Background: RNAP from several species of Archaea and Eukarya is predicted to contain Fe-S clusters.
Results: We show here that Subunit D of RNAP from M. acetivorans contains two redox-active and oxygen-labile [4Fe-4S] clusters.
Conclusion: The [4Fe-4S] clusters are not required for D-L heterodimer formation but affect the stability of the heterodimer.
Significance: The [4Fe-4S] clusters may regulate the assembly and activity of RNAP in an oxidant-dependent manner.
Keywords: Archaea, Iron-Sulfur Protein, Oxidative Stress, Redox Regulation, RNA Polymerase, Transcription, Methanogen
Abstract
Subunit D of multisubunit RNA polymerase from many species of archaea is predicted to bind one to two iron-sulfur (Fe-S) clusters, the function of which is unknown. A survey of encoded subunit D in the genomes of sequenced archaea revealed six distinct groups based on the number of complete or partial [4Fe-4S] cluster motifs within domain 3. Only subunit D from strictly anaerobic archaea, including all members of the Methanosarcinales, are predicted to bind two [4Fe-4S] clusters. We report herein the purification and characterization of Methanosarcina acetivorans subunit D in complex with subunit L. Expression of subunit D and subunit L in Escherichia coli resulted in the purification of a D-L heterodimer with only partial [4Fe-4S] cluster content. Reconstitution in vitro with iron and sulfide revealed that the M. acetivorans D-L heterodimer is capable of binding two redox-active [4Fe-4S] clusters. M. acetivorans subunit D deleted of domain 3 (DΔD3) was still capable of co-purifying with subunit L but was devoid of [4Fe-4S] clusters. Affinity purification of subunit D or subunit DΔD3 from M. acetivorans resulted in the co-purification of endogenous subunit L with each tagged subunit D. Overall, these results suggest that domain 3 of subunit D is required for [4Fe-4S] cluster binding, but the [4Fe-4S] clusters and domain 3 are not required for the formation of the D-L heterodimer. However, exposure of two [4Fe-4S] cluster-containing D-L heterodimer to oxygen resulted in loss of the [4Fe-4S] clusters and subsequent protein aggregation, indicating that the [4Fe-4S] clusters influence the stability of the D-L heterodimer and therefore have the potential to regulate the assembly and/or activity of RNA polymerase in an oxidant-dependent manner.
Introduction
DNA-dependent RNA polymerase (RNAP)2 is a multisubunit enzyme that is essential in all cellular organisms where it acts in concert with a vast array of gene-specific activators, repressors, and general transcription factors to modulate the synthesis of RNA. The core catalytic component of RNAP is conserved in all three domains of life (Bacteria, Archaea, and Eukarya). The diversity in RNAPs lies in the number of RNAP complexes and their subunit composition. Eukaryotes have three separate nuclear RNAPs (Pol I, II, and III) that transcribe non-overlapping subsets of genes (1, 2). Each eukaryotic RNAP contains at least 10 subunits. In addition to the three RNAPs, higher plants possess Pol IV (NRPD1) and Pol V (NRPE1) that transcribe non-coding RNAs involved in RNA silencing (3). Archaea and Bacteria contain a single RNAP that transcribes all genes. Bacterial RNAP is the least complex, containing only the core component comprising five subunits (α2ββ′ω). Archaeal RNAP consists of 12–13 subunits (RpoA-P) depending on phyla, similar to the subunit composition of eukaryotic RNAPs. Archaeal RNAP is most similar to eukaryotic Pol II, which consists of 12 subunits (Rpb1–12). The similarities between archaeal and eukaryotic RNAP were confirmed following the recent elucidation of the structures of archaeal RNAP (4–6). One of the more striking features of the Sulfolobus solfataricus RNAP structure was the presence of an iron-sulfur (Fe-S) cluster in subunit D. Fe-S clusters are cofactors typically found in metabolic enzymes where they function in the transfer of electrons but also have catalytic, sensing, and structural roles (7). The function of Fe-S cluster(s) in RNAP is unknown.
Subunit D of archaeal RNAP contains three domains. Domains 1 and 2 are conserved in the homologous Rpb3 subunit of Pol II, AC40 subunit of Pol I/III, and bacterial α subunit (8). Domain 1 is divided into three regions and is involved in dimerization with subunit L (RpoL), the initial step in the assembly of RNAP (9, 10). Domain 2 is involved in the interaction with additional RNAP subunits and general transcription factors. Domain 2 of S. solfataricus subunit D contains four cysteines that form two intramolecular disulfides (4). However, these cysteines are not conserved in all RpoD/Rpb3/AC40/α subunits, and the four cysteines of domain 2 in yeast Rpb3 chelate a zinc ion (11). Domain 3 of S. solfataricus subunit D contains six cysteine residues, four of which are found in a typical [4Fe-4S] cluster binding motif. The structure of the D-L heterodimer from S. solfataricus revealed a [3Fe-4S] cluster present in domain 3 of subunit D ligated by Cys-183, Cys-203, and Cys-209 (4). The cluster is likely a [4Fe-4S] cluster in vivo with Cys-206 serving as the fourth ligand. The two remaining cysteines of domain 3 form an intramolecular disulfide. The [4Fe-4S] cluster binding motif is not restricted to S. solfataricus subunit D but is found in D and Rpb3/AC40 subunits in various species of Archaea and Eukarya. However, all bacterial α subunits lack the residues of domain 3 and therefore are not predicted to bind Fe-S clusters (8).
Previous phylogenetic analysis indicated that there are four groups of archaeal D subunits based on the number of complete [4Fe-4S] cluster binding motifs in domain 3 (8). A re-examination here of the cysteine content in subunit D domain 3 encoded in the genomes of sequenced archaea in the NCBI database indicates that the classification should be expanded to six distinct groups based on complete or partial [4Fe-4S] cluster motifs (see “Results”). Conservation of the [4Fe-4S] cluster binding motif(s) in RNAP in multiple archaeal lineages and in some eukaryotes suggests that the clusters serve an important role(s). The Fe-S clusters have been postulated to function in regulation of the de novo assembly of RNAP in which the presence of the cluster(s) is required for the formation of the D-L heterodimer (8), the first step in the assembly of RNAP in archaea.
Only strictly anaerobic archaea contain RNAP subunit D with two [4Fe-4S] cluster motifs. However, not every anaerobic archaeon possesses an RNAP subunit D with [4Fe-4S] cluster motifs. For example, all methane-producing archaea (methanogens) are strict anaerobes, but there is extensive diversity in the properties of domain 3 of subunit D. Moreover, the ability of subunit D to bind two [4Fe-4S] clusters has not been documented. We report herein that subunit D from the methanogen Methanosarcina acetivorans binds two [4Fe-4S] clusters, consistent with the presence of two [4Fe-4S] cluster binding motifs, and that the clusters are similar to the [4Fe-4S] clusters in ferredoxin. Expression studies combined with mutational analysis also demonstrated that although domain 3 of subunit D is required for [4Fe-4S] cluster binding the [4Fe-4S] clusters and amino acid residues of domain 3 are not required for the initial association of subunit D with subunit L. However, the [4Fe-4S] clusters are oxygen-labile, and oxygen-dependent loss of the [4Fe-4S] clusters impacts the structural stability of the D-L heterodimer. Implications for the regulation of RNAP assembly and activity by the [4Fe-4S] clusters in methanogens are presented.
EXPERIMENTAL PROCEDURES
Homology Model of M. acetivorans D-L Heterodimer
A structural model of the D-L heterodimer from M. acetivorans was generated by SWISS-MODEL (12). The Protein Data Bank coordinates of the S. solfataricus D-L heterodimer (Protein Data Bank code 2PA8) and ferredoxin-like domain of Archaeoglobus fulgidus adenylylsulfate reductase (Protein Data Bank code 1JNR) were used as templates for the generation of the structural model. The model generated for domain 3, including the two [4Fe-4S] clusters, was manually traced onto domain 3 of the S. solfataricus structure by Coot (13).
Plasmid Construction and Mutagenesis
For co-expression of M. acetivorans subunits D (RpoD) and L (RpoL) in Escherichia coli, the plasmid pMaRpoDL was constructed. Primers were used to amplify rpoL from M. acetivorans C2A genomic DNA, resulting in a product with NdeI and XhoI sites at the 5′- and 3′-ends, respectively. The PCR product was digested with NdeI and XhoI and ligated to pET21a that had been similarly digested. The resulting plasmid (pMaRpoL) contained rpoL fused with a C-terminal six-histidine (His) tag under the control of the T7 promoter. The gene encoding rpoD was amplified from M. acetivorans C2A genomic DNA using a forward primer with an NdeI site and a reverse primer containing a BamHI site. The PCR product was digested with NdeI and BamHI and ligated with pET21a, which was digested with the same enzymes. The resultant plasmid, pMaRpoD, contained an untagged rpoD gene under the control of the T7 promoter. pMaRpoD was digested with BglII and BamHI. The roughly 1-kb fragment containing the T7 promoter and rpoD was gel-extracted, purified, and ligated with BglII-digested pMaRpoL, creating the pMaRpoDL plasmid. pMaRpoDL contains both rpoD and rpoL, each regulated by a separate T7 promoter. Importantly, only subunit L contains a His tag, designated subunit L(His). E. coli DH5α was used as the parent strain for all manipulations and was grown in Luria broth or agar containing appropriate antibiotics at 37 °C.
PCR amplification was used to remove the domain 3 amino acid residues 171–221 from M. acetivorans subunit D, the region that encompasses both [4Fe-4S] cluster motifs. Primers RpoDFeS2KORev (5′-AAT GGT AAT TAC AGG CAT GTT TTT G-3′) and RpoDFeS1KOFor (5′-GTG GAC TTC TAT GAA AAC TCT TTT G-3′) were 5′-phosphorylated and used to amplify pMaRpoDL minus domain 3 of subunit D. The ends of the amplified product were blunt-ligated forming pDL408. In-frame deletion of the codons encoding amino acid residues 171–221 of subunit D was confirmed by DNA sequencing. Subunit D harboring the deletion was designated subunit DΔD3. Plasmid pDL408 was used for co-expression of subunit DΔD3 with subunit L(His).
Expression and Purification of Recombinant M. acetivorans D-L Heterodimer
All recombinant proteins were expressed in E. coli Rosetta (DE3) (pLacI) cells containing pRKISC, which increases the level of iron-sulfur cluster biogenesis proteins (14). Unless stated otherwise, cells were grown in Terrific broth containing 50 μg/ml ampicillin, 17 μg/ml chloramphenicol, and 5 μg/ml tetracycline at 37 °C with shaking at 250 rpm. Once an optical density at 600 nm of 0.5–0.7 was reached, the growth temperature was adjusted to 25 °C, 0.2 mm ferrous ammonium sulfate was added to the medium, and the culture was induced with the addition of 0.5 mm isopropyl β-d-thiogalactopyranoside. The cells were harvested by centrifugation 16 h after induction.
All subsequent purification procedures were performed anaerobically within an anaerobic chamber (Coy Laboratory Products) containing an atmosphere of 95% N2 and 5% H2. Approximately 10 g of cells was suspended in 35 ml of 20 mm Tris-HCl (pH 8) containing 500 mm NaCl, 2 mm benzamidine, and a few crystals of DNase I. The cells were lysed by two passages through a French pressure cell at over 110 megapascal. The lysate was centrifuged at 70,000 × g for 35 min at 4 °C. The supernatant containing the expressed protein(s) was filtered (pore size, 0.45 μm) and applied at a flow rate of 0.5 ml min−1 to a column containing 5 ml of Ni2+-agarose resin (Genscript). The column was then washed with 25 ml of 20 mm Tris-HCl (pH 8), 500 mm NaCl followed by a 25-ml wash with the same buffer containing 10 mm imidazole. Bound protein(s) was eluted from the column by the addition of 15 ml of 20 mm Tris-HCl (pH 8), 500 mm NaCl containing 250 mm imidazole. The eluate was concentrated using a Vivacell concentrator (Sartorius) with a 10,000-molecular weight cutoff under nitrogen flow inside the anaerobic chamber. The concentrated protein was desalted into 50 mm HEPES (pH 7.5) containing 150 mm NaCl using a PD-10 column (GE Healthcare). The desalted protein was either frozen under N2 at −80 °C or immediately processed by size exclusion chromatography.
Size exclusion chromatography was performed using a Sephacryl HiPrep S-200 gel filtration fast protein liquid chromatography column (GE Healthcare) with a Biologic LP system (Bio-Rad) within an anaerobic chamber. Samples were analyzed with a running buffer of 50 mm HEPES (pH 7.5) containing 150 mm NaCl and 2 mm DTT at a flow rate of 0.5 ml min−1. The column was calibrated with the following proteins having known molecular masses: β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), carbonic anhydrase (29 kDa), and cytochrome c (12.4 kDa). Samples (2 ml) containing 50–75 mg of total protein were loaded onto the column for each run.
D-L(His) proteins were reconstituted with iron and sulfide similarly to that described (15). All additions and incubations were done under anaerobic conditions at 4 °C. Briefly, the eluate containing subunit D and subunit L(His) from the Ni2+-agarose resin was concentrated and desalted into 50 mm Tris-HCl (pH 8.0). Approximately 50 mg of total protein was diluted in 50 ml of 50 mm Tris-HCl (pH 8.0) containing 2 mm β-mercaptoethanol in a 125-ml serum bottle. After 30 min, ferrous ammonium sulfate was added dropwise to a final concentration of 120 mm. After another incubation for 30 min, sodium sulfide was added dropwise to a final concentration of 120 mm. The protein/iron/sulfide mixture was incubated for 16 h at 4 °C. The protein was then concentrated using a stirred cell concentrator (Millipore) with a 10,000-molecular weight cutoff and subsequently desalted into 50 mm HEPES (pH 7.5) containing 150 mm NaCl and analyzed by size exclusion chromatography.
Analytical Methods
Protein concentrations were determined by the method of Bradford (16) using bovine serum albumin as a standard. The concentration of the D/L(His) heterodimer was also determined using the calculated extinction coefficient at 280 nm of 21,890 mm−1 cm−1. The results from both methods were in good agreement (<5% deviation). The iron and acid-labile sulfide contents of D-L(His) heterodimers were determined as described previously (17). The number of thiols in D-L(His) heterodimers was determined using Ellman's reagent (dithionitrobenzoate (DTNB)) (18). In brief, thiol measurements were performed in 100 mm phosphate buffer (pH 8) with 0.2 mm DTNB in both the presence and absence of 6 m guanidine hydrochloride. The absorbance at 412 nm was measured after room temperature incubation for 15 min. Extinction coefficients at 412 nm of 14,150 and 13,700 mm−1 cm−1 were used for sulfhydryl content determination in buffer or buffer containing guanidine hydrochloride, respectively. Mass spectrometry of purified proteins was performed at the University of Arkansas Statewide Mass Spectrometry facility (Fayetteville, AR). SDS-PAGE and Western blotting were performed by standard procedures. The anti-RpoDL antibody was raised in rabbit against recombinantly purified D-L(His) heterodimer.
Continuous wave electron paramagnetic resonance (EPR) spectra were measure at X-band (9-GHz) frequency on a Bruker EMX spectrometer fitted with the ER-4119-HS high sensitivity perpendicular-mode cavity. General EPR conditions were as follows: microwave frequency, 9.385 GHz; field modulation frequency, 100 kHz; field modulation amplitude, 0.6 millitesla. Sample-specific conditions are indicated in the figure legends.
Cloning, Expression, and Purification of Subunit D and DΔD3 with N-terminal His Tag in M. acetivorans
PCR was used to amplify rpoD from M. acetivorans C2A genomic DNA. The forward primer for the amplification contained the sequence for an NdeI restriction site and an N-terminal His tag (5′-ATT AAG GCA TAT GCA TCA TCA TCA TCA TCA TAC GAT GGA AGT AGA CAT TCT-3′), whereas the reverse primer contained a HindIII restriction site (5′-GGT GGT AAG CTT TCA GAG CTG GTC CAG AAT TGC-3′). The PCR product was digested with NdeI and HindIII and ligated with similarly digested pJK027A (19). The resulting plasmid was named pDL516. To construct a similar plasmid carrying the RpoDΔD3 deletion, the same procedure listed here was performed except pDL408 was used as a PCR template with the resulting plasmid named pDL409.
M. acetivorans strain WWM73 was transformed with pDL516 and pDL409 as described previously (20). Successful integration of the plasmid was determined as described (19), and the resulting strains were named DJL30 (His-subunit D (His-D)) and DJL31 (His-DΔD3). These strains allow for the tetracycline-regulated chromosomal expression of His-D or His-DΔD3. Cultures of DJL30 or DJL31 were grown anaerobically in HS medium (21) supplemented with methanol and sodium sulfide in the presence and absence of 100 μg/ml tetracycline at 37 °C. The cells were harvested by centrifugation at an optical density at 600 nm of 0.6 under anaerobic conditions.
All purification steps were performed in an anaerobic environment using either lysis buffer (10 mm Tris-HCl (pH 8), 1 m KCl, 10 μm ZnCl2, 10 mm imidazole, 15% glycerol), wash buffer (10 mm Tris-HCl (pH 8), 0.1 m KCl, 10 μm ZnCl2, 10 mm imidazole, 15% glycerol), or elution buffer (10 mm Tris-HCl (pH 8), 0.1 m KCl, 10 μm ZnCl2, 200 mm imidazole, 15% glycerol). Cell pellets were thawed and suspended on ice in lysis buffer supplemented with a few crystals of DNase I and benzamidine HCl. Cells were lysed by sonication, and clarified lysates were obtained after a 35-min centrifugation step at 70,000 × g at 4 °C. The soluble fraction was passed through a 0.45-μm filter and slowly loaded onto a Ni2+-agarose column pre-equilibrated with lysis buffer. The column was washed with 10 column volumes of lysis buffer followed by 20 column volumes of wash buffer. Protein was eluted from the column with the addition of 3 column volumes of elution buffer. Eluates were stored anaerobically in sealed vials at −80 °C.
RESULTS
Archaeal RNAP Subunit D Classification and Diversity of Methanogen Subunit D
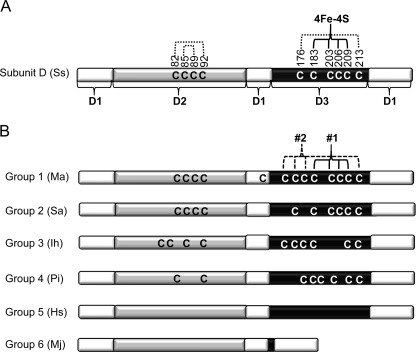
A survey of RNAP D subunits from the 99 sequenced archaeal genomes currently in the NCBI database reveals six distinct groups based on the absent, complete, or partial [4Fe-4S] cluster motifs of domain 3. The domain organization and cysteine residue content of a representative subunit D from each group is shown in Fig. 1 with S. solfataricus subunit D shown as a reference. A complete list of subunit D from individual species and their grouping is listed in supplemental Table S1: briefly, group 1, complete [4Fe-4S] cluster 1 and 2 motifs (restricted to strict anaerobes); group 2, a complete [4Fe-4S] cluster 1 motif; group 3, a complete [4Fe-4S] cluster 2 motif; group 4, partial [4Fe-4S] cluster 1 and/or 2 motif; group 5, absence of [4Fe-4S] cluster motif cysteines; group 6, lack of amino acid residues of domain 3 (only Methanococcales). The corresponding subunit Rpb3 in Pol II or AC40 Pol I/III in certain eukaryotes contains only [4Fe-4S] cluster binding motif 1 and therefore falls in archaeal group 2.
FIGURE 1.
Domain architecture and diversity of cysteine residue content of subunit D of archaeal RNAP. A, domain architecture and cysteine content of subunit D from S. solfataricus (Ss) (4). The regions comprising domains 1, 2, and 3 (D1–D3) are indicated and shaded differently. The cysteine residues that function as ligands to a [4Fe-4S] cluster are indicated by 4Fe-4S. Cysteine residues documented to participate in disulfide bonds are indicated by dotted lines. B, diagram depicting the six distinct groups of archaeal subunit D based on the number of complete or partial [4Fe-4S] cluster motifs within domain 3. The domain architecture is the same as that depicted in A. The two [4Fe-4S] cluster motifs in group 1 subunit D are labeled 1 and 2 with 1 corresponding to the single [4Fe-4S] cluster motif in S. solfataricus subunit D. Representative members of each group are shown: Ma, M. acetivorans; Sa, Sulfolobus acidocaldarius; Ih, Ignicoccus hospitalis; Pi, Pyrobaculum islandicum; Hs, Halobacterium salinarum; Mj, Methanocaldococcus jannaschii.
All members of an order typically encode a subunit D within the same group, indicating that the presence or absence of the clusters is correlated with particular taxonomic lineages. For example, all Halobacteriales encode a group 5 subunit D, one that is devoid of any cysteine residues. Interestingly, group 1 subunit D is found only in strictly anaerobic archaea but is not a universal feature of strict anaerobes. For example, all methanogenic archaea are strict anaerobes and are only capable of growth by methanogenesis using a limited number of substrates, including acetate, H2/CO2, and methylated compounds (22). However, despite the uniform mode of growth and nutritional requirements, there is extensive diversity in the cysteine residue content of subunit D, in particular the number of [4Fe-4S] cluster motifs (supplemental Fig. S1). Subunit D from all sequenced methanogens, except for members of the Methanococcales, contains four cysteines in domain 2. Members of Methanosarcinales, Methanomicrobiales, and Methanobacteriales encode subunit D that contains one cysteine residue in domain 1. The majority of methanogens encode subunit D with a domain 3 predicted to bind at least one [4Fe-4S] cluster. All members of the Methanosarcinales and Methanomicrobiales encode a group 1 subunit D, whereas members of Methanobacteriales encode a group 1 or 2 subunit D. All species of the Methanococcales encode subunit D that lacks the amino acid residues of domain 3 and are the only members of group 6. The sole species of Methanopyrales encodes a group 4 subunit D. Thus, methanogens provide a unique opportunity to examine the effect of the presence or absence of the [4Fe-4S] clusters within a single group of organisms and correlate results obtained with the metabolism and environment of particular species.
Homology Model of Group 1 Subunit D from M. acetivorans in Complex with Subunit L
The genome of M. acetivorans contains a single rpoD gene encoding a 266-amino acid protein with 32% identity to the 265-amino acid subunit D from S. solfataricus. However, in the original annotation of the M. acetivorans genome, rpoD was incorrectly identified as two separate genes (NTO2MA1356–7) (23). Based on the x-ray crystal structure of the S. solfataricus D-L heterodimer (4), a homology model of the M. acetivorans subunit D in complex with subunit L was generated (Fig. 2).
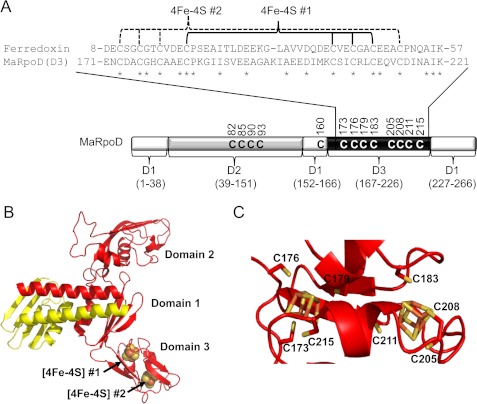
FIGURE 2.
Domain 3 of subunit D of RNAP from M. acetivorans is predicted to bind two [4Fe-4S] clusters similar to two-[4Fe-4S] cluster ferredoxin. A, schematic of the domain architecture of M. acetivorans subunit D, including an amino acid alignment of domain 3 (MaRpoD (D3)) to two-[4Fe-4S] cluster ferredoxin (MA0431) from M. acetivorans. Conserved residues are indicated by an asterisk, including the cysteine residues postulated to bind the two [4Fe-4S] clusters. D1–D3, domains 1–3. B, homology model of the heterodimer of M. acetivorans subunit D (red) and subunit L (yellow) D-L heterodimer. The two [4Fe-4S] clusters in domain 3 are represented by sphere models. C, close-up view of domain 3 of subunit D showing putative cysteine ligands to each [4Fe-4S] cluster. The two [4Fe-4S] clusters in domain 3 are represented in stick models.
Similar to S. solfataricus subunit D, M. acetivorans subunit D contains three domains (Fig. 2A). However, there are differences in the cysteine content of each domain between S. solfataricus and M. acetivorans D subunits. Domain 1 makes up the dimerization interface with subunit L. M. acetivorans subunit D contains a single cysteine residue (Cys-160) in domain 1 unlike that of S. solfataricus. Domain 2 of M. acetivorans subunit D contains four cysteine residues similar to S. solfataricus; however, there is a difference in the spacing of the cysteines. Domain 3 of M. acetivorans subunit D contains eight cysteine residues that comprise the two [4Fe-4S] cluster motifs (group 1 subunit D), whereas domain 3 of S. solfataricus subunit D contains six cysteines (group 2 subunit D). Moreover, a 50-amino acid region of M. acetivorans subunit D domain 3 (residues 171–221) has 39% identity to the predicted 60-amino acid two-[4Fe-4S] ferredoxin encoded by ma0431 in the M. acetivorans genome (Fig. 2A). Based on the homology model of the M. acetivorans D-L heterodimer, neither domain 2 nor domain 3 is in contact with subunit L, indicating that they are likely unnecessary for dimerization with subunit L. In addition, all members of the Methanoccocales completely lack the amino acid residues of domain 3, suggesting that domain 3 is likely not necessary for dimerization with subunit L in methanogens.
Expression and Purification of Recombinant M. acetivorans D-L Heterodimer
To determine the capacity of M. acetivorans subunit D to bind Fe-S clusters and whether the clusters are required for the interaction and stable association of subunit D with subunit L, each subunit was separately expressed with a C-terminal His tag in E. coli. Subunit L(His) expressed in aerobically grown E. coli was found in the soluble fraction of cell lysates and was purified to homogeneity using Ni2+ affinity and size exclusion chromatography (data not shown). However, subunit D(His) expressed in aerobically grown E. coli was found only in inclusion bodies in the insoluble fraction of cell lysates. Expression of untagged subunit D or subunit D with a glutathione S-transferase (GST) tag also resulted in the formation of inclusion bodies (data not shown). The formation of inclusion bodies indicates that subunit D is improperly folded, potentially as a result of the absence of subunit L or incomplete Fe-S cluster incorporation. The lack of Fe-S clusters incorporated into subunit D may be a result of limiting cellular levels of iron, sulfur, or iron-sulfur biogenesis machinery during protein expression in E. coli. However, subunit D expressed in anaerobically grown E. coli containing pRKISC, which encodes iron-sulfur cluster biogenesis proteins and has been shown to increase the cluster content in recombinant Fe-S proteins (24), combined with the addition of supplemental iron still resulted in the protein being found in insoluble inclusion bodies (data not shown).
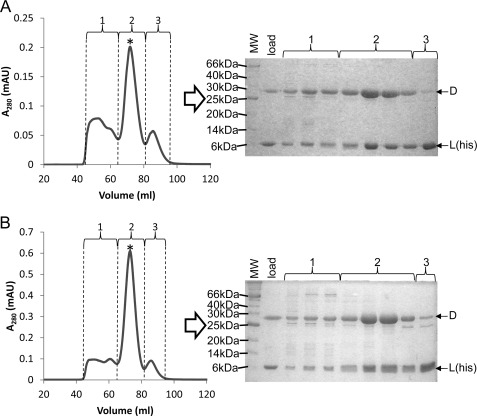
Subunit D was subsequently co-expressed with subunit L(His) in aerobically grown E. coli. Size exclusion chromatography and SDS-PAGE analyses of imidazole-eluted protein from the Ni2+-agarose loaded with cell lysate from E. coli co-expressing subunit D and subunit L(His) revealed that subunit D co-purifies with subunit L(His) (Fig. 3A). Three major peaks were detected, all of which contained subunit D and/or subunit L(His). Based on the elution profile of known molecular mass standards, the elution volume of peak 1 is consistent with D-L(His) complexes larger than a heterodimer (molecular mass, >45 kDa). The elution volume of peak 3 is consistent with the molecular weight of subunit L(His) (molecular mass, 11.1 kDa). The elution volume of the major peak (peak 2) is consistent with a heterodimer of D-L(His) (molecular mass, 40.1 kDa). The presence of higher molecular weight D-L(His) complexes as well as subunit L(His) monomers may be a result of differences in expression levels of subunit D and subunit L(His). Co-purification of untagged subunit D with subunit L(His) combined with the insolubility of subunit D in the absence of subunit L under the same conditions indicates that subunit D is likely unstable or improperly folded when not associated with subunit L.
FIGURE 3.
Purification of recombinant M. acetivorans D-L(His) and D-L(His)-FeS heterodimers. A, anaerobic size exclusion chromatography of eluate (∼50 mg of total protein) from Ni2+ affinity chromatography of E. coli cell lysate containing M. acetivorans subunit L(His) and subunit D. B, same as A except that the eluate was reconstituted with iron and sulfide prior to loading onto the size exclusion column (see “Experimental Procedures” for details). For each purification, samples from fractions containing peaks 1, 2, and 3 were analyzed by SDS-PAGE as indicated. The asterisk indicates the peak that is consistent with a the molecular mass of a D-L(His) heterodimer (40.2 kDa). mAU, milliabsorbance units.
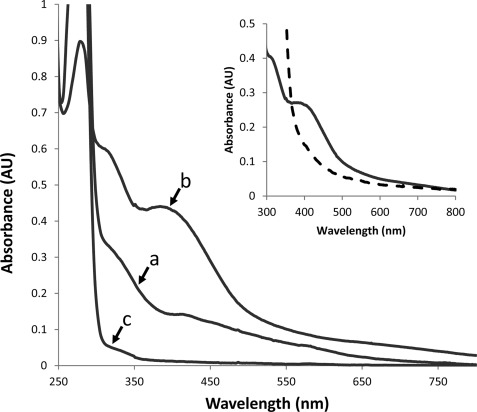
Purified D-L(His) heterodimer was red-brown in color, indicative of the presence of a chromophore. The UV-visible spectrum of the D-L(His) heterodimer contained absorbance maxima centered at 320 and 390 nm in addition to 280 nm (Fig. 4), consistent with the presence of Fe-S clusters (25). Although M. acetivorans subunit D is predicted to contain two [4Fe-4S] clusters, the A390/A280 ratio is low, and the experimentally determined iron and acid-labile sulfide contents of as-purified D-L(His) heterodimer is substantially less than predicted (Table 1). The ability to purify D-L(His) heterodimer, which lacks two [4Fe-4S] clusters, indicates that the presence of both clusters is not required for the interaction and dimerization of M. acetivorans subunit D with subunit L. Attempts to increase the cluster content by co-expression of subunit D with subunit L(His) in anaerobically grown E. coli containing pRKISC and supplementation of the medium with iron did not significantly increase the cluster content of as-purified D-L(His) heterodimer (data not shown).
FIGURE 4.
UV-visible spectra of purified D-L(His), D-L(His)-FeS, and DΔD3/L(His) heterodimers. a, D-L(His) (67.5 μm); b, D-L(His)-FeS (13.5 μm); c, DΔD3/L(His) (67.5 μm). The inset shows the D-L(His)-FeS heterodimer before (solid line) and after (dashed line) the addition of sodium dithionite (10 mm). AU, absorbance units.
TABLE 1.
Comparison of properties of the M. acetivorans D-L(His), D-L(His)-FeS, and DΔD3-L(His) heterodimers
| Heterodimer | A390/A280 | ϵ390 | Irona | Sulfideb | Thiolc | Thiold, denatured |
|---|---|---|---|---|---|---|
| mm−1cm−1 | ||||||
| D-L(His) | 0.06 | 2.2 ± 0.08 | 1.1 ± 0.1 | 1.1 ± 0.05 | 13.8 ± 0.4 | 14.5 ± 0.7 |
| D-L(His)-FeS | 0.50 | 33.2 ± 0.6 | 8.3 ± 0.2 | 7.7 ± 0.8 | 16.2 ± 0.3 | 21.5 ± 1.6 |
| DΔD3-L(His) | 0.01 | 0.2 ± 0.01 | NDe | ND | 5.9 ± 0.7 | 5.4 ± 1.5 |
a nmol of iron/nmol of DL heterodimer.
b nmol of acid-labile sulfide/nmol of DL heterodimer.
c nmol of thiol/nmol of DL heterodimer.
d Measured in buffer containing 6 m guanidine HCl.
e ND, not determined.
Reconstitution of [4Fe-4S] Clusters in Recombinant M. acetivorans D-L(His) Heterodimer
To determine the full capacity and type of Fe-S clusters within M. acetivorans subunit D, in vitro reconstitution of the eluate from the Ni2+-agarose column containing a mixture of subunit D and subunit L(His) was performed by incubation with iron and sulfide as described previously (15). The elution profile from size exclusion chromatography of the Fe/S-reconstituted sample was identical to that of the non-reconstituted sample (Fig. 3). However, the major peak (peak 2) corresponding to the D-L(His) heterodimer was of greater intensity than that of non-reconstituted D-L(His) heterodimer despite loading a similar amount of protein onto the column. The fractions containing peak 2 from the Fe/S-reconstituted sample were dark brown in color, indicative of increased Fe-S cluster content. These fractions were collected and concentrated, and the sample was designated as the D-L(His)-FeS heterodimer. The D-L(His)-FeS heterodimer had a substantial increase in the A390/A280 ratio and extinction coefficient at 390 nm compared with the D-L(His) heterodimer (Table 1), consistent with an increase in Fe-S cluster content. Additionally, compared with the non-reconstituted D-L(His) heterodimer, the D-L(His)-FeS heterodimer contained 8 times the amount of iron and acid-labile sulfide (Table 1), consistent with the incorporation of two [4Fe-4S] clusters into subunit D. Importantly, this experimental protocol is reproducible as additional preparations of Fe/S-reconstituted D-L(His) heterodimer had similar levels of iron and acid-labile sulfide (data not shown). These data support that M. acetivorans subunit D coordinates two [4Fe-4S] clusters.
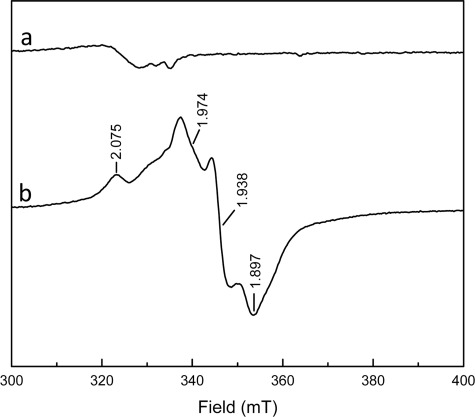
The D-L(His)-FeS heterodimer showed a broad absorption band centered around 390 nm and a second peak centered around 300 nm similar to the non-reconstituted heterodimer (Fig. 4). The similarity of domain 3 of M. acetivorans subunit D to two-[4Fe-4S] ferredoxin, which functions in electron transfer (26, 27), indicates that the [4Fe-4S] clusters in subunit D may have the capacity to participate in oxidation-reduction reactions. The addition of the reductant dithionite to the D-L(His)-FeS heterodimer under anaerobic conditions resulted in a decrease of the absorption band at 390 nm, consistent with reduction of the clusters. EPR spectroscopic analyses of the D-L(His)-FeS heterodimer in the absence of reductant did not reveal any significant signals (Fig. 5). The lack of a g = 2.01 EPR signal under these conditions indicates that the D-L(His)-FeS heterodimer does not contain [3Fe-4S]+ clusters, which is consistent with the elemental analyses (Table 1). Addition of dithionite to the sample of D-L(His)-FeS resulted in the appearance of EPR signals with g values at 2.075, 1.974, 1.938, and 1.897 (Fig. 5). The complex gav = 1.94 is consistent with the presence of two [4Fe-4S]+ clusters as is seen with ferredoxins harboring two spin-coupled [4Fe-4S] clusters (28, 29). The complexity is caused by spin-spin coupling of the two clusters in close vicinity of each other. Taken together, these results support that the D-L(His)-FeS heterodimer contains two redox-coupled [4Fe-4S]2+/+ clusters that can possibly participate in one-electron transfers. EPR spectroscopic analyses of the non-reconstituted D-L(His) heterodimer did not produce an EPR signal in the as-purified state or after the addition of dithionite, which may be attributed to insufficient clusters in the samples analyzed. Attempts to obtain more concentrated samples of non-reconstituted D-L(His) for EPR analysis resulted in the precipitation of the protein (data not shown).
FIGURE 5.
EPR spectra of D-L(His)-FeS heterodimer before (a) and after the addition of dithionite (b). The sample contained 150 μm D-L(His)-FeS in 50 mm HEPES, 50 mm NaCl (pH 7.5). EPR conditions were as follows: microwave frequency, 9.385 GHz; microwave power incident to the cavity, 2.0 milliwatts; field modulation frequency, 100 kHz; microwave amplitude, 0.6 millitesla (mT); temperature, 8 K.
Domain 3 of Subunit D Is Required for [4Fe-4S] Cluster Binding but Is Not Required for Interaction with Subunit L
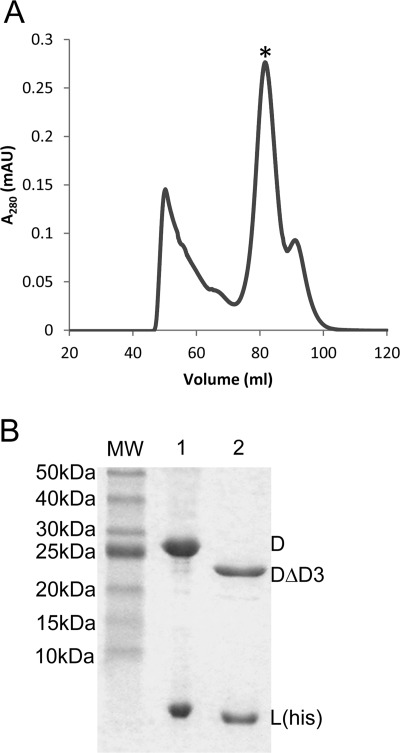
To determine whether domain 3 of subunit D is required for [4Fe-4S] cluster binding and dimerization with subunit L, a variant of subunit D deleted of amino acid residues 171–221 (domain 3) was generated. This variant (subunit DΔD3) was co-expressed with subunit L(His) and subjected to the same purification procedure as that used for wild-type subunit D. The elution profile from the size exclusion column for the sample containing subunit DΔD3 and subunit L(His) was similar to that observed for purification of the wild-type D-L(His) heterodimer (Fig. 6). However, the DΔD3/L(His) heterodimer eluted at a greater elution volume, consistent with the expected smaller size of the DΔD3/L(His) heterodimer (35 kDa) compared with wild-type heterodimer (40 kDa). The sizes of D subunits and the presence of subunit L(His) in each heterodimer were confirmed by SDS-PAGE (Fig. 6). The ability of subunit DΔD3 to co-purify with subunit L(His) and form a stable heterodimer reveals that domain 3 is not required for the interaction of subunit D with subunit L. Moreover, deletion of domain 3 from subunit D resulted in a heterodimer devoid of [4Fe-4S] clusters as revealed by UV-visible spectroscopy and elemental analyses (Fig. 4 and Table 1). These results support that the eight cysteine residues in domain 3 are necessary for the binding of the two [4Fe-4S] clusters in M. acetivorans subunit D.
FIGURE 6.
Co-purification of M. acetivorans subunit DΔD3 with subunit L(His) after co-expression in E. coli. A, anaerobic size exclusion chromatography of eluate (∼50 mg of total protein) from Ni2+ affinity chromatography of E. coli cell lysate containing M. acetivorans subunits L(His) and DΔD3. The asterisk indicates the peak that is consistent with a the molecular mass of a heterodimer of DΔD3-L(His) (34.8 kDa). B, SDS-PAGE analysis of the purified D-L(His) (lane 1) and DΔD3-L(His) (lane 2) heterodimers, confirming the smaller molecular mass of DΔD3 (23.3 kDa) compared with D (28.7 kDa). mAU, milliabsorbance units.
Formation of M. acetivorans D-L Heterodimer Does Not Require Disulfides in Subunit D
M. acetivorans subunit D contains 13 cysteine residues (Fig. 2). M. acetivorans subunit L has one cysteine residue. Each cysteine may be in the free reduced state (thiol), participate in disulfide bonding, or serve to coordinate a cofactor (e.g. [4Fe-4S] cluster). The structure of the S. solfataricus D-L heterodimer revealed that each cysteine that is not a ligand to the [4Fe-4S] cluster participates in disulfide bonding, suggesting that disulfides may be required for the formation and stability of the heterodimer (4). Thus, the number of thiols in purified M. acetivorans D-L(His) heterodimers was quantified using the thiol-specific reagent DTNB to infer the presence or absence of disulfides (18).
The purified D-L(His) heterodimers were first incubated in non-denaturing buffer containing DTNB under anaerobic conditions. Incubation of the D-L(His) heterodimer with DTNB resulted in the release of TNB corresponding to 14 thiols (Table 1), indicating that all 14 cysteine residues are in the reduced state. The DΔD3/L(His) heterodimer reacted with DTNB under non-denaturing conditions to release TNB corresponding to six thiols, consistent with the deletion of the eight cysteine residues of domain 3. There was no significant difference in the reactivity of each heterodimer with DTNB under denaturing conditions, indicating that each cysteine is readily accessible to DTNB in the folded state of the heterodimer. This result is consistent with the model showing that the domain 2 and 3 cysteine residues are relatively surface-exposed within the heterodimer (Fig. 2). The cysteine residues of domain 2 of Rbp3 from yeast have been shown to coordinate a zinc ion instead of participating in disulfide bonding. However, the reaction of Cys-82, Cys-85, Cys-90, and Cys-93 of domain 2 of M. acetivorans subunits D and DΔD3 with DTNB under both non-denaturing and denaturing conditions suggests that these residues do not bind zinc or participate in disulfides under the conditions examined.
The quantitation of thiols in the D-L(His)-FeS heterodimer was more variable. Incubation of D-L(His)-FeS with DTNB under non-denaturing conditions produced TNB corresponding to 16 thiols (Table 1). The number of thiols detected in the D-L(His)-FeS heterodimer is likely a combination of TNB produced by thiols and by sulfide released from the [4Fe-4S] clusters as the reaction of one sulfide anion with DTNB yields two TNB anions (30). The addition of denaturant resulted in an increase in the number of thiols detected, consistent with an increase in sulfide released from the [4Fe-4S] clusters. These data indicate that the cysteine residues of the purified M. acetivorans D-L heterodimers are either in the thiol state or participate in coordination of the [4Fe-4S] clusters, indicating that disulfides are not required for dimerization of M. acetivorans subunit D with subunit L.
[4Fe-4S] Clusters of M. acetivorans Subunit D Are Oxygen-labile and Affect Structural Stability of D-L Heterodimer
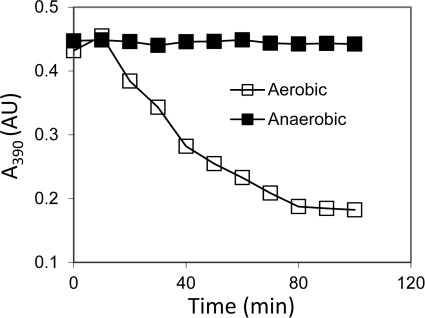
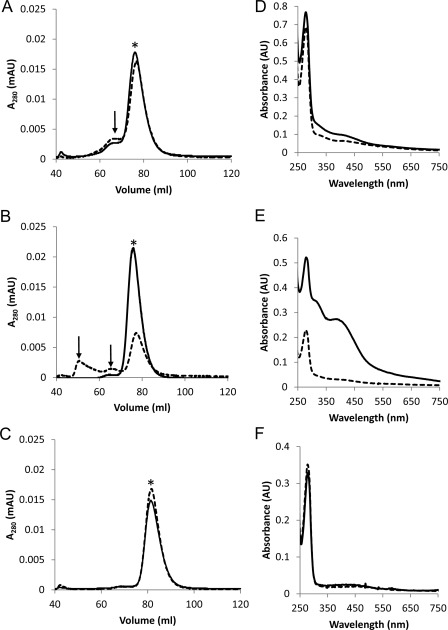
The Fe-S clusters in proteins from methanogens are often oxygen-labile, including the [4Fe-4S] clusters in ferredoxin from Methanosarcina (28). The exposure of the D-L(His)-FeS heterodimer to air resulted in a time-dependent decrease in the absorbance at 390 nm, whereas no significant decrease at 390 nm was seen when the heterodimer was kept anaerobic (Fig. 7). To determine whether destruction of the clusters due to oxygen exposure affects the stability of the D-L(His) heterodimer, oxygen-induced structural changes were examined by size exclusion chromatography. Each purified heterodimer was incubated under nitrogen or air for 1 h and then subjected to size exclusion chromatography under anaerobic conditions (Fig. 8, A–C). For each sample, the fractions corresponding to heterodimer were collected and concentrated, and the UV-visible spectrum was recorded (Fig. 8, D and E). The elution profile of the non-reconstituted D-L(His) heterodimer following anaerobic incubation resulted in a major heterodimer peak and a minor peak, which was consistent with a larger molecular weight aggregate of D-L(His). Exposure to oxygen resulted in a very slight increase in the population of the minor, non-heterodimeric species. Importantly, oxygen did not cause disassociation of subunit D and subunit L as indicated by the lack of peaks that corresponded to D or L monomers. Compared with the collected, anaerobically incubated heterodimer, a decrease in absorbance in the 300–420 nm range of the collected aerobically incubated heterodimer was observed, consistent with some loss of the small amount of Fe-S cluster present in the non-reconstituted D-L(His) heterodimer.
FIGURE 7.
Effect of oxygen on stability of the [4Fe-4S] clusters in purified D-L(His)-FeS heterodimer. The loss of the [4Fe-4S] clusters in purified D-L(His)-FeS (20 μm) incubated under N2 or in air in 50 mm HEPES, 50 mm NaCl (pH 7.5) was monitored by measuring A390 over time. AU, absorbance units.
FIGURE 8.
Effect of oxygen on stability of D-L(His), D-L(His)-FeS, and DΔD3-L(His) heterodimers. Each heterodimer was incubated anaerobically or aerobically in 50 mm HEPES, 50 mm NaCl (pH 7.5) and analyzed by size exclusion chromatography under anaerobic conditions using a running buffer of 50 mm HEPES (pH 7.5), 150 mm NaCl. Elution profiles of each heterodimer after anaerobic (solid line) or aerobic incubation (dashed line) are shown in A–C. A, D-L(His), 1.1 mg of each; B, D-L(His)-FeS, 0.8 mg of each; C, DΔD3/L(His), 1.5 mg of each. For each elution profile, an asterisk indicates the heterodimer peak, and arrows indicate D-L(His) aggregates. The UV-visible spectra of concentrated fractions containing heterodimer from anaerobic samples (solid line) and aerobic samples (dashed line) are shown in D–F. D, D-L(His), 26 μm each; E, D-L(His)-FeS, 10 μm each; F, DΔD3-L(His),18 μm each. AU, absorbance units; mAU, milliabsorbance units.
The elution profile of the anaerobically incubated D-L(His)-FeS heterodimer displayed a single heterodimer peak. Although the non-reconstituted D-L(His) heterodimer exhibited some aggregation after anaerobic incubation, the lack of this aggregation by the D-L(His)-FeS suggests that the heterodimer is more stable (i.e. has less structural heterogeneity) when both [4Fe-4S] clusters are present. In contrast, a substantial increase in the structural heterogeneity of the D-L(His)-FeS heterodimer was observed following aerobic incubation as indicated by the appearance of two additional peaks in the elution profile (Fig. 8B). The increased structural heterogeneity is a result of oxygen-induced [4Fe-4S] cluster loss in the D-L(His)-FeS sample as revealed by the UV-visible spectrum of the collected heterodimer sample (Fig. 8E). The most likely explanation for the increased aggregation observed in the D-L(His)-FeS heterodimer, compared with the D-L(His) heterodimer, upon oxygen exposure is iron-catalyzed production of reactive oxygen species, leading to amino acid oxidation and protein aggregation. The presence of only a heterodimer peak in the elution profile of both anaerobically incubated and aerobically incubated DΔD3/L(His) heterodimer demonstrates that domain 3 is involved in the formation of the D-L(His) aggregates. These results suggest that the presence or absence of domain 3 along with the [4Fe-4S] clusters affects the structural stability of the D-L heterodimer. Overall, these data support that the presence of both [4Fe-4S] clusters is important for the integrity of domain 3 and serves to stabilize the D-L heterodimer.
Domain 3 of Subunit D Is Not Required for Formation of D-L Heterodimer within M. acetivorans
Co-purification of subunit DΔD3 with subunit L(His) after co-expression in E. coli along with the complete lack of domain 3 in subunit D from all members of the Methanococcales indicates that domain 3 of subunit D is not required for the formation of the D-L heterodimer within M. acetivorans. To test this hypothesis, two merodiploid strains of M. acetivorans were constructed. In addition to expressing native subunit D, these strains contain a second rpoD gene that encodes an N-terminally His-tagged subunit D and is under the control of a tetracycline-dependent promoter. Strain DJL30 expresses His-D, and strain DJL31 expresses His-subunit DΔD3 (His-DΔD3). Importantly, subunit L was not tagged or overexpressed in these strains. Western blot analysis with anti-D-L antibodies of lysates from induced DJL30 cells did not reveal an increase in the concentration of subunit D when compared with uninduced cells (data not shown). The inability to detect an increase in the cellular levels of subunit D in the merodiploid His-D expression strain is likely due to the degradation of unassembled subunits by proteases, which has been seen in yeast (31). This result is consistent with His-D competing with native D for association with endogenous subunit L and assembly into RNAP with unassembled His-D or D being degraded. Inducible expression was seen in the DJL31 strain where a smaller immunoreactive protein (His-DΔD3) was detected only when DJL31 cells were grown in the presence of tetracycline (data not shown). Strains DJL30 and DJL31 grown in the absence and presence of tetracycline exhibited a similar growth rate and cell yield (data not shown), indicating that the expression of subunit D containing an affinity tag or the presence of subunit DΔD3 is not detrimental to M. acetivorans.
To verify expression and to determine whether His-D and His-DΔD3 can associate with native subunit L, clarified cell lysates from strains DJL30 and DJL31 grown in the presence or absence of tetracycline were used for Ni2+ affinity purification. Imidazole eluates from the columns were analyzed by Western blot using anti-D-L antibodies. A band consistent with subunit D or DΔD3 was only detected in the eluate from lysate of cells grown in the presence of tetracycline (Fig. 9), consistent with inducible expression of His-D and His-DΔD3 in strains DJL30 and DJL31, respectively. Moreover, endogenous subunit L (untagged) co-purified with both subunit His-D and His-DΔD3, revealing that domain 3 of subunit D is not required for the formation of the D-L heterodimer within M. acetivorans (Fig. 9). The additional immunoreactive proteins in the eluates are degradation products of unassembled His-D and His-DΔD3, similar to the appearance of Rpb3 degradation products when affinity-tagged Rpb3 is expressed in yeast (31–33).
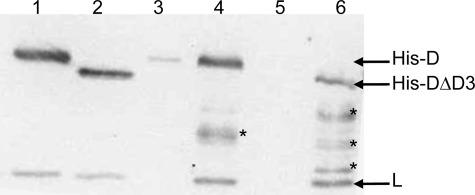
FIGURE 9.
Co-purification of endogenous subunit L with His-tagged subunit D or subunit DΔD3 expressed within M. acetivorans. Shown is a Western blot of a 15% SDS-polyacrylamide gel with anti-D-L antibodies. Lane 1, recombinant D-L(His) heterodimer (50 ng); lane 2, recombinant DΔD3-L(His) heterodimer (150 ng); lane 3, imidazole eluate from a Ni2+-agarose column loaded with cell lysate of DJL30 grown in the absence of tetracycline; lane 4, imidazole eluate from a Ni2+-agarose column loaded with cell lysate of DJL30 grown in the presence of tetracycline; lane 5, imidazole eluate from a Ni2+-agarose column loaded with cell lysate of DJL31 grown in the absence of tetracycline; lane 6, imidazole eluate from a Ni2+-agarose column loaded with cell lysate of DJL31 grown in the presence of tetracycline. The asterisks indicate subunit His-D degradation products.
DISCUSSION
The results presented herein reveal the ability of subunit D of RNAP from M. acetivorans to bind two redox-active and oxygen-labile [4Fe-4S] clusters. Given that all archaeal species harboring group 1 subunit D are strictly anaerobic, similar to M. acetivorans, it is highly likely that all group 1 subunit D contain two [4Fe-4S] clusters. However, the presence of a group 1 subunit D is not a universal feature of strictly anaerobic archaea. In fact, there is extensive diversity in the properties of subunit D within the five orders of methanogens, indicating that the [4Fe-4S] clusters are not essential to the function of RNAP in methanogens. The presence of one or two [4Fe-4S] clusters in subunit D of archaeal RNAP likely imparts some advantage to those species that have acquired or retained this feature within their RNAP. Other DNA processing and repair enzymes, including helicase, primase, glycosylases, and endonucleases, bind [4Fe-4S] clusters (34), indicating that these cofactors are a common, but not universal, feature of DNA processing enzymes. In many of these enzymes, the [4Fe-4S] cluster is important to the structural stability of the enzyme and serves to monitor the redox state of the cell and modulate enzyme activity. For example, the DNA damage-inducible protein DinG, a DNA helicase, contains a [4Fe-4S] cluster that regulates DinG helicase activity (35). The [4Fe-4S] clusters in RNAP may serve a similar role. Based on previous structure-function studies of bacterial α N-terminal domain (36, 37), eukaryotic Rpb3 (32, 38), and archaeal D of RNAPs (2) along with results obtained with M. acetivorans subunit D, a plausible function for RNAP clusters is to regulate the de novo assembly of RNAP or to modulate the postassembly activity of RNAP. A proposed model for the possible role(s) of the [4Fe-4S] clusters in M. acetivorans subunit D is presented in Fig. 10.
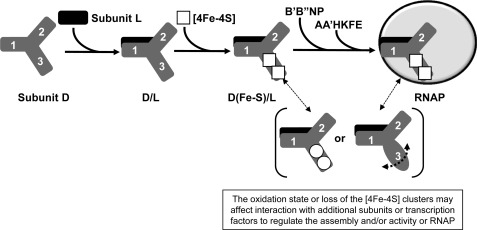
FIGURE 10.
Model depicting potential roles of subunit D [4Fe-4S] clusters in modulating assembly and/or activity of M. acetivorans RNAP. The three domains of subunit D are represented by 1, 2, and 3. A change in the reduction state of each [4Fe-4S] cluster is indicated by a circle versus a square. The dashed semicircle arrow depicts the increased flexibility of domain 3 upon loss of the [4Fe-4S] clusters.
Iron is used as a cofactor in numerous metabolic enzymes in virtually all organisms. This is especially true in anaerobes, including methanogens. Methanogens are predicted to contain the largest number of [4Fe-4S] proteins, many of which are oxygen-labile (39). Many enzymes directly involved in methanogenesis, including hydrogenases, carbon-monoxide dehydrogenase, and heterodisulfide reductase, contain Fe-S clusters (40, 41). These enzymes are rapidly inactivated in the presence of oxygen due to the oxidation and/or destruction of the Fe-S clusters, resulting in a decrease in the energy available for biosynthesis. Moreover, two-[4Fe-4S] ferredoxin is a key electron transfer protein that interacts with a number of redox partners, including carbon-monoxide dehydrogenase (40). In M. acetivorans and other organisms, the [4Fe-4S] clusters in ferredoxin-like domain 3 of subunit D may impart the ability to directly integrate global gene expression with the metabolic state of the cell.
One plausible role for the [4Fe-4S] clusters is to regulate the assembly of RNAP in response to cellular conditions, and this is supported by previous studies documenting archaeal subunit D, bacterial α subunit, and eukaryotic Rpb3 as key subunits that initiate the assembly of RNAP (9, 10, 37, 38). The D/α/Rpb3 subunits are not directly involved in binding of template DNA and synthesis of RNA. Instead, one function of these subunits is to serve as a scaffold for the assembly and proper interaction of the catalytic subunits. In bacteria, the assembly of RNAP is initiated by α subunit dimerization, and it is assembled in the following order: αI + αII → αI/II → αI/IIβ → αI/IIββ′ → αI/IIββ′σ (36). The determinants for α dimerization are located within the bacterial α N-terminal domain. The β and β′ subunits comprise the catalytic core. The assembly of eukaryotic Pol II is initiated by Rpb3 dimerization with Rpb11 followed by addition of Rpb2 to form the assembly subcomplex Rpb2-Rpb3-Rpb11, which is analogous to αI/IIβ. The catalytic subunit Rpb1 and the auxiliary subunits Rpb4–10 and Rbp12 then assemble to form complete Pol II. Similarly, the assembly of archaeal RNAP is initiated by the formation of a heterodimer of subunits D and L followed by the addition of subunits N and P, forming the DLNP assembly platform (42). A stable BDLNP subcomplex that is competent in DNA binding and transcription factor interaction has also been identified, indicating that subunit B is likely added prior to the addition of the core A′ and A″ subunits and auxiliary HKFE subunits (10). Subunit B is split into two polypeptides (B′ and B″) in some Euryarchaeota, including M. acetivorans (42).
The ability to purify the M. acetivorans D-L heterodimer, which lacks the [4Fe-4S] clusters, reveals that the clusters are not required for initial D-L heterodimer formation. However, the presence and redox state of the [4Fe-4S] clusters in domain 3 of subunit D may influence the interaction of subunit D with additional assembly or catalytic subunits. Moreover, the instability of recombinant subunit D when expressed in the absence of subunit L indicates that subunit L serves to stabilize subunit D and that dimerization occurs prior to [4Fe-4S] cluster incorporation (Fig. 10). Based on the S. solfataricus RNAP structure and previous interaction studies with Pyrococcus furiosus RNAP, the D-L heterodimer directly contacts subunits B, N, and P (4, 10). In particular, domain 3 of S. solfataricus subunit D is in close proximity to subunit B (B′ in M. acetivorans RNAP), indicating that the conformational state of domain 3 could impact the interaction between the D-L heterodimer and the B subunit. The lack of cluster incorporation or loss of incorporated clusters likely increases the flexibility of domain 3, which could prevent favorable interaction of the D-L heterodimer with subunits B, N, and P (Fig. 10).
Because M. acetivorans is a strict anaerobe and exposure to oxygen shuts down metabolism (17, 43), one possible mechanism to globally conserve energy to maintain critical functions during times of iron depletion or oxidative stress would be to decrease production of RNA to minimum levels adequate for cellular maintenance. The lack of cluster incorporation, the oxidation of the clusters, or oxidative loss of incorporated clusters within domain 3 of subunit D may prevent the de novo assembly of RNAP. Thus, the [4Fe-4S] clusters in RNAP may be used to sense metabolic factors within the cell, such as iron/sulfur levels and intracellular redox state, which are influenced by environmental factors (e.g. nutrients and oxygen). The reduction potential and oxidative/reductive stability of the clusters is likely tuned to the metabolism of each particular species. For example, the single [4Fe-4S] cluster in S. solfataricus subunit D, an aerobic archaeon, is quite stable in air compared with the two [4Fe-4S] clusters in subunit D from M. acetivorans, a strict anaerobe. One possible explanation for this difference is the presence of the disulfide bond, instead of a second cluster, in domain 3 of S. solfataricus subunit D (4), that is absent from M. acetivorans subunit D. The disulfide may serve to stabilize the cluster in S. solfataricus subunit D such that the cluster is only lost under more extreme oxidative conditions. In addition to preventing the assembly of RNAP, oxidation or loss of the clusters in assembled RNAP may induce dispersion and subsequent inactivation of RNAP.
An alternative function of the clusters may be to regulate RNAP activity whereby the clusters serve as a recognition element necessary for optimal protein-protein interactions with general or gene-specific transcription factors. Importantly, subunit D is on the periphery of archaeal RNAP (as is Rpb3 on eukaryotic Pol II) in a position to interact with general or specific transcription factors (4, 44). The α subunit of bacterial RNAP is also the primary subunit that interacts with specific transcription factors to regulate promoter-dependent gene expression in bacteria (36). Moreover, Rpb3 has been shown to directly interact with a number of transcription factors (45, 46). An Rpb3 mutagenesis study in yeast identified Rpb3 mutants with a temperature-sensitive defect in activator-dependent transcription, indicating that Rpb3 contains determinants for interaction with transcription factors (47). More recently, a direct interaction of Rpb3 with the Med17 subunit of the Mediator complex was demonstrated using an in vivo cross-linking approach (48). Mediator is a large multisubunit complex conserved in eukaryotes that is required for transcription of most Pol II-transcribed genes. Mediator also serves to link specific regulatory proteins with the Pol II transcription complex. Therefore, Rpb3 is important for assembly and the direct interaction with general and specific transcription factors. Proteins that interact with subunit D of archaeal RNAP have not been identified, but given the high degree of similarity to Rpb3, it is highly likely that subunit D contains regions required for interaction with transcription factors.
In M. acetivorans, two [4Fe-4S] cluster-containing domain 3 of subunit D may function as a recognition element for the interaction with regulatory proteins needed to recruit RNAP to specific promoters or to control transcription initiation rates. Furthermore, given the rapid loss of the [4Fe-4S] clusters in the purified D-L(FeS) heterodimer upon exposure to air, the clusters may be used to sense the redox state of the cell to direct changes in gene expression. However, there may be differences in the stability of the [4Fe-4S] clusters in fully assembled RNAP. For example, the [4Fe-4S] cluster in S. solfataricus RNAP was more recalcitrant to removal by chelators as compared with the [4Fe-4S] cluster in D-L heterodimer. As seen with other DNA-interacting proteins, changes in the redox state of the [4Fe-4S] clusters in M. acetivorans RNAP may be enough to induce structural changes in domain 3 that alter the affinity for interacting partners. Oxidized RNAP may be recruited to the promoters of genes involved in response to stress. To test this hypothesis, it will be necessary to purify M. acetivorans RNAP and compare the properties of the [4Fe-4S] clusters with those of clusters in the D-L heterodimer. It is possible that the clusters affect both the assembly and activity of RNAP. For example, exposure of M. acetivorans to oxygen and loss of the clusters from the D-L heterodimer may prevent assembly and oxidation, or loss of the clusters from RNAP may direct RNAP to the promoters of genes needed for cellular maintenance. Using the established M. acetivorans genetic system combined with in vitro approaches, it will be possible to test both the assembly and activity function of the [4Fe-4S] clusters. For example, the ability to generate the M. acetivorans strain capable of expressing subunit DΔD3 without a negative phenotype indicates that it will be possible to use this mutant to determine subunit D-interacting partners that are dependent on domain 3.
CONCLUSIONS
Our results from a survey of RNAP subunit D from sequenced archaea reveal extensive diversity in the cysteine content and number of [4Fe-4S] cluster motifs, dividing archaeal RNAPs into six distinct groups. Subunit D from species of an individual order typically falls within a single group, indicating that subunit D may be used as an additional marker for phylogenetic analyses. For example, all members of the Methanosarcinales encode a group 1 subunit D, whereas all Methanococcales encode group 6 subunit D. We have also demonstrated that subunit D from M. acetivorans is capable of binding two redox-active and oxygen-labile [4Fe-4S] clusters. Data obtained from recombinant studies reveal that the clusters and domain 3 are not required for the formation of the D-L heterodimer, and this was supported by in vivo studies revealing that domain 3 is not required for D-L heterodimer formation within M. acetivorans. Overall, these results suggest that the clusters are not essential to RNAP, consistent with a regulatory role. Given the extreme sensitivity of the clusters to oxygen and the cluster-induced structural changes of the D-L heterodimer, a potential function is in the redox-dependent regulation of RNAP assembly and/or activity.
Supplementary Material
Acknowledgments
We thank Professor Y. Takahashi for providing pRKISC, Bill Metcalf for M. acetivorans strains and plasmids, Mack Ivey for critical reading of the manuscript, and Liz Karr for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant 1P30RR031154-02 from the Centers of Biomedical Research Excellence, National Center for Research Resources (to D. J. L.). This work was also supported by National Science Foundation Grant MCB1121292 (to D. J. L.), the Arkansas Biosciences Institute (to D. J. L.), and the Institute for Fermentation, Osaka (to A. H.).

This article contains supplemental Fig. S1 and Table S1.
- RNAP
- RNA polymerase
- DΔD3
- subunit D deleted of domain 3
- DTNB
- dithionitrobenzoate
- TNB
- thionitrobenzoate
- D-L(His)
- purified D-L heterodimer
- D-L(His)-FeS
- purified D-L heterodimer reconstituted with Fe/S
- DΔD3-L(His)
- purified DΔD3-L heterodimer
- His-D
- six-histidine-tagged subunit D
- His-DΔD3
- six-histidine-tagged subunit DΔD3
- Pol
- polymerase.
REFERENCES
- 1. Ebright R. H. (2000) RNA polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J. Mol. Biol. 304, 687–698 [DOI] [PubMed] [Google Scholar]
- 2. Werner F. (2008) Structural evolution of multisubunit RNA polymerases. Trends Microbiol. 16, 247–250 [DOI] [PubMed] [Google Scholar]
- 3. Haag J. R., Pikaard C. S. (2011) Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 12, 483–492 [DOI] [PubMed] [Google Scholar]
- 4. Hirata A., Klein B. J., Murakami K. S. (2008) The x-ray crystal structure of RNA polymerase from Archaea. Nature 451, 851–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Korkhin Y., Unligil U. M., Littlefield O., Nelson P. J., Stuart D. I., Sigler P. B., Bell S. D., Abrescia N. G. (2009) Evolution of complex RNA polymerases: the complete archaeal RNA polymerase structure. PLoS Biol. 7, e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kusser A. G., Bertero M. G., Naji S., Becker T., Thomm M., Beckmann R., Cramer P. (2008) Structure of an archaeal RNA polymerase. J. Mol. Biol. 376, 303–307 [DOI] [PubMed] [Google Scholar]
- 7. Johnson D. C., Dean D. R., Smith A. D., Johnson M. K. (2005) Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74, 247–281 [DOI] [PubMed] [Google Scholar]
- 8. Hirata A., Murakami K. S. (2009) Archaeal RNA polymerase. Curr. Opin. Struct. Biol. 19, 724–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eloranta J. J., Kato A., Teng M. S., Weinzierl R. O. (1998) In vitro assembly of an archaeal D-L-N RNA polymerase subunit complex reveals a eukaryote-like structural arrangement. Nucleic Acids Res. 26, 5562–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goede B., Naji S., von Kampen O., Ilg K., Thomm M. (2006) Protein-protein interactions in the archaeal transcriptional machinery: binding studies of isolated RNA polymerase subunits and transcription factors. J. Biol. Chem. 281, 30581–30592 [DOI] [PubMed] [Google Scholar]
- 11. Spåhr H., Calero G., Bushnell D. A., Kornberg R. D. (2009) Schizosaccharomyces pombe RNA polymerase II at 3.6-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 106, 9185–9190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 13. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 14. Takahashi Y., Nakamura M. (1999) Functional assignment of the ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster involved in the assembly of Fe-S clusters in Escherichia coli. J. Biochem. 126, 917–926 [DOI] [PubMed] [Google Scholar]
- 15. Cruz F., Ferry J. G. (2006) Interaction of iron-sulfur flavoprotein with oxygen and hydrogen peroxide. Biochim. Biophys. Acta 1760, 858–864 [DOI] [PubMed] [Google Scholar]
- 16. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 17. Lessner D. J., Ferry J. G. (2007) The archaeon Methanosarcina acetivorans contains a protein disulfide reductase with an iron-sulfur cluster. J. Bacteriol. 189, 7475–7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ellman G. L. (1959) Tissue sulfhydryl groups. Arch. Biochem. Biophys. 82, 70–77 [DOI] [PubMed] [Google Scholar]
- 19. Guss A. M., Rother M., Zhang J. K., Kulkarni G., Metcalf W. W. (2008) New methods for tightly regulated gene expression and highly efficient chromosomal integration of cloned genes for Methanosarcina species. Archaea 2, 193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Metcalf W. W., Zhang J. K., Apolinario E., Sowers K. R., Wolfe R. S. (1997) A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc. Natl. Acad. Sci. U.S.A. 94, 2626–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sowers K. R., Boone J. E., Gunsalus R. P. (1993) Disaggregation of Methanosarcina spp. and growth as single cells at elevated osmolarity. Appl. Environ. Microbiol. 59, 3832–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zinder S. (1993) in Methanogenesis (Ferry J. G., ed) pp. 128–206, Chapman and Hall, New York [Google Scholar]
- 23. Galagan J. E., Nusbaum C., Roy A., Endrizzi M. G., Macdonald P., FitzHugh W., Calvo S., Engels R., Smirnov S., Atnoor D., Brown A., Allen N., Naylor J., Stange-Thomann N., DeArellano K., Johnson R., Linton L., McEwan P., McKernan K., Talamas J., Tirrell A., Ye W., Zimmer A., Barber R. D., Cann I., Graham D. E., Grahame D. A., Guss A. M., Hedderich R., Ingram-Smith C., Kuettner H. C., Krzycki J. A., Leigh J. A., Li W., Liu J., Mukhopadhyay B., Reeve J. N., Smith K., Springer T. A., Umayam L. A., White O., White R. H., Conway de Macario E., Ferry J. G., Jarrell K. F., Jing H., Macario A. J., Paulsen I., Pritchett M., Sowers K. R., Swanson R. V., Zinder S. H., Lander E., Metcalf W. W., Birren B. (2002) The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12, 532–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nakamura M., Saeki K., Takahashi Y. (1999) Hyperproduction of recombinant ferredoxins in Escherichia coli by coexpression of the ORF1-ORF2-iscS-iscU-iscA-hscB-hs cA-fdx-ORF3 gene cluster. J. Biochem. 126, 10–18 [DOI] [PubMed] [Google Scholar]
- 25. Sweeney W. V., Rabinowitz J. C. (1980) Proteins containing 4Fe-4S clusters: an overview. Annu. Rev. Biochem. 49, 139–161 [DOI] [PubMed] [Google Scholar]
- 26. Terlesky K. C., Ferry J. G. (1988) Purification and characterization of a ferredoxin from acetate-grown Methanosarcina thermophila. J. Biol. Chem. 263, 4080–4082 [PubMed] [Google Scholar]
- 27. Terlesky K. C., Ferry J. G. (1988) Ferredoxin requirement for electron transport from the carbon monoxide dehydrogenase complex to a membrane-bound hydrogenase in acetate-grown Methanosarcina thermophila. J. Biol. Chem. 263, 4075–4079 [PubMed] [Google Scholar]
- 28. Clements A. P., Kilpatrick L., Lu W. P., Ragsdale S. W., Ferry J. G. (1994) Characterization of the iron-sulfur clusters in ferredoxin from acetate-grown Methanosarcina thermophila. J. Bacteriol. 176, 2689–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Prince R. C., Adams M. W. (1987) Oxidation-reduction properties of the two Fe4S4 clusters in Clostridium pasteurianum ferredoxin. J. Biol. Chem. 262, 5125–5128 [PubMed] [Google Scholar]
- 30. Crack J. C., Green J., Le Brun N. E., Thomson A. J. (2006) Detection of sulfide release from the oxygen-sensing [4Fe-4S] cluster of FNR. J. Biol. Chem. 281, 18909–18913 [DOI] [PubMed] [Google Scholar]
- 31. Mitobe J., Mitsuzawa H., Ishihama A. (2001) Functional analysis of RNA polymerase II Rpb3 mutants of the fission yeast Schizosaccharomyces pombe. Curr. Genet. 39, 210–221 [DOI] [PubMed] [Google Scholar]
- 32. Kimura M., Ishihama A. (2000) Involvement of multiple subunit-subunit contacts in the assembly of RNA polymerase II. Nucleic Acids Res. 28, 952–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Svetlov V., Nolan K., Burgess R. R. (1998) Rpb3, stoichiometry and sequence determinants of the assembly into yeast RNA polymerase II in vivo. J. Biol. Chem. 273, 10827–10830 [DOI] [PubMed] [Google Scholar]
- 34. Genereux J. C., Boal A. K., Barton J. K. (2010) DNA-mediated charge transport in redox sensing and signaling. J. Am. Chem. Soc. 132, 891–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ren B., Duan X., Ding H. (2009) Redox control of the DNA damage-inducible protein DinG helicase activity via its iron-sulfur cluster. J. Biol. Chem. 284, 4829–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ebright R. H., Busby S. (1995) The Escherichia coli RNA polymerase α subunit: structure and function. Curr. Opin. Genet. Dev. 5, 197–203 [DOI] [PubMed] [Google Scholar]
- 37. Ishihama A. (1981) Subunit of assembly of Escherichia coli RNA polymerase. Adv. Biophys. 14, 1–35 [PubMed] [Google Scholar]
- 38. Kimura M., Ishiguro A., Ishihama A. (1997) RNA polymerase II subunits 2, 3, and 11 form a core subassembly with DNA binding activity. J. Biol. Chem. 272, 25851–25855 [DOI] [PubMed] [Google Scholar]
- 39. Major T. A., Burd H., Whitman W. B. (2004) Abundance of 4Fe-4S motifs in the genomes of methanogens and other prokaryotes. FEMS Microbiol. Lett. 239, 117–123 [DOI] [PubMed] [Google Scholar]
- 40. Ferry J. G. (2003) in Biochemistry and Physiology of Anaerobic Bacteria (Ljungdahl L. G., Adams M. W., Barton L. L., Ferry J. G., Johnson M. K., eds) pp. 143–156, Springer-Verlag, New York [Google Scholar]
- 41. Lessner D. J. (2009) in eLS, John Wiley & Sons, Ltd., Chichester, U.K. [Google Scholar]
- 42. Werner F. (2007) Structure and function of archaeal RNA polymerases. Mol. Microbiol. 65, 1395–1404 [DOI] [PubMed] [Google Scholar]
- 43. Brioukhanov A. L., Netrusov A. I., Eggen R. I. (2006) The catalase and superoxide dismutase genes are transcriptionally up-regulated upon oxidative stress in the strictly anaerobic archaeon Methanosarcina barkeri. Microbiology 152, 1671–1677 [DOI] [PubMed] [Google Scholar]
- 44. Cramer P., Bushnell D. A., Kornberg R. D. (2001) Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292, 1863–1876 [DOI] [PubMed] [Google Scholar]
- 45. De Angelis R., Iezzi S., Bruno T., Corbi N., Di Padova M., Floridi A., Fanciulli M., Passananti C. (2003) Functional interaction of the subunit 3 of RNA polymerase II (RPB3) with transcription factor-4 (ATF4). FEBS Lett. 547, 15–19 [DOI] [PubMed] [Google Scholar]
- 46. Oufattole M., Lin S. W., Liu B., Mascarenhas D., Cohen P., Rodgers B. D. (2006) Ribonucleic acid polymerase II binding subunit 3 (Rpb3), a potential nuclear target of insulin-like growth factor binding protein-3. Endocrinology 147, 2138–2146 [DOI] [PubMed] [Google Scholar]
- 47. Tan Q., Linask K. L., Ebright R. H., Woychik N. A. (2000) Activation mutants in yeast RNA polymerase II subunit RPB3 provide evidence for a structurally conserved surface required for activation in eukaryotes and bacteria. Genes Dev. 14, 339–348 [PMC free article] [PubMed] [Google Scholar]
- 48. Soutourina J., Wydau S., Ambroise Y., Boschiero C., Werner M. (2011) Direct interaction of RNA polymerase II and mediator required for transcription in vivo. Science 331, 1451–1454 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.