Abstract
A DNA double strand break (DSB) is a highly toxic lesion, which can generate genetic instability and profound genome rearrangements. However, DSBs are required to generate diversity during physiological processes such as meiosis or the establishment of the immune repertoire. Thus, the precise regulation of a complex network of processes is necessary for the maintenance of genomic stability, allowing genetic diversity but protecting against genetic instability and its consequences on oncogenesis. Two main strategies are employed for DSB repair: homologous recombination (HR) and non-homologous end-joining (NHEJ). HR is initiated by single-stranded DNA (ssDNA) resection and requires sequence homology with an intact partner, while NHEJ requires neither resection at initiation nor a homologous partner. Thus, resection is an pivotal step at DSB repair initiation, driving the choice of the DSB repair pathway employed. However, an alternative end-joining (A-EJ) pathway, which is highly mutagenic, has recently been described; A-EJ is initiated by ssDNA resection but does not require a homologous partner. The choice of the appropriate DSB repair system, for instance according the cell cycle stage, is essential for genome stability maintenance. In this context, controlling the initial events of DSB repair is thus an essential step that may be irreversible, and the wrong decision should lead to dramatic consequences. Here, we first present the main DSB repair mechanisms and then discuss the importance of the choice of the appropriate DSB repair pathway according to the cell cycle phase. In a third section, we present the early steps of DSB repair i.e., DSB signaling, chromatin remodeling, and the regulation of ssDNA resection. In the last part, we discuss the competition between the different DSB repair mechanisms. Finally, we conclude with the importance of the fine tuning of this network for genome stability maintenance and for tumor protection in fine.
Keywords: DNA double strand break, Homologous recombination, Non homologous end joining, alternative end-joining, Resection, chromatin remodeling, genetic instability, genome rearrangements
Introduction
The coordination of a complex network of metabolic pathways is essential for the maintenance, duplication, and transmission of the genome. This network controls the DNA damage response (DDR) pathway, associating DNA replication, recombination and repair with cell cycle control. A defect in any of the participants in these pathways results in genetic instability and tumor predisposition.
Understanding the control of genome integrity is essential in cancer studies at the cognitive, diagnostic and therapeutic levels. Indeed, tumor initiation requires an initial event that modifies/alters the genetic information. It is therefore notable that the presence of DNA breaks and activation of the DDR have been observed in the pre-cancerous stages of untreated cells. This activation of the DDR pathway is considered the response to spontaneous endogenous replicative stress [1,2]. Indeed, replication forks are routinely arrested by a wide variety of endogenous stresses including bulging regions in the DNA, regions of DNA/RNA hybrids and endogenous cell metabolism, which produces reactive oxygen species (ROS) [3]. ROS generate damaged bases, which can block replication progression and oxidize DNA replication or repair proteins, thus inactivating them [4-6]. In addition, genetic instability, which also favors tumor progression, is a hallmark of cancer cells. First, DNA damaging agents are generally carcinogenic, and second, many inherited syndromes affecting the DDR predispose to genetic instability and cancer [7]. Furthermore, at present, of the 12 germline mutations that confer a familial predisposition to breast cancer, 11 directly affect genes involved in the DDR, particularly those at the replication/recombination interface. In particular, the most frequently mutated genes are BRCA1 and BRCA2, which play key roles in homologous recombination [8-11]. The 12th mutated gene, PTEN, also affects homologous recombination, either directly through the regulation of the RAD51 gene or indirectly through the regulation of the AKT1 signaling pathway [9,12-16]. This over-representation of genes involved in a specific metabolic pathway emphasizes the importance of the DDR in the etiology of breast cancer. In addition, the DDR can also be altered in sporadic breast cancer (which represents 80 to 90% of all breast cancers) through the dysregulation of the AKT1/PTEN signaling pathway because AKT1 has been found to be up regulated in 40 to 60% of sporadic breast cancers [9,13,17,18].
Thus, a profound knowledge of the intrinsic processes governing genome integrity maintenance and transmission is essential to understand cancer etiology from its genesis and progression up to metastatic invasion. Furthermore, DNA is a widely used target to cure cancer. Indeed, treatments based on ionizing radiation, DNA damaging chemicals, and synthetic lethality of DNA metabolism pathways are efficient and promising strategies used in cancer therapy. Consequently, understanding the DDR is also important for the development of innovative and personalized approaches to improve therapeutic strategies for killing cancer cells while preserving adjacent healthy tissue.
A DNA double strand break (DSB) is a particular lesion that is highly toxic and can generate genetic rearrangements; therefore, it is at the crossroads between genetic variability and instability. DSBs can be generated by exogenous stresses, such as ionizing radiation (IR), and endogenous stresses, such as endogenous oxidative or replication stresses. Indeed, prolonged replication fork arrest leads to DSB accumulation[19,20]. Replication forks can be arrested by exogenous chemotherapeutic agents (e.g., interstrand crosslinking agents and topoisomerase inhibitors) or by a variety of endogenous stresses as discussed above [3].
However, in some common tightly regulated physiological processes, such as meiosis and immune repertoire generation, DSBs are used to promote genetic diversity. In these cases, DSBs are controlled by cellular enzymes, but the repair mechanisms use the same systems as those used for the repair of stress-induced breaks. Moreover, while different molecular mechanisms compete or collaborate for DSB repair [21-25], the choice of the appropriate repair pathway, notably according to the cell cycle phases, is essential to maintain genome stability [26]. Therefore, precise regulation is necessary to control the equilibrium between genetic stability and diversity while avoiding genetic instability.
Two main strategies allow the repair of DSBs:homologous recombination (HR) and nonhomologous end joining (NHEJ). HR requires sequence homology and is initiated by 5’ to 3’ single-stranded DNA (ssDNA) resection; the resulting 3’ ssDNA invades then an intact homologous duplex, driving the pairing of homologous sequences and strand exchange. In contrast, canonical NHEJ (C-NHEJ) is dependent upon neither sequence homology nor ssDNA resection. Thus, resection appears to be an essential step in determining the DSB repair pathway employed, favoring HR initiation. Recently, an alternative end-joining (A-EJ) pathway has been described that, like cannonical C-NHEJ, canonical C-NHEJ, it does not require sequence homology, but it is initiated by ssDNA resection in a manner similar to HR [27-29]. Importantly, A-EJ appears to be a major source of translocations [30-35]. Therefore, controlling the initiation of resection is a pivotal step determining the DSB repair pathway employed.
Here, we will first briefly present the different DSB repair systems and discuss the importance of the choice of the DSB repair process according to the cell cycle stage. Second, we will describe the early molecular steps of DSB repair leading to an essential process for determining the choice of the repair pathway used: single-stranded DNA resection.
The different DSB repair pathways
Homologous Recombination
The products of HR are gene conversion (i.e., the non-reciprocal exchange of genetic information) and crossing over (i.e., the reciprocal exchange of adjacent sequences). The proposed DSB repair model [36] accounts for the two products of HR: gene conversion associated with and without crossover (Figure 1A). HR is initiated by single-stranded DNA (ssDNA) resection, which generates a 3’ tail coated with the ssDNA binding protein RPA. Afterward, the BRCA2/PALB2 complex removes RPA from the ssDNA and loads the pivotal HR protein RAD51, which then promotes the invasion of a homologous double-stranded DNA (dsDNA), leading to strand exchange. The process of DNA strand exchange tolerates some differences between the molecules involved, thereby creating a hybrid dsDNA molecule called a heteroduplex, which carries mismatches. Nevertheless, a minimal length of perfect homology, i.e., the MEPS (Minimal Efficient Processing Segment) is necessary for efficient HR initiation. In mammalian cells, this distance corresponds to 200-250 bp [37-39]. According to the direction of mismatch repair, genetic information may or may not be converted to match the original strand, leading to gene conversion. In addition, the 3’ end of the invading strand allows DNA synthesis initiation by copying the recipient molecule; a sequence absent from the invading strand can be recopied, leading to a non-reciprocal transfer of genetic information from the invaded molecule to the invading molecule and therefore to gene conversion. Strand exchange also creates cruciform structures called Holliday junctions. The orientation of the resolution of the Holliday junction, may result in the exchange of adjacent sequences (i.e., crossing over). The result of HR is therefore gene conversion with or without a crossover (Figure 1A).
Figure 1.
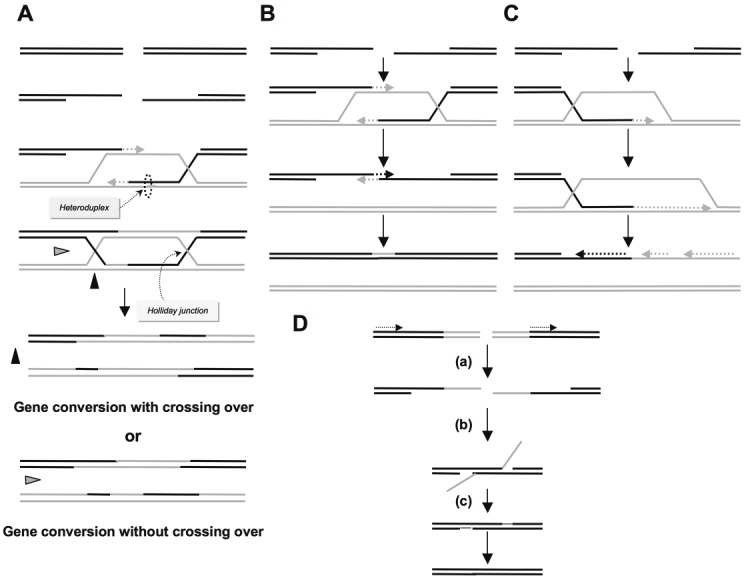
A) The double-strand break repair model. Resection of ssDNA generates single-stranded 3’tails that can invade a homologous double-stranded DNA, forming a D-loop (displacement loop). Synthesis of DNA is primed from the 3’end of the invasive strand. The D-loop can hybridize with the second single-stranded 3’end. DNA synthesis repairs the break and can therefore fill gaps. Cruciform junctions called Holliday junctions are formed and can migrate. Resolution of these junctions can occur in two different orientations (black or grey triangles) resulting in either gene conversion associated with crossing over or gene conversion without crossing over. B) Synthesis-dependent strand annealing (SDSA). Initiation is similar to that of the previous model, with a single-strand resection, invasion of the homologous double-stranded DNA and DNA synthesis, but the invading strand dehybridizes and reanneals at the other end of the injured molecule; no Holliday junction is thus formed. This model only accounts for gene conversion without crossovers. C) Break-induced replication (BIR). Initiation is similar to that of the previous models, with a single-strand resection, invasion of the homologous double-stranded DNA and DNA synthesis, but the synthesis continues over longer distances on the chromosome arms and even can reach the end of the chromosome. Here again, there is neither resolution of Holliday junctions nor crossover. D) Single-strand annealing (SSA). A double-strand break occurs between two homologous sequences in tandem in the same orientation (dotted arrows) (a). A single-strand resection then reveals two complementary DNA strands that can hybridize (b). If the tandem sequences are in opposite orientations, the revealed single strands of DNA are not complementary but are identical; they therefore cannot hybridize. In this process, the break does not need to be in or near a region of homology. (c) Resolution of this intermediate and filling the gap of the single strand completes the repair of the double-strand break, leading to the deletion of the intergenic sequences between the initial repetitions.
Other models of HR exist (for a review, see [40], including synthesis-dependent strand annealing (SDSA) and break-induced replication (BIR). These two models do not involve resolution of the Holliday junction but are initiated by common steps: ssDNA resection followed by invasion and exchange of a homologous DNA strand (Figures 1B and 1C). In particular, BIR seems to be the mechanism underlying the ALT system that allows telomere maintenance in the absence of telomerase [41]. Finally, in the last model, single-strand annealing (SSA), can occur between two sequences in tandem, and it is initiated by ssDNA resection (Figure 1D); however, contrary to the models previously described, this step is not followed by invasion of the DNA duplex and strand exchange. When the two sequences are in direct orientation, the ssDNA sequences revealed are complementary and can hybridize, forming a branched structure. Resolution of the intermediate structure inevitably leads to a deletion at the repair junction; SSA is a non-conservative process. Moreover, SSA cannot occur between inverted repeat sequences because the strands revealed by resection are not complementary but identical. Notably, if two breaks simultaneously occur in ectopic homologous sequences, SSA can generate translocations [42].
Non-homologous End Joining (NHEJ)
In contrast to HR, repair by canonical NHEJ (C-NHEJ) is not based on an intact partner sharing homologous sequences to repair broken ends. In this process, double-stranded DNA ends are recognized by the KU80/KU70 heterodimer followed by recruitment of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs). Accessory factors that allow the processing of DNA structures at double-stranded ends that impede repair, e.g., 3’-phosphoglycolates, are also recruited to the break, and notably include the Artemis, PALF (PNK and APTX-like FHA protein) and PNKP (Polynucleotide kinase/phosphatase) proteins. The ultimate NHEJ repair steps involve the rejoining of DNA ends by DNA ligase IV, which is assisted by its cofactors XRCC4 and Cernunnos/XLF (Figure 2A; recently reviewed in [43]).
Figure 2.

A. The different stages of canonical NHEJ and AE-J. After the formation of DSBs, the heterodimer Ku70/Ku80 interacts with the ends of the damaged DNA and promotes the recruitment of DNA-PKcs and Artemis. Artemis processes the ends of the DNA to make them compatible for enzymatic ligation by the complex Cernunnos-XLF/XRCC4/Ligase IV complex at the final step. B/ A-EJ: Alternatively, DNA ends that are not protected by Ku70/Ku80 are degraded. It is proposed that a single strand DNA resection reveals complementary microhomologies (2 to 4 nt and more), which can anneal; filling in of the single strand gap or nick complete the end-joining. A-EJ (for Alternative end-joining) is always mutagenic, with deletion at the junctions and frequently (but not systematically) uses microhomologie distant from the DSB. A-EJ is independent on Ku80, Xrcc4, Ligase IV, and is dependent on Parp1, Ligase III. The nuclease activity of MRE11 favors A-EJ.
Thus, in addition to the differences in sequence homology requirements, HR and NHEJ mechanistically differ at the initiation steps. The ssDNA resection machinery is crucial for the initiation of all HR models (see Figure 1), and while it is not required for NHEJ (see Figure 2), long resection may even impair it because it renders DNA ends incompatible for repair by the KU80/70 heterodimer. Thus, controlling ssDNA resection is the pivotal step for the choice between HR and NHEJ for DSB repair.
Alternative End Joining (A-EJ)
Recently, a new DSB repair process mechanistically and genetically distinct from both HR and C-NHEJ (the KU/LigaseIV pathway) has emerged. This alternative end-joining process was called different names: A-EJ (Alternative end-joining), B-NHEJ (Backup NHEJ), and MMEJ (Micro-homology Mediated End Joining). Although this pathway is not fully characterized and may correspond to different processes, we will refer to it here as A-EJ. A-EJ is highly mutagenic and frequently uses (but not always) micro-homologies (2 to 8 nt) that are distant from the break, while micro-homologies at the break site can be used by C-NHEJ [28,31]. In the proposed model, A-EJ is initiated by ssDNA resection that reveals complementary micro-homologies that can anneal, creating branched intermediate structures in a manner similar to SSA; the resolution of this intermediate structure results in deletions at the repair junctions (Figure 2B). Importantly, in human cell lines, it appears that many A-EJ events do not reveal the use of microhomologies, suggesting a repair model in which the occurring DNA end synapse does not involve strand pairing.
A-EJ shares the initial resection step with HR; however, it importantly differs from HR because it requires neither extended resection nor extended sequence homology. Indeed, less than a 50 bp resection is sufficient for A-EJ, while HR requires longer ssDNA stretches compatible with the MEPS. Moreover, A-EJ is independent of KU80 and XRCC4 and thus varies from C-NHEJ [28,31,35,44,45]. PARP-1, XRCC1, DNA ligase III, PNK and WRN have been implicated in this process [46-51]. Other studies demonstrated that MRE11 was implicated in both C-NHEJ and A-EJ and that its nuclease activity favors mutagenic A-EJ [29,52,53]. In addition, CtIP and NBS1 have also been shown to play a role at the initiation of A-EJ [29,54-57]. Finally, a role for ERCC1 has been reported [58]; however, its role has been challenged in another study [54]. The differences in the substrates used in these studies may explain these discrepancies.
Finally A-EJ has been described in C-NHEJ deficient mice in the course of physiological processes such as class switch recombination and V(D)J recombination [35,44,45].
Moreover, in addition to deletions at the resealed junctions, A-EJ appears to be a major source of DNA translocations induced by DSB [30-35].
A sequential model for the choice of the DSB repair process has been proposed [29]. In this model, the initiation (or not) of ssDNA resection (through the nuclease activity of MRE11) will orientate DSB repair either to C-NHEJ or A-EJ and HR; the size of the resection, which is associated with the cell cycle phase, will then direct the DSB repair to either HR or A-EJ (Figure 3). The appropriate choice of DSB repair pathway is essential for genome stability maintenance. In fact, the cell cycle phase is also pivotal for the choice between HR and NHEJ (as discussed below). Thus, DNA damage recognition and ssDNA resection control appear to be major steps for the choice between the DSB repair pathways used (see Figure 3).
Figure 3.

Model for sequential steps for DSB repair. The MRN complex is involved at early step of DSB signaling and can activate both C-NHEJ and A-EJ. Binding of Ku80/Ku70 and C-NHEJ protects from ssDNA resection leading to a conservative DSB repair outcome. Short ssDNA resection allows A-EJ but not homologous recombination. Long ssDNA resection allows A-EJ, but homologous recombination requires the activation of the subsequent steps such as the loading of RAD51 on the ssDNA.
The importance of the choice of the appropriate DSB repair pathway according to the cell cycle
Efficient and faithful DSB repair is essential for the maintenance of genome stability and cell survival, thus the choice of the appropriate repair pathway is pivotal. Cell signaling pathways, chromatin modifications and the early steps of break processing play a major role in directing repair towards HR or NHEJ.
Resection is essential for HR initiation, but it must be tightly controlled for several reasons. First, A-EJ, which is also initiated by resection, is an error-prone pathway at the repair junction, and it is also involved in chromosomal translocation [30-35]; thus, to maintain genome stability, A-EJ should be repressed. Second, SSA is a non-conservative DSB repair process, which is initiated by ssDNA resection. Genome stability maintenance should limit SSA. Third, although HR is required for genomic stability maintenance, it represents a double-edged sword. Repeated sequences dispersed throughout the genome can participate in HR. Crossing-over between repeated sequences can lead to profound genomic rearrangements (Figure 4A) including amplifications, deletions, inversions, translocations, and gene conversions (with an heteroallele or pseudogene) and can result in the loss of functional alleles (Figure 4B), as discussed elsewhere [9,26]. Moreover, unresolved HR intermediates are toxic and/or generate genetic instability [59]. Thus, to avoid the excessive unscheduled initiation of HR events, HR must also be tightly controlled. Cell signaling can allow the restriction of excessive HR initiation, as proposed for AKT1 [9,13]; in addition, helicases, have been shown to destabilize abortive HR intermediates [60-63]. These observations emphasize that HR is tightly constrained.
Figure 4.
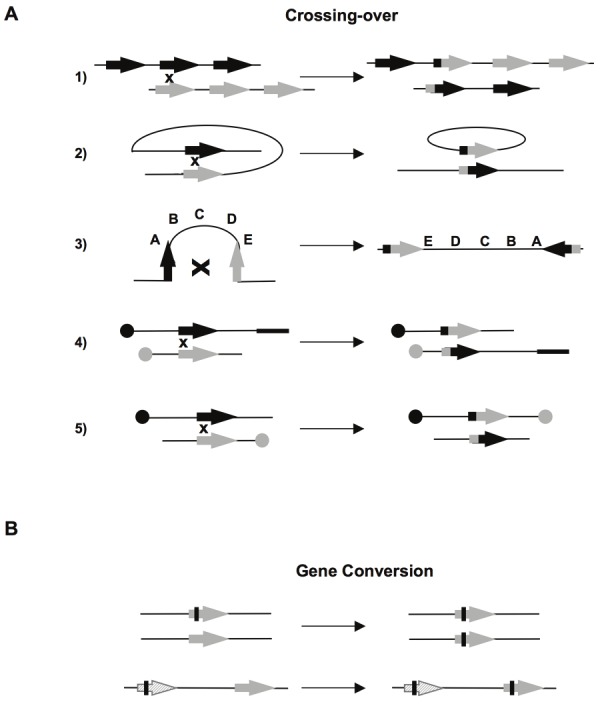
A) Chromosomal rearrangements resulting from crossovers (CO). (1) CO between repetitions on two chromosomes or unequal sister chromatids exchange results in amplification of one molecule and deletion of the other. (2) Intra-chromosomal CO between two direct repeat sequence, results in excision of the intervening sequence. (3) CO between two inverted repeat sequences leads to inversion of the internal fragment. (4) and (5) Inter-chromosomal CO. According to the orientation of the sequences with respect to the centromers (black or grey circles), the process generates a translocation (4) or one dicentric and one acentric chromosome (5). B) Genetic modifications resulting from gene conversion between two heteroalleles, leading to a loss of heterozygosity (upper panel). Lower panel: Gene conversion between one pseudogene (hatched), which often contains nonsense mutations (black bar), and one gene, leading to the inactivation of the latter.
NHEJ is active throughout the entire cell cycle and competes for the repair of DSBs with HR in S phase. Thus, double-strand breaks resulting from prolonged replication blockage can be addressed by HR or non-homologous end joining (NHEJ) [20,64,65]. However, unlike doublestrand breaks produced by enzymes and ionizing radiation, breaks produced by replication stops have only one double-stranded end (see Figure 5A). Ligation of the two ends of the double strand (which are therefore far apart) by NHEJ inevitably leads to chromosomal rearrangement (Figure 5B). In contrast, during S phase, HR can use the intact sister chromatid (Figure 5B). Because the two chromatids have identical sequences, the genetic impact is minimal. There is a potential risk for unequal sister chromatid exchange or crossovers with sequences of different chromosomes. However, the close proximity of the sister chromatids (particularly due to cohesins) favors the use of sister chromatids as templates for the repair of double-strand breaks [66]. In addition, in somatic cells, gene conversion without crossover is favored, limiting the risks associated with crossovers [66,67]. Of note, in the G1 phase, the sister chromatid is not present, and HR would thus be forced to use one homologous ectopic partner, jeopardizing genome integrity. This inhibition of HR in G1 allows the maintenance of genome stability; indeed, several mechanisms regulate HR to constraint its activity in the S and G2 phases (as discussed below). In addition, p53, which inhibits HR [68], is stimulated upon genotoxic stresses, blocking cells at the G1/S transition; one can thus propose that one role of p53 is to inhibit HR when cells are arrested in the G1 phase.
Figure 5.
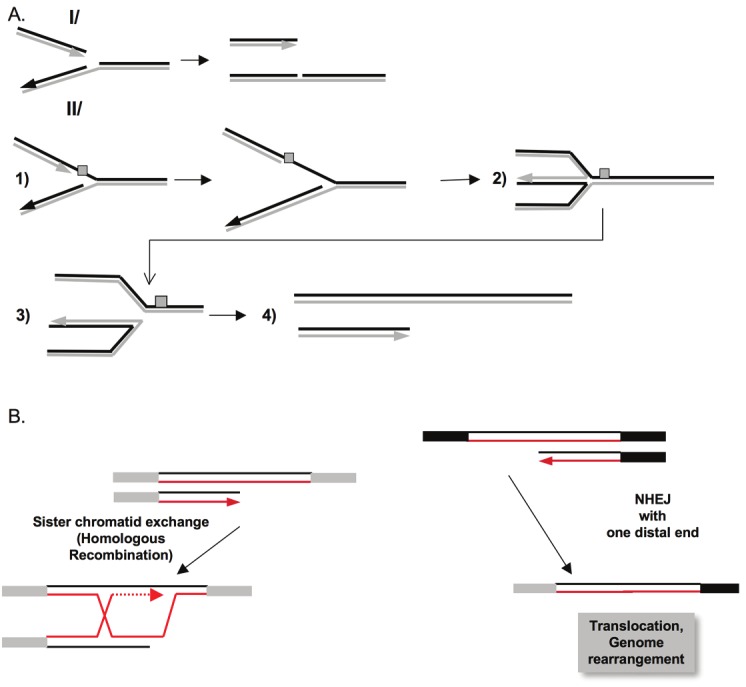
A/ I/ A replication fork reaching a single-stranded gap in the template, results in a collapsed fork. Recombination with the sister chromatid allows the restarting of the fork via a process similar to BIR (see Figure 1). II/ (1) When the fork reaches a blocking lesion (grey scare), fork reversion forms a structure called “chicken foot” (2); (3) the cruciform structure can be resolved (in the manner of a Holliday junction), resulting in a double end (4). Note that in both case single double strand ends are generated, a situation different from DSB induced by endonucleases or IR in which two close double strand ends are generated, favoring end-joining. B) Recombination with the sister chromatid (Left panel) allows replication to restart by a BIR-like mechanism, maintaining genome stability. In such situation, NHEJ, which join distant single-ended double strand ends, resulting thus in genetic exchange (Right panel).
Thus, the choice of the DSB repair pathway strongly impacts genome stability maintenance; this essential choice is dependent of early DSB repair steps, which are discussed below.
Early steps of DNA DSB recognition and repair; signaling and resection
DSB signaling
At an early step, double-strand break recognition is accomplished by the MRN complex (MRE11, RAD50 and NBS1), which acts in an activation loop together with ATM kinase to signal the existence of a DSB. It remains unclear whether the KU heterodimer competes with MRN for binding to damaged DNA extremities or whether an interplay exists between these two complexes for the protection of DNA ends, which involves the displacement of one of the latter to correctly direct towards the appropriate repair pathway. PARP-1 is a prime candidate to mediate the choice between the two players [69]. The rapid phosphorylation of the H2AX histones surrounding the break follows ATM activation. This process leads to the recruitment of MDC1, which then stabilizes MRN at the break site and thereby enhances ATM-mediated signaling. The phosphorylation of MDC1 by ATM leads to the recruitment of two E3 ubiquitin ligases RNF8 and RNF168, which are essential for chromatin remodeling. The subsequent relaxation of the chromatin then exposes epigenetic markers, such as methylated histone H4, which leads to the recruitment of 53BP1 and RAP80-BRCA1 to damage sites (for review, see [70]. Of note, KAP-1, an ATM effector, is required for the remodeling of heterochromatin, allowing DNA repair in such closed regions of the genome [71-73].
Chromatin remodeling
The nucleosome is a barrier that blocks the access of enzymes involved in transcription, replication and repair to DNA. Chromatin remodeling, by way of the post-translational modifications of histones, occurs during the detection and repair of damaged DNA. One class of enzymes alters histone-histone and histone-DNA interactions through serine/threonine phosphorylation, acetylation, ubiquitination, sumoylation and the methylation of lysine and arginine residues [74,75]. The second class of enzymes uses energy released by ATP hydrolysis to modify histone organization, thereby impeding the movement and repositioning of nucleosomes [74,76,77]. This step is summarized in Figure 6.
Figure 6.
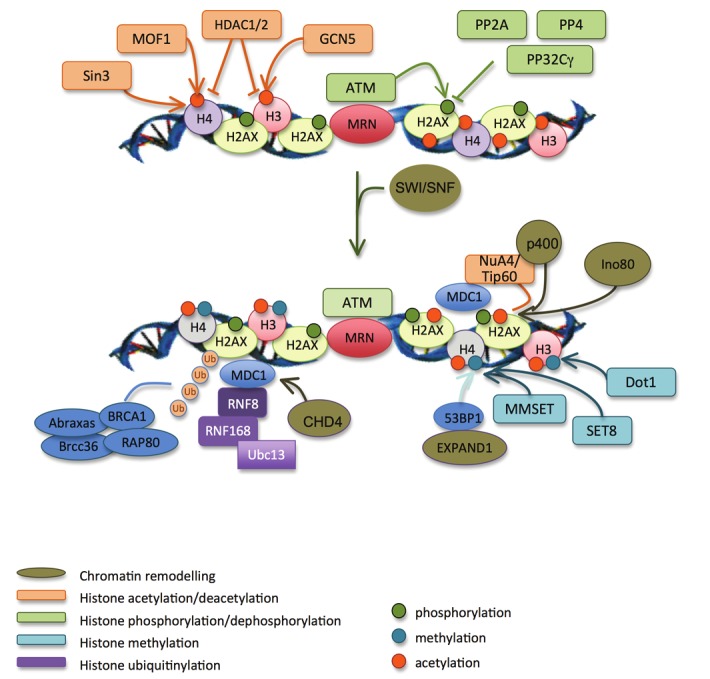
Chromatin remodelling in response to DSBs. H2AX (phosphoryltaion by ATM) and histone H3 (acetylation by GCN5) are modified at low levels on DSB flanking regions. This helps the recruitment of MDC1. GCN5 then binds to firstly phosphorylated H2AX, which increases histones acetylation and facilitates the recruitment of SWI/SNF chromatin remodelling factors. They increase the accessibility of chromatin to chromatin remodelling and repair factors. Sin3 and MOF1 acetylate histone H4, which also helps the efficient recruitment of MDC1, BRCA1 and 53BP1. MDC1 recruit RNF8 and RNF168 that mediate the ubiquitylation of histones. RAP80, through its ubiquitin-binding domain, recruits BRCA1 and the other members of the ABRAXAS complex at DSBs. These reactions are amplified upon the chromatin relaxation operated by NuA4, Tip60, Ino80, CHD4 and P400. MDC1 also recruits MMSET and SET8 methyltransferases that methylate histone H4. This, with histones ubiquitylation mediated by RNF8 and RNF168, mediates the chromatin relaxation by EXPAND.
The importance of histone H2AX phosphorylation
Phosphorylation of serine-139 at the C-terminus of H2AX is a crucial chromatin modification in response to double-stranded DNA breakage [78-81]. This phosphorylation is performed by the PI3 kinases ATM, ATR and DNA-PK [82-84] and begins at the break and extends beyond 2 Mb in higher eukaryotes [85]. The phosphorylation of H2AX is accompanied by the acetylation of histone H3 by GCN5 [86]. These two events facilitate the recruitment of the SWI/SNF ATPase BRG1, which presumably increases the accessibility of DSB flanking chromatin and thereby amplifies γ-H2AX and acetylated H3 signal. It has been suggested that H2AX phosphorylation stabilizes the interaction between repair proteins such as 53BP1, BRCA1 and NBS1 at the break site and facilitates the downstream accumulation of other DDR proteins [87-89]. Phosphorylation of γ-H2AX is recognized by the BRCT (BRCA1 carboxy-terminal) domain of MDC1 (mediator of DNA damage checkpoint) [90,91]. This complex regulates the level of H2AX phosphorylation and is an early step that allows the downstream accumulation of key protein complexes such as MRE11-RAD50-NBS1 and the ubiquitin ligase UBC13/RNF8 [92-95] and of chromatin remodeling factors such as EXPAND1, CHD4 and the NuA4 and Ino80 complexes [16,96-99]. The NuA4 complex consists of two types of chromatin remodelers, p400 and Tip60, and a histone acetyltransferase (HAT). The Ino80 complex has various activities including a 3’-5’ helicase, nucleosome sliding and DNA-dependent ATPase [100]. The Ino80 complex has also been implicated in the generation of ssDNA by participating in 3’-5’ DNA resection. Indeed, Ino80 mutants demonstrate a defect in the recruitment of Mre11 nuclease and the formation of ssDNA [99]. It is important to note that these chromatin remodeling factors are necessary for H2AX phosphorylation [101].
During repair, MDC1 opposes the action of the phosphatases via its interaction with H2AX [91] and this interaction is stabilized by the acetylation of H4 by MOF1. However, when repair is completed, histone γ-H2AX must be dephosphorylated to regain the original structure. In mammals, the phosphatases PP2A, PP4 and PP2C gamma participate in the dephosphorylation of histone H2AX [102-104].
Other posttranslational modifications of histones
In addition to phosphorylation, histones undergo other modifications during DNA repair. Histone H3, which is methylated at lysine 79 (H3K79me) by Dot1 methyltransferase, is recognized by the Tudor domain of the 53BP1 protein after damage [105,106]. 53BP1 also directly binds to histone H4 that is methylated at lysine 20 (H4K20me) [107] by MMSET and SET8 [108,109].
In response to ionizing radiation, H2A and H2AX are both polyubiquitinated by the UBC13/RNF8 ubiquitin ligase complex, which occurs at the beginning of the ubiquitination process, and the RNF168 ubiquitin ligase, which amplifies this phenomenon [92,110,111].
Rap80 acts as a mediator by recognizing polyubiquitin chains generated by RNF8 and in directing polyubiquitinated proteins to the site of the break. Rap80 also interacts with the Abraxas complex, which is composed of BRCA1, Abraxas and Brcc36 [93,112,113]. Thus, RNF8 plays a major role in the recruitment of RAP80, BRCA1, Abraxas, BRCC6 and 53BP1 after irradiation. RNF8 is also necessary for the accumulation of H2AX and MDC1 proteins at the repair sites at the earliest stages. The proposed hypothesis is that RNF8 restructures chromatin at the breakage site facilitating the accumulation near the lesion of factors that act later in repair (such as 53BP1 and BRCA1) and therefore favor the maintenance of the integrity of the genome.
The acetylation of histone lysine residues can facilitate repair protein access to the site of the damage. Indeed, changes in the dynamics of histone acetylation have been observed during HR and play a key role in post-HR viability [114]. The balance between the acetyltransferase (i.e., HATs) and deacetylase (i.e., HDACs) activities control histone acetylation. Transient acetylation of histones H3 and H4 has been found at the sites of double-strand breaks (van Attikum and Gasser, 2005, Cell Cycle, 4, 1011-4). In mammals, the Tip60 complex is recruited to the break where it acetylates H4 and its cofactor TRRAP [115]. Moreover, Tip60 also promotes γ-H2AX ubiquitination [115]. In addition, H2AX acetylation by the Tip60-UBC13 complex causes its detachment from chromatin. It therefore seems that the sequential acetylation and ubiquitination of H2AX affect histone dynamics at the break site. Recently, it has been shown that the changes in nucleosome and chromatin structure induced by p400 favor the ubiquitination of chromatin and the accumulation of BRCA1 and 53BP1 at the damaged site [116]. Moreover, an interaction between Tip60 and MRN was described [117].
The role of chromatin in repair pathway choice
The deacetylation of H3K56 and H4K16 residues by HDAC1 and HDAC2 is proposed to favor NHEJ repair [118] through the maintenance of a compact chromatin structure during repair, which impedes HR and thus promotes NHEJ. However, the opposite has also been reported. Indeed, the acetylation of histone H3 facilitates ssDNA resection and thereby the initiation of HR [119]. In addition, the hyperacetylation of histone H4 by sin3 provokes a defect in NHEJ [120]. Moreover, DNA-PK can favor NHEJ by phosphorylating the histone acetyltransferase GCN5 in vivo, which inhibits its HAT activity [121].
Analogous to methylations, H3K36 dimethylation on residues that are different from those leading to the recruitment of 53BP1 to chromatin favors NHEJ by increasing the recruitment of KU70 and NBS1 [122].
ssDNA resection and its regulation during the cell cycle
Two-step model for resection
An essential step for initiating HR is ssDNA resection, leading to the production of a single-stranded 3’ tail (Figure 1). This step determines the choice of mechanism for double-strand break repair, and different factors, including 53BP1 and RAP80, have been shown to protect DNA ends from resection [123-125,139].
Recent studies in S. cerevisiae demonstrated that the resection necessary for the initiation of HR is a two-step process. In the first step, limited resection occurs, involving the action of the Mre11, Rad50 and Xrs2 (MRX) protein complex and the Sae2 protein (functional homolog of human CtIP). These proteins are able to remove up to 100 bp from the ends of the double-stranded break [126]. The single-stranded end formed is a preferable substrate for the Exo1 exonuclease or the Sgs1 RecQ helicase/Dna2 endonuclease complex. These enzymes perform a more processive recession, allowing the production of an outgoing 3’ single-stranded DNA of appropriate length to allow the invasion of an intact homologous molecule [127,128].
In mammals, studies have suggested a similar two-step model; however, the mechanism has yet to be determined, particularly concerning the second step of extended resection (for a review, see [129]). The role of the MRN complex in the initiation of resection during HR in mammalian cells is well established. The protein CtIP, a partner of the MRN complex, interacts with the FHA domains of NBS1 and stimulates the endonuclease activity of MRE11. Importantly, no nuclease activity has been assigned to mammalian CtIP, which is unlike the yeast Sae2 protein. Despite this fact, CtIP plays a key role in the stimulation of resection in mammals, and its activity is tightly regulated at multiple levels during the cell cycle.
In agreement with the two-step model, the first initiation of resection should be followed by the concerted action of nucleases and helicases. The involvement of Exo1 in HR was shown by Bolderson et al. [130]. Another study demonstrated that Exo1 recruitment depends on MRE11 and CtIP [131], and CtIP can inhibit the Exo1 exonuclease activity in vitro. The involvement of BLM (a RecQ helicase mutated in Bloom Syndrome) in the resection process was also suggested, but these data remain controversial. One study demonstrated that BLM and Exo1 act in parallel in independent pathways [132]. Other data demonstrated a specific interaction between these two proteins. BLM could stimulate the exonuclease activity of Exo1 in vitro [133] or the affinity of Exo1 towards DNA ends [134], suggesting their cooperation during resection. The latter in vitro study demonstrated the involvement of the two independent machineries in the resection of ssDNA, involving either Exo1 or Dna2 [134]. BLM was also proposed to unwind the double helix to allow DNA2 to perform endonucleolytic cleavage of DNA. In this scenario, the RPA protein would be necessary to ensure proper directionality of the resection in the 5’-3’ direction because DNA2 possesses both 5’-3’ and 3’-5’ nuclease activities. However, it should be noted that an active role of BLM in HR initiation is highly inconsistent with the phenotype of cells from patients with Bloom Syndrome who demonstrate the trait of hyper-recombination but not HR deficiency. It is possible that one of the other RecQ helicases (i.e., RecQ1, WRN, RecQ4, or RecQ5) is a part of the resection machinery in vivo.
Regulation of resection during cell cycle
Because HR is the mechanism that should be privileged during replicative and post-replicative stages of the cell cycle and should be avoided in the G1 phase, resection needs to be restricted to the S and G2 stages of the cell cycle.
The phosphorylation of the Ser327 residue on CtIP by Cyclin Dependent Kinase 1 (CDK1) kinases allows the association of CtIP with BRCA1 and the MRN complex [135,136]. This leads to the stimulation of resection not only through MRE11 activation but also through displacing factors antagonizing resection such as 53BP1 [124]. CtIP can also be ubiquitinylated by BRCA1, but the function of this modification remains unclear [137]. It is speculated that it would give CtIP the necessary advantage to replace two other BRCA1 interacting partners, RAP80/Abraxas, whose role is to impair resection, and thereby favor ssDNA appearance [138,139]. A role for SIRT6 in promoting resection has also been proposed at two levels: first, SIRT6 deacetylates CtIP, which favors resection [140] and second, it also stimulates PARP-1 activity at damage sites upon stress, which stimulates both HR and A-EJ [141].
The ATM kinase seems to contribute to the choice between resection and the protection of DNA ends. It has been shown that in G1 lymphocytes, ATM-dependent H2AX phosphorylation impedes CtIP-mediated resection. Furthermore, MDC1 recruited to the break in an ATM-dependent manner, binds to the FHA domains of NBS1, thus competing with CtIP and impairing resection [142]. Paradoxically, ATM has also been proposed to stimulate limited resection in G1 through CtIP and KAP1 phosphorylation, but this could be explained by the necessity to process the breaks blocked by bulky adducts that impede proper repair [72,143].
Competition and the choice between the different DSB repair mechanisms
The choice for resection or NHEJ?
In yeast, it has been shown that the presence of KU80/70 restricts the access of nucleases to DNA extremities, particularly Exo1, thus inhibiting early steps of HR. This recent study also suggests that Sae2 (the functional ortholog of CtIP) would be necessary for the displacement of the KU heterodimer from DNA ends [144]. In S. pombe, the nuclease activity of Mre11 and Ctp1 (CtIP) are necessary for liberating the ssDNA tail from the presence of KU and the MRN complex to allow HR repair [145]. Recently, the RNF8 E3 ligase has been proposed to regulate the removal of KU80 from DSBs. In the absence of RNF8, the prolonged presence of KU at break sites impeded repair by NHEJ [146]. In human cells, the repair defect observed in FANCD2 deficient cells could be restored by the depletion of KU [147]. Taken together, these data suggest that some proteins could antagonize the binding of KU to DNA ends, particularly in S phase when HR needs to be promoted, and particularly upon replicative stress, when collapsed replication forks require restarting by the HR machinery. Nevertheless, the sequence of events leading to KU displacement necessitates further investigation.
In the absence of KU, HR is stimulated due to increased resection [24,148,149]. However, the resection stimulation by itself may be not sufficient to induce HR events. Indeed, in Xenopus extracts, an uncoupling between the early resection steps and the RAD51 filament formation has been proposed [150]. Therefore, as expected, HR is tightly regulated at multiple levels: at the resection steps at a minimum and at the assembly of an active RAD51 filament; these two steps may be disconnected, although RAD51 filament assembly requires resection.
Another key player of NHEJ, DNA-PKcs, is able to impede HR solely by its physical presence because the absence of DNA-PKcs leads to an increase in HR, but its chemical inhibition impedes DSB repair in general for NHEJ and HR [21].
A recent study proposes that in G2, despite the presence of sister chromatids, NHEJ is the first mechanism to act, and this is possibly due to the abundance of its key proteins and the chromatin status [151]. If NHEJ is unable to accomplish repair, then resection would channel repair towards HR. However, HR is inhibited in late G2 by the AKT1 signaling pathway [116].
A-EJ vs. HR and NHEJ
HR and A-EJ are both initiated by ssDNA resection. A-EJ events arise from limited resection, which is insufficient to accomplish HR repair, or occurs during non-replicative stages of the cell cycle as a result of break processing by nucleases. Resection is mediated in both cases by the same set of proteins (i.e., MRE11 in cooperation with CtIP). In S phase, the obvious choice is to guide the repair process towards the error-free HR pathway by ensuring extended resection that is compatible with MEPS and the activation of the factors required by HR. In contrast, in G1, break processing can impede C-NHEJ, leading to the appearance of error-prone events and no possibility of faithful repair.
Conclusions
The initiation of DSB repair represents an essential challenge for maintaining genome integrity and for tumor protection in fine.
HR appears to be strictly controlled at several levels (at least resection and RAD51 filament assembly), which, in addition, may be disconnected, increasing the security to avoid inappropriate HR. An efficient generation of RPA-coated ssDNA is not sufficient to lead to the appearance of HR events due to the lack of RAD51 filament formation and ATR signaling [150]. Moreover, Cdk1, which stimulates resection through CtIP phosphorylation, would also be responsible for preventing the assembly of the RAD51 filament on resected DNA. Thus, these two steps are uncoupled, and the occurrence of HR events requires that all factors implicated in both the early and late steps of HR should be available and properly activated.
Interestingly, it has recently been shown that HR repair at heterochromatic loci necessitates the shuttling of HR intermediates towards adjacent sites to avoid associated instability, i.e., with recombination between repeated sequences [152]. The initial resection steps are allowed inside heterochromatin domains but the progression of repair is blocked until the recombination intermediates are displaced to loci outside of heterochromatin. HR can then be completed at the periphery without interference from repetitive DNA sequences. These data further underline the important role of the chromatin environment during DSB repair and adds another level of regulation of HR events.
Altogether, the initiation of DSB repair represents an essential challenge for maintaining genome integrity and for tumor protection in fine. In this context, controlling the initial step of DSB repair, particularly resection initiation according to the cell cycle stage, is an essential step that may be irreversible, and thus, the wrong decision may lead to dramatic consequences.
Acknowledgements
This work was supported by the Association for Research against Cancer (ARC) and the Institut National du Cancer (INCa).
References
- 1.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 2.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA Jr, Kastrinakis NG, Levy B, Kletsas D, Yoneta A, Herlyn M, Kittas C, Halazonetis TD. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 3.Hyrien O. Mechanisms and consequences of replication fork arrest. Biochimie. 2000;82:5–17. doi: 10.1016/s0300-9084(00)00344-8. [DOI] [PubMed] [Google Scholar]
- 4.Bravard A, Vacher M, Gouget B, Coutant A, de Boisferon FH, Marsin S, Chevillard S, Radicella JP. Redox regulation of human OGG1 activity in response to cellular oxidative stress. Mol Cell Biol. 2006;26:7430–7436. doi: 10.1128/MCB.00624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montaner B, O'Donovan P, Reelfs O, Perrett CM, Zhang X, Xu YZ, Ren X, Macpherson P, Frith D, Karran P. Reactive oxygen-mediated damage to a human DNA replication and repair protein. EMBO Rep. 2007;8:1074–1079. doi: 10.1038/sj.embor.7401084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallace SS. Biological consequences of free radical-damaged DNA bases. Free Radic Biol Med. 2002;33:1–14. doi: 10.1016/s0891-5849(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 7.Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 8.Erkko H, Pylkas K, Karppinen SM, Winqvist R. Germline alterations in the CLSPN gene in breast cancer families. Cancer Lett. 2008;261:93–97. doi: 10.1016/j.canlet.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Guirouilh-Barbat J, Wilhelm T, Lopez BS. AKT1/BRCA1 in the control of homologous recombination and genetic stability: the missing link between hereditary and sporadic breast cancers. Oncotarget. 2010;1:691–699. doi: 10.18632/oncotarget.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh T, King MC. Ten genes for inherited breast cancer. Cancer Cell. 2007;11:103–105. doi: 10.1016/j.ccr.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM, Roeb W, Agnew KJ, Stray SM, Wickramanayake A, Norquist B, Pennington KP, Garcia RL, King MC, Swisher EM. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci USA. 2011;108:18032–18037. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McEllin B, Camacho CV, Mukherjee B, Hahm B, Tomimatsu N, Bachoo RM, Burma S. PTEN loss compromises homologous recombination repair in astrocytes: implications for glioblastoma therapy with temozolomide or poly(ADP-ribose) polymerase inhibitors. Cancer Res. 2010;70:5457–5464. doi: 10.1158/0008-5472.CAN-09-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plo I, Laulier C, Gauthier L, Lebrun F, Calvo F, Lopez BS. AKT1 inhibits homologous recombination by inducing cytoplasmic retention of BRCA1 and RAD51. Cancer Res. 2008;68:9404–9412. doi: 10.1158/0008-5472.CAN-08-0861. [DOI] [PubMed] [Google Scholar]
- 14.Plo I, Lopez B. AKT1 represses gene conversion induced by different genotoxic stresses and induces supernumerary centrosomes and aneuploidy in hamster ovary cells. Oncogene. 2009;28:2231–2237. doi: 10.1038/onc.2009.85. [DOI] [PubMed] [Google Scholar]
- 15.Saal LH, Gruvberger-Saal SK, Persson C, Lovgren K, Jumppanen M, Staaf J, Jonsson G, Pires MM, Maurer M, Holm K, Koujak S, Subramaniyam S, Vallon-Christersson J, Olsson H, Su T, Memeo L, Ludwig T, Ethier SP, Krogh M, Szabolcs M, Murty VV, Isola J, Hibshoosh H, Parsons R, Borg A. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat Genet. 2008;40:102–107. doi: 10.1038/ng.2007.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP, Yin Y. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 17.Sun M, Wang G, Paciga JE, Feldman RI, Yuan ZQ, Ma XL, Shelley SA, Jove R, Tsichlis PN, Nicosia SV, Cheng JQ. AKT1/PKBalpha kinase is frequently elevated in human cancers and its constitutive activation is required for oncogenic transformation in NIH3T3 cells. Am J Pathol. 2001;159:431–437. doi: 10.1016/s0002-9440(10)61714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang H, Wen YY, Zhao R, Lin YL, Fournier K, Yang HY, Qiu Y, Diaz J, Laronga C, Lee MH. DNA damage-induced protein 14-3-3 sigma inhibits protein kinase B/Akt activation and suppresses Akt-activated cancer. Cancer Res. 2006;66:3096–3105. doi: 10.1158/0008-5472.CAN-05-3620. [DOI] [PubMed] [Google Scholar]
- 19.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saintigny Y, Delacote F, Vares G, Petitot F, Lambert S, Averbeck D, Lopez BS. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 2001;20:3861–3870. doi: 10.1093/emboj/20.14.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen C, Kurimasa A, Brenneman MA, Chen DJ, Nickoloff JA. DNA-dependent protein kinase suppresses double-strand break-induced and spontaneous homologous recombination. Proc Natl Acad Sci USA. 2002;99:3558–3563. doi: 10.1073/pnas.052545899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belmaaza A, Chartrand P. One-sided invasion events in homologous recombination at double-strand breaks. Mutat Res. 1994;314:199–208. doi: 10.1016/0921-8777(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 23.Delacote F, Han M, Stamato TD, Jasin M, Lopez BS. An xrcc4 defect or Wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells. Nucleic Acids Res. 2002;30:3454–3463. doi: 10.1093/nar/gkf452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richardson C, Jasin M. Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol Cell Biol. 2000;20:9068–9075. doi: 10.1128/mcb.20.23.9068-9075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delacote F, Lopez BS. Importance of the cell cycle phase for the choice of the appropriate DSB repair pathway, for genome stability maintenance: the trans-S double-strand break repair model. Cell Cycle. 2008;7:33–38. doi: 10.4161/cc.7.1.5149. [DOI] [PubMed] [Google Scholar]
- 27.Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 2000;28:2585–2596. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guirouilh-Barbat J, Rass E, Plo I, Bertrand P. Lopez BS Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc Natl Acad Sci USA. 2007;104:20902–20907. doi: 10.1073/pnas.0708541104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 30.Boboila C, Jankovic M, Yan CT, Wang JH, Wesemann DR, Zhang T, Fazeli A, Feldman L, Nussenzweig A, Nussenzweig M, Alt FW. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc Natl Acad Sci USA. 2010;107:3034–3039. doi: 10.1073/pnas.0915067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, Lopez BS. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 32.Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol. 2010;17:410–416. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, Greenman CD, Jia M, Latimer C, Teague JW, Lau KW, Burton J, Quail MA, Swerdlow H, Churcher C, Natrajan R, Sieuwerts AM, Martens JW, Silver DP, Langerod A, Russnes HE, Foekens JA, Reis-Filho JS, van 't Veer L, Richardson AL, Borresen-Dale AL, Campbell PJ, Futreal PA, Stratton MR. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weinstock DM, Brunet E, Jasin M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol. 2007;9:978–981. doi: 10.1038/ncb1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, Rajewsky K, Alt FW. IgH class switching and translocations use a robust nonclassical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 36.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 37.Liskay RM, Letsou A, Stachelek JL. Homology requirement for efficient gene conversion between duplicated chromosomal sequences in mammalian cells. Genetics. 1987;115:161–167. doi: 10.1093/genetics/115.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez BS, Corteggiani E, Bertrand-Mercat P, Coppey J. Directional recombination is initiated at a double strand break in human nuclear extracts. Nucleic Acids Res. 1992;20:501–506. doi: 10.1093/nar/20.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rubnitz J, Subramani S. The minimum amount of homology required for homologous recombination in mammalian cells. Mol Cell Biol. 1984;4:2253–2258. doi: 10.1128/mcb.4.11.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 42.Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- 43.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soulas-Sprauel P, Le Guyader G, Rivera-Munoz P, Abramowski V, Olivier-Martin C, Goujet-Zalc C, Charneau P, de Villartay JP. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J Exp Med. 2007;204:1717–1727. doi: 10.1084/jem.20070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, Shah S, Brandt VL, Meek K, Roth DB. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 46.Audebert M, Salles B, Weinfeld M, Calsou P. Involvement of polynucleotide kinase in a poly (ADP-ribose) polymerase-1-dependent DNA double-strand breaks rejoining pathway. J Mol Biol. 2006;356:257–265. doi: 10.1016/j.jmb.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 47.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 48.Robert I, Dantzer F, Reina-San-Martin B. Parp1 facilitates alternative NHEJ, whereas Parp2 suppresses IgH/c-myc translocations during immunoglobulin class switch recombination. J Exp Med. 2009;206:1047–1056. doi: 10.1084/jem.20082468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sallmyr A, Tomkinson AE, Rassool FV. Up-regulation of WRN and DNA ligase IIIalpha in chronic myeloid leukemia: consequences for the repair of DNA double-strand breaks. Blood. 2008;112:1413–1423. doi: 10.1182/blood-2007-07-104257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simsek D, Brunet E, Wong SY, Katyal S, Gao Y, McKinnon PJ, Lou J, Zhang L, Li J, Rebar EJ, Gregory PD, Holmes MC, Jasin M. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Windhofer F, Iliakis G. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 52.Dinkelmann M, Spehalski E, Stoneham T, Buis J, Wu Y, Sekiguchi JM, Ferguson DO. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat Struct Mol Biol. 2009;16:808–813. doi: 10.1038/nsmb.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-HEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deriano L, Stracker TH, Baker A, Petrini JH, Roth DB. Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol Cell. 2009;34:13–25. doi: 10.1016/j.molcel.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee-Theilen M, Matthews AJ, Kelly D, Zheng S, Chaudhuri J. CtIP promotes microhomology-mediated alternative end joining during classs-switch recombination. Nat Struct Mol Biol. 2011;18:75–79. doi: 10.1038/nsmb.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, Jasin M. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat Struct Mol Biol. 2011;18:80–84. doi: 10.1038/nsmb.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ahmad A, Robinson AR, Duensing A, van Drunen E, Beverloo HB, Weisberg DB, Hasty P, Hoeijmakers JH, Niedernhofer LJ. ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol Cell Biol. 2008;28:5082–5092. doi: 10.1128/MCB.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gangloff S, Soustelle C, Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- 60.Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O'Neil NJ, Petalcorin MI, Martin JS, Collis SJ, Cantor SB, Auclair M, Tissenbaum H, West SC, Rose AM, Boulton SJ. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA repair. Annu Rev Genet. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 63.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, Jasin M, Vogel H, Sung P, Luo G. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Couedel C, Mills KD, Barchi M, Shen L, Olshen A, Johnson RD, Nussenzweig A, Essers J, Kanaar R, Li GC, Alt FW, Jasin M. Collaboration of homologous recombination and nonhomologous end-joining factors for the survival and integrity of mice and cells. Genes Dev. 2004;18:1293–1304. doi: 10.1101/gad.1209204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mills KD, Ferguson DO, Essers J, Eckersdorff M, Kanaar R, Alt FW. Rad54 and DNA Ligase IV cooperate to maintain mammalian chromatid stability. Genes Dev. 2004;18:1283–1292. doi: 10.1101/gad.1204304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liskay RM, Stachelek JL, Letsou A. Homologous recombination between repeated chromosomal sequences in mouse cells. Cold Spring Harb Symp Quant Biol. 1984;49:183–189. doi: 10.1101/sqb.1984.049.01.021. [DOI] [PubMed] [Google Scholar]
- 68.Bertrand P, Saintigny Y, Lopez BS. p53's double life: transactivation-independent repression of homologous recombination. Trends Genet. 2004;20:235–243. doi: 10.1016/j.tig.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 69.Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, Poirier GG. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 70.Stewart GS. Solving the RIDDLE of 53BP1 recruitment to sites of damage. Cell Cycle. 2009;8:1532–1538. doi: 10.4161/cc.8.10.8351. [DOI] [PubMed] [Google Scholar]
- 71.Goodarzi AA, Kurka T, Jeggo PA. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol. 2011;18:831–839. doi: 10.1038/nsmb.2077. [DOI] [PubMed] [Google Scholar]
- 72.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 73.Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 74.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 75.Marmorstein R, Berger SL. Structure and function of bromodomains in chromatin-regulating complexes. Gene. 2001;272:1–9. doi: 10.1016/s0378-1119(01)00519-4. [DOI] [PubMed] [Google Scholar]
- 76.Lusser A, Kadonaga JT. Chromatin remodeling by ATP-dependent molecular machines. Bioessays. 2003;25:1192–1200. doi: 10.1002/bies.10359. [DOI] [PubMed] [Google Scholar]
- 77.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 78.Fillingham J, Keogh MC, Krogan NJ. Gamma-H2AX and its role in DNA double-strand break repair. Biochem Cell Biol. 2006;84:568–577. doi: 10.1139/o06-072. [DOI] [PubMed] [Google Scholar]
- 79.Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 81.Sedelnikova OA, Pilch DR, Redon C, Bonner WM. Histone H2AX in DNA damage and repair. Cancer Biol Ther. 2003;2:233–235. doi: 10.4161/cbt.2.3.373. [DOI] [PubMed] [Google Scholar]
- 82.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 83.Stiff T, O'Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–2396. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 84.Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem. 2001;276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- 85.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brand M, Moggs JG, Oulad-Abdelghani M, Lejeune F, Dilworth FJ, Stevenin J, Almouzni G, Tora L. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 2001;20:3187–3196. doi: 10.1093/emboj/20.12.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ward IM, Minn K, Jorda KG, Chen J. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J Biol Chem. 2003;278:19579–19582. doi: 10.1074/jbc.C300117200. [DOI] [PubMed] [Google Scholar]
- 88.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 89.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 90.Lee MS, Edwards RA, Thede GL, Glover JN. Structure of the BRCT repeat domain of MDC1 and its specificity for the free COOH-terminal end of the gamma-H2AX histone tail. J Biol Chem. 2005;280:32053–32056. doi: 10.1074/jbc.C500273200. [DOI] [PubMed] [Google Scholar]
- 91.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 92.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 93.Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci USA. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008;9:795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Melander F, Bekker-Jensen S, Falck J, Bartek J, Mailand N, Lukas J. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol. 2008;181:213–226. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Cote J. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 97.Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 98.Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010;29:3130–3139. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 100.Osley MA, Tsukuda T, Nickoloff JA. ATP-dependent chromatin remodeling factors and DNA damage repair. Mutat Res. 2007;618:65–80. doi: 10.1016/j.mrfmmm.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, Imbalzano AN, Kwon J. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. Embo J. 2006;25:3986–3997. doi: 10.1038/sj.emboj.7601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chowdhury D, Keogh MC, Ishii H, Peterson CL, Buratowski S, Lieberman J. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 103.Chowdhury D, Xu X, Zhong X, Ahmed F, Zhong J, Liao J, Dykxhoorn DM, Weinstock DM, Pfeifer GP, Lieberman J. A PP4-phosphatase complex dephosphorylates gamma-H2AX generated during DNA replication. Mol Cell. 2008;31:33–46. doi: 10.1016/j.molcel.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kimura H, Takizawa N, Allemand E, Hori T, Iborra FJ, Nozaki N, Muraki M, Hagiwara M, Krainer AR, Fukagawa T, Okawa K. A novel histone exchange factor, protein phosphatase 2Cgamma, mediates the exchange and dephosphorylation of H2A-H2B. J Cell Biol. 2006;175:389–400. doi: 10.1083/jcb.200608001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huyen Y, Zgheib O, Ditullio RA Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 106.Wysocki R, Javaheri A, Allard S, Sha F, Cote J, Kron SJ. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol Cell Biol. 2005;25:8430–8443. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell. 2010;40:364–376. doi: 10.1016/j.molcel.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, Bergsagel PL, Wang L, You Z, Lou Z. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470:124–128. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, Lukas J, Lukas C. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 111.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, Oldreive C, Wildenhain J, Tagliaferro A, Pelletier L, Taubenheim N, Durandy A, Byrd PJ, Stankovic T, Taylor AM, Durocher D. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 112.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 113.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25:4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, Kimura H, Ikura M, Nishikubo S, Ito T, Muto A, Miyagawa K, Takeda S, Fishel R, Igarashi K, Kamiya K. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol. 2007;27:7028–7040. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xu N, Hegarat N, Black EJ, Scott MT, Hochegger H, Gillespie DA. Akt/PKB suppresses DNA damage processing and checkpoint activation in late G2. J Cell Biol. 2010;190:297–305. doi: 10.1083/jcb.201003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chailleux C, Tyteca S, Papin C, Boudsocq F, Puget N, Courilleau C, Grigoriev M, Canitrot Y, Trouche D. Physical interaction between the histone acetyl transferase Tip60 and the DNA double-strand breaks sensor MRN complex. Biochem J. 2010;426:365–371. doi: 10.1042/BJ20091329. [DOI] [PubMed] [Google Scholar]
- 118.Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Qin S, Parthun MR. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol Cell Biol. 2002;22:8353–8365. doi: 10.1128/MCB.22.23.8353-8365.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jazayeri A, McAinsh AD, Jackson SP. Saccharomyces cerevisiae Sin3p facilitates DNA double-strand break repair. Proc Natl Acad Sci USA. 2004;101:1644–1649. doi: 10.1073/pnas.0304797101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Barlev NA, Poltoratsky V, Owen-Hughes T, Ying C, Liu L, Workman JL, Berger SL. Repression of GCN5 histone acetyltransferase activity via bromodomain-mediated binding and phosphorylation by the Ku-DNA-dependent protein kinase complex. Mol Cell Biol. 1998;18:1349–1358. doi: 10.1128/mcb.18.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M, Radhakrishnan K, Lee SH, Nickoloff JA, Hromas R. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci USA. 2011;108:540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, Haffty BG, Tommiska J, Blomqvist C, Drapkin R, Adams DJ, Nevanlinna H, Bartek J, Tarsounas M, Ganesan S, Jonkers J. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, Xu X, Deng CX, Finkel T, Nussenzweig M, Stark JM, Nussenzweig A. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cao L, Xu X, Bunting SF, Liu J, Wang RH, Cao LL, Wu JJ, Peng TN, Chen J, Nussenzweig A, Deng CX, Finkel T. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol Cell. 2009;35:534–541. doi: 10.1016/j.molcel.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mimitou EP, Symington LS. Nucleases and helicases take center stage in homologous recombination. Trends Biochem Sci. 2009;34:264–272. doi: 10.1016/j.tibs.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 128.Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair (Amst) 2009;8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bolderson E, Tomimatsu N, Richard DJ, Boucher D, Kumar R, Pandita TK, Burma S, Khanna KK. Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res. 2010;38:1821–1831. doi: 10.1093/nar/gkp1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Eid W, Steger M, El-Shemerly M, Ferretti LP, Pena-Diaz J, Konig C, Valtorta E, Sartori AA, Ferrari S. DNA end resection by CtIP and exonuclease 1 prevents genomic instability. EMBO Rep. 2010;11:962–968. doi: 10.1038/embor.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci USA. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen L, Nievera CJ, Lee AY, Wu X. Cell cycle-dependent complex formation of BRCA1. CtIP.MRN is important for DNA double-strand break repair. J Biol Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 136.Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 Cterminal domains. Mol Cell Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yu X, Fu S, Lai M, Baer R, Chen J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 2006;20:1721–1726. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Coleman KA, Greenberg RA. The BRCA1-RAP80 Complex Regulates DNA Repair Mechanism Utilization by Restricting End Resection. J Biol Chem. 2011;286:13669–13680. doi: 10.1074/jbc.M110.213728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hu Y, Scully R, Sobhian B, Xie A, Shestakova E, Livingston DM. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev. 2011;25:685–700. doi: 10.1101/gad.2011011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329:1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 141.Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Helmink BA, Tubbs AT, Dorsett Y, Bednarski JJ, Walker LM, Feng Z, Sharma GG, McKinnon PJ, Zhang J, Bassing CH, Sleckman BP. H2AX prevents CtIP-mediated DNA end resection and aberrant repair in G1-phase lymphocytes. Nature. 2011;469:245–249. doi: 10.1038/nature09585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li S, Ting NS, Zheng L, Chen PL, Ziv Y, Shiloh Y, Lee EY, Lee WH. Functional link of BRCA1 and ataxia telangiectasia gene product in DNA damage response. Nature. 2000;406:210–215. doi: 10.1038/35018134. [DOI] [PubMed] [Google Scholar]
- 144.Mimitou EP, Symington LS. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010;29:3358–3369. doi: 10.1038/emboj.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Langerak P, Mejia-Ramirez E, Limbo O, Russell P. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 2011;7:e1002271. doi: 10.1371/journal.pgen.1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Feng L, Chen J. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol. 2012 doi: 10.1038/nsmb.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010;329:219–223. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- 148.Liang F, Jasin M. Ku80-deficient cells exhibit excess degradation of extrachromosomal DNA. J Biol Chem. 1996;271:14405–14411. doi: 10.1074/jbc.271.24.14405. [DOI] [PubMed] [Google Scholar]
- 149.Liang F, Romanienko PJ, Weaver DT, Jeggo PA, Jasin M. Chromosomal double-strand break repair in Ku80-deficient cells. Proc Natl Acad Sci USA. 1996;93:8929–8933. doi: 10.1073/pnas.93.17.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Peterson SE, Li Y, Chait BT, Gottesman ME, Baer R, Gautier J. Cdk1 uncouples CtIP-dependent resection and Rad51 filament formation during M-phase double-strand break repair. J Cell Biol. 2011;194:705–720. doi: 10.1083/jcb.201103103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shibata A, Conrad S, Birraux J, Geuting V, Barton O, Ismail A, Kakarougkas A, Meek K, Taucher-Scholz G, Lobrich M, Jeggo PA. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J. 2011;30:1079–1092. doi: 10.1038/emboj.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]


