Background: STAT3 suppresses carcinogenesis of intestinal tumors in Apc min mice.
Results: STAT3 suppresses expression of SNAI in intestinal epithelium by regulating GSK3β activity.
Conclusion: STAT3 induces degradation of SNAI by promoting GSK3β activity and thereby suppresses adenoma-to-adenocarcinoma transition in Apc min mice.
Significance: Our data provide a new insight into the role of STAT3 in colorectal cancer biology.
Keywords: Carcinogenesis, Cell Invasion, Colorectal Cancer, EMT, STAT3, Apcmin, GSK3β, SNAI
Abstract
STAT3 was recently reported to suppress tumor invasion in Apcmin/+ mice. We investigated the mechanisms by which STAT3 inhibits intestinal epithelial tumors using Apcmin/+/Stat3IEC-KO mice (intestinal epithelial cell (IEC)-specific deletion of STAT3 in the Apcmin/+ background) to determine the role of STAT3 in carcinogenesis in vivo as well as colorectal cancer cell lines in vitro. To inhibit invasion of IEC tumors, STAT3 functions as a molecular adaptor rather than a transcription factor. Accordingly, the tumors in Apcmin/+/Stat3IEC-KO mice undergo adenoma-to-carcinoma transition and acquire an invasive phenotype. Similarly, STAT3 knockdown in a colorectal cell line enhances IEC invasion. We demonstrate that STAT3 down-regulates SNAI (Snail-1) expression levels and hence suppresses epithelial-mesenchymal transition of colorectal cancer cells. Mechanistically, STAT3 facilitates glycogen synthase kinase (GSK) 3β-mediated degradation of SNAI by regulating phosphorylation of GSK3β. Our data identified a new role for STAT3 in the adenoma-to-carcinoma sequence of intestinal tumors.
Introduction
STAT3 is a multifunctional transcription factor that is activated by various growth factors (e.g. epidermal growth factor and hepatocyte growth factor) and cytokines (e.g. IL-6 and IL-10). Consequently, STAT3 plays a key role in many biological processes such as cell growth, apoptosis, and inflammation (1). STAT3 is phosphorylated on a tyrosine residue (Tyr-705) by an upstream kinase JAK2 (Janus kinase 2). The phosphorylated protein forms either a homo- or heterodimer with other STAT proteins, then translocates to the nucleus and transcribes target genes. Rare nontranscriptional activities of STAT3 have also been reported. For example, STAT3 functions as an adaptor protein connecting IFNAR1 (interferon α receptor 1) and the p85 regulatory subunit of PI3K (phosphoinositide 3-kinase) (2), and it inhibits stathmin that depolymerizes microtubules (3). Recently, STAT3 was shown to inhibit intestinal epithelial cell (IEC)4 tumor invasion in Apcmin/+ mice (4), but the underlying mechanisms remain unclear.
The development of human colon adenomas is initially induced by mutations in APC or β-catenin, and its transition to carcinoma is followed by sequential genetic mutations in K-Ras, SMAD2 or SMAD4, and p53. Whereas deletion of p53 does not provoke malignant transformation in Apcmin/+ mice (5), at least two genes were found critical for adenoma-to-adenocarcinoma transition. Tumors in Smad4+/−/Apcmin/+ mice become malignant, showing an extensive stromal cell proliferation and submucosal invasion (6), and Ephb3−/−/Apcmin/+ mice develop carcinomas that invade the muscle layer (7). However, the mechanisms by which these genes suppress IEC carcinogenesis are not fully understood.
The invasion of epithelial tumors to the surrounding tissues is associated with epithelial-mesenchymal transition (EMT), a process in which the epithelial tumor cells lose their epithelial phenotype and acquire a mesenchymal phenotype. Epithelial cells that have undergone EMT display reduced intercellular interactions but increased motility and invasiveness. Several transcriptional repressors were identified as inducers of EMT, including SNAI, Slug (Snail-2), E47, Twist, and the Zeb factors (8). Main targets of these proteins are cell adhesion molecules such as E-cadherin and claudins (CLDNs) that link epithelial cells together (8–11).
Glycogen synthase kinase 3β (GSK3β) performs multiple cellular functions (12). As an important regulator of metastasis, it suppresses the level of SNAI by being phosphorylated on two different sites: phosphorylation on the first site leads to ubiquitination and proteasomal degradation of SNAI whereas that on the second site controls the subcellular localization of SNAI (13). GSK3β is inactivated by phosphorylation by several different kinases at the Ser-9 residue (12, 14) but also can be regulated by phosphorylation at other residues: it is inactivated by phosphorylation at Thr-390 (15) and is activated by phosphorylation at Tyr-216 (16, 17). In addition, GSK3β activity is modulated by protein complex formation mediated by GSK3β-binding proteins and its subcellular localization (18). GSK3β is also a key regulator of β-catenin (19, 20), an effector molecule for the Wnt signaling pathway. Constitutive activation of β-catenin due to a mutation in the APC gene is the major cause of colorectal cancers (CRCs) (21) and induces the multiple intestinal neoplasia (min) phenotype in Apcmin/+ mice (21–23)
Here, we identified a novel adaptor function for STAT3 in IECs, which has a broad implication to CRC. Our data show that in a nontranscriptional fashion, STAT3 suppresses expression of SNAI, a major driving force in EMT and in the invasion of CRC cells. STAT3 interacts with GSK3β and negatively regulates phosphorylation of GSK3β, thereby inducing degradation of SNAI.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Anti-STAT3, anti-β-catenin, anti-phospho-β-catenin, anti-GSK3β, anti-phospho-GSK3β (Ser-9), anti-SNAI, anti-Slug, anti-Zeb-1, anti-matrix metalloproteinase 7 (MMP7), anti-MMP9, and anti-E-cadherin antibodies were obtained from Cell Signaling Technologies (Danvers, MA). Anti-β-actin came from Sigma. Anti-ubiquitin, anti-MMP14, and anti-MMP15 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-claudin and anti-occludin antibodies were obtained from Lab Vision (Fremont, CA). Anti-vimentin and anti-fibronectin antibodies were obtained from BD Biosciences. Anti-axin antibody is from Invitrogen. The GFP-STAT3 plasmid was generously provided by Nancy C. Reich (24). Myc-GSK3β-CA plasmid (13) was obtained from Addgene (Cambridge MA). LiCl was obtained from Sigma and SB216763 from EMD Biosciences (Darmstadt, Germany).
Isolation of IECs from Mouse Intestines
Cell Culture
Culture conditions of IEC cells have been described previously (25). HCA-7 cells were cultured in low glucose DMEM, and HCT116 and RKO cells in high glucose DMEM (Mediatech, Manassas, VA).
Cell Invasion Assay
The cell invasion assay was performed with a basement membrane-coated CytoSelectTM 24-well cell invasion assay kit according to the manufacturer's instructions (Cell Biolabs, San Diego, CA). IECs (1 × 105 cells/well) were plated into each well 3 days after siRNA transfection and cultured for 2 days. The number of invaded cells per 4 high powered fields/well was visually counted using a microscope. The results are the average of two independent experiments, and the statistical analysis was performed by one-way ANOVA.
Quantitative PCR (qPCR), Immunoblotting, Immunohistochemistry, and Immunoprecipitation
These procedures were performed as described previously (25, 26). PCR primers are shown in Table 1.
TABLE 1.
PCR primers
| Gene names | Forward primer | Reverse primer |
|---|---|---|
| h-GAPDH | 5′-CAT GTT CGT CAT GGG TGT GAA CCA-3′ | 5′-AGT GAT GGC ATG GAC TGT GGT CAT-3′ |
| h-SNAI | 5′-TAC AGC GAG CTG CAG GAC TCT AAT-3′ | 5′-AGG ACA GAG TCC CAG ATG AGC ATT-3′ |
Cell Proliferation Assay in Mice
Mice were administered BrdU intraperitoneally (2 mg/mouse) and killed 2 h after the injection. BrdU incorporation (proliferation) in paraffinized colonic tissues and the TUNEL assay in paraffinized colonic tissues were performed and analyzed according to the manufacturer's instructions (BD Biosciences).
siRNA-mediated Knockdown
Knockdown in IECs was performed with Nucleofector (Amaxa, Germany). Nontargeting siRNA #2 (luciferase targeting siRNA) from Dharmacon was used as a control. STAT3 siRNA (5′-CAACATGTCATTTGCTGAA-3′), GSK3β siRNA (5′-TCCGAGGAGAACCCAATGTTTCGTATA-3′), and SNAI siRNA (Applied Biosystems, Cat. No. s13186) were used.
Generation of STAT3IEC-KO Mice
To generate STAT3 deficiency specifically in IEC in mice, Stat3flox/flox mice (a gift from Shizou Akira, Osaka University, Osaka, Japan) were crossed to villin-Cre mice (on the C57BL/6J background; The Jackson Laboratory, Bar Harbor, ME), to create Stat3IEC-KO mice. Genotyping was performed as described (27). Apcmin/+ mice were obtained from The Jackson Laboratory. Apcmin/+/Stat3IEC-KO mice were generated by crossing Apcmin/+ mice with Stat3IEC-KO mice. All animal protocols received prior approval by the Institutional Animal Care and Use Committee.
RESULTS
STAT3 Suppresses Invasion of Intestinal Tumors in Apcmin/+ Mice
To determine the role of STAT3 in intestinal tumorigenesis, we conditionally deleted STAT3 in IEC (STAT3IEC-KO) in Apcmin/+ mice (supplemental Fig. 1, A and B). At 20 weeks of age, the tumor count in Apcmin/+/Stat3IEC-KO mice was comparable with that in Apcmin/+ mice (supplemental Fig. 1C). In addition, proliferation of IECs in normal or tumor IEC was not significantly affected by STAT3 deletion (supplemental Fig. 1D).
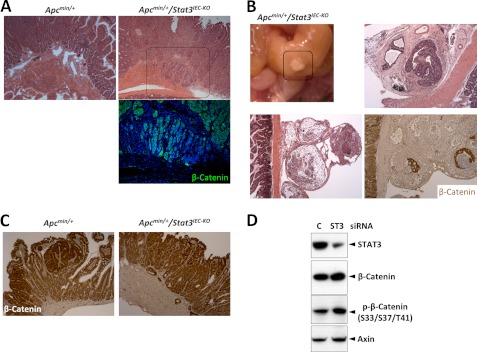
Whereas the IEC tumors in Apcmin/+ mice were strictly confined to the mucosal crypts, those in the Apcmin/+/Stat3IEC-KO mice invaded the mucosal stroma, submucosa, and muscle (Fig. 1A). Such invasive tumors were found in all of the Apcmin/+/Stat3IEC-KO mice examined at 20 weeks (n = 20) of age and were confirmed by β-catenin immunostaining (Fig. 1A). At 25 weeks of age, the IEC tumors in Apcmin/+/Stat3Δ1EC mice penetrated through the intestinal serosa (Fig. 1B). Although some of the tumors penetrating serosa were necrotic, live tumor cells (β-catenin-positive) were also found surrounded by stromal tissues (Fig. 1B). However, STAT3 deletion did not significantly affect the levels of β-catenin in tumor cells (Fig. 1C). In a human colorectal cell line, STAT3 knockdown slightly enhanced the levels of β-catenin, phospho-β-catenin, and axin (a target of β-catenin), indicating that it can suppress the β-catenin pathway (Fig. 1D). Taken together, these findings validate that STAT3 in this animal model inhibits invasion of IEC tumors without affecting tumor induction (4).
FIGURE 1.
STAT3 suppresses tumor invasion in Apcmin/+ mice. A, deletion of STAT3IEC induces invasion of tumors in Apcmin/+ mice. H&E (upper panel) and β-catenin (lower panel) staining of the small intestines shows the invasion of tumors in Apcmin/+/Stat3IEC-KO mice. B, tumors in Apcmin/+/Stat3IEC-KO mice penetrate the intestinal walls into the peritoneum. The boxed area in the upper left panel was analyzed by H&E and β-catenin staining. C, STAT3 does not regulate expression of β-catenin in Apcmin/+ mice. Expression of β-catenin in the small intestines was measured by immunohistochemistry. D, STAT3 suppresses activation of β-catenin in a CRC cell line. HCT116 cells were transfected with GFP or GFP-STAT3-WT (ST3), and the levels of the indicated proteins were measured by immunoblotting.
STAT3 Suppresses EMT
Because EMT often precedes tumor invasion, we next investigated whether STAT3 deletion induces EMT in IEC tumors. Expression of E-cadherin or occludin was largely unaffected by STAT3 deletion (Fig. 2A), but expression of CLDN-3 and CLDN-5 was markedly decreased in tumor cells of Apcmin/+/Stat3IEC-KO mice compared with those in Apcmin/+ mice (Fig. 2B). Vimentin was expressed mainly in mesenchymal cells and rarely in tumor cells in Apcmin/+ mice; however, all of the tumor cells in the Apcmin/+/Stat3IEC-KO mice expressed vimentin at a high level (Fig. 2C). Fibronectin was also up-regulated in the tumor cells of Apcmin/+/Stat3IEC-KO mice whereas it was not detectable in tumor from Apcmin/+ mice (Fig. 2D). Interestingly fibronectin in Apcmin/+/Stat3IEC-KO was observed only in IEC tumor cells around the necrotic regions (Fig. 2D). Collectively, these data highly suggest that STAT3 suppresses EMT in IEC tumor cells in Apcmin/+ mice.
FIGURE 2.
STAT3 suppresses EMT in Apcmin/+ mice. A, STAT3 does not regulate expression of E-cadherin in Apcmin/+ mice. Expression of E-cadherin in the small intestines was measured by immunohistochemistry. B, STAT3IEC maintains the expression of CLDNs in Apcmin/+ mice. Expression of the indicated proteins was measured by immunoblotting with the isolated tumor cells from the small intestines. C, STAT3 suppresses the expression of mesenchymal marker vimentin in Apcmin/+ mice. Expression of vimentin was measured by confocal imaging in the small intestines (lower panel original magnification, ×100). D, STAT3 suppresses the expression of mesenchymal marker fibronectin in Apcmin/+ mice. Expression of vimentin was measured by confocal imaging in the small intestines. Note that most of the fibronectin expression in Apcmin/+/Stat3IEC-KO mice is found in tumor cells around the necrotic regions. E, STAT3 suppresses the expression of MMP14 in Apcmin/+ mice. Expression of MMP14 was measured by immunohistochemistry in the small intestines.
STAT3 Suppresses Expression of MMP14
We next checked the expression of MMPs, which are important for tumor invasion (28, 29). Whereas MMP7 expression in tumor cells was significantly lower in Apcmin/+/Stat3IEC-KO mice than that in Apcmin/+ mice, MMP9 or MMP15 (MT2-MMP) expression was minimal and observed mostly in nonepithelial cells in both strains of mice (supplemental Fig. 2). In contrast, MMP14 (MT1-MMP) expression in tumor cells was highly induced in Apcmin/+/Stat3IEC-KO mice but undetectable in Apcmin/+ mice (Fig. 2E). Interestingly, MMP14, a membrane type metalloproteinase, is particularly efficient in hydrolyzing basement membranes (30, 31), and its expression can be induced by SNAI in carcinomas (30).
STAT3 Suppresses Tumor Invasion via Down-regulation of SNAI
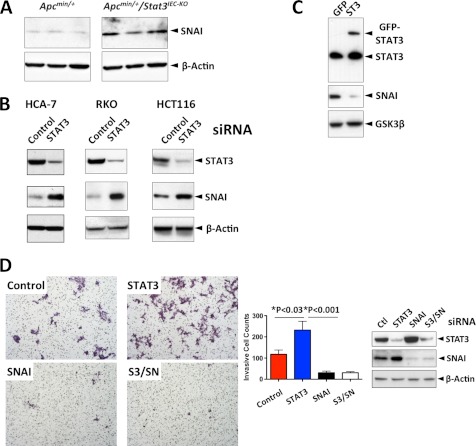
To examine further the suppressive effect of STAT3 on EMT, known regulators of EMT were examined. Whereas expression of Slug and Zeb-1 were barely detectable, and their levels were not significantly different in IEC tumor cells from both strains of mice (supplemental Fig. 3), SNAI expression was significantly elevated in Apcmin/+/Stat3IEC-KO tumor cells (Fig. 3A). To investigate further the role of STAT3 in SNAI expression and the role of SNAI in invasion, we used CRC cell lines in the subsequent studies. In three different CRC cell lines, HCT116, HCA-7, and RKO, STAT3 knockdown increased the expression of SNAI (Fig. 3B). Conversely, ectopic expression of STAT3-WT suppressed expression of SNAI (Fig. 3C).
FIGURE 3.
STAT3 inhibits tumor invasion by suppressing expression of SNAI. A, STAT3 suppresses expression of SNAI in Apcmin/+ mice. Expression of the indicated proteins was measured by immunoblotting from the isolated tumor cells from the small intestines. B, STAT3 suppresses expression of SNAI in human CRC lines. STAT3 was silenced by transfection of siRNA, and expression of the indicated proteins in the indicated CRC lines was measured by immunoblotting 3 days after transfection. C, STAT3 suppresses expression of SNAI in a human CRC cell line. HCT116 cells were transfected with either GFP or GFP-STAT3-WT, and the levels of the indicated proteins were measured by immunoblotting. D, SNAI mediates the invasion of the STAT3-depleted HCT116 cells. Cancer cell invasion assay was performed as described under “Experimental Procedures” 3 days after transfection of the indicated siRNA (S3, STAT3; SN, SNAI). The results are the average of two independent experiments (n = 6). The levels of the indicated proteins on the bottom right were measured by immunoblotting. Error bars, S.D.
We next tested whether the elevated SNAI was responsible for the invasive phenotype of STAT3-depleted CRC cells. STAT3, SNAI, or STAT3 with SNAI was silenced in HCT116 cells, and the transfected cells were subjected to an in vitro cell invasion assay. STAT3 knockdown significantly enhanced invasiveness compared with control. In addition, SNAI knockdown alone or a combination of STAT3 and SNAI knockdown significantly curtailed the invasion (Fig. 3D). Thus, these data indicate that SNAI promotes invasion and that STAT3 down-regulates SNAI and thereby suppresses invasion of CRC cells.
STAT3 Promotes GSK3β Activity to Induce Degradation of SNAI
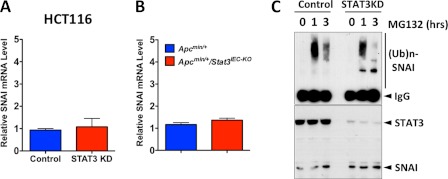
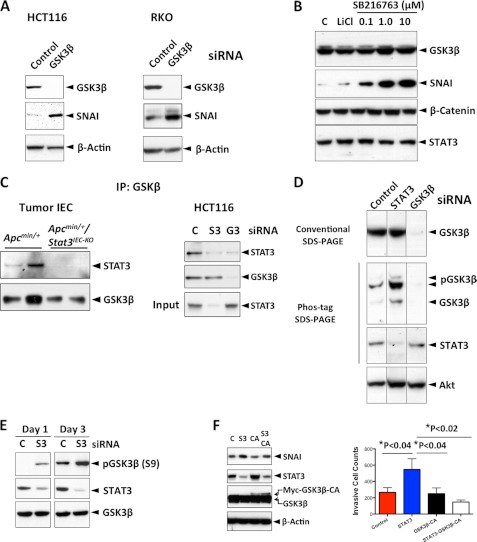
We next studied how STAT3 regulates SNAI expression. STAT3 deletion or knockdown in IEC did not significantly affect the transcription of SNAI (Fig. 4, A and B). We therefore examined whether STAT3 regulated SNAI via post-translational modification. SNAI in HCT116 cells was constitutively ubiquitinated and degraded by proteasomes (Fig. 4C). STAT3 knockdown significantly diminished the levels of SNAI ubiquitination, even though the levels of SNAI were higher than control (Fig. 4C). Because GSK3β phosphorylates SNAI, which leads to ubiquitination and proteasomal degradation (13), we investigated whether STAT3 is involved in the ubiquitin-mediated proteasomal degradation of SNAI via GSK3β. First, we found that GSK3β also down-regulates SNAI in CRC cell lines (Fig. 5A) and that inhibition of GSK3β activity with pharmacological inhibitors significantly increases SNAI (Fig. 5B). In addition, STAT3 forms a complex with GSK3β (Fig. 5C). Because STAT3 deletion or knockdown did not significantly affect the expression of GSK3β (Fig. 5C), we examined whether STAT3 regulates activation (or phosphorylation) of GSK3β by Phos-tag SDS-PAGE (13, 32). Whereas there was no noticeable difference in GSK3β expression level between control and STAT3-deleted cells in a traditional SDS-PAGE analysis, the putative phosphorylated (retarded in mobility) GSK3β level was much higher in STAT3-deleted cells compared with that in control cells (Fig. 5D). STAT3 knockdown significantly increased the phosphorylation level on Ser9 residue of GSK3β (Fig. 5E). This result indicated that STAT3 negatively affects phosphorylation of GSK3β and suggested that STAT3 regulates SNAI expression and tumor invasion by promoting GSK3β activity. Therefore, we tested whether a constitutively active GSK3β (GSK3β-CA) mutant can reverse the effects of STAT3 depletion in HCT116 cells. Indeed, expression of GSK3β-CA reversed the increased SNAI expression and the increased invasiveness induced by STAT3 depletion (Fig. 5F). Taken together, these data provide evidence for a nontranscriptional and inhibitory role of STAT3 in IEC tumor invasion.
FIGURE 4.
STAT3 regulates SNAI at the post-transcriptional level. A, STAT3 does not regulate transcription of SNAI. The mRNA levels of SNAI were measured by qPCR in HCT116 cells 3 days after siRNA transfection (n = 4). B, STAT3 does not regulate transcription of SNAI. The mRNA levels of SNAI were measured by qPCR in the indicated tumor cells (n = 4). C, STAT3 promotes ubiquitin-mediated degradation of SNAI in IECs. HCT116 cells were transfected with either control or STAT3 siRNA, treated with MG132 (10 μm) as indicated, SNAI was immunoprecipitated, and the gel was immunoblotted for ubiquitin (upper panel (Ub)n, polyubiquitin). The levels of STAT3 and SNAI were measured by immunoblotting (lower panel).
FIGURE 5.
STAT3 promotes GSK3β activity to induce degradation of SNAI. A, GSK3β suppresses SNAI expression in CRC lines. The indicated protein levels were measured by immunoblotting 3 days after siRNA transfection. B, GSK3β suppresses SNAI via its kinase activity. HCT116 cells were treated with either LiCl (4 mm) or SB216763, GSK3β inhibitors as indicated for 4 h, and the indicated proteins were measured by immunoblotting. C, STAT3 interacts with GSK3β. GSK3β in isolated mouse tumor cells or in HCT116 cells was immunoprecipitated and immunoblotted for STAT3 (S3, STAT3; G3, GSK3β). D, STAT3 regulates phosphorylation levels of GSK3β. Lysates from HCT116 cells transfected with the indicated siRNA were divided and separated by either traditional SDS-PAGE or Phos-tag SDS-PAGE, and the indicated proteins were measured by immunoblotting. E, STAT3 inhibits phosphorylation at Ser-9 of GSK3β. HCT116 cells were transfected with the indicated siRNA (C, control; S3, STAT3), total cell lysates were collected 1 or 3 days after transfection, and the levels of indicated proteins were measured by immunoblotting. F, GSK3β-CA reverses the effects of STAT3 depletion. HCT116 cells were transfected with control siRNA (C), STAT3 siRNA (S3), GSK3β-CA plasmid, or STAT3 siRNA (S3) plus GSK3β-CA plasmid. Cell invasion assay (n = 3) and immunoblotting were performed 3 days after transfection.
DISCUSSION
Although no naturally occurring mutations in STAT3 have been identified as a cause of any human cancer, it is considered as a strong promoter of carcinogenesis. STAT3 is activated in multiple solid and hematologic human malignancies, and its role in carcinogenesis has been validated in animal models (1, 33). For example, STAT3 was found to be crucial in initiation and progression of skin carcinoma (34), and the growth and survival of lymphomas are dependent on STAT3 (35). Recently, constitutively activating STAT3 mutations were identified in human hepatocellular adenomas, where the Tyr-640 mutant homodimerizes independently of upstream signals and is hypersensitive to IL-6 stimulation (36). In CRC, STAT3 activation (phosphorylation) was considerably up-regulated during adenoma-to-carcinoma progression (37, 38), but the precise role of STAT3 in CRC has not been yet defined.
Our data provide the mechanisms by which STAT3 suppresses tumor invasion both in an animal model and in CRC cell lines. STAT3 in IEC tumors of Apcmin/+ mice inhibits expression of an EMT inducer SNAI and mesenchymal markers, such as vimentin and fibronectin. Similarly, in CRC cell line HCT116, STAT3 suppresses invasiveness by limiting expression of SNAI. Overexpression of a dominant negative STAT3 mutant in a CRC cell line was reported to cause down-regulation of E-cadherin (39). Although SNAI is widely known as a suppressor of E-cadherin, our data and another recent report (4) show that its expression is not significantly affected by STAT3 deletion in IEC. However, overexpression of SNAI did not suppress E-cadherin in a breast cancer cell line, whereas a mutant form (6SA) was able to do so (13). Therefore, it is possible that Ser-6 of SNAI may be phosphorylated in STAT3-deficient tumor cells.
Like STAT3, GSK3β is also a multifunctional protein involved in a wide range of physiological activities such as metabolism, cell development, and body pattern formation (12). It also plays a key role in Wnt signaling whose dysregulation is the most common cause of human CRC (2, 20). Our data show that STAT3 regulates GSK3β activity to control SNAI expression. We also found that STAT3 inhibits activation of β-catenin in a CRC cell line, but the mechanisms behind this phenomenon are yet to be investigated.
Interestingly, STAT3 in IECs is required for tumor induction in models of colitis-associated cancer. In colitis-associated cancer, STAT3 activation in IEC regulates cell survival and cell cycle progression through its transcriptional activation of downstream targets such as Bcl-XL, c-Myc, and cyclin D1 (40–42). This colitis-associated cancer model employs a chemical mutagen, azoxymethane, and three cycles of the chemical irritant, dextran sodium sulfate, to induce colorectal tumors. Despite the observation that Stat3IEC-KO mice are much more susceptible to dextran sodium sulfate-induced colitis (41), it appears that STAT3 in IEC in Apc min model is necessary for their malignant transformation.
Overall, our data document the restraining role of STAT3 on the acquisition of an invasive phenotype of IEC tumors and explain the molecular basis that controls this process. We therefore propose that the inhibition of STAT3 in patients with CRC especially in advanced stages should be practiced with caution.
Supplementary Material
Acknowledgements
We thank Jennifer Meerloo for assistance in confocal imaging at the University of California at San Diego Neuroscience Microscopy Shared Facility funded by National Institutes of Health Grant P30 NS047101 through the NINDS.
This work was supported, in whole or in part, by National Institutes of Health Grants A1068685, A1095623, DK35108, and DK080506. This work was also supported by a grant from Crohns and Colitis Foundation of America.

This article contains supplemental Figs. 1–3.
- IEC
- intestinal epithelial cell
- CA
- constitutively active
- CLDN
- claudin
- CRC
- colorectal cancer
- EMT
- epithelial-mesenchymal transition
- GSK3β
- glycogen synthase kinase 3β
- min
- multiple intestinal neoplasia
- MMP
- matrix metalloproteinase
- qPCR
- quantitative PCR
- SNAI
- Snail-1
- APC
- adenomatous polyposis coli.
REFERENCES
- 1. Yu H., Pardoll D., Jove R. (2009) STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer 9, 798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pfeffer L. M., Mullersman J. E., Pfeffer S. R., Murti A., Shi W., Yang C. H. (1997) STAT3 as an adapter to couple phosphatidylinositol 3-kinase to the IFNAR1 chain of the type I interferon receptor. Science 276, 1418–1420 [DOI] [PubMed] [Google Scholar]
- 3. Ng D. C., Lin B. H., Lim C. P., Huang G., Zhang T., Poli V., Cao X. (2006) Stat3 regulates microtubules by antagonizing the depolymerization activity of stathmin. J. Cell Biol. 172, 245–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Musteanu M., Blaas L., Mair M., Schlederer M., Bilban M., Tauber S., Esterbauer H., Mueller M., Casanova E., Kenner L., Poli V., Eferl R. (2010) Stat3 is a negative regulator of intestinal tumor progression in Apc(Min) mice. Gastroenterology 138, 1003–1011.e1–5 [DOI] [PubMed] [Google Scholar]
- 5. Halberg R. B., Katzung D. S., Hoff P. D., Moser A. R., Cole C. E., Lubet R. A., Donehower L. A., Jacoby R. F., Dove W. F. (2000) Tumorigenesis in the multiple intestinal neoplasia mouse: redundancy of negative regulators and specificity of modifiers. Proc. Natl. Acad. Sci. U.S.A. 97, 3461–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takaku K., Oshima M., Miyoshi H., Matsui M., Seldin M. F., Taketo M. M. (1998) Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Apc genes. Cell 92, 645–656 [DOI] [PubMed] [Google Scholar]
- 7. Batlle E., Bacani J., Begthel H., Jonkeer S., Gregorieff A., van de Born M., Malats N., Sancho E., Boon E., Pawson T., Gallinger S., Pals S., Clevers H. (2005) EphB receptor activity suppresses colorectal cancer progression. Nature 435, 1126–1130 [DOI] [PubMed] [Google Scholar]
- 8. Peinado H., Olmeda D., Cano A. (2007) Snail, Zeb, and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer 7, 415–428 [DOI] [PubMed] [Google Scholar]
- 9. Carrozzino F., Soulié P., Huber D., Mensi N., Orci L., Cano A., Féraille E., Montesano R. (2005) Inducible expression of Snail selectively increases paracellular ion permeability and differentially modulates tight junction proteins. Am. J. Physiol. Cell Physiol. 289, C1002–1014 [DOI] [PubMed] [Google Scholar]
- 10. Ikenouchi J., Matsuda M., Furuse M., Tsukita S. (2003) Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J. Cell Sci. 116, 1959–1967 [DOI] [PubMed] [Google Scholar]
- 11. Martínez-Estrada O. M., Cullerés A., Soriano F. X., Peinado H., Bolós V., Martínez F. O., Reina M., Cano A., Fabre M., Vilaró S. (2006) The transcription factors Slug and Snail act as repressors of claudin-1 expression in epithelial cells. Biochem. J. 394, 449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Plyte S. E., Hughes K., Nikolakaki E., Pulverer B. J., Woodgett J. R. (1992) Glycogen synthase kinase-3: functions in oncogenesis and development. Biochim. Biophys. Acta 1114, 147–162 [DOI] [PubMed] [Google Scholar]
- 13. Zhou B. P., Deng J., Xia W., Xu J., Li Y. M., Gunduz M., Hung M. C. (2004) Dual regulation of Snail by GSK-3β-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat. Cell Biol. 6, 931–940 [DOI] [PubMed] [Google Scholar]
- 14. Cross D. A., Alessi D. R., Cohen P., Andjelkovich M., Hemmings B. A. (1995) Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378, 785–789 [DOI] [PubMed] [Google Scholar]
- 15. Thornton T. M., Pedraza-Alva G., Deng B., Wood C. D., Aronshtam A., Clements J. L., Sabio G., Davis R. J., Matthews D. E., Doble B., Rincon M. (2008) Phosphorylation by p38 MAPK as an alternative pathway for GSK3β inactivation. Science 320, 667–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hartigan J. A., Johnson G. V. (1999) Transient increases in intracellular calcium result in prolonged site-selective increases in tau phosphorylation through a glycogen synthase kinase 3β-dependent pathway. J. Biol. Chem. 274, 21395–21401 [DOI] [PubMed] [Google Scholar]
- 17. Lesort M., Jope R. S., Johnson G. V. (1999) Insulin transiently increases tau phosphorylation: involvement of glycogen synthase kinase-3β and Fyn tyrosine kinase. J. Neurochem. 72, 576–584 [DOI] [PubMed] [Google Scholar]
- 18. Schaffer B., Wiedau-Pazos M., Geschwind D. H. (2003) Gene structure and alternative splicing of glycogen synthase kinase 3β (GSK-3β) in neural and non-neural tissues. Gene 302, 73–81 [DOI] [PubMed] [Google Scholar]
- 19. Peifer M., Pai L. M., Casey M. (1994) Phosphorylation of the Drosophila adherens junction protein Armadillo: roles for wingless signal and zeste-white 3 kinase. Dev. Biol. 166, 543–556 [DOI] [PubMed] [Google Scholar]
- 20. Yost C., Torres M., Miller J. R., Huang E., Kimelman D., Moon R. T. (1996) The axis-inducing activity, stability, and subcellular distribution of β-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 10, 1443–1454 [DOI] [PubMed] [Google Scholar]
- 21. Fearon E. R. (2011) Molecular genetics of colorectal cancer. Annu. Rev. Pathol. 6, 479–507 [DOI] [PubMed] [Google Scholar]
- 22. Bilger A., Shoemaker A. R., Gould K. A., Dove W. F. (1996) Manipulation of the mouse germ line in the study of Min-induced neoplasia. Semin. Cancer Biol. 7, 249–260 [DOI] [PubMed] [Google Scholar]
- 23. Fearon E. R., Vogelstein B. (1990) A genetic model for colorectal tumorigenesis. Cell 61, 759–767 [DOI] [PubMed] [Google Scholar]
- 24. Liu L., McBride K. M., Reich N. C. (2005) STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-α3. Proc. Natl. Acad. Sci. U.S.A. 102, 8150–8155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee J., Mo J. H., Katakura K., Alkalay I., Rucker A. N., Liu Y. T., Lee H. K., Shen C., Cojocaru G., Shenouda S., Kagnoff M., Eckmann L., Ben-Neriah Y., Raz E. (2006) Maintenance of colonic homeostasis by distinctive apical TLR9 signalling in intestinal epithelial cells. Nat. Cell Biol. 8, 1327–1336 [DOI] [PubMed] [Google Scholar]
- 26. Lee S. H., Hu L. L., Gonzalez-Navajas J., Seo G. S., Shen C., Brick J., Herdman S., Varki N., Corr M., Lee J., Raz E. (2010) ERK activation drives intestinal tumorigenesis in Apcmin/+ mice. Nat. Med. 16, 665–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kobayashi M., Kweon M. N., Kuwata H., Schreiber R. D., Kiyono H., Takeda K., Akira S. (2003) Toll-like receptor-dependent production of IL-12p40 causes chronic enterocolitis in myeloid cell-specific Stat3-deficient mice. J. Clin. Invest. 111, 1297–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kessenbrock K., Plaks V., Werb Z. (2010) Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 141, 52–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zucker S., Vacirca J. (2004) Role of matrix metalloproteinases (MMPs) in colorectal cancer. Cancer Metastasis Rev. 23, 101–117 [DOI] [PubMed] [Google Scholar]
- 30. Hotary K. B., Allen E. D., Brooks P. C., Datta N. S., Long M. W., Weiss S. J. (2003) Membrane type I matrix metalloproteinase usurps tumor growth control imposed by the three-dimensional extracellular matrix. Cell 114, 33–45 [DOI] [PubMed] [Google Scholar]
- 31. Ota I., Li X. Y., Hu Y., Weiss S. J. (2009) Induction of a MT1-MMP and MT2-MMP-dependent basement membrane transmigration program in cancer cells by Snail1. Proc. Natl. Acad. Sci. U.S.A. 106, 20318–20323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kinoshita E., Kinoshita-Kikuta E. (2011) Improved Phos-tag SDS-PAGE under neutral pH conditions for advanced protein phosphorylation profiling. Proteomics 11, 319–323 [DOI] [PubMed] [Google Scholar]
- 33. Catlett-Falcone R., Landowski T. H., Oshiro M. M., Turkson J., Levitzki A., Savino R., Ciliberto G., Moscinski L., Fernández-Luna J. L., Nuñez G., Dalton W. S., Jove R. (1999) Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity 10, 105–115 [DOI] [PubMed] [Google Scholar]
- 34. Chan K. S., Sano S., Kiguchi K., Anders J., Komazawa N., Takeda J., DiGiovanni J. (2004) Disruption of Stat3 reveals a critical role in both the initiation and the promotion stages of epithelial carcinogenesis. J. Clin. Invest. 114, 720–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chiarle R., Simmons W. J., Cai H., Dhall G., Zamo A., Raz R., Karras J. G., Levy D. E., Inghirami G. (2005) Stat3 is required for ALK-mediated lymphomutagenesis and provides a possible therapeutic target. Nat. Med. 11, 623–629 [DOI] [PubMed] [Google Scholar]
- 36. Pilati C., Amessou M., Bihl M. P., Balabaud C., Nhieu J. T., Paradis V., Nault J. C., Izard T., Bioulac-Sage P., Couchy G., Poussin K., Zucman-Rossi J. (2011) Somatic mutations activating STAT3 in human inflammatory hepatocellular adenomas. J. Exp. Med. 208, 1359–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kusaba T., Nakayama T., Yamazumi K., Yakata Y., Yoshizaki A., Nagayasu T., Sekine I. (2005) Expression of p-STAT3 in human colorectal adenocarcinoma and adenoma: correlation with clinicopathological factors. J. Clin. Pathol. 58, 833–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Park J. K., Hong R., Kim K. J., Lee T. B., Lim S. C. (2008) Significance of p-STAT3 expression in human colorectal adenocarcinoma. Oncol. Rep. 20, 597–604 [PubMed] [Google Scholar]
- 39. Rivat C., De Wever O., Bruyneel E., Mareel M., Gespach C., Attoub S. (2004) Disruption of STAT3 signaling leads to tumor cell invasion through alterations of homotypic cell-cell adhesion complexes. Oncogene 23, 3317–3327 [DOI] [PubMed] [Google Scholar]
- 40. Bollrath J., Phesse T. J., von Burstin V. A., Putoczki T., Bennecke M., Bateman T., Nebelsiek T., Lundgren-May T., Canli O., Schwitalla S., Matthews V., Schmid R. M., Kirchner T., Arkan M. C., Ernst M., Greten F. R. (2009) gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell cycle progression during colitis-associated tumorigenesis. Cancer Cell 15, 91–102 [DOI] [PubMed] [Google Scholar]
- 41. Grivennikov S., Karin E., Terzic J., Mucida D., Yu G. Y., Vallabhapurapu S., Scheller J., Rose-John S., Cheroutre H., Eckmann L., Karin M. (2009) IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 15, 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grivennikov S. I., Karin M. (2010) Dangerous liaisons: STAT3 and NF-κB collaboration and cross-talk in cancer. Cytokine Growth Factor Rev. 21, 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.