Background: Search for novel avidins with reduced oligomeric state revealed an intriguing dimeric protein.
Results: Structures of shwanavidin from Shewanella denitrificans identified features accountable for high affinity and psychrophilic properties.
Conclusion: Phe-43 and disulfide bridge are responsible for the high affinity compensating the lack of intermonomeric interaction in tetrameric avidins.
Significance: Proteins from extremophile organisms provide excellent platforms toward the development of novel biotechnological utilizations.
Keywords: Biotin, Circular Dichroism (CD), Isothermal Titration Calorimetry, Structural Biology, X-ray Crystallography, High Affinity Systems, Avidin, Biotin Interaction, Psychrophilic Adaptation, Thermostability
Abstract
Shwanavidin is an avidin-like protein from the marine proteobactrium Shewanella denitrificans, which exhibits an innate dimeric structure while maintaining high affinity toward biotin. A unique residue (Phe-43) from the L3,4 loop and a distinctive disulfide bridge were shown to account for the high affinity toward biotin. Phe-43 emulates the function and position of the critical intermonomeric Trp that characterizes the tetrameric avidins but is lacking in shwanavidin. The 18 copies of the apo-monomer revealed distinctive snapshots of L3,4 and Phe-43, providing rare insight into loop flexibility, binding site accessibility, and psychrophilic adaptation. Nevertheless, as in all avidins, shwanavidin also displays high thermostability properties. The unique features of shwanavidin may provide a platform for the design of a long sought after monovalent form of avidin, which would be ideal for novel types of biotechnological application.
Introduction
Proteins belonging to the avidin family are recognized for their remarkable affinity toward d-biotin (1). During the past 4 decades, the avidin-biotin system has been utilized extensively and has developed into an essential biotechnological tool in diverse fields of research and analysis (2, 3). Historically, the first avidin to be documented was from the egg white of chicken and other avian species (1). Later, streptavidins from a few Streptomyces species (4), and, more recently, isolated examples of avidin-like proteins were discovered in other prokaryotic (5–8) and eukaryotic (fungal) sources (9). Despite the sporadic but wide distribution of high affinity biotin-binding avidin-like proteins, their precise function in nature is essentially unknown.
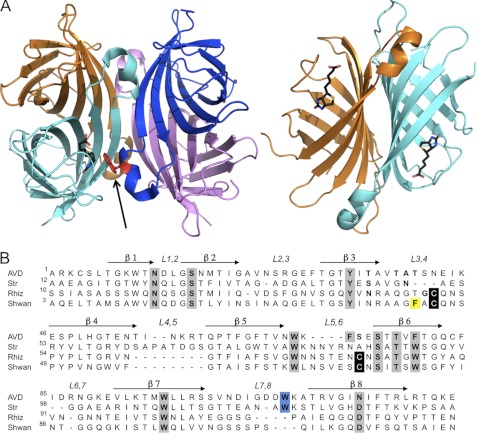
Avidins are usually homotetrameric proteins with four biotin-binding sites, and their high affinity characteristics are closely dependent on their quaternary assembly (Fig. 1A). The monomeric subunit of all avidins consists of a highly similar topology comprising an eight-stranded antiparallel β-barrel. The tertrameric assembly is regarded as a dimer of dimers (10), each monomer of which forms three types of interactions defined as 1-2, 1-3, and 1-4 (11). The 1-2 interaction includes a critical tryptophan residue, located in the L7,8 loop (connecting β7 and β8), which is inserted into the biotin-binding site of an adjacent monomer and plays an important role in both high affinity biotin binding and tetrameric integrity (12–14). The dissociation of the avidins into dimers or monomers results in a dramatic decrease (7–8 orders of magnitude) in affinity toward biotin (13–15). The 1-4 interface forms the largest contact area (∼1500 Å2) with numerous intermonomeric interactions (Fig. 1A), whereas the 1-3 interface involves only 3–4 amino acids contributed by each monomer and exhibits the smallest intermolecular contact surface (∼210 Å2).
FIGURE 1.
Structures of tetrameric avidins. A, tetrameric assembly of egg white avidin (left), with respective monomers shown in magenta, blue, cyan, and orange. The biotin ligand is shown in black in the binding site of the cyan monomer (for clarity). The Trp residue (shown in red) from an adjacent monomer (blue) contributes to the high affinity of biotin binding and is indicated by an arrow. The 1-4 sandwich-like monomer-monomer interaction (right) forms numerous stabilizing interactions (orange and cyan). All molecular graphics figures were generated using PyMOL (47). B, multiple sequence alignment of avidin (AVD), streptavidin (Str), rhizavidin (Rhiz), and shwanavidin (Shwan). The eight β-strands forming the tertiary structure are indicated by arrows, and the loops connecting the strands are labeled. Residues participating in biotin binding are highlighted in gray, cysteines participating in the disulfide bridge in the new binding site are in black, and the Trp residues from the L7,8 contributing to the biotin binding in tetrameric avidins are highlighted in blue. Phe-43 from the L3,4 loop compensates for the lack of the intermonomeric Trp residue and is highlighted in yellow. Shwanavidin exhibits 46, 50, and 61% sequence similarity and 26, 33, and 47% identity compared with avidin, streptavidin, and rhizavidin, respectively.
Another essential component for biotin binding is the L3,4 loop connecting strands β3 and β4. This loop is disordered in the apo forms of avidin and streptavidin, but becomes ordered upon biotin binding, leaving merely a small portion of the biotin carboxylate available to the solvent (11, 16). Additionally, upon biotin binding the L3,4 loop forms several polar interactions with biotin thus stabilizing its conformation (11, 16). In addition to the interactions with the L3,4 loop, there are important hydrophobic and polar interactions that accommodate the biotin molecule into the highly complementary contours of the binding site. In the tetrameric avidins the biotin ligand inhabits a hydrophobic box, formed by either 3 or 4 conserved aromatic residues, which is then sealed by the additional Trp contributed by a neighboring monomer. The biotin molecule is also stabilized by an intricate network of H-bond interactions in which its bicyclic segment and the carboxylate moiety are highly conserved throughout the avidin family (11, 17).
Biotechnological considerations have fueled the quest for additional avidin-like proteins that would exhibit divergent oligomeric states that could lead to the development of high affinity mono- and divalent avidins of smaller size consisting of a single polypeptide chain (18). Several attempts have been conducted to achieve these types of avidin, such as dual-chain and circular permutated avidins and streptavidins; yet, the resulting affinity of the product was compromised (19–22). In addition, a monovalent tetrameric assembly of streptavidin was obtained with one biotin-binding site available, yet its overall size remained as large as other tetrameric avidins (23).
The recently discovered rhizavidin from Rhizobium etli is the first known dimeric member of the avidin family (1-4 interaction) (6). The divalent form maintains high affinity toward biotin, thus providing an initial molecular platform toward the development of a minimized high affinity avidin. The 1-2 and 1-3 monomer-monomer interactions are lacking in rhizavidin, and the crucial Trp from the L7,8 loop is redundant and missing in the sequence (12, 13, 24). The crystal structure of rhizavidin provided indications of how the high affinity is maintained in the absence of the critical Trp residue (6, 25). In this context, a disulfide bridge connecting L3,4 and L5,6 contributes to the rigidity of the loops and also forms interactions with biotin; thus, the bridge was assumed to contribute to the high affinity (25).
Rhizavidin is the first member of the avidin family found to have a unique dimeric structure. We were thus interested to discover additional dimeric (or preferably monomeric) forms of avidin, to identify the diverse features contributing to the high affinity that could be further utilized to form high affinity biotin-binding monomers. In the advent of recent genome-sequencing efforts, several novel avidin-like genes have been detected, particularly in various bacteria, that could potentially exhibit high affinity biotin-binding properties. One of these sequences, identified from the Gram-negative bacterium Shewanella denitrificans (26), is termed herein shwanavidin. Whereas rhizavidin from R. etli and streptavidin from Streptomyces avidinii inhabit the soil rhizosphere (27), S. denitrificans is found at 120–130 m depths of the Baltic Sea at low temperatures (28), and shwanavidin exhibits a high degree of sequence similarity to other avidins (Fig. 1B). Interestingly, the protein was shown to be expressed in the parent bacterium.2
Because shwanavidin and rhizavidin appear to form a unique subgroup in the avidin family, we aimed to determine its biochemical and structural properties. Indeed, similar to rhizavidin, shwanavidin was found to be a thermostable dimer in solution, maintaining high affinity toward biotin. However, the crystal structure of shwanavidin in the apo- and biotin-complexed forms revealed a unique feature in the binding site that contributes to the high affinity and differs significantly from that of rhizavidin. The roles of the disulfide bridge and Phe-43 in biotin binding were examined via mutagenesis and structural analysis and were found to be critical in maintaining the high affinity. Intriguingly, despite its thermostable properties, shwanavidin maintains its high affinity biotin-binding function under psychrophilic conditions. The repulsive interactions of the unique Phe-43 residue with solvent results in high flexibility of a segment of the L3,4 loop, thereby permitting biotin binding at low temperatures.
EXPERIMENTAL PROCEDURES
Shwanavidin Gene and Plasmid Construction
The shwanavidin sequence was discovered using BLAST in the UniProtKB data base via the ExPASy web site (29) as well as the NCBI BLASTp (30, 31). Shwanavidin exhibits 50 and 61% sequence similarity, 33 and 47% identity compared with streptavidin and rhizavidin, respectively. Running the BLASTn (30, 31) search with shwanavidin gene sequence versus the NCBI data base (32) did not result in any significant hits, not even for other Shewanella species. The full-length shwanavidin sequence contained 172 amino acids, where the first 42 residues were assumed to be part of a signal peptide. The last 10 residues, which also corresponds to the streptavidin C-terminal cleavage and rhizavidin flexible C′ terminus, were omitted from the synthetic gene leaving only 122 residues (residues number 43–162 from the original sequence, with 2 additional residues (Met-Ala) at the N′ terminus, resulting from two restriction sites now numbered 1–122). A synthetic gene of shwanavidin was constructed based on its amino acid sequence as available in the UniProtKB data base with codon usage suitable for Escherichia coli expression (by GenScript). The shwanavidin gene was cloned into a pHis2 parallel plasmid (33) using restriction enzymes NdeI and XhoI to obtain a core protein consisting of residues 43–162 with Met-Ala at the N terminus (referred to henceforth as shwanavidin). Furthermore, a His-tagged protein (His-shwanavidin), with an additional 24 residues at the N′ terminus, comprising the His6 tag and a tobacco etch virus protease cleavage site between the His6 and the core protein, was cloned into the vector using restriction sites NcoI and XhoI.
C45A/C74A and F43A Mutations
Site-directed mutagenesis of C45A/C74A and F43A mutants were carried out using the QuikChange kit with Pfu ultra DNA polymerase (Stratagene). The reaction was performed on the shwanavidin gene subcloned into pHis2 parallel (33) downstream and in-frame to the His6-tobacco etch virus protease cleavage site coding sequence. The procedure included PCR using specific complementary primers (supplemental Table 1) followed by a DpnI digestion. The elongation step was set to 12 min at 68 °C.
Structure Determination of Shwanavidin
Expression, purification, crystallization, and data collection of shwanavidin (wild type and mutants) are described in supplemental Experimental Procedures. The native apo structure was solved by molecular replacement using PHASER (34, 35) implemented in the CCP4i suite (36). The search models included monomer and dimer from a Swiss-Model (37, 38) homology model of shwanavidin based on the rhizavidin structure (25). Matthew's coefficient indicated the presence of 8–14 shwanavidin molecules in the asymmetric unit (Vm = 2.84–1.89 Å3/Da, 56.7–35% solvent content). The resulting PHASER solution at the resolution range of 50.0–3.0 Å using a dimeric model resulted in a dodecamer in the asymmetric unit (Vm = 1.89 Å3/Da; 35% solvent). The structure was initially refined using the rigid body protocol in REFMAC (39) resulting in an R values of 39.7% and Rfree of 40.1%. The structure was further refined to the resolution of 50–1.45 Å using REFMAC5, and solvent molecules were added utilizing ARP/wARP (40), after several iterative cycles of refinement and model building utilizing COOT (41) (Table 1).
TABLE 1.
Data collection and refinement statistics
| Parameter | Shwanavidin apo-Form-A | Shwanavidin apo-Form-B | Shwanavidin-biotin complex | Shwanavidin apo-F43A | Shwanavidin F43A-biotin complex |
|---|---|---|---|---|---|
| ESRF beamline | ID 23-1 | ID 29 | ID 23-1 | ID 29 | ID 29 |
| Wavelength (Å) | 0.972 | 0.979 | 0.875 | 0.9736 | 0.9736 |
| Space group | P21 | P1 | P212121 | R3 (H3) | P43212 |
| Unit cell parameters (Å) | a + 67.5, b + 110.6, c + 84.2, β + 93.9° | a + 43.3, b + 63.5, c + 67.7, α + 103.8°, β + 107.3°, γ + 103.8° | a + 47.7, b + 56.0, c + 86.6 | a + 113.93, c + 83.43 | a + 63.21, c + 65.83 |
| Resolution rangea (Å) (last resolution shell) | 50–1.40 (1.42–1.40) | 50.0–1.07 (1.09–1.07) | 50.0–1.45 (1.48–1.45) | 50–1.15 (1.17–1.15) | 50.0–1.5 (1.53–1.5) |
| Unique reflections | 236,160 | 257,028 | 37,616 | 143,125 | 21,973 |
| Redundancy | 2.4 | 3.0 | 4.3 | 3.0 | 6.9 |
| Rsym(I)b | 3.5 (52.0) | 5.8 (53.9) | 8.0 (58.5) | 6.6 (73.0) | 5.5 (93.8) |
| Completeness | 97.5 (95.5) | 92.5 (80.5) | 99.8 (99.8) | 99.7 (99.9) | 99.2 (99.9) |
| I/σ | 24.9 (1.6) | 35.3 (1.5) | 16.7 (1.5) | 25.8 (1.8) | 40.5 (2.15) |
| Number of protein atoms | 11,029 | 5,513 | 1,890 | 3,678 | 890 |
| Number of ligand atoms | 32 | 16 | |||
| Number of solvent/formate atoms | 1,374/33 | 851 | 197 | 546 | 109 |
| R factor | 0.158 | 0.161 | 0.162 | 0.146 | 0.18 |
| Rfreec | 0.193 | 0.187 | 0.190 | 0.171 | 0.212 |
| Average B factor (Å2) | |||||
| Protein | 16.7 | 13.9 | 10.8 | 12.04 | 24.08 |
| Biotin | 8.6 | 10.58 | |||
| Solvent | 31.6 | 29.7 | 27.8 | 28.83 | 37.62 |
| Formate | 30.5 | ||||
| R.m.s.d. from ideal | |||||
| Bond length | 0.011 | 0.010 | 0.012 | 0.012 | 0.014 |
| Bond angle | 1.43 | 1.38 | 1.55 | 1.54 | 1.603 |
| Ramachandran plot (PROCHECK) (%) | |||||
| Favored | 87.8 | 87.0 | 87.8 | 87.9 | 88.8 |
| Allowed | 12.2 | 12.8 | 12.2 | 12.1 | 11.2 |
| Generously allowed | 0.0 | 0.2 | 0.0 | 0.0 | 0.0 |
| Disallowed | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
a Outer shell resolution range.
b Rsym(I) + Σ|I − <I>|/ΣI.
c Test set consists of 5% for all data.
Another data set of a different crystal form of apo-shwanavidin was collected to the maximal resolution of 1.07 Å (termed Form-B). The structure of the Form-B triclinic shwanavidin was solved by PHASER using the shwanavidin refined Form-A dimer as the search model at the resolution range of 50–3.5 Å. The solution indicated the presence of six shwanavidin monomers in the asymmetric unit having a Vm value of 1.96 Å3/Da with 37.2% solvent content. The structure was refined in a similar protocol described for the Form-A apo-shwanavidin (Table 1). Crystals of the shwanavidin-biotin complex belonged to the orthorhombic space group P212121 with cell parameters a = 47.7 Å, b = 56.0 Å, c = 86.6 Å. The structure of the biotin complex was solved by PHASER (50–4 Å) using the shwanavidin refined monomer as the search model. The solution indicated the presence of two shwanavidin monomers in asymmetric unit (Vm = 1.99 Å3/Da, 35% solvent). Difference electron density maps clearly indicated the presence of biotin molecules in the corresponding binding sites.
The apo-F43A crystals belonged to the rhombohedral space group R3(H3) with cell parameters a = 113.9 Å, c = 83.4 Å, and the biotin-complex F43A crystals belonged to the tetragonal P43212 space group; both structures were solved by PHASER using the Form-A solved structure as the search model. The solution for the apo-F43A mutant indicated the presence of four shwanavidin monomers in asymmetric unit (Vm = 2 Å3/Da, 38.7% solvent). For the biotin complex the solution indicated one shwanavidin monomer in asymmetric unit (Vm = 1.23 Å3/Da, 51.4% solvent). The inherent dimer was generated by a 2-fold symmetry. The difference electron density maps clearly indicated the presence of a biotin molecule in the binding site.
Affinity and Thermostability Measurements
CD spectra of the shwanavidin and its mutants were conducted using a JASCO J-810 spectrophotometer using SpectraManager software. Proteins were initially dialyzed overnight against a solution containing 10 mm NaH2PO4 buffer. Protein concentration was then determined using spectrophotometer, and a 7-μl saturated biotin solution was added to 200 μl of 14 μm protein solutions in 10 mm phosphate buffer and incubated for 30 min on ice. Samples with and without biotin were placed in 0.2-cm quartz cells (Starna, Atascadero, CA), and spectra were recorded in the wavelength range λ = 190–260 nm, with five accumulations for each measurement and a data pitch of 0.1 nm. Background CD spectra of the appropriate buffer (with and without biotin) were recorded and subtracted from each spectrum. Thermal melts were performed by measuring CD spectra at the temperature range of 4–95 °C with a heating rate of 60 °C/h. CD signal was recorded every 0.1 °C, and the CD spectrometer was set to operate with a spectral bandwidth of 1 nm and a response time of 1 s.
The CD melting curve was measured at the wavelength of 233 nm (apo forms) and 234 nm (biotin-complex forms) where a maximum difference between the native and unfolded forms appeared. Melting temperatures (Tm) were calculated using Origin 8.1 software. Normalized Tm curves of all shwanavidin forms were then fitted to the Boltzman model, and its midpoint was calculated as the Tm. For each sample, a measurement between λ values of 190 and 260 nm was measured at 20 °C upon pre- and postheating.
The melting temperatures of native shwanavidin, in the apo- and biotin-complexed forms, were analyzed by differential scanning calorimetry (DSC).3 The experiments were performed both in the absence and presence of biotin, in a protein concentration of 1 mg/ml (76 μm), and shwanavidin-biotin (monomer) ratio of 3:1, in 50 mm sodium phosphate, 1 m NaCl, pH 7. The thermograms (25–130 °C, 0.92 °C/min) were collected with a VP-DSC calorimeter from MicroCal (Northampton, MA). Data analysis was carried out using Origin 7.0 software (MicroCal) to calculate Tm. Because the process was nonreversible, ΔH could not be calculated.
The affinity measurements of the wild-type shwanavidin toward biotin were first conducted using the isothermal titration calorimetry (ITC) method, using a VP-ITC MicroCalorimeter instrument (MicroCal). Shwanavidin was dialyzed overnight against a sodium phosphate buffer (50 mm NaH2PO4, 150 mm NaCl, pH 7). After dialysis, the protein was diluted to a concentration of 12 μm. d-Biotin was diluted to a 10-fold concentration using the same buffer. The measurements were conducted at 25 °C using 10-μl titration aliquots. For comparison, 8 μm rhizavidin and 5 μm streptavidin were measured against a 10-fold concentration of d-biotin at the same buffer. Another set of Kd measurements was conducted at 4 °C. Thermal titration data were fitted to a single binding site model per monomer.
The affinity of shwanavidin and its mutants toward biotin and 2-iminobiotin was measured by the surface plasmon resonance (SPR) method using a Biacore 3000 optical biosensor instrument (Biacore AB, Uppsala, Sweden). A sensor chip was prepared by activating the surface of CM5 chip (carboxymethyl dextran-derivatized surface; Biacore AB) with a mixture of 0.4 m EDC and 0.1 m N-hydroxysuccinimide (NHS) followed by attachment of ethylenediamine. In the resultant chip one channel was treated by injecting a mixture of 0.4 m EDC and 10 mm 2-iminobiotin, and another with NHS-biotin as previously described (25). For the affinity measurements of 2-iminobiotin, 80–800 nm shwanavidin and 5–500 nm rhizavidin were injected over sensor surfaces in 1 m NaCl, 50 mm Na2CO3, pH 11.0 buffer. Proteins were exchanged with the buffer prior to analysis. Surfaces were regenerated between measurements by injecting 20 μl of 0.5 m acetic acid, pH 3, followed by washing with the measurement buffer. For biotin measurements, shwanavidin F43A (70 nm–2 μm), shwanavidin C45A/C74A (10–300 nm), and rhizavidin C50A/C79A (0.5–5 μm) mutants were injected over the NHS-biotin channel sensor (flow rate 20 μl/min) in 1 m NaCl, 50 mm sodium phosphate buffer, pH 7, for 240 s. Surface was regenerated between measurements by injecting 20 μl of 7.2 m urea containing 0.2 m NaOH, followed by wash with the measurement buffer for 600 s. Data analysis was performed using Biaevaluation 3.2 RC1 software, and the entire data set was fitted globally. A period of 20 s from the beginning of each injection was included in the association phase fit, and the dissociation rate analysis was performed for a 60-s period, starting immediately after the end of the injection.
A qualitative affinity assessment toward 2-iminobiotin was also conducted using an open 2-imminobiotin column. Purified shwanavidin and its F43A and C45A/C74A mutants (0.2 mg/ml) were incubated for 1.5 h with 2-iminobiotin beads (Pierce) in 50 mm sodium carbonate, 1 m NaCl buffer, pH 11, and eluted by 0.1 m acetic acid. The unbound, washed, and eluted (bound) fractions were uploaded on SDS-PAGE for analysis.
RESULTS
Structure of Shwanavidin
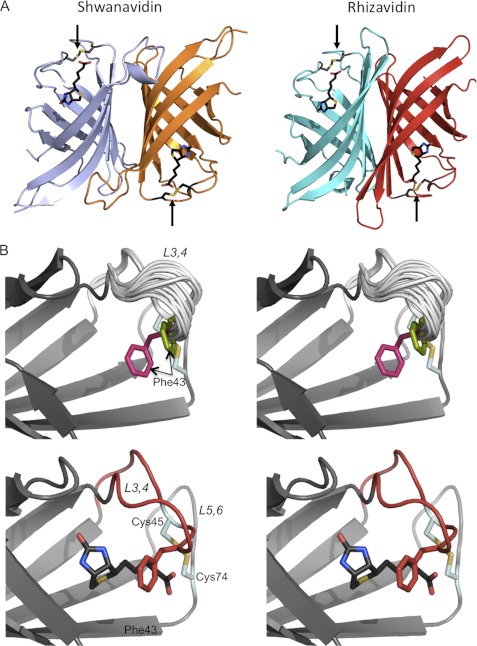
The resolved crystal structures reveal a molecule displaying a 1–4 dimer highly similar to that of rhizavidin (r.m.s.d. of 1.21 Å for 186 Cα pairs) (25) (Fig. 2A and Table 1). Gel filtration analysis clearly supported the presence of shwanavidin as a dimer in solution (supplemental Fig. 1). The inherent shwanavidin dimer is stabilized by numerous interactions with an overall contact surface area of ∼1680 Å2/monomer. As in all avidins, the tertiary structure of shwanavidin consists of an eight-antiparallel β-stranded β-barrel. A distinctive feature in shwanavidin, thus far observed only in the structure of rhizavidin, is a disulfide bridge between Cys-45 and Cys-74 linking the L3,4 and L5,6 loops, respectively. Additionally, conserved Cys residues were also found in homologous positions in sequences of other proteobacteria avidin-like proteins, whose structural characteristics have yet to be studied (5, 7, 8).
FIGURE 2.
Structure of shwanavidin. A, ribbon presentations of the dimeric structures of shwanavidin (left) and rhizavidin (right) in complex with biotin (shown as black sticks). The two structures are very similar, forming an innate dimer consistent with the 1-4 dimeric form of the tetrameric avidins. The binding sites and the corresponding biotin (black) carboxylates are located at opposite positions. The disulfide bridges (colored dark gray, sulfurs in yellow) are shown in stick presentation indicated by arrows. B, stereo presentations of the vicinity of the biotin-binding site of apo- (top) and biotin-complexed (bottom) shwanavidin. For the apo-shwanavidin the superimposed 18 models emphasize the conformational flexibility of the L3,4 loop (gray) especially between residues 40 ad 44 N-terminal to the disulfide bridge. The two most extreme conformations of Phe-43 are indicated in magenta and green and indicated by arrows. The portion of the L3,4 loop, N-terminal to the disulfide bridge, adopts a similar conformation in all monomers; for clarity, only one conformation of the L5,6 loop is presented. In the biotin complex (biotin shown in black), the L3,4 loop (shown in red) adopts a stabilized conformation while forming hydrophobic interactions with biotin. The side chain of Phe-43 (red) now seals the biotin almost completely in its binding site, making most of it unavailable to solvent.
The L3,4 loop is considered to be critical for biotin binding (16, 42–44). In the apo forms of avidin and streptavidin the L3,4 loop is disordered and becomes ordered upon biotin binding by forming several polar interaction with the ligand (11, 16). In shwanavidin the L3,4 loop has the same size as avidin and rhizavidin, highly similar in composition to the latter (Fig. 1B). The main difference in the L3,4 between shwanavidin and rhizavidin is the presence of a Phe-43 residue rather than a Thr. For apo-shwanavidin, the A and B crystal forms provided 18 distinct copies of the monomer, which is an infrequent occurrence in crystal structure analysis (Table 1). Upon superposition of the 18 shwanavidin monomers, it is apparent that the L3,4 loop exhibits different conformational states which could represent incremental snapshots of its flexibility in solution (Fig. 2B). Interestingly, only about half of the loop is highly flexible. The position of Cys-45, part of the disulfide bridge, is similar in all models. In contrast, the 5 residues immediately N-terminal (residues 40–44) of Cys-45 display extremely high variability, where the maximal Cα distance between the two most extreme conformations for Phe-43 is 2.43 Å (r.m.s.d. of 2.02 Å for Cαs of residues 40–44) (Fig. 2B).
Biotin Binding Site
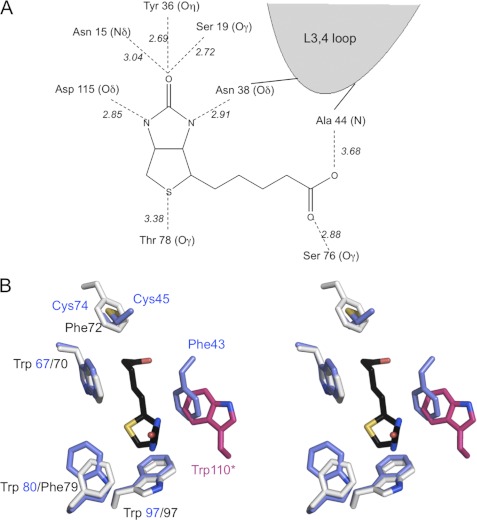
Upon biotin binding, the L3,4 loop adopts a distinct conformation while forming polar and hydrophobic interactions with biotin (Fig. 3). The polar interactions of shwanavidin with biotin consist of a canonical network of H-bond interactions largely identical in most avidins (Fig. 3). The biotin bicyclic ring forms six conserved polar interactions. The biotin ureido ring carbonyl group is stabilized by three H-bond interactions with Asn-15 Nδ, Ser-19 Oγ, and Tyr-36 Oη. The tetrahydrothiophene ring sulfur forms a polar interaction with Thr-78 Oγ. One oxygen of the biotin carboxylate moiety is stabilized by H-bond interactions with Ala-44 N, whereas the other interacts with Ser-76 Oγ. In addition to maintaining the rigidity of the L3,4 and L5,6 loops, the disulfide bridge also contributes to the stabilization of biotin in the binding site, similar to that also observed for rhizavidin. The disulfide bridge is located in a position equivalent to Phe-72 in avidin, where the latter participates in the extended hydrophobic box of the biotin-binding site (Fig. 3). The disulfide bridge is also stabilized by an H-bond interaction of Cys-45 S with Tyr-50 Oη (3.2Å), where the latter residue is conserved in all avidins exhibiting 2 Cys residues in the same positions.
FIGURE 3.
Biotin-binding site. A, schematic representation of the hydrogen-bonding network in the shwanavidin complex with biotin. The biotin ring system forms an identical network of H-bond interactions as in other avidins where five are formed with the uriedo ring and one with the tetrahydrothiophene sulfur. Furthermore, the L3,4 loop contributes a single H-bond interaction with one of the biotin ureido nitrogens. Each of the biotin carboxylate oxygens forms a single H-bond interaction with Ser-76 and main chain nitrogen of Ala-44. B, aromatic residues and the disulfide bridge involved in biotin binding in shwanavidin. Shown is stereo presentation of superimposed residues involved in forming the hydrophobic box accommodating biotin in shwanavidin (light blue) and avidin (light gray). For clarity, the biotin molecule (black) from the shwanavidin complex is shown. The disulfide bridge between the L3,4 and L5,6 loops in shwanavidin accommodates the precise position as the avidin Phe-72 side chain ring. Phe-43 from the L3,4 loop accommodates a highly similar position as Trp-110 of avidin (shown in magenta) contributed by an adjacent monomer. This configuration of aromatic residues in shwanavidin leaves the biotin molecule almost completely unavailable to solvent.
In addition to the polar interactions, there are critical hydrophobic interactions consisting mainly of conserved aromatic residues forming the canonical hydrophobic box. In this regard, there are three Trp residues (Trp-67, Trp-80, and Trp-97) in shwanavidin, which are also conserved in rhizavidin (25) (Fig. 3). Unlike rhizavidin, however, there is a significant addition to the framework of the aromatic box in the binding site. The Phe-43, contributed by the L3,4 loop and unique to shwanavidin, represents an additional feature of the hydrophobic box and seals almost completely the ligand in its binding site (Fig. 4). The interaction of Phe-43 with biotin also contributes to the overall stability of the L3,4 loop. Incredibly, the uniqueness of Phe-43 is far more pronounced because its side chain is situated almost precisely where the Trp residues from L7,8 in tetrameric avidins are positioned (Figs. 3 and 4). Phe-43 thus compensates for the lack of the Trp residue and maintains a more complete configuration of the hydrophobic box compared with that of the tetrameric avidins.
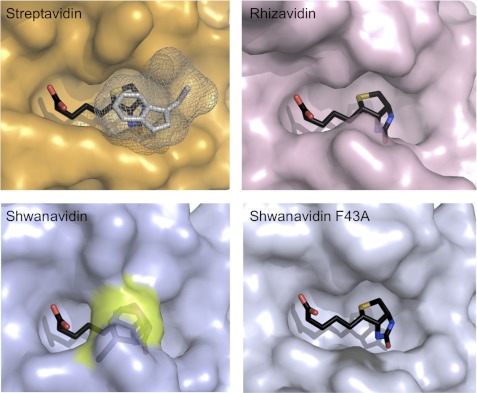
FIGURE 4.
Biotin availability in the binding sites. Surface presentation of the biotin-complexed shwanavidin (light blue), rhizavidin (light pink), and streptavidin (orange) is shown. In streptavidin, the Trp-120 (shown in gray mesh) contributed by a neighboring monomer seals the biotin ligand almost completely. In rhizavidin and shwanavidin an equivalent Trp is lacking due to the dimeric structure. Phe-43 in shwanavidin (shown in the transparent surface and highlighted in yellow) emulates almost exactly the position of the intermonomeric Trp of the tetrameric avidins and caps the biotin molecule in the binding site, rendering the biotin molecule essentially unavailable to solvent. Upon mutating Phe-43 to Ala, the binding site of shwanavidin becomes largely exposed to solvent thus substantiating its low affinity for biotin.
Structure of F43A Mutant
To examine the role of Phe-43 in biotin binding, we mutated this residue into alanine. Biophysical studies of this mutant (see below) indicated that the resultant affinity indeed decreased substantially (Kd ∼ 10−8 m), yet the overall structural features remained remarkably similar to those of the wild-type molecule. However, there are notable differences in the L3,4 loop which affect the biotin-binding site and consequently the affinity. Unlike wild-type shwanavidin, the entire L3,4 loop region of the mutant is well ordered in the apo form. Upon biotin binding, only a small conformational change is observed in L3,4 compared with the wild-type protein. The substitution of Phe-43 by Ala leaves biotin far more available to solvent (Fig. 4). Furthermore, unlike in the wild-type shwanavidin, the distance between one of the biotin carboxylic oxygens to Ala-44 N (4.7 Å) indicates that the H-bond interactions available in the wild-type protein could not be formed (data not shown), thereby decreasing stabilization of the biotin ligand in the binding site (Fig. 4). The remaining polar and hydrophobic interactions between the protein and biotin are identical to those of the wild-type protein.
Affinity and Thermostability Characteristics of Shwanavidin
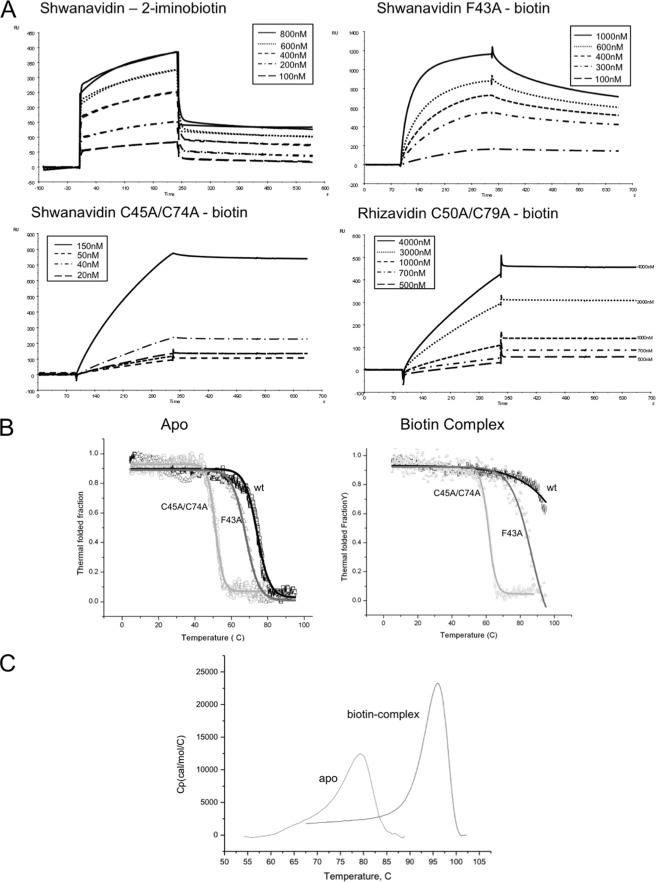
ITC measurements of the interaction between shwanavidin and biotin revealed an extremely high affinity similar to that observed for rhizavidin and streptavidin. The data were fitted to a single set of one site and resulted in a single data point for the Ka slope, indicating that the affinities exceed the capacity of this method (supplemental Fig. 2). Hence, further affinity assays were conducted using SPR as described previously for rhizavidin (25). The binding of rhizavidin to the biotin-coated chip essentially resulted in irreversible binding, even in the presence of 7.2 m urea, and affinity assays were therefore performed using 2-iminobiotin at pH 11.0 where its affinity toward the avidins is maximal. The affinity of shwanavidin toward 2-iminobiotin is of the same order of magnitude (10−7 m) as that reported for rhizavidin (Table 2). Both the F43A and C45A/C74A mutants were unable to bind 2-iminobiotin even at high concentrations. However, the mutants all bound biotin at lower affinities, compared with wild-type shwanavidin, with Kd values of 10−8 and 10−9 m for F43A and C45A/C74A, respectively. Similarly, the disulfide bridge mutant (C50A/C79A) of rhizavidin also exhibited decreased affinity for biotin (Kd = 3.1 × 10−8 m) (Fig. 5A and Table 2).
TABLE 2.
Affinity and thermostability properties of rhizavidin and shwanavidin
FIGURE 5.
Binding and thermostability properties. A, SPR sensograms showing the association and dissociation of wild-type shwanavidin to 2-iminobiotin. Because the shwanavidin F43A mutant and the disulfide bridge mutants of shwanavidin and rhizaividin did not bind 2-iminobiotin, SPR measurements were conducted using biotin. Protein samples were injected over the chip surface for 240 s at a flow rate of 20 μl/min and then washed with elution buffer without protein for 300 s. Shwanavidin sensograms are similar to those reported previously for rhizavidin (25). The disulfide bridge mutants from shwanavidin and rhizavidin show similar sensograms, although in rhizavidin the Kd is 1 order of magnitude higher than that of shwanavidin. RU, response unit. B, CD thermograms of shwanavidin wild type and mutants (F43A and C45A/C74A). Data were normalized to the thermostable fraction, ranging between 1 (natively folded protein) and 0 (unfolded). Sigmoidal plots of the folded state versus temperature in λ values of 233 and 234 nm for the apo- and biotin-complexed proteins, respectively, clearly show the shift in thermostability upon mutating Phe-43 and the disulfide bridges. C, DSC thermograms of shwanavidin as a function of temperature measured in the absence and presence of biotin in 1 m NaCl, 50 mm NaH2PO4, pH 7. The Tm values of 77.2 and 95.2 °C were obtained at the highest point in the thermograms for the apo- and biotin-complexed shwanavidin, respectively, indicating a higher stability upon biotin binding.
To assess further the affinity of the shwanavidin and rhizavidin mutants toward 2-iminobiotin, the two dimeric forms of avidin were incubated with 2-iminobiotin beads at high pH values (11). Whereas wild-type shwanavidin bound to the column and was eluted by reducing the pH, the mutants failed to bind to the resin, indicating very low affinity toward 2-iminobiotin, in agreement with the SPR results. Wild-type shwanavidin could thus be purified using a one-step protocol, whereas the mutants required a different purification strategy.
The thermal stability characteristics of shwanavidin and its mutants were initially evaluated using circular dichroism (CD). The measurements were performed on shwanavidin (wild type and mutants) in their apo- and biotin-complexed forms, using rhizavidin as a control. All samples were measured within the temperature range of 4–95 °C in 10 mm sodium phosphate buffer, pH 4.8. The Tm value for apo-shwanavidin was determined to be 74.2 °C, whereas the biotin complex remained stable to 95 °C, hence its thermostability curve could not be determined (Fig. 5B and Table 2). Thus, biotin has a stabilizing effect on shwanavidin, and further measurements of the apo- and biotin complex were thus performed using DSC. DSC measurements revealed the Tm of the apo- and biotin-complexed shwanavidin to be 77.9 and 95.2 °C, respectively (Fig. 5C and Table 2). These results are in agreement with previously reported values for rhizavidin and other avidins (6, 45), clearly validating shwanavidin as a thermostable protein (Table 2).
The CD measurements for the shwanavidin mutants (F43A and C45A/74A) displayed a dramatic decrease in thermostability properties. For the apo-F43A mutant, a Tm value of 67.9 °C was evaluated, where the biotin-complexed protein exhibited higher stability with a Tm value of 83.8 °C. The double C45A/C74A mutant of shwanavidin had a more dramatic effect on thermal stability with Tm values of 51.1 and 61.3 °C for the apo- and biotin-complexed molecules, respectively (Fig. 5B and Table 2).
DISCUSSION
The search for novel avidins with unique characteristics has led us to the discovery of a bacterial sequence bearing features consistent with a high propensity to bind biotin. Shwanavidin from the marine bacterium, S. denitrificans, is the second known innate homodimeric avidin-like protein (after rhizavidin) which maintains high affinity toward biotin. Rhizavidin and shwanavidin are highly similar in structure and biotin-binding site, yet the latter has an exceptional structural feature, a unique Phe-43 residue located on the L3,4 loop, which emulates the precise position of the crucial intermonomeric Trp in tetrameric avidins (Figs. 3 and 4) (11, 13). The intramonomeric Phe-43 seals the biotin molecule almost completely, thus mimicking the binding-site framework in tetrameric avidins.
Intriguingly, the two crystal forms of apo-shwanavidin provide 18 copies of its monomer, which collectively provide a unique insight into the structural flexibility of residues 40–44 of the L3,4 loop. Mutating Phe-43 into alanine resulted in a striking decrease in affinity toward biotin and eliminated binding to 2-iminobiotin. In addition, the L3,4 loop in the F43A mutant is far less flexible and exhibits a similar conformation in the two subunits. Hence, the presence of solvent-exposed Phe-43 appears to be unfavorable, probably due to its repulsive interactions with solvent molecules leading to multiple conformations. Upon biotin binding, Phe-43 forms hydrophobic interactions with biotin resulting in conformational stability. Removal of Phe-43 in the F43A mutant substantially increases the availability of biotin to solvent, resulting in lower affinity for the ligand (Fig. 4 and Table 2). A similar phenomenon was observed upon mutating the analogous Trp-120 in streptavidin to Ala, whereby the affinity decreased from 10−14 to 10−7 m (46). Mutating the intermonomeric Trp into Lys in avidin and streptavidin resulted in the dissociation into dimers in solution and decreased affinity toward biotin (∼10−8 m) (12).
One of the essential biotin-binding characteristics of both rhizavidin and shwanavidin is the disulfide bridge, which connects loops L3,4 and L5,6. The disulfide bridge of both proteins resides in a similar position, forming almost identical interactions with biotin (25) (Fig. 3). Its role in the high affinity biotin binding was clearly established via mutagenesis of the corresponding Cys residues into Ala. The resulting affinity toward biotin was substantially decreased for both rhizavidin and shwanavidin. Moreover, their binding capacities toward 2-iminobiotin were completely abolished. Hence, removal of the disulfide bridge likely increases the flexibility of the L3,4 and L5,6 loops and the respective degrees of freedom, resulting in a higher loss of entropy and weaker binding of biotin. The apparent increase in flexibility of the unrestrained L3,4 loop renders the bound biotin molecule more available to solvent, thus increasing the off rate of the ligand. In addition, the higher flexibility and entropy could explain the decrease in the corresponding Tm values of the mutant. The high flexibility in the L3,4 and L5,6 loops could also serve as a rationale as to why crystals of the C45A/C74A mutant could not be obtained.
The dimeric rhizavidin and shwanavidin show extremely similar characteristics in their overall structural topology and functional properties. Each originates from a different host bacterium which inhabits a distinctive environment. Whereas rhizavidin from R. etli is found in the soil rhizosphere habitat (27), shwanavidin is found in the deep Gotland basin region of the Baltic Sea, at a depth of 120–130 m (26). Furthermore, both rhizavidin and shwanavidin are unique proteins in their families, where no other species of Shewanella or Rhizobium is known to exhibit another avidin-like sequence in its genome. If ancestral forms of the two bacteria occupied the same environment, acquisition of these avidin-like molecules could reflect horizontal gene transfer event(s). However, because the environments are so distinct and different, other explanations such as convergent or divergent evolution resulting in environmental adaptation could also be possible. In this regard, shwanavidin might have gone through adaptive changes for the low temperature environment. Psychrophilic proteins are reported to have a higher ligand-accessible binding site compared with their mesophilic homologs (48). Flexible loops in the vicinity of the ligand-binding site are one way to increase its availability. Hence, the presence of solvent-exposed Phe-43 in the L3,4 loop results in entropic destabilization of the 40–44 segment of the loop. The observed flexibility in the loop increases the availability of the binding site to the ligand (49) (Fig. 2). After biotin binding, the loop forms stabilizing interactions with biotin, thus adopting a defined conformation. The presence of Phe-43 could thus reflect adaptation to the cold environment of shwanavidin while maintaining its function (48).
Natural evolutionary adaptations of avidin-like proteins have revealed many of the subtle molecular determinants that generated high affinity biotin-binding properties in their dimeric forms. Together with rhizavidin, the high affinity of dimeric shwanavidin provides an excellent platform toward the design of monovalent high affinity biotin-binding systems. Moreover, continued searches of the expanding genomic data bases could also lead to the discovery of a monomeric homolog that maintains high affinity toward biotin. The adaptability of the thermostable shwanavidin into a cold environment also extends its potential utilization in biotechnological applications over a large temperature range.
Accession Codes
The coordinates and the corresponding structure factors of the solved structure are available in the RCSB-PDB data base with the entry codes of 3SZI, 3SZH, 3SZJ, 3T2X, 3T2W, for the shwanavidin apo-form A, form B, biotin complex, apo-F43A, and F43A biotin complex, respectively.
Supplementary Material
Acknowledgments
We thank Prof. Manfred Höfle, Helmholtz Centre for Infection Research, Braunschweig, Germany, for providing the S. denitrificans strain OS217; the staff of ESRF for upgrading and maintaining the first class facility; Shahar Sukenik for assistance with the CD measurements; and Dr. Aharon Rabinkof and Dr. Irina Shin for assistance in conducting the SPR, ITC, and DSC measurements.

This article contains supplemental Table 1, Figs. 1 and 2, Experimental Procedures, and additional references.
The atomic coordinates and structure factors (codes 3SZI, 3SZH, 3SZJ, 3T2X, and 3T2W) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
A. Meir and O. Livnah, unpublished data.
- DSC
- differential scanning calorimetry
- ITC
- isothermal titration calorimetry
- NHS
- N-hydroxysuccinimide
- r.m.s.d.
- root mean square deviation
- SPR
- surface plasmon resonance.
REFERENCES
- 1. Green N. M. (1975) Avidin. Adv. Protein Chem. 29, 85–133 [DOI] [PubMed] [Google Scholar]
- 2. Wilchek M., Bayer E. A. (1989) Avidin-biotin technology ten years on: has it lived up to its expectations? Trends Biochem. Sci. 14, 408–412 [DOI] [PubMed] [Google Scholar]
- 3. Bayer E. A., Wilchek M. (1990) Application of avidin-biotin technology to affinity-based separations. J. Chromatogr. 510, 3–11 [DOI] [PubMed] [Google Scholar]
- 4. Bayer E. A., Kulik T., Adar R., Wilchek M. (1995) Close similarity among streptavidin-like, biotin-binding proteins from Streptomyces. Biochim. Biophys. Acta 1263, 60–66 [DOI] [PubMed] [Google Scholar]
- 5. Helppolainen S. H., Määttä J. A., Halling K. K., Slotte J. P., Hytönen V. P., Jänis J., Vainiotalo P., Kulomaa M. S., Nordlund H. R. (2008) Bradavidin II from Bradyrhizobium japonicum: a new avidin-like biotin-binding protein. Biochim. Biophys. Acta 1784, 1002–1010 [DOI] [PubMed] [Google Scholar]
- 6. Helppolainen S. H., Nurminen K. P., Määttä J. A., Halling K. K., Slotte J. P., Huhtala T., Liimatainen T., Ylä-Herttuala S., Airenne K. J., Närvänen A., Jänis J., Vainiotalo P., Valjakka J., Kulomaa M. S., Nordlund H. R. (2007) Rhizavidin from Rhizobium etli: the first natural dimer in the avidin protein family. Biochem. J. 405, 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nordlund H. R., Hytönen V. P., Laitinen O. H., Kulomaa M. S. (2005) Novel avidin-like protein from a root nodule symbiotic bacterium, Bradyrhizobium japonicum. J. Biol. Chem. 280, 13250–13255 [DOI] [PubMed] [Google Scholar]
- 8. Sardo A., Wohlschlager T., Lo C., Zoller H., Ward T. R., Creus M. (2011) Burkavidin: a novel secreted biotin-binding protein from the human pathogen Burkholderia pseudomallei. Protein Expr. Purif. 77, 131–139 [DOI] [PubMed] [Google Scholar]
- 9. Takakura Y., Tsunashima M., Suzuki J., Usami S., Kakuta Y., Okino N., Ito M., Yamamoto T. (2009) Tamavidins: novel avidin-like biotin-binding proteins from the tamogitake mushroom. FEBS J. 276, 1383–1397 [DOI] [PubMed] [Google Scholar]
- 10. Kurzban G. P., Bayer E. A., Wilchek M., Horowitz P. M. (1991) The quaternary structure of streptavidin in urea. J. Biol. Chem. 266, 14470–14477 [PubMed] [Google Scholar]
- 11. Livnah O., Bayer E. A., Wilchek M., Sussman J. L. (1993) Three-dimensional structures of avidin and the avidin-biotin complex. Proc. Natl. Acad. Sci. U.S.A. 90, 5076–5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Laitinen O. H., Airenne K. J., Marttila A. T., Kulik T., Porkka E., Bayer E. A., Wilchek M., Kulomaa M. S. (1999) Mutation of a critical tryptophan to lysine in avidin or streptavidin may explain why sea urchin fibropellin adopts an avidin-like domain. FEBS Lett. 461, 52–58 [DOI] [PubMed] [Google Scholar]
- 13. Freitag S., Le Trong I., Chilkoti A., Klumb L. A., Stayton P. S., Stenkamp R. E. (1998) Structural studies of binding site tryptophan mutants in the high affinity streptavidin-biotin complex. J. Mol. Biol. 279, 211–221 [DOI] [PubMed] [Google Scholar]
- 14. Sano T., Vajda S., Smith C. L., Cantor C. R. (1997) Engineering subunit association of multisubunit proteins: a dimeric streptavidin. Proc. Natl. Acad. Sci. U.S.A. 94, 6153–6158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pazy Y., Eisenberg-Domovich Y., Laitinen O. H., Kulomaa M. S., Bayer E. A., Wilchek M., Livnah O. (2003) Dimer-tetramer transition between solution and crystalline states of streptavidin and avidin mutants. J. Bacteriol. 185, 4050–4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weber P. C., Ohlendorf D. H., Wendoloski J. J., Salemme F. R. (1989) Structural origins of high affinity biotin binding to streptavidin. Science 243, 85–88 [DOI] [PubMed] [Google Scholar]
- 17. Eisenberg-Domovich Y., Hytönen V. P., Wilchek M., Bayer E. A., Kulomaa M. S., Livnah O. (2005) High-resolution crystal structure of an avidin-related protein: insight into high affinity biotin binding and protein stability. Acta Crystallogr. D Biol. Crystallogr. 61, 528–538 [DOI] [PubMed] [Google Scholar]
- 18. Lim K. H., Huang H., Pralle A., Park S. (2011) Engineered streptavidin monomer and dimer with improved stability and function. Biochemistry 50, 8682–8691 [DOI] [PubMed] [Google Scholar]
- 19. Freitag S., Le Trong I., Klumb L., Stayton P. S., Stenkamp R. E. (1997) Structural studies of the streptavidin binding loop. Protein Sci. 6, 1157–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nordlund H. R., Hytönen V. P., Hörhä J., Määttä J. A., White D. J., Halling K., Porkka E. J., Slotte J. P., Laitinen O. H., Kulomaa M. S. (2005) Tetravalent single-chain avidin: from subunits to protein domains via circularly permuted avidins. Biochem. J. 392, 485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nordlund H. R., Laitinen O. H., Hytönen V. P., Uotila S. T., Porkka E., Kulomaa M. S. (2004) Construction of a dual chain pseudotetrameric chicken avidin by combining two circularly permuted avidins. J. Biol. Chem. 279, 36715–36719 [DOI] [PubMed] [Google Scholar]
- 22. Chivers C. E., Crozat E., Chu C., Moy V. T., Sherratt D. J., Howarth M. (2010) A streptavidin variant with slower biotin dissociation and increased mechanostability. Nat. Methods 7, 391–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Howarth M., Chinnapen D. J., Gerrow K., Dorrestein P. C., Grandy M. R., Kelleher N. L., El-Husseini A., Ting A. Y. (2006) A monovalent streptavidin with a single femtomolar biotin binding site. Nat. Methods 3, 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sano T., Smith C. L., Cantor C. R. (1997) Expression and purification of recombinant streptavidin-containing chimeric proteins. Methods Mol. Biol. 63, 119–128 [DOI] [PubMed] [Google Scholar]
- 25. Meir A., Helppolainen S. H., Podoly E., Nordlund H. R., Hytönen V. P., Määttä J. A., Wilchek M., Bayer E. A., Kulomaa M. S., Livnah O. (2009) Crystal structure of rhizavidin: insights into the enigmatic high-affinity interaction of an innate biotin-binding protein dimer. J. Mol. Biol. 386, 379–390 [DOI] [PubMed] [Google Scholar]
- 26. Brettar I., Christen R., Höfle M. G. (2002) Shewanella denitrificans sp. nov., a vigorously denitrifying bacterium isolated from the oxic-anoxic interface of the Gotland Deep in the central Baltic Sea. Int. J. Syst. Evol. Microbiol. 52, 2211–2217 [DOI] [PubMed] [Google Scholar]
- 27. González V., Santamaría R. I., Bustos P., Hernández-González I., Medrano-Soto A., Moreno-Hagelsieb G., Janga S. C., Ramírez M. A., Jiménez-Jacinto V., Collado-Vides J., Dávila G. (2006) The partitioned Rhizobium etli genome: genetic and metabolic redundancy in seven interacting replicons. Proc. Natl. Acad. Sci. U.S.A. 103, 3834–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brettar I., Christen R., Höfle M. G. (2012) Analysis of bacterial core communities in the central Baltic by comparative RNA-DNA-based fingerprinting provides links to structure-function relationships. Isme J. 6, 195–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A. (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T. L. (2009) BLAST+: architecture and applications. BMC Bioinformatics 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jain E., Bairoch A., Duvaud S., Phan I., Redaschi N., Suzek B. E., Martin M. J., McGarvey P., Gasteiger E. (2009) Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics 10, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T. L. (2008) NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sheffield P., Garrard S., Derewenda Z. (1999) Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 15, 34–39 [DOI] [PubMed] [Google Scholar]
- 34. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zwart P. H., Afonine P. V., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., McKee E., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Storoni L. C., Terwilliger T. C., Adams P. D. (2008) Automated structure solution with the PHENIX suite. Methods Mol. Biol. 426, 419–435 [DOI] [PubMed] [Google Scholar]
- 36. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 37. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 38. Kiefer F., Arnold K., Künzli M., Bordoli L., Schwede T. (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37, D387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vagin A. A., Steiner R. A., Lebedev A. A., Potterton L., McNicholas S., Long F., Murshudov G. N. (2004) REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr 60, 2184–2195 [DOI] [PubMed] [Google Scholar]
- 40. Morris R. J., Perrakis A., Lamzin V. S. (2003) ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 374, 229–244 [DOI] [PubMed] [Google Scholar]
- 41. Emsley P., Cowtan K. (2004) COOT: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 42. Livnah O., Bayer E. A., Wilchek M., Sussman J. L. (1993) The structure of the complex between avidin and the dye, 2-(4′-hydroxyazobenzene) benzoic acid (HABA). FEBS Lett. 328, 165–168 [DOI] [PubMed] [Google Scholar]
- 43. Chivers C. E., Koner A. L., Lowe E. D., Howarth M. (2011) How the biotin-streptavidin interaction was made even stronger: investigation via crystallography and a chimaeric tetramer. Biochem. J. 435, 55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Teulon J. M., Delcuze Y., Odorico M., Chen S. W., Parot P., Pellequer J. L. (2011) Single and multiple bonds in (strept)avidin-biotin interactions. J. Mol. Recognit. 24, 490–502 [DOI] [PubMed] [Google Scholar]
- 45. González M., Argaraña C. E., Fidelio G. D. (1999) Extremely high thermal stability of streptavidin and avidin upon biotin binding. Biomol. Eng. 16, 67–72 [DOI] [PubMed] [Google Scholar]
- 46. Chilkoti A., Tan P. H., Stayton P. S. (1995) Site-directed mutagenesis studies of the high affinity streptavidin-biotin complex: contributions of tryptophan residues 79, 108, and 120. Proc. Natl. Acad. Sci. U.S.A. 92, 1754–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 48. Feller G., Gerday C. (2003) Psychrophilic enzymes: hot topics in cold adaptation. Nat. Rev. Microbiol. 1, 200–208 [DOI] [PubMed] [Google Scholar]
- 49. Casanueva A., Tuffin M., Cary C., Cowan D. A. (2010) Molecular adaptations to psychrophily: the impact of “omic” technologies. Trends Microbiol. 18, 374–381 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.