Background: Human Tudor staphylococcal nuclease (Tudor-SN) is involved in the snRNP assembly.
Results: The efficient formation of Tudor-SN·SmB complex requires binding orientation of the methylated ligand and the specific binding pocket.
Conclusion: Tudor-SN takes part in regulating pre-mRNA splicing via the recruitment of U5 snRNP and the association of Sm protein.
Significance: The mechanism underlying the involvement of Tudor-SN in regulating snRNP biogenesis was revealed.
Keywords: Protein Domains, Protein-Protein Interactions, Ribonuclear Protein (RNP), RNA Splicing, Spliceosome, Sm Protein, Tudor-SN
Abstract
Human Tudor staphylococcal nuclease (Tudor-SN) is composed of four tandem repeats of staphylococcal nuclease (SN)-like domains, followed by a tudor and SN-like domain (TSN) consisting of a central tudor flanked by two partial SN-like sequences. The crystal structure of the tudor domain displays a conserved aromatic cage, which is predicted to hook methyl groups. Here, we demonstrated that the TSN domain of Tudor-SN binds to symmetrically dimethylarginine (sDMA)-modified SmB/B′ and SmD1/D3 core proteins of the spliceosome. We demonstrated that this interaction ability is reduced by the methyltransferase inhibitor 5-deoxy-5-(methylthio)adenosine. Mutagenesis experiments indicated that the conserved amino acids (Phe-715, Tyr-721, Tyr-738, and Tyr-741) in the methyl-binding cage of the TSN domain are required for Tudor-SN-SmB interaction. Furthermore, depletion of Tudor-SN affects the association of Sm protein with snRNAs and, as a result, inhibits the assembly of uridine-rich small ribonucleoprotein mediated by the Sm core complex in vivo. Our results reveal the molecular basis for the involvement of Tudor-SN in regulating small nuclear ribonucleoprotein biogenesis, which provides novel insight related to the biological activity of Tudor-SN.
Introduction
Tudor staphylococcal nuclease (Tudor-SN),4 also known as SND1 (staphylococcal nuclease domain containing 1) or p100, has been identified as a ubiquitous protein present in humans, cattle (1), rats (2), zebrafish (3), Tetrahymena thermophila (4), rice (5), peas (6), and many other species. Tudor-SN is a multifunctional protein implicated in a variety of cellular processes, such as gene transcription, pre-mRNA splicing, formation of stress granules, as well as the RNA-induced silencing complex in which small RNAs are complexed with ribonucleoproteins to ensure an RNAi-mediated gene (7–14). Combination of the modeled three-dimensional structures and x-ray crystallography indicates that the full-length structure of Tudor-SN resembles a stick with a hook, where SN-like domains form the stick and the tudor domain makes up the hook (11). It indicates that different domains of Tudor-SN protein may recruit different protein complexes to play different roles. In line with this concept, we have demonstrated that Tudor-SN functions as a transcriptional co-activator of STAT6 via interaction with the SN-like domains (12, 15), whereas the TSN domain is involved in the spliceosome assembly and accelerates the kinetics of precursor-messenger RNA (pre-mRNA) splicing (13). However, the precise molecular mechanism underlying the involvement of Tudor-SN in pre-mRNA processing has not been fully elucidated.
The pre-mRNA splicing process is essential for the successful execution of eukaryotic gene expression and mediates the production of mature mRNA through excision of introns and ligation of exons in pre-mRNAs by the spliceosome machinery. The spliceosome consists of five conserved snRNPs formed by an ordered binding of specific protein complexes onto metabolically stable U snRNAs, including U1, U2, U5, and base-paired U4/U6, which are highly abundant in eukaryotic cells (16–19). The biogenesis of U snRNPs is a stepwise process, and the hallmark of which is the formation of an Sm protein ring consisting of seven polypeptides (B/B′, D1, D2, D3, E, F, and G) around the U snRNAs (20–22). Notably, arginine residues in SmB/B′, SmD1, and SmD3 proteins can be methylated by protein-arginine methyltransferases (23–27). There are two general types (i.e. type I and type II) of protein-arginine methyltransferase responsible for protein arginine methylation. Type I methyltransferases (PRMT1, -3, -4, and -6) mainly generate monomethylarginine and asymmetrical dimethylarginine, whereas type II methyltransferases (PRMT5, -7, and -9) predominantly generate monomethylarginine as well as symmetrical dimethylarginine (sDMA) (24–27). sDMA is detected on both nuclear and cytoplasmic Sm proteins, whereas asymmetrical dimethylarginine is only identified on nuclear Sm proteins (28). The tudor domain has previously been shown to be able to bind methylated proteins. For example, sDMA-modified Sm proteins are recognized by the tudor domains of SMN and SPF30 (29–31).
The formation of the spliceosome complex is a dynamic process of snRNP particles occurring on pre-mRNA. The U5 snRNP binds the 5′ and 3′ splice sites of exons, allowing the spliceosome to “tether” the exons at the 5′ splice site, and it intermediately carries out the first catalytic step and then aligns with the 3′ splice site of exon for the second step. Prp8, U5–116, and hBrr2 are three key components of U5 snRNP that interact extensively with each other and play essential roles in the formation, activation, and remodeling of the spliceosome. We reported earlier that purified Tudor-SN could accelerate the first catalytic step of splicing, but it did not affect the overall level of splicing (13). The association of Tudor-SN- and U5 snRNP-specific proteins may explain the phenomenon. However, it cannot explain how Tudor-SN could associate with different U snRNAs, such as U1, U2, U4, U5, and U6 snRNAs.
The tudor domain in Tudor-SN shows a high level of homology to the tudor domain of the SMN protein (13). SMN protein associates with the spliceosomal Sm proteins via its tudor domain and plays essential roles in the assembly of U snRNPs (32, 33). Thus, it is possible that Tudor-SN protein participates in snRNP assembly through similar interaction mechanisms, which led us to investigate the relationship between the TSN domain and Sm proteins. In addition, surface electrostatic potential plots of tudor domain in Tudor-SN protein also reveal negatively charged surfaces, which are involved in recognition and binding of methylation marks (11). The aim of this study was thus to investigate the molecular mechanisms of the Tudor-SN protein in the pre-mRNA splicing process. In this study, we demonstrate that the interaction of Tudor-SN and snRNPs involves the efficient association of Sm protein, apart from the interaction with U5 snRNPs.
EXPERIMENTAL PROCEDURES
Cell Culture and Plasmid Construction
HeLa cells and COS-7 cells were cultured as reported previously (12). COS-7 cells were transiently transfected with expression plasmids by electroporation with a Bio-Rad gene pulser at 220 V/950 microfarads. siRNA was transfected into HeLa cells using Lipofectamine RNAi MAX (Invitrogen), according to the manufacturer's instructions. Tudor-SN siRNA was generated as reported previously (14), and SMN siRNA was purchased from Santa Cruz Biotechnology. pCMV6-AC-GFP-SMN plasmid was purchased from OriGene Technologies. The pSG5-Tudor-SN expression plasmid containing the full-length cDNA of human Tudor-SN and the GST-TSN plasmid containing the TSN domain (640–885 amino acids) of Tudor-SN protein were constructed as described previously (12). GST-Tudor was constructed by cloning PCR products corresponding to amino acids 678–769 of human TSN into pGEXT-4T-1 vector with EcoRI and NotI. GST-SN1 (26–184 amino acids), SN2 (186–341 amino acids), SN3 (342–504 amino acids), or SN4 (509–673 amino acids) were constructed by cloning PCR products corresponding to amino acids into pGEX-4T-1 vector with EcoRI and NotI. GST-TSN mutant constructs (F715A, Y721A, F715A/Y721A, Y741A, Y738A, and Y738A/Y741A) were generated as described previously (11). All PCR products were sequenced.
Glycerol Density Gradient Analysis
The total cell lysates (TCLs) of HeLa cells were harvested with Nonidet P-40 lysis buffer (50 mm Tris-HCl (pH 7.6), 300 mm NaCl, 0.1 mm EDTA, 0.5% Nonidet P-40, 20% glycerol, 0.1 mm sodium orthovanadate, 1 mm sodium butyrate). The TCLs were layered on a 10-ml gradient of 10–30% glycerol and centrifuged at 24,000 rpm (Beckman SW-41Ti rotor) for 20 h at 4 °C. All glycerol solutions were prepared with 25 mm Tris-HCl buffer (pH 7.5), 1.5 mm EGTA, 15 mm MgCl2, 50 mm NaCl, 0.1 mm NaF (pH 7.2), 1 mm DTT, 0.04% Nonidet P-40. and 10–30% glycerol. Eighteen fractions of 500 μl each were collected from top to bottom. Each fraction was run on SDS-PAGE and analyzed by Western blotting with corresponding antibodies. The extracted snRNAs from each fraction were also analyzed by Northern blotting with U1, U2, U4, U5, and U6 snRNA probes as described previously (34). Radiolabeled probes of U1, U2, U4, U5, and U6 snRNA were made by in vitro transcription of the linearized snRNA plasmids (34).
RNA-binding Protein Immunoprecipitation (RIP) Assays
HeLa cells (2.0 × 107) were incubated with 1% formaldehyde for 10 min at 37 °C to cross-link RNA protein. Then the glycine (125 mm, pH 7.0) was added to quench the cross-linking. Total cell lysates were harvested with SDS-lysis buffer (Upstate Biotechnology), supplemented with protease inhibitor mixture (Roche Applied Science) and RiboLock Ribonuclease inhibitor (MBI E00381) for 10 min on ice. Then the lysates were incubated with corresponding antibodies conjugated with protein G Dynabeads (Invitrogen) or protein A-agarose (Upstate Biotechnology) in the binding buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 167 mm NaCl, 16.7 mm Tris (pH 8.1), 400 units/ml MBL RiboLock Ribonuclease inhibitor and Roche Applied Science protease inhibitors mixture) at 4 °C overnight with head-over-tail rotation. The precipitated complexes were sequentially washed with buffer containing 500 mm NaCl, 150 mm NaCl, TE buffer (pH 8.0) and then incubated with the elution buffer (1% SDS, 0.1 m NaHCO3, 400 units/ml MBL RiboLock Ribonuclease inhibitor) at 4 °C for 15 min and at 65 °C for 2 h with addition of 5 m NaCl.
The bound RNAs were isolated using TRIzol reagent (Invitrogen) and used for the first-strand cDNA synthesis with reverse transcriptase M-MLV(RNase H−) and random hexamer primers (Takara, Japan), according to the manufacturer's protocol. The quantitative real time PCR (Q-PCR) assay was performed to detect the presence of precipitated U1, U2, U4, U5, and U6 snRNA with the specific primers (35). Amplification specificity was detected by gel electrophoresis and dissociation curve analysis. For the Q-PCR, the RIP fold changes were calculated validly using the ΔΔCt (cycle time) method (36, 37). The ΔCt value of bound snRNAs with either experimental (anti-Y12 or anti-TMG) or control (anti-IgG) beads was normalized to the Input fraction Ct value in the same assay. Then the ΔΔCt value was calculated by subtracting the ΔCt(normalized anti-IgG control RIP) from the ΔCt(normalized Y12 or anti-TMG RIP). Fold change value (experimental RIP versus control RIP) was determined by raising 2 to the power of the negative ΔΔCt.
GST Pulldown Assays
GST pulldown assays were performed as described previously (12). GST alone or GST fusion proteins were produced in Escherichia coli BL21 bacteria and bound to glutathione-Sepharose 4B beads (Amersham Biosciences) according to the manufacturer's instructions. The presence of GST fusion protein in the lysate was separated by SDS-PAGE, followed by Coomassie Blue staining. TCLs of COS-7 cells or HeLa cells were collected with Nonidet P-40 lysis buffer (20 mm Tris-HCl (pH 7.6), 100 mm NaCl, 10% glycerol, 1% Nonidet P-40, 2 mm EDTA, and 50 mm NaF) supplemented with protease inhibitor mixture (P8340, Sigma). Protein concentrations of TCLs were measured using the protein assay system from Bio-Rad. TCLs of HeLa cells were treated with or without 100 μl of RNase mixture (MBI Fermentas) at 37 °C for 30 min. The information on the RNase mixture system used in protein binding assay is shown in supplemental Table S1. The bead-bound fusion proteins were incubated with TCLs overnight at 4 °C with head-over-tail rotation and then washed five times with binding buffer containing 75 mm NaCl. The bound proteins were separated by SDS-PAGE and blotted with monoclonal anti-SmB (Clone 12F5, Sigma) or anti-Sm (Y12, ab3138, Abcam).
Co-immunoprecipitation Assays and Antibodies
HeLa or COS-7 cells were incubated with or without the methyltransferase inhibitor, 5-deoxy-5-(methylthio)adenosine (250 μm, MTA, Sigma) for 20 h. The total cell lysates of HeLa or COS-7 cells were collected and then incubated with corresponding antibodies conjugated with either protein G Dynabeads (Invitrogen) or protein A-agarose (Upstate Biotechnology) at 4 °C for 12 h with head-over-tail rotation. Bound proteins were subjected to SDS-PAGE and detected by blotting with corresponding antibodies.
Antibodies
The following antibodies were used: anti-trimethylguanosine (TMG)-agarose beads and anti-TMG-cap antibody (K121, Calbiochem); mouse monoclonal anti-Tudor-SN antibody and rabbit polyclonal anti-IgG antibody (Santa Cruz Biotechnology); monoclonal anti-SmB antibody (Clone 12F5, Sigma); anti-Sm (Y12, Abcam); mouse monoclonal anti-FLAGM2 coupled with agarose and anti-His-agarose (H-0767, Sigma); goat anti-Tudor-SN (C-17, Santa Cruz Biotechnology); rabbit polyclonal anti-sDMA antibody (anti-dimethylarginine, symmetric, SYM10, Upstate Biotechnology); and rabbit anti-SMN (H-195, Santa Cruz Biotechnology). The rabbit polyclonal anti-Tudor-SN antibody was generated against TSN domain (amino acids 640–885) of Tudor-SN.
In Vitro Splicing Assays and Native Gel Spliceosome Assembly Analysis
[32P]UTP-labeled adenovirus-splicing substrates was produced by in vitro transcription with T7 RNA polymerase (Promega) using the Adenovirus Major Late (AdML) plasmid linearized with HindIII as template and purified as described previously (13, 38). The wild- type TSN proteins or TSN with single or double amino acid substitutions were purified as described previously (11). The in vitro splicing reactions, containing 40% (v/v) HeLa extracts, 2 mm MgCl2, 10 mm DTT, 20 mm creatine phosphate, and 2 mm ATP, were supplemented with different purified proteins and preincubated at 30 °C for 10 min on ice, followed by addition of 10,000 cpm of the AdML pre-mRNA. The splicing reactions were incubated at 30 °C for different time points and stopped by placing the reaction on ice. Heparin (1 mg/ml final concentration) was added to the splicing reactions prior to loading. Native gel electrophoresis on 4% acrylamide was performed as described previously (39) and visualized by autoradiography.
Statistical Analysis
All experiments were repeated at least three times. The results were presented as means ± S.E. and compared by Independent-Samples Student's t test or one-way analysis of variance using the SPSS 16.0 software. p value less than 0.05 was considered as statistical significance.
RESULTS
Tudor-SN Is in the Complex with Spliceosomal snRNP in Vivo
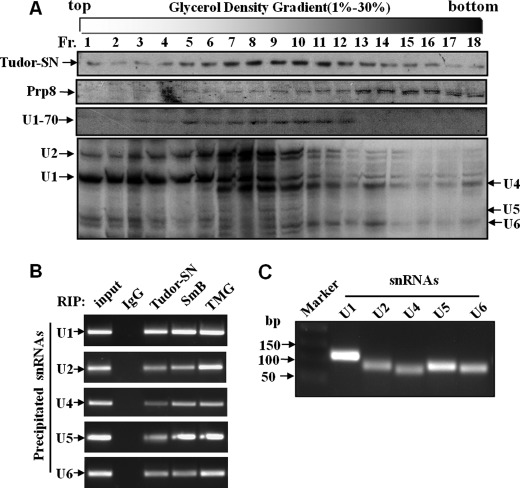
We previously demonstrated that Tudor-SN interacts with U1, U2, U4, U5, U6, and U7 snRNAs in vitro but not with 7SK snRNA (13). To further consolidate the in vivo association of Tudor-SN and U snRNAs, we performed glycerol gradient analysis (Fig. 1A) and RIP assays (Fig. 1, B and C). The total cell lysates of HeLa cells were fractionated by 10–30% glycerol density gradient followed by Western or Northern blotting analysis. The antibodies against Tudor-SN, U1–70, or Prp8 protein were used respectively. U1–70, also known as snRNP70, is a specific component of U1 snRNP that can directly interact with a stem-loop of U1 snRNA (40). Prp8 is a highly conserved member of U5 snRNP-specific proteins (41). As shown in Fig. 1A, the endogenous Tudor-SN proteins distributed throughout the fractions, especially peaked in fractions from 6 to 11. U1–70 proteins (fractions 3–12) and U1, U2 snRNAs (fractions 1–12 in particular) were found in the same fractions as Tudor-SN. Prp8, Tudor-SN, and U4, U5, and U6 snRNAs were detected simultaneously in the same fractions from 6 to 18. The peaked distribution of all the U snRNAs was in the same fraction (fractions 6–10) as Tudor-SN. All these data provided the evidence that Tudor-SN could exist in the same complex with different U snRNPs in vivo.
FIGURE 1.
Analysis of Tudor-SN and U snRNA profiles by glycerol gradient sedimentation. A, U snRNP complexes from HeLa cells were analyzed by glycerol gradient sedimentation. The total lysates of HeLa cells were sedimented through 10–30% glycerol gradients. The distribution of Tudor-SN, Prp8, and U1–70 in each fraction was analyzed by SDS-PAGE and immunoblotted with specific antibodies as indicated. Fraction (Fr.) 1 is at the top of the gradient. The extracted snRNAs from fractions were analyzed by Northern blotting with U1, U2, U4, U5, and U6 snRNA probes and visualized by autoradiography. Positions of snRNAs are indicated on the left and right. B, Tudor-SN and SmB associate with U1, U2, U4, U5, and U6 snRNA in vivo. RNA-binding protein immunoprecipitation assay was performed with total cell lysates of HeLa cells with formaldehyde and glycine treatment. The TCLs were incubated with bead-bound anti-Tudor-SN, anti-SmB, anti-TMG-cap, and rabbit IgG antibody as indicated. The co-precipitated snRNAs was extracted and reverse-transcribed to cDNA with random hexamer primers. The U1, U2, U4, U5, and U6 snRNAs were generated by PCR analysis. C, U1, U2, U4, U5, and U6 snRNAs in the TCLs of HeLa cells for RIP assays were extracted and reverse-transcribed to cDNA with random hexamer primers and then were generated by PCR assay.
Moreover, the RIP assay was performed to verify the association of Tudor-SN and U snRNAs. The RNA-protein complexes in HeLa cells were precipitated with anti-Tudor-SN antibody, anti-IgG as negative control, and anti-TMG-cap or anti-SmB as positive control. The RNA extracted from the precipitated complex was detected by reverse transcription-PCR analysis. As shown in Fig. 1B, U1, U2, U4, U5, and U6 snRNAs can be detected in anti-Tudor-SN, anti-TMG-cap, or anti-SmB precipitation, whereas no PCR products were detected in IgG-negative control. Fig. 1C indicated the PCR products of different U snRNA. These results confirmed the in vivo association of Tudor-SN with U1, U2, U4/6, and U5 snRNAs.
Tudor-SN Interacts with SmB/B′ and SmD1/D3 Proteins
U5 snRNP is an integral component of the spliceosome, containing a group of specific proteins, such as Prp8 and U5–116. These proteins and U5 snRNA can be cross-linked to the sites of pre-mRNA transcript and play crucial roles in the spliceosome complex formation and pre-mRNA splicing (41, 42). Our previous (13) and present results (shown in supplemental Figs. S1 and S2) demonstrate that Tudor-SN and U5–116 interact with different domains of Prp8 to form a stable complex. However, to some extent, these data cannot explain why Tudor-SN interacts with not only U5 snRNA but also other U snRNAs such as U1 and U2 snRNAs.
The structure of the tudor domain of Tudor-SN is similar to that of SMN, which has been shown to bind U1, U2, U5, and U4/U6 snRNPs via interaction with Sm proteins. Therefore, it is possible that Tudor-SN protein participates in snRNP assembly through the similar mechanism. We thus performed co-immunoprecipitation and GST pulldown assays to detect the association of Tudor-SN, SmB/B′, and SmD1/D3 proteins, which are the key proteins in the Sm complex. Y12 antibody (Abcam) and anti-SmB antibody (Sigma) were used in the present study. Y12 antibody can recognize cross-reactive epitopes on the B/B′ and D polypeptides of Sm and has been utilized to detect the SmB/B′ and SmD1/D3 proteins in many studies (43–46). To further confirm the interaction between Tudor-SN and SmB, the anti-SmB antibody specifically against cellular SmB protein was used as well. COS-7 cells were transfected with mammalian expression vector encoding full-length Tudor-SN tagged with the FLAG epitope. The total cell lysates were immunoprecipitated with anti-FLAG antibody or anti-His antibody as negative control and then detected with anti-FLAG (Fig. 2A, upper panel) or anti-SmB antibody (Fig. 2A, lower panel). We found that the ectopically expressed Tudor-SN protein was efficiently co-precipitated with the endogenous SmB proteins (Fig. 2A). Furthermore, the association of endogenous Tudor-SN with SmB/B′ or SmD1/D3 was also detected in HeLa cells (Fig. 2C), and the interaction of SMN and SmB/B′ or SmD1/D3 was performed as positive control (Fig. 2B). As shown in Fig. 2, B and C, the total cell lysates of HeLa cells were immunoprecipitated with anti-Tudor-SN or SMN antibody. The endogenous SmB/B′ and SmD1/D3 proteins were co-immunoprecipitated with both SMN (Fig. 2B) and Tudor-SN protein (Fig. 2C). These results demonstrate that Tudor-SN protein, the same as SMN, interacts with SmB/B′ and SmD1/D3 proteins in vivo.
FIGURE 2.
Tudor-SN interacts with SmB/B′ and SmD1/D3 via the tudor domain. A, ectopically expressed Tudor-SN interacts with SmB. COS-7 cells were transfected with the pSG5-Tudor-SN-FLAG plasmid. The total cell lysates of the transfected COS-7 cells were used in immunoprecipitation (IP) with anti-FLAG or anti-His-agarose beads as control. The co-precipitated proteins were subjected to SDS-PAGE and blotted with anti-FLAG (upper panel) or anti-SmB (lower panel) antibody. Approximately 10% of the TCLs were included as input. B and C, endogenous Tudor-SN or SMN interacts with SmB/B′ and SmD1/D3. TCLs of HeLa cells were immunoprecipitated with rabbit anti-Tudor-SN, anti-SMN, or anti-IgG as control. Bound proteins were subjected to SDS-PAGE and blotted with mouse anti-Tudor-SN, anti-SMN, anti-SmB, or Y12 antibody. Approximately 10% of the TCLs were included as input. D, mapping the interaction domain of Tudor-SN with SmB. TCLs of HeLa cells were incubated with equal amounts of GST and GST fusion proteins containing the isolated SN domains or the TSN domain as indicated in the upper panel. Bound proteins were separated by SDS-PAGE and immunoblotted with anti-SmB antibody (middle panel). The expression levels of the GST fusion proteins were visualized by Coomassie Blue staining (lower panel). Approximately 10% of the TCLs were included as input. E, GST pulldown assay was also performed with GST fusion proteins containing TSN or tudor domain alone. Bound proteins were detected with anti-SmB (upper panel) or Y12 antibody (2 middle panels), and the GST fusion proteins were visualized by Coomassie Blue staining (lower panel).
To determine the interaction domain of Tudor-SN and SmB, GST fusion proteins containing different domains of Tudor-SN (SN1, SN2, SN3, SN4, TSN, and tudor), as indicated in the upper panel of Fig. 2D, were incubated with total cell lysate of HeLa cells. Compared with SN1, SN2, SN3, and SN4 domains, the TSN domain strongly interacted with SmB (middle panel in Fig. 2D). We further identified that the isolated tudor domain alone (without the flanking SN-sequence) could sufficiently interact with SmB/B′ and SmD1/D3 proteins (Fig. 2E). These data indicate that the tudor domain mediates the association of Tudor-SN and Sm proteins.
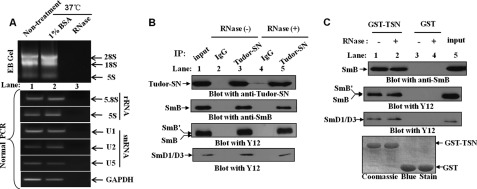
To exclude the possibility that Tudor-SN and SmB association is mediated by RNAs, GST pulldown assay and co-immunoprecipitation experiments were also carried out in the presence of RNase mixture (detailed in supplemental Table S1), which cleaves RNAs. The total cell lysate of HeLa cells was treated with or without RNase mixture at 37 °C for 30 min; the RNA was then extracted, and the existence of rRNA (28 S, 18 S, and 5 S) in the total cell lysate was detected directly by agarose gel separation (Fig. 3A, upper panel, lanes 1 and 2). After RNase treatment, there was no detectable RNA in the sample (Fig. 3A, upper panel, lane 3). Furthermore, reverse transcription-PCR assays were performed to detect the existence of rRNA (5.8 S and 5 S), snRNAs (U1, U2, and U5), and mRNA of GAPDH in different samples. As shown in the lower panel of Fig. 3A, there was no detectable PCR product in the RNase-treated sample (lane 3), compared with the blank control (lane 1) or negative control (lane 2). These data indicate that the RNase mixture successfully degraded the RNAs in the total cell lysate. Then the RNase-treated or nontreated HeLa TCLs were incubated with bead-bound GST fusion protein or immunoprecipitated with rabbit anti-Tudor-SN or anti-IgG as control. Bound proteins were subjected to SDS-PAGE and blotted with mouse anti-Tudor-SN, anti-SmB, or Y12 antibodies. The results in Fig. 3B revealed that even after RNase treatment, the SmB/B′ and SmD1/D3 proteins could efficiently precipitate with Tudor-SN. In addition, the GST pulldown assays demonstrated the consistent result that GST-TSN fusion protein could bind to SmB/B′ and SmD1/D3 proteins even after the removal of RNAs from the lysates (Fig. 3C). These data suggest that Tudor-SN and SmB/B′/SmD1/D3 proteins form a stable complex independent of RNA. As SmB/B′ and SmD1/D3 proteins are sufficient to bind the Sm core site of U1, U2, U4, and U5 snRNAs (20–22), it is possible that Tudor-SN binds snRNAs indirectly via the association of the Sm core complex.
FIGURE 3.
Tudor-SN interacts with SmB/B′ and SmD1/D3 in RNA-free lysates. A, TCLs of HeLa cells were incubated in the presence (+) or absence (−) of RNase mixture (120 μg of RNase A; 300 units of RNase Ti; 50 units of RNase H; 300 units of RNase I) or 1% BSA (Sigma) for 30 min at 37 °C. The RNAs were extracted and separated by EB-agarose gel electrophoresis. The 28 S, 18 S, and 5 S rRNAs are shown. rRNAs (5S and 5.8S), snRNAs (U1, U2, and U5), and GAPDH mRNAs were also detected using normal PCR assay. The primer sequences of rRNAs and GAPDH are shown in the supplemental Table S2. B, Tudor-SN interacts with SmB/B′ and SmD1/D3 in an RNase-resistant manner in vivo. The RNase treated or nontreated TCLs were immunoprecipitated with rabbit anti-Tudor-SN or anti-IgG as control. Bound proteins were subjected to SDS-PAGE and blotted with mouse anti-Tudor-SN (upper panel), anti-SmB (middle panel), or Y12 antibody (2 lower panels). 10% of the TCLs was included as input. C, TSN domain binds endogenous SmB/B′ and SmD1/D3 in an RNase-resistant manner. TCLs of HeLa cells (±RNase) were incubated with equal amounts of GST or GST-TSN fusion protein. Bound proteins were resolved by SDS-PAGE and blotted with anti-SmB (upper panel) or Y12 antibody (2 middle panels). The expression levels of the GST fusion proteins were visualized by Coomassie Blue staining (lower panel). Approximately 10% of the TCLs were also included as a control. IP, immunoprecipitated.
Tudor-SN Associates with Dimethylated Sm Proteins
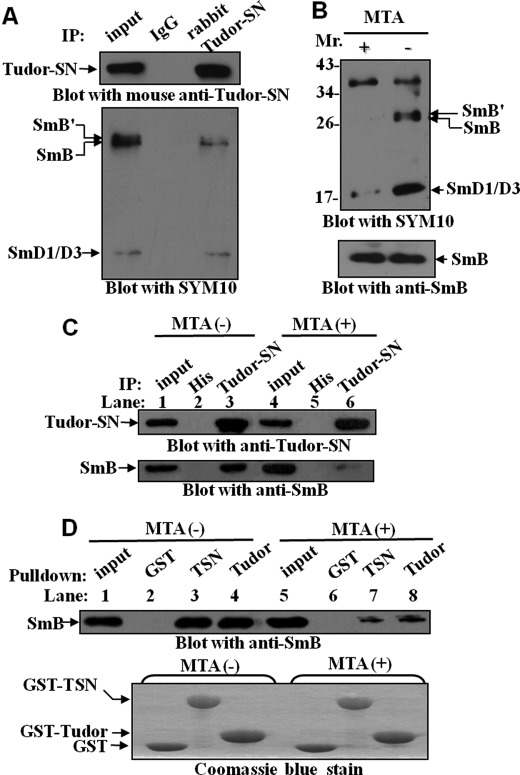
The arginines in the RG-rich C-terminal domains of SmB/B′ protein could be post-translationally modified by symmetrical dimethylation (43). Recent NMR study demonstrated that the extended tudor domain of Drosophila melanogaster Tudor-SN could bind symmetrically dimethylated putative ligands derived from the C-terminal tails of Sm proteins (47). Notably, the crystal structure of the tudor domain of D. melanogaster is similar to that of the human Tudor-SN, which also contains a conserved cage that potentially hooks methylated ligands (11). We therefore investigated whether methylation of SmB/B′ and SmD1/D3 is required for the efficient association with Tudor-SN. It was reported that SYM10 antibody recognizes proteins containing multiple symmetrically dimethylated arginines (sDMA), especially SmB/B′ and SmD1/D3 (45, 48). Hence, a co-immunoprecipitation experiment was performed with anti-Tudor-SN antibody or anti-IgG antibody as negative control and then detected with anti-Tudor-SN (Fig. 4A, upper panel) or SYM10 antibody (Fig. 4A, lower panel). The results suggested that the Tudor-SN protein efficiently precipitated the sDMA-modified SmB/B′ and SmD1/D3. To further verify this point, the MTA (a general methylation inhibitor) was used to de-methylate arginines. HeLa cells were grown in the presence or absence of 250 μm MTA for 20 h. The results in Fig. 4B indicated that MTA treatment reduced more than 75% of the methylated epitopes (SmB/B′ and SmD1/D3) recognized by SYM10 (upper panel), whereas the levels of cellular SmB remained equivalent (lower panel). Furthermore, the MTA-treated lysates were immunoprecipitated with anti-Tudor-SN antibody (Fig. 4C) or incubated with bead-bound GST fusion proteins (Fig. 4D). After MTA treatment (Fig. 4C), less SmB was co-immunoprecipitated with endogenous Tudor-SN protein (lane 6) compared with nontreated cells (lane 3). Consistently, in the lysate with MTA treatment (Fig. 4D), less SmB precipitated with GST-TSN (lane 7) or GST-tudor (lane 8) fusion protein, compared with the untreated lysate (lanes 3 and 4), although the SmB protein levels in the lysates without (lane 1) or with MTA (lane 5) treatment were the same. All these data indicate that methylation modification of SmB is required for the efficient interaction with Tudor-SN.
FIGURE 4.
Tudor-SN interacts with sDMA-modified SmB/B′ and SmD1/D3 proteins. A, endogenous Tudor-SN interacts with sDMA-SmB/B′ and SmD1/D3. TCLs of HeLa cells were immunoprecipitated (IP) with rabbit anti-Tudor-SN or anti-IgG as control. Bound proteins were subjected to SDS-PAGE and blotted with mouse anti-Tudor-SN (upper panel) or anti-sDMA antibody (SMY10) (lower panel). Approximately 10% of the TCLs were included as input. B, total cell lysates were prepared from HeLa cells cultured in the presence (+) or absence (−) of the methylation inhibitor MTA (250 μm) for 20 h and then immunoblotted with SMY10 antibody. The filter was stripped and re-blotted with anti-SmB to assess the reduction in methylation level. C, same HeLa TCLs (+/− MTA) were immunoprecipitated with anti-Tudor-SN antibody or anti-His- agarose beads as negative control. The precipitated proteins were separated by SDS-PAGE and blotted with either anti-Tudor-SN (upper panel) or anti-SmB (lower panel) antibody. D, total cell lysates from +/− MTA-treated HeLa cells were incubated with GST, GST-TSN, or GST-Tudor fusion proteins. The amount of bound SmB protein was visualized by immunoblotting with anti-SmB antibody (upper panel). The GST fusion proteins were visualized by Coomassie Blue staining (lower panel).10% of the TCLs was included as input.
Intact Aromatic Cage in TSN Domain Is Required for the Efficient Association of Sm Proteins
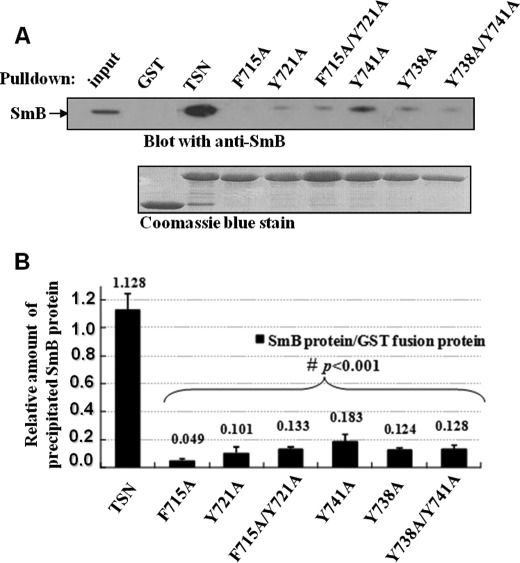
Previously, we demonstrated that the TSN domain of the Tudor-SN protein possesses a conserved aromatic cage composed of three tyrosine residues (Tyr-721, Tyr-738, and Tyr-741) and a phenylalanine residue (Phe-715) (11). Mutagenesis studies indicated that the single point mutation of Tyr-721, Tyr-738, Tyr-741, or Phe-715 or the double point mutation (Y738A/Y741A or Y721A/F715A) of the aromatic cage diminished binding ability of TSN to snRNAs (11). As we know that Tudor-SN itself cannot recognize the TMG-cap structure, the co-existence of Tudor-SN and snRNA is not due to their direct interaction, rather it is mediated by other proteins such as the Sm complex. To further verify this point, we performed GST pulldown assays to investigate the association of GST fusion proteins containing different mutants of TSN with the SmB proteins from total cell lysate of HeLa cells. As shown in Fig. 5, the wild-type TSN efficiently interacted with SmB proteins, whereas the SmB binding ability of the TSN with single or double mutants was significantly reduced (p < 0.001, n = 3), especially the F715A and Y721A mutants. These data imply that the intact cage structure of the TSN domain is a prerequisite for the efficient association of Tudor-SN and SmB. Consistent with the data, we previously demonstrated the TSN domain with single or double mutants could not efficiently associate with U snRNPs (11).
FIGURE 5.
TSN mutants affect the SmB binding. A, mutations of TSN affect the binding ability to SmB. TCLs of HeLa cells were incubated with GST and GST fusion proteins containing wild-type (WT) TSN or different mutants of TSN as indicated. The precipitated proteins were resolved by SDS-PAGE and immunoblotted with anti-SmB antibody (upper panel). The expression levels of GST and different GST fusion proteins were determined by Coomassie Blue staining (lower panel). 10% of the TCLs were included as input. B, band density was digitized with TotalLab software and then analysis of variance was used for statistical analysis. Significant differences were indicated as follows: #, p < 0.001 versus control (n = 3). The level of bound SmB proteins was normalized against the corresponding GST fusion proteins.
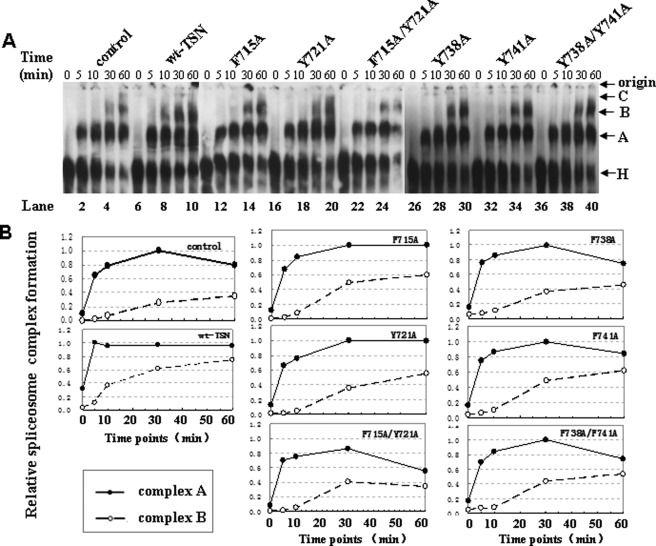
Moreover, the native gel analysis was performed to investigate the effect of intact cage structure of the TSN domain on in vitro spliceosome assembly. We analyzed the formation of pre-spliceosomal complex A and fully assembled spliceosomal complex B with in vitro transcripted AdML pre-mRNA as substrates. Fig. 6A demonstrates a time course analysis of spliceosomal complex formation. The reactions were either kept on ice (0 min) or incubated at 30 °C for 5, 10, 30, or 60 min prior to gel analysis. Fig. 6B shows a time course quantitative analysis of spliceosomal complex A and B formation and normalized each set by setting the highest value of complex A to 1. As shown in Fig. 6A, in the control reaction (lanes 1–5), complex A was first detected at the 5-min time point, and complex B was observed after 30 min of incubation. We also carried out the same native gel analysis using the purified GST or BSA as control, which demonstrated the same results (data not shown). The addition of a different purified single or double mutated TSN domain (Fig. 6A, lanes 11–40) presented the same pattern in in vitro spliceosome assembly as the controls. However, the addition of the purified wild-type TSN protein affected the formation of complexes (Fig. 6A, lanes 6–10), especially the transition from complex A to B. We reproducibly observed that complex B appeared as early as 5 min after the reaction (Fig. 6A, lane 7) and gave strong signal after 10 min (lane 8), although no obvious complex B was observed in the control and TSN mutations after 10 min of reaction. These results further imply that the intact structure of Tudor-SN is required for snRNP assembly.
FIGURE 6.
TSN proteins with mutations have no ability to promote the kinetics of in vitro spliceosome formation. A, in vitro splicing reactions were performed at different time points with the addition of indicated purified proteins followed by native gel analysis of spliceosome complex formation. The gel was visualized by autoradiography. The bands corresponding to the H, A, B, and C complexes as well as the gel origin are indicated on the right. B, quantitative analysis of spliceosome formation. The vertical axis of coordinate represents the density value of complex A and B. The horizontal axis of coordinate represents the different time points (0, 5, 10, 30, and 60 min) for the spliceosome assembly. The intensities of complex A and B were determined by PhosphorImager and normalized by setting the highest value of complex A to 1.
Effect of Tudor-SN Protein on Recruitment of Sm to snRNP in Vivo
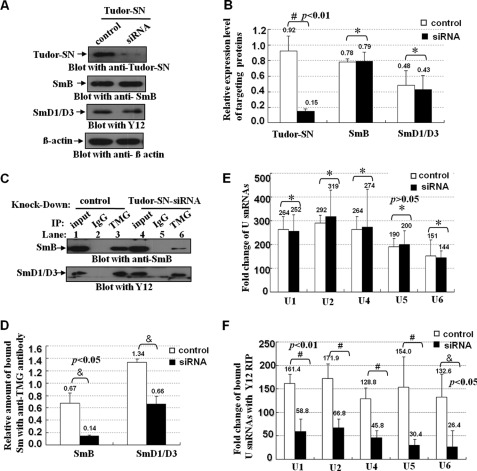
The Sm core complex is responsible for the recruitment of U snRNPs to specific U snRNA, although the trimethylguanosine (TMG)-cap is present in U1, U2, U4, and U5 snRNAs (49). Thus, using the antibody specifically to recognize the TMG-cap structure could efficiently precipitate the spliceosomal snRNPs components, including Sm core proteins. If Tudor-SN-SmB-D1-D3 interactions play important roles in the U snRNP formation, knockdown of Tudor-SN would affect the snRNA binding ability of Sm complex proteins. We first transfected the HeLa cells with Tudor-SN siRNA to knock down the endogenous Tudor-SN protein. As shown in Fig. 7, A and B, the endogenous Tudor-SN protein level was significantly reduced (#, p < 0.01, n = 5) with Tudor-SN mRNA-specific siRNAs duplex, although no detectable change of the endogenous SmB and SmD1/D3 proteins compared with the scrambled siRNA control (Fig. 7, A and B, *, p > 0.05, n = 5). The total cell lysates of different samples were immunoprecipitated with anti-TMG antibody or anti-IgG antibody as negative control to capture the snRNP complex, and then the precipitated SmB and SmD1/D3 proteins were detected by blotting with anti-SmB or Y12 antibody, although the bound U1, U2, U4, U5, and U6 snRNAs were determined with quantitative real time PCR (Q-PCR) assay. As shown in Fig. 7C, SmB and SmD1/D3 proteins were precipitated with anti-TMG-cap antibody (lanes 3 and 6) but not with the anti-IgG antibody (lanes 2 and 4); however, a lesser amount of Sm proteins was precipitated (Fig. 7C, lane 6) when knockdown of endogenous Tudor-SN protein was compared with the scrambled control (lane 3). The statistical analysis indicates significant differences (Fig. 7D, &, p < 0.05, n = 3) between the Tudor-SN knockdown and the scrambled control samples. To rule out the possibility of a previous difference in the accumulation of U snRNAs with anti-TMG-cap Dynabeads, we isolated and quantitated the bound U1, U2, U4/6, and U5 snRNA by RIP-Q-PCR assay. Fig. 7E demonstrated that equivalent amounts of U snRNAs were purified in both samples (*, p > 0.05, n = 3). As shown in Fig. 7F, the amount of precipitated U snRNAs in the lysate with knockdown of Tudor-SN was significantly reduced, compared with scrambled control. The statistical analysis indicates significant differences (Fig. 7F, #, p < 0.01; &, p < 0.05, n = 3). All these data demonstrated that knockdown of Tudor-SN protein affected the recruitment of SmB and SmD1/D3 to spliceosomal U snRNPs in vivo.
FIGURE 7.
Knockdown of TSN reduces the recruitment of Sm to the U snRNP in vivo. A, HeLa cells were transfected with the Tudor-SN siRNA or scramble siRNA (control). The total cell lysates of different samples were blotted with anti-Tudor-SN (upper panel), anti-SmB (1st middle panel), Y12 (2nd middle panel), or anti-β-actin antibody (lower panel) to detect the protein level of corresponding proteins. B, band density was digitized with TotalLab software and then Independent-Samples Student's t Test was performed. Significant difference was indicated: #, p < 0.01 versus control (n = 5); *, p > 0.05 (n = 5). The expression level of targeting protein (Tudor-SN, SmB, and SmD1/D3) in HeLa cells was normalized against the β-actin. C, total lysates of different samples were immunoprecipitated (IP) with anti-TMG-cap antibody or anti-IgG coupled with protein G Dynabeads as negative control. The co-precipitated proteins were subjected to SDS-PAGE and blotted with anti-SmB (upper panel) or Y12 antibody (lower panel). D, amount of SmB and SmD1/D3 proteins immunoprecipitated with anti-TMG-cap antibody was normalized against the total input Sm proteins. Band density was measured and then Independent-Samples Student's t Test was used for statistical analysis. Significant difference was indicated: &, p < 0.05 versus control group (n = 3). E, RNAs in the total cell lysates of different samples were isolated and reverse-transcribed to cDNA with random hexamer primers, and we then performed the quantitative real time PCR assay to detect the relative fold changes of U1, U2, U4, U5, and U6 snRNA. The fold changes were analyzed with Independent-Samples Student's t test. *, p > 0.05 (n = 3). F, total lysates of different samples were immunoprecipitated with Y12 Dynabeads or anti-IgG Dynabeads as control. The bound RNAs were extracted and reverse-transcribed to cDNA, and we then performed the quantitative real time PCR assay to detect the relative fold changes of precipitated U1, U2, U4, U5, and U6 snRNA. The RIP fold changes were analyzed with Independent-Samples Student's t test. Significant difference was indicated as follows: #, p < 0.01; &, p < 0.05, versus control (n = 3).
SMN and Tudor-SN Have Similar Function in Loading Sm Protein onto snRNP
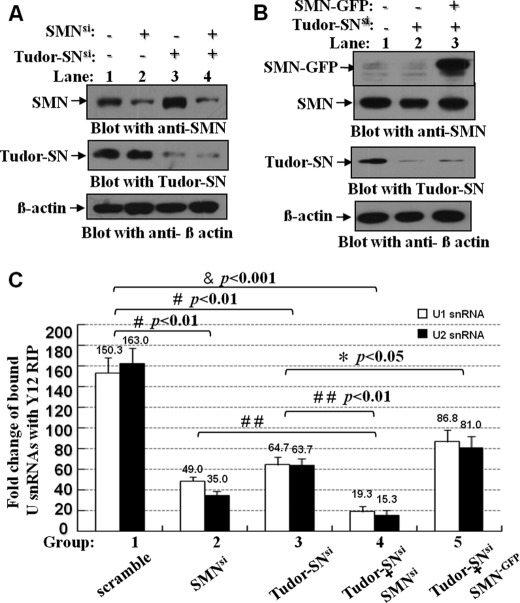
This study suggests that Tudor-SN functions in loading Sm protein onto snRNP, which is also attributed to the SMN protein. Because Tudor-SN and SMN possess the highly similar structure and function, we therefore investigate whether SMN could restore the defect function of Tudor-SN in recruiting Sm protein onto snRNP. The endogenous Tudor-SN or SMN protein was either knocked down alone (Tudor-SNsi or SMNsi) or both were knocked down (SMNsi/Tudor-SNsi) with transfection of Tudor-SN siRNA or SMN siRNA, respectively, into HeLa cells. After 72 h, cells were harvested, and the prepared lysates were examined for the Tudor-SN or SMN protein level. As shown in Fig. 8A, the respective endogenous SMN (lane 2) or Tudor-SN protein (lane 3) or both together (lane 4) were 75–85% decreased compared with the control protein β-actin.
FIGURE 8.
Ectopically expressed SMN restored the reduced U snRNP assembly caused by depletion of endogenous Tudor-SN. A and B, HeLa cells were transfected with the Tudor-SN siRNA and SMN siRNA (A) or mammalian expression plasmids containing full-length SMN tagged with GFP epitope (SMN-GFP) as indicated (B). The total cell lysates of different samples were blotted with anti-SMN (upper panel), anti-Tudor-SN (middle panel), or anti-β-actin antibody (lower panel) to detect the protein level of corresponding proteins. C, total lysates of different samples were immunoprecipitated with Y12 Dynabeads or anti-IgG Dynabeads as control. The bound RNAs were isolated and reverse-transcribed to cDNA with random hexamer primers, and we then performed the quantitative real time PCR assay to detect the relative fold changes of precipitated U1 and U2 snRNA. The RIP fold changes were analyzed with analysis of variance. Significant difference was indicated as follows: &, <0.001; #, p < 0.01 versus nontreatment group (n = 3), *, p < 0.05; # #, p < 0.01 versus Tudor-SNsi or SMNsi group (n = 3).
Meanwhile, SMN protein was also ectopically expressed in the HeLa cells with 80–85% depletion of Tudor-SN (Tudor-SNsi/SMN−GFP) (Fig. 8B, lane 3) by transfecting SMN-GFP expression plasmid. In Fig. 8B, the upper band is the SMN-GFP fusion protein, and the lower band is the endogenous SMN protein. Then the RIP-Q-PCR assay was conducted with immunoprecipitation with Y12 antibody in the corresponding lysates to examine the amount of the bound U1 and U2 snRNAs in the snRNP complex. As shown in Fig. 8C, compared with scrambled control, single knockdown of either endogenous SMN (SMNsi) or Tudor-SN (Tudor-SNsi) reduced the in vivo recruitment of Sm to U1 and U2 snRNAs (#, p < 0.01, n = 3). An even smaller amount of precipitated U1 and U2 snRNAs was detected in the lysate with knockdown of both SMN and Tudor-SN proteins (SMNsi/Tudor-SNsi), compared with the single SMNsi or Tudor-SNsi (##, p < 0.01, n = 3) or scrambled control (&, p < 0.001, n = 3). The ectopic expression of SMN protein in the HeLa cells with depletion of endogenous Tudor-SN (Tudor-SNsi/SMN−GFP) partially restored the reduced association of Sm with U1 and U2 snRNAs (*, p < 0.05, n = 3). These data suggest that Tudor-SN and SMN proteins play similar roles in facilitating the loading of Sm core protein to snRNPs.
DISCUSSION
Spliceosome is composed of five snRNPs and a range of non-snRNP-associated protein factors. The ordered assembly of Sm proteins and the recruitment of different specific snRNP proteins to specific U snRNA are critical initial steps in the complex process of snRNP biogenesis (20–22). The recognition of pre-mRNA involves U1 snRNP binding to the 5′ end splice site of the pre-mRNA, and U2 snRNP associates with the branch point sequence of pre-mRNA and other non-snRNP-associated factors to form complex A. The U4/U5/U6 tri-snRNP is recruited to the assembling spliceosome to form complex B, and following several rearrangements, the spliceosome complex is activated for catalysis (41).
In our previous and present study, we have demonstrated that Tudor-SN associates with U1, U2, U4, U5, U6, and U7 snRNAs but not 7SK snRNA (13). This suggests that two common features of snRNP, Sm core protein and trimethylated cap structure, are required for the interaction of Tudor-SN, for 7SK missed both features. However, the hyper-methylation of the snRNA cap structure is dependent on the recognition and association with Sm core proteins. Therefore, we hypothesize two possibilities that Tudor-SN interacts with TMG-capped snRNA as follows: 1) Tudor-SN protein binds Sm proteins anchoring on the U1, U2, U4, U5, and U6 snRNAs; 2) alternatively, Tudor-SN may recognize the TMG structure of snRNAs in the spliceosomal complex. The efficient co-immunoprecipitation of Tudor-SN with anti-TMG-cap antibody excludes the possibility that the TMG-cap structure of snRNAs is the target of Tudor-SN (13). Therefore, in this study, we speculate about the association of Tudor-SN and Sm proteins.
The tudor domain is a 60-amino acid structure motif that is generally found in proteins with putative functions in RNA or protein binding. In this study, we demonstrate that the interaction of Tudor-SN and Sm proteins (SmB and SmD1/D3) is mediated by the tudor domain, and the formation of a high affinity Tudor-SN·SmB·D1·D3 complex requires binding orientation of the methylated ligand and the presented specific binding pocket of the tudor domain, for example, the GST pulldown assay demonstrated that the tudor domain of Tudor-SN protein could efficiently interact with SmB and SmD1/D3 proteins. Meanwhile, the co-immunoprecipitation assay also indicated that either ectopically expressed or endogenous Tudor-SN protein could efficiently associate with SmB and SmD1/D3 proteins. However, when the Sm proteins are de-methylated by MTA, they could not sufficiently interact with Tudor-SN or recognize U snRNAs. The Drosophila and human tudor domains are structurally similar, specifically the corresponding aromatic cages. Shaw et al. (11) reported that the tudor structure of human Tudor-SN is implicated in recognition and binding of methyl groups. The TSN of Drosophila can also bind with symmetrically dimethylated putative ligands derived from the C-terminal tails of Sm proteins (47), which support our present data that sDMA modification of SmB/B′ and SmD1/D3 is required for the efficient association of human Tudor-SN.
Recent structure analysis of Tudor-SN in different species (11, 47, 50) has shown that the tudor domain also associates with other methylated peptides. For example, Liu et al. (50) reported that the TSN domain of Drosophila Tudor-SN could bind PIWIL1 in an arginine methylation-dependent manner. In PIWIL1 binding, both the N- and C-terminal SN parts of the TSN domain were essential for the ligand binding, whereas in our study the tudor core domain alone was sufficient to associate with SmB. Tudor domains of other proteins have also been shown to bind to methylated protein and regulate protein-protein interaction. For instance, the tudor domain of 53BP1 (51) and JMJD2A (52) can bind methylated histone tails, but not the tudor domain of Drosophila Tudor-SN protein (47). Therefore, preference for different sDMAs or methyl-lysine modification of the ligands may contribute to the distinct biological functions of different tudor domain-containing proteins.
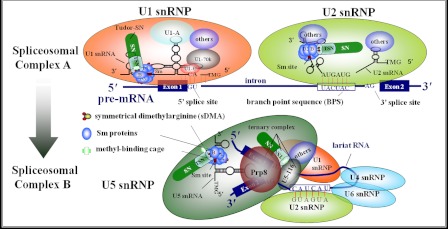
Concerning the role of the SMN protein in snRNP biogenesis, it is quite similar to Tudor-SN protein. SMN interacts with the Sm core proteins by binding to the sDMA-rich C-terminal domains of SmB, SmD1, and SmD3. Here, we speculate that the tudor domain of Tudor-SN protein participates in snRNP through Sm protein interaction. Moreover, the efficient association of Tudor-SN, SmB/B′, and SmD1/D3 in the RNA-free lysate strongly supports the idea that the Sm·Tudor-SN complex formation is not mediated by RNA. The assembled snRNPs could be immunoprecipitated with anti-TMG-coated beads, which recognize the cap structure present on the mature Sm-snRNAs. Using this assay, we further demonstrate that knockdown of endogenous Tudor-SN or SMN reduced the amount of Sm proteins anchored on the U snRNAs and impacted the level of snRNA TMG capping in vivo. Furthermore, depletion of both Tudor-SN and SMN significantly impairs the association of the Sm core complex to the snRNA, although ectopically expressed SMN could restore the impaired association of the Sm to the snRNA caused by knockdown of endogenous Tudor-SN protein. All these data indicate that Tudor-SN, as well as SMN, is required for the recruitment of the Sm core complex to U1, U2, U4, U5, and U6 snRNAs. In addition, Tudor-SN and U5–116 could form a stable complex via interaction with different domains of Prp8 protein, even in a high salt concentration condition (shown in supplemental Figs. S1 and S2). Therefore, there are two possibilities to explain our finding that Tudor-SN takes part in snRNP assembly (Fig. 9) as follows: one is the efficient binding of Tudor-SN with the Sm proteins, and the recruitment to the U snRNAs, and the other one is via the association of U5 snRNP.
FIGURE 9.
Model demonstrates two roles of Tudor-SN in regulating pre-mRNA splicing, particularly the formation of spliceosomal complex A and the transition from complex A to B. Tudor-SN preferentially interacts with symmetrically dimethylated SmB/B′, SmD1, and SmD3 proteins, which facilitates the recruitment of Sm proteins to the U snRNAs and the formation of spliceosomal complex A. However, Tudor-SN takes part in U5 snRNP via the association with Prp8 and U5–116, which may facilitate the transition from complex A to complex B.
However, in the tudor domain of Tudor-SN protein, mutations of the conserved aromatic residues Phe-715, Tyr-721, Tyr-738, and Tyr-741, which form the rectangle aromatic cage, disrupt methyl binding and consequently impair the association of SmB. This explains our previous observation that mutation of TSN proteins exhibits weaker interaction with U1, U2, U5, and U4/U6 snRNAs (13). Mutation of key amino acids within certain multifunctional proteins frequently alters protein-protein interactions, influences various biogenesis pathways, and even contributes to the pathogenesis of certain diseases. For instance, in type I spinal muscular atrophy, the point mutations E134K (53) and W92S (54) have previously been shown to affect the interaction of SMN with Sm proteins. Thus, it is possible that specific point mutation(s) in the TSN domain may be involved in the pathogenesis of related human diseases.
Interestingly, the crystal structure indicates that the SN domain of Tudor-SN could capture nucleic acids (7), and we reported earlier that the recombinant SN domain of Tudor-SN protein totally blocked the in vitro splicing reaction and the spliceosome complex formation (13). The possible explanation is that ectopically expressed SN domain may interact with pre-mRNA and occupy the splicing sites. As a result, U snRNP could not efficiently recognize the splicing sites. We hypothesize that SN and TSN domain functions are coordinated in regulating pre-mRNA splicing. In general, the TSN domain is responsible for the taking part of the U snRNP, and the SN domain may help the Tudor-SN-containing U snRNP complex to anchor on pre-mRNA and then facilitate the recognition and interaction with the splicing site. More experiments need to be done to confirm this hypothesis.
The stepwise snRNP biogenesis pathway begins with the transcription of small nuclear RNAs in the nucleus, followed by their export to the cytoplasm, where the major Sm core assembly of the snRNPs occurs. Once in the nucleus, mature snRNPs carry out the process of pre-mRNA splicing (55, 56). The multifunctional Tudor-SN protein is present in both the nucleus and cytoplasm (1, 3, 9–11, 13); however, it remains unclear whether a transport mechanism of Tudor-SN protein between the cytoplasm and nucleus exists during snRNP assembly and pre-mRNA splicing processes. Further studies need to be conducted to unravel these open questions.
Supplementary Material
Acknowledgments
We thank X. M. Sun for technical assistance and Dr. M. Frilander from the University of Helsinki for the reagents.
This work was supported by Project 863 of the Ministry of Science and Technology of China Grants 2007AA02Z115, NSFC 31125012, 90919032, 30970582, 30670441, 31100967, and 31170830, 973 Program Grant 2009CB918903, Specialized Fund for the Doctoral Program of Higher Education Grant 20091202110001, China Postdoctoral Science Foundation Grant 2011M500529, Tianjin Educational Committee Foundation Grant 2008ZD01, and Tianjin Municipal Science and Technology Commission Grant 08ZCGHHZ01900.
- SN
- staphylococcal nuclease-like domain
- TSN
- tudor and SN-like domain
- U snRNPs
- uridine-rich small ribonucleoprotein
- sDMA
- symmetrical dimethylarginine
- TCL
- total cell lysate
- Q-PCR
- quantitative real time PCR
- TMG
- trimethylguanosine
- snRNP
- small nuclear ribonucleoprotein
- RIP
- RNA-binding protein immunoprecipitation
- MTA
- 5-deoxy-5-(methylthio)adenosine
- SMN
- survival of motor neuron.
REFERENCES
- 1. Broadhurst M. K., Lee R. S., Hawkins S., Wheeler T. T. (2005) The p100 EBNA-2 co-activator. A highly conserved protein found in a range of exocrine and endocrine cells and tissues in cattle. Biochim. Biophys. Acta 1681, 126–133 [DOI] [PubMed] [Google Scholar]
- 2. Rodríguez L., Ochoa B., Martínez M. J. (2007) NF-Y and Sp1 are involved in transcriptional regulation of rat SND p102 gene. Biochem. Biophys. Res. Commun. 356, 226–232 [DOI] [PubMed] [Google Scholar]
- 3. Zhao C. T., Shi K. H., Su Y., Liang L. Y., Yan Y., Postlethwait J., Meng A.M. (2003) Two variants of zebrafish p100 are expressed during embryogenesis and regulated by Nodal signaling. FEBS Lett. 543, 190–195 [DOI] [PubMed] [Google Scholar]
- 4. Howard-Till R. A., Yao M. C. (2007) Tudor nuclease genes and programmed DNA rearrangements in Tetrahymena thermophila. Eukaryot. Cell 6, 1795–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sami-Subbu R., Choi S. B., Wu Y., Wang C., Okita T. W. (2001) Identification of a cytoskeleton-associated 120-kDa RNA-binding protein in developing rice seeds. Plant Mol. Biol. 46, 79–88 [DOI] [PubMed] [Google Scholar]
- 6. Abe S., Sakai M., Yagi K., Hagino T., Ochi K., Shibata K., Davies E. (2003) A Tudor protein with multiple SNc domains from pea seedlings. Cellular localization, partial characterization, sequence analysis, and phylogenetic relationships. J. Exp. Bot. 54, 971–983 [DOI] [PubMed] [Google Scholar]
- 7. Li C. L., Yang W. Z., Chen Y. P., Yuan H. S. (2008) Structural and functional insights into human Tudor-SN, a key component linking RNA interference and editing. Nucleic Acids Res. 36, 3579–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Paukku K., Kalkkinen N., Silvennoinen O., Kontula K. K., Lehtonen J. Y. (2008) p100 increases AT1R expression through interaction with AT1R 3′-UTR. Nucleic Acids Res. 36, 4474–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Caudy A. A., Ketting R. F., Hammond S. M., Denli A. M., Bathoorn A. M., Tops B. B., Silva J. M., Myers M. M., Hannon G. J., Plasterk R. H. (2003) A micrococcal nuclease homologue in RNAi effector complexes. Nature 425, 411–414 [DOI] [PubMed] [Google Scholar]
- 10. Gao X., Ge L., Shao J., Su C., Zhao H., Saarikettu J., Yao X., Yao Z., Silvennoinen O., Yang J. (2010) Tudor-SN interacts with and co-localizes with G3BP in stress granules under stress conditions. FEBS Lett. 584, 3525–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shaw N., Zhao M., Cheng C., Xu H., Saarikettu J., Li Y., Da Y., Yao Z., Silvennoinen O., Yang J., Liu Z. J., Wang B. C., Rao Z. (2007) The multifunctional human p100 protein “hooks” methylated ligands. Nat. Struct. Mol. Biol. 14, 779–784 [DOI] [PubMed] [Google Scholar]
- 12. Yang J., Aittomäki S., Pesu M., Carter K., Saarinen J., Kalkkinen N., Kieff E., Silvennoinen O. (2002) Identification of p100 as a co-activator for STAT6 that bridges STAT6 with RNA polymerase II. EMBO J. 21, 4950–4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang J., Välineva T., Hong J., Bu T., Yao Z., Jensen O. N., Frilander M. J., Silvennoinen O. (2007) Transcriptional co-activator protein p100 interacts with snRNP proteins and facilitates the assembly of the spliceosome. Nucleic Acids Res. 35, 4485–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Välineva T., Yang J., Palovuori R., Silvennoinen O. (2005) The transcriptional co-activator protein p100 recruits histone acetyltransferase activity to STAT6 and mediates interaction between the CREB-binding protein and STAT6. J. Biol. Chem. 280, 14989–14996 [DOI] [PubMed] [Google Scholar]
- 15. Dong L., Zhang X., Fu X., Zhang X., Gao X., Zhu M., Wang X., Yang Z., Jensen O. N., Saarikettu J., Yao Z., Silvennoinen O., Yang J. (2011) PTB-associated splicing factor (PSF) functions as a repressor of STAT6-mediated Ig ϵ gene transcription by recruitment of HDAC1. J. Biol. Chem. 286, 3451–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Will C. L., Lührmann R. (2001) Spliceosomal UsnRNP biogenesis, structure, and function. Curr. Opin. Cell Biol. 13, 290–301 [DOI] [PubMed] [Google Scholar]
- 17. Kambach C., Walke S., Nagai K. (1999) Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Curr. Opin. Struct Biol. 9, 222–230 [DOI] [PubMed] [Google Scholar]
- 18. Jurica M. S., Moore M. J. (2003) Pre-mRNA splicing. Awash in a sea of proteins. Mol. Cell 12, 5–14 [DOI] [PubMed] [Google Scholar]
- 19. Nagai K., Muto Y., Pomeranz Krummel D. A., Kambach C., Ignjatovic T., Walke S., Kuglstatter A. (2001) Structure and assembly of the spliceosomal snRNPs. Novartis Medal Lecture. Biochem. Soc. Trans. 29, 15–26 [DOI] [PubMed] [Google Scholar]
- 20. Achsel T., Stark H., Lührmann R. (2001) The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proc. Natl. Acad. Sci. U.S.A. 98, 3685–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kambach C., Walke S., Young R., Avis J. M., de la Fortelle E., Raker V.A., Lührmann R., Li J., Nagai K. (1999) Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 96, 375–387 [DOI] [PubMed] [Google Scholar]
- 22. Stark H., Dube P., Lührmann R., Kastner B. (2001) Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature 409, 539–542 [DOI] [PubMed] [Google Scholar]
- 23. Boisvert F. M., Chénard C. A., Richard S. (2005) Protein interfaces in signaling regulated by arginine methylation. Sci. STKE 2005, re2. [DOI] [PubMed] [Google Scholar]
- 24. Lee J. H., Cook J. R., Yang Z. H., Mirochnitchenko O., Gunderson S. I., Felix A. M., Herth N., Hoffmann R., Pestka S. (2005) PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. J. Biol. Chem. 280, 3656–3664 [DOI] [PubMed] [Google Scholar]
- 25. Meister G., Eggert C., Bühler D., Brahms H., Kambach C., Fischer U. (2001) Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr. Biol. 11, 1990–1994 [DOI] [PubMed] [Google Scholar]
- 26. Cheng D., Côté J., Shaaban S., Bedford M. T. (2007) The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol. Cell 25, 71–83 [DOI] [PubMed] [Google Scholar]
- 27. Gonsalvez G. B., Tian L., Ospina J. K., Boisvert F. M., Lamond A. I., Matera A. G. (2007) Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins. J. Cell Biol. 178, 733–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miranda T. B., Khusial P., Cook J. R., Lee J. H., Gunderson S. I., Pestka S., Zieve G. W., Clarke S. (2004) Spliceosome Sm proteins D1, D3, and B/B′ are asymmetrically dimethylated at arginine residues in the nucleus. Biochem. Biophys. Res. Commun. 323, 382–387 [DOI] [PubMed] [Google Scholar]
- 29. Sprangers R., Groves M. R., Sinning I., Sattler M. (2003) High resolution x-ray and NMR structures of the SMN Tudor domain. Conformational variation in the binding site for symmetrically dimethylated arginine residues. J. Mol. Biol. 327, 507–520 [DOI] [PubMed] [Google Scholar]
- 30. Côté J., Richard S. (2005) Tudor domains bind symmetrical dimethylated arginines. J. Biol. Chem. 280, 28476–28483 [DOI] [PubMed] [Google Scholar]
- 31. Tripsianes K., Madl T., Machyna M., Fessas D., Englbrecht C., Fischer U., Neugebauer K. M., Sattler M. (2011) Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat. Struct. Mol. Biol. 18, 1414–1420 [DOI] [PubMed] [Google Scholar]
- 32. Kolb S. J., Battle D. J., Dreyfuss G. (2007) Molecular functions of the SMN complex. J. Child Neurol. 22, 990–994 [DOI] [PubMed] [Google Scholar]
- 33. Bühler D., Raker V., Lührmann R., Fischer U. (1999) Essential role for the Tudor domain of SMN in spliceosomal U snRNP assembly. Implications for spinal muscular atrophy. Hum. Mol. Genet. 8, 2351–2357 [DOI] [PubMed] [Google Scholar]
- 34. Pessa H. K., Ruokolainen A., Frilander M. J. (2006) The abundance of the spliceosomal snRNPs is not limiting the splicing of U12-type introns. RNA 12, 1883–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G. (2008) SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133, 585–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real time quantitative PCR and the 2−ΔΔC(T)) Method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 37. Chakrabarti S. K., James J. C., Mirmira R. G. (2002) Quantitative assessment of gene targeting in vitro and in vivo by the pancreatic transcription factor Pdx1. Importance of chromatin structure in directing promoter binding. J. Biol. Chem. 277, 13286–13293 [DOI] [PubMed] [Google Scholar]
- 38. Zhou Z., Licklider L. J., Gygi S. P., Reed R. (2002) Comprehensive proteomic analysis of the human spliceosome. Nature 419, 182–185 [DOI] [PubMed] [Google Scholar]
- 39. Frilander M. J., Meng X. (2005) Proximity of the U12 snRNA with both the 5′ splice site and the branch point during early stages of spliceosome assembly. Mol. Cell. Biol. 25, 4813–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Surowy C. S., van Santen V. L., Scheib-Wixted S. M., Spritz R. A. (1989) Direct sequence-specific binding of the human U1–70K ribonucleoprotein antigen protein to loop I of U1 small nuclear RNA. Mol. Cell. Biol. 9, 4179–4186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Grainger R. J., Beggs J. D. (2005) Prp8 protein: at the heart of the spliceosome. RNA 11, 533–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brenner T. J., Guthrie C. (2006) Assembly of Snu114 into U5 snRNP requires Prp8 and a functional GTPase domain. RNA 12, 862–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brahms H., Meheus L., de Brabandere V., Fischer U., Lührmann R. (2001) Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B′ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA 7, 1531–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sleeman J. E., Ajuh P., Lamond A. I. (2001) snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J. Cell Sci. 114, 4407–4419 [DOI] [PubMed] [Google Scholar]
- 45. Boisvert F. M., Cote J., Boulanger M. C., Cleroux P., Bachand F., Autexier C., Richard S. (2002) Symmetrical dimethylarginine methylation is required for the localization of SMN in Cajal bodies and pre-mRNA splicing. J. Cell Biol. 159, 957–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brahms H., Raymackers J., Union A., de Keyser F., Meheus L., Lührmann R. (2000) The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J. Biol. Chem. 275, 17122–17129 [DOI] [PubMed] [Google Scholar]
- 47. Friberg A., Corsini L., Mourão A., Sattler M. (2009) Structure and ligand binding of the extended Tudor domain of D. melanogaster Tudor-SN. J. Mol. Biol. 387, 921–934 [DOI] [PubMed] [Google Scholar]
- 48. Boisvert F. M., Côté J., Boulanger M. C., Richard S. (2003) A proteomic analysis of arginine-methylated protein complexes. Mol Cell. Proteomics 2, 1319–1330 [DOI] [PubMed] [Google Scholar]
- 49. Andersen J., Zieve G. W. (1991) Assembly and intracellular transport of snRNP particles. BioEssays 13, 57–64 [DOI] [PubMed] [Google Scholar]
- 50. Liu K., Chen C., Guo Y., Lam R., Bian C., Xu C., Zhao D. Y., Jin J., MacKenzie F., Pawson T., Min J. (2010) Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain. Proc. Natl. Acad. Sci. U.S.A. 107, 18398–18403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huyen Y., Zgheib O., Ditullio R. A., Jr., Gorgoulis V.G., Zacharatos P., Petty T. J., Sheston E. A., Mellert H. S., Stavridi E. S., Halazonetis T. D. (2004) Methylated lysine 79 of histone H3 targets 53BP1 to DNA double strand breaks. Nature 432, 406–411 [DOI] [PubMed] [Google Scholar]
- 52. Huang Y., Fang J., Bedford M. T., Zhang Y., Xu R. M. (2006) Recognition of histone H3 lysine-4 methylation by the double Tudor domain of JMJD2A. Science 312, 748–751 [DOI] [PubMed] [Google Scholar]
- 53. Selenko P., Sprangers R., Stier G., Bühler D., Fischer U., Sattler M. (2001) SMN Tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Biol. 8, 27–31 [DOI] [PubMed] [Google Scholar]
- 54. Kotani T., Sutomo R., Sasongko T. H., Sadewa A. H., Gunadi, Minato T., Fujii E., Endo S., Lee M. J., Ayaki H., Harada Y., Matsuo M., Nishio H. (2007) A novel mutation at the N-terminal of SMN Tudor domain inhibits its interaction with target proteins. J. Neurol. 254, 624–630 [DOI] [PubMed] [Google Scholar]
- 55. Guthrie C. (2010) From the ribosome to the spliceosome and back again. J. Biol. Chem. 285, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tkacz I. D., Gupta S. K., Volkov V., Romano M., Haham T., Tulinski P., Lebenthal I., Michaeli S. (2010) Analysis of spliceosomal proteins in Trypanosomatids reveals novel functions in mRNA processing. J. Biol. Chem. 285, 27982–27999 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.