Abstract
Epithelial ovarian cancer is a malignancy with high rate of death due to an advanced disease at diagnosis and frequent relapse after chemotherapy. Nowadays, there is a lack of knowledge for clear risk factors and predictive and/or prognostic genetic markers although genomic alterations such as mutations in p53, PTEN, BRCA1/BRCA2, HER2, KRAS and PI3K genes have been associated to this pathology. A genomic variant in the 3’ untraslated region of cancer related gene KRAS, is able to disrupt the let-7 miRNA binding site. The SNP, commonly named KRAS-LCS6, determines the substitution of the more abundant T-allele to a G-allele which was observed to increase the KRAS expression and in turn to activate the downstream pathway at higher levels if compared to the T-allele. In this study we assessed the role of the KRAS-LCS6 polymorphism (rs61764370) in 97 early (stages I and II) and 232 advanced (stages III and IV) ovarian cancer patients in order to associate this SNP to any physiopathological characteristic of the patients cohort, including progression free survival and overall survival, with a follow up data longer than ten years. Our data indicate that KRAS-LCS6 polymorphism is not relevant in ovarian cancer, in fact, in our cohort of patients, is not associated to any outcome or physiopathological characteristic.
Keywords: KRAS, let-7, ovarian cancer, LCS6, miRNA, rs61764370
Introduction
Epithelial ovarian cancer (EOC) is one of the deadliest cancer worldwide, with poor survival rates as the majority of cases are diagnosed in late stages. Worldwide more than 200,000 new cases of ovarian cancer were diagnosed every year and more than 100,000 cases are lethal. More than 80% of patients at the time of diagnosis present late stage malignancies with a survival rate less than 30% at 5 years [1,2]. It is well known that there are some risk factors for ovarian cancer including age over 55, familiar history of breast or ovarian cancer and hormone replacement therapy but it is also known that EOC is a heterogeneous disease showing many different genomic alterations such as mutations in p53, PTEN, BRCA1/BRCA2, HER2, KRAS and PI3K genes [3].
RAS family members are important proteins able to regulate cell growth, survival and differentiation by activation of downstream effectors. Three distinct genes encode for the three different proteins H-, K-, and NRAS but KRAS is the most frequently mutated in human cancer [4]. Genetic analysis of ovarian cancers demonstrated that low grade malignancies (typically stage I and II) harbor KRAS gene mutations, whereas late stages (III and IV) rarely display mutations in this gene [5-8]. Gene amplification is another mechanism able to deregulate KRAS in ovarian cancer, in fact, about 11% of ovarian tumors present KRAS amplification [9].
In addition to gene mutation and amplification, KRASactivity can be altered by the short RNA molecule (miRNA) let-7 [10]. It has been shown that let-7 binds to its specific site in the 3′-UTR of KRASmRNA and induces KRASdown regulation [10]. A single-nucleotide polymorphism (SNP) in the KRASlet-7 complementary site (KRAS-LCS6) has been identified and demonstrated to affect KRASexpression. The KRAS-LCS6 (rs61764370) SNP determines the change of the ancestral T-allele to a G-allele which was observed to increase the KRASexpression and in turn to activate the downstream pathway compared to the ancestral allele [11]. The KRAS-LCS6 variant is relatively uncommon, in fact, is almost absent in Native Americans and in East Asians, is very rare in Africans and has a minor allele frequency of about 7% in the European populations [11].
The moderate smoker population harboring the G-allele has been shown to have an increased lung cancer risk [11] but not a reduced survival [12]. The KRAS-LCS6 was also associated to higher cancer risk for triple-negative breast cancer [13] and reduced survival in oral cancer patients [14]. On the contrary, the KRAS-LCS6 SNP was associated to a better outcome in early stage colorectal cancer, but this feature was lost in advanced stages of this disease [15]. Wild-type KRASpatients with metastatic colorectal cancer also seem to better respond to cetuximab monotherapy if the infrequent variant is present [16]. The question for ovarian cancer is still uncertain given that published papers result in opposite conclusions [17,18].
In this paper, we assess the role of the KRAS-LCS6 polymorphism as a biomarker of outcome and response to platinum-based chemotherapy in EOC.
Materials and methods
Patients and samples collection
Biopsies and blood specimens were collected at the Clinic of Obstetrics and Gynecology, San Gerardo Hospital (Monza, Italy). Fresh tumors tissues were minced and kept frozen with blood samples at -80°C. The collection and use of the samples was approved by the local scientific ethical committee and patients gave their written informed consent.
Genotyping
DNA from blood was extracted using Maxwell 16 DNA Purification Kit (Promega, Milan, Italy). The rs61764370 SNP was genotyped using TaqMan SNP Genotyping assay (Applied Biosystems, Monza, Milan), based on Real Time PCR technique (ABI 7900, Applied Biosystems). The PCR was carried out in 384-wells plate with a reaction volume of 5 μL containing genomic DNA (10 ng), 2x TaqMan Genotyping Master Mix (Applied Biosystems), 40x MGB probes and primers. Primers and probe sequences (MGB probes specifically designed for Allelic Discrimination) are property of Applied Biosystems. Thermal cycle conditions were 95°C for 10 minutes and 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Completed PCR plates were analyzed using the Allelic Discrimination Sequence Detection Software (Applied Biosystems).
Copy number variation
DNA from tumors was extracted using Maxwell 16 DNA Purification Kit (Promega, Milan, Italy). The KRAS gene copy number was assessed using TaqMan Copy Number assay (Applied Biosystems, Monza, Milan), based on Real Time PCR technique (ABI 7900, Applied Biosystems). TERT copy number was used as reference gene. The PCR was carried out in 384-wells plate with a reaction volume of 10 μL containing genomic DNA (10 ng), 2x TaqMan Genotyping Master Mix (Applied Biosystems), 40x MGB probes and primers. Primers and probe sequences (MGB probes specifically designed for copy number analysis) are property of Applied Biosystems. Thermal cycle conditions were 95°C for 10 minutes and 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Completed PCR plates were analyzed using the Copy Caller 2.0 Software (Applied Biosystems).
Mutational status of KRAS
The KRAS mutational status in exon 2 was determined in tumor specimens. Genomic DNA, obtained as previously described, was PCR amplified with the following primers: Fw 5’-CTTAAGCGTCGATGGAGGAG and Rw: 5’-AGAATGGTCCTGCACCAGTAA. Amplification was performed in a thermocycler (TC-510, Techna) with 35 cycles at 95°C for 1’; 60°C for 30”; 72°C for 1’ Sequencing was performed by Primm (Milan, Italy).
Statistical methods
A consecutive cohort of patients with ovarian cancer for which biological material was available was identified and retrospectively enrolled in this monocentric study. Baseline covariate distributions were summarized using descriptive statistics (median and range for continuous variables; absolute and percentage frequencies for categorical variables); nonparametric tests (Wilcoxon-Mann-Whitney test for continuous covariates and Fisher’s exact test for categorical covariates) were used to detect statistical association. Progression Free Survival (PFS) was defined as the time from the date of diagnosis up to the date of first progression or death from any cause, whichever came first. Subjects who have not progressed or died while on study were censored at the last disease assessment date. Overall survival (OS) was defined as the time from the date of diagnosis up to the date of death from any cause. Subjects who have not died while on study were censored at the last follow-up. Survival curves were estimated with the Kaplan-Meier method. Cox proportional hazards models were used for univariate and multivariate analysis to estimate and test demographic characteristics, clinical features, and biological parameters for their associations with PFS and OS. Results were expressed as Hazard Ratios (HRs) and their 95% confidence intervals (95%CIs). Statistical analyses were carried out using SAS version 9.1 (SAS Institute, Cary, NC).
Results
Clinical and histopathological characteristics
From September 1979 to December 2004 356 patients with ovarian cancer were identified. 27 (7.6%) patients with borderline grading were not considered for the study; out of the remaining patients 82 (24.9%) had FIGO stage I, 15 (4.6%) had FIGO stage II, 206 (62.6%) patients had FIGO stage III and 26 (7.9%) had FIGO stage IV disease.
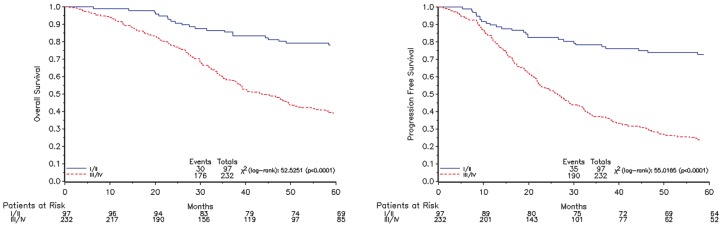
After an initial comparison between stage I and II no statistical differences were highlighted between these stages (early stage) (Table 1). The comparison between stage III and IV (late stage) suggested a difference between these stages when compared for histotype (Table 1). Overall, the estimate risk of the two populations detected considering progression free survival (PFS, Table 5) and overall survival (OS, Table 6) as endpoints was not statistically different; therefore we decided to group these two categories of patients.
Table 1.
Comparison within stages according to clinical and histopathological characteristics
| Early stage (p-value) | Late stage (p-value) | |
|---|---|---|
| Age at diagnosis | 0.221 | 0.844 |
| Grading | 0.128 | 0.565 |
| Histotype | 0.247 | 0.005 |
| Residual tumor | - | 0.827 |
| Adjuvant therapy | 0.571 | - |
| KRAS status | 1.000 | - |
| KRAS gene copy number | 0.775 | 0.157 |
| KRAS-LCS6 SNP | 1.000 | 0.421 |
Table 5.
Prognostic evaluation of clinical and histopatological characteristics of late stages: Progression Free Survival
| Progression Free Survival - Univariated analysis | |||||
|---|---|---|---|---|---|
| HR | Lower 95% CI | Upper 95% CI | p-value | ||
| Age at diagnosis | 1.02 | 1.01 | 1.03 | p <0.001 | |
| Stage | III | 1 | |||
| IV | 1.33 | 0.86 | 2.05 | 0.206 | |
| Grading | G1 | 1 | |||
| G2 | 2.14 | 1.05 | 4.34 | 0.035 | |
| G3 | 2.13 | 1.08 | 4.20 | 0.029 | |
| Histotype | Serous | 1 | |||
| Endometroid | 1.18 | 0.72 | 1.92 | 0.516 | |
| Clear cell | 1.21 | 0.66 | 2.24 | 0.535 | |
| Mucinous | 1.90 | 0.93 | 3.87 | 0.078 | |
| Undifferentiated | 1.01 | 0.42 | 2.47 | 0.977 | |
| Other | 0.30 | 0.04 | 2.13 | 0.227 | |
| Residual tumor | < 2 cm | 1 | |||
| ≥ 2 cm | 1.83 | 1.33 | 2.51 | p <0.001 | |
| KRAS gene copy number | Disomy | 1 | |||
| Amplification | 1.11 | 0.78 | 1.59 | 0.556 | |
| Deletion | 1.69 | 0.99 | 2.86 | 0.054 | |
| KRAS-LCS6 polymorphism | T/G-G/G | 1 | |||
| T/T | 1.04 | 0.72 | 1.51 | 0.831 | |
| Progression Free Survival - Multivariated analysis | |||||
| HR | Lower 95% CI | Upper 95% CI | p-value | ||
| KRAS-LCS6 polymorphism | T/G-G/G | 1 | |||
| T/T | 1.19 | 0.80 | 1.76 | 0.383 | |
Table 6.
Prognostic evaluation of clinical and histopatological characteristics of late stages: Overall Survival
| Overall Survival - Univariated analysis | |||||
|---|---|---|---|---|---|
| HR | Lower 95% CI | Upper 95% CI | p-value | ||
| Age at diagnosis | 1.02 | 1.01 | 1.04 | p <0.001 | |
| Stage | III | 1 | |||
| IV | 1.40 | 0.90 | 2.20 | 0.136 | |
| Grading | G1 | 1 | |||
| G2 | 3.27 | 1.40 | 7.62 | 0.006 | |
| G3 | 2.86 | 1.26 | 6.50 | 0.012 | |
| Histotype | Serous | 1 | |||
| Endometroid | 1.08 | 0.64 | 1.85 | 0.767 | |
| Clear cell | 1.41 | 0.76 | 2.60 | 0.278 | |
| Mucinous | 2.19 | 1.02 | 4.68 | 0.044 | |
| Undifferentiated | 1.23 | 0.50 | 2.99 | 0.656 | |
| Other | 0.34 | 0.05 | 2.44 | 0.283 | |
| Residual tumor | < 2 cm | 1 | |||
| ≥ 2 cm | 2.29 | 1.63 | 3.21 | p <0.001 | |
| KRAS gene copy number | Disomy | 1 | |||
| Amplification | 0.98 | 0.68 | 1.42 | 0.912 | |
| Deletion | 1.34 | 0.76 | 2.37 | 0.317 | |
| KRAS-LCS6 polymorphism | T/G-G/G | 1 | |||
| T/T | 0.91 | 0.62 | 1.33 | 0.635 | |
| Overall Survival - Multivariated analysis | |||||
| HR | Lower 95% CI | Upper 95% CI | p-value | ||
| KRAS-LCS6 polymorphism | T/G-G/G | 1 | |||
| T/T | 0.98 | 0.66 | 1.46 | 0.922 | |
For the stage I/II population the median age at diagnosis was 52.1 years (range: 16.5 – 81.8 years); 82 (84.5%) patients had FIGO stage I; the predominant histology was serous subtype (35 patients, 36.1%) and poorly differentiated grade (43 patients, 44.3%). 64 patients (66.0%) received an adjuvant cisplatin-based therapy. For these stages mutation in KRAS gene was found in 15 (15.5%) patients, while 22 patients (22.7%) presented KRAS gene copy number variation. After a median follow-up of 10.0 years 67 (69.1%) patients were alive, 62 (92.5%) of them were progression-free. Age at diagnosis was the only baseline covariate statistically associated to PFS (HR=1.04, 95%CI: 1.00-1.07; p-value=0.024); age at diagnosis (HR=1.04, 95% CI: 1.01-1.08; p-value=0.019), cancer grading (HR(G3vsG1)=4.39, 95%CI: 1.01-19.10; p-value=0.049) were baseline covariates statistically associated to OS.
For the stage III/IV population the median age at diagnosis was 54.7 years (range: 13.2 – 79.1 years; 206 (88.8%) patients had FIGO stage III disease; residual tumor size was more than 2 cm in 151 (65.1%) patients; the predominant histology was serous subtype (n.of patients:180, 77.6%) and poorly differentiated grade (n.of patients: 154, 66.4%). All patients received platinum based therapy. Sixty-one (26.4%) patients presented KRAS gene copy variation while the mutational status of KRAS was not evaluated for this group of patients given the literature data demonstrating a very low rate of mutation for this gene. After a median follow-up of 11.3 years, 56 (24.1%) patients were alive, 42 (75.0%) of them were progression-free. Age at diagnosis (HR=1.02, 95% CI: 1.01-1.03; p-value<0.001), residual tumor size (HR=1.83, 95%CI: 1.33-2.51; p-value=<0.001) and grading (HR(G2vsG1)=2.14, 95%CI: 1.05-4.34; p-value=0.035 / HR(G3vsG1)=2.13, 95%CI: 1.08-4.20; p-value=0.029) were the baseline covariates statistically correlated to PFS, globally or at least comparing single risk factor categories; the correlations detected considering OS as endpoint were age at diagnosis (HR=1.02, 95%CI: 1.01-1.04; p-value=<0.001), residual tumor size (HR=2.29, 95%CI: 1.63-3.21; p-value=<0.001), histotype (HR(MUCINOUSvsSEROUS)=2.19, 95%CI: 1.02-4.68; p-value=0.044) and grading (HR(G2vsG1)=3.27, 95%CI: 1.40-7.62; p-value=0.006 / HR(G3vsG1)=2.86, 95%CI: 1.26-6.50; p-value=0.012).
Clinical and histopathological characteristics of the populations are summarized in Table 2. Survival estimates are plotted in Figure 1 and risk estimates of baseline covariates are reported in Tables 3, 4, 5 and 6.
Table 2.
Clinical and histopatological characteristics
| Early stage | Late stage | ||||
|---|---|---|---|---|---|
| N | % | N | % | ||
| Patients | 97 | 232 | |||
| Age at diagnosis | Median | 52.1 | 54.7 | ||
| Range | 16.5-81.8 | 13.2-79.1 | |||
| Stage | I | 82 | 84.5 | 206 | 88.8 |
| II | 15 | 15.5 | 26 | 11.2 | |
| Grading | G1 | 19 | 19.6 | 15 | 6.5 |
| G2 | 35 | 36.1 | 63 | 27.1 | |
| G3 | 43 | 44.3 | 154 | 66.4 | |
| Histotype | Serous | 35 | 36.1 | 180 | 77.6 |
| Endometroid | 24 | 24.7 | 21 | 9.0 | |
| Clear cell | 19 | 19.6 | 13 | 5.6 | |
| Mucinous | 12 | 12.4 | 9 | 3.9 | |
| Undifferentiated | 2 | 2.1 | 6 | 2.6 | |
| Other | 5 | 5.1 | 3 | 1.3 | |
| Residual tumor | < 2 cm | Absent | 81 | 34.9 | |
| ≥ 2 cm | 151 | 65.1 | |||
| Adjuvant therapy | Platinum-based | 64 | 66.0 | 232 | 100.0 |
| No therapy | 33 | 34.0 | |||
| KRAS status | Wild-type | 82 | 84.5 | Not evaluated | |
| Mutated | 15 | 15.5 | |||
| KRAS gene copy number | Disomy | 75 | 77.3 | 170 | 73.6 |
| Amplification | 15 | 15.5 | 45 | 19.5 | |
| Deletion | 7 | 7.2 | 16 | 6.9 | |
| KRAS – LCS6 polymprphism | T/G-G/G | 18 | 18.6 | 41 | 17.7 |
| T/T | 79 | 81.4 | 191 | 82.3 | |
Figure 1.
survival estimate plots for OS (left) and PFS (right) according to the disease stage.
Table 3.
Prognostic evaluation of clinical and histopatological characteristics of early stages: Progression Free Survival
| Progression Free Survival - Univariated analysis | |||||
|---|---|---|---|---|---|
| HR | Lower 95% CI | Upper 95% CI | p-value | ||
| Age at diagnosis | 1.04 | 1.00 | 1.07 | 0.024 | |
| Stage | I | 1 | |||
| II | 1.24 | 0.54 | 2.85 | 0.606 | |
| Grading | G1 | 1 | |||
| G2 | 1.95 | 0.63 | 5.97 | 0.245 | |
| G3 | 2.63 | 0.89 | 7.77 | 0.081 | |
| Histotype | Serous | 1 | |||
| Endometroid | 0.46 | 0.17 | 1.26 | 0.132 | |
| Clear cell | 1.15 | 0.49 | 2.71 | 0.756 | |
| Mucinous | 0.59 | 0.17 | 2.04 | 0.404 | |
| Undifferentiated | 6.88 | 1.46 | 32.37 | 0.01 | |
| Other | 0.76 | 0.17 | 3.34 | 0.717 | |
| First line therapy | No therapy | 1 | |||
| Platinum-based | 0.79 | 0.40 | 1.58 | 0.510 | |
| KRAS status | Wild-type | 1 | |||
| Mutated | 0.85 | 0.33 | 2.19 | 0.736 | |
| KRAS gene copy number | Disomy | 1 | |||
| Amplification | 1.48 | 0.64 | 3.44 | 0.357 | |
| Deletion | 1.22 | 0.37 | 4.04 | 0.748 | |
| KRAS-LCS6 polymorphism | T/G-G/G | 1 | |||
| T/T | 1.08 | 0.45 | 2.60 | 0.865 | |
| Progression Free Survival - Multivariated analysis | |||||
| HR | Lower 95% CI | Upper 95% CI | p-value | ||
| KRAS-LCS6 polymorphism | T/G-G/G | 1 | |||
| T/T | 1.34 | 0.50 | 3.57 | 0.564 | |
Table 4.
Prognostic evaluation of clinical and histopatological characteristics of early stages: Overall survival
| Overall Survival - Univariated analysis | |||||
|---|---|---|---|---|---|
| HR | Lower 95% CI | Upper 95% CI | p-value | ||
| Age at diagnosis | 1.04 | 1.01 | 1.08 | 0.019 | |
| Stage | I | 1 | |||
| II | 1.37 | 0.56 | 3.40 | 0.492 | |
| Grading | G1 | 1 | |||
| G2 | 3.35 | 0.74 | 15.11 | 0.116 | |
| G3 | 4.39 | 1.01 | 19.10 | 0.049 | |
| Histotype | Serous | 1 | |||
| Endometroid | 0.81 | 0.28 | 2.37 | 0.699 | |
| Clear cell | 1.89 | 0.74 | 4.80 | 0.182 | |
| Mucinous | 1.06 | 0.29 | 3.89 | 0.930 | |
| Undifferentiated | 7.68 | 1.65 | 35.81 | 0.009 | |
| Other | 0.67 | 0.09 | 5.21 | 0.699 | |
| First line therapy | No therapy | 1 | |||
| Platinum-based | 0.63 | 0.30 | 1.33 | 0.227 | |
| KRAS status | Wild-type | 1 | |||
| Mutated | 1.18 | 0.45 | 3.11 | 0.737 | |
| KRAS gene copy number | Disomy | 1 | |||
| Amplification | 1.41 | 0.57 | 3.49 | 0.455 | |
| Deletion | 0.38 | 0.05 | 2.86 | 0.350 | |
| KRAS-LCS6 polymorphism | T/G-G/G | 1 | |||
| T/T | 2.13 | 0.64 | 7.05 | 0.215 | |
| Overall Survival - Multivariated analysis | |||||
| HR | Lower 95% CI | Upper 95% CI | p-value | ||
| KRAS-LCS6 polymorphism | T/G-G/G | 1 | |||
| T/T | 3.11 | 0.72 | 13.43 | 0.129 | |
Correlation between KRAS-LCS6 polymorphism and clinical, histopathological characteristics of ovarian cancer patients
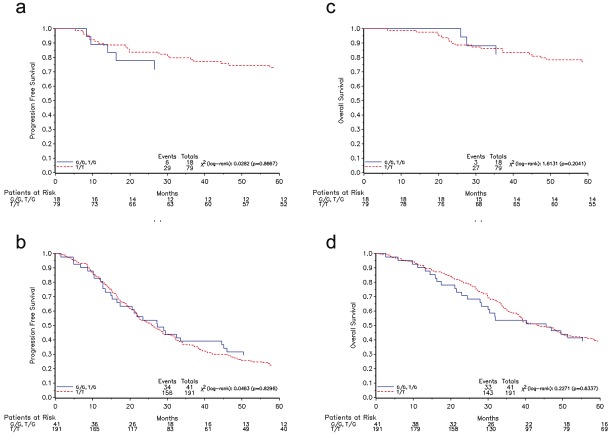
For both the stage I/II and stage III/VI populations the predominant status of the polymorphic site LCS6 was T/T (n.of patients: 79, 81.4% and 191, 82.3% for early and advanced stages, respectively). After the analysis of literature, we decided to evaluate the role of KRAS-LCS6 polymorphism as G containing (G/G plus G/T) and Tonly (T/T) population. The G containing variants were present in about 18% of the whole population (18.6% in early stage and 17.7% in late stage) and were not statistically significantly correlated to any clinical and histopathological characteristic such as age at diagnosis, residual tumor size, histotype, grading and response to therapy in both early and late stages. This SNP was also not correlated to the other KRAS features such as mutational status and gene amplification. Moreover, the genotype was not able to predict any prognosis tested as PFS and OS and plotted as Kaplan-Meier curves (Figure 2). Correlation between KRAS features and clinical and histopathological characteristics of patient are reported in Table 7 whereas relative risk estimates are reported in Tables 3, 4, 5 and 6.
Figure 2.
Kaplan-Meier plots for PFS (left panels) and OS (right panels) in early (upper panels) and late (lower panels) stages according to the LCS6 polymorphism
Table 7.
Correlations between clinical and histopathological parameters and genotypes
| Early Stages | ||||||
|---|---|---|---|---|---|---|
| T/G-G/G | T/T | p-value | ||||
| N | % | N | % | |||
| Age at diagnosis | Patients | 18 | 79 | 0.792 | ||
| Median | 52.5 | 52.1 | ||||
| Range | 30.8-70.8 | 16.5-81.8 | ||||
| Stage | I | 15 | 83.3 | 67 | 84.8 | 1.000 |
| II | 3 | 16.7 | 12 | 15.2 | ||
| Grading | G1 | 3 | 16.7 | 16 | 20.3 | 1.000 |
| G2 | 7 | 38.9 | 28 | 35.4 | ||
| G3 | 8 | 44.4 | 35 | 44.3 | ||
| Histotype | Serous | 8 | 44.4 | 27 | 34.2 | 0.801 |
| Endometroid | 6 | 33.3 | 18 | 22.8 | ||
| Clear cell | 3 | 16.7 | 16 | 20.3 | ||
| Mucinous | 1 | 5.6 | 11 | 13.9 | ||
| Undifferentiated | - | - | 2 | 2.5 | ||
| Other | - | - | 5 | 6.3 | ||
| Adjuvant therapy | Platinum-based | 12 | 66.7 | 27 | 65.8 | 1.000 |
| No therapy | 6 | 33.3 | 52 | 34.2 | ||
| KRAS status | Wild-type | 14 | 82.4 | 66 | 84.6 | 0.728 |
| Mutated | 3 | 17.6 | 12 | 15.4 | ||
| KRAS gene copy number | Disomy | 13 | 72.2 | 62 | 78.5 | 0.720 |
| Amplification | 4 | 22.2 | 11 | 13.9 | ||
| Deletion | 1 | 5.6 | 6 | 7.6 | ||
| Late Stages | ||||||
| T/G-G/G | T/T | p-value | ||||
| N | % | N | % | |||
| Age at diagnosis | Patients | 41 | 191 | 0.502 | ||
| Median | 57.9 | 54.5 | ||||
| Range | 20.6-77.7 | 13.2-79.1 | ||||
| Stage | III | 35 | 85.4 | 171 | 89.5 | 0.421 |
| IV | 6 | 14.6 | 20 | 10.5 | ||
| Grading | G1 | 4 | 9.8 | 11 | 5.8 | 0.462 |
| G2 | 9 | 22.0 | 54 | 28.2 | ||
| G3 | 28 | 68.2 | 126 | 66.0 | ||
| Histotype | Serous | 32 | 78.0 | 148 | 77.5 | 0.624 |
| Endometroid | 2 | 4.9 | 19 | 9.9 | ||
| Clear cell | 3 | 7.3 | 10 | 5.2 | ||
| Mucinous | 3 | 7.3 | 6 | 3.1 | ||
| Undifferentiated | 1 | 2.4 | 5 | 2.6 | ||
| Other | - | - | 3 | 1.6 | ||
| Residual tumor | < 2 cm | 13 | 31.7 | 68 | 35.6 | 0.719 |
| ≥ 2 cm | 28 | 68.3 | 123 | 64.4 | ||
| KRAS gene copy number | Disomy | 34 | 82.9 | 136 | 71.6 | 0.056 |
| Amplification | 3 | 7.3 | 42 | 22.1 | ||
| Deletion | 4 | 9.8 | 12 | 6.3 | ||
Discussion
KRAS is an important factor associated with poor prognosis and decreased response in several human tumors. Mutation in KRAS gene, for example, is determinant in colon cancer patients for the selection of treatment with the EGFR inhibitor cetuximab [19].
KRAS mutation has been found in several tumors [20-22] and KRAS amplification has been found in ovarian cancer [9,20]. In addition, a polymorphic site in the 3’UTR of KRAS gene has been detected. The base change in the polymorphic site alters the binding of the let-7 miRNA to the KRAS sequence, resulting in altered expression of KRAS protein and its downstream effectors [10]. The presence of the less abundant G allele variant, is associated to an increased risk in some human tumors including lung and breast [11-13]. As for response to treatment, the data are less clear and, depending on the tumor type and the stage, the outcome seems to be different [14,16]. Controversial is also the role of the polymorphism in ovarian cancer [17,18,23].
We here analyzed, in a relatively large cohort of patients with a follow up data longer than ten years, the role of KRAS-LCS6 SNP in determining response of ovarian cancer patients to platinum based therapy. The distribution of polymorphism in the group of patients analyzed, is in line with the reported values present in the literature [24], and no differences were found between early and late stage ovarian cancer. The presence of a G allele, which associates with an increased expression of KRAS, does not modify the response of patients to platinum based therapy. We also evaluated for the first time the presence of G allele together with KRAS gene mutation and gene copy number variation.
Given the importance of determining new biomarkers able to discriminate between responders or non responders to a given therapy, our data would suggest that KRAS gene alterations, either mutation, gene copy number variation or KRAS let-7 complementary site SNP variation, do not impact on response to therapy in ovarian cancer patients. We also assessed the risk grouping the “KRAS altered” population (mutated, gene amplified and G-containing KRAS let-7 complementary site SNP) against wild-type patients: again the HR of KRAS altered population was very close to 1 and no statistical difference between the two cohorts was highlighted (Table 8).
Table 8.
Risk in “KRAS altered” ovarian cancer population
| Progression Free Survival | ||||
|---|---|---|---|---|
| HR | Lower 95% CI | Upper 95% CI | p-value | |
| Whole cohort | 0.98 | 0.75 | 1.28 | 0.860 |
| Early stages | 0.92 | 0.47 | 1.84 | 0.809 |
| Late stages | 1.04 | 0.78 | 1.40 | 0.724 |
| Overall Survival | ||||
| HR | Lower 95% CI | Upper 95% CI | p-value | |
| Whole cohort | 0.95 | 0.72 | 1.26 | 0.719 |
| Early stages | 0.84 | 0.39 | 1.78 | 0.642 |
| Late stages | 1.05 | 0.77 | 1.42 | 0.768 |
We have recently found that KRAS gene alteration reduces the response to platinum based first line therapy in NSCLC [25] and Garassino M et al., submitted. We do not have a clear explanation for the different response to platinum based therapy in NSCLC and EOC in relation to KRAS alterations. One possibility could be that KRAS gene alterations are more frequent in NSCLC [22] than in EOC where KRAS mutations are present in a relatively low percentage and only in low grade tumors [7]. This would imply that changes in KRAS downstream signalling are relevant for tumor initiation and progression in NSCLC and to a lesser extent in EOC, hence alterations in its function have a different impact on the aggressiveness of the two tumors. NSCLC is strongly associated with smoking habits, and KRAS mutations are frequent in smokers relative to non smoker population [26]. This could account for the different role of KRAS in the two malignancies.
Whatever the reason, our data suggest that KRAS-LCS6 polymorphism and more in general KRAS gene alterations do not impact on EOC response to platinum therapy. It will be interesting to analyze, in the future, the role, if any, of KRAS in determining response of EOC patients to new emerging targeted therapies (such as anti-angiogenic therapy) which are likely to become new standard therapies in the next years for this malignancy.
Acknowledgements
The generous contributions of the Nerina and Mario Mattioli Foundation are gratefully acknowledged.
References
- 1.Ferlay J, Parkin DM, Steliarova-Foucher E. Estimates of cancer incidence and mortality in Europe in 2008. Eur J Cancer. 2010;46:765–781. doi: 10.1016/j.ejca.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Gayther SA, Pharoah PD. The inherited genetics of ovarian and endometrial cancer. Curr Opin Genet Dev. 2010;20:231–238. doi: 10.1016/j.gde.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 5.Vereczkey I, Serester O, Dobos J, Gallai M, Szakacs O, Szentirmay Z, Toth E. Molecular characterization of 103 ovarian serous and mucinous tumors. Pathol Oncol Res. 2011;17:551–559. doi: 10.1007/s12253-010-9345-8. [DOI] [PubMed] [Google Scholar]
- 6.Matulonis UA, Hirsch M, Palescandolo E, Kim E, Liu J, van Hummelen P, MacConaill L, Drapkin R, Hahn WC. High throughput interrogation of somatic mutations in high grade serous cancer of the ovary. PLoS One. 2011;6:e24433. doi: 10.1371/journal.pone.0024433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singer G, Oldt R 3rd, Cohen Y, Wang BG, Sidransky D, Kurman RJ, Shih Ie M. Mutations in BRAF and KRAS characterize the development of low-grade ovarian serous carcinoma. J Natl Cancer Inst. 2003;95:484–486. doi: 10.1093/jnci/95.6.484. [DOI] [PubMed] [Google Scholar]
- 8.Sieben NL, Macropoulos P, Roemen GM, Kolkman-Uljee SM, Jan Fleuren G, Houmadi R, Diss T, Warren B, Al Adnani M, De Goeij AP, Krausz T, Flanagan AM. In ovarian neoplasms, BRAF, but not KRAS, mutations are restricted to lowgrade serous tumours. J Pathol. 2004;202:336–340. doi: 10.1002/path.1521. [DOI] [PubMed] [Google Scholar]
- 9.Hanrahan AJ, Schultz N, Westfal ML, Sakr RA, Giri DD, Scarperi S, Janikariman M, Olvera N, Stevens EV, She QB, Aghajanian C, King TA, Stanchina ED, Spriggs DR, Heguy A, Taylor BS, Sander C, Rosen N, Levine DA, Solit DB. Genomic complexity and AKT dependence in serous ovarian cancer. Cancer Discov. 2012;2:56–67. doi: 10.1158/2159-8290.CD-11-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 11.Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, Muller RU, Straka E, Su L, Burki EA, Crowell RE, Patel R, Kulkarni T, Homer R, Zelterman D, Kidd KK, Zhu Y, Christiani DC, Belinsky SA, Slack FJ, Weidhaas JB. A SNP in a let-7 microRNA complementary site in the KRAS 3' untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson HH, Christensen BC, Plaza SL, Wiencke JK, Marsit CJ, Kelsey KT. KRAS mutation, KRAS-LCS6 polymorphism, and non-small cell lung cancer. Lung Cancer. 2010;69:51–53. doi: 10.1016/j.lungcan.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paranjape T, Heneghan H, Lindner R, Keane FK, Hoffman A, Hollestelle A, Dorairaj J, Geyda K, Pelletier C, Nallur S, Martens JW, Hooning MJ, Kerin M, Zelterman D, Zhu Y, Tuck D, Harris L, Miller N, Slack F, Weidhaas J. A 3'-untranslated region KRAS variant and triple-negative breast cancer: a case-control and genetic analysis. Lancet Oncol. 2011;12:377–386. doi: 10.1016/S1470-2045(11)70044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen BC, Moyer BJ, Avissar M, Ouellet LG, Plaza SL, McClean MD, Marsit CJ, Kelsey KT. A let-7 microRNA-binding site polymorphism in the KRAS 3' UTR is associated with reduced survival in oral cancers. Carcinogenesis. 2009;30:1003–1007. doi: 10.1093/carcin/bgp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smits KM, Paranjape T, Nallur S, Wouters KA, Weijenberg MP, Schouten LJ, van den Brandt PA, Bosman FT, Weidhaas JB, van Engeland M. A Let-7 MicroRNA SNP in the KRAS 3'UTR Is Prognostic in Early-Stage Colorectal Cancer. Clin Cancer Res. 2011;17:7723–7731. doi: 10.1158/1078-0432.CCR-11-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W, Winder T, Ning Y, Pohl A, Yang D, Kahn M, Lurje G, Labonte MJ, Wilson PM, Gordon MA, Hu-Lieskovan S, Mauro DJ, Langer C, Rowinsky EK, Lenz HJ. A let-7 microRNA-binding site polymorphism in 3'-untranslated region of KRAS gene predicts response in wildtype KRAS patients with metastatic colorectal cancer treated with cetuximab monotherapy. Ann Oncol. 2011;22:104–109. doi: 10.1093/annonc/mdq315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pharoah PD, Palmieri RT, Ramus SJ, Gayther SA, Andrulis IL, Anton-Culver H, Antonenkova N, Antoniou AC, Goldgar D, Beattie MS, Beckmann MW, Birrer MJ, Bogdanova N, Bolton KL, Brewster W, Brooks-Wilson A, Brown R, Butzow R, Caldes T, Caligo MA, Campbell I, Chang-Claude J, Chen YA, Cook LS, Couch FJ, Cramer DW, Cunningham JM, Despierre E, Doherty JA, Dork T, Durst M, Eccles DM, Ekici AB, Easton D, Fasching PA, de Fazio A, Fenstermacher DA, Flanagan JM, Fridley BL, Friedman E, Gao B, Sinilnikova O, Gentry-Maharaj A, Godwin AK, Goode EL, Goodman MT, Gross J, Hansen TV, Harnett P, Rookus M, Heikkinen T, Hein R, Hogdall C, Hogdall E, Iversen ES, Jakubowska A, Johnatty SE, Karlan BY, Kauff ND, Kaye SB, Chenevix-Trench G, Kelemen LE, Kiemeney LA, Kjaer SK, Lambrechts D, Lapolla JP, Lazaro C, Le ND, Leminen A, Leunen K, Levine DA, Lu Y, Lundvall L, Macgregor S, Marees T, Massuger LF, McLaughlin JR, Menon U, Montagna M, Moysich KB, Narod SA, Nathanson KL, Nedergaard L, Ness RB, Nevanlinna H, Nickels S, Osorio A, Paul J, Pearce CL, Phelan CM, Pike MC, Radice P, Rossing MA, Schildkraut JM, Sellers TA, Singer CF, Song H, Stram DO, Sutphen R, Lindblom A, Terry KL, Tsai YY, van Altena AM, Vergote I, Vierkant RA, Vitonis AF, Walsh C, Wang-Gohrke S, Wappenschmidt B, Wu AH, Ziogas A, Berchuck A, Risch HA. The role of KRAS rs61764370 in invasive epithelial ovarian cancer: implications for clinical testing. Clin Cancer Res. 2011;17:3742–3750. doi: 10.1158/1078-0432.CCR-10-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ratner ES, Keane FK, Lindner R, Tassi RA, Paranjape T, Glasgow M, Nallur S, Deng Y, Lu L, Steele L, Sand S, Muller RU, Bignotti E, Bellone S, Boeke M, Yao X, Pecorelli S, Ravaggi A, Katsaros D, Zelterman D, Cristea MC, Yu H, Rutherford TJ, Weitzel JN, Neuhausen SL, Schwartz PE, Slack FJ, Santin AD, Weidhaas JB. A KRAS variant is a biomarker of poor outcome, platinum chemotherapy resistance and a potential target for therapy in ovarian cancer. Oncogene 2011. doi: 10.1038/onc.2011.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J. Clin. Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 20.Sasaki H, Hikosaka Y, Kawano O, Moriyama S, Yano M, Fujii Y. Evaluation of Kras gene mutation and copy number gain in non-small cell lung cancer. J Thorac Oncol. 2011;6:15–20. doi: 10.1097/JTO.0b013e31820594f0. [DOI] [PubMed] [Google Scholar]
- 21.Adelstein BA, Dobbins TA, Harris CA, Marschner IC, Ward RL. A systematic review and metaanalysis of KRAS status as the determinant of response to anti-EGFR antibodies and the impact of partner chemotherapy in metastatic colorectal cancer. Eur J Cancer. 2011;47:1343–1354. doi: 10.1016/j.ejca.2011.03.031. [DOI] [PubMed] [Google Scholar]
- 22.Riely GJ, Marks J, Pao W. KRAS mutations in non-small cell lung cancer. Proc Am Thorac Soc. 2009;6:201–205. doi: 10.1513/pats.200809-107LC. [DOI] [PubMed] [Google Scholar]
- 23.Ratner E, Lu L, Boeke M, Barnett R, Nallur S, Chin LJ, Pelletier C, Blitzblau R, Tassi R, Paranjape T, Hui P, Godwin AK, Yu H, Risch H, Rutherford T, Schwartz P, Santin A, Matloff E, Zelterman D, Slack FJ, Weidhaas JB. A KRAS-variant in ovarian cancer acts as a genetic marker of cancer risk. Cancer Res. 2010;70:6509–6515. doi: 10.1158/0008-5472.CAN-10-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.NCBI. http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=61764370. [Google Scholar]
- 25.Marabese M, Rulli E, Bettini A, Garassino MC, Longo F, Moscetti L, Pavese I, Lauricella C, Broggini M, Farina G. KRAS mutational status impact progression-free survival of patients treated with platinum-based chemotherapy in NSCLC. Molecular Cancer Therapeutics. 2011;10:B77. [Google Scholar]
- 26.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]