Abstract
Myc protein plays a fundamental role in regulation of cell cycle, proliferation, differentiation and apoptosis by modulating the expression of a large number of targets. Here we report the transactivation ability of the human Myc protein to activate the SUMO-activating enzyme SAE1 transcription. We found that Myc activates SAE1 transcription via direct binding to canonical E-Boxes sequences located close to the SAE1 transcription start site. A recent report has highlighted the crucial role of the SAE gene expression in Myc mediated oncogenesis. Our study adds new insight in this context since we show here that Myc directly activates SAE1 transcription, suggesting that Myc oncogenic activity which depends on SAE1 is ensured by Myc itself through direct binding and transcriptional activation of SAE1 expression.
Keywords: Myc, SUMOylation, SAE1, transcription
Introduction
c-Myc is a regulatory gene that codes for a basic helix-loop-helix transcription factor (Myc) that can both activate and repress transcription. Myc activates transcription of dedicated target genes as component of the Myc/Max complex, which binds to specific DNA sequences, termed E-box [1,2]. Oncogenic activation of Myc promotes regulation of a plethora of genes in the cell and the delicate balance of pro and anti tumorigenic properties of its targets defines the fate of the cells. Understanding the Myc-driven transcriptional program represent one of the most challenging aspect to decipher the molecular mechanisms underlying Myc-induced tumorigenesis [2,3].
Here we report a novel and important target of Myc activity. We found that SAE1 transcription is activated by Myc through recruitment to E-box sequences located at SAE1 transcriptional start site. SAE1 in association with SAE2 form a critical component of the sole SUMO-activating enzyme necessary for SUMO conjugation to proteins [4]. SAE1 together with its partner SAE2/ UBA2 has been recently shown to be necessary for the tumorigenicity of Myc-dependent cancer cells in vitro and in vivo [5], and most importantly gene expression analysis of Myc-high human breast cancer suggested that low SAE1/2 abundance in the tumors correlates with longer survival of the patients [5]. The findings reported here, showing that Myc directly activates SAE1 transcription, add new insight in this context, suggesting that Myc oncogenic activity which depends on SAE1 is ensured by Myc itself through direct binding and transcriptional activation of SAE1 expression.
Materials and methods
Cell culture and drugs
RAT1 cells expressing a 4-hydroxytamoxifen (OHT)-inducible MycER chimera and RAT-Myc-/- cells were cultured in DMEM medium supplemented with 10% fetal calf serum and MycER induction was carried out with OHT (1μM) as recently described [6,7]. Cells were made quiescent by contact inhibition followed by serum removal for two days. To induce entry into the cycle, synchronized Go arrested cells were treated alone or OHT plus serum or serum alone as indicated in the text and harvested at the indicated times. Human retinal hT-RPE-MycER cell lines in which the inducible Myc-estrogen receptor fusion transgene (MycER protein) is activated upon treatment with tamoxifen (OHT) were grown and OHT-treated as recently described [8].
Transfections and siRNA
To carry out transient transfection experiments in RAT-Myc-/- and RAT-MycER cells, we used MicroPoRATor Digital Bio Technology, a pipette-type electroporation [6,7]. Indicated plasmids, DNAs or siRNA were introduced into each 3X106-dissociated cells in 100-μl volume according to manufacturer’s instructions. For siRNA treatments, ON-TARGETplus SMARTpool (L-003282-00-0005) Myc; and ON-TARGETplus Non-targeting pool (D-001810-10-5) were obtained from Dharmacon.
mRNA quantification by qPCR
cDNA was prepared from total RNA with the Quantitect Reverse Transcription Kit (Qiagen) according to manufactory instructions. Each sample was assayed in triplicate. The qPCR data were normalized to the expression of the housekeeping beta-glucuronidase (GUS) gene and after normalization the data were presented as fold change relative to the 0 point [6].
Quantitative chromatin immunoprecipitation (qChIP)
qChIP experiments were performed essentially as described [6,7]. For qPCR 3μl out of 150μl of immunoprecipitated DNA was used with primers described in Supplementary Data. ACHR promoter amplicon was used as negative control in all experiments. Normal serum and input DNA values were used to subtract/normalize the values from qChIP samples by using: % Input = 2DCtx3; DCt = Ct(input)-Ct(cIP). qRT-PCR and qChIP data are presented as means of three at least independent biological experiments each analyzed by triplicate (± SD).
Results and discussion
We have recently shown that the proapoptotic BBC3/PUMA (p53 up-regulated modulator of apoptosis) gene is a Myc target and through recruitment to an E-box binding site on its promoter, Myc cooperates with the PI3/AKT pathway to repress FOXO3a mediated activation of PUMA expression [7]. In the course of this study we were intrigued to observe a Myc responsive RNAPII peak in the second intron of PUMA locus in condition in which the PUMA expression was inhibited (data not shown). This finding prompted us to investigate the presence of actively expressed genes responsive to Myc activation in the region downstream of PUMA. Exploring the genomic region downstream of PUMA locus we found the presence of SUMO-activating enzyme SAE1 gene whose transcription proceeds in the opposite strand to PUMA.
To determine the expression of SAE1 in the presence of hyper-activated Myc protein, we used two Myc-inducible cell lines: the rat fibroblast RAT-MycER and the human retinal hT-RPE-MycER cell lines in which the inducible Myc-estrogen receptor fusion transgene (MycER protein) is activated upon treatment with tamoxifen (OHT) [6-8].
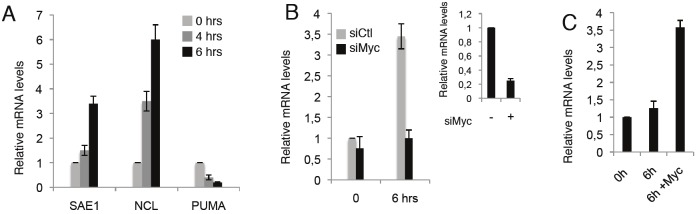
As shown in Figure 1A, in RAT-MycER, SAE1 expression as well as the known Nucleolin (NCL) Myc target, increase in response to Myc activation while the levels of expression of PUMA are inhibited as previously described [6]. To determine the contribution of Myc in the SAE1 activation we
Figure 1.
Myc activates SAE1 expression. (A) mRNA expression levels of SAE1, NCL and PUMA were quantified by qRT–PCR in quiescent cells (0) and after 4 and 6hrs of treatment with serum and OHT. (B) Myc expression was inhibited with specific siRNA (siMyc) and siRNA non-targeting (siCtl) was used as scrambled RNAs. SAE1 mRNAs expression levels were quantified by qRT–PCR in quiescent cells (0) and 6hrs of treatment with serum and OHT. The efficiency of Myc silencing by siRNA treatments measured by qRT–PCR is shown on the right. (C) RAT-Myc-/- cells as well as cells transfected with a Myc expression vector were serum deprived for 48 hrs. SAE1 mRNA levels were evaluated by qRT– PCR 6 hrs after serum induction. Values were compared to quiescent cells (0).
silenced Myc expression by transfection of specific siRNAs in the RAT-MycER cells and we found that Myc silencing inhibits activation of SAE1 transcription (Figure 1B). To further determine the contribution of Myc in SAE1 activation, we used the isogenic RAT-Myc-/- cells and measured SAE1 expression levels in starved versus serum-induced cells. As shown in Figure 1C, SAE1 expression slightly increases upon serum addiction while the expression was significantly higher when a Myc expression vector was introduced by transfection into RAT-Myc-/- cells. Collectively these results suggest that SAE1 expression is indeed regulated by Myc.
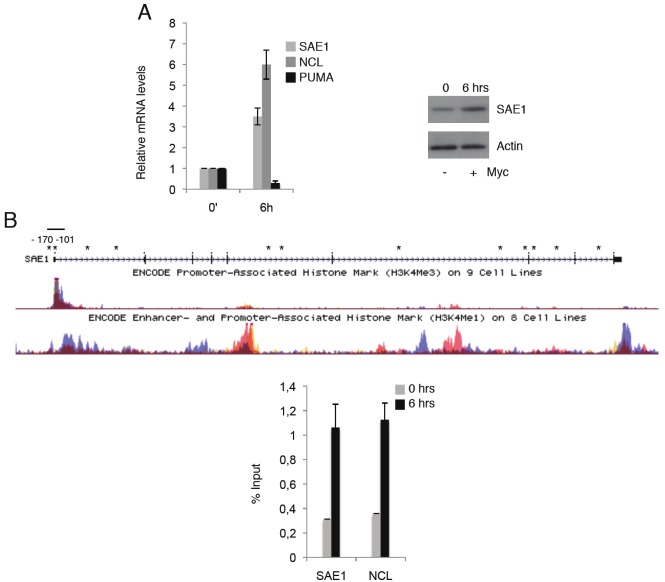
SAE1 sequence is extremely conserved between rat and human suggesting an evolutionary fundamental role in cell physiology. In this respect we analyzed SAE1 expression in the retinal human Myc inducible cell line hT-RPE-MycER [8] and, as shown in Figure 2A, Myc activation results in induction
Figure 2.
(A) hT-RPE-MycER cells were synchronized by 2 days of growth factors deprivation. SAE1, NCL and PUMA mRNAs expression levels were quantified by qRT–PCR in synchronized (0) and cells treated with growth factor + OHT (6hrs). All mRNA levels were normalized to β-glucuronidase (GUS) mRNA levels and all values represent the average of at least three independent experiments. Error bars indicate SD. Protein expression is shown in immunoblots of whole-cell extracts with anti-SAE1 and actin for loading control. (B) An adapted UCSC genome browser view of SAE1 genomic sequence displaying H3K4me3 and H3K4me1 levels and putative E-box (asterisks). Myc occupancy on SAE1 and NCL chromatin in synchronized (0) and Myc induced hT-RPE-MycER cells for 6hrs is shown. ChIP-enriched DNA was quantified by real-time PCR analysis using amplicon covering the region containing the Myc binding E-boxes as shown in Figure 2A.
of SAE1 expression at levels comparable to the known Nucleolin (NCL), a well defined Myc target. To explore the role of Myc in SAE1 activation we scanned its genomic sequences for putative Myc binding sites. Several E-boxes were found, Figure 2B, and we focalized our attention on two closely associated E-boxes in position close to the TSS site (-170 and -101). Most importantly these E-boxes are located in a region with high level of H3K4Me3, a prerequisite for Myc binding (Figure 2B) [9,10]. To assess Myc occupancy on SAE1 we carried out qChIP analysis in the RPE cells, which were growth factors-deprived for 2 days (0) and treated for 6hrs with OHT for Myc activation. Myc-immunoprecipitated chromatin was analyzed using amplicons spanning the SAE1 and NCL E-boxes and the qChIP data (Figure 2B) show that Myc is recruited on the E-box sites at the SAE1 promoter with efficiency comparable to recruitment on NCL. Although we cannot exclude the presence of additional E-box binding sites that might contribute to Myc activation, we can conclude that SAE1 is a bona-fide positively regulated Myc target through a direct binding to the E-boxes close to SAE1 TSS site.
While our studies on Myc role on SAE1 expression were in progress it has been reported the identification of genes vital to support Myc-addicted tumors through a genome wide genetic screen for Myc-synthetic lethal (MySL) shRNAs in human mammary epithelial cells [5]. The most significant candidates that have been isolated in this study are the SAE1 and SAE2 genes whose products associate to form the heterodimer that is a critical component of the SUMO activating enzyme needed for SUMO conjugation to proteins [4]. The authors found that SAE was required for growth of Myc dependent tumors and that the low expression of SAE1/2 in human breast cancers with high Myc expression levels correlates with longer survival of the patients. SAE depleted cells with high levels of Myc expression were found impaired in a correct mitotic spindle formation and consequently committed to apoptosis highlighting the crucial role of the SAE gene expression in Myc mediated oncogenesis.
Our study adds new insight in this context since we show here that Myc directly activates SAE1 transcription, suggesting that Myc oncogenic activity which depends on SAE1 is ensured by Myc itself through direct binding and transcriptional activation of SAE1 expression. Thus, recruitment of Myc on SAE1 modulates its expression supporting Myc oncogenic program by activation of Sumoylation dependent Myc-switchers (SMS genes).
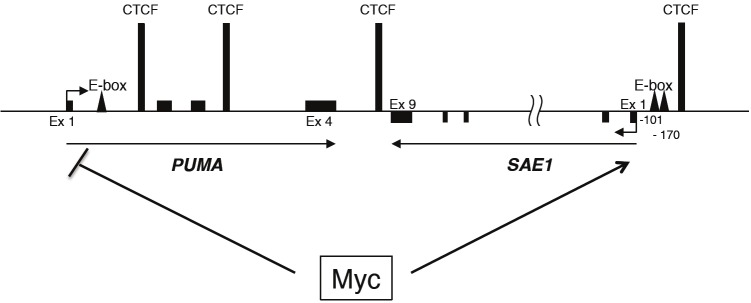
Intriguing is the fact that the PUMA and SAE1 are adjacent on the human chromosome 19 and that their expression has been always found inversely correlated. While Myc cooperates with PI3/AKT pathway to repress PUMA transcription, hyper activation of Myc activates SAE1 transcription. An extensive analysis of chromatin domains at the PUMA locus and surrounding region including SAE1, has been recently provided by the Espinosa’s laboratory [11,12]. It has been identified a peculiar transcriptional organization of the locus with the presence of the insulator protein CTCF (CCCTC-binding factor) and the associated cohesin complex at intragenic PUMA as well as at intergenic regions between PUMA and SAE1 (Figure 3). Since CTCF functions involve the formation of ‘chromatin loops’ and these loops seems to be regulated in a signaling specific manner [13], it is possible that Myc binding might induce an intergenic gene looping between PUMA and SAE1 leading to the divergent Myc-mediated transcription control of SAE1 and PUMA loci. Further investigations will be necessary to validate these speculative hypotheses.
Figure 3.
A general schematic of SAE1/PUMA locus showing CTCF binding sites and identified Myc binding sites (E-box).
Acknowledgments
We thank L Lania for stimulating discussions and I Iaccarino for providing the hT-RPE-MycER cell line. This work was supported by a grant from MIUR (PRIN) to B.M.
References
- 1.Wasylishen AR, Penn LZ. Myc: The beauty and the beast. Genes and Cancer. 2010;1:532–541. doi: 10.1177/1947601910378024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 3.Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hay RT. SUMO. Mol Cell. 2005;18:1–7. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, Rao M, Yu P, Dominguez-Vidana R, Liang AC, Solimini NL, Bernardi RJ, Yu B, Hsu T, Golding I, Luo J, Osborne CK, Creighton CJ, Hilsenbeck SG, Schiff R, Shaw CA, Elledge SJ, Westbrook TF. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335:348–353. doi: 10.1126/science.1212728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amente S, Zhang J, Lubrano Lavadera M, Lania L, Avvedimento EV, Majello B. Myc and PI3K/ AKT signaling cooperatively repress FOXO3-adependent PUMA and GADD45a gene expression. Nucleic Acids Res. 2011;39:9498–9507. doi: 10.1093/nar/gkr638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amente S, Bertoni A, Morano A, Lania L, Avvedimento EV, Majello B. LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription. Oncogene. 2010;29:3691–3702. doi: 10.1038/onc.2010.120. [DOI] [PubMed] [Google Scholar]
- 8.Alfano D, Votta G, Schulze A, Downward J, Caputi M, Stoppelli MP, Iaccarinoi I. Modulation of cellular migration and survival by c-Myc through the downregulation of Urokinase (uPA) and uPA receptor. Mol Cell Biol. 2010;30:1838–1851. doi: 10.1128/MCB.01442-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall' Olio V, Zardo G, Nervi C, Bernard L, Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 10.Martinato F, Cesaroni M, Amati B, Guccione E. Analysis of Myc-induced histone modifications on target chromatin. PLoS One. 2008;3:e3650. doi: 10.1371/journal.pone.0003650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomez NP, Espinosa JM. Disparate chromatin landscapes and kinetics of inactivation impact differential regulation of p53 target genes. Cell Cycle. 2010;9:3428–3437. doi: 10.4161/cc.9.17.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez NP, Espinosa JM. Gene-specific repression of the p53 target gene PUMA via intragenic CTCF-Cohesin binding. Genes Dev. 2010;24:1022–1034. doi: 10.1101/gad.1881010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesion. Proc Natl Acad Sci USA. 2010;107:3651–3756. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]