Background: Plants produce glycoside hydrolase inhibitor protein to protect cell walls.
Results: The inhibition mechanism for a xyloglucanase-inhibitor of a fungal xyloglucanase is revealed by crystal structures.
Conclusion: Xyloglucanase inhibitor protein distinguishes specific structural features of a glycosyl hydrolase to protect the plant cell wall from degradation.
Significance: Understanding the mechanism of xyloglucanase inhibition is key to comprehending how plants defend themselves against microbes that express glycosyl hydrolases.
Keywords: Cell Wall, Crystal Structure, Glycoside Hydrolases, Plant Defense, Protein Complexes, Inhibitor Protein
Abstract
Microorganisms such as plant pathogens secrete glycoside hydrolases (GHs) to digest the polysaccharide chains of plant cell walls. The degradation of cell walls by these enzymes is a crucial step for nutrition and invasion. To protect the cell wall from these enzymes, plants secrete glycoside hydrolase inhibitor proteins (GHIPs). Xyloglucan-specific endo-β-1,4-glucanase (XEG), a member of GH family 12 (GH12), could be a great threat to many plants because xyloglucan is a major component of the cell wall in most plants. Understanding the inhibition mechanism of XEG by GHIP is therefore of great importance in the field of plant defense, but to date the mechanism and specificity of GHIPs remain unclear. We have determined the crystal structure of XEG in complex with extracellular dermal glycoprotein (EDGP), a carrot GHIP that inhibits XEG. The structure reveals that the conserved arginines of EDGP intrude into the active site of XEG and interact with the catalytic glutamates of the enzyme. We have also determined the crystal structure of the XEG-xyloglucan complex. These structures show that EDGP closely mimics the XEG-xyloglucan interaction. Although EDGP shares structural similarity to a wheat GHIP (Triticum aestivum xylanase inhibitor-IA (TAXI-IA)) that inhibits GH11 family xylanases, the arrangement of GH and GHIP in the XEG-EDGP complex is distinct from that in the xylanase-TAXI-IA complex. Our findings imply that plants have evolved structures of GHIPs to inhibit different GH family members that attack their cell walls.
Introduction
Plant cell walls are composed of various polysaccharides such as cellulose, hemicellulose, and pectin. In most plant cell wall models, cellulose microfibrils are linked via hemicellulose. This cellulose-hemicellulose network provides tensile strength and acts as a physical barrier against microorganisms such as invading pathogens. To penetrate and utilize plant cell walls nutritionally, microorganisms secrete hydrolases for cell wall degradation. These enzymes, which include endoglucanases, xylanases, and polygalacturonases, are classified into glycoside hydrolase (GH)2 families in the CAZY data base (1). In response to pathogenic attack, plants produce glycoside hydrolase inhibitor proteins (GHIPs) against the cell wall-degrading enzymes (2, 3).
Extracellular dermal glycoprotein (EDGP) from carrot is one such GHIP. EDGP shows inhibitory activity toward the xyloglucan-specific endo-β-1,4-glucanase (XEG) from the fungus Aspergillus aculeatus (4). EDGP is alternatively called XEG inhibitor protein (XEGIP). XEG belongs to GH family 12 (GH12) and specifically cleaves xyloglucan, which consists of a β-linked glucose backbone substituted with xylose side chains (5). Xyloglucan is a major hemicellulose in most plants (6), and thus xyloglucanases such as XEG are a great threat to plants because the degradation of hemicellulose causes great damage. The inhibition of XEG by EDGP is an important component of the plant defense system. Proteins homologous to EDGP have been identified in various plants, and several of these proteins have been characterized. Tomato XEGIP inhibits XEG by forming an associated 1:1 complex (7). Tobacco Necturin IV (NEC IV) also inhibits XEG (8). In contrast, the homologous protein from wheat, TAXI-IA (Triticum aestivum xylanase inhibitor-IA), inhibits a GH family 11 (GH11) xylanase from fungus, ANXI (Aspergillus niger xylanase I) (9–11). Interestingly, the homologous protein from soybean, basic 7S globulin (Bg7S), lacks inhibitory activity for either GH11 or GH12 enzymes (12). The crystal structures of the ANXI-TAXI-IA complex and Bg7S have been determined (10, 12). However, not only the inhibition mechanism of XEG but also the mechanism underlying family-specific inhibition by GHIP have remained unclear.
In this work, we have determined the crystal structures of XEG, the XEG-xyloglucan complex, EDGP, and the XEG-EDGP complex. The structure of the XEG-xyloglucan complex provides a structural basis of specific recognition of xyloglucan by XEG. The structure of the XEG-EDGP complex reveals how GHIP recognizes the active site of GH12 and inhibits its activity. Surprisingly, the arrangement of GH and GHIP in the XEG-EDGP complex is distinct from that in the ANXI-TAXI-IA complex. Our findings clarify the mechanism of family-specific inhibition of GH12 and GH11 by EDGP homologous GHIPs.
EXPERIMENTAL PROCEDURES
Preparation of EDGP and XEG
The preparation of EDGP and XEG has been described previously (12, 13). In brief, EDGP was purified from carrot callus culture medium. The carrot callus was grown for 2–3 weeks at 298 K in Murashige-Skoog medium containing 1 mg/liter 2,4-dichlorophenoxyacetic acid. The protein was purified using HiTrap SP (GE Healthcare). The cDNA encoding XEG was obtained by PCR-based gene synthesis (14) and inserted into pGEX6P-I vector (GE Healthcare) at the BamHI-XhoI site. N-terminal GST-fused XEG was expressed in Escherichia coli BL21. The protein was purified using glutathione-Sepharose 4B resin (GE Healthcare), a HiTrap Q HP column (GE Healthcare), and a HiLoad Superdex 75 26/60 column (GE Healthcare).
Enzyme Activity Assay
The activity of XEG wild-type or mutants was measured using p-hydroxybenzoic acid hydrazide (PAHBAH) method (15). The reaction mixture contained 50 mm sodium acetate, pH 4.6, 100 mm NaCl, 5 mg/ml xyloglucan from tamarind seeds (DS Pharma), and 100 ng of XEG in the absence or presence of 5 μg of EDGP. The final reaction volume was 20 μl. The reaction mixtures were incubated at room temperature for 15 min, and then the amount of reducing sugar was measured.
Crystallographic Analyses
All crystals were obtained by the hanging-drop vapor diffusion method at 293 K. Hexagonal crystals of XEG were obtained under 0.1 m sodium acetate, pH 5.5, and 1.5 m ammonium sulfate. The co-crystal of XEG-xyloglucan was obtained under 0.1 m sodium acetate, pH 4.6, 25% PEG 3000, and 5 mg/ml digested xyloglucan. XEG used in crystallization with xyloglucan was wild type. The preparation of digested xyloglucan has been described previously (16). Hexagonal crystals of EDGP were obtained under 0.2 m ammonium acetate, 0.1 m sodium acetate, pH 4.6, and 30% PEGMME2000. Iodine-derived crystals of EDGP were prepared by the vaporizing iodine labeling method (17). A droplet of iodine solution (0.67 m KI and 0.47 m I2) was placed next to the crystallization droplet containing EDGP crystals. After 20 min, the iodine solution was removed and the crystallization droplet was incubated for a further 20 h.
To crystallize the XEG-EDGP complex, XEG protein with an N-terminal truncation, XEG(8–224), was prepared. The XEG-EDGP complex was prepared by mixing EDGP and XEG(8–224) in an equal molar ratio. Monoclinic crystals of the XEG-EDGP complex were obtained under 0.24 m Morpheus alcohols (Molecular Dimensions), 0.1 m Morpheus Buffer system 3, pH 8.5 (Molecular Dimensions), 30% Morpheus EOD_P8K (Molecular Dimensions), and 9% dextran sulfate.
X-ray diffraction data for crystals of XEG, XEG-xyloglucan, EDGP native, EDGP derivative, and XEG-EDGP were collected in-house at UltraX18 (Rigaku), Photon Factory (PF) BL-17A, SPring-8 BL-41XU, PF NE-3A, and SPring-8 BL-32XU, respectively. All diffraction data were processed using the program HKL2000 (18). The XEG structure was solved by the molecular replacement method with the program MOLREP (19) using the endo-β-1,4-glucanase from Hypocrea jecorina as a search model (Protein Data Bank (PDB) ID code 1OA2). The EDGP structure was solved by the SIRAS method using the programs SOLVE and RESOLVE (20). The EDGP-XEG complex structure was solved by the molecular replacement method with the program MOLREP (19) using EDGP and XEG structures as search models. Model building was performed with the program COOT (21). Structure refinement was performed with the programs CNS (22) and REFMAC (23). The geometries of the final structures were validated with the program PROCHECK (24). Data collection and refinement statistics are given in Table 1. Final coordinates and structure factors have been deposited in the Protein Data Bank Japan (PDBj).
TABLE 1.
Data collection and refinement statistics
| Parameters | XEG | XEG-xyloglucan | EDGP | EDGP deriv | XEG-EDGP |
|---|---|---|---|---|---|
| Data collection | |||||
| Wavelength (Å) | 1.5418 | 1.0000 | 0.8000 | 1.900 | 1.0000 |
| Space group | P65 | P212121 | P62 | P62 | C2 |
| a (Å) | 93.0 | 62.6 | 130.1 | 130.1 | 249.0 |
| b (Å) | 93.0 | 79.3 | 130.1 | 130.1 | 51.7 |
| c (Å) | 62.0 | 80.4 | 44.5 | 44.5 | 143.2 |
| α (°) | 90 | 90 | 90 | 90 | 90 |
| β (°) | 90 | 90 | 90 | 90 | 122.2 |
| γ (°) | 120 | 90 | 120 | 120 | 90 |
| Resolution (Å) | 50.0–1.90 | 50.0–1.20 | 50–0.92 | 50.0–2.40 | 50.0–2.70 |
| Observed reflections | 134,050 | 743,148 | 1,283,262 | 343,185 | 121,934 |
| Unique reflections | 24,103 | 120,607 | 272,047 | 16,855 | 40,308 |
| R-merge (%) | 8.3 (31.6) | 6.9 (31.7) | 7.5 (32.6) | 6.4 (21.8) | 7.8 (38.4) |
| Completeness (%) | 99.6 (98.8) | 95.8 (75.2) | 91.6 (74.3) | 98.2 (95.2) | 91.9 (83.2) |
| <I>/σ<I> | 14.8 (7.0) | 12.8 (3.3) | 12.2 (2.0) | 19.2 (11.3) | 12.4 (3.3) |
| Refinement | |||||
| Resolution (Å) | 1.90 | 1.20 | 0.95 | 2.70 | |
| Refined reflections | 22,833 | 114,328 | 239,679 | 37,707 | |
| Free reflections | 1,142 | 5,716 | 11,983 | 1,885 | |
| R (%) | 18.0 | 12.3 | 12.6 | 25.2 | |
| R-free (%) | 21.2 | 16.2 | 14.5 | 33.9 | |
| Root mean square deviation | |||||
| Bond length (Å) | 0.005 | 0.015 | 0.016 | 0.015 | |
| Bond angles (°) | 0.951 | 1.637 | 1.782 | 1.823 | |
| Ramachandran plot | |||||
| Most favored (%) | 93.7 | 92.7 | 89.3 | 82.3 | |
| Additional allowed (%) | 5.8 | 7.3 | 10.1 | 16.4 | |
| Generously allowed (%) | 0.5 | 0.0 | 0.3 | 0.8 | |
| Disallowed (%) | 0.0 | 0.0 | 0.3 | 0.5 | |
| Protein Data Bank ID code | 3VL8 | 3VL9 | 3VLA | 3VLB | |
Pulldown Assay
GST-fused XEG and its mutants were overexpressed in E. coli BL21 and purified by glutathione-Sepharose 4B resin. Next, EDGP and GST-fused XEG were incubated with glutathione-Sepharose 4B beads equilibrated with a buffer containing 50 mm sodium acetate, pH 4.6, and 100 mm NaCl for 1 h at 25 °C. The beads were washed with the above buffer and eluted with a buffer containing 100 mm Tris-HCl, pH 9.0, 200 mm NaCl, and 50 mm reduced glutathione. The GST tags were cleaved by HRV3C protease. The proteins in solution were analyzed by SDS-PAGE with Coomassie Brilliant Blue staining. Band intensities were calculated by the program ImageJ (National Institutes of Health).
Figure Preparation
Protein structures were prepared with the program PyMOL (DeLano Scientific). All of the figures were modified with the programs PHOTOSHOP and ILLUSTRATOR (Adobe Systems).
RESULTS
Structure of XEG-Xyloglucan Complex
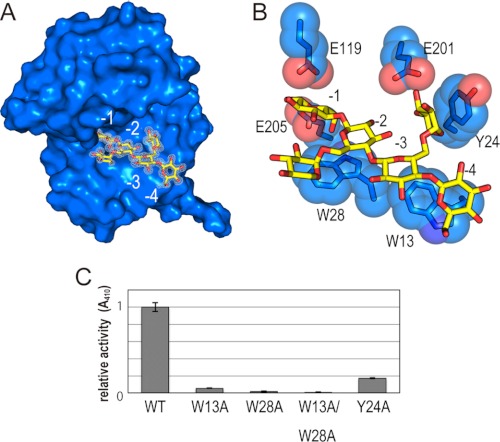
We determined the crystal structures of XEG and XEG in complex with xyloglucan at 1.9 and 1.2 Å resolution, respectively. The structure of XEG bound to xyloglucan could be superimposed on that of XEG, with a root mean square deviation value of 0.6 Å for comparable Cα atoms. XEG adopts a β-jelly roll fold, as observed in other enzymes of the family GH12 (25). A cleft runs across the surface of the protein, and the xyloglucan-binding subsites are located within this cleft. The hydrolysis reaction of the GH12 enzymes proceeds with a two-step retaining mechanism catalyzed by two glutamate residues: one acts as the nucleophile and the other as the acid/base (25). The putative nucleophile and acid/base in XEG are Glu119 and Glu205, respectively. The electron density map clearly shows binding of xyloglucan within the cleft, where the β-1,4-glucose backbone is bound to the −1 to −4 subsites, and α-1,6-xylose residues branching from the glucose moieties in subsites −2 and −3 are also observed in the electron density (Fig. 1A). Trp13 and Trp28 form hydrophobic interactions with the glucose moieties of the β-1,4-glucan chain in the −4 and −2 subsites, respectively (Fig. 1B). In addition to these hydrophobic interactions, the catalytic Glu119 and Glu205 residues form hydrogen bonds with oxygen atoms of the glucose moiety in the −1 subsite.
FIGURE 1.
Recognition of xyloglucan by XEG. A, structure of XEG-xyloglcan complex. XEG and xyloglucan are represented by a surface and stick models, respectively. The observed electron density of xyloglucan is shown by white mesh (2 Fo − Fc, 1σ). B, detailed interactions between XEG and xyloglucan are shown by stick and sphere models. Carbon of XEG, carbon of xyloglucan, oxygen, and nitrogen are colored light blue, yellow, red, and blue, respectively. C, activities of the XEG mutants were estimated relative to that of wild-type (WT).
The XEG structure shows the structural basis for specific recognition of xyloglucan by the enzyme. Tyr24 stacks against the xylose side chain linked to the glucose moieties in the −3 subsite, and Glu201 interacts with the O5 oxygen atoms of xylose (Fig. 1B). To investigate which residues are responsible for XEG activity, we prepared XEG mutants and measured their xyloglucanase activity (Fig. 1C). An XEG mutant with a W13A, W28A, or W13A/W28A substitution(s) lacked almost all xyloglucanase activity, clearly indicating that these tryptophans, which are involved in the hydrophobic interaction with the glucose backbone, are essential for this activity. Remarkably, a Y24A substitution also markedly decreased the activity of XEG. Tyr24 of XEG is not conserved in the GH12 enzyme from A. aculeatus, FI-CMCase (26). FI-CMCase hydrolyzes carboxylmethylated β-1,4-glucan, a model substrate of cellulase, and cleaves xyloglucan with approximately half of the activity of XEG. These results suggest that a stacking interaction of a hydrophobic nature between Tyr24 and the xylose side chain is crucial for xyloglucan recognition. A previous study has revealed the crystal structure of a GH12 xyloglucanase from Bacillus licheniformis (BlXG12) in complex with xyloglucan (27). In this structure, tryptophan residues are involved in interactions with glucose backbone just as we observe with XEG. However, stacking interactions between an aromatic residue and a xylose residue were not observed. This is consistent with the fact that the BlXG12 also possesses glucanase activity to digest carboxymethyl cellulose (CMC) (27).
Structure of EDGP
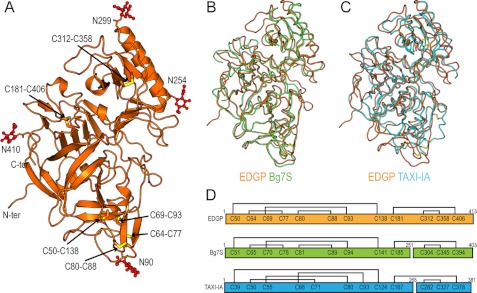
The crystal structure of carrot EDGP was determined at 0.95 Å resolution (Fig. 2A). In the structure, the N-terminal glutamine is converted to a pyroglutamic acid (28). EDGP adopts a pepsin-like fold that is β-rich with several α-helices and is roughly divided by a center cleft comprising the active site (29). Despite the structural similarity to pepsin, one of the catalytic aspartates in pepsin is replaced by Ser271 in EDGP, and thus EDGP lacks protease activity. Consistent with this, other GHIPs also lack the catalytic aspartate. EDGP has six disulfide bonds, and these supposedly stabilize the tertiary structure of EDGP in the extracellular environment (Fig. 2, A and D). EDGP has four putative N-linked glycosylation sites: Asn90, Asn254, Asn299, and Asn410 (28). In each putative N-linked glycosylation site, the electron density map indicated at least one sugar moiety linked to the asparagine (Fig. 2A).
FIGURE 2.
Crystal structure of EDGP. A, overall structure of EDGP is represented by a ribbon model. The disulfide bonds are shown by stick models, in which sulfur atoms are colored yellow. N-Linked glycans are represented by ball and stick models and colored red. B, superimposed structures of EDGP (orange) and Bg7S (light green) are represented as wire models. C, superimposed structures of EDGP (orange) and TAXI-IA (cyan) are represented as wire models. D, schematic drawing shows disulfide bonds in EDGP, Bg7S, and TAXI-IA.
The crystal structures of homologous proteins from wheat (TAXI-IA) and soybean (Bg7S) have been reported previously (10, 12). The overall structure of EDGP is similar to those of Bg7S and TAXI-IA. The root mean square deviation value for Cα atoms comparable with Bg7S (PDB ID code 3AUP) and TAXI-IA (PDB ID code 1T6E) is 1.6 and 2.4 Å, respectively (Fig. 2, B and C). EDGP, Bg7S, and TAXI-IA all have six disulfide bonds. Although the patterns of disulfide formation are conserved in EDGP and Bg7S, they differ from that in TAXI-IA (Fig. 2D). Bg7S and TAXI-IA undergo post-translational cleavage in their internal regions, whereas EDGP does not (Fig. 2D). The variation in post-translational modification of GHIPs is of interest, although the functional associations of these modifications in GHIPs remains unclear.
Structure of XEG-EDGP Complex, and Its Interaction
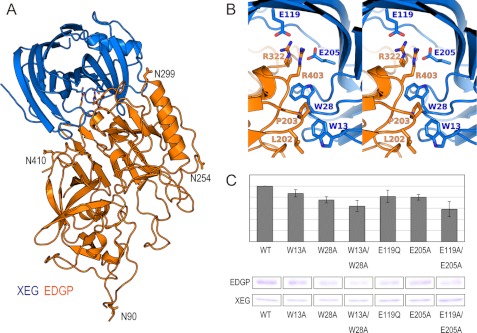
The crystal structure of XEG in complex with EDGP was determined at 2.7 Å resolution (Fig. 3A). The XEG-EDGP complex structure shows that EDGP completely covers the active cleft of XEG. The averaged root mean square deviations of comparable Cα atoms between each native structure (XEG and EDGP) and the XEG-EDGP complex are 0.8 and 0.7 Å, respectively, suggesting no substantial conformational changes occur in EDGP upon the binding of XEG. The calculated buried solvent-accessible surface area is 2046 Å2 in the complex, comparable with that of the ANXI-TAXI-IA complex (1998 Å2). We did not observe sugar chains at all of the N-linked glycosylation sites of EDGP because of ambiguity in the electron density map. In the XEG-EDGP structure, Arg322 and Arg403 of EDGP insert into the active cleft of XEG and form an electrostatic interaction with the catalytic residues, Glu119 and Glu205 (Fig. 3B). Hydrophobic interactions are made between the aliphatic moiety of Arg403 of EDGP and Trp28 of XEG, Leu202 and Pro203 of EDGP and Trp13 of XEG (Fig. 3B). To identify the amino acid residues of XEG responsible for the interaction with EDGP, we performed a pulldown assay using XEG mutants and EDGP (Fig. 3C). We did not observe a marked reduction in the interaction with EDGP with single or double mutations. However, mutations in residues involved in the interaction with EDGP weakened the binding ability, suggesting that both electrostatic and hydrophobic interactions are important in the interaction between XEG and EDGP (Fig. 3, B and C).
FIGURE 3.
Crystal structure of the XEG-EDGP inhibition complex and interactions between EDGP and XEG. A, overall structure of inhibition complex of EDGP (orange) and XEG (light blue) shown by a ribbon model. B, stereo view of the detailed interactions between EDGP (orange) and XEG (light blue). C, binding activity of XEG mutants with EDGP (upper panel) estimated by band intensity of pulldown assay (bottom panel).
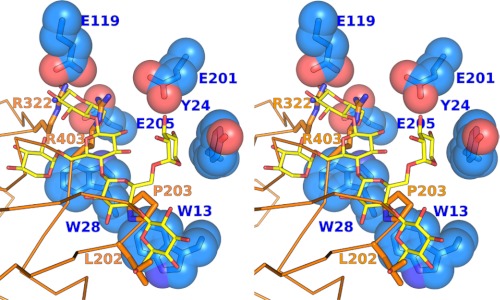
Structural overlay of the XEG-EDGP inhibition complex and the XEG-xyloglucan complex revealed the strategy employed by EDGP for inhibition (Fig. 4). Namely, the guanidium moieties of Arg322 and Arg403 of EDGP in the inhibition complex are located in the −1 subsite of XEG, and the aliphatic moiety of the side chain of Arg403 is located in the −2 subsite of XEG. Furthermore, Leu202 and Pro203 of EDGP, which are conserved in most plants, are located in the −4 subsite of XEG. These findings indicate that EDGP mimics the interaction between XEG and xyloglucan observed in the structure of the XEG-xyloglucan complex. Interestingly, EDGP does not interact with Tyr24 of XEG, which is involved in xylose recognition (Fig. 1B). The lack of any interaction with Tyr24 of XEG might be explained by our unpublished data that EDGP also inhibits the glucanase activity of FI-CMCase, which lacks this tyrosine residue. The EDGP complex with FI-CMCase has been crystallized (13) and solved.3
FIGURE 4.
Comparison between XEG-EDGP and XEG-xyloglucan interactions. Stereo view shows XEG-EDGP interactions, with xyloglucan superimposed from the structure of XEG-xyloglucan. XEG (light blue), EDGP (orange), and xyloglucan (yellow) are shown by stick and sphere, stick and wire, and stick models, respectively.
DISCUSSION
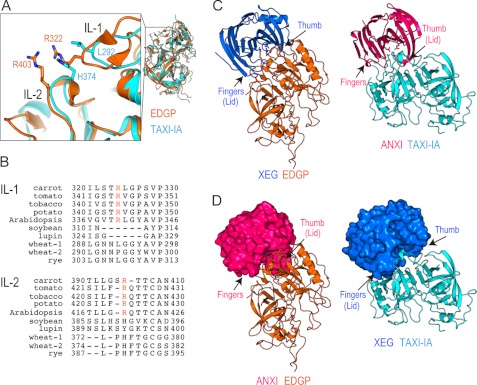
The crystal structure of the XEG-xyloglucan complex provides a structural basis for understanding the specific recognition of xyloglucan by XEG. The structure of the XEG-EDGP inhibition complex reveals details of the inhibition mechanism, in which two arginines located in inhibition loops 1 and 2 (IL-1 and IL-2) of EDGP intrude into the catalytic cleft of XEG and mimic the interactions formed between XEG and xyloglucan (Figs. 4 and 5A). The two arginines in IL-1 and IL-2 are conserved in most GHIPs, including homologous proteins from tomato, tobacco, potato, and Arabidopsis (Fig. 5B). Of these, tomato and tobacco GHIPs inhibit GH12 enzymes (7, 8). Furthermore, Leu202 and Pro203 (which contact the −4 subsite of the enzyme active site) are also conserved in GHIPs of most plants. Although the inhibitory activity of GHIP from potato or Arabidopsis has not been reported, our results suggest that these GHIPs might inhibit GH12 enzymes by means of their conserved arginines, leucine, and proline. In the GH11-TAXI-IA complex, Leu292 in IL-1 and His374 in IL-2 are involved in target-binding interactions. These residues are conserved in GHIPs of some monocots of the order Poales, including grasses, whose hemicellulose is xylan. Thus, rye GHIP might inhibit GH11 enzymes instead of GH12.
FIGURE 5.
Diverse mechanisms of target recognition of GH12 and GH11 by GHIP. A, close-up view (left) around IL-1 and IL-2 of the superimposed structures (right) of EDGP (orange) and TAXI-IA (cyan). B, sequence alignment of IL-1 and IL-2 of EDGP with homologous proteins from various plants, tomato (UniProt ID; Q8GT67), tobacco (Q3KU27), potato (Q7XJE7), Arabidopsis (Q8LF70), soybean (P13917), lupin (Q42369), wheat-1 (Q8H0K8), wheat-2 (Q53IQ4), and rye (Q6KE41). Conserved arginines are colored orange. C, comparison of structures between XEG-EDGP (left panel) and ANXI-TAXI-IA (right panel). Complexes are represented by ribbon models. D, putative model structure of an ANXI-EDGP (left panel) or XEG-TAXI-IA (right panel) complex.
Structural overlays of EDGP and TAXI-IA in the GH-GHIP complexes reveal very similar positioning of IL-1 and IL-2 in these complexes (Fig. 5A), indicating that EDGP and TAXI-IA use similar regions to interact with their corresponding GHs. Although the overall structures of XEG and EDGP are comparable with those of ANXI and TAXI-IA (Fig. 2C), the arrangement of GH and GHIP in the XEG-EDGP complex is distinct from that in the ANXI-TAXI-IA complex (Fig. 5C). There is a small but significant difference between the XEG and ANXI structures. The structure of GH displaying the β-jelly roll fold including GH11 and GH12 has been likened to a right hand, with a thumb, palm, and fingers (30). The thumb or fingers forms a lid over the active site cleft in both ANXI and XEG (Fig. 5C). The putative model structure of an ANXI-EDGP or XEG-TAXI-IA complex built by superimposition of the GH structures clearly shows a steric clash between the lid of GH and GHIP (Fig. 5D). This indicates an intrinsic difference in the lid structures of ANXI and XEG which is conserved in each member of the GH11 and GH12 families. Therefore, our findings imply that plants have evolved structures of GHIPs to inhibit specific GH families of enzymes that attack their cell walls. In addition to TAXI, wheat has xylanase inhibitor protein I, which possesses two independent enzyme-binding sites and is able to inhibit both GH10 and GH11 xylanases (31). Plants employ related proteins and mechanisms for target recognition of GH to protect their cell walls using conserved patterns of interacting residues.
Acknowlegments
We thank the beamline staff of SPring-8 and PF for data collection and Dr. H. Shiota, Yokohama City University, for supplying carrot callus, and Dr. J. R. H. Tame, Yokohama City University for English corrections.
This work was supported by a Grant-in-Aid for Scientific Research (KAKENHI), the National Project on Protein Structural and Functional Analyses Protein 3000 Project, and the Targeted Proteins Research Program from Ministry of Education, Culture, Sports, Science, and Technology in Japan (MEXT) (to T. S., M. S., and H. H.).
The atomic coordinates and structure factors (codes 3VL8, 3VL9, 3VLA, and 3VLB) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
T. Yoshizawa, T. Shimizu, H. Hirano, M. Sato, and H. Hashimoto, unpublished data.
- GH
- glycoside hydrolase
- ANXI
- Aspergillus niger xylanase I
- Bg7S
- basic 7S globulin
- EDGP
- extracellular dermal glycoprotein
- GHIP
- GH inhibitor protein
- IL
- inhibition loop
- PDB
- Protein Data Bank
- TAXI-IA
- Triticum aestivum xylanase inhibitor-IA
- XEG
- xyloglucan-specific endo-β-1,4-glucanase
- XEGIP
- XEG inhibitor protein
- FI-CMCase
- FI-carboxymethyl cellulose.
REFERENCES
- 1. Henrissat B. (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Juge N. (2006) Plant protein inhibitors of cell wall degrading enzymes. Trends Plant Sci. 11, 359–367 [DOI] [PubMed] [Google Scholar]
- 3. Lagaert S., Beliën T., Volckaert G. (2009) Plant cell walls: protecting the barrier from degradation by microbial enzymes. Semin. Cell Dev. Biol. 20, 1064–1073 [DOI] [PubMed] [Google Scholar]
- 4. Shang C., Sassa H., Hirano H. (2005) The role of glycosylation in the function of a 48-kDa glycoprotein from carrot. Biochem. Biophys. Res. Commun. 328, 144–149 [DOI] [PubMed] [Google Scholar]
- 5. Pauly M., Andersen L. N., Kauppinen S., Kofod L. V., York W. S., Albersheim P., Darvill A. (1999) A xyloglucan-specific endo-β-1,4-glucanase from Aspergillus aculeatus: expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology 9, 93–100 [DOI] [PubMed] [Google Scholar]
- 6. Hayashi T. (1989) Xyloglucans in the primary cell wall. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40, 139–168 [Google Scholar]
- 7. Qin Q., Bergmann C. W., Rose J. K., Saladie M., Kolli V. S., Albersheim P., Darvill A. G., York W. S. (2003) Characterization of a tomato protein that inhibits a xyloglucan-specific endoglucanase. Plant J. 34, 327–338 [DOI] [PubMed] [Google Scholar]
- 8. Naqvi S. M., Harper A., Carter C., Ren G., Guirgis A., York W. S., Thornburg R. W. (2005) Nectarin IV, a potent endoglucanase inhibitor secreted into the nectar of ornamental tobacco plants: isolation, cloning, and characterization. Plant Physiol. 139, 1389–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gebruers K., Brijs K., Courtin C. M., Fierens K., Goesaert H., Rabijns A., Raedschelders G., Robben J., Sansen S., Sørensen J. F., Van Campenhout S., Delcour J. A. (2004) Properties of TAXI-type endoxylanase inhibitors. Biochim. Biophys. Acta 1696, 213–221 [DOI] [PubMed] [Google Scholar]
- 10. Sansen S., De Ranter C. J., Gebruers K., Brijs K., Courtin C. M., Delcour J. A., Rabijns A. (2004) Structural basis for inhibition of Aspergillus niger xylanase by Triticum aestivum xylanase inhibitor-I. J. Biol. Chem. 279, 36022–36028 [DOI] [PubMed] [Google Scholar]
- 11. Fierens K., Gils A., Sansen S., Brijs K., Courtin C. M., Declerck P. J., De Ranter C. J., Gebruers K., Rabijns A., Robben J., Campenhout S., Volckaert G., Delcour J. A. (2005) His374 of wheat endoxylanase inhibitor TAXI-I stabilizes complex formation with glycoside hydrolase family 11 endoxylanases. FEBS J. 272, 5872–5882 [DOI] [PubMed] [Google Scholar]
- 12. Yoshizawa T., Shimizu T., Yamabe M., Taichi M., Nishiuchi Y., Shichijo N., Unzai S., Hirano H., Sato M., Hashimoto H. (2011) Crystal structure of basic 7S globulin, a xyloglucan-specific endo-β-1,4-glucanase inhibitor protein-like protein from soybean lacking inhibitory activity against endo-β-glucanase. FEBS J. 278, 1944–1954 [DOI] [PubMed] [Google Scholar]
- 13. Yoshizawa T., Shimizu T., Hirano H., Sato M., Hashimoto H. (2011) Purification, crystallization and x-ray diffraction study of extracellular dermal glycoprotein from carrot and the inhibition complex that it forms with an endo-β-glucanase from Aspergillus aculeatus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 830–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoover D. M., Lubkowski J. (2002) DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 30, e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lever M. (1972) A new reaction for colorimetric determination of carbohydrates. Anal. Biochem. 47, 273–279 [DOI] [PubMed] [Google Scholar]
- 16. Martinez-Fleites C., Guerreiro C. I., Baumann M. J., Taylor E. J., Prates J. A., Ferreira L. M., Fontes C. M., Brumer H., Davies G. J. (2006) Crystal structures of Clostridium thermocellum xyloglucanase, XGH74A, reveal the structural basis for xyloglucan recognition and degradation. J. Biol. Chem. 281, 24922–24933 [DOI] [PubMed] [Google Scholar]
- 17. Miyatake H., Hasegawa T., Yamano A. (2006) New methods to prepare iodinated derivatives by vaporizing iodine labelling (VIL) and hydrogen peroxide VIL (HYPER-VIL). Acta Crystallogr. D Biol. Crystallogr. 62, 280–289 [DOI] [PubMed] [Google Scholar]
- 18. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 19. Vagin A., Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 20. Terwilliger T. (2004) SOLVE and RESOLVE: automated structure solution, density modification and model building. J. Synchrotron Radiat. 11, 49–52 [DOI] [PubMed] [Google Scholar]
- 21. Emsley P., Cowtan K. (2004) COOT: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 22. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 23. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 24. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structure. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 25. Sandgren M., Ståhlberg J., Mitchinson C. (2005) Structural and biochemical studies of GH family 12 cellulases: improved thermal stability, and ligand complexes. Prog. Biophys. Mol. Biol. 89, 246–291 [DOI] [PubMed] [Google Scholar]
- 26. Kanda T., Wakabayashi K., Nisizawa K. (1976) Synergistic action of two different types of endocellulase components from Irpex lacteus (Polyporus tulipiferae) in the hydrolysis of some insoluble celluloses. J Biochem. 79, 997–1005 [DOI] [PubMed] [Google Scholar]
- 27. Gloster T. M., Ibatullin F. M., Macauley K., Eklöf J. M., Roberts S., Turkenburg J. P., Bjørnvad M. E., Jørgensen P. L., Danielsen S., Johansen K. S., Borchert T. V., Wilson K. S., Brumer H., Davies G. J. (2007) Characterization and three-dimensional structures of two distinct bacterial xyloglucanases from families GH5 and GH12. J. Biol. Chem. 282, 19177–19189 [DOI] [PubMed] [Google Scholar]
- 28. Shang C., Shibahara T., Hanada K., Iwafune Y., Hirano H. (2004) Mass spectrometric analysis of posttranslational modifications of a carrot extracellular glycoprotein. Biochemistry 43, 6281–6292 [DOI] [PubMed] [Google Scholar]
- 29. Browner M. F., Smith W. W., Castelhano A. L. (1995) Matrilysin-inhibitor complexes: common themes among metalloproteases. Biochemistry 34, 6602–6610 [DOI] [PubMed] [Google Scholar]
- 30. Törrönen A., Harkki A., Rouvinen J. (1994) Three-dimensional structure of endo-1,4-β-xylanase II from Trichoderma reesei: two conformational states in the active site. EMBO J. 13, 2493–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Payan F., Leone P., Porciero S., Furniss C., Tahir T., Williamson G., Durand A., Manzanares P., Gilbert H. J., Juge N., Roussel A. (2004) The dual nature of the wheat xylanase protein inhibitor XIP-I: structural basis for the inhibition of family 10 and family 11 xylanases. J. Biol. Chem. 279, 36029–36037 [DOI] [PubMed] [Google Scholar]