Abstract
Phagocytosis is an evolutionarily ancient, receptor-driven process, by which phagocytic cells recognize invading microbes and destroy them after internalization. The phagocytosis receptor Eater is expressed exclusively on Drosophila phagocytes and is required for the survival of bacterial infections. In a recent study, we explored how Eater can defend fruit flies against different kinds of bacteria. We discovered that Eater bound to certain types of bacteria directly, while for others bacterial binding was dependent on prior disruption of the bacterial envelope. Similar to phagocytes, antimicrobial peptides and lysozymes are ancient components of animal immune systems. Our results suggest that cationic antimicrobial peptides, as well as lysozymes, can facilitate Eater binding to live Gram-negative bacteria. Both types of molecules promote surface-exposure of bacterial ligands that otherwise would remain buried and hidden under an outer membrane. We propose that unmasking ligands for phagocytic receptors may be a conserved mechanism operating in many animals, including humans. Thus, studying a Drosophila phagocytosis receptor may advance our understanding of innate immunity in general.
Keywords: antimicrobial peptides, cecropin A, Eater, Gram-negative bacteria, innate immunity, lysozyme, pattern recognition, pattern recognition receptor, phagocytic receptor, phagocytosis
The Drosophila Phagocyte Receptor Eater and Recognition of Pathogenic Bacteria
Most animals possess mobile cells that survey tissues and body surfaces for foreign intruders and, once recognition occurs, quickly react by ingesting and destroying them. These phagocytes are part of the innate immune system, an evolutionarily old form of immunity. The innate immune system uses a limited set of germline-encoded receptors to fend off a multitude of microbes, which differ in surface and composition. Drosophila melanogaster has emerged as an attractive model system to study how animals meet this challenge.1,2 The concept of pattern recognition, originally proposed by Charles Janeway,3,4 provides an elegant solution to the problem: by recognizing evolutionarily conserved, or structurally and functionally constrained molecules of microbial origin (patterns), a host can prevail despite using a limited set of receptors (termed pattern recognition receptors).
Scavenger receptors are one group of molecules that participate in pattern recognition.5 They form a superfamily of structurally unrelated molecules with emerging functions in vertebrate and invertebrate immunity. Many of these receptors remain poorly understood. They are prevalent on endothelial cells and phagocytes and display a preference for multiple, polyanionic ligands.6 In 2005 we identified Eater, a Drosophila receptor with scavenger receptor properties and involved in bacterial phagocytosis.7 Eater is expressed exclusively in phagocytic hemocytes and their precursors. Genetic analysis showed that flies deficient in Eater expression were immunocompromised and succumbed more readily to bacterial infections with a range of diverse bacteria.7-11
Unexpectedly, we found that the ectodomain of Eater consisted mostly of EGF-like repeats, an extracellular protein domain implicated, thus far, mostly in protein–protein interactions. However, the N-terminal part of Eater (comprising four EGF-like or NIM repeats)7,12,13 was able to bind directly to heat-inactivated bacteria,7 establishing Eater as a bona-fide pattern recognition receptor. In a recent study,14 we pursued these initial observations to better understand how a single phagocytosis receptor can defend a host organism against an array of different microbes. In particular, we wondered whether signaling to a transcriptional response might contribute to Eater’s protective role and how Eater interacts with live bacteria in vivo.
Binding and Internalizing Rather Than Sensing and Signaling?
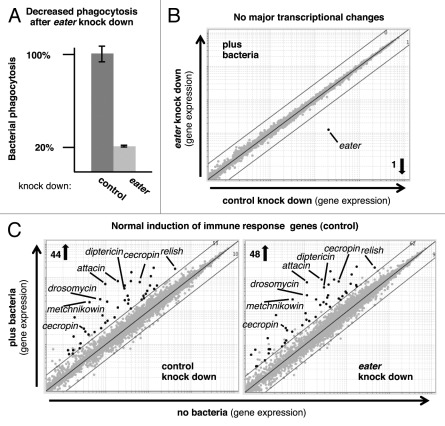
Inducible and constitutive responses in the Drosophila fat body or barrier epithelia are mediated by two well characterized immune signaling pathways, Toll and Immune deficiency (Imd), which display striking similarities to mammalian NFkB signaling.1,2 In eater null flies, as well as in the phagocytic cell line S2, signaling via these pathways seems to be intact in the absence of eater-expression, as judged by the transcriptional induction of selected examples of immune reporter genes, mainly antimicrobial peptides.7 To determine whether Eater is involved in the transcriptional regulation of other genes that are induced after bacterial binding and phagocytosis, we performed a comprehensive survey of transcriptional responses in S2 cells in which eater-expression was knocked down by RNAi. Figure 1A shows that reduced eater-expression caused a profound decrease in the phagocytosis of heat-inactivated bacteria. We monitored genome-wide transcriptional changes at 30, 90 and 180 min of bacterial phagocytosis (GEO accession GSE31564). As can be seen in Figure 1B, eater knock down was specific; only eater transcripts were significantly reduced. In response to bacterial phagocytosis, no other significant transcriptional changes could be detected when comparing control and eater knock down cells (Fig. 1B). Similar results were obtained at all time points. We found a normal induction of immune response genes, including a battery of antimicrobial peptides, in eater knock down cells at 90 and 180 min of phagocytosis, although induction appeared slightly weaker at 30 min (Fig. 1C and data not shown).
Figure 1. Knock down of eater mRNA in Drosophila S2 cells leads to reduced bacterial phagocytosis in the absence of major transcriptional changes. (A) S2 cells were analyzed for bacterial phagocytosis using a mixture of heat-inactivated, fluorescently labeled Gram-positive and Gram-negative bacteria (Staphylococcus aureus and Serratia marcescens, respectively). Error bars indicate SD (B and C) Gene expression microarray analysis of S2 cells after RNAi-mediated knock down and exposure to bacteria. Black dots indicate significant expression changes of > 3-fold (FDR ≤ 0.05; mean of three independent repeat experiments). (B) Genome-wide transcriptional changes after 90 min of exposure to bacteria. One transcript (eater) was reduced significantly by > 3-fold. Similar results were obtained at 30 and 180 min. (C) Genes significantly upregulated by > 3-fold after 90 min of exposure to bacteria were strongly enriched for immune response functions (DAVID enrichment scores of 7.92 and 7.66). Overlap between eater knock down and control samples: 93%.
These data illustrate that Eater may work differently from the upstream pattern recognition receptors in the Toll and Imd pathways, which are expressed in several immune tissues and act as sensors whose activation gets translated into a transcriptional response.1,2 Binding of microbial components to these sensors induces a variety of genes that are strongly enriched in immune functions (illustrated in Fig. 1C; DAVID enrichment scores above 7). By contrast, our data suggest that bacterial binding by Eater does not significantly alter this transcriptional response in phagocytes, since it remained largely unchanged when eater expression was strongly reduced (on average more than 10-fold; Fig. 1B and C). The main role of the phagocyte-specific pattern recognition receptor Eater may, therefore, lie in binding and internalization of microbial particles rather than in sensing and signaling to induce a transcriptional response.
Unmasking of Eater Ligands by Conserved Immune Effectors
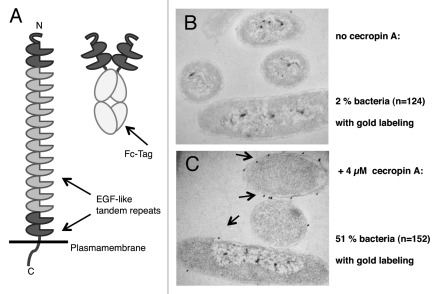
Typically, bacterial phagocytosis and phagocyte binding experiments are performed with inactivated bacteria, in order to avoid confounding effects on phagocyte physiology resulting from lytic or otherwise toxic molecules that may be secreted by live bacteria. However, the surface of live bacteria resembles more closely the molecular landscape that is encountered by phagocytic receptors in vivo.15-17 In a recent study,14 we decided to carry out Eater binding assays with live bacteria. To that end, we engineered a soluble Eater receptor, Eater-Fc, by grafting the N-terminal 199 amino acids of Eater onto an antibody Fc-tag (Fig. 2A).
Figure 2. Eater-Fc binding to live Gram-negative bacteria is dependent on prior exposure of bacteria to a cationic antibacterial peptide. (A, left) The ectodomain of the native Eater receptor consists mainly of tandem EGF-like repeats. N- and C-terminal tandem repeats (dark gray) show higher variability than internal repeats (light gray). Eater-Fc (right): the N-terminal 199 amino acids of Eater were fused to an IgG1-Fc tag to generate a soluble fusion protein. (B and C) Pre-formed complexes of biotinylated Eater-Fc with 15 nm gold–streptavidin conjugate were used to reveal binding of Eater to live E. coli in the absence (B) or presence (C) of cecropin A. Electron micrographs, magnification 60,000x.
With the help of Eater-Fc, we were able to show that Eater binds directly to live Gram-positive bacteria such as Staphylococcus aureus and Enterococcus faecalis.14 To our surprise, Eater-Fc could not react with live Gram-negative bacteria (Fig. 2B).14 It turned out that the surface of Gram-negative bacteria required disruption of the bacterial outer membrane before Eater-Fc binding could take place: heat- or ethanol-inactivation, but not inactivation with formaldehyde rendered bacteria accessible for Eater.14
How might this work in vivo? How can Eater ligands on Gram-negative bacteria become exposed in an infected host? Antimicrobial peptides are evolutionarily ancient and occur in virtually all the animal epithelia and barrier tissues. Furthermore, many antimicrobial peptides possess membrane-disrupting activities,18,19 and it was reported that cationic peptides can enhance phagocytosis of Gram-negative bacteria.20 We used live Escherichia coli as a model system to test whether pre-treating with a cationic peptide might render a Gram-negative bacterial surface accessible for Eater-Fc binding. Figure 2C illustrates that this was the case: incubation of E. coli with the evolutionarily conserved, cationic antimicrobial peptide cecropin A lead to exposure of Eater-binding determinants.14 Our data raise the possibility that some antimicrobial peptides may possess a previously unknown function.21 They may contribute to the in vivo processing of the bacterial surface in order to unmask phagocytosis receptor ligands that otherwise would be hidden under an outer membrane.
What may be the nature of the ligands that are recognized by Eater? We found that Eater-Fc was able to bind to polymeric insoluble peptidoglycan,14 a conserved cell wall component that is nearly ubiquitous in bacteria.22 However, Eater did not bind equally well to all types of polymeric peptidoglycan. For example, Eater did not bind to the actinobacterium Micrococcus luteus, neither to live nor inactivated bacterial particles or to M. luteus-derived polymeric peptidoglycan.14 M. luteus peptidoglycan differs in important ways from the peptidoglycans of many other bacteria. On the one hand, the crosslinking peptide stems and peptide bridges within the polymer are distinct.23 On the other hand, it is not associated with a class of negatively charged molecules, the teichoic acids that are abundant in the cell walls of S. aureus and E. faecalis,17 and instead contains teichuronic acids and lipomannan.17,24
Open Questions
One rather interesting possibility is that Eater might recognize a combination of different polyanionic molecules on different types of bacteria, or even on the same bacterium. In this respect, it may perhaps behave like a “dirty drug.” This concept was suggested for cationic antimicrobial peptides, which are able to bind multiple polyanionic targets with moderate affinity.16,21,25 Different experimental approaches will be required to address this possibility.
Further experiments will also be necessary to confirm a role for antimicrobial peptides in ligand processing and to firmly establish a biologically relevant cooperation between antimicrobial peptides and phagocytic receptors. It will be important to demonstrate that this mechanism operates not only on laboratory E. coli strains such as the one used in our study14 and shown in Figure 2, but also on fully virulent, smooth Gram-negative bacteria that carry full length lipopolysaccharide chains. Our attempts to use smooth type Serratia marcescens, Pseudomonas aeruginosa or E. coli in conjunction with cecropin A were unsuccessful. Yet, two membrane-disrupting treatments, heat-inactivation or ethanol-fixation, both efficiently exposed Eater ligands on these bacteria.14 This finding led us to speculate that antimicrobial peptides may not be the only molecules that participate in the process of ligand unmasking in vivo.
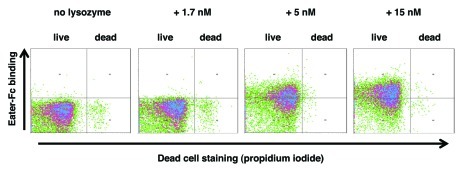
Similar to antimicrobial peptides, lysozyme is a ubiquitous and evolutionarily conserved antibacterial molecule. Lysozyme possesses a membrane-lytic activity, which is independent of its enzymatic function as a peptidoglycan-degrading muramidase.26,27 We were curious, whether lysozyme might be able to promote Eater-binding to Gram-negative bacteria. Figure 3 shows that incubation of live E. coli with lysozyme led to Eater-Fc binding in a concentration-dependent way. It is remarkable that Eater ligands were exposed on live bacteria at low nM concentrations, which are below lysozyme levels in many tissues (e.g., 9 µM in mouse lung)28 and the bactericidal concentrations of lysozyme (micromolar range).26,29 Our data suggest that multiple immune mechanisms exist in vivo that can lead to exposure of previously hidden bacterial determinants, and that these could be important for efficient phagocytosis. Furthermore, antibacterial peptides and lysozyme may act synergistically.30
Figure 3. Lysozyme promotes Eater-Fc binding to live E. coli. Flow cytometry analysis of binding of 200 nM Eater-Fc to lysozyme-treated E. coli. Lysozyme efficiently unmasked Eater ligands on live bacteria in a concentration-dependent way; a plateau was reached at 15 nM. Lysozyme concentrations up to 1.5 µM did not affect bacterial viability under these conditions (≥ 95% live E. coli). Data shown are representative of four independent runs.
Conclusions
In vivo efficiency of pathogen clearance by phagocytes matters, because bacteria can outcompete host defenses by rapid replication. This is especially important in the initial stages of infection or in tissues that are constantly exposed to an influx of external pathogens, such as the non-inflamed lung.31-33 It is conceivable, that an animal host uses multiple synergistic ways to cope with this problem. Phagocytes, antimicrobial peptides and lysozymes are well poised to take part in this process. Our data raise the possibility that these ancient immune components act not only as parallel forces, but also by directly cooperating and synergizing in the preparation and processing of ligands for immune receptors. By studying how a Drosophila phagocytosis receptor interacts with live bacteria, we may have uncovered a more general mechanism by which the innate immune system maximizes its use of a limited number of germ-line encoded receptors in order to counter a great variety of potential pathogens.
Materials and Methods
Phagocytosis and eater RNAi knock down experiments were performed as described.14 Quantitative real time PCR (qPCR) was used to determine the abundance of eater mRNA in S2 cells (30–60 copies per cell). Eater knock down was found to reduce eater mRNA levels by 5- to 10-fold (n = 8; average: 8-fold reduction) when measured by qPCR and 16- to 25-fold when measured by microarray (n = 12; average: 19-fold reduction). Gene expression data were generated using Affymetrix GeneChips (Drosophila Genome Array 2.0) and GenePattern 2.0 software.34 Gene expression data in MIAME-compliant form and details of the experimental design are available from the GEO public repository (GEO accession GSE31564). To identify “enriched biological themes,” gene lists were analyzed with DAVID (v6.7) using the “Functional Annotation Clustering” module with “medium” stringency and Affymetrix Drosophila Genome Array 2.0 as background.35,36 Eater-Fc binding assays were performed as described using E. coli DH10B/TOP10.14 Neutrophil lysozyme was purchased from Athens Research and Technology, Inc., reconstituted with water at 200 µg/ml (12 µM), aliquoted and stored at -80°C. Lysozyme was diluted in phosphate-buffered saline at pH 7.2 to the indicated concentrations and incubated with washed, live E. coli for 30 min at 25°C.
Acknowledgments
We gratefully acknowledge funding by NIH/NIAID through grant P01 AI44220. We thank Janice Lee for expert technical help and our colleagues in the Ausubel laboratory and the Department of Molecular Biology for support, in particular Julia Dewdney and Yuval Tabach for advice with microarray analysis and Read Pukkila-Worley for comments on the manuscript. EM was performed by Mary McKee, Microscopy Core, Center for Systems Biology/Program in Membrane Biology, which is partially supported by NIH Center for the Study of Inflammatory Bowel Disease Grant DK43351 and Boston Area Diabetes and Endocrinology Research Center Award DK57521.
Footnotes
Previously published online: www.landesbioscience.com/journals/fly/article/18497
References
- 1.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 2.Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila systemic immune response: Sensing and signalling during bacterial and fungal infections. Nat Rev Immunol. 2007;7:862–74. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- 3.Janeway CA., Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54:1–13. doi: 10.1101/SQB.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Janeway CA, Jr., Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 5.Areschoug T, Gordon S. Scavenger receptors: role in innate immunity and microbial pathogenesis. Cell Microbiol. 2009;11:1160–9. doi: 10.1111/j.1462-5822.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- 6.Greaves DR, Gordon S. Thematic review series: the immune system and atherogenesis. Recent insights into the biology of macrophage scavenger receptors. J Lipid Res. 2005;46:11–20. doi: 10.1194/jlr.R400011-JLR200. [DOI] [PubMed] [Google Scholar]
- 7.Kocks C, Cho JH, Nehme N, Ulvila J, Pearson AM, Meister M, et al. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell. 2005;123:335–46. doi: 10.1016/j.cell.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 8.Charroux B, Royet J. Elimination of plasmatocytes by targeted apoptosis reveals their role in multiple aspects of the Drosophila immune response. Proc Natl Acad Sci USA. 2009;106:9797–802. doi: 10.1073/pnas.0903971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Defaye A, Evans I, Crozatier M, Wood W, Lemaitre B, Leulier F. Genetic ablation of Drosophila phagocytes reveals their contribution to both development and resistance to bacterial infection. J Innate Immun. 2009;1:322–34. doi: 10.1159/000210264. [DOI] [PubMed] [Google Scholar]
- 10.Nehme NT, Quintin J, Cho JH, Lee J, Lafarge MC, Kocks C, et al. Relative roles of the cellular and humoral responses in the Drosophila host defense against three gram-positive bacterial infections. PLoS ONE. 2011;6:e14743. doi: 10.1371/journal.pone.0014743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Limmer S, Haller S, Drenkard E, Lee J, Yu S, Kocks C, et al. Pseudomonas aeruginosa RhlR is required to neutralize the cellular immune response in a Drosophila melanogaster oral infection model. Proc Natl Acad Sci USA. 2011;108:17378–83. doi: 10.1073/pnas.1114907108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurucz E, Markus R, Zsamboki J, Folkl-Medzihradszky K, Darula Z, Vilmos P, et al. Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr Biol. 2007;17:649–54. doi: 10.1016/j.cub.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 13.Somogyi K, Sipos B, Penzes Z, Kurucz E, Zsamboki J, Hultmark D, et al. Evolution of genes and repeats in the Nimrod superfamily. Mol Biol Evol. 2008;25:2337–47. doi: 10.1093/molbev/msn180. [DOI] [PubMed] [Google Scholar]
- 14.Chung YS, Kocks C. Recognition of pathogenic microbes by the Drosophila phagocytic pattern recognition receptor Eater. J Biol Chem. 2011;286:26524–32. doi: 10.1074/jbc.M110.214007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peschel A, Sahl HG. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat Rev Microbiol. 2006;4:529–36. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]
- 17.Weidenmaier C, Peschel A. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol. 2008;6:276–87. doi: 10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- 18.Hancock RE. Alterations in outer membrane permeability. Annu Rev Microbiol. 1984;38:237–64. doi: 10.1146/annurev.mi.38.100184.001321. [DOI] [PubMed] [Google Scholar]
- 19.Boman HG, Hultmark D. Cell-free immunity in insects. Annu Rev Microbiol. 1987;41:103–26. doi: 10.1146/annurev.mi.41.100187.000535. [DOI] [PubMed] [Google Scholar]
- 20.Sawyer JG, Martin NL, Hancock RE. Interaction of macrophage cationic proteins with the outer membrane of Pseudomonas aeruginosa. Infect Immun. 1988;56:693–8. doi: 10.1128/iai.56.3.693-698.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hancock RE, Scott MG. The role of antimicrobial peptides in animal defenses. Proc Natl Acad Sci USA. 2000;97:8856–61. doi: 10.1073/pnas.97.16.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salton RJ. Immunochemistry of bacterial antigens. In: Atassi MZ, van Oss CJ, Absolom DR, eds. Molecular Immunology. New York, NY: Marcel Dekker, Inc., 1984:91-116. [Google Scholar]
- 23.Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–77. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Powell DA, Duckworth M, Baddiley J. A membrane-associated lipomannan in micrococci. Biochem J. 1975;151:387–97. doi: 10.1042/bj1510387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hale JD, Hancock RE. Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert Rev Anti Infect Ther. 2007;5:951–9. doi: 10.1586/14787210.5.6.951. [DOI] [PubMed] [Google Scholar]
- 26.Ibrahim HR, Matsuzaki T, Aoki T. Genetic evidence that antibacterial activity of lysozyme is independent of its catalytic function. FEBS Lett. 2001;506:27–32. doi: 10.1016/S0014-5793(01)02872-1. [DOI] [PubMed] [Google Scholar]
- 27.Ibrahim HR, Thomas U, Pellegrini A. A helix-loop-helix peptide at the upper lip of the active site cleft of lysozyme confers potent antimicrobial activity with membrane permeabilization action. J Biol Chem. 2001;276:43767–74. doi: 10.1074/jbc.M106317200. [DOI] [PubMed] [Google Scholar]
- 28.Cappuccino JG, Reilly HC, Winston S. Elevation of lysozyme in extracts of kidneys and spleens from tumor-bearing animals. Cancer Res. 1962;22:850–6. [PubMed] [Google Scholar]
- 29.Markart P, Faust N, Graf T, Na CL, Weaver TE, Akinbi HT. Comparison of the microbicidal and muramidase activities of mouse lysozyme M and P. Biochem J. 2004;380:385–92. doi: 10.1042/BJ20031810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan H, Hancock RE. Synergistic interactions between mammalian antimicrobial defense peptides. Antimicrob Agents Chemother. 2001;45:1558–60. doi: 10.1128/AAC.45.5.1558-1560.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rabinovitch M. Professional and non-professional phagocytes: an introduction. Trends Cell Biol. 1995;5:85–7. doi: 10.1016/S0962-8924(00)88955-2. [DOI] [PubMed] [Google Scholar]
- 32.Speert DP. Phagocytosis of Pseudomonas aeruginosa by macrophages: receptor-ligand interactions. Trends Microbiol. 1993;1:217–21. doi: 10.1016/0966-842X(93)90135-E. [DOI] [PubMed] [Google Scholar]
- 33.Markart P, Korfhagen TR, Weaver TE, Akinbi HT. Mouse lysozyme M is important in pulmonary host defense against Klebsiella pneumoniae infection. Am J Respir Crit Care Med. 2004;169:454–8. doi: 10.1164/rccm.200305-669OC. [DOI] [PubMed] [Google Scholar]
- 34.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 35.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]