Background: CPEB and PUF proteins collaborate to control mRNA expression by binding to elements in the 3′-UTR.
Results: A conserved region of PUF proteins is required for PUF/CPEB interactions in nematodes and humans.
Conclusion: PUF proteins from diverse species utilize a common surface to interact with diverse protein partners.
Significance: Understanding molecular recognition between RNA-binding proteins is crucial for describing their roles in biology.
Keywords: RNA, RNA-binding Protein, RNA Turnover, RNA-binding Proteins, RNA-Protein Interaction
Abstract
Members of the PUF (Pumilio and FBF) and CPEB (cytoplasmic polyadenylation element-binding) protein families collaborate to regulate mRNA expression throughout eukaryotes. Here, we focus on the physical interactions between members of these two families, concentrating on Caenorhabditis elegans FBF-2 and CPB-1. To localize the site of interaction on FBF-2, we identified conserved amino acids within C. elegans PUF proteins. Deletion of an extended loop containing several conserved residues abolished binding to CPB-1. We analyzed alanine substitutions at 13 individual amino acids in FBF-2, each identified via its conservation. Multiple single point mutations disrupted binding to CPB-1 but not to RNA. Position Tyr-479 was particularly critical as multiple substitutions to other amino acids at this position did not restore binding. The complex of FBF-2 and CPB-1 repressed translation of an mRNA containing an FBF binding element. Repression required both proteins and was disrupted by FBF-2 alleles that failed to bind CPB-1 or RNA. The equivalent loop in human PUM2 is required for binding to human CPEB3 in vitro, although the primary sequences of the human and C. elegans PUF proteins have diverged in that region. Our findings define a key region in PUF/CPEB interactions and imply a conserved platform through which PUF proteins interact with their protein partners.
Introduction
Every step in the life of an mRNA is controlled. Sequences present in the mRNA specify regulation through RNA-binding proteins (1). Collaborations among regulatory factors assembled onto an mRNA determine the location, timing, and quantity of protein that an mRNA produces (2, 3). These regulatory proteins typically bind to elements in the 3′-untranslated region (UTR). These multiprotein complexes are a dominant theme in mRNA control, particularly during early development (4, 5). Here, we examine the interaction between two collaborating families of mRNA regulatory proteins: the CPEB6 (cytoplasmic polyadenylation element-binding) and PUF (Pumilio and FBF) proteins. These two families of proteins bind one another and contribute to the control of learning, pattern formation, and meiotic maturation (6–16).
CPEBs are conserved among metazoans and are key players in mRNA control (10). They bind U-rich elements designated CPEs (cytoplasmic polyadenylation elements) via zinc knuckles and RNA recognition motif domains (17). CPEB proteins regulate translation, localization, and poly(A) tail length and can either activate or repress their targets (10). Among the proteins identified in CPEB-containing complexes are PARN, GLD-2, symplekin, maskin, eIF4E-T, and PUF proteins (18–20). In vertebrates, both CPEB and PUF proteins contribute to control of cyclin B1 mRNA during oocyte maturation (7, 9, 19–24). CPEBs physically associate with PUF proteins in many animal species, including Caenorhabditis elegans, Aplysia, Drosophila, and Xenopus (6–9). CPEB proteins are critical in very diverse biological contexts, from synaptic plasticity to the cell cycle, cancer progression, and cellular senescence (10, 24–26).
PUF proteins are widespread throughout eukaryotes and commonly bind 3′-UTR sequences containing a UGU motif (27). The PUF protein fold is stringently conserved and consists of eight repeats of three-helix bundles, termed PUF repeats; together these eight repeats form a crescent (28–32). The concave face of the crescent binds an extended single-stranded RNA through a modular form of recognition. Typically, each PUF repeat recognizes a single base along the concave surface of the protein. However, in the structure of FBF-2, two bases in the binding site flip away from the protein (see Fig. 1A) (28). PUFs commonly function as mRNA repressors and promote deadenylation and mRNA decay; however, they also can enhance translation and promote subcellular localization (33–38). PUFs recruit multiple protein partners, in addition to CPEB, including CCR4·NOT complex, Nanos (NOS), Brain Tumor (BRAT), Argonaute (AGO), and the regulatory poly(A) polymerase complex consisting of GLD-2 and GLD-3 (31, 36, 39–42). These interactions are crucial as they dictate whether a transcript will be properly localized, degraded, translated, or repressed. Metazoan PUF proteins have important roles in controlling stem cell maintenance, pattern formation, learning, and memory (11–15, 43).
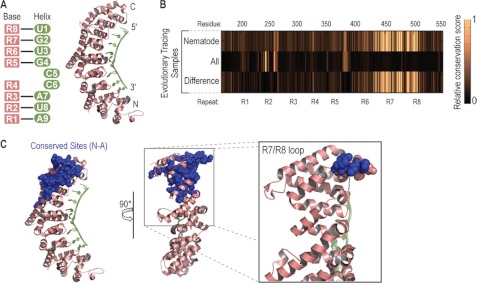
FIGURE 1.
Identification of conserved sites in FBF-2. A, recognition of RNA by PUFs is highly modular; a schematic and the structure of FBF-2 (28) are shown. PUF repeats (pink) specify recognition of RNA bases (green). B, identification of conserved residues in nematode PUFs, all PUFs, and a subtraction of the two sets. Conservation was calculated using Evolutionary Tracing (46). C, an extended loop along one side of the PUF repeats contains conserved sites. Inset, the R7/R8 loop residues (479–485) are shown as spheres; two of residues in this region were identified as conserved sites using Evolutionary Tracing (blue spheres).
In C. elegans, two nearly identical PUF proteins, FBF-1 and FBF-2 (collectively referred to as FBF), are required for stem cell maintenance, oocyte fate specification, and normal spermatogenesis (12, 13, 44). One of the four C. elegans CPEBs, CPB-1, interacts physically with FBF and is also required for normal spermatogenesis (44). Therefore, FBF and CPB-1 share a function in controlling spermatogenesis, although their roles in this process are not yet clear.
Here, we demonstrate that a discrete molecular interface governs the interaction between C. elegans CPB-1 and FBF-2. Furthermore, we show that CPB-1 enhances repression of translation by FBF-2 in vitro. The site of the interaction is broadly conserved; analogous regions of the C. elegans and human PUF proteins are required for their interactions with CPEBs of the same species. We suggest that a protein interaction platform located near the C-terminal segment of the PUF repeats mediates interactions between PUFs and multiple protein partners across eukaryotic species.
EXPERIMENTAL PROCEDURES
Evolutionary Tracing
Alignments for homologous nematode PUF proteins were detected based on similarity to FBF-2 using BLASTP (www.ncbi.nlm.nih.gov/BLAST). Similarly, Pum2 was used to detect PUF proteins across distantly related organisms. Ungapped protein alignments were generated using ClustalW and supplied to the Evolutionary Trace server. Heat maps and difference values were calculated using MATLAB (MathWorks).
Protein Purification
Recombinant FBF-2 (residues 164–575), CPB-1(1–80), and CPB-1(19–80) were produced from a pET-22b(+) (Novagen) vector. Proteins were purified using the TALON metal affinity resin (Clontech). Human CPEB3 (440–698) and PUM2 (456–1064) were expressed in pGex6P-1 (GE Healthcare).
Translation Assays
Purified PCR products encoding CPB-1 and FBF-2 were used as templates for T7 transcription reactions (Ambion). FBF-2 and CPB-1 proteins were generated from 50 ng of transcribed mRNA using rabbit reticulocyte translation extracts (Promega). Reactions were allowed to proceed for 90 min. 1 μl of the newly synthesized protein sample was added to the second reaction containing reporter RNAs. Reporter RNAs were transcribed from the pYC2 plasmid using primers that generated the candidate regulatory elements directly after the stop codon. Renilla luciferase was transcribed from pSP65-Ren (45). Individual reactions were assembled as described previously and assayed using the Dual-Luciferase assay system (Promega) measured with a 96-well Synergy 2 plate reader (45).
Affinity Chromatography
In the experiments assaying association of CPB-1 and FBF-2 (see Fig. 2C), a solution containing 15 μm recombinant polyhistidine-tagged CPB-1 (19–80) was added to FBF-2 in 20 mm Tris-HCl (pH7.5), 100 mm NaCl, 16 mm imidazole, and 2.5 mm DTT. Binding reactions were incubated at 25 °C for 3 h prior to immobilization on His SpinTrap columns (GE Healthcare). In assays of the interaction between human PUF and CPEB proteins (see Fig. 6), GST-CPEB3 was bound to glutathione-agarose at a concentration of ∼1 μg/binding reaction. GST was removed from purified PUM2 using PreScission (GE Healthcare). An equal amount (1 μg) of PUM2 or PUM2Δ984–989 was added to CPEB3 and allowed to incubate at 4 °C for 1 h in the presence of RNase A and T1 (TNEMN150 buffer (50 mm Tris-HCl (pH 8), 0.5% (v/v) Nonidet P40, 1 mm EDTA, 2 mm MgCl2, and 150 mm NaCl)). The final volume of sample was adjusted to 200 μl using TNEMN150. The resin was pelleted and washed five times with 0.5 ml of TNEMN150 prior to analysis by SDS-PAGE.
FIGURE 2.
Analysis of a mutant in FBF-2 that lacks the R7/R8 loop. A, schematic of the yeast two-hybrid assay. Binding assays of a wild-type and a mutant form of FBF-2 and wild-type CPB-1 are shown. The YBZ-1 cell line was transformed with plasmids expressing the indicated GAL4 activation domain (AD) or DNA binding domain (LexA) fusions indicated. B, RNA binding assayed in the yeast three-hybrid system. Interactions between a high affinity binding site from the gld-1 FBEa and wild-type or mutant version of FBF-2 are shown. Error bars in A and B indicate S.D. C, an affinity chromatography-based assay of FBF-2 binding to CPB-1. CPB-1 was immobilized on resin and incubated with FBF-2. Unbound FBF-2 was washed away using a series of wash steps, and the specifically bound protein eluted at high concentrations of imidazole. Ni-NTA, nickel-nitrilotriacetic acid. D, FBF-2 eluted at high concentrations of imidazole. In the absence of CPB-1, FBF-2 is weakly associated with the resin. However, in the presence of CPB-1, FBF-2 is eluted at high concentrations of imidazole. The deletion mutant behaves in an identical fashion to FBF-2 in the absence of CPB-1.
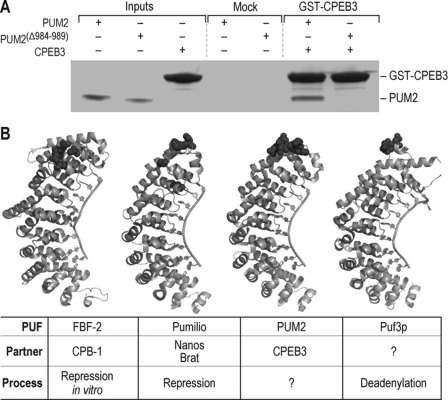
FIGURE 6.
Identification of a conserved site of interaction between PUFs and CPEBs. A, affinity chromatography of human CPEB3 was conducted with either wild-type PUM2 or mutant PUM2Δ984–989. Wild-type PUM2 specifically associates with CPEB3 and not a mock GST alone control. However, PUM2Δ984–989 failed to interact with either. B, a broadly conserved interface mediates protein-protein interactions throughout PUF proteins. The sites of mutations that disrupt specific interactions are shown as spheres (40, 55).
Yeast Two- and Three-hybrid Assays
These experiments were conducted as described with minor adjustments (44). Yeast two- and three-hybrid experiments were conducted in strains L40 Ura− and YBZ-1, respectively. CPB-1 (residues 40–80) was overexpressed from p414TEF and fused to an SV40 nuclear localization signal. Luminescence data were collected using the β-Glo reagent (Promega) using the Dual-Luciferase assay system (Promega) measured with a 96-well Synergy 2 plate reader. Average luminescence values were corrected for cell density as determined by optical density at 600 nm.
RESULTS
Conservation in FBF-2
We sought to identify the amino acid residues comprising the interface between C. elegans FBF-2 and CPB-1. To find candidate sites in FBF-2 that might mediate its interactions with other proteins, we calculated conservation at each amino acid residue in its RNA binding domain using a computational method, Evolutionary Tracing (46). Scores were determined using a multiple sequence alignment of a wide array of either distantly related eukaryotic PUFs or more closely related nematode PUF proteins (Fig. 1B). Amino acids involved in RNA binding were identified through structural analysis and were excluded from the analysis (28). We hypothesized that conserved sites across distantly related taxa likely were involved in protein stability and folding. Therefore, we subtracted conservation scores derived from a comparison of all organisms from those of the nematode group. The remaining set of residues was conserved specifically among nematodes and unlikely to be involved in RNA binding. The majority of conserved sites clustered near the C-terminal region of FBF-2. Several of these residues reside in a loop linking PUF repeats 7 and 8, referred to as the R7/R8 loop (Fig. 1C).
Analysis of an Extended Loop in FBF-2
To determine whether the R7/R8 loop was required for the interaction between CPB-1 and FBF-2, we generated a deletion mutant spanning residues 479–485. We first performed a two-hybrid assay in which either wild-type or mutant FBF-2 GAL4 activation domain fusions were introduced into yeast expressing a peptide derived from CPB-1 fused to a DNA binding domain (Fig. 2A). CPEBs bind RNA using two RNA recognition motif domains and two zinc-chelating segments. However, a region of CPB-1 (residues 1–80) outside the RNA binding domain is sufficient to bind tightly to FBF-2 (44). This peptide was used in the two-hybrid assays. Levels of expression of β-galactosidase, produced from the LacZ reporter gene, were used to quantify FBF-2 binding to the CPB-1 peptide. Deletion of the R7/R8 loop impaired the interaction of CPB-1 (Fig. 2A) in the two-hybrid assay. As a control, we examined binding of the same mutant to a high affinity RNA binding element (the gld-1 FBF binding element, FBEa) using a yeast three-hybrid assay (Fig. 2B). In electrophoretic mobility shift assays (EMSAs), the apparent dissociation constants for RNA binding to the wild-type and R7/R8 loop deletion mutant proteins were 0.4 and 0.7 nm, respectively, indicating that the R7/R8 loop deletion did not disrupt folding of the protein (data not shown). We conclude that the R7/R8 loop is specifically required for the interaction of FBF-2 with CPB-1, and not for binding of FBF-2 to RNA.
To test whether the physical binding interaction of purified FBF-2 and CPB-1 proteins was compromised by deletion of the loop, we devised a series of affinity chromatography experiments (Fig. 2C). A recombinant version of CPB-1 containing the minimal portion sufficient for interaction with FBF-2 was immobilized on nickel-nitrilotriacetic acid-agarose using a hexahistidine affinity tag. After removal of unbound CPB-1, purified samples containing FBF-2 were added to form the protein complex. FBF-2 was then eluted from the resin through a series of wash steps using increasing concentrations of imidazole. Wild-type FBF-2 was eluted together with CPB-1 at high concentrations of imidazole (Fig. 2D). In contrast, the FBF-2 R7/R8 loop deletion mutant was eluted at low imidazole concentrations and behaved in an identical manner to FBF-2 without any immobilized CPB-1. The deletion mutant protein still bound RNA, indicating that it was not misfolded (supplemental Fig. S1).
Screen to Identify CPB-1 Binding Site on FBF-2
We sought to determine which specific amino acids among the set of highly conserved sites in FBF-2 (identified in Fig. 1) were required for binding CPB-1. To do so, we developed a screen that combined yeast two- and three-hybrid assays (Fig. 3A). Alanine substitutions in the residues conserved throughout nematodes were introduced into FBF-2 fused to the Gal4 activation domain. These were analyzed following the protocol we used for the loop deletion mutant. Each variant was examined for CPB-1 binding in two-hybrid assays (Fig. 3B) and for RNA binding in three-hybrid assays (Fig. 3C). β-Galactosidase activity levels were used as a proxy for binding affinity in both assays. The ratio of β-galactosidase levels obtained from the two assays was used to identify mutations that specifically disrupted binding to CPB-1 but not to RNA (Fig. 3D). Of the 13 mutations analyzed, several did not significantly reduce either CPB-1 or RNA binding; others disrupted RNA binding and were excluded from further consideration. Six mutations, E474A, D488A, A489G, L444A, M472A, and Y479A, disrupted the interaction between FBF-2 and CPB-1 without significantly affecting binding to RNA. A single alanine substitution at position Tyr-479 reduced the level of LacZ reporter activation more than 10-fold. This tyrosine is situated in the loop required for CPB-1 binding (Fig. 4A).
FIGURE 3.
Identification of interaction-defective mutants in FBF-2 using targeted screen. A, logic of the experiment. B and C, measurements of CPB-1 binding in a yeast two-hybrid assay (B) and RNA binding in a yeast three-hybrid assay (C). D, the ratio of LacZ values obtained from the assays in B and C was used to identify mutations that specifically disrupted binding to CPB-1 but not to RNA (black bars). Error bars in B–D indicate S.D.
FIGURE 4.
Clustered interaction-defective alleles and biochemical analysis of key residue, Tyr-479. A, neutral mutations (gray), interaction-defective mutants (purple), and the most defective point mutant (blue) are shown as spheres in the structure of FBF-2 (28). Inset, the single largest loss of interaction point mutant, Y479A, is adjacent to the other loss of interaction mutants. B, additional mutations at position 479 fail to interact with CPB-1 in yeast two-hybrid assays. C, however, RNA binding is uncompromised in yeast three-hybrid assays. Error bars in B and C indicate S.D.
The six residues required for CPB-1 binding are adjacent to one another in the three-dimensional structure of FBF-2 (Fig. 4A) spanning repeats 7 and 8. Mutations that had little or no effect generally lie outside this region. Thus, a discrete surface in FBF-2 appeared to mediate the interaction with CPB-1.
A Conserved Tyrosine Is Required for CPB-1 Binding
To better understand the CPB-1 binding site on FBF-2, we focused on the most severely impaired mutant, Y479A. Single point mutations were introduced at position 479. None restored the ability of FBF-2 to bind CPB-1 to more than 10% of the wild-type level of LacZ activity (Fig. 4B). The most active mutant was Y479G, and the least active was Y479P. The Y479F mutant failed to restore substantial activity, suggesting that the phenolic hydroxyl group was important for CPB-1 binding. To test whether any of the mutant proteins were compromised in RNA binding, we again used yeast three-hybrid assays (Fig. 4C). All the proteins bound RNA as well as wild type. Similarly, in EMSA assays, purified Y479A protein bound RNA as well as wild-type protein in vitro with equilibrium dissociation constants of 0.3 and 0.4 nm, respectively (data not shown). We conclude that Tyr-479 is a critical component of the CPB-1 binding site in FBF-2.
CPB-1 Enhances Repression by FBF-2 in Vitro
CPEB and PUF proteins bind to the 3′-UTRs of target mRNAs and can repress or activate translation (21, 33–37, 47, 48). The biological effects of their interaction have not been directly examined or recapitulated in vitro, nor, to our knowledge, has purified CPEB been shown to affect translation in vitro. To evaluate the effects of the CPB-1·FBF-2 complex on translation, we conducted a series of in vitro translation experiments. Proteins FBF-2 and CPB-1 were translated in vitro, yielding similar amounts of protein for all three FBF proteins analyzed: wild-type FBF, CPB-1 binding-defective (CPBdef, Y479A), and RNA binding-defective (RNAdef, H326A) (supplemental Fig. S1A). To assay translational activity, two reporter mRNAs were then added (Fig. 5A). The 3′-UTR of the firefly luciferase reporter contained a putative FBE derived from the major sperm protein SSP-10 (AUUGUGAAUUG), whereas the Renilla reporter did not contain an FBE. The ratio of firefly to Renilla luciferase activities was used to quantify repression mediated through the FBE (45). The value obtained in a mock control reaction containing only CPB-1 was used to normalize the data. As a control, we examined translation of the Renilla reporter over a broad range of firefly luciferase reporter concentrations (supplemental Fig. S1B). We found that translation in rabbit reticulocyte lysate was not limiting for production of either reporter over a 150-fold range of concentrations.
FIGURE 5.
CPB-1 is required for translational repression of FBE-containing reporter by FBF-2. A, schematic of the cell-free repression assay of translation. Two rounds of translation were conducted. In the first, CPB-1 and FBF-2 proteins were in vitro translated from 50 ng of mRNA. In the second round, newly synthesized proteins were incubated with two reporter RNAs. The firefly luciferase reported contained a 3′-UTR PUF binding element derived from the 3′-UTR of SSP-10. The second Renilla reporter was used as a control. These were incubated for 90 min, and activities of both reporters were quantified. B, the translation of the firefly reporter is repressed only in the presence of FBF-2 and CPB-1. As controls, CPB-1 binding-defective (CPBdef) (Y479A), an RNA binding-defective (RNAdef) (H326A) form of FBF-2, and an FBF-2 binding defective mutant of CPB-1 (FBFdef, L40A) were assayed. FBFdef, FBF-binding defective. FF/Ren, firefly/Renilla. C, the mechanism utilized by FBF-2 in the presence of CPB-1 is not dependent on deadenylation as similar results are obtained on a reporter lacking a poly(A) tail. D, schematic of a modified yeast three-hybrid assay. CPB-1 is expressed with an SV40 nuclear localization sequence (NLS). The effects on FBF-2 binding to RNA are assayed using Lac-Z expression. E, expression of CPB-1 alters binding of FBF-2 to the ssp-10 RNA but not an empty vector or high affinity gld-1a site. Error bars in B, C, and E indicate S.D.
We tested repression by FBF-2 alone or in the presence of the CPB-1 peptide (Fig. 5, B and C). Repression by wild-type FBF-2 was negligible. However, the mixture of CPB-1 and FBF-2 specifically reduced the level of firefly luciferase activity (Fig. 5, B and C). CPB-1-mediated repression was eliminated by point mutations in FBF that prevented its interaction either with CPB-1 or with RNA.
Reporters that possessed a 29-nucleotide poly(A) tail (Fig. 5B) or no poly(A) tail (Fig. 5C) behaved identically. To test whether the CPB-1·FBF-2 complex influenced mRNA stability, we determined the quantity of both reporters prior to and after the translation assays (supplemental Fig. S1C). The FBF-2 and CPB-1 proteins did not alter the stability of the firefly reporter mRNA (supplemental Fig. S1, D and E). These findings suggest that the mechanism of repression by the CPB-1·FBF-2 complex was not dependent on deadenylation.
Similarly, mutant versions of CPB-1 that failed to bind FBF-2 in two hybrid assays also abolished repression in vitro in the presence of FBF-2 (data not shown). We conclude that the physical interaction between CPB-1 and FBF-2 is required for translational repression in vitro.
CPB-1 Affects RNA specificity of FBF-2
To determine whether CPB-1 influences binding of FBF-2 to the ssp-10 FBE, we devised a modified yeast three-hybrid assay (Fig. 5D). In these experiments, binding of FBF-2 to RNA is assayed in the presence or absence of CPB-1 containing a nuclear localization sequence. CPB-1 enhanced LacZ expression 17.5-fold for the ssp-10 FBE (Fig. 5E). However, there was only a modest 1.6-fold stimulation on the high affinity gld-1a site.
A Conserved Site Mediates Human PUF/CPEB Interaction
CPEBs physically associate with PUF proteins in many animal species, including humans, yet the amino acid sequence in the R7/R8 loop region has diverged (6). We reasoned that the site of interaction might nonetheless be constrained. To test this idea, we removed the loop between repeats 7 and 8repeats 7 and 8 from human PUM2, guided by comparisons of the crystal structures of human PUM2 and C. elegans FBF-2 (29, 49). We purified recombinant wild-type PUM2 or mutant PUM2Δ984–989. These two proteins were used in affinity chromatography experiments with recombinant human CPEB3 (Fig. 6A). Wild-type PUM2 interacted with CPEB3. The interaction was RNA-independent because it persisted after treatment with a mixture of purified RNase A and T1. However, the human PUM2 deletion protein failed to bind CPEB3. Both forms of PUM2 bound RNA containing the PUM binding site in vitro (supplemental Fig. S2). We conclude that the loop linking repeats 7 and 8 plays a conserved role in mediating interactions between PUFs and CPEBs.
DISCUSSION
The prevalence and importance of multiprotein complexes are dominant themes in mRNA control. We have focused here on the interactions between two well characterized and biologically important protein families: the PUF and CPEB proteins. Guided by conservation and structural analysis, we identified a loop between PUF repeats 7 and 8 that was required for the interaction of C. elegans FBF-2 and CPB-1. A single substitution in that region, Y479A, disrupted CPB-1 binding and FBF-mediated repression in vitro (Figs. 3D and 5, B and C).
Might the same region of PUF proteins be required for interactions with CPEBs in other organisms? Mouse CPEB3 is expressed in the hippocampus (50) and represses mRNAs to which it is tethered in neuronal cells (51). Guided by structural comparisons, we constructed a deletion mutant in human PUM2 that lacked the segment of FBF-2 that bound CPB-1 (Fig. 6B) (28, 49). The PUM2 deletion mutant was defective in binding to human CPEB3. We conclude that the site of interaction between PUFs and CPEBs is conserved from worms to humans.
We propose that the binding site we have identified is a common platform for interactions between PUF proteins and multiple protein partners (Fig. 6B). The FBF-2/CPB-1 interaction parallels that of Drosophila Pumilio and its protein partner, BRAT (40, 52). Recruitment of BRAT through the combined action of two RNA-binding proteins, Nanos and Pumilio, is required for the regulation of hunchback mRNA (53, 54). Several mutations in Pumilio disrupt its interaction with Nanos and assembly of the ternary complex with BRAT (Fig. 6B) (52). On the basis of these data, a model validated by mutational analysis was proposed for the binding site of BRAT on Pumilio (39). The proposed binding site resides in the C-terminal region directly over repeats 7 and 8. These observations are consistent with the notion that a single region of the PUF domain serves as a hub in a network of protein/protein interactions.
The Saccharomyces cerevisiae Pumilio orthologue, Puf3p, may possess an analogous interaction platform. Puf3p represses COX17 mRNA and promotes its deadenylation and decay (34). A deletion that spans PUF repeats 7 and 8 abrogated regulation, whereas deletion of a loop joining repeats six and seven did not (Fig. 6B) (55). The protein that interacts with this region of Puf3p has not been identified.
We suggest that the region we have identified is an ancient site of collaboration maintained across the PUF family of RNA-binding proteins. However, it is not the only site through which PUF proteins recruit protein partners. For instance, mutations that impair the interaction between PUM2 and AGO reside in the center portion of the protein along the convex surface (42). It remains to be seen whether residues in the PUM2 R7/R8 loop are also involved despite their distance (>20 Å) from the known interaction-defective mutations.
Conservation of interactions between molecules despite sequence divergence at their interface may at first appear enigmatic but is common (56, 57). Sites can diverge in sequence during evolution through neutral mutations at an interface with redundant contacts. Understanding the evolutionary divergence of the PUF-CPEB interface will require analysis of the structures of the complexes. Multiple structures spanning diverse species that represent intermediates during the divergence process may reveal the evolutionary trajectory of this conserved interaction surface.
The CPB-1·FBF-2 complex repressed translation of an FBE-containing reporter in vitro. The repression activity resides exclusively in the complex as neither protein alone is active. Two models could account for the mechanism by which CPB-1 exerts its effects. In the first, CPB-1 enhances the affinity of FBF-2 for certain RNAs. In this fashion, CPB-1 would be required for repression at one site, but not another. Alternatively, CPB-1 could promote an interaction with an unknown third partner that mediates repression. In this view, recruitment of the CPB-1 peptide to the RNA by FBF-2 causes repression of that mRNA in cis.
The Evolutionary Trace method is a potentially powerful means to identify regions that mediate protein/protein interactions, and it yielded interaction-defective alleles of FBF-2. The method requires having a structure that can be used for comparison. An analogous approach led us to amino acids that mediate binding of human PUM2 to AGO/eEF1A, which lie in a different region of the PUF protein (42). We note that Evolutionary Trace revealed three residues, Asn-1105, Gly-1107, and Pro-1108, that are conserved among vertebrate PUF proteins and lie in the loop 7/8 region that mediates binding of human PUM2 to CPEB3.
Our data imply a novel activity for the collaborations among protein partners. Full-length CPEB proteins possess RNA binding motifs that enable binding to specific sequences (CPEs) in 3′-UTRs. In Xenopus oocytes, the activity of a noncanonical CPE is enhanced by an appropriately spaced PUF binding site (8). These data suggest that the two bound proteins interact with one another while they are bound to the RNA. Our studies show that a segment of CPB-1 that lacks its RNA binding regions alters repression even when not bound directly to the target RNA. We infer that CPEB proteins can influence translation through their PUF partners even without binding RNA. Such actions in vivo would be revealed by RNA targets that associate with CPEB yet lack its binding site. Such mRNAs are common in RNA-binding protein immunoprecipitation-microarray profiling of PUF proteins and may indicate effects on specificity due to protein partners.
3′-UTRs are repositories for regulatory elements that dictate assembly of multiprotein complexes. These complexes bind others that are not associated with the RNA, such as the Ccr4·Not complex (58). Understanding how these regulatory assemblies form and exert their effects on translation requires dissection of the interfaces involved. Here, we have presented such an analysis of PUF/CPEB interactions. The data reveal that the two proteins interact and repress translation through a discrete interface, the location of which is conserved among PUF proteins.
Supplementary Material
Acknowledgments
We thank Laura Vanderploeg for help preparing the figures and Melanie A. Preston for careful reading of the manuscript.

This article contains supplemental Figs. S1 and S2.
- CPEB
- cytoplasmic polyadenylation element binding
- CPE
- cytoplasmic polyadenylation element
- PUF
- Pumilio and FBF
- FBF
- fem-3 mRNA-binding factor
- FBE
- FBF binding element
- BRAT
- Brain Tumor.
REFERENCES
- 1. Valencia-Sanchez M. A., Liu J., Hannon G. J., Parker R. (2006) Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20, 515–524 [DOI] [PubMed] [Google Scholar]
- 2. Gebauer F., Hentze M. W. (2004) Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 5, 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Besse F., Ephrussi A. (2008) Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat. Rev. Mol. Cell Biol. 9, 971–980 [DOI] [PubMed] [Google Scholar]
- 4. Osborne H. B., Richter J. D. (1997) Translational control by polyadenylation during early development. Prog. Mol. Subcell. Biol. 18, 173–198 [DOI] [PubMed] [Google Scholar]
- 5. Wickens M., Kimble J., Strickland S. (1996) Translational control of developmental decisions in Translational Control (Hershey J., Mathews M., Sonenberg N., eds) pp. 411–450, Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 6. Richter J. D. (2007) CPEB: a life in translation. Trends Biochem. Sci 32, 279–285 [DOI] [PubMed] [Google Scholar]
- 7. Nakahata S., Katsu Y., Mita K., Inoue K., Nagahama Y., Yamashita M. (2001) Biochemical identification of Xenopus Pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a Nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element-binding protein. J. Biol. Chem. 276, 20945–20953 [DOI] [PubMed] [Google Scholar]
- 8. Piqué M., López J. M., Foissac S., Guigó R., Méndez R. (2008) A combinatorial code for CPE-mediated translational control. Cell 132, 434–448 [DOI] [PubMed] [Google Scholar]
- 9. Nakahata S., Kotani T., Mita K., Kawasaki T., Katsu Y., Nagahama Y., Yamashita M. (2003) Involvement of Xenopus Pumilio in the translational regulation that is specific to cyclin B1 mRNA during oocyte maturation. Mech. Dev. 120, 865–880 [DOI] [PubMed] [Google Scholar]
- 10. Richter J. D., Lasko P. (2011) Translational control in oocyte development. Cold Spring Harb. Perspect. Biol. 3, a002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dubnau J., Chiang A. S., Grady L., Barditch J., Gossweiler S., McNeil J., Smith P., Buldoc F., Scott R., Certa U., Broger C., Tully T. (2003) The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr. Biol. 13, 286–296 [DOI] [PubMed] [Google Scholar]
- 12. Zhang B., Gallegos M., Puoti A., Durkin E., Fields S., Kimble J., Wickens M. P. (1997) A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390, 477–484 [DOI] [PubMed] [Google Scholar]
- 13. Crittenden S. L., Bernstein D. S., Bachorik J. L., Thompson B. E., Gallegos M., Petcherski A. G., Moulder G., Barstead R., Wickens M., Kimble J. (2002) A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417, 660–663 [DOI] [PubMed] [Google Scholar]
- 14. Suh N., Crittenden S. L., Goldstrohm A., Hook B., Thompson B., Wickens M., Kimble J. (2009) FBF and its dual control of gld-1 expression in the Caenorhabditis elegans germline. Genetics 181, 1249–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cho S., Rogers K. W., Fay D. S. (2007) The C. elegans glycopeptide hormone receptor ortholog, FSHR-1, regulates germline differentiation and survival. Curr. Biol. 17, 203–212 [DOI] [PubMed] [Google Scholar]
- 16. Costa-Mattioli M., Sossin W. S., Klann E., Sonenberg N. (2009) Translational control of long-lasting synaptic plasticity and memory. Neuron 61, 10–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hake L. E., Mendez R., Richter J. D. (1998) Specificity of RNA binding by CPEB: requirement for RNA recognition motifs and a novel zinc finger. Mol. Cell. Biol. 18, 685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Radford H. E., Meijer H. A., de Moor C. H. (2008) Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim. Biophys. Acta 1779, 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mendez R., Richter J. D. (2001) Translational control by CPEB: a means to the end. Nat. Rev. Mol. Cell Biol. 2, 521–529 [DOI] [PubMed] [Google Scholar]
- 20. Minshall N., Reiter M. H., Weil D., Standart N. (2007) CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J. Biol. Chem. 282, 37389–37401 [DOI] [PubMed] [Google Scholar]
- 21. Groisman I., Huang Y. S., Mendez R., Cao Q., Theurkauf W., Richter J. D. (2000) CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell 103, 435–447 [DOI] [PubMed] [Google Scholar]
- 22. Tay J., Hodgman R., Richter J. D. (2000) The control of cyclin B1 mRNA translation during mouse oocyte maturation. Dev. Biol. 221, 1–9 [DOI] [PubMed] [Google Scholar]
- 23. Kadyrova L. Y., Habara Y., Lee T. H., Wharton R. P. (2007) Translational control of maternal cyclin B mRNA by Nanos in the Drosophila germline. Development 134, 1519–1527 [DOI] [PubMed] [Google Scholar]
- 24. Standart N., Minshall N. (2008) Translational control in early development: CPEB, P-bodies, and germinal granules. Biochem. Soc. Trans. 36, 671–676 [DOI] [PubMed] [Google Scholar]
- 25. Burns D. M., Richter J. D. (2008) CPEB regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Genes Dev. 22, 3449–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ortiz-Zapater E., Pineda D., Martínez-Bosch N., Fernández-Miranda G., Iglesias M., Alameda F., Moreno M., Eliscovich C., Eyras E., Real F. X., Méndez R., Navarro P. (2012) Key contribution of CPEB4-mediated translational control to cancer progression. Nat. Med. 18, 83–90 [DOI] [PubMed] [Google Scholar]
- 27. Wickens M., Bernstein D. S., Kimble J., Parker R. (2002) A PUF family portrait: 3′-UTR regulation as a way of life. Trends Genet. 18, 150–157 [DOI] [PubMed] [Google Scholar]
- 28. Wang Y., Opperman L., Wickens M., Hall T. M. (2009) Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein. Proc. Natl. Acad. Sci. U.S.A. 106, 20186–20191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang X., McLachlan J., Zamore P. D., Hall T. M. (2002) Modular recognition of RNA by a human Pumilio homology domain. Cell 110, 501–512 [DOI] [PubMed] [Google Scholar]
- 30. Wang X., Zamore P. D., Hall T. M. (2001) Crystal structure of a Pumilio homology domain. Mol. Cell 7, 855–865 [DOI] [PubMed] [Google Scholar]
- 31. Edwards T. A., Pyle S. E., Wharton R. P., Aggarwal A. K. (2001) Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell 105, 281–289 [DOI] [PubMed] [Google Scholar]
- 32. Zhu D., Stumpf C. R., Krahn J. M., Wickens M., Hall T. M. (2009) A 5′ cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs. Proc. Natl. Acad. Sci. U.S.A. 106, 20192–20197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wharton R. P., Struhl G. (1991) RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell 67, 955–967 [DOI] [PubMed] [Google Scholar]
- 34. Olivas W., Parker R. (2000) The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 19, 6602–6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wreden C., Verrotti A. C., Schisa J. A., Lieberfarb M. E., Strickland S. (1997) Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development 124, 3015–3023 [DOI] [PubMed] [Google Scholar]
- 36. Goldstrohm A. C., Seay D. J., Hook B. A., Wickens M. (2007) PUF protein-mediated deadenylation is catalyzed by Ccr4p. J. Biol. Chem. 282, 109–114 [DOI] [PubMed] [Google Scholar]
- 37. Hook B. A., Goldstrohm A. C., Seay D. J., Wickens M. (2007) Two yeast PUF proteins negatively regulate a single mRNA. J. Biol. Chem. 282, 15430–15438 [DOI] [PubMed] [Google Scholar]
- 38. Kaye J. A., Rose N. C., Goldsworthy B., Goga A., L'Etoile N. D. (2009) A 3′-UTR Pumilio-binding element directs translational activation in olfactory sensory neurons. Neuron 61, 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Edwards T. A., Wilkinson B. D., Wharton R. P., Aggarwal A. K. (2003) Model of the Brain Tumor-Pumilio translation repressor complex. Genes Dev. 17, 2508–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sonoda J., Wharton R. P. (1999) Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 13, 2704–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eckmann C. R., Crittenden S. L., Suh N., Kimble J. (2004) GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics 168, 147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Friend K., Campbell Z. T., Cooke A., Kroll-Conner P., Wickens M. P., Kimble J. (2012) A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nat. Struct. Mol. Biol. 19, 176–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ariz M., Mainpal R., Subramaniam K. (2009) C. elegans RNA-binding proteins PUF-8 and MEX-3 function redundantly to promote germline stem cell mitosis. Dev. Biol. 326, 295–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Luitjens C., Gallegos M., Kraemer B., Kimble J., Wickens M. (2000) CPEB proteins control two key steps in spermatogenesis in C. elegans. Genes Dev. 14, 2596–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chritton J. J., Wickens M. (2010) Translational repression by PUF proteins in vitro. RNA 16, 1217–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rajagopalan L., Pereira F. A., Lichtarge O., Brownell W. E. (2009) Identification of functionally important residues/domains in membrane proteins using an evolutionary approach coupled with systematic mutational analysis. Methods Mol. Biol. 493, 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mendez R., Hake L. E., Andresson T., Littlepage L. E., Ruderman J. V., Richter J. D. (2000) Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature 404, 302–307 [DOI] [PubMed] [Google Scholar]
- 48. Barnard D. C., Ryan K., Manley J. L., Richter J. D. (2004) Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119, 641–651 [DOI] [PubMed] [Google Scholar]
- 49. Lu G., Hall T. M. (2011) Alternate modes of cognate RNA recognition by human PUMILIO proteins. Structure 19, 361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Theis M., Si K., Kandel E. R. (2003) Two previously undescribed members of the mouse CPEB family of genes and their inducible expression in the principal cell layers of the hippocampus. Proc. Natl. Acad. Sci. U.S.A. 100, 9602–9607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huang Y. S., Kan M. C., Lin C. L., Richter J. D. (2006) CPEB3 and CPEB4 in neurons: analysis of RNA binding specificity and translational control of AMPA receptor GluR2 mRNA. EMBO J. 25, 4865–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sonoda J., Wharton R. P. (2001) Drosophila Brain Tumor is a translational repressor. Genes Dev. 15, 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hülskamp M., Schröder C., Pfeifle C., Jäckle H., Tautz D. (1989) Posterior segmentation of the Drosophila embryo in the absence of a maternal posterior organizer gene. Nature 338, 629–632 [DOI] [PubMed] [Google Scholar]
- 54. Irish V., Lehmann R., Akam M. (1989) The Drosophila posterior-group gene nanos functions by repressing hunchback activity. Nature 338, 646–648 [DOI] [PubMed] [Google Scholar]
- 55. Houshmandi S. S., Olivas W. M. (2005) Yeast Puf3 mutants reveal the complexity of Puf-RNA binding and identify a loop required for regulation of mRNA decay. RNA 11, 1655–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Haag E. S. (2007) Compensatory versus pseudocompensatory evolution in molecular and developmental interactions. Genetica 129, 45–55 [DOI] [PubMed] [Google Scholar]
- 57. Haag E. S., Molla M. N. (2005) Compensatory evolution of interacting gene products through multifunctional intermediates. Evolution 59, 1620–1632 [PubMed] [Google Scholar]
- 58. Goldstrohm A. C., Wickens M. (2008) Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 9, 337–344 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.