Background: CD154, an immuno-inflammatory molecule, binds to four receptors.
Results: CD154 differentially binds its various receptors and is capable of simultaneously interacting with multiple ones, inducing synergistic responses in monocytes.
Conclusion: The simultaneous engagement of CD154 receptors can create a cross-talk between them.
Significance: Concomitant binding of CD154 to multiple receptors is greatly significant in therapies of CD154-related diseases.
Keywords: Cell Signaling, Immunology, Inflammation, Monocytes, Mutagenesis, α5β1, αIIbβ3, CD154, CD40
Abstract
In addition to its classical CD40 receptor, CD154 also binds to αIIbβ3, α5β1, and αMβ2 integrins. Binding of CD154 to these receptors seems to play a key role in the pathogenic processes of chronic inflammation. This investigation was aimed at analyzing the functional interaction of CD154 with CD40, αIIbβ3, and α5β1 receptors. We found that the binding affinity of CD154 for αIIbβ3 is ∼4-fold higher than for α5β1. We also describe the generation of sCD154 mutants that lost their ability to bind CD40 or αIIbβ3 and show that CD154 residues involved in its binding to CD40 or αIIbβ3 are distinct from those implicated in its interaction to α5β1, suggesting that sCD154 may bind simultaneously to different receptors. Indeed, sCD154 can bind simultaneously to CD40 and α5β1 and biologically activate human monocytic U937 cells expressing both receptors. The simultaneous engagement of CD40 and α5β1 activates the mitogen-activated protein kinases, p38, and extracellular signal-related kinases 1/2 and synergizes in the release of inflammatory mediators MMP-2 and -9, suggesting a cross-talk between these receptors.
Introduction
CD154, also known as CD40 ligand (CD40L) or gp39, is a type II transmembrane protein that belongs to the tumor necrosis factor (TNF) superfamily, expressed on a variety of activated hematopoietic and nonhematopoietic cells (1, 2). CD154 expression on T cells is triggered primarily by T cell receptor signaling and is regulated by CD28-dependent and -independent pathways. In platelets and a subpopulation of T cells, CD154 is stored and rapidly translocates to the cell membrane following cellular activation (3, 4). In addition to membrane-bound CD154 (mCD154), soluble CD154 (sCD154), which originates from a matrix metalloproteinase-dependent proteolytic cleavage of mCD154, has been found at high levels in several inflammatory conditions (5–7). Both mCD154 and sCD154 are found to be as a homotrimers (8), and such property is an absolute requirement for its biological activity.
For almost 2 decades, CD40 was considered the sole receptor for CD154 (1). CD40 is a type I transmembrane protein and a member of the TNF receptor superfamily. It is expressed on the surface of many immune and nonimmune cells, including B-lymphocytes, monocytes/macrophages, dendritic cells, platelets, epithelial cells, as well as endothelial cells (2, 9, 10). Most biological functions of CD154 have been attributed to its direct interaction with CD40. However, studies using CD154−/− and CD40−/− mice have suggested that CD154 may also bind other receptors (11). In support of this hypothesis, it has been demonstrated that sCD154 interacts with αIIbβ3 (GPIIb/IIIa), an integrin expressed on the megakaryocyte-platelet lineage, mast cells, and hematopoietic progenitor cells (12). CD154−/− mice exhibit increased bleeding times and unstable thrombi, thus highlighting the physiological relevance of the CD154/αIIbβ3 interaction (12).
Based on the above observations, and on earlier studies showing that CD154 induces CD40-independent cytokine production in human monocytes (13, 14), we hypothesized that CD154 may bind to yet another unknown receptor on human monocytes. In support of our hypothesis, we reported that sCD154 binds to CD40-negative human monocytic U937 cells, a binding that is mediated by the α5β1 integrin (15). Binding of sCD154 to α5β1 induced translocation of α5β1 to the Triton X-100-insoluble fraction in U937 cells (CD40−, α5β1+), as well as rapid activation of the mitogen-activated protein kinase (MAPK)-extracellular signal regulated kinase (ERK1/2) and IL-8 gene expression (15). Additionally, the leukocyte αMβ2 integrin (also named Mac-1) has also been identified as a receptor for CD154 (16, 17). This integrin was found to be implicated in atherosclerosis development via its role in leukocyte adhesion to the endothelial layer and their subsequent trans-endothelial migration (18). Taken together, these results clearly indicate that CD154 interacts with four different receptors (CD40, αIIbβ3, α5β1, and αMβ2) of biological relevance.
Although the three newly described receptors (αIIbβ3, α5β1, and αMβ2) all belong to the integrin superfamily, CD154 binds to each of them in a specific manner. CD154 binds to the active and inactive forms of αIIbβ3 (12), whereas its binding to αMβ2 is mainly mediated by the active form of the integrin (16). Activation of α5β1, which increases its ability to interact with fibronectin, completely abolished CD154 binding (15). In this investigation, we present data showing the affinity of CD154 to α5β1 and demonstrate that CD154 residues involved in its interaction with α5β1 are distinct from those implicated in its interaction with CD40 and αIIbβ3. According to the topological location of the residues involved in sCD154 binding to CD40 and αIIbβ3, these findings may indicate that sCD154 simultaneously binds different receptors (CD40, α5β1, and/or αIIbβ3). Simultaneous interaction of CD154 with CD40 and α5β1 on CD40-transfected U937 cells generates a cross-talk between these receptors and induces rapid p38 and ERK1/2 activation, which lead to enhanced MMP-2 and MMP-9 secretion.
EXPERIMENTAL PROCEDURES
Cells
The myelomonocytic cell line U937 (ATCC, Manassas, VA) and the B cell lymphoma cell line BJAB (from Dr. J. Menezes, Sainte-Justine Hospital, Montréal, Quebec, Canada) were maintained in RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum (FBS), l-glutamine, penicillin, and streptomycin (Wisent, St-Bruno, Quebec, Canada).
Plasmids and Mutagenesis
The pMTBip/V5-His-(A) vector was purchased from Invitrogen. pMTBip/V5-His-(A)-sCD154 wild type (sCD154WT) was generated by subcloning sCD154WT at BglII and XhoI sites within the pMTBip/V5-His-(A) vector. Prior to cloning, the 450-bp sequence encoding the soluble form of wild type CD154 (Met-113 to Leu-261) was first amplified by PCR from pSecTag2A/CD154, using sCD154 primers (Table 1) containing a BglII and XhoI restriction site, respectively. The cDNA encoding mutants of sCD154 (D117E, R203A, Y145A, and R203A/Y145A (R/Y)) were generated by PCR overlap extension methods using the primers described in Table 1. The PCR product was then cloned in pMTBip/V5-His-(A) at BglII and XhoI. Nucleotide sequence was confirmed by ABI DNA sequencing.
TABLE 1.
Primers for cloning CD154 wild type and mutants
| Sequence name | Primer sequence |
|---|---|
| sCD154wt | Forward, 5′-C GGG AGA TCT ATG CAA AAA GGT GAT-3′ |
| Reverse, 5′-TAG ACT CGA GTT TGA GTA AGC C- 3′ | |
| sCD154-D117E | Forward, 5′-C GGG AGA TCT ATG CAA AAA GGT GAA-3′ |
| Reverse, 5′-TAG ACT CGA GTT TGA GTA AGC C-3′ | |
| sCD154-R203A | Forward, 5′-GAG TAA GAT TGC CTC GAA TCT-3′ |
| Reverse, 5′-AGA TTC GAG GCA ATC TTA CTC-3′ | |
| sCD154-Y145A | Forward, 5′-GAA AAA GGA GCC TAC ACC ATG-3′ |
| Reverse, 5′-CAT GGT GTA GGC TCC TTT TTC-3′ |
Cell Culture, Transfection, and Protein Purification
Schneider's (S2) cells (1.5–3 × 106 cells/ml) were cultured in complete DES Schneider's Drosophila medium (Invitrogen), supplemented with 10% heat-inactivated FBS (Wisent Inc., St-Bruno, Quebec, Canada), 100 units of penicillin G sodium, 100 μg/ml streptomycin sulfate, and 0.25 μg/ml amphotericin B as fungicide (Invitrogen) at 26 °C without CO2. To produce recombinant soluble CD154 (rsCD154) containing different point mutations, cells were co-transfected with pMT-BiP/V5–6× HisA carrying the sCD154 DNA sequence and pCoHygro vectors (Invitrogen) at a weight ratio of 19:1, respectively, by calcium phosphate. The clones were then selected by 300 μg/ml hygromycin B (Wisent Inc., St-Bruno, Quebec, Canada). The expression of rsCD154 was tested by seeding 1–2 × 106 cells in a 100-mm Petri dish followed by induction with 100 mm copper sulfate for 24 h. Cell debris was removed, and Western blot analysis of the generated supernatant was performed. A stable population of hygromycin-resistant S2 cells was obtained after a 20-day period. The selected cultures were initiated to a large scale-up in serum-free medium (Drosophila-SFM) (Invitrogen) supplemented with antibiotics and antimycotic without hygromycin B. Upon induction, the proteins released in the supernatant were recovered by centrifugation at 3000 × g for 20 min. The samples were then filtered through a 0.22-μm filter and concentrated to one-twentieth of the initial volume with a Centricon concentrator. The hexahistidine tag within the recombinant sCD154 protein was used for purification with the HisTrap-FF column (GE Healthcare). Purity of all recombinants, CD154 mutants as well as recombinants obtained from commercial sources, was assessed by Coomassie Blue staining.
Reagents and Antibodies
Anti-CD154 hybridoma 5C8 (IgG2a) and anti-CD40 hybridoma G28.5 (IgG1) antibodies were obtained from ATCC. The isotype controls anti-TSST-1 hybridoma 2H8 (IgG1) and anti-SEB hybridoma 8C12 (IgG2a) antibodies were developed in our laboratory. The following antibodies (Abs)3 were purchased: goat anti-mouse IgG-fluorescein isothiocyanate (FITC) Ab (Sigma); rabbit anti-phospho-p38 Abs, anti-p38 Abs, rabbit anti-phospho-ERK1/2, anti-ERK1/2, and JBS5 anti-α5β1 Abs (Cell Signaling Technology, Inc., Beverly, MA); and goat anti-rabbit IgG-HRP Ab and goat anti-mouse IgG-HRP Ab (Santa Cruz Biotechnology, Santa Cruz, CA). Soluble α5β1 and αIIbβ3 were produced as described previously (19, 20). Recombinant soluble CD40-Fc was from R&D Systems (Minneapolis, MN). Avidin was procured from Sigma, and the Alexa Fluor-488 labeling kit came from Molecular Probes (Molecular Probes, Eugene, OR). Alexa Fluor-488 labeling of rsCD154 (rsCD154-A) and avidin (Avidin-A) was performed according to the manufacturer's instructions.
BIAcore Analysis
The affinity and kinetic analyses of the interactions between rsCD154 and α5β1 and between rsCD154 and αIIbβ3 were determined at 25 °C using the surface plasmon resonance detections with a BIAcore 3000 instrument (GE Healthcare). Briefly, rsCD154 was immobilized (∼980 response units) onto a CM4 sensor chip (GE Healthcare), as described for other systems (21). The concentrations used for the injected analyte samples are α5β1 (0.63 μm to 80 nm) and αIIbβ3 (0.18 μm to 23 nm), with 2-fold dilutions. Association and dissociation rates (Kon/Koff), as well as the dissociation constant (KD), were obtained by global fitting of the surface plasmon resonance data from multiple concentrations to a simple 1:1 Langmuir binding model, using the BIAevaluation software Version 4.1.
Flow Cytometric Analysis
Cell surface analysis was performed with specific mAbs or Alexa-labeled rsCD154-A, as described previously (22). Washed cells were then analyzed on a FACSort. For competition binding assay, cells (2 × 105 cells/100 ml) were incubated in binding assay medium (RPMI 1640, HEPES 10 mm, bovine serum albumin (BSA 1%)) containing 20 ng of labeled rsCD154-A or Alexa-labeled avidin-A in the presence or absence of a 10-fold molar excess of unlabeled rsCD154, sCD40-Fc, α5β1, or αIIbβ3 for 1 h at 37 °C in a humidified incubator (5% CO2 atmosphere). Washed cells were analyzed on a FACSort (BD Biosciences). For binding analysis, cells were incubated with sCD154 (CD154WT, CD154 D117E, CD154 R203A, CD154 Y145A, or and CD154 R203A/Y145A) in staining medium for 30 min at 4 °C for BJAB cells and at 37 °C for U937 cells. Washed cells were then incubated with biotinylated anti-CD154 poly-Ab for 30 min at 4 °C, followed by streptavidin-phycoerythrin (Sigma) for 30 min. Sample were then washed and analyzed on a FACSort.
Cell Stimulation
U937 and BJAB cells were incubated in serum-free medium for 4 h at 37 °C and stimulated with rsCD154WT or mutants at different concentrations (15–500 ng/5 × 105 cells/ml) for 1, 2, 5, or 10 min at 37 °C. The stimulation was stopped by the addition of heated/warmed 2× SDS sample buffer containing 10% 2-mercaptoethanol, and a mixture of protease (Roche Applied Science) and phosphatase inhibitors (Sigma) inhibitors. After boiling for 7 min, cell lysates were separated by SDS-PAGE for Western blot analysis.
Immunoblot Analysis
Polyvinylidene fluoride (PVDF) membranes were blocked in 5% skim milk in Tris saline, pH 7.5, 0.15% Tween 20 (Fisher) for 1 h at room temperature. The phosphorylation of p38 and ERK1/2 was assessed by immunoblotting using phospho-specific Abs according to the manufacturer's instructions. Membranes were stripped (62 mm Tris-HCl, pH 6.8, 2% SDS, 100 mm 2-mercaptoethanol, 30 min, 50 °C) and reprobed with antibodies recognizing total p38 or ERK1/2. Antigen-antibody complexes were revealed by ECL (GE Healthcare).
Analysis of MMP-2 and -9 Secretion by Zymography
U937 cells (106 cells/ml) were stimulated with 100 or 500 ng/ml of anti-CD40 mAb G28.5, anti-α5β1 mAb JBS5, or with both mAbs in RPMI serum-free medium at 37 °C for 48 h. The presence of MMP-2 and -9 was assessed by gelatin zymography (23). Briefly, 45 μl of the different supernatants were analyzed at 4 °C on an 8% SDS-polyacrylamide gel containing 2 mg/ml gelatin (Sigma). Gels were then washed twice for 15 min with 2.5% Triton X-100 wash buffer to extract SDS and allow gelatinases to re-nature within the gel. Thereafter, the gel was incubated overnight in enzyme assay buffer (38 mm Trizma hydrochloride, pH 7.5, 13 mm CaCl2 anhydride, 0.05% NaN3) at 37 °C to allow development of enzyme activity bands. Following incubation, gels were washed and stained with 0.2% Coomassie Brilliant Blue (methanol/acetic acid/water, 4:1:6) for 1 h on a shaker at room temperature. Gels were then destained in 40% methanol with 10% acetic acid until detection of gelatinase activity, appearing as clear bands against a dark background. Finally, the gel was scanned, and the density of the bands was measured (Bio-Rad GS-800 and QuantityOne Analysis Software).
RESULTS
CD154 Molecules Bind to Their Receptors with Different Affinity
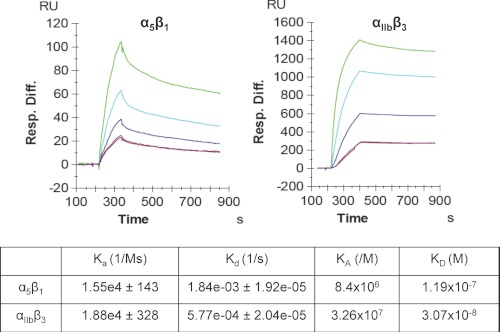
The ability of a ligand to bind several receptors is not unique for CD154, as it has been reported for various cytokines (24) and different cell surface molecules such as CD28/CTLA4:B7.1/B7.2 (25), BAFF/APRIL:BAFF-R/BCMA/TACI (26), and PD-1:PD-L1/PD-L2 (27). Although CD28 and CTLA-4 bind to the same receptors, B7.1 and B7.2, CTLA-4 shows a higher affinity to B7.1 and B7.2 receptors than does CD28. The binding affinity of CD154 to its receptors and the nature of this interaction are not well known. To determine the binding affinity of CD154/receptors, we used the surface plasma resonance approach by immobilizing soluble purified CD154 and injecting increasing concentrations of soluble α5β1 and αIIbβ3. We found that the binding affinity of CD154 for αIIbβ3 (3.07 × 10−8 m) is ∼4-fold higher than for α5β1 (1.19 × 10−7 m) (Fig. 1).
FIGURE 1.
Binding affinity and kinetic analysis of the interactions between sCD154 and α5β1 and between sCD154 and αIIbβ3, as determined by the surface plasmon resonance approach. Soluble purified sCD154 was immobilized (980 response units) onto biosensor chips (GE Healthcare) and α5β1 (630, 315, 157, and 78.75 nm) and αIIbβ3 (180, 90, 45, and 23 nm) at different concentrations were injected. Sensograms were fitted to a 1:1 binding model to obtain on- and off-rates (Kon and Koff). KD was calculated as koff/kon.
Binding of CD154 to BJAB Cells Is Inhibited by sCD40, whereas Its Binding to U937 Cells Is Inhibited by α5β1 and αIIbβ3
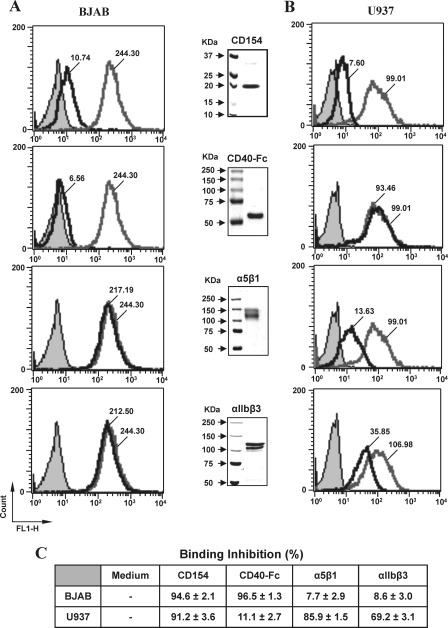
To verify the possible concomitant binding of sCD154 to two receptors and determine the nature of the interaction of CD154 with various receptors, we first investigated the ability of soluble CD40-Fc, α5β1, or αIIbβ3 to modulate the binding of sCD154 to CD40 or α5β1 expressed on the cell surface. For this purpose, BJAB cells (a B cell line expressing only CD40) and U937 cells (a monocytic cell line expressing only α5β1) were used as models. Soluble CD154-Alexa was added to BJAB or U937 cells alone or in the presence of unlabeled sCD154, sCD40-Fc, α5β1, or αIIbβ3. Purity of these recombinant proteins was also determined by Coomassie Blue staining (Fig. 2, middle blots). After incubation, washed cells were analyzed by flow cytometry. Our results indicate that the binding of sCD154-Alexa to BJAB cells was only inhibited by sCD40-Fc (94.6 ± 2.1% inhibition, Fig. 2, A and C). In contrast, binding of sCD154-Alexa to U937 cells was unaffected by sCD40-Fc (11.1 ± 2.7% inhibition) but significantly blocked by α5β1 (85.9 ± 1.5% inhibition) and, to a lesser extent, by αIIbβ3 (69.2 ± 3.1% inhibition) (Fig. 2, B and C). Competition analysis in the presence of unlabeled sCD154 (10-fold molar excess) was also assessed to show binding specificity of sCD154-Alexa (Fig. 2, A and B, top panels). As expected, binding to both BJAB and U937 cells was completely block by unlabeled CD154 (94.6 ± 2.1% inhibition and 91.2 ± 3.6% inhibition, respectively). Taken together, these results strongly suggest that sCD154 may concomitantly bind CD40 and α5β1 or CD40 and αIIbβ3. Hence, it is highly likely that the residues involved in its interaction with CD40 are different from those implicated in its interaction with α5β1 and αIIbβ3.
FIGURE 2.
CD154 binding to U937 cells is inhibited by soluble α5β1 and αIIbβ3, whereas binding to BJAB cells is inhibited by sCD40. BJAB (A) or U937 cells (B) were incubated for 1 h with labeled rsCD154-Alexa (200 ng/ml) in the absence (Media) or presence of 2 μg/ml of sCD154, sCD40-Fc, α5β1, or αIIbβ3. For competition analysis with CD154, cells were preincubated with 10-fold molar excess of uncoupled sCD154 (2 μg/ml) prior to incubation with labeled rsCD154-Alexa (200 ng/ml), although CD40-Fc, α5β1, and αIIbβ3 were co-incubated with labeled rsCD154-Alexa prior to incubation with cells. Cells were then washed and analyzed by flow cytometry for CD154 binding. Filled gray plots indicate avidin-Alexa staining (negative control); empty gray plots show the binding of rsCD154-Alexa in the absence of competitors; and empty black plots represent rsCD154-Alexa binding in the presence of unlabeled competitors (CD154, CD40-Fc, α5β1, or αIIbβ3). Values within plots represent the mean fluorescence intensity for rsCD154-Alexa binding in the absence or presence of unlabeled competitors. Middle blots show the expression of the recombinant competitors. Proteins (2 μg total) were resolved on 6% (CD40-Fc, α5β1, and αIIbβ3) or 12% (CD154) SDS-polyacrylamide gels and stained with Coomassie Blue (R-250). Data presented in C show the mean percentage of inhibition (±S.E.) in the presence of unlabeled competitors. All results shown are representative of four independent experiments.
Distinct CD154 Residues Are Involved in CD40, α5β1, and αIIbβ3 Binding
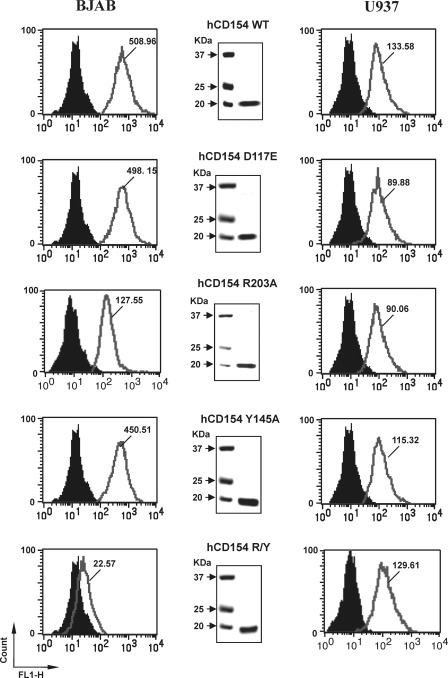
The CD154 residues involved in CD40 binding have been initially studied by site-directed mutagenesis (28–30) and more recently by co-crystal analysis (31). However, with respect to the CD154/αIIbβ3 interaction, structural analysis has only been analyzed by site-directed mutagenesis (12). According to these analysis, CD154 residues involved in CD40 binding are centered on at least two clusters of residues (Lys-143, Lys-145, Lys-146 and Arg-203, Gln-220) (28–31), whereas the CD154/αIIbβ3 interaction is mediated by the RGD residues of murine CD154 and the KGD residues of human CD154 (at positions 115–117) (12). Residue Asp-117 seems to play a major role in the CD154/αIIbβ3 interaction, because substitution of Asp-117 by alanine or glutamic acid abolishes the interaction (12). The RGD and KGD motifs of α5β1 do not appear to mediate CD154 binding, because activation of α5β1 with Mn2+ or DTT abolished binding (15). Therefore, the ability of αIIbβ3 to inhibit the binding of sCD154-Alexa to α5β1 on U937 cells, as observed in the above experiments, might be due to hindrance and/or binding competition for shared residues between both receptors. To verify these hypotheses and to investigate whether CD154 residues involved in CD40 or αIIbβ3 binding are distinct from those involved in binding to α5β1, recombinant sCD154 mutants have been generated, purified, and used for binding and functional analysis. These CD154 mutants include the following: 1) D117A or D117E, which lost the ability to interact with αIIbβ3, and 2) Y145A, R203A, or Y145A/R203A double mutant, which abolished CD154 binding to CD40. Binding of sCD154WT or mutants to BJAB and U937 cells was assayed using flow cytometry, as described above. Our results indicate that all sCD154 mutants were capable of binding to U937 cells (Fig. 3). In contrast, as expected, sCD154 R203A/Y145A mutants lost their ability to bind to BJAB cells, thereby confirming that CD154 residues involved in the α5β1 interaction are distinct from those required for binding to CD40 and αIIbβ3. Similar results were also generated by ELISAs using receptor-bound plates in the presence of CD154 and its mutants (data not shown).
FIGURE 3.
Binding of CD154 to α5β1 is independent of residues Tyr-145, Arg-203, and Asp-117. BJAB or U937 cells were incubated with rsCD154 (250 ng/sample) and wild type or mutant (hCD154 D117E, hCD154 R203A, hCD154 Y145A, and hCD154 R203A/Y145A) for 30 min at 4 °C for BJAB cells and 30 min at 37 °C for U937 cells. Washed cells were then incubated with biotinylated anti-CD154 polyclonal Ab for 30 min at 4 °C, followed by streptavidin-phycoerythrin for 30 min and washed and analyzed by flow cytometry. Filled black plots indicate the cells incubated with streptavidin-phycoerythrin (negative control), as compared with the empty gray plots that represent the binding of rsCD154 to BJAB and U937 cells. Values within plots represent the mean fluorescence intensity for rsCD154 binding (wild type or mutants). Middle plots show the expression of the recombinant competitors. Proteins (2 μg total) were resolved on 12% SDS-polyacrylamide gels and stained with Coomassie Blue (R-250). All results shown are representative of four independent experiments.
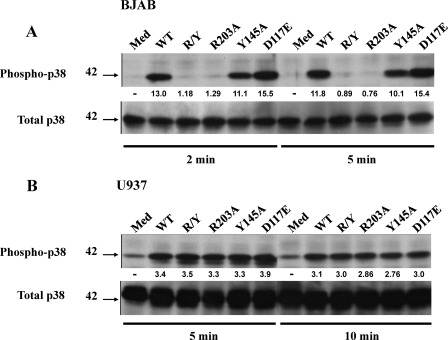
CD154 Mutants Maintain Their Ability to Induce p38 MAPK Phosphorylation in U937 Cells but Not in BJAB Cells
To further support the hypotheses drawn from the above experiments, we investigated the effect of these sCD154 mutants in cellular activation. U937 and BJAB cells were stimulated with sCD154WT or its derived mutants. In contrast to sCD154WT and to a lesser extent sCD154 Y145A that induced p38 phosphorylation in BJAB cells, sCD154 R203A and R203A/Y145A failed to stimulate such responses in these cells (Fig. 4A). However, sCD154WT as well as all sCD154 mutants described above induced p38 phosphorylation in U937 cells (Fig. 4B). Altogether, these results confirmed that CD154 residues involved in the α5β1 interaction are distinct from those required for binding to CD40 and αIIbβ3.
FIGURE 4.
CD154 mutants (R203A and R203A/Y145A) induce p38 MAPK phosphorylation in U937 cell, but not in CD40+ BJAB cells. BJAB (A) or U937 (B) cell lines were stimulated at 37 °C with 250 ng of sCD154WT or each of its mutants for the indicated time points. Cell lysates were separated by SDS-PAGE and analyzed by immunoblot using an anti-phospho-p38-specific antibody. Membranes were stripped and re-hybridized with an anti-p38 specific antibody. Medium (Med) was used as negative control. The values below the blots represent arbitrary units of increase in phosphorylation relative to total protein levels, as measured by densitometry using the Quantity One analysis software (Bio-Rad). Blots are representative of three independent experiments.
Simultaneous Binding of CD154 to α5β1 and CD40 Induces a Cross-talk between Both Receptors
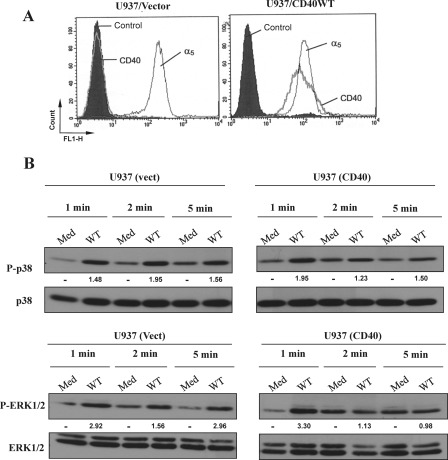
Our recent studies clearly indicated that engagement of α5β1 with sCD154 on human monocytic U937 cells triggers ERK1/2 and p38 MAPK phosphorylation, which leads to IL-8 gene expression (15). Here, we investigated the outcome of sCD154WT binding to cells expressing both CD40 and α5β1. Vector-transfected U937 (CD40−/α5β1+) or CD40-transfected U937 cell lines (CD40+/α5β1+) (Fig. 5A) were stimulated with sCD154WT, and the phosphorylation of the MAPK p38 and ERK1/2 was analyzed (Fig. 5B). Stimulation of vector-transfected U937 cells with sCD154WT induced phosphorylation of both p38 and ERK1/2 at 1 min that persisted up to 5 min. However, stimulation of U937 cells expressing both CD40 and α5β1 with sCD154WT induced phosphorylation of p38 and ERK1/2 that reached maximal responses at 1 min and quickly decreased at 2 and 5 min (Fig. 5B).
FIGURE 5.
Simultaneous ligation of CD154 to CD40 and α5β1 on CD40-transfected U937 cells modulates CD154-induced MAPK signaling. A, U937 cells were transfected with an empty vector (U937/vector) or with CD40WT (U937/CD40WT) and analyzed for cell surface expression of CD40 and α5β1 by flow cytometry. Cells were analyzed using specific anti-CD40 mAbs (G28.5) or anti-α5β1 mAbs (JBS5). B, vector (vect)-transfected or CD40-transfected U937 cells were stimulated at 37 °C with CD154 WT (250 ng/sample) for the indicated time points. Medium (Med) was used as a negative control. Cells were lysed, and total cell lysates were analyzed by immunoblot using anti-phospho-p38-specific and anti-phospho-ERK1/2-specific Abs. Anti-p38 and anti-ERK1/2 total antibodies were used to control the sample loading. The values below the blots represent arbitrary units of increase in phosphorylation relative to total protein levels, as measured by densitometry using the Quantity One analysis software (Bio-Rad). Results are representative of three independent experiments.
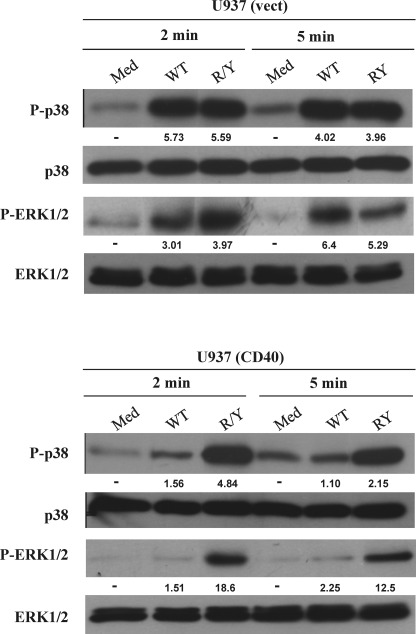
To delineate the mechanisms involved in the above observations, both U937 cell lines were activated with sCD154WT or sCD154 R/Y mutant. As with sCD154WT, stimulation of vector-transfected U937 cells with sCD154 R/Y mutant induced phosphorylation of both p38 and ERK1/2 at 2 min that persisted up to 5 min. (Fig. 6). However, in contrast to sCD154WT, which decreased phosphorylation of p38 and ERK1/2 at 2 and 5 min in CD40-transfected U937 cells, sCD154 R/Y mutant stimulated CD40-transfected U937 cells with persistent phosphorylation of p38 and ERK1/2 up to 5 min (Fig. 6). There are two possible explanations for the rapidly decreased phosphorylation of p38 and ERK1/2 at 2 and 5 min upon simultaneous binding of sCD154 to CD40 and α5β1. Soluble CD154WT could ligate simultaneously CD40 and α5β1 without cross-linking the receptors or sCD154 could cross-link two different receptors on the cell surface. In both cases, signals induced via each receptor could modulate each other; modulation was reflected by changes in the kinetics of the activation of the MAPK p38 and ERK1/2 pathways. Taken together, these results strongly suggest a cross-talk between the CD154 receptors when ligated with their ligand.
FIGURE 6.
Simultaneous binding of CD154WT to both receptors alters p38 and ERK1/2 phosphorylation at 2 and 5 min. CD40-transfected U937 cells and its control (vector (vect)) were stimulated with CD154WT (WT) or with sCD154 R/Y mutant at 37 °C for the indicated time. Medium (Med) was used as negative control. Cells were lysed, and total cell lysates were analyzed by immunoblot using specific Abs against phosphorylated p38, total p38, phosphorylated ERK1/2, and total ERK1/2. The values below the blots represent arbitrary units of an increase in phosphorylation relative to total protein levels, as measured by densitometry using the Quantity One analysis software (Bio-Rad). Blots are representative of three independent experiments.
Simultaneous Ligation of α5β1 and CD40 on U937 Cells with Specific Abs Alters the Activation of MAPKs
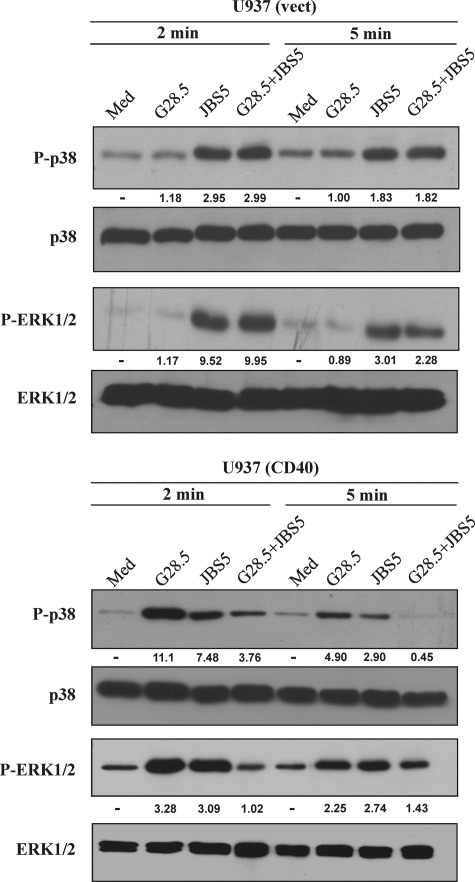
To investigate the first possibility, vector-transfected and CD40-transfected U937 cells were stimulated with the specific anti-CD40 mAb G28.5 or with the anti-α5β1 mAb JBS5, alone or in combination. Our results showed that engaging α5β1 or CD40 individually was capable of inducing phosphorylation of p38 and ERK1/2 in U937 cells, indicating that CD40 expression on these cells is functionally linked to the MAPK pathway, similarly to that on BJAB cells. In contrast, the combination of both mAbs inhibited the phosphorylation of p38 and ERK1/2 at 2 and 5 min (Fig. 7). Although our results are in support of the first possibility, they do not rule out the possible cross-linking of two receptors by sCD154WT.
FIGURE 7.
Simultaneous engagement of CD40 and α5β1 with specific antibodies alters p38 and ERK1/2 phosphorylation in CD40-transfected U937. Transfected U937 cell lines were stimulated at 37 °C with 500 ng of anti-CD40 mAb (G28.5) or anti-α5β1 mAb (JBS5), alone or in combination, as indicated. Cells were lysed, and total cell lysates were analyzed by immunoblot using specific Abs as mentioned above in Fig. 5. Anti-total p38 and anti-total ERK1/2 Abs were used to control the sample loading. The values below the blots represent arbitrary units of an increase in phosphorylation relative to total protein levels, as measured by densitometry using the Quantity One analysis software (Bio-Rad). Blots are representative of three independent experiments.
Simultaneous Ligation of CD40 and α5β1 with Specific Abs Synergizes in MMP-2 and -9 Secretion by U937-transfected Cells
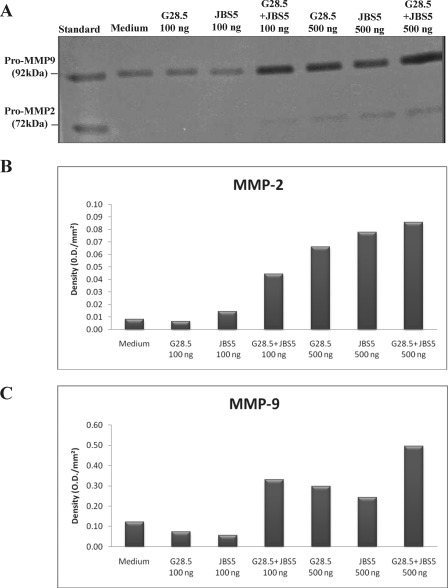
It is well established that ligation of CD40 or α5β1 on human monocytes or monocytic cell lines leads to the secretion of several inflammatory proteins, such as MMP-2 and -9 (32). In addition, the expression and secretion of these two metalloproteinases are known to be mediated by p38-dependent pathways (33–35). To further assess the cellular response that could be induced in our U937 cells, we analyzed the release of MMP-2 and -9. For this purpose, cells were stimulated with different concentrations of specific anti-CD40 mAb G28.5 and anti-α5β1 mAb JBS5 alone or in combination. Supernatants were collected after 48 h of stimulation and analyzed for MMP-2 and -9 secretions by gelatin zymography. When the specific Abs were used alone at a suboptimal concentration (100 ng/ml), no detectable MMP-2 or -9 secretion was observed, whereas the combination of both mAbs induced a synergistic secretion of both MMP-2 and -9 (Fig. 8). Under optimal concentrations (500 ng/ml), each mAb induced a significant up-regulation of matrix metalloproteinases, and their combination leads to an additive response (Fig. 8). Together, these results confirm that both CD40 and α5β1 are linked to functional activities in monocytic U937 cells and that simultaneous stimulation of both receptors, even under suboptimal conditions, can induce a synergistic cellular response.
FIGURE 8.
Simultaneous ligation of CD40 and α5β1 with specific antibodies synergizes in MMP-2 and MMP-9 secretion in U937 cells. CD40-transfected U937 cells were analyzed at base line (medium) or upon stimulation with anti-CD40 (G28.5) or anti-α5β1 (JBS5), alone or in combination, for 48 h at 37 °C. 45 μl of the different supernatants were analyzed at 4 °C on an 8% SDS-polyacrylamide gel containing 2 mg/ml gelatin. Gelatin zymography of MMP-2 and MMP-9 is represented in A. Densitometry measurements of MMP-2 and MMP-9 secreted by U937 cells are represented in B and C, respectively. This figure is representative of three independent experiments.
DISCUSSION
CD40 was initially thought to be the sole receptor for CD154. However, studies from different groups showed that CD154 also interacts with three members of the integrin family, α5β1, αIIbβ3, and αMβ2 (12, 15, 16). The nature of these interactions and their biological responses remain fairly uncharacterized. In this study, by using the BIAcore approach we determined and compared the binding affinity between CD154 and α5β1 and between CD154 and other receptors. We also showed that the CD154 residues involved in its interaction with α5β1 are different from those implicated in CD154 interaction with CD40 and αIIbβ3. We further studied the functional outcome of simultaneous binding of sCD154 to two receptors.
In general, high affinity binding of ligands to receptors is often physiologically important when some of the binding energy can be used to cause a conformational change in the receptor, resulting in altered behavior of an associated ion channel or enzyme. High affinity binding implies that a relatively low concentration of a ligand is adequate to maximally occupy a ligand-binding site and trigger a physiological response. Low affinity binding implies that a relatively high concentration of a ligand is required before the binding site is maximally occupied, and the maximum physiological response to the ligand is achieved. The binding affinity of CD154 for CD40 has been previously evaluated by plasmon resonance on BIAcore biosensors and appears to be in the range of 10–30 nm (36). Here, we found similar binding affinities for αIIbβ3 (30 nm), as reported by Andre et al. (12). Interestingly, our binding data indicate that the affinity of CD154 for α5β1 is around 120 nm, which is ∼4-fold lower than the affinity of CD154 for CD40 and αIIbβ3. These results do not undermine the importance of α5β1 in CD154-driven reactions, given that this integrin can be expressed at higher levels than CD40 on the cell surface, as is the case for monocytes/macrophages and T-lymphocytes, which are of great importance in inflammatory diseases. Moreover, integrin signaling and activity are known to mediate crucial aspects of inflammation, indicating that even though CD154 shows lower binding affinities for α5β1, this interaction may be of utmost importance in certain conditions. In accordance with this argument, it has recently been shown that the binding affinity of CD154 for αMβ2 is in the range of 200 nm (37), which would make αMβ2 the lowest affinity receptor for CD154. Nevertheless, the CD154/αMβ2 interaction appears crucial for leukocyte recruitment and atherogenesis in mice, thus supporting the concept that each receptor may mediate a particular element of CD154-driven inflammation, regardless of their binding affinities.
With the TNF-β/TNF receptor interaction in mind, Bajorath et al. (29) have identified the CD40 and CD154 residues that are important for receptor-ligand binding, using structure-based sequence alignments, a molecular model of CD154, site-directed mutagenesis, and receptor-ligand binding assays. They identified residues in CD154 (Lys-143 and Tyr-145) and CD40 (Tyr-82, Asp-84, and Asn-86) involved in CD154/CD40 interactions (29). The same group has extended their mutagenesis analysis of the CD154/CD40 interaction, where they identified additional CD154 (Tyr-146, Arg-203, and Gln-220) and CD40 (Glu-74 and Glu-117) residues that contribute to such interaction. They also explored the importance of the CD154 residue Tyr-145 in the CD154/CD40 interaction by conservatively replacing this residue with Phe (30). These findings are in harmony with the recent co-crystal analysis of An et al. (31), and our current results show the importance of Tyr-145 and Arg-203 residues in binding of CD154 to CD40. Andre et al. (31) reported that binding of αIIbβ3 to CD154 is mediated through the RGD (murine) or KGD (human) sequence present on CD154 (12). Residue Asp-117 of CD154 seems to be the major binding player (12), and substitution of Asp-117 to Glu or Ala was shown to diminish integrin binding activity (38). Our results showed that this substitution failed to affect the binding of CD154 mutants to α5β1 expressed in U937 cells. This is supported by our earlier study showing that divalent cation (Mn2+), which induces activation of α5β1 and increases its ability to interact with fibronectin, inhibited CD154 binding to α5β1 (15). Indeed, our studies also suggested that distinct CD154 residues are implicated in the interaction of CD154 with CD40, αIIbβ3, α5β1, and probably αMβ2. Here, we disregarded the comparison between CD154 residues implicated in CD154/CD40, CD154/α5β1, or CD154/αIIbβ3 interactions and those involved in CD154/αMβ2 binding for two reasons. First, in contrast to α5β1, only the active form of αMβ2 binds to CD154 (16), and second, the residues implicated in the binding of CD154 to αMβ2 are still unknown and will be the subject of further investigations by our group.
Interestingly, sCD154 can simultaneously bind to α5β1 and CD40, as shown by our cell binding and activation experiments. The simultaneous binding of sCD154 to α5β1 and CD40 confirms that sCD154 interacts with α5β1 outside the CD40-binding site. These results raise the possibility that sCD154 trimers may serve as molecular bridges between CD40 and α5β1 from different cell types, triggering as such signal transduction in these cells. Alternatively, sCD154 may cross-link these two receptors (or perhaps three receptors) on the same cell, thus inhibiting, modulating, or inducing signal-related events via both receptors. This phenomenon is potentially of great importance when evaluating the role of CD154 in cells expressing more than one CD154 receptor and should be carefully monitored when designing therapeutic strategies directed against these cells. Indeed, our results have shown that simultaneous ligation of sCD154 to CD40 and α5β1 induces maximal p38 and ERK1/2 MAPK phosphorylation faster than ligation of each receptor alone. One of the interesting findings of this study is the synergistic response observed with respect to MMP-2 and MMP-9 secretion when both receptors were ligated. These data suggest new biological mechanisms of interaction between CD154 and its receptors, as other ligands function differently upon binding to one or more of their receptors. For example, CD28, a ligand of at least two members of the B7 family, acts as an activating molecule, whereas CTLA-4, another ligand of the B7 family, acts as an inhibitory molecule, and such responses seem to be influenced by the affinity of ligand/receptor interaction (39, 40). In addition, APRIL and TACI interact with Syndecan-1 on the surface of multiple myeloma cells to form an essential survival loop (41). The efficiency of many cell-surface receptors is sometimes dependent on the rate of binding soluble or surface-attached ligands (42).
In conclusion, this study describes the differential binding of CD154 to its various receptors and suggests that CD154 can interact with all of its receptors simultaneously. These findings bear an intriguing importance with regard to anti-CD154 therapies. Indeed, CD154 interfering agents, in particular anti-CD154 antibodies, have been used for the treatment or prevention of various CD154-associated diseases such as systemic lupus erythematosus, arthritis, and multiple sclerosis (43, 44). However, the use of such therapies has been accompanied with thromboembolic complications, possibly resulting from the interaction of the antibody with the platelet Fc receptor or the formation of unstable thrombi, as described earlier (45). These observations press the need for the development of novel therapeutic strategies aiming at inhibiting a particular CD154/receptor interaction implicated in a given cellular response, without affecting other CD154 interactions. To address this issue, we generated CD154 mutants that lost the ability to specifically bind CD40 or αIIbβ3, without affecting binding to α5β1. We are currently also investigating CD154 residues implicated in its binding to α5β1 and Mac-1. These mutants will specifically interfere with the binding of endogenous CD154 to a specific receptor, while preserving its capacity to bind and activate other CD154 receptors, thus reducing undesired complications associated with anti-CD154 therapies.
Acknowledgments
We thank Angelic Bellemare for the generation of CD154 mutants and CD154 vectors. We thank the Immunology Core at the Wadsworth Center for BIAcore binding studies. We also thank Dr. Ghada Hassan for critical reading of the manuscript.
This work was supported by the Canadian Institutes of Health Research and the Canadian Arthritis Network.
- Ab
- antibody.
REFERENCES
- 1. van Kooten C., Banchereau J. (1997) Functional role of CD40 and its ligand. Int. Arch. Allergy Immunol. 113, 393–399 [DOI] [PubMed] [Google Scholar]
- 2. Schönbeck U., Libby P. (2001) The CD40/CD154 receptor/ligand dyad. Cell. Mol. Life Sci. 58, 4–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hermann A., Rauch B. H., Braun M., Schrör K., Weber A. A. (2001) Platelet CD40 ligand (CD40L)-subcellular localization, regulation of expression, and inhibition by clopidogrel. Platelets 12, 74–82 [DOI] [PubMed] [Google Scholar]
- 4. André P., Nannizzi-Alaimo L., Prasad S. K., Phillips D. R. (2002) Platelet-derived CD40L. The switch-hitting player of cardiovascular disease. Circulation 106, 896–899 [DOI] [PubMed] [Google Scholar]
- 5. Menchén L., Marín-Jiménez I., Arias-Salgado E. G., Fontela T., Hernández-Sampelayo P., Rodríguez M. C., Butta N. V. (2009) Matrix metalloproteinase 9 is involved in Crohn's disease-associated platelet hyperactivation through the release of soluble CD40 ligand. Gut 58, 920–928 [DOI] [PubMed] [Google Scholar]
- 6. Matthies K. M., Newman J. L., Hodzic A., Wingett D. G. (2006) Differential regulation of soluble and membrane CD40L proteins in T cells. Cell. Immunol. 241, 47–58 [DOI] [PubMed] [Google Scholar]
- 7. Danese S., Fiocchi C. (2005) Platelet activation and the CD40/CD40 ligand pathway. Mechanisms and implications for human disease. Crit. Rev. Immunol. 25, 103–121 [DOI] [PubMed] [Google Scholar]
- 8. Hsu Y. M., Lucci J., Su L., Ehrenfels B., Garber E., Thomas D. (1997) Heteromultimeric complexes of CD40 ligand are present on the cell surface of human T lymphocytes. J. Biol. Chem. 272, 911–915 [DOI] [PubMed] [Google Scholar]
- 9. Noelle R. J. (1996) CD40 and its ligand in host defense. Immunity 4, 415–419 [DOI] [PubMed] [Google Scholar]
- 10. Noelle R. J., Mackey M., Foy T., Buhlmann J., Burns C. (1997) CD40 and its ligand in autoimmunity. Ann. N.Y. Acad. Sci. 815, 384–391 [DOI] [PubMed] [Google Scholar]
- 11. Mehlhop P. D., van de Rijn M., Brewer J. P., Kisselgof A. B., Geha R. S., Oettgen H. C., Martin T. R. (2000) CD40L, but not CD40, is required for allergen-induced bronchial hyper-responsiveness in mice. Am. J. Respir. Cell Mol. Biol. 23, 646–651 [DOI] [PubMed] [Google Scholar]
- 12. André P., Prasad K. S., Denis C. V., He M., Papalia J. M., Hynes R. O., Phillips D. R., Wagner D. D. (2002) CD40L stabilizes arterial thrombi by an α3 integrin-dependent mechanism. Nat. Med. 8, 247–252 [DOI] [PubMed] [Google Scholar]
- 13. Ribbens C., Dayer J. M., Chizzolini C. (2000) CD40-CD40 ligand (CD154) engagement is required but may not be sufficient for human T helper 1 cell induction of interleukin-2- or interleukin-15-driven, contact-dependent, interleukin-1β production by monocytes. Immunology 99, 279–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lindmark E., Tenno T., Siegbahn A. (2000) Role of platelet P-selectin and CD40 ligand in the induction of monocytic tissue factor expression. Arterioscler. Thromb. Vasc. Biol. 20, 2322–2328 [DOI] [PubMed] [Google Scholar]
- 15. Léveillé C., Bouillon M., Guo W., Bolduc J., Sharif-Askari E., El-Fakhry Y., Reyes-Moreno C., Lapointe R., Merhi Y., Wilkins J. A., Mourad W. (2007) CD40 ligand binds to α5β1 integrin and triggers cell signaling. J. Biol. Chem. 282, 5143–5151 [DOI] [PubMed] [Google Scholar]
- 16. Zirlik A., Maier C., Gerdes N., MacFarlane L., Soosairajah J., Bavendiek U., Ahrens I., Ernst S., Bassler N., Missiou A., Patko Z., Aikawa M., Schönbeck U., Bode C., Libby P., Peter K. (2007) CD40 ligand mediates inflammation independently of CD40 by interaction with Mac-1. Circulation 115, 1571–1580 [DOI] [PubMed] [Google Scholar]
- 17. Sandilands G. P., McCrae J., Hill K., Perry M., Baxter D. (2006) Major histocompatibility complex class II (DR) antigen and costimulatory molecules on in vitro and in vivo activated human polymorphonuclear neutrophils. Immunology 119, 562–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sotiriou S. N., Orlova V. V., Al-Fakhri N., Ihanus E., Economopoulou M., Isermann B., Bdeir K., Nawroth P. P., Preissner K. T., Gahmberg C. G., Koschinsky M. L., Chavakis T. (2006) Lipoprotein(a) in atherosclerotic plaques recruits inflammatory cells through interaction with Mac-1 integrin. FASEB J. 20, 559–561 [DOI] [PubMed] [Google Scholar]
- 19. Stupack D. G., Cheresh D. A. (2002) Get a ligand, get a life. Integrins, signaling, and cell survival. J. Cell Sci. 115, 3729–3738 [DOI] [PubMed] [Google Scholar]
- 20. Krokhin O. V., Cheng K., Sousa S. L., Ens W., Standing K. G., Wilkins J. A. (2003) Mass spectrometric based mapping of the disulfide bonding patterns of integrin α chains. Biochemistry 42, 12950–12959 [DOI] [PubMed] [Google Scholar]
- 21. Li H., Zhao Y., Guo Y., Li Z., Eisele L., Mourad W. (2007) Zinc induces dimerization of the class II major histocompatibility complex molecule that leads to cooperative binding to a superantigen. J. Biol. Chem. 282, 5991–6000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Léveillé C., Chandad F., Al-Daccak R., Mourad W. (1999) CD40 associates with the MHC class II molecules on human B cells. Eur. J. Immunol. 29, 3516–3526 [DOI] [PubMed] [Google Scholar]
- 23. Abou-Saleh H., Théorêt J. F., Yacoub D., Merhi Y. (2005) Neutrophil P-selectin-glycoprotein-ligand-1 binding to platelet P-selectin enhances metalloproteinase 2 secretion and platelet-neutrophil aggregation. Thromb. Haemost. 94, 1230–1235 [DOI] [PubMed] [Google Scholar]
- 24. Groppe J., Hinck C. S., Samavarchi-Tehrani P., Zubieta C., Schuermann J. P., Taylor A. B., Schwarz P. M., Wrana J. L., Hinck A. P. (2008) Cooperative assembly of TGF-β superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol. Cell 29, 157–168 [DOI] [PubMed] [Google Scholar]
- 25. Greenwald R. J., Freeman G. J., Sharpe A. H. (2005) The B7 family revisited. Annu. Rev. Immunol. 23, 515–548 [DOI] [PubMed] [Google Scholar]
- 26. Bossen C., Schneider P. (2006) BAFF, APRIL and their receptors: structure, function, and signaling. Semin. Immunol. 18, 263–275 [DOI] [PubMed] [Google Scholar]
- 27. Keir M. E., Butte M. J., Freeman G. J., Sharpe A. H. (2008) PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 26, 677–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bajorath J. (1998) Detailed comparison of two molecular models of the human CD40 ligand with an x-ray structure and critical assessment of model-based mutagenesis and residue mapping studies. J. Biol. Chem. 273, 24603–24609 [DOI] [PubMed] [Google Scholar]
- 29. Bajorath J., Chalupny N. J., Marken J. S., Siadak A. W., Skonier J., Gordon M., Hollenbaugh D., Noelle R. J., Ochs H. D., Aruffo A. (1995) Identification of residues on CD40 and its ligand, which are critical for the receptor-ligand interaction. Biochemistry 34, 1833–1844 [DOI] [PubMed] [Google Scholar]
- 30. Bajorath J., Marken J. S., Chalupny N. J., Spoon T. L., Siadak A. W., Gordon M., Noelle R. J., Hollenbaugh D., Aruffo A. (1995) Analysis of gp39/CD40 interactions using molecular models and site-directed mutagenesis. Biochemistry 34, 9884–9892 [DOI] [PubMed] [Google Scholar]
- 31. An H. J., Kim Y. J., Song D. H., Park B. S., Kim H. M., Lee J. D., Paik S. G., Lee J. O., Lee H. (2011) Crystallographic and mutational analysis of the CD40-CD154 complex and its implications for receptor activation. J. Biol. Chem. 286, 11226–11235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hassan G. S., Merhi Y., Mourad W. M. (2009) CD154 and its receptors in inflammatory vascular pathologies. Trends Immunol. 30, 165–172 [DOI] [PubMed] [Google Scholar]
- 33. Park J. E., Chang W. Y., Cho M. (2009) Induction of matrix metalloproteinase-9 by galectin-7 through p38 MAPK signaling in HeLa human cervical epithelial adenocarcinoma cells. Oncol. Rep. 22, 1373–1379 [DOI] [PubMed] [Google Scholar]
- 34. Hou X., Han Q. H., Hu D., Tian L., Guo C. M., Du H. J., Zhang P., Wang Y. S., Hui Y. N. (2009) Mechanical force enhances MMP-2 activation via p38 signaling pathway in human retinal pigment epithelial cells. Graefes Arch. Clin. Exp. Ophthalmol. 247, 1477–1486 [DOI] [PubMed] [Google Scholar]
- 35. Grabellus F., Worm K., Schmid K. W. (2007) Induction of the matrix metalloproteinase-2 activation system in arteries by tensile stress. Involvement of the p38 MAP kinase pathway. Pathol. Res. Pract. 203, 135–143 [DOI] [PubMed] [Google Scholar]
- 36. Khandekar S. S., Silverman C., Wells-Marani J., Bacon A. M., Birrell H., Brigham-Burke M., DeMarini D. J., Jonak Z. L., Camilleri P., Fishman-Lobell J. (2001) Determination of carbohydrate structures N-linked to soluble CD154 and characterization of the interactions of CD40 with CD154 expressed in Pichia pastoris and Chinese hamster ovary cells. Protein Expr. Purif. 23, 301–310 [DOI] [PubMed] [Google Scholar]
- 37. Wolf D., Hohmann J. D., Wiedemann A., Bledzka K., Blankenbach H., Marchini T., Gutte K., Zeschky K., Bassler N., Hoppe N., Rodriguez A. O., Herr N., Hilgendorf I., Stachon P., Willecke F., Duerschmied D., von zur Muhlen C., Soloviev D. A., Zhang L., Bode C., Plow E. F., Libby P., Peter K., Zirlik A. (2011) Binding of CD40L to Mac-1's I-domain involves the EQLKKSKTL motif and mediates leukocyte recruitment and atherosclerosis but does not affect immunity and thrombosis in mice. Circ. Res. 109, 1269–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Masuta Y., Kato K., Tomihara K., Nakamura K., Sasaki K., Takahashi S., Hamada H. (2007) Gene transfer of noncleavable cell surface mutants of human CD154 induces the immune response and diminishes systemic inflammatory reactions. J. Immunother. 30, 694–704 [DOI] [PubMed] [Google Scholar]
- 39. Alegre M. L., Fallarino F. (2006) Mechanisms of CTLA-4-Ig in tolerance induction. Curr. Pharm. Des. 12, 149–160 [DOI] [PubMed] [Google Scholar]
- 40. Lazetic S., Leong S. R., Chang J. C., Ong R., Dawes G., Punnonen J. (2002) Chimeric co-stimulatory molecules that selectively act through CD28 or CTLA-4 on human T cells. J. Biol. Chem. 277, 38660–38668 [DOI] [PubMed] [Google Scholar]
- 41. Moreaux J., Sprynski A. C., Dillon S. R., Mahtouk K., Jourdan M., Ythier A., Moine P., Robert N., Jourdan E., Rossi J. F., Klein B. (2009) APRIL and TACI interact with syndecan-1 on the surface of multiple myeloma cells to form an essential survival loop. Eur. J. Haematol. 83, 119–129 [DOI] [PubMed] [Google Scholar]
- 42. Robert P., Limozin L., Pierres A., Bongrand P. (2009) Biomolecule association rates do not provide a complete description of bond formation. Biophys. J. 96, 4642–4650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Howard L. M., Miller S. D. (2004) Immunotherapy targeting the CD40/CD154 costimulatory pathway for treatment of autoimmune disease. Autoimmunity 37, 411–418 [DOI] [PubMed] [Google Scholar]
- 44. Toubi E., Shoenfeld Y. (2004) The role of CD40-CD154 interactions in autoimmunity and the benefit of disrupting this pathway. Autoimmunity 37, 457–464 [DOI] [PubMed] [Google Scholar]
- 45. Boumpas D. T., Furie R., Manzi S., Illei G. G., Wallace D. J., Balow J. E., Vaishnaw A. (2003) A short course of BG9588 (anti-CD40 ligand antibody) improves serologic activity and decreases hematuria in patients with proliferative lupus glomerulonephritis. Arthritis Rheum. 48, 719–727 [DOI] [PubMed] [Google Scholar]