Background: Retroviruses and retrotransposons integrate with poorly understood preferences.
Results: Depending on in vitro reaction conditions, Ty3 retroelement integrase was targeted by a transcription factor or by Ty3 inverted-repeat terminal sequence.
Conclusion: Retroelement targeting can be reproduced in a defined system.
Significance: Biochemical models will enable improved understanding of retrovirus integration preferences and gene therapy vector safety.
Keywords: DNA Enzymes, Integrase, Protein DNA-Interaction, Retrovirus, RNA polymerase III, Yeast, Retrotransposon
Abstract
The Saccharomyces cerevisiae long terminal repeat retrotransposon Ty3 integrates within one or two nucleotides of the transcription initiation sites of genes transcribed by RNA polymerase III. In this study the minimal components required to re-constitute position-specific strand transfer by Ty3 integrase are defined. Ty3 integrase targeted by a synthetic fusion of RNA polymerase III transcription factor IIIB subunits, Brf1 and TBP, mediated position-specific strand transfer of duplex oligonucleotides representing the ends of the Ty3 cDNA. These results further delimit the TFIIIB domains targeted by the Ty3 element and show that IN is the Ty3 component sufficient in vitro to target integration. These results underscore the commonality of protein interactions that mediate transcription and retrotransposon targeting. Surprisingly, in the presence of MnCl2, strand transfer was TFIIIB-independent and targeted sequences resembling the Ty3 terminal inverted repeat.
Introduction
Retroviruses display preferential patterns of integration in eukaryotic genomes, reflecting influences of host transcription factors and effects of chromatin components and DNA modification, sequence, and structure on activity of the preintegration complex of integrase (IN)2 and cDNA known as the intasome (1–3). Understanding these integration biases, particularly in the case of retroviruses, is complicated both by the complexity of the target and the animal genomes themselves. For example, although it is known that lens epithelium-derived growth factor (LEDGF) is required for efficient integration of HIV-1 (4–5) and that the interaction between IN and LEDGF maps to the C-terminal end of the catalytic core domain (6), mechanistic details of how LEDGF tethers the intasome to the target DNA remain elusive.
The relatively subtle integration preferences of retroviruses contrast with the striking preferences of some retrotransposons in lower eukaryotes and plants (7–9). For example, in Saccharomyces cerevisiae, the copia-like LTR retrotransposon Ty5 is targeted to heterochromatic DNA by interactions between the IN C-terminal domain and the Sir4 silencing protein (10–11) and copia-like Ty1 and gypsy-like Ty3 LTR retrotransposons target the 5′-flanking regions of Pol III-transcribed genes (12–13). In Dictystelium discoideum, the non-LTR retrotransposon TRE5 targets 5′-flanking regions of tRNA genes and the TRE5 ORF1 protein interacts with components of the Pol III transcription factor TFIIIB (14). The Schizosaccharomyces pombe gypsy-like Tf1 interacts with a subset of transcription factors to target RNA Pol II promoters (15).
Ty3 is distinguished by the precision of its integration within a few bases of Pol III TSS (13). However, despite this unusual insertion specificity, Ty3 has substantial structural and functional similarity to retroviruses (16). For example, cells expressing Ty3 accumulate VLPs containing processed Ty3 proteins and cDNA and the Ty3 IN has a conserved core domain that contains residues conserved among retroviral integrases, including the D, DX35E catalytic motif of polynucleotide esterases. Although the amino- and carboxyl-terminal domains of IN proteins are generally less-well conserved, they contain a zinc finger and a GPY/F motif, respectively. These motifs are also found in the Ty3 IN protein (17). Similar to retroviral cDNA, the Ty3 cDNA has LTRs and terminates with two “extra” bp at each end, which are endonucleolytically removed from the 3′-ends by Ty3 IN prior to strand transfer (18). Based on the retrovirus model, the resulting 3′-hydroxyls mediate SN2 nucleophilic attacks at staggered positions in the duplex chromosomal DNA. These positions are offset by 5 nts so that concerted strand transfer generates the characteristic 5-bp direct repeats flanking the ends of Ty3 insertions. These similarities to retroviruses coupled with precise targeting make Ty3 an attractive model for probing the mechanisms by which targeting proteins might interact with the retroelement intasome. However, a biochemically defined in vitro system that recapitulates the natural specificity of any retroelement including Ty3 has been lacking. We describe such a system here and use it to investigate Ty3 substrate and target sequences that influence integration.
EXPERIMENTAL PROCEDURES
Recombinant DNA Constructions
Plasmids were constructed using standard molecular biology procedures (19) unless otherwise noted. Details of plasmid constructions, plasmids and sequences of oligonucleotides used for constructions are provided in supplemental Experimental Procedures and supplemental Tables S1 and S2, respectively. Constructs were verified by DNA sequence analysis (Genewiz Inc., La Jolla, CA).
Proteins
S. cerevisiae strain BY4741 was induced to express galactose-regulated Ty3 from pDLC201 (20), and VLPs were harvested as previously described (21). Triple fusion protein (Brf11–382-TBP61–240-Brf1439–596, TFP) was expressed in bacterial strain Rosetta (DE3) pLysS (EMD Biosciences. San Diego, CA) and was purified essentially as described (22).
Recoded Ty3 IN (23) was cloned to allow expression of a C-terminal His6-tagged protein under control of the lac promoter (pKN2412). Expression was induced in Rosetta (DE3) pLysS according to standard procedures. Extracts were enriched for IN by affinity chromatography using His60 Ni Superflow. IN was further purified using anion exchange chromatography over DEAE Sephadex A-25. Details of protein purifications are provided in supplemental Experimental Procedures.
In Vitro Integration and Strand-Transfer Assays
In vitro integration using VLPs was performed as described previously (24). Either TFIIIB or TFP were mixed with target plasmids on ice for 30 min before VLPs were added, and samples were incubated at 16 °C for 15 min. Strand-transfer reactions were performed in buffer R (20 mm HEPES pH 7.5, 70 mm NaCl, 0.1% Nonidet P-40, 7.5% DMSO, 5 mm DTT) supplemented with MgCl2 or MnCl2 cofactors. Generally, samples contained 50 fmol of target plasmid, 250 fmol of duplex DNA, 250 fmol of TFP, and 1000 fmol of IN in a total volume of 40 μl. Reactions were incubated at 24 °C for 1 h, and DNA products were extracted as described previously (24).
PCR was performed to amplify fragments diagnostic of strand transfer. For VLP integrations, one tenth of the DNA products were combined with primers 242 and 411, which anneal within the SNR6 gene and at the downstream end of the internal domain of Ty3, respectively (25). In the PCR reactions monitoring strand-transfer products of duplex DNA substrates, primer HH1707, which anneals at the first half of the DNA substrates, was substituted for primer 411. Control PCR reactions amplified a segment of the target plasmid. Products were resolved by electrophoresis on non-denaturing 8% polyacrylamide gel or 1.5% agarose gel and visualized by staining with ethidium bromide. To determine strand-transfer sites, DNA fragments were extracted from the gel, cloned into pCR2.1 and sequenced.
Protein DNA Binding Assay
A 57-bp 32P-labeled TATA-containing DNA probe was labeled, and EMSA was performed as described previously (22).
RESULTS AND DISCUSSION
A Synthetic Brf1 and TBP Fusion Protein Supports Ty3 Position-specific Integration
SNR6 is transcribed by Pol III, but is distinguished in yeast from some other Pol III templates by the presence of an upstream TATA box. TFIIIB composed of TBP, Bdp1, and Brf1 functions to dock Pol III and enhance duplex opening at the position of transcription initiation (26). In vitro TFIIIB binds DNA via interactions between TBP and the SNR6 TATA element and these interactions are sufficient to support TFIIIC-independent transcription initiation (27–28). On a template containing heteroduplex DNA at the transcription initiation site, Brf1 and TBP alone are sufficient to support transcription initiation (29). Function of TFIIIB subunits Brf1 and TBP in transcription initiation can be substituted by a structure-based fusion of the conserved domain of TBP flanked by segments of Brf1 (Brf11–382-TBP61–240-Brf1439–596) referred to as TFP (22).
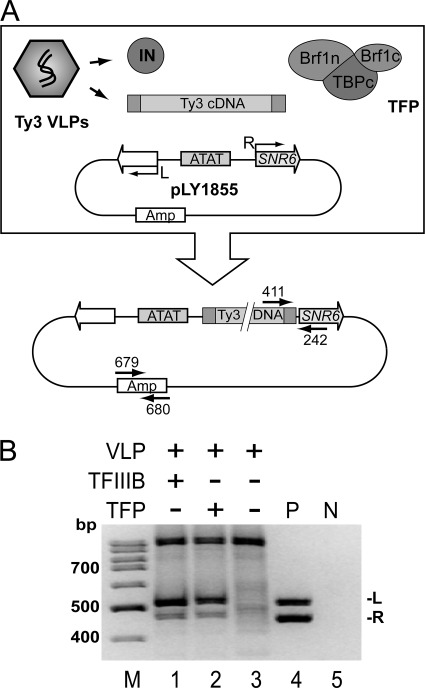
A particulate fraction containing Ty3 VLPs isolated from yeast extracts by sucrose gradient centrifugation can provide active IN and substrate cDNA (30). Ty3 VLP-mediated cDNA strand transfer differs from Pol III transcription initiation in that it can be targeted by Brf1 and TBP without introduction of heteroduplex DNA at the TSS (24). However, the requirement of Ty3 for TFIIIB and TFIIIC for integration at most tRNAs and for TFIIIB or even Brf1 and TBP at SNR6 complicates identification of interactions key to targeting. In order to better define the activities required for Ty3 strand-transfer targeting, we examined whether TFP could replace TBP and Brf1 as was found for Pol III transcription. The TATA box upstream of SNR6 can bind TBP in either orientation and thus mediate bi-directional transcription initiation at upstream (SNR6 distal) and downstream (SNR6 proximal) sites (31). A related variant on plasmid pLY1855 supports Ty3 integration at both initiation sites (25). This target plasmid was combined with bacterially-expressed TFIIIB (TBP, Brf1, and Bdp1) or TFP and Ty3 VLPs were added as the source of integration activity and cDNA (25). Strand transfer was assayed using a PCR primed by cDNA- and target plasmid-specific oligonucleotides (Fig. 1A). Products consistent with TFIIIB bound to the TATA box in each orientation were observed in positive control reactions containing TFIIIB and VLPs (Fig. 1B, lane 1) (25) and in a test reaction containing TFP (Fig. 1B, lane 2), but not in reactions containing only VLPs (Fig. 1B, lane 3). Therefore, non-conserved TBP residues 1–60 and Brf1 region 383–438, which contains HR I and the HRI-II spacer, both of which are lacking in TFP, are dispensable for targeting Ty3 strand transfer to Pol III TSS.
FIGURE 1.
Identification of minimal in vitro targeted strand transfer components. A, in vitro strand-transfer assay. Reactions contained VLPs as a source of Ty3 cDNA and IN and TFIIIB or TFP (Brf11–382-TBP61–240-Brf1439–596). A functionally symmetric TATA box (ATAT) binds TBP in either orientation allowing bidirectional transcription initiation (L and R arrows). Strand-transfer products were detected in PCR primed from within Ty3 and SNR6 (primers 411 and 242, respectively). DNA recovery was monitored with PCR using primers annealed to the backbone of the target plasmid (primers 679 and 680). B, TFP substitutes for TFIIIB in targeting Ty3 strand transfer. Strand-transfer reactions with test reactants (lanes 1–3), positive control (P) plasmids with a Ty3 LTR at leftward (pLY1842) and rightward TSS (pDLC370) (lane 4), or negative control (N) containing water alone (lane 5) were used to template PCR using primers 411 and 242. PCR products were separated on a nondenaturing 8% polyacrylamide gel. Products amplified from leftward (L) and rightward (R) TSS are indicated.
Recombinant IN Is Sufficient to Mediate Position-specific Integration
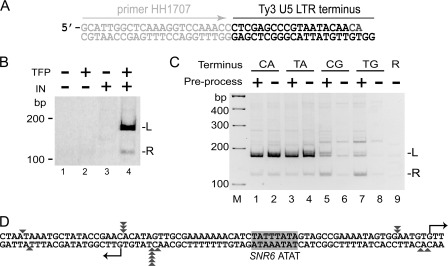
A remaining major limitation in defining the Ty3 components required for targeting was the requirement for a complex VLP fraction as the source of both IN and cDNA. Although it might be anticipated that IN would directly mediate specificity, recent findings in the retrovirus system indicate that domains within some retroviral Gag proteins have the capacity to influence integration patterns (2). A system was therefore developed in which recombinant IN and duplex oligonucleotides were substituted for VLPs. These strategies were previously used to reconstitute the retroviral strand-transfer reaction (32), although the greater size of the Ty3 IN complicated direct adoption of those protocols. The portion of POL3 encoding the 61-kDa Ty3 IN was tagged with 6× His and recoded for bacterial expression (23). Wt IN and a catalytic site mutant (D225E/E261D) derivative (18) were expressed in Escherichia coli. These recombinant IN proteins were purified by nickel affinity chromatography. A duplex oligonucleotide containing 23 nt with complementarity to a PCR primer followed by 20 nt representing the downstream (U5) end of the unprocessed Ty3 LTR and a non-transferred, complementary strand of 45 nt were introduced into the in vitro strand-transfer reaction to substitute for unprocessed VLP cDNA. An identical substrate lacking two nts from the U5 3′-end (“pre-processed”) was also tested (Fig. 2A). IN, duplex pre-processed substrates, SNR6 DNA and TFP were combined in the strand-transfer reaction. Products of this reaction were used to template PCR primed with oligonucleotides complementary to the substrate and plasmid target. The reactions including wt IN generated fragments of the size expected for Ty3 strand transfer at the divergent TSS (Fig. 2B, lane 4; Fig. 2C, lanes 1 and 2); the D22E/E261D mutant IN failed to generate these products (data not shown). Sequence analysis of four independent reactions identified eleven distinct joints of targeted strand transfers. The majority of joints were distributed within one or two nt of the TSS on the template strand or offset upstream by five nt on the nontemplate strand (Fig. 2D). A similar amount of product was generated in reactions using unprocessed duplexes (Fig. 2C, compare lanes 1 and 2). In addition, sequence analysis of strand-transfer products of the blunt substrate showed that the junction occurred at the terminal CA, so that strand transfer was preceded by removal of two nt from the 3′-end of the duplex (data not shown). These assays demonstrated for the first time that Ty3 IN is the sole Ty3 protein required to process 3′ extra nts and target strand transfer to the Pol III transcription initiation site.
FIGURE 2.
Recombinant Ty3 IN mediates targeted strand transfer with duplex oligonucleotide substrates. A, duplex oligonucleotide substrate. The duplex oligonucleotide substrate for strand transfer: left end (light gray): 23 bp sequence complementary to PCR primer HH1707; right end (black): 20/22 nt sequence of Ty3 LTR U5 terminus lacking the two 3′-nt removed by processing (preprocessed). B, recombinant IN mediates strand transfer of duplex Ty3 LTR U5 substrate. Duplex preprocessed oligonucleotide substrate (A) was incubated in the absence and presence of TFP and recombinant IN as described under “Experimental Procedures.” PCR was performed on the products using primers HH1707 and 242 and products were fractionated as described in the Fig. 1 legend. C, recombinant IN mediates strand transfer of unprocessed and preprocessed substrates and is sensitive to substrate sequence. Strand-transfer assays were as in B except that substrates were either preprocessed (+) or blunt-end (−) and the terminal 2 nt of the Ty3 IR (5′-CA-3′) were varied as shown. R, DNA substrate in which random 22 nt replaced Ty3 U5 sequence (oligonucleotides HH1798 and HH1799). D, target sites of strand transfer. Products of four strand-transfer reactions (B, lane 4) were cloned and sequenced. Strand-transfer joints between Ty3 U5 and target sequence are shown (triangles). Multiple triangles at the identical site in all three cases represent two independent reactions. Eleven independent sites were identified.
Ty3 IN Strand Transfer Is Sensitive to Mutations in the cDNA Inverted Repeat Dinucleotide
Terminal IR are a signature feature of integrated transposons and retroviruses with TG/CA being virtually universally conserved. Upstream of the conserved dinucleotide the two ends can have distinct sequences and in vitro evolution of IN substrates has shown that additional variation is possible in the absence of requirements for replication (33). As discussed above, in the cDNA the IR copies are flanked on the outside ends by 2 “extra” bp which are removed during integration. Ty3 has a terminal 8-bp IR and 2 extra bp (plus strand, 5′-gaTGTTGTAT-3′ … ATACAACAcc-3′). U5 oligonucleotide substrates substituted in the outside ends of the IR (CA, wt; TA, CG, TG, mutants) and a duplex oligonucleotide in which the terminal Ty3 sequence was randomized, were assayed for strand transfer (Fig. 2C). This assay showed little difference in activity among reactions using blunt or pre-processed substrates with IR sequences ending in wt CA or mutant TA (Fig. 2C, lanes 1–4). Strand-transfer products were not generated from the randomized oligonucleotide substrate (Fig. 2C, lane 9). Significantly less strand transfer was observed for processed and blunt substrates with IR ending in G, rather than wt A (Fig. 2C, lanes 5–8). In addition, among the latter templates, more strand transfer was observed for preprocessed substrates indicating that processing was sensitive to mutations of the terminal “A” (Fig. 2C, lanes 5–8). In the case of in vitro relative rate assays of HIV-1 and Ty1 and Tf1 retrotransposon IN proteins, 3′-end processing of the two extra nt was blocked by mutations in the IR terminal “A” and was greatly reduced by changes in the conserved penultimate IR “C” (34) for Ty1 (35) and Tf1 (36). Although these strand-transfer assays combined with PCR detection are unlikely to be as sensitive to perturbation as real time enzymatic assays, they showed that Ty3 IN activity is sensitive to changes in the terminal IR.
TFP Mediates IN Association with Target DNA
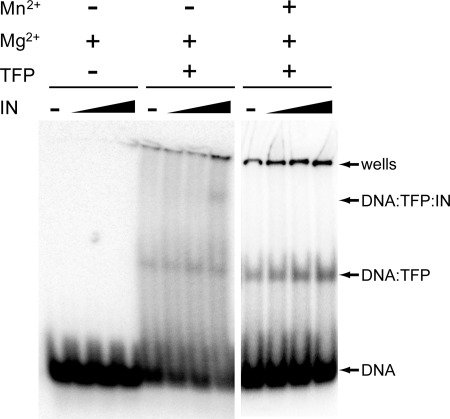
Retroviral IN proteins display robust in vitro strand-transfer activity in the absence of host targeting factors. In the case of Ty3 IN, strand-transfer assays did not show evidence of a default nonspecific pathway. Nonetheless, this activity would yield more diffuse products in our assay and therefore be more difficult to detect than specific strand transfer. Therefore, the ability of IN to interact with target DNA was reinvestigated using a more direct assay. A 57-bp duplex oligonucleotide DNA containing the SNR6 TATA element was used to represent the target DNA. An identical duplex was previously used to measure binding of TFP specifically to TATA-containing DNA (22). Over a range of IN concentrations, no interaction between IN and the target DNA was observed (Fig. 3, left panel). In contrast, as reported previously, addition of TFP alone retarded mobility of the SNR6 target duplex (22). In the presence of TFP, supershifting of the TATA-containing duplex was proportional to the amount of IN (Fig. 3, middle panel). However, this interaction was weak for both wt IN and a catalytic site mutant (data not shown). Overall, these results support a model in which the Ty3 intasome interactions with Pol III promoters is mediated by direct interaction of Ty3 IN with Brf1 and TBP components of TFIIIB. This model is similar to what has been proposed for targeting of Ty5 (37) and Tf1 (15) integration by IN tethering to target-bound proteins.
FIGURE 3.
Ty3 IN interacts with target DNA via interactions mediated by TFP. Protein-32P-labeled DNA complexes were formed with 0.3 nm 57-bp TATA box-containing probe (see “Experimental Procedures”: EMSA), 12.5 nm TFP (middle and right panels), and increasing amounts of IN (0, 12.5, 25, 50 nm), and were analyzed on a 4% native polyacrylamide gel. Binding buffers contained 7 mm MgCl2 (all panels), and MnCl2-containing reactions were 10 mm (right panel). IN was titrated in each series (− and wedge).
In the Presence of MnCl2, Strand Transfer Is TFP-independent and Sequence Specific
In vitro substitution of the natural MgCl2 metal cofactor with MnCl2 in the case of HIV-1 IN reduces specificity for cDNA termini (34) and enhances activity in disintegration assays (38). To test the effect of MnCl2 on the association of Ty3 IN with its target, MgCl2 was either supplemented or substituted with MnCl2 in the strand-transfer reactions. PCR analysis of products of MnCl2-containing reactions showed surprisingly that strand transfer was dependent upon IN, but independent of TFP (Fig. 4A). Strand transfer was not observed for the randomized oligonucleotide substrate in the presence of MnCl2, indicating that it required specific interactions with IN (data not shown). Reactions containing VLPs showed only a low level of non-targeted products in the presence of MnCl2 (supplemental Fig. S1). In the presence of MnCl2, TFP shifted the TATA-containing probe indicating that MnCl2 does not produce TFP-independent strand transfer by disruption of TFP binding (Fig. 3, right panel). However, the IN supershift was no longer observed, suggesting that the presence of MnCl2 affected the interaction between TFP and IN.
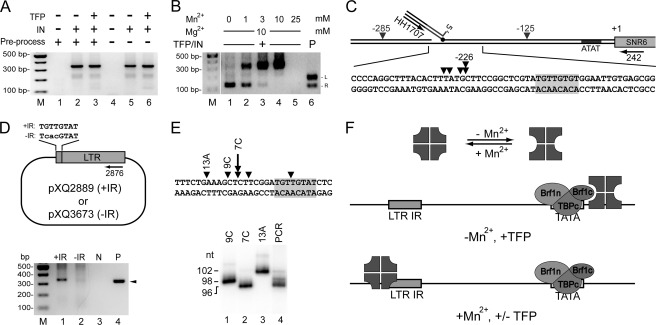
FIGURE 4.
Ty3 IN mediates sequence-specific targeting in the presence of MnCl2. A, Ty3 IN strand-transfer in the presence of MnCl2 is TFP independent. Strand transfer reactions and analysis were performed as described in the Fig. 2 legend except that 10 mm MnCl2 replaced MgCl2 and PCR products were resolved on a 1.5% agarose gel. B, appearance of novel strand-transfer product is dependent upon MnCl2. Reactions as described in A performed in the presence of 10 mm MgCl2 and MnCl2 concentrations from 0 to 25 mm (lanes 1–5). Plasmids pXQ3659 and pXQ3660 were used as control for strand-transfer positions under MgCl2-containing conditions (lane 6). PCR products were analyzed as in A. C, MnCl2-dependent strand transfer targets vicinity of sequences similar to Ty3 IR. PCR reactions were performed as described above (A, lane 3), and products were sequenced and labeled in diagram as described in the Fig. 2 legend. Target plasmid Ty3 IR (5′-TGTTGTAT-3′/5′-ATACAACA-3′)-like sequence, shaded box. D, MnCl2-dependent strand-transfer targets Ty3 IR. Upper panel, plasmids containing Ty3 LTR lacking the downstream inverted repeat and wt (+IR) (pXQ2889) or mutated (−IR) (pXQ3673) IR were tested for targeting activity. Lower panel, reaction conditions and assay were as described above (A, lane 3) with pre-processed Ty3 U5 duplex oligonucleotide substrate. Insertions were analyzed using the same forward primer (HH1707) and a reverse primer (XQ2876) annealed downstream of the Ty3 IR. Control PCR used pXQ3661 containing a Ty3 U5 end upstream of the IR sequence as template (P). DNA recovery was monitored by PCR using primers XQ2876 and XQ2877 (data not shown). PCR products were analyzed as in A. E, strand transfer is imprecise and proximal to the outside end of the Ty3 IR. PCR amplicons (D, lane 1) were cloned and sequenced allowing identification of products with strand transfers at positions −7(C), −9(C), and −13(A). These clones (lanes 9C, 7C, and 13A) and the total original reaction products (as described for D, lane 1) were used to template 32P-labeling PCR using primers XQ2876/XQ3135. Products were resolved on a 10% denaturing polyacrylamide gel. F, model for differential sequence or transcription complex targeting by Ty3 IN. Ty3 IN adopts different states in the presence or absence of MnCl2 (top). In the absence of MnCl2, TFP recruits IN (middle). IN conformation in MnCl2 prevents interaction with TFP and allows association of IN subunits not interacting with substrate to interact with target (bottom).
The PCR amplicon from products generated in the presence of MnCl2 concentrations greater than 10 mm in the presence or absence of TFP was ∼300 bp (Fig. 4, A and B). Experiments were performed in which MnCl2 or MgCl2 was increased in the absence or presence of the other metal cation and the products were amplified using PCR (Fig. 4B and data not shown). In high MgCl2 and low MnCl2 bands representing products of strand transfers flanking the TFP binding site were observed as previously described. However increasing MnCl2 correlated with increasing amounts of higher molecular weight products including a major product of about 300 bp and decreasing amounts of lower molecular weight products (Fig. 4B). Since these products were clearly discrete from previously observed targeted strand-transfer products, products of three independent reactions were cloned and submitted for sequencing. This analysis showed strand transfer mainly within a small region. Among the six sites revealed by sequencing, four occurred within a 5-bp region from −231 to −226 upstream of SNR6 and the others occurred at positions −285 and −125 (Fig. 4C). One possibility was that strand transfer at a secondary TFP-binding site occurred in the presence of MnCl2. However, inspection failed to identify TATA-like sequences, and insertions were independent of TFP (Fig. 4A). Instead, the sequence (5′-TGTTGTGT-3′/3′-ACAACACA-5′) resembling the terminal IR sequence of Ty3 (5′-TGTTGTAT-3′/3′-ACAACATA-5′) was identified between −213 and −205 upstream of SNR6. The four clustered positions of strand transfer occurred 13 to 18 nt upstream of the 5′-end of this sequence (Fig. 4C).
The strand-transfer products recovered in the vicinity of plasmid sequences resembling the Ty3 IR suggested that IN might confer sequence specificity to strand transfer under some conditions. To directly test whether Ty3 IN targeted Ty3 IR-like sequences, a plasmid containing an isolated Ty3 LTR truncated at the downstream end to remove one IR (pXQ2889) was used as a target (Fig. 4D). Strand-transfer assays were performed using MnCl2 as the cation and the preprocessed U5 oligonucleotide duplex substrate. PCR templated by products of this reaction showed dominant fragments of about 300 bp (Fig. 4D, lane 1). The mixed PCR products were cloned and sequences of six clones were determined. This analysis showed strand-transfer joints at positions −13, −9, and −7 relative to the outside end of the target Ty3 LTR (5′-TGTTGTAT-3′). To assess the distribution of target sites more completely, cloned strand-transfer products at these positions were used as templates to obtain 32P-labeled markers. Migration of these markers was compared with that of products of an independent strand-transfer reaction. Comparison of the distribution of PCR fragments templated by products of the total strand-transfer reaction to the sizes of the sequenced standards showed that there was a narrow distribution of strand transfers ∼5–13 bp from the upstream end of the plasmid-borne LTR (Fig. 4E, lane 4). To further test the dependence of the strand-transfer reaction on the Ty3 IR, the target plasmid was modified in the Ty3 IR from TGTTGTAT to TCACGTAT to produce plasmid pXQ3673 (Fig. 4D). In contrast to PCR templated by the reaction using the wt IR target, PCR of the reaction containing the mutated IR failed to generate a product (Fig. 4D, lane 2). Control PCR reactions monitoring DNA recovery showed no difference in plasmid recovery between the two sets of samples (data not shown). Although it appears that in MgCl2, in the absence of a targeting factor, strand transfer does not occur or is extremely inefficient, this may be because the PCR assay is less sensitive to detection of highly distributed products. In the presence of MnCl2, strand transfer was independent of TFP and concentrated near Ty3 IR-like sequences. Thus, IN strand transfer activity per se does not depend upon the presence of a targeting transcription factor. U5 strand transfer was only observed upstream of the Ty3 IR. This is consistent with asymmetric targeting by the 8-bp sequence to regions which in a chromosomal context would lie outside of Ty3.
The experiments in which the position of TFP was shifted by IN in the presence of Mg2+ but not in the presence of Mn2+, together with the redirection of strand transfer in the presence of MnCl2 suggested that IN interacts directly with IR-containing DNA in the presence of MnCl2. However, gel shift experiments similar to those which detected weak TFP-mediated IN association with TATA-containing target probe failed to identify detectable binding to IR-containing 50-mers (data not shown).
The possibility that the presence of MnCl2 enhances weak sequence-specific interactions is intriguing. The crystal structure of the primate foamy virus intasome (39) showed a dimer of dimers with the catalytic site at the dimer-dimer interface; residues interacting with the donor IR mapped to the catalytic core and C-terminal domains of the interface. If we assume a similar structure for Ty3 IN, outer subunits might be available to participate in targeting. We speculate that in the presence of MgCl2, they interact preferentially with the TFIIIB complex, whereas in the presence of MnCl2, this interaction is disfavored and IR-interacting residues mediate interactions (Fig. 4F). Although this activity is interesting in terms of intasome structure-function, it may have minimal in vivo significance. In vivo, integration into Pol III initiation sites clearly dominates (40) and the concentration of MnCl2 required for IR targeting was significantly greater than the reported physiologic concentration (41).
In summary, the involvement of multiple retroelement and host proteins and poorly-defined insertion preferences complicate elucidation of retroelement targeting. This study reconstitutes precise retroelement targeting in vitro for the first time and delimits the retroelement and host components responsible. Intriguingly, our studies showed that both protein-targeted and IR sequence-targeted modes of strand transfer can occur in vitro. We propose that outer intasome subunits not involved in strand transfer are available for target interaction.
Supplementary Material
Acknowledgments
We thank B. Irwin for technical support and helpful discussions. We thank K. Nguyen for providing the IN expression plasmid. We thank G. A. Kassavetis and E. P. Geiduschek, University of California, San Diego, for providing TFIIIB and TBP, Brf1, and TFP expression plasmids and for many helpful discussions.
This work was supported, in whole or in part, by funds from the National Institutes of Health Public Service Grant GM33281 and NSF Grant ID 0450159 (to S. S.).

This article contains supplemental Fig. S1, Tables S1 and S2, and Experimental Procedures.
- IN
- integrase
- EMSA
- electrophoretic mobility shift assay
- HR
- homology region
- IR
- inverted repeats
- LEDGF
- lens epithelium-derived growth factor
- LTR
- long terminal repeat
- Pol III
- RNA polymerase III
- TSS
- transcription start sites
- VLPs
- virus-like particles
- TFP
- Brf1n-TBPc-Brf1c triple fusion protein.
REFERENCES
- 1. Bushman F., Lewinski M., Ciuffi A., Barr S., Leipzig J., Hannenhalli S., Hoffmann C. (2005) Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 3, 848–858 [DOI] [PubMed] [Google Scholar]
- 2. Delelis O., Zamborlini A., Thierry S., Saïb A. (2010) Chromosomal tethering and proviral integration. Biochim. Biophys. Acta 1799, 207–216 [DOI] [PubMed] [Google Scholar]
- 3. Van Maele B., Busschots K., Vandekerckhove L., Christ F., Debyser Z. (2006) Cellular co-factors of HIV-1 integration. Trends Biochem. Sci. 31, 98–105 [DOI] [PubMed] [Google Scholar]
- 4. Maertens G., Cherepanov P., Pluymers W., Busschots K., De Clercq E., Debyser Z., Engelborghs Y. (2003) LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278, 33528–33539 [DOI] [PubMed] [Google Scholar]
- 5. Llano M., Saenz D. T., Meehan A., Wongthida P., Peretz M., Walker W. H., Teo W., Poeschla E. M. (2006) An essential role for LEDGF/p75 in HIV integration. Science 314, 461–464 [DOI] [PubMed] [Google Scholar]
- 6. Cherepanov P., Ambrosio A. L., Rahman S., Ellenberger T., Engelman A. (2005) Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc. Natl. Acad. Sci. U.S.A. 102, 17308–17313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beauregard A., Curcio M. J., Belfort M. (2008) The take and give between retrotransposable elements and their hosts. Annu. Rev. Genet. 42, 587–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Winckler T., Szafranski K., Glöckner G. (2005) Transfer RNA gene-targeted integration: an adaptation of retrotransposable elements to survive in the compact Dictyostelium discoideum genome. Cytogenet Genome Res. 110, 288–298 [DOI] [PubMed] [Google Scholar]
- 9. Peterson-Burch B. D., Nettleton D., Voytas D. F. (2004) Genomic neighborhoods for Arabidopsis retrotransposons: a role for targeted integration in the distribution of the Metaviridae. Genome Biol. 5, R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zou S., Wright D. A., Voytas D. F. (1995) The Saccharomyces Ty5 retrotransposon family is associated with origins of DNA replication at the telomeres and the silent mating locus HMR. Proc. Natl. Acad. Sci. U.S.A. 92, 920–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu Y., Dai J., Fuerst P. G., Voytas D. F. (2003) Controlling integration specificity of a yeast retrotransposon. Proc. Natl. Acad. Sci. U.S.A. 100, 5891–5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Devine S. E., Boeke J. D. (1996) Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 10, 620–633 [DOI] [PubMed] [Google Scholar]
- 13. Chalker D. L., Sandmeyer S. B. (1992) Ty3 integrates within the region of RNA polymerase III transcription initiation. Genes Dev. 6, 117–128 [DOI] [PubMed] [Google Scholar]
- 14. Chung T., Siol O., Dingermann T., Winckler T. (2007) Protein interactions involved in tRNA gene-specific integration of Dictyostelium discoideum non-long terminal repeat retrotransposon TRE5-A. Mol. Cell Biol. 27, 8492–8501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leem Y. E., Ripmaster T. L., Kelly F. D., Ebina H., Heincelman M. E., Zhang K., Grewal S. I., Hoffman C. S., Levin H. L. (2008) Retrotransposon Tf1 is targeted to Pol II promoters by transcription activators. Mol. Cell 30, 98–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sandmeyer S. B., Aye M., Menees T. M. (2002) in Saccharomyces cerevisiae. in Mobile DNA II (Craig N. L., Craigie R., Martin G., Lambowitz A. M. eds), pp. 663–682, ASM Press, Washington D. C [Google Scholar]
- 17. Malik H. S., Eickbush T. H. (1999) Modular evolution of the integrase domain in the Ty3/Gypsy class of LTR retrotransposons. J. Virol. 73, 5186–5190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kirchner J., Sandmeyer S. B. (1996) Ty3 integrase mutants defective in reverse transcription or 3′-end processing of extrachromosomal Ty3 DNA. J. Virol. 70, 4737–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. (2007) Current Protocols in Molecular Biology, John Wiley and Sons, Inc., New York [Google Scholar]
- 20. Hansen L. J., Chalker D. L., Sandmeyer S. B. (1988) Ty3, a yeast retrotransposon associated with tRNA genes, has homology to animal retroviruses. Mol. Cell Biol. 8, 5245–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clemens K., Larsen L., Zhang M., Kuznetsov Y., Bilanchone V., Randall A., Harned A., Dasilva R., Nagashima K., McPherson A., Baldi P., Sandmeyer S. (2011) The Ty3 Gag3 spacer controls intracellular condensation and uncoating. J. Virol. 85, 3055–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kassavetis G. A., Soragni E., Driscoll R., Geiduschek E. P. (2005) Reconfiguring the connectivity of a multiprotein complex: fusions of yeast TATA-binding protein with Brf1, and the function of transcription factor IIIB. Proc. Natl. Acad. Sci. U.S.A. 102, 15406–15411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Larsen L. S., Wassman C. D., Hatfield G. W., Lathrop R. H. (2008) Computationally optimized DNA assembly of synthetic genes. Int. J. Bioinform Res. Appl. 4, 324–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yieh L., Hatzis H., Kassavetis G., Sandmeyer S. B. (2002) Mutational analysis of the transcription factor IIIB-DNA target of Ty3 retroelement integration. J. Biol. Chem. 277, 25920–25928 [DOI] [PubMed] [Google Scholar]
- 25. Yieh L., Kassavetis G., Geiduschek E. P., Sandmeyer S. B. (2000) The Brf and TATA-binding protein subunits of the RNA polymerase III transcription factor IIIB mediate position-specific integration of the gypsy-like element, Ty3. J. Biol. Chem. 275, 29800–29807 [DOI] [PubMed] [Google Scholar]
- 26. Kassavetis G. A., Kumar A., Letts G. A., Geiduschek E. P. (1998) A post-recruitment function for the RNA polymerase III transcription-initiation factor IIIB. Proc. Natl. Acad. Sci. U.S.A. 95, 9196–9201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Margottin F., Dujardin G., Gérard M., Egly J. M., Huet J., Sentenac A. (1991) Participation of the TATA factor in transcription of the yeast U6 gene by RNA polymerase C. Science 251, 424–426 [DOI] [PubMed] [Google Scholar]
- 28. Kassavetis G. A., Geiduschek E. P. (2006) Transcription factor TFIIIB and transcription by RNA polymerase III. Biochem. Soc. Trans. 34, 1082–1087 [DOI] [PubMed] [Google Scholar]
- 29. Kassavetis G. A., Letts G. A., Geiduschek E. P. (1999) A minimal RNA polymerase III transcription system. EMBO J. 18, 5042–5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kirchner J., Connolly C. M., Sandmeyer S. B. (1995) Requirement of RNA polymerase III transcription factors for in vitro position-specific integration of a retroviruslike element. Science 267, 1488–1491 [DOI] [PubMed] [Google Scholar]
- 31. Whitehall S. K., Kassavetis G. A., Geiduschek E. P. (1995) The symmetry of the yeast U6 RNA gene's TATA box and the orientation of the TATA-binding protein in yeast TFIIIB. Genes Dev. 9, 2974–2985 [DOI] [PubMed] [Google Scholar]
- 32. Bushman F. D., Craigie R. (1991) Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc. Natl. Acad. Sci. U.S.A. 88, 1339–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou H., Rainey G. J., Wong S. K., Coffin J. M. (2001) Substrate sequence selection by retroviral integrase. J. Virol. 75, 1359–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Esposito D., Craigie R. (1998) Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interaction. EMBO J. 17, 5832–5843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sharon G., Burkett T. J., Garfinkel D. J. (1994) Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol. Cell Biol. 14, 6540–6551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hizi A., Levin H. L. (2005) The integrase of the long terminal repeat-retrotransposon Tf1 has a chromodomain that modulates integrase activities. J. Biol. Chem. 280, 39086–39094 [DOI] [PubMed] [Google Scholar]
- 37. Xie W., Gai X., Zhu Y., Zappulla D. C., Sternglanz R., Voytas D. F. (2001) Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol. Cell Biol. 21, 6606–6614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hickman A. B., Palmer I., Engelman A., Craigie R., Wingfield P. (1994) Biophysical and enzymatic properties of the catalytic domain of HIV-1 integrase. J. Biol. Chem. 269, 29279–29287 [PubMed] [Google Scholar]
- 39. Maertens G. N., Hare S., Cherepanov P. (2010) The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature 468, 326–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Qi X., Daily K., Nguyen K., Wang H., Mayhew D., Rigor P., Forouzan S., Johnston M., Mitra R. D., Baldi P., Sandmeyer S. (2012) Retrotransposon profiling of RNA polymerase III initiation sites. Genome Res. 22, 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lange H. C., Heijnen J. J. (2001) Statistical reconciliation of the elemental and molecular biomass composition of Saccharomyces cerevisiae. Biotechnol. Bioeng. 75, 334–344 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.