Background: Glutaminase C (GAC) is down-regulated by diphenylarsinic acid (DPAA).
Results: DPAA promotes the conformational changes of GAC, and unfolded GAC is degraded by mitochondrial Lon protease.
Conclusion: Destruction of tetrameric GAC by DPAA may be a trigger of the proteolysis process induced by Lon protease.
Significance: Lon protease plays an important role in preventing the accumulation of chemically damaged proteins.
Keywords: Mitochondria, Molecular Chaperone, Protease Inhibitor, Protein Assembly, Protein Chemical Modification, Protein Degradation, Protein Denaturation, Diphenylarsinic acid, Glutaminase C, Lon Protease
Abstract
Glutaminase C (GAC), a splicing variant of the kidney-type glutaminase (KGA) gene, is a vital mitochondrial enzyme protein that catalyzes glutamine to glutamate. Earlier studies have shown that GAC proteins in the human hepatocarcinoma cell line, HepG2, were down-regulated by diphenylarsinic acid (DPAA), but the mechanism by which DPAA induced GAC protein down-regulation remained poorly understood. Here, we showed that DPAA promoted GAC protein degradation without affecting GAC transcription and translation. Moreover, DPAA-induced GAC proteolysis was mediated by mitochondrial Lon protease. DPAA insolubilized 0.5% Triton X-100-soluble GAC protein and promoted the accumulation of insoluble GAC in Lon protease knockdown cells. DPAA destroyed the native tetrameric GAC conformation and promoted an increase in the unassembled form of GAC when DPAA was incubated with cell extracts. Decreases in the tetrameric form of GAC were observed in cells exposed to DPAA, and decreases occurred prior to a decrease in total GAC protein levels. In addition, decreases in the tetrameric form of GAC were observed independently with Lon protease. Mitochondrial heat shock protein 70 is known to be an indispensable protein that can bind to misfolded proteins, thereby supporting degradation of proteins sensitive to Lon protease. When cells were incubated with DPAA, GAC proteins that can bind with mtHsp70 increased. Interestingly, the association of mtHsp70 with GAC protein increased when the tetrameric form of GAC was reduced. These results suggest that degradation of native tetrameric GAC by DPAA may be a trigger in GAC protein degradation by Lon protease.
Introduction
Diphenylarsinic acid (DPAA2 (V)) was detected in ground water after a poisonous incident in Kamisu, Japan. Patients showed central nervous system dysfunctions, such as ataxic gait, titubation, scanning speech, and myoclonus as cerebellar and brain symptoms, and visual impairments, insomnia, nightmares, memory impairment, and mental retardation in some children as cerebral symptoms (1).
An approach to define the toxic target molecules of DPAA(V) by a high throughput analysis of proteins from cultured human cells demonstrated down-regulation of glutaminase C (GAC) (2). Glutaminase (EC 3.5.1.2) is an enzyme that catalyzes the hydrolysis of glutamine to glutamate and plays a fundamental role in several physiological processes, such as renal ammoniagenesis, hepatic ureagenesis, and the synthesis of glutamate and GABA for neurotransmission in the brain (3). GAC is an isozyme, an alternatively spliced mRNA that encodes a protein whose C-terminal region differs from kidney-type glutaminase (KGA). GAC was identified and characterized in the human kidney (4, 5), rat kidney, and pig renal cells (6, 7).
In the central nervous system, knock-out mice for the Gls1 gene, which encodes for brain/kidney phosphate-activated glutaminase, have impaired glutamatergic synaptic transmission, altered breathing, disorganized goal-directed behavior, and death shortly after birth (8). Moreover, decreases in glutamate release from neurons were also associated with disruptions in hippocampal short term memory (9). Therefore, we thought that glutaminase plays an important role in neuronal function, and clarification of the down-regulation mechanism of GAC by DPAA is helpful for a therapeutic approach.
Recently, Gao et al. (10) reported that c-Myc oncogenic transcription factor, which is known to regulate microRNAs and stimulate cell proliferation, transcriptionally repressed miR-23a and miR23b, resulting in greater expression of their target protein, mitochondrial glutaminase (GLS1), in human cancer cells. These observations suggested the possibility that GAC was translationally regulated in tumor cells expressing c-Myc.
The human KGA gene is located in chromosome 2, and its transcript is translated in the cytosol as a preprotein containing an N-terminal amino acid signal sequence that directs its import to the mitochondrial matrix. Upon import, the presequence is processed by mitochondrial peptidases to yield a mature form. Matured proteins are associated with each other to form a tetrameric complex (11–14).
In the mammalian mitochondrial matrix, three ATP-dependent proteolytic complexes, mammalian homologues of bacterial FtsH, ClpXP, and Lon, have been identified. Like the cytosolic 26 S proteasome, these proteases were considered to be members of the AAA+ (ATPases associated with a wide variety of cellular activities) protein superfamily (15–17). FtsH, which is a membrane-bound zinc metalloprotease identified in bacteria, is localized in the mammalian mitochondrial inner membrane and plays a role in the turnover of inner membrane-embedded proteins (15, 18, 19). ClpP holoenzymes, which have also been identified in bacteria, consist of soluble two-component systems composed of the same proteolytic component (ClpP) and different ATPase chaperon components (ClpA or ClpX). Different from bacterial ClpAP and ClpXP, which recognize and degrade the SsrA-tagged substrate, the mammalian ClpA homologue has not been identified, and the roles of ClpXP and its substrate are not fully understood because a covalent modification like SsrA tag or ubiquitin that targets specific proteins for degradation has not been discovered in mammalian mitochondria (15, 20–22).
Among these proteases, Lon is associated with protein quality control because Lon protease exhibits ATP-dependent degradation of proteins that are damaged, mutated, or display non-native conformations. Lon is a soluble homo-oligomeric protease located in the bacterial cytoplasm and mammalian mitochondrial matrix (23–27). Recent reports have shown that Lon protease played an important role in the turnover of native steroidogenic acute regulatory protein and termination of steroidogenic acute regulatory activity in cells related to steroid synthesis (28, 29).
In this study, to understand the overall mechanisms of down-regulation of GAC induced by DPAA, GAC was investigated at the levels of transcription, translation, and post-translation, respectively, in cultured human cells. The results showed that GAC was post-translationally regulated by DPAA, and mitochondrial Lon protease plays an important role in this regulation.
EXPERIMENTAL PROCEDURES
Materials
DPAA was purchased from Wako Pure Chemicals (Osaka, Japan). MG132, lactacystin, and epoxomicin were purchased from Peptide Institute, Inc. (Osaka, Japan). Polyclonal anti-KGA/GAC antiserum was prepared as described previously (2). Mouse monoclonal anti-β-actin antibody, rabbit polyclonal anti-voltage-dependent anion channel 1 (VDAC1) antibody, and mouse monoclonal anti-mtHsp70 antibody were purchased from Abcam plc (Cambridge, UK). Mouse monoclonal anti-peroxiredoxin III antibody was purchased from LabFrontier Co. Ltd. (Gyeonggi-do, Korea). Rabbit polyclonal anti-LONP1 was purchased from Sigma. Mouse monoclonal anti CLPP antibody was purchased from Abnova Corp. (Taipei, Taiwan). Mouse monoclonal anti-HSP60 antibody was purchased from Santa Cruz Biotechnology, Inc., and peroxidase-conjugated anti-rabbit IgG and anti-mouse IgG were purchased from Jackson ImmunoResearch. ECL Plus Western blotting detection system and protein A-Sepharose beads were purchased from GE Healthcare. EXPRE35S35S protein labeling mix was purchased from PerkinElmer Life Sciences.
Cell Culture
Cells of the human hepatocarcinoma cell line, HepG2, were cultured in Dulbecco's modified Eagle's medium (DMEM) (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) supplemented with 10% (v/v) fetal bovine serum (FBS) (Biowest, Nuaillé, France) under standard culture conditions at 37 °C, 5% CO2 in humidified air.
Quantitative Real Time PCR Analysis
Total RNA was extracted using the GeneElute mammalian total RNA miniprep kit (Sigma) according to the manufacturer's instructions. cDNA was prepared using the Superscript III first-stranded synthesis system (Invitrogen). The primers used were as follows: GAC-F, 5′-GGTCTCCTCCTCTGGATAAGATGG-3′, and GAC-R, 5′-GATGTCCTCATTTGACTCAGGTGAC-3′; GAPDH-F, 5′-CCACCCATGGCAAATTCCATGGCA-3′, GAPDH-R, 5′-TCTAGACGGCAGGTCAGGTCCACC-3′. Reaction mix was prepared using SYBR Premix Ex TaqII (Takara Bio Inc., Shiga, Japan) and analyzed by the ABI 7500 real time PCR system (Applied Biosystems). All PCRs were performed in triplicate.
Uptake of [35S]Methionine into Newly Translated GAC Protein
HepG2 cells were cultured for 24 h in DPAA-containing medium, then washed, and incubated in 22 μCi of [35S]methionine-containing starvation medium (cysteine and methionine-free medium (Invitrogen) supplemented with 10% (v/v) dialyzed FBS (Invitrogen)) for 1 h. After thorough washings with PBS, cells were harvested and suspended in ice-cold RIPA buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS) containing protease inhibitor mixture (Sigma) and kept on ice for 20 min. DNA and cellular debris were removed by centrifugation (15 min at 17,000 × g), and the protein content in the supernatant was determined and then subjected to an immunoprecipitation assay. 35S-Labeled GAC levels were analyzed with a Fuji Bio-Imaging analyzer (BAS-1000, Fuji Photo Film Co., Tokyo, Japan).
Pulse-Chase Analysis
Pulse-chase analysis was performed as described previously (28). HepG2 cells were cultured for 24 h and then incubated for 1 h in starvation medium (as described above). The medium was exchanged with 22 μCi of [35S]methionine-containing starvation medium and then incubated for 2 h. After removal of the radioactive medium, cells were washed and further incubated with 500 μm DPAA-containing medium for the indicated times. 35S-Labeled GAC proteins were collected by immunoprecipitation and analyzed with a Fuji Bio-Imaging analyzer.
Immunoprecipitation
Anti-GAC/KGA antiserum was incubated with 400 μg of the cell lysate for 2 h in a revolving tube carousel at 4 °C. After this incubation, protein A-Sepharose beads were added to each sample, followed by a 4-h incubation in a revolving tube carousel at 4 °C. Samples were centrifuged at 17,000 × g for 30 s at 4 °C; pelleted beads were washed three times, and the immune complex was released with 30 μl of SDS-PAGE sample buffer and 5 min of boiling.
Preparation of Total Cell Extract, Soluble Fraction, and Insoluble Membrane Fraction
To prepare the soluble fraction and insoluble membrane fraction, HepG2 cells were fractionated by differential extract buffer as described previously (30). For the soluble fraction, HepG2 cells were suspended in Cell lysis buffer (10 mm phosphate buffer, pH 7.4, 10% ethylene glycol, 0.5% Triton X-100, and standard protease inhibitors (Sigma)) and separated by centrifugation at 17,000 × g for 15 min at 4 °C, and the supernatant was collected. Pellets, which were washed twice with Cell lysis buffer, were resuspended in whole cell extract buffer (10 mm phosphate buffer, pH 7.4, 10% ethylene glycol, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate, and standard protease inhibitors) and then sonified to obtain the insoluble membrane fraction. To prepare the total cell extract, cell pellets were suspended in whole cell extract buffer and then sonified. Protein contents of each fraction and extract were determined by the Coomassie Plus Protein Assay Reagent (Pierce) using BSA as the standard.
Immunoblotting
Protein samples were electrophoretically separated using 10% SDS-PAGE and then transferred onto a PVDF membrane. The membrane was blocked overnight at 4 °C with 5% skim milk dissolved in TBS-T (20 mm Tris-HCl, pH 7.5, and 500 mm NaCl (TBS), containing 0.1% Tween 20). The membrane was then incubated with antibody for 6–16 h at 4 °C with gentle rocking. The blot was washed twice with TBS-T for 10 min and once for 5 min, and thereafter, the membrane was incubated with 1:20,000 diluted anti-rabbit IgG or anti-mouse IgG coupled to horseradish peroxidase (Jackson ImmunoResearch) for 2 h at 4 °C. The blot was washed with TBS-T, and the membrane was developed using the ECL Plus immunoblotting detection system (GE Healthcare) and then analyzed with the LAS-3000 mini luminoimage analyzer (Fuji Photo Film Co.).
RNAi Experiments
siRNAs targeting human LONP (target sequence, GCGUCUUUCUAAAGAGAGA), human CLPP (target sequence, GGGCCAAGCCACAGACAUU), or negative control siRNA (sequence, AUCCGCGCGAUAGUACGUA) were purchased from B-Bridge International, Inc. Transfection of the siRNAs into HepG2 cells was performed using the LipofectamineTM RNAiMAX reagent (Invitrogen) according to the manufacturer's instructions. Forty eight hours after siRNA transfection, cells were cultured in fresh growth medium for 40 h and then incubated with 500 μm DPAA-containing medium. After treatment with DPAA for the indicated time period, cells were harvested, and the expression levels of each protein were examined by immunoblot analyses.
Blue Native Gel Electrophoresis
HepG2 cells were lysed in 80 μl of ice-cold DDM buffer (1% n-dodecyl-β-d-maltoside, 50 mm BisTris, pH 7.2, 0.5 m 6-aminocaproic acid, 50 mm NaCl, 10% glycerol, 0.001% Ponceau S, and standard protease inhibitor) and 20 μl of ice-cold CHAPS buffer (5% CHAPS, 50 mm BisTris, pH 7.2, 0.5 m 6-aminocaproic acid, 50 mm NaCl, 10% glycerol, 0.001% Ponceau S, and standard protease inhibitor). Insolubilized material was removed by centrifugation for 30 min at 20,000 × g. After addition of 6.5 μl of sample buffer (0.5% (w/v) Coomassie Brilliant Blue G-250, 10 mm BisTris, pH 7.2, 50 mm 6-aminocaproic acid, 10% glycerol, 0.1 mm PMSF) to 13.1 μl of supernatant, 10 μl of the sample (protein concentration of 2 μg/μl) was analyzed directly by BN-PAGE. In short, a 4–16% polyacrylamide gradient gel (Invitrogen) was run in a cold room with pre-chilled dark blue cathode buffer (50 mm BisTris, 50 mm Tricine, pH 6.8, 0.02% Coomassie Brilliant Blue G-250) for 50 min at 150 V. The dark blue cathode buffer was changed for light blue cathode buffer (Coomassie Brilliant Blue G-250 changed to 0.002%) and continued to electrophoresis for 90–100 min until the dye front was leaked from the bottom of the gel. The same buffer without Coomassie Brilliant Blue G-250, was used for electrophoresis as an anode buffer. For Western blotting, the blue native gel was soaked in blot buffer (20 mm Bicine, 25 mm BisTris, 1 mm EDTA, pH 7.2) for 20 min, and denatured proteins were transferred to PVDF membranes (Bio-Rad) in the same buffer using the semi-dry blotting technique. Immunodecoration was according to standard procedures and was visualized by the ECL method (GE Healthcare).
RESULTS
Effect of DPAA on GAC Transcription
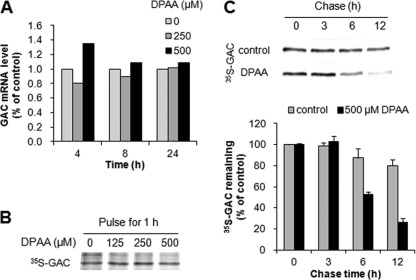
To understand the mechanism of GAC protein down-regulation by DPAA, we determined GAC mRNA levels after treatment with DPAA. HepG2 cells were incubated with 0, 250, and 500 μm DPAA for 4, 8, and 24 h; the total RNA was then extracted, and GAC mRNA levels were determined by quantitative real time PCR. As shown in Fig. 1A, no significant changes in GAC mRNA levels were observed in HepG2 cells incubated with various concentrations of DPAA for the specified times.
FIGURE 1.
Effects of DPAA on GAC mRNA expression, translation, and GAC proteolysis. A, GAC mRNA expression levels after treatment with DPAA. GAC mRNA levels were analyzed by quantitative real time PCR in HepG2 cells that were incubated with 0, 250, and 500 μm DPAA for 4, 8, and 24 h. The reproducibility of results was confirmed by separate experiments. B, newly translated GAC protein levels after treatment with DPAA. HepG2 cells were cultured for 24 h in 0–500 μm DPAA-containing culture medium, which was then replaced with [35S]methionine-containing medium and pulsed for 1 h, followed by immunoprecipitation of GAC, PAGE, and autoradiography. The reproducibility of results was confirmed by separate experiments. C, proteolysis of GAC protein by DPAA. Upper panel, HepG2 cells were pulsed for 2 h with [35S]methionine and chased at the indicated times with or without 500 μm DPAA-containing chase medium, followed by immunoprecipitation of GAC, PAGE, and autoradiography. Lower panel, a quantitative phosphoimager analysis of results. Data are presented as the percentage of labeling at time 0 of the chase (mean ± S.D., n = 3).
Effect of DPAA on GAC Translation
A recent study indicated that GAC protein levels were post-transcriptionally regulated by microRNAs, and their translation was inhibited by these microRNAs (10). Therefore, we investigated the effects of DPAA on GAC translation. The translational levels of GAC were assessed by [35S]methionine uptake in newly translated GAC proteins after treatment with DPAA. As shown in Fig. 1B, no significant changes in [35S]methionine-labeled GAC (35S-GAC) were observed by DPAA.
Effect of DPAA on GAC Proteolysis
To evaluate the effects of DPAA on GAC protein degradation, GAC protein turnover in HepG2 cells was investigated by pulse-chase analysis. For pulse labeling, HepG2 cells were incubated in [35S]methionine-containing medium for 2 h, and then the remaining 35S-GAC proteins were chased in the control and 500 μm DPAA-containing chase medium for up to 12 h. As shown in Fig. 1C, 80.1% of 35S-GAC proteins remained in HepG2 cells when incubated in the control chase medium for 12 h, whereas in the presence of DPAA, 35S-GAC levels decreased time-dependently, and only 25.9% of 35S-GAC proteins remained after 12 h of chase.
Effects of Proteasome Inhibitors on DPAA-induced GAC Proteolysis
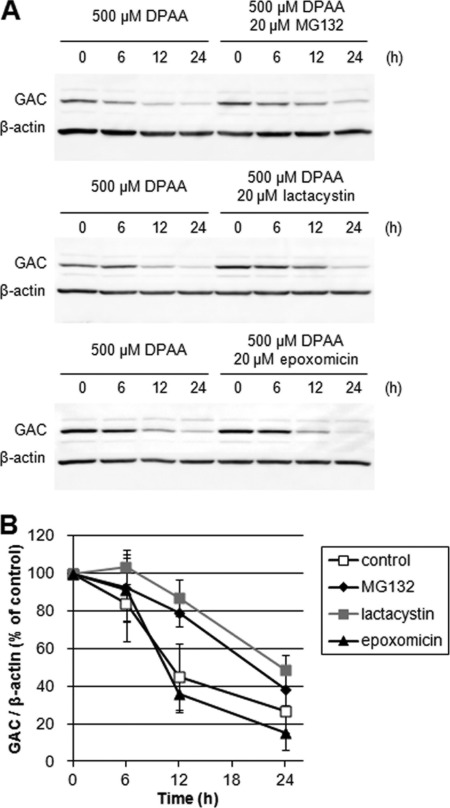
As DPAA promoted the ubiquitination of cellular proteins (supplemental Fig. S1), we hypothesized that GAC proteolysis may be mediated by the ubiquitin-proteasome system. To clarify this hypothesis, HepG2 cells were pretreated with proteasome inhibitors, and cellular GAC protein levels were determined after DPAA treatment. Each proteasome inhibitor was added to the medium 2 h before addition of 500 μm DPAA, and incubation was conducted for the indicated times. As shown in Fig. 2A (upper and middle panels), DPAA-induced decreases in GAC proteins at the time point of 12 h were partially blocked by pretreatment with MG132 and lactacystin. After a 12-h treatment with DPAA, only 44.8% of GAC proteins remained in the cells, whereas ∼80% of GAC proteins remained in cells pretreated with MG132 or lactacystin (Fig. 2B). In contrast, pretreatment with epoxomicin had no effect on DPAA-induced degradation of GAC (Fig. 2A, lower panels), and about 40% of GAC proteins remained in cells (Fig. 2B). To define the inhibitory effects of epoxomicin on proteasomes, the ubiquitination level of cellular proteins was evaluated by immunoblot analysis. Ubiquitination levels were significantly increased by epoxomicin treatment, and the effect was the same as the ubiquitination level of MG132 and lactacystin treatment (data not shown). Therefore, we concluded that epoxomicin could fully inhibit the proteasome.
FIGURE 2.
Effects of 26 S proteasome inhibitors on DPAA-induced GAC degradation. A, immunoblot pattern of GAC expression levels in HepG2 cells after treatment by DPAA with or without proteasome inhibitor. HepG2 cells were preincubated for 2 h in each proteasome inhibitor (lactacystin, MG132, and epoxomicin)-containing medium, after which DPAA was added and the culture continued for the indicated time. A cell extract was prepared, and immunoblot analysis was performed as described under “Experimental Procedures.” B, quantitative analysis of GAC protein levels. GAC protein levels are presented as the percentage of levels at 0 h (mean ± S.D., n = 3).
Involvement of Lon Protease in DPAA-induced GAC Proteolysis
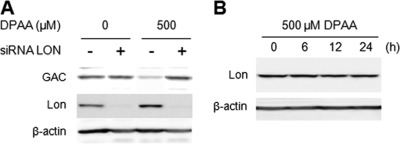
Previous studies showed that MG132 and lactacystin had inhibitory effects on Lon protease in addition to the inhibition of proteasome, although epoxomicin did not (28, 29). Therefore, partial inhibition by MG132 and lactacystin of DPAA-induced GAC degradation may be due to inhibition of Lon protease. To determine the involvement of Lon protease in GAC degradation caused by DPAA, the effects of siRNA Lon on the induction of GAC degradation were investigated. Double-stranded short oligonucleotides for human Lon protease mRNA (siRNA LON) were synthesized and transfected into HepG2 cells. After a 48-h transfection of siRNA LON in HepG2 cells, cellular Lon protease levels significantly decreased, and this decrease lasted for the following 72 h (data not shown). Thus, HepG2 cells transfected transiently with siRNA LON for 48 h and then incubated for 40 h in normal culture medium were used as “Lon protease knockdown cells.” As shown in Fig. 3A, GAC protein levels were significantly decreased by treatment with DPAA in control siRNA-transfected cells, whereas GAC protein levels in Lon protease knockdown cells were not changed by treatment with DPAA. In contrast, in the absence of DPAA treatment, GAC protein levels were not significantly changed in both control and Lon protease knockdown cells.
FIGURE 3.
Effect of Lon protease on DPAA-induced GAC degradation. A, GAC protein levels in Lon protease knockdown cells after treatment with or without DPAA. Double-stranded oligonucleotides for Lon protease (siRNA LON +) or negative control oligonucleotides (siRNA LON −) were transiently transfected into HepG2 cells for 48 h, and then the medium was replaced with fresh medium. After 40 h of culture, 500 μm DPAA was added, and incubation was continued for 12 h. Cellular GAC and Lon protease protein levels were determined by immunoblot analysis as described under “Experimental Procedures.” The reproducibility of results was confirmed by separate experiments. B, changes in Lon protease protein levels after treatment with DPAA. HepG2 cells were cultured in medium containing 500 μm DPAA for up to 24 h, and Lon protease levels were examined by immunoblot analysis. β-Actin was indicated as the loading control. The reproducibility of results was confirmed by separate experiments.
Effects of DPAA on Lon Expression
To determine whether the promotion of GAC degradation by DPAA is due to Lon protease up-regulation, Lon protein expression levels were investigated. HepG2 cells were incubated with 500 μm DPAA for up to 24 h, and then cellular Lon protein levels were determined by immunoblot analysis. As shown in Fig. 3B, no changes in Lon protein levels were observed for up to 24 h after treatment with DPAA.
Changes in Subcellular Distribution of GAC Protein after Treatment with DPAA
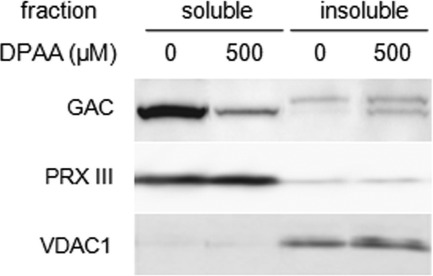
Lon protease preferentially degrades misfolded or oxidatively modified proteins to prevent the formation of insoluble aggregates (23–25, 31, 32). Likewise, protein modification caused by DPAA was investigated in terms of changes in GAC protein solubility. HepG2 cells were treated with 500 μm DPAA for 12 h and then soluble and insoluble cellular proteins were separated by 0.5% Triton X-100-containing buffer as described previously (30). Separation of soluble and insoluble cellular compartments was evaluated by peroxiredoxin III (PRX III). PRX III is known as a soluble mitochondria matrix protein used as a soluble protein marker, and voltage-dependent anion channel-1 (VDAC1) is a mitochondrial outer membrane-embedded protein used as an insoluble membrane fraction marker. As shown in Fig. 4, regardless of the presence or absence of DPAA treatment, PRX III- and VDAC1-positive bands were detected in soluble and insoluble membrane fractions, respectively. In control cells, GAC proteins were detected in the soluble fraction but not the insoluble membrane fraction. However, GAC protein levels in the soluble fractions were significantly decreased in cells treated with DPAA and slightly increased in the insoluble membrane fraction.
FIGURE 4.
Changes in the subcellular distribution of GAC proteins after treatment with DPAA. After treatment with 500 μm DPAA for 12 h, soluble and insoluble cellular proteins in HepG2 cells were prepared as described under “Experimental Procedures.” Contamination of the soluble or insoluble membrane fractions was checked by immunoblots using antibodies against PRX III, which is a mitochondrial soluble matrix protein, and voltage-dependent anion channel-1 (VDAC1), which is a component of porin that is embedded in the mitochondrial outer membrane. The reproducibility of results was confirmed by separate experiments.
Subcellular Distribution of GAC Protein in Lon Protease Knockdown Cells
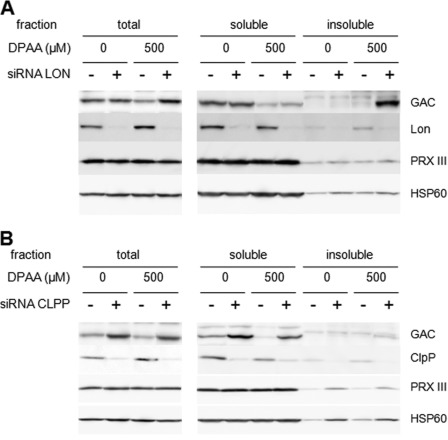
To understand the role of Lon protease in GAC degradation, subcellular distribution of the GAC protein in Lon protease knockdown cells was investigated. As shown in Fig. 5A, in the absence of DPAA, most GAC proteins were detected in the soluble fraction, and their expression levels decreased in both control and Lon protease knockdown cells when cells were treated with DPAA. However, insoluble GAC proteins were significantly increased in Lon protease knockdown cells after treatment with DPAA. Both PRX III and mitochondrial heat-shock protein 60 (HSP60) levels were not significantly changed in control and Lon protease knockdown cells after treatment with DPAA.
FIGURE 5.
Subcellular distribution of GAC protein in DPAA-treated Lon protease or ClpP protease knockdown cells. A, subcellular distribution of GAC protein in Lon protease knockdown cells after treatment with DPAA. Double-stranded oligonucleotides for Lon protease (siRNA LON +) or negative control oligonucleotides (siRNA LON −) were transiently transfected into HepG2 cells for 48 h and then the medium was replaced with fresh medium. After a 40-h incubation, 500 μm DPAA was added and incubated further for 12 h. Total, soluble, and insoluble cellular extracts of HepG2 cells were prepared as described under “Experimental Procedures.” GAC and other mitochondrial protein (PRX III and HSP60) levels in each fraction were determined by immunoblot analysis as described under “Experimental Procedures.” The reproducibility of results was confirmed by separate experiments. B, subcellular distribution of GAC protein in ClpP protease knockdown cells after treatment with DPAA. Double-stranded oligonucleotides for ClpP protease (siRNA CLPP +) or negative control oligonucleotides (siRNA CLPP −) were transiently transfected into HepG2 cells for 48 h and then the medium was replaced with fresh medium. After a 40-h incubation, 500 μm DPAA was added and incubated further for 12 h. Total, soluble, and insoluble cellular extracts of HepG2 cells were prepared as described under “Experimental Procedures.” GAC and other mitochondrial protein (PRX III and HSP60) levels in each fraction were determined by immunoblot analysis as described under “Experimental Procedures.” The reproducibility of results was confirmed by separate experiments.
Effect of ClpP Protease on GAC Protein Degradation
To clarify the effects of ClpP protease on GAC degradation, ClpP knockdown cells were constructed, and GAC protein expression and their subcellular distribution were investigated (Fig. 5B). GAC protein levels in the total cell extract were significantly higher in ClpP protease knockdown cells than those of control and Lon protease knockdown cells. To determine the increase in cellular GAC content, subcellular distribution of GAC protein was investigated. In the soluble fraction, GAC proteins were significantly increased in ClpP knockdown cells but not in the insoluble membrane fraction. In the presence of DPAA, soluble GAC proteins were slightly decreased by ClpP protease knockdown. However, unlike Lon protease knockdown cells, no significant increase in insoluble GAC proteins was observed in ClpP protease knockdown cells after treatment with DPAA. PRX III and HSP60 protein levels in insoluble membrane fractions were slightly increased in ClpP protease knockdown cells. However, PRX III and HSP60 protein levels in both the total cellular fraction and soluble fraction were not significantly affected by both ClpP protease knockdown and DPAA treatment.
Effect of DPAA on Native GAC Conformation
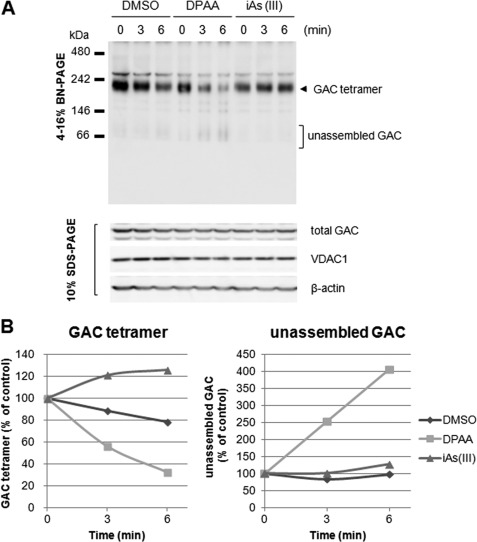
To understand in detail the mechanism of DPAA-induced changes in the solubility of GAC proteins, conformational changes in GAC proteins were investigated by BN-PAGE analysis. In a normal state, immunodecoration with antibodies directed against GAC identified an abundant complex of ∼232 kDa (Fig. 6A, upper panel). The same samples were denatured by SDS and applied to SDS-PAGE. GAC migrated at the monomer position (∼58 kDa, Fig. 6A, lower panel). These observations indicated that native GAC existed in a tetrameric form. Native tetrameric GAC was decreased, and low molecular weight GAC increased time-dependently when DPAA was added and incubated with cell extracts prepared from HepG2 cells at 25 °C (Fig. 6B). However, when the cell extract was incubated with equimolar inorganic arsenicals (iAs(III)) instead of DPAA, both tetrameric and low molecular weight GAC levels were not significantly changed. These results indicated that DPAA directly destructed the native tetrameric GAC complex to accumulate the unassembled one.
FIGURE 6.
DPAA directly destroys the native tetrameric GAC and forms unassembled GAC. A, BN-PAGE analysis of native GAC protein after incubation with DPAA. HepG2 cell extracts were prepared and incubated with 14.8 mm DPAA or NaAsO2 for the indicated time points at 25 °C. Native GAC proteins were determined with 4–16% BN-PAGE and immunoblot analysis as described under “Experimental Procedures.” Total GAC, VDAC1, and β-actin levels in cell extracts were determined by SDS-PAGE and immunoblot analysis. B, changes in tetrameric and unassembled forms of GAC after incubation with DPAA. Each form of GAC protein prior to incubation at 25 °C was set to 100%.
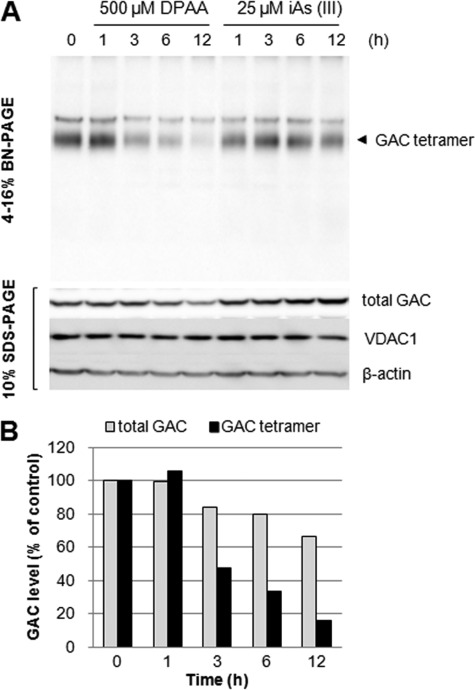
We further examined tetrameric GAC levels in HepG2 cells incubated with DPAA for the indicated time points. As shown in Fig. 7A, native tetrameric GAC levels when incubated with DPAA were decreased at 3 h, and this continued for up to 12 h. Quantification of tetrameric GAC levels in cells treated with DPAA revealed that 85% of tetrameric GAC was decreased for up to 12 h (Fig. 7B). However, when the same samples were analyzed with SDS-PAGE and total GAC levels in HepG2 cells were examined, ∼80% of GAC remained for 3 h, and more than 60% of GAC remained even after 12 h of DPAA treatment over control levels (Fig. 7, A, lower panel, and B). Tetrameric and total GAC levels remained constant when cells were incubated with iAs(III) for up to 12 h (Fig. 7A, upper panel).
FIGURE 7.
Native tetrameric GAC in HepG2 cells treated with DPAA rapidly disappears before total cellular GAC degradation. A, BN-PAGE analysis of native GAC protein in cells treated with DPAA. HepG2 cells were incubated with 500 μm DPAA or 25 μm NaAsO2 for the indicated time points, and native tetrameric GAC levels were determined with 4–16% BN-PAGE and immunoblot analysis as described under “Experimental Procedures.” Total GAC, VDAC1, and β-actin levels in cell extracts were determined by SDS-PAGE and immunoblot analysis. B, quantitative analysis of total and tetrameric GAC protein levels in HepG2 cells treated with DPAA. GAC levels in cells cultured without DPAA medium were set to 100%.
Effect of Lon Protease on Native GAC Protein Degradation
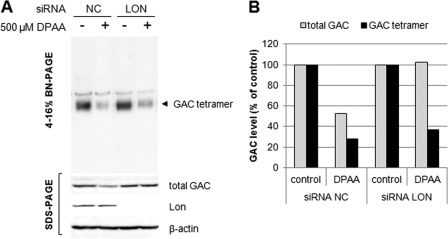
We investigated tetrameric GAC levels in Lon protease knockdown cells. As shown in Fig. 8A, tetrameric GAC levels were significantly decreased in both control and Lon protease knockdown cells. Approximately 70% of tetrameric GAC was decreased after 12 h of treatment with DPAA. However, 50 and 100% of total GAC proteins remained in control and Lon protease knockdown cells, respectively, after 12 h of treatment with DPAA (Fig. 8B).
FIGURE 8.
DPAA promotes native tetrameric GAC destruction in both control and Lon protease knockdown cells. A, tetrameric GAC levels in Lon protease knockdown cells before and after treatment with DPAA. Double-stranded oligonucleotides for Lon protease (siRNA LON) or negative control oligonucleotides (siRNA NC) were transiently transfected into HepG2 cells for 48 h, and then the medium was replaced with fresh medium. After a 40-h incubation, 500 μm DPAA was added and incubated for 12 h. Native tetrameric GAC was determined by BN-PAGE and immunoblot analysis. Total GAC, Lon, and β-actin levels were determined by SDS-PAGE and immunoblot analysis as described under “Experimental Procedures.” B, quantity of total and tetrameric GAC protein levels in Lon protease knockdown HepG2 cells treated with DPAA. GAC levels in cells cultured without DPAA medium were set to 100%.
Effect of DPAA on the Interaction of GAC and mtHsp70
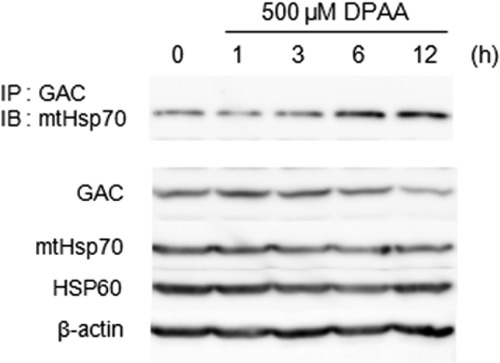
mtHsp70, which is known as a molecular chaperon protein, has an essential role for the maintenance of misfolded proteins in a soluble state and for the process of proteolysis of PIM1 protease, a yeast homologue of mammalian Lon (33, 34). If unassembled GAC is proteolyzed by the Lon protease, mtHsp70 may recognize the unassembled GAC as a misfolded protein. To address the role of mtHsp70 in the process of Lon protease-mediated GAC proteolysis, the association of mtHsp70 and GAC was determined by the immunoprecipitation method. HepG2 cells, treated with DPAA for the indicated time points, were collected and cellular GAC proteins were immunoprecipitated by anti-GAC antibody, and immunoblot analysis was performed using anti-mtHsp70 antibody. As shown in Fig. 9, GAC protein associated with mtHsp70 was increased 3–6 h after incubation with 500 μm DPAA and then leveled off for up to 12 h. However, total cellular GAC levels were time-dependently decreased by DPAA. In contrast, total cellular mtHsp70 protein levels remained constant with DPAA treatment.
FIGURE 9.
PDAA promotes the interaction of GAC with mtHsp70. HepG2 cells were incubated with DPAA for the indicated time points, and immunoprecipitation (IP) was performed as described under “Experimental Procedures.” Each protein level in cell extracts and immunoprecipitants was determined by SDS-PAGE and immunoblot (IB) analysis.
DISCUSSION
This study was conducted in human hepatocarcinoma HepG2 cells to understand the mechanism of DPAA-induced GAC down-regulation. As a first step approach, GAC mRNA expression, translation, and GAC proteolysis were investigated in cells exposed to DPAA. Results revealed the promotion of GAC protein degradation by DPAA (Fig. 1). In addition, GAC protein degradation in DPAA-treated HepG2 cells was partially blocked by proteasome inhibitors, such as MG132 and lactacystin, but not by epoxomicin (Fig. 2), and the blocking effects of MG132 and lactacystin may be attributable to the action of mitochondrial Lon protease (26, 28, 29, 35, 36). Indeed, DPAA-induced GAC protein turnover was completely blocked by the knockdown of Lon protease (Fig. 3A), suggesting that GAC was regulated post-translationally in cells exposed to DPAA and that GAC is a substrate of Lon protease.
Lon protease in mammals is a hexameric homoprotein complex located within the mitochondrial matrix and displays a serine protease activity that is stimulated by ATP (17). Lon mRNA and protein were increased in response to various stresses, such as oxidants, hypoxia, heat shock, and endoplasmic reticulum stress to remove damaged proteins from the mitochondria (37–41). Hence, we postulated that the promotion of GAC protein degradation by Lon protease may be due to up-regulation of Lon protease resulting from some stress response caused by DPAA. However, as shown in Fig. 3B, Lon protein levels were not significantly increased by DPAA treatment. We further examined mitochondrial ATP-dependent protease activities, which reflect the total mitochondrial proteases, including Lon and other mitochondrial matrix proteases such as ClpXP, and we confirmed that DPAA did not affect these protease activities (data not shown). These observations suggested that GAC protein degradation by DPAA was not attributable to the increase in Lon protease.
Lon protease preferentially degrades misfolded, unassembled, or oxidatively damaged proteins to prevent their accumulation and subsequent formation of an insoluble aggregate (27, 40–42). Therefore, we hypothesized that GAC protein may be modified by DPAA, and the modified GAC may be a target in Lon protease-mediated proteolysis. To test this hypothesis, changes in GAC protein solubility were investigated. The GAC protein, which was normally detected in the soluble fraction, accumulated in the insoluble membrane fraction after treatment with DPAA (Fig. 4). In addition, insoluble GAC proteins were markedly increased in Lon protease knockdown cells (Fig. 5A). In contrast, soluble GAC proteins in Lon protease knockdown cells decreased to the same level as those seen in parental cells after treatment with DPAA (Fig. 5A). However, DPAA-dependent insoluble GAC accumulation was not observed in knockdown cells of ClpP, a protease subunit of ClpXP protease in mitochondria. These observations suggested that DPAA may promote the soluble GAC protein modification, and the modified GAC protein was recognized and degraded by Lon protease.
How do DPAA promote the accumulation of GAC that is prone to aggregate? A conformational change of GAC in cells treated with DPAA was investigated by the BN-PAGE technique. In normal cell extracts, GAC-positive signals were abundant around ∼232 kDa. When the same samples were denatured by SDS and applied to SDS-PAGE, GAC migrated at the monomer position (∼58 kDa). These results indicated that GAC proteins are associated with each other to form a tetrameric complex as observed in KGA (14). Native tetrameric GAC proteins were destroyed, and unassembled forms of GAC were accumulated when DPAA was added and incubated with cell extract (Fig. 6A). Moreover, decreases in tetrameric GAC were observed prior to a decrease in total cellular GAC when HepG2 cells were treated with DPAA (Fig. 7). In addition, decreases in tetrameric GAC were also observed in both control and Lon protease knockdown cells after treatment with DPAA (Fig. 8). These observations indicated that DPAA-promoted tetrameric GAC destructions were independent of Lon protease, and Lon protease may play a role in proteolysis for the unassembled GAC. The mechanism of substrate recognition and binding of Lon protease remains to be elucidated. However, Major et al. (43) reported that it is not only protein that is not successfully folded in the native conformation but also protein that has not formed a complex due to the lack of prosthetic groups, such as metals, that may be a target for Lon protease. Consequently, they proposed that the overall tertiary structure of protein is one of the major determinants of susceptibility to Lon/Pim1-dependent degradation. Moreover, von Janowsky et al. (44) proposed that hydrophobic loops exposed at the surface of substrate proteins were essential for Lon protease in recognizing substrates. Although detailed structural changes in GAC after treatment with DPAA remain obscure, GAC proteins that interacted with mtHsp70 were increased when DPAA treatment was prolonged (Fig. 9). Interestingly, the mtHsp70 that interacted with GAC increased when tetrameric GAC destruction was apparently observed (Fig. 7). Wagner et al. (33) reported that mtHsp70 played an essential role in the maintenance of misfolded proteins in a soluble state at the proteolysis process of misfolded mitochondrial proteins by PIM1 proteases. Consequently, unassembled GAC itself or further modified GAC may be recognized with mtHsp70 and degraded by Lon protease.
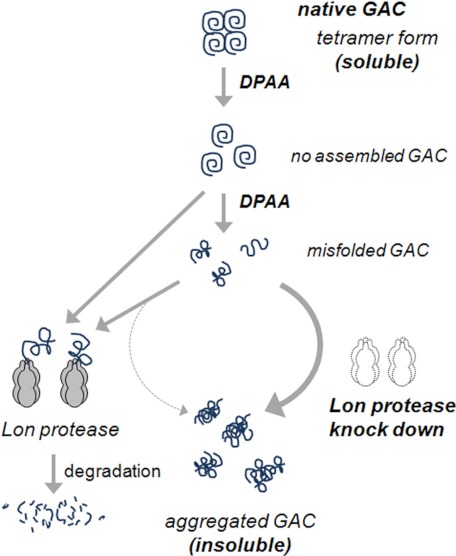
Taken together, we proposed a model for the mechanism of down-regulation of GAC by DPAA (Fig. 10). A native GAC protein, which is newly translated and imported into the mitochondrial matrix space, exists in a soluble tetrameric form. When cells were exposed to DPAA, the tetrameric GAC complex was destroyed and subsequently accumulated unassembled GAC. Unassembled GAC itself and/or further modified GAC were prone to form insoluble aggregates; however, they were recognized with mtHsp70 and degraded by Lon protease to prevent the formation of insoluble aggregates. We concluded that destruction of tetrameric GAC by DPAA may be a trigger of the proteolysis process induced by Lon protease, and this is the cause of GAC protein down-regulation induced by DPAA.
FIGURE 10.
Hypothetical illustration of DPAA-induced GAC protein degradation. In the normal state, GAC proteins assembled each other and formed the soluble tetrameric complex. In the presence of DPAA, tetrameric GAC was destroyed, and unassembled GAC was formed. It is obscure whether DPAA promoted unassembled GAC modifications; however, unassembled GAC itself or further modified ones may be degraded by mitochondrial Lon protease. If functional Lon protease was lost, GAC proteins may accumulate and form an insoluble aggregate.
Supplementary Material
This work was supported by Grant-in-aid for Young Scientists (B) 20790113 from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan.

This article contains supplemental Fig. S1.
- DPAA
- diphenylarsinic acid
- GAC
- glutaminase C
- KGA
- kidney-type glutaminase
- mtHsp70
- mitochondrial heat shock protein 70
- BN-PAGE
- blue native-PAGE
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Ishii K., Tamaoka A., Otsuka F., Iwasaki N., Shin K., Matsui A., Endo G., Kumagai Y., Ishii T., Shoji S., Ogata T., Ishizaki M., Doi M., Shimojo N. (2004) Diphenylarsinic acid poisoning from chemical weapons in Kamisu, Japan. Ann. Neurol. 56, 741–745 [DOI] [PubMed] [Google Scholar]
- 2. Kita K., Suzuki T., Ochi T. (2007) Down-regulation of glutaminase C in human hepatocarcinoma cell by diphenylarsinic acid, a degradation product of chemical warfare agents. Toxicol. Appl. Pharmacol. 220, 262–270 [DOI] [PubMed] [Google Scholar]
- 3. Curthoys N. P., Watford M. (1995) Regulation of glutaminase activity and glutamine metabolism. Annu. Rev. Nutr. 15, 133–159 [DOI] [PubMed] [Google Scholar]
- 4. Elgadi K. M., Meguid R. A., Qian M., Souba W. W., Abcouwer S. F. (1999) Cloning and analysis of unique human glutaminase isoforms generated by tissue-specific alternative splicing. Physiol. Genomics 1, 51–62 [DOI] [PubMed] [Google Scholar]
- 5. Holcomb T., Taylor L., Trohkimoinen J., Curthoys N. P. (2000) Isolation, characterization, and expression of a human brain mitochondrial glutaminase cDNA. Mol. Brain Res. 76, 56–63 [DOI] [PubMed] [Google Scholar]
- 6. Roberg B., Torgner I. A., Laake J., Takumi Y., Ottersen O. P., Kvamme E. (2000) Properties and submitochondrial localization of pig and rat renal phosphate-activated glutaminase. Am. J. Physiol. Cell Physiol. 279, C648–C657 [DOI] [PubMed] [Google Scholar]
- 7. Porter L. D., Ibrahim H., Taylor L., Curthoys N. P. (2002) Complexity and species variation of the kidney-type glutaminase gene. Physiol. Genomics 9, 157–166 [DOI] [PubMed] [Google Scholar]
- 8. Masson J., Darmon M., Conjard A., Chuhma N., Ropert N., Thoby-Brisson M., Foutz A. S., Parrot S., Miller G. M., Jorisch R., Polan J., Hamon M., Hen R., Rayport S. (2006) Mice lacking brain/kidney phosphate-activated glutaminase have impaired glutamatergic synaptic transmission, altered breathing, disorganized goal-directed behavior, and die shortly after birth. J. Neurosci. 26, 4660–4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sullivan J. M. (2000) Cellular and molecular mechanisms underlying learning and memory impairments produced by cannabinoids. Learn Mem. 7, 132–139 [DOI] [PubMed] [Google Scholar]
- 10. Gao P., Tchernyshyov I., Chang T. C., Lee Y. S., Kita K., Ochi T., Zeller K. I., De Marzo A. M., Van Eyk J. E., Mendell J. T., Dang C. V. (2009) c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458, 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aledo J. C., Gómez-Fabre P. M., Olalla L., Márquez J. (2000) Identification of two human glutaminase loci and tissue-specific expression of the two related genes. Mamm. Genome 11, 1107–1110 [DOI] [PubMed] [Google Scholar]
- 12. Srinivasan M., Kalousek F., Curthoys N. P. (1995) In vitro characterization of the mitochondrial processing and the potential function of the 68-kDa subunit of renal glutaminase. J. Biol. Chem. 270, 1185–1190 [DOI] [PubMed] [Google Scholar]
- 13. Srinivasan M., Kalousek F., Farrell L., Curthoys N. P. (1995) Role of the N-terminal 118 amino acids in the processing of the rat renal mitochondrial glutaminase precursor. J. Biol. Chem. 270, 1191–1197 [DOI] [PubMed] [Google Scholar]
- 14. Haser W. G., Shapiro R. A., Curthoys N. P. (1985) Comparison of the phosphate-dependent glutaminase obtained from rat brain and kidney. Biochem. J. 229, 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ogura T., Wilkinson A. J. (2001) AAA+ superfamily ATPases. Common structure-diverse function. Genes Cells 6, 575–597 [DOI] [PubMed] [Google Scholar]
- 16. Gottesman S. (2003) Proteolysis in bacterial regulatory circuits. Annu. Rev. Cell Dev. Biol. 19, 565–587 [DOI] [PubMed] [Google Scholar]
- 17. Kuzela S., Goldberg A. L. (1994) Mitochondrial ATP-dependent protease from rat liver and yeast. Methods Enzymol. 244, 376–383 [DOI] [PubMed] [Google Scholar]
- 18. Banfi S., Bassi M. T., Andolfi G., Marchitiello A., Zanotta S., Ballabio A., Casari G., Franco B. (1999) Identification and characterization of AFG3L2, a novel paraplegin-related gene. Genomics 59, 51–58 [DOI] [PubMed] [Google Scholar]
- 19. Coppola M., Pizzigoni A., Banfi S., Bassi M. T., Casari G., Incerti B. (2000) Identification and characterization of YME1L1, a novel paraplegin-related gene. Genomics 66, 48–54 [DOI] [PubMed] [Google Scholar]
- 20. Yu A. Y., Houry W. A. (2007) ClpP. A distinctive family of cylindrical energy-dependent serine proteases. FEBS Lett. 581, 3749–3757 [DOI] [PubMed] [Google Scholar]
- 21. Kang S. G., Dimitrova M. N., Ortega J., Ginsburg A., Maurizi M. R. (2005) Human mitochondrial ClpP is a stable heptamer that assembles into a tetradecamer in the presence of ClpX. J. Biol. Chem. 280, 35424–35432 [DOI] [PubMed] [Google Scholar]
- 22. Kang S. G., Ortega J., Singh S. K., Wang N., Huang N. N., Steven A. C., Maurizi M. R. (2002) Functional proteolytic complexes of the human mitochondrial ATP-dependent protease, hClpXP. J. Biol. Chem. 277, 21095–21102 [DOI] [PubMed] [Google Scholar]
- 23. Chung C. H., Goldberg A. L. (1981) The product of the lon (capR) gene in Escherichia coli is the ATP-dependent protease, protease La. Proc. Natl. Acad. Sci. U.S.A. 78, 4931–4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rotanova T. V., Melnikov E. E., Khalatova A. G., Makhovskaya O. V., Botos I., Wlodawer A., Gustchina A. (2004) Classification of ATP-dependent proteases Lon and comparison of the active sites of their proteolytic domains. Eur. J. Biochem. 271, 4865–4871 [DOI] [PubMed] [Google Scholar]
- 25. Tsilibaris V., Maenhaut-Michel G., Van Melderen L. (2006) Biological roles of the Lon ATP-dependent protease. Res. Microbiol. 157, 701–713 [DOI] [PubMed] [Google Scholar]
- 26. Lee I., Suzuki C. K. (2008) Functional mechanics of the ATP-dependent Lon protease. Lessons from endogenous protein and synthetic peptide substrates. Biochim. Biophys. Acta 1784, 727–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gur E., Sauer R. T. (2008) Recognition of misfolded proteins by Lon, a AAA+ protease. Gene Dev. 22, 2267–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Granot Z., Geiss-Friedlander R., Melamed-Book N., Eimerl S., Timberg R., Weiss A. M., Hales K. H., Hales D. B., Stocco D. M., Orly J. (2003) Proteolysis of normal and mutated steroidogenic acute regulatory proteins in the mitochondria. The fate of unwanted proteins. Mol. Endocrinol. 17, 2461–2476 [DOI] [PubMed] [Google Scholar]
- 29. Granot Z., Kobiler O., Melamed-Book N., Eimerl S., Bahat A., Lu B., Braun S., Maurizi M. R., Suzuki C. K., Oppenheim A. B., Orly J. (2007) Turnover of mitochondrial steroidogenic acute regulatory (StAR) protein by Lon protease. The unexpected effect of proteasome inhibitors. Mol. Endocrinol. 21, 2164–2177 [DOI] [PubMed] [Google Scholar]
- 30. Zhao Q., Wang J., Levichkin I. V., Stasinopoulos S., Ryan M. T., Hoogenraad N. J. (2002) A mitochondrial specific stress response in mammalian cells. EMBO J. 21, 4411–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shineberg B., Zipser D. (1973) The ion gene and degradation of β-galactosidase nonsense fragments. J. Bacteriol. 116, 1469–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kowit J. D., Goldberg A. L. (1977) Intermediate steps in the degradation of a specific abnormal protein in Escherichia coli. J. Biol. Chem. 252, 8350–8357 [PubMed] [Google Scholar]
- 33. Wagner I., Arlt H., van Dyck L., Langer T., Neupert W. (1994) Molecular chaperones cooperate with PIM1 protease in the degradation of misfolded proteins in mitochondria. EMBO J. 13, 5135–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Savel'ev A. S., Novikova L. A., Kovaleva I. E., Luzikov V. N., Neupert W., Langer T. (1998) ATP-dependent proteolysis in mitochondria. m-AAA protease and PIM1 protease exert overlapping substrate specificities and cooperate with the mtHsp70 system. J. Biol. Chem. 273, 20596–20602 [DOI] [PubMed] [Google Scholar]
- 35. Bayot A., Basse N., Lee I., Gareil M., Pirotte B., Bulteau A. L., Friguet B., Reboud-Ravaux M. (2008) Toward the control of intracellular protein turnover. Mitochondrial Lon protease inhibitors versus proteasome inhibitors. Biochimie 90, 260–269 [DOI] [PubMed] [Google Scholar]
- 36. Frase H., Hudak J., Lee I. (2006) Identification of the proteasome inhibitor MG262 as a potent ATP-dependent inhibitor of the Salmonella enterica serovar Typhimurium Lon protease. Biochemistry 45, 8264–8274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hori O., Ichinoda F., Tamatani T., Yamaguchi A., Sato N., Ozawa K., Kitao Y., Miyazaki M., Harding H. P., Ron D., Tohyama M., M Stern D., Ogawa S. (2002) Transmission of cell stress from endoplasmic reticulum to mitochondria. Enhanced expression of Lon protease. J. Cell Biol. 157, 1151–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fukuda R., Zhang H., Kim J. W., Shimoda L., Dang C. V., Semenza G. L. (2007) HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 129, 111–122 [DOI] [PubMed] [Google Scholar]
- 39. Ngo J. K., Davies K. J. (2009) Mitochondrial Lon protease is a human stress protein. Free Radic. Biol. Med. 46, 1042–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Guillon B., Bulteau A. L., Wattenhofer-Donzé M., Schmucker S., Friguet B., Puccio H., Drapier J. C., Bouton C. (2009) Frataxin deficiency causes up-regulation of mitochondrial Lon and ClpP proteases and severe loss of mitochondrial Fe-S proteins. FEBS J. 276, 1036–1047 [DOI] [PubMed] [Google Scholar]
- 41. Bota D. A., Davies K. J. (2002) Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat. Cell Biol. 4, 674–680 [DOI] [PubMed] [Google Scholar]
- 42. Bayot A., Gareil M., Rogowska-Wrzesinska A., Roepstorff P., Friguet B., Bulteau A. L. (2010) Identification of novel oxidized protein substrates and physiological partners of the mitochondrial ATP-dependent Lon-like protease Pim1. J. Biol. Chem. 285, 11445–11457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Major T., von Janowsky B., Ruppert T., Mogk A., Voos W. (2006) Proteomic analysis of mitochondrial protein turnover: identification of novel substrate proteins of the matrix protease pim1. Mol. Cell. Biol. 26, 762–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. von Janowsky B., Knapp K., Major T., Krayl M., Guiard B., Voos W. (2005) Structural properties of substrate proteins determine their proteolysis by the mitochondrial AAA+ protease Pim1. Biol. Chem. 386, 1307–1317 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.