Background: The molecular basis for sequence-dependent variation in DNA repair is poorly understood.
Results: A systematic study of lesion encounter by MutM reveals major differences in stacking of the target oxoG.
Conclusion: Sequence-dependent changes in base stacking may contribute to lesion extrusion and repair.
Significance: This is the first structural study of sequence context effects on lesion recognition for a DNA repair enzyme.
Keywords: Base Excision Repair, DNA Damage, DNA Repair, DNA-Protein Interaction, Structural Biology, DNA Glycosylase, Disulfide Cross-linking, Lesion Recognition
Abstract
MutM, a bacterial DNA-glycosylase, plays a critical role in maintaining genome integrity by catalyzing glycosidic bond cleavage of 8-oxoguanine (oxoG) lesions to initiate base excision DNA repair. The task faced by MutM of locating rare oxoG residues embedded in an overwhelming excess of undamaged bases is especially challenging given the close structural similarity between oxoG and its normal progenitor, guanine (G). MutM actively interrogates the DNA to detect the presence of an intrahelical, fully base-paired oxoG, whereupon the enzyme promotes extrusion of the target nucleobase from the DNA duplex and insertion into the extrahelical active site. Recent structural studies have begun to provide the first glimpse into the protein-DNA interactions that enable MutM to distinguish an intrahelical oxoG from G; however, these initial studies left open the important question of how MutM can recognize oxoG residues embedded in 16 different neighboring sequence contexts (considering only the 5′- and 3′-neighboring base pairs). In this study we set out to understand the manner and extent to which intrahelical lesion recognition varies as a function of the 5′-neighbor. Here we report a comprehensive, systematic structural analysis of the effect of the 5′-neighboring base pair on recognition of an intrahelical oxoG lesion. These structures reveal that MutM imposes the same extrusion-prone (“extrudogenic”) backbone conformation on the oxoG lesion irrespective of its 5′-neighbor while leaving the rest of the DNA relatively free to adjust to the particular demands of individual sequences.
Introduction
Oxidative damage to DNA, resulting primarily from attack by the electrophilic byproducts of aerobic respiration, represents a significant source of endogenous genotoxicity in cells. A particularly mutagenic event is the oxidation of guanine to 7,8-dihydro-8-oxoguanine (8-oxoguanine (oxoG)4; see Fig. 1A) due to the high propensity of oxoG to mispair with adenine during processive DNA replication and thereby giving rise to G:C → T:A transversion mutations (1). In bacteria, removal of oxoG is catalyzed by MutM, a bifunctional DNA glycosylase/lyase that excises oxoG from the oxoG:C base pair (2). In eukaryotes from fungi to humans, the same function is carried out by the structurally non-homologous enzyme Ogg1 (3, 4). A major question confronting the field of DNA repair is how these repair enzymes locate and detect rare lesions such as oxoG in the presence of an astronomical (106-107-fold) excess of undamaged bases, with only thermal diffusion as their available means of propulsion. The search process is further complicated by the fact that all DNA glycosylases acting on monomeric lesions, including MutM and Ogg1, catalyze base-excision only after extruding the substrate nucleobase entirely from the DNA helix and inserting it into an extrahelical active site on the enzyme (2, 5–7). Although obligate extrusion of helix-destabilizing lesions would seem to pose little structural or energetic challenge to the enzyme, being as these lesions undergo spontaneous extrusion at an appreciable rate, the situation is considerably more difficult with oxoG, which has little effect on the structure and energetics of duplex DNA (8–11). Thus, with the possible exception of differences in structural dynamics, all MutM and Ogg1 have as signposts are the two atoms that differ between oxoG and its closest structural relative, G, those being, respectively, O versus H at C8 and NH versus N at the 7-position (see Fig. 1A) (12).
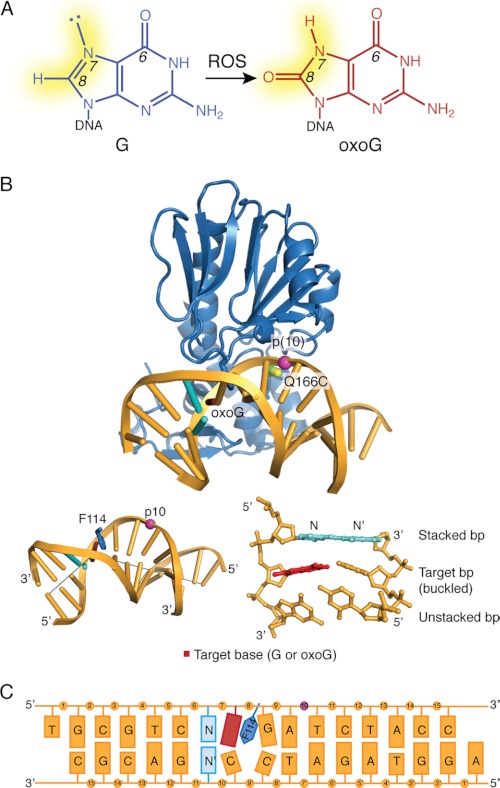
FIGURE 1.
A, formation of oxoG from guanine (G) by reactive oxygen species (ROS) is shown. The changes in atom substituents at N7 and C8 are highlighted in yellow. B, the structure of MutM cross-linked to an intrahelical oxoG (PDB code 3GP1) is shown. The cross-linking sites on the protein and DNA are as indicated. A view of the base-stacking to the target base is shown in the bottom right (with the stacked base pair shown in cyan); insertion of Phe114 into the duplex and the resulting buckle in the DNA (with gray lines marking the helical axis flanking the target base) is shown at the bottom left. C, shown is a schematic diagram of the DNA used in this study. The target base, shown in red, was either G or oxoG; base N is the 5′-stacking neighbor fg and was varied between A, T, and G. The modified phosphate group is shown in purple.
A substantial body of structural and supporting biochemical and computational information has provided a rich picture of the events after extrusion of the lesion from DNA and leading up to the moment of catalysis (13–22). Only quite recently, however, did the first structural snapshots become available of MutM at the stage of its initial encounter with a fully intrahelical, base-paired oxoG lesion; indeed, these structures provided the first view of any DNA glycosylase encountering an intrahelical lesion in duplex DNA (19). The most striking feature of these structures is the muscular but localized inspection of the DNA helix see (Fig. 1B), with the DNA exhibiting a pronounced bend localized entirely at the site of the target base pair, with the amino acid side chain of Phe114 being intercalated on the 3′-side of the lesion, and with the target base pair being drastically buckled (19). Sequence-matched structures having the oxoG lesion swapped for an undamaged G, a change of only two atoms throughout the entire protein-DNA interface, showed unambiguously that MutM detects the presence of an intrahelical oxoG lesion (13, 19). Specifically, the bending and buckling induced by MutM causes the 8-oxo carbonyl of oxoG to clash with the DNA backbone at the lesion-containing site, thereby inducing movement of the backbone along a trajectory coincident with that of base extrusion; in other words, the presence of oxoG creates an extrudogenic conformation in the DNA backbone. The C8-H atom of G, being smaller and less electron-rich, induces no such conformational change. Computational simulations based on these structures confirmed that the activation barrier for extrusion of oxoG is lower by ∼7 kcal/mol than with G (19). Additional sequence-matched sets of structures, changing from set to set the base pair immediately 5′ or 3′ to the target pair (oxoG:C or G:C), revealed that in all cases the oxoG structure showed the same extrudogenic conformation. Some conformational heterogeneity, however, was seen among the non-lesion-containing (G) structures, specifically when the 5′-base pair was changed. These hints of sequence dependence on the ability of MutM to induce deformation of the DNA backbone provided the impetus for this study.
It has long been known that oxoG is more mutagenic in some sequences than in others, and evidence suggests that repair efficiency may contribute to this behavior. For example, although MutM can remove oxoG from all sequence contexts, the enzyme repairs oxoG more efficiently from pyrimidine-rich sequences than purine-rich sequences (23). Similarly, in vivo studies varying the sequence flanking known G → T hotspots, which result from replication of an oxoG:A base pair, in the supF gene showed higher mutation frequencies when flanked by purine bases on either side of the hotspot site (24, 25). These data are consistent with statistical analysis of the sequence context surrounding G → T transversion mutations in the Escherichia coli lacI, human p53, and human factor IX genes, which show an overrepresentation of a 5′-GNA-3′ sequence flanking the mutated base (N) (23). The authors went so far as to propose that these sequences minimize lesion-induced distortion of the DNA, allowing oxoG to escape detection by repair enzymes (23). Studies using plasmids containing a site-specific oxoG placed in different sequence contexts showed significantly higher mutation frequencies (>90% G → T transversions) when oxoG was placed in a GC-rich stretch of DNA (26).
In this study we have focused our initial investigation of sequence-dependent structural changes on the 5′-base pair flanking the lesion. This choice follows directly from the structures of MutM interrogating fully base-paired DNA, which showed the 3′-base pair to be fully unstacked from the target base pair through the intercalation of Phe114 (see Fig. 1B). The 5′-base pair not only retains its stacking interaction with the target base pair in these structures but is forced into an even more intimate association through buckling of the target base pair in the 5′-direction. Below we describe a complete set of sequence-matched structures in which we have completed a systematic variation of the 5′-base pair to include all four possible permutations, with either G or oxoG being targeted for extrusion by the enzyme. Here we report the results of this comprehensive analysis of sequence variation on intrahelical recognition by MutM, and we discuss the implications for sequence-dependence in DNA repair by this enzyme.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Mutant MutM proteins were overexpressed and purified as previously described (19). MutM was expressed in E. coli BL21 (DE3) plysS cells and grown at 37 °C until A600 reached 0.5–0.7. Protein expression was induced by the addition of 0.5 mm isopropyl-β-d-thiogalactopyranoside (Invitrogen) and 0.05 mm ZnCl2. The cells were allowed to grow for 4–5 h at 30 °C, harvested by ultracentrifugation, resuspended in 50 mm Na2HPO4, pH 8.0, 500 mm NaCl, and 0.1% β-mercaptoethanol, flash-frozen in liquid nitrogen, and stored at −80 °C.
Thawed cells were supplemented with 1 mm PMSF and one Complete, EDTA-free Protease Inhibitor Mixture tablet (Roche Applied Science), lysed by sonication, and clarified by centrifugation at 14,000 rpm for 20 min. The clarified lysate was diluted 1:5 with buffer A (20 mm Tris, pH 7.4, and 0.1% β-mercaptoethanol) and loaded onto a 20-ml SFF column (GE Healthcare). The protein was purified using a linear gradient from 0.1 to 0.5 m NaCl over 10 column volumes followed by size-exclusion chromatography (Superdex75, GE Healthcare) into 1 m NaCl, 10 mm Tris, pH 7.4, 5 mm β-mercaptoethanol. Unused protein was stored by adding glycerol to 20% final concentration, flash-frozen, and stored at −80 °C.
DNA Synthesis and Purification
All DNA substrates were synthesized using solid phase synthesis on an ABI 392 DNA synthesizer using standard reagents and protocols (Applied Biosystems, Glen Research). For the cross-linker-modified oligos, the protocol was modified to include a 3′-H-phosphonate (Glen Research) in the coupling step at the site of the backbone modification. After coupling, the disulfide tether was incorporated onto the backbone by an oxidation step with carbon tetrachloride (Sigma) and the diamine disulfide (free base). All oligos were purified according to the same protocol. The oligonucleotides were deprotected and cleaved from the CPG resin using ammonium hydroxide at 55 °C for 8–10 h. The ammonium hydroxide was removed using a SpeedVac, and the oligos were purified using the crush-and-soak method (27). The oligos were desalted using hydrophobic chromatography (Sep pak columns, Waters chromatography) and then evaporated to dryness using a SpeedVac. The oligos were dissolved in 10 mm Tris, pH 8.0, and their appropriate masses were verified using MALDI-TOF mass spectrometry. The lesion-containing strand was annealed to the complementary strand in a 1:1.2 ratio in 1× NaTE (50 mm NaCl, 10 mm Tris-HCl, 1 mm EDTA, pH 8.0).
Preparative Cross-linking Reactions
All complexes were made by cross-linking the corresponding DNA substrates to E3Q Q166C V222P MutM. Reactions were set up using 10 μm cross-linker-containing duplex substrate with 20 μm protein in degassed cross-linking buffer (50 mm NaCl and 20 mm Tris, pH 7.4). The reactions were purged with argon and kept rotating at 4 °C for 2–3 days.
The reactions were purified using a Mono Q column (GE Healthcare) with a linear gradient from 0.1 to 0.6 m NaCl in buffer A (20 mm Tris pH 7.4, degassed) over 30 column volumes. Fractions were pooled, and buffer was exchanged into degassed cross-linking buffer and concentrated to ∼200–250 μm for crystallization experiments. The concentration was determined by the A260 of the DNA in the complex.
Crystallization, Data Collection, and Structure Determination
Crystallization drops were set up at 4 °C using the hanging drop vapor diffusion method in a 1:1 or 1:2 ratio of protein-DNA complex to reservoir solution. The complexes crystallized in 12–18% PEG 8000, 100 mm sodium cacodylate, pH 7.0, and 5% glycerol. Crystals appeared with 2–3 days and were allowed to grow to size over 1–2 weeks before briefly soaking in cryoprotectant solution containing 18% PEG 8000, 100 mm sodium cacodylate, pH 7.0, and 25% glycerol followed by flash-freezing in liquid nitrogen. Diffraction data were collected at the 24ID-E beamline at Argonne Photon Source. Data were processed using the HKL2000 program suites (28).
Protein coordinates from the isomorphous structure of MutM cross-linked to undamaged DNA (PDB code 3GPX) were used as the initial model for refinement (19). Rigid body fitting, energy minimization, and simulated annealing in PHENIX resulted in a partial model (29). The DNA (including the disulfide tether) was built into the Fo − Fc map using the strong density for the cross-link to determine the register. Manual readjustment of protein side chains and DNA bases were done in COOT followed by successive rounds of simulated annealing, energy minimization, and individual B-factor refinement (30, 31). After all visible protein and DNA atoms were built and the Rfree dropped below 28%, water molecules were added to the model using automated water picking and manual inspection of the difference map in PHENIX. TLS refinement was also included during refinement, with manual selection of TLS groups (two groups total; the protein and then both DNA strands were treated as one TLS group) (32, 33). Electron density for residues 217–237 (comprising the oxoG-capping loop) was not visible, and these residues were omitted from the model. Protein side chains were truncated in instances where density was not observed. Data collection and refinement statistics are shown in Tables 1–2 and supplemental Table S1.
TABLE 1.
Data collection and refinement statistics for set 1 NpGo structures
| Data collection | GpGo | ApGo | TpGo |
|---|---|---|---|
| Radiation source | APS-24-IDE | APS-24-IDE | APS-24-IDE |
| Resolution (Å) | 50-1.85 | 50-1.80 | 50-1.70 |
| Unique reflections | 38,874 | 40,545 | 49,297 |
| Completeness (%)a | 99.4 (99.1) | 96.8 (97.6) | 99.1 (98.4) |
| Redundancya | 4.4 (4.6) | 4.9 (4.8) | 6.5 (6.0) |
| Rmergea,b | 0.052 (0.476) | 0.055 (0.524) | 0.055 (0.578) |
| 〈I/σ〉a | 32.5 (3.1) | 31.5 (2.8) | 44.1 (3.6) |
| Space group | P212121 | P212121 | P212121 |
| Unit cell dimensions | a + 45.42 | a + 45.25 | a + 45.28 |
| b + 93.42 | b + 93.59 | b + 93.94 | |
| c + 104.54 | c + 104.23 | c + 104.13 | |
| Refinement and model | |||
| Resolution (Å) | 32.7-1.87 | 32.7-1.80 | 32.7-1.70 |
| Rworka,c (%) | 18.6 (20.9) | 18.9 (21.0) | 19.1 (18.9) |
| Rfreea,c (%) | 21.6 (26.8) | 21.1 (25.4) | 20.7 (21.4) |
| Mean B-factors | |||
| Protein | 27.63 | 27.47 | 27.03 |
| Water | 40.31 | 39.43 | 39.70 |
| Root mean square deviation from ideality | |||
| Bond lengths (Å) | 0.007 | 0.006 | 0.007 |
| Bond angles (°) | 1.283 | 1.217 | 1.287 |
| Ramachandran plotd (%) | |||
| Most favored | 95.3 | 94.4 | 93.9 |
| Additionally allowed | 4.2 | 5.2 | 5.6 |
| Generously allowed | 0.5 | 0.5 | 0.9 |
| PDB ID | 3U6D | 3U6C | 3U6E |
a Values in parentheses refer to the highest resolution shell.
b Rmerge + Σ|I − 〈I〉|/Σ〈I〉, where I is the observed intensity.
c Rwork + Σ|Fo − Fc|/Σ|Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively. Rfree was calculated based on 5% data randomly selected and omitted throughout structure refinement (50).
d Values calculated using PROCHECK (34).
TABLE 2.
Data collection and refinement statistics for Set 1 NpG structures
| Data collection | GpG | ApG | TpG |
|---|---|---|---|
| Radiation source | APS-24-IDE | APS-24-IDE | APS-24-IDE |
| Resolution (Å) | 50-1.60 | 50-1.90 | 50-1.77 |
| Unique reflections | 58,218 | 34,913 | 43,744 |
| Completeness (%)a | 99.9 (100.0) | 97.4 (97.8) | 99.6 (99.2) |
| Redundancya | 4.6 (4.8) | 4.5 (4.4) | 4.6 (4.3) |
| Rmergea,b | 0.053 (0.561) | 0.053 (0.226) | 0.059 (0.572) |
| I/Σa | 29.5 (2.4) | 38.7 (6.7) | 32.9 (2.9) |
| Space group | P212121 | P212121 | P212121 |
| Unit cell dimensions | a + 45.31 | a + 45.31 | a + 45.24 |
| b + 93.86 | b + 94.64 | b + 94.62 | |
| c + 104.01 | c + 103.26 | c + 103.11 | |
| Refinement and model | |||
| Resolution (Å) | 32.7-1.60 | 32.7-1.90 | 32.7-1.77 |
| Rworka,c (%) | 19.3 (23.3) | 18.3 (16.9) | 17.9 (20.5) |
| Rfreea,c (%) | 20.5 (12.7) | 19.9 (17.7) | 19.4 (17.2) |
| Mean B-factors | |||
| Protein | 24.83 | 26.61 | 24.45 |
| Water | 40.62 | 38.73 | 39.01 |
| Root mean square deviation from ideality | |||
| Bond lengths (Å) | 0.005 | 0.007 | 0.007 |
| Bond angles (°) | 1.268 | 1.309 | 1.331 |
| Ramachandran plotd (%) | |||
| Most favored | 94.4 | 94.4 | 93.0 |
| Additionally allowed | 5.2 | 5.2 | 6.6 |
| Generously allowed | 0.5 | 0.5 | 0.5 |
| PDB ID | 3U6P | 3U6O | 3U6S |
a Values in parentheses refer to the highest resolution shell.
b Rmerge + Σ|I − 〈I〉|/Σ〈I〉; where I is the observed intensity.
c Rwork + Σ|Fo − Fc|/Σ|Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively. Rfree was calculated based on 5% data randomly selected and omitted throughout structure refinement (50).
d Values were calculated using PROCHECK (34).
Model statistics and validation were carried out using PROCHECK and Molprobity (34, 35). Analysis of the DNA conformation including base-step parameters, sugar puckers, and torsion angles were done using 3DNA (36). The figure were made using PyMOL Version 1.3 (37).
oxoG Cleavage Assays
The duplex DNA substrate used for the cleavage assays was identical in sequence to the oligonucleotides used for crystallization but did not contain an N-ethylthio tether. The oxoG-containing strands were 5′-end-labeled using T4 polynucleotide kinase (New England Biolabs) and [γ-32P] ATP (PerkinElmer Life Sciences) and then annealed with a 1.1-fold excess of the complementary strand. Single turnover cleavage reactions were carried out using 100 nm DNA duplex and 150 nm wild-type MutM in a standard reaction buffer of 150 mm NaCl, 50 mm Tris-HCl, pH 7.4, and 10 mm MgCl2 at room temperature. Aliquots of the reaction were removed periodically and quenched with an equal amount of 100 mm dithiothreitol in 95% formamide and 1× Tris borate/EDTA buffer, subjected to denaturing urea-PAGE, and then visualized on a phosphorimaging plate. Quantification of the cleavage product was done using ImageQuant TL (GE Healthcare).
RESULTS
Experimental Strategy
Our previous studies on the inspection of an intrahelical target base pair in duplex DNA by MutM employed a sequence denoted IC3 and its congener EC3, which differed only in whether the target nucleobase (Fig. 1C, red) was G or oxoG, respectively. This particular sequence contains the nucleobase C stacked over the target nucleobase on the 5′-side (i.e. Fig. 1C, N = C; N′ = G) (13, 19). We systematically varied this 5′-stacking nucleobase N to include all other possibilities (N = G, A, and C), while varying the target nucleobase as either G or oxoG; in all cases, complementary nucleobases (N′) were incorporated on the paired DNA strand (Fig. 1B; Tables 1 and 2). The four NpG structures contain a target G at the target site (red in Fig. 1C), whereas the four corresponding sequence-matched NpGo structures contain a target oxoG. With the exception of the N/N′ base pair, the DNA sequences remain identical to the published IC3/EC3 sequence. As in our previous study, the MutM/DNA complexes were stabilized using a disulfide cross-linking system designed and validated for MutM (13). This system uses as cross-linking components a variant of MutM bearing an engineered Cys at position 166 in MutM (Q166C mutation) and duplex DNA containing a two-carbon thiol tether attached to DNA at the 10th phosphate (p10) on the lesion-containing strand (Fig. 1, B and C).
The protein used in these studies also included the V222P mutation, which is necessary to prevent formation of an extrahelical lesion recognition complex in the NpGo series of structures (19). Although this mutation is unnecessary for obtaining the intrahelical state with the NpG series, it was maintained in that series to minimize extraneous alterations and provide a consistent comparison between the NpG and NpGo series of structures. Thus, the only point of difference between each sequence-matched NpG and NpGo structure is the two-atom change in the target nucleobase (Fig. 1A).
Sequence-independent Features of MutM-DNA Interaction
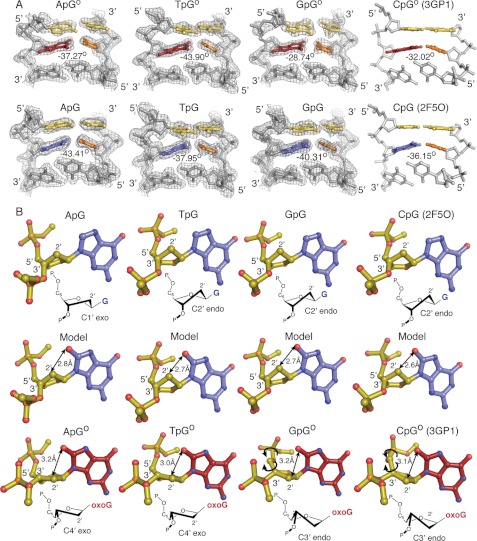
All the newly determined structures presented here show a global architecture very similar to each other and to the previously published IC3 and EC3 structures (PDB IDs 3GP1 and 2F5O, respectively). Superpositions using only Cα atoms between each NpGo structure to the EC3 structure (corresponding to the CpGo state) and between each NpG structure with the IC3 structure (corresponding to the CpG state) yielded an average root mean square deviation 0.20 and 0.25 Å, respectively (19). Both the target base pair and the base pair 3′ to the target base are buckled via insertion of Phe114, whereas the base pair 5′ to the target base remains planar and stacked with the target base pair (Fig. 2A). At the target base, comparisons between the sequence-matched sets of NpG and NpGo structures reveal several stereotypic changes in the sugar pucker and backbone conformation that, as previously described for IC3 and EC3, appear to be driven by the need to accommodate the O8 atom present in oxoG but replaced in G by the smaller H8 atom. Specifically, the presence of O8 is associated with a consistent rearrangement of the sugar pucker from the C2′ endo conformation, found in the NpG structures, to the C4′ exo or C3′ endo conformation in the NpGo structures (Fig. 2B). Maintaining a C2′ endo pucker conformation of oxoG would result in a steric clash between the C2′ atom and the O8; however, the change in pucker shifts the C2′ carbon away from the O8, preventing the unfavorable contact. Similarly, rotations in the phosphodiester and C4′-C5′ bonds reposition the phosphate backbone away from the O8, relieving the repulsive interactions between the phosphate oxygens and the O8. These changes in conformation are localized to the immediate vicinity of the oxoG, with the remainder of the DNA backbone being nearly unchanged between each NpGo structure (supplemental Fig. S1A). This study thus confirms and extends the previous conclusions that MutM detects the presence of an oxoG lesion by grasping the DNA in such a way as to force a steric and electronic clash between the oxoG O8 atom and the backbone of that same residue; the clash is avoided through an adjustment in backbone conformation that lowers the kinetic barrier for extrusion of the target nucleoside from DNA (19).
FIGURE 2.
A, buckling of the target base pair and the 3′-neighboring base pair is shown. The target base is shown as red for oxoG and blue for G, with the cytosine base-pairing partner shown in orange. The 5′-stacking neighbor base pair to the target base is shown in yellow. Buckle angles were calculated using 3DNA. The 2 Fo − Fc maps for the new structures reported in this manuscript are contoured to 1.0 σ. B, steric clashes resulting from the O8 carbonyl are shown. Double-headed arrows indicate the distance between the O8 atom and the C2′ carbon when modeled into a NpG structure or in the NpGo structure. The sugar pucker for each structure is as indicated. Single-headed arrows indicate rotations in the phosphodiester bonds.
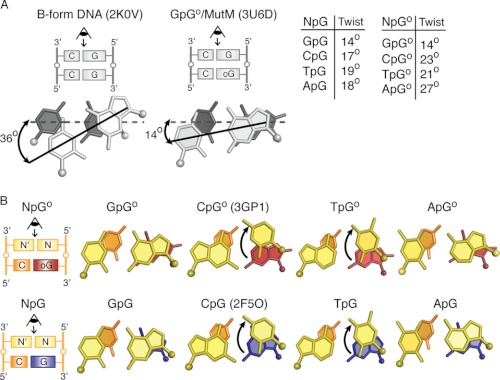
Sequence-specific Effects on Base Stacking
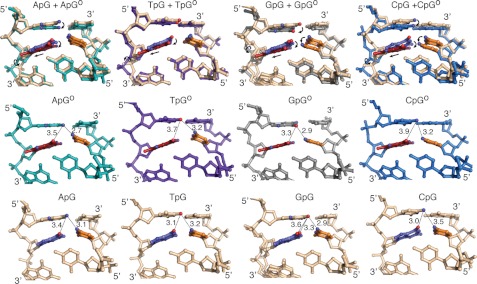
The base pair step between the target base and the 5′-stacking neighbor is significantly underwound in all the NpG and NpGo structures, as judged by analysis of base step parameters using 3DNA (36). This step in the NpG and NpGo structures had an average twist value of 17.1 ± 1.8° and 21.6 ± 5.5°, respectively, as compared with 36° for B-form DNA (Fig. 3A). The decrease in rotation of the base pairs along the helix axis significantly increases the extent of base pair stacking relative to that in canonical B-form DNA (38), as judged by inspection of helix projections of target base and the 5′-stacking neighbor (Fig. 3); that these base-pairs are indeed in π-π contact in all cases is evident in views perpendicular to the helix axis (Fig. 3B). The extent of DNA unwinding is greatest in the GpG and GpGo structures, which have helical twist values of 14.7° and 14.4°, respectively. In both of these structures a view down the helical axis shows nearly complete overlap between the 5′-G and the target base, an extent of stacking rarely observed for DNA in any other context. Similar views for the other structures in each set show different degrees of overlap that vary in magnitude depending on the twist angle and the identity of the 5′-base (Fig. 3).
FIGURE 3.
A, shown is the effect of Twist angle on base-stacking. The target base pair is shown in dark gray, and the 5′-base pair shown in light gray. The dotted line is drawn along the base-pairing axis of the target base pair, whereas the solid line indicates the base-pairing axis for the stacked base pair. A table of Twist angles for each structure is shown to the right. The angles correspond to Twist values for that base step as calculated by 3DNA. B, shown is a base-stacking diagram for oxoG (Go) and G-containing structures. Arrows indicate movement of 5′ base. PDB codes for previously published structures are indicated.
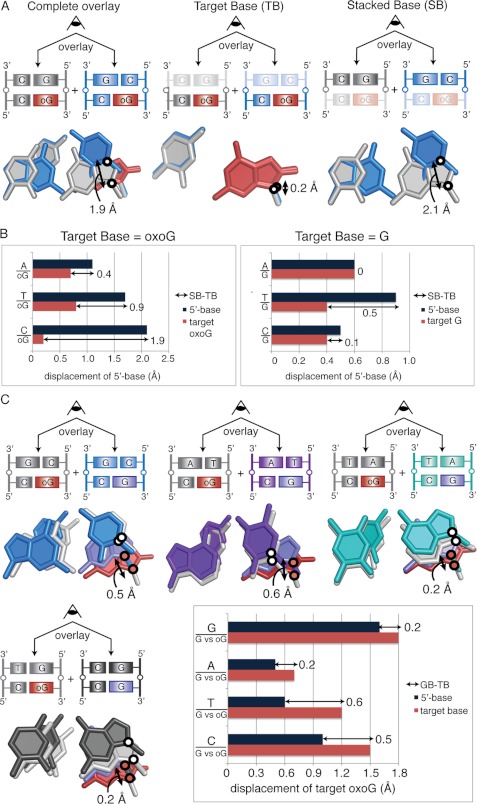
To facilitate comparisons between the various dinucleotide sequences, we decided to superimpose each on a common reference structure. Any member of the sequence-matched quartet of structures could have served as a valid reference, but we chose GpG and GpGo as reference structures because these exhibit the least extent of helical twist and consequently have the greatest extent of stacking. Thus, each of the NpG structures was superimposed on GpG and the NpGo structures were superimposed on GpGo. We selected the glycosidic nitrogen atom of the stacked base (SB) and target base (TB) as our positional marker, because this nitrogen atom is the one invariant structural feature of all nucleobases. One caveat of this analysis is that the distances measured encompass displacement in all three dimensions, such as movement in the direction of the helical axis in addition to movements in the plane of the base. However, the majority of the 5′-N bases in each alignment remain nearly coplanar with the 5′-G in the reference structures (supplemental Fig. S1C), allowing us to treat the net displacement as representative of translational motion in the plane of the base pair. As illustrated in Fig. 4A for the superposition of CpGo and GpGo, the displacement of the target base (TB, middle panel) is quite modest (0.2 Å) compared with that of the stacked base (SB, right-hand panel) (2.1 Å). The bar graphs in Fig. 4B show the displacements of SB (dark blue bars) and TB (red bars) for the quartet of sequence-matched structures having a target Go (left panel) and a target G (right panel). Several trends become evident from this analysis. First, considering that the margin of error of the superpositions is within the range of a few tenths of Å, the target base pair shows only minor sequence-dependent positional variation. The stacked base, on the other hand, shows considerable sequence-dependent positional variation for pyrimidines stacked over a target Go but much less variation for purines stacked over a target Go. Specifically, the 5′-pyrimidines in the CpGo and TpGo structures are significantly offset from the GpGo state, with net displacements of 1.9 and 0.9 Å, respectively; in contrast, the ApGo structure is offset by only 0.4 Å from the GpGo state. With a target G, neither stacked purines nor pyrimidines show pronounced sequence-dependent displacement. When the stacked base is a purine, SB and TB tend to move in lock step. Although Fig. 4B does not capture the directionality of the displacement, inspection of the structures (refer to Fig. 3) reveals that indeed SB and TB are being displaced in the same direction in comparing GpGo with ApGo and in comparing GpG with ApG. This is at least suggestive that conservation of stacking interactions is more important for sequences having a purine base stacked over the target base than for those having a stacked pyrimidine.
FIGURE 4.
A, calculation of net displacement of the 5′-stacking neighbor is shown. The CpGo structure is shown in blue, and the GpGo structure is shown in gray. Black circles mark the position of the glycosidic nitrogens used for the measurements. Each base pair is viewed down the helical axis. Measurement of the displacement of the SB is shown to the far right and of the TB is in the middle. Subtraction of TB from SB results in the net displacement of the stacked base, shown to the far left. B, the distances between glycosidic nitrogens measured for each NpGo aligned with GpGo pair are shown for NpGo (left) and NpG structures (right). C, shown is net displacement of oxoG compared with G in the NpG versus NpGo structures. White circles mark the glycosidic nitrogens on the 5′-base-stacking partner, and red circles mark the glycosidic nitrogens on the target base. Overlays are shown for CpG/Go (blue), TpG/Go (purple), ApG/Go (teal), and GpG/Go (gray) alignments. The net displacement was calculated using the average of the distance between oxoG and G from both conformations. The net displacement of the target base is calculated by subtracting the movement of 5′-base (dark blue bars) from the movement of the target base (pink bars).
Sequence-specific Displacement of oxoG toward Minor Groove
Next we analyzed the response of duplex structure to changing the target base, G versus Go, in an otherwise invariant sequence. In this analysis we designated each NpG structure as the reference point for a fully intrahelical, nonextruded target base and measured the displacement of each NpGo structure with respect to the position of the corresponding sequence-matched NpG structure. Each NpG structure was aligned to a sequence-matched NpGo structure, and the displacement between glycosidic nitrogens for the target G and oxoG was measured (GB). To correct for global variations in the alignment of the two structures, the distance between glycosidic nitrogens in the 5′-N bases was also measured (NB). The net displacement of the oxoG (Do) from the G state was calculated as Do = GB-NB.
The results of this analysis are shown in Fig. 4C. Interestingly, GB ranges from 0.6 to 1.5 Å across all NpGo structures, with the oxoG shifting stereotypically in the direction of the minor groove. However, in the structures having a stacked 5′-purine (i.e. GpGo versus GpG and ApGo versus ApG), the 5′-neighbor also shifts toward the minor groove by a distance nearly equal to that of the target base, resulting in only a 0.2 Å net displacement of the oxoG from the G state. Stated another way, even though the presence of oxoG causes a displacement of the target nucleobase by altering the DNA backbone conformation, a 5′-purine neighbor moves along with the oxoG, such that the 5′-purine/target-G versus 5′-purine/target-oxoG stacking interactions remain nearly unchanged. In contrast, 5′-neighboring pyrimidines (TpGo and CpGo structures) do not move with the oxoG; hence, the oxoG shears with respect to the neighbor in these dinucleotides. This leads to a net displacement of 0.6 and 0.5 Å for the TpGo and CpGo structures, respectively, possibly facilitating extrusion of oxoG as it shifts toward the minor groove. These sequence-specific observations suggest that differences in base-stacking between oxoG and the 5′-base can influence the position of the oxoG (with respect to the neighboring bases) and accentuate differences between the G- and oxoG-bound states.
Sequence Dependence of Buckling of Target Base Pair
As shown in Fig. 2A, insertion of Phe114 at the target site significantly buckles the target base pair and the base pair 3′ to the target base, with buckle angles ranging from 36 to 43° for the NpG set and 28 to 43° for the NpGo set. This buckling thrusts the major groove edge of the target base pair, in particular the O6 from the target base and N4 from the opposing cytosine, toward the 5′-stacking neighbor. Measurement of the distances between the target base pair and the 5′-N base are shown in Fig. 5. Interestingly, these measurements indicate that the target G is within 3.0–3.4 Å of the 5′-N base in the NpG set, whereas oxoG remains 3.3–3.9 Å away from the 5′-N base in the NpGo set. The larger distances in the NpGo set are consistent with the previous observation that oxoG:C target base pairs in our structures are generally less buckled (and, therefore, more planar relative to the neighboring base pairs) than G:C pairs from corresponding structures.
FIGURE 5.
Measurements of distance between O6 from the target base (G or oxoG) and exocyclic atom at C6 (A and G) or C4 (T and C) position of the 5′-neighbor. Distance from the 5′-neighbor to the N4 of the opposing C is also shown. The target base and its base-stacking partner are consistently closer in distance in the G-bound state compared with the oxoG-bound state. Additionally, alignments between the oxoG-bound and G-bound states show how the position of the target oxoG responds to changes in the backbone conformation. Rotation of the backbone away from the state seen in the G-bound structure pulls the target oxoG into a more planar position, reducing the buckle angle of the target oxoG base pair compared with the buckle for the target G base pair.
In the NpGo set, the O6 atom of the target oxoG appears to make to an electrostatically favorable interaction with the 5′-A (N6-H) and 5′-C (N4-H) but makes an electrostatically repulsive interaction with the 5′-G (O6) and 5′-T (O4). The repulsion between oxoG and the 5′-T is mitigated by the large distance between the two bases (3.7 Å) but not in the case of 5′-G, with only 3.3 Å separating the two oxygen atoms.
Experimental Measurement of oxoG Repair Rates
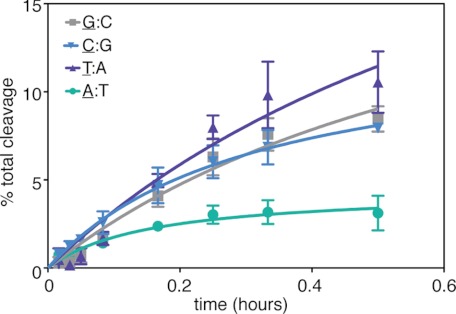
To determine whether the sequence-dependent structural differences observed in this study were also manifest in functional terms, we measured the rates of MutM-catalyzed base-excision of an oxoG lesion for the four 5′ sequence-permuted oligonucleotides characterized structurally in this study. Again, these substrates differ only in the identity of the base pair flanking oxoG:C on the 5′-side. The substrate with T:A stacked on the target base pair was cleaved fastest, whereas the A:T substrate was cleaved slowest, the difference being ∼3-fold (Fig. 6). The C:G and G:C substrates were cleaved at about the same rate as each other, intermediate between A:T and T:A. These rates correlate roughly with the extent of base stacking between oxoG and the 5′-neighbor observed crystallographically, with the exception of G:C, which is cleaved faster than expected on the basis of stacking alone.
FIGURE 6.
OxoG cleavage assay using substrates in which the 5′-neighbor is varied between all four bases. The underlined nucleobase denotes the base that is stacked on the oxoG. The 5′-A substrate shows a markedly slower rate of oxoG repair compared with the 5′-T, 5′-C, and 5′-G substrates.
Effect of cross-linker
To determine the potential effects of cross-linking position on these backbone conformations, we solved an additional set of NpGo structures using a different cross-linking position on the DNA (p11). Crystals for these complexes diffracted to 1.9–2.1 Å (supplemental Table S1). The sequence flanking the oxoG remains the same as the first set: the 3′-base to the oxoG remains a G, whereas the 5′-stacking neighbor is varied between A, T, and C (a previously published structure with 5′-G cross-linked at p11 completes the set) (19). Consistent with our previous observations, structures with a 5′-pyrimidine have less overlap with the target oxoG, whereas structures with a 5′-purine remain stacked to the oxoG (supplemental Fig. S3). This second set of structures confirms that our previous observations are independent of cross-linking position and are determined solely by the identity of the base 5′ to the oxoG.
DISCUSSION
Previous work from our laboratory has shown that MutM locates intrahelical oxoG residues and discriminates them from G by kinetically controlled extrusion into the DNA minor groove (19, 20). High resolution structures of MutM captured in the act of interrogating an undamaged G or encountering an oxoG lesion have revealed that the protein performs a muscular interrogation of the DNA helix with pronounced bending of the DNA localized to the interrogation site and with full insertion of an intercalating residue (Phe114) into the helix on the 3′ side of the lesion. Insertion of Phe114 severely buckles the target base pair, thrusting it toward the base pair on the 5′-side of the lesion, which nevertheless remains stacked with the target base pair. The concerted bending and buckling of DNA causes the substituent at the 8-position of the target nucleobase to project toward both the 2′-CH2 group of its own sugar moiety and toward the 5′-phosphate. When the C-8 substituent is an electropositive H, no steric or electronic clash is engendered with the 2′-CH2 or 5′-phosphate, and the sugar retains a normal 2′-endo pucker. However, when the 8-substituent is a larger, electronegative O, a severe clash develops with the 2′-CH2 and the 5′-phosphate. To avoid this clash, the sugar pseudorotates to the 4′-exo conformation. Molecular dynamics simulations showed that the adoption of this alternative backbone conformation lowered the activation barrier for breakage of the target base pair and extrusion of the target nucleobase from DNA by 7 kcal/mol (19). Based on these observations, we concluded that MutM locates intrahelical oxoG residues by a kinetically controlled process involving formation of an extrudogenic conformation at oxoG residues (19). Given that the target base pair in these structures remains in intimate contact with its 5′-neighbor, we had to consider the possibility that this extrudogenic switch in going from oxoG to G might be a property of the particular sequence employed in our prior study. Furthermore, even if the extrudogenic switch turned out to be general, the rate of extrusion could well be influenced by the 5′-neighbor, and this could be one factor that influences the known fact that the mutagenicity of lesions is influenced by the surrounding sequence. Given the present lack of experimental methods that can distinguish the difference between base pair breathing rates and extrusion rates, computational molecular dynamics simulations provide the only reliable means of accessing the energetic barrier to extrusion of a target nucleobase from DNA. Such simulations are most informative when performed from a starting point of an experimental structure that is as close as possible to the actual species of interest. Therefore, we undertook this study to assess the generality of the extrudogenic conformation model and to provide a complete set of experimental structures in which the 5′-neighboring base pair was exhaustively varied in the presence of either a G:C or oxoG:C target base pair. Although not emphasized here in detail, we also solved a sequence-matched series of structures in which MutM was repositioned along the length of the DNA helix so as to examine the system in a different crystal packing environment. MutM is the only DNA repair system for which intrahelical lesion recognition has been demonstrated and elucidated at the molecular level, and this analysis was intended to broaden and deepen our understanding of that recognition event. To our knowledge this study is the most comprehensive structural investigation of sequence -dependence in substrate recognition for any catalytic DNA-binding protein.
A key finding of this study is that, without exception, all of the sequence-matched structures determined here show the same stereotypic extrudogenic switch in going from a target G to oxoG. In addition to the aforementioned backbone changes, we further note that the oxoG nucleobase in the NpGo structures is invariably extended farther toward the minor groove than G in the NpG structures, and this again favors extrusion into the minor groove. We also note that the hydrogen-bonding distances for the target oxoG:C base pair (supplemental Fig. S6) are consistently longer in our structures than in the sequence-matched structures having a target G:C, whereas no such difference is seen in the absence of MutM (8), again indicative that MutM is an active participant in wrenching oxoG residues from the DNA helical stack.
There are numerous caveats in attempting to reconcile our x-ray structures with DNA repair rates. First, the kinetics of extrusion of a nucleobase from DNA involves both the rate of association with that site and the rate of extrusion, and our structures provide little insight into the former. Secondly, the base-excision kinetics also entail multiple conformational events after initial ejection of the target nucleobase, for example, ordering and closure of the oxoG-capping loop, and again these late events are not easily gleaned from our structures. Finally, even if one focuses solely on the rate of extrusion, our structures reveal a complex ensemble of sequence-dependent differences in stacking with the 5′-neighbor and in other interactions resulting from buckling of the target base pair, among other more subtle differences. All other things being equal, one would expect that stacking of the target G with its 5′-neighbor would stabilize the intrahelical conformation of oxoG and thereby pose a kinetic impediment to extrusion. In our structures, oxoG is substantially destacked from 5′-pyrimidines T and C, and indeed, these are repaired rapidly. Furthermore, oxoG is extensively stacked with a 5′-A, and this substrate is repaired the most slowly. However, the correlation of repair rates with stacking breaks down for a 5′-G, which despite being the most extensively stacked is repaired at the same rate as the destacked 5′-C. Thus, although base-stacking at the pre-extrusion stage is most likely a contributor to overall repair rates, other factors are clearly at play. By way of speculation, it could be that the GpGo structure has destabilizing interactions that weigh against the stabilization provided by its extensive stacking; one obvious culprit could be the unfavorable electrostatic interaction between the O6 atoms of the 5′-G and the target oxoG. Future computational simulations will help to define the specific interactions that govern target extrusion in these different sequence contexts.
Variations in base stacking at a lesion site have been proposed to aid in lesion recognition for various DNA repair enzymes (38–40). Many DNA repair enzymes such as MutM, Ogg1, AlkA, photolyase, and T4 endonuclease V enforce a distinctive bend in the DNA at the lesion site that destacks the target base pair from one of its neighboring base pairs (the 3′-base pair in the case of MutM) (15, 41–45). In the pre-extrusion state seen in our structures, we show that the base stacking conformation near the target oxoG is unique to the MutM-bound state (compared with base stacking in B-form DNA), providing the first structural basis for sequence-specific differences in the conformation of an intrahelical lesion base during lesion recognition.
The influence of sequence context on lesion recognition and repair has significant biological consequences. Previous work has demonstrated that poor repair efficiency is an important factor in the formation of mutational hotspots in the genome, suggesting that the prevalence of oxoG hotspots at purine-rich sequences in the DNA may be due to diminished repair efficiency at those sites (46). Our structures show that adjacent 5′-purine bases share a significant base stacking interaction with oxoG, but the particular sequence used in our study does not show a clear correlation between the extent of stacking for a 5′-G. It would be of interest to determine experimental structures of MutM encountering oxoG in a known mutational sequence to see whether any clear structural basis for this effect emerges. Given the unique characteristics of a GG stack, further work is necessary to explore the effect of guanine-rich sequences on oxoG repair efficiency.
Our work with MutM has implications for understanding the effect of sequence on oxoG repair in humans as well. Currently available crystal structures of Ogg1, the functional equivalent of MutM in humans, in complex with DNA containing an extrahelical oxoG shows striking similarities to the extrahelical MutM structures (44). Both proteins introduce sharp bends into the DNA at the lesion site (80° for Ogg1, 75° for MutM), inserting residues into the duplex to extrude oxoG into the active site. Closer examination of the base pairs flanking the extruded oxoG in the Ogg1 complex indicate that the base pair 3′ to the oxoG is slightly buckled, whereas the base pair 5′ to the oxoG remains planar (47–49). The unperturbed state of the 5′ base pair to the oxoG suggests that lesion recognition by Ogg1 may also be susceptible to the effects of sequence context; however, these hypotheses await confirmation by crystal structures of Ogg1 bound to intrahelical oxoG-containing DNA.
Supplementary Material
Acknowledgments
We are grateful to staff from the 24-ID beamline of the Argonne Photon Source for assistance in data collection. We thank Kwangho Nam, Danaya Pakotiprapha, and Brian Bowman for constructive feedback during the writing of this manuscript. We also thank members of the Verdine group for helpful advice.
This work was supported, in whole or in part, by National Institutes of Health Grants GM044853 and CA100742.

This article contains supplemental Table S1 and Figs. S1–S6.
The atomic coordinates and structure factors (codes 3U6C, 3U6D, 3U6E, 3U6P, 3U6O, 3U6S) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- oxoG
- 8-oxoguanine
- SB
- stacked base
- TB
- target base.
REFERENCES
- 1. Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature 362, 709–715 [DOI] [PubMed] [Google Scholar]
- 2. Michaels M. L., Miller J. H. (1992) The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 174, 6321–6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nash H. M., Bruner S. D., Schärer O. D., Kawate T., Addona T. A., Spooner E., Lane W. S., Verdine G. L. (1996) Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr. Biol. 6, 968–980 [DOI] [PubMed] [Google Scholar]
- 4. van der Kemp P. A., Thomas D., Barbey R., de Oliveira R., Boiteux S. (1996) Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine. Proc. Natl. Acad. Sci. U.S.A. 93, 5197–5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barnes D. E., Lindahl T. (2004) Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 38, 445–476 [DOI] [PubMed] [Google Scholar]
- 6. Fromme J. C., Verdine G. L. (2004) Base excision repair. Adv. Protein Chem. 69, 1–41 [DOI] [PubMed] [Google Scholar]
- 7. Grollman A. P., Moriya M. (1993) Mutagenesis by 8-oxoguanine. An enemy within. Trends Genet 9, 246–249 [DOI] [PubMed] [Google Scholar]
- 8. Bowman B. R., Lee S., Wang S., Verdine G. L. (2008) Structure of the E. coli DNA glycosylase AlkA bound to the ends of duplex DNA. A system for the structure determination of lesion-containing DNA. Structure 16, 1166–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lipscomb L. A., Peek M. E., Morningstar M. L., Verghis S. M., Miller E. M., Rich A., Essigmann J. M., Williams L. D. (1995) X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine. Proc. Natl. Acad. Sci. U.S.A. 92, 719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oda Y., Uesugi S., Ikehara M., Nishimura S., Kawase Y., Ishikawa H., Inoue H., Ohtsuka E. (1991) NMR studies of a DNA containing 8-hydroxydeoxyguanosine. Nucleic Acids Res. 19, 1407–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Plum G. E., Grollman A. P., Johnson F., Breslauer K. J. (1995) Influence of the oxidatively damaged adduct 8-oxodeoxyguanosine on the conformation, energetics, and thermodynamic stability of a DNA duplex. Biochemistry 34, 16148–16160 [DOI] [PubMed] [Google Scholar]
- 12. Cheng X., Kelso C., Hornak V., de los Santos C., Grollman A. P., Simmerling C. (2005) Dynamic behavior of DNA base pairs containing 8-oxoguanine. J. Am Chem. Soc. 127, 13906–13918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Banerjee A., Santos W. L., Verdine G. L. (2006) Structure of a DNA glycosylase searching for lesions. Science 311, 1153–1157 [DOI] [PubMed] [Google Scholar]
- 14. Coste F., Ober M., Carell T., Boiteux S., Zelwer C., Castaing B. (2004) Structural basis for the recognition of the FapydG lesion (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by formamidopyrimidine-DNA glycosylase. J. Biol. Chem. 279, 44074–44083 [DOI] [PubMed] [Google Scholar]
- 15. Fromme J. C., Verdine G. L. (2002) Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM. Nat. Struct. Biol. 9, 544–552 [DOI] [PubMed] [Google Scholar]
- 16. Fromme J. C., Verdine G. L. (2003) DNA lesion recognition by the bacterial repair enzyme MutM. J. Biol. Chem. 278, 51543–51548 [DOI] [PubMed] [Google Scholar]
- 17. Gilboa R., Zharkov D. O., Golan G., Fernandes A. S., Gerchman S. E., Matz E., Kycia J. H., Grollman A. P., Shoham G. (2002) Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J. Biol. Chem. 277, 19811–19816 [DOI] [PubMed] [Google Scholar]
- 18. Pereira de Jésus K., Serre L., Zelwer C., Castaing B. (2005) Structural insights into abasic site for Fpg-specific binding and catalysis. Comparative high resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA. Nucleic Acids Res 33, 5936–5944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qi Y., Spong M. C., Nam K., Banerjee A., Jiralerspong S., Karplus M., Verdine G. L. (2009) Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme. Nature 462, 762–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qi Y., Spong M. C., Nam K., Karplus M., Verdine G. L. (2010) Entrapment and structure of an extrahelical guanine attempting to enter the active site of a bacterial DNA glycosylase, MutM. J. Biol. Chem. 285, 1468–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Serre L., Pereira de Jésus K., Boiteux S., Zelwer C., Castaing B. (2002) Crystal structure of the Lactococcus lactis formamidopyrimidine-DNA glycosylase bound to an abasic site analogue-containing DNA. EMBO J. 21, 2854–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sugahara M., Mikawa T., Kumasaka T., Yamamoto M., Kato R., Fukuyama K., Inoue Y., Kuramitsu S. (2000) Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8. EMBO J. 19, 3857–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hatahet Z., Zhou M., Reha-Krantz L. J., Morrical S. W., Wallace S. S. (1998) In search of a mutational hotspot. Proc. Natl. Acad. Sci. U.S.A. 95, 8556–8561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kamiya H., Murata-Kamiya N., Koizume S., Inoue H., Nishimura S., Ohtsuka E. (1995) 8-Hydroxyguanine (7,8-dihydro-8-oxoguanine) in hot spots of the c-Ha-ras gene. Effects of sequence contexts on mutation spectra. Carcinogenesis 16, 883–889 [DOI] [PubMed] [Google Scholar]
- 25. Watanabe T., Nunoshiba T., Kawata M., Yamamoto K. (2001) An in vivo approach to identifying sequence context of 8-oxoguanine mutagenesis. Biochem. Biophys. Res. Commun. 284, 179–184 [DOI] [PubMed] [Google Scholar]
- 26. Pastoriza-Gallego M., Armier J., Sarasin A. (2007) Transcription through 8-oxoguanine in DNA repair-proficient and Csb(−)/Ogg1(−) DNA repair-deficient mouse embryonic fibroblasts is dependent upon promoter strength and sequence context. Mutagenesis 22, 343–351 [DOI] [PubMed] [Google Scholar]
- 27. Maxam A. M., Gilbert W. (1977) A new method for sequencing DNA. Proc. Natl. Acad. Sci. U.S.A. 74, 560–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 29. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 31. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 32. Painter J., Merritt E. A. (2006) TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 109–111 [Google Scholar]
- 33. Painter J., Merritt E. A. (2006) Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 34. Laskowski R. A., Macarthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 35. Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. (2004) MOLPROBITY. Structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res 32, W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lu X. J., Olson W. K. (2003) 3DNA. A software package for the analysis, rebuilding, and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res 31, 5108–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. DeLano W. L. (2010) The PyMOL Molecular Graphics System, Version 1.3r1, Schrodinger, LLC, New York [Google Scholar]
- 38. Cooper V. R., Thonhauser T., Puzder A., Schröder E., Lundqvist B. I., Langreth D. C. (2008) Stacking interactions and the twist of DNA. J. Am Chem. Soc. 130, 1304–1308 [DOI] [PubMed] [Google Scholar]
- 39. Yang W. (2006) Poor base stacking at DNA lesions may initiate recognition by many repair proteins. DNA Repair 5, 654–666 [DOI] [PubMed] [Google Scholar]
- 40. Yang W. (2008) Structure and mechanism for DNA lesion recognition. Cell Res. 18, 184–197 [DOI] [PubMed] [Google Scholar]
- 41. Vassylyev D. G., Kashiwagi T., Mikami Y., Ariyoshi M., Iwai S., Ohtsuka E., Morikawa K. (1995) Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate. Structural basis for damaged DNA recognition. Cell 83, 773–782 [DOI] [PubMed] [Google Scholar]
- 42. Lau A. Y., Schärer O. D., Samson L., Verdine G. L., Ellenberger T. (1998) Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA. Mechanisms for nucleotide flipping and base excision. Cell 95, 249–258 [DOI] [PubMed] [Google Scholar]
- 43. Hollis T., Ichikawa Y., Ellenberger T. (2000) DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J. 19, 758–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bruner S. D., Norman D. P., Verdine G. L. (2000) Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature 403, 859–866 [DOI] [PubMed] [Google Scholar]
- 45. Mees A., Klar T., Gnau P., Hennecke U., Eker A. P., Carell T., Essen L. O. (2004) Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair. Science 306, 1789–1793 [DOI] [PubMed] [Google Scholar]
- 46. Feng Z., Hu W., Chen J. X., Pao A., Li H., Rom W., Hung M. C., Tang M. S. (2002) Preferential DNA damage and poor repair determine ras gene mutational hotspot in human cancer. J. Natl. Cancer Inst. 94, 1527–1536 [DOI] [PubMed] [Google Scholar]
- 47. Banerjee A., Yang W., Karplus M., Verdine G. L. (2005) Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature 434, 612–618 [DOI] [PubMed] [Google Scholar]
- 48. Banerjee A., Verdine G. L. (2006) A nucleobase lesion remodels the interaction of its normal neighbor in a DNA glycosylase complex. Proc. Natl. Acad. Sci. U.S.A. 103, 15020–15025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Radom C. T., Banerjee A., Verdine G. L. (2007) Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations. J. Biol. Chem. 282, 9182–9194 [DOI] [PubMed] [Google Scholar]
- 50. Brunger A. (1993) Acta Crystallogr. D 49, 24–36 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.