Backgound: p53 is accumulated and activated in response to influenza virus infection.
Results: p53 accumulation results from protein stabilization, which is associated with compromised Mdm2-mediated p53 ubiquitination. Viral nucleoprotein binds to p53 and impairs Mdm2-mediated p53 ubiquitination.
Conclusion: p53 stabilization results from compromised Mdm2-mediated p53 ubiquitination.
Significance: First time to demonstrate the mechanism of p53 stabilization in influenza virus-infected cells.
Keywords: Host-Pathogen Interactions, Influenza Virus, p53, Protein Stability, Ubiquitination, Murine Double Minute 2, Nucleoprotein
Abstract
Influenza A virus (IAV) induces apoptosis of infected cells. In response to IAV infection, p53, a tumor suppressor involved in regulating apoptosis and host antiviral defense, accumulates and becomes activated. This study was undertaken to examine the mechanism of p53 accumulation in IAV-infected cells. Here we show that p53 accumulation in IAV-infected cells results from protein stabilization, which was associated with compromised Mdm2-mediated ubiquitination of p53. In IAV-infected cells, p53 was stabilized and its half-life was remarkably extended. The ladders of polyubiquitinated p53 were not detectable in the presence of the proteasome inhibitor MG132 and were less sensitive to proteasome-mediated degradation. IAV infection did not affect the abundance of Mdm2, a major ubiquitin E3 ligase responsible for regulating p53 ubiquitination and degradation, but weakened the interaction between p53 and Mdm2. Viral nucleoprotein (NP) was able to increase the transcriptional activity and stability of p53. Furthermore, NP was found to associate with p53 and to impair the p53-Mdm2 interaction and Mdm2-mediated p53 ubiquitination, demonstrating its role in inhibiting Mdm2-mediated p53 ubiquitination and degradation.
Introduction
Influenza A virus (IAV)2 is a member of the Orthomyxoviridae family of RNA viruses and is highly infective in humans, causing ∼500,000 deaths worldwide per year (1), and many animal species. IAV can escape host immunity via antigenic drift and antigenic shift, leading to the emergence of new virulent strains, such as the pandemic H1N1 2009 strain (2) and the highly pathogenic H5N1 strain (3), that represent a serious threat to global public health. Understanding the factors that contribute to host antiviral defenses, virus replication and disease pathogenesis is critical to the development of better therapeutic strategies to combat IAV infection.
IAV is a cytolytic virus that induces apoptosis in numerous cell types (4). Apoptosis has been considered to be a host defense mechanism against viral infection in multi-cellular organisms (5). Previous studies suggested that apoptosis inhibits the replication of IAV (6). However, several groups have recently linked the apoptotic response in IAV-infected cells to the efficiency of virus replication (7, 8).
Tumor suppressor p53, a major cellular defense against tumor development, plays an important role in mediating apoptosis in response to stress, including virus infection (9). Several studies have demonstrated that p53 is accumulated and activated in IAV-infected cells and that it is essential for the induction of apoptosis in these cells (4, 10, 11). p53 is tightly regulated and maintained at low levels in unstressed cells. However, the levels of p53 increase dramatically in response to various types of stress including virus infection. The activation of p53 is generally regulated at the post-translational level. Although the precise mechanisms are not fully understood, they are generally thought to involve posttranslational modifications such as ubiquitination, acetylation, phosphorylation, sumoylation, neddylation, methylation, and glycosylation (12). Among these post-translational modifications, ubiquitination has been established to play a major role in p53 regulation (13).
Mdm2 is a RING finger domain-containing protein that exhibits E3 ubiquitin-protein ligase activity. It was first identified to regulate p53 stability and was later shown to be the major factor that effectively regulates p53 ubiquitination through its E3 ligase activity (13). In unstressed cells, Mdm2 binds to p53 and functions as an ubiquitin E3 ligase that maintains p53 at low levels by continuous proteasomal degradation (12). In response to stress, a decrease in Mdm2 protein levels, and (or) the interaction between Mdm2 and p53, leads to p53 stabilization and enhancement of p53 transcriptional activity (14).
Besides its well established role in protection against the development of cancer, p53 has recently been implicated in host antiviral defense (15–18). In response to IAV infection, p53 is accumulated and activated, and consequently transactivates its target genes responsible for regulating apoptosis and the innate immune response (10, 11). The inhibition of p53 activity in IAV-infected cells leads to increased viral titers (4). In addition, p53-mediated apoptosis may cause tissue damage and increase susceptibility to infection by bacterial pathogens, playing an important role in disease pathogenesis (19, 20). Therefore, exploring the basis of p53 activation would contribute to our understanding of its role in IAV infection and disease pathogenesis, and may aid the development of new therapeutic strategies. In this study, we investigated the mechanisms of p53 accumulation in IAV-infected cells.
EXPERIMENTAL PROCEDURES
Viruses and Cells
Influenza A/Swine/Jiangsu/2/2006 (H3N2 subtype), A/Swine/Guangdong/96/06 (H1N1 subtype), and A/Swine/Gangxi/7/07 (H9N2 subtype) viruses were propagated in the allantoic cavities of 9-day-old embryonated specific-pathogen-free chicken eggs or in Madin-Darby canine kidney (MDCK) cells. MDCK, Vero, CV-1, and p53-null H1299 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen), penicillin (100 units/ml), and streptomycin (100 mg/ml) in an atmosphere containing 5% CO2. For viral infection, cells were washed with phosphate-buffered saline (PBS) and infected with IAV at a multiplicity of infection (MOI) of 5 (for Vero, CV-1 and H1299 cells) or 1 (for MDCK cells) at 37 °C. After a 1 h adsorption period, the inocula were removed, and the cells were incubated with DMEM containing 1% FBS and 1 μg/ml TPCK trypsin at 37 °C. For mock-infection, the procedure was performed in an identical fashion to the viral infection using PBS as the inoculum.
Antibodies
The commercial antibodies used were an anti-p53 monoclonal antibody (DO-1, Santa Cruz Biotechnology, Santa Cruz, CA), an anti-p53 polyclonal antibody (FL-393, Santa Cruz Biotechnology), an anti-Mdm2 monoclonal antibody (SMP14, Santa Cruz Biotechnology), an anti-Mdm2 polyclonal antibody (c-18, Santa Cruz Biotechnology), an anti-GFP monoclonal antibody (ab1218, Abcam, Cambridge, MA), an anti-p300 polyclonal antibody (C-20, Santa Cruz Biotechnology), an anti-β-actin monoclonal antibody (AC-15, Sigma), an anti-Flag monoclonal antibody (M2, Sigma), an anti-His polyclonal antibody (sc-803, Santa Cruz Biotechnology), a horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (sc-2004, Santa Cruz Biotechnology) and goat anti-mouse IgG antibody (sc-2005, Santa Cruz Biotechnology). An anti-NS1 polyclonal antibody was generated in our laboratory.3
Drug Administration
To block protein synthesis of cells, cycloheximide (CHX) (Sigma), an inhibitor of protein biosynthesis, was added at 100 μg/ml to the media 16 h postinfection for Vero cells, 8 h postinfection for MDCK cells, and incubated for the indicated periods at 37 °C. To determine the effect of the proteasome inhibitor MG132 on the stability of p53, MG132 (Merck & Co, Whitehouse Station, NJ) was added at 10 μm to the media 12 h postinfection, and CHX was subsequently added at 100 μg/ml to the media 16 h postinfection, and incubated for the indicated periods at 37 °C. To detect the ubiquitination of p53 in IAV-infected cells, MG132 was added at 10 μm to the media 12 h postinfection and incubated for 6 h at 37 °C. To determine the effect of the Mdm2 inhibitor TIBC on p53 ubiquitination, the cells were treated with TIBC (Sigma) alone at 10 μm, or MG132 alone at 10 μm, or a combination of both at 12 h postinfection, and incubated for 6 h at 37 °C.
Plasmids and Transfection
p53-luciferase reporter plasmid (p53-Luc) and Renilla luciferase pRL-TK plasmid were purchased from Stratagene (San Diego, CA) and Promega (Madison, WI), respectively. Plasmid engineered to express GFP-tagged human p53 (GFP-p53) was constructed by inserting the full-length human p53 cDNA into pEGFP-C1 vector (BD Biosciences Clontech, Mountain View, CA). Plasmids engineered to express Flag-tagged human Mdm2 (Flag-Mdm2), viral NP (Flag-NP), viral M1 (Flag-M1), viral M2 (Flag-M2), viral NS1 (Flag-NS1), or viral NS2 (Flag-NS2) were constructed by inserting the indicated gene into p3xFLAG-CMV-7.1 vector (Sigma). Plasmid engineered to express His-tagged ubiquitin (His-ubiquitin) was constructed by inserting the ubiquitin cDNA into pcDNA3.1/His vector (Invitrogen). Plasmid engineered to express human p53 (pcDNA-p53) was constructed by inserting human p53 cDNA into pcDNA3(+) vector (Invitrogen). Cells were transfected using LipofectamineTM 2000 (Invitrogen), according to the manufacturer's protocol.
Reporter Gene Assay
MDCK cells grown in 35-mm plates were transiently co-transfected with 0.75 μg of p53-Luc reporter plasmid and 0.075 μg of control plasmid Renilla luciferase pRL-TK and incubated for 12 h. The transfectants were then mock-infected or infected with influenza A/Swine/Jiangsu/2/2006 at a MOI of 1 and incubated for the indicated periods. To analyze the effect of viral proteins on p53 transcriptional activity, Vero cells were transfected with the indicated expression plasmids in the presence of the p53-Luc reporter plasmid and the control plasmid Renilla luciferase pRL-TK. Transfectants were harvested 24 h post-transfection for luciferase assays. The luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega), according to the manufacturer's protocol. The firefly luciferase activity of individual cell lysates was normalized to Renilla luciferase activity.
Immunoprecipitation
H1299 cells grown in 100 mm plates were transiently transfected with 12 μg of GFP-p53 construct and incubated for 12 h. The transfectants were then mock-infected or infected with influenza A/Swine/Jiangsu/2/2006 at a MOI of 5 and incubated for 20 h. The cells were lysed in Nonidet P-40 lysis buffer (1% Nonidet P-40, 50 mm Tris, pH 8.0, 5 mm EDTA, 150 mm NaCl, 2 mg/ml leupeptin, 2 mg/ml aprotinin, 1 mm phenylmethanesulfonyl fluoride) and precleared by incubation with protein G-agarose beads (Sigma) for 1 h at 4 °C. The lysates were then incubated with anit-p53 or anti-Mdm2 antibodies on a rotating wheel overnight at 4 °C. Protein G-agarose beads were then added, and the mixtures were further incubated on a rotating wheel for 2 h at 4 °C. The agarose beads were pelleted and washed three times in Nonidet P-40 lysis buffer. Antibody-antigen complexes bound to the beads were eluted in SDS-PAGE sample buffer by boiling, resolved by SDS-PAGE, and analyzed by Western blot analysis with the appropriate antibodies. To analyze interaction between p53 and viral nucleoprotein (NP), H1299 cells were transiently co-transfected with plasmid GFP-p53 and Flag-NP and incubated for 24 h. The transfectants were lysed in the Nonidet P-40 lysis buffer and subjected to immunoprecipitation assay.
Ubiquitination Assay and Purification of His-tagged Ubiquitin Conjugates
H1299 cells grown in 100 mm plate were transiently co-transfected with plasmid His-ubiquitin (8 μg), pcDNA-p53 (4 μg) and Flag-Mdm2 (4 μg) in the presence or absence of plasmid Flag-NP (2 or 4 μg) and harvested 24 h post-transfection. The transfectants were treated with 15 μm MG132 for 3 h before harvest. The purification of His-tagged ubiquitin conjugates was performed as described previously (21). Briefly, the cells were lysed in a lysis buffer (6 m guanidinium-HCl, 0.1 m Na2HPO4-NaH2PO4, 0.01 m Tris-HCl pH 8.0, 5 mm imidazole, 10 mm β-mercaptoethanol). The lysates were mixed with Ni2+-NTA-agarose beads (Qiagen, Hilden, Germany) and incubated on a rotating wheel for 12 h at 4 °C. The beads were then washed as described previously (21) and incubated with an elution buffer (200 mm imidazole, 0.15 m Tris-HCl, pH 6.7, 30% glycerol, 0.72 m β-mercaptoethanol, 5% SDS) for 20 min at room temperature. The eluates were mixed with SDS-PAGE sample buffer and analyzed by Western blot analysis with the appropriate antibodies.
Bimolecular Fluorescence Complementation (BiFC) Assay
To construct expression plasmids for BiFC assay, plasmid expressing Venus fluorescent protein (VFP) was generated based on plasmid pEYFP-N1 (BD Biosciences Clontech) as described previously (22). Sequences encoding the N- (amino acids 1–173) and C- (amino acids 174–239) terminal fragments of VFP were fused by a short linker to human p53 (p53-VN) and Mdm2 gene (Mdm2-VC), respectively (23). The sequences encoding all fusion proteins were verified by DNA sequencing. H1299 cells grown in 35-mm plates were transiently co-transfected with plasmid p53-VN (0.7 μg), Mdm2-VC (0.7 μg), Flag-NP (1 or 2 μg), and pECFP vector (0.4 μg) expressing cyan fluorescent protein (CFP) (BD Biosciences Clontech) and incubated for 13 h. The fluorescence emission was imaged using a fluorescence microscopy, and the BiFC/CFP ratio analysis was similarly performed as described previously (24).
Western Blot Analysis
Western blot analysis was performed as described previously (25).
RESULTS
Accumulation and Activation of p53 Protein in IAV-infected cells
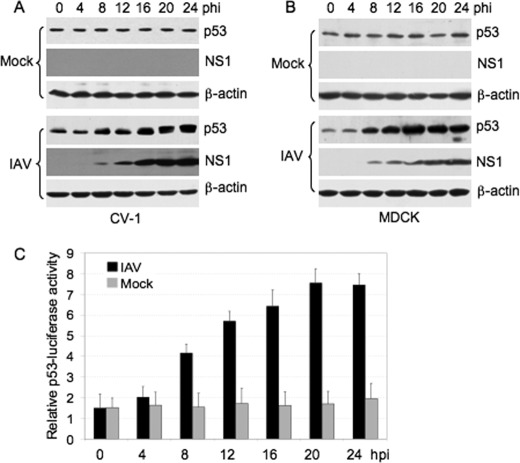
Three strains of IAV (influenza A/Swine/Jiangsu/2/2006 (H3N2 subtype), A/Swine/Guangdong/96/06 (H1N1 subtype) and A/Swine/Gangxi/7/07 (H9N2 subtype)) were used in this study. Because similar results were observed with all IAV strains, we only show the results obtained from strain A/Swine/Jiangsu/2/2006. Cells were infected with IAV at the indicated MOI and intracellular p53 was monitored by Western blot analysis. Viral nonstructural protein 1 (NS1) was used as an indicator of virus replication. Mock-infected cells revealed a relatively constant amount of p53 (Fig. 1, A and B, mock panels), whereas in the parallel cultures of IAV-infected cells, p53 levels increased (Fig. 1, A and B, IAV panels), consistent with the findings of previous studies (4, 10, 11).
FIGURE 1.
Accumulation and activation of p53 in IAV-infected cells. CV-1 (A) and MDCK (B) cells were mock-infected (mock) or infected with influenza A/Swine/Jiangsu/2/2006 (IAV) at a MOI of 5 and 1, respectively. The cells were collected at various time points (hours) postinfection (hpi) and subjected to Western blot analysis using the indicated antibodies. β-actin was used as a protein loading control. C, MDCK cells were transiently co-transfected with a p53-Luc reporter plasmid and a control plasmid Renilla luciferase pRL-TK, 12 h prior to infection. The transfectants were then mock-infected or infected with influenza A/Swine/Jiangsu/2/2006 at a MOI of 1. Cells were collected at the indicated times and subjected to luciferase activity analysis. The firefly luciferase activity of individual cell lysates was normalized to Renilla luciferase activity. Results are presented as the means ± S.E. from three independent experiments. The black and gray bars indicate IAV- and mock-infected cells, respectively.
To determine whether the increased p53 protein levels correlated with increased transcriptional activity, MDCK cells were transiently co-transfected with a p53-Luc reporter plasmid which contained the luciferase reporter gene driven by a basic promoter element joined to 14 repeats of the p53 binding site, and a control plasmid, Renilla luciferase pRL-TK, prior to IAV infection. The transfectants were then mock-infected or infected with IAV (MOI 1). The p53 luciferase activities of the IAV- or mock-infected cells were measured and normalized to Renilla luciferase activity. The activation of p53 was detected with kinetics similar to the pattern of p53 accumulation in IAV-infected cells, but not in mock-infected cells (Fig. 1C), suggesting that p53 was activated.
Proteasome-mediated Degradation of p53 Was Inhibited in IAV-infected Cells
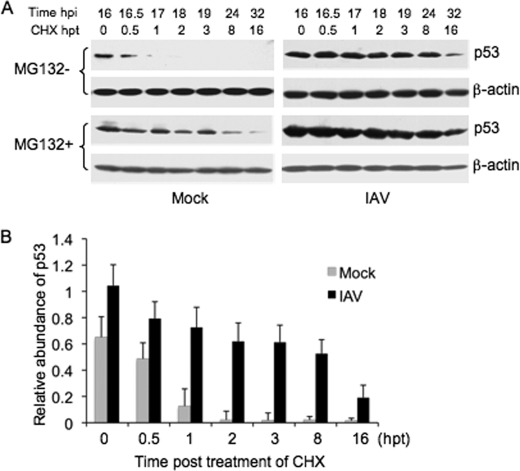
Previous studies demonstrated that the increase in p53 protein in IAV-infected cells was independent of increased transcription (4, 11) and it is known that certain types of viral infection affect the stability of p53 protein (26, 27). Therefore, in this study, we analyzed p53 stability in IAV-infected cells. To test whether the accumulation of p53 was dependent on the regulation of protein stability, CHX, an inhibitor of protein biosynthesis in eukaryotic cells, was administrated at 100 μg/ml to IAV- and mock-infected cells to block protein synthesis. The decay of p53 protein was monitored by Western blot analysis (Fig. 2A, MG132- panels). The half-life of p53 was ∼0.6 h in mock-infected cells, whereas in IAV-infected cells, it was remarkably extended (Fig. 2B). In contrast, the stability of β-actin showed no significant difference between the mock- and IAV-infected cells. These results indicated that p53 protein was stabilized in IAV-infected cells.
FIGURE 2.
Stabilization of p53 in IAV-infected cells. A, Vero cells were mock-infected (mock) or infected with influenza A/Swine/Jiangsu/2/2006 (IAV) at a MOI of 5. The cells were first mock-treated (MG132− panels) or treated with 10 μm of MG132 (MG132+ panels) at 12 h postinfection (hpi) and subsequently exposed to 100 μg/ml of CHX at 16 hpi. The cells were collected at the indicated times (hours) post-treatment (hpt) and subjected to Western blot analysis using the indicated antibodies. B, change in abundance of p53 after CHX treatment in MG132-untreated cells (MG132− panels) was determined by densitometric analysis and normalized to β-actin. The relative abundance of p53 at each hpt was plotted. Results are presented as the means ± S.E. from three independent experiments. The black and gray bars indicate the IAV- and mock-infected cells, respectively.
It is known that p53 is predominantly degraded through the ubiquitin-proteasome pathway (28). MG132 is an inhibitor of proteasome-mediated proteolysis (29) and has been used extensively as a tool for ubiquitin-proteasome pathway studies. Administration of MG132 (10 μm) to mock-infected cells resulted in an increase in the stabilization of p53 compared with MG132-untreated cells (Fig. 2A, mock panels), suggesting that ubiquitin-proteasome-mediated degradation was responsible for the short lifespan of p53 in mock-infected cells. In contrast, the p53 lifespan in IAV-infected cells in the presence of MG132 was similar to that in the absence of MG132 (Fig. 2A, IAV panels), suggesting that MG132 had no significant effect on the p53 lifespan in IAV-infected cells. Taken together, these results suggested that the proteasome-mediated degradation of p53 was substantially inhibited in IAV-infected cells.
Mdm2-mediated p53 Ubiquitination Was Impaired in IAV-infected Cells
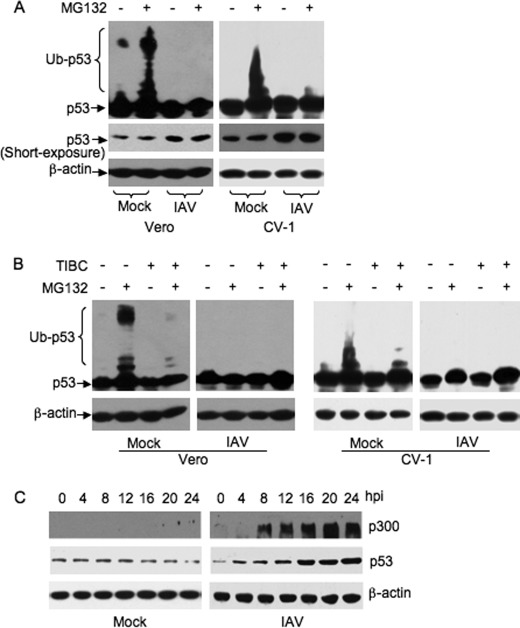
p53 is targeted for proteasome-mediated degradation by ubiquitination (13). To examine whether the stabilization of p53 in IAV-infected cells was a result of compromised p53 ubiquitination, cells were first infected with IAV and were then mock-treated or treated with MG132 (10 μm). The cells were harvested 6 h after MG132 treatment and subjected to Western blot analysis (Fig. 3A). The slowly migrating p53 bands corresponding to polyubiquitinated p53 (ladders of polyubiquitinated p53) were detected in the mock-infected cells treated with MG132, but not in the mock-treated cells (Fig. 3A, mock lanes). The ladders of polyubiquitinated p53 were not detectable in the absence of MG132, presumably because of the rapid degradation of ubiquitinated p53 (30). In contrast, the ladders of polyubiquitinated p53 were not detected in IAV-infected cells in the presence or absence of MG132 (Fig. 3A, IAV lanes). These results suggested that the ubiquitination of p53 was inhibited in IAV-infected cells.
FIGURE 3.
Detection of p53 ubiquitination and p300 in IAV-infected cells. Vero and CV-1 cells were mock-infected (mock) or infected with influenza A/Swine/Jiangsu/2/2006 (IAV) at a MOI of 5. A, cells were treated with 10 μm of MG132 or mock-treated at 12 h postinfection. The cell lysates were prepared 6 h after MG132 treatment and subjected to Western blot analysis using the indicated antibodies. To detect the ladders of polyubiquitinated p53 (Ub-p53), films were exposed ∼30 min longer than for the short-exposure p53 films. B, cells were treated with 10 μm TIBC alone, 10 μm MG132 alone, or a combination of both, or mock-treated, at 12 h postinfection. The cell lysates were prepared 6 h after treatment and subjected to Western blot analysis using the indicated antibodies. C, CV-1 cells were collected at the indicated time points postinfection (hpi) and subjected to Western blot analysis using the indicated antibodies.
Although Mdm2 is the major ubiquitin E3 ligase responsible for regulating p53 ubiquitination, Mdm2-independent ubiquitination also occurs and results in similar regulation of p53 (13). To determine whether Mdm2 was the major ubiquitin E3 ligase responsible for regulating p53 ubiquitination in the cells used in this study, we administrated 5 μm TIBC, a cell-permeable Mdm2 inhibitor that binds strongly to Mdm2 and irreversibly disrupts Mdm2-p53 protein complexes (27, 31), and measured its effect on p53 ubiquitination in mock- and IAV-infected cells. The ladders of polyubiquitinated p53 were detected in mock-infected cells in the presence of MG132 (Fig. 3B, mock panels), consistent with the results illustrated in Fig. 3A. However, when the mock-infected cells were treated with a combination of TIBC and MG132, only attenuated ladders of polyubiquitinated p53 were detectable (Fig. 3B, mock panels). The ladders of polyubiquitinated p53 were not detected in IAV-infected cells even in the presence of MG132 (Fig. 3B, IAV panels), confirming our findings shown in Fig. 3A. Taken together, these results demonstrated that Mdm2 was the major ubiquitin E3 ligase catalyzing p53 ubiquitination in the cells tested and that Mdm2-mediated p53 ubiquitination is inhibited in IAV-infected cells.
Mdm2 usually catalyzes p53 mono-ubiquitination, which is not a substrate for proteasome degradation (32). p300 is an ubiquitin conjugation factor E4 ligase that adds additional ubiquitin molecules to mono-ubiquitinated p53 (33). To determine whether IAV infection reduced cellular levels of p300 and consequently polyubiquitination of p53, we examined the abundance of p300 in IAV-infected cells by Western blot analysis. As shown in Fig. 3C, p300 was undetectable in mock-infected cells (mock panel), whereas p300 was detected at 8 h postinfection, and gradually increased in a pattern similar to that of p53 accumulation in IAV-infected cells (IAV panel). These results revealed that a lack of abundance of p300 was not a contributory factor in the compromised level of p53 ubiquitination.
Interaction between p53 and Mdm2 Was Weakened in IAV-infected Cells
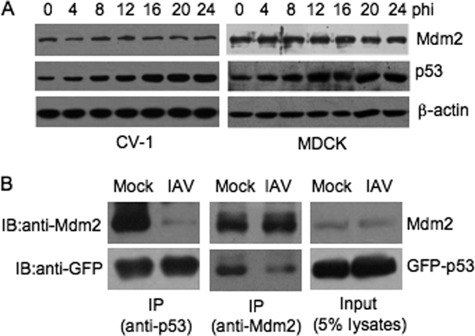
The decrease in p53 ubiquitination in IAV-infected cells could result from several different mechanisms that are responsible for the inhibition of the ubiquitin pathway. Because Mdm2 was the major ubiquitin E3 ligase in the cells used in this study (Fig. 3), we speculated that Mdm2 was involved in the down-regulation of p53 ubiquitination in IAV-infected cells. We first examined the cellular abundance of Mdm2 by Western blot analysis because a decrease in Mdm2 protein levels leads to p53 stabilization (14). In contrast to the observed gradual increase in abundance of p53, Mdm2 abundance was relatively constant. No remarkable changes in Mdm2 protein levels were detected during IAV infection (Fig. 4A), suggesting that the compromised p53 ubiquitination was not related to decreased abundance of Mdm2. In addition, the sequestration of Mdm2 to a specific cellular compartment, which results in p53 and Mdm2 being localized in distinct subcellular compartments and the consequent impairment of Mdm2-mediated p53 ubiquitination (14), was not observed by immunofluorescent analysis (data not shown).
FIGURE 4.
Detection of the abundance of Mdm2 and the interaction between p53 and Mdm2 in IAV-infected cells. A, CV-1 and MDCK cells were infected with influenza A/Swine/Jiangsu/2/2006 (IAV) at a MOI of 5 and 1, respectively. The cells were collected at the indicated time points (hours) postinfection (hpi) and subjected to Western blot analysis using the indicated antibodies. B, H1299 cells were transiently transfected with a GFP-fused p53 construct (GFP-p53) 12 h prior to IAV infection. The transfectants were then mock-infected (mock) or infected with influenza A/Swine/Jiangsu/2/2006 (IAV) at a MOI of 5 and incubated for 20 h. The cell lysates were prepared and subjected to co-immunoprecipitation using either anti-p53 antibody (FL393) (left panels) or anti-Mdm2 antibody (SMP14) (middle panels). The co-immunoprecipitates were then immunoblotted with anti-GFP and anti-Mdm2 antibodies. The cell lysates (5% of the amount used for immunoprecipitation) were included as a loading control (right panels). IP, immunoprecipitation. IB, Western blot.
Mdm2-mediated p53 ubiquitination is dependent on the binding of Mdm2 to p53 (34). Disruption of the interaction between p53 and Mdm2 results in de-ubiquitination and stabilization of p53 (14). We therefore examined whether the interaction between p53 and Mdm2 was impaired during IAV infection by transiently transfecting H1299 cells with a GFP-fused p53 construct (GFP-p53) 12 h prior to IAV infection. The transfectants were then infected with IAV (MOI 5) and harvested 20 h postinfection and used in a co-immunoprecipitation assay with anti-p53 or anti-Mdm2 antibody. The advantage of this approach is that it distinguishes the exogenously expressed GFP-p53 from the immunoglobulin G heavy chain that migrates close to native p53. The abundance of GFP-p53 or Mdm2 in mock-infected cells was similar to that in IAV-infected cells (Fig. 4B, input panel). The amount of Mdm2 co-immunoprecipitated from the lysates of IAV-infected cells by anti-p53 antibody, however, was less than that from the lysates of mock-infected cells (Fig. 4B, anti-p53 panel). Similarly, the amount of GFP-p53 co-immunoprecipitated from the lysates of IAV-infected cells by anti-Mdm2 antibody was less than that from the lysates of mock-infected cells (Fig. 4B, anti-Mdm2 panel). These results indicated that the interaction between p53 and Mdm2 was weakened in IAV-infected cells.
Viral Nucleoprotein Increased the Transcriptional Activity and Stability of p53 and Impaired p53 Ubiquitination
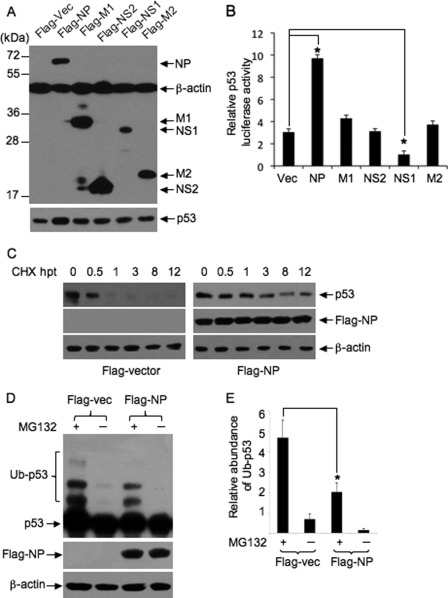
It is known that viral proteins of certain viruses are capable of impairing Mdm2-mediated p53 ubiquitination and subsequently inducing p53 stabilization (9, 35). To identify viral protein(s) responsible for the stabilization of p53 in IAV-infected cells, several viral proteins, such as nucleoprotein (NP), NS1, nonstructural protein 2 (NS2), matrix protein 1 (M1), and matrix protein 2 (M2), were transiently expressed as Flag-tagged fusion proteins in Vero cells in the presence of p53-Luc reporter plasmid, and their effect on p53 transcriptional activity was determined by luciferase assay. The expression of Flag-tagged proteins was confirmed by Western blot analysis (Fig. 5A). The expression of Flag-M1, -M2, and -NS2 had no significant effect on p53 luciferase activity, whereas the expression of Flag-NP significantly increased p53 luciferase activity (∼3-fold), compared with Flag-vector (Fig. 5B). The p53 luciferase activity was reduced significantly in the cells transfected with Flag-NS1, which was consistent with our previous observation (36). These data suggested that viral NP increased the transcriptional activity of p53.
FIGURE 5.
Analysis of the effect of viral NP on p53 stability. Vero cells were transiently transfected with plasmid Flag-NP, Flag-M1, Flag-NS2, Flag-NS1, Flag-M2 or Flag-vector (Vec) in the presence of p53-Luc reporter plasmid and incubated for 24 h. A, protein expression was detected by Western blot analysis using the indicated antibodies. B, p53 luciferase activity of lysates prepared from the transfectants was analyzed. Results are presented as the means ± S.E. from three independent experiments. *, p < 0.05 compared with cells transfected with the empty vector. C, Vero cells were transiently transfected with plasmid Flag-NP or Flag-vector and incubated for 24 h. The transfectants were treated with CHX at 100 μg/ml for the indicated times (hours) post-treatment (hpt) and subjected to Western blot analysis using the indicated antibodies. D, Vero cells were transiently transfected with plasmid Flag-NP or Flag-vector (Vec) and incubated for 24 h. The transfectants were treated with 10 μm of MG132 or mock-treated for 6 h and subjected to Western blot analysis using the indicated antibodies. To detect the ladders of polyubiquitinated p53 (Ub-p53), films were exposed for longer. E, change in abundance of polyubiquitinated p53 (Ub-p53) in the transfectants was determined by densitometric analysis and normalized to β-actin. The relative abundance of Ub-p53 was plotted. Results are presented as the means ± S.E. from three independent experiments. *, p < 0.05 compared with cells transfected with Flag-vector.
Because NP is able to increase the transcriptional activity of p53 (Fig. 5B), we detected the stability of p53 in Flag-NP-transfected cells. The expression of Flag-NP resulted in an increase in p53 protein levels in the transfected cells compared with other transfectants (Fig. 5A). When the transfectants were treated with the protein biosynthesis inhibitor CHX, p53 lifespan was extended remarkably in Flag-NP-transfected cells, compared with that in Flag-vector-transfected cells (Fig. 5C), suggesting that p53 was stabilized in Flag-NP-transfected cells. We next examined whether the stabilization of p53 in Flag-NP-transfected cells was a result of compromised p53 ubiquitination. The Flag-NP-transfected cells were treated with MG132 (10 μm) and the ubiquitinated p53 was detected by Western blot analysis (Fig. 5D). Ladders of polyubiquitinated p53 were detected in Flag-vector-transfected cells treated with MG132, whereas, the intensity of these ladders was significantly attenuated in Flag-NP-transfected cells (Fig. 5, D and E), suggesting that p53 ubiquitination was impaired by viral NP expression.
Mdm2-mediated p53 Ubiquitination Was Attenuated by Viral Nucleoprotein
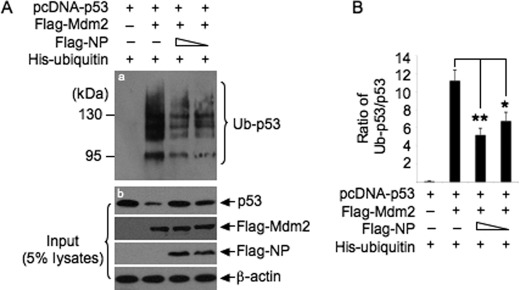
Given that Mdm2 is the major ubiquitin E3 ligase responsible for regulating p53 ubiquitination in the cells used in our experiments (Fig. 3), we detected whether viral NP is able to affect Mdm2-mediated p53 ubiquitination. H1299 cells were transiently transfected with a combination of plasmids expressing human p53 (pcDNA-p53), Flag-tagged Mdm2 (Flag-Mdm2), or Flag-NP in the presence of plasmid expressing His-ubiquitin. Ubiquitinated p53 in the transfectants was pulled down using Ni2+-NTA-agarose beads and detected by Western blot analysis with anti-p53 antibody (DO-1). The co-expression of pcDNA-p53 with Flag-Mdm2 resulted in an increase of ubiquitinated p53 (Fig. 6A, panel a) and a decrease of non-ubiquitinated p53 (Fig. 6A, panel b) at the protein level, compared with the expression of pcDNA-p53 alone, suggesting that Mdm2 promotes the ubiquitination and subsequent degradation of p53. However, when pcDNA-p53 was co-expressed with Flag-Mdm2 and Flag-NP, the levels of ubiquitinated p53 were decreased and the levels of non-ubiquitinated p53 were increased, compared with those in the cells transfected with a combination of pcDNA-p53 and Flag-Mdm2 (Fig. 6A). The change in abundance of ubiquitinated p53 in the transfectants was further determined by densitometric analysis. A significant decrease in ubiquitinated p53 protein levels was detected in the cells transfected with a combination of pcDNA-p53, Flag-Mdm2, and Flag-NP, compared with the cells transfected with a combination of pcDNA-p53 and Flag-Mdm2 (Fig. 6B). These data suggested that Mdm2-mediated p53 ubiquitination was attenuated by viral NP expression.
FIGURE 6.
Analysis of the effect of viral NP on Mdm2-mediated p53 ubiquitination. H1299 cells were transiently transfected with a combination of the indicated plasmids and incubated for 24 h. The transfectants were treated with 15 μm MG132 for 3 h before harvest. A, ubiquitinated p53 (Ub-p53) in the transfectants was pulled down using Ni2+-NTA-agarose beads and detected by Western blot analysis with anti-p53 antibody (DO-1) (panel a). The protein expression in the transfectants was detected by Western blot analysis using the indicated antibodies (input panels). B, change in abundance of Ub-p53 in the transfectants was determined by densitometric analysis. The ratio between ubiquitinated and non-ubiquitinated p53 (Ub-p53/p53) was plotted. Results are presented as the means ± S.E. from three independent experiments. *, p < 0.05, **, p < 0.01 compared with cells transfected with pcDNA-p53, Flag-Mdm2, and His-ubiquitin.
Viral Nucleoprotein Binds to p53 and Impairs Interaction between p53 and Mdm2
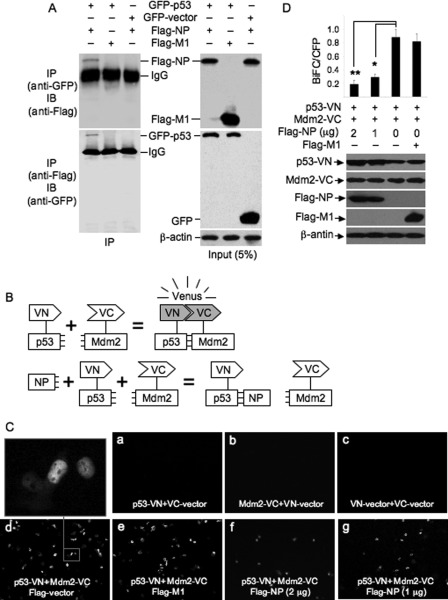
To explore the mechanism of how viral NP impairs Mdm2-mediated p53 ubiquitination, we first detected whether viral NP binds to p53. H1299 cells were transiently co-transfected with plasmids expressing GFP-p53 and Flag-NP, and subjected to a co-immunoprecipitation assay with anti-GFP or anti-Flag antibodies. Plasmid expressing Flag-M1 and GFP-vector were used as control. The protein expression in the transfectants was confirmed by Western blot analysis (Fig. 7A, right panels). GFP-p53 immunoprecipitated Flag-NP, but did not pulldown Flag-M1 (Fig. 7A, left panels). A reverse co-immunoprecipitation assay was performed using anti-Flag antibody. Flag-NP, but not Flag-M1, immunoprecipitated GFP-p53. No GFP was immunoprecipitated by anti-Flag antibody (Fig. 7A, left panels). We also performed a co-immunoprecipitation assay to detect whether viral NP interacts with Mdm2. No interaction between viral NP and Mdm2 was detected (data not shown). These data suggested that viral NP was able to interact with p53.
FIGURE 7.
Detection of viral NP-p53 interaction and BiFC analysis of the effect of viral NP on interaction between p53 and Mdm2. A, H1299 cells were transiently transfected with a combination of the indicated plasmids and incubated for 16 h. The cell lysates prepared from the transfectants were subjected to co-immunoprecipitation assay using anti-GFP and anti-Flag antibodies. The immunoprecipitates were immunoblotted with the indicated antibodies (left panels). The cell lysates were included as a loading control (right panels). IP, immunoprecipitation. IB, Western blot. B, schematic illustration of BiFC assay of interaction between p53 and Mdm2. VN and VC are the N- and C-terminal fragments of Venus fluorescent protein, respectively. NP, viral nucleoprotein. C, H1299 cells were transiently transfected with a combination of the indicated plasmids and incubated for 13 h. The fluorescence emission was detected using a fluorescence microscope. D, H1299 cells were transiently transfected with a combination of the indicated plasmids and incubated for 13 h. pECFP vector expressing cyan fluorescent protein (CFP) was co-transfected as transfection control. The protein expression in the transfectants was detected by Western blot analysis using the indicated antibodies. The BiFC efficiencies (BiFC/CFP) in the transfectants were quantified by normalizing the number of Venus-fluorescence-positive cells to the number of CFP-positive cells. The number of cells in ten randomly selected visual fields was counted. Results are presented as the means ± S.E. from three independent experiments. *, p < 0.05; **, p < 0.01 compared with cells transfected with p53-VN and Mdm2-VC.
Because viral NP interacts with p53, we performed a BiFC assay, which allows detection of protein-protein interaction in vivo without disturbance of the cellular environment (24), to detect whether viral NP interferes with the interaction between p53 and Mdm2. In this assay, the N-terminal region (amino acids 1–173) defined as VN and the C-terminal region (amino acids 174–239) defined as VC of VFP, which were fused by a short linker to the C terminus of human p53 (p53-VN) and Mdm2 (Mdm2-VC), respectively, can complement and reconstitute a functional fluorophore because of the interaction between p53 and Mdm2. In contrast, the functional fluorophore cannot be reconstituted if viral NP impairs the interaction between p53 and Mdm2 (Fig. 7B). H1299 cells were transiently transfected with a combination of expression plasmids (Fig. 7, C and D) and the fluorescence emission was imaged using a fluorescence microscopy. No fluorescence emission was detected in the cells transfected with a combination of p53-VN and non-fused VC (VC-vector), Mdm2-VC and non-fused VN (VN-vector), or VN-vector and VC-vector (Fig. 7C, panels a, b, and c). However, the cells co-transfected with plasmids expressing p53-VN and Mdm2-VC emitted fluorescence (Fig. 7C, panel d), suggesting that the interaction between p53-VN and Mdm2-VC occurred. The co-expression of p53-VN and Mdm2-VC, or Flag-M1 and p53-VN and Mdm2-VC resulted in a large number of cells with strong fluorescence (Fig. 7C, panels d and e). In contrast, only a small number of cells emitted fluorescence when Flag-NP was co-expressed with p53-VN and Mdm2-VC (Fig. 7C, panels f and g). The BiFC efficiencies (BiFC/CFP) were significantly reduced in the cells transfected with Flag-NP, p53-VN, and Mdm2-VC, compared with the cells transfected with p53-VN and Mdm2 (Fig. 7D). Taken together, these data indicated that viral NP interacted with p53 and impaired the interaction between p53 and Mdm2.
DISCUSSION
Activation of p53 is generally regulated at the post-translational level. Its activation comprises three basic steps: stabilization, sequence-specific DNA binding, and transactivation of target genes (9). In response to IAV infection, p53 is accumulated and activated (4, 10, 11). In this study, we confirmed that IAV infection results in the accumulation and activation of p53 (Fig. 1) and found that the accumulation of p53 was a result of protein stabilization (Fig. 2). Protein decay studies in CHX-treated cells indicated that the p53 lifespan was remarkably extended in IAV-infected cells, compared with mock-infected cells. The view that p53 accumulation results from its stabilization in IAV-infected cells was further supported by the observation that the ladders of polyubiquitinated p53 were not detected in IAV-infected cells, even in the presence of MG132 (Fig. 3, A and B).
Although the precise mechanisms underlying p53 stabilization are not fully understood, they are generally thought to involve post-translational modifications (12). Ubiquitination, one form of post-translational modification, is well established to play a major role in regulating p53 stabilization (13). The ubiquitin-proteasome pathway was intact in the cells used in this study (Fig. 2), however, we found that the ladders of polyubiquitinated p53 were not detectable in IAV-infected cells (Fig. 3, A and B), suggesting that p53 ubiquitination was inhibited during IAV infection. We therefore consider that the compromised ubiquitination of p53 contributes to its stabilization in IAV-infected cells.
Mdm2 is the major ubiquitin E3 ligase responsible for regulating p53 ubiquitination (13). We therefore speculated that Mdm2 was involved in down-regulation of p53 ubiquitination and found that Mdm2-mediated p53 ubiquitination is decreased in IAV-infected cells (Fig. 3B). It is known that a decrease in Mdm2 protein levels, disruption of the interaction between p53 and Mdm2, or sequestration of Mdm2 to a specific cellular compartment, can lead to p53 stabilization and enhancement of p53 transcriptional activity (14). However, neither a lack of abundance (Fig. 4A) nor altered subcellular distribution of Mdm2 (data not shown) was observed in IAV-infected cells compared with mock-infected cells, suggesting that the compromised ubiquitination of p53 did not result from decreased Mdm2 abundance or sequestration of Mdm2 to a specific cellular compartment. However, when we analyzed the binding affinity between p53 and Mdm2, a weakened interaction between p53 and Mdm2 was observed (Fig. 4B). These results suggested that the compromised p53 ubiquitination was a result of the weakened interaction between p53 and Mdm2. To exclude the possibility that the compromised p53 ubiquitination resulted from a lack of abundance of p300, an ubiquitin conjugation factor E4 ligase that adds additional ubiquitin molecules to mono-ubiquitinated p53 (33), we examined p300 abundance in IAV-infected cells. In contrast to the undetectable level of p300 in mock-infected cells, p300 was detected at 8 h postinfection in IAV-infected cells with gradually increasing levels (Fig. 3C). These results revealed that the compromised p53 ubiquitination was not due to a lack of abundance of p300.
It is known that viral proteins of certain viruses are capable of impairing Mdm2-mediated p53 ubiquitination and subsequently inducing p53 stabilization (9, 35). We analyzed the effect of several viral proteins of IAV on the transcriptional activity of p53 and found that viral NP was able to increase the transcriptional activity and stability of p53 (Fig. 5, B and C). Furthermore, NP was found to associate with p53 (Fig. 7A) and to impair the Mdm2-mediated p53 ubiquitination (Fig. 6) and the interaction between p53 and Mdm2 (Fig. 7, C and D). These findings demonstrated that viral NP is involved in the activation and stabilization of p53. NP is a multifunctional protein that interacts with a wide variety of viral and cellular macromolecules and plays an important role at many steps of the virus life cycle, such as viral RNA transcription, replication, and packaging (37). Although the activation of p53 during IAV infection has been described (4, 10, 11), the viral components responsible for p53 activation have not been identified. We first demonstrated that the viral NP is involved in p53 activation and stabilization. In addition, in contrast to viral NP that increases the transcriptional activity of p53, viral NS1 was found to inhibit the transcriptional activity of p53 (36, 38). It will be interesting to know how these two proteins cooperatively regulate p53 activity during IAV infection.
In conclusion, we observed that p53 was stabilized and its half-life was remarkably extended in IAV-infected cells. The stabilization of p53 was a result of compromised Mdm2-mediated ubiquitination of p53, which was associated with a weakened interaction between p53 and Mdm2. Viral NP was able to increase the transcriptional activity and stability of p53. Furthermore, NP was found to associate with p53 and to impair the interaction between p53 and Mdm2 and Mdm2-mediated p53 ubiquitination, showing its involvement in inhibiting Mdm2-mediated p53 ubiquitination and degradation.
It should be mentioned here that the precise mechanisms underlying Mdm2-mediated p53 stabilization are not fully understood (13), and the mechanisms responsible for the stabilization and activation of p53 in IAV-infected cells are likely to be complex. The findings presented here gain some insight into these mechanisms. It is possible that other cellular protein(s) or viral protein(s) are involved in the stabilization and activation of p53.
Acknowledgments
We thank the Key Open Laboratory of Animal Parasitology, Ministry of Agriculture of China, for the provision of laboratory equipment.
This work was sponsored by the National Natural Science Foundation of China (No. 30970141 and No. 81171547) and the Natural Science Foundation of Shanghai (No. 10JC1417300).
Z. Ma, unpublished data.
- IAV
- influenza A virus
- NP
- nucleoprotein
- Mdm2
- murine double minute 2
- TIBC
- trans-4-iodo, 4′-boranyl-chalcone
- MDCK
- Madin-Darby canine kidney
- MOI
- multiplicity of infection
- CHX
- cycloheximide
- BiFC
- bimolecular fluorescence complementation.
REFERENCES
- 1. Smith D. J., Lapedes A. S., de Jong J. C., Bestebroer T. M., Rimmelzwaan G. F., Osterhaus A. D., Fouchier R. A. (2004) Mapping the antigenic and genetic evolution of influenza virus. Science 305, 371–376 [DOI] [PubMed] [Google Scholar]
- 2. Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team (2009) Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360, 2605–2615 [DOI] [PubMed] [Google Scholar]
- 3. Centers for Disease Control and Prevention (1998) Update: isolation of avian influenza A(H5N1) viruses from humans–Hong Kong, 1997–1998. MMWR Morb. Mortal. Wkly. Rep. 46, 1245–1247 [PubMed] [Google Scholar]
- 4. Turpin E., Luke K., Jones J., Tumpey T., Konan K., Schultz-Cherry S. (2005) Influenza virus infection increases p53 activity: role of p53 in cell death and viral replication. J. Virol. 79, 8802–8811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Koyama A. H., Adachi A., Irie H. (2003) Physiological significance of apoptosis during animal virus infection. Int. Rev. Immunol. 22, 341–359 [DOI] [PubMed] [Google Scholar]
- 6. Kurokawa M., Koyama A. H., Yasuoka S., Adachi A. (1999) Influenza virus overcomes apoptosis by rapid multiplication. Int. J. Mol. Med. 3, 527–530 [DOI] [PubMed] [Google Scholar]
- 7. Wurzer W. J., Planz O., Ehrhardt C., Giner M., Silberzahn T., Pleschka S., Ludwig S. (2003) Caspase 3 activation is essential for efficient influenza virus propagation. EMBO J. 22, 2717–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McLean J. E., Datan E., Matassov D., Zakeri Z. F. (2009) Lack of Bax prevents influenza A virus-induced apoptosis and causes diminished viral replication. J. Virol. 83, 8233–8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Collot-Teixeira S., Bass J., Denis F., Ranger-Rogez S. (2004) Human tumor suppressor p53 and DNA viruses. Rev. Med. Virol. 14, 301–319 [DOI] [PubMed] [Google Scholar]
- 10. Zhirnov O. P., Klenk H. D. (2007) Control of apoptosis in influenza virus-infected cells by up-regulation of Akt and p53 signaling. Apoptosis 12, 1419–1432 [DOI] [PubMed] [Google Scholar]
- 11. Shen Y., Wang X., Guo L., Qiu Y., Li X., Yu H., Xiang H., Tong G., Ma Z. (2009) Influenza A virus induces p53 accumulation in a biphasic pattern. Biochem. Biophys. Res. Commun. 382, 331–335 [DOI] [PubMed] [Google Scholar]
- 12. Brooks C. L., Gu W. (2006) p53 ubiquitination: Mdm2 and beyond. Mol. Cell 21, 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee J. T., Gu W. (2010) The multiple levels of regulation by p53 ubiquitination. Cell Death. Differ. 17, 86–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marine J. C., Lozano G. (2010) Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death. Differ. 17, 93–102 [DOI] [PubMed] [Google Scholar]
- 15. Takaoka A., Hayakawa S., Yanai H., Stoiber D., Negishi H., Kikuchi H., Sasaki S., Imai K., Shibue T., Honda K., Taniguchi T. (2003) Integration of interferon-α/β signaling to p53 responses in tumor suppression and antiviral defense. Nature 424, 516–523 [DOI] [PubMed] [Google Scholar]
- 16. Munoz-Fontela C., Garcia M. A., Garcia-Cao I., Collado M., Arroyo J., Esteban M., Serrano M., Rivas C. (2005) Resistance to viral infection of super p53 mice. Oncogene 24, 3059–3062 [DOI] [PubMed] [Google Scholar]
- 17. Muñoz-Fontela C., Macip S., Martínez-Sobrido L., Brown L., Ashour J., García-Sastre A., Lee S. W., Aaronson S. A. (2008) Transcriptional role of p53 in interferon-mediated antiviral immunity. J. Exp. Med. 205, 1929–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dharel N., Kato N., Muroyama R., Taniguchi H., Otsuka M., Wang Y., Jazag A., Shao R. X., Chang J. H., Adler M. K., Kawabe T., Omata M. (2008) Potential contribution of tumor suppressor p53 in the host defense against hepatitis C virus. Hepatology 47, 1136–1149 [DOI] [PubMed] [Google Scholar]
- 19. Ramphal R., Small P. M., Shands J. W., Jr., Fischlschweiger W., Small P. A., Jr. (1980) Adherence of Pseudomonas aeruginosa to tracheal cells injured by influenza infection or by endotracheal intubation. Infect. Immun. 27, 614–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brydon E. W., Morris S. J., Sweet C. (2005) Role of apoptosis and cytokines in influenza virus morbidity. FEMS Microbiol. Rev. 29, 837–850 [DOI] [PubMed] [Google Scholar]
- 21. Xirodimas D., Saville M. K., Edling C., Lane D. P., Laín S. (2001) Different effects of p14ARF on the levels of ubiquitinated p53 and Mdm2 in vivo. Oncogene. 20, 4972–4983 [DOI] [PubMed] [Google Scholar]
- 22. Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., Miyawaki A. (2002) A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87–90 [DOI] [PubMed] [Google Scholar]
- 23. Shyu Y. J., Liu H., Deng X., Hu C. D. (2006) Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. BioTechniques 40, 61–66 [DOI] [PubMed] [Google Scholar]
- 24. Hu C. D., Chinenov Y., Kerppola T. K. (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 9, 789–798 [DOI] [PubMed] [Google Scholar]
- 25. Qiu Y., Shen Y., Li X., Ding C., Ma Z. (2008) Molecular cloning and functional characterization of a novel isoform of chicken myeloid differentiation factor 88 (MyD88). Dev. Comp. Immunol. 32, 1522–1530 [DOI] [PubMed] [Google Scholar]
- 26. Takemoto M., Mori Y., Ueda K., Kondo K., Yamanishi K. (2004) Productive human herpesvirus 6 infection causes aberrant accumulation of p53 and prevents apoptosis. J. Gen. Virol. 85, 869–879 [DOI] [PubMed] [Google Scholar]
- 27. Chen Z., Knutson E., Wang S., Martinez L. A., Albrecht T. (2007) Stabilization of p53 in human cytomegalovirus-initiated cells is associated with sequestration of HDM2 and decreased p53 ubiquitination. J. Biol. Chem. 282, 29284–29295 [DOI] [PubMed] [Google Scholar]
- 28. Vogelstein B., Lane D., Levine A. J. (2000) Surfing the p53 network. Nature 408, 307–310 [DOI] [PubMed] [Google Scholar]
- 29. Saito Y., Tsubuki S., Ito H., Kawashima S. (1990) The structure-function relationship between peptide aldehyde derivatives on initiation of neurite outgrowth in PC12h cells. Neurosci. Lett. 120, 1–4 [DOI] [PubMed] [Google Scholar]
- 30. Yang Y., Li C. C., Weissman A. M. (2004) Regulating the p53 system through ubiquitination. Oncogene 23, 2096–2106 [DOI] [PubMed] [Google Scholar]
- 31. Kumar S. K., Hager E., Pettit C., Gurulingappa H., Davidson N. E., Khan S. R. (2003) Design, synthesis, and evaluation of novel boronic-chalcone derivatives as antitumor agents. J. Med. Chem. 46, 2813–2815 [DOI] [PubMed] [Google Scholar]
- 32. Moll U. M., Petrenko O. (2003) The MDM2-p53 interaction. Mol. Cancer Res. 1, 1001–1008 [PubMed] [Google Scholar]
- 33. Grossman S. R., Deato M. E., Brignone C., Chan H. M., Kung A. L., Tagami H., Nakatani Y., Livingston D. M. (2003) Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science 300, 342–344 [DOI] [PubMed] [Google Scholar]
- 34. Xirodimas D. P., Stephen C. W., Lane D. P. (2001) Cocompartmentalization of p53 and Mdm2 is a major determinant for Mdm2-mediated degradation of p53. Exp. Cell Res. 270, 66–77 [DOI] [PubMed] [Google Scholar]
- 35. Yang M. R., Lee S. R., Oh W., Lee E. W., Yeh J. Y., Nah J. J., Joo Y. S., Shin J., Lee H. W., Pyo S., Song J. (2008) West Nile virus capsid protein induces p53-mediated apoptosis via the sequestration of HDM2 to the nucleolus. Cell Microbiol. 10, 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang X., Shen Y., Qiu Y., Shi Z., Shao D., Chen P., Tong G., Ma Z. (2010) The non-structural (NS1) protein of influenza A virus associates with p53 and inhibits p53-mediated transcriptional activity and apoptosis. Biochem. Biophys. Res. Commun. 395, 141–145 [DOI] [PubMed] [Google Scholar]
- 37. Portela A., Digard P. (2002) The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J. Gen. Virol. 83, 723–734 [DOI] [PubMed] [Google Scholar]
- 38. Li W., Wang G., Zhang H., Xin G., Zhang D., Zeng J., Chen X., Xu Y., Cui Y., Li K. (2010) Effects of NS1 variants of H5N1 influenza virus on interferon induction, TNFalpha response and p53 activity. Cell. Mol. Immunol. 7, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]