Background: Allosteric ligands targeting cholecystokinin receptors are needed.
Results: Stereochemically distinct iodinated 1,4-benzodiazepine antagonists of type 1 and 2 cholecystokinin receptors dock to analogous intramembranous pockets that have distinct shape and molecular determinants.
Conclusion: The geometry of the binding pockets and specific residue interactions are unique for each receptor.
Significance: The predictive power of these insights should be useful in the discovery of lead compounds and in their refinement.
Keywords: G Protein-coupled Receptors (GPCR), Homology Modeling, Site-specific Mutagenesis, Peptide Hormones, Receptor Structure-Function
Abstract
Allosteric binding pockets in peptide-binding G protein-coupled receptors create opportunities for the development of small molecule drugs with substantial benefits over orthosteric ligands. To gain insights into molecular determinants for this pocket within type 1 and 2 cholecystokinin receptors (CCK1R and CCK2R), we prepared a series of receptor constructs in which six distinct residues in TM2, -3, -6, and -7 were reversed. Two novel iodinated CCK1R- and CCK2R-selective 1,4-benzodiazepine antagonists, differing only in stereochemistry at C3, were used. When all six residues within CCK1R were mutated to corresponding CCK2R residues, benzodiazepine selectivity was reversed, yet peptide binding selectivity was unaffected. Detailed analysis, including observations of gain of function, demonstrated that residues 6.51, 6.52, and 7.39 were most important for binding the CCK1R-selective ligand, whereas residues 2.61 and 7.39 were most important for binding CCK2R-selective ligand, although the effect of substitution of residue 2.61 was likely indirect. Ligand-guided homology modeling was applied to wild type receptors and those reversing benzodiazepine binding selectivity. The models had high predictive power in enriching known receptor-selective ligands from related decoys, indicating a high degree of precision in pocket definition. The benzodiazepines docked in similar poses in both receptors, with C3 urea substituents pointing upward, whereas different stereochemistry at C3 directed the C5 phenyl rings and N1 methyl groups into opposite orientations. The geometry of the binding pockets and specific interactions predicted for ligand docking in these models provide a molecular framework for understanding ligand selectivity at these receptor subtypes. Furthermore, the strong predictive power of these models suggests their usefulness in the discovery of lead compounds and in drug development programs.
Introduction
With the recent solution of a series of class A G protein-coupled receptor (GPCR)3 structures (1–7), the promise of structure-based rational design and refinement of receptor-active drugs is coming closer to fruition. This is particularly true for drugs that target the highly conserved and spatially constrained helical bundle region of receptors in this family. The type 1 and type 2 cholecystokinin (CCK) receptors (CCK1R and CCK2R) are closely related, physiologically important members of this family that have distinct patterns of structural selectivity both for natural peptide ligands and for non-peptidyl small molecule ligands (8–10). This makes them potentially useful targets for a chimeric approach to defining the structural basis of ligand binding and thereby contributing to drug development.
These receptors possess 53% sequence identity, including 69% identity within predicted transmembrane helical segments. They control a variety of gastrointestinal and behavioral functions. Type 1 CCK receptors are expressed on gallbladder smooth muscle, pancreatic cells, enteric neurons, and central nervous system nuclei, whereas type 2 CCK receptors are expressed on gastric parietal cells and diffuse regions of the brain (11, 12). There have been extensive efforts to develop drugs to target these molecules for the treatment of obesity, anxiety, gastric hypersecretory states, and even particular types of cancer (13–15).
Although both types of CCK receptors bind and are activated by CCK and gastrin peptides, the molecular basis for peptide binding to these receptors appears to be distinct. Whereas the CCK1R requires the carboxyl-terminal heptapeptide-amide, including the sulfated tyrosine residue in position 27 (using the numbering scheme of CCK-33 first isolated), for high affinity binding and potent action, the CCK2R requires only the carboxyl-terminal tetrapeptide that is shared between CCK and gastrin peptides (16, 17). Whereas naturally occurring CCK peptides eight or more residues in length bind with high affinity and are potent activators of both the CCK1R and the CCK2R, gastrin and shorter CCK peptides are high affinity ligands and potent activators of the CCK2R only (18). The basis for CCK binding to CCK1R is particularly well defined, based on direct photoaffinity labeling spatial approximation constraints at seven positions within CCK-8, which show determinants distributed throughout the extracellular loop and amino-terminal tail regions but not within the predicted transmembrane domain bundle (12, 19). In contrast, at the CCK2R, the carboxyl-terminal end of the peptide may be sited closer to the helical bundle (12, 20, 21).
Selective, small molecule ligands have been developed for both CCK1R and CCK2R (22, 23). It is now clear, based on limited receptor mutagenesis, photoaffinity labeling, and pharmacological manipulations, that these ligands bind to an allosteric site within the intramembranous helical bundle that is distinct from the orthosteric CCK peptide-binding site of the CCK1R (24–27). However, the molecular basis for ligand selectivity between the two subtypes of CCK receptors remains unclear. Using the photo-cross-linking point as an anchor, crude homology modeling predicts up to 20 transmembrane residues that could be involved in the formation of this pocket (26). Of these amino acids, only six differ between CCK1R and CCK2R (residues 2.61, 3.28, 3.29, 6.51, 6.52, and 7.39, using the nomenclature of Ballesteros and Weinstein (28)).
The current work utilized a focused chimeric receptor approach targeting these six residues and took advantage of a unique pair of subtype-selective radioiodinatable 1,4-benzodiazepine ligands that could be used as direct probes of this intramembranous interhelical ligand-binding site (29). Direct binding analysis was performed for an extensive series of chimeric CCK1R/CCK2R constructs to determine the contributions of each of these residues in small molecule ligand selectivity and to demonstrate the reversal of selectivity via distinct combinations of substitutions. The allosteric nature of the small molecule binding of key chimeric constructs was also demonstrated directly using a peptide ligand dissociation assay. Ligand-directed homology modeling was utilized to generate predictive receptor models in which the geometry of the binding pockets and distinct residue interactions predicted for ligand docking provided a molecular framework for understanding allosteric ligand selectivity at the two receptor subtypes.
EXPERIMENTAL PROCEDURES
Materials
Costar 96-well V-bottom assay plates were from Corning (Corning, NY); Ham's F-12 medium and other tissue culture supplements were from Invitrogen; fetal clone II cell culture medium supplement was from HyClone Laboratories (Logan, UT). All other reagents were analytical grade.
Benzodiazepines
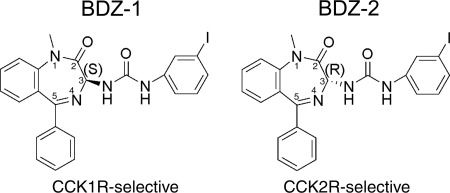
The reagents that are the focus of this work are the 1,4-benzodiazepine antagonists of CCK receptors that differ only in the stereochemistry (S or R) of their 3-position side chains while exhibiting selective binding to either the type 1 cholecystokinin receptor (CCK1R) (BDZ-1, identified previously as compounds 5 and 9 (29)) or the type 2 cholecystokinin receptor (CCK2R) (BDZ-2, identified previously as compounds 3 and 7 (29)). The 1,4-benzodiazepines with an S-orientation, (S)-1-(3-iodophenyl)-3-(1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)urea, represent the BDZ-1 radioligand with incorporation of radioactive iodine (125I) and BDZ-1 with non-radioactive 127I, whereas those with an R-orientation, (R)-1-(3-iodophenyl)-3-(1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)urea, represent the BDZ-2 radioligand with 125I and BDZ-2 with 127I (Fig. 1) (29). These were prepared by oxidative iodination of precursor compounds (S)-1-(1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-3(3(trimethylstannyl)phenyl)urea and (R)-1-(1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-3(3(trimethylstannyl)phenyl)urea, respectively, as we described previously (29). These compounds have been fully characterized chemically and pharmacologically (29) and have the advantage of providing radioligands for competition binding assays with direct binding to the allosteric site within the helical bundle of CCK receptors.
FIGURE 1.
Chemical structure of CCK receptor ligands. Shown are the structures of BDZ-1 and BDZ-2, representing benzodiazepine ligands selective for binding to the CCK1R and CCK2R. These correspond to compounds that were described previously (29): compounds 5 and 9, which are selective for CCK1R, and compounds 3 and 7, which are selective for CCK2R, with the first compound of each pair being non-radioactive and incorporating 127I and the other in the radioiodinated form.
Peptide Ligands
Synthetic peptides, human gastrin-17 and cholecystokinin octapeptide (CCK-8 and CCK-26–33), were from Bachem AG (Bubendorf, Switzerland) and Peninsula Laboratories (Belmont, CA), respectively. The CCK-like peptide radioligand 125I-d-Tyr-Gly-[(Nle28,31)CCK-26–33] was synthesized, purified to homogeneity on reversed-phase HPLC, and radioiodinated in our laboratory as we described previously (30).
Preparation of Human CCK Receptor Constructs
Human CCK1R and CCK2R constructs were inserted into the eukaryotic expression vectors pcDNA3.1zeo (Invitrogen) and pcDNA3, respectively (31). Primers were designed to introduce mutations in transmembrane (TM) segments TM2 (position 2.61), TM3 (positions 3.28 and 3.29), TM6 (positions 6.51 and 6.52), or TM7 (position 7.39) of CCK1R and CCK2R constructs using the QuikChangeTM site-directed mutagenesis kit (Stratagene, La Jolla, CA). Each construct introduced the reciprocal residue into the specified position of the other receptor of the pair of subtypes, type 1 or type 2, in the CCK receptors. The CCK1R variant with residues changed in positions 3.28/3.29 and 6.51/6.52 was prepared by ligation of the individual constructs, CCK1R 3.28/3.29 and CCK1R 6.51/6.52. These were digested with BspEI and XhoI with appropriate fragments ligated together. The CCK2R variant with residues changed in positions 3.28/3.29 and 6.51/6.52 was created by the ligation of the individual constructs, CCK2R 3.28/3.29 and CCK2R 6.51/6.52, after digestion with BsrGI and HindIII. All constructs had their sequences confirmed by automated dye-terminator DNA sequencing.
Cell Culture
Wild type and mutant CCK1R and CCK2R constructs were transiently expressed in COS-1 cells. Cells were plated in 100-mm tissue culture dishes in Dulbecco's modified Eagle's medium supplemented with fetal clone II at a density of 0.5 × 106 cells/plate, and 3 μg of cDNA from each construct was introduced by the DEAE-dextran method (32). Cells were maintained in a humidified atmosphere of 5% CO2 at 37 °C. Select constructs were also studied as stably expressed in Chinese hamster ovary (CHO) cells, when higher levels of receptor expression were necessary to characterize radioligand binding. These lines were established as we described previously (33).
Membrane Preparation
Receptor-bearing membranes were isolated from confluent COS-1 cells 72 h after transfection using a previously described protocol that involves sucrose density centrifugation (33). All procedures were carried out on ice or at 4 °C. Membranes were suspended in Krebs-Ringers-HEPES (KRH) medium (25 mm HEPES, pH 7.4, 104 mm NaCl, 5 mm KCl, 2 mm CaCl2,1 mm KH2PO4, and 1.2 mm MgSO4) supplemented with 0.01% soybean trypsin inhibitor and 1 mm phenylmethylsulfonyl fluoride and were stored at −80 °C until use.
Receptor Binding Assays
Binding of the radioiodinated benzodiazepines, BDZ-1 radioligand and BDZ-2 radioligand, to the wild type and each of the mutant CCK receptors was assessed using filtration assays. In brief, binding reactions were performed in KRH medium supplemented with 0.2% bovine serum albumin and 0.01% soybean trypsin inhibitor. Receptor-bearing membranes (1–20 μg of protein) were mixed with the BDZ radioligands (0.5–1.0 pm; ∼10,000 cpm/well) in a low retention polypropylene plate for 1 h at room temperature in the presence or absence of increasing concentrations of the non-radioactive BDZ compounds (the respective 127I-labeled analogues of the BDZ radioligands). Nonspecific binding was defined using competition with the analogous non-radioactive BDZ compound at 1 μm concentration. The receptor-bound fractions were separated from the free radioligand by filtration using Unifilter-96 GF/B filter mats in a FilterMate Harvester (PerkinElmer Life Sciences). The plate was then washed six times with wash buffer (0.9% NaCl and 0.2% bovine serum albumin), air-dried overnight, and counted on a TopCount® NXTTM counter (Packard) after the addition of 30 μl of MicroScintTM-O (PerkinElmer). All assays were performed in duplicate in at least three independent experiments. Radioligand binding data were analyzed using the Ligand program (34) and were plotted using the nonlinear least-squares curve-fitting routine in Prism (GraphPad 4.0, San Diego, CA).
Binding assays using the CCK-like radioligand 125I-d-Tyr-Gly-[(Nle28,31)CCK-26–33] were also performed, following the techniques established previously (31). These assays were performed in intact cells to establish cell surface expression and in membranes of select receptor constructs to determine the structural specificity of peptide binding. In peptide binding experiments, nonspecific binding was determined in the presence of 1 μm unlabeled CCK.
Assays to quantify the rates of dissociation of receptor-bound CCK-like radioligand in the absence or presence of small molecule ligands were used as one determinant of the allosteric mode of action, following the procedure described previously in detail (27).
Molecular Modeling
All molecular modeling was conducted using a stochastic global energy optimization procedure in internal coordinate mechanics (ICM) (35) with the ICM-Pro package, version 3.7-2 (MolSoft LLC, San Diego, CA). This procedure consisted of three iterative steps: (a) random conformational change of a dihedral angle according to the biased probability Monte Carlo method (36); (b) local minimization of all free dihedral angles; and (c) acceptance or rejection of the new conformation based on the Metropolis criterion at the simulation temperature, usually at 600 K (37).
Specifically, a ligand-guided homology modeling method was used (38). In this procedure, a simple naive homology model was first generated using the ICM standard homology modeling function (39). Four distinct x-ray structures of class A GPCRs were attempted as templates for the standard threading method: β-2 adrenergic receptor, 2RH1; dopamine 3 receptor, 3PBL; A2a adenosine receptor, 3EML; and CXCR-4 receptor, 3ODU. A seed ligand was placed within the ligand pocket, which was predicted by the ICM PocketFinder algorithm (40). A distance restraint was set between the ligand and an anchor residue on the receptor to limit the size of the sampling space and to keep the ligand within the pocket during the modeling process. The receptor was then subjected to cycles of Monte Carlo side chain sampling and backbone minimization, which was followed by loop modeling. After each cycle, multiple receptor conformations were generated.
Molecular models were evaluated by docking a structurally diverse set of test ligands selected from the ChEMBL database (41) based on available data for CCK receptor binding affinity. For CCK1R, the ligand set contained 53 positive ligands with pKi > 6 and 115 decoys with pKi < 6 against CCK1R. For CCK2R, the ligand set contained 76 positive ligands with pKi > 8 and 171 decoys with pKi < 8 against CCK2R. The best model was selected by the composite score,
 |
where SCOREfinal is the final score of the model and SCOREICM is the median ICM docking score of the positive compounds, to encourage discrimination of positive compounds by rewarding better ligand-receptor interactions rather than by penalizing decoys. FHB is the fraction of positive compounds forming hydrogen bond contacts with the corresponding anchor residue. SCOREcluster is the ICM docking pose clustering score, to encourage more consistent docking poses. NSQ_AUC is the normalized square root AUC (38) defined by the equation,
 |
where AUC* is similar to area under the receiver-operating characteristic (ROC) curve (42), except the x axis is replaced by the square root of the percentage of false positives. Compounds that form a hydrogen bond contact with the anchor are presumably docked correctly and are prioritized in the AUC* calculation. The NSQ_AUC has an advantage over the traditional AUC, because NSQ_AUC is more sensitive to initial enrichment and therefore is more relevant in a virtual screening setting where only the top 1% of the ligands or fewer are selected.
After the initial sampling and evaluation, the best model was selected. The seed ligand was redocked into the receptor, generating multiple models with different docking poses. Each of these models was then refined by side chain sampling and backbone minimization. All final models were then re-evaluated by docking the entire test ligand set, and the best model was selected. For the model of the CCK1R mutant construct in which the benzodiazepine selectivity was reversed, the final model of CCK1R was used as the initial template, changing the following residues to their counterparts present in the same positions of CCK2R: N2.61T, T3.28V, T3.29S, I6.51V, F6.52Y, and L7.39H. For the model of the CCK2R mutant construct in which the benzodiazepine selectivity was reversed, the analogous approach using the final model of CCK2R as initial template did not yield an acceptable, high quality molecular model. Instead, for this mutant construct, the final model of CCK1R was used as the initial template, changing all residues to those of CCK2R except for Ile6.51, Phe6.52, and Leu7.39, the key residues in these positions in CCK1R. The naive models were then subjected to the ligand-guided modeling method by using the test ligand set for the opposite receptor for docking and selection.
RESULTS
In the current work we focused on the six residues lining the predicted allosteric pocket of the CCK1R that are different in the two CCK receptor subtypes (Fig. 2). These exist within TM2, TM3, TM6, and TM7, with one residue in each of TM2 (2.61) and TM7 (7.39) and two adjacent residues in each of TM3 (3.28/3.29) and TM6 (6.51/6.52). We have changed these residues to the corresponding residues in the opposite CCK receptor subtype using TM segment groups and typically refer to them according to the TM segment or segments changed in the chimeric CCK1R/CCK2R receptor constructs.
FIGURE 2.
Primary structures of receptor constructs used in this work. Shown are the aligned sequences and proposed topology of human CCK1R and CCK2R, with the TM segments enclosed in boxes. The 20 amino acids proposed to align the intrahelical small molecule-binding pocket (residues 2.61, 2.64, 2.65, 3.28, 3.29, 3.32, 3.36, 5.42, 6.48, 6.51, 6.52, 6.54, 6.55, 6.58, 7.35, 7.36, 7.38, 7.39, 7.42, and 7.43, using the nomenclature of Ballesteros and Weinstein (28)) are shaded, with those residues that are different in the CCK1R and the CCK2R shown in bold and labeled in italics. The residues of interest exist within transmembrane segments TM2, TM3, TM6, and TM7, with one residue in each of TM2 (2.61) and TM7 (7.39) and two adjacent residues in each of TM3 (3.28/3.29) and TM6 (6.51/6.52). We mutated these residues in TM segment groups and generally refer to them according to the TM segment changed in the chimeric CCK1R/CCK2R constructs.
Characterization of Ligand Binding to Chimeric Receptor Constructs Involving Single TM Segments
Competition binding was performed using each of the selective benzodiazepine radioligands with the wild type CCK receptors and the single TM segment chimeric constructs (Fig. 3 and Tables 1 and 2). Each of the wild type and single region mutants was synthesized and trafficked normally to the cell surface where they were shown in intact cell assays to bind saturably at least one of the radioligands, including the benzodiazepines and the CCK-like radioligand (data not shown). Only the wild type CCK1R and chimeric constructs based on the CCK1R structure bound the type 1-selective BDZ-1 radioligand in a saturable manner (Fig. 3A). Loss of function, with reduced binding affinity for BDZ-1, was observed for each of the CCK1R constructs when replacing the unique residues in TM6 (6.51 and 6.52), TM3 (3.28 and 3.29), and TM7 (7.39), whereas replacing residue 2.61 in TM2 had no significant negative impact (affinity of 3.6 ± 0.7 nm; not different from that of wild type CCK1R, 2.3 ± 0.3 nm) (Fig. 3A).
FIGURE 3.
Receptor binding characterization of wild type and single TM chimeric CCK1R/CCK2R constructs. Competition binding assays were performed using each of the selective benzodiazepine radioligands, BDZ-1 radioligand and BDZ-2 radioligand, with the wild type CCK receptors and the single TM segment chimeric receptor constructs. Data for the CCK1R-based constructs are shown in the upper set of panels (A–D), and data for the CCK2R-based constructs are shown in the lower set of panels (E and F). In each set of panels, the use of each radioligand is shown in a row, with the homologous competition curves in the left column and the heterologous competition curves in the right column. No saturable binding of BDZ-1 radioligand was observed for the single region CCK2R mutants involving TM2, TM3, TM6, or TM7. No saturable binding of BDZ-2 radioligand was observed for the single region mutant of CCK2R involving TM2 or the single region mutants of CCK1R involving TM3, TM6, or TM7. Data for wild type receptors are shown with dashed lines.
TABLE 1.
Benzodiazepine ligand binding to CCK1R and CCK1R-based mutant constructs
NBD, no demonstrable saturable binding; NDB^, no demonstrable saturable binding of BDZ-1 or BDZ-2 radioligands when expressed transiently in COS-1 cells or stably in CHO cells; however, saturable CCK-like radioligand binding confirmed the expression of these constructs on the cell membrane. *, p < 0.05; **, p < 0.001; ***, p < 0.0001 relative to values with wild type CCK1R.
| CCK1R receptor constructs | Abbreviation | CCK1Ra |
|||||
|---|---|---|---|---|---|---|---|
| BDZ-1 radioligand binding, homologous competition Ki | BDZ-1 radioligand binding, heterologous competition Ki | BDZ-2 radioligand binding, homologous competition Ki | BDZ-2 radioligand binding, heterologous competition Ki | BDZ-1 radioligand binding, homologous competition Bmax | BDZ-2 radioligand binding, homologous competition Bmax | ||
| nm | nm | nm | nm | pmol/mg | pmol/mg | ||
| CCK1R-WT | 2.3 ± 0.3 | 197.4 ± 31.7 | NDB | NDB | 19.2 ± 2.5 | NDB | |
| N2.61T | CCK1R TM2 | 3.6 ± 0.7 | 36.2 ± 10.5** | 14.4 ± 1.3*** | 2.4 ± 0.4*** | 61.2 ± 7.2*** | 23.8 ± 1.2*** |
| T3.28V, T3.29S | CCK1R TM3 | 14.5 ± 1.8*** | 259.3 ± 119.1 | NDB | NDB | 33.6 ± 7.5* | NDB |
| I6.51V, F6.52Y | CCK1R TM6 | 10.8 ± 2.2*** | 238.7 ± 29.0 | NDB | NDB | 14.1 ± 0.2 | NDB |
| L7.39H | CCK1R TM7 | 11.4 ± 0.6*** | 104.2 ± 7.9 | NDB | NDB | 46.1 ± 5.3*** | NDB |
| N2.61T + T3.28V, T3.29S | CCK1R TM(2,3) | 17.2 ± 6.7** | 26.6 ± 13.6** | 14.9 ± 1.9*** | 25.5 ± 10.5** | 23.4 ± 6.2 | 38.7 ± 8.3*** |
| N2.61T + I6.51V, F6.52Y | CCK1R TM(2,6) | 32.5 ± 8.7** | 18.1 ± 11.0** | 22.7 ± 7.5** | 23.8 ± 1.9** | 14.8 ± 3.7 | 19.7 ± 1.7*** |
| N2.61T + L7.39H | CCK1R TM(2,7) | 11.0 ± 2.3*** | 4.5 ± 0.2** | 7.1 ± 2.2* | 25.0 ± 4.0** | 33.5 ± 10.0 | 43.4 ± 1.5*** |
| T3.28V, T3.29S + I6.51V, F6.52Y | CCK1R TM(3,6) | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ |
| T3.28V, T3.29S + L7.39H | CCK1R TM(3,7) | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ |
| I6.51V, F6.52Y + L7.39H | CCK1R TM(6,7) | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ |
| N2.61T + T3.28V, T3.29S + I6.51V, F6.52Y | CCK1R TM(2,3,6) | NDB | NDB | 6.4 ± 0.5** | 81.8 ± 4.6 | NDB | 21.1 ± 2.6*** |
| N2.61T + T3.28V, T3.29S + L7.39H | CCK1R TM(2,3,7) | 19.5 ± 1.4*** | 5.1 ± 0.7** | 32.9 ± 4.4*** | 250.3 ± 31.1*** | 20.0 ± 3.6 | 48.1 ± 9.8*** |
| N2.61T + I6.51V, F6.52Y + L7.39H | CCK1R TM(2,6,7) | 36.7 ± 4.7*** | 37.6 ± 9.3* | 37.7 ± 10.1*** | 623.0 ± 67.17*** | 12.4 ± 2.7 | 50.9 ± 15.5*** |
| T3.28V, T3.29S + I6.51V, F6.52Y + L7.39H | CCK1R TM(3,6,7) | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ |
| N2.61T + T3.28V, T3.29S + I6.51V, F6.52Y + L7.39H | CCK1R TM(2,3,6,7) | NDB | NDB | 2.1 ± 0.3 | 86.9 ± 7.1 | NDB | 15.6 ± 1.6*** |
a Values are expressed as means ± S.E.
TABLE 2.
Benzodiazepine ligand binding to CCK2R and CCK2R-based mutant constructs
NDB, no demonstrable saturable binding; NDB^, no demonstrable saturable binding of BDZ-1 or BDZ-2 radioligands when expressed transiently in COS-1 cells or stably in CHO cells; however, saturable CCK-like radioligand binding confirmed the expression of these constructs on the cell membrane, except for CCK2R TM(2,3,7) and CCK2R TM(2,3,6,7). #, CCK2R TM(3,7) binding was assessed in a stably transfected CHO cell line, because it had inadequate saturable binding for characterization when expressed transiently in COS-1 cells. *, p < 0.05; **, p < 0.001; ***, p < 0.0001 relative to values with wild type CCK2R.
| CCK2R receptor constructs | Abbreviation | CCK2Ra |
|||||
|---|---|---|---|---|---|---|---|
| BDZ-2 radioligand binding, homologous competition Ki | BDZ-2 radioligand binding, heterologous competition Ki | BDZ-1 radioligand binding, homologous competition Ki | BDZ-1 radioligand binding, heterologous competition Ki | BDZ-2 radioligand binding, homologous competition Bmax | BDZ-1 radioligand binding, homologous competition Bmax | ||
| nm | nm | nm | nm | pmol/mg | pmol/mg | ||
| CCK2R-WT | 3.4 ± 0.4 | 99.9 ± 12.2 | NDB | NDB | 3.0 ± 0.4 | NDB | |
| T2.61N | CCK2R TM2 | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ |
| V3.28T, S3.29T | CCK2R TM3 | 3.1 ± 0.9 | 154.5 ± 60.0 | NDB | NDB | 4.6 ± 0.9 | NDB |
| V6.51I, Y6.52F | CCK2R TM6 | 2.3 ± 0.6 | 20.2 ± 1.3** | NDB | NDB | 1.2 ± 0.3* | NDB |
| H7.39L | CCK2R TM7 | 10.3 ± 3.9* | 30.4 ± 10.4** | NDB | NDB | 1.4 ± 0.4* | NDB |
| T2.61N + V3.28T, S3.29T | CCK2R TM(2,3) | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ |
| T2.61N + V6.51I, Y6.52F | CCK2R TM(2,6) | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ |
| T2.61N + H7.39L | CCK2R TM(2,7) | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ |
| V3.28T, S3.29T + V6.51I, Y6.52F | CCK2R TM(3,6) | 3.7 ± 0.3 | 28.3 ± 7.2** | NDB | NDB | 1.5 ± 0.1* | NDB |
| V3.28T, S3.29T + H7.39L | CCK2R TM(3,7) | 34.7 ± 15.0**# | 41.1 ± 2.4*# | 39.5 ± 10.4***# | 30.0 ± 5.4**# | 8.1 ± 2.9*# | 7.7 ± 3.0*# |
| V6.51I, Y6.52F + H7.39L | CCK2R TM(6,7) | NDB | NDB | 4.1 ± 1.3 | 23.0 ± 10.3** | NDB | 2.4 ± 0.5*** |
| T2.61N + V3.28T, S3.29T + V6.51I, Y6.52F | CCK2R TM(2,3,6) | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ |
| T2.61N + V3.28T, S3.29T + H7.39L | CCK2R TM(2,3,7) | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ |
| T2.61N + V6.51I, Y6.52F + H7.39L | CCK2R TM(2,6,7) | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ |
| V3.28T, S3.29T + V6.51I, Y6.52F + H7.39L | CCK2R TM(3,6,7) | NDB | NDB | 9.0 ± 3.1* | 59.6 ± 22.2* | NDB | 3.0 ± 0.9*** |
| T2.61N + V3.28T, S3.29T + V6.51I, Y6.52F + H7.39L | CCK2R TM(2,3,6,7) | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ | NDB^ |
a Values are expressed as means ± S.E.
The type 2-selective BDZ-2 radioligand bound saturably to the wild type CCK2R and to selected chimeric constructs based on that structure but not to the CCK1R (Fig. 3, C and E). Loss of function, with reduced binding affinity for BDZ-2, was observed for the CCK2R constructs when replacing the unique residues in TM2 (2.61) and TM7 (7.39), whereas replacing those in TM3 and TM6 had no significant negative impact on binding affinity. Of particular interest, gain of function was observed for the CCK1R TM2 construct, with this receptor construct gaining the ability to saturably bind BDZ-2 radioligand and for BDZ-2 to compete for its binding of the BDZ-1 radioligand.
Characterization of Ligand Binding to Chimeric Receptor Constructs Involving Multiple TM Segments
The multiple TM segment chimeric constructs were also analyzed using the full complement of competition binding studies with each of the selective benzodiazepine radioligands (Figs. 4 and 5 and Tables 1 and 2). There was evidence of normal receptor folding, trafficking, and cell surface expression for all of the constructs based on the CCK1R structure, with all constructs that did not bind the benzodiazepine radioligands demonstrated to saturably bind the CCK-like radioligand (data not shown). In contrast, for the chimeric constructs based on the CCK2R structure, two of the 15 constructs (CCK2R TM(2,3,7) and CCK2R TM(2,3,6,7)) did not saturably bind radiolabeled benzodiazepines or CCK, likely reflecting misfolding and intracellular trapping of these constructs within a biosynthetic compartment. It is also notable that CCK2R TM(3,7) and CCK2R TM(3,6,7) constructs, which trafficked normally to the cell surface where they bound BDZ-2 radioligand, did not exhibit saturable binding of the CCK radioligand.
FIGURE 4.
Receptor binding characterization of multiple TM CCK1R-based chimeric constructs. Competition binding assays were performed using each of the selective benzodiazepine radioligands, BDZ-1 radioligand and BDZ-2 radioligand, with the wild type CCK1R and multiple TM segment chimeric receptor constructs. The use of each radioligand is shown in a row (BDZ-1 in A and B; BDZ-2 in C and D), with the homologous competition curves in the left column (A and C) and the heterologous competition curves in the right column (B and D). No detectable binding was observed for homologous and heterologous binding of BDZ-1 radioligand with CCK1R chimeric constructs TM(3,6), TM(3,7), TM(6,7), TM(2,3,6), TM(3,6,7), and TM(2,3,6,7). No detectable binding was observed for homologous and heterologous binding of BDZ-2 radioligand with CCK1R chimeric constructs TM(3,6), TM(3,7), TM(6,7), and TM(3,6,7). Data for wild type receptors are shown with dashed lines.
FIGURE 5.
Receptor binding characterization of multiple TM CCK2R-based chimeric constructs. Competition binding assays were performed using each of the selective benzodiazepine radioligands, BDZ-1 radioligand and BDZ-2 radioligand, with the wild type CCK2R and multiple TM segment chimeric receptor constructs. The use of each radioligand is shown in a row (BDZ-1 in A and B; BDZ-2 in C and D), with the homologous competition curves in the left column (A and C) and the heterologous competition curves in the right column (B and D). Data for wild type receptors are shown with dashed lines. No detectable binding was observed for homologous and heterologous binding of BDZ-1 radioligand with CCK2R chimeric constructs TM(2,3), TM(2,6), TM(2,7), TM(6,7), TM(2,3,6), TM(2,3,7), TM(2,6,7), and TM(2,3,6,7). No detectable binding was observed for homologous and heterologous binding of BDZ-2 radioligand with CCK2R chimeric constructs TM(2,3), TM(2,6), TM(2,7), TM(6,7), TM(2,3,6), TM(2,3,7), TM(2,6,7), TM(3,6,7), and TM(2,3,6,7).
The competition binding curves for the CCK1R-selective BDZ-1 radioligand are shown in Fig. 4, with the homologous competition on the left (Fig. 4, A and C) and the heterologous competition on the right (Fig. 4, B and D). Of interest, in the homologous competition curves, adding the chimeric changes to CCK1R in TM7, TM3, or TM6 to TM2 reduced the affinities of binding BDZ-1 from what was observed with TM2 only. Adding the chimeric changes in TM6 and TM7 to the CCK2R bound the BDZ-1 radioligand with an affinity (4.1 ± 1.3 nm) that did not differ from that of wild type CCK1R (2.3 ± 0.3 nm) (p > 0.05). Thus, TM6 and TM7 residues were capable of both loss of function at the CCK1R and gain of function at the CCK2R, supporting the critical importance of these residues for BDZ-1 binding to the CCK1R.
The competition binding curves for the CCK2R-selective BDZ-2 radioligand are shown in Fig. 5, with the homologous competition on the left (Fig. 5, A and C) and the heterologous competition on the right (Fig. 5, B and D). In the homologous competition curves, adding the chimeric changes in TM3 and TM6 to CCK2R (3.7 ± 0.3 nm) or TM2, TM3, TM6, and TM7 to CCK1R (2.1 ± 0.3 nm) resulted in high affinity binding that did not differ from that of wild type CCK2R (3.4 ± 0.4 nm) (p > 0.05). Several of the combinations were able to enhance the apparent affinity in the heterologous competition binding for this radioligand.
Of note, some binding characteristics of a few select mutants were anomalous (Tables 1 and 2). Although there was general accord in the estimated number of binding sites with the two BDZ radioligands (when significant binding could be measured), there were three exceptions to this pattern. The N2.61T mutant (CCK1R TM2) exhibited an ∼3-fold greater number of binding sites with the BDZ-1 radioligand than with the BDZ-2 radioligand, suggesting that the BDZ-2 radioligand bound only a subset of these receptors that were detected with the BDZ-1 radioligand. The reverse was observed for the CCK1R TM(2,3,7) and CCK1R TM(2,6,7) constructs, with a 2–4-fold greater number of binding sites observed with the BDZ-2 radioligand than with the BDZ-1 radioligand. This suggests that the radioligands may have sampled distinct populations of receptors that do not readily interchange.
CCK Peptide Binding Selectivity
Fig. 6 shows the CCK peptide binding selectivity in the chimeric constructs in which benzodiazepine selectivity was most effectively reversed, the CCK1R TM(2,3,6,7) construct with CCK2R-like benzodiazepine selectivity and the CCK2R TM(6,7) construct with CCK1R-like benzodiazepine selectivity. In both cases, the peptide selectivity of the parent construct was maintained. Note also that competitive binding curves were also performed using BDZ-1 and BDZ-2. The relevant high affinity benzodiazepine ligand was able to fully inhibit saturable CCK-like radioligand binding to each of these constructs. Based on this set of observations, it was not possible to distinguish competitive inhibition of peptide ligand binding from the influence of negative cooperativity of allosteric ligands.
FIGURE 6.
CCK peptide binding to wild type (A and B) and key chimeric CCK1R/CCK2R constructs (C and D). Competition binding assays using a CCK-like peptide radioligand were performed to evaluate CCK peptide binding selectivity in the chimeric receptor constructs in which the benzodiazepine selectivity was most effectively reversed, the CCK1R TM(2,3,6,7) construct with CCK2R benzodiazepine selectivity (C) and the CCK2R TM(6,7) construct with CCK1R benzodiazepine selectivity (D). In both cases, the peptide selectivity of the parent construct was maintained. Also shown in each panel are curves with dashed lines reflecting the abilities of the high affinity benzodiazepine ligands, BDZ-1 and BDZ-2, to inhibit the saturable binding of the CCK radioligand.
Allosteric Nature of Small Molecule Ligand Binding
The possible allosteric nature of the binding of the benzodiazepine ligands to these receptors was examined using a classical orthosteric radioligand dissociation assay as described previously (27). In this assay, the ability of a second ligand (here BDZ-1 or BDZ-2) to alter the rate of dissociation of the radioligand provides definitive evidence for the allosteric nature of the small molecule binding. Fig. 7 shows that the CCK2R and each of the key chimeric receptors that effectively reversed their benzodiazepine ligand selectivity displayed small molecule-induced changes in orthosteric ligand dissociation. Of note, CCK1R exhibited no difference in these two dissociation curves. This does not rule out an allosteric mode of action for this ligand at this receptor but shows only that its binding did not modify the kinetics of orthosteric ligand dissociation. It is noteworthy that the orthosteric ligand dissociation was quite rapid at this receptor, and a small change might not have been detectable under the assay conditions. However, there are strong independent data to support an allosteric mode of action of BDZ-1 at the CCK1R, specifically the inability of CCK to fully compete for BDZ-1 binding to this receptor in both a model receptor-bearing cell line and natural receptor-expressing gallbladder cells (29).
FIGURE 7.
Dissociation of CCK radioligand saturably bound to the wild type (A and B) and key chimeric CCK1R/CCK2R constructs (C and D). Control curves with dashed lines represent dissociation in the absence of benzodiazepine ligands, whereas the second curve in each panel represents dissociation of the CCK radioligand in the presence of the relevant high affinity benzodiazepine ligand, BDZ-1 or BDZ-2 (10 nm). Shown in parentheses are the Koff values for each condition. These curves were significantly different from each other for the CCK2R, CCK2R TM(6,7) and CCK1R TM(2,3,6,7) constructs (p < 0.05) but were not different for the two conditions studied at the CCK1R receptor.
Molecular Modeling
The best template for the molecular modeling of both the CCK1R and the CCK2R was found to be the A2a adenosine receptor, 3EML. This resulted in the highest scores for the ultimate models (described below). In this approach, an anchor residue is critical for the initial docking of the ligands during the modeling process. For the CCK1R, Asn6.55 was initially selected as the anchor residue, because it is a polar residue in close proximity to Ile6.51 and Phe6.52 (shown to be critically important in the mutagenesis studies) that is potentially capable of forming a specific hydrogen bond with the benzodiazepine ligand. This residue had also been utilized effectively as the anchor residue in previous small molecule ligand docking to the A2a adenosine receptor (43). This tether turned out to be quite effective, yielding a strong, credible, and discriminating model. For the CCK2R, Thr2.61 was initially selected as the anchor residue, because it had appropriate chemical characteristics and was shown to be functionally important in the mutagenesis studies. However, after extensive trials, using this anchor we were unable to obtain a reasonably high scoring model that had significant enrichment. This supports an indirect functional effect of mutation of this residue in CCK2R. Switching the anchor for the CCK2R modeling to Asn6.55, however, produced a final model that exhibited excellent enrichment and selectivity properties.
The molecular models of BDZ-1 docked with the CCK1R and BDZ-2 docked with the CCK2R are shown in Fig. 8. The benzodiazepine ligands in both of these models were docked in pockets in analogous positions high in the intramembranous helical bundles. Both ligands displayed similar poses for their C3 urea substituents and benzo rings; however, the C5 phenyl rings and N1 methyl groups exhibited different orientations and spatial approximations with the receptors because of their differing stereochemistry at C3. The benzodiazepine C3 urea substituents of both ligands pointed upward toward the top of the TM5–TM6 region and were flanked partially by ECL2 and ECL3, and the benzo rings were located close to TM3. The C2 carbonyl of BDZ-1 in the CCK1R model was predicted to make a hydrogen bond contact with Asn333 (Asn6.55), with the N1 methyl group pointed down toward the bottom of the ligand pocket and the C5 phenyl ring pointed toward the TM2–TM7 region. In contrast, for BDZ-2 in the CCK2R model, N4 of the benzodiazepine was predicted to make a hydrogen bond contact with Asn353 (Asn6.55), the N1 methyl group pointed toward the TM2–TM7 region, and the C5 phenyl ring pointed down toward the bottom of the pocket.
FIGURE 8.
Docking of benzodiazepine ligands into pockets in CCK1R and CCK2R. The best docking poses of BDZ-1 in the CCK1R model and BDZ-2 in the CCK2R model are shown. The view shown in A is from the extracellular side of the receptors (top view) looking down through extracellular loop 2, which has been cut away. The view in B is from the membrane side of the receptors with helices 4 and 5 cut away. CCK1R and CCK2R are displayed in ribbon and stick representation and are colored orange and green, respectively. BDZ-1 and BDZ-2 are displayed in stick representation, and the carbon atoms are colored orange and green, respectively. Hydrogen bonds are displayed as spheres and colored according to the receptor and ligand. The BDZ compounds are docked in similar poses in both receptor models. Both compounds have their C3 urea substituents pointing upward toward the top of the TM5–TM6 region and are flanked partially by ECL2 and ECL3. Because of the differences in the stereochemistry at C3, the benzodiazepine cores are flipped in the two compounds. For BDZ-1 in the CCK1R model, the C2 carbonyl makes a hydrogen bond contact with Asn333 (Asn6.55), and the N1 methyl group points down toward the floor of the ligand pocket. The benzo ring is positioned next to TM3 and the C5 phenyl ring points toward the TM2–TM7 region. In contrast, for BDZ-2 in the CCK2R model, N4 of the benzodiazepine makes a hydrogen bond contact with Asn353 (Asn6.55) and the C5 phenyl ring points down toward the bottom of the pocket. The benzo ring is located close to TM3, and the N1 methyl group points toward the TM2–TM7 region.
This also resulted in differing shapes of the binding pockets in the two receptors, with the volumes between TM3 and TM5, TM3 and TM6, and TM6 and TM7 being larger for the CCK2R than the CCK1R and the pocket in the CCK1R protruding further into the extracellular loop region than the pocket in the CCK2R (Fig. 9). The different shapes of these pockets help to explain the receptor subtype selectivity imposed by differing stereochemistry at position 3 of the benzodiazepines.
FIGURE 9.
Intramembranous small molecule binding pockets of CCK1R and CCK2R models. The view in A is from the extracellular side of the receptors (top view) looking down through extracellular loop 2, which has been cut away. The view in B is from the membrane side of the receptors with helices 4 and 5 cut away. The ligand binding pockets of CCK1R and CCK2R are displayed as a wire mesh, colored orange and green, respectively. The ligands and receptor side chains are color-coded as described in the legend for Fig. 8. The pockets are similar in overall shape, but CCK2R has a larger volume between TM3 and TM5, TM3 and TM6, and TM6 and TM7. The CCK1R pocket protrudes further into the extracellular loop region compared with the CCK2R pocket.
The receptor residues in contact with the ligands, BDZ-1 docked at the CCK1R and BDZ-2 docked at the CCK2R, are illustrated in Fig. 10. This again emphasizes the differences in the determinants for docking the two ligands to these two receptors. Of note, the residue in position 2.61 within the helical bundle of CCK1R was in contact with BDZ-1, whereas the analogous residue in CCK2R did not exhibit substantial contact with BDZ-2. This was also true for the residues in positions 6.51 and 7.43. Intrahelical residues in positions 3.36, 6.48, and 7.35 of CCK2R were found to be in contact with BDZ-2, whereas the analogous residues in CCK1R did not exhibit substantial contact with BDZ-1. Selected residues within extracellular loops 2 and 3 of both receptors were also shown to establish contact with their respective ligands, as indicated in Fig. 10.
FIGURE 10.
CCK1R and CCK2R residues predicted to be in direct contact with benzodiazepine ligands based on molecular modeling. Shown are alignments of the predicted transmembrane segments (enclosed in boxes) and associated extracellular loops (ECL) of CCK1R and CCK2R. Residues that are predicted to be in direct contact with the corresponding ligand, defined as having 10% or more of its solvent-accessible area covered upon ligand binding, are shaded. The most conserved TM residues are identified by TM number and position number (_.50) based on the nomenclature of Ballesteros and Weinstein (28). The residues that were mutated in the current work are also shown in italics. The binding patches in the TM bundle that are common to both receptors include residues 3.29, 3.32, 5.38, 5.42, 6.52, 6.55, 6.58, 6.59, and 7.39. TM bundle residues that are specific for BDZ-1 in CCK1R are Asn2.61, Tyr3.30, Gly3.33, Ile6.51, and Tyr7.43. For CCK2R, the only TM bundle residue specific for BDZ-2 binding was Ile7.35.
The NSQ_AUC scores, reflecting the performance of these final models, are shown in Table 3. The NSQ_AUC values can range from 100 to −50, with a score of 0 indicating that the model cannot differentiate positives from decoys and a score of 100 indicating that the model prefers the positives and excludes the decoys. The CCK1R model had NSQ_AUC scores of 66.5 and 13.9 for CCK1R ligands and CCK2R ligands, respectively, indicating that the model has a much better ability to differentiate CCK1R ligands from decoys than to differentiate CCK2R ligands from decoys. Conversely, the CCK2R model had NSQ_AUC scores of 20.6 and 61.5 for CCK1R ligands and CCK2R ligands, respectively, supporting its preference for CCK2R ligands. For the key mutant models, the selectivity scores were reversed, as would be predicted. For the model of the mutant CCK1R (N2.61T, T3.28V, T3.29S, I6.51V, F6.52Y, and L7.39H), having experimental CCK2R-like benzodiazepine selectivity, the scores were 25.6 and 64.3 for CCK1R and CCK2R ligands, respectively. This supports its reversal of selectivity, with its being better at selecting CCK2R ligands from decoys. For the model of the mutant CCK2R (V6.51I, Y6.52F, and H7.39L), having experimental CCK1R-like benzodiazepine selectivity, the scores were 63.4 and 16.9 for CCK1R and CCK2R ligands, respectively. This supports its reversal of selectivity, with its being better at selecting CCK1R ligands from decoys.
TABLE 3.
Docking performance of wild type and key mutant CCK receptor models
| Model | NSQ_AUC for CCK1R ligandsa | NSQ_AUC for CCK2R ligandsa |
|---|---|---|
| CCK1R | 66.5 | 13.9 |
| CCK2R | 20.6 | 61.5 |
| CCK1R → CCK2R | 25.6 | 64.3 |
| CCK2R → CCK1R | 63.4 | 16.9 |
a NSQ_AUC values range from 100 to −50, where a score of 100 indicates a model with the maximal ability to differentiate positives from decoys; a score of 0 indicates a random model with no preference for positives or decoy.
ROC curves plotting the rate of finding true positives versus the rate of finding false positives for these models are shown in Fig. 11. These curves for individual models show the preference of each model for a particular ligand set. As expected, the wild type CCK1R (Fig 11A) and mutant CCK2R (Fig 11C) models preferred CCK1R ligands over CCK2R ligands, whereas the CCK2R (Fig 11B) and mutant CCK1R (Fig 11D) models preferred CCK2R ligands over CCK1R ligands.
FIGURE 11.
ROC curves for CCK1R and CCK2R models and their most informative mutant constructs with BDZ compounds. The curves for an individual model show the preference of that model for a particular ligand set. Blue curves, 53 CCK1R ligands versus 115 decoys. Red curves, 76 CCK2R ligands versus 171 decoys. As expected, the wild type CCK1R (A) and mutant CCK2R (V6.51I, Y6.52F, and H7.39L) (C) models preferred CCK1R ligands over CCK2R ligands, whereas the CCK2R (B) and mutant CCK1R (N2.61T, T3.28V, T3.29S, I6.51V, F6.52Y, and L7.39H) (D) models preferred CCK2R ligands over CCK1R ligands.
The top two panels of Fig. 12 illustrate the comparisons of the docking poses of BDZ-1 at the wild type CCK1R and the chimeric CCK2R TM(6,7) construct, which reverses its benzodiazepine selectivity, and the docking poses of BDZ-2 at the wild type CCK2R and the chimeric CCK1R TM(2,3,6,7) construct, which reverses its benzodiazepine selectivity. The poses of BDZ-1 are identical, whereas those of BDZ-2 are quite similar within the primary pharmacophore of the benzodiazepine and most divergent for the C3 urea substituent that structure-activity series have shown to be extraneous to action. The lower two panels of Fig. 12 illustrate the high degree of similarity between the docking of BDZ-1 and the Merck benzodiazepine antagonist L364,718, at the CCK1R and that between the docking of BDZ-2 and the Merck antagonist L365,260 at the CCK2R.
FIGURE 12.
Comparisons of docking poses for benzodiazepine ligands. The view is from the extracellular side of the receptors (top view) looking down through extracellular loop 2, which has been cut away. A shows the wild type CCK1R versus chimeric CCK2R construct reversing benzodiazepine selectivity. Shown are the structures of BDZ-1 docked at the wild type CCK1R (orange) and CCK2R TM(6,7) (gray). Mutated residues V6.51I, V6.52F, and H7.39L are displayed as gray sticks. This ligand is shown to dock in exactly the same pose in both models. B shows wild type CCK2R versus chimeric CCK1R construct reversing benzodiazepine selectivity. Shown are the structures of BDZ-2 docked at the wild type CCK2R (green ribbon) and CCK1R TM(2,3,67) (blue ribbon). Mutated residues N2.61T, T3.28V, T3.29S, I6.51V, F6.52Y, and L7.39H are displayed as blue sticks. C shows how the docking of L364,718 (cyan stick) relates to that of BDZ-1 (orange stick) at CCK1R (orange ribbon). Both ligands dock in the same poses. D shows how the docking of L365,260 (red stick) relates to that of BDZ-2 (green stick) at the CCK2R (green ribbon). Both ligands dock in the same pose.
DISCUSSION
Understanding the molecular basis for ligand binding and activation of receptors can contribute substantially to the development of receptor-active drugs. The selectivity of binding and spectrum of action of such drugs are quite important. In recent years, it has become clear that drugs acting at allosteric sites that are distinct from the sites of action of natural hormonal ligands can gain additional levels of selectivity of binding, exhibit subsets of the spectrum of action of natural agonists, and even modulate the actions of natural agonist ligands (44, 45). Thus, allosteric ligands can provide unique therapeutic opportunities.
The model ligands that were the focus of the current work were closely related 1,4-benzodiazepine antagonists that differ only in the stereochemistry (S or R) of their 3-position side chains while exhibiting highly selective binding to either CCK1R or CCK2R, respectively (29). The current experimental approach included the use of two distinct radioiodinated ligands that bind directly to the small molecule ligand-binding pockets of interest in these receptors and the extensive mutagenesis of residues that represent candidates for contributing to the selectivity of binding of these ligands. Within this pocket, there are only six residues that differ between these two receptors, and these were systematically modified individually and in all combinations of groupings.
The initial effort to modify the residues within each TM segment individually, by exchanging the residue present in that position in the CCK1R or CCK2R, provided some useful insights. Indeed, the characteristics of binding these ligands were affected by some of these changes, supporting the proposed binding to the intramembranous pocket within the helical bundle (24–27). Of interest, the patterns of residue changes that had effects were distinct for the two ligands and for the two receptors. Reduced saturable binding of BDZ-1 radioligand was observed for CCK1R constructs with changes in TM6 (residues 6.51 and 6.52), TM3 (residues 3.28 and 3.29), or TM7 (residue 7.39), and reduced saturable binding of BDZ-2 radioligand was observed for CCK2R constructs with changes in TM2 or TM7 (residue 2.61 or 7.39, respectively). TM7 residue 7.39 appeared to be important for the selectivity of both receptors, whereas the other residues that caused loss of function were specific for each receptor.
Other publications have also described the importance of some of these residues for the binding of other benzodiazepine ligands to CCK receptors (25–27, 46, 47). Those studies, however, were more indirect, utilizing the orthosteric radioligand based on the natural peptide that is known to bind to extracellular loop and tail regions rather than a radioligand that binds directly to the intramembranous docking site of these small molecules. Two of the residues identified as important in the current work (residues 6.51 and 7.39) have also been identified as important for the binding of the benzodiazepine antagonist L364,718 to the CCK1R (25, 46, 47). This is helpful in confirming the more general relevance of these residues for small molecule ligand binding to this receptor. Insights into binding determinants of the CCK2R have also been reported. Effects on CCK peptide radioligand binding were described for an extensive set of mutations of transmembrane residues in CCK2R, replacing 58 divergent amino acids present in CCK2R with corresponding residues in CCK1R. Of note, eight changes (R1.35Q, T2.61N, S3.29T, S5.39H, V6.51I, Y6.52F, T6.56A, and H7.39L) shifted the ability of natural peptide agonists, CCK and gastrin, or benzodiazepine antagonists, L364,718 and L365,260, to compete for the binding of the peptide radioligand by more than 2.5-fold relative to that at the wild type CCK2R (48). Mutations decreasing the binding of L365,260, the CCK2R-selective benzodiazepine antagonist, included T2.61N, S5.39H, and H7.39L. Mutations increasing the binding of L364,718, the CCK1R-selective benzodiazepine antagonist, included R1.35Q, S3.29T, S5.39H, V6.51I, and Y6.52F. This, too, provides further evidence for the general relevance of this pocket for small molecule docking while emphasizing the ability of changes in many of these residues to affect indirectly the binding of different types of ligands. It is particularly noteworthy that an earlier study following the effects on CCK radioligand binding reported a critical effect of CCK2R residue 6.51 on benzodiazepine L365,260 binding (46), but the current study in which a benzodiazepine radioligand was utilized observed no such effect.
Although loss-of-function mutations provide information on the importance of specific TM residues/combinations of residues for maintenance of binding, such data can be difficult to interpret because of the potential for both loss of direct binding contacts and conformational effects on the binding pocket propagated through changes to interhelical packing. Gain-of-function observations are potentially more informative, although here, too, the effects can be indirect. The most impressive gain of function was produced by replacing the residue in TM2 at position 2.61 in the CCK1R with that present in CCK2R. This resulted in the ability of this CCK1R TM2 construct to saturably bind the BDZ-2 radioligand, the only construct not based on CCK2R structure to be capable of binding this ligand. Indeed, the most impressive loss of function of the CCK2R series of constructs was observed with the replacement of this residue. Thus, residue 2.61 appears to be both sufficient and necessary for high affinity binding of BDZ-2. However, using this residue as a tether in ligand docking was not effective in molecular modeling, and it likely reflects an indirect, rather than a direct effect on BDZ-2 docking. Presumably, the effect of this residue is through its impact on interhelical packing by changing the geometry of the actual ligand binding pocket or the kinetics or stability of the intermediate conformations leading to stable ligand binding. Although these single site modifications provided initial insights into the determinants for benzodiazepine ligand binding and selectivity, supporting the distinct nature of the molecular determinants, the effects of each construct were not robust enough to be confident of the distinct poses optimal for each ligand.
There is also direct evidence that small molecules bind to this region of the CCK1R, provided by photoaffinity labeling studies with benzophenone derivatives of 1,5-benzodiazepines, representing both antagonists and agonists of CCK1R (26). Although a segment of the receptor covalently labeled with the antagonist in this series was localized, such observations can only provide general insights into the region of labeling and does not elucidate the specific pose of the docked ligand.
Combining the chimeric receptor modifications confirmed the insights gained from the single TM segment chimeric constructs and provided better global understanding of the nature of the small molecule-binding pocket. BDZ-1 binding was most affected by the residues in TM6 and TM7, with manifestations of loss of function of the CCK1R when mutated and gain of function when these residues were introduced into the CCK2R. The binding affinity of BDZ-1 was not different for CCK2R TM(6,7) than for the wild type CCK1R. BDZ-2 binding was most affected by the residues in TM2 and TM7, with manifestations of loss of function of the CCK2R when mutated and gain of function when these residues were introduced into the CCK1R. CCK1R TM(2,7) gained a high binding affinity of BDZ-2 that was only slightly lower than that for wild type CCK2R. Introducing all six of the distinct residues from CCK2R into CCK1R resulted in further improvement in BDZ-2 binding affinity, not different from that of wild type CCK2R.
Another important finding in this work was that the selectivity of the allosteric ligand binding site can be modified independently of the orthosteric ligand binding to the CCK receptors. Indeed, the peptide binding selectivities were retained for these receptors while completely reversing their benzodiazepine binding selectivities. The demonstration of benzodiazepine-induced modification of the rate of dissociation of bound orthosteric peptide ligand at the key chimeric receptor constructs further supports the allosteric nature of the small molecule-binding sites as totally distinct from the peptide-binding sites for these receptors.
Of interest, the data were different for the type 2 CCK receptor, where the benzodiazepine selectivity could be changed while retaining the peptide binding selectivity characteristics of that receptor only when modifying the distinct residues in both TM6 and TM7. Adding the other distinct CCK1R residues present in TM2 and TM3 to the CCK2R resulted in disruption of the folding, trafficking, and/or binding of benzodiazepine and/or CCK-like radioligands. It is particularly interesting that the CCK2R TM(3,6,7) construct was folded normally and expressed on the cell surface, where its benzodiazepine binding was demonstrated as having reversed its selectivity to reflect normal high affinity binding of the CCK1R-selective BDZ-1, although not being capable of binding CCK. This may reflect the proposed differences in the modes of peptide docking with CCK1R and CCK2R (12, 20, 49). Unlike the CCK1R, where CCK is believed to bind to cell surface loops and amino-terminal tail regions, in the CCK2R, the peptide is believed to be directed more toward the helical bundle, where these mutations may interfere with CCK peptide binding.
Molecular modeling was guided by the experimental data generated in the current project. This has resulted in models that have selectivity compatible with extensive sets of potential small molecule ligands. These are substantially different from those models that have been proposed previously in the literature. In 2005, Archer-Lahlou et al. (50) proposed a computational model of the benzodiazepine devazepide docked with the CCK1R based entirely on indirect mutagenesis data. This model predicts that the C2 carbonyl group of the benzodiazepine ring forms a hydrogen bond with Arg336 in ECL3 and the N4 group interacts with Asn333 (Asn6.55) in TM6. The comparison of the Archer-Lahlou CCK1R model docked with devazepide (50) with the current CCK1R model docked with BDZ-1 shows that these antagonists occupy a similar region within the intramembranous helical bundle but are docked in distinctly different orientations.
The molecular models of CCK1R and CCK2R that were developed in the current work are very instructive and useful. They clearly illustrate the different shapes of the small molecule docking sites in these closely related receptors that were influenced by a very small number of distinct residues within the transmembrane segments comprising the helical bundle. The benzodiazepine C3 urea substituents of both docked ligands were directed upward, with their benzo rings directed toward TM3 of both the CCK1R and the CCK2R. The differing stereochemistry of these ligands at their benzodiazepine C3 positions resulted in a substantial change in the orientation of their benzodiazepine pharmacophores. At the CCK1R, the benzodiazepine C5 phenyl ring was directed toward TM2 and TM7, whereas at the CCK2R this ring was directed down toward the base of the pocket. Similarly, at the CCK1R, the benzodiazepine N1 methyl group was directed down toward the base of the pocket, whereas at the CCK2R this group was directed toward TM2 and TM7. In both receptors, TM7 residue 7.39 appeared to play an important direct role in ligand docking. In the CCK1R, it was in contact with the C5 phenyl group of the benzodiazepine, whereas in the CCK2R, it was in contact with the N1 methyl group of the benzodiazepine. Asn6.55, which is conserved in both receptors, also appeared to play important roles, forming a hydrogen bond with the C2 carbonyl of the benzodiazepine in the CCK1R and forming a hydrogen bond with the N4 of the benzodiazepine in the CCK2R. Although residue 2.61 appears to play a critically important role in the CCK2R, this is most likely indirect, as it does not make substantial contact with the benzodiazepine ligand in this model. It should be noted that, again based on an interpretation of indirect mutagenesis data, residue 2.61 in the CCK2R has been suggested to play an anchoring role for relatively large non-peptide ligands based on a dibenzobicyclo[2.2.2]octane skeleton (JB93,182 and JB93,242), but these are not structurally similar to benzodiazepines and may more closely reflect peptide binding to this receptor (51).
The currently proposed molecular models highlight the differential spatial geometry of the intramembranous interhelical binding pockets and the distinct residue interactions predicted for high affinity ligand binding. The compatibility of these models with a large number of small molecule ligands and their ability to discriminate subtype selectivity of these ligands are very exciting. These models will likely have high predictive value, both for the rational design and development of new drugs and for the refinement of existing lead compounds for this effort.
Acknowledgment
We thank Dr. F. Gao for performing preliminary experiments relevant to this project.
This work was supported, in whole or in part, by National Institutes of Health Grants DK32878 (to L. J. M.) and U54 GM094618 and U01 GM 094612 (to R. A.). This work was also supported by a grant from the Mayo Clinic (to L. J. M.) and by Grant 519461 from the Australian National Health and Medical Research Council (NHMRC) (to P. M. S.).
- GPCR
- G protein-coupled receptor
- AUC
- area under the curve
- BDZ
- benzodiazepine
- CCK
- cholecystokinin
- CCK1R
- type 1 cholecystokinin receptor
- CCK2R
- type 2 cholecystokinin receptor
- ICM
- internal coordinate mechanics
- NSQ
- normalized square root
- ROC
- receiver-operating characteristic
- TM
- transmembrane.
REFERENCES
- 1. Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C. (2007) High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Choe H. W., Kim Y. J., Park J. H., Morizumi T., Pai E. F., Krauss N., Hofmann K. P., Scheerer P., Ernst O. P. (2011) Crystal structure of metarhodopsin II. Nature 471, 651–655 [DOI] [PubMed] [Google Scholar]
- 3. Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Crystal structure of rhodopsin: A G protein-coupled receptor. Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 4. Rasmussen S. G., Choi H. J., Rosenbaum D. M., Kobilka T. S., Thian F. S., Edwards P. C., Burghammer M., Ratnala V. R., Sanishvili R., Fischetti R. F., Schertler G. F., Weis W. I., Kobilka B. K. (2007) Crystal structure of the human β2 adrenergic G-protein-coupled receptor. Nature 450, 383–387 [DOI] [PubMed] [Google Scholar]
- 5. Rosenbaum D. M., Cherezov V., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Yao X. J., Weis W. I., Stevens R. C., Kobilka B. K. (2007) GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science 318, 1266–1273 [DOI] [PubMed] [Google Scholar]
- 6. Warne T., Serrano-Vega M. J., Baker J. G., Moukhametzianov R., Edwards P. C., Henderson R., Leslie A. G., Tate C. G., Schertler G. F. (2008) Structure of a β1-adrenergic G-protein-coupled receptor. Nature 454, 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu B., Chien E. Y., Mol C. D., Fenalti G., Liu W., Katritch V., Abagyan R., Brooun A., Wells P., Bi F. C., Hamel D. J., Kuhn P., Handel T. M., Cherezov V., Stevens R. C. (2010) Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 330, 1066–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chang R. S., Lotti V. J., Chen T. B., Kunkel K. A. (1986) Characterization of the binding of [3H]-(+/−)-L-364,718: a new potent, nonpeptide cholecystokinin antagonist radioligand selective for peripheral receptors. Mol. Pharmacol. 30, 212–217 [PubMed] [Google Scholar]
- 9. Gaisano H. Y., Klueppelberg U. G., Pinon D. I., Pfenning M. A., Powers S. P., Miller L. J. (1989) Novel tool for the study of cholecystokinin-stimulated pancreatic enzyme secretion. J. Clin. Invest. 83, 321–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lotti V. J., Chang R. S. (1989) A new potent and selective non-peptide gastrin antagonist and brain cholecystokinin receptor (CCK-B) ligand: L-365,260. Eur. J. Pharmacol. 162, 273–280 [DOI] [PubMed] [Google Scholar]
- 11. Dufresne M., Seva C., Fourmy D. (2006) Cholecystokinin and gastrin receptors. Physiol. Rev. 86, 805–847 [DOI] [PubMed] [Google Scholar]
- 12. Miller L. J., Gao F. (2008) Structural basis of cholecystokinin receptor binding and regulation. Pharmacol. Ther. 119, 83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Berna M. J., Tapia J. A., Sancho V., Jensen R. T. (2007) Progress in developing cholecystokinin (CCK)/gastrin receptor ligands that have therapeutic potential. Curr. Opin. Pharmacol. 7, 583–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cawston E. E., Miller L. J. (2010) Therapeutic potential for novel drugs targeting the type 1 cholecystokinin receptor. Br. J. Pharmacol. 159, 1009–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morton M., Prendergast C., Barrett T. D. (2011) Targeting gastrin for the treatment of gastric acid-related disorders and pancreatic cancer. Trends Pharmacol. Sci. 32, 201–205 [DOI] [PubMed] [Google Scholar]
- 16. Eysselein V. E., Eberlein G. A., Hesse W. H., Schaeffer M., Grandt D., Williams R., Goebell H., Reeve J. R., Jr. (1990) Molecular variants of cholecystokinin after endogenous stimulation in humans: a time study. Am. J. Physiol. Gastrointest. Liver Physiol. 258, G951–G957 [DOI] [PubMed] [Google Scholar]
- 17. Rehfeld J. F., Sun G., Christensen T., Hillingsø J. G. (2001) The predominant cholecystokinin in human plasma and intestine is cholecystokinin-33. J. Clin. Endocrinol. Metab. 86, 251–258 [DOI] [PubMed] [Google Scholar]
- 18. Dockray G. J., Varro A., Dimaline R., Wang T. (2001) The gastrins: their production and biological activities. Annu. Rev. Physiol. 63, 119–139 [DOI] [PubMed] [Google Scholar]
- 19. Miller L. J., Lybrand T. P. (2002) Molecular basis of agonist binding to the type A cholecystokinin receptor. Pharmacol. Toxicol. 91, 282–285 [DOI] [PubMed] [Google Scholar]
- 20. Anders J., Blüggel M., Meyer H. E., Kühne R., ter Laak A. M., Kojro E., Fahrenholz F. (1999) Direct identification of the agonist binding site in the human brain cholecystokinin-B receptor. Biochemistry 38, 6043–6055 [DOI] [PubMed] [Google Scholar]
- 21. Harikumar K. G., Pinon D. I., Miller L. J. (2006) Fluorescent indicators distributed throughout the pharmacophore of cholecystokinin provide insights into distinct modes of binding and activation of type A and B cholecystokinin receptors. J. Biol. Chem. 281, 27072–27080 [DOI] [PubMed] [Google Scholar]
- 22. Freidinger R. M. (1989) Non-peptide ligands for peptide receptors. Trends Pharmacol. Sci. 10, 270–274 [DOI] [PubMed] [Google Scholar]
- 23. Aquino C. J., Armour D. R., Berman J. M., Birkemo L. S., Carr R. A., Croom D. K., Dezube M., Dougherty R. W., Jr., Ervin G. N., Grizzle M. K., Head J. E., Hirst G. C., James M. K., Johnson M. F., Miller L. J., Queen K. L., Rimele T. J., Smith D. N., Sugg E. E. (1996) Discovery of 1,5-benzodiazepines with peripheral cholecystokinin (CCK-A) receptor agonist activity. 1. Optimization of the agonist “trigger.” J. Med. Chem. 39, 562–569 [DOI] [PubMed] [Google Scholar]
- 24. Kopin A. S., Beinborn M., Lee Y. M., McBride E. W., Quinn S. M. (1994) The CCK-B/gastrin receptor. Identification of amino acids that determine nonpeptide antagonist affinity. Ann. N.Y. Acad. Sci. 713, 67–78 [DOI] [PubMed] [Google Scholar]
- 25. Jagerschmidt A., Guillaume-Rousselet N., Vikland M. L., Goudreau N., Maigret B., Roques B. P. (1996) His-381 of the rat CCKB receptor is essential for CCKB versus CCKA receptor antagonist selectivity. Eur. J. Pharmacol. 296, 97–106 [DOI] [PubMed] [Google Scholar]
- 26. Hadac E. M., Dawson E. S., Darrow J. W., Sugg E. E., Lybrand T. P., Miller L. J. (2006) Novel benzodiazepine photoaffinity probe stereoselectively labels a site deep within the membrane-spanning domain of the cholecystokinin receptor. J. Med. Chem. 49, 850–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gao F., Sexton P. M., Christopoulos A., Miller L. J. (2008) Benzodiazepine ligands can act as allosteric modulators of the Type 1 cholecystokinin receptor. Bioorg. Med. Chem. Lett. 18, 4401–4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ballesteros J. A., Weinstein H. (1995) Integrated Methods for the Construction of Three-dimensional Models and Computational Probing of Structure-Function Relations in G Protein-coupled Receptors. Methods Neurosci. 25, 366–428 [Google Scholar]
- 29. Akgün E., Körner M., Gao F., Harikumar K. G., Waser B., Reubi J. C., Portoghese P. S., Miller L. J. (2009) Synthesis and in vitro characterization of radioiodinatable benzodiazepines selective for type 1 and type 2 cholecystokinin receptors. J. Med. Chem. 52, 2138–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Powers S. P., Pinon D. I., Miller L. J. (1988) Use of N,O-bis-Fmoc-d-Tyr-ONSu for introduction of an oxidative iodination site into cholecystokinin family peptides. Int. J. Pept. Protein Res. 31, 429–434 [DOI] [PubMed] [Google Scholar]
- 31. Cheng Z. J., Harikumar K. G., Holicky E. L., Miller L. J. (2003) Heterodimerization of type A and B cholecystokinin receptors enhance signaling and promote cell growth. J. Biol. Chem. 278, 52972–52979 [DOI] [PubMed] [Google Scholar]
- 32. Ulrich C. D., Ferber I., Holicky E., Hadac E., Buell G., Miller L. J. (1993) Molecular cloning and functional expression of the human gallbladder cholecystokinin A receptor. Biochem. Biophys. Res. Commun. 193, 204–211 [DOI] [PubMed] [Google Scholar]
- 33. Hadac E. M., Ghanekar D. V., Holicky E. L., Pinon D. I., Dougherty R. W., Miller L. J. (1996) Relationship between native and recombinant cholecystokinin receptors: role of differential glycosylation. Pancreas 13, 130–139 [DOI] [PubMed] [Google Scholar]
- 34. Munson P. J., Rodbard D. (1980) Ligand: a versatile computerized approach for characterization of ligand-binding systems. Anal. Biochem. 107, 220–239 [DOI] [PubMed] [Google Scholar]
- 35. Abagyan R., Totrov M., Kuznetsov D. (1994) ICM: A new method for protein modeling and design. Applications to docking and structure prediction from the distorted native conformation. J. Comput. Chem. 15, 488–506 [Google Scholar]
- 36. Abagyan R., Totrov M. (1994) Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J. Mol. Biol. 235, 983–1002 [DOI] [PubMed] [Google Scholar]
- 37. Metropolis N., Rosenbluth A. W., Rosenbluth M. N., Teller A. H., Teller E. (1953) Equation of state calculations by fast computing machines. J. Chem. Phys. 21, 1087–1092 [Google Scholar]
- 38. Katritch V., Rueda M., Lam P. C., Yeager M., Abagyan R. (2010) GPCR 3D homology models for ligand screening: lessons learned from blind predictions of adenosine A2a receptor complex. Proteins 78, 197–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cardozo T., Totrov M., Abagyan R. (1995) Homology modeling by the ICM method. Proteins 23, 403–414 [DOI] [PubMed] [Google Scholar]
- 40. An J., Totrov M., Abagyan R. (2005) Pocketome via comprehensive identification and classification of ligand binding envelopes. Mol. Cell Proteomics 4, 752–761 [DOI] [PubMed] [Google Scholar]
- 41. Overington J. (2009) ChEMBL. An interview with John Overington, team leader, chemogenomics at the European Bioinformatics Institute Outstation of the European Molecular Biology Laboratory (EMBL-EBI). Interview by Wendy A. Warr. J. Comput. Aided Mol. Des. 23, 195–198 [DOI] [PubMed] [Google Scholar]
- 42. Truchon J. F., Bayly C. I. (2007) Evaluating virtual screening methods: good and bad metrics for the “early recognition” problem. J. Chem. Inf. Model. 47, 488–508 [DOI] [PubMed] [Google Scholar]
- 43. Katritch V., Jaakola V. P., Lane J. R., Lin J., Ijzerman A. P., Yeager M., Kufareva I., Stevens R. C., Abagyan R. (2010) Structure-based discovery of novel chemotypes for adenosine A(2A) receptor antagonists. J. Med. Chem. 53, 1799–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kenakin T., Miller L. J. (2010) Seven transmembrane receptors as shape-shifting proteins: the impact of allosteric modulation and functional selectivity on new drug discovery. Pharmacol. Rev. 62, 265–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Keov P., Sexton P. M., Christopoulos A. (2011) Allosteric modulation of G protein-coupled receptors: a pharmacological perspective. Neuropharmacology 60, 24–35 [DOI] [PubMed] [Google Scholar]
- 46. Beinborn M., Lee Y. M., McBride E. W., Quinn S. M., Kopin A. S. (1993) A single amino acid of the cholecystokinin-B/gastrin receptor determines specificity for non-peptide antagonists. Nature 362, 348–350 [DOI] [PubMed] [Google Scholar]
- 47. Mantamadiotis T., Baldwin G. S. (1994) The seventh transmembrane domain of gastrin/CCK receptors contributes to non-peptide antagonist binding. Biochem. Biophys. Res. Commun. 201, 1382–1389 [DOI] [PubMed] [Google Scholar]
- 48. Kopin A. S., McBride E. W., Quinn S. M., Kolakowski L. F., Jr., Beinborn M. (1995) The role of the cholecystokinin-B/gastrin receptor transmembrane domains in determining affinity for subtype-selective ligands. J. Biol. Chem. 270, 5019–5023 [DOI] [PubMed] [Google Scholar]
- 49. Dong M., Liu G., Pinon D. I., Miller L. J. (2005) Differential docking of high-affinity peptide ligands to type A and B cholecystokinin receptors demonstrated by photoaffinity labeling. Biochemistry 44, 6693–6700 [DOI] [PubMed] [Google Scholar]
- 50. Archer-Lahlou E., Tikhonova I., Escrieut C., Dufresne M., Seva C., Pradayrol L., Moroder L., Maigret B., Fourmy D. (2005) Modeled structure of a G-protein-coupled receptor: the cholecystokinin-1 receptor. J. Med. Chem. 48, 180–191 [DOI] [PubMed] [Google Scholar]
- 51. Foucaud M., Marco E., Escrieut C., Low C., Kalindjian B., Fourmy D. (2008) Linking non-peptide ligand binding mode to activity at the human cholecystokinin-2 receptor. J. Biol. Chem. 283, 35860–35868 [DOI] [PubMed] [Google Scholar]