Background: β-Catenin and EGFR are often overexpressed and activated in human cancers, but the relation between them is not completely understood.
Results: β-Catenin targets EGFR gene for increased expression in GSK3β-inactivated prostate cancer cells.
Conclusion: GSK3β inactivation initiates β-catenin-EGFR cross-talk leading to enhanced survival and proliferation of prostate cancer cells.
Significance: Targeting the β-catenin-EGFR pathway may be a novel therapeutic approach for prostate cancer.
Keywords: Epidermal Growth Factor Receptor (EGFR), Proliferation, Protein Kinase A (PKA), Signaling, STAT3, Cross-talk, GSK3β, β-catenin-Ser552
Abstract
Wnt/β-catenin and EGFR pathways are important in cancer development and often aberrantly activated in human cancer. However, it is very important to understand the mechanism responsible for this activation and the relation between them. Here, we report the mechanism of EGFR expression by transcriptionally active β-catenin in GSK3β-inactivated prostate cancer cells that eventually leads to its enhanced proliferation and survival. Expressions of β-catenin and EGFR are elevated in various cancers specifically in prostate cancer cells, DU145. When GSK3β is inactivated in these cells, β-catenin gets stabilized, phosphorylated at Ser-552 by protein kinase A, accumulates in the nucleus, and regulates the expression of its target genes that include EGFR. Chromatin immunoprecipitation (ChIP) and promoter analysis revealed that the EGFR promoter gets occupied by transcriptionally active β-catenin when elevated in GSK3β-inactivated cells. This phenomenon not only leads to increased expression of EGFR but also initiates the activation of its downstream molecules such as ERK1/2 and Stat3, ultimately resulting in up-regulation of multiple genes involved in cell proliferation and survival.
Introduction
In canonical Wnt signaling, β-catenin is the key effector molecule, which has been implicated in a dual role in the cellular context. First identified as a major structural component of the cell-cell adherence junctions, it links members of the cadherin family of transmembrane adhesion proteins to the actin cytoskeleton (1). Disruption of the β-catenin destruction complex (consisting of APC, axin, and GSK3β)2 formation by inactivating GSK3β results in β-catenin stabilization. This facilitates its translocation into the nucleus where it acts as a transcriptional activator of T-cell factor (TCF)/lymphoid enhancer factor-mediated gene expression that controls an array of target genes, like c-jun, fra-1, c-myc, cyclin D1, etc., as well as genes involved in developmental stages and adult tissue homeostasis (2, 3). There are studies reporting aberrant activation of this signaling playing an important role in the development of colorectal cancer (4). Activating mutations in β-catenin, as well as inactivating mutations of its regulators such as APC, have been discovered in multiple human cancers (5–7). Recent studies suggest that phosphorylation of β-catenin by several kinases can modulate not only its stability but also its transcriptional activity (8–12). Several lines of evidence in human and animal models also indicate that aberrant activation of β-catenin plays an important role in the development of prostate cancer (PCa) (13, 14).
Similarly, deregulation of EGF receptor (EGFR) is often associated with carcinogenesis, which can be caused by its overexpression, gene amplification, mutations, or deletions (15). Increased expression of EGFR family members EGFR (Erb-B1) and Her2 (Erb-B2) have been found in majority of human cancers that include PCa, and this is associated with a more aggressive clinical outcome (16). Overexpression of either EGFR or Her2 leads to transformation of NIH-3T3 cells (17–18). Increased expressions of EGFR and its ligands (EGF and TGF-α) have been described in prostate tumors (19), and autocrine activation of EGFR signaling regulates the growth of androgen-independent PCa cell line, DU145 (20). It has also been reported that EGFR expression is elevated in androgen-independent prostate tumors (21).
Although there is an expansive evidence of literature deciphering the central role of β-catenin and EGFR in the architecture of intracellular signaling networks, little is known about their cross-talk, which may be involved in the regulation of cell proliferation and survival for tumorigenesis. In the present study, we investigate the role of β-catenin during GSK3β suppression and the mechanism of cross-talk with EGFR pathway in inducing cell proliferation and survival in PCa. The current data support an important role of β-catenin in enhanced survival and proliferation of PCa cells via up-regulation of EGFR gene expression that leads to activation of multiple downstream pathways.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, and Drug Treatments
The human cell lines HEK293 (embryonic kidney), MCF7 and MDA-MB-231 (breast cancer), DU145 and PC3 (prostate cancer), and HCT116 (colon cancer) used in this study were obtained from ATCC. Cells were cultured in DMEM supplemented with 10% heat-inactivated FBS, 2000 units/liter penicillin and 2 mg/liter streptomycin (Invitrogen). All cells were maintained at 37 °C in a humid incubator with 5% CO2. Transfections of different DNA constructs were performed using either Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions or calcium phosphate protocol as described previously (22). The final concentrations of the following inhibitors used in this study were LiCl (20 mm), PD153035 (10 μm), LY294002 (20 μm), U0126 (15 μm), and H89 (10 μm) (Calbiochem) unless otherwise mentioned, and incubation times are shown in the figure legends.
Plasmids and Luciferase Reporter Assay
Control pGZ21dx vector and full-length β-catenin in pBI expression vector (pBI-β-catenin) were kind gifts from Dr. K. M. Yamada and Dr. B. Alman, respectively. Full-length human EGFR promoter was amplified from genomic DNA using flanking primers containing KpnI sites and inserted into pGL3-basic plasmid to generate pGL3-EGFR. We have analyzed the EGFR promoter to find the putative TCF4 site using TRANSFAC software, and the point mutant was generated by changing the sequence −797GCTTCAAAGT−787 to −797GCTTCACCGT−787 at the TCF4 site in the EGFR promoter using the QuikChange XL site-directed mutagenesis kit (Stratagene). SuperTopFlash-TCF4 luciferase reporter (under the control of eight TCF4 consensus sites) was procured from Addgene. All constructs were verified by sequencing (sequences of all the primers used for cloning and subcloning are given in supplemental Table S1). To examine the effects of LiCl on EGFR promoter activity, HEK293 cells were transiently transfected with firefly luciferase constructs (1 μg) of pGL3-EGFR or pGL3-EGFR-Mut along with Renilla luciferase vector (pRL-TK) using calcium phosphate protocol. After 36 h, cells were kept in serum-free medium in the presence or absence of LiCl for another 6 h before being harvested for determination of luciferase activity, which was measured in a VICTOR X multilabel plate reader (PerkinElmer Life Sciences). The efficiency of transfection was normalized with the Renilla luciferase expression. Luciferase activity of cell lysates was determined luminometrically using the Dual-Luciferase assay system (Promega) as specified by the manufacturer. Quantification was based on three independent experiments.
Immunoblotting and Immunoprecipitation
For immunoblotting, cells were washed twice with phosphate-buffered saline (PBS) and lysed on ice using Tris lysis buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40 (Nonidet P-40), 10% glycerol + protease inhibitor mixture Set V, Calbiochem). Cytoplasmic and nuclear extracts were prepared using buffers of composition 150 mm NaCl, 1.5 mm MgCl2,10 mm KCl, 10 mm HEPES for cytoplasmic extracts, and 420 mm NaCl, 1.5 mm MgCl2, 10 mm HEPES, 0.2 mm EDTA, 25% glycerol for nuclear extracts, respectively. Thirty or fifty microgram protein equivalent lysates were separated by SDS-PAGE and subjected to immunoblotting. For immunoprecipitation experiments, cells were lysed on ice using immunoprecipitation buffer (50 mm HEPES, pH 7.2, 250 mm NaCl, 10% glycerol, 1% Nonidet P-40, 1.0 mm EDTA, 0.5 mm DTT, 10 mm PMSF, and protease inhibitor mixture Set V). After preclearing with protein A-Sepharose beads (GE Healthcare), 1 mg of total protein was subjected to immunoprecipitation as described previously (22). The following antibodies were used: EGFR, β-catenin, Mcl-1, PARP, CDC6, cyclin A, GAPDH, β-actin, α-tubulin, lamin B (Santa Cruz Biotechnology) and Stat3, phospho-Stat3-Tyr705, phospho-β-catenin-Ser552, GSK3β, phospho-GSK3β-Ser9, cyclin D1, ERK1/2, phospho-ERK1/2-Thr202/Tyr204, AKT, phospho-AKT-Ser473, phospho-PKA (phospho-PKAα/β-Thr197), Bcl-xL, proliferating cell nuclear antigen, CDC25A, and cyclin B (Cell Signaling Technology).
Quantitative PCR (qPCR)
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. For each sample, 2 μg of RNA was converted to cDNA using the high capacity reverse transcription kit (Applied Biosystems). and 100 ng of cDNA was subsequently used for qPCR analysis using Power SYBR Green Master Mix on 7500 Fast real time PCR system (Applied Biosystems). In all experiments, 18 S rRNA served as the internal control (normalization), and calibrator controls were chosen appropriately. Sequences of all the primers used in qPCR are given in supplemental Table S1.
Chromatin Immunoprecipitation (ChIP) Assay
DU145 cells were cross-linked with 1% formaldehyde for 12 min at room temperature. The reaction was quenched with glycine at a final concentration of 0.125 m and successively washed three times with PBS. The cells were then resuspended in ChIP lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl, pH 8.1, protease inhibitor mixture Set V) and sonicated to an average size of 200–1000 bp using a Misonix Ultrasonic XL-2000 liquid processor following an established protocol (23). Briefly, the precleared sonicated chromatin (25 μg) was incubated for 12 h at 4 °C with either 3 μg of anti-β-catenin polyclonal antibody (Santa Cruz Biotechnology) or normal rabbit IgG followed by pulldown with protein A-Sepharose beads, which were preblocked with 3% BSA. The beads were successively washed with low salt buffer (0.1% SDS, 1% Triton X-100, 0.15 m NaCl, 2 mm EDTA, 20 mm Tris-HCl, pH 8.1) and then with high salt buffer (0.1% SDS, 1% Triton X-100, 0.5 m NaCl, 2 mm EDTA, 20 mm Tris-HCl, pH 8.1), LiCl buffer (0.25 m LiCl, 1% sodium deoxycholate, 1% Nonidet P-40, 1 mm EDTA, 10 mm Tris-HCl, pH 8.1), and finally Tris-EDTA buffer (1 mm EDTA, 10 mm Tris-HCl, pH 8.1) twice for 5 min each at 4 °C. The precipitated chromatin was eluted by incubation of the beads with elution buffer (1% SDS, 0.1 m NaHCO3) at room temperature for 20 min, de-cross-linked by incubation at 65 °C for 4 h in the presence of 200 mm NaCl, extracted with phenol-chloroform, and precipitated using standard ammonium acetate protocol. ChIP experiments were performed by normal PCR using suitable primers (see supplemental Table S1), Reactions were processed for one cycle at 94 °C for 3 min, and then 40 cycles at 94 °C for 1 min, 60 °C for 30 s, and at 72 °C for 30 s using Qiagen's Top Taq master mix.
Immunofluorescence Microscopy
DU145 cells were fixed and permeabilized with 3.7% paraformaldehyde and 0.5% Triton X-100, respectively. Cells were blocked with 3% BSA (in PBS) before incubation with primary antibodies overnight at 4 °C followed by secondary antibody for 1 h at room temperature under dark conditions (24). Confocal microscopy images were taken with Nikon A1 confocal microscope using NIS-Elements software. The antibodies were used against the following proteins: phospho-β-catenin-Ser552, phospho-Stat3-Tyr705 (Cell Signaling Technology), TCF4 (Santa Cruz Biotechnology); AF488 or AF594 (Molecular Probes). Mounting medium containing DAPI was purchased from Vector Laboratories. ImageJ software was used to merge the images.
RNAi Knockdown
Human β-catenin shRNA (pLKO.1 puro shRNA β-catenin) was purchased from Addgene (construct no. 18803). Control shRNA (non-targeting to any known genomic loci) was constructed according to the standard protocol (25). Both sense and antisense oligonucleotides were synthesized by IDT, annealed, and inserted into pLKO.1 expression vector (Addgene) between AgeI and EcoRI sites. All constructs were verified by sequencing. HiPerfect reagent (Qiagen) was used for transfection of 3–4 μg of shRNA constructs.
FACS Analysis
Cells were trypsinized, pelleted, washed, and resuspended in PBS followed by fixation with ice-cold 70% ethanol and kept for 45 min on ice. After washing twice with PBS, cells were resuspended in propidium iodide/RNase staining buffer (BD Biosciences) and incubated in the dark for 15 min at ambient temperature. Cell cycle distribution was examined by flow cytometry using a FACS Array flow cytometer (BD Biosciences) and analyzed using BD FACSDiva software (version 6.1.2).
Histology and Immunohistochemistry (IHC)
Tumor sections were processed following standard methods. Histological features were examined using routine H&E staining. For immunohistochemistry, antigen retrieval was done using standard citrate buffer protocol. The following antibodies were used: phospho-GSK3β-Ser9, β-catenin, EGFR, phospho-ERK1/2, phospho-Stat3-Tyr705, and Ki67 (Cell Signaling Technology, Santa Cruz Biotechnology, and Biogenex).
RESULTS
EGFR and β-Catenin Are Overexpressed in Prostate Cancer
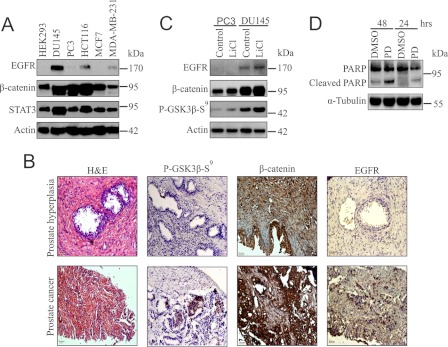
EGFR and Wnt/β-catenin pathways are activated in most of the human cancers. To understand their involvement in cancer initiation and progression, it was important to investigate the expression pattern of EGFR and β-catenin in various established human cancer cell lines. First, we tested the expression of these two key molecules along with Stat3, an important oncoprotein, in several cell lines that include human embryonic kidney (HEK293), prostate cancer (DU145, PC3), colon cancer (HCT116), and breast cancer (MCF7, MDA-MB231) by immunoblotting. It was found that EGFR and β-catenin as well as Stat3 were highly up-regulated in metastatic prostate cancer cell line, DU145 as compared with the others (Fig. 1A). Up-regulation of β-catenin in HCT116 may be due to APC mutation (4).
FIGURE 1.
EGFR is highly expressed along with β-catenin in PCa tissue and DU145 cell line that provide survival. A, whole cell lysates (WCLs) were prepared from HEK293, DU145, PC3, HCT116, MCF7, and MDA-MB231 cells and checked for the expression of EGFR, β-catenin, and Stat3 by immunoblotting. B, IHC was performed in 10 patient samples each of both human PHp and PCa to see the status of phospho-GSK3β-Ser9 (P-GSK3β-S9), β-catenin, and EGFR. Representative histological images of adjacent tumor sections stained with H&E are also provided. The images were captured at a magnification of 20×. C, twenty four-h serum-starved PC3 and DU145 (SS-PC3, SS-DU145) cells were incubated with or without LiCl for 6 h in serum-free media. Cells were lysed and examined for the levels of phospho-GSK3β-Ser9, EGFR, and β-catenin by immunoblotting. D, SS-DU145 cells were treated with or without PD153035 (PD) for 24 and 48 h. WCLs were analyzed for poly ADP ribose polymerase cleavage. DMSO, dimethyl sulfoxide.
Several reports suggest that both EGFR and Wnt/β-catenin pathways are important for tumor development and are up-regulated highly in DU145 cells (6, 26). It is also known that multiple growth signals can induce inactivation of GSK3β by its phosphorylation at Ser9 (27, 28). Thus, it was interesting to find the status of β-catenin in prostate cancer patient samples. Our IHC analysis showed a distinct nuclear staining of β-catenin and increased phosphorylation of GSK3β at Ser9 in PCa. The same tissue samples were then investigated for EGFR, which depicted its elevated expression in PCa compared with PHp (Fig. 1B). Statistically significant up-regulation of β-catenin and EGFR in 2385 prostate cancer patient samples has been reported independently (29–31). Thus, it can be hypothesized that there is a positive correlation between nuclear β-catenin and EGFR. Next, the effect of GSK3β inactivation was tested on these molecules. DU145 and PC3 cells were serum-starved (SS) for 24 h before treatment with LiCl for 6 h to inactivate GSK3β. The result suggested that not only β-catenin but also EGFR expression was increased due to the inactivation of GSK3β by phosphorylation at Ser9 in DU145 cells and this response was highly correlated with IHC data of patient samples (Fig. 1C).
As EGFR was overexpressed upon LiCl treatment, we next focused on the involvement of EGFR signaling in the survival of DU145 cells. For this, SS-DU145 cells were treated with PD153035 for the indicated time periods. PARP cleavage was detected as early as 24 h (Fig. 1D). The result indicates that EGFR signaling plays a central role in the survival of these cells because EGFR inhibitor-treated cells undergo apoptosis. Altogether, the findings indicate that EGFR is overexpressed along with β-catenin in prostate tumor samples as well as the PCa cell line DU145, and this signaling is important for the survival of these cells.
GSK3β Inhibition Promotes Stabilization and Nuclear Accumulation of β-Catenin by PKA-mediated Phosphorylation at Ser552
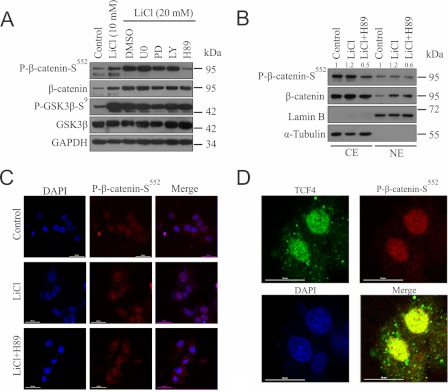
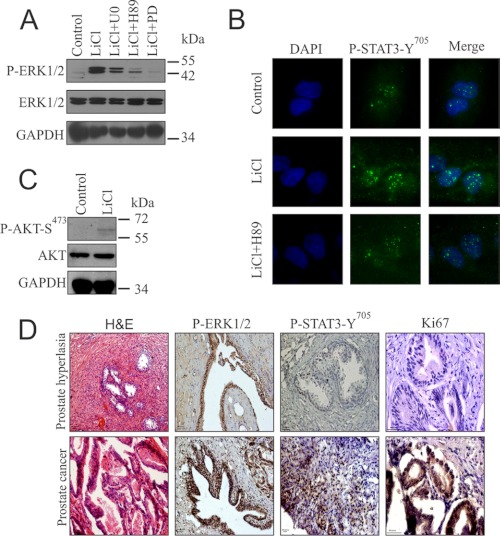
To investigate the possible mechanism of EGFR expression upon GSK3β inactivation, we focused on the transcriptional activation and nuclear accumulation of β-catenin. β-Catenin can be phosphorylated on residues Ser552 and Ser675 by protein kinase B and PKA (10–11). We checked whether any of these phosphorylations was present in LiCl-treated cells and the kinase involved in this context. We treated the SS-DU145 cells with specific inhibitors U0126, LY294002, H89, and PD153035 targeting MEK, PI3K (for protein kinase B/AKT), PKA, and EGFR, respectively. We observed that LiCl treatment resulted in inactivation of GSK3β (phospho-GSK3β-Ser9) leading to a dose-dependent phosphorylation of β-catenin at Ser552 (Fig. 2A). Interestingly, PKA but not protein kinase B seems to play the major role in this phosphorylation event. Next, we examined the distribution of phospho-β-catenin-Ser552 in untreated and treated SS-DU145 cells. Results showed an increased level of β-catenin and its phosphorylated form (phospho-β-catenin-Ser552) in the nucleus when compared with the untreated cells and that was reduced by PKA inhibition (Fig. 2B). To further confirm this important finding, we visualized the localization pattern of phospho-β-catenin-Ser552 after treating the cells with LiCl in the presence or absence of H89 (Fig. 2C). LiCl-treated cells showed higher nuclear accumulation of phospho-β-catenin-Ser552, which was abrogated in the presence of H89 thus corroborating our findings from immunoblot.
FIGURE 2.
GSK3β inhibition leads to PKA-mediated phosphorylation of β-catenin resulting in its increased nuclear accumulation and colocalization with TCF4. A, SS-DU145 cells were pretreated with U0126 (U0), PD153035 (PD), LY294002 (LY), and H89 for 1 h before treatment with LiCl for 24 h in serum-free medium. WCLs were probed for GSK3β, phospho-GSK3β-Ser9, β-catenin, and phospho-β-catenin-Ser552. GAPDH was kept as a loading control. B and C, SS-DU145 cells were treated with or without H89 in the presence of LiCl for 6 h in serum-free medium. Dimethyl sulfoxide (DMSO)-treated cells were kept as a control. B, cytoplasmic extracts (CE) and nuclear extracts (NE) were prepared and immunoblotted with antibodies against β-catenin and phospho-β-catenin-Ser552. Densitometric analysis was performed using ImageJ software, and the relative values were calculated for phospho-β-catenin-Ser552 with respective controls (α-tubulin (cytoplasmic extracts) or lamin-B (nuclear extracts)). C, SS-DU145 cells immunostained with β-catenin (primary) and AF594 (secondary; red) antibodies and visualized under a confocal fluorescence microscope. Images were captured with DAPI-stained nuclei. D, DU145 cells were treated with LiCl for 6 h before staining with primary antibodies against TCF4 and phospho-β-catenin-Ser552, followed by secondary antibodies conjugated to AF488 (TCF4; green) or AF594 (phospho-β-catenin-Ser552; red) and visualized under a confocal fluorescence microscope. Images were captured with DAPI-stained nuclei at 60× magnification.
To explore the possibility for the involvement of phospho-β-catenin-Ser552, which transactivates TCF4-dependent gene expression, a confocal immunofluorescence study was done to demonstrate their colocalization in the nucleus, and as expected, it was found that they do colocalize in LiCl-treated cells (Fig. 2D). These results clearly indicate that β-catenin not only gets stabilized but also accumulates in the nucleus and co-localizes with TCF4 in GSK3β-inactivated cells.
Association of PKA with β-Catenin Leads to Its Phosphorylation and Increased Trans-activation Potential
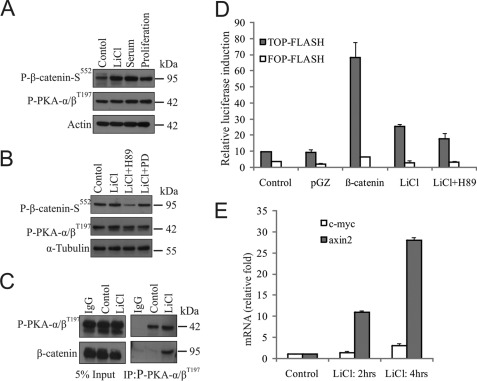
PDK1 phosphorylates PKA at Thr197 residue for its full activation (32). Because we have found increased Ser552 phosphorylation of β-catenin upon LiCl treatment is due to PKA, we have tested whether any upstream molecule regulates the level of activated PKA in DU145 cells. We examine this possibility in these cells under different conditions. The level of phospho-β-catenin-Ser552 was increased in LiCl-treated cells when compared with serum-starved control without any significant change of activated PKA (phospho-PKAα/β-Thr197) level (Fig. 3A). Furthermore, we examined this phosphorylation of β-catenin in the presence of either H89 or PD153035 to see the possible involvement of EGFR-PKA pathway (Fig. 3B). Our result indicates that PKA but not EGFR is involved in this regulation. We speculated that for this phosphorylation, β-catenin should physically interact with phospho-PKAα/β-Thr197. To confirm their association, a coimmunoprecipitation assay was performed, and their interaction was indeed confirmed (Fig. 3C).
FIGURE 3.
β-Catenin associates with phospho-PKA for its phosphorylation and transcriptional activation in GSK3β-inactivated cells. A and B, SS-DU145 cells were treated with either LiCl or 10% serum for 24 h (A); in the presence of LiCl alone or with either H89 or PD153035 (PD) for 6 h (B) in serum-free medium. Untreated SS-DU145 and proliferating cells were kept for respective controls. WCLs were prepared and checked for the levels of phospho-β-catenin-Ser552 and phospho-PKAα/β-Thr197 by immunoblotting. C, WCLs of SS-DU145 cells treated with or without LiCl for 6 h were immunoprecipitated with antibodies against either normal IgG or anti-phospho-PKAα/β-Thr197. Co-immunoprecipitated proteins were probed for β-catenin and phospho-PKAα/β-Thr197. D, luciferase activities were determined in lysates prepared from 36 h post-transfected cells. Values represent the ratio of TOP-Flash versus FOP-Flash firefly luciferase activities normalized previously to Renilla luciferase activity. The average (± S.D.) was calculated from triplicate plates, and experiments were repeated twice with similar results. E, SS-DU145 cells were treated with LiCl for 2 and 4 h or kept untreated. cDNAs were synthesized, and qPCR was performed to check the expression levels of c-myc and axin-2 transcripts.
Next, to examine the activity of nuclear phospho-β-catenin-Ser552, we performed a transcriptional assay using TOP-Flash/FOP-Flash system for β-catenin/T-cell factor (TCF) dependent transcription. Results showed that TCF-dependent transcription is increased upon either β-catenin overexpression or by LiCl treatment, which was reduced when treated with the PKA inhibitor H89 (Fig. 3D). To further support the data, quantitative RT-PCR analysis was performed with β-catenin target genes c-myc and axin-2, which resulted in increased expression of ∼3-fold and ∼28-fold, respectively (Fig. 3E). Henceforth, it can be stated that upon LiCl treatment β-catenin associates with PKA for its phosphorylation, which is stabilized and accumulated in the nucleus and is transcriptionally active in these cells.
β-Catenin/TCF-dependent Transactivation of EGFR Promoter Leads to Its Increased Expression in GSK3β-inactivated Cells
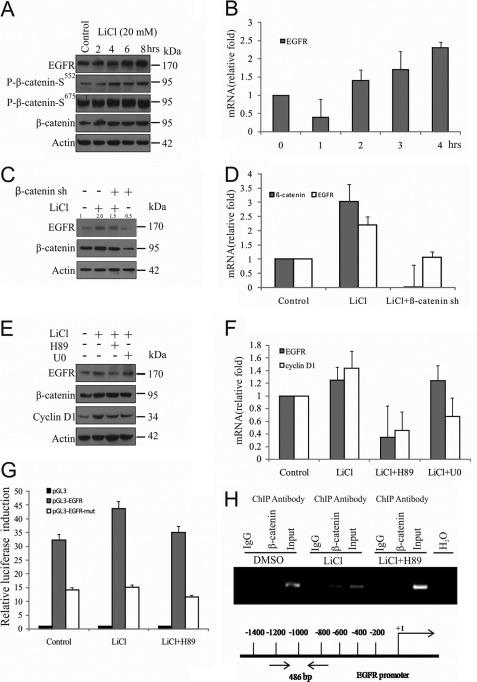
To evaluate whether nuclear accumulation of phospho-β-catenin-Ser552 causes increased EGFR expression, SS-DU145 cells were treated with LiCl for the indicated time periods, and EGFR status was examined at both protein and mRNA levels. Results indicate that both protein and mRNA levels of EGFR were up-regulated with increased accumulation of phospho-β-catenin-Ser552 (Fig. 4, A and B). The β-catenin mediated up-regulation of EGFR expression also is reverse-corroborated by knockdown of β-catenin using β-catenin shRNA in the presence of LiCl followed by immunoblotting and qRT-PCR analysis (Fig. 4, C and D). Here, we have also noticed that the β-catenin mRNA level was increased in LiCl-treated cells, indicating the additional transcriptional regulation of β-catenin through a positive feedback loop, which has been reported previously (33, 34). To rule out the possible involvement of MAPK pathway in EGFR gene expression in this context, SS-DU145 cells were pretreated with U0126 and H89, where cyclin D1 expression was kept as a positive control for MAPK-dependent expression. This result clearly shows that EGFR gene expression is regulated by PKA but not by the MAPK pathway (Fig. 4, E and F).
FIGURE 4.
GSK3β inhibition augments β-catenin-induced EGFR overexpression. A and B, SS-DU145 cells were treated with LiCl for the indicated time periods. Induction of EGFR expression was examined by immunoblotting (protein), qPCR (mRNA), and compared with control. C and D, cells were transfected with β-catenin shRNA or control (scrambled) shRNA (sh) before serum starvation for 24 h followed by treatment with or without LiCl for another 6 h. WCLs and cDNAs were prepared and checked for β-catenin and EGFR expression by immunoblotting and qPCR, respectively. In qPCR, relative fold changes were calculated with respect to untreated control (value, 1.0), and 18 S rRNA was used as an internal control. Also densitometric analysis was performed for immunoblotting, and the relative values were calculated for EGFR with respect to control. E and F, SS-DU145 cells were pretreated with or without H89 or U0126 for 1 h before LiCl treatment for a further 6 h. WCLs and cDNAs were prepared and checked for cyclin D1 and EGFR expression by immunoblotting and qPCR, respectively. G, HEK293 cells were transfected with EGFR-pGL3 luciferase reporter plasmid. After 36 h of treatment, including the last 6 h with LiCl, WCLs were prepared, and luciferase activity was measured in triplicates. Results were presented in terms of fold change, and the values represent the mean ± S.D. from three independent transfections. H, cross-linked chromatin was immunoprecipitated using ChIP grade antibodies as indicated from SS-DU145 cells treated with LiCl in the presence or absence of H89 for 6 h. Untreated cells were kept as a control. DNA was isolated and PCR-amplified. Input, DNA isolated from cross-linked chromatin without immunoprecipitation. DMSO, dimethyl sulfoxide.
To further assess the impact of phospho-β-catenin-Ser552 on expression of EGFR, analysis of EGFR promoter activity was carried out, which revealed that pGL3-EGFR-WT was increased upon LiCl treatment and reduced by H89 treatment in HEK293 cells but did not activate pGL3-EGFR-Mut, in which the putative β-catenin-binding site was mutated. Our results also showed a 50% reduction of luciferase activity in mutant promoter when compared with the wild type (Fig. 4G). In support of this finding, we performed ChIP assay with the primer set as indicated to confirm the presence of β-catenin on the EGFR promoter where two consecutive TCF4 sites exist. The result indicates that β-catenin occupies the EGFR promoter when it is accumulated in the nucleus of LiCl-treated cells, and this association is abolished in presence of H89, indicating the importance of phospho-β-catenin-Ser552 in EGFR gene expression (Fig. 4H). Collectively, these results strongly support that transcriptionally activated nuclear β-catenin up-regulates EGFR gene expression and initiate a signaling cross-talk in DU145 cells when GSK3β remains in an inactivated condition.
Enhanced Expression of EGFR Leads to Increased Activation of Its Downstream Pathways
EGFR overexpression in GSK3β-inactivated PCa cells is involved in increased activation of its downstream pathways that include MAPK (ERK1/2), AKT, and Stat3. LiCl-treated cells in the absence or presence of the specific inhibitors as indicated showed increased activation of ERK1/2, which was reduced by the used inhibitors (Fig. 5A). Stat3 is a highly oncogenic member of Stat family of transcription factors and is activated constitutively by phosphorylation at Tyr705 in most of the human cancer cells through either IL-6 or EGF. In support of the EGFR activation, our immunofluorescence study in the presence and absence of H89 showed increased accumulation of activated Stat3 in LiCl-treated cells, which was inhibited when treated in combination with H89 (Fig. 5B). But we did not find any significant change in the basal level of AKT activation, which may be due to the stringent negative regulation of AKT activation by wild type PTEN in DU145 cells (Fig. 5C).
FIGURE 5.
Increased activation of ERK1/2 and Stat3 in GSK3β-inactivated cells and PCa tissues. A, SS-DU145 cells were preincubated with either dimethyl sulfoxide, U0126 (U0), PD153035 (PD), or H89 for 1 h before treatment with LiCl for 6 h. WCLs were analyzed for ERK1/2 and phospho-ERK1/2. B, SS-DU145 cells were treated with or without H89 in the presence of LiCl, and untreated cells were kept as control before immunostaining with phospho-Stat3 (primary) and AF488 (green) antibodies and visualized under a confocal fluorescence microscope. The green image was merged with DAPI. C, SS-DU145 cells were treated with or without LiCl for 6 h, and the lysates were probed for AKT and phospho-AKT. D, IHC was performed in 10 patient samples each of both human PHp and PCa to see the status of phospho-ERK1/2, phospho-Stat3, and Ki67. Representative histological images of adjacent tumor sections stained with H&E were also provided. The images were captured at a magnification of 20× except for Ki67, which was taken at a magnification of 40×.
Increased activation of ERK1/2 and Stat3 along with EGFR overexpression in GSK3β inactivated SS-DU145 cells prompted us to check whether the same is true in prostate cancer patient samples where GSK3β gets inactivated by its phosphorylation at Ser9 as shown in Fig. 1B. IHC analysis using antibody against phospho-ERK1/2 and phospho-Stat3-Tyr705 on human PCa samples revealed increased activation of both the proteins and nuclear accumulation of activated Stat3 (Fig. 5D). To summarize, the results highlight that GSK3β inactivation leads to increased activation of both ERK1/2 and Stat3 in prostate cancer cell lines and patient samples.
β-Catenin-EGFR Cross-talk Initiated by GSK3β Inactivation Enhances Survival and Proliferation of Prostate Cancer Cells
Stat3 is involved in survival, proliferation, and differentiation in different context in both normal and cancer cells (35). It was interesting for us to discern the role of activated Stat3 under GSK3β-inactivated conditions. For this, we directly examined the status of Stat3 target genes Mcl-1 and Bcl-xL (Bcl-2 prosurvival family members) and found increased expressions of these genes at both the protein and mRNA (Fig. 6, A and B).
FIGURE 6.
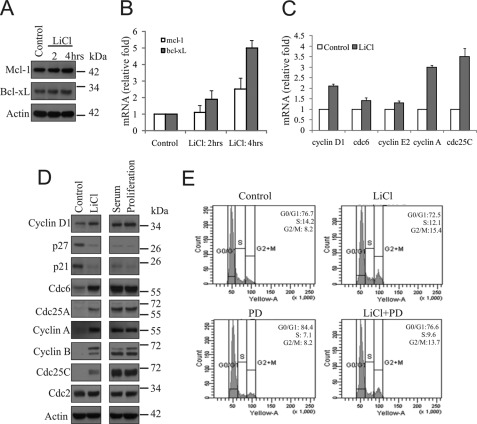
β-Catenin-EGFR cross-talk leads to enhanced survival and proliferation of PCa cells. A and B, SS-DU145 cells were treated with LiCl for the indicated time periods. Expression of Mcl-1 and Bcl-xL were examined by immunoblotting for protein and qPCR for mRNA and compared with untreated control. C, SS-DU145 cells were treated with or without LiCl for 24 h, and the expressions of cyclin D1, cdc6, cyclin E2, cyclin A, and cdc25C were examined by qPCR analysis, and the relative fold changes were compared with untreated control normalized to 1.0. D, SS-DU145 cells were treated with either LiCl or 10% serum in serum-free medium for 24 h. Untreated SS-DU145 and proliferating cells were kept as controls. WCLs were prepared and checked for the levels of cyclin D1, p27, p21, Cdc6, Cdc25A, cyclin A, cyclin B, Cdc25C, and Cdc2 by immunoblotting. Actin served as the loading control. E, SS-DU145 cells were treated with LiCl alone or in the presence or absence of PD153035 (PD) for 24 h. Untreated SS-DU145 cells were kept as a control. Cells were harvested by trypsinization, stained with PI solution, and finally analyzed by FACS.
There are several reports deciphering the importance of GSK3β in cell physiology (28, 36–38). Thus, we investigated the importance of these molecular consequences in GSK3β-inactivated cells and their implication on cell cycle progression. In fact, we found increased levels of cyclin D1, Cdc6, cyclin E2, cyclin A, and Cdc25c transcripts by qRT-PCR in SS-DU145 cells when treated with LiCl for 24 h (Fig. 6C). We further looked for the expression levels of some important proteins involved in different phases of the cell cycle by immunoblotting. We found down-regulation of these proteins in SS-DU145 cells, whereas their expressions were induced upon LiCl treatment, a phenomenon that was comparable with serum-treated or proliferating cells (Fig. 6D).
Finally, the cell cycle analysis revealed an increased amount of proliferative cells (S-G2-M phase cells) in LiCl treatment, when compared with SS cells alone, which can be well explained by the fact that multiple cell cycle regulator genes were shown to be up-regulated under GSK3B-inactivated conditions (Fig. 6E). Collectively, all these results indicate that GSK3β inactivation by LiCl treatment induces the expressions of Bcl-2 family prosurvival as well as multiple cell cycle regulatory genes, which synergistically lead to increased survival and proliferation of DU145 cells.
DISCUSSION
PCa is currently the most frequently diagnosed malignancy in men and the second leading cause of cancer related death in Western populations (39). On the other hand, there are several reports that are suggestive of Wnt-independent stabilization of β-catenin leads to the enhancement of β-catenin trans-activation and tumorigenesis (40). Activation of the β-catenin pathway has been observed in PCa patients, and a recent study showed that 32% of these patients at advanced stages carried mutations in the β-catenin gene, highlighting the important role of β-catenin in the development of PCa (7, 14, 41). The hallmark of β-catenin activity in cells is nuclear localization and its transcriptional up-regulation. Here, we also have confirmed this phenomenon by observing strong nuclear localization in prostate cancer samples. EGFR has been known to be overexpressed in many human cancer cell types, including androgen-independent PCa cells (43, 44). Thus, there arises the obvious task to identify the molecular mechanisms that might lead to the up-regulation of these key molecules there by promoting prostate cancer growth and tumorigenesis. The IHC data from the current study demonstrates that phosphorylation of GSK3β is highly correlated with the up-regulated expressions of both β-catenin and EGFR in PCa compared with PHp.
The present study describes three major findings: (i) GSK3β inhibition promotes β-catenin stabilization and its PKA-dependent phosphorylation of Ser552 and Ser675 residues in DU145 cells. (ii) Phosphorylation of β-catenin at Ser552 by PKA promotes its transcriptional activity, including EGFR gene expression. (iii) Increased expression of EGFR through phospho-β-catenin-Ser552 may facilitate the activation of ERK1/2 and Stat3, which provide a molecular mechanism how β-catenin promotes survival and proliferation of PCa cells.
It has also been reported that PKA regulates the post-translational stabilization of β-catenin by phosphorylation of either Ser552 or Ser675. These phosphorylations are implicated in enhancing the transcriptional activity of β-catenin by facilitating its interaction with a transcriptional co-activator, CREB-binding protein (9, 10, 45). In this study, we demonstrate that the inhibition of GSK3β by LiCl leads to PKA-mediated phosphorylation of β-catenin at Ser552 and its consequent nuclear accumulation. In this context, nuclear translocation of β-catenin may be due to inhibition of its β-transducin repeat-containing protein (βTRCP)-mediated degradation. Here, we also show that phospho-β-catenin-Ser552 co-localizes with TCF4 in the nucleus of GSK3β-inactivated cells, which strengthens the previous view that phospho-β-catenin-Ser552 is involved directly in transcription (10). Our data proves that several known targets of β-catenin such as cyclin D1, c-myc, and axin-2 were up-regulated. We also found increased expression of the EGFR gene that is inhibited by H89, which strengthens the importance of nuclear phospho-β-catenin-Ser552 level in these cells. Our data indicates that β-catenin occupies the EGFR promoter in LiCl-treated cells, which is in agreement with a previous report in transgenic animal system. However, the TCF4 binding sequence used previously (WWCAAWG) is different from the current consensus sequence, (A/T)TCAAAG and was used for ChIP analysis (46–48).
Recently, it has been shown that Wnt signaling is involved in regulation of the EGFR pathway (46, 49, 50). Our results show that by inhibiting GSK3β with LiCl, the level of phospho-β-catenin-Ser552 increases, which further induces the expression of EGFR in a time-dependent manner, indicating a strong cross-talk between these two pathways, a novel mechanism in PCa cells. We had also observed that the overexpressed EGFR activates both ERK1/2 and Stat3 in GSK3β-inhibited PCa cells, which supports the claim that persistently activated EGFR is an important upstream physiological controller of these molecules. In this context, the critical difference in signaling between normal and cancer cells may arise due to the β-catenin-mediated cross-talk with EGFR that leads to aberrant activation of its downstream pathways. Our IHC analysis revealed that EGFR expression is up-regulated significantly in PCa samples, and its expression pattern seems to overlap with nuclear phospho-Stat3-Tyr705, when compared with PHp samples.
Furthermore, we have identified the up-regulation of anti-apoptotic family of Stat3 target genes as well as cell cycle regulatory proteins in GSK3β-inhibited PCa cells. Thus, in our results, we have established that β-catenin when phosphorylated at Ser552 is a key factor in promoting EGFR expression and EGFR-dependent proliferation and survival. All of these results strongly suggest that activation of β-catenin signaling may be associated with the progression of the disease through signaling cross-talk. Thus, our study can open a new avenue in the combinatorial therapeutic approach which is currently under development in cancer treatment (42, 51, 52).
Acknowledgments
We thank Dr. K. M. Yamada (Chief, Laboratory of Cell and Developmental Biology, National Institute of Health/NIDCR) and Dr. B. Alman (Canada Research Chair; Head, Division of Orthopaedic Surgery; Department of Surgery, University of Toronto, Canada) for providing us with the pGZ21dx plasmid and pBI-β-catenin construct respectively.
This work was supported by grants provided by Department of Science & Technology (SR/SO/HS-0126/2007) and Council of Scientific and Industrial Research (EMPOWER: OLP-2) (to M. K. G.).

This article contains supplemental Table S1.
- GSK3β
- glycogen synthase kinase 3 β
- PCa
- prostate cancer
- SS-DU145
- serum-starved DU145
- qPCR
- quantitative real time PCR
- PHp
- prostate hyperplasia
- TCF
- T-cell factor
- IHC
- immunohistochemistry
- PKA
- protein kinase A
- CREB
- cAMP-responsive element-binding protein
- WCL
- whole cell lysate
- APC
- Adenomatous polyposis coli.
REFERENCES
- 1. Ozawa M., Baribault H., Kemler R. (1989) The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J. 8, 1711–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huber A. H., Weis W. I. (2001) The structure of the β-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by β-catenin. Cell 105, 391–402 [DOI] [PubMed] [Google Scholar]
- 3. Huelsken J., Behrens J. (2002) The Wnt signaling pathway. J. Cell Sci. 115, 3977–3978 [DOI] [PubMed] [Google Scholar]
- 4. Morin P. J., Sparks A. B., Korinek V., Barker N., Clevers H., Vogelstein B., Kinzler K. W. (1997) Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science 275, 1787–1790 [DOI] [PubMed] [Google Scholar]
- 5. Voeller H. J., Truica C. I., Gelmann E. P. (1998) β-Catenin mutations in human prostate cancer. Cancer Res. 58, 2520–2523 [PubMed] [Google Scholar]
- 6. Gerstein A. V., Almeida T. A., Zhao G., Chess E., Shih I. e. M., Buhler K., Pienta K., Rubin M. A., Vessella R., Papadopoulos N. (2002) APC/CTNNB1 (β-catenin) pathway alterations in human prostate cancers. Genes Chromosomes Cancer 34, 9–16 [DOI] [PubMed] [Google Scholar]
- 7. Chesire D. R., Ewing C. M., Sauvageot J., Bova G. S., Isaacs W. B. (2000) Detection and analysis of β-catenin mutations in prostate cancer. Prostate 45, 323–334 [DOI] [PubMed] [Google Scholar]
- 8. Daugherty R. L., Gottardi C. J. (2007) Phosphoregulation of β-catenin adhesion and signaling functions. Physiology 22, 303–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fang D., Hawke D., Zheng Y., Xia Y., Meisenhelder J., Nika H., Mills G. B., Kobayashi R., Hunter T., Lu Z. (2007) Phosphorylation of β-catenin by AKT promotes β-catenin transcriptional activity. J. Biol. Chem. 282, 11221–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Taurin S., Sandbo N., Qin Y., Browning D., Dulin N. O. (2006) Phosphorylation of β-catenin by cyclic AMP-dependent protein kinase. J. Biol. Chem. 281, 9971–9976 [DOI] [PubMed] [Google Scholar]
- 11. Verheyen E. M., Gottardi C. J. (2010) Regulation of Wnt/β-catenin signaling by protein kinases. Dev. Dyn. 239, 34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao J., Yue W., Zhu M. J., Sreejayan N., Du M. (2010) AMP-activated protein kinase (AMPK) cross-talks with canonical Wnt signaling via phosphorylation of β-catenin at Ser552. Biochem. Biophys. Res. Commun. 395, 146–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chesire D. R., Isaacs W. B. (2002) Ligand-dependent inhibition of β-catenin/TCF signaling by androgen receptor. Oncogene 21, 8453–8469 [DOI] [PubMed] [Google Scholar]
- 14. Bierie B., Nozawa M., Renou J. P., Shillingford J. M., Morgan F., Oka T., Taketo M. M., Cardiff R. D., Miyoshi K., Wagner K. U., Robinson G. W., Hennighausen L. (2003) Activation of β-catenin in prostate epithelium induces hyperplasias and squamous transdifferentiation. Oncogene 22, 3875–3887 [DOI] [PubMed] [Google Scholar]
- 15. Gullick W. J. (1991) Prevalence of aberrant expression of the epidermal growth factor receptor in human cancers. Br. Med. Bull. 47, 87–98 [DOI] [PubMed] [Google Scholar]
- 16. Di Lorenzo G., Tortora G., D'Armiento F. P., De Rosa G., Staibano S., Autorino R., D'Armiento M., De Laurentiis M., De Placido S., Catalano G., Bianco A. R., Ciardiello F. (2002) Expression of epidermal growth factor receptor correlates with disease relapse and progression to androgen independence in human prostate cancer. Clin. Cancer Res. 8, 3438–3444 [PubMed] [Google Scholar]
- 17. Di Fiore P. P., Pierce J. H., Fleming T. P., Hazan R., Ullrich A., King C. R., Schlessinger J., Aaronson S. A. (1987) Overexpression of the human EGF receptor confers an EGF-dependent transformed phenotype to NIH-3T3 cells. Cell 51, 1063–1070 [DOI] [PubMed] [Google Scholar]
- 18. Di Fiore P. P., Pierce J. H., Kraus M. H., Segatto O., King C. R., Aaronson S. A. (1987) erbB-2 is a potent oncogene when overexpressed in NIH-3T3 cells. Science 237, 178–182 [DOI] [PubMed] [Google Scholar]
- 19. Djakiew D. (2000) Dysregulated expression of growth factors and their receptors in the development of prostate cancer. Prostate 42, 150–160 [DOI] [PubMed] [Google Scholar]
- 20. Connolly J. M., Rose D. P. (1991) Autocrine regulation of DU145 human prostate cancer cell growth by epidermal growth factor-related polypeptides. Prostate 19, 173–180 [DOI] [PubMed] [Google Scholar]
- 21. Shah R. B., Ghosh D., Elder J. T. (2006) Epidermal growth factor receptor (ErbB1) expression in prostate cancer progression: Correlation with androgen independence. Prostate 66, 1437–1444 [DOI] [PubMed] [Google Scholar]
- 22. Ghosh M. K., Sharma P., Harbor P. C., Rahaman S. O., Haque S. J. (2005) PI3K-AKT pathway negatively controls EGFR-dependent DNA-binding activity of Stat3 in glioblastoma multiforme cells. Oncogene 24, 7290–7300 [DOI] [PubMed] [Google Scholar]
- 23. Ghosh M. K., Harter M. L. (2003) A viral mechanism for remodeling chromatin structure in G0 cells. Mol. Cell 12, 255–260 [DOI] [PubMed] [Google Scholar]
- 24. Sha J., Ghosh M. K., Zhang K., Harter M. L. (2010) E1A interacts with two opposing transcriptional pathways to induce quiescent cells into S phase. J. Virol. 84, 4050–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sarbassov D. D., Sabatini D. M. (2005) Redox regulation of the nutrient-sensitive raptor-mTOR pathway and complex. J. Biol. Chem. 280, 39505–39509 [DOI] [PubMed] [Google Scholar]
- 26. Festuccia C., Muzi P., Millimaggi D., Biordi L., Gravina G. L., Speca S., Angelucci A., Dolo V., Vicentini C., Bologna M. (2005) Molecular aspects of gefitinib antiproliferative and proapoptotic effects in PTEN-positive and PTEN-negative prostate cancer cell lines. Endocr. Relat. Cancer 12, 983–998 [DOI] [PubMed] [Google Scholar]
- 27. O'Brien W. T., Klein P. S. (2009) Validating GSK3 as an in vivo target of lithium action. Biochem. Soc. Trans. 37, 1133–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mishra R. (2010) Glycogen synthase kinase 3β: Can it be a target for oral cancer. Mol. cancer 9, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jaggi M., Johansson S. L., Baker J. J., Smith L. M., Galich A., Balaji K. (2005) Aberrant expression of E-cadherin and β-catenin in human prostate cancer. Urol. Oncol. 23, 402–406 [DOI] [PubMed] [Google Scholar]
- 30. Whitaker H. C., Girling J., Warren A. Y., Leung H., Mills I. G., Neal D. E. (2008) Alterations in β-catenin expression and localization in prostate cancer. The Prostate 68, 1196–1205 [DOI] [PubMed] [Google Scholar]
- 31. Schlomm T., Kirstein P., Iwers L., Daniel B., Steuber T., Walz J., Chun F. H., Haese A., Kollermann J., Graefen M., Huland H., Sauter G., Simon R., Erbersdobler A. (2007) Clinical significance of epidermal growth factor receptor protein overexpression and gene copy number gains in prostate cancer. Clin. Cancer Res. 13, 6579–6584 [DOI] [PubMed] [Google Scholar]
- 32. Moore M. J., Kanter J. R, Jones K. C., Taylor S. S. (2002) Phosphorylation of the catalytic subunit of protein kinase A. Autophosphorylation versus phosphorylation by phosphoinositide-dependent kinase-1. J. Biol. Chem. 277, 47878–47884 [DOI] [PubMed] [Google Scholar]
- 33. Li Q., Dashwood W. M., Zhong X., Al-Fageeh M., Dashwood R. H. (2004) Cloning of the rat β-catenin gene (Ctnnb1) promoter and its functional analysis compared with the Catnb and CTNNB1 promoters. Genomics 83, 231–242 [DOI] [PubMed] [Google Scholar]
- 34. Bandapalli O. R., Dihlmann S., Helwa R., Macher-Goeppinger S., Weitz J., Schirmacher P., Brand K. (2009) Transcriptional activation of the β-catenin gene at the invasion front of colorectal liver metastases. J. Pathol. 218, 370–379 [DOI] [PubMed] [Google Scholar]
- 35. Yu H., Jove R. (2004) The STATs of cancer–new molecular targets come of age. Nat. Rev. Cancer 4, 97–105 [DOI] [PubMed] [Google Scholar]
- 36. Wang Q., Zhou Y., Wang X., Evers B. M. (2006) Glycogen synthase kinase-3 is a negative regulator of extracellular signal-regulated kinase. Oncogene 25, 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu C., Kim N. G., Gumbiner B. M. (2009) Regulation of protein stability by GSK3 mediated phosphorylation. Cell Cycle 8, 4032–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Beurel E., Jope R. S. (2006) The paradoxical pro- and anti-apoptotic actions of GSK3 in the intrinsic and extrinsic apoptosis signaling pathways. Prog. Neurobiol. 79, 173–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jemal A., Siegel R., Ward E., Hao Y., Xu J., Murray T., Thun M. J. (2008) Cancer statistics, 2008. CA. Cancer J. Clin. 58, 71–96 [DOI] [PubMed] [Google Scholar]
- 40. Lu Z., Hunter T. (2004) Wnt-independent β-catenin transactivation in tumor development. Cell Cycle 3, 571–573 [PubMed] [Google Scholar]
- 41. Yardy G. W., Brewster S. F. (2005) Wnt signaling and prostate cancer. Prostate Cancer Prostatic Dis. 8, 119–126 [DOI] [PubMed] [Google Scholar]
- 42. Das T., Sa G., Saha B., Das K. (2010) Multifocal signal modulation therapy of cancer: Ancient weapon, modern targets. Mol. Cell Biochem. 336, 85–95 [DOI] [PubMed] [Google Scholar]
- 43. Luwor R., Taylor L., Wang B., Zhu H. J. (January 23, 2008) Tumor-associated EGFR overexpression specifically activates Stat3 and Smad7 resulting in desensitization of TGF-β signaling. Nature Precedings, hdl:10101/npre.2008.1535.1 [Google Scholar]
- 44. Lorenzo G. D., Bianco R., Tortora G., Ciardiello F. (2003) Involvement of growth factor receptors of the epidermal growth factor receptor family in prostate cancer development and progression to androgen independence. Clin. Prostate Cancer 2, 50–57 [DOI] [PubMed] [Google Scholar]
- 45. He X. C., Yin T., Grindley J. C., Tian Q., Sato T., Tao W. A., Dirisina R., Porter-Westpfahl K. S., Hembree M., Johnson T., Wiedemann L. M., Barrett T. A., Hood L., Wu H., Li L. (2007) PTEN-deficient intestinal stem cells initiate intestinal polyposis. Nat. Genet. 39, 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tan X., Apte U., Micsenyi A., Kotsagrelos E., Luo J. H., Ranganathan S., Monga D. K., Bell A., Michalopoulos G. K., Monga S. P. (2005) Epidermal growth factor receptor: A novel target of the Wnt/β-catenin pathway in liver. Gastroenterology 129, 285–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hatzis P., van der Flier L. G., van Driel M. A., Guryev V., Nielsen F., Denissov S., Nijman I. J., Koster J., Santo E. E., Welboren W., Versteeg R., Cuppen E., van de Wetering M., Clevers H., Stunnenberg H. G. (2008) Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol. Cell Biol. 28, 2732–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bottomly D., Kyler S. L., McWeeney S. K., Yochum G. S. (2010) Identification of β-catenin binding regions in colon cancer cells using ChIP-Seq. Nucleic Acids Res. 38, 5735–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hu T., Li C. (2010) Convergence between Wnt-β-catenin and EGFR signaling in cancer. Mol. Cancer 9, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Musgrove E. A. (2004) Wnt signaling via the epidermal growth factor receptor: A role in breast cancer? Breast Cancer Res. 6, 65–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Siddique H. R., Mishra S. K., Karnes R. J., Saleem M. (2011) Lupeol, a novel androgen receptor inhibitor: Implications in prostate cancer therapy. Clin. Cancer Res. 17, 5379–5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saia G., Zhang M., Depalo V., Lautenschlager T., Chakravarti A. (2007) Molecular and genetic profiling of prostate cancer: Implications for future therapy. Curr. Cancer Ther. Rev. 3, 25–36 [Google Scholar]