Plasticity in developmental programming has evolved to provide the best chances of survival and reproductive success to the organism under changing environments. The window of developmental plasticity extends from preconception to early childhood, and involves epigenetic responses to environmental changes, which exert their effects during life history phase-transitions. This review provides a comprehensive presentation of translational epigenetics as it pertains to child health, growth, and maturation. Identifying the epigenetic consequences of fetal programming creates potential applications in clinical practice: the development of epigenetic biomarkers for early diagnosis of disease, the ability to identify susceptible individuals at risk for adult diseases, and the development of novel preventive and curative measures that are based on diet and/or novel epigenetic drugs.
Abstract
Plasticity in developmental programming has evolved in order to provide the best chances of survival and reproductive success to the organism under changing environments. Environmental conditions that are experienced in early life can profoundly influence human biology and long-term health. Developmental origins of health and disease and life-history transitions are purported to use placental, nutritional, and endocrine cues for setting long-term biological, mental, and behavioral strategies in response to local ecological and/or social conditions. The window of developmental plasticity extends from preconception to early childhood and involves epigenetic responses to environmental changes, which exert their effects during life-history phase transitions. These epigenetic responses influence development, cell- and tissue-specific gene expression, and sexual dimorphism, and, in exceptional cases, could be transmitted transgenerationally. Translational epigenetic research in child health is a reiterative process that ranges from research in the basic sciences, preclinical research, and pediatric clinical research. Identifying the epigenetic consequences of fetal programming creates potential applications in clinical practice: the development of epigenetic biomarkers for early diagnosis of disease, the ability to identify susceptible individuals at risk for adult diseases, and the development of novel preventive and curative measures that are based on diet and/or novel epigenetic drugs.
I. Introduction
Each living organism has two histories that determine its biology: an evolutionary history whose duration is in the hundreds of thousands of years, and a developmental history that starts at the time of its conception. Developmental history of an organism is associated with the appearance of new structures that cannot be explained in terms of its developmental programming. The ability of the genotype to produce different phenotypes in response to different environments is termed “plasticity.” The time of maximal plasticity appears to be during development. However, heritable phenotypic variation at a later stage is also possible because of the individual's capability to respond to environmental cues. This ability of the organism to facilitate change is termed “adaptability” (1), and the expressions of suites of genes, particularly during development or life-history transitions, probably underlie the fundamental plasticity of an organism (2).
Trait variability, irrespective of whether it is molecular, cellular, physiological, morphological, or behavioral, is the leading edge of evolution. Plasticity in developmental programming has evolved to provide the best chances of survival and reproductive success to the organism. It was recently appreciated that the life-history evolutionary theory is a powerful tool for understanding child growth and development from an evolutionary perspective (3) (Fig. 1). By applying this theory to developmental data, adaptive growth- and metabolic-related strategies for transition from one life-history phase to the next and the timing of such transitions (inherent adaptive plasticity) have evolved.
Fig. 1.
Preadult periods of adaptive plasticity in the transition between life-history phases (double arrows). Prenatal growth affects adult health and disease. The transition from infancy to childhood confers a predictive adaptive response that determines adult height. The transition from childhood to juvenility bestows an adaptive response that resolves adult body composition and metabolic consequences. The transition from juvenility to adolescence establishes longevity and the age of reproduction and fecundity. IC, Infancy-childhood (transition).
The environmental conditions that are experienced in early life can profoundly influence human biology and long-term health. Early-life nutrition and stress are among the best documented examples of such conditions because they influence the adult risk of developing metabolic diseases, such as type 2 diabetes mellitus (T2D), and cardiovascular diseases (4). Individuals who are born small-for-gestational age (SGA) have an increased risk of cardiovascular morbidity and mortality when they are adults (4–7). This epidemiological evidence is now supported by an extensive experimental literature in animals [see Gluckman et al. (8)]. Accordingly, cardiovascular morbidity can now be considered to be, in part, a prenatal and pediatric disease. Evidence on the importance of prenatal and early postnatal growth for later morbidity suggests the existence of a link between developmental responses to early environments and adult biology. These associations are grounded in functional relationships and are broadly consistent with life-history evolution theory. Moreover, they complement current research on the impact of early-life environments on disease occurrence and susceptibility in later life.
A. Developmental origins of health and disease (DOHaD)
Interest in developmental plasticity and its relationship to human health arose from the results of early epidemiological and subsequent clinical and experimental studies that identified a relationship between early cues (often measured using birth weight as the surrogate marker) and the later risk of developing metabolic and other diseases (5, 7, 9–16). This relationship is the basis of the DOHaD phenomenon, and the inevitable association between immediate and predictive adaptive responses best models the original birth weight-disease relationships (7). There is a growing consensus that this association is broader than that of grossly disturbed early growth. Indeed, the relationships between the maternal state and later phenotypic changes of pathophysiological relevance can be independent of the birth weight (17).
The DOHaD phenomenon is an example of developmental plasticity, through which alternative phenotypes (morphs) are generated from a specific genotype by adjusting the developmental program in response to persistent environmental cues (8, 18–24). Such phenotypic variation is considered to be anticipatory of later conditions and is termed a “predictive adaptive response” that the organism induces with the expectation of a future (fitness) benefit. The recognition that environmental cues can profoundly influence development encourages the appearance of functional morphs in the population (a postgenomic interpretation of phenotype). Two examples of such developmental plasticity are often cited: the appearance of the “helmeted” morph in the freshwater crustacean Daphnia in response to an increased presence of predators (25), and the marked shifts in morphology and behavior of desert locusts in response to increased population density (26). In mammals, such adaptive plasticity is typified by the fetal meadow vole, which determines the thickness of its postnatal coat in utero in response to maternally derived signals of day length, which are used as an indicator of the season (27). The DOHaD phenomenon challenges the simplistic interpretation of phenotype as a deterministic fixed outcome of the genotype, an interpretation that has dominated much of developmental and evolutionary biology thinking in the 20th century. When phenotype is viewed as “the expression of a given genotype under its particular environmental influences” (28), the arguments of the early evolutionary biologists, such as Schmalhausen (564) and Waddington (565), are reignited. The DOHaD phenomenon also sits comfortably with emerging notions in modern molecular biology.
Mismatch arises when our evolution as a species and our development as an individual do not leave us well-matched to an “evolutionarily novel” world (20, 23). Metabolic disease is an example of a mismatch (Fig. 2). The individual variation in the sensitivity of mismatch can be explained in part by genomic variation, and in part by developmental plasticity. Although we have yet to fully understand the overnutrition pathway, medicine is reengaging with development. As a result, a new developmental synthesis is evolving where the weights of genetics, development, and the ancestral, intergenerational, and current environments in disease causation are more balanced than previously thought.
Fig. 2.
The match-mismatch paradigm of metabolic disease. The developing organism senses maternally transmitted environmental cues, such as undernutrition, during prenatal and early postnatal life. Developmental plasticity in response to these cues modifies the default trajectory defined by the inherited fetal genome and epigenome according to whether the environment is perceived as adequate (dark background) or deprived (light background), resulting in adjustment of metabolic set points. If the eventual mature environment, whether adequate or deprived, matches the prediction, then the risk of metabolic disease in later life is low. If there is a mismatch between the predicted and actual mature environments, particularly if the mature environment is richer than anticipated, then the risk of metabolic disease is enhanced. [Reproduced from P. D. Gluckman et al.: Am J Hum Biol 19:1–19, 2007 (23). © 2006 Wiley-Liss, Inc.; reprinted with permission from John Wiley & Sons, Inc.]
B. Plasticity in developmental programming
Environments change continuously, and a species adapts its phenotype to the prevailing environment, even when the environmental change is disruptive or even catastrophic. A species is considered to be well adapted and fit in evolutionary terms when it can survive to reproduce and display relative phenotypic consistency across many generations. Phenotype stability is most likely to occur when the species has adapted to a normative range of environments that remains relatively stable on generational time scales. Numerous plants and animals utilize phenotypic variation as a means to maintain fitness (reproductive success) under the challenge of a changing environment, and phenotypic variations might occur on a rapid time basis, such as in acclimatization, or over many generations, such as in natural selection.
Almost all organisms exist within an environment that can change rapidly, and those species with a relatively fixed phenotype may not be able to respond sufficiently quickly to survive an unexpected environmental change. Maintaining this flexibility results in polyphenisms (alternative phenotypes in different environments, such as in metamorphosis). Adaptive plasticity enables a species to respond to an environmental change to survive and reproduce and may manifest itself as polyphenism or as a continuous variation in traits. In evolutionary terms, plastic and developmental responses in early life enable an organism to adjust its phenotype so that it can survive in the environment in which it will grow and reproduce. However, not all developmental responses to environmental cues have an adaptive basis. When the cue is severe or novel, the outcome may be disruptive and may result in teratogenesis, disease, or death (23, 24).
The time scale and persistence of an environmental change can impact upon the phenotype because both can trigger phenotypic shifts. These shifts are also adaptive responses and have survival and/or reproductive value. Many organisms maintain a degree of phenotypic plasticity to live their life to its full potential in a constantly changing environment. The predictability of environmental changes is also an important determinant of the degree of adaptive flexibility of a species. In some instances, the environmental change is highly predictable, and an adapted species exists as a limited range of subtle but distinct and definable phenotypes. Adaptive plasticity of an organism is associated with immediate adaptive responses (forecasting or predicting), which are concerned with its immediate survival with no consideration for the long-term consequences. These adaptive responses adjust the developmental phenotype and comprise a set of processes that can be triggered by a wide range of environmental cues to promote lifetime fitness. Recognition of an environmental cue often occurs during sensitive periods in the life span of a species, namely the prenatal period and/or during transitions between life-history phases. Recognition of an environmental cue also enables the organism to adapt or acclimatize to an environment change and creates future trajectories in its development. Adaptive responses override the canalization of development and the inheritance of acquired characteristics (the constancy of the wild-type phenotype under varying developmental conditions) (29) to maintain developmental robustness (23). The resultant adaptive advantage depends on the fidelity of the cue about the future state of the environment. High-fidelity cues enable the organism to optimize its adaptation or fit to the anticipated environment. Low-fidelity cues carry a fitness disadvantage, although the impact will depend on the extent of mismatch between the predicted and actual future environment. According to the Darwinian theory of natural selection, the surviving phenotypes are better adapted to the prevailing conditions than the alternative forms (morphs)—postdevelopmental determination of the best-fit phenotype. One such cue is the energy/nutrition supply, which can cause a shift in the growth trajectory of subsequent generations (maternal prediction). Another example is population density, which is sometimes used as a surrogate indicator and predictor of future nutritional supply.
Two types of adaptive responses or plasticity exist (23). The first type is the anticipatory or predictive adaptive responses, where the developing organism forecasts the future environment and then adjusts its phenotypic trajectory accordingly. The second type is the immediate adaptive responses that promote short-term maternal or fetal survival with some advantages in later life (developmental plasticity). Because these two types of adaptive responses come with a significant cost, individual members of a species make a cost-benefit analysis to determine the true value of an adaptive response. Within the adaptive responses, the organism may engage in a trade-off between phenotypic changes to ensure its short-term survival at the expense of a long-term advantage. Hence, trade-offs occur because energy needs to be allocated to meet the different metabolic and physiological demands of a developing organism. Therefore, trade-offs can often manifest themselves as longevity, as an alternative to reduced survival of the juveniles. Such is the consequence of embryonic fetal development when it occurs in a deprived intrauterine environment as a result of a limited transplacental nutrient supply. In response, the fetus protects the development of its heart and brain at the expense of other organs, and somatic growth is retarded. Underlying developmental plasticity is the fundamental premise that the physiology of an individual is driven by the induction of a particular developmental program, which is influenced by the prevailing environment during critical developmental periods (18). To improve the chances of survival at birth, the offspring are small and have high rates of morbidity and mortality. Intrauterine growth restriction (IUGR) is an example of an immediate cryptically maladaptive response to the environment (19, 20, 22). Under severe situations in polytocous species, which produce numerous offspring in a single birth, adaptation may be driven by maternal interests that compromise some of the offspring. However, this adaptation seems unlikely to happen in slow-reproducing, single-offspring species (8, 23, 24).
When the environmental cue is subtle in early life, often no immediate adaptive responses occur because the cue is interpreted as a surrogate predictor of the later reproductive environment. Because developmental plasticity is limited by temporal constraints, the interpretation of this response can create a situation where it is evolutionarily advantageous for the fetus to adjust its phenotypic development to create a “better-matched” postnatal phenotype. Although such processes are robustly selected across taxa, there is a high risk of low fidelity of the prediction in mammals. In fitness terms, such adjustments may not have much impact because health and fitness are distinct concepts. However, for humans who live beyond the peak reproduction period, the health consequences, rather than the reproductive consequences, become the primary concern. For example, the occurrence of metabolic disease is more likely to occur when the nutritional status in adulthood differs markedly from that experienced and predicted during development and the expectation of a poor environment is not subsequently met. Life-history theory argues that energy- and stress-related cues experienced by the mother are likely to be the primary environmental triggers of developmental plastic responses (30). The predictive responses are primarily induced by subtle cues, and the immediate adaptive responses are induced by more obvious cues. However, both types of responses could coexist when the immediate adaptive responses are induced.
C. Plasticity in phase transitions of human life history
The secular trends in child growth and puberty are dazzling examples of such adaptation (31). European men are now 13 cm taller than they were 150 yr ago. This range of plasticity in growth over approximately six generations is not long enough to result from changes in the DNA sequence. Over the same six generations, the age of menarche in Western countries has decreased by 4 yr. This reduction has a fitness advantage on the fecundity span in an environment that is rich in energy resources and demonstrates plasticity in the maturation of the hypothalamic-pituitary-gonadal axis. As a consequence of constantly changing life conditions and environment, today's children may be stunted in growth or be tall, adapt their body composition and energy metabolism, and modulate their longevity, fertility, and fecundity. The signals of energy balance that modulate this plasticity are both intrinsic (internal) and extrinsic (environmental). The internal signals include leptin, the GH-IGF-I axis, ghrelin, thyroid hormones, insulin, and the cortisone-cortisol shuttle (11β-hydroxysteroid dehydrogenases), whereas the environmental signals include prenatal and postnatal nutrition, stressors, endocrine-disrupting chemicals (EDCs), and light.
Human growth and development are orchestrated processes of well-recognized and predictable events with five overlapping, yet distinct, preadult life-history phases: the prenatal, infantile, childhood, juvenile, and pubertal growth phases (Fig. 1). The transition periods between these phases are sensitive windows of developmental plasticity, and there is now some evidence that the features of transition from one phase to the next are transmitted transgenerationally (32). With decreasing sensitivity, the transitions between phases are periods of adaptive plasticity, and the multifactorial regulation of growth during each phase mirrors the interplay between genetic, hormonal, environmental, and psychosocial factors.
Four adaptive processes influence human phenotype, and each operates on a different time scale (33). The first process involves changes in gene sequence and frequency in a population or species, and this process occurs over several hundred thousand years. The second process is modification of homozygosity of the population, and this process occurs over several hundred years and numerous generations. The third process is adaptive phenotypic plasticity, and this process occurs over the entire life span of the individual, and may be carried forward for three to four generations. The fourth process is short-term acclimatization that can last several months or years. In response to environmental cues, especially those that relate to energy resources, a life-history phase can be added or deleted and can have its duration, intensity, and onset time altered (3). Thus, the timing of infancy-childhood transition (ICT) adaptively adjusts an individual's size to the prevailing environment in response to environmental cues (3). Hochberg and Albertsson-Wikland (34) have previously reported that the ICT is a major determinant of final adult height and a delayed ICT is the most common cause of idiopathic short stature. The transition from juvenility to adolescent-related puberty and the growth spurt is a function of maturation of the hypothalamic-pituitary-gonadal axis. Poor quality of life during this transition delays fecundity and increases longevity (35). Hence, a series of control mechanisms must exist to enable 1) the GH-IGF-I axis to dominate as the child transits into childhood; 2) adrenarche at the onset of juvenility; and 3) an abrupt increase in sex hormones at initiation of puberty (3).
As already noted, an organism distributes its energy resources during its life by timed allocations toward growth, self-maintenance, reproduction, and raising offspring to independence to avoid death (33, 36). Whereas the environment at any one geographical location may vary slowly, nutritional conditions may change rapidly. Evolution has provided organisms with the mechanisms to adapt to such extremes. Humans can also use sociocultural adjustments to fill the gaps when the changes occur faster than the evolutionary time scale. This can be seen when one examines the evolution of hominid life history from Australopithecus afarensis to Homo sapiens. In humans, the duration of infancy has been shortened and that of childhood has been prolonged, and these two phases are followed by a relatively short juvenility and late adolescence to increase fitness (36–38). The overall result of this strategy is increased body size and longevity, and reproduction at a later age, compared with other primates. This strategy has been very successful for humans, who can thrive and propagate in extremely diverse environments that encompass the entire range of geographic latitudes and altitudes.
An important environmental cue for infants and young children is the caregiving behavior of their parents, which can be used as a predictive indicator of the security of their environment. The resultant attachment patterns are transmitted transgenerationally (39, 40). The degree of security that is experienced during childhood sets development on alternative pathways and adaptively shapes the individual's future reproductive strategy. A secure attachment will result in a reproductive strategy that is based on late maturation, a commitment to a long-term relationship, and a large investment in parenting. In terms of evolutionary developmental biology, which studies the developmental mechanisms that control body shape and form and the alterations in gene expression and function that lead to changes in body shape and pattern (41), the expected response to a secure environment will include investment in large body size (42, 43). This example of transgenerational phenotypic plasticity contrasts that of an insecure attachment and a small parental investment that involves a large number of children; the response is a compromise in body size, early reproduction, and short-term mating.
Child growth and body composition display a vast range of adaptive plasticity. Short-term plasticity in the various child growth phases and transitions suggests that epigenetic mechanisms determine the extent of adaptive plasticity during growth in response to environmental cues. In light of these new findings, this article considers the utility of life-history theory and the links between epigenetics, developmental programming, and plasticity in early growth and nutrition. Current research in child health strives to identify mechanisms that underlie plasticity in developmental programming and life-history transitions. Developmental programming and life-history transitions are purported to use nutritional or endocrine cues for setting long-term biological strategies in response to local ecological and/or social conditions (18, 23, 44). Rapid changes in nutrition during one's lifetime can then lead to “mismatch” and metabolic disease (20). It has been further proposed that intergenerational influences on nutrition and growth stabilize the nutritional signals that are received in utero to increase the reliability of an intrauterine cue as a predictive signal (44). It is now also known that the effects of hormones, stress, and drugs during embryogenesis can not only influence the subsequent behavioral phenotype of the individual, but can also modify the individual's response to adult experiences (2).
In his recent review on phenotypic plasticity and the epigenetics of human disease, Feinberg (45) argues that epigenetic changes are involved in normal development and human disease. He proposes that the term “epigenetic disease” be used to describe defects in the epigenome that are known to lead to disease. These defects include changes in the localized or global density of DNA methylation, incorrect histone modifications, or altered distribution or function of chromatin-modifying proteins that, in turn, lead to aberrant gene expression. According to Feinberg, defects in phenotypic plasticity or the cell's ability to change its behavior in response to internal or external environmental cues are the underlying theme of epigenetic disease. Feinberg proposes that this theme can also be applied to common diseases with late-onset phenotypes that involve interactions between the epigenome, the genome, and the environment.
The almost exponential expansion in our understanding of epigenetic regulation now provides mechanistic insights to developmental plasticity and the molecular relationships between the environment and the response of genes. This article (45) proposes that phenotypic plasticity is the manifestation of adaptive programming and that “softly inherited” epigenetic mechanisms may underlie phenotypic plasticity and adaptive programming. The article also reviews the evolving idea of plasticity in developmental programming with respect to human life history and transitions between life-history phases and proposes that epigenetics provides a molecular mechanism for programming that links genes, the prenatal environment, intrauterine growth, and subsequent susceptibility to disease. For this purpose, the epigenetic basis of plasticity is reviewed in the setting of early nutritional experiences and developmental programming. The notion that the phenotype of the placenta and its ability to support fetal growth are established at the time of conception, or even implantation, is also explored. This notion is further discussed in the light of increasing knowledge that the placenta and the fetus continue to adapt throughout pregnancy in response to the prevailing environmental conditions.
II. Epigenetic Programming and Developmental Plasticity
Epigenetics has evolved very quickly from the study of an obscure collection of diverse phenomena to become one of the most exciting topics in contemporary biology. It is a rapidly expanding field of study in which the molecular mechanisms of seemingly unrelated normal processes, such as paramutation in maize, position effect variegation (PEV) in the fruit fly, and genomic imprinting and X-chromosomal inactivation in mammals, are now recognized as evolutionarily conserved epigenetic processes. In medicine, epigenetics has become the new frontier.
The exact definition of epigenetics is controversial (46, 47), and discussion on its definition is beyond the scope of this review. Irrespective of its definition, epigenetics is important for understanding gene function and expression because expression profiles are influenced by epigenetic modifications, and the epigenetic regulation of gene expression is essential for the normal growth, development, and aging of higher organisms (45). Epigenetics also underlies genomic imprinting, programming, and reprogramming in early life and the increased susceptibility to disease in later life. In this section, we will describe the epigenome and the epigenetic machinery to provide an overview of the components of the epigenome and the processes that the epigenetic machinery uses to influence and/or modulate gene expression, programming, and disease susceptibility, which are topics that will be discussed in detail in the various sections of this review.
The term “epigenetic landscape” is widely used when discussing epigenetics (48). It describes the range of epigenetic marks that are acquired during the developmental course of a cell, or an embryo, or its parts to take specific trajectories that lead to different cellular or organismal fates (lineage commitment) in response to the environmental cues (49, 50). Initially, several possible pathways are available because the cell is pluripotent, and early embryonic cells follow one of these pathways. However, the further a cell or embryo travels down any one developmental pathway or trajectory, the more difficult it becomes for it to move into an alternative one (canalization). The choice of a developmental trajectory can be influenced by exposure of the mother to an environmental cue, which, in turn, alters the nature of the mother-offspring interaction and is capable of inducing a shift in the developmental trajectory of the offspring (environmental programming). Reprogramming between generations is a corollary of Waddington's (565) idea and refers to the zygote regaining its totipotency so that it can go down all the possible pathways as a new organism develops. This section will also discuss these concepts, as well as presenting an overview of the heritability of epigenetic change and the determination of phenotype.
A. The epigenome
Our genomes constitute more than just the DNA blueprint. DNA is packaged as chromatin, and to fit within the nucleus of the cell, DNA is very tightly coiled and bundled into three-dimensional chromosomal structures. In eukaryotes, DNA is wrapped around an octamer of histone proteins that consists of two copies of the core histones, H2A, H2B, H3, and H4. These core particles or nucleosomes are the basic unit of chromatin, which can then be assembled further into higher-order chromatin structures. The nucleosome compacts the DNA-histone complex, and the degree of compaction creates an added layer of regulatory control of the genome.
The compact histone-DNA configuration is maintained by electrostatic bonds between positively charged histones and negatively charged DNA, and changes in the patterns of these bonds regulate gene expression (51). Nucleosomes also carry covalent modifications on their core histones and on the DNA. These epigenetic modifications can determine whether parts of chromosomes are tightly or loosely packaged, which in turn influences whether a gene is switched “on” or “off.” It is now recognized that epigenetic information is crucial for the dynamic interpretation of genetic information so that the correct genes are expressed at the right time during critical cell fate decisions.
The term “epigenome” refers to the global epigenetic patterns that are characteristic of an organism. Changes in epigenetic information during the life span can occur by modifying the epigenetic marks on the DNA and/or histone proteins without altering the underlying DNA sequence, whereas changes in genetic information occur by altering the underlying DNA sequence. Epigenetic mechanisms of gene regulation are relevant throughout development from when the sperm first meets the egg, through early lineage decisions, to fetal development and postnatal life. Epigenetic patterns that were acquired during development are, in most cases, stable in somatic cells and during adult life. However, somatic epigenetic patterns need to be “reset” or “reprogrammed” in germ cells and also in early embryos to achieve developmental pluripotency. Reprogramming normally results in differences in some epigenetic marks on chromosomes that are inherited from eggs, when compared with those that are inherited from sperm. The most striking example of such epigenetic asymmetry is “genomic imprinting,” which occurs in mammals. There are nearly 100 genes that are subject to imprinting in humans and mice (52, 53). These genes are marked epigenetically in the germ line, and this process results in only one of the parental copies being expressed after fertilization (discussed in Section IV.A). Epigenetic regulation also underlies X-chromosome inactivation, a phenomenon through which one of the two X-chromosomes is inactivated in every cell of the body, and in the silencing of transposable elements, thereby preventing insertional mutagenesis. X-inactivation and genomic imprinting are discussed in detail in Section IV.C.
Almost all of the different cell types that make up an organism share an identical genotype, yet each cell type has well-defined, individual, and stable profiles of gene expression. Subsets of the 20,000–25,000 genes of the human genome are active in different tissues because of their regulation by different sets of transcription factors and epigenetic modifications (54, 55). Thus, the epigenetic marks that accumulate in a differentiated cell will differ from those in pluripotent cells and will also be distinct from those of other differentiated lineages (Fig. 3). The set of epigenetic patterns, the “epigenotype,” is specific to each cell type and influences its fate, irrespective of whether the cell is derived from the liver, brain, or bone. Remarkably, during cell division, committed cells acquire the same epigenotype as their parent cells. Therefore, epigenetic events create a memory of cell identity to sustain genomic function that includes, among others, maintenance of cell identity after differentiation (Fig. 3) (56).
Fig. 3.
Top panel, DNA vs. chromatin. The genome is the invariant DNA sequence of an individual. The epigenome is the overall chromatin composition, which indexes the entire genome in any given cell. It varies according to cell type and response to the internal and external signals that it receives. Lower panel, Epigenome diversification occurs during development in multicellular organisms as differentiation proceeds from a single stem cell (the fertilized embryo) to more committed cells. Reversal of differentiation or transdifferentiation requires the reprogramming of the cell's epigenome. [Fig. 3 and its legend have been reproduced with permission from C. D. Allis et al.: Epigenetics, Chap 3, Cold Spring Harbor Laboratory Press, Woodbury, NY, 2007 (561). © 2007 CSHL Press.]
Epigenetic states, however, also have an inherent flexibility because they can undergo regulated change in response to particular stimuli to modulate gene expression as the need arises. For example, this flexibility is evident during the development of stem cells into particular organ systems, reprogramming events in germ cells and early embryo to restore pluripotency, and in the response to external environmental factors, such as diet and environmental chemicals. On the other hand, the functional states of nondividing quiescent cells with a long lifetime, such as neurons, require an epigenetic mechanism for their quiescence and longevity. If these naturally occurring epigenetic processes occur improperly, major adverse health and behaviors can ensue. Epigenetic modifications, therefore, can render the genome functionally flexible and adaptable, but at the same time render it vulnerable in many ways. The vulnerability of the genome and underlying epigenetic mechanisms of gene expression in various settings are discussed in Section VIII.C–F.
B. The epigenetic marks
The traditional view of epigenetic modifications as static on/off switches in the control of gene expression is now being altered by the idea that these marks are dynamic. Because some environmental factors or cues can act on the epigenetic machinery to bring about either short-term or long-term outcomes, this next section will briefly review the key features of the epigenetic marks and associated machinery in the context of developmentally regulated genes and plasticity throughout mammalian life.
1. DNA methylation
The most comprehensively studied epigenetic mark is DNA cytosine methylation (m5C). In mammals, nearly all m5C is found at sites where cytosines are followed by guanines (CpG dinucleotides) (57). The mammalian genome is greatly depleted of CpG dinucleotides, which accounts for only about 1% of all DNA bases (57). However, the promoter regions of around 60% of all human genes contain small stretches of DNA with a relatively high CpG content (57). Although these promoter-region “CpG islands” are largely unmethylated, promoter-region methylation of islands is frequently correlated with transcriptional silencing (57–59). Cytosine methylation is required for the allele-specific expression of imprinted genes, the transcriptional repression of retrotransposons (mobile genetic elements that are transposed through RNA intermediates) in germ cells and somatic cells, X-chromosome inactivation in females, and stable silencing of some pluripotency-associated genes during differentiation.
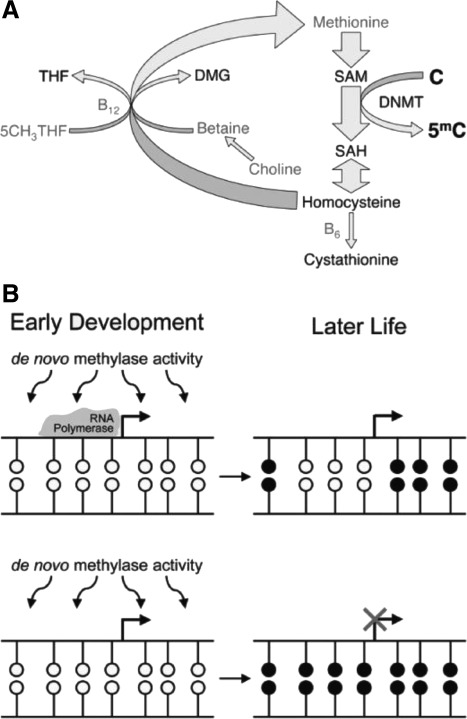
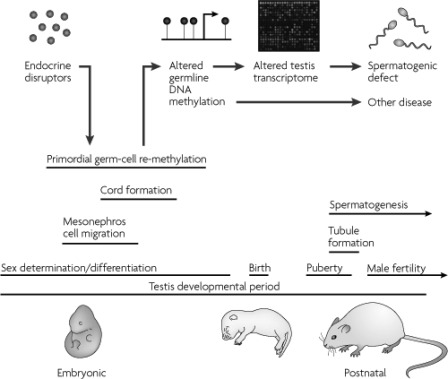
In the mammalian genome, methyl groups are placed on DNA by a group of highly conserved proteins called DNA methyltransferases (DNMTs). The de novo DNMTs (DNMT3A, DNMT3B, and their cofactor DNMT3-like) establish methylation patterns early in development (60). Although there is increasing evidence that DNMT3A and DMNT3B are also involved in the maintenance of DNA methylation (61), the activity of the maintenance DNMT, DNMT1, ensures that DNA methylation patterns are stably maintained during adult life. DNMT1 preserves methylation patterns throughout cell divisions by adding methyl groups to hemimethylated CpG dinucleotides (60). However, widespread losses of DNA methylation are observed during the epigenetic “reprogramming” that occurs in primordial germ cells (PGCs) and the early embryo during particular developmental windows (Fig. 4) (62, 63). DNA methylation can be lost either passively by blocking methylation of newly synthesized DNA during DNA replication or actively by unknown mechanisms, which possibly involve DNA repair (64). The erasure of methylation marks at imprinted genes in the germ line is a key developmental event so that gender-specific methylation is imposed subsequently during germ cell development (65). Also crucial for development is the ability of imprinted genes to maintain their methylation marks throughout early embryo reprogramming to ensure the inheritance of parental-specific epigenetic information (Fig. 4) (66).
Fig. 4.
Alterations in methylation status during development. During embryonic development and gonadal sex determination, primordial germ cells undergo genome-wide demethylation, which erases previous parental-specific methylation marks that regulate imprinted gene expression. In the male germ line, paternal methylation marks in imprinted genes are laid down in developing gonocytes that will develop into spermatogonia. The female germ line establishes maternal methylation marks in imprinted genes at a later stage. After fertilization, the paternal genome is actively demethylated, whereas the maternal genome undergoes passive demethylation (176). Genome-wide remethylation occurs on both parental genomes before implantation. However, imprinted genes maintain their methylation marks throughout this reprogramming, allowing for the inheritance of parental-specific monoallelic expression in somatic tissues throughout adulthood. [Reprinted with permission from R. L. Jirtle and M. K. Skinner: Nat Rev Genet 8:253–262, 2007 (62). © 2007 Macmillan Publishers Ltd.]
It is now recognized that DNA methylation contributes to specifying cell fates and maintenance of cell identity. Pluripotency transcription factors, such as OCT4 and NANOG, are expressed in embryonic stem cells (ESCs) but are silenced by DNA methylation and histone modifications during the differentiation of these cells (63). Conversely, the transcription factor gene, Elf5, is methylated and silenced in the embryonic lineage but hypomethylated and expressed in the trophoblast lineage (67). The function of this type of epigenetic marking of Elf5 is to reinforce the trophoblast-specific transcriptional circuit and fixation of the lineage fate (embryonic vs. extraembryonic) (68). Lastly, the results of several genome-wide studies have shown that methylation patterns differ between tissues; a gene might be methylated in one tissue but unmethylated in another, thereby constituting the so-called tissue-specific differentially methylated regions (69–71). Collectively, these examples support the notion that DNA methylation plays a key role in tissue differentiation by maintaining the transcriptional silence of genes whose expression is not required in specific cell lineages.
2. Histone modifications
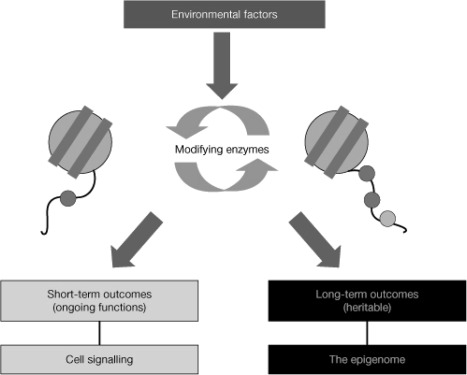
Each core histone has an end-amino-terminal tail that protrudes from the nucleosome and can be subjected to a diverse array of covalent posttranslational modifications (72). These modifications include acetylation of lysines, methylation of lysines and arginines, ubiquitylation and sumoylation (small ubiquitin-like modification) of lysines, and phosphorylation of serines and threonines. Histone modifications recruit and bind critical DNA-regulatory proteins, and these processes ultimately lead to changes in DNA transcription, replication, recombination, and repair. Histone modifications constitute signals that are read alone or in combination with other marks on the same or neighboring histones, and the resultant codes are referred to as the “histone code.” Thus, histone modifications are recruitment signals for protein effectors that exert a series of diverse functional effects with short-term and long-term outcomes (Fig. 5).
Fig. 5.
Histone modifications can generate both short-term and long-term outcomes. The amino-terminal tails of all eight core histones protrude through the DNA and are exposed on the nucleosome surface, where they are subject to an enormous range of enzyme-catalyzed modifications of specific amino-acid side chains, including acetylation of lysines, methylation of lysines and arginines, and phosphorylation of serines and threonines. Histone tail modifications are put in place by modifying and demodifying enzymes whose activities can be modulated by environmental and intrinsic signals. Modifications may function in short-term, ongoing processes (such as transcription, DNA replication, and repair) and in more long-term functions (as determinants of chromatin conformation, for example heterochromatin formation, or as heritable markers that both predict and are necessary for future changes in transcription). Short-term modifications are transient and show rapidly fluctuating levels. Long-term, heritable modifications need not necessarily be static; in theory, they still show enzyme-catalyzed turnover, but the steady-state level must be relatively consistent. [Reprinted with permission from B. M. Turner: Nat Cell Biol 9:2–6, 2007 (562). © 2007 Macmillan Publishers Ltd.]
Chromatin is generally compartmentalized into two main domain types: heterochromatin, which is condensed and gene-poor, and euchromatin, which is decondensed and gene-rich (72). These domains have different patterns of histone modifications and are associated with different modes of nucleosome packaging, higher-order structure, and nuclear organization. A link between heterochromatin formation and gene silencing has been inferred from the loss of gene activity on the inactive X-chromosome (Xi) and in PEV in Drosophila and other organisms. PEV occurs when a gene that is normally “euchromatic” is juxtaposed with heterochromatin by transposition or rearrangement; the resulting variegating phenotype indicates that the gene has been silenced in a proportion of the cells. In general, heterochromatin is associated with repressive histone marks and DNA methylation, whereas euchromatin is associated with active histone marks (72).
Histone acetylation is restricted to conserved lysines across the core histones, despite being one of the most prevalent of all the histone modifications (73). It is generally considered that a mark of open, active chromatin domains corresponds to actively transcribed genes with high levels of acetylation at their promoter regions, transcription start sites, CpG islands, and functional regulatory elements (74). The levels of acetylation across chromatin are determined by histone acetyltransferases (HATs), which catalyze the addition of acetyl moieties to the lysine residues, and histone deacetylases (HDACs), which remove the acetyl group from the lysine residues (75). The balance of the activities of these two enzymes determines the state of histone acetylation, which in turn can influence the level of expression of the underlying genes (73). When HDACs remove the acetyl groups from histone lysines, a positive charge is restored to the lysine residue, thereby condensing the structure of nucleosomes (76). Nucleosomes that contain highly charged hypoacetylated histones bind tightly to the phosphate backbone of DNA, thereby inhibiting transcription, presumably because transcription factors, regulatory complexes, and RNA polymerase do not have access to the DNA. This closed chromatin structure commonly precludes transcription factor binding to DNA and underscores the importance of enzymes that modify histone-DNA interactions. On the other hand, HATs catalyze the acetylation of selected positively charged amino acids, such as lysine, on the protruding histone tails, of which histone H3 or H4 is the most common. Acetylation of K9 residues on the end-amino-terminal tails of H3 histones (H3K9ac), for example, neutralizes the positive charge of the histone tail and decreases histone's affinity to negatively charged DNA, and generates a more open DNA conformation. This results in “relaxing” of the DNA, which is wrapped around the octamer of histone residues. Transcription factors and the transcription apparatus can then access the DNA, and expression of the corresponding genes is facilitated (77). Importantly, many transcriptional regulators and factors possess intrinsic HAT activity (73). Consistent with the role of acetylation in transcriptional activation, deacetylation is generally associated with gene silencing. Indeed, HDACs are generally considered as transcriptional corepressors (78).
Compared with histone acetylation, histone methylation is considerably more complex (79). It can occur on conserved lysine and arginine residues and across all four histone proteins. Up to three methyl moieties can be applied to the lysine amino group (monomethylation, dimethylation, and trimethylation), whereas arginine can be either monomethylated or dimethylated. Histone methyltransferases are the enzymes that are responsible for the addition of methyl groups to either lysine or arginine residues. Until recently, it was believed that histone methylation represented a more permanent, stable modification because the global turnover of this mark was lower than of the highly dynamic acetylation mark (67). However, the recent identification of enzymes that are capable of removing methyl groups from histones has shown that this mark may be equally dynamic (80). In contrast to acetylation that affects the charge of the residue, and thereby directly impacts on histone-histone or histone-DNA binding, the role of methylation is likely to be solely orchestrated through the recruitment of additional regulatory factors. Therefore, methyl marks have the potential to influence gene expression in opposing ways that depend on both the location and the timing of the mark. For example, trimethylation of lysine 4 on histone H3 (H3K4me3) is generally perceived as an active modification and occurs preferentially at active promoters, whereas methylation of H3K9 (H3K9me) is detected at the promoter of inactive genes and yet is deposited in the coding regions of active genes (81). H3K27me3 is strongly associated with gene silencing via unique interactions with the Polycomb group (PcG) proteins, which are discussed in Section II.B.3.a.
The regulatory potential of histone modifications is substantial in view of the large number of different histone modifications and the extensive cross-regulation that may occur between certain histone marks. The cross talk between histone marks is a fundamental concept of the histone code hypothesis, which predicts that combinatorial sets of histone marks act in concert to regulate the chromatin structure. There are various ways by which this cross-regulation can occur. Different marks can antagonize each other on the same residue. For example, when a lysine residue is acetylated, it cannot also be methylated, and only one level of methylation (namely mono-, di-, or trimethylation) can be present (82). Another level of cross talk involves removal or recruitment of a protein complex by an adjacent modification, as is the case for phosphorylation of serine 10 on histone H3, which is necessary to disrupt the binding of the heterochromatin protein 1 (HP1) to H3K9me3 (83). Alternatively, an enzyme complex can be affected by multiple histone modifications: H3K9ac and H3K14ac enhance the binding of the general transcription factor TFIID to H3K4me3, and asymmetric dimethylation of H3R2 prevents it (84). Cross talk can also involve trans-histone effects, where modifications on different histone proteins can regulate each other. For example, monoubiquitylation of K120 is required for di- and trimethylation of H3K4 (85, 86). Lastly, cross talk can ultimately determine the final transcriptional output through specific trans-acting effects of developmentally regulated noncoding RNAs (ncRNAs) on transcription where modifications on different histone proteins can regulate one another (87–89).
As we acquire more knowledge about histone marking systems, the accepted concept of classifying histone marks as either active or inactive is being challenged. Indeed, marks that were originally thought to be “active” can be found within silent genes and vice versa; H3K9me3 is found both in silent heterochromatin and at some active genes (90). Therefore, it now seems prudent to consider that a single type of histone posttranslational modification does not dictate a single outcome. Instead, it seems that a combination and enrichment of histone posttranslational modifications define different chromatin domains with specific functional outcomes. Although it is widely accepted that chromatin has a crucial role in the inheritance of transcriptional regulation, it is still unclear how histone modifications are reproduced after DNA replication and are transmitted from one cell generation to the next. Several mechanisms have been proposed by which “new” and “old” histones, of which the latter carry their original posttranslational modifications, are distributed after DNA replication. These include random, semiconservative, and asymmetric modes of histone distribution (72, 91). The genome-wide epigenetic reprogramming that occurs both in the germ line and embryo extends beyond DNA methylation and also involves histone marks, histone exchange, and the use of histone variants (Fig. 4) (66, 68, 91–93). The mechanistic aspects of reprogramming are under intense investigation and are beyond the scope of this review [for further reading, see Morgan et al. (94), Hayashi and Surani (95), Hemberger et al. (68), Popp et al. (96), Ray-Gallet and Almouzni (97), and Xu et al. (98)].
3. Noncoding RNAs
Almost all of the genome is transcribed, yet only a small proportion of it (1.2%) encodes proteins. One explanation for this phenomenon is the existence of a large repertoire of short and long ncRNAs that includes many new RNAs, in addition to the well-known groups of rRNAs, small nuclear RNAs, and tRNAs. It has become clear that these newly discovered ncRNAs are functional and central to complex genetic phenomena in eukaryotes that include transcriptional and posttranscriptional gene silencing, X-inactivation, genomic imprinting, and germ cell reprogramming, all of which involve epigenetic processes.
a. Short ncRNAs.
The fundamental biological role for short regulatory RNAs was demonstrated in the late 1990s by the discovery of RNA interference (RNAi) (99). RNAi is a process through which exposure to double-stranded RNA leads to silencing of homologous genes, most often posttranscriptionally. This phenomenon was originally thought to be restricted to exogenous double-stranded RNAs, but it soon became clear that animals and plants produce an array of small RNAs, which include endogenous small interfering RNAs, microRNAs (miRNAs), and P-element-induced wimpy testes (PIWI)-interacting RNAs (piRNAs), and this repertoire is continually increasing. The three major small RNA silencing pathways identified thus far seem to be involved in both posttranscriptional gene silencing through RNA degradation or translation arrest and chromatin-dependent gene-silencing pathways, which in turn, also appear to occur through both transcriptional and cotranscriptional gene silencing. A detailed examination of the function and biogenesis of small RNAs is beyond the scope of this review and has recently been covered in detail in several excellent reviews by Moazed (100) and Taft (101).
miRNAs are small ncRNAs that regulate gene expression at the posttranscriptional level by either degradation or translational repression of a target mRNA (102). They are generated from hairpin precursors by the successive actions of the RNAse III enzymes, Drosha and Dicer, which are located in the nucleus and cytoplasm, respectively. Most miRNAs seem to act exclusively in the cytoplasm, where they mediate mRNA degradation or translational arrest. Some of the first miRNAs that were discovered, such as the let-7 family of miRNAs, are master regulators of developmental differentiation, both in early embryos and adult tissues (103, 104). The let-7 targets are “canonical,” in that the miRNA seed “sequence” (eight nucleotides long) binds to target the 3′ untranslated region of mRNA and represses translation. The principal targets of let-7 family members, of which there are 11 in vertebrates, are cell cycle regulators, oncofetal genes, pluripotency factors, and components of the miRNA biogenesis pathway. Importantly, let-7 biogenesis and gene regulation are characterized by a series of autoregulatory feedback loops (105). For example, let-7 targets the pluripotency factor LIN28, which can in turn bind to the conserved loop of the primary let-7 transcript (pri-let-7) to directly inhibit the cleavage steps by the nuclear RNase, Drosha. Consistent with the central role of let-7 miRNAs in developmental regulation, genetic variants of the LIN28B locus have recently been associated with the timing of human pubertal growth and development (106) (see discussion in Section III.C).
In plants and in fission yeast, short regulatory ncRNAs often work in concert with various components of the cell's chromatin and DNA methylation machinery to achieve stable silencing (107). Although endogenous small interfering RNAs or other classes of small RNAs that mediate transcriptional gene silencing have yet to be characterized in mammals, the results of recent studies suggest that small RNA-directed epigenetic processes exist in mammals. Indeed, human miRNAs have been found to guide chromatin remodeling by inducing heterochromatin formation at promoters (108, 109). For example, Kim et al. (109) recently reported that miR-320, a conserved miRNA, can direct the association of the RNAi protein, Argonaute-1; the PcG protein, Ezh2; and H3K27me3 at the promoter of the cell cycle gene, POLR3D. Furthermore, RNA-directed DNA methylation, which is a conserved mechanism for control of gene expression, has been recently described in mammals, where it appears to be restricted to germ cells (110). Members of the PIWI clade of proteins and associated piRNAs are involved in the repression of retrotransposons and are essential for gametogenesis (111). Mutations in the mouse family members of PIWI proteins, MIWI2 or MILI, result in demethylation of the LINE-1 retrotransposon and intracisternal particle (IAP) transposable elements in the testis (112). This finding suggests that piRNAs, directly or indirectly, mediate changes in DNA methylation. However, the mechanisms by which they trigger de novo methylation are at present unclear, but this may involve demethylation of H3K4 (60).
b. Long ncRNAs.
It is estimated that at least 80% of transcriptional activity in mammals corresponds to long ncRNAs (lncRNAs) (113), which are generally more than 2 kb long, although some are more than 100 kb. lncRNAs are spliced RNAs that contain canonical polyadenylation signals. Several lncRNAs have been found to be associated with chromatin modifying complexes (89, 114). A primary role of lncRNAs appears to be the regulation of protein-coding gene expression through modulation of chromatin states or through direct effects on gene transcription. For example, Rinn et al. (115) recently identified a 2.2-kb lncRNA, which they termed HOTAIR, residing in the HOXC locus in an antisense orientation. HOTAIR represses transcription across different chromosomes (in trans) by maintaining a transcriptional silent chromosomal domain that spans 40 kb of the HOXD locus through PcG protein-mediated repressive H3K27me3. In another study, Feng et al. (116) recently demonstrated that the lncRNA Evf-2, which is partially encoded by the Dlx-5/6 ultraconserved region, stably complexes with the transcription factor Dlx-2 to increase the transcriptional activity of the Dlx-5/6 enhancer, which in turn regulates Dlx-5/6 expression. Of note, Evf-2-deficient mice show reduced numbers of GABAergic interneurons in early postnatal hippocampus and dentate gyrus and reduced synaptic inhibition in adulthood (117). From these results, Bond et al. (117) suggested that ncRNA-dependent balanced gene regulation in the embryonic brain is critical for proper formation of GABA-dependent neuronal circuitry in the adult brain. Such findings provide additional evidence that lncRNAs are one of many critical factors in the developing embryo that influence GABAergic interneuron function in adults (118). The findings from these studies are relevant to the etiologies of adult mental health disorders because these results show that lncRNA-dependent processes are fundamental to the development of the central nervous system. Moreover, the findings suggest that adult mental disorders, in the absence of apparent physiological deficits, may be the result of altered embryonic development, a topic that is discussed in Section VIII.E.
lncRNAs are also associated with genomic imprinting and X-chromosome inactivation (see Sections III.C and IV.C). These two epigenetic phenomena have been proposed to share some mechanistic features [see review by Reik and Lewis (119)]. Several imprinted gene clusters use lncRNAs as the main epigenetic mechanism to silence their adjacent genes, possibly by establishing nuclear domains with repressive histone modifications (120–122). Another lncRNA, X inactive-specific transcript gene (Xist), which is transcribed specifically from the inactive X-chromosome, coats the chromosome in cis to help create a repressive environment with recruitment of histone modifications and DNA methylation (123) (see also Section IV.C).
Disruption in the expression of small RNAs and lncRNAs has been linked to human disease. For example, miRNAs are frequently found aberrantly expressed in a variety of cancers, central nervous system disorders, and cardiovascular disease (101). Furthermore, microdeletions of the small nucleolar RNA clusters, HBII-85 and HBII-52, on chromosome 15q11-q13 result in Prader-Willi syndrome, an imprinting disorder that is characterized by hyperphagia, hypogonadism, and cognitive impairment (124) (see Section III.C).
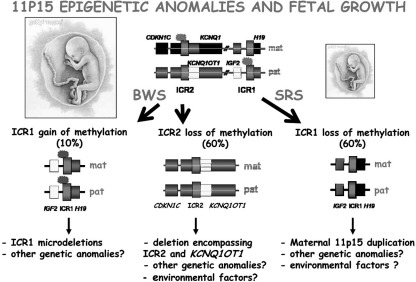
Dysregulation of lncRNAs is also a primary feature in many cancer types, Alzheimer's disease, spinocerebellar ataxia type 8, and the Beckwith-Wiedemann syndrome (BWS) (see Section III.C), among other diseases. Interestingly, the results of genome-wide association studies (GWAS) are beginning to identify ncRNAs as novel disease loci (101). This is perhaps not surprising because the genome is highly transcribed from intergenic regions and many disease variants map far from genes; thus, the likelihood of interrupted lncRNAs is high (101).
C. Reading the epigenetic marks and developmental and physiological consequences
Epigenetic information is conveyed in mammals by synergistic interaction between mitotically heritable patterns of DNA methylation, histone modifications, and various DNA-binding proteins (49, 125). Cross talk between DNA methylation and histone modifications occurs, and this cross talk is mediated by methyl-binding or histone-binding proteins (125), which decipher the regulatory information that is encoded in the DNA methylation and histone marks. The methyl CpG-binding domain protein family is a highly conserved family of DNA-binding proteins with a common sequence motif (126, 127). This family of proteins is widely believed to decode information that is encoded in DNA methylation patterns into an appropriate functional state by recruiting HDACs, for example, to effect gene silencing (128, 129). As mentioned previously, modified histones are recognition sites for effector proteins. For example, the spreading of repressive chromatin can be achieved by H3K9me3, which is recognized by HP1, to recruit the lysine methyltransferase Suv39h1 and the DNMTs (129). This process facilitates further H3K9me3 marking, HP1 binding, and DNA methylation on the adjacent nucleosomes and results in the spreading of chromatin domains.
PcG protein complexes are another group of proteins that can modify histones. The PcG system, which was originally shown to repress developmental Hox genes in Drosophila melanogaster, is important for the stability of the transcriptional program during development and maintenance of stem cell pluripotency. PcG proteins are repressors of target genes (130, 131). The catalytic component of PRC (polycomb repressive complex) 2, Ezh2, catalyzes the trimethylation of histone H3K27 (H3K27me3), which in turn recruits PRC1 via its chromodomain-containing components and facilitates histone H2A ubiquitination and chromatin condensation (132). In ESCs, PcG proteins suppress cell fate-specific genes to keep stem cells in a pluripotent state. Indeed, genes such as Dlx, Pax, Six, and Hox, which are required during development and for differentiation, are held repressed in pluripotent ESCs by induced H3K27 methylation (131, 133). Upon differentiation, the reduced recruitment of PcG proteins activates their target genes (134), and the trithorax group proteins may be involved in this activation by substituting the PcG proteins on the target genes (130).
Another prominent way by which histone modifications can influence developmental gene expression was revealed from the results of epigenomic studies on murine and human ESCs (133, 135). Using chromatin immunoprecipitation, Bernstein et al. (135) showed that developmental genes that are repressed in ESCs but are required for later differentiation are marked by bivalent chromatin, which contains both inactivating (H3K27me3) and activating (H3K4me3) marks (133, 135, 136). These bivalent chromatin domains render genes poised for activation, and therefore reflect the cell state and lineage potential (135). Importantly, several genes that are not marked by either H3K27me3 or H3K4me3 tend to be marked by DNA methylation in a complementary mechanism to histone modifications that ensures heritable gene repression (137, 138).
Emerging evidence suggests that the distribution of DNA methylation may be a major determinant of the chromatin landscape by controlling histone modifications and histone variant deposition (138–140). In accordance with this notion, H3K4me and DNA methylation have been shown to be inversely correlated (138, 141). The results of recent functional studies raise the possibility that H3K4me needs to be removed by KDM1B lysine-demethylase so that some DNA methylation imprints can be established in germ cells (142). This finding is consistent with previous observations that DNMT3L recognizes histone H3 tails that are unmethylated at lysine 4 and induces de novo DNA methylation by recruitment or activation of DNMT3A (139). In contrast to H4K4me, H3K9me is found to be highly coincident with DNA methylation. Knockdowns of enzymes that catalyze these modifications impact DNA methylation levels at defined loci, and knockdown of DNMT1 results in altered levels of these marks (143–145).
Many specialized sets of nuclear proteins, which are not involved in chromatin modifications per se, are also critical for epigenetic regulation (146, 147). These include chromatin remodeling complexes, which are thought to modify chromatin accessibility by sliding or ejecting nucleosomes, and enhancer-blocking insulator proteins, such as CTCF, are thought to form a chromatin barrier that protects a gene from neighboring transcriptional influences (148). In addition, specialized histone variants, such as H3.3 and H2A.Z, introduce variation into the chromatin template and often carry their own modifications (81, 149, 150).
The establishment of links between external signals and the epigenetic machinery with specific physiological outcomes is an area of increasing importance because of its impact on developmental programming and child health. The roles of histone demethylases in the context of whole body physiology are now being uncovered. The histone demethylase, Jhdm2a, for instance, was recently identified as a crucial regulator of the genes that are involved in energy expenditure and fat storage (151). This finding suggests that Jhdm2a may be a key factor in obesity and metabolic syndrome. Sirtuins also represent exciting new avenues of research on developmental programming. Sirtuins are proteins that possess either HDAC or monoribosyltransferase activity and are found in a variety of organisms that range from bacteria to humans (152). Several sirtuins are class III NAD+-dependent deacetylases with key roles as metabolic sensors and mediators of survival for stressed cells. They regulate chromatin structure and function by targeting histones, in particular H4K16ac, as well as other nonhistone chromatin proteins (152, 153). Importantly, their activity conveys information about the state of cellular metabolism to chromatin as part of the adaptive response to environmental stimuli. The underlying mechanism of the signaling action of glucose, fatty acids, insulin, and other metabolites and hormones to chromatin is a fundamental question at the cellular level and is discussed in several parts of Section VIII. Recently, a new mechanism that links glucose metabolism to chromatin modification and global transcriptional control via the enzyme ATP-citrate lysate and production of acetyl-coenzyme A was proposed (154). Acetyl-coenzyme A was shown to be a nuclear substrate for HATs, thereby providing additional evidence for a glucose-to-gene link (154–156).
D. Epigenetics as a molecular mechanism for developmental origins of disease
There are three intriguing and fascinating facets about the epigenetic state that are important to discuss in the context of developmental programming and child health. First, epigenetic states can be paradoxically both reversible and heritable. Second, epigenetic states can be both heritable across cell divisions in somatic cells and potentially “inherited” across several generations. Third, epigenetic states can be both carriers of “memory” of early-life experiences and “triggers” of disease susceptibility in later life.
1. Heritability and reversibility
As previously discussed, it is well established that epigenetic marks are stably propagated during mitotic divisions and contribute to cell lineage determination and differentiation. Different cell types have their own “unique” epigenotype as a result of distinct epigenetic programs that are faithfully maintained through cellular heritability. The process by which cells acquire epigenetic marks that are important for cell specification is generally referred to as epigenetic programming.
We now know that epigenetic states can be switched from being “stable” to being “flexible. The most striking example of this flexibility is that which occurs during “natural” epigenetic reprogramming, when epigenetic information is erased from the genome during periods of development (see Sections II.E, IV.A, and VIII.F). To the best of our knowledge, only germ cells and early embryos have been shown to be able to “reset” or “reprogram” the epigenetic marks on a genome-wide scale. Therefore, it is reasonable to ask why reprogramming is needed and confined to specific periods and embryonic types. Epigenetic reprogramming in PGCs is important for the erasure of genomic imprints and possibly for the control of transposon silencing. Furthermore, it has the potential of erasing “epimutations” that could otherwise be inherited across generations, and reprogramming of PGCs could also limit the amount of epigenetic information that is passed onto subsequent generations. In general terms, the global resetting of epigenetic marks is thought to achieve developmental potency to allow the return of developmental pluripotency to embryonic cells. It is thus not surprising that the reprogramming that occurs in the early embryo is crucial for erasing the gametic gene expression programs and restoring totipotency to the zygote to form an entire organism. Importantly, epigenetic reprogramming can also occur in “artificial” experimental systems in which differentiated cells are converted into inducible pluripotent stem cells (157), and this reprogramming has important implications for the emerging field of regenerative medicine.
Epigenetic information can pass over to the next generation (158) (see Section II.E). An important example is that of IAP transposon insertions that can alter the expression of the neighboring endogenous genes depending on the methylation status of the IAP. IAP elements seem to resist methylation reprogramming in PGCs and during preimplantation development, thereby potentially enabling the expression state of the associated genes to be inherited across generations (159). Another mechanistic example of “spillover” of epigenetic information is epigenetic asymmetry between parental alleles at imprinted loci. Indeed, some methylated DNA sequences at imprinted genes in mature gametes are protected from demethylation at or after fertilization (Fig. 4) (160). As a result, the transmission of epigenetic information to the subsequent generations is made possible only when epigenetic states are not completely erased during the normal reprogramming in germs cells and the early embryo. Epigenetic inheritance is relatively common in plants, but it is still unclear how widespread it is and whether it has a role in phenotypic variation and evolution in mammals (see Section II.E). Nonetheless, it is fascinating that it is important to erase epigenetic marks between generations, while simultaneously having a need to maintain certain epigenetic marks between generations, such as at certain retrotransposons. Remarkably, there is growing evidence that epigenetic marks may escape erasure between generations, thereby leading to multigenerational influences on inheritance and phenotype (see Section II.E).
2. Defining epigenetic programming
Reprogramming is the most dramatic example of a dynamic epigenetic state. As mentioned before, it refers to the resetting of epigenetic marks to achieve developmental potency. Other more “subtle” examples of epigenetic flexibility are also seen at developmental genes (see Sections II.C and IV.A). Indeed, the genes that are required later in development are transiently held in a repressed state by histone modifications, which are highly dynamic marks and easily reversed when expression of these genes is needed. This flexibility contrasts with long-term repression that is brought about by DNA methylation and associated histone modifications and is observed in genes that are crucial for pluripotency during differentiation and at imprinted genes and transposons.
An exciting topic for future research will be the full characterization of the dynamic epigenome to establish the “flexibility” of the epigenome beyond reprogramming and the developmental program. As previously defined, epigenetic programming is associated with acquisition of marks that are important for cell specification and long-term stability. DNA methylation has long been considered the most stable epigenetic mark and is thus critical for epigenetic programming. However, it was recently reported that certain promoters might be actively methylated and demethylated during transcriptional cycling in differentiated somatic cells (161–163). Therefore, it is tempting to speculate that gene-specific resetting of epigenetic marks (or “gene-specific reprogramming”) may occur in particular cell types, for example, in adult stem cells, and in tissues with a high cellular turnover. It is also important to realize that certain epigenetic marks can be removed before a cell divides or within few cell divisions (or short-term flexibility), whereas others can be maintained for many divisions (long-term stability) (Fig. 5). Short-term flexibility of epigenetic marks is particularly important to allow appropriate responses to acute environmental cues (Fig. 5).
As our knowledge about the flexibility of epigenetic mechanisms increases, epigenetic programming should perhaps be viewed as more than just the acquisition of marks that define cell types and maintain cellular memory. Instead, epigenetic programming should be thought of as being inclusive: the dynamic epigenomic program that operates beyond the early embryo and throughout the lifetime, from the establishment of epigenetic marks in the embryo that specify lineages to the intrinsic responses to environmental factors and aging. Therefore, this definition of epigenetic programming will be used in the various discussions of this review. Inherent in this definition is the concept of epigenetic misprogramming, which refers to abnormal epigenetic programming that can be caused by either intrinsic or extrinsic (environmental) factors.
3. Cellular memory of early-life experiences and disease risk
The DOHaD hypothesis proposes that some disorders, such as T2D and cardiovascular disease, can result from an imbalance between the environments that are experienced in utero, in early infancy, and later in life (Fig. 2). More recently, it has been proposed that the memory of the fetal history and adaptive responses in aging organs and cells may be mediated through epigenetic mechanisms of gene regulation (Fig. 6) (164, 165). The key feature of the “epigenotype model” of DOHaD is largely based on the finding that the environment can modulate epigenetic states. Indeed, there is mounting evidence that links environmental stimuli and the epigenome [reviewed by Jirtle and Skinner (62) and Jaenisch and Bird (166)]. Examples include normal physiological responses to cold exposure in plants, behavioral programming by maternal care in rats (see Section VIII.D), and divergence between monozygotic (MZ) twin pairs and between genetically identical inbred mice (see Sections V and X). Furthermore, the results of several studies have shown that environmental influences in early life can induce permanent alterations in the epigenotype and determine adult phenotypes and disease susceptibility [reviewed by Skinner et al. (167)]. Such embryonic exposures include suboptimal nutrition, glucocorticoids, and EDCs, with transgenerational effects being reported in some of these studies (168–170) (see also Sections VI.B and VIII.C–F).
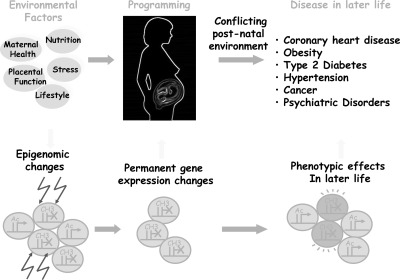
Fig. 6.
The epigenotype model of developmental origins of disease. Environmental factors acting in early life have consequences that become manifest as an altered disease risk in later life. The period of life in which external factors can influence biology extends from conception to the neonatal period and early infancy. It has been suggested that the baby receives from its mother a forecast of the environment it will encounter after birth and modifies its metabolism, whole body physiology, and growth trajectory appropriately to maximize its chances of survival postnatally. However, these adaptations become detrimental if the conditions after birth are not the same as the ones encountered during early life. These adaptations include metabolic and endocrine changes that may lead to lifelong changes in the function and structure of the body—a concept that has been termed programming. The molecular mechanisms by which a phenomenon that occurs in utero has a phenotypic consequence many years later are likely to involve epigenetic mechanisms of gene regulation. Epigenetic marks can be modulated by environmental factors, are heritable, and perpetuate gene-expression changes that underlie programming and may contribute to the onset of disease in later life. Ac, Histone acetylation/active genes; CH3, DNA methylation/silent genes. [Reprinted with permission from I. Sandovici et al.: Epigenetics, Horizon Scientific Press/Caister Academic Press, Norfolk, UK, 2008 (563). © with permission from the publisher]
Epigenetic misprogramming may occur throughout development, during which particular developmental time windows may be associated with specific outcomes. For example, induced changes that affect reprogramming in the germ line may alter the resetting of the normal chromatin state of the affected genes in subsequent generations and result in transgenerational disease (see Section VIII.F). Also, environmentally induced changes that affect programming during early development, and especially during cell differentiation, may have a greater, more widespread effect than those that occur during less “plastic” times in development. A still unanswered question is the identity of those genes that are likely to be involved in the enhanced susceptibility when they are epigenetically deregulated by environmental factors. Many approaches are being followed to identify these genes using genome-wide approaches in MZ and dizygotic (DZ) twins and inbred mouse strains to study the environmental impact on the chromosome machinery without the confounding effect of genetic variation. At this point in time, there are only a few described examples of epigenetic targets of the environment, and they include transposable elements, metastable epialleles, and a small number of developmental transcription factors and imprinted genes (see Sections II.E, III.D, and V). The outcome of the many ongoing epigenome screens is eagerly awaited, in particular those that are trying to look at the unexplored area of ncRNAs and how they might impact on the programming of disease.
E. Heritability of epigenetic changes and the determination of phenotype
The approximate composition of chromosomes is 50% DNA and 50% protein (mainly histones). During the last 50 yr, most research effort has been directed toward investigating the DNA molecule and its nucleotides. In more recent times, the research effort has been directed toward the protein content and function of DNA, and their roles in epigenetic phenomena. The outbred nature of the human species poses challenges for the conduct of research into epigenetic causes of phenotypic variation. One way to overcome this problem is to study MZ twins and look for epigenetic marks that are variable between twins who are discordant for disease (see Section V). These studies are not easy to conduct because of the need to recruit sufficient numbers of MZ twin pairs to enable statistical validity. Other barriers that need to be overcome include ethical review, duration, cost, and, more importantly, the exact interpretation of DNA methylation and other epigenetic marks.
The epigenetic machinery, as described in Section II.B, is mitotically heritable to establish cell type-specific gene expression during development. Once established, epigenetic marks are maintained, in most cases, with high fidelity as cells proliferate throughout life. However, some epigenetic marks can sometimes be meiotically inherited. Although it is obvious that reprogramming of the genome in the sperm and oocyte cells is essential, some chromosomal components, such as telomeres, centromeres, and transposable elements (retrotransposons), probably remain in their original state. Therefore, it is important to know the extent of transgenerational transmission of epigenetic marks. Assuming a 10–15% loss of global DNA methylation in a single life span (171), complete gene repression or “epigenetic collapse” would occur after five to 10 generations if there was transgenerational transmission of all epigenetic marks. Therefore, a repair or restorative system is needed to erase epigenetic marks. In this regard, Teixeira et al. (172) recently reported that genetically induced epigenetic alterations can be transmitted to the next generation in plants but are corrected in successive generations by posttranscriptional gene silencing and RNAi. However, it should not be overlooked that reprogramming could be life-threatening, or even lethal, for the developing organism. Lastly, it is also important to know whether the transmission of epigenetic marks occurs on a genome-wide scale, or whether only stable locus-specific marks are transmitted.
The observed heritability of some epigenetic marks in animals (173) and humans (174, 175) raises the possibility that part of the epigenotype can be transmitted from one generation to the next generation. If this is true, how are epigenetic signatures transmitted to the next generation? One possibility is that the process depends on certain genotypes that directly affect the epigenetic machinery, such as the DMNTs. In this case, the epigenotype of the offspring would resemble that of the progenitors due to the direct action of the epigenotype-associated inherited genotype. The dependence on genotype is supported by the association between methylation at the IGF2/H19 locus with single nucleotide polymorphisms (SNPs) in cis in DZ twins (174). Another possibility is that epigenetic marks can be directly transmitted to the next generation. This idea is supported by the results of the study of Kaminsky et al. (175) in which they found that epigenetic differences between outbred mice were not significantly associated with variation in the DNA sequence. Because MZ twins develop from the same zygote and should possess similar epigenomes at the time of blastocyst splitting, Kaminsky et al. (175) proposed that DZ twins have more epigenetic differences than MZ twins because they originate from different zygotes that carry two different epigenetic profiles. It is possible that genetic and direct transmission of epigenetic marks occurs simultaneously (i.e., some DNA regions are directed by one mechanism and others by a different mechanism). The molecular mechanisms that underlie the heritability of epigenetic marks are still unclear. Indeed, most genomic DNA methylation is erased during embryonic development (62, 176), which implies that other epigenetic mechanisms, in addition to DNA demethylation, must participate in the reprogramming process.
The influence of environmental factors on the epigenetic marking of genes and the heritability of epigenetic marks are of particular interest to childhood growth and development. Irrespective of the method of inheritance of epigenetic phenomena and the problems of assessing epigenetic patterns in humans, there are some epidemiological data that support the concept of transgenerational inheritance of environmental effects. The results from the Dutch famine birth cohort studies stand out because they provide good evidence of this phenomenon (177–181). The infants of pregnant women who experienced famine due to the Dutch Hunger Winter during World War II weighed less than expected when maternal undernutrition occurred during the third trimester. This effect was passed on to the second- and third-born infants in the second generation (F2) from those who were exposed to the famine during the first trimester of pregnancy. The results of these studies also showed that these children were more susceptible to diabetes mellitus, obesity, cardiovascular disease, and other health problems. Until very recently, there was no molecular evidence of epigenetic correlates with these phenomena. Heijmans et al. (182) recently reported that individuals who were prenatally exposed to Dutch famine have, six decades later, less DNA methylation of the imprinted IGF2 gene compared with their unexposed, same-sex siblings. The association was specific for periconceptional exposure, thereby reinforcing the notion that very early mammalian development is a crucial period for establishing and maintaining epigenetic marks. These data are the first to provide evidence that early-life environmental conditions can cause epigenetic changes in humans that persist throughout life.
Using data from the Overkalix and Avon Longitudinal Study of Parents and Children (ALSPAC) cohort, Pembrey et al. (35) supported the existence in humans of sex-specific, male-line transgenerational responses by showing that smoking by the father during his childhood correlated with a higher body mass index (BMI) in his male offspring. Pembrey et al. (35) hypothesized that these transgenerational transmissions were mediated by epigenetic events on the Y-chromosome. In doing so, they added an entirely new dimension to the study of gene-environment interactions in development and health. Such data are challenging to interpret, and other mechanisms or even other explanations are possible.
Some genes can be sensitive to environmental factors because their activity is dependent on their epigenetic state. Such alleles of mammalian genes with such characteristics are termed “metastable epialleles” and are named so as to distinguish them from traditional alleles [for clinical examples, see review by Dolinoy et al. (183)]. These alleles differ from SNPs in that they have epimutations, rather than point mutations. The term “metastable” is used to describe the state of permanency of the change: they are not as stable as point mutations, and they can change more rapidly. These alleles display more plasticity than traditional alleles. At this stage, it is unclear how common these alleles are, but an appreciation of their existence will aid in their identification. DNA methylation often correlates with other regional chromatin features, such as histone acetylation and methylation. Accordingly, DNA methylation is used as an overall indicator of locus-specific epigenetic alterations that regulate gene expression. Most genomic regions undergo developmentally programmed establishment of epigenetic regulation and show little interindividual variability in DNA methylation. Conversely, developmental establishment of DNA methylation at metastable epialleles occurs probabilistically and results in dramatic interindividual differences in epigenetic regulation (184).
Although the epigenetic states, once established, usually last for the lifetime of the individual, some can change during that lifetime depending on genetic, environmental, or stochastic factors (185) (see Section V). Therefore, the epigenetic state is a record of the environmental history of the individual. The epigenetic state is also labile (see Section II.A), and phenotypic mosaicism exists between cells (variegation) and between individuals (variable expressivity). The establishment of the epigenetic state that occurs during early embryogenesis is a probabilistic event that in some cases is influenced by whether the allele is carried on paternal or maternal alleles (see Sections III.C and IV.A and D). In addition, the epigenetic state determines whether these alleles are dominant. Some mammalian genes display variable expressivity in the absence of genetic heterogeneity. A litter of isogenic mice will display variable expressivity and variegation during early development. This variegation occurs because daughter cells remembered the epigenetic state of their founder cell. Assuming that methylation of the gene promoter is the underlying mechanism, one can speculate that methylation of the promoter occurred during the early development of the founder cell and was passed onto subsequent generations of offspring cells through cell division in clonal patches.
Phenotypic variation among genetically identical individuals exists, even when the environmental influences are controlled, and is called “intangible variation” or “developmental noise” in genetic textbooks. Intangible variation results from the stochastic establishment of epigenetic modifications to the DNA nucleotide sequence in early development (54, 186). These modifications, which involve DNA methylation and chromatin remodeling, result in alterations in gene expression that, in turn, affect the phenotype of the organism. Random mutagenesis of the genome can be used to identify genes that are involved in epigenomic programming to understand the mechanisms that underlie establishment of the epigenome and its reprogramming during development. Transgenes that are known to variegate have been used to develop a mutagenesis screen to determine protein expression levels in red blood cells using green fluorescent protein as the reporter and fluorescence-activated cell sorting analysis (187). In this way, dominant and recessive screens can be developed for identifying modifications in metastable epialleles. For example, the PEV of eye color in Drosophila has been extensively used to define genes when investigating epigenetic reprogramming (188).
Using this strategy, Whitelaw and colleagues (187) carried out a “sensitized” N-ethyl-N-nitrosourea mutagenesis screen in mice that were carrying a variegating transgene to identify genes that modify the epigenetic state. They screened 1000 first-generation (F1) mutant offspring for dominant mutations and identified 10 mutants or modifiers of murine metastable epialleles (Mommes). In most cases, they were homozygous lethal, and this finding indicates the obligate requirement for those genes (187). They have since characterized seven of the underlying mutations, of which two are novel (189, 190). The others have been mapped to between 1- and 3-cM intervals. All mutations that have been tested so far affect expression at epigenetically sensitive loci and include the agouti viable yellow allele. Interestingly, the mutations in a number of cases show both paternal and maternal effects; namely, the wild-type offspring from heterozygous mutant parents were different from the wild-type offspring from wild-type parents (191). Heterozygosity for the mutations was associated with mild abnormalities in phenotypes. These results highlight the essential role of epigenetic reprogramming in early development (189). These studies have been extended, and another 12 Mommes have been identified after screening of another 1000 F1 mice. This project has the potential to identify many more novel genes involved in epigenetic phenomena and to produce loss of function (hypomorphs) and gain of function (hypermorphs) versions of known modifiers of the epigenetic state. Mice that are haploinsufficient for such proteins show a range of subtle phenotypes that include obesity and behavioral abnormalities. Viable mouse strains with mutations in epigenetic modifier genes enable investigation into the role of epigenetics in maintaining genome stability. These mutant lines are therefore a valuable resource to study the role of epigenetics in gene/environment interactions. For example, the risk of a poor outcome after a gestational exposure to an environmental pollutant or a nutrient deficiency may be greater for a MommeD heterozygous individual than for a MommeD homozygous individual.
To summarize, we know that epigenetic regulation of gene expression is essential for normal growth, development, and the aging of higher organisms (45) and that epigenetic dysregulation of gene expression is causally linked with various pathologies, such as cancer (192). Regrettably, our current understanding of epigenetic marks and the epigenetic machinery is incomplete. Understanding the code of epigenetic marks and the underlying processes of their writing, reading, and erasure is a “work in progress.” The epigenome is not static, and we are now beginning to appreciate that the epigenome is both stable and labile. As we will now discuss throughout this review, the dynamic and heritable nature of the epigenome is important for understanding the underlying mechanisms of metabolic programming, how programming influences intrauterine and postnatal growth, and the origins of disease in later life.
III. Human Growth and Developmental Programming
A. Plasticity in human growth
Postnatal growth in body weight and stature can be assessed by three measures: growth velocity, attained body size, and the timing or “tempo” of growth, which is a measure of how rapidly an individual achieves its growth potential. Human growth rates differ markedly between individuals, particularly during the most rapid phases of growth, which occur during infancy and adolescence (193, 194).
Human growth demonstrates both “elasticity” and “plasticity” (or long-term programming) during the different growth periods. The concepts of growth elasticity and plasticity arose from the results of studies in experimental animals that date back to the 1960s in which the influence of nutrition on growth was investigated. The results of these studies demonstrated that there are critical time windows in which the outcome of a programmed growth trajectory can be changed. McCance and Widdowson (195, 196) were the first to report this phenomenon when they showed that the exact timing of undernourishment in the growth phase can exert either a permanent or transient effect on final body size. When rats are transiently undernourished (food-restricted) in very early postnatal life, they remained smaller throughout later life than control rats that are not undernourished. In contrast, rats that are transiently undernourished during later growth phases show catch-up growth after the period of undernutrition and attain the same adult weights as the control rats.
Although human growth may be impacted by severe acute or chronic diseases, there is growing awareness that growth rates, and in particular the tempo of growth, may have marked influences on the subsequent risks for morbidity and mortality and, hence, reproductive fitness. Birth weight is strongly correlated with perinatal mortality and is the single strongest predictor of infant survival. Neonates who are born at term and weigh between 1500 and 2500 g (<10th percentile) have a 5- to 30-fold increase in perinatal morbidity and mortality when compared with neonates whose birth weights lie between the 10th and 90th percentiles. The strength of the correlation between birth weight and perinatal mortality depends on gestational age (the lower the birth weight, the higher the rate of neonatal mortality for the estimated gestational age) (197) and also on factors that are unrelated to gestational age. This low birth weight association with neonatal mortality is echoed in adult life with the development of later disease and mortality (15).
In postnatal life, there is growing evidence that the “natural variations” in body size and growth rate may have major relevance, not only on adult height but also more importantly on infant and childhood survival and reproductive fitness (198). Pygmies are an “extreme” example of the interplay between postnatal growth and development, survival, and reproductive fitness. Their characteristic small adult size does not appear to have evolved through any positive selection for short stature, but rather as the result of a life-history trade-off between the fertility benefits of large body size against the costs of late growth cessation in a setting of extremely high childhood and early adult mortality (199).
In Western settings, rapid weight gain during early postnatal life is associated with increased risks for disease. For example, Ong et al. (193) showed that children who showed catch-up growth between birth and 2 yr of age were fatter and had more central fat distribution at 5 yr when compared with children with normal early growth. Ekelund et al. (200) examined the independent associations between weight gain during infancy (0–6 months) or early childhood (3–6 yr) with components of the metabolic syndrome in young adults in a prospective cohort study in 128 individuals from birth to 17 yr. They concluded that rapid weight gain during infancy (0–6 months), but not during early childhood (3–6 yr), predicted the clustered metabolic risk at age 17 yr.
Infant feeding type and feeding patterns can also influence growth trajectories and disease risk. Compared with formula feeding, breast feeding is associated with slower infant weight gain and lower later obesity risk. The results of several meta-analyses suggest that breast feeding has a protective effect, especially in SGA and preterm infants (201). Experimental evidence from several randomized control trials of nasogastric feeding of breast milk and various nutrient formulae for 4 wk showed long-term differences on adiposity levels and the later propensity to cardiovascular disease (202–206). Precocious puberty that is associated with rapid weight gain and growth, particularly during infancy, also has implications for future life events. Ong et al. (207) have shown that an early age of menarche confers increased risk for disease, such as obesity, T2D, and hypertension, and death from cardiovascular disease and cancer in later life (208).
Finally, the mechanisms that signal and regulate early catch-up growth in the postnatal period may mediate or modify the associations between small size at birth and risks for disease in adulthood. The combination of low birth weight and a subsequent high BMI is related to the increased incidence of T2D in later life. Using longitudinal data that were collected from 8760 individuals who were born in Helsinki between 1934 and 1944, Eriksson et al. (209) reported that the large differences in the incidence of T2D were associated with growth rates in utero, weight gain in infancy, and the age at adiposity rebound. These observations have implications for the early origins of both obesity and cardiovascular disease, in that programmable windows of human obesity may exist during the periods of greatest weight velocity. However, current evidence has failed as yet to agree on the specific programmable windows during postnatal growth and development for later disease risks (200, 202, 209).
B. Epigenetic regulation of human growth
If environment can influence long-term growth trajectories in infants and children and their later life outcomes, how do epigenetic changes influence growth at the molecular level? Human growth is highly heritable. The results of twin studies estimate that genetic factors account for 80% of the variations in adult height and approximately 80% of the variations in the timing of the adolescent peak height velocity (210). Traditionally, pediatricians and endocrinologists have had a hormone-centric view of the regulation of growth and development. Growth velocity is regulated by insulin, GH, IGF-I, and, particularly during fetal life, IGF-II. The results of recent studies (211–213) have described the BWS and the Silver-Russell syndrome (SRS) as growth disorders due to defective IGF2 imprinting. The specific molecular defects and consequent prenatal and postnatal growth phenotypes are discussed in detail in Section III.D. Rare mutations in the genes that regulate the GH-IGF-I axis underlie some causes of prenatal and postnatal growth failure and may also retard intrauterine and subsequent growth. These genes include the GH receptor (GHR) gene (214, 215), the STAT5B gene (216, 217), IGF1 (218, 219), and the IGFIR gene (220, 221). Although these growth factors and sex steroids are the downstream factors that drive the growth process, they do not convey messages of when the various growth stages should start or stop, how such timings are influenced by environmental factors, or how variations in growth might be related to long-term disease risks.
Adult height is the summation of the velocity of childhood growth and also the duration of the various growth phases, and it reflects the combined influence of multiple genetic factors. The results of several recent GWAS have begun to reveal the multitude of biological pathways that contribute to the normal variation in adult height. A major surprise has been the absence of genes that are involved in the traditional hormone regulatory pathways (222). Rather, newly identified genetic loci for adult height highlight several targets for let-7 miRNAs, chromatin remodeling proteins, and Hedgehog signaling as important regulators of human stature (222). The expression of let-7 miRNAs is tightly correlated with the onset of adult development in many animals, and this relationship suggests that let-7 miRNAs function as evolutionarily conserved regulators of developmental timing (223) (see also Section II.B.3.a).
Recently, the results of four GWAS identified that the location of common genetic markers for the timing of menarche in girls coincides with the height-related gene, LIN28B (6q21) (106, 224–226), which is a potent and specific regulator of let-7 miRNA processing. This gene is the first common variant to be associated with the timing of human growth and maturation in both boys and girls (106). Dysregulation of mRNA expression by the let-7 miRNAs is considered to be critical for the activity of LIN28B in development; Viswanathan et al. (227) reported that the LIN28B protein product selectively blocks the processing of pri-let-7 miRNAs in embryonic cells by blocking microprocessor-mediated cleavage of pri-let-7 miRNAs. More recent studies have described the roles of let-7 miRNAs and the LIN-28 protein in controlling cell self-renewal and cell differentiation (228). Collectively, these findings indicate a remarkable parallel in the molecular regulation of both cellular differentiation and whole organism maturation. Further investigations are now needed to explore how LIN28B and let-7 miRNAs might signal the effects of early-life environmental exposures on subsequent height trajectory, the tempo of growth and timing of puberty, and long-term disease risks (229).
C. Intrauterine growth and imprinted genes
Human growth is a complex event that requires the programmed contribution and interaction of many operators. Key participants in the growth pathways are regulated epigenetically to a certain extent, and correct genomic imprinting is essential for mammalian ontogenesis. Somatic maintenance of imprints throughout development is a highly complex process that involves not only the allelic DNA methylation at imprinting control regions (ICRs), but also covalent histone modifications and nonhistone proteins (230–233) (see Sections II.B and III.A).
Errors in the mechanisms for resetting and maintaining genomic imprints lead to imprinting defects with or without nucleotide sequence abnormalities (234). In humans, dysregulation of imprinting mechanisms has been linked to fetal and postnatal growth, neurological development, and behavior (21, 235). BWS (OMIM 130650) and SRS (OMIM 180860) are two human growth disorders that exhibit opposite phenotypes. BWS is characterized by pre- and postnatal overgrowth, macroglossia, abnormal wall defects, hemihyperplasia, and an increased risk of childhood tumors (236, 237). SRS, in contrast, first described by Silver et al. (238) and Russell (239), is characterized by pre- and postnatal growth retardation, a prominent forehead, relative macrocephaly, body asymmetry with hemihypoplasia, feeding difficulties, and a BMI less than −2 sd score (213).
Both growth disorders are often caused by abnormalities in DNA methylation at the 11p15 region, which encompasses many imprinted genes that encode key growth regulators, such as the IGF2 gene. BWS has been reported to result from loss of methylation (LOM) at the centromeric KCNQ1OT1 region on the maternal allele or gain of methylation (GOM) at the telomeric IGF2/H19 region on the maternal allele (236). Conversely, LOM at the telomeric IGF2/H19 domain on the paternal allele has been demonstrated in SRS (211, 213, 240–243) (Fig. 7). Aberrant imprinting at different loci and uniparental disomies (the loss of one parental allele and duplication of the opposite parental allele origin) are also the cause of several other diseases. For example, aberrant LOM at imprinted loci on chromosomes 6q24 and 11p15.5 has been described in transient neonatal diabetes mellitus (244–248).
Fig. 7.
The two imprinted domains of the 11p15 chromosomal region are under the control of two ICRs. The reciprocal imprinting of the maternally (mat) expressed H19 and the paternally (pat) expressed IGF2 depends on an ICR1 located upstream from the H19 gene that acts as an insulator. The repressor factor CTCF (CCCTC-binding factor) binds to the unmethylated maternal copy of the ICR and prevents the IGF2 gene promoter from interacting with enhancers downstream from the H19 gene. This results in transcriptional silencing of the maternal IGF2 allele. On the paternal allele, the ICR is methylated, and CTCF binding is prevented. This leads to IGF2 transcription on the paternal allele and silencing of the H19 gene. The centromeric KCNQ1 domain produces a noncoding RNA (antisense KCNQ1OT1 RNA) that silences many of the genes in this domain. Paternally expressed genes are represented as white boxes, maternally expressed genes as black boxes, and nonexpressed genes as gray boxes. BWS is associated with a variety of genetic and epigenetic defects within the imprinted 11p15 region. Most patients (70%) exhibit an epigenetic defect. Ten percent of BWS patients display an imprinting defect at the IGF2-H19 domain (aberrant GOM at the maternal copy of the ICR), which results in silencing of the maternal H19 gene and a biallelic expression of the IGF2 gene. The majority of the BWS patients exhibit a LOM at the ICR of the KCNQ1 domain. Loss of methylation at this ICR results in activation of the normally silent maternal allele of KCNQ1OT1 and CDKN1C silencing. In SRS, the mirror phenotype of BWS, a loss of imprinting at the IGF2–H19 domain was identified: the paternal allele switches to a maternal epigenotype, and this results in biallelic expression of H19 and loss of IGF2 expression. Genetic and environmental factors could induce these epigenetic anomalies.
The recognition that disrupted imprinting underlies these syndromes comes from the results of studies on Igf2-knockout mice (65, 249–251). In humans, BWS and SRS are caused by abnormal DNA methylation at the 11p15 region that encompasses many imprinted genes, including the IGF2 gene (212, 252). As already mentioned, LOM at the ICR of the KCNQ1 domain and GOM at the ICR of the neighboring IGF2/H19 domain on the maternal allele occur in individuals with BWS. The latter epigenetic defect is associated with a high risk of cancer in such individuals (236, 240, 253–257). Epigenetic changes in the 11p15 region also occur in individuals with SRS, and this syndrome could be perceived as a molecular mirror of BWS (211–213). In these SRS patients, LOM at the IGF2/H19 ICR occurs on the paternal allele. These human 11p15 imprinting anomalies (LOM and GOM) probably occur in the postfertilization period because of the presence of mosaic patterns of imprinting abnormalities and because such imprinting defects are seen exclusively in the affected twin within MZ twin pairs who are discordant for these syndromes (211, 236).
D. Postnatal developmental epigenomics
Realization that developmental plasticity extends into the postnatal period led Waterland and Garza (164) in 1995 to propose “metabolic imprinting” as the biological phenomenon that putatively underlies the associations between nutritional experiences in early life and later diseases. The term is intended to encompass those adaptive responses to early-life nutritional challenges that are characterized by: 1) a susceptibility that is limited to a critical ontogenic window early in development; 2) a persistent effect that lasts through adulthood; 3) a specific and measurable outcome (that may differ quantitatively among individuals); and 4) a dose-response or threshold relation between a specific exposure and outcome.
The mouse agouti gene encodes a paracrine signaling molecule that promotes follicular melanocytes to produce the yellow pheomelanin pigment rather than black eumelanin pigment. As a result, the hair coat in these mice is yellow. There are several agouti gene mutations, such as lethal yellow and viable yellow, in which the agouti gene is deregulated and expressed ectopically. In addition to causing a yellow coat, deregulation of the agouti gene has pleiotropic effects that include adult-onset obesity, increased tumor susceptibility, and premature infertility. Waterland et al. (258, 259) have shown that food supplementation with a methyl donor to female mice before and during pregnancy permanently increases tissue-specific DNA methylation at the agouti viable yellow (Avy) and axin-fused (AxinFu) alleles in the offspring (the latter controls dorsal-ventral axis development through the Wnt signaling pathway, which describes a network of proteins that are involved in embryogenesis and cancer). This is an example of an epigenetic effect within one generation because Waterland and his colleagues did not find evidence of a memory of methyl donor supplementation across generations. Accordingly, they concluded that stochastic establishment of epigenotype at metastable epialleles is, in general, labile to methyl donor nutrition, and such influences are not limited to early embryonic development. Such findings in mice may have direct relevance to the wide variation of the human phenotype (see also Sections VIII.C and F and IX.A).
Extensive data indicate that epigenetic dysregulation can contribute to obesity (260). To test the hypothesis that maternal obesity induces transgenerational amplification of obesity, Waterland et al. (261) passed the Avy allele through obese Avy/a females for three generations in two separate, but contemporaneous populations of mice. One population was fed a standard rat diet, and the second population was fed a methyl-supplemented diet during development to assess the cumulative effects on coat color and body weight. They reported that the genetic tendency for obesity in Avy mice was progressively exacerbated when the Avy allele was passed through successive generations of obese Avy/a female mice, and this transgenerational amplification of body weight was prevented by a promethylation dietary supplement. Importantly, the effect of methyl supplementation on body weight was independent of epigenetic changes at the Avy locus.
From these data, Waterland and Michels (55) proposed two general mechanisms to explain the early postnatal environmental influence on the developmental establishment of DNA methylation: the supply of dietary methyl donors and/or activity of DNA methyltransferases can induce either hyper- or hypomethylation at metastable epialleles and alterations in transcriptional activity of specific genes during ontogenic periods when DNA methylation is being established (Fig. 8).
Fig. 8.
Potential mechanisms for environmental influences on developmental establishment of DNA methylation. A, Nutritional or other stimuli that affect either the efficiency of one-carbon metabolism or the activity of DNMT1 could alter the developmental establishment of DNA methylation at metastable epialleles. Flux through the transmethylation/remethylation pathway is dependent upon nutrients including folate, vitamins B12 and B6, choline, betaine, and methionine. B, Transcriptional activity during critical developmental periods can impair de novo methylation. Any nutritional or other environmental exposure that activates gene transcription during periods of de novo CpG methylation can permanently imprint transcriptional competence by preventing hypermethylation. Methylated CpG sites are shown as “filled lollipops.” Although a gene promoter region is shown here, similar effects could occur at any genomic region contributing to transcriptional regulation, such as a distal enhancer. 5CH3THF, 5-Methyl tetrahydrofolate; SAH, S-adenosylhomocysteine; DMG, dimethyl glycine. [Reprinted with permission from R. A. Waterland and K. B. Michels: Annu Rev Nutr 27:363–388, 2007 (55). © Annual Reviews.]
To summarize, we are becoming increasingly aware that epigenetic changes have important relevance to growth. However, we do not yet know how these processes interact with the hormonal axes or growth plates to regulate the timing and extent of growth. We need to understand which specific types of epigenetic changes influence growth and the timing of postnatal development, as well as the precise genetic loci in each tissue that these epigenetic changes affect. With this kind of information, one can begin to explore whether nutritional or other exposures create or alter epigenetic modifications during key periods of growth and thereby exert their long-term programming effects. Some of these concepts are discussed in Section VIII.
IV. Genomic Imprinting, X-Inactivation, and Childhood Disease
A. Epigenetic mechanisms in mammalian genomic imprinting
Monoallelic gene expression is achieved by genomic imprinting and X-inactivation, both of which are mechanisms that depend on DNA methylation and histone modifications. Genomic imprinting refers to the differential expression where either the maternal or the paternal copy of a gene is expressed (monoallelic expression or functional hemizygosity) (65, 262). In genomic imprinting, the activity of a gene is reversibly modified, depending on the sex of the parent that transmits it, and leads to unequal expression of the maternal and paternal alleles in the offspring (263). It is a form of non-Mendelian inheritance and is believed to have evolved in mammals to regulate, in part, the dosage of developmentally important genes. Genomic imprints are established upon passage of the genome through either the female or the male germ line. They are fully acquired in sperm (paternal imprints) and in mature oocytes (maternal imprints). After fertilization, these parental imprints are maintained throughout development in all somatic cells and tissues. The allelic expression of imprinted genes that are mediated by the parental imprints can be cell type- or tissue-specific (66). In the developing PGCs of the embryo, imprints are erased so that new imprints can be established at a later developmental stage according to the sex of the embryo (52). The expression of imprinted genes is made additionally complex when, exceptionally, erasure and resetting of the imprint are not entirely complete in a single generation (234).
As mentioned earlier, more than 100 imprinted autosomal genes have been identified to date in mammals, and many of these participate in the regulation of growth and cellular proliferation, whereas others influence behavior (52, 53). Imprinting is important in mammals because imprinted genes affect intrauterine and postnatal growth and behavior, such as IGF2 (11p15) (242). There are two critical time periods in epigenetic reprogramming: gametogenesis and early preimplantation development. Early embryonic maintenance is particularly critical because this process is sensitive to environmental factors. Major reprogramming takes place in PGCs to erase parental imprints and restore totipotency. Imprint marks are then reestablished later during spermatogenesis or oogenesis. Upon fertilization, genome-wide demethylation occurs and is followed by a wave of de novo methylation, both of which are resisted by imprinted loci (Figs. 4, 8, and 9).
Fig. 9.
Parental imprints are established during oogenesis or spermatogenesis at sequence elements that control the imprinted expression (the ICRs). After fertilization of the egg by the sperm, these imprints are maintained throughout development. DNA methylation (lollipops) is the most consistent hallmark of imprints. Two examples of ICRs are depicted: one with paternally derived (ICR1) and one with maternally derived (ICR2) DNA methylation. [Reprinted from K. Delaval and R. Feil: Curr Opin Genet Dev 14:188–195, 2004 (52), with permission from Elsevier.]
Almost all known imprinted genes are clustered in large chromosomal domains whose organization is similar in humans and mice. The parental allele-specific gene expression at these domains is mediated by ICRs, and the parental imprints are established at these regions (Fig. 9). The precise nature of these parental imprints is not yet fully understood, but DNA methylation is a hallmark of genomic imprinting at all ICRs. DNA methylation is not the only epigenetic modification found at ICRs (52). Results from chromatin studies show that the chromatin is compacted by repressive histone modifications on the DNA-methylated allele. On the opposite parental allele, where there is no DNA methylation, there are histone modifications, which are typical for an open chromatin structure. In this way, differential DNA methylation and the associated chromatin features at ICRs convey the allelic expression of imprinted genes at imprinted gene clusters (52, 235, 252).
There are at least three ways through which differential DNA methylation at ICRs can result in the silencing of one of the two parental alleles at close-by genes (264). The simplest way is through direct silencing of a promoter, where the methylated allele is nontranscribed. One of the best-studied imprinted genes is the IGF2 gene on human chromosome 11p15.5. IGF2 is expressed from the paternally inherited allele during fetal development and after birth (52, 252). The allelic repression of this gene is regulated by an ICR that is located upstream of the H19 gene and 90 kb from the IGF2 gene. This CpG-rich regulatory region is marked by DNA methylation on its paternally inherited copy only. This paternal methylation is acquired during spermatogenesis and is maintained during development in all the somatic lineages. The mechanism of imprinting at the H19 ICR involves formation of an insulator on the unmethylated allele, which prevents the IGF2 promoter from interacting with downstream enhancers and consequently prevents IGF2 expression from the maternal chromosome (265, 266). Another well-characterized mechanism of imprinting is DNA methylation-dependent repression of a lncRNA transcript on one of the parental alleles. Consequently, the ncRNA transcript is expressed only from the unmethylated allele of the ICR. At some imprinted domains, the ncRNA recruits PcG proteins, such as EZH2 and EED, and histone methyltransferases to the locus. Enrichment of repressive histone modifications to the region consequently silences the surrounding genes. This mechanism is thought to be involved in the imprinting of both the KCNQ1 and IGF2R domains (267, 268). Imprinted expression at these domains is tissue-specific, and several of the genes are imprinted in the placenta only (269). The ncRNA-mediated recruitment of PcG proteins is important for this placenta-specific imprinting (270), which also requires the histone methyltransferase G9a (also called KMT1C) (120, 271). Not all genes of the KCNQ1 domain are imprinted in the human placenta. Monk et al. (270) reported that imprinting in the placenta at the IGF2R domain in humans is polymorphic, thereby potentially increasing the susceptibility of IGF2R locus to the effects of epigenetic perturbation in some pregnancies. Another example is the growth factor receptor-binding protein 10 (GRB10), which is a potent growth inhibitor (272). In the mouse, the Grb10 gene displays a complex tissue-specific imprinting pattern that is controlled by different promoters. Some tissues show expression from the maternal chromosome, and others show expression from the paternal chromosome (273, 274). Whereas Grb10 is expressed from the maternal allele in most tissues, its expression in the brain is only from the paternal allele, and its maternal and paternal expressions are initiated from different promoter regions (275). In addition, Sanz et al. (275) have shown that the tissue differences in the imprinted expression of this gene in mice and humans are due not to the acquisition of an imprint mark, but rather to differences in the reading of this mark. Specifically, this ICR is methylated on the maternally inherited allele in both humans and mice. Whereas this maternal imprint in the mouse conveys maternal allele-specific expression in many mesodermal and endodermal tissues that include the placenta, maternal expression in humans is detected in the villous trophoblast of the placenta only. However, in both species, the ICR conveys paternal allele-specific expression in the brain (276).
B. Assisted reproductive technologies and genomic imprinting
As described in the previous section, the somatic maintenance of imprints throughout development is a highly complex process that not only involves the allelic DNA methylation at ICRs but is also associated with covalent histone modifications and nonhistone proteins (231–233, 275). Different types of environmental stress, particularly those that result from transferring cells and embryos from their natural environment to in vitro conditions, can interfere with the somatic maintenance of imprints. Assisted reproductive technology (ART) is a stressor of special interest because in vitro culture of embryonic cells and embryos can perturb the imprints at ICRs in different model systems, thereby affecting fetal growth and development (235, 277–284). Such findings have implications in humans because of the association between ART and increased incidence of BWS (231, 281, 285–287), SRS (288), and Angelman syndrome, which is a severe neurodevelopmental condition characterized by microcephalus, absence of speech, severe mental retardation, and frequent laughing (279, 289, 290).
In humans, it has been suggested that ART may favor imprinting alterations at the imprinted KCNQ1 domain [LOM at the maternally methylated ICR (281, 291) and the IGF2/H19 ICR (211, 213, 288, 292)] in SRS. ART has also been associated with imprinting anomalies at multiple other loci (288, 293, 294). Therefore, it is presumed that ART interferes with the acquisition or maintenance of maternal methylation marks and results in unfaithful maintenance of DNA methylation marks after fertilization due to dysregulation of trans-acting regulatory factors. It remains to be discovered what precisely gives rise to the altered DNA methylation patterns at ICRs (the “epimutations”), but ART could possibly affect proteins that recruit the DNA methylation machinery to the ICRs during each cell cycle. Because not all cells and tissues are affected to the same extent, the mosaic distribution of these human epimutations suggests that imprinting is perturbed after fertilization due to a failure to maintain the differential methylation marks during preimplantation development (281, 293).
In sheep and cattle, epigenetic changes have been shown to be involved in large offspring syndrome (295). Affected animals exhibit various phenotypes that include large size at birth. In both species, the syndrome is caused by the in vitro exposure of embryos to various unnatural environments between fertilization and the blastocyst stage. Large offspring syndrome is often related to the loss of imprinting of the Igf2-receptor gene which ensures internalization and degradation of IGF2 and exerts an antiproliferative function (296). In vitro preimplantation procedures in mice are also responsible for fetal overgrowth due to the abnormal expression of various imprinted genes, and in particular those genes that are located at distal chromosome 7 (H19 and Igf2 genes), which is orthologous to the human 11p15 region.
In the mouse, Kcnq1 domain paternal repression is found at several genes in the placenta only. Interestingly, the allelic repression does not involve acquisition of DNA methylation at the promoters of these genes. Rather, the paternal gene silencing in the placenta involves repressive H3K9me and H3K27me (297, 298). This repressive histone methylation becomes established on the paternal chromosome already during early development, a process that requires a ncRNA, which is expressed from the ICR on the paternal allele. This ncRNA recruits PCR2, which methylates histone H3 tails at lysine 27 (297). Recently, it was reported that repressive H3K9Me at the KCNQ1 domain and some other imprinted domains is controlled by the histone methyltransferase G9a (Kmt1c) (271). Also, this histone methyltransferase is recruited to the chromatin by the same ncRNA (120).
Finally, based on the association between ART and the increased incidence of BWS, Miles et al. (299) hypothesized that subtle differences, which manifest as differences in the phenotype and hormonal profiles in midchildhood, may also exist in the previously underinvestigated in vitro fertilization (IVF) population. After analyzing data from 69 IVF and 71 naturally conceived control children, they reported that the prepubertal IVF children were taller when adjusted for parents' heights, had higher serum IGF-II and IGF-I levels, had altered ratios between serum IGF-II and IGF-binding protein 3 levels, and had a different lipid profile when compared with those in the naturally conceived children. Based on these preliminary findings, they speculated that these differences in stature, serum growth factors, and lipid metabolism may be due to subtle epigenetic alteration of imprinted genes and/or other genes that are involved in growth and development. This hypothesis remains to be tested. Although ART in humans is associated with exposure to multiple environmental factors, no specific aspect of the procedures has been implicated as a unique cause of the alterations in genomic imprinting and the risk of congenital abnormalities. This inability to identify a unique cause of the alterations in genomic imprinting is quite amazing, despite the birth of approximatley 4 million IVF children worldwide and the accumulation of substantial data from birth registries. Future studies should aim to identify the causal factor(s) and emphasize the need to monitor the health of IVF children through childhood into adulthood.
Important insights into the epigenetic changes that can occur with ART have come from somatic cell nuclear transfer (cloning) experiments in mice and domestic animals. Here, zygotes are cultured to the blastocyst stage after introducing a nucleus of a somatic cell into an enucleated oocyte and are then implanted into the uterus. This in vitro culturing step frequently leads to altered DNA methylation at ICRs and hence, perturbed imprinted gene expression and phenotype when fetal calf serum is added to the culture medium (278, 285, 300). In cloning experiments in experimental animals, the uterine environment is also an important factor that influences the development of the cloned embryo until birth [reviewed in Loi et al. (301)]. Importantly, results from experiments in cattle and mice show that phenotypic alterations due to cloning become corrected in the naturally conceived offspring of cloned animals (302). This finding suggests that altered DNA methylation patterns become largely corrected upon transmission through the germ line and do not affect the phenotype across multiple generations (303).
C. X-inactivation, imprinted genes, and the Turner syndrome
In humans and other eutherians, sex is determined by the X- and Y-chromosomes, which evolved from an ordinary pair of autosomes by the acquisition of a sex-determining gene or mutation on one of the two autosomes. Over time, the sex chromosomes diverged more and more from each other by accumulating sex-linked mutations and genes, and the only chromosomal part that is still shared between the two sex chromosomes is the pseudoautosomal region. This chromosomal region is where crossover in meiosis can occur between the different sex chromosomes and recombination of the sex-specific region outside the pseudoautosomal region is suppressed (304).
The lack of recombination between the X- and Y-chromosomes led to the loss and differentiation of genes on the Y-chromosome and a single copy of most X-linked genes in males (305, 306). One outcome of the evolution of genetic sex determination and distinct sex chromosomes was an imbalance of gene dosage between autosomes and sex chromosomes and between males and females. To protect the organism against the deleterious effects of X-chromosome monosomy, two mechanisms of dosage compensation evolved: X-chromosome up-regulation, which equalizes the gene dosage between the X-chromosome and the autosomes; and X-chromosome inactivation, which silences one X-chromosome in females to equalize the gene dosage between the sexes (305, 306).
The term “X-inactivation” is used to describe the initial transition from a transcriptionally active to an inactive state and also the subsequent stable maintenance of the silent state (307). In female mammals, most genes in one X-chromosome are silenced as a result of random X-chromosome. In X-inactivation of extraembryonic tissues in the mouse, the paternal X-chromosome (Xp) is transcriptionally silenced, and the maternal X-chromosome (Xm) is transcriptionally active (306–308).
X-inactivation is a remarkable example of epigenetic inheritance in which the silencing of more than 1000 genes occurs by packaging DNA into transcriptionally inactive chromatin through a process that is able to distinguish between one of an essentially identical pair of chromosomes. The process of X-inactivation displays plasticity: the Xi is reactivated during oogenesis, thereby permitting inactivation in the next generation. One of the differences between Xi and the active X-chromosome (Xa) and autosomes in differentiated cells is the unique combination of epigenetic features of Xi that include histone modifications, DNA methylation, late replication timing, and a peripheral nuclear location.
Critical for the inactivation process is the X-inactivation center (XIC), which is a multifunctional domain on the X-chromosome to be inactivated, and its crucial component, the Xist gene (308). The Xist gene is crucial for both imprinted X-inactivation from either of the parental chromosome and random X-inactivation and gene silencing in eutherians because it encodes a nontranslated RNA that coats Xi (see Section II.B.3.b). The mechanism of choosing which X-chromosome will remain active and which will be inactivated is complex and is regulated by multiple elements in the XIC. Although Xist is able to initiate inactivation, it does not appear to be sufficient for recapitulating the entire X-inactivation process. The elements that are involved in the number of X-chromosomes relative to autosomes (count) and the selection of the diploid set that will remain active (choice) lie outside the Xist gene (309). Although the mechanism of count is currently unclear, Morey et al. (309) proposed that H3K4me within Xist may be functionally implicated in the counting process (see Section II.B.3.b).
Monoallelic expression of most of genes on the X-chromosome in females is determined mostly by random X-inactivation and partly by imprinted X-inactivation. X-chromosome inactivation involves features that are common to autosomal imprinted genes. However, it is poorly understood how Xist RNA and other epigenetic modifications are directed to sites along the inactive X-chromosome and how inactivation spreads in cis over the silenced X-chromosome (310). One critical unresolved question on the mechanism of imprinted X-inactivation is the nature and origin of the imprinted gene(s). Is gene imprinting established exclusively in the maternal or the paternal germ line, or are the two different maternal and paternal gene imprints necessary? At this stage, there are no exact answers to these questions, but the results of studies in mice suggest the existence of an epigenetic switch at the XIC, which underlies the molecular aspects of chromosome-wide silencing (306). In addition, the inheritance of imprinted epigenetic marks on Xp and the imprinting of Xist on Xm appear to be linked to ensure faithful imprinted X-inactivation (306).
The epigenetic mechanisms of X-inactivation are now beginning to be understood. Reik and Lewis (119) recently proposed that the processes of X-inactivation and genomic imprinting are mechanistically similar and are thought to have evolved together when the evolution of the placenta exerted selective pressure to imprint growth-related genes. These authors also proposed that ncRNAs and histone modifications were adopted for the imprinting of growth suppressors on the X-chromosome and autosomes. The initiation of X-inactivation is thought to be tightly correlated with early differentiation events during development (311); Xp undergoes imprinted inactivation from the cleavage stages onward, well before cellular differentiation. In another study, Silva et al. (312) have shown that the recruitment of PRCs to the Xi occurs in both imprinted and random X-inactivation, which occurs in the embryo proper. Localization of these repressive complexes to Xi occurs very early, at the onset of Xist expression, but becomes less pronounced as differentiation and development progress. In addition, the PcG complex is required to establish H3K27me on Xi, which in turn is required to stabilize the Xi chromatin structure. Lastly, Patrat et al. (313) recently reported their findings from a systematic, single-cell transcriptional analysis that they performed to examine the activity of the Xp for a panel of X-linked genes throughout early preimplantation development in the mouse. From the results of this analysis, they concluded that imprinted X-inactivation in mice is far less concerted than previously thought and highlighted the epigenetic diversity that underlies the dosage compensation process during early mammalian development.
Between 15 and 25% of female X-linked genes escape X-inactivation, and these genes are potential contributors to sexually dimorphic traits, phenotypic variability among females heterozygous for X-linked conditions, and clinical abnormalities in patients with abnormal X-chromosomes (310). There is a remarkable degree of expression heterogeneity linked to the X-chromosome (310). This heterogeneity has been attributed to: 1) the inactivation “escape” of about 15% of X-linked genes; 2) the dramatic differences in the proportion of escape genes between different regions of the X-chromosome; and 3) variable patterns of inactivation and expression of 10% of X-linked genes from some inactive X-chromosomes.
Turner syndrome (TS) is thought to be caused by haploinsufficiency of the escape genes and arises from a complete and/or partial monosomy of the X-chromosome as a result of loss of either part or all of a second X- or Y-chromosome (314). Between 60 and 80% of TS individuals have an intact Xm (45,Xm) (315–317). Because TS can manifest in nonmosaic or mosaic forms with or without the presence of a normal 46,XX karyotype, or occasionally the 46,XY karyotype, its phenotypic spectrum is broad, and it encompasses both physical and neurocognitive features (318). Evidence exists for X-linked parent-of-origin effects in TS individuals because phenotypic and cognitive profiles differ between 45,Xm and 45,Xp individuals. X-Imprinting effects in TS are found for cognitive function and social cognition, statural growth, visceral adiposity, and lipid metabolism (263, 317–323).
Individuals with TS have been used to investigate the impact of putative X-linked imprinted genes on growth and neurocognitive development. Whereas implementation and conduct of these studies in humans are difficult, genetically engineered 39,XO mice have now been developed to study X-chromosome allele-specific expression (Xa vs. Xi): the Xp mouse (324), and the Xm mouse (325–328). Using such mice, Raefski and O'Neill (329) identified a cluster of X-linked genes that contains at least three genes that show transcriptional repression of paternal alleles. They also established that the imprinting of these three genes was independent of X-chromosome inactivation and has a dynamic and complex pattern of tissue and stage specificity. In addition to these genetically engineered mice, some nonanimal methods have been developed to study various aspects of X-chromosome allele-specific expression. Fibroblasts from women, where about 15% of X-linked genes escape inactivation, have been used to study polymorphisms in genes of interest (310); somatic mouse-human cell hybrids that contain either Xa or Xi have been used to study transcription of genes of interest, which are normalized to a known gene that always escapes inactivation (pseudoautosomal gene) (310); and human ESCs have been used to study the early lethality of 45,X embryos (330).
Both human and murine data support the notion that the TS phenotype could be modulated by imprinted loci, particularly with respect to growth and neurocognition. Nevertheless, conflicting data on the basis of TS in human studies still exist due to unaccounted confounding variables, different test measures, small sample sizes, and statistical bias. Therefore, studies on TS in humans should focus on quantifiable variables and on the underlying physiological and genetic mechanisms. These studies would complement future studies in the genetically engineered mice and cell systems whose aims could be to investigate the underlying and diverse epigenetic mechanisms that are associated with X-inactivation and the variability of the TS phenotype.
V. The Role of Epigenetics in Aging
Understanding the links between epigenetics, the DOHaD phenomenon, and age-related diseases has emerged as an exciting research topic because epigenetic factors are now known to mediate, at least in part, the relationship between the genome and the environment. Focusing specifically on the relationship between epigenetics and aging, an active role for epigenetics in aging must meet two prior conditions: there must be specific epigenetic changes, and the epigenetic changes must be functionally associated with the aging phenotype. One of the theories of aging claims that aging is the progressive decay of the potential of adult stem cells to maintain correct tissular homeostasis (331, 332). The variation in the life span of a species seems to be more strongly affected by the accumulation of molecular errors over time that compromise adult stem cell function than by specific genetic programs (332, 333). These molecular alterations can occur at both the genetic and epigenetic levels, and age-dependent accumulation of epigenetic marks depends on the genotype (intrinsic factors), the environment (extrinsic factors), and stochastic factors (185). Young adult stem cells are pluripotent and consequently participate in tissue regeneration. As these cells grow older, their genome is marked epigenetically, and this marking may be accompanied by a loss in their ability to participate in tissue regeneration. Genotypes with a low efficiency for repairing genetic and/or epigenetic defects or maintaining epigenetic stability in response to harmful environmental exposures can accelerate the accumulation of molecular alterations at the genetic and epigenetic levels, which in turn can accelerate the aging process. In contrast, genotypes with a high resilience to genetic and/or epigenetic defects or maintaining epigenetic stability in response to harmful environmental exposures can slow the accumulation of molecular alterations at the genetic and epigenetic levels, which in turn can delay the aging process (185).
Epigenetic variation, as illustrated by genomic methylation patterns, is dynamic because it changes over time and during the aging process. Time-associated epigenetic variation was first observed more than 40 yr ago by Berdyshev et al. (334), who reported that spawning humpbacked salmon showed a global decrease in 5-methyldeoxycytidine levels with age. In a more detailed follow-up study, Vanyushin et al. (335) reported a global loss of m5C in DNA in the brain, heart, and spleen; no m5C changes in the liver and lungs; and modest m5C increases in the kidneys of rats with aging. Based on these results, Vanyushin et al. (335) proposed that DNA methylation may be one of the regulatory mechanisms of gene activity and the observed changes in m5C could be responsible for the process of aging. These early findings were confirmed by Wilson et al. (336), who reported gradual loss of DNA methylation with aging in different mouse tissues and human bronchial epithelial cells. Wilson et al. (336, 337) also provided convincing evidence that the reduction in DNA methylation was unrelated to the proliferation rate of the cells and could not be ascribed to the dilution effect of cell division. Age-dependent global hypomethylation has since been demonstrated in humans and other mammals (338, 339). In addition, specific loci are known to become hypermethylated during aging in mammals. Examples include hypermethylated clusters of ribosomal DNA in the liver and germ cells of old rats (340) and hypermethylated CpG islands in the promoters of tumor suppressor genes, lysyl oxidase, p16INK4a, runt-related transcription factor, and tumor promotor TPA-inducible gene 1 in various human tissues (171). Intriguingly, global DNA hypomethylation and aberrant promoter hypermethylation also occur in cancer (192, 341). Such findings lend support to the notion of age-related loss of normal epigenetic patterns as a mechanism for the late onset of many human diseases (342).
The relative importance of genetic and nongenetic components in aging can be estimated from the results of twin studies (174, 175, 339, 343). The underlying rationale of twin studies is that MZ twins are identical genetically, whereas DZ twins on average share 50% of their segregating genes and are as genetically different or similar as ordinary siblings (344). Despite being genetically identical, MZ twin pairs vary in a wide range of anthropomorphic features and also in their susceptibility to disease (344). The cause of phenotypic discordance in MZ twins has been traditionally attributed to unique exposure to postnatal environmental factors of each sibling, namely the nonshared environment. Nevertheless, there is increasing evidence that postzygotic genetic, epigenetic, and prenatal environmental factors may contribute to the phenotypic discordance in MZ twins (344). In fact, MZ twins display numerous epigenetic differences, and in some cases, these differences are associated with specific behavioral and physical features (344). Recently, Fraga et al. (343) analyzed the global epigenetic differences in different-aged MZ twins and showed that elderly MZ twin pairs, who lived apart from their own families, exhibited numerous phenotypic differences. Moreover, these elderly MZ twin pairs have more epigenetic differences than young and phenotypically similar MZ twin pairs who lived in the same household with their parents. In agreement with other reports (345–347), Fraga et al. (343) also found that MZ twins have significantly different gene-expression phenotypes, although most of the epigenetic changes occurred in nonfunctional and repetitive DNA elements. Overall, the results from these different-aged MZ twin studies suggest that intraindividual epigenetic changes do occur over time.
This notion has been recently corroborated by the results of a longitudinal study in which successive DNA samples were collected more than 10 yr apart in two populations, each with more than 100 individuals (339). In this study, Bjornsson et al. (339) measured global DNA methylation in two samples that were collected, on average, 11 yr apart from 111 individuals of an Icelandic cohort and, on average, 16 yr apart from 126 individuals of a Utah cohort. They reported that the change in DNA methylation over time was greater than 10% in 29% of the Icelandic individuals. The family-based Utah sample also displayed similar intraindividual changes in DNA methylation over time, as well as familial clustering of the methylation change. In addition, families that showed the greatest global DNA methylation loss also have the greatest loss of gene-specific methylation. From these results, the authors concluded that changes in DNA methylation occur over time and proposed that the maintenance of DNA methylation may be under genetic control. The results of other studies provide further support for the existence of epigenetic differences between twin pairs. For example, DZ twins have more differences in genome-wide (175) and locus-specific (174) DNA methylation than MZ twins. Collectively, these findings suggest that the maintenance of epigenetic marks with aging is genetically regulated.
Environmental exposures affect time-associated epigenetic variation [reviewed by Feinberg (45)]. Using smoking as an example, Belinsky et al. (348) determined the prevalence of aberrant promoter methylation of the p16, the O(6)-methylguanine DNA methyltransferase, the death-associated protein (DAP) kinase, and the Ras effector homolog (RASSFIA) genes in nonmalignant bronchial epithelial cells from current and former smokers in a hospital-based, case control study of lung cancer. They also determined the relationship between loss of heterozygosity at 9p and p16 methylation in bronchial epithelium and the prevalence for methylation of these four genes in sputum from cancer-free, current, and former smokers. From their results, Belinsky et al. (348) concluded that aberrant promoter hypermethylation of the p16 gene, and to a lesser extent the DAP kinase gene, frequently occurs in the bronchial epithelium of lung cancer patients and cancer-free controls and persists after smoking cessation. Moreover, the strong association between methylation of the p16 gene in the bronchial epithelium and the corresponding primary tumor led them to suggest that inactivation of the p16 gene, although itself is not transforming, is likely to be permissive for the acquisition of additional genetic and epigenetic changes that lead to lung cancer.
Genetic or environmental effects cannot explain all the epigenetic changes, such as the differences in DNA methylation, that have been reported in isogenic animals that live under the same environmental conditions (349). Isogenic laboratory animals that are maintained under identical environmental conditions also exhibit marked phenotypic differences (350), of which life span is one (351). Such phenotypic variability is thought to be due to stochastic factors, which are independent of the environment and can contribute to random biological variability (350).
Hereditary, environmental, and stochastic factors determine the accumulation of epigenetic variation over time, but their relative contribution to the phenotypic outcome is unclear because little data are available. For example, Ronn et al. (352) reported that age influences the extent of DNA methylation and the expression of OXPHOS genes, a set of genes that influence oxidative phosphorylation in muscles. However, they were unable to determine whether the age-associated changes in DNA methylation were due to hereditary, environmental, and/or stochastic factors.
The finding of concordance of some psychological aptitudes between MZ twins that were reared either apart or together (353) suggests that stochastic events are more important than hereditary or environmental factors if these aptitudes depend on epigenetic factors. In contrast, results from studies of large cohorts of MZ twins that are discordant for cancer, which is one of the best known epigenetic-dependent diseases (354), have shown that environmental factors have an important role in the etiology of this disease (355). It is possible that the influence of one factor, be it environmental, genetic, or stochastic, is different in different genomic regions, such as the coding elements and repeated sequences. Consistent with this notion, most of the environment-related changes in phenotypic expression between MZ twins preferentially occur in heterochromatic, gene-poor regions (346, 347), which are the regions where most epigenetic differences are found in environmentally dependent, phenotypically discordant MZ twins (343). In contrast, the IGF2/H19 locus, whose epigenetic variation depends primarily on genetic factors, is resistant to age-related changes in DNA methylation (174).
The functional role of epigenetic alterations that occur over time depends on the genomic region that is affected by these changes. Although genome-wide (175, 343) approaches suggest that epigenetic differences between MZ twins occur frequently outside the functional coding elements, the discordant expression phenotypes of MZ twins (347) and the relationship between environmentally dependent epigenetic marks and cancer (45) suggest that epigenetic differences can have significant functional implications. In this regard, Mill et al. (356) assessed the methylation status of two CpG sites in the promoter region of the COMT gene in 12 MZ twin-pairs who were discordant for birth weight but were clinically normal. They found that the extent of DNA methylation at the two CpG sites was highly correlated, but there was considerable variation in the concordance of methylation levels between MZ twin-pairs, which explains the incomplete phenotypic concordance. Differences in DNA methylation have also been found in MZ twins discordant for the caudal duplication syndrome, which is a rare family of developmental defects in which structures derived from the embryogenic cloaca and notochord are variously duplicated at the AXIN1 gene, which has been implicated in this syndrome (357). In addition, differential methylation of the X-chromosome has been proposed as a possible source of discordance among female MZ twin-pairs with a bipolar disorder (358).
In conclusion, epigenetic states can change over time, and this epigenetic variation depends on hereditary, environmental, and stochastic factors (Fig. 10). Future studies are now needed to: 1) quantify the contributions of each component to epigenetic variation over time; 2) determine the molecular mechanism involved in the transmission of epigenetic patterns between generations; and 3) assess their functional role and the DNA regions in which they occur. The application of the new technologies of ultra-deep sequencing to large cohorts of accurately phenotypically annotated MZ and DZ twins should generate enough epigenome-wide information to gain insights into the functional relevance of the epigenetic changes that occur during aging and to determine the contributions of the genetic, environmental, and stochastic factors to their establishment.
Fig. 10.
A model of how genetic and epigenetic factors can affect aging. Young adult stem cells present no alterations in either the genetic or epigenetic levels, and so there is proper stem cell function and, consequently, tissue regeneration. Genotypes of low efficiency in repairing genetic or epigenetic (represented as lollipops over the structure of the DNA) defects or in maintaining epigenetic stability accompanied by harmful environmental exposures can accelerate the accumulation of molecular alterations at the genetic and the epigenetic levels, which in turn can accelerate the aging process. On the other hand, genotypes that are highly efficient in repairing genetic and epigenetic defects and in maintaining epigenetic stability accompanied by harmless environmental exposures can slow the accumulation of molecular alterations at the genetic and epigenetic levels, which, in turn, can delay the aging process. [Reprinted from F. M. Fraga: Curr Opin Immunol 21:446–453, 2009 (185). © with permission from Elsevier.]
VI. Tissue-Specific Epigenetic Changes
There is now much evidence that tissue-specific epigenetic patterns exist across chromosomal regions. These patterns are conserved across individuals because patterns in the same tissue from different donors are strongly correlated. Indeed, DNA methylation profiles of the same tissue correlate better across individuals than those of different tissues from the same individual (359). Interestingly, the DNA methylation profiles across various regions of the brain are strongly correlated. This strong correlation suggests that the shared methylation pattern of these tissues was established in a common precursor cell type, and that the functions of these cells are sufficiently similar to be reflected in a similar pattern of epigenetic modifications.
Over the past 30 yr, research has established that type 1 diabetes mellitus (T1D) is a family of disorders in which glucose homeostasis is disrupted due to loss of tolerance to β-cell autoantigens. As a result, progressive and selective destruction of insulin-secreting β-cells of the islets of Langerhans occurs by a multigenic process in which numerous immune and β-cell defects associate to drive the diabetogenic process. This process has been extensively studied in the nonobese diabetic mouse, a spontaneous model of T1D. Findings from the nonobese diabetic mouse and other genetically engineered T1D mice have been instrumental in understanding the complexity of this disease and deciphering its autoimmune basis. From these studies, proinsulin, glutamic acid decarboxylase, phogrin (1A-2β), IA-2, carboxypeptidase E, and many other β-cell autoantigens have been identified as targets of autoimmunity in T1D. The results of other studies have established that the thymus also contributes to the development of T1D autoimmunity (360, 361).
Recently, Concannon et al. (362) reviewed the genetic basis for T1D, whose complex polygenic etiology has been gradually teased out. The combination of heritability estimates from family studies and the specific results of linkage studies and GWAS clearly reveals that T1D susceptibility has a major genetic basis. A recent GWAS meta-analysis identified at least 40 loci that are associated with the risk of T1D in humans (363). The concordance rates for T1D in MZ twin pairs are around 30–35%, and such figures have been used to simply allocate the contributions of genetic vs. environmental factors (362, 364). However, disease onset can occur at different ages in each of the twins and sometimes with a 30- to 40-yr interval (365). This long period before disease onset in MZ twins was also found by Redondo et al. (366), who investigated the apparent discordance of T1D in MZ twins. They reported that 65% of the discordant unaffected twins do eventually develop the disease. Epigenetic mechanisms could, therefore, be relevant in mediating the effects of environment on disease risk and also on the timing of disease onset.
Nontraditional, apparently epigenetic inheritance does occur in T1D, but its basis is still unknown. Akesson et al. (367) recently suggested that not only the inherited haplotypes, but also the noninherited haplotypes, may influence the risk of the disease. They investigated the risk of T1D in 563 children with the disease and 286 nondiabetic children according to the human leukocyte antigen (HLA) haplotypes, which can be classified as noninherited from either the maternal side (NIMA) or paternal side (NIPA). They found no difference in the frequency of the positively associated haplotypes between the NIMA and NIPA individuals. They also reported that NIMA, but not NIPA, was associated with the risk of T1D.
The notion that maternal microchimerism (maternal cells in the circulation and tissues of her offspring) might affect growth and development, or may contribute to disease or tissue repair in the progeny and persist into adult life in healthy subjects, arises from observations in children with severe combined immunodeficiency. Maternal microchimerism as a mechanism of nontraditional inheritance of T1D has been investigated by Nelson et al. (368). To identify and quantify maternal microchimerism, they first developed a panel of quantitative PCR assays that targeted nontransmitted, nonshared maternal-specific HLA alleles. They then assayed maternal microchimerism levels, which were expressed as the genome equivalent per 100,000 tested cells, in the peripheral blood from 172 individuals, of which 94 had T1D, 54 were unaffected siblings, and 24 were unrelated healthy subjects. Maternal microchimerism levels were significantly higher in the T1D individuals than those in the unaffected siblings and healthy subjects. The differences between the groups were evident, irrespective of HLA genotypes. However, for individuals with the T1D-associated haplotype, maternal microchimerism was found more often when the haplotype was transmitted paternally (70%), when compared with that found when transmitted maternally (14%). From these results, they concluded that maternal microchimerism may contribute to islet β-cell autoimmunity in a mother's progeny because T1D individuals have higher levels of maternal microchimerism in their circulation than unaffected siblings and healthy individuals.
Given the variable heritability of T1D, investigation has now shifted to environmental causes to explain the heterogeneous phenotype of the disease. Viruses (congenital rubella, Coxsackie B, mumps, echovirus, cytomegalovirus, Epstein-Barr virus, retrovirus, rotavirus, parvovirus B19), bacteria in the gut microbiota (369), diet [cow's milk; decreased vitamin C, D, and E intake; early introduction of cereals (370), potatoes, carrots, fruit, berries, N-nitroso compounds, and increased caloric intake], and psychosocial factors have all been implicated as environmental causes (371).
Miao et al. (372) examined histone methylation patterns in blood cells from T1D individuals and found significant increases in H3K9me2 patterns in a subset of genes in lymphocytes, but not in monocytes. They also found increased H3K9me2 in the promoter of one of the candidate T1D susceptibility genes, CLTA4, and two high-scoring networks of genes. Many genes in the two high-scoring networks have been previously identified as known T1D candidate genes and are associated with several autoimmune and inflammation-related processes and molecules, such as TGF-β, nuclear factor-κB (NF-κB), p38 MAPK, Toll-like receptors, and IL-6 (372). In another study, El-Osta et al. (373) reported transient hyperglycemia-induced long-lasting epigenetic changes in the promoter of the NF-κB subunit p65 in aortic endothelial cells, both in vitro and in nondiabetic mice. Furthermore, El-Osta et al. (373) reported that the epigenetic and gene expression changes persisted for at least 6 d of subsequent normal glycemia, as did the NF-κB-induced increases in the expressions of monocyte chemoattractant protein 1 and vascular cell adhesion molecule 1. The results of these two studies show that even hyperglycemia has epigenetic consequences.
One known epigenetic mechanism in transient neonatal diabetes, a rare form of nonpermanent diabetes mellitus in newborns, is LOM at the TNDM locus on chromosome 6q24. Loss of this epigenetic mark in the mesodermal lineage leads to the prune belly sequence, which is a syndrome of abdominal muscle hypoplasia, urinary tract abnormalities, and cryptorchidism (244). Laborie et al. (374) investigated a family with transient neonatal diabetes and prune belly sequence that included one set of MZ twins. The twin with both transient neonatal diabetes and prune belly sequence had extensive LOM at the TNDM locus, as well as at the IGF2R, DIRAS3, and PEG1 loci, whereas the healthy MZ twin and other family members had normal methylation. Therefore, the LOM at the loci that are associated with both transient neonatal diabetes and prune belly sequence may indicate a generalized maternal hypomethylation syndrome.
A potential for inducing epigenetic modifications in cell therapy for T1D has been raised by the finding that endocrine pancreatic cell lineages can be prompted to become endocrine cells by treatment with HDAC inhibitors (375, 376). This approach as a treatment for T1D clearly merits further investigation, and epigenetic drug therapy is discussed in Section IX.A of this review.
VII. Sexual Dimorphism of Gene Expression and Epigenetics
Many tissues exhibit sexual dimorphism for a substantial proportion of the genes that they express (377, 378). Sexual dimorphism has been explained traditionally by the regulatory pathways that underlie sexual development of the gonads, brain, and other organs, and the impact of lifelong fluctuations in the circulating level of sex hormones. Sensitivity to specific environmental challenges for each sex also exists during gametogenesis and developmental programming and throughout the individual's life (378). Because environmental factors can influence epigenetic marking during particular spatiotemporal windows of life in a sex-related manner, it is therefore not surprising that the sexes differ in their sensitivity to environmental challenges throughout an individual's life. There are many examples of sex differences on the effects of prenatal and early postnatal life exposures and the risks of subsequent metabolic disease (378–384). These sex differences could be attributed to the properties of the sex chromosomes, the different regulatory pathways that underlie the sexual development of most organs, and the lifelong fluctuating impact of sex hormones. In fact, sex-specific gene expression appears to be under the control of sex-specific epigenetic marks. For example, modifications of histone H3 are sexually dimorphic in the developing mouse brain, and patterns of acetylation, but not methylation, are masculinized in females by testosterone in utero (385).
A. Sex chromosomes and the hormonal basis of sexual dimorphism
Mammalian sex determination is initiated by the presence or absence of the testis-determining SRY gene on the Y-chromosome and is expressed only in Sertoli cells in a very narrow spatiotemporal window, namely between the sixth and seventh week of gestation. This male factor induces the differentiation of testes and the secretion of those hormones that are responsible for male secondary sexual differentiation (386). This does not mean that female development occurs by default; the results of recent studies suggest the existence of both X and Y sex-chromosomal mechanisms of sex determination (387). In addition, sex-determining genes on the sex chromosomes are thought to contribute to the development of nongonadal organs in secondary sexual development and to the development of organs beyond the reproductive system, such as the brain (387).
All male cells possess a single X-chromosome of maternal origin and a Y-chromosome of paternal origin. Female cells comprise two X-chromosomes, one of which is silenced by X-inactivation. This can be either the Xm or the Xp, thereby defining two populations of cells in females. In about half of the female cells, Xm is inactivated, whereas Xp is inactivated in the other female cells. Overall, gene expression in a given female tissue is approximately the sum of the gene expression profiles of these two cell populations. Several classes of genes may be expressed in a sexually dimorphic manner, depending on their origin and location on the X- and Y-chromosomes. Y-chromosome-encoded genes are expressed solely in males, and those X-chromosome-encoded genes that escape (or partially escape) X-inactivation will be more highly expressed in females. In addition, some genes on the X-chromosome can be imprinted in such a manner that either the maternal or the paternal copy is expressed. This expression is independent from the random X-inactivation mechanism that controls the allelic repression of most X-linked genes. This can generate differences in expression levels between males and females. For instance, X-linked imprinted genes that are expressed from Xp will be expressed solely in females because males have only an Xm.
It has been proposed that the dimorphism between male and female fetuses could start before the formation of the gonads (387). Recently, it was found that the response of male and female cells to chemical exposure to either ethanol or camptothecin was sexually dimorphic. These sexually dimorphic responses apparently occurred at fetal stages that preceded the production of sex hormones and could therefore be directly attributed to a sex chromosome effect (388). Thus, cells can differ according to sex, irrespective of their history of exposure to sex hormones. From the results of this experiment, one can conclude that sex chromosomes are crucial for establishing sexual dimorphism.
B. Sexual dimorphism of gene expression in the liver
Gene expression in somatic cells and tissues can be influenced by external factors, such as the extracellular hormonal milieu. A good example of hormonal regulation is the effect of GH on gene expression in the liver, which leads to sex differences in many metabolic processes, such as steroid and fatty acid metabolism, cholesterol homeostasis, and drug metabolism (389). Important sex differences also characterize responses to various hepatic stresses in both rodent models and humans. For example, alcohol-induced liver fibrosis is more prevalent in women than in men, whereas sepsis- and hepatitis virus-induced liver fibrosis, hepatic ischemia/reperfusion injury, and hepatocellular carcinoma are more prevalent in men than in women; and these sex differences are, at least, in part due to hormonal factors (389, 390).
Liver sex differences are best studied for the major hepatic steroid- and drug-metabolizing cytochrome P450 enzymes (CYPs), where male-female differences in the levels of individual CYP mRNAs range from about 2-fold to as high as 1000-fold in both rats and mice (389). Sex differences in the expression of CYPs and other enzymes of steroid and drug metabolism are much smaller in the human liver, but nevertheless can have substantial impact on physiological and pathophysiological processes (389). In rats, liver sex differences emerge at puberty when the onset of a pulsatile pattern of pituitary GH secretion first appears in males and a near-continuous pattern of GH secretion emerges in females (391–393). These sex differences in plasma GH patterns are programmed by the action of gonadal hormones on the hypothalamus during the early neonatal period; they are first seen at puberty, continue into adulthood, and then decline with senescence (394). The sex-dependent patterns of pituitary GH secretion, in turn, dictate the sex differences in the expression of many liver-expressed genes (395, 396). Plasma GH patterns are also gonadal hormone-regulated and sex-dependent in humans (397, 398). These plasma GH patterns regulate the sex differences in liver gene expression at the level of initiation of gene transcription (399, 400).
The results from mouse knockout studies have identified two nuclear factors that are required for the observed sex differences in liver gene expression: signal transducer and activator of transcription 5b (STAT5b), and hepatocyte nuclear factor (HNF) 4α (401, 402). STAT5b protein is rapidly activated by each incoming plasma GH pulse in the adult male rat liver; this in turn gives rise to a pulsatile pattern of nuclear STAT5b activity that parallels the pulsatile pattern of plasma GH stimulation. In contrast, STAT5b activity in the adult female liver is maintained at a low, but persistent, level by the more continuous plasma GH pattern (403–405). Thus, although there are no sex differences in liver STAT5b mRNA or protein levels, liver STAT5b DNA-binding activity shows major sex differences. These findings led to the proposal by Waxman et al. (403) that STAT5b maintains male liver gene expression through its stimulatory effects. As revealed by microarray analysis of STAT5b-deficient mouse liver, these stimulatory effects were shown to impact approximately 90% of the genes in the mouse liver that show male-predominant expression (389). On the other hand, STAT5b exerts its inhibitory effects on approximately 60% of the genes that show female-predominant expression in the mouse liver (389). Indeed, STAT5b deficiency in male mouse liver primarily, but not exclusively, affects those genes that show sex differences in expression in wild-type mice. Further support for this model is provided by the results of recent studies that have shown that STAT5 protein (primarily STAT5b) binds dynamically to its chromatin binding sites in the liver, cycling on and off with each plasma GH pulse, with marked sex differences in STAT5 DNA binding occurring at low-affinity, but not at high-affinity, STAT5 sites (406).
A second nuclear factor, HNF4α, is a liver-enriched transcription factor that is required for hepatic expression of many of the same sex-dependent genes that are targets of GH and STAT5b. However, it is not precisely clear how HNF4α contributes to sexual dimorphism of the liver insofar as it also regulates many liver-expressed genes that do not show sex differences in their expression. One possible mechanism is suggested by the discovery that HNF4α and GH-activated STAT5b are both required for the expression of several other transcription factors, whose expression shows sex differences in mouse and rat liver (407) and might, in turn, control the expression of downstream sex-dependent target genes via a GH-regulated transcriptional regulatory network (407).
Support for the involvement of chromatin features in the regulation of genes that show sex differences in the liver comes from the discovery of short genomic regions that show sex-dependent and GH-regulated differences in chromatin accessibility (“hypersensitivity sites”) in liver tissue, as probed using the enzyme DNA nuclease (DNase) I. Thus, increased hypersensitivity to DNase I cleavage in the male liver tissue, compared with that of female liver tissue, is seen in the promoter regions of two male-specific genes, C4a/Slp, sex-limited protein, and Cyp2c11, which catalyzes testosterone hydroxylation (408, 409). Correspondingly, female-specific DNase I hypersensitivity sites have been identified adjacent to the female-specific Cyp2c12, a steroid sulfate hydroxylase (410) (Fig. 11). DNase hypersensitive chromosomal regions, such as these, have increased access to transcription factors and other DNA-binding proteins and include promoters, enhancers, silencers, and insulators. These findings of sex differences in DNase hypersensitivity are indicative of a sex-specific liver chromatin organization, which is presumably established and/or maintained by the sexually dimorphic patterns of pituitary GH secretion that emerge at puberty and through their downstream signaling, which leads directly to the sex-dependent patterns of nuclear STAT5b activity (403–405). Further studies are now needed to determine whether these GH- and sex-regulated differences in chromatin accessibility are a general characteristic of sex-specific genes in mammalian liver and how these sex differences in the epigenome are functionally linked to the sexual dimorphism of liver gene expression. Differential factors that regulate liver GH responsiveness are also indicated by the finding that intrinsic sex differences in early GH pulse responsiveness characterize 45 individual genes that show male-predominant expression; these genes are rapidly induced by GH (within 30–90 min) in the livers of hypophysectomized male mice, but not in the livers of hypophysectomized female mice (411). The persistence of these sex differences in GH responsiveness several weeks after hypophysectomy could be the result of epigenetic programming, which could be mediated by hormonal exposure at an earlier point in time, i.e., before hypophysectomy.
Fig. 11.
Epigenetic regulation of sex-specific CYPs. Female-specific Cyp genes are proposed to be repressed in the male liver, and male-specific Cyp genes are proposed to be repressed in the female liver by packaging in heterochromatin. Continuous GH is proposed to activate female-specific genes, such as Cyp3a genes, by a mechanism that involves the local conversion of heterochromatin to euchromatin, which enables the binding of transcription factors (TF) that activate CYP gene expression. This process could involve the loss of DNA CpG methylation and/or loss of chromatin marks that are associated with repressed chromatin, such as histone H3 lysine 27 trimethylation, which is typically found in genes in a compact chromatin structure and is associated with a stable, inactive heterochromatic state. [Reprinted from D. J. Waxman and M. G. Holloway: Mol Pharmacol 76:215–228, 2009 (389), with permission from the publisher.]
VIII. Phenotypic Plasticity and Environmental Programming
A. Epigenetics and environmental programming
As noted earlier, there is now a large body of evidence indicating that epigenetic events mediate developmental plasticity and that chromatin modifications may be transmitted transgenerationally to influence the development and behavior of subsequent generations, especially when they are acquired during development and transitions between life-history phases. Indeed, the initial data that gave support to the DOHaD phenomenon were gathered by experimental replication (412, 413). Only a few environmental influences have been shown to cause DNA sequence changes that could explain altered gene expression or increases in disease frequency in a particular region (414). Evidence is also accumulating that different environments are able to alter gene expression and change the phenotype by modifying the epigenome [see review by Gluckman et al. (8)]. Such findings add further support for the likely role of epigenetic mechanisms in developmental plasticity. Moreover, when environmentally induced epigenetic adaptations occur at crucial stages of life, they can potentially change behavior, disease susceptibility, and survival (62). For example, Barber et al. (415) showed that radiation could induce transgenerational germ-line instability in mice that persisted for at least two generations. This finding raises important issues of risk evaluation in humans and highlights the existence of an alternate pathway for disease etiology that does not involve a change in the DNA sequence.
Jirtle and Skinner (62) proposed that three kinds of genomic targets are susceptible to gene-expression changes owing to environmental perturbations of epigenetic marks: the promoter regions of certain housekeeping genes, transposable elements that lie adjacent to genes with metastable epialleles, and regulatory elements of imprinted genes. Both DNA methylation and histone modifications are markedly altered in the promoter regions of tumor suppressor genes and oncogenes in human cancer. The importance of epigenetics in the etiology of cancer is not within the scope of this review but has been discussed extensively elsewhere [for example, see Esteller (192)]. The second genomic target is genes with metastable epialleles, which are loci that can be epigenetically modified in a variable and reversible manner such that a distribution of phenotypes occurs from genetically identical cells (see Sections II.B and IV.D). As discussed in Section IV.A, the allelic expression of the third genomic target, imprinted genes in the present generation, may depend on the parental environment in which these genes resided in the previous generation.
It is therefore not surprising that the influence of environmental factors on the epigenetic marking of genes and the heritability of epigenetic marks are of particular interest to childhood growth and development. The resultant alterations in gene expression could have consequences for cellular function and health of the individual throughout the life span. Diet, hormones, and social and lifestyle factors (all of which will be discussed in this section) have all been shown to influence the epigenotype and may exert profound effects on many aspects of child and adult health and disease susceptibility in later life (62, 167, 416). In animals, chromatin transitions at genes have been linked to: 1) circadian, sleep-wake, and rest-activity rhythms; 2) the hunger-satiety cycles; and 3) the major components of energy homeostasis and thermogenesis (383). Because chromatin modifications are both dynamic and labile (see Section II.B), epigenetic modulation or modification of gene expression is possible because epigenetic plasticity extends beyond birth. For example, maternal nutrition can influence gene expression of the growing fetus in utero and in the developing progeny after its birth (170, 382, 417, 418), and DNA methylation can be prenatally manipulated by hormonal stimuli (168). Diet also can affect the gene expression by altering the extent of DNA methylation in gene promoters and histone acetylation in the chromatin structure (286, 419). Epigenetic marks that accumulate during aging can lead to dysregulation of gene expression, which are important in tumorigenesis and the onset of age-related diseases (420–422). Maternal behavior has long-term effects on the methylation of the glucocorticoid receptor (GR) gene in the hippocampus of the offspring (423). Lastly, EDCs can exert transgenerational effects (TGEs) on gene expression and DNA methylation patterns in the progeny that can last for as long as four generations (62, 168, 424, 425) and are discussed in Section VIII.F. The flexibility of epigenetic marks makes it possible for all these above-mentioned influences to alter existing DNA methylation patterns, create new histone marks, or modify the chromatin structure during a particular spatiotemporal window, sometimes in a sex-specific manner (377, 426).
B. Environmental factors and epigenetic processes
A wide range of environmental factors and compounds may influence long-term disease risks, despite only transient exposures at specific earlier periods of development [reviewed by Gluckman and Hanson (24)]. Epigenetic modifications have been proposed as a plausible link between the alterations in gene expression, environment, and disease phenotypes and susceptibilities (62). Epigenetic mechanisms result in stable regulation of gene expression without alterations to the DNA sequence and trigger initiation and/or maintenance of cell-specific transcriptional profiles. Indeed, the precise control of transcription is achieved by modulating the chromatin structure and three-dimensional organization of the nuclear architecture and genome (see Section II). Because epigenetic programming and reprogramming can lead to stable changes in lineage specification, deviation of cell-type determination by either amplifying or decreasing the number of specific cell subtypes in early life can lead to disease and/or changes in disease susceptibility in adulthood (375), and this process can be potentially manipulated or even reversed by appropriate epigenetic drugs (427) (see Section IX.A).
Abnormal maternal behavior, inadequate maternal feeding, and exposure to deleterious environmental compounds during critical periods of life (periconception and fetal and infant development) can change developmental trajectories. Some epigenetic marks may originate from a previous generational experience and increase disease susceptibility in the offspring (62). Epigenetic changes that failed to be erased in the germ line or early embryo may be transmitted to the next generation in a sex-specific manner and exert a TGE (186, 428–433) (Fig. 12). The results of early studies on TGEs assumed that they were the result of the epigenetic malprogramming of somatic processes. However, paternal or maternal germ line epigenetic inheritance could also account for the TGEs (168, 184, 434). Moreover, both somatic and germ line effects may be sexually dimorphic and can affect both mitochondrial and nuclear DNA through the maternal line (435) (Fig. 12). Therefore, the phenotype of an individual is the result of lifelong remodeling of the epigenome due to a complex interaction between the genotype and the ancestral and current environments.
Fig. 12.
Sexual dimorphism in the modes of transmission and its effects on the offspring in successive generations. The sex specificity of these effects operates at three different levels: 1) the maternal transmission during pregnancy and postnatal periods; 2) the sex of the parent who transmits the consequences of a stimulus exposure via the germline; and 3) the sex of the offspring who displays the maternal effect or paternal and/or maternal germline TGEs. [Reprinted from A. Gabory et al.: Mol Cell Endocrinol 304:8–18, 2009 (378), with permission from Elsevier.]
There are at least three signaling pathways that can transduce signals from the extracellular environment to the epigenetic machinery (378, 436). These include the traditional membrane receptor signaling cascade, ligand activation of nuclear receptors (NRs) by small lipophilic molecules, and metabolic activators and inhibitors of the epigenetic machinery. The consequences of stimulating each of these pathways can lead to altered tissue-, stage-, sex-, and age-specific epigenetic landscapes (Fig. 13). Chemical and nonchemical environmental factors, such as drugs, food, toxins, social cues, and cultural factors can have specific impacts on the epigenetic machinery that depend on their access to chromatin (Fig. 13).
Fig. 13.
The three signaling pathways transduce environmental signals from the cell membrane to the chromatin structure in epigenetic programming of the genome: 1) activation or inhibition of the chromatin epigenetic machinery by metabolites of these substrates; 2) activation of nuclear receptors by ligands; and 3) traditional membrane receptor signaling cascades. [Reprinted from A. Gabory et al.: Mol Cell Endocrinol 304:8–18, 2009 (378), with permission from Elsevier.]
In the first signaling pathway, specific environmental factors, the aging process, and the actions of sex hormones may influence the chromatin modifying enzymes (437, 438). After their passive or active entry across the cell membrane, exogenous and/or endogenous substrates undergo cell-specific metabolism. Folates and methionine are precursors for the biosynthesis of S-adenosyl methionine (SAM), which is the principal methyl donor for DNA and histone methylation. Thus, agents that modulate one-carbon metabolism or directly affect the intracellular and/or nuclear levels of SAM can affect epigenetic programming (439). Such agents, including some bioactive constituents in foods, such as sulforaphane (SFN) in broccoli, and beverages, such as resveratrol in red wine, or drugs, such as valproic acid and trichostatin A, are HDAC inhibitors (see Section IX.A). Surprisingly, some drugs have been shown to cause DNA demethylation, even in the presence of the DNA methylation inhibitor 5-azaC, thus illustrating the complex relationship between histone modifications and DNA methylation processes (440) (discussed in Section II). Thus, endogenous or exogenous compounds may lead to the alteration of a critical balance of the chromatin remodeling enzymes, not only for specific sets of dysregulated genes, but also at the whole genome level.
In the second pathway, some compounds specifically bind to NRs, which provide direct links between signaling molecules, epigenetic remodeling, and transcriptional response. Their action involves several mechanisms (441). NRs may be present in the nucleus or cytoplasm where they bind to their ligand. When this binding occurs in the cytoplasm, they may undergo several modifications before being translocated to the nucleus where they bind to their specific response elements (REs). Some environmental compounds, such as EDCs, may bind to estrogen and testosterone receptors to cause the same (or slightly different) effect as the natural ligand (see Section VIII.F). Other NRs, such as peroxisome proliferator-activated receptors (PPARs) and RXRs (retinoid X receptors), are already dimerized in the nucleus on their RE at the promoter of target genes. Their binding to a complex of corepressors and HDACs prevents transcription of these genes in the absence of PPAR or RXR ligands. Upon binding with their natural polyunsaturated fatty acid ligands or drugs such as fibrates, allosteric rearrangement leads to the recruitment of coactivators and chromatin remodeling factors to form a transcription-prone chromatin complex that activates or inhibits chromatin-modifying enzymes. Appropriate modifications of the epigenetic marks at PPAR or RXR RE in target gene promoters modulate the expression of genes in a tissue-specific manner depending on the presence of the appropriate cofactor(s) (442).
The third pathway comprises traditional membrane receptor signaling cascades (443, 444). The basic idea, which was proposed by Szyf (440) and co-workers, is that behavioral exposures stimulate signaling pathways in the brain, which in turn activates sequence-specific factors that recruit or direct HATs to specific targets that enhance the probability of DNA demethylation. Such a mechanism provides a conduit through which both social and behavioral experiences, as well as endogenous and environmental chemicals, could affect the epigenome, and thus gene expression and function. It is possible, depending on the type of ligand or spatiotemporal conditions, that different pathways could be activated. The maintenance of DNA methylation patterns is dependent on the preservation of the balance of several factors, such as DNMTs, DNA demethylases, HATs, HDACs, histone methyltransferases, and histone demethylases. Extra- or intracellular signaling pathways could trigger the activation or suppression of one or some of these enzymes, which, for example, could change loci-specific histone acetylation and tilt the balance toward DNA demethylation and a change in the expression state of a particular gene.
C. Nutrition, epigenetics, and programming in early life
The placenta evolved in eutherian mammals to provide nutrients to the developing fetus, and fetal growth and survival depend on its integrity (445). To fulfill this physiological role as a nutrient sensor and supplier, the placenta follows a carefully orchestrated developmental program during gestation.
Many of the detrimental events that occur in the fetus are “secondary” phenomena due to a wide range of causes, which include preeclampsia and other hypertensive disorders, abnormal development of the placenta and its vasculature, and viral infections (50). IUGR is a term that is used to describe slow fetal growth and has many causes [see Section I.A and reviews by Gluckman and others (19, 20, 22)]. IUGR is closely linked to placental development and function, and undernutrition during pregnancy reduces birth weight and programs the adult phenotype for growth and body composition, with consequences for morbidity and life expectancy (446, 447) (see also Sections I.A and II). In fact, placental insufficiency and an abnormal uterine environment are two of the known environmental factors that predispose the developing fetus to epigenetic misprogramming, which in turn increases susceptibility of the offspring to disease in later life (16, 382, 433, 448–450) (see Section II.D).
Several concepts are now emerging to explain the consequences of IUGR, and these concepts were recently summarized by Borowicz and Reynolds (446). The first concept, which will be discussed in this section, is the idea that the placenta can be epigenetically programmed in response to a maternal stressor, such as maternal nutrient restriction, and that epigenetic programming in the placenta can lead in turn to altered nutrient transport to the fetus, and hence to fetal growth restriction. The second concept is the idea that a compensatory increase in placental function, such as an increase in nutrient transport, occurs in response to undernutrition during pregnancy. The third concept is the idea that the placental response to maternal stress is variable and complex because the response depends on the type of stressor, such as the maternal age and the maternal environmental stress. The fourth concept is that altered placental angiogenesis, which can include a compensatory increase in vascularity in some cases, is also an important component of placental programming. Of these four concepts, the first concept, epigenetic programming in the placenta, will be discussed in the next two sections, and it focuses on the effects of dietary modifications and hormones on the phenotype and gene expression in early life. In these two sections, the discussion on the role of epigenetics in the plasticity and epigenetic regulation of human growth will be an extension of the discussions in Section III.A of this review.
There are basically two causes of epigenetic programming in the placenta: nutritional programming (which was introduced in Section III.D and will be discussed in this subsection and Sections VIII.F and IX.A), and endocrine programming (which will be discussed in Section VIII.D). Epigenetic programming of the placenta (placental programming) can result in structural and functional changes in genes, cells, tissues, and even whole organs (451). These changes may be isolated or widespread events with either discrete or cumulative effects on development depending on the nature and timing of the programming stimulus (21). The consequences of placental programming depend on whether the developmental deficit is the inadvertent outcome of an insult that acts as mutagen or a specific adaptation to an environmental challenge that is designed to maximize survival to reproductive age (8, 22, 23). With mutagenesis, the structural and functional deficits are permanent and invariably detrimental to long-term survival. In contrast, the physiological adaptations made in response to suboptimal intrauterine conditions may improve viability in the short to medium term, but at the risk of later morbidity (discussed in Section I). Placental programming also has consequences for the next generation, and this topic is discussed in Section VIII.F.
Focusing on the effects of nutrition on placental programming, the effects are mediated through changes in placental gene expression, which includes imprinted genes (447, 452). As already discussed in Section IV.A, imprinted genes are expressed monoallelically according to their parental origin. Although not well understood, imprinted genes seem to have a disproportionately important influence on placental development (453–455). For example, Coan et al. (447) recently reported that undernutrition of pregnant mice resulted in decreased expression of the placental-specific transcript of the Igf2 gene, although methylation of its promoter was unaffected. There is also increasing evidence that nonimprinted genes can be nutritionally programmed in utero. This programming involves changes in DNA modifications, particularly at promoter regions, which are related to altered gene expression in adulthood (456, 457). Whether these changes in gene expression are mediated through alterations in the placenta is unknown because it has also been shown that feeding a protein-restricted diet to pregnant mice during the first 3.5 d of the pregnancy before blastocyst implantation can induce nutritional programming and an altered phenotype of the offspring (458).
Lillycrop and colleagues (459–461) fed either a control or a protein-restricted diet to pregnant rats and mice and then measured the hepatic expressions of the GR and PPARα, as well as other markers of glucose homeostasis and β-oxidation in the offspring at different times after birth. GR and PPARα are NR proteins (whose signaling pathway to chromatin was discussed in Section VIII.B) and play key roles in glucose and lipid metabolism, two processes that are dysregulated in the offspring of protein-restricted mothers. The results of these experiments showed that modest changes to maternal intake of macronutrients during pregnancy induce stable changes to the epigenetic regulation of GR and PPARα in the livers of the juvenile (459, 461) and adult offspring (460). Maternal protein restriction also induced the down-regulation of the maintenance DNMT, DNMT1 (462). This finding suggested that hypomethylation of the PPARα and GR promoters may result from the decreased expression of DNMT1, and hence the progressive loss of methyl groups from CpG dinucleotides after mitosis, rather than active demethylation. Thus, altered gene methylation may provide a causal mechanism to explain how maternal diet can induce stable changes in gene expression within the offspring and may represent a fundamental mechanism for altering the phenotype.
The finding that maternal protein restriction induces down-regulation of DNMT1 and the hypomethylation of the GR110 promoter suggests that DNMT1 mRNA expression may be related to the level of methylation of the hepatic GR promoter. To determine whether DNMT1 expression was also related to the level of methylation of the GR1-CTotal promoter, which shows 70.6% homology with the rat GR110 promoter (463), in fetal human tissue, Lillycrop et al. (462) investigated DNA methylation and DNMT expression in human umbilical cords that were collected from 15 term infants whose birth weights were within the normal range. They found that DNMT1 expression significantly predicted 49% of the variation in GR1-CTotal methylation, whereas DNMT3A expression was not related to GR1-CTotal methylation (462). Thus, methylation of human GR appears to be associated with the capacity of DNMT1 to maintain methylation of CpG dinucleotides, rather than the capacity for DNA methylation de novo. These findings are consistent not only with findings in rats, but also with the hypothesis that the induction of different phenotypes in humans by prenatal nutrition may involve variations in DNMT1 expression and, in turn, DNA methylation. These findings lend support to the notion that there are critical periods during embryogenesis and early postnatal life when epigenetic processes are susceptible to perturbations by maternal nutrition.
There are studies that explored the “rescue” of aberrant phenotypes in utero or the reversibility of induced phenotypic effects (see Section III.D). Lillycrop et al. (459) showed that supplementation of the protein-restricted diet with folic acid, a methyl donor cofactor, during pregnancy prevented changes to the methylation status of the GR and PPARα promoters and led to the normalization of GR and PPARα expression. Their result is consistent with those of Jackson et al. (464) and Brawley et al. (465), who were also able to prevent induction of an altered metabolic phenotype in the offspring of rats who had been fed either a folic acid- or glycine-enriched protein-restricted diet. In addition, Burdge et al. recently reported that folic acid supplementation in the peripubertal period did not normalize the effect of the maternal protein-restricted diet on the phenotype or epigenotype of the offspring (466). These findings imply that the timing of the nutritional intervention is an important factor for determining the phenotypic outcome. The apparently contrasting effects of increased folic acid exposure before birth or after weaning may reflect differences in the supply of folate to the offspring. Before birth, maternal physiology may buffer or modulate supply of folate to the offspring. After weaning, the offspring receive folic acid directly, and so the exposure of the juvenile animals may be greater than fetuses. Furthermore, the results of the postweaning folic acid supplementation study showed that the juvenile-pubertal period is another period of plasticity when specific nutrient intakes may alter the phenotype of the offspring through epigenetic changes in specific genes. Folic acid supplementation in the juvenile-pubertal period may therefore provide a window of opportunity for appropriate nutritional interventions to reverse the effects of a poor prenatal environment.
There are now also several published reports on attenuation, and even reversal of the adverse effects of high-fat, high-carbohydrate (HC), or low-protein diets on the neonatal phenotype by maternal nutritional interventions in rodents (467). Srinivasan and colleagues (468, 469) showed that artificial rearing of female rat pups on a HC milk formula resulted in chronic hyperinsulinemia and adult-onset obesity (HC phenotype), and that the maternal HC phenotype was transmitted to their progeny. Using this model, Srinivasan et al. (470) then tested the idea that the fetal adaptations that predisposed the progeny for the expression of the HC phenotype in adulthood and the transfer of the HC phenotype to the progeny could be reversed by maternal food restriction. For this purpose, they modified the intrauterine environment of HC rats by pair-feeding pregnant rats the identical amount of food that was consumed by age-matched control rats from the time of their weaning. This mild dietary restriction reversed the HC phenotype and also prevented the development of the HC phenotype in the progeny. From these results, they concluded that: 1) malprogramming of the progeny of hyperinsulinemic-obese HC female rats for the expression of the HC phenotype is initiated in utero; and 2) normalization of the maternal environment by mild food restriction in HC female rats resulted in a normal phenotype in their progeny.
Junien and colleagues (382) investigated whether altering fat intake in mothers during the periconceptual, gestational, and lactational periods by feeding them a high-fat diet (HFD) that induced obesity could be used to modify fetal/neonatal metabolic programming to prevent the development of the postnatal metabolic phenotype. To this end, they crossed F1 obese female mice with T2D mice that were fed a HFD with F1 lean males that had been fed a normal rodent chow. The HFD of these F1 females was then switched to the normal chow before mating and during the gestational and lactational periods, and all second-generation (F2) mice were fed a HFD for 5 months. Sensitivity or resistance to the HFD differed significantly between generations and sexes. The proportions of the F1 and F2 males that developed hyperphagia, obesity, and T2D were similar. In contrast, the proportion of F2 female mice that were hyperphagic and obese and developed T2D was significantly lower than that of the F1 (57 vs. 83%). In other words, the proportion of the F2 female offspring that were resistant to the effects of the HFD was significantly higher than that of the F1 (43 vs. 17%). Despite having free access to the HFD, these “resistant” F2 female mice displayed a “satiety phenotype”; they were lean, no longer hyperphagic, and had normal plasma glucose levels and insulin sensitivity, despite being mildly hypercholesterolemic and glucose intolerant (382). These results suggest that feeding rats a diet with an appropriate fatty acid profile before mating and during the gestational and lactational periods in the setting of maternal obesity interfered with fetal or neonatal programming of the metabolic syndrome.
In another study, Howie et al. (418) reported recently on the long-term impact of a moderately HFD during preconception and/or pregnancy and lactation on postnatal growth and metabolism of the rats from birth to adulthood. The major findings of this study were: 1) the offspring of dams that were fed a HFD during pregnancy and lactation were smaller than normal; 2) a postweaning HFD increased adiposity in all treatment groups, but the offspring whose dams were fed a HFD during pregnancy and lactation had elevated adiposity compared with controls regardless of postweaning diet; 3) this increased adiposity was accompanied by hyperinsulinemia and hyperleptinemia; and 4) maternal preconceptual diet did not impact offspring adiposity. From these results, the authors suggested that the diet in pregnancy and lactation, but not preconception, is an important influence on the long-term health of the offspring.
Lastly, Mao et al. (471) recently reported contrasting effects of different maternal diets on sexually dimorphic gene expression in the murine placenta. In their study, they examined the impact of diet and fetal sex on placental gene expression in mice that were fed a very high-fat, low-fat, or chow diet of intermediate caloric density, and then extracted and analyzed placental RNA by microarray on embryonic day (E) 12.5. First, they found that the changes in the expression of 1972 genes were more than 2-fold in at least one of the three treatment groups. Second, they reported that the placentae that were attached to female pups (female placentae) demonstrated more striking alterations in gene expression in response to the maternal diet than that found in the placentae that were attached to male pups (male placentae). Third, they reported that each diet provided a distinctive signature of sexually dimorphic genes; the expression of the sexually dimorphic genes from female placentae was generally higher than those from male placentae. Fourth, they found that the expression of other genes, which are normally considered as characteristic of kidney function, were affected by diet and included genes that regulated ion balance and chemoreception. Fifth, they found that transcript levels of many known olfactory receptor genes in the placenta, which may allow the placenta to sense odorant molecules and other minor dietary component genes, were influenced by diet and the sex of the fetus. From these results, they concluded that gene expression in the murine placenta is adaptive and could be influenced by the maternal diet. Moreover, they found that the placenta exhibits pronounced sexual dimorphism; the female placentae were more sensitive to nutritional perturbations than the male placentae.
The results of the rodent studies in which the maternal diet was manipulated and induced the metabolic phenotype and its underlying epigenetic changes in the offspring may have implications for humans. From these results, it is also now evident that future studies should investigate the effects of maternal nutrition on the epigenetic regulation of gene expression in placental programming to understand how the maternal diet influences the health of the resulting offspring during the life span. The metabolic phenotype, which includes obesity, T2D, and other metabolic disorders, belongs to the group of the fastest-growing health problems worldwide, and a substantial body of evidence indicates that lifestyle factors contribute to the increased prevalence of obesity and T2D. The risk of spontaneous abortions and congenital malformations of neonates is higher in diabetic women than nondiabetic women, and poor diabetic control increases these risks (209, 472). As a result, a vicious cycle develops; prenatal development in a diabetic milieu favors the development of T2D in the offspring in later life (473). In the context of a worldwide epidemic of obesity, the increasing prevalence of excessive body weight and obesity due to imbalanced nutrition (25% of women in France and 50% of women in the United States) creates an abnormal uterine environment in women of childbearing age. Therefore, making pregnant women, especially overweight pregnant women, aware of 1) their diet and/or nutrition and their importance during pregnancy; 2) their body composition and the consequences of excessive weight; and 3) the subsequent effects of diet, nutrition, body composition, and weight on the phenotype and health of their babies may help disrupt the vicious cycle of the mother-offspring transmission of adult metabolic phenotype.
D. The effect of hormones on epigenetic gene regulation in early life
Hormones have an important role in regulating normal growth and development in utero. Undernutrition, hypoxemia, and stress can alter both maternal and fetal concentrations of many hormones that include GH, IGFs, insulin, glucocorticoids, catecholamines, leptin, thyroid hormones, placental eicosanoids, sex steroids, and placental lactogen. Some of the epigenetic effects of these hormones have already been discussed in this review. The epigenetic regulation of imprinted genes that are important for growth regulation, such as IGF2, and the role of let-7 miRNAs have been discussed in Sections III.B and IV.A of this review. In addition, the consequences of dysregulation of imprinting mechanisms at the 11p15 region that encompasses many imprinted genes and underlies the development of BWS and SRS in humans have been discussed in Section IV.B of this review. In addition, IGF2 and its epigenetic regulation have been recently reviewed in depth [for example, see reviews by Gicquel and Le Bouc (242, 252) and Chao and D'Amore (474)]. From the results of studies using Igf2 knockout mice, evidence is emerging that imprinted genes and hormones have central roles in controlling both the fetal demand for and the placental supply of maternal nutrients during mammalian development [see reviews by Reik, Constancia, and others (269, 452, 453, 475, 476)]. We also refer readers to several recent reviews on the epigenetic effects of other pregnancy-related hormones by Fowden and Forhead (477, 478) and Pasca and Penn (479).
In view of these referrals to other sections of the review and published reviews, the discussion of this section will focus on the epigenetic effects of glucocorticoids, which have widespread programming effects in utero and mediate the programming effects of nutritional and other environmental challenges during pregnancy (480–482). Specifically, glucocorticoids act at cellular and molecular levels and can induce changes in tissue accretion and differentiation by direct and indirect mechanisms. Many of these effects are mediated by genes, and comprehensive lists of the genes that are influenced by glucocorticoids can be found in the various reviews of Fowden and colleagues (451, 455, 481, 483–487).
The epigenetic effects of glucocorticoids have been reported. Thomassin et al. (488) reported that glucocorticoid administration caused DNA demethylation associated with increased gene expression of a hepatic aminotransferase in rats during the perinatal period. In another study, Drake et al. (489) showed that fetal exposure to the synthetic glucocorticoid, dexamethasone, during the last quarter of pregnancy in rats exerted a TGE on the key hepatic gluconeogenic enzyme, phosphoenolpyruvate carboxylase, and the subsequent development of hyperinsulinemia and hyperglycemia that persisted into the F2 without further treatment of the F1 animals. In another study, Weaver (490) reported that the levels of methylation at the 5′-end of the GR gene promoter in the hippocampus were inversely proportional to the extent to which rat pups were licked, groomed, and nursed by their mothers, and that the reduced level of methylation at the GR promoter correlated with higher GR transcription. Collectively, the results of such studies provide support for the notion that glucocorticoids are intimately linked with fetal programming of adult pathophysiology (482) (see also Section VIII.E).
Although a complete understanding of the epigenetic actions of glucocorticoids on the placental and fetal genome and its TGE on the hypothalamic-pituitary-adrenal (HPA) axis, and the fetal “programming” of adult pathophysiology is still required, antenatal glucocorticoid administration has significant implications in humans because multiple courses of synthetic glucocorticoids are currently recommended for various conditions. Preterm delivery occurs in approximately 10% of all pregnancies, and prenatal exposure to synthetic glucocorticoids is very efficacious in reducing the incidence of respiratory distress syndrome in these babies. Despite the beneficial therapeutic effect of antenatally administered synthetic glucocorticoids, its TGE is different in males and females with respect to the risk of developing cardiovascular disease and the metabolic phenotype in later life (482, 489, 491–493). Moreover, the sex differences of the TGEs that occur in later life after the antenatally administered synthetic glucocorticoids are exacerbated by the type of synthetic glucocorticoid and the timing of their administration (494). From the results of such studies, there is now more awareness of the potential consequences of repeated antenatal administration of glucocorticoids on the health of male and female infants. This increased awareness is echoed in a recent article by Newnham and Jobe (495) who wrote, “Unless further evidence of both benefit and safety of repeated courses is provided, it would be prudent for clinicians at this time to confine their use of antenatal corticosteroids to single-course treatment.”
E. Epigenetics of mental health and behavior
Following on from the discussion in Section II.B.3.b, which presented evidence on the relationship between epigenetics and mental health disease, the discussion in this section will present information on the role of the postnatal environment in generating vulnerabilities to chronic mental disease (496–498) and behavior (499, 500). The long-term effects of maternal behavior in the rat, as well as other mammals, on the stress responsiveness and behavior of the offspring during adulthood are well documented (501–503). The results of these studies have shown that early handling and deprivation can exert effects on spontaneous open-field behavior, acoustic startle, and the endocrine stress response when they become adults. In addition, these adult rats exhibited enhanced active avoidance and their stress hormone responses were reduced. Permanent changes in emotional and neuroendocrine reactivity have been observed in rodents after a variety of experiences, even minor ones, during postnatal life. In addition, it is well documented that stressful events that occur prenatally, and even at preimplantation, can have permanent consequences on behavior in later life (504–508).
Epigenetic programming occurs in response to rat maternal care, and the epigenetic programming of DNA methylation and histone acetylation is different in the low and high maternal care offspring (509, 510). Specifically, the nature and amount of care that a pup receives from its mother modulates its reaction to stress later in life largely through effects on the GR in the hippocampus (509). This maternal effect is transgenerational or heritable only insofar as its manifestation depends upon the pup's experience in the first week of life.
Maternal behavior also triggers a signaling pathway that involves the serotonin receptor, an increase in cAMP, and recruitment of the transcription factor nerve growth factor-inducible factor A. In turn, the HAT CREB-binding protein, and the methylated DNA binding protein, methyl-CpG-binding domain 2, are recruited to activate the GR gene promoter (511). Using this pathway to study the epigenetic consequences of maternal care in rats, Weaver et al. (511) demonstrated that DNA methylation patterns are dynamic: the methylation and demethylation of genes could be reversed during adulthood, although DNA methylation was established early in life. They observed differences in DNA methylation and histone acetylation in the regulatory regions of the GR exon 17 promoter in the hippocampus of the offspring from high- and low-caring nursing rats. They also showed differences in DNA methylation and histone acetylation that appeared to cover wide regions of chromosome 18 in adult rats that had experienced high and low maternal care when they were pups. In adult rats, they also reported that hippocampal gene expression was significantly altered and that the extent of the change was a function of the early-life maternal care that they had received. Therefore, differences in hippocampal gene expression due to early-life experiences determined the HPA stress response in the developing offspring and were maintained throughout their lives (510–513). They also reported that high maternal care resulted in hypomethylation in some brain regions and hypermethylation in other regions and was reversed by inhibiting histone acetylation (513). When an HDAC inhibitor was injected into the brains of adult offspring of low-caring mothers, they reported reversal of the epigenetic programming of the hippocampal GR promoter. Moreover, the treatment reestablished stress responsivity and open field behaviors that were indistinguishable from the adult offspring of high-caring mothers. When methionine, which is also an inhibitor of active demethylation (514), was injected into the brains of adult offspring of high-caring mothers, they reported: 1) DNA methylation; 2) down-regulation of GR; 3) heightened stress responsivity; and 4) open field behaviors that were indistinguishable from the adult offspring of low-caring mothers. These findings suggest that variations in maternal behavior can directly program the rudimentary defensive responses to stress through epigenetic mechanisms. These findings are supported by results of recent studies in which Miller et al. (515, 516) reported that the dynamic response of DNA methylation in neurons is involved in fear conditioning.
These challenging findings in rats have been extended to humans by identifying an association between early childhood adversity and epigenetic marks in later life (423, 517, 518). McGowan et al. (518) investigated the extent of DNA methylation in the promoter of genes that encoded rRNA genes in the brains of suicide victims. Suicide victims who had experienced childhood abuse had higher overall rRNA gene methylation and expressed less rRNA than other victims with no history of child abuse. From these results, the authors concluded that the difference in methylation was driven by an environmental factor, namely child abuse, rather than by a genetic variation (518). More recently, McGowan et al. (518) compared the GR exon 1f promoter of suicide victims who were or were not abused as children. They report site-specific differences in DNA methylation in the GR exon between suicide victims who experienced or did not experience social adversity in early life. These differences in DNA methylation were associated with reduced expression of the GR gene (423).
These data are the first demonstration of differences in DNA methylation states that were possibly triggered by an early-life exposure to social adversity. Moreover, these epigenetic imprints have functional consequences that result in reduced expression of a key regulator of the HPA stress response. Thus, early-life adversity might have lasting impact on gene regulation and results in susceptibility to mental health problems in later life. Moreover, the idea that epigenetic processes are involved in mental health disease adds a new dimension to the impact of early-life experience on disease susceptibility in later life and points to the possibilities of prediction, early diagnosis, and new therapeutic approaches to treating these diseases.
F. Transgenerational actions of endocrine-disrupting chemicals
EDCs are exogenous environmental chemicals that mimic or block the actions of endogenous hormones (2). To date, the vast majority of known EDCs are those that activate the parts of the endocrine system that are associated with the steroid/retinoid/thyroid superfamily of receptors, and within this superfamily, receptors that are related to the hormone estrogen are usually stimulated.
We are becoming increasingly aware of the role of environmental factors in disease susceptibility. Because the genome is evolutionarily and chemically stable, these environmental influences regulate genome activity independent of DNA sequence changes. An additional consideration for environmental influences on disease etiology is the developmental stage of exposure. Exposures during a crucial time of development can alter genome activity associated with the differentiation-linked programming of cells or organ systems. This altered program and gene expression profile can then promote an abnormal physiology and disease at the later adult stage of development (167).
Exposure of the germ cells during this critical period to environmental toxicants, such as EDCs, can reprogram the germ line (168, 519), and an epigenetic transgenerational phenotype can develop. When postnatal and/or adult exposures to environmental toxicants occur, gametogenesis can be affected with potential reprogramming of the germ line. Although epigenetic effects on gametogenesis have been described (520–523), no TGEs have been observed. In contrast, the transmission of a permanently altered germ line epigenome can promote a transgenerational inheritance of corresponding phenotypes to subsequent generations and progeny (168).
Much of our knowledge on the TGEs of EDCs has come from the studies of Skinner and colleagues (168, 424, 425, 519, 524–528). They used rodents to examine the TGEs of two chemical pesticides with endocrine disruptor activity, namely vinclozolin, which is a fungicide with antiandrogenic properties, and methoxychlor, a replacement for dichlorodiphenyltrichloroethane, whose metabolites have estrogenic, antiestrogenic, and antiandrogenic activities. They reported that transiently exposing pregnant rats between E8 and E15 to either methoxychlor or vinclozolin promoted a spermatogenic defect that was characterized by increased apoptosis and decreased cell number and motility in the adult F1 (168). They also reported that this spermatogenic defect was carried over four generations. The preliminary data suggest that this defect is linked to altered DNA methylation of the male germ line and the induction of new imprinted-like DNA methylation sites. This altered sperm epigenome also impacted on the genome activity of other developing tissues and cell types through the paternal genome (528). Interestingly, the expression of over 200 genes was altered in the embryonic testis, and this altered transcriptome was present in generations F1-F3. Altered transgenerational transcriptomes were also identified in several other tissues and cell types, with each tissue and cell type having a unique set of differentially expressed genes. The next step in understanding this TGE is to establish the functional relationships between the differential DNA methylation and transcriptome effects (2). In addition to detecting the male testis disorder, Skinner and colleagues (424, 527) also reported a TGE on the development of other disease states as the animals aged, such as tumor development, prostate disease, kidney disease, and immune abnormalities, as well as on the pregnancies and the onset of disease in female adults (425). In fact, the phenotype of the young adult rats resembled one of an aged animal, which suggests that vinclozolin accelerated the aging process (528). Skinner and colleagues (528, 529) also recently reported TGEs on behaviors, such as sexual selection and anxiety, due to vinclozolin exposure.
Based on these results, two potential epigenetic mechanisms of action for EDCs have been proposed (62) (Fig. 14). The first mode occurs during active development of a specific organ when the epigenome and transcriptome are progressing through a cascade of developmental stages to establish the adult organ transcriptome and physiology. The second mode occurs during reprogramming of the epigenome of the germ line and promotes an abnormal epigenome (168, 424, 519). In both mechanisms, the transgenerational epigenetic mechanism of action of an EDC occurs at the time of sex determination to cause altered epigenetic programming of the germ line. As a result, the transcriptomes of developing organs are altered in such a way as to induce various adult disease states transgenerationally.
Fig. 14.
A model for endocrine-disruptor-induced epigenetic transgenerational disease. Endocrine-disruptor action reprograms the epigenome of the developing germ cell during embryonic sex determination, leading to genes and other DNA sequences with altered DNA methylation. These changes are proposed to alter the transcriptomes of the testis and other organs, thereby promoting adult pathologies, some of which are inherited transgenerationally. Epigenetic mechanisms might therefore have a role in the induction of adult-onset disease through environmental exposures early in development. [Reprinted with permission from R. L. Jirtle and M. K. Skinner: Nat Rev Genet 8:253–262, 2007 (62). © 2007 Macmillan Publishers Ltd.]
Other EDCs of potential importance include phytoestrogens, which are naturally occurring estrogenic chemicals, and are present in high levels in plant-based diets. One such phytoestrogen is genistein, an isoflavone that is found mainly in soybeans and is widely consumed worldwide (see also Sections IV.D and VIII.C). Concern has been raised over the potential estrogenic effects of genistein in soy-based infant milk formulae on fetal development and the long-term consequences on female reproductive performance, adiposity, and cancer risk (530, 531). These concerns originate from the results of studies of McLachlan and colleagues (2, 532–534), who proposed that developmental reprogramming by early-life estrogenic exposures may be linked to cancer in later life due to altered epigenetic memory. At the heart of this concern is the lifetime isoflavone exposure profile, despite the low and variable potency of soy isoflavones to bind and activate estrogen receptors, and their ability to act as selective estrogen receptor modulators.
Jirtle and colleagues (530) reported that maternal dietary genistein supplementation of mice during gestation, at levels comparable with humans consuming high-soy diets, shifted the coat color of heterozygous viable yellow agouti (Avy/a) offspring toward pseudo-agouti. This marked phenotypic change was significantly associated with increased cytosine methylation at a retrotransposon upstream of the transcription start site of the Agouti gene. Because the extents of DNA methylation were similar in endodermal, mesodermal, and ectodermal tissues, they concluded that genistein acts during early embryonic development. They also concluded that in utero dietary genistein protected the offspring from obesity because genistein-induced hypermethylation persisted into adulthood. Since the publication of this study, other studies, such as those of Cederroth et al. (535, 536), have confirmed the beneficial effects of soy and/or genistein on adiposity and body composition. For example, Cederroth et al. (535, 536) reported that male mice fed from conception to adulthood with a high soy-containing diet had reduced body weight and adiposity and decreased glucose intolerance. In a recent follow-up study, they showed that eating a high soy-containing diet during gestation, lactation, or after weaning could alter body composition, glucose tolerance, and blood pressure in adult individuals independently of adipose gain (537). In addition to these animal studies, the Arkansas Children's Nutrition Center conducted a prospective longitudinal study that compared the growth, development, and health of breastfed children and soy formula-fed children from birth to age 6 yr (531). After 5 yr, the growth of soy formula-fed children was normal. Although this study reported that no adverse effects occurred with eating high levels of soybean, further studies with longer follow-up and wider assessment of the outcomes after consuming high-soy diets are needed.
Although the actions of most EDCs will likely involve alterations in the somatic cell epigenome and will not promote a transgenerational phenotype, one can speculate that some component of adult-onset disease will involve the actions of EDCs on the germ line to promote transgenerational inheritance. The suggestion that EDCs can reprogram the germ line and induce epigenetic transgenerational disease is a new paradigm and should be considered in disease states that have a familial inheritance that does not follow normal genetic mechanisms (2).
IX. Perspectives in Clinical Epigenetics
Epidemiological evidence provides strong support that environmental exposures early in development may influence susceptibility to disease in later life. Furthermore, some of these environmental effects could rarely be passed on to subsequent generations. In this review (Section VIII.C–F), we have described the experimental evidence from animal studies that supports the notion that epigenetic modifications provide a plausible link between the environment and alterations in gene expression that might lead to disease phenotypes. Although differences between humans and other species in regard to epigenomic regulation and, most importantly, imprinting require caution in extrapolating the findings from one species to another, there is an increasing body of evidence from animal studies that supports a role of environmental epigenetics in disease susceptibility (62, 167). In this review, we have also emphasized that the growing fetus has “environmentally sensitive” periods for building the epigenome. In this section, we will discuss therapeutic targeting of the epigenetic mechanisms of these sensitive periods and the potential use of epigenetic biomarkers to identify fetuses at risk for developing an abnormal physiology or phenotype and disease at the later adult stage of development.
A. Drug and dietary targeting of epigenetic mechanisms
As discussed in Sections II.B and C, genomic programming is accomplished by DNA methylation and changes to the chromatin architecture during cellular differentiation, and critical periods of gestation and early life is a highly organized process. The resultant epigenetic patterns are dynamic, and the epigenome is sculpted continuously by the complex chromatin machineries throughout life in response to external cues. In contrast to genetic information, which is highly stable, epigenetic information is flexible and therefore potentially reversible. Therefore, if one could modify epigenetic patterns pharmacologically or by nonpharmacological means, it would be possible to alter deleterious gene expression programs (440). The idea that epigenetic states could be prevented or reversed has immensely important implications for the potential of interventions to override the effects of early-life adversity on health and behavior.
Selective DNA methylation inhibitors and DNMT1 modulators are available experimentally but are not in clinical use. However, there are drugs in current clinical use that influence DNA demethylation. Procainamide is a widely used antiarrhythmic drug that inhibits DNMT activity and promotes DNA hypomethylation. Hydralazine, a peripheral vasodilator that is used to treat some types of hypertension, and valproic acid, a widely used antiepileptic and mood stabilizer, are now known to cause DNA demethylation. Because an effect on DNA methylation is one of the actions of these drugs, concerns that other drugs in current clinical use might also affect DNA methylation patterns have been raised. Accordingly, it has been suggested that future drug safety tests should now include measures of DNA demethylation (440, 538).
The DOHaD hypothesis has been much discussed in this review. Acceptance of this hypothesis has been the main reason for advocating maternal supplementation with the dietary methyl donors and cofactors, such as folic acid, vitamin B12, betaine, and choline, to optimize DNA methylation and gene expression (55) (see Sections III.D and VIII.C). However, it is now known that eating cruciferous vegetables is also associated with epigenetic modifications because naturally occurring HDAC inhibitors are normal constituents in these foods. For example, diallyl disulfide in garlic and SFN in broccoli are class I and class II HDAC inhibitors (419, 539). There are many other known or putative diet-derived HDAC inhibitors, such as butyrate, which is derived from the fermentation of dietary fiber and is the primary metabolic fuel for colonic epithelial cells. Of interest are the results of the study of Myzak et al. (540), who measured the level of HDAC inhibition in peripheral blood mononuclear cells in healthy volunteers after ingestion of a single dose of SFN-rich broccoli sprouts. Each participant consumed 68 g (1 cup) of broccoli sprouts, and blood was collected over the next 48 h. They reported that HDAC activity was inhibited as early as 3 h after broccoli sprout intake and returned to normal after 24 h. There was strong induction of histone H3 and H4 acetylation coincident with HDAC inhibition at 3 and 6 h, and histone hyperacetylation was evident for at least 48 h. Overall, the level of HDAC inhibition and histone hyperacetylation was equal to or greater than that achieved with the clinically used HDAC inhibitor, vorinostat. Other dietary constituents can alter HDAC activity through other mechanisms. For example, resveratrol, a polyphenol constituent of red wine, can activate human SIRT1, which is member of the family of sirtuin proteins that are essential for gene silencing (541) (see Section II.C). Lastly, the major polyphenol in green tea, epigallocatechin-3-gallate, has been reported to inhibit DNMT and reactivate methylation-silenced genes in human esophageal squamous cell carcinoma cell lines (542).
To summarize, much research is still required to “connect the dots” between diet, epigenetically altered genes, child growth and development, and adult disease. With increasing attention now being put on the effect of diet and nutritional supplements on the placental and fetal epigenome in pregnancy, we should begin to pay more attention to the constituents of the maternal diet and their potential effects on these two epigenomes. Optimizing the nutritional environment to which an individual is exposed during development has the potential to improve the health of the global population. On the assumption that nutraceuticals (a food or naturally occurring food supplement thought to have a beneficial effect on human health) and food and beverage constituents can cross the placental barrier and/or be secreted unchanged into milk, the saying that “you are what you eat” still holds true, but it now has the following caveat “you are what your mother ate and did when she was pregnant with or nursing you.”
B. Epigenetic biomarkers
Biomarkers are biometric measurements that provide information about the biological condition of the subject that is being tested and provide varied information depending on the category of interest. Accordingly, biomarkers fall into several categories, and there are now biomarkers of exposure, biomarkers of susceptibility, and biomarkers of response (543). The clinical potential of a biomarker must satisfy three criteria: ease of clinical measurement, provision of new information, and therapeutic value and utility (544). To these ends, biomarkers have been used as a screen for the presence of a disease, an indicator of the disease process and its clinical course, a measure of disease severity, and a method of confirming the diagnosis. Biomarkers have also been used to detect the predisposition to disease, as well as to predict clinical responses to therapies. The ideal biomarker should have the following characteristics: 1) highly sensitive; 2) highly specific; 3) cost effective, rapid, and simple; 4) noninvasive; and 5) accurate, with a standard reference range (545).
The epigenetic consequences of placental and fetal programming create a potential opportunity for the development of epigenetic biomarkers for use in child and adult health and practice. Such epigenetic biomarkers could be used for early diagnosis of disease, identification of individuals at high risk of diseases, and the monitoring of responses to preventive or curative interventions (546–548).
As shown in Fig. 8, allele-specific CpG methylation is the most comprehensively studied epigenetic mark and one of the hallmarks of genomic imprinting. Accordingly, patterns of DNA methylation and histone modification of imprinted genes that are associated with development also have potential clinical use. Methylation-specific amplification (MSA) can identify genes that are differentially methylated in cancers and novel tumor suppressor and drug resistance genes (549, 550). Shen et al. (59) recently advanced this technique by combining MSA with oligo-based microarray hybridization for rapid methylation profiling and genome-wide analysis of CpG methylation. Based on these techniques, Tycko and colleagues (551) developed a SNP chip-based method for combined genetic and epigenetic profiling, which they called a methylation-sensitive SNP array for identifying sequence-dependent allele-specific DNA methylation. SNP arrays are used for methylation profiling to produce bar code readouts of the methylation status at thousands of SNP-tagged loci (552). Using this method, they surveyed DNA from 12 normal tissue samples that included peripheral blood leukocytes, kidney, brain, lung, placenta, and buccal cells at 50 and 250K resolution. Based on their results, they claimed that the recurrent phenomenon of sequence-dependent allele-specific methylation (ASM) has practical implications for mapping and interpreting associations of noncoding SNPs and haplotypes with human phenotypes.
As noted in Section VIII.D, the epigenetic lability of imprinted genes underlies placental programming. Tycko et al. (553) used ASM to develop a method for measuring DNA methylation patterns in placental tissue. They were able to distinguish specific patterns of DNA methylation in the human placenta from other human organs and uncovered other differences that distinguish normal placental tissue from hydatidiform moles. Their method seems to be unique because it revealed not only net gains and losses in DNA methylation when comparing two biological samples, but also differences in ASM.
Fetal DNA circulates in maternal plasma, and this discovery has enabled new approaches for noninvasive prenatal diagnosis of disease and monitoring using maternal blood samples (554). Although still in its infancy, the molecular characterization of plasma nucleic acids for distinguishing Y-chromosomal DNA sequences from the background maternal plasma DNA shows great potential in prenatal diagnosis. Using this approach, Chim et al. (555) investigated the potential clinical utility of the maspin (SERPINB5) tumor suppressor gene, a gene that is expressed in the placenta, for the noninvasive prenatal assessment of the developing fetus. They used bisulfite DNA sequencing to determine the methylation status of the maspin gene promoter in placental tissues and paired maternal blood cells from pregnant women. They found that the maspin gene promoter was hypomethylated in placental tissues and densely methylated in maternal blood cells. Using real-time quantitative methylation-specific PCR, they reported that the unmethylated maspin sequences that were detected in maternal plasma in all three trimesters of pregnancy were cleared within 24 h after delivery. Of particular interest, the maternal plasma concentration of unmethylated maspin sequences was elevated almost 6-fold in preeclampsia, when compared with non-preeclamptic pregnancies. From these results, Chim et al. (555) proposed that hypomethylated maspin DNA could be used as a universal marker for fetal DNA in maternal plasma to diagnose pregnancy-associated disorders, irrespective of fetal gender and genetic polymorphisms.
The future clinical utility of epigenetic biomarkers and techniques, such as methylation specific amplification microarray, in routine clinical practice will require rapid, quantitative, accurate, and cost-effective techniques and objective criteria for selection of the suitable genes. An additional consideration is the choice of tissue; epigenetic marks are cell- and tissue-specific, although preliminary evidence suggests that some conclusions can be drawn from the results of studies in germ and blood cells. Notwithstanding the social, ethical, and regulatory aspects on the use of epigenetic biomarkers for prenatal diagnosis, there can be little doubt that the introduction of epigenetic biomarkers will have considerable impact on prenatal diagnosis and the management of the developing fetus. The speed at which these biomarkers will become clinically useful tools depends on our rate of acquiring knowledge on the relationship between epigenetic marks and the linked consequences on placental and environmental programming on the fetal phenotype. Until such knowledge is acquired through research, epigenetic biomarkers are not yet appropriate for routine clinical use.
X. Future Directions: Identifying the Needs and Opportunities for Advancing Epigenetic Research in Child Health
The end-target of translational research is the patient, with the goal to improve medical care. Traditionally, translational research has followed a one-dimensional sequence of events: discovery of a new mechanism and a potential target in an experimental or basic research setting; development of a biomarker; validation of its utility in the clinical setting; and its eventual introduction into clinical practice. Translational epigenetic research in child health must be seen as a reiterative process that ranges from research in the basic sciences to preclinical research and pediatric clinical research. Wide knowledge gaps still exist in our current understanding of the epigenetic machinery, despite the increasing use of numerous experimental systems. As a result, we still do not know whether some of the epigenetic mechanisms that have been identified thus far using these experimental systems are operative in humans and other eutherians. For example, the short ncRNAs often work in concert with various components of the cell's chromatin and DNA methylation machinery to achieve stable silencing in fission yeast and plants (107) (see Section II.B.3.a).
Hereditary, environmental, and stochastic factors determine the accumulation of epigenetic variation over time, but their relative contribution to the phenotypic outcome and the extent of stochastic epigenetic reprogramming that is required to alter human phenotypes is not known because few data are available (see Sections II.E and V). Although we do not also fully understand the underlying mechanism of programming and reprogramming, a reasonable hypothesis would be that an inadequate set of epigenetic modifiers increases the risk of somatic epimutations, or mutations, or both.
If the environment (epigenetic events) can influence growth and developmental trajectories during preadult life-history stages and later life outcomes, how do epigenetic events influence the transition from one life-history stage to the next, growth, and puberty at the molecular level? Growth and puberty are regulated by insulin, GH, the IGFs, and the sex hormones. These hormones drive the rate of growth and development, but it is unclear what determines the timing of the different phases of developmental events and the quantity of growth. At the target tissues for these hormones, we need to first identify the specific types of epigenetic changes that occur in each tissue, as well as the precise genetic loci in each tissue that are affected by these epigenetic changes. The cell type specificity and tissue specificity of chromatin regulation are great challenges for future human studies into epigenetic regulation of gene expression and the role of epigenetics in disease susceptibility.
Epigenetic mechanisms potentially play an important role in the DOHaD phenomenon. Environmental influences during embryonic and early-life development can permanently alter epigenetic gene regulation, which in turn can result in imprinting and reprogramming of the epigenome and influence disease susceptibility in later life. The mechanisms by which cues about nutrient availability in the uterus and postnatal environment are transmitted to the offspring and by which different stable phenotypes are induced are still unknown. The genetic control of the regulation of placental supply and fetal demand for maternal nutrients is not fully understood, and many of the detrimental events that occur in the fetus could possibly be due to epigenetic misprogramming.
As stated in Section VIII.F, epigenetic transgenerational disease is a new paradigm in disease etiology that has not been considered previously. In fact, research into epigenetic transgenerational disease is now one of the new topics that is undertaken to understand the etiology of disease states that have a familial inheritance but do not follow normal genetic mechanisms. Epigenetic epidemiology provides a basis for future studies into the relationships between early-life exposures, epigenetic mechanisms, and adult disease, and epigenomics will accelerate the discovery of human loci at which epigenetic regulation is correlated with early environmental exposures.
Whereas it is generally held that the mouse is neurologically immature at birth, relative to the human, it is possible that the timing of specific developmental mechanisms is conserved from mouse to human (556). Almost all current epigenetic research is conducted in laboratory animals or cell systems. Finding the most appropriate animal models to study the epigenetic machinery is critical to advancing our knowledge for child health. For this purpose, inbred mice can be very useful to allow the investigator to control for genetic factors and the environment. Inbred littermates are a powerful tool to study epigenetics because they are homozygous at every allele. Any observed phenotypic change in these mice is epigenetic because genetic noise has been eliminated by definition. Therefore, a litter of inbred mice is similar to a set of MZ twins. By manipulating the genetics of the mouse in a controlled environment, gene-environment interactions and transgenerational epigenetic inheritance can be studied. Moreover, the mice can be used to seek explanations for complex and sporadic disease. In this regard, work from a number of laboratories suggests that the establishment of epigenetic states can be influenced by the environment, and epigenetic states are involved in the developmental origins of some disease states. Agouti viable yellow mice are particularly useful because changes in their coat color can be used to quantify phenotypic variation among inbred littermates. For example, alterations in the DNA methylation levels have now been reported in agouti viable yellow mice, which were given 10% alcohol before fertilization and during early pregnancy (557). Because specific loci on the epigenome of these mice are particularly sensitive to nutritional influences, these findings underscore the role of epigenetic mechanisms in the development of fetal alcohol syndrome.
Because no other animal has a similar preadult life history to that of humans, an obvious question is whether the findings from any experimental animal can be extrapolated to humans. Many tissues can be sampled in humans by noninvasive and minimally invasive methods, including red blood cells, T cells, sperm, placentae, umbilical tissue and blood, and fetal cells and/or fetal DNA in the maternal circulation, foreskin, urine, cord blood, nails, and hair. Obviously, the lineage of the specimens is highly important when studying epigenetic mechanisms. For example, foreskin derives from endoderm and cord blood from the mesoderm. Appropriate specimen collection includes the need for robust protocols for sample preparation, storage, and retrieval. In addition, accurate phenotyping of the donor is crucial to ensure the reliability of the data from any past, current, or future cohort studies. Finally, the available options for noninvasive sample collection overcome some, but not all, of the stringent ethical requirements for conducting experiments in humans.
As described in Section II of this review, the epigenetic machinery is complex, and our appreciation of its complexity continues to grow with our increasing knowledge. Future studies are now needed to: 1) establish the contribution of each component to epigenetic variation over time; 2) determine the molecular mechanism involved in the possible transmission of epigenetic patterns between generations; and 3) assess their functional role and the DNA regions in which they occur. The burgeoning complexity of the epigenetic machinery leads to two issues: selection of genetic locus to study an epigenetic mechanism, and the validity and interpretation of the resultant epigenetic mark. Although this review has not discussed chromatin techniques in detail, readers should be aware of the development of next-generation sequencing vs. the microarray in epigenetic research for ultimate clinical use in humans (558).
Focusing specifically on the needs and opportunities in child health, we need better phenotypic assessments than those currently available to define study populations and, in particular, to distinguish between IUGR infants and other SGA infants. The SGA infant embraces two different phenotypes: first, a small infant who has been a small fetus throughout pregnancy with a normal fetal growth rate; and second, an IUGR infant with reduced fetal growth rate. To distinguish between these two phenotypes, an accurate classification for identifying each phenotype is critical for the clinician to: 1) reduce avoidable perinatal morbidity and mortality; and 2) more accurately assess the risk of developing disease in later life. Although birth weight and length are easily obtainable, they are inadequate indices to fully phenotype SGA and IUGR infants, even with additional information on ethnicity, sex, and parental size. Birth weight and length are sometimes crudely used as indicators of fetal growth and nutrition but are measures of attained size, rather than measures of fetal growth rate. Epigenetic biomarkers have the potential to greatly improve the phenotyping of these subsets of infants, and this is an example of the need for such a biomarker in child health. Animal models will clearly inform the identification of suitable candidate biomarkers and selection of the most appropriate animal model of IUGR. Nijland et al. (559) used a nonhuman primate model of IUGR to investigate epigenetic modifications in gluconeogenesis in response to fetal malnutrition. They induced maternal nutrient restriction in pregnant baboons by limiting their caloric intake to 70% of controls' ad libitum intake. Although this difference in caloric intake did not significantly lower fetal weight at 0.9 of gestation, fetuses from the nutrient-restricted mothers were thinner. In addition, fetuses from the nutrient-restricted mothers had an increase in hepatic phosphoenolpyruvate carboxylase 1 in both expression and immunoreactive protein and hypomethylation of the promoter in their liver. These data emphasize that analysis of fetal weight may not be sufficient to fully classify IUGR or the fetal response to intrauterine nutrient deprivation, and more refined measures are needed to classify the pathological process of IUGR, such as morphological asymmetry or genetic profiling assays (560). Although nonhuman primates were used by Nijland et al. (559), most investigations on the effects of maternal nutrient restriction on the fetus have been done in rodents for ethical reasons and cost, among others. Choice of the IUGR model is also crucial when rodents are used as the model, as demonstrated by the recent publication by Shahkhalili et al. (467), who compared two rodent models of IUGR, maternal food restriction and dexamethasone exposure, for early postnatal catch-up growth and later development of glucose intolerance and obesity. Their finding that prenatal food restriction is a more sensitive model than the dexamethasone-exposed model to study the consequences of IUGR should also be considered in the light of the discussion on placental programming by glucocorticoids, which was covered in Section VIII.D.
Epigenetic information also has the potential to help in all phases of the developing individual during his/her life history (Fig. 1). Epigenetic information could be used to indicate the fetus' experience of prenatal nutrition and maternal health. It could also be used to monitor infant nutrition and the role of supplementary vitamins and dietary methyl donors (folic acid), and the quality of bonding and attachment, and possibly help to predict the effectiveness of GH treatment or other interventions in SGA infants who remain short.
The topics that have been discussed and ideas that have been presented in this review indicate that the epigenetic program that is established early in life in response to certain maternal behaviors has the potential to be manipulated or even potentially reversed by social and cognitive interventions, drugs, and diet. Epigenetic information that is collected during childhood could potentially represent a stable indicator of the child's psychological and behavioral experiences that include neglect or even abuse and might provide insights into risks for mental and physical health. These results are very intriguing because they hint of the existence of an epigenetic link between nurture and nature.
Lastly, epigenetic information that is collected during the transition to juvenility could potentially be used to inform the prevention of obesity and the metabolic phenotype. The focus of many studies in prenatal programming of adult health is restricted to specific narrow windows of embryonic development or, at the most, to a single life-history stage. As a result, these studies have not considered the cumulative nature of critical experiences throughout life history. To incorporate these considerations, careful thought must be given on ways to include an additional dimension, namely the social environment in which the individual lives and which itself has a genetic basis. This type of research, as an adjunct to existing epidemiological methods, requires future close collaboration between epigeneticists and clinical scientists.
Acknowledgments
The authors thank the three anonymous reviewers for their constructive comments and Dr. Arieh Bomzon, Consulwrite (www.consulwrite.com), for his nonexpert scientific input and editorial assistance.
This manuscript was prepared from the presentations at the New Inroads for Child Health (NICHe) conference held May 2009 in Marstrand, Sweden. The NICHe conference was supported by Växthuset Foundation for Children, the European Society for Pediatric Endocrinology, and an educational grant from Pfizer Endocrine Care.
Disclosure Summary: All authors were invited speakers at the NICHe conference. K.A.-W., P.B., J.-C.C., M.C., C.L.D., M.F., Z.H., C.J., K.L., K.O., R.S., R.A.W., and E.W. have nothing to declare. R.F. receives research funds from the Agence National de la Recherche, the Institut National du Cancer, and the Association for International Cancer Research. Y.L.B. receives basic and clinical research funds from INSERM, the Université Pierre et Marie Curie-Paris 6 (UPMC), the French Research Agency (ANR), the French National Ministry of Health (PHRC), and the Biomedecine Agency. M.Sk., M.Sz., and D.J.W. receive research funding from the United States National Institutes of Health. A.S. receives research funds from the New Zealand government.
Footnotes
- ART
- Assisted reproductive technology
- ASM
- allele-specific methylation
- BMI
- body mass index
- BWS
- Beckwith-Wiedemann syndrome
- CpG
- cytosine and guanine linked by a phosphate moiety
- CYP
- cytochrome P450 enzyme
- DNase
- DNA nuclease
- DNMT
- DNA methyltransferase
- DOHaD
- developmental origins of health and disease
- DZ
- dizygotic
- E
- embryonic day
- EDC
- endocrine-disrupting chemical
- ESC
- embryonic stem cell
- F1
- first generation
- F2
- second generation
- GOM
- gain of methylation
- GR
- glucocorticoid receptor
- GRB10
- growth factor receptor-binding protein 10
- GWAS
- genome-wide association study or studies
- HAT
- histone acetyltransferase
- HC
- high-carbohydrate
- HDAC
- histone deacetylase
- HFD
- high-fat diet
- HLA
- human leukocyte antigen
- HNF
- hepatocyte nuclear factor
- HP1
- heterochromatin protein 1
- HPA
- hypothalamic-pituitary-adrenal
- IAP
- intracisternal particle
- ICR
- imprinting control region
- ICT
- infancy-childhood transition
- IUGR
- intrauterine growth restriction
- IVF
- in vitro fertilization
- lncRNA
- long ncRNA
- LOM
- loss of methylation
- m5C
- DNA cytosine methylation
- miRNA
- microRNA
- Momme
- modifier of murine metastable epiallele
- MSA
- methylation-specific amplification
- MZ
- monozygotic
- ncRNA
- noncoding RNA
- NF-κB
- nuclear factor-κB
- NIMA
- noninherited maternal antigen
- NIPA
- noninherited paternal antigen
- NR
- nuclear receptor
- PcG
- polycomb group (proteins)
- PEV
- position effect variegation
- PGC
- primordial germ cell
- piRNA
- PIWI-interacting RNA
- PIWI
- P-element-induced wimpy testes
- PPAR
- peroxisome proliferator-activated receptor
- PRC
- polycomb repressive complex
- RE
- response element
- RNAi
- RNA interference
- RXR
- retinoid X receptor
- SAM
- S-adenosyl methionine
- SFN
- sulforaphane
- SGA
- small-for-gestational age
- SNP
- single nucleotide polymorphism
- SRS
- Silver-Russell syndrome
- STAT5b
- signal transducer and activator of transcription 5b
- T1D
- type 1 diabetes mellitus
- T2D
- type 2 diabetes mellitus
- TGE
- transgenerational effect
- TS
- Turner syndrome
- Xa
- active X-chromosome
- Xi
- inactive X-chromosome
- XIC
- X-inactivation center
- Xist
- X inactive-specific transcript (gene)
- Xm
- maternal X-chromosome
- Xp
- paternal X-chromosome.
References
- 1. Bateson P. 2005. The return of the whole organism. J Biosci 30:31–39 [DOI] [PubMed] [Google Scholar]
- 2. Crews D, McLachlan JA. 2006. Epigenetics, evolution, endocrine disruption, health, and disease. Endocrinology 147:S4–S10 [DOI] [PubMed] [Google Scholar]
- 3. Hochberg Z. 2009. Evo-devo of child growth II: human life history and transition between its phases. Eur J Endocrinol 160:135–141 [DOI] [PubMed] [Google Scholar]
- 4. Barker DJ. 1995. Fetal origins of coronary heart disease. BMJ 311:171–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barker DJ. 1992. The fetal origins of adult hypertension. J Hypertens 10:S39–S44 [PubMed] [Google Scholar]
- 6. Barker DJP. 1995. The fetal and infant origins of disease. Eur J Clin Invest 25:457–463 [DOI] [PubMed] [Google Scholar]
- 7. Barker DJ. 1998. In utero programming of chronic disease. Clin Sci 95:115–128 [PubMed] [Google Scholar]
- 8. Gluckman PD, Hanson MA, Cooper C, Thornburg KL. 2008. Effect of in utero and early-life conditions on adult health and disease. N Engl J Med 359:61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Forsdahl A. 1977. Are poor living conditions in childhood and adolescence an important risk factor for arteriosclerotic heart disease? Br J Prev Soc Med 31:91–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barker DJ, Osmond C. 1986. Infant mortality, childhood nutrition, and ischaemic heart disease in England and Wales. Lancet 327:1077–1081 [DOI] [PubMed] [Google Scholar]
- 11. Barker DJ. 1992. Fetal growth and adult disease. BJOG 99:275–276 [DOI] [PubMed] [Google Scholar]
- 12. Barker DJ, Martyn CN. 1992. The maternal and fetal origins of cardiovascular disease. J Epidemiol Community Health 46:8–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Law CM, Barker DJ, Osmond C, Fall CH, Simmonds SJ. 1992. Early growth and abdominal fatness in adult life. J Epidemiol Community Health 46:184–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barker DJ. 1995. The fetal origins of adult disease. Proc R Soc Lond B Biol Sci 262:37–43 [DOI] [PubMed] [Google Scholar]
- 15. Barker DJP. 1997. Fetal nutrition and cardiovascular disease in later life. Br Med Bull 53:96–108 [DOI] [PubMed] [Google Scholar]
- 16. Godfrey KM, Barker DJ. 2001. Fetal programming and adult health. Public Health Nutr 4:611–624 [DOI] [PubMed] [Google Scholar]
- 17. Gale CR, Javaid MK, Robinson SM, Law CM, Godfrey KM, Cooper C. 2007. Maternal size in pregnancy and body composition in children. J Clin Endocrinol Metab 92:3904–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bateson P, Barker D, Clutton-Brock T, Deb D, D'Udine B, Foley RA, Gluckman P, Godfrey K, Kirkwood T, Lahr MM, McNamara J, Metcalfe NB, Monaghan P, Spencer HG, Sultan SE. 2004. Developmental plasticity and human health. Nature 430:419–421 [DOI] [PubMed] [Google Scholar]
- 19. Gluckman PD, Hanson MA. 2004. Maternal constraint of fetal growth and its consequences. Semin Fetal Neonatal Med 9:419–425 [DOI] [PubMed] [Google Scholar]
- 20. Gluckman PD, Hanson MA. 2004. The developmental origins of the metabolic syndrome. Trends Endocrinol Metab 15:183–187 [DOI] [PubMed] [Google Scholar]
- 21. McMillen IC, Robinson JS. 2005. Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiol Rev 85:571–633 [DOI] [PubMed] [Google Scholar]
- 22. Gluckman PD, Lillycrop KA, Vickers MH, Pleasants AB, Phillips ES, Beedle AS, Burdge GC, Hanson MA. 2007. Metabolic plasticity during mammalian development is directionally dependent on early nutritional status. Proc Natl Acad Sci USA 104:12796–12800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gluckman PD, Hanson MA, Beedle AS. 2007. Early life events and their consequences for later disease: a life history and evolutionary perspective. Am J Hum Biol 19:1–19 [DOI] [PubMed] [Google Scholar]
- 24. Gluckman PD, Hanson MA. 2004. Developmental origins of disease paradigm: a mechanistic and evolutionary perspective. Pediatr Res 56:311–317 [DOI] [PubMed] [Google Scholar]
- 25. Agrawal AA, Laforsch C, Tollrian R. 1999. Transgenerational induction of defences in animals and plants. Nature 401:60–63 [Google Scholar]
- 26. Anstey ML, Rogers SM, Ott SR, Burrows M, Simpson SJ. 2009. Serotonin mediates behavioral gregarization underlying swarm formation in desert locusts. Science 323:627–630 [DOI] [PubMed] [Google Scholar]
- 27. Lee TM, Spears N, Tuthill CR, Zucker I. 1989. Maternal melatonin treatment influences rates of neonatal development of meadow vole pups. Biol Reprod 40:495–502 [DOI] [PubMed] [Google Scholar]
- 28. Sultan SE. 2003. Commentary: the promise of ecological developmental biology. J Exp Zool B Mol Dev Evol 296:1–7 [DOI] [PubMed] [Google Scholar]
- 29. Waddington CH. 1942. Canalization of development and the inheritance of acquired characters. Nature 150:563–565 [DOI] [PubMed] [Google Scholar]
- 30. Vickers MH, Breier BH, McCarthy D, Gluckman PD. 2003. Sedentary behavior during postnatal life is determined by the prenatal environment and exacerbated by postnatal hypercaloric nutrition. Am J Physiol Regul Integr Comp Physiol 285:R271–R273 [DOI] [PubMed] [Google Scholar]
- 31. Arcaleni E. 2006. Secular trend and regional differences in the stature of Italians, 1854–1980. Econ Hum Biol 4:24–38 [DOI] [PubMed] [Google Scholar]
- 32. Stein AD, Barnhart HX, Wang M, Hoshen MB, Ologoudou K, Ramakrishnan U, Grajeda R, Ramirez-Zea M, Martorell R. 2004. Comparison of linear growth patterns in the first three years of life across two generations in Guatemala. Pediatrics 113:e270–e275 [DOI] [PubMed] [Google Scholar]
- 33. Müller GB. 2007. Evo-devo: extending the evolutionary synthesis. Nat Rev Genet 8:943–949 [DOI] [PubMed] [Google Scholar]
- 34. Hochberg Z, Albertsson-Wikland K. 2008. Evo-devo of infantile and childhood growth. Pediatr Res 64:2–7 [DOI] [PubMed] [Google Scholar]
- 35. Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjöström M, Golding J. 2006. Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet 14:159–166 [DOI] [PubMed] [Google Scholar]
- 36. Bogin B, Silva MI, Rios L. 2007. Life history trade-offs in human growth: adaptation or pathology? Am J Hum Biol 19:631–642 [DOI] [PubMed] [Google Scholar]
- 37. Pratt JH, Manatunga AK, Li W. 1994. Familial influences on the adrenal androgen excretion rate during the adrenarche. Metabolism 43:186–189 [DOI] [PubMed] [Google Scholar]
- 38. Hochberg Z. 2008. Juvenility in the context of life history theory. Arch Dis Child 93:534–539 [DOI] [PubMed] [Google Scholar]
- 39. Belsky J, Fearon RM. 2002. Early attachment security, subsequent maternal sensitivity, and later child development: does continuity in development depend upon continuity of caregiving? Attach Hum Dev 4:361–387 [DOI] [PubMed] [Google Scholar]
- 40. Del Giudice M. 2009. Sex, attachment, and the development of reproductive strategies. Behav Brain Sci 32:1–67 [DOI] [PubMed] [Google Scholar]
- 41. Goodman CS, Coughlin BC. 2000. The evolution of evo-devo biology. Proc Natl Acad Sci USA 97:4424–4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu YX, Jalil F, Karlberg J. 1998. Growth stunting in early life in relation to the onset of the childhood component of growth. J Pediatr Endocrinol Metab 11:247–260 [DOI] [PubMed] [Google Scholar]
- 43. Liu Y, Albertsson-Wikland K, Karlberg J. 2000. Long-term consequences of early linear growth retardation (stunting) in Swedish children. Pediatr Res 47:475–480 [DOI] [PubMed] [Google Scholar]
- 44. Kuzawa CW. 2007. Developmental origins of life history: growth, productivity, and reproduction. Am J Hum Biol 19:654–661 [DOI] [PubMed] [Google Scholar]
- 45. Feinberg AP. 2007. Phenotypic plasticity and the epigenetics of human disease. Nature 447:433–440 [DOI] [PubMed] [Google Scholar]
- 46. Haig D. 2004. The (dual) origin of epigenetics. Cold Spring Harb Symp Quant Biol 69:67–70 [DOI] [PubMed] [Google Scholar]
- 47. Bird A. 2007. Perceptions of epigenetics. Nature 447:396–398 [DOI] [PubMed] [Google Scholar]
- 48. Waddington CH. 1957. The strategy of the genes: a discussion of some aspects of theoretical biology. London: Allen and Unwin [Google Scholar]
- 49. Goldberg AD, Allis CD, Bernstein E. 2007. Epigenetics: a landscape takes shape. Cell 128:635–638 [DOI] [PubMed] [Google Scholar]
- 50. Hemberger M. 2007. Epigenetic landscape required for placental development. Cell Mol Life Sci 64:2422–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Grunstein M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349–352 [DOI] [PubMed] [Google Scholar]
- 52. Delaval K, Feil R. 2004. Epigenetic regulation of mammalian genomic imprinting. Curr Opin Genet Dev 14:188–195 [DOI] [PubMed] [Google Scholar]
- 53. Feil R, Berger F. 2007. Convergent evolution of genomic imprinting in plants and mammals. Trends Genet 23:192–199 [DOI] [PubMed] [Google Scholar]
- 54. Rakyan VK, Preis J, Morgan HD, Whitelaw E. 2001. The marks, mechanisms and memory of epigenetic states in mammals. Biochem J 356:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Waterland RA, Michels KB. 2007. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr 27:363–388 [DOI] [PubMed] [Google Scholar]
- 56. Kelley RL, Meller VH, Gordadze PR, Roman G, Davis RL, Kuroda MI. 1999. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 98:513–522 [DOI] [PubMed] [Google Scholar]
- 57. Illingworth RS, Bird AP. 2009. CpG islands—‘A rough guide’. FEBS Lett 583:1713–1720 [DOI] [PubMed] [Google Scholar]
- 58. Ching TT, Maunakea AK, Jun P, Hong C, Zardo G, Pinkel D, Albertson DG, Fridlyand J, Mao JH, Shchors K, Weiss WA, Costello JF. 2005. Epigenome analyses using BAC microarrays identify evolutionary conservation of tissue-specific methylation of SHANK3. Nat Genet 37:645–651 [DOI] [PubMed] [Google Scholar]
- 59. Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, Shu J, Chen X, Waterland RA, Issa JP. 2007. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet 3:2023–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ooi SK, O'Donnell AH, Bestor TH. 2009. Mammalian cytosine methylation at a glance. J Cell Sci 122:2787–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jones PA, Liang G. 2009. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet 10:805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jirtle RL, Skinner MK. 2007. Environmental epigenomics and disease susceptibility. Nat Rev Genet 8:253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Reik W. 2007. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447:425–432 [DOI] [PubMed] [Google Scholar]
- 64. Gehring M, Reik W, Henikoff S. 2009. DNA demethylation by DNA repair. Trends Genet 25:82–90 [DOI] [PubMed] [Google Scholar]
- 65. Reik W, Walter J. 2001. Genomic imprinting: parental influence on the genome. Nat Rev Genet 2:21–32 [DOI] [PubMed] [Google Scholar]
- 66. Reik W, Dean W, Walter J. 2001. Epigenetic reprogramming in mammalian development. Science 293:1089–1093 [DOI] [PubMed] [Google Scholar]
- 67. Ng RK, Dean W, Dawson C, Lucifero D, Madeja Z, Reik W, Hemberger M. 2008. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat Cell Biol 10:1280–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hemberger M, Dean W, Reik W. 2009. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nat Rev Mol Cell Biol 10:526–537 [DOI] [PubMed] [Google Scholar]
- 69. Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schübeler D. 2005. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37:853–862 [DOI] [PubMed] [Google Scholar]
- 70. Rakyan VK, Down TA, Thorne NP, Flicek P, Kulesha E, Gräf S, Tomazou EM, Bäckdahl L, Johnson N, Herberth M, Howe KL, Jackson DK, Miretti MM, Fiegler H, Marioni JC, Birney E, Hubbard TJ, Carter NP, Tavaré S, Beck S. 2008. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs). Genome Res 18:1518–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462:315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Margueron R, Reinberg D. 2010. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet 11:285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kouzarides T. 2007. Chromatin modifications and their function. Cell 128:693–705 [DOI] [PubMed] [Google Scholar]
- 74. Roh TY, Cuddapah S, Zhao K. 2005. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev 19:542–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kuo MH, Allis CD. 1998. Roles of histone acetyltransferases and deacetylases in gene regulation. BioEssays 20:615–626 [DOI] [PubMed] [Google Scholar]
- 76. Davie JR, Chadee DN. 1998. Regulation and regulatory parameters of histone modifications. J Cell Biochem Suppl 30–31:203–213 [PubMed] [Google Scholar]
- 77. Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293:1074–1080 [DOI] [PubMed] [Google Scholar]
- 78. Haberland M, Montgomery RL, Olson EN. 2009. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet 10:32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhang Y, Reinberg D. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 15: 2343–2360 [DOI] [PubMed] [Google Scholar]
- 80. Cloos PA, Christensen J, Agger K, Helin K. 2008. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev 22:1115–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823–837 [DOI] [PubMed] [Google Scholar]
- 82. Latham JA, Dent SYR. 2007. Cross-regulation of histone modifications. Nat Struct Mol Biol 14:1017–1024 [DOI] [PubMed] [Google Scholar]
- 83. Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438:1116–1122 [DOI] [PubMed] [Google Scholar]
- 84. Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. 2007. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131:58–69 [DOI] [PubMed] [Google Scholar]
- 85. Li S, Shang Y. 2007. Regulation of SRC family coactivators by post-translational modifications. Cell Signal 19:1101–1112 [DOI] [PubMed] [Google Scholar]
- 86. Shilatifard A. 2008. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol 20:341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lau NC, Lai EC. 2005. Diverse roles for RNA in gene regulation. Genome Biol 6:315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. 2009. RNA regulation of epigenetic processes. BioEssays 31:51–59 [DOI] [PubMed] [Google Scholar]
- 89. Mercer TR, Dinger ME, Mattick JS. 2009. Long non-coding RNAs: insights into functions. Nat Rev Genet 10:155–159 [DOI] [PubMed] [Google Scholar]
- 90. Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. 2005. Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol Cell 19:381–391 [DOI] [PubMed] [Google Scholar]
- 91. Probst AV, Dunleavy E, Almouzni G. 2009. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol 10:192–206 [DOI] [PubMed] [Google Scholar]
- 92. Santos F, Peters AH, Otte AP, Reik W, Dean W. 2005. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev Biol 280:225–236 [DOI] [PubMed] [Google Scholar]
- 93. Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA. 2008. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 452:877–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Morgan HD, Santos F, Green K, Dean W, Reik W. 2005. Epigenetic reprogramming in mammals. Hum Mol Genet 14:R47–R58 [DOI] [PubMed] [Google Scholar]
- 95. Hayashi K, Surani MA. 2009. Resetting the epigenome beyond pluripotency in the germline. Cell Stem Cell 4:493–498 [DOI] [PubMed] [Google Scholar]
- 96. Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. 2010. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463:1101–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ray-Gallet D, Almouzni G. 2010. Mixing or not mixing. Science 328:56–57 [DOI] [PubMed] [Google Scholar]
- 98. Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. 2010. Partitioning of histone H3–H4 tetramers during DNA replication-dependent chromatin assembly. Science 328:94–98 [DOI] [PubMed] [Google Scholar]
- 99. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811 [DOI] [PubMed] [Google Scholar]
- 100. Moazed D. 2009. Small RNAs in transcriptional gene silencing and genome defence. Nature 457:413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. 2010. Non-coding RNAs: regulators of disease. J Path 220:126–139 [DOI] [PubMed] [Google Scholar]
- 102. Kuehbacher A, Urbich C, Dimmeler S. 2008. Targeting microRNA expression to regulate angiogenesis. Trends Pharmacol Sci 29:12–15 [DOI] [PubMed] [Google Scholar]
- 103. Büssing I, Slack FJ, Grosshans H. 2008. let-7 MicroRNAs in development, stem cells and cancer. Trends Mol Med 14:400–409 [DOI] [PubMed] [Google Scholar]
- 104. Roush S, Slack FJ. 2008. The let-7 family of microRNAs. Trends Cell Biol 18:505–516 [DOI] [PubMed] [Google Scholar]
- 105. Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. 2008. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol 10:987–993 [DOI] [PubMed] [Google Scholar]
- 106. Ong KK, Elks CE, Li S, Zhao JH, Luan J, Andersen LB, Bingham SA, Brage S, Smith GD, Ekelund U, Gillson CJ, Glaser B, Golding J, Hardy R, Khaw KT, Kuh D, Luben R, Marcus M, McGeehin MA, Ness AR, Northstone K, Ring SM, Rubin C, Sims MA, Song K, Strachan DP, Vollenweider P, Waeber G, Waterworth DM, Wong A, Deloukas P, Barroso I, Mooser V, Loos RJ, Wareham NJ. 2009. Genetic variation in LIN28B is associated with the timing of puberty. Nat Genet 41:729–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Verdel A, Vavasseur A, Le Gorrec M, Touat-Todeschini L. 2009. Common themes in siRNA-mediated epigenetic silencing pathways. Int J Dev Biol 53:245–257 [DOI] [PubMed] [Google Scholar]
- 108. Gonzalez S, Pisano DG, Serrano M. 2008. Mechanistic principles of chromatin remodeling guided by siRNAs and miRNAs. Cell Cycle 7:2601–2608 [DOI] [PubMed] [Google Scholar]
- 109. Kim DH, Saetrom P, Snøve O, Jr, Rossi JJ. 2008. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA 105:16230–16235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Bayne EH, Allshire RC. 2005. RNA-directed transcriptional gene silencing in mammals. Trends Genet 21:370–373 [DOI] [PubMed] [Google Scholar]
- 111. Aravin AA, Hannon GJ, Brennecke J. 2007. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318:761–764 [DOI] [PubMed] [Google Scholar]
- 112. Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12:503–514 [DOI] [PubMed] [Google Scholar]
- 113. Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316:1484–1488 [DOI] [PubMed] [Google Scholar]
- 114. Wilusz JE, Sunwoo H, Spector DL. 2009. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 23:1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. 2007. Functional demarcation of active and silent chromatin domains in human HOX Loci by noncoding RNAs. Cell 129:1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. 2006. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev 20:1470–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD. 2009. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci 12:1020–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Hsieh J, Eisch AJ. 2010. Epigenetics, hippocampal neurogenesis, and neuropsychiatric disorders: unraveling the genome to understand the mind. Neurobiol Dis 39:73–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Reik W, Lewis A. 2005. Co-evolution of X-chromosome inactivation and imprinting in mammals. Nat Rev Genet 6:403–410 [DOI] [PubMed] [Google Scholar]
- 120. Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. 2008. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322:1717–1720 [DOI] [PubMed] [Google Scholar]
- 121. Terranova R, Yokobayashi S, Stadler MB, Otte AP, van Lohuizen M, Orkin SH, Peters AH. 2008. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev Cell 15:668–679 [DOI] [PubMed] [Google Scholar]
- 122. Redrup L, Branco MR, Perdeaux ER, Krueger C, Lewis A, Santos F, Nagano T, Cobb BS, Fraser P, Reik W. 2009. The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development 136:525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Chaumeil J, Le Baccon P, Wutz A, Heard E. 2006. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev 20:2223–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Sahoo T, del Gaudio D, German JR, Shinawi M, Peters SU, Person RE, Garnica A, Cheung SW, Beaudet AL. 2008. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet 40:719–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Ikegami K, Ohgane J, Tanaka S, Yagi S, Shiota K. 2009. Interplay between DNA methylation, histone modification and chromatin remodeling in stem cells and during development. Int J Dev Biol 53:203–214 [DOI] [PubMed] [Google Scholar]
- 126. Fatemi M, Wade PA. 2006. MBD family proteins: reading the epigenetic code. J Cell Sci 119:3033–3037 [DOI] [PubMed] [Google Scholar]
- 127. Dhasarathy A, Wade PA. 2008. The MBD protein family—reading an epigenetic mark? Mutat Res 647:39–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Brunmeir R, Lagger S, Seiser C. 2009. Histone deacetylase 1 and 2-controlled embryonic development and cell differentiation. Int J Dev Biol 53:275–289 [DOI] [PubMed] [Google Scholar]
- 129. Sasai N, Defossez PA. 2009. Many paths to one goal? The proteins that recognize methylated DNA in eukaryotes. Int J Dev Biol 53:323–334 [DOI] [PubMed] [Google Scholar]
- 130. Ringrose L, Paro R. 2004. Epigenetic regulation of cellular memory by the Polycomb and trithorax group proteins. Annu Rev Genet 38:413–443 [DOI] [PubMed] [Google Scholar]
- 131. Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441:349–353 [DOI] [PubMed] [Google Scholar]
- 132. Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D. 2008. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol 28:2718–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. 2006. Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8:532–538 [DOI] [PubMed] [Google Scholar]
- 134. Tiwari VK, McGarvey KM, Licchesi JD, Ohm JE, Herman JG, Schübeler D, Baylin SB. 2008. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol 6:2911–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315–326 [DOI] [PubMed] [Google Scholar]
- 136. Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448:553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G. 2008. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell 2:160–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454:766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448:714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. 2008. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456:125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Ooi SL, Henikoff S. 2007. Germline histone dynamics and epigenetics. Curr Opin Cell Biol 19:257–265 [DOI] [PubMed] [Google Scholar]
- 142. Ciccone DN, Chen T. 2009. Histone lysine methylation in genomic imprinting. Epigenetics 4:216–220 [DOI] [PubMed] [Google Scholar]
- 143. Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 13:1192–1200 [DOI] [PubMed] [Google Scholar]
- 144. Espada J, Ballestar E, Fraga MF, Villar-Garea A, Juarranz A, Stockert JC, Robertson KD, Fuks F, Esteller M. 2004. Human DNA methyltransferase 1 is required for maintenance of the histone H3 modification pattern. J Biol Chem 279:37175–37184 [DOI] [PubMed] [Google Scholar]
- 145. Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S, Yang HW, Lam LL, Mager DL, Schübeler D, Tachibana M, Shinkai Y, Lorincz MC. 2008. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J 27:2691–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Kim JK, Samaranayake M, Pradhan S. 2009. Epigenetic mechanisms in mammals. Cell Mol Life Sci 66:596–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Ho L, Crabtree GR. 2010. Chromatin remodelling during development. Nature 463:474–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Bushey AM, Dorman ER, Corces VG. 2008. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell 32:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. 2006. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell 24:309–316 [DOI] [PubMed] [Google Scholar]
- 150. Morrison AJ, Shen X. 2009. Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat Rev Mol Cell Biol 10:373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Tateishi K, Okada Y, Kallin EM, Zhang Y. 2009. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature 458:757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Haigis MC, Guarente LP. 2006. Mammalian sirtuins—emerging roles in physiology, aging, and calorie restriction. Genes Dev 20:2913–2921 [DOI] [PubMed] [Google Scholar]
- 153. Vaquero A. 2009. The conserved role of sirtuins in chromatin regulation. Int J Dev Biol 53:303–322 [DOI] [PubMed] [Google Scholar]
- 154. Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. 2009. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324:1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. 2006. Nucleocytosolic acetyl-coenzyme A synthetase is required for histone acetylation and global transcription. Mol Cell 23:207–217 [DOI] [PubMed] [Google Scholar]
- 156. Rathmell JC, Newgard CB. 2009. A glucose-to-gene link. Science 324:1021–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676 [DOI] [PubMed] [Google Scholar]
- 158. Whitelaw NC, Whitelaw E. 2008. Transgenerational epigenetic inheritance in health and disease. Curr Opin Genet Dev 18:273–279 [DOI] [PubMed] [Google Scholar]
- 159. Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W. 2003. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35:88–93 [DOI] [PubMed] [Google Scholar]
- 160. Edwards CA, Ferguson-Smith AC. 2007. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol 19:281–289 [DOI] [PubMed] [Google Scholar]
- 161. Kangaspeska S, Stride B, Métivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G. 2008. Transient cyclical methylation of promoter DNA. Nature 452:112–115 [DOI] [PubMed] [Google Scholar]
- 162. Métivier R, Gallais R, Tiffoche C, Le Péron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, Gannon F, Salbert G. 2008. Cyclical DNA methylation of a transcriptionally active promoter. Nature 452:45–50 [DOI] [PubMed] [Google Scholar]
- 163. Guibert S, Forne T, Weber M. 2009. Dynamic regulation of DNA methylation during mammalian development. Epigenomics 1:81–98 [DOI] [PubMed] [Google Scholar]
- 164. Waterland RA, Garza C. 1999. Potential mechanisms of metabolic imprinting that lead to chronic disease. Am J Clin Nutr 69:179–197 [DOI] [PubMed] [Google Scholar]
- 165. Ozanne SE, Constância M. 2007. Mechanisms of disease: the developmental origins of disease and the role of the epigenotype. Nat Clin Pract Endocrinol Metab 3:539–546 [DOI] [PubMed] [Google Scholar]
- 166. Jaenisch R, Bird A. 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 33:245–254 [DOI] [PubMed] [Google Scholar]
- 167. Skinner MK, Manikkam M, Guerrero-Bosagna C. 2010. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol Metab 21:214–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Anway MD, Cupp AS, Uzumcu M, Skinner MK. 2005. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 308:1466–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Park JH, Stoffers DA, Nicholls RD, Simmons RA. 2008. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest 118:2316–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Wolff GL, Kodell RL, Moore SR, Cooney CA. 1998. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J 12:949–957 [PubMed] [Google Scholar]
- 171. Calvanese V, Lara E, Kahn A, Fraga MF. 2009. The role of epigenetics in aging and age-related diseases. Ageing Res Rev 8:268–276 [DOI] [PubMed] [Google Scholar]
- 172. Teixeira FK, Heredia F, Sarazin A, Roudier F, Boccara M, Ciaudo C, Cruaud C, Poulain J, Berdasco M, Fraga MF, Voinnet O, Wincker P, Esteller M, Colot V. 2009. A role for RNAi in the selective correction of DNA methylation defects. Science 323:1600–1604 [DOI] [PubMed] [Google Scholar]
- 173. Morgan HD, Sutherland HG, Martin DI, Whitelaw E. 1999. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet 23:314–318 [DOI] [PubMed] [Google Scholar]
- 174. Heijmans BT, Kremer D, Tobi EW, Boomsma DI, Slagboom PE. 2007. Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum Mol Genet 16:547–554 [DOI] [PubMed] [Google Scholar]
- 175. Kaminsky ZA, Tang T, Wang SC, Ptak C, Oh GH, Wong AH, Feldcamp LA, Virtanen C, Halfvarson J, Tysk C, McRae AF, Visscher PM, Montgomery GW, Gottesman II, Martin NG, Petronis A. 2009. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet 41:240–245 [DOI] [PubMed] [Google Scholar]
- 176. Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. 2000. Demethylation of the zygotic paternal genome. Nature 403:501–502 [DOI] [PubMed] [Google Scholar]
- 177. Hart N. 1993. Famine, maternal nutrition and infant mortality: a re-examination of the Dutch hunger winter. Popul Stud 47:27–46 [DOI] [PubMed] [Google Scholar]
- 178. Stein AD, Ravelli AC, Lumey LH. 1995. Famine, third-trimester pregnancy weight gain, and intrauterine growth: the Dutch famine birth cohort study. Hum Biol 67:135–150 [PubMed] [Google Scholar]
- 179. Lumey LH, Stein AD. 1997. In utero exposure to famine and subsequent fertility: the Dutch famine birth cohort study. Am J Public Health 87:1962–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Lumey LH, Stein AD. 1997. Offspring birth weights after maternal intrauterine undernutrition: a comparison within sibships. Am J Epidemiol 146:810–819 [DOI] [PubMed] [Google Scholar]
- 181. Ravelli AC, van der Meulen JH, Michels RP, Osmond C, Barker DJ, Hales CN, Bleker OP. 1998. Glucose tolerance in adults after prenatal exposure to famine. Lancet 351:173–177 [DOI] [PubMed] [Google Scholar]
- 182. Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. 2008. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA 105:17046–17049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Dolinoy DC, Das R, Weidman JR, Jirtle RL. 2007. Metastable epialleles, imprinting, and the fetal origins of adult diseases. Pediatr Res 61:30R–37R [DOI] [PubMed] [Google Scholar]
- 184. Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, Luu KV, Whitelaw E. 2003. Transgenerational inheritance of epigenetic states at the murine AxinFu allele occurs after maternal and paternal transmission. Proc Natl Acad Sci USA 100:2538–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Fraga MF. 2009. Genetic and epigenetic regulation of aging. Curr Opin Immunol 21:446–453 [DOI] [PubMed] [Google Scholar]
- 186. Whitelaw NC, Whitelaw E. 2006. How lifetimes shape epigenotype within and across generations. Hum Mol Genet 15:R131–R137 [DOI] [PubMed] [Google Scholar]
- 187. Blewitt ME, Vickaryous NK, Hemley SJ, Ashe A, Bruxner TJ, Preis JI, Arkell R, Whitelaw E. 2005. An N-ethyl-N-nitrosourea screen for genes involved in variegation in the mouse. Proc Natl Acad Sci USA 102:7629–7634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Hsieh J, Fire A. 2000. Recognition and silencing of repeated DNA. Annu Rev Genet 34:187–204 [DOI] [PubMed] [Google Scholar]
- 189. Ashe A, Morgan DK, Whitelaw NC, Bruxner TJ, Vickaryous NK, Cox LL, Butterfield NC, Wicking C, Blewitt ME, Wilkins SJ, Anderson GJ, Cox TC, Whitelaw E. 2008. A genome-wide screen for modifiers of transgene variegation identifies genes with critical roles in development. Genome Biol 9:R182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Blewitt ME, Gendrel AV, Pang Z, Sparrow DB, Whitelaw N, Craig JM, Apedaile A, Hilton DJ, Dunwoodie SL, Brockdorff N, Kay GF, Whitelaw E. 2008. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet 40:663–669 [DOI] [PubMed] [Google Scholar]
- 191. Chong S, Vickaryous N, Ashe A, Zamudio N, Youngson N, Hemley S, Stopka T, Skoultchi A, Matthews J, Scott HS, de Kretser D, O'Bryan M, Blewitt M, Whitelaw E. 2007. Modifiers of epigenetic reprogramming show paternal effects in the mouse. Nat Genet 39:614–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Esteller M. 2008. Epigenetics in cancer. N Engl J Med 358:1148–1159 [DOI] [PubMed] [Google Scholar]
- 193. Ong KK, Ahmed ML, Emmett PM, Preece MA, Dunger DB. 2000. Association between postnatal catch-up growth and obesity in childhood: prospective cohort study. BMJ 320:967–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Parent AS, Teilmann G, Juul A, Skakkebaek NE, Toppari J, Bourguignon JP. 2003. The timing of normal puberty and the age limits of sexual precocity: variations around the world, secular trends, and changes after migration. Endocr Rev 24:668–693 [DOI] [PubMed] [Google Scholar]
- 195. McCance RA, Widdowson EM. 1962. Nutrition and growth. Proc R Soc Lond B Biol Sci 156:326–335 [Google Scholar]
- 196. McCance RA, Widdowson EM. 1974. The determinants of growth and form. Proc R Soc Lond B Biol Sci 185:1–17 [DOI] [PubMed] [Google Scholar]
- 197. Wilcox AJ, Skjaerven R. 1992. Birth weight and perinatal mortality: the effect of gestational age. Am J Public Health 82:378–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Jolley CD. 2003. Failure to thrive. Curr Probl Pediatr Adolesc Health Care 33:183–206 [DOI] [PubMed] [Google Scholar]
- 199. Migliano AB, Vinicius L, Lahr MM. 2007. Life history trade-offs explain the evolution of human pygmies. Proc Natl Acad Sci USA 104:20216–20219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Ekelund U, Ong KK, Linné Y, Neovius M, Brage S, Dunger DB, Wareham NJ, Rössner S. 2007. Association of weight gain in infancy and early childhood with metabolic risk in young adults. J Clin Endocrinol Metab 92:98–103 [DOI] [PubMed] [Google Scholar]
- 201. Owen CG, Martin RM, Whincup PH, Smith GD, Cook DG. 2005. Effect of infant feeding on the risk of obesity across the life course: a quantitative review of published evidence. Pediatrics 115:1367–1377 [DOI] [PubMed] [Google Scholar]
- 202. Singhal A, Farooqi IS, Cole TJ, O'Rahilly S, Fewtrell M, Kattenhorn M, Lucas A, Deanfield J. 2002. Influence of leptin on arterial distensibility: a novel link between obesity and cardiovascular disease? Circulation 106:1919–1924 [DOI] [PubMed] [Google Scholar]
- 203. Singhal A, Farooqi IS, O'Rahilly S, Cole TJ, Fewtrell M, Lucas A. 2002. Early nutrition and leptin concentrations in later life. Am J Clin Nutr 75:993–999 [DOI] [PubMed] [Google Scholar]
- 204. Singhal A, Wells J, Cole TJ, Fewtrell M, Lucas A. 2003. Programming of lean body mass: a link between birth weight, obesity, and cardiovascular disease? Am J Clin Nutr 77:726–730 [DOI] [PubMed] [Google Scholar]
- 205. Singhal A, Cole TJ, Fewtrell M, Deanfield J, Lucas A. 2004. Is slower early growth beneficial for long-term cardiovascular health? Circulation 109:1108–1113 [DOI] [PubMed] [Google Scholar]
- 206. Singhal A, Cole TJ, Fewtrell M, Kennedy K, Stephenson T, Elias-Jones A, Lucas A. 2007. Promotion of faster weight gain in infants born small for gestational age: is there an adverse effect on later blood pressure? Circulation 115:213–220 [DOI] [PubMed] [Google Scholar]
- 207. Ong KK, Northstone K, Wells JC, Rubin C, Ness AR, Golding J, Dunger DB. 2007. Earlier mother's age at menarche predicts rapid infancy growth and childhood obesity. PLoS Med 4:e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Lakshman R, Forouhi NG, Sharp SJ, Luben R, Bingham SA, Khaw KT, Wareham NJ, Ong KK. 2009. Early age at menarche associated with cardiovascular disease and mortality. J Clin Endocrinol Metab 94:4953–4960 [DOI] [PubMed] [Google Scholar]
- 209. Eriksson JG, Forsén T, Tuomilehto J, Osmond C, Barker DJ. 2003. Early adiposity rebound in childhood and risk of type 2 diabetes in adult life. Diabetologia 46:190–194 [DOI] [PubMed] [Google Scholar]
- 210. Wehkalampi K, Silventoinen K, Kaprio J, Dick DM, Rose RJ, Pulkkinen L, Dunkel L. 2008. Genetic and environmental influences on pubertal timing assessed by height growth. Am J Hum Biol 20:417–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Gicquel C, Rossignol S, Cabrol S, Houang M, Steunou V, Barbu V, Danton F, Thibaud N, Le Merrer M, Burglen L, Bertrand AM, Netchine I, Le Bouc Y. 2005. Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver-Russell syndrome. Nat Genet 37:1003–1007 [DOI] [PubMed] [Google Scholar]
- 212. Rossignol S, Netchine I, Le Bouc Y, Gicquel C. 2008. Epigenetics in Silver-Russell syndrome. Best Pract Res Clin Endocrinol Metab 22:403–414 [DOI] [PubMed] [Google Scholar]
- 213. Netchine I, Rossignol S, Dufourg MN, Azzi S, Rousseau A, Perin L, Houang M, Steunou V, Esteva B, Thibaud N, Demay MC, Danton F, Petriczko E, Bertrand AM, Heinrichs C, Carel JC, Loeuille GA, Pinto G, Jacquemont ML, Gicquel C, Cabrol S, Le Bouc Y. 2007. 11p15 Imprinting center region 1 loss of methylation is a common and specific cause of typical Russell-Silver syndrome: clinical scoring system and epigenetic-phenotypic correlations. J Clin Endocrinol Metab 92:3148–3154 [DOI] [PubMed] [Google Scholar]
- 214. Amselem S, Sobrier ML, Dastot F, Duquesnoy P, Duriez B, Goossens M. 1996. Molecular basis of inherited growth hormone resistance in childhood. Baillieres Clin Endocrinol Metab 10:353–369 [DOI] [PubMed] [Google Scholar]
- 215. Amit T, Youdim MB, Hochberg Z. 2000. Does serum growth hormone (GH) binding protein reflect human GH receptor function? J Clin Endocrinol Metab 85:927–932 [DOI] [PubMed] [Google Scholar]
- 216. Kofoed EM, Hwa V, Little B, Woods KA, Buckway CK, Tsubaki J, Pratt KL, Bezrodnik L, Jasper H, Tepper A, Heinrich JJ, Rosenfeld RG. 2003. Growth hormone insensitivity associated with a STAT5b mutation. N Engl J Med 349:1139–1147 [DOI] [PubMed] [Google Scholar]
- 217. Hwa V, Little B, Adiyaman P, Kofoed EM, Pratt KL, Ocal G, Berberoglu M, Rosenfeld RG. 2005. Severe growth hormone insensitivity resulting from total absence of signal transducer and activator of transcription 5b. J Clin Endocrinol Metab 90:4260–4266 [DOI] [PubMed] [Google Scholar]
- 218. Woods KA, Camacho-Hübner C, Savage MO, Clark AJ. 1996. Intrauterine growth retardation and postnatal growth failure associated with deletion of the insulin-like growth factor I gene. N Engl J Med 335:1363–1367 [DOI] [PubMed] [Google Scholar]
- 219. Walenkamp MJ, Karperien M, Pereira AM, Hilhorst-Hofstee Y, van Doorn J, Chen JW, Mohan S, Denley A, Forbes B, van Duyvenvoorde HA, van Thiel SW, Sluimers CA, Bax JJ, de Laat JA, Breuning MB, Romijn JA, Wit JM. 2005. Homozygous and heterozygous expression of a novel insulin-like growth factor-I mutation. J Clin Endocrinol Metab 90:2855–2864 [DOI] [PubMed] [Google Scholar]
- 220. Abuzzahab MJ, Schneider A, Goddard A, Grigorescu F, Lautier C, Keller E, Kiess W, Klammt J, Kratzsch J, Osgood D, Pfäffle R, Raile K, Seidel B, Smith RJ, Chernausek SD. Intrauterine Growth Retardation (IUGR) Study Group 2003. IGF-I receptor mutations resulting in intrauterine and postnatal growth retardation. N Engl J Med 349:2211–2222 [DOI] [PubMed] [Google Scholar]
- 221. Domené HM, Bengolea SV, Martínez AS, Ropelato MG, Pennisi P, Scaglia P, Heinrich JJ, Jasper HG. 2004. Deficiency of the circulating insulin-like growth factor system associated with inactivation of the acid-labile subunit gene. N Engl J Med 350:570–577 [DOI] [PubMed] [Google Scholar]
- 222. Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, Sanna S, Eyheramendy S, Voight BF, Butler JL, Guiducci C, Illig T, Hackett R, Heid IM, Jacobs KB, Lyssenko V, Uda M, Boehnke M, Chanock SJ, Groop LC, Hu FB, Isomaa B, Kraft P, Peltonen L, Salomaa V, Schlessinger D, Hunter DJ, Hayes RB, Abecasis GR, Wichmann HE, Mohlke KL, Hirschhorn JN. 2008. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet 40:584–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Tennessen JM, Thummel CS. 2008. Developmental timing: let-7 function conserved through evolution. Curr Biol 18:R707–R708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. He C, Kraft P, Chen C, Buring JE, Pare G, Hankinson SE, Chanock SJ, Ridker PM, Hunter DJ, Chasman DI. 2009. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet 41:724–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Perry JR, Stolk L, Franceschini N, Lunetta KL, Zhai G, McArdle PF, Smith AV, Aspelund T, Bandinelli S, Boerwinkle E, Cherkas L, Eiriksdottir G, Estrada K, Ferrucci L, Folsom AR, Garcia M, Gudnason V, Hofman A, Karasik D, Kiel DP, Launer LJ, van Meurs J, Nalls MA, Rivadeneira F, Shuldiner AR, Singleton A, Soranzo N, Tanaka T, Visser JA, Weedon MN, Wilson SG, Zhuang V, Streeten EA, Harris TB, Murray A, Spector TD, Demerath EW, Uitterlinden AG, Murabito JM. 2009. Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nat Genet 41:648–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Sulem P, Gudbjartsson DF, Rafnar T, Holm H, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Alexandersen P, Feenstra B, Boyd HA, Aben KK, Verbeek AL, Roeleveld N, Jonasdottir A, Styrkarsdottir U, Steinthorsdottir V, Karason A, Stacey SN, Gudmundsson J, Jakobsdottir M, Thorleifsson G, Hardarson G, Gulcher J, Kong A, Kiemeney LA, Melbye M, Christiansen C, Tryggvadottir L, Thorsteinsdottir U, Stefansson K. 2009. Genome-wide association study identifies sequence variants on 6q21 associated with age at menarche. Nat Genet 41:734–738 [DOI] [PubMed] [Google Scholar]
- 227. Viswanathan SR, Daley GQ, Gregory RI. 2008. Selective blockade of microRNA processing by Lin28. Science 320:97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Nimmo RA, Slack FJ. 2009. An elegant miRror: microRNAs in stem cells, developmental timing and cancer. Chromosoma 118:405–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Hartge P. 2009. Genetics of reproductive lifespan. Nat Genet 41:637–638 [DOI] [PubMed] [Google Scholar]
- 230. Delaval K, Govin J, Cerqueira F, Rousseaux S, Khochbin S, Feil R. 2007. Differential histone modifications mark mouse imprinting control regions during spermatogenesis. EMBO J 26:720–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Pannetier M, Feil R. 2007. Epigenetic stability of embryonic stem cells and developmental potential. Trends Biotechnol 25:556–562 [DOI] [PubMed] [Google Scholar]
- 232. Hirasawa R, Feil R. 2008. A KRAB domain zinc finger protein in imprinting and disease. Dev Cell 15:487–488 [DOI] [PubMed] [Google Scholar]
- 233. Mackay DJ, Callaway JL, Marks SM, White HE, Acerini CL, Boonen SE, Dayanikli P, Firth HV, Goodship JA, Haemers AP, Hahnemann JM, Kordonouri O, Masoud AF, Oestergaard E, Storr J, Ellard S, Hattersley AT, Robinson DO, Temple IK. 2008. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat Genet 40:949–951 [DOI] [PubMed] [Google Scholar]
- 234. Jiang YH, Bressler J, Beaudet AL. 2004. Epigenetics and human disease. Annu Rev Genomics Hum Genet 5:479–510 [DOI] [PubMed] [Google Scholar]
- 235. Arnaud P, Feil R. 2005. Epigenetic deregulation of genomic imprinting in human disorders and following assisted reproduction. Birth Defects Res C Embryo Today 75:81–97 [DOI] [PubMed] [Google Scholar]
- 236. Gaston V, Le Bouc Y, Soupre V, Burglen L, Donadieu J, Oro H, Audry G, Vazquez MP, Gicquel C. 2001. Analysis of the methylation status of the KCNQ1OT and H19 genes in leukocyte DNA for the diagnosis and prognosis of Beckwith-Wiedemann syndrome. Eur J Hum Genet 9:409–418 [DOI] [PubMed] [Google Scholar]
- 237. Cooper WN, Luharia A, Evans GA, Raza H, Haire AC, Grundy R, Bowdin SC, Riccio A, Sebastio G, Bliek J, Schofield PN, Reik W, Macdonald F, Maher ER. 2005. Molecular subtypes and phenotypic expression of Beckwith-Wiedemann syndrome. Eur J Hum Genet 13:1025–1032 [DOI] [PubMed] [Google Scholar]
- 238. Silver HK, Kiyasu W, George J, Deamer WC. 1953. Syndrome of congenital hemihypertrophy, shortness of stature, and elevated urinary gonadotrophins. Pediatrics 12:368–376 [PubMed] [Google Scholar]
- 239. Russell A. 1954. A syndrome of “intra-uterine” dwarfism recognizable at birth with cranio-facial dysostosis, disproportionately short arms and other anomalies (5 examples). Proc R Soc Med 47:1040–1044 [PubMed] [Google Scholar]
- 240. Schneid H, Seurin D, Vazquez MP, Gourmelen M, Cabrol S, Le Bouc Y. 1993. Parental allele specific methylation of the human insulin-like growth factor II gene and Beckwith-Wiedemann syndrome. J Med Genet 30:353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Reik W, Brown KW, Schneid H, Le Bouc Y, Bickmore W, Maher ER. 1995. Imprinting mutations in the Beckwith-Wiedemann syndrome suggested by an altered imprinting pattern in the IGF2-H19 domain. Hum Mol Genet 4:2379–2385 [DOI] [PubMed] [Google Scholar]
- 242. Gicquel C, Le Bouc Y. 2006. Hormonal regulation of fetal growth. Horm Res 65(Suppl 3):28–33 [DOI] [PubMed] [Google Scholar]
- 243. Eggermann T, Eggermann K, Schönherr N. 2008. Growth retardation versus overgrowth: Silver-Russell syndrome is genetically opposite to Beckwith-Wiedemann syndrome. Trends Genet 24:195–204 [DOI] [PubMed] [Google Scholar]
- 244. Mackay DJ, Hahnemann JM, Boonen SE, Poerksen S, Bunyan DJ, White HE, Durston VJ, Thomas NS, Robinson DO, Shield JP, Clayton-Smith J, Temple IK. 2006. Epimutation of the TNDM locus and the Beckwith-Wiedemann syndrome centromeric locus in individuals with transient neonatal diabetes mellitus. Hum Genet 119:179–184 [DOI] [PubMed] [Google Scholar]
- 245. Cassidy SB. 1997. Prader-Willi syndrome. J Med Genet 34:917–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Hoveyda N, Shield JP, Garrett C, Chong WK, Beardsall K, Bentsi-Enchill E, Mallya H, Thompson MH. 1999. Neonatal diabetes mellitus and cerebellar hypoplasia/agenesis: report of a new recessive syndrome. J Med Genet 36:700–704 [PMC free article] [PubMed] [Google Scholar]
- 247. Clayton-Smith J, Laan L. 2003. Angelman syndrome: a review of the clinical and genetic aspects. J Med Genet 40:87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Kotzot D. 2004. Advanced parental age in maternal uniparental disomy (UPD): implications for the mechanism of formation. Eur J Hum Genet 12:343–346 [DOI] [PubMed] [Google Scholar]
- 249. DeChiara TM, Efstratiadis A, Robertson EJ. 1990. A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature 345:78–80 [DOI] [PubMed] [Google Scholar]
- 250. DeChiara TM, Robertson EJ, Efstratiadis A. 1991. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64:849–859 [DOI] [PubMed] [Google Scholar]
- 251. Kido Y, Nakae J, Hribal ML, Xuan S, Efstratiadis A, Accili D. 2002. Effects of mutations in the insulin-like growth factor signaling system on embryonic pancreas development and β-cell compensation to insulin resistance. J Biol Chem 277:36740–36747 [DOI] [PubMed] [Google Scholar]
- 252. Gicquel C, El-Osta A, Le Bouc Y. 2008. Epigenetic regulation and fetal programming. Best Pract Res Clin Endocrinol Metab 22:1–16 [DOI] [PubMed] [Google Scholar]
- 253. Schneid H, Vazquez MP, Vacher C, Gourmelen M, Cabrol S, Le Bouc Y. 1997. The Beckwith-Wiedemann syndrome phenotype and the risk of cancer. Med Pediatr Oncol 28:411–415 [DOI] [PubMed] [Google Scholar]
- 254. Engel JR, Smallwood A, Harper A, Higgins MJ, Oshimura M, Reik W, Schofield PN, Maher ER. 2000. Epigenotype-phenotype correlations in Beckwith-Wiedemann syndrome. J Med Genet 37:921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Bliek J, Maas SM, Ruijter JM, Hennekam RC, Alders M, Westerveld A, Mannens MM. 2001. Increased tumour risk for BWS patients correlates with aberrant H19 and not KCNQ1OT1 methylation: occurrence of KCNQ1OT1 hypomethylation in familial cases of BWS. Hum Mol Genet 10:467–476 [DOI] [PubMed] [Google Scholar]
- 256. Weksberg R, Nishikawa J, Caluseriu O, Fei YL, Shuman C, Wei C, Steele L, Cameron J, Smith A, Ambus I, Li M, Ray PN, Sadowski P, Squire J. 2001. Tumor development in the Beckwith-Wiedemann syndrome is associated with a variety of constitutional molecular 11p15 alterations including imprinting defects of KCNQ1OT1. Hum Mol Genet 10:2989–3000 [DOI] [PubMed] [Google Scholar]
- 257. DeBaun MR, Niemitz EL, McNeil DE, Brandenburg SA, Lee MP, Feinberg AP. 2002. Epigenetic alterations of H19 and LIT1 distinguish patients with Beckwith-Wiedemann Syndrome with cancer and birth defects. Am J Hum Genet 70:604–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Waterland RA, Jirtle RL. 2003. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 23:5293–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Waterland RA, Dolinoy DC, Lin JR, Smith CA, Shi X, Tahiliani KG. 2006. Maternal methyl supplements increase offspring DNA methylation at Axin fused. Genesis 44:401–406 [DOI] [PubMed] [Google Scholar]
- 260. Waterland RA. 2008. Epigenetic epidemiology of obesity: application of epigenomic technology. Nutr Rev 66:S21–S23 [DOI] [PubMed] [Google Scholar]
- 261. Waterland RA, Travisano M, Tahiliani KG, Rached MT, Mirza S. 2008. Methyl donor supplementation prevents transgenerational amplification of obesity. Int J Obes (Lond) 32:1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Ferguson-Smith AC, Surani MA. 2001. Imprinting and the epigenetic asymmetry between parental genomes. Science 293:1086–1089 [DOI] [PubMed] [Google Scholar]
- 263. Skuse DH, James RS, Bishop DV, Coppin B, Dalton P, Aamodt-Leeper G, Bacarese-Hamilton M, Creswell C, McGurk R, Jacobs PA. 1997. Evidence from Turner's syndrome of an imprinted X-linked locus affecting cognitive function. Nature 387:705–708 [DOI] [PubMed] [Google Scholar]
- 264. Monk D, Wagschal A, Arnaud P, Müller PS, Parker-Katiraee L, Bourc'his D, Scherer SW, Feil R, Stanier P, Moore GE. 2008. Comparative analysis of human chromosome 7q21 and mouse proximal chromosome 6 reveals a placental-specific imprinted gene, TFPI2/Tfpi2, which requires EHMT2 and EED for allelic-silencing. Genome Res 18:1270–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Murrell A, Heeson S, Reik W. 2004. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet 36:889–893 [DOI] [PubMed] [Google Scholar]
- 266. Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. 2006. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci USA 103:10684–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Sleutels F, Zwart R, Barlow DP. 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415:810–813 [DOI] [PubMed] [Google Scholar]
- 268. Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM. 2006. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev 20:1268–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Wagschal A, Feil R. 2006. Genomic imprinting in the placenta. Cytogenet Genome Res 113:90–98 [DOI] [PubMed] [Google Scholar]
- 270. Monk D, Arnaud P, Apostolidou S, Hills FA, Kelsey G, Stanier P, Feil R, Moore GE. 2006. Limited evolutionary conservation of imprinting in the human placenta. Proc Natl Acad Sci USA 103:6623–6628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Wagschal A, Sutherland HG, Woodfine K, Henckel A, Chebli K, Schulz R, Oakey RJ, Bickmore WA, Feil R. 2008. G9a Histone methyltransferase contributes to imprinting in the mouse placenta. Mol Cell Biol 28:1104–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Charalambous M, Smith FM, Bennett WR, Crew TE, Mackenzie F, Ward A. 2003. Disruption of the imprinted Grb10 gene leads to disproportionate overgrowth by an Igf2-independent mechanism. Proc Natl Acad Sci USA 100:8292–8297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Arnaud P, Monk D, Hitchins M, Gordon E, Dean W, Beechey CV, Peters J, Craigen W, Preece M, Stanier P, Moore GE, Kelsey G. 2003. Conserved methylation imprints in the human and mouse GRB10 genes with divergent allelic expression suggests differential reading of the same mark. Hum Mol Genet 12:1005–1019 [DOI] [PubMed] [Google Scholar]
- 274. Hikichi T, Kohda T, Kaneko-Ishino T, Ishino F. 2003. Imprinting regulation of the murine Meg1/Grb10 and human GRB10 genes; roles of brain-specific promoters and mouse-specific CTCF-binding sites. Nucl Acids Res 31:1398–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Sanz LA, Chamberlain S, Sabourin JC, Henckel A, Magnuson T, Hugnot JP, Feil R, Arnaud P. 2008. A mono-allelic bivalent chromatin domain controls tissue-specific imprinting at Grb10. EMBO J 27:2523–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Monk D, Arnaud P, Frost J, Hills FA, Stanier P, Feil R, Moore GE. 2009. Reciprocal imprinting of human GRB10 in placental trophoblast and brain: evolutionary conservation of reversed allelic expression. Hum Mol Genet 18:3066–3074 [DOI] [PubMed] [Google Scholar]
- 277. Dean W, Bowden L, Aitchison A, Klose J, Moore T, Meneses JJ, Reik W, Feil R. 1998. Altered imprinted gene methylation and expression in completely ES cell-derived mouse fetuses: association with aberrant phenotypes. Development 125:2273–2282 [DOI] [PubMed] [Google Scholar]
- 278. Khosla S, Dean W, Brown D, Reik W, Feil R. 2001. Culture of preimplantation mouse embryos affects fetal development and the expression of imprinted genes. Biol Reprod 64:918–926 [DOI] [PubMed] [Google Scholar]
- 279. Cox GF, Bürger J, Lip V, Mau UA, Sperling K, Wu BL, Horsthemke B. 2002. Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am J Hum Genet 71:162–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. DeBaun MR, Niemitz EL, Feinberg AP. 2003. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet 72:156–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Gicquel C, Gaston V, Mandelbaum J, Siffroi JP, Flahault A, Le Bouc Y. 2003. In vitro fertilization may increase the risk of Beckwith-Wiedemann syndrome related to the abnormal imprinting of the KCNQ1OT gene. Am J Hum Genet 72:1338–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Mann MR, Chung YG, Nolen LD, Verona RI, Latham KE, Bartolomei MS. 2003. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol Reprod 69:902–914 [DOI] [PubMed] [Google Scholar]
- 283. Maher ER, Brueton LA, Bowdin SC, Luharia A, Cooper W, Cole TR, Macdonald F, Sampson JR, Barratt CL, Reik W, Hawkins MM. 2003. Beckwith-Wiedemann syndrome and assisted reproduction technology (ART). J Med Genet 40:62–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Halliday J, Oke K, Breheny S, Algar E, J Amor D. 2004. Beckwith-Wiedemann syndrome and IVF: a case-control study. Am J Hum Genet 75:526–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Khosla S, Dean W, Reik W, Feil R. 2001. Epigenetic and experimental modifications in early mammalian development. Part II: Culture of preimplantation embryos and its long-term effects on gene expression and phenotype. Hum Reprod Update 7:419–427 [DOI] [PubMed] [Google Scholar]
- 286. Feil R. 2006. Environmental and nutritional effects on the epigenetic regulation of genes. Mutat Res 600:46–57 [DOI] [PubMed] [Google Scholar]
- 287. Lim D, Bowdin SC, Tee L, Kirby GA, Blair E, Fryer A, Lam W, Oley C, Cole T, Brueton LA, Reik W, Macdonald F, Maher ER. 2009. Clinical and molecular genetic features of Beckwith-Wiedemann syndrome associated with assisted reproductive technologies. Hum Reprod 24:741–747 [DOI] [PubMed] [Google Scholar]
- 288. Azzi S, Rossignol S, Steunou V, Sas T, Thibaud N, Danton F, Le Jule M, Heinrichs C, Cabrol S, Gicquel C, Le Bouc Y, Netchine I. 2009. Multilocus methylation analysis in a large cohort of 11p15-related foetal growth disorders (Russell Silver and Beckwith Wiedemann syndromes) reveals simultaneous loss of methylation at paternal and maternal imprinted loci. Hum Mol Genet 18:4724–4733 [DOI] [PubMed] [Google Scholar]
- 289. Ørstavik KH, Eiklid K, van der Hagen CB, Spetalen S, Kierulf K, Skjeldal O, Buiting K. 2003. Another case of imprinting defect in a girl with Angelman syndrome who was conceived by intracytoplasmic sperm injection. Am J Hum Genet 72:218–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Ludwig M, Katalinic A, Gross S, Sutcliffe A, Varon R, Horsthemke B. 2005. Increased prevalence of imprinting defects in patients with Angelman syndrome born to subfertile couples. J Med Genet 42:289–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Chang AS, Moley KH, Wangler M, Feinberg AP, Debaun MR. 2005. Association between Beckwith-Wiedemann syndrome and assisted reproductive technology: a case series of 19 patients. Fertil Steril 83:349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Svensson J, Björnståhl A, Ivarsson SA. 2005. Increased risk of Silver-Russell syndrome after in vitro fertilization? Acta Paediatr 94:1163–1165 [DOI] [PubMed] [Google Scholar]
- 293. Rossignol S, Steunou V, Chalas C, Kerjean A, Rigolet M, Viegas-Pequignot E, Jouannet P, Le Bouc Y, Gicquel C. 2006. The epigenetic imprinting defect of patients with Beckwith–Wiedemann syndrome born after assisted reproductive technology is not restricted to the 11p15 region. J Med Genet 43:902–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Bliek J, Verde G, Callaway J, Maas SM, De Crescenzo A, Sparago A, Cerrato F, Russo S, Ferraiuolo S, Rinaldi MM, Fischetto R, Lalatta F, Giordano L, Ferrari P, Cubellis MV, Larizza L, Temple IK, Mannens MM, Mackay DJ, Riccio A. 2009. Hypomethylation at multiple maternally methylated imprinted regions including PLAGL1 and GNAS loci in Beckwith-Wiedemann syndrome. Eur J Hum Genet 17:611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Young LE, Sinclair KD, Wilmut I. 1998. Large offspring syndrome in cattle and sheep. Rev Reprod 3:155–163 [DOI] [PubMed] [Google Scholar]
- 296. Young LE, Fernandes K, McEvoy TG, Butterwith SC, Gutierrez CG, Carolan C, Broadbent PJ, Robinson JJ, Wilmut I, Sinclair KD. 2001. Epigenetic change in IGF2R is associated with fetal overgrowth after sheep embryo culture. Nat Genet 27:153–154 [DOI] [PubMed] [Google Scholar]
- 297. Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R. 2004. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet 36:1296–1300 [DOI] [PubMed] [Google Scholar]
- 298. Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W. 2004. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet 36:1291–1295 [DOI] [PubMed] [Google Scholar]
- 299. Miles HL, Hofman PL, Peek J, Harris M, Wilson D, Robinson EM, Gluckman PD, Cutfield WS. 2007. In vitro fertilization improves childhood growth and metabolism. J Clin Endocrinol Metab 92:3441–3445 [DOI] [PubMed] [Google Scholar]
- 300. Young LE, Schnieke AE, McCreath KJ, Wieckowski S, Konfortova G, Fernandes K, Ptak G, Kind AJ, Wilmut I, Loi P, Feil R. 2003. Conservation of IGF2–H19 and IGF2R imprinting in sheep: effects of somatic cell nuclear transfer. Mech Dev 120:1433–1442 [DOI] [PubMed] [Google Scholar]
- 301. Loi P, Ptak G, Feil R. 2009. Epigenetic mechanisms in mammals and their effects on cloning procedures. In: Schon I, Martens K, van Dijk P. eds. Lost sex: the evolutionary biology of parthenogenesis. Heidelberg: Springer; 559–580 [Google Scholar]
- 302. Kasai K, Sano F, Miyashita N, Watanabe S, Nagai T. 2007. Comparison of the growth performances of offspring produced by a pair of cloned cattle and their nuclear donor animals. J Reprod Dev 53:135–142 [DOI] [PubMed] [Google Scholar]
- 303. Thuan NV, Kishigami S, Wakayama T. 2010. How to improve the success rate of mouse cloning technology. J Reprod Dev 56:20–30 [DOI] [PubMed] [Google Scholar]
- 304. Graves JA. 2006. Sex chromosome specialization and degeneration in mammals. Cell 124:901–914 [DOI] [PubMed] [Google Scholar]
- 305. Charlesworth B. 1996. The evolution of chromosomal sex determination and dosage compensation. Curr Biol 6:149–162 [DOI] [PubMed] [Google Scholar]
- 306. Payer B, Lee JT. 2008. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet 42:733–772 [DOI] [PubMed] [Google Scholar]
- 307. Lucchesi JC, Kelly WG, Panning B. 2005. Chromatin remodeling in dosage compensation. Annu Rev Genet 39:615–651 [DOI] [PubMed] [Google Scholar]
- 308. Chow JC, Yen Z, Ziesche SM, Brown CJ. 2005. Silencing of the mammalian X chromosome. Annu Rev Genomics Hum Genet 6:69–92 [DOI] [PubMed] [Google Scholar]
- 309. Morey C, Navarro P, Debrand E, Avner P, Rougeulle C, Clerc Ph. 2004. The region 3′ to Xist mediates X chromosome counting and H3 Lys-4 dimethylation within the Xist gene. EMBO J 23:594–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Carrel L, Willard HF. 2005. X-Inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434:400–404 [DOI] [PubMed] [Google Scholar]
- 311. Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. 2004. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303:644–649 [DOI] [PubMed] [Google Scholar]
- 312. Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N. 2003. Establishment of histone H3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell 4:481–495 [DOI] [PubMed] [Google Scholar]
- 313. Patrat C, Okamoto I, Diabangouaya P, Vialon V, Le Baccon P, Chow J, Heard E. 2009. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc Natl Acad Sci USA 106:5198–5203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Cheng MK, Nguyen DK, Disteche CM. 2006. Dosage compensation of the X chromosome and Turner syndrome. Int Congr Ser 1298:3–8 [Google Scholar]
- 315. Lorda-Sanchez I, Binkert F, Hinkel KG, Moser H, Rosenkranz W, Maechler M, Schinzel A. 1992. Uniparental origin of sex chromosome polysomies. Hum Hered 42:193–197 [DOI] [PubMed] [Google Scholar]
- 316. Mathur A, Stekol L, Schatz D, MacLaren NK, Scott ML, Lippe B. 1991. The parental origin of the single X chromosome in Turner syndrome: lack of correlation with parental age or clinical phenotype. Am J Hum Genet 48:682–686 [PMC free article] [PubMed] [Google Scholar]
- 317. Sagi L, Zuckerman-Levin N, Gawlik A, Ghizzoni L, Buyukgebiz A, Rakover Y, Bistritzer T, Admoni O, Vottero A, Baruch O, Fares F, Malecka-Tendera E, Hochberg Z. 2007. Clinical significance of the parental origin of the X chromosome in Turner syndrome. J Clin Endocrinol Metab 92:846–852 [DOI] [PubMed] [Google Scholar]
- 318. Hamelin CE, Anglin G, Quigley CA, Deal CL. 2006. Genomic imprinting in Turner syndrome: effects on response to growth hormone and on risk of sensorineural hearing loss. J Clin Endocrinol Metab 91:3002–3010 [DOI] [PubMed] [Google Scholar]
- 319. Reiss AL, Mazzocco MM, Greenlaw R, Freund LS, Ross JL. 1995. Neurodevelopmental effects of X monosomy: a volumetric imaging study. Ann Neurol 38:731–738 [DOI] [PubMed] [Google Scholar]
- 320. Nijhuis-van der Sanden MW, Eling PA, Otten BJ. 2003. A review of neuropsychological and motor studies in Turner Syndrome. Neurosci Biobehav Rev 27:329–338 [DOI] [PubMed] [Google Scholar]
- 321. Lawrence K, Kuntsi J, Coleman M, Campbell R, Skuse D. 2003. Face and emotion recognition deficits in Turner syndrome: a possible role for X-linked genes in amygdala development. Neuropsychology 17:39–49 [PubMed] [Google Scholar]
- 322. Kesler SR, Blasey CM, Brown WE, Yankowitz J, Zeng SM, Bender BG, Reiss AL. 2003. Effects of X-monosomy and X-linked imprinting on superior temporal gyrus morphology in Turner syndrome. Biol Psychiatry 54:636–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Bondy CA. 2006. Genomic imprinting in Turner syndrome. Int Congr Ser 1298:21–25 [Google Scholar]
- 324. Burgoyne PS, Ojarikre OA, Turner JMA. 2002. Evidence that postnatal growth retardation in XO mice is due to haploinsufficiency for a non-PAR X gene. Cytogenet Genome Res 99:252–256 [DOI] [PubMed] [Google Scholar]
- 325. Burgoyne PS, Mahadevaiah S, Mittwoch U. 1985. A reciprocal autosomal translocation which causes male sterility in the mouse also impairs oogenesis. J Reprod Fertil 75:647–652 [DOI] [PubMed] [Google Scholar]
- 326. Burgoyne PS, Baker TG. 1985. Perinatal oocyte loss in XO mice and its implications for the aetiology of gonadal dysgenesis in XO women. J Reprod Fertil 75:633–645 [DOI] [PubMed] [Google Scholar]
- 327. Hultcrantz M, Stenberg AE, Fransson A, Canlon B. 2000. Characterization of hearing in an X,0 ‘Turner mouse’. Hear Res 143:182–188 [DOI] [PubMed] [Google Scholar]
- 328. Lynn PM, Davies W. 2007. The 39,XO mouse as a model for the neurobiology of Turner syndrome and sex-biased neuropsychiatric disorders. Behav Brain Res 179:173–182 [DOI] [PubMed] [Google Scholar]
- 329. Raefski AS, O'Neill MJ. 2005. Identification of a cluster of X-linked imprinted genes in mice. Nat Genet 37:620–624 [DOI] [PubMed] [Google Scholar]
- 330. Urbach A, Benvenisty N. 2009. Studying early lethality of 45,XO (Turner's Syndrome) embryos using human embryonic stem cells. PLoS One 41:e4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331. Fraga MF, Esteller M. 2007. Epigenetics and aging: the targets and the marks. Trends Genet 23:413–418 [DOI] [PubMed] [Google Scholar]
- 332. Sharpless NE, DePinho RA. 2007. How stem cells age and why this makes us grow old. Nat Rev Mol Cell Biol 8:703–713 [DOI] [PubMed] [Google Scholar]
- 333. Kirkwood TB. 2005. Understanding the odd science of aging. Cell 120:437–447 [DOI] [PubMed] [Google Scholar]
- 334. Berdyshev GD, Korotaev GK, Boiarskikh GV, Vaniushin BF. 1967. Nucleotide composition of DNA and RNA from somatic tissues of humpback salmon and its changes during spawning. Biokhimiia 32:988–993 [PubMed] [Google Scholar]
- 335. Vanyushin BF, Nemirovsky LE, Klimenko VV, Vasiliev VK, Belozersky AN. 1973. The 5-methylcytosine in DNA of rats. Gerontologia 19:138–152 [PubMed] [Google Scholar]
- 336. Wilson VL, Smith RA, Ma S, Cutler RG. 1987. Genomic 5-methyldeoxycytidine decreases with age. J Biol Chem 262:9948–9951 [PubMed] [Google Scholar]
- 337. Wilson CB, Merkenschlager M. 2006. Chromatin structure and gene regulation in T cell development and function. Curr Opin Immunol 18:143–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338. Fuke C, Shimabukuro M, Petronis A, Sugimoto J, Oda T, Miura K, Miyazaki T, Ogura C, Okazaki Y, Jinno Y. 2004. Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Ann Hum Genet 68:196–204 [DOI] [PubMed] [Google Scholar]
- 339. Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, Yu W, Rongione MA, Ekström TJ, Harris TB, Launer LJ, Eiriksdottir G, Leppert MF, Sapienza C, Gudnason V, Feinberg AP. 2008. Intra-individual change over time in DNA methylation with familial clustering. JAMA 299:2877–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340. Oakes CC, Smiraglia DJ, Plass C, Trasler JM, Robaire B. 2003. Aging results in hypermethylation of ribosomal DNA in sperm and liver of male rats. Proc Natl Acad Sci USA 100:1775–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341. Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JP. 1998. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res 58:5489–5494 [PubMed] [Google Scholar]
- 342. Bjornsson HT, Fallin MD, Feinberg AP. 2004. An integrated epigenetic and genetic approach to common human disease. Trends Genet 20:350–358 [DOI] [PubMed] [Google Scholar]
- 343. Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suñer D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M. 2005. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA 102:10604–10609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344. Poulsen P, Esteller M, Vaag A, Fraga MF. 2007. The epigenetic basis of twin discordance in age-related diseases. Pediatr Res 61:38R–42R [DOI] [PubMed] [Google Scholar]
- 345. Cheung VG, Conlin LK, Weber TM, Arcaro M, Jen KY, Morley M, Spielman RS. 2003. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet 33:422–425 [DOI] [PubMed] [Google Scholar]
- 346. Sharma A, Sharma VK, Horn-Saban S, Lancet D, Ramachandran S, Brahmachari SK. 2005. Assessing natural variations in gene expression in humans by comparing with monozygotic twins using microarrays. Physiol Genomics 21:117–123 [DOI] [PubMed] [Google Scholar]
- 347. Choi JK, Kim SC. 2007. Environmental effects on gene expression phenotype have regional biases in the human genome. Genetics 175:1607–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348. Belinsky SA, Palmisano WA, Gilliland FD, Crooks LA, Divine KK, Winters SA, Grimes MJ, Harms HJ, Tellez CS, Smith TM, Moots PP, Lechner JF, Stidley CA, Crowell RE. 2002. Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res 62:2370–2377 [PubMed] [Google Scholar]
- 349. Vogt G, Huber M, Thiemann M, van den Boogaart G, Schmitz OJ, Schubart CD. 2008. Production of different phenotypes from the same genotype in the same environment by developmental variation. J Exp Biol 211:510–523 [DOI] [PubMed] [Google Scholar]
- 350. Gärtner K. 1990. A third component causing random variability beside environment and genotype. A reason for the limited success of a 30 year long effort to standardize laboratory animals? Lab Anim 24:71–77 [DOI] [PubMed] [Google Scholar]
- 351. Johnson TE. 1990. Increased life-span of age-1 mutants in Caenorhabditis elegans and lower Gompertz rate of aging. Science 249:908–912 [DOI] [PubMed] [Google Scholar]
- 352. Rönn T, Poulsen P, Hansson O, Holmkvist J, Almgren P, Nilsson P, Tuomi T, Isomaa B, Groop L, Vaag A, Ling C. 2008. Age influences DNA methylation and gene expression of COX7A1 in human skeletal muscle. Diabetologia 51:1159–1168 [DOI] [PubMed] [Google Scholar]
- 353. Bouchard TJ, Jr, Lykken DT, McGue M, Segal NL, Tellegen A. 1990. Sources of human psychological differences: the Minnesota Study of Twins Reared Apart. Science 250:223–228 [DOI] [PubMed] [Google Scholar]
- 354. Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. 2000. Environmental and heritable factors in the causation of cancer: analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 343:78–85 [DOI] [PubMed] [Google Scholar]
- 355. Hoover RN. 2000. Cancer: nature, nurture, or both. N Engl J Med 343:135–136 [DOI] [PubMed] [Google Scholar]
- 356. Mill J, Dempster E, Caspi A, Williams B, Moffitt T, Craig I. 2006. Evidence for monozygotic twin (MZ) discordance in methylation level at two CpG sites in the promoter region of the catechol-O-methyltransferase (COMT) gene. Am J Med Genet B Neuropsychiatr Genet 141B:421–425 [DOI] [PubMed] [Google Scholar]
- 357. Oates NA, van Vliet J, Duffy DL, Kroes HY, Martin NG, Boomsma DI, Campbell M, Coulthard MG, Whitelaw E, Chong S. 2006. Increased DNA methylation at the AXIN1 gene in a monozygotic twin from a pair discordant for a caudal duplication anomaly. Am J Hum Genet 79:155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358. Rosa A, Picchioni MM, Kalidindi S, Loat CS, Knight J, Toulopoulou T, Vonk R, van der Schot AC, Nolen W, Kahn RS, McGuffin P, Murray RM, Craig IW. 2008. Differential methylation of the X-chromosome is a possible source of discordance for bipolar disorder female monozygotic twins. Am J Med Genet B Neuropsychiatr Genet 147B:459–462 [DOI] [PubMed] [Google Scholar]
- 359. De Bustos C, Ramos E, Young JM, Tran RK, Menzel U, Langford CF, Eichler EE, Hsu L, Henikoff S, Dumanski JP, Trask BJ. 2009. Tissue-specific variation in DNA methylation levels along human chromosome 1. Epigenetics Chromatin 2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360. Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D. 2002. Projection of an immunological self shadow within the thymus by the Aire protein. Science 298:1395–1401 [DOI] [PubMed] [Google Scholar]
- 361. Faideau B, Lotton C, Lucas B, Tardivel I, Elliott JF, Boitard C, Carel JC. 2006. Tolerance to proinsulin-2 is due to radioresistant thymic cells. J Immunol 177:53–60 [DOI] [PubMed] [Google Scholar]
- 362. Concannon P, Rich SS, Nepom GT. 2009. Genetics of type 1A diabetes. N Engl J Med 360:1646–1654 [DOI] [PubMed] [Google Scholar]
- 363. Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, Plagnol V, Pociot F, Schuilenburg H, Smyth DJ, Stevens H, Todd JA, Walker NM, Rich SS. The Type 1 Diabetes Genetics Consortium 2009. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 41:703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364. Hemminki K, Li X, Sundquist J, Sundquist K. 2009. Familial association between type 1 diabetes and other autoimmune and related diseases. Diabetologia 52:1820–1828 [DOI] [PubMed] [Google Scholar]
- 365. Alper CA, Husain Z, Larsen CE, Dubey DP, Stein R, Day C, Baker A, Beyan H, Hawa M, Ola TO, Leslie RD. 2006. Incomplete penetrance of susceptibility genes for MHC-determined immunoglobulin deficiencies in monozygotic twins discordant for type 1 diabetes. J Autoimmun 27:89–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366. Redondo MJ, Jeffrey J, Fain PR, Eisenbarth GS, Orban T. 2008. Concordance for islet autoimmunity among monozygotic twins. N Engl J Med 359:2849–2850 [DOI] [PubMed] [Google Scholar]
- 367. Akesson K, Carlsson A, Ivarsson SA, Johansson C, Weidby BM, Ludvigsson J, Gustavsson B, Lernmark A, Kockum I. 2009. The non-inherited maternal HLA haplotype affects the risk for type 1 diabetes. Int J Immunogenet 36:1–8 [DOI] [PubMed] [Google Scholar]
- 368. Nelson JL, Gillespie KM, Lambert NC, Stevens AM, Loubiere LS, Rutledge JC, Leisenring WM, Erickson TD, Yan Z, Mullarkey ME, Boespflug ND, Bingley PJ, Gale EAM. 2007. Maternal microchimerism in peripheral blood in type 1 diabetes and pancreatic islet β cell microchimerism. Proc Natl Acad Sci USA 104:1637–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369. Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, Gordon JI, Chervonsky AV. 2008. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455:1109–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370. Norris JM, Barriga K, Klingensmith G, Hoffman M, Eisenbarth GS, Erlich HA, Rewers M. 2003. Timing of initial cereal exposure in infancy and risk of islet autoimmunity. JAMA 290:1713–1720 [DOI] [PubMed] [Google Scholar]
- 371. Peng H, Hagopian W. 2006. Environmental factors in the development of type 1 diabetes. Rev Endocr Metab Disord 7:149–162 [DOI] [PubMed] [Google Scholar]
- 372. Miao F, Smith DD, Zhang L, Min A, Feng W, Natarajan R. 2008. Lymphocytes from patients with type 1 diabetes display a distinct profile of chromatin histone H3 lysine 9 dimethylation. Diabetes 57:3189–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373. El-Osta A, Brasacchio D, Yao D, Pocai A, Jones PL, Roeder RG, Cooper ME, Brownlee M. 2008. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med 205:2409–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374. Laborie LB, Mackay DJ, Temple IK, Molven A, Søvik O, Njølstad PR. 2010. DNA hypomethylation, transient neonatal diabetes, and prune belly sequence in one of two identical twins. Eur J Pediatr 169:207–213 [DOI] [PubMed] [Google Scholar]
- 375. Haumaitre C, Lenoir O, Scharfmann R. 2008. Histone deacetylase inhibitors modify pancreatic cell fate determination and amplify endocrine progenitors. Mol Cell Biol 28:6373–6383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376. Haumaitre C, Lenoir O, Scharfmann R. 2009. Directing cell differentiation with small-molecule histone deacetylase inhibitors: the example of promoting pancreatic endocrine cells. Cell Cycle 8:536–544 [DOI] [PubMed] [Google Scholar]
- 377. Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, Drake TA, Lusis AJ. 2006. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res 16:995–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378. Gabory A, Attig L, Junien C. 2009. Sexual dimorphism in environmental epigenetic programming. Mol Cell Endocrinol 304:8–18 [DOI] [PubMed] [Google Scholar]
- 379. Flanagan DE, Moore VM, Godsland IF, Cockington RA, Robinson JS, Phillips DIW. 2000. Fetal growth and the physiological control of glucose tolerance in adults: a minimal model analysis. Am J Physiol Endocrinol Metab 278:E700–E706 [DOI] [PubMed] [Google Scholar]
- 380. Sugden MC, Holness MJ. 2002. Gender-specific programming of insulin secretion and action. J Endocrinol 175:757–767 [DOI] [PubMed] [Google Scholar]
- 381. Wilcoxon JS, Schwartz J, Aird F, Redei EE. 2003. Sexually dimorphic effects of maternal alcohol intake and adrenalectomy on left ventricular hypertrophy in rat offspring. Am J Physiol Endocrinol Metab 285:E31–E39 [DOI] [PubMed] [Google Scholar]
- 382. Gallou-Kabani C, Vigé A, Gross MS, Boileau C, Rabes JP, Fruchart-Najib J, Jais JP, Junien C. 2007. Resistance to high-fat diet in the female progeny of obese mice fed a control diet during the periconceptual, gestation, and lactation periods. Am J Physiol Endocrinol Metab 292:E1095–E1100 [DOI] [PubMed] [Google Scholar]
- 383. Gallou-Kabani C, Vigé A, Junien C. 2007. Lifelong circadian and epigenetic drifts in metabolic syndrome. Epigenetics 2:137–146 [DOI] [PubMed] [Google Scholar]
- 384. Owens JA, Thavaneswaran P, De Blasio MJ, McMillen IC, Robinson JS, Gatford KL. 2007. Sex-specific effects of placental restriction on components of the metabolic syndrome in young adult sheep. Am J Physiol Endocrinol Metab 292:E1879–E1889 [DOI] [PubMed] [Google Scholar]
- 385. Tsai HW, Grant PA, Rissman EF. 2009. Sex differences in histone modifications in the neonatal mouse brain. Epigenetics 4:47–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386. Wilhelm D, Koopman P. 2006. The makings of maleness: towards an integrated view of male sexual development. Nat Rev Genet 7:620–631 [DOI] [PubMed] [Google Scholar]
- 387. Blecher SR, Erickson RP. 2007. Genetics of sexual development: a new paradigm. Am J Med Genet A 143A:3054–3068 [DOI] [PubMed] [Google Scholar]
- 388. Penaloza C, Estevez B, Orlanski S, Sikorska M, Walker R, Smith C, Smith B, Lockshin RA, Zakeri Z. 2009. Sex of the cell dictates its response: differential gene expression and sensitivity to cell death inducing stress in male and female cells. FASEB J 23:1869–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389. Waxman DJ, Holloway MG. 2009. Sex differences in the expression of hepatic drug metabolizing enzymes. Mol Pharmacol 76:215–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390. Yokoyama Y, Nimura Y, Nagino M, Bland KI, Chaudry IH. 2005. Current understanding of gender dimorphism in hepatic pathophysiology. J Surg Res 128:147–156 [DOI] [PubMed] [Google Scholar]
- 391. Tannenbaum GS, Martin JB. 1976. Evidence for an endogenous ultradian rhythm governing growth hormone secretion in the rat. Endocrinology 98:562–570 [DOI] [PubMed] [Google Scholar]
- 392. Edén S. 1979. Age- and sex-related differences in episodic growth hormone secretion in the rat. Endocrinology 105:555–560 [DOI] [PubMed] [Google Scholar]
- 393. Robinson IC, Gevers EF, Bennett PA. 1998. Sex differences in growth hormone secretion and action in the rat. Growth Horm IGF Res 8(Suppl B):39–47 [DOI] [PubMed] [Google Scholar]
- 394. Jansson JO, Edén S, Isaksson O. 1985. Sexual dimorphism in the control of growth hormone secretion. Endocr Rev 6:128–150 [DOI] [PubMed] [Google Scholar]
- 395. Mode A, Gustafsson JA, Jansson JO, Edén S, Isaksson O. 1982. Association between plasma level of growth hormone and sex differentiation of hepatic steroid metabolism in the rat. Endocrinology 111:1692–1697 [DOI] [PubMed] [Google Scholar]
- 396. Waxman DJ, Pampori NA, Ram PA, Agrawal AK, Shapiro BH. 1991. Interpulse interval in circulating growth hormone patterns regulates sexually dimorphic expression of hepatic cytochrome P450. Proc Natl Acad Sci USA 88:6868–6872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397. Veldhuis JD. 1998. Neuroendocrine control of pulsatile growth hormone release in the human: relationship with gender. Growth Horm IGF Res 8:49–59 [DOI] [PubMed] [Google Scholar]
- 398. Veldhuis JD, Bowers CY. 2003. Human GH pulsatility: an ensemble property regulated by age and gender. J Endocrinol Invest 26:799–813 [DOI] [PubMed] [Google Scholar]
- 399. Mode A, Gustafsson JA. 2006. Sex and the liver—a journey through five decades. Drug Metab Rev 38:197–207 [DOI] [PubMed] [Google Scholar]
- 400. Waxman DJ, O'Connor C. 2006. Growth hormone regulation of sex-dependent liver gene expression. Mol Endocrinol 20:2613–2629 [DOI] [PubMed] [Google Scholar]
- 401. Clodfelter KH, Holloway MG, Hodor P, Park SH, Ray WJ, Waxman DJ. 2006. Sex-dependent liver gene expression is extensive and largely dependent upon signal transducer and activator of transcription 5b (STAT5b): STAT5b-dependent activation of male genes and repression of female genes revealed by microarray analysis. Mol Endocrinol 20:1333–1351 [DOI] [PubMed] [Google Scholar]
- 402. Holloway MG, Miles GD, Dombkowski AA, Waxman DJ. 2008. Liver-specific hepatocyte nuclear factor-4α deficiency: greater impact on gene expression in male than in female mouse liver. Mol Endocrinol 22:1274–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403. Waxman DJ, Ram PA, Park SH, Choi HK. 1995. Intermittent plasma growth hormone triggers tyrosine phosphorylation and nuclear translocation of a liver-expressed, Stat 5-related DNA binding protein. J Biol Chem 270:13262–13270 [DOI] [PubMed] [Google Scholar]
- 404. Choi HK, Waxman DJ. 1999. Growth hormone, but not prolactin, maintains low-level activation of STAT5a and STAT5b in female rat liver. Endocrinology 140:5126–5135 [DOI] [PubMed] [Google Scholar]
- 405. Choi HK, Waxman DJ. 2000. Plasma growth hormone pulse activation of hepatic JAK-STAT5 signaling: developmental regulation and role in male-specific liver gene expression. Endocrinology 141:3245–3255 [DOI] [PubMed] [Google Scholar]
- 406. Laz EV, Sugathan A, Waxman DJ. 2009. Dynamic in vivo binding of STAT5 to growth hormone-regulated genes in intact rat liver. Sex-specific binding at low- but not high-affinity STAT5 sites. Mol Endocrinol 23:1242–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407. Laz EV, Holloway MG, Chen CS, Waxman DJ. 2007. Characterization of three growth hormone-responsive transcription factors preferentially expressed in adult female liver. Endocrinology 148:3327–3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408. Hemenway C, Robins DM. 1987. DNase I-hypersensitive sites associated with expression and hormonal regulation of mouse C4 and Slp genes. Proc Natl Acad Sci USA 84:4816–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409. Ström A, Eguchi H, Mode A, Legraverend C, Tollet P, Strömstedt PE, Gustafsson JA. 1994. Characterization of the proximal promoter and two silencer elements in the CYP2C11 gene expressed in rat liver. DNA Cell Biol 13:805–819 [DOI] [PubMed] [Google Scholar]
- 410. Endo M, Takahashi Y, Sasaki Y, Saito T, Kamataki T. 2005. Novel gender-related regulation of CYP2C12 gene expression in rats. Mol Endocrinol 19:1181–1190 [DOI] [PubMed] [Google Scholar]
- 411. Wauthier V, Sugathan A, Meyer RD, Dombkowski AA, Waxman DJ. 2010. Intrinsic sex differences in the early growth hormone responsiveness of sex-specific genes in mouse liver. Mol Endocrinol 24:667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412. Vickers MH, Gluckman PD, Coveny AH, Hofman PL, Cutfield WS, Gertler A, Breier BH, Harris M. 2005. Neonatal leptin treatment reverses developmental programming. Endocrinology 146:4211–4216 [DOI] [PubMed] [Google Scholar]
- 413. Burdge GC, Lillycrop KA, Jackson AA, Gluckman PD, Hanson MA. 2008. The nature of the growth pattern and of the metabolic response to fasting in the rat are dependent upon the dietary protein and folic acid intakes of their pregnant dams and post-weaning fat consumption. Br J Nutr 99:540–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414. Li E. 2002. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet 3:662–673 [DOI] [PubMed] [Google Scholar]
- 415. Barber R, Plumb MA, Boulton E, Roux I, Dubrova YE. 2002. Elevated mutation rates in the germ line of first- and second-generation offspring of irradiated male mice. Proc Natl Acad Sci USA 99:6877–6882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416. Waterland RA, Jirtle RL. 2004. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition 20:63–68 [DOI] [PubMed] [Google Scholar]
- 417. Dolinoy DC, Huang D, Jirtle RL. 2007. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci USA 104:13056–13061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418. Howie GJ, Sloboda DM, Kamal T, Vickers MH. 2009. Maternal nutritional history predicts obesity in adult offspring independent of postnatal diet. J Physiol 587:905–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419. Dashwood RH, Ho E. 2007. Dietary histone deacetylase inhibitors: from cells to mice to man. Semin Cancer Biol 17:363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420. Raptis S, Bapat B. 2006. Genetic instability in human tumors. In: Bignold LP. ed. Cancer: cell structures, carcinogens and genomic instability. Basel, Boston, Berlin: Birkhauser Verlag; 303–320 [Google Scholar]
- 421. De Flora S, Izzotti A. 2009. Modulation of genomic and postgenomic alterations in noncancer diseases and critical periods of life. Mutat Res 667:15–26 [DOI] [PubMed] [Google Scholar]
- 422. Maslov AY, Vijg J. 2009. Genome instability, cancer and aging. Biochim Biophys Acta 1790:963–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423. McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonté B, Szyf M, Turecki G, Meaney MJ. 2009. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci 12:342–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424. Anway MD, Leathers C, Skinner MK. 2006. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology 147:5515–5523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425. Nilsson EE, Anway MD, Stanfield J, Skinner MK. 2008. Transgenerational epigenetic effects of the endocrine disruptor vinclozolin on pregnancies and female adult onset disease. Reproduction 135:713–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426. Wauthier V, Waxman DJ. 2008. Sex-specific early growth hormone response genes in rat liver. Mol Endocrinol 22:1962–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427. Szyf M, McGowan P, Meaney MJ. 2008. The social environment and the epigenome. Environ Mol Mutagen 49:46–60 [DOI] [PubMed] [Google Scholar]
- 428. Levin BE, Govek E. 1998. Gestational obesity accentuates obesity in obesity-prone progeny. Am J Physiol 275:R1374–R1379 [DOI] [PubMed] [Google Scholar]
- 429. Armitage JA, Khan IY, Taylor PD, Nathanielsz PW, Poston L. 2004. Developmental programming of the metabolic syndrome by maternal nutritional imbalance: how strong is the evidence from experimental models in mammals? J Physiol 561:355–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430. Khan I, Dekou V, Hanson M, Poston L, Taylor P. 2004. Predictive adaptive responses to maternal high-fat diet prevent endothelial dysfunction but not hypertension in adult rat offspring. Circulation 110:1097–1102 [DOI] [PubMed] [Google Scholar]
- 431. Armitage JA, Taylor PD, Poston L. 2005. Experimental models of developmental programming: consequences of exposure to an energy rich diet during development. J Physiol 565:3–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432. Das UN. 2005. Pathophysiology of metabolic syndrome X and its links to the perinatal period. Nutrition 21:762–773 [DOI] [PubMed] [Google Scholar]
- 433. Nathanielsz PW, Poston L, Taylor PD. 2007. In utero exposure to maternal obesity and diabetes: animal models that identify and characterize implications for future health. Obstet Gynecol Clin North Am 34:201–212, vii–viii [DOI] [PubMed] [Google Scholar]
- 434. Campbell JH, Perkins P. 1988. Transgenerational effects of drug and hormonal treatments in mammals: a review of observations and ideas. Prog Brain Res 73:535–753 [DOI] [PubMed] [Google Scholar]
- 435. Taylor PD, McConnell J, Khan IY, Holemans K, Lawrence KM, Asare-Anane H, Persaud SJ, Jones PM, Petrie L, Hanson MA, Poston L. 2005. Impaired glucose homeostasis and mitochondrial abnormalities in offspring of rats fed a fat-rich diet in pregnancy. Am J Physiol Regul Integr Comp Physiol 288:R134–R139 [DOI] [PubMed] [Google Scholar]
- 436. Lelièvre SA. 2009. Contributions of extracellular matrix signaling and tissue architecture to nuclear mechanisms and spatial organization of gene expression control. Biochim Biophys Acta 1790:925–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437. Ke X, Lei Q, James SJ, Kelleher SL, Melnyk S, Jernigan S, Yu X, Wang L, Callaway CW, Gill G, Chan GM, Albertine KH, McKnight RA, Lane RH. 2006. Uteroplacental insufficiency affects epigenetic determinants of chromatin structure in brains of neonatal and juvenile IUGR rats. Physiol Genomics 25:16–28 [DOI] [PubMed] [Google Scholar]
- 438. Xiao Y, Word B, Starlard-Davenport A, Haefele A, Lyn-Cook BD, Hammons G. 2008. Age and gender affect DNMT3a and DNMT3b expression in human liver. Cell Biol Toxicol 24:265–272 [DOI] [PubMed] [Google Scholar]
- 439. Vaissière T, Hung RJ, Zaridze D, Moukeria A, Cuenin C, Fasolo V, Ferro G, Paliwal A, Hainaut P, Brennan P, Tost J, Boffetta P, Herceg Z. 2009. Quantitative analysis of DNA methylation profiles in lung cancer identifies aberrant DNA methylation of specific genes and its association with gender and cancer risk factors. Cancer Res 69:243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440. Szyf M. 2009. Epigenetics, DNA methylation, and chromatin modifying drugs. Annu Rev Pharmacol Toxicol 49:243–263 [DOI] [PubMed] [Google Scholar]
- 441. Gronemeyer H, Gustafsson JA, Laudet V. 2004. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov 3:950–964 [DOI] [PubMed] [Google Scholar]
- 442. Sharma RP. 2005. Schizophrenia, epigenetics and ligand-activated nuclear receptors: a framework for chromatin therapeutics. Schizophr Res 72:79–90 [DOI] [PubMed] [Google Scholar]
- 443. McGowan PO, Meaney MJ, Szyf M. 2008. Diet and the epigenetic (re)programming of phenotypic differences in behavior. Brain Res 1237:12–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444. Patra SK, Szyf M. 2008. DNA methylation-mediated nucleosome dynamics and oncogenic Ras signaling. FEBS J 275:5217–5235 [DOI] [PubMed] [Google Scholar]
- 445. Watson ED, Cross JC. 2005. Development of structures and transport functions in the mouse placenta. Physiology 20:180–193 [DOI] [PubMed] [Google Scholar]
- 446. Borowicz P, Reynolds LP. 2010. ‘Placental programming’: more may still be less. J Physiol 588:393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447. Coan PM, Vaughan OR, Sekita Y, Finn SL, Burton GJ, Constancia M, Fowden AL. 2010. Adaptations in placental phenotype support fetal growth during undernutrition of pregnant mice. J Physiol 588:527–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448. Bertram CE, Hanson MA. 2001. Animal models and programming of the metabolic syndrome: type 2 diabetes. Br Med Bull 60:103–121 [DOI] [PubMed] [Google Scholar]
- 449. Junien C, Nathanielsz P. 2007. Report on the IASO Stock Conference 2006: early and lifelong environmental epigenomic programming of metabolic syndrome, obesity and type II diabetes. Obes Rev 8:487–502 [DOI] [PubMed] [Google Scholar]
- 450. Armitage JA, Poston L, Taylor PD. 2008. Developmental origins of obesity and the metabolic syndrome. Front Horm Res 36:73–84 [DOI] [PubMed] [Google Scholar]
- 451. Fowden AL, Giussani DA, Forhead AJ. 2006. Intrauterine programming of physiological systems: causes and consequences. Physiology 21:29–37 [DOI] [PubMed] [Google Scholar]
- 452. Angiolini E, Fowden A, Coan P, Sandovici I, Smith P, Dean W, Burton G, Tycko B, Reik W, Sibley C, Constancia M. 2006. Regulation of placental efficiency for nutrient transport by imprinted genes. Placenta 27(Suppl A):S98–S102 [DOI] [PubMed] [Google Scholar]
- 453. Reik W, Constância M, Fowden A, Anderson N, Dean W, Ferguson-Smith A, Tycko B, Sibley C. 2003. Regulation of supply and demand for maternal nutrients in mammals by imprinted genes. J Physiol 547:35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454. Coan PM, Burton GJ, Ferguson-Smith AC. 2005. Imprinted genes in the placenta—a review. Placenta 26:S10–S20 [DOI] [PubMed] [Google Scholar]
- 455. Fowden AL, Sibley C, Reik W, Constancia M. 2006. Imprinted genes, placental development and fetal growth. Horm Res 65:50–58 [DOI] [PubMed] [Google Scholar]
- 456. Rees WD, Hay SM, Brown DS, Antipatis C, Palmer RM. 2000. Maternal protein deficiency causes hypermethylation of DNA in the livers of rat fetuses. J Nutr 130:1821–1826 [DOI] [PubMed] [Google Scholar]
- 457. Hanson MA, Gluckman PD. 2008. Developmental origins of health and disease: new insights. Basic Clin Pharmacol Toxicol 102:90–93 [DOI] [PubMed] [Google Scholar]
- 458. Kwong WY, Wild AE, Roberts P, Willis AC, Fleming TP. 2000. Maternal undernutrition during the preimplantation period of rat development causes blastocyst abnormalities and programming of postnatal hypertension. Development 127:4195–4202 [DOI] [PubMed] [Google Scholar]
- 459. Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. 2005. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr 135:1382–1386 [DOI] [PubMed] [Google Scholar]
- 460. Burdge GC, Slater-Jefferies J, Torrens C, Phillips ES, Hanson MA, Lillycrop KA. 2007. Dietary protein restriction of pregnant rats in the F0 generation induces altered methylation of hepatic gene promoters in the adult male offspring in the F1 and F2 generations. Br J Nutr 97:435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461. Lillycrop KA, Phillips ES, Torrens C, Hanson MA, Jackson AA, Burdge GC. 2008. Feeding pregnant rats a protein-restricted diet persistently alters the methylation of specific cytosines in the hepatic PPARα promoter of the offspring. Br J Nutr 100:278–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462. Lillycrop KA, Slater-Jefferies JL, Hanson MA, Godfrey KM, Jackson AA, Burdge GC. 2007. Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. Br J Nutr 97:1064–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463. Turner JD, Muller CP. 2005. Structure of the glucocorticoid receptor (NR3C1) gene 5′ untranslated region: identification, and tissue distribution of multiple new human exon 1. J Mol Endocrinol 35:283–292 [DOI] [PubMed] [Google Scholar]
- 464. Jackson AA, Dunn RL, Marchand MC, Langley-Evans SC. 2002. Increased systolic blood pressure in rats induced by a maternal low-protein diet is reversed by dietary supplementation with glycine. Clin Sci (Lond) 103:633–639 [DOI] [PubMed] [Google Scholar]
- 465. Brawley L, Torrens C, Anthony FW, Itoh S, Wheeler T, Jackson AA, Clough GF, Poston L, Hanson MA. 2004. Glycine rectifies vascular dysfunction induced by dietary protein imbalance during pregnancy. J Physiol 554:497–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466. Burdge GC, Lillycrop KA, Phillips ES, Slater-Jefferies JL, Jackson AA, Hanson MA. 2009. Folic acid supplementation during the juvenile-pubertal period in rats modifies the phenotype and epigenotype induced by prenatal nutrition. J Nutr 139:1054–1060 [DOI] [PubMed] [Google Scholar]
- 467. Shahkhalili Y, Moulin J, Zbinden I, Aprikian O, Macé K. 2010. Comparison of two models of intrauterine growth restriction for early catch-up growth and later development of glucose intolerance and obesity in rats. Am J Physiol Regul Integr Comp Physiol 298:R141–R146 [DOI] [PubMed] [Google Scholar]
- 468. Patel MS, Srinivasan M. 2002. Metabolic programming: causes and consequences. J Biol Chem 277:1629–1632 [DOI] [PubMed] [Google Scholar]
- 469. Srinivasan M, Laychock SG, Hill DJ, Patel MS. 2003. Neonatal nutrition: metabolic programming of pancreatic islets and obesity. Exp Biol Med Maywood 228:15–23 [DOI] [PubMed] [Google Scholar]
- 470. Srinivasan M, Aalinkeel R, Song F, Mitrani P, Pandya JD, Strutt B, Hill DJ, Patel MS. 2006. Maternal hyperinsulinemia predisposes rat fetuses for hyperinsulinemia, and adult-onset obesity and maternal mild food restriction reverses this phenotype. Am J Physiol Endocrinol Metab 290:E129–E134 [DOI] [PubMed] [Google Scholar]
- 471. Mao J, Zhang X, Sieli PT, Falduto MT, Torres KE, Rosenfeld CS. 2010. Contrasting effects of different maternal diets on sexually dimorphic gene expression in the murine placenta. Proc Natl Acad Sci USA 107:5557–5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472. Eriksson UJ. 2009. Congenital anomalies in diabetic pregnancy. Semin Fetal Neonatal Med 14:85–93 [DOI] [PubMed] [Google Scholar]
- 473. Dabelea D, Hanson RL, Lindsay RS, Pettitt DJ, Imperatore G, Gabir MM, Roumain J, Bennett PH, Knowler WC. 2000. Intrauterine exposure to diabetes conveys risks for type 2 diabetes and obesity: a study of discordant sibships. Diabetes 49:2208–2211 [DOI] [PubMed] [Google Scholar]
- 474. Chao W, D'Amore PA. 2008. IGF2: Epigenetic regulation and role in development and disease. Cytokine Growth Factor Rev 19:111–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475. Constância M, Kelsey G, Reik W. 2004. Resourceful imprinting. Nature 432:53–57 [DOI] [PubMed] [Google Scholar]
- 476. Constância M, Angiolini E, Sandovici I, Smith P, Smith R, Kelsey G, Dean W, Ferguson-Smith A, Sibley CP, Reik W, Fowden A. 2005. Adaptation of nutrient supply to fetal demand in the mouse involves interaction between the Igf2 gene and placental transporter systems. Proc Natl Acad Sci USA 102:19219–19224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477. Fowden AL, Forhead AJ. 2009. Hormones as epigenetic signals in developmental programming. Exp Physiol 94:607–625 [DOI] [PubMed] [Google Scholar]
- 478. Fowden AL, Forhead AJ. 2009. Endocrine regulation of feto-placental growth. Horm Res 72:257–265 [DOI] [PubMed] [Google Scholar]
- 479. Pasca AM, Penn AA. 2010. The placenta: the lost neuroendocrine organ. NeoReviews 11:e64–e77 [Google Scholar]
- 480. Bertram CE, Hanson MA. 2002. Prenatal programming of postnatal endocrine responses by glucocorticoids. Reproduction 124:459–467 [DOI] [PubMed] [Google Scholar]
- 481. Fowden AL, Forhead AJ. 2004. Endocrine mechanisms of intrauterine programming. Reproduction 127:515–526 [DOI] [PubMed] [Google Scholar]
- 482. Seckl JR, Holmes MC. 2007. Mechanisms of disease: glucocorticoids, their placental metabolism and fetal ‘programming’ of adult pathophysiology. Nat Clin Pract Endocrinol Metab 3:479–488 [DOI] [PubMed] [Google Scholar]
- 483. Fowden AL. 1995. Endocrine regulation of fetal growth. Reprod Fertil Dev 7:351–363 [DOI] [PubMed] [Google Scholar]
- 484. Fowden AL, Li J, Forhead AJ. 1998. Glucocorticoids and the preparation for life after birth: are there long-term consequences of the life insurance? Proc Nutr Soc 57:113–122 [DOI] [PubMed] [Google Scholar]
- 485. Fowden AL, Sibley C, Reik W, Constancia M. 2006. Imprinted genes, placental development and fetal growth. Horm Res 65(Suppl 3):50–58 [DOI] [PubMed] [Google Scholar]
- 486. Fowden AL, Ward JW, Wooding FP, Forhead AJ, Constancia M. 2006. Programming placental nutrient transport capacity. J Physiol 572:5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487. Fowden AL, Sferruzzi-Perri AN, Coan PM, Constancia M, Burton GJ. 2009. Placental efficiency and adaptation: endocrine regulation. J Physiol 587:3459–3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488. Thomassin H, Flavin M, Espinás ML, Grange T. 2001. Glucocorticoid-induced DNA demethylation and gene memory during development. EMBO J 20:1974–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489. Drake AJ, Walker BR, Seckl JR. 2005. Intergenerational consequences of fetal programming by in utero exposure to glucocorticoids in rats. Am J Physiol Regul Integr Comp Physiol 288:R34–R38 [DOI] [PubMed] [Google Scholar]
- 490. Weaver IC. 2009. Epigenetic effects of glucocorticoids. Semin Fetal Neonatal Med 14:143–150 [DOI] [PubMed] [Google Scholar]
- 491. Liu L, Li A, Matthews SG. 2001. Maternal glucocorticoid treatment programs HPA regulation in adult offspring: sex-specific effects. Am J Physiol Endocrinol Metab 280:E729–E739 [DOI] [PubMed] [Google Scholar]
- 492. O'Regan D, Kenyon CJ, Seckl JR, Holmes MC. 2004. Glucocorticoid exposure in late gestation in the rat permanently programs gender-specific differences in adult cardiovascular and metabolic physiology. Am J Physiol Endocrinol Metab 287:E863–E870 [DOI] [PubMed] [Google Scholar]
- 493. Kapoor A, Matthews SG. 2008. Prenatal stress modifies behavior and hypothalamic-pituitary-adrenal function in female guinea pig offspring: effects of timing of prenatal stress and stage of reproductive cycle. Endocrinology 149:6406–6415 [DOI] [PubMed] [Google Scholar]
- 494. Dunn E, Kapoor A, Leen J, Matthews SG. 2010. Prenatal synthetic glucocorticoid exposure alters hypothalamic-pituitary-adrenal regulation and pregnancy outcomes in mature female guinea pigs. J Physiol 588:887–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495. Newnham JP, Jobe AH. 2009. Should we be prescribing repeated courses of antenatal corticosteroids? Semin Fetal Neonatal Med 14:157–163 [DOI] [PubMed] [Google Scholar]
- 496. Power C, Hertzman C, Matthews S, Manor O. 1997. Social differences in health: life-cycle effects between ages 23 and 33 in the 1958 British birth cohort. Am J Public Health 87:1499–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497. Hertzman C, Power C, Matthews S, Manor O. 2001. Using an interactive framework of society and lifecourse to explain self-rated health in early adulthood. Soc Sci Med 53:1575–1585 [DOI] [PubMed] [Google Scholar]
- 498. Power C, Jefferis BJ, Manor O, Hertzman C. 2006. The influence of birth weight and socioeconomic position on cognitive development: does the early home and learning environment modify their effects? J Pediatr 148:54–61 [DOI] [PubMed] [Google Scholar]
- 499. Nagin D, Tremblay RE. 1999. Trajectories of boys' physical aggression, opposition, and hyperactivity on the path to physically violent and nonviolent juvenile delinquency. Child Dev 70:1181–1196 [DOI] [PubMed] [Google Scholar]
- 500. Broidy LM, Nagin DS, Tremblay RE, Bates JE, Brame B, Dodge KA, Fergusson D, Horwood JL, Loeber R, Laird R, Lynam DR, Moffitt TE, Pettit GS, Vitaro F. 2003. Developmental trajectories of childhood disruptive behaviors and adolescent delinquency: a six-site, cross-national study. Dev Psychol 39:222–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501. Pryce CR, Feldon J. 2003. Long-term neurobehavioural impact of the postnatal environment in rats: manipulations, effects and mediating mechanisms. Neurosci Biobehav Rev 27:57–71 [DOI] [PubMed] [Google Scholar]
- 502. de Kloet ER, Sibug RM, Helmerhorst FM, Schmidt MV, Schmidt M. 2005. Long-term neurobehavioural impact of the postnatal environment in rats: manipulations, effects and mediating mechanisms. Neurosci Biobehav Rev 29:271–281 [DOI] [PubMed] [Google Scholar]
- 503. Spivey J, Barrett D, Padilla E, Gonzalez-Lima F. 2008. Mother-infant separation leads to hypoactive behavior in adolescent Holtzman rats. Behav Processes 79:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504. Fride E, Weinstock M. 1989. Alterations in behavioral and striatal dopamine asymmetries induced by prenatal stress. Pharmacol Biochem Behav 32:425–430 [DOI] [PubMed] [Google Scholar]
- 505. Hockman CH. 1961. Prenatal maternal stress in the rat: its effects on emotional behavior in the offspring. J Comp Physiol Psychol 54:679–684 [DOI] [PubMed] [Google Scholar]
- 506. Thompson WR, Quinby S. 1964. Prenatal maternal anxiety and offspring behavior: parental activity and level of anxiety. J Genet Psychol 105:359–371 [DOI] [PubMed] [Google Scholar]
- 507. Suchecki D, Palermo Neto J. 1991. Prenatal stress and emotional response of adult offspring. Physiol Behav 49:423–426 [DOI] [PubMed] [Google Scholar]
- 508. Glover V, O'Connor TG, O'Donnell K. 2010. Prenatal stress and the programming of the HPA axis. Neurosci Biobehav Rev 35:17–22 [DOI] [PubMed] [Google Scholar]
- 509. Meaney MJ. 2001. Maternal care, gene expression, and the transmission of individual differences in stress reactivity across generations. Annu Rev Neurosci 24:1161–1192 [DOI] [PubMed] [Google Scholar]
- 510. Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. 2004. Epigenetic programming by maternal behavior. Nat Neurosci 7:847–854 [DOI] [PubMed] [Google Scholar]
- 511. Weaver IC, D'Alessio AC, Brown SE, Hellstrom IC, Dymov S, Sharma S, Szyf M, Meaney MJ. 2007. The transcription factor nerve growth factor-inducible protein A mediates epigenetic programming: altering epigenetic marks by immediate-early genes. J Neurosci 27:1756– 1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512. Weaver IC, Champagne FA, Brown SE, Dymov S, Sharma S, Meaney MJ, Szyf M. 2005. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci 25:11045–11054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513. Weaver IC, Meaney MJ, Szyf M. 2006. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci USA 103:3480–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514. Detich N, Hamm S, Just G, Knox JD, Szyf M. 2003. The methyl donor S-adenosylmethionine inhibits active demethylation of DNA: a candidate novel mechanism for the pharmacological effects of S-adenosylmethionine. J Biol Chem 278:20812–20820 [DOI] [PubMed] [Google Scholar]
- 515. Miller CA, Sweatt JD. 2007. Covalent modification of DNA regulates memory formation. Neuron 53:857–869 [DOI] [PubMed] [Google Scholar]
- 516. Miller CA, Campbell SL, Sweatt JD. 2008. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol Learn Mem 89:599–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517. Brown SE, Szyf M. 2007. Epigenetic programming of the rRNA promoter by MBD3. Mol Cell Biol 27:4938–4952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518. McGowan PO, Sasaki A, Huang TC, Unterberger A, Suderman M, Ernst C, Meaney MJ, Turecki G, Szyf M. 2008. Promoter-wide hypermethylation of the ribosomal RNA gene promoter in the suicide brain. PLoS One 3:e2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519. Anway MD, Skinner MK. 2006. Epigenetic transgenerational actions of endocrine disruptors. Endocrinology 147:S43–S49 [DOI] [PubMed] [Google Scholar]
- 520. Trasler JM. 1998. Origin and roles of genomic methylation patterns in male germ cells. Semin Cell Dev Biol 9:467–474 [DOI] [PubMed] [Google Scholar]
- 521. McLaren A. 2003. Primordial germ cells in the mouse. Dev Biol 262:1–15 [DOI] [PubMed] [Google Scholar]
- 522. Allegrucci C, Thurston A, Lucas E, Young L. 2005. Epigenetics and the germline. Reproduction 129:137–149 [DOI] [PubMed] [Google Scholar]
- 523. Flanagan JM, Popendikyte V, Pozdniakovaite N, Sobolev M, Assadzadeh A, Schumacher A, Zangeneh M, Lau L, Virtanen C, Wang SC, Petronis A. 2006. Intra- and interindividual epigenetic variation in human germ cells. Am J Hum Genet 79:67–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524. Cupp AS, Skinner MK. 2001. Actions of the endocrine disruptor methoxychlor and its estrogenic metabolite on in vitro embryonic rat seminiferous cord formation and perinatal testis growth. Reprod Toxicol 15:317–326 [DOI] [PubMed] [Google Scholar]
- 525. Cupp AS, Uzumcu M, Suzuki H, Dirks K, Phillips B, Skinner MK. 2003. Effect of transient embryonic in vivo exposure to the endocrine disruptor methoxychlor on embryonic and postnatal testis development. J Androl 24:736–745 [DOI] [PubMed] [Google Scholar]
- 526. Anway MD, Skinner MK. 2008. Transgenerational effects of the endocrine disruptor vinclozolin on the prostate transcriptome and adult onset disease. Prostate 68:517–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527. Anway MD, Rekow SS, Skinner MK. 2008. Transgenerational epigenetic programming of the embryonic testis transcriptome. Genomics 91:30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528. Skinner MK, Anway MD, Savenkova MI, Gore AC, Crews D. 2008. Transgenerational epigenetic programming of the brain transcriptome and anxiety behavior. PLoS One 3:e3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529. Crews D, Gore AC, Hsu TS, Dangleben NL, Spinetta M, Schallert T, Anway MD, Skinner MK. 2007. Transgenerational epigenetic imprints on mate preference. Proc Natl Acad Sci USA 104:5942–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530. Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. 2006. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect 114:567–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531. Badger TM, Gilchrist JM, Pivik RT, Andres A, Shankar K, Chen JR, Ronis MJ. 2009. The health implications of soy infant formula. Am J Clin Nutr 89:1668S–1672S [DOI] [PubMed] [Google Scholar]
- 532. Li S, Hursting SD, Davis BJ, McLachlan JA, Barrett JC. 2003. Environmental exposure, DNA methylation, and gene regulation: lessons from diethystilbestrol-induced cancers. Ann NY Acad Sci 983:161–169 [DOI] [PubMed] [Google Scholar]
- 533. Li S, Washburn KA, Moore R, Uno T, Teng C, Newbold RR, McLachlan JA, Negishi M. 1997. Developmental exposure to diethylstilbestrol elicits demethylation of estrogen-responsive lactoferrin gene in mouse uterus. Cancer Res 57:4356–4359 [PubMed] [Google Scholar]
- 534. Li S, Hansman R, Newbold R, Davis B, McLachlan JA, Barrett JC. 2003. Neonatal diethylstilbestrol exposure induces persistent elevation of c-fos expression and hypomethylation in its exon-4 in mouse uterus. Mol Carcinog 38:78–84 [DOI] [PubMed] [Google Scholar]
- 535. Cederroth CR, Vinciguerra M, Gjinovci A, Kühne F, Klein M, Cederroth M, Caille D, Suter M, Neumann D, James RW, Doerge DR, Wallimann T, Meda P, Foti M, Rohner-Jeanrenaud F, Vassalli JD, Nef S. 2008. Dietary phytoestrogens activate AMP-activated protein kinase with improvement in lipid and glucose metabolism. Diabetes 57:1176–1185 [DOI] [PubMed] [Google Scholar]
- 536. Cederroth CR, Vinciguerra M, Kühne F, Madani R, Doerge DR, Visser TJ, Foti M, Rohner-Jeanrenaud F, Vassalli JD, Nef S. 2007. A phytoestrogen-rich diet increases energy expenditure and decreases adiposity in mice. Environ Health Perspect 115:1467–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537. Cederroth CR, Nef S. 2009. Fetal programming of adult glucose homeostasis in mice. PLoS One 4:e7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538. Szyf M. 2001. Towards a pharmacology of DNA methylation. Trends Pharmacol Sci 22:350–354 [DOI] [PubMed] [Google Scholar]
- 539. Dashwood RH, Myzak MC, Ho E. 2006. Dietary HDAC inhibitors: time to rethink weak ligands in cancer chemoprevention? Carcinogenesis 27:344–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540. Myzak MC, Tong P, Dashwood WM, Dashwood RH, Ho E. 2007. Sulforaphane retards the growth of human PC-3 xenografts and inhibits HDAC activity in human subjects. Exp Biol Med (Maywood) 232:227–234 [PMC free article] [PubMed] [Google Scholar]
- 541. Borra MT, Smith BC, Denu JM. 2005. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem 280:17187–17195 [DOI] [PubMed] [Google Scholar]
- 542. Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, Welsh W, Yang CS. 2003. Tea polyphenol (−)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res 63:7563–7570 [PubMed] [Google Scholar]
- 543. Amacher DE. 2010. The discovery and development of proteomic safety biomarkers for the detection of drug-induced liver toxicity. Toxicol Appl Pharmacol 245:134–142 [DOI] [PubMed] [Google Scholar]
- 544. Morrow DA, de Lemos JA. 2007. Benchmarks for the assessment of novel cardiovascular biomarkers. Circulation 115:949–952 [DOI] [PubMed] [Google Scholar]
- 545. Thomas JC, Vohra RS, Beer S, Bhatti K, Ponnambalam S, Homer-Vanniasinkam S. 2009. Biomarkers in peripheral arterial disease. Trends Cardiovasc Med 19:147–151 [DOI] [PubMed] [Google Scholar]
- 546. Castro-Chavez F, Yechoor VK, Saha PK, Martinez-Botas J, Wooten EC, Sharma S, O'Connell P, Taegtmeyer H, Chan L. 2003. Coordinated upregulation of oxidative pathways and downregulation of lipid biosynthesis underlie obesity resistance in perilipin knockout mice. Diabetes 52:2666–2674 [DOI] [PubMed] [Google Scholar]
- 547. Kutlu B, Cardozo AK, Darville MI, Kruhøffer M, Magnusson N, Ørntoft T, Eizirik DL. 2003. Discovery of gene networks regulating cytokine-induced dysfunction and apoptosis in insulin-producing INS-1 cells. Diabetes 52:2701–2719 [DOI] [PubMed] [Google Scholar]
- 548. Roy S, Rink C, Khanna S, Phillips C, Bagchi D, Bagchi M, Sen CK. 2004. Body weight and abdominal fat gene expression profile in response to a novel hydroxycitric acid-based dietary supplement. Gene Expr 11:251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549. Ueki T, Toyota M, Sohn T, Yeo CJ, Issa JP, Hruban RH, Goggins M. 2000. Hypermethylation of multiple genes in pancreatic adenocarcinoma. Cancer Res 60:1835–1839 [PubMed] [Google Scholar]
- 550. Youssef EM, Chen XQ, Higuchi E, Kondo Y, Garcia-Manero G, Lotan R, Issa JP. 2004. Hypermethylation and silencing of the putative tumor suppressor tazarotene-induced gene 1 in human cancers. Cancer Res 64:2411–2417 [DOI] [PubMed] [Google Scholar]
- 551. Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, Li K, Murty VV, Schupf N, Vilain E, Morris M, Haghighi F, Tycko B. 2008. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet 40:904–908 [DOI] [PubMed] [Google Scholar]
- 552. Pomraning KR, Smith KM, Freitag M. 2009. Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods 47:142–150 [DOI] [PubMed] [Google Scholar]
- 553. Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, Li K, Murty VV, Schupf N, Vilain E, Morris M, Haghighi F, Tycko B. 2008. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet 40:904–908 [DOI] [PubMed] [Google Scholar]
- 554. Dennis Lo YM, Chiu RW. 2007. Prenatal diagnosis: progress through plasma nucleic acids. Nat Rev Genet 8:71–77 [DOI] [PubMed] [Google Scholar]
- 555. Chim SS, Tong YK, Chiu RW, Lau TK, Leung TN, Chan LY, Oudejans CB, Ding C, Lo YM. 2005. Detection of the placental epigenetic signature of the maspin gene in maternal plasma. Proc Natl Acad Sci USA 102:14753–14758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556. Bouret SG, Draper SJ, Simerly RB. 2004. Trophic action of leptin on hypothalamic neurons that regulate feeding. Science 304:108–110 [DOI] [PubMed] [Google Scholar]
- 557. Kaminen-Ahola N, Ahola A, Maga M, Mallitt KA, Fahey P, Cox TC, Whitelaw E, Chong S. 2010. Maternal ethanol consumption alters the epigenotype and the phenotype of offspring in a mouse model. PLoS Genet 6:e1000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558. Hurd PJ, Nelson CJ. 2009. Advantages of next-generation sequencing versus the microarray in epigenetic research. Brief Funct Genomic Proteomic 8:174–183 [DOI] [PubMed] [Google Scholar]
- 559. Nijland MJ, Mitsuya K, Li C, Ford S, McDonald TJ, Nathanielsz PW, Cox LA. 2010. Epigenetic modification of fetal baboon hepatic phosphoenolpyruvate carboxykinase following exposure to moderately reduced nutrient availability. J Physiol 588:1349–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560. Green AS, Limesand SW. 2010. Remembering development—epigenetic responses to fetal malnutrition. J Physiol 588:1379–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 561. Allis CD, Jenuwein T, Reinberg D. 2007. Overview and concepts. In: Allis CD, Jenuwein T, Reinberg D, Caparros M-L. eds. Epigenetics. Chap 3 Woodbury, NY: Cold Spring Harbor Laboratory Press; 23–61 [Google Scholar]
- 562. Turner BM. 2007. Defining an epigenetic code. Nat Cell Biol 9:2–6 [DOI] [PubMed] [Google Scholar]
- 563. Sandovici I, Smith N, Ozanne S, Constancia M. 2008. The dynamic epigenome: the impact of the environment on epigenetic regulation of gene expression and developmental programming. In: Tost J. ed. Epigenetics. Chap 15 Norfolk, UK: Horizon Scientific Press/Caister Academic Press; 344–370 [Google Scholar]
- 564. Levit GS, Hossfeld U, Olsson L. 2006. From the “modern synthesis” to cybernetics: Ivan Ivanovich Schmalhausen (1884–1963) and his research program for a synthesis of evolutionary and developmental biology. J Exp Zool B Mol Dev Evol 306:89–106 [DOI] [PubMed] [Google Scholar]
- 565. Waddington CH. 1975. The evolution of an evolutionist. Ithica, NY: Cornell University Press [Google Scholar]