Background: mTORC1 integrates diverse signals including stress to control cell growth.
Results: JNK phosphorylates Raptor, a component of mTORC1, and activates mTORC1 kinase upon osmotic stress.
Conclusion: mTORC1 is regulated by JNK during osmotic stress.
Significance: Our findings provide the JNK-Raptor relationship as a potential mechanism by which stress activates mTORC1 signaling pathway.
Keywords: Jun N-terminal Kinase (JNK), mTOR, mTOR Complex (mTORC), Phosphorylation, Stress, Osmolarity, Raptor
Abstract
mTOR complex 1 (mTORC1) is a multiprotein complex that integrates diverse signals including growth factors, nutrients, and stress to control cell growth. Raptor is an essential component of mTORC1 that functions to recruit specific substrates. Recently, Raptor was suggested to be a key target of regulation of mTORC1. Here, we show that Raptor is phosphorylated by JNK upon osmotic stress. We identified that osmotic stress induces the phosphorylation of Raptor at Ser-696, Thr-706, and Ser-863 using liquid chromatography-tandem mass spectrometry. We found that JNK is responsible for the phosphorylation. The inhibition of JNK abolishes the phosphorylation of Raptor induced by osmotic stress in cells. Furthermore, JNK physically associates with Raptor and phosphorylates Raptor in vitro, implying that JNK is responsible for the phosphorylation of Raptor. Finally, we found that osmotic stress activates mTORC1 kinase activity in a JNK-dependent manner. Our findings suggest that the molecular link between JNK and Raptor is a potential mechanism by which stress regulates the mTORC1 signaling pathway.
Introduction
The mammalian target of rapamycin (mTOR)2 is an evolutionarily conserved protein kinase that has key roles in several fundamental cellular processes (1, 2). In cells, mTOR exists as two functionally distinct multiprotein complexes: mTOR complex 1 (mTORC1) and 2 (mTORC2) (1). These two complexes have different sensitivities to the allosteric inhibitor rapamycin and distinct roles in cells. Rapamycin strongly and specifically inhibits mTORC1, which acts as a central controller of cell growth by regulating several biosynthetic pathways, including protein synthesis, ribosome biogenesis, and lipid biosynthesis (1, 3). Genetic and pharmacological studies have demonstrated that abnormal hyperactivation of mTORC1 induces cell growth and proliferation, and aberrantly elevated mTORC1 activity has consistently been found in various human diseases such as cancer and diabetes (4, 5).
mTORC1 needs to be tightly regulated by a variety of intra- and extracellular signals to control cell growth accurately. Indeed, mTORC1 senses and integrates diverse signals, including growth factors, nutrient availability, energy status, and stress (6). Signaling by growth factors such as insulin and epidermal growth factor (EGF) activates mTORC1; consequently, complex metazoans are able to maintain homeostasis of organ and organism size (6). Simultaneously, mTORC1 activity is highly sensitive to nutrient and energy levels because mTORC1-induced biosynthetic pathways consume much energy and nutrients (6). Consequently, uncontrolled mTORC1 activation during energy or nutrient deprivation causes cellular apoptosis (7, 8). mTORC1 is also regulated by several stresses, including hypoxia, DNA damage, and oxidative stress, enabling cells to adapt in these stresses (9). At the molecular level, many of these signals converge on TSC1/TSC2 and small GTPase Rheb to regulate mTORC1 (10). Rheb directly binds to and enhances mTORC1 kinase activity (11, 12). TSC1/TSC2 complex negatively regulates Rheb via its GTPase-accelerating protein activity and thereby inhibits mTORC1 (13). Several kinases including Akt, Erk, and 5′-AMP-activated protein kinase (AMPK) were reported to phosphorylate and thereby modulate the GTPase-accelerating protein (GAP) activity of TSC1/TSC2 to regulate mTORC1 (10). TSC1/TSC2 and Rheb is essential for the regulation of mTORC1; however, recent studies have shown that components of mTORC1 including Raptor can function as molecular sensors for mTORC1 (12, 14–18).
Raptor, a defining component of mTORC1, is essential for mTORC1 activity (19, 20). It functions as a scaffold protein that recruits mTORC1-specific substrates (21). Recently, several studies have suggested Raptor as an important signal acceptor of mTORC1. Energy deprivation causes Raptor phosphorylation via 5′-AMP-activated protein kinase, thereby inhibiting mTORC1 (14). The Ras-mediated oncogenic signal also regulates mTORC1 via Raptor phosphorylation by p90 ribosomal S6 kinase (RSK) and Erk (15, 16). Recent findings also revealed that mTORC1 is regulated via cdc2-dependent Raptor phosphorylation during mitosis (17, 18). Therefore, mTORC1 activity is closely related with the status of Raptor phosphorylation.
In this study, we demonstrate that osmotic stress also regulates mTORC1 via Raptor phosphorylation. We found that Raptor is phosphorylated at several proline-directed sites upon osmotic stress and identified JNK as being responsible for the phosphorylation and activation of mTORC1 under osmotic stress. Our findings reveal a novel mechanism of mTORC1 regulation by osmotic stress and suggest that the molecular link between JNK and Raptor has a potential role in the stress regulation of mTORC1.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
All chemicals were purchased from Sigma unless stated otherwise. SP600125, U0126, SB203580, roscovitine, rapamycin, and calyculin A were obtained from Merck (Darmstadt, Germany). Antibodies for mTOR, Raptor, rictor, phospho-mTOR (Ser-2448), phospho-S6K (Thr-389), phospho-S6 (Ser-240/244), phospho-TSC2 (Thr-1462), phospho-Akt (Ser-473), phospho-RSK (Thr-573), phospho-Erk1/2, phospho-AMPK (Thr-172), JNK, and phospho-c-Jun (Ser-63) were acquired from Cell Signaling Technology (Beverly, MA). Anti-phospho-Raptor (Ser-863) and anti-phospho-Raptor (Thr-706) were purchased from Millipore (Billerica, MA). Anti-phospho-Raptor (Ser-696) was kindly provided by Dr. Diane C. Fingar (University of Michigan Medical School, Ann Arbor, MI).
Constructs
HA-Raptor and GST-S6K1 were kindly provided by Dr. David M. Sabatini (Massachusetts Institute of Technology (MIT), Cambridge, MA). Full-length Raptor cDNA obtained by PCR was subcloned into pFLAG-CMV2 (Sigma). FLAG-Raptor S696A/T706A/S863A and FLAG-Raptor S863A were generated by site-directed mutagenesis using Pyrobest DNA polymerase (Takara Bio, Kyoto, Japan). FLAG-JNK1 was a gift from Dr. Eui-Ju Choi (Korea University, Seoul, Korea). GST-JNK1 was generated by subcloning full-length JNK1 cDNA into pGEX-4T-1 (Amersham Biosciences). FLAG-MKK7-JNK1 wild type (WT) and the APF mutant were obtained from Dr. Roger Davis (University of Massachusetts Medical School, Worcester, MA) via Addgene (Cambridge, MA).
Cell Culture and Transfection
HEK293 cells were maintained in Dulbecco's modified Eagle's medium (Cambrex, Charles City, IA) containing 10% fetal bovine serum (Cambrex). Transfection was performed using the Lipofectamine reagent (Invitrogen) according to the manufacturer's instructions. For RNA interference, cells were transfected with 20 nm JNK1/2 siRNA or control siRNA using Lipofectamine. The JNK1/2 and control siRNA sequences were 5′-AAAGAAUGUCCUACCUUCUTT-3′ and 5′-UUCUUCGAACGUGUCACGUTT-3′, respectively. HEK293 cells were transfected and serum-starved for 24 h before stimulation with 0.5 m sorbitol and lysis.
Cell Lysis, Immunoprecipitation, and Immunoblotting
Cells washed twice with ice-cold phosphate-buffered saline (PBS) were lysed with CHAPS lysis buffer (40 mm HEPES, pH 7.5, 0.3% CHAPS, 120 mm NaCl, 1 mm EDTA, 10 mm pyrophosphate, 10 mm glycerophosphate, 50 mm NaF, 1.5 mm Na2VO3, 1 mm phenylmethylsulfonyl fluoride (PMSF), and 10 μg/ml leupeptin). The soluble fractions of the cell lysates were isolated by centrifugation at 14,000 rpm for 15 min. For immunoprecipitation, anti-FLAG M2 beads (Sigma) were added to the lysates and incubated at 4 °C with gentle agitation. The immunoprecipitates were washed four times with lysis buffer. Whole-cell lysates or immunoprecipitates were subjected to SDS-PAGE and immunoblotting. All immunoblots were detected by enhanced chemiluminescence (ECL System; Amersham Biosciences).
In Vitro Kinase Assay
The mTORC1 kinase assay was performed as described by Sancak et al. (12). Briefly, mTORC1 was immunoprecipitated with anti-FLAG M2 beads from FLAG-Raptor-transfected cells. The immunoprecipitates were washed twice in 25 mm HEPES (pH 7.5), 20 mm KCl. The kinase assay was performed for 30 min at 30 °C in mTORC1 kinase assay buffer (25 mm HEPES, pH 7.5, 50 mm KCl, 10 mm MgCl2, 250 μm ATP) and 150 ng of 4E-BP1 (Stratagene, La Jolla, CA) or GST-S6K1. The reactions were stopped by boiling in gel loading buffer and then analyzed by SDS-PAGE and immunoblotting.
For the JNK assay, HEK293 cells transfected with FLAG-Raptor WT, S696A/T706A/S863A mutant, or S863A mutant were serum-starved for 24 h and then lysed with Triton X-100 lysis buffer (40 mm HEPES, pH 7.5, 1% Triton X-100, 120 mm NaCl, 10 mm pyrophosphate, 10 mm glycerophosphate, 50 mm NaF, 1.5 mm Na2VO3, 1 mm PMSF, and 10 μg/ml leupeptin). Then, the FLAG-Raptor proteins were immobilized with anti-FLAG-M2 beads. To purify activated JNK1, FLAG-JNK1-transfected HEK293 cells were serum-starved for 24 h and then stimulated with 0.5 m sorbitol for 1 h to activate JNK. The cells were lysed with Triton X-100 lysis buffer. FLAG-JNK1 was isolated from the lysate with anti-FLAG M2 beads and eluted with 100 μg/ml 3×FLAG peptide (Sigma). The JNK assay was performed by incubating purified active JNK1 and immunoprecipitated FLAG-Raptor proteins in JNK assay buffer (20 mm HEPES, pH 7.4, 10 mm MgCl2, 0.5 mm DTT, 100 μm ATP) at 30 °C for 60 min.
Protein Purification and in Vitro Binding Analysis
GST-S6K1 expression construct was transfected into HEK293 cells, and after 24 h, serum-starved for 24 h. The cells were treated with 20 nm rapamycin for 1 h prior to cell lysis. GST-S6K1 was purified using glutathione-Sepharose bead (GE Healthcare) and eluted with 50 mm reduced glutathione (GSH).
GST and GST-tagged JNK1 proteins were purified from Escherichia coli strain BL21 containing the appropriate constructs. Expression was induced by adding 0.1 mm isopropyl-β-d-thiogalactopyranoside at 25 °C for 4 h. After the cells were harvested, the GST-tagged proteins were purified using glutathione-Sepharose beads and then eluted with 50 mm reduced GSH. In vitro binding between JNK and Raptor was examined with a GST pulldown assay after incubation with GST-tagged proteins and HEK293 lysates. HEK293 cells were lysed with Triton X-100 lysis buffer. The soluble fractions isolated by centrifugation at 14,000 rpm for 15 min were incubated with purified GST or GST-JNK1 at 4 °C for 4 h with gentle agitation. Then, glutathione-Sepharose beads were added, and the incubation was continued for an additional 30 min. After incubation, the beads were washed four times with lysis buffer. Bound Raptor was detected by immunoblotting.
CIP Assay
The FLAG-Raptor immunoprecipitates were washed twice in calf intestinal phosphatase buffer (50 mm Tris, pH 7.9, 10 mm MgCl2, 1 mm dithiothreitol) and then incubated with 1 μl of 10 units/μl calf intestinal phosphatase (New England Biolabs, Ipswich, MA) in 50 μl of calf intestinal phosphatase buffer at 37 °C for 1 h. The reactions were stopped by boiling in gel loading buffer.
Mass Spectrometry
To identify the phosphorylation sites in Raptor, FLAG-Raptor was immunoprecipitated from HEK293 cells after stimulation with 0.5 m sorbitol and then subjected to SDS-PAGE. The Coomassie Blue-stained gel band corresponding to Raptor was subjected to in-gel trypsin digestion. The digested peptides were analyzed using a linear trap quadrupole XL mass spectrometer (Thermo Fisher Scientific,) equipped with a nano-LC system (Eksigent, Dublin, CA) for nano-flow chromatography.
The peptides were trapped with a trapping column (inner diameter 75 μm, length 3 cm, particle size 5 μm) for preconcentration and desalting using solvent A (99.9% distilled water, 0.1% formic acid). Then, the trapped peptides were eluted from the trapping column using a mobile phase gradient (solvent B, 99.9% acetonitrile, 0.1% formic acid) directly onto a reversed phase analytical column (length 10 cm, inner diameter 75 μm) packed with C18 (particle size 5 μm) and eluted separately. The gradient began at 1% solvent B for 9 min for preconcentration and desalting and then was ramped to 40% solvent B for 80 min and finally to 80% solvent B for 20 min at a flow rate of 250 nl/min. The peptides were separated using a reversed phase analytical column (length 10 cm, inner diameter 75 μm) packed with C18 (5 μm). For preconcentration and desalting, the peptides were left in the trapping column (length 3 cm, inner diameter 75 μm) packed with C18 (5 μm) for 9 min and then eluted from the precolumn directly to a reversed phase analytical column using a mobile phase gradient (99.9% acetonitrile, 0.1% formic acid). The eluent was introduced into the linear trap quadrupole mass spectrometer from a nano-ion source with a 1.9-kV electrospray voltage. The analysis method consisted of a full MS scan with a range of 400–1800 m/z followed by data-dependent MS/MS on the five most intense ions from the full MS scan.
The raw data from the linear trap quadrupole were searched using the International Protein Index human FASTA database (version 3.63) with the MASCOT search engine (version 2.2.04) selecting Decoy database analysis. A peptide mass tolerance of 2.0 Da and MS/MS tolerance of 0.8 Da were allowed with tryptic specificity. The database search conditions considered two missed cleavages, variable modification of phosphorylation of Ser, Thr, and Tyr, oxidation of methionine, and carbamidomethylation of cysteine as a fixed modification.
RESULTS
Osmotic Stress Induces Raptor Phosphorylation
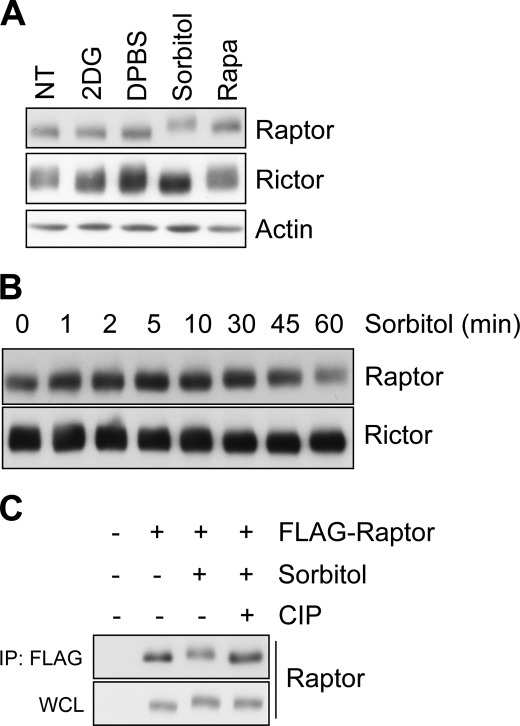
To explore the stresses affecting the posttranslational modification of Raptor, we examined the mobility of Raptor in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Among several stressors, we found that osmotic stress induced by high osmolarity sorbitol treatment retarded the mobility of Raptor (Fig. 1A). An upshift of Raptor was clearly observed after 30 min of sorbitol treatment (Fig. 1B). Then, we wanted to determine which modification causes the mobility retardation. First, we suspected phosphorylation because several studies have reported on mobility retardation of phosphorylated Raptor (17, 18, 22). To examine this issue, the modified Raptor was incubated with calf intestinal phosphatase in vitro. We found that calf intestinal phosphatase treatment reversed the osmotic stress-induced mobility shift of Raptor, indicating that the upshift is due to the phosphorylation of Raptor (Fig. 1C). Therefore, we concluded that Raptor is strongly phosphorylated in response to osmotic stress.
FIGURE 1.
Osmotic stress induces Raptor phosphorylation. A, HEK293 cells were serum-starved overnight and exposed for 60 min to 25 mm 2-deoxyglucose (2DG) for glucose starvation, 45 min to Dulbecco's PBS (DPBS) for amino acid withdrawal, 60 min to 0.5 m sorbitol to induce osmotic stress, or 30 min to 20 nm rapamycin (Rapa). The lysate was resolved by SDS-PAGE and subjected to immunoblotting with the indicated antibody. NT, not treated. B, HEK293 cells were serum-starved overnight and exposed to 0.5 m sorbitol for the indicated times. C, HEK293 cells transfected with FLAG-Raptor were serum-starved overnight and stimulated with 0.5 m sorbitol for 60 min. The immunoprecipitates (IP) were incubated with or without calf intestinal phosphatase (CIP) and then subjected to immunoblotting with Raptor antibody. WCL, whole-cell lysates.
Multiple Sites of Raptor Are Phosphorylated in Response to Osmotic Stress
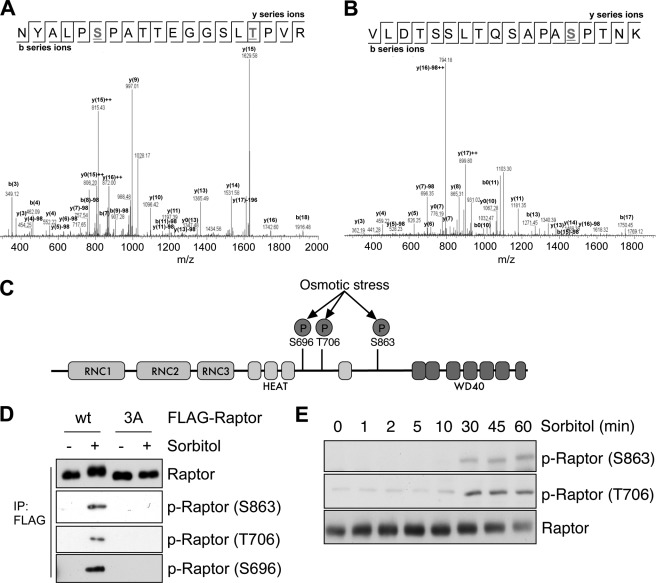
To identify the residues of Raptor phosphorylated by osmotic stress, Raptor immunoprecipitated from HEK293 cells was challenged to osmotic stress, digested with trypsin, and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Several phosphorylation sites were identified, including Ser-696, Thr-706, Ser-863, Ser-859, and Ser-877, from both samples and controls (Fig. 2, A and B, and data not shown). To determine the residues phosphorylated by osmotic stress, we compared the MS/MS spectral count of each phosphopeptide. We found that the spectra of NYALP(pS)PATTEGGSL(pT)PVR for Ser(P)-696 and Thr(P)-706 and VLDTSSLTQSAPA(pS)PTNK (where pS and pT are phospho-Ser and phospho-Thr, respectively) for Ser(P)-863 were more identified in the sorbitol-treated sample than in the control sample, indicating that osmotic stress induces the phosphorylation at Ser-696, Thr-706, and Ser-863. All of the identified phosphorylation sites are located between HEAT repeats and WD40 domains (Fig. 2C). To confirm that these sites are responsible for the osmotic stress-induced phosphorylation, we examined whether mutation of the identified sites affects the mobility shift of Raptor induced by sorbitol treatment. Mutation of Ser-696/Thr-706/Ser-863 to nonphosphorylatable alanine (S696A/T706A/S863A) did not show the band shift with sorbitol treatment, whereas WT Raptor did (Fig. 2D). We also examined the phosphorylation level with phospho-specific antibodies against Ser-696, Thr-706, and Ser-863. The phospho-specific antibodies recognized only WT Raptor and not S696A/T706A/S863A mutant Raptor, and osmotic stress increased the phosphorylation of WT Raptor at Ser-696, Thr-706, and Ser-863 (Fig. 2D). In addition, we also tried to examine the phosphorylation of endogenous Raptor with phospho-specific antibodies. Although the phosphorylation at Ser-696 was unable to be detected because the antibody is too weak to detect the phosphorylation of endogenous Raptor, the phosphorylation of Raptor at Ser-863 and Thr-706 was detected beginning 30 min after sorbitol treatment when the upshift of Raptor began to appear (Fig. 2E). Taken together, we concluded that osmotic stress induces the Raptor phosphorylation at Ser-696, Thr-706, and Ser-863.
FIGURE 2.
Multiple sites of Raptor are phosphorylated in response to osmotic stress. A, the MS/MS spectrum of the peptide with phospho-Ser-696 and phospho-Thr-706. B, the MS/MS spectrum of the peptide with phospho-Ser-863. C, a schematic representation of the Raptor domains and phosphorylated sites (P) induced by osmotic stress. D, FLAG-Raptor WT or S696A/T706A/S863A mutant (3A) was expressed and exposed to 0.5 m sorbitol for 60 min. The level of phosphorylation was determined by the mobility shift in SDS-PAGE or phospho-specific antibodies. p-Raptor, phospho-Raptor. E, lysates from Fig. 1B were subjected to immunoblotting with the indicated phospho-Raptor antibodies.
JNK Pathway Mediates Osmotic Stress-induced Raptor Phosphorylation
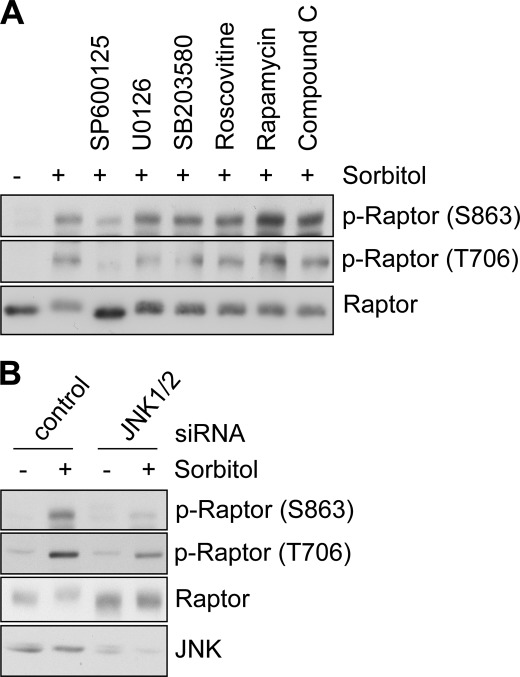
Next, we tried to identify the upstream kinase responsible for Raptor phosphorylation upon osmotic stress. We selected several kinases as candidates, including kinases known to phosphorylate Raptor (14–18). Proline-directed kinases were also selected because the phosphorylated residues that we identified are followed by proline. Then, we examined whether the inhibition of each kinase affected the osmotic stress-induced mobility shift of Raptor. Of the inhibitors, SP600125, which inhibits JNK, was found to totally block the mobility retardation of Raptor in osmotic stress. Consistently, SP600125 resulted in the inhibition of Raptor phospho-Ser-863 and phospho-Thr-706 (Fig. 3A). To confirm that JNK is the responsible kinase, we also examined the effect of JNK1/2 knockdown on Raptor phosphorylation. We found that JNK1/2 knockdown also inhibited the Raptor mobility shift and phosphorylation induced by osmotic stress (Fig. 3B). As JNK is known to be activated by osmotic stress (23), we concluded that JNK is activated upon osmotic stress and subsequently induces Raptor phosphorylation.
FIGURE 3.
JNK pathway mediates osmotic stress-induced Raptor phosphorylation. A, HEK293 cells were serum-starved overnight, pretreated with 20 μm SP600125, 10 μm U0126, 10 μm SB203580, 10 μm Roscovitine, 20 nm rapamycin, or 10 μm Compound C for 30 min, and then exposed to 0.5 m sorbitol for an additional 60 min. The lysates were immunoblotted with the indicated antibodies. p-Raptor, phospho-Raptor. B, HEK293 cells transfected with control or JNK1/2 siRNA were serum-starved overnight and exposed to 0.5 m sorbitol for 60 min. The lysates were immunoblotted with the indicated antibodies.
JNK Physically Associates with and Phosphorylates Raptor
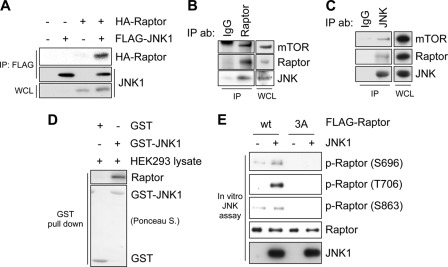
The fact that inhibition of the JNK pathway blocks the osmotic stress-induced Raptor phosphorylation suggests that JNK phosphorylates Raptor directly. First, we examined whether JNK interacts with Raptor. HA-tagged Raptor was co-immunoprecipitated with FLAG-tagged JNK1 (Fig. 4A). Endogenous JNK was also co-immunoprecipitated with endogenous Raptor (Fig. 4B). In addition, we readily detected the interaction between Raptor from HEK293 cell lysate and bacterially purified GST-JNK1 in a GST pulldown experiment (Fig. 4C). Because we prepared the HEK293 cell lysate with lysis buffer containing Triton X-100 detergent, which disrupts the interaction between Raptor and mTOR during lysis, the result indicates that Raptor interacted with JNK directly, not via the other mTORC1 components. Finally, we performed an in vitro JNK kinase assay to examine whether JNK could phosphorylate Raptor directly. Activated JNK1 was found to induce the phosphorylation of WT Raptor at Ser-696, Thr-706, and Ser-863 in vitro, whereas JNK could not phosphorylate S696A/T706A/S863A mutant Raptor (Fig. 4D). Taken together, we concluded that JNK interacts with and phosphorylates Raptor directly.
FIGURE 4.
JNK physically associates with and phosphorylates Raptor. A, HA-Raptor or FLAG-JNK1 was expressed in HEK293 cells. The lysates were immunoprecipitated (IP) with anti-FLAG and immunoblotted with Raptor and FLAG antibodies. WCL, whole-cell lysates. B, the presence of JNK in Raptor immunoprecipitates was determined by immunoblotting with JNK antibody (IP ab). C, the presence of Raptor in JNK immunoprecipitates was determined by immunoblotting with Raptor antibody (IP ab). D, purified GST or GST-JNK1 was incubated with HEK293 lysates prepared using Triton X-100 detergent, and then the GST pulldown assay was performed, and bound Raptor was detected by immunoblotting with Raptor antibody. The amount of GST or GST-JNK1 was shown with Ponceau S staining. E, immunoprecipitated FLAG-Raptor WT or S696A/T706A/S863A (3A) was incubated with purified activated JNK1 in a kinase reaction and then subjected to SDS-PAGE and immunoblotting with phospho-Raptor (p-Raptor) antibodies.
JNK Alone Sufficiently Activates mTORC1 Signaling Pathway
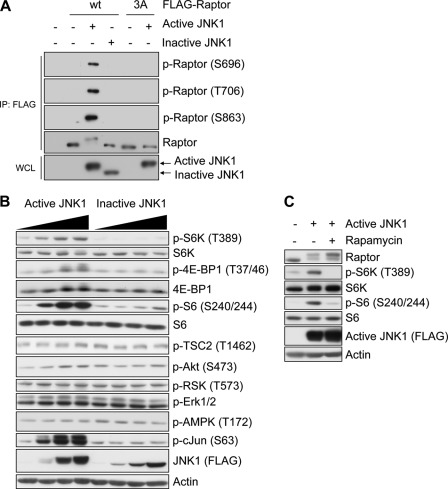
Next, we wanted to determine the role of JNK-mediated Raptor phosphorylation in osmotic stress, which perturbs several signaling pathways, as well as JNK. Therefore, to examine the JNK-specific effect on mTORC1, we used a constitutively active JNK1 construct. Constitutively active JNK1 (called active JNK1) is fused to an upstream kinase MKK7. Therefore, MKK7 can phosphorylate JNK1 constitutively; consequently, JNK1 can be activated without any signal (24, 25). As a negative control, we used an MKK7-JNK1 fusion construct containing JNK1 mutant in which the dual TPY phosphorylation site was replaced with APF (called inactive JNK1) (25). We found that the expression of active JNK1 brings about a phosphorylation-induced mobility shift of Raptor and phosphorylation at Ser-696, Thr-706, and Ser-863, confirming that JNK1 can phosphorylate these sites in cells (Fig. 5A). We monitored the phosphorylation status of several signaling proteins that are upstream regulators or downstream effectors of mTORC1. Notably, the expression of active JNK1 dramatically increased the phosphorylation of S6K, 4E-BP1, and S6, which are markers of mTORC1 activity, whereas inactive JNK1 did not affect their phosphorylation (Fig. 5B), indicating that JNK activation leads to up-regulated mTORC1 activity in cells. Rapamycin treatment completely blocked the JNK-enhanced S6K phosphorylation, consistent with JNK functioning upstream from mTORC1 (Fig. 5C). The expression of active JNK1 had no effect on known upstream regulators, such as the Akt, Erk, and 5′-AMP-activated protein kinase pathways (Fig. 5B), which indicates that the induction of mTORC1 activation by JNK is independent of those pathways. Therefore, we concluded that JNK activation results in Raptor phosphorylation at Ser-696, Thr-706, and Ser-863, along with mTORC1 activation in cells.
FIGURE 5.
JNK alone sufficiently activates mTORC1 signaling pathway. A, HEK293 cells were transfected with FLAG-Raptor WT or S696A/T706A/S863A (3A) mutant and FLAG-MKK7-JNK1 (active JNK1) or FLAG-MKK7-JNK1 mutant (inactive JNK1) and serum-starved overnight. The FLAG-Raptor proteins were immunoprecipitated (IP) and immunoblotted with the indicated antibodies. WCL, whole-cell lysates; p-Raptor, phospho-Raptor. B, active JNK1 or inactive JNK1 was expressed at several levels. The phosphorylation (P) levels of downstream effectors and upstream regulators of mTORC1 were monitored by immunoblotting with the indicated antibodies. C, HEK293 cells were transfected with active JNK1 and serum-starved for 24 h and then treated with 20 nm rapamycin for 30 min before lysis. The lysates were subjected to immunoblotting with the indicated antibodies.
Osmotic Stress Increases mTORC1 Kinase Activity
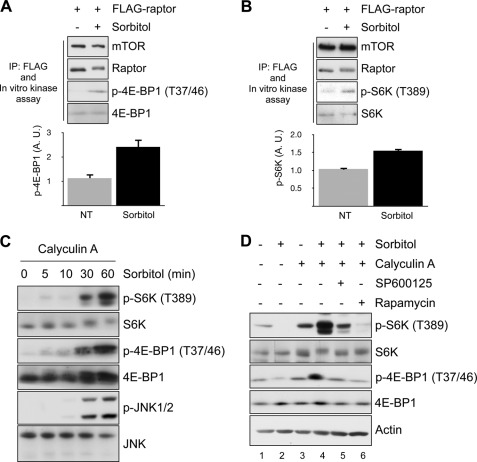
The phosphorylation sites identified in this study have already been reported to be phosphorylated by mTOR or Erk in mitogenic signals, including insulin and EGF, and the phosphorylation is positively associated with mTORC1 activity (16, 22). Indeed, when we purified mTORC1 from cells treated with sorbitol and compared mTORC1 activity with an in vitro kinase assay, mTORC1 from sorbitol-treated cells showed higher activity than control cells (Fig. 6, A and B). This result was unexpected because osmotic stress is thought to inhibit mTORC1 as osmotic stress completely abolishes S6K and 4E-BP1 phosphorylation. We also found that sorbitol treatment induces the dephosphorylation of S6K and 4E-BP1 (supplemental Fig. 1). Therefore, a discrepancy exists between the change in S6K and 4E-BP1 phosphorylation in a cell and the results of the in vitro kinase assay (Fig. 6, A and B). Interestingly, we found that dephosphorylation of S6K was completed within 5 min of sorbitol treatment, whereas Raptor phosphorylation and JNK activation started at 30 min, indicating that these two events are independent (Fig. 1B, supplemental Fig. 1). From these results, we speculated that the possibility exists that the dephosphorylation of S6K is not due to the inactivation of mTORC1. Calyculin A-sensitive phosphatase is reportedly responsible for the inhibition of S6K upon osmotic stress (26). It has been reported that 4E-BP1 is also dephosphorylated by calyculin A-sensitive phosphatase (27). Therefore, we reasoned that upon osmotic stress, S6K and 4E-BP1 are dephosphorylated by the strong phosphatase activity despite the activation of mTORC1. To examine this possibility, we pretreated cells with calyculin A to exclude the effect of phosphatase. Surprisingly, under the calyculin A-treated condition, the phosphorylation of S6K and 4E-BP1 was strongly induced by sorbitol treatment, whereas sorbitol treatment strongly decreased the phosphorylation of S6K and 4E-BP1 without calyculin A, supporting our hypothesis (Fig. 6, C and D). Osmotic stress-induced S6K and 4E-BP1 phosphorylation in the calyculin A-treated condition was completely inhibited by rapamycin, confirming that the increase of phosphorylation is mediated by mTORC1 (Fig. 6D). In addition, the induction of S6K and 4E-BP1 phosphorylation occurred 30 min after sorbitol treatment when JNK phosphorylation started to be shown, implying that S6K and 4E-BP1 phosphorylation is JNK activity-dependent (Fig. 6C). Finally, we found that JNK inhibition reversed the phosphorylation of S6K and 4E-BP1 to the level of phosphorylation before sorbitol treatment (Fig. 6D, lanes 3 and 5), indicating that JNK is responsible for the activation of mTORC1 by sorbitol treatment. Taken together, these data suggest that osmotic stress induces mTORC1 activation and JNK mediates the activation.
FIGURE 6.
Osmotic stress enhances mTORC1 kinase activity. A and B, HEK293 cells transfected with FLAG-Raptor were serum-starved overnight and exposed to osmotic stress. mTORC1 was immunoprecipitated (IP) with FLAG antibody and the in vitro kinase assay was performed with purified 4E-BP1 and S6K, respectively. The bar graph at the bottom shows the quantification of 4E-BP1 and S6K1 phosphorylation, respectively. A. U., arbitrary units. p-4E-BP1, phospho-4E-BP1; p-S6K, phospho-S6K. C, HEK293 cells were pretreated with 100 nm calyculin A for 30 min and then exposed to 0.5 m sorbitol for the indicated times. The cell lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. D, HEK293 cells were pretreated with 100 nm calyculin A and 20 μm SP600125 or 20 nm rapamycin where indicated and then exposed to 0.5 m sorbitol for an additional 60 min. The cell lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
DISCUSSION
Understanding how cell growth is controlled with diverse environmental perturbations is important because many diseases are caused by the uncontrolled regulation of cell growth under such circumstances (4). As a critical integrator of environmental inputs into cell growth, mTORC1 has been studied intensively in numerous environmental contexts. However, how osmotic stress, one such environmental perturbation, affected the mTORC1 signaling pathway at a molecular level remains unclear. Here, we demonstrated that osmotic stress regulates mTORC1 kinase activity via JNK-mediated Raptor phosphorylation. We found that osmotic stress induces strong Raptor phosphorylation and determined that Raptor Ser-696, Thr-706, and Ser-863 are phosphorylated. We also found that JNK interacts with Raptor directly and phosphorylates these sites. Finally, we found that the kinase activity of mTORC1 is up-regulated in osmotic stress.
We demonstrated that osmotic stress induces Raptor phosphorylation at three sites, Ser-696, Thr-706, and Ser-863, as demonstrated in previous studies. mTOR phosphorylates Raptor at Ser-696, Thr-706, and Ser-863 with the insulin signal (22). Erk can also phosphorylate Raptor at Ser-696 and Ser-863 in Ras-dependent mTORC1 activation (16). In addition, cdc2 phosphorylates Raptor at Ser-696 and Thr-706 during mitosis (17, 18). Recently, arsenite was shown to induce Raptor phosphorylation at Ser-863 mediated by p38 (28). Here, we found that upon osmotic stress, JNK is the predominant kinase inducing Raptor phosphorylation. This was demonstrated by blocking mTOR, Erk, p38, and cdc2 with specific inhibitors that did not affect or partially affected osmotic stress-induced Raptor phosphorylation and the electrophoretic mobility shift, whereas JNK inhibition using SP600125 fully blocked the Raptor phosphorylation upon osmotic stress. Consistently, JNK knockdown reduced Raptor phosphorylation, confirming that the JNK pathway is responsible for Raptor phosphorylation in osmotic stress (Fig. 3). Furthermore, we demonstrated that JNK directly phosphorylates Raptor at three sites. During the preparation of this manuscript, Fujishita et al. (29) also reported that JNK can phosphorylate the Ser-863 site of Raptor in intestinal tumorigenesis. Here, we claim that JNK also phosphorylates Raptor Ser-696 and Thr-706 as well as Ser-863. We found that constitutively active JNK still induced the mobility shift of Raptor that had its Ser-863 site mutated to alanine, indicating the existence of additional phosphorylation (supplemental Fig. 2A). In agreement, the expression of active JNK1 resulted in increased phosphorylation of three sites in Raptor (Fig. 5A). Finally, we demonstrated that JNK could phosphorylate Raptor at three sites in vitro (Fig. 4E, supplemental Fig. 2B).
In this study, we also demonstrated that mTORC1 kinase activity was enhanced under osmotic stress. The Raptor phosphorylation sites that we identified are reported to have a positive role in mTORC1. Therefore, our results suggest that mTORC1 is in an active state under osmotic stress. Consistently, we demonstrated that mTORC1 kinase activity increased when measured with an in vitro kinase assay (Fig. 6). Contrary to our results, however, the notion that osmotic stress suppresses mTORC1 has been widely accepted because S6K, an established effector of mTORC1, is completely inhibited upon osmotic stress. Calyculin A-sensitive phosphatase has been suggested to mediate the inhibition of S6K upon osmotic stress (26). However, no evidence exists that mTORC1 is required for the inhibition of S6K. Therefore, to resolve this discrepancy, we hypothesized that S6K inhibition during osmotic stress is independent of mTORC1. To test this hypothesis, we excluded the effect of phosphatase by using calyculin A. We found that osmotic stress induces S6K phosphorylation in the absence of phosphatase activity, indicating the activation of mTORC1. Evidence supporting our conclusion has appeared in previous studies. First, Inoki et al. (13) showed that the expression of the constitutively active mutant of Rheb, which is a potent activator of mTORC1, does not rescue the inhibition of S6K by osmotic stress, whereas it restores the phosphorylation of S6K under other stress conditions that inactivate mTORC1, implying that the dephosphorylation of S6K is not dependent on mTORC1 inhibition under osmotic stress. Second, Kim et al. (19) showed that mTORC1 kinase activity in vitro does not change with a 10-min sorbitol treatment, although cellular S6K phosphorylation disappears completely under that condition. The inconsistency between Kim et al. (19) and our study regarding the in vitro mTOR activity may be due to the difference in the length of the sorbitol treatment. A 10-min sorbitol treatment did not induce Raptor phosphorylation (Fig. 2E); therefore, the mTORC1 activity may not be affected at that time. Third, the osmotic stress in 3T3-L1 adipocytes induces IRS-1 Thr-307 phosphorylation in a rapamycin-sensitive manner, indicating that mTORC1 activity increases with osmotic stress in adipocytes (30).
Here, we also showed that JNK is responsible for the activation of mTORC1 upon osmotic stress (Figs. 5 and 6B). JNK kinase is activated by a variety of cellular stresses, especially including UV and reactive oxygen species (31). Interestingly, several studies have shown that UV and reactive oxygen species are stresses that induce the activation of mTORC1 (32, 33). Therefore, this raises several interesting possibilities. First, the possibility exists that JNK-activating stressors activate mTORC1. Therefore, one must determine whether JNK mediates mTORC1 activation under those stress conditions. Second, mTORC1 may also have active roles in apoptosis because JNK activation induces apoptosis. Unregulated mTORC1 activation has been reported to induce apoptosis under nutrient-starved conditions (7, 8). Constitutive mTORC1 activity has been shown to induce p53 activation by stimulating p53 translation in response to energy starvation (8). Therefore, JNK may utilize mTORC1 activity to induce apoptosis efficiently. Further investigation is required to better understand the role of mTORC1 in stress and apoptosis.
Although most mammalian cells maintained in an internal environment are exposed to very limited changes in osmolality, increasing evidence supports the importance of the osmotic stress response in various tissues, including kidney and lymphoid tissues (34). The renal medulla is exposed to extremely hyperosmotic stress in the process of reabsorbing water in the kidneys to concentrate urine. Lymphoid tissues, including the thymus, spleen, and liver, can also be hyperosmolar relative to blood (35). Therefore, the role of osmotic stress regulation of mTORC1 in these tissues remains to be investigated.
Supplementary Material
Acknowledgments
We thank Dr. David M. Sabatini for providing HA-Raptor and Dr. Eui-ju Choi for FLAG-JNK1. We also thank Dr. Diane C. Fingar for providing the Ser(P)-696-Raptor antibody.
This work was supported by the Global Frontier Project (Grant NRF-MIAXA002-2010-0029764) and Global Research Network Program (Grant KRF-2008-220-C00036).

This article contains supplemental Figs. 1 and 2.
- mTOR
- mammalian target of rapamycin
- mTORC
- mTOR complex
- CIP
- calf intestinal phosphatase
- S6K
- p70 ribosomal S6 kinase
- Raptor
- regulatory associated protein of mTOR
- JNK
- c-Jun N-terminal Kinase.
REFERENCES
- 1. Wullschleger S., Loewith R., Hall M. (2006) TOR signaling in growth and metabolism. Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 2. Jacinto E., Hall M. (2003) Tor signaling in bugs, brain and brawn. Nat. Rev. Mol. Cell Biol. 4, 117–126 [DOI] [PubMed] [Google Scholar]
- 3. Ma X. M., Blenis J. (2009) Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10, 307–318 [DOI] [PubMed] [Google Scholar]
- 4. Zoncu R., Efeyan A., Sabatini D. M. (2011) mTOR: from growth signal integration to cancer, diabetes, and aging. Nat. Rev. Mol. Cell Biol. 12, 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guertin D. A., Sabatini D. M. (2007) Defining the role of mTOR in cancer. Cancer Cell 12, 9–22 [DOI] [PubMed] [Google Scholar]
- 6. Sengupta S., Peterson T. R., Sabatini D. M. (2010) Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell 40, 310–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Choo A. Y., Kim S. G., Vander Heiden M. G., Mahoney S. J., Vu H., Yoon S. O., Cantley L. C., Blenis J. (2010) Glucose addiction of TSC null cells is caused by failed mTORC1-dependent balancing of metabolic demand with supply. Mol. Cell 38, 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee C. H., Inoki K., Karbowniczek M., Petroulakis E., Sonenberg N., Henske E. P., Guan K. L. (2007) Constitutive mTOR activation in TSC mutants sensitizes cells to energy starvation and genomic damage via p53. EMBO J. 26, 4812–4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reiling J. H., Sabatini D. M. (2006) Stress and mTORture signaling. Oncogene 25, 6373–6383 [DOI] [PubMed] [Google Scholar]
- 10. Huang J., Manning B. (2008) The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem. J. 412, 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Long X., Lin Y., Ortiz-Vega S., Yonezawa K., Avruch J. (2005) Rheb binds and regulates the mTOR kinase. Curr. Biol. 15, 702–713 [DOI] [PubMed] [Google Scholar]
- 12. Sancak Y., Thoreen C. C., Peterson T. R., Lindquist R. A., Kang S. A., Spooner E., Carr S. A., Sabatini D. M. (2007) PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell 25, 903–915 [DOI] [PubMed] [Google Scholar]
- 13. Inoki K., Li Y., Xu T., Guan K. L. (2003) Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17, 1829–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gwinn D. M., Shackelford D. B., Egan D. F., Mihaylova M. M., Mery A., Vasquez D. S., Turk B. E., Shaw R. J. (2008) AMPK phosphorylation of Raptor mediates a metabolic checkpoint. Mol. Cell 30, 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carrière A., Cargnello M., Julien L. A., Gao H., Bonneil E., Thibault P., Roux P. P. (2008) Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated Raptor phosphorylation. Curr. Biol. 18, 1269–1277 [DOI] [PubMed] [Google Scholar]
- 16. Carriere A., Romeo Y., Acosta-Jaquez H. A., Moreau J., Bonneil E., Thibault P., Fingar D. C., Roux P. P. (2011) ERK1/2 phosphorylate Raptor to promote Ras-dependent activation of mTOR complex 1 (mTORC1). J. Biol. Chem. 286, 567–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ramírez-Valle F., Badura M. L., Braunstein S., Narasimhan M., Schneider R. J. (2010) Mitotic Raptor promotes mTORC1 activity, G2/M cell cycle progression, and internal ribosome entry site-mediated mRNA translation. Mol. Cell Biol. 30, 3151–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gwinn D. M., Asara J. M., Shaw R. J. (2010) Raptor is phosphorylated by cdc2 during mitosis. PLoS One 5, e9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) mTOR interacts with Raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 20. Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., Hidayat S., Tokunaga C., Avruch J., Yonezawa K. (2002) Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110, 177–189 [DOI] [PubMed] [Google Scholar]
- 21. Schalm S. S., Fingar D. C., Sabatini D. M., Blenis J. (2003) TOS motif-mediated Raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr. Biol. 13, 797–806 [DOI] [PubMed] [Google Scholar]
- 22. Foster K. G., Acosta-Jaquez H. A., Romeo Y., Ekim B., Soliman G. A., Carriere A., Roux P. P., Ballif B. A., Fingar D. C. (2010) Regulation of mTOR complex 1 (mTORC1) by Raptor Ser-863 and multisite phosphorylation. J. Biol. Chem. 285, 80–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huangfu W. C., Omori E., Akira S., Matsumoto K., Ninomiya-Tsuji J. (2006) Osmotic stress activates the TAK1-JNK pathway while blocking TAK1-mediated NF-κB activation: TAO2 regulates TAK1 pathways. J. Biol. Chem. 281, 28802–28810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zheng C., Xiang J., Hunter T., Lin A. (1999) The JNKK2-JNK1 fusion protein acts as a constitutively active c-Jun kinase that stimulates c-Jun transcription activity. J. Biol. Chem. 274, 28966–28971 [DOI] [PubMed] [Google Scholar]
- 25. Lei K., Nimnual A., Zong W. X., Kennedy N. J., Flavell R. A., Thompson C. B., Bar-Sagi D., Davis R. J. (2002) The Bax subfamily of Bcl2-related proteins is essential for apoptotic signal transduction by c-Jun NH2-terminal kinase. Mol. Cell Biol. 22, 4929–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parrott L. A., Templeton D. J. (1999) Osmotic stress inhibits p70/85 S6 kinase through activation of a protein phosphatase. J. Biol. Chem. 274, 24731–24736 [DOI] [PubMed] [Google Scholar]
- 27. Peterson R. T., Desai B. N., Hardwick J. S., Schreiber S. L. (1999) Protein phosphatase 2A interacts with the 70-kDa S6 kinase and is activated by inhibition of FKBP12-rapamycinassociated protein. Proc. Natl. Acad. Sci. U.S.A. 96, 4438–4442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu X. N., Wang X. K., Wu S. Q., Lu J., Zheng M., Wang Y. H., Zhou H., Zhang H., Han J. (2011) Phosphorylation of Raptor by p38β participates in arsenite-induced mammalian target of rapamycin complex 1 (mTORC1) activation. J. Biol. Chem. 286, 31501–31511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fujishita T., Aoki M., Taketo M. M. (2011) JNK signaling promotes intestinal tumorigenesis through activation of mTOR complex 1 in Apc(Δ716) mice. Gastroenterology 140, 1556–1563.e6 [DOI] [PubMed] [Google Scholar]
- 30. Gual P., Gonzalez T., Grémeaux T., Barres R., Le Marchand-Brustel Y., Tanti J. F. (2003) Hyperosmotic stress inhibits insulin receptor substrate-1 function by distinct mechanisms in 3T3-L1 adipocytes. J. Biol. Chem. 278, 26550–26557 [DOI] [PubMed] [Google Scholar]
- 31. Weston C. R., Davis R. J. (2007) The JNK signal transduction pathway. Curr. Opin. Cell Biol. 19, 142–149 [DOI] [PubMed] [Google Scholar]
- 32. Zhang Y., Dong Z., Nomura M., Zhong S., Chen N., Bode A. M., Dong Z. (2001) Signal transduction pathways involved in phosphorylation and activation of p70S6K following exposure to UVA irradiation. J. Biol. Chem. 276, 20913–20923 [DOI] [PubMed] [Google Scholar]
- 33. Huang C., Li J., Ke Q., Leonard S. S., Jiang B. H., Zhong X. S., Costa M., Castranova V., Shi X. (2002) Ultraviolet-induced phosphorylation of p70S6K at Thr-389 and Thr-421/Ser-424 involves hydrogen peroxide and mammalian target of rapamycin but not Akt and atypical protein kinase C. Cancer Res. 62, 5689–5697 [PubMed] [Google Scholar]
- 34. Ho S. N. (2006) Intracellular water homeostasis and the mammalian cellular osmotic stress response. J. Cell. Physiol. 206, 9–15 [DOI] [PubMed] [Google Scholar]
- 35. Go W. Y., Liu X., Roti M. A., Liu F., Ho S. N. (2004) NFAT5/TonEBP mutant mice define osmotic stress as a critical feature of the lymphoid microenvironment. Proc. Natl. Acad. Sci. U.S.A. 101, 10673–10678 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.