Abstract
The pathophysiology of type 2 diabetes mellitus (DM) is varied and complex. However, the association of DM with obesity and inactivity indicates an important, and potentially pathogenic, link between fuel and energy homeostasis and the emergence of metabolic disease. Given the central role for mitochondria in fuel utilization and energy production, disordered mitochondrial function at the cellular level can impact whole-body metabolic homeostasis. Thus, the hypothesis that defective or insufficient mitochondrial function might play a potentially pathogenic role in mediating risk of type 2 DM has emerged in recent years. Here, we summarize current literature on risk factors for diabetes pathogenesis, on the specific role(s) of mitochondria in tissues involved in its pathophysiology, and on evidence pointing to alterations in mitochondrial function in these tissues that could contribute to the development of DM. We also review literature on metabolic phenotypes of existing animal models of impaired mitochondrial function. We conclude that, whereas the association between impaired mitochondrial function and DM is strong, a causal pathogenic relationship remains uncertain. However, we hypothesize that genetically determined and/or inactivity-mediated alterations in mitochondrial oxidative activity may directly impact adaptive responses to overnutrition, causing an imbalance between oxidative activity and nutrient load. This imbalance may lead in turn to chronic accumulation of lipid oxidative metabolites that can mediate insulin resistance and secretory dysfunction. More refined experimental strategies that accurately mimic potential reductions in mitochondrial functional capacity in humans at risk for diabetes will be required to determine the potential pathogenic role in human insulin resistance and type 2 DM.
The aim of this review is to critically examine the emerging hypothesis that defective or insufficient mitochondrial function might play a pathogenic role in mediating the complex pathophysiology of type 2 diabetes mellitus (DM). We summarize (1) current evidence for the specific role(s) of mitochondria in key metabolic tissues, (2) data suggesting alterations in mitochondrial function in DM, and (3) metabolic phenotypes in animal models of impaired mitochondrial function. We conclude that while the association between impaired mitochondrial function and DM is strong, a causal pathogenic relationship remains uncertain. However, alterations in mitochondrial oxidative activity may directly impact adaptive responses to overnutrition and/or inactivity, causing an imbalance between oxidative capacity and nutrient load, and initiation of a vicious cycle of insulin resistance and secretory dysfunction.
- I. Type 2 Diabetes Pathogenesis
- A. Risk factors associated with type 2 diabetes
- II. General Overview of Mitochondrial Biology
- A. The dynamic morphology of mitochondria
- B. Mechanisms that control mitochondrial density and capacity
- III. Role of Mitochondria in Tissue-Specific Contexts
- A. Muscle
- B. Adipose tissue
- C. Liver
- D. Pancreatic β-cells
- IV. Experimental Strategies to Explore the Relationship between Mitochondrial Function and DM
- A. PGC-1 α and β overexpression
- B. PGC-1 knockout models
- C. Other mitochondrial function defects
V. Conclusions
I. Type 2 Diabetes Pathogenesis
Type 2 diabetes mellitus (DM) in the United States and around the world has reached epidemic proportions. At present, 17.9 million people in the United States have been diagnosed with diabetes, with an additional 5.7 million undiagnosed (1). Together, this encompasses 8% of the population, and thus, diabetes is a major public health issue. In addition, current data indicate that 57 million Americans suffer from prediabetes (defined as fasting blood glucose between 100 and 125 mg/dl) (1). Diabetes disproportionately affects specific ethnic populations, with risk increased 1.8-fold in African-Americans, 1.7-fold in Mexican-Americans, and 2.2-fold in Native Americans. In addition to the major health consequences to individuals, including higher risk of death, heart disease, stroke, kidney disease, blindness, amputations, neuropathy, and pregnancy-related complications, diabetes and its complications result in a total cost of $174 billion in the United States (2). By far, the largest proportion is derived from type 2 DM, which accounts for more than 90% of diabetes. Unfortunately, the incidence of diabetes has more than doubled in the past 25 yr, with 1.6 million new cases diagnosed in adults in 2007 (2) and a projected increase of 165% from 2000 to 2050 (4).
Intimately linked with the rise in diabetes prevalence is the burgeoning epidemic of obesity around the world, particularly in developed societies (5). In 2004, 17% of children in the United States between ages 2 and 19 yr were overweight, and 32% of adults over age 20 were obese (6). Both obesity and related inactivity are likely to contribute to the pathogenesis of diabetes because the incidence of diabetes can be reduced by modest weight loss and exercise (7,8,9). In light of these findings, an important public health goal should be to understand the complex pathophysiology of diabetes and to identify and target specific mechanisms to prevent DM in at-risk individuals.
A. Risk factors associated with type 2 diabetes
Multiple physiological abnormalities can be found in individuals with established type 2 DM, defined on the basis of elevations in fasting and/or postprandial glucose (2). These include insulin resistance in muscle and adipose tissue, β-cell dysfunction leading to impaired insulin secretion, increased hepatic glucose production, abnormal secretion and regulation of incretin hormones, and altered balance of central nervous system pathways controlling food intake and energy expenditure. Given this diverse constellation of abnormalities in multiple tissues and the secondary consequences of established hyperglycemia and hyperlipidemia, it is difficult to identify the primary events that lead to the development of diabetes. To address this key clinical and scientific question, it is important not only to determine abnormalities associated with established disease, but also to identify underlying metabolic characteristics preceding the onset of disease in at-risk individuals.
Risk factors for the development of and/or progression of type 2 DM include: 1) genetics (10,11,12,13,14,15,16), exemplified by the high risk of type 2 DM in particular ethnic groups (17) and the high concordance rates in monozygotic twin pairs (18); and 2) both prenatal and postnatal environmental factors, including suboptimal intrauterine environment (19,20), low birth weight (19,21), obesity (22,23), inactivity (24), gestational diabetes (25), and advancing age (26). Several longitudinal studies have indicated that insulin resistance, measured as reduced insulin-stimulated glucose disposal during the hyperinsulinemic euglycemic clamp or by iv glucose tolerance testing, is common in high-risk individuals years before the onset of type 2 DM (27,28,29). However, insulin resistance is not predictive of diabetes in individuals without a family history of diabetes, indicating that additional unidentified factors are necessary for disease progression (30).
Multiple mechanisms have recently emerged as potential causes of insulin resistance and/or diabetes progression, among them impaired mitochondrial capacity and/or function; altered insulin signaling due to cellular lipid accumulation, proinflammatory signals, and endoplasmic reticulum stress; and reduced incretin-dependent and -independent β-cell insulin secretion. In this review, we will focus on a critical assessment of the evidence linking mitochondrial function to diabetes pathogenesis, at both a cellular and whole-body level.
II. General Overview of Mitochondrial Biology
Mitochondria are double-membrane organelles that serve multiple essential cellular functions (Fig. 1) mediated by thousands of mitochondrial-specific proteins encoded by both the nuclear and mitochondrial genomes (31,32). Although mitochondria are most often recognized for their role in generating the majority of cellular ATP via oxidative phosphorylation (OXPHOS), other essential metabolic functions include the generation by the tricarboxylic acid (TCA) cycle of numerous metabolites that function in cytosolic pathways, oxidative catabolism of amino acids, ketogenesis, ornithine cycle activity (“urea cycle”), the generation of reactive oxygen species (ROS) with important signaling functions (33,34), the control of cytoplasmic calcium (35,36), and the synthesis of all cellular Fe/S clusters, protein cofactors essential for cellular functions such as protein translation and DNA repair (37). The rate-limiting first step in steroidogenesis also occurs in mitochondria, thus linking mitochondrial function to endocrine homeostasis (38,39,40,41). This multiplicity of organelle functions explains the variability in pathophysiology, severity, and age of onset of the increasing number of diseases recognized to arise from primary or secondary alterations in specific mitochondrial pathways (37,42,43,44).
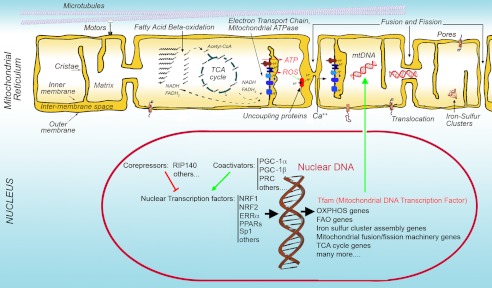
Figure 1.
Basic structural and functional features of the mitochondrial reticulum (illustrated from left to right). The mitochondrial reticulum is composed of an inner and outer membrane, between which lies the intermembrane space, and a matrix contained within the inner membrane. The surface of the inner membrane is folded into cristae. The organization and distribution of the mitochondrial reticulum is controlled by interactions with cytoskeletal elements such as microtubules. The matrix contains the enzymatic machinery for fatty acid β oxidation, which generates acetyl-CoA from acyl chains, and reducing equivalents in the form of NADH and FADH2 in the process. Acetyl-CoA fuels the TCA cycle, which also produces NADH and FADH2. These donate electrons to the ETC, leading to the generation of a proton gradient across the inner mitochondrial membrane. Dissipation of this gradient through the mitochondrial ATPase generates ATP. Delay of electron transport by the ETC results in the production of ROS, which can activate UCPs that dissipate the proton gradient without producing ATP. The electrochemical gradient also causes cytoplasmic Ca++ to enter the matrix, buffering cytoplasmic Ca++ levels and promoting TCA cycle flux. Mitochondria are also crucial in the generation of iron-sulfur clusters that form the prosthetic group of numerous proteins involved in multiple cellular pathways. The mitochondrial reticulum undergoes continuous fusion and fission reactions that involve both the inner and outer mitochondrial membranes, allowing redistribution of matrix content, such as mtDNA, within the reticulum. The proteins that compose all mitochondrial machineries are encoded both by mtDNA and by nuclear DNA. The master transcription factor operating on mtDNA is TFAM, which is encoded in the nuclear genome. The expression of mitochondrial genes in the nucleus is driven by numerous transcription factors, which are in turn controlled by specific coactivators and corepressors that respond to cellular energy demands.
A. The dynamic morphology of mitochondria
In the thin sections observed by electron microscopy and shown in most textbooks, mitochondria appear as discrete, small, bean-shaped, double-membrane organelles. However, more recent studies based on light microscopy in live cells have revealed that mitochondria exist as a reticulum that is in continuous communication through dynamic fusion and fission events, moving actively to different regions of the cell through interactions with the cytoskeleton (Fig. 2). The mitochondrial reticulum is composed of an outer and an inner membrane, between which is the intermembrane space, and a matrix limited by the inner membrane (Fig. 1). The area of the inner membrane can be greater than that of the outer membrane due to the presence of cristae, inner membrane invaginations that contain all the transmembrane proteins of the electron transport chain (ETC) as well as the mitochondrial ATPase (45,46,47). The mitochondrial matrix contains the components of the TCA cycle and of the β-oxidation pathway, which provide reduced nicotinamide adenine dinucleotide (NADH) and reduced flavin adenine dinucleotide (FADH2) to the ETC.
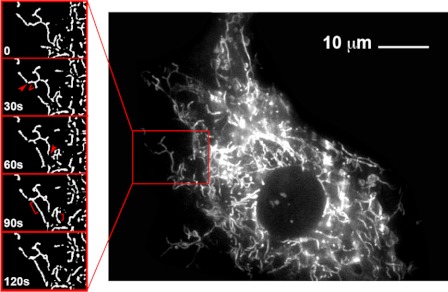
Figure 2.
Visualization of the dynamic nature of the mitochondrial reticulum in a cultured muscle cell. C2C12 mouse myoblasts were stained with MitoTracker green and imaged in culture at 37 C. Images of a segment of the cell captured at 30-sec intervals are shown on the left. The comparison between successive frames reveals scission events (arrowheads), branching events (V), and fusion events (brackets) occurring at 30-sec intervals.
The ETC is composed of four large multisubunit complexes (complexes I to IV) with more than 85 individual gene products. The ETC transports electrons from donors (NADH at complex I, FADH2 at complex II) to a final acceptor, molecular oxygen, forming H2O at complex IV. The transport of electrons is accompanied by release of large amounts of free energy, most of which is harnessed for the translocation of protons from the matrix to the intermembrane space; the remainder is dissipated as heat (Fig. 3). The energy contained in the proton electrochemical gradient generated by the ETC is then coupled to ATP production as protons flow back into the matrix through the mitochondrial ATPase. Thus, OXPHOS results from electron transport, the generation of a proton gradient, and subsequent proton flux coupled to the mitochondrial ATPase. Each of these steps can vary in efficiency; for example, the exact stoichiometry between electron flow and proton pumping, or between proton pumping and ATP synthesis varies depending on the probability of loss of electrons from the ETC before reaching complex IV and on non-ATPase-coupled proton leak through the inner mitochondrial membrane [e.g., via uncoupling proteins (UCPs)].
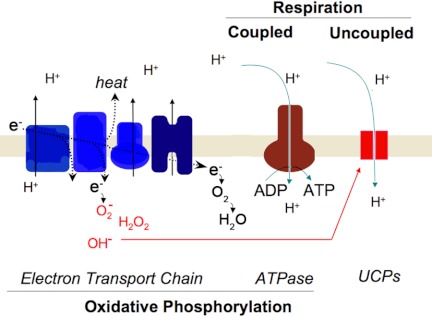
Figure 3.
Coupled and uncoupled respiration. Electrons derived from reduced donors NADH and FADH2 are transported within the ETC to molecular oxygen, producing water. The flow of electrons within the ETC is coupled to translocation of protons due to the large amount of free energy released during electron transport. The remainder of this free energy is released as heat. The proton gradient thus produced is dissipated through the mitochondrial ATPase, and the consequent decrease in free energy drives ATP synthesis. This process is known as OXPHOS, or coupled respiration. Under circumstances where NADH and FADH2 are available, but movement of electrons down the respiratory chain is slow, some of those electrons will be released from the respiratory chain and reduce molecular oxygen, forming the superoxide anion O2−, hydrogen peroxide, and the hydroxyl radical OH−. These are the main ROS formed at steady state. Accumulation of ROS activates UCPs, which dissipate the proton gradient without producing ATP, resulting in uncoupled respiration.
The high electronegative potential generated by the proton gradient also drives the rapid entry of Ca++ into the mitochondrial matrix, buffering its concentration in the cytoplasm. In the mitochondrial matrix, Ca++ can stimulate flux through the Krebs cycle by stimulating dehydrogenase activities (36). The exit of Ca++ from the matrix is driven by electroneutral exchange with Na+ or H+.
The ETC is also a potent source of ROS. Loss of electrons from the ETC can result in reduction of oxygen to form O2−, which can be dismutated to H2O2 and subsequently converted to the hydroxyl radical, OH−. These three products constitute the major ROS formed during respiration. As the name implies, these species are highly reactive, and acute, very high elevations, or more chronic elevations can be extremely damaging to the cell. ROS generation is more likely to occur when the proton gradient is large and electron carriers are highly reduced, e.g., when ADP is rate-limiting for ATP production or when availability of O2 is limiting. Uncoupling proteins are considered to be natural regulators of this process, responding to and controlling ROS production by mitigating the formation of a large proton gradient.
The mitochondrial matrix also contains the circular mitochondrial DNA (mtDNA) molecule, which encodes for 37 genes (13 of which are subunits of the ETC). Translation of these proteins occurs within the mitochondrial matrix, utilizing mtDNA-encoded rRNA and tRNA.
Mitochondrial fission and fusion allow the transcriptional products of mtDNA, as well as multiple metabolites generated in the mitochondrial matrix, to be shared within the entire mitochondrial reticulum. Although the molecular machinery of mitochondrial fusion and fission has been elucidated (48), it has only recently been established that mitochondrial fusion and fission also contribute multiple other mitochondrial functions, including the control of cellular calcium handling, ROS production, and energetic output (49,50,51). Moreover, human diseases arising from mutations in conserved elements of the mitochondrial fusion machinery have been identified, such as Charcot-Marie-Tooth type 2A caused by mutations in mitofusin 2, and autosomal dominant optic atrophy, caused by mutations in optic atrophy 1 (49). A role for mitochondrial fusion machinery in metabolic control has also been suggested by the findings that mitofusin 2 levels are controlled during muscle development and are reduced in both obesity and type 2 DM in parallel with insulin resistance (52).
B. Mechanisms that control mitochondrial density and capacity
The term mitochondrial biogenesis is often used to describe the generation of more mitochondria in response to increased energy demands, or the multiplication of mitochondria necessary for cell growth and division. However, the copy number of specific mitochondrial proteins and the functional capacity of each distinct mitochondrial pathway may be very variable between different tissues and between different physiological conditions. Thus, the term mitochondrial biogenesis can be ambiguous because multiple parameters, including mtDNA copy number, mitochondrial density, levels of specific mitochondrial proteins, and mitochondrial functional output may vary independently of each other. For example, the proliferation of mitochondria occurring to sustain hyperplastic growth is probably very different from that occurring to support hypertrophic growth in any given tissue, and the regulatory mechanisms controlling these adaptive changes are likely to be distinct.
1. Transcriptional control mechanisms
Although we know very little about specific mechanisms that control different modalities of mitochondrial biogenesis, it is clear that these mechanisms require coordination between the nuclear and mitochondrial genomes. Transcription of the mitochondrial genome is under the control of a single transcription factor, Tfam, which is encoded by the nuclear genome. In turn, Tfam expression is regulated by the transcription factors NRF (nuclear respiratory factor)-1 and NRF-2, which specifically activate numerous nuclear-encoded genes involved in mitochondrial respiration (53,54). Thus, through NRF-stimulated expression of Tfam, the transcription of the mitochondrial genome is stimulated in coordination with that of nuclear-encoded mitochondrial genes. The expression of many other mitochondrial genes is controlled by additional nuclear transcription factors, including peroxisome proliferator-activated receptor (PPAR) α, PPARδ, estrogen-related receptor (ERR) α/γ, and Sp1, which can induce expression of mitochondrial genes in a tissue-dependent and physiological context-dependent manner (55).
A high level of transcriptional coordination is required to ensure coupling of mitochondrial activity to other metabolic activities within the cell and to mediate appropriate parallel changes in all components of multiprotein complexes. This coordination is accomplished through the action of transcriptional coactivators and corepressors. The best studied coactivators of mitochondrial gene transcription are members of the PPARγ coactivator (PGC) family, including PGC-1α, PGC-1β (56,57), and PPRC, a related serum-responsive coactivator (58). These respond to cellular energy-requiring conditions such as cell growth, hypoxia, glucose deprivation, and exercise (55) to activate transcription factors promoting mitochondrial remodeling and/or biogenesis, thus restoring cellular energetics. For example, PCG-1α is highly expressed in muscle, liver, and brown fat, and expression is further increased in these tissues in response to exercise, fasting, and cold exposure, respectively. Although PGC-1α and -β do not appear to be required for mitochondrial biogenesis during development (59), they are necessary for the expression of the full complement of proteins of mitochondrial OXPHOS and fatty acid β-oxidation pathways in muscle and brown adipose tissue (59,60,61,62,63,64,65,66,67,68,69). Moreover, PGC-1α and PGC-1β are crucial for the rapid bursts in mitochondrial proliferation that accompany perinatal heart and brown adipose tissue development (59). These data support the concept that mitochondrial adaptation to specific energy needs is mediated by PGC-1α and PGC-1β; by contrast, mitochondrial expansion during cell proliferation is more likely to depend on serum-responsive coactivators such as PPRC (70).
The role of corepressors in the transcriptional control of energy metabolism genes is less extensively studied. However, evidence in cultured cells and in mouse models points to a critical role of the corepressor RIP140 in controlling important aspects of mitochondrial energy metabolism in both adipose tissue and muscle (71,72,73,74,75). RIP140 suppresses UCP1 through interaction with specific enhancer elements and also suppresses expression of genes involved in β-oxidation and respiratory chain assembly. RIP140 also interacts directly with many of the transcription factors coactivated by PGC-1α (76). The mechanisms that control the balance between PGC-1 coactivators and RIP140 and other corepressors are not clear but are likely to represent key regulatory mechanisms of energetic adaptation.
2. Posttranscriptional control mechanisms
The expansion of the mitochondrial reticulum requires not only the expression of genes encoding mitochondrial proteins but also the import of these into the mitochondrial space (77,78,79,80) and the coordinated expansion of mitochondrial membranes. Mitochondrial inner and outer membranes have distinct lipid compositions that differ from that of other membrane-bound organelles and from the plasma membrane. Specific features of mitochondrial membranes are their relative lack of cholesterol and the high content of cardiolipin, which is unique to mitochondrial membranes and essential for the proper assembly and function of the respiratory chain (81,82,83). Mitochondrial lipids are most likely synthesized in the endoplasmic reticulum (the primary site of lipid biosynthesis in eukaryotic cells) and transferred to mitochondria via as-yet unidentified mechanisms. However, recent work has identified mechanisms regulating the synthesis of cardiolipin and phosphatidylethanolamine in mitochondria inner membranes via the action of mitochondrial prohibitins (84). In addition, cardiolipin synthesis requires the mitochondrial translocator assembly and maintenance protein Tam41, revealing a mechanism for the coordination of protein import and mitochondrial membrane lipid assembly (85).
The area and composition of the mitochondrial inner and outer membranes must be tailored to accommodate the specific components of mitochondria from different cells and tissues, which are each likely to have optimal lipid composition and density. This essential requirement for specific lipid composition is underscored by the morphological and functional alterations in mitochondria seen in Barth syndrome, a disorder arising from mutations in a lipid acyltransferase, tafazzin (41,86). The resulting alterations in cardiolipin structure cause profound changes in the assembly and distribution of respiratory chain components within mitochondrial cristae (84,87,88). Interestingly, lymphoblasts from patients with Barth syndrome can produce ATP at normal levels but display an expanded mitochondrial reticulum (89). These observations underscore the existence of mechanisms that can compensate in part for specific mitochondrial deficiencies.
Given the complex and dynamic structure of mitochondria and the diversity and physiological importance of their multiple functions, assessing the role of mitochondria in human pathology requires a comprehensive characterization not only of mitochondrial structure and abundance, but also of the pathways that compensate for suboptimal mitochondrial capacity and functional output—which may then modify disease severity and progression. In the following sections, we will critically analyze the findings that have suggested a role for mitochondrial function in the establishment of diabetes risk and the gaps in our knowledge that must be filled to determine the merits of this hypothesis.
III. Role of Mitochondria in Tissue-Specific Contexts
A. Muscle
1. Role of mitochondria in muscle
Mitochondria are particularly important for skeletal muscle function, given the high oxidative demands imposed on this tissue by intermittent contraction. Mitochondria play a critical role in ensuring adequate levels of ATP needed for contraction by the muscle sarcomere. This high-level requirement for ATP by sarcomeres has likely contributed to the distinct subsarcolemmal and sarcomere-associated populations of mitochondria in muscle. Moreover, muscle cells must maintain metabolic flexibility, defined as the ability to rapidly modulate substrate oxidation as a function of ambient hormonal and energetic conditions. For example, healthy muscle tissue predominantly oxidizes lipid in the fasting state, as evidenced by low respiratory quotient (RQ), with subsequent transition to carbohydrate oxidation (increased RQ) during the fed state. Availability of fuels, particularly lipids, and capacity to oxidize them within mitochondria are also critical for sustained exercise. Thus, mitochondrial functional capacity is likely to directly affect muscle metabolic function and, because of its large contribution to total body mass, to have a significant impact on whole-body metabolism. This possibility is supported by the findings of increased mitochondrial content in skeletal muscle in an individual with hypermetabolism and resistance to weight gain (Luft syndrome) (90).
2. Potential mechanisms by which impaired muscle mitochondrial oxidative function could result in insulin resistance
Skeletal muscle is the largest insulin-sensitive organ in humans, accounting for more than 80% of insulin-stimulated glucose disposal. Thus, insulin resistance in this tissue has a major impact on whole-body glucose homeostasis. Indeed, multiple metabolic defects have been observed in muscle from insulin-resistant but normoglycemic subjects at high risk for diabetes development, including: 1) reduced insulin-stimulated glycogen synthesis (27,91,92); 2) alterations in insulin signal transduction (93); and 3) increased muscle lipid accumulation (94). Although it remains unclear whether any of these defects play a causal role in insulin resistance, intramyocellular lipid excess strongly correlates with the severity of insulin resistance, even after correction for the degree of obesity (94), and has been observed in muscles of multiple fiber types (95). Moreover, lipid excess has been linked experimentally to induction of insulin resistance (96) and alterations in insulin signal transduction (97,98,99).
Thus, one possible mechanism by which impaired mitochondrial function might contribute to insulin resistance is via altered metabolism of fatty acids. Increased tissue lipid load, as with obesity, and/or sustained inactivity, may lead to the accumulation of fatty acyl coenzyme A (CoA), diacylglycerols, ceramides, products of incomplete oxidation, and ROS, all of which have been linked experimentally to reduced insulin signaling and action (96,97,98,99,100,101,102). Additional mechanisms potentially linking impaired mitochondrial oxidative function to insulin resistance include: 1) reduced ATP synthesis for energy-requiring functions such as insulin-stimulated glucose uptake; 2) abnormalities in calcium homeostasis (necessary for exercise-induced glucose uptake) (103,104,105); and 3) reduced ATP production during exercise (106), potentially contributing to reduced aerobic capacity, muscle fatigue, and decreased voluntary exercise over time—further feeding a vicious cycle of inactivity-fueled insulin resistance.
3. Evidence for reduced muscle mitochondrial oxidative function in DM
An important early clue suggesting that muscle mitochondrial oxidative dysfunction may be associated with insulin resistance in humans was the series of observations by Simoneau and Kelley that obesity is associated with reductions in citrate synthase, malate dehydrogenase, carnitine palmitoylotransferase 1 (CPT1), and cytochrome oxidase (COX) activity in the fasting state (107,108) and with parallel increases in activity of the glycolytic enzymes hexokinase and phosphofructokinase (109). Moreover, oxidative activity (e.g., citrate synthase, acyl CoA dehydrogenase) is a robust correlate of insulin sensitivity, even better than either im triglycerides or long-chain fatty acyl CoA (110). Furthermore, leg balance studies demonstrated that obesity-linked insulin resistance and diabetes are both associated with reduced fasting lipid oxidation, as indicated by higher RQ, as well as inability to suppress lipid oxidation and switch to carbohydrate oxidation in response to meals/insulin stimulation (111), a state termed “metabolic inflexibility” (112). Impaired flexibility also correlates with intramyocellular accumulation of lipids (107), and 24-h RQ can predict subsequent weight gain (110,113). Together, these data suggest that an intrinsic defect in multiple components of oxidative metabolism, or altered regulation, may contribute to the development of both obesity and insulin resistance.
The diminished capacity for appropriate regulation of oxidative metabolism observed in the above studies could be linked to reduced mitochondrial function due to: 1) abnormal mitochondrial density and/or in vivo function; and/or 2) intrinsic defects in oxidative metabolism of lipids or other substrates. Multiple studies suggest that human insulin resistance is indeed accompanied by impaired in vivo mitochondrial oxidative function—in turn linked, at least in part, to reduced mitochondrial density. Ritov et al. (114) demonstrated that the enzymatic activity of OXPHOS complex I, as assessed by the activity of rotenone-sensitive NADH:O2 oxidoreductase, was reduced by about 40% in skeletal muscle biopsy samples from individuals with type 2 DM and by 20% in obese individuals. Similarly, Boushel et al. (115) found modest reductions in ADP and succinate-stimulated oxygen consumption in permeabilized muscle fibers from obese individuals with type 2 DM. In each of these studies, differences in oxidative capacity did not remain after normalization for mitochondrial mass by citrate synthase activity or mtDNA content, respectively, suggesting that reduced mitochondrial mass might be a major contributor. This possibility is consistent with electron microscopy demonstrating diminished mitochondrial size in obesity and diabetes (116), particularly in subsarcolemmal fractions (114). Interestingly, this fraction is also characterized by even greater reductions in OXPHOS activity (114).
Nuclear magnetic resonance (NMR) spectroscopy has also been used to assess mitochondrial function in vivo, with studies finding similar reductions in oxidative function in both insulin resistance and type 2 DM. For example, rates of mitochondrial OXPHOS in offspring of type 2 diabetic subjects, as assessed by 31P spectroscopy, are reduced by 30% in the fasting state (117), and TCA cycle flux, modeled using rates of 4-13C-glutamate enrichment during infusion of 13C-acetate, is reduced by 30% (118). The magnitude of these changes is strikingly similar to the 38% lower muscle mitochondrial density, assessed by electron microscopy, in this same population—again suggesting that decreased mitochondrial density might be an important factor in reduced oxidative capacity in individuals with a family history of diabetes.
Alterations in intrinsic function of mitochondria have also been identified in isolated mitochondria from humans with insulin resistance and DM. Mogensen et al. (119) observed decreases in maximal ADP-stimulated respiration (state 3, malate and pyruvate as substrates) in mitochondria isolated from obese subjects with DM as compared with obesity alone; these differences persisted even after normalization to citrate synthase activity. Thus, these data suggest that in addition to decreased mitochondrial density, there is an additional intrinsic defect(s) in TCA, OXPHOS, membrane potential, or adenine nucleotide transporters in mitochondria of individuals with established diabetes.
Such underlying functional defects may be subtle at baseline but may be unmasked during acute energetic stress. For example, short-term exercise normally increases ATP synthesis rates. However, this adaptive response is completely mitigated/abolished in nonobese first-degree relatives of type 2 diabetics—despite normal basal ATP synthesis rates (106). Similarly, insulin-stimulated ATP synthesis is reduced by more than 90% in nonobese first-degree relatives of type 2 diabetics (120), more than would be expected from the 30% decrease in mitochondrial density and oxidative function observed in the same population. Because these short-term experimental protocols (several hours in duration at most) would not be expected to alter mitochondrial density, DNA content, or number, these data strongly suggest that inability to appropriately modulate oxidative function in response to the prevailing energetic environment is a signature of insulin resistance and diabetes risk.
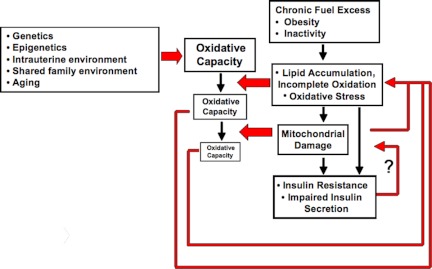
Analysis of global gene expression patterns has also demonstrated a 20–30% reduction in mRNA expression levels for multiple nuclear-encoded genes of the OXPHOS pathway in humans with type 2 DM (121,122,123). Importantly, similar reductions in OXPHOS gene expression have been observed in some, but not all, populations of insulin-resistant, but completely normoglycemic, individuals (122,124).a These differences may reflect population-specific differences in obesity, physical fitness, or ethnicity. Interestingly, a recent study of Asian Indian subjects found no correlation between changes in OXPHOS gene expression and insulin resistance (125). In these individuals, expression of OXPHOS and TCA cycle genes, mtDNA content, and ATP production rates were actually higher in both nondiabetic and diabetic individuals compared with Northern European controls, despite lower overall insulin sensitivity. However, circulating triglycerides were significantly elevated in both nondiabetic and diabetic individuals of Asian Indian origin (125). These results also raise the question of whether levels of OXPHOS gene expression and function must be considered relative to the oxidative fuel load in an individual. For example, high OXPHOS expression in the population mentioned above may still be inadequate for appropriate and complete oxidation of a chronic high load of circulating lipids, whereas lower OXPHOS levels may be sufficient under conditions of a low circulating lipid load (see Fig. 5).
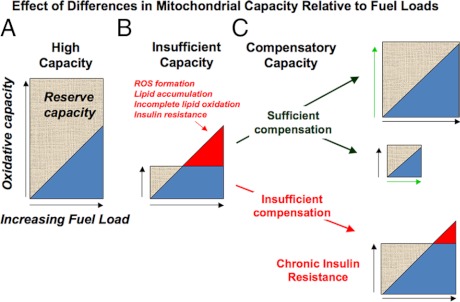
Figure 5.
Dynamic relationship between oxidative activity and fuel load leading to development of disease. In this diagram, oxidative capacity refers to the ability to generate energy in response to varying energy requirements, and balance is indicated by oxidative capacity equaling or exceeding fuel loads. A, High oxidative activity can ensue from high-energy requirements (chronic exercise, high metabolic rate). Individuals with a high oxidative capacity will have high tolerance to large fuel loads. B, Low oxidative activity can ensue from a lack of energy requirement (sedentary lifestyle) or inability to generate energy (mitochondrial myopathies, intrauterine exposures, genetics) as seen in the failure of ATP synthesis in DM patients in response to exercise. Individuals with low oxidative capacity are intolerant to moderate-high fuel loads that lead to ROS generation, lipid accumulation, incomplete oxidation, and acute insulin resistance. C, Insufficient oxidative capacity can be resolved by compensatory mechanisms that increase oxidative activity (e.g., exercise, top right) decrease fuel load (weight loss, middle right), or resolve maladaptive patterns of oxidative stress. Insufficient compensation results in chronic insulin resistance (bottom right).
Such data also highlight the importance of considering additional aspects of oxidative mitochondrial function beyond OXPHOS expression or capacity. For example, primary myotubes isolated from obese humans with type 2 DM display reduced basal lipid oxidation and insulin-stimulated glucose oxidation with no differences in OXPHOS gene expression (126). Thus, defects in lipid oxidation in DM can be significant contributors to disordered oxidative metabolism even in the absence of detectable alterations in OXPHOS gene expression or function.
4. Factors affecting OXPHOS gene expression in muscle
Several conditions associated with susceptibility to insulin resistance, including obesity, lipid accumulation, and aging, have all been associated with reduced nuclear-encoded OXPHOS gene expression. Reduced OXPHOS gene expression has been observed in response to genetic and nutritional obesity (127), short-term high-fat feeding (even in humans) (128), lipid infusion (129), and lipid loading of myotubes (127). However, these responses are not observed in all studies of high-fat feeding; in fact, some studies demonstrate that high-fat feeding is associated with increased numbers of mitochondrial protein and DNA content, potentially mediated by chronic fatty acid activation of PPAR nuclear receptors (130,131,132). Similarly, relatively short-term reductions in serum fatty acids and intracellular fatty acyl CoA levels mediated by acipimox treatment in healthy humans are associated with reduced expression of nuclear-encoded mitochondrial oxidative genes—in parallel with enhanced insulin sensitivity (294). Together, these seemingly disparate data suggest that genetic background (127), age at dietary intervention, specific dietary lipid composition, and duration of diet may be important variables to consider when analyzing the interaction between OXPHOS gene expression and diet. Moreover, alterations in OXPHOS gene expression may be a secondary response to an underlying primary defect in oxidative metabolism, reflecting attempts to compensate for reductions in mitochondrial capacity (increased OXPHOS expression), or the deleterious effects of lipid overload and accumulation on transcription of OXPHOS genes (decreased OXPHOS expression), or a mixture of both. Additionally, because OXPHOS gene expression is coordinately regulated, patterns of differential OXPHOS expression may be more readily detectable in disease states, yet not necessarily mirror other aspects of mitochondrial oxidative capacity.
Reduced physical fitness is associated with reduced muscle OXPHOS gene expression. In humans, maximal oxygen uptake is robustly correlated with OXPHOS gene expression (133). Similarly, in rats bred for low aerobic capacity over multiple generations, expression of several OXPHOS genes is markedly reduced, even in the absence of obesity (134). Conversely, OXPHOS expression can be increased with exercise training (133,135), a potent insulin sensitizer.
Genetic and epigenetic modifications may also contribute to reduced expression of OXPHOS genes in type 2 DM. For example, expression of COX7A1, a complex IV gene down-regulated in type 2 DM, is heritable (50–72% heritability, as assessed by analysis in monozygotic and dizygotic twins), indicating a strong genetic or shared familial environmental contribution (136). Similar patterns are observed for the complex I gene NDUFB6 (137) and the ATP synthase component ATP5O (138). Indeed, expression of nuclear-encoded OXPHOS genes is significantly more concordant between monozygotic twins than expected and is the top-ranking gene set for concordance in pathway analysis of global gene expression. Mediators of mitochondrial biogenesis, including ERRα, may contribute to the strong heritability of OXPHOS components.b Interestingly, epigenetic mechanisms may also contribute to these patterns because reduced expression parallels increased DNA methylation of both the COX7A1 promoter (136) and NDUFB6 (137,139).
Aging is also linked to impaired oxidative function (140) in parallel with reductions in OXPHOS gene expression, including COX7A1, NDUFB6, and ATP50 (136,137,138). It is unclear at this time whether this is a direct effect of aging per se or related to reduced physical fitness, increased tissue lipid accumulation, or other factors accompanying typical patterns of aging. Genetic polymorphisms may also influence age-dependent reductions in expression (137).
A key question is whether the changes in OXPHOS gene expression observed in type 2 DM are secondary features of the diabetes metabolic environment such as hyperglycemia or insulin resistance. Reductions in OXPHOS gene expression in patients with established type 2 DM can be partially normalized by insulin treatment (123). Expression of multiple OXPHOS genes is also markedly reduced in mice made insulin deficient by treatment with the β-cell toxin streptozotocin, and can be normalized by insulin (141). Similarly, withdrawal of insulin in individuals with type 1 diabetes reduces muscle OXPHOS gene expression and ATP production rates (142). Short-term experimental induction of acute hyperglycemia in humans does not fully mirror this pattern of gene expression (143), suggesting that the response to insulin deficiency is not completely due to resultant hyperglycemia. Moreover, experimental insulin therapy does not modulate mitochondrial respiration (144), so mechanisms linking insulin action with OXPHOS gene expression remain unclear.
Changes in the levels of OXPHOS and other oxidative genes must occur in response to cellular energetic and metabolic needs, and in a coordinated manner that ensures the stoichiometric assembly of the products of distinct genes into functional complexes. As in other tissues, the coordination of OXPHOS gene expression in muscle is mediated in part by the action of coactivators and corepressors. PGC-1α has been recognized as an important coactivator in skeletal muscle, contributing to fiber type determination, glucose uptake, and oxidative capacity (see Section IV. A). Moreover, alterations in muscle PGC-1α and -β mRNA expression are observed in humans with insulin resistance—being reduced by nearly 50% in muscle from individuals with diabetes (122,145) and in some populations of normoglycemic insulin-resistant humans (121,124,137). In turn, PGC-1α expression may also be reduced as a consequence of promoter methylation (146) or caused by insulin itself (145), obesity (126), and sustained lipid exposure (126). For example, saturated fatty acids reduce PGC-1α promoter transcriptional activity and expression in cultured myotubes, in parallel with reduced OXPHOS expression and O2 consumption (127). PGC-1 activity can also be modulated at the level of translation and by posttranscriptional changes, including inhibitory GCN5-mediated acetylation (147) and stimulatory sirtuin 1 mediated deacetylation (148). These multiple modes of PGC-1α regulation are likely to have evolved from the need to adapt mitochondrial energy metabolism in response to increasingly diverse inputs.
In summary, insulin resistance has been associated with alterations in skeletal muscle mitochondrial oxidative function and its transcriptional regulatory pathways. However, several lines of evidence suggest that this may not be a causal relationship in all situations. First, oxidative dysfunction is not observed in all insulin resistant individuals (125). Second, oxidative activity is determined by the need to generate energy to meet cellular demands, e.g., contraction and ion transport; thus oxidative capacity is not likely to be limiting in the resting state in muscle (3). Rather, alterations in relative utilization of substrates, an imbalance between fuel load and cellular energy requirements, and/or differential thresholds for generation of or resolution of oxidative stress in this setting may contribute to differential susceptibility to insulin resistance in muscle. These concepts are examined more fully in the conclusion (Section V).
B. Adipose tissue
1. Roles of mitochondria in adipose tissue
The role of adipose tissue mitochondria is most apparent in brown adipose tissue, where flux through the ETC generates heat in the process of thermogenesis, a potentially important mechanism regulating systemic metabolism even in adult humans (149,150,151,152). In this tissue, electron transport is greatly accelerated due to tissue-specific expression of the mitochondrial UCP1. UCP1 hinders the establishment of, or dissipates, a proton gradient of sufficient magnitude to sustain the synthetic activity of the mitochondrial ATPase (150,153,154,155), thus driving continuous accelerated electron transport. UCP1-mediated uncoupling alone, however, cannot fully account for the large thermogenic capacity of brown adipocytes in the absence of mechanisms that ensure continuous substrate delivery to the ETC. Thus, brown adipocyte mitochondria also contain high levels of CPT1b, which is critical for the entry of fatty acids into the mitochondria for β-oxidation. β-Oxidation, in turn, generates large amounts of reducing equivalents for the ETC.
White adipocytes have been described to contain low levels of mitochondria, which is indeed the case when compared with brown adipocytes or muscle. However, mitochondrial density increases dramatically, and mitochondrial remodeling occurs during white adipocyte differentiation (156,157,158), suggesting that mitochondrial functions are required to support the multiple biological roles of mature white adipocytes. Interestingly, a recent compendium of mitochondrial proteins from 14 different mouse tissues indicates that white adipocyte mitochondria contain a more diverse protein repertoire than mitochondria from heart, skeletal muscle, or brain (31). Thus, white adipocyte mitochondria appear to be equipped for a broader array of functions compared with mitochondria in tissues that must sustain rapid bursts of energy-requiring processes. Among the mitochondrial functions that may be relevant for white adipose tissue function are the anaplerotic generation of metabolic intermediates for fatty acid synthesis and esterification (159), the maintenance of a robust pathway for the folding and secretion of high abundance circulating proteins such as adiponectin (160), and interactions between mitochondrial function and components of the insulin signaling pathway (161).
2. Potential mechanisms by which impaired adipose tissue mitochondrial oxidative capacity could result in insulin resistance
The large capacity of brown adipose tissue mitochondria to oxidize fatty acids results in a measurable impact on whole-body metabolism; increased brown adipose tissue abundance correlates negatively with fuel storage and weight gain in rodents, and vice versa (162). The role of brown adipose tissue in human metabolism has typically been thought to be minor. However, recent work has led to reconsideration of this notion, noting that humans possess adipose tissue depots that are cold-sensitive and hypermetabolic, as assessed by their very high uptake of labeled glucose (152,163). Such depots appear to be less active as a function of aging and/or obesity (151,164,165,166,167). Thus, impaired mitochondrial capacity in brown adipose tissue might be functionally linked to impaired thermogenesis and energy expenditure, and thus increased susceptibility to obesity-linked insulin resistance.
The relevance of white adipocyte mitochondria to whole-body metabolism and metabolic disease may depend on the extent to which mitochondrial respiratory capacity and/or the total mass of white adipose tissue would be sufficient to impact circulating free fatty acid levels. White adipocytes display a high degree of plasticity (168), and regional differences in metabolic activity can be linked to varying mitochondria densities (169). Higher mitochondrial density and even UCP1 can be induced in response to pharmacological or genetic alterations of white adipocytes (170,171,172,173,174,175,176,177), suggesting that white adipose tissue could potentially be induced to acquire more oxidative metabolic phenotypes, promoting increased fuel consumption and thus energy expenditure. Whether respiratory chain uncoupling mediated through the induction of UCP1 in white adipocytes alone could reduce free fatty acid release, or whether an additional increase in mitochondrial oxidative capacity would be required, is debated (178,179,180,181,182).
Gain-of-function studies in mice where ectopic expression of UCPs mitigate diet-induced obesity support the notion that uncoupling could be sufficient (183,184). However, UCP1 expression in adipocytes driven by the aP2 promoter failed to significantly raise resting metabolic rate (185). Moreover, in cultured adipocytes, ectopic expression of UCP1 impairs fatty acid synthesis (186,187). These results suggest that, in the absence of mechanisms to ensure continuously elevated fuel oxidation, such as those present in brown fat, uncoupling of white adipose tissue mitochondria may decrease ATP levels and impair anabolic flux (183).
In addition to effects on fuel utilization, decreased mitochondrial capacity in adipocytes may also alter adipocyte insulin sensitivity and/or function due to the high energetic requirements for fatty acid storage, adipokine secretion (160), insulin signaling (161), and glucose uptake. Interestingly, in cultured adipocytes, impairment of respiratory chain function through depletion of Tfam during adipocyte differentiation results in impaired insulin-stimulated glucose transport (161); data in animal models are necessary to determine the physiological relevance of this finding.
3. Evidence for reduced adipose tissue mitochondrial capacity in DM
White adipocyte mitochondrial content is decreased in both rodent and human obesity (177,188,189,190,191) and correlates with insulin resistance that accompanies obesity. In humans, white adipocyte mtDNA copy number is inversely correlated with age and BMI and directly correlated with basal and insulin-induced lipogenesis (192). Thus, reduced mtDNA content could reduce adipocyte capacity for lipid storage, promoting ectopic lipid accumulation in peripheral tissues such as muscle and liver. In parallel, expression of nuclear-encoded OXPHOS genes is down-regulated in visceral adipose tissue of humans with type 2 DM (193). Administration of thiazolidinediones induces changes in mitochondrial content and remodeling in white adipocytes concomitantly with an improvement in insulin sensitivity (170,173,177,190,194,195,196,197,198). Mitochondrial levels in white adipocytes are also increased in response to adrenergic stimulation, β-3 agonists, and CB1 blockade in mice (195,199,200), again in parallel with enhanced insulin sensitivity.
Whether changes in mitochondrial density are a cause or consequence of changes in insulin sensitivity is unclear. However, some evidence suggests that lack of insulin signaling does not reduce mitochondrial capacity in adipose tissue. For example, mice with adipose tissue-specific ablation of the insulin receptor (FIRKO mice) display high levels of mitochondrial genes involved in fatty acid oxidation and OXPHOS over the lifespan of the animals (201). Thus, mechanisms that induce and maintain active mitochondria in adipocytes can bypass defects in insulin signaling, and indeed, insulin signaling may repress mitochondrial gene expression and/or function.
4. Factors affecting mitochondrial OXPHOS expression and function in adipose tissue
The genetic program leading to brown adipose tissue development, and potentially to the high abundance of mitochondria, is initiated by the zinc-finger protein PRDM16 (202,203,204). Current reports support the hypothesis that brown adipocytes and myocytes share a common cellular lineage, potentially explaining their similarity with regard to containing mitochondria specialized in fuel oxidation. In addition, the transcriptional coactivators PGC-1α and -1β (56) play a critical role in the expansion of the mitochondrial reticulum and in the induction of UCP1 and the brown adipose tissue thermogenic program during the perinatal period (59).
Adipocyte mitochondrial density and OXPHOS activity can be regulated in response to factors that affect lipid metabolism. For example, Toh et al. (176) and Nishino, et al. (205) find that mice deficient in Fsp27, a lipid droplet protein that promotes lipid storage in white and brown adipocytes, have increased whole-body energy expenditure, resistance to diet-induced obesity, and enhanced insulin sensitivity. This apparent paradoxical result (high insulin sensitivity despite deficiency in lipid storage), appears to be due to the increased mitochondrial density and activity in white adipocytes, which are brown-like in their increased capacity to oxidize large quantities of fatty acids. Nitric oxide production by the endothelial nitric oxide synthase has also been linked to enhanced adipose tissue mitochondrial biogenesis and prevention of high-fat diet-induced obesity (200). Conversely, both genetic and diet-induced obesity result in decreased mitochondrial density and OXPHOS activity in adipose tissue (127,177,189,190,191), potentially contributing to adipose tissue dysfunction and exacerbation of insulin resistance. The mechanisms whereby obesity results in a reduction in adipose mitochondrial density are not known but could be mediated by decreased expression of PGC-1α, as observed in obese humans (206).
C. Liver
The liver plays a central, unique role in carbohydrate, protein, and fat metabolism. It is critical for maintaining glucose homeostasis (1) during fuel availability, via storage of glucose as glycogen or conversion to lipid for export and storage in adipose tissue, and (2) in the fasting state, via catabolism of glycogen, synthesis of glucose from noncarbohydrate sources such as amino acids (gluconeogenesis), and ketogenesis. In turn, these responses are regulated by the key hormones insulin and glucagon, which modulate signaling pathways and gene expression, leading to inhibition or stimulation of glucose production, respectively.
Recent human data have highlighted the importance of disordered hepatic metabolism, including inappropriately increased hepatic glucose production, hyperlipidemia, and lipid accumulation, in both obesity and type 2 DM (207). Similarly, rodent data also support an important role for the liver in diabetes pathogenesis. For example, liver-specific insulin receptor knockout (LIRKO) mice develop insulin resistance, glucose intolerance, impaired insulin suppression of hepatic glucose production, and altered patterns of hepatic gene expression (208). Interestingly, these mice are also dyslipidemic and susceptible to atherosclerosis (209).
1. Role of mitochondria in liver
Given the diverse array of unique metabolic functions centered in the liver, it is not surprising that ultrastructure and function of hepatic mitochondria are distinct from that of muscle. Electron microscopy demonstrates that mitochondrial area is 44% lower in liver than in heart (210) with smaller size, fewer cristae, and lower matrix density. Protein expression of multiple OXPHOS components and Tfam (expressed per milligram of protein) and citrate synthase activity are also lower in liver (e.g., 7% that of cardiac muscle) (211). Similarly, patterns of gene expression are distinct in liver (32). Functionally, isolated hepatic mitochondria have relative reductions in OXPHOS proteins, respiratory chain cytochromes, and maximal activity of complexes III and IV (211). Despite lower OXPHOS capacity, state 3 respiration and respiratory control ratio are equivalent in liver and muscle, indicating differences in relative substrate concentrations and lower “excess capacity” in liver. Recent application of 31P NMR to the liver in humans demonstrates that rates of ATP synthesis are 3-fold higher in liver than in muscle (212). By contrast, the content of mtDNA, expressed either per gram of tissue or per mitochondrion, is actually higher in liver than in other tissues. Together, these data again emphasize differences in protocols assessing mitochondrial abundance, capacity, and function and highlight tissue diversity of mitochondrial structure and function, which may contribute to tissue-specific disease susceptibility.
2. Potential mechanisms by which impaired hepatic mitochondrial function could influence hepatic insulin sensitivity
Impairments in mitochondrial number and/or oxidative function could potentially affect multiple cellular functions within hepatocytes, both directly (e.g., reduced ATP generation, alterations in oxidative stress, reduced capacity for fatty acid oxidation) and indirectly, via effects on energy-requiring processes, including gluconeogenesis, synthesis of urea, bile acids, cholesterol, and proteins, and detoxification. Because accumulation of lipid within hepatocytes is a key marker of insulin resistance in humans (207) and a major contributor to nonalcoholic fatty liver disease, nonalcoholic steatohepatitis (NASH), and cirrhosis, we will first consider relationships between hepatic lipid metabolism and insulin resistance, and in Section III.C.3 will review evidence linking DM and hepatic steatosis to alterations in fatty acid metabolism or more global mitochondrial dysfunction.
Hepatic lipid accumulation may result when adipose lipid storage capacity is exceeded, as in obesity or adipocyte dysfunction (e.g., lipodystrophy) (213). Alternatively, lipid accumulation may reflect an additional imbalance between de novo hepatic lipogenesis and mitochondrial oxidative metabolism. Although the relative roles of each of these possibilities is incompletely understood, hepatic lipid accumulation is associated with obesity in humans, particularly central (abdominal) in location (214,215), and in parallel with low adiponectin levels (216). Interestingly, hepatic lipid accumulation is also a robust predictor of not only hepatic, but also muscle and adipose insulin sensitivity [better than intraabdominal fat, body mass index (BMI), or other obesity measures] (217,218). Conversely, modest weight loss (about 8 kg) normalizes intrahepatic lipid in subjects with type 2 DM, in parallel with normalization of hepatic insulin sensitivity, even in the absence of changes in intramyocellular lipid accumulation or circulating adipocytokines (215).
Although these data highlight an intimate relationship between obesity, intrahepatic lipid metabolism, and insulin sensitivity in humans, mechanisms responsible for these links remain unclear. One possibility is that excessive hepatic lipid accumulation may play a central, pathogenic role in insulin resistance. Support for this hypothesis comes from experimental lipid loading, which can induce hepatic insulin resistance. Transgenic mice expressing lipoprotein lipase in the liver have a 2-fold increase in hepatic triglyceride content and are insulin resistant (219). At a cellular level, incubation of hepatocytes with saturated long-chain fatty acids induces insulin resistance by reducing insulin-stimulated tyrosine phosphorylation of the insulin receptor and its downstream substrates (220,221). These effects in the liver appear to be mediated via reduced expression of the insulin receptor (221). Although these effects could be mediated by accumulation of fatty acyl CoA, diacylglycerols, and ceramides (as in muscle; Section III.A), it is intriguing that effects of fatty acids in liver cells can be prevented by inhibition of CPT1, indicating a critical role for mitochondrial oxidation in inducing lipid-mediated insulin resistance, perhaps via products of incomplete oxidation and/or generation of ROS (220). Fatty acids can also alter expression and/or function of key regulatory transcription factors in the liver (e.g., PGC-1β, PPARα, hepatic nuclear factor 4α) (127,222,223,224) or posttranscriptional regulation of mRNA stability (225). Fatty acid-induced reductions in insulin receptor number and function in the liver (211) may also reduce hepatic insulin clearance (226), causing systemic hyperinsulinemia, itself a contributor to both insulin resistance and reduced mitochondrial function (214,227,228).
A second possibility is that hepatic insulin resistance itself contributes to alterations in mitochondrial oxidative capacity. Indeed, a recent paper demonstrated that mice with hepatic insulin resistance due to deletions of the major insulin receptor substrates (IRS-1 and IRS-2) have impaired mitochondrial function and biogenesis, as demonstrated by reduced NADH oxidation, reduced ATP production rates, reduced numbers of mitochondria per cell, reduced fatty acid oxidation, and increased hepatic triglyceride accumulation (229). Mitochondrial dysfunction was reversed by deletion of Foxo1. These data indicate that normal insulin signaling, which inhibits Foxo1, is required for maintenance of normal mitochondrial function in this model. It remains unclear whether additional components of the in vivo environment, such as glucose intolerance and hyperinsulinemia, contribute to mitochondrial dysfunction in these mice. However, more broadly, these data indicate that hepatic insulin resistance can cause mitochondrial dysfunction, at least in mice.
3. Evidence for impaired liver mitochondrial function in diabetes and NASH
Although human liver studies have been limited due to lack of tissue biopsy samples from otherwise healthy individuals, two groups have examined hepatic gene expression related to mitochondrial function in both obesity and type 2 DM (230,231,232). In the first (232), severe obesity (mean BMI 52 kg/m2) was associated with reduced expression of seven of 25 genes encoding OXPHOS genes; expression of these genes was inversely correlated with hepatic lipid accumulation and paralleled by reduced expression of PGC-1α and genes known to be regulated by thyroid hormone. Similar patterns were observed in obese subjects with established type 2 DM. Interestingly, reduced expression of OXPHOS genes (e.g., COX7C, ATP5C1) was also observed in mice fed a high-fat diet and normalized by acute therapy with thyroid hormone T3—suggesting that functional hepatic thyroid hormone resistance could contribute to reduced expression of mitochondrial oxidative genes in this context (232).
In contrast, studies in Japanese individuals with established DM and modest obesity (BMI 27 kg/m2) observed a modestly increased expression of multiple genes within all complexes of OXPHOS complexes, in parallel with BMI and insulin resistance (measured by homeostasis model assessment of insulin resistance, HOMA-IR) (231). Up-regulation of these OXPHOS genes was also positively associated with expression of several genes linked to mitochondrial biogenesis (e.g., PGC-1β, ERRα, NRF, thyroid hormone receptor) and both ROS generation (e.g., NADPH oxidase) and attenuation (e.g., glutathione peroxidase). Thus, increased ROS related to increased fatty acid oxidation and/or hyperglycemia might contribute to up-regulation of OXPHOS gene expression in coexisting obesity and type 2 DM. Although these two data sets appear to be discordant (i.e., obesity-linked down-regulation of mitochondrial oxidative gene expression in the first, and up-regulation in the second), several differences in the study population may account for these findings: 1) much greater degree of adiposity and hepatic steatosis in the first; 2) differences in ethnicity (Caucasian-Americans vs. Japanese); and 3) differences in insulin sensitivity and glycemia (insulin sensitive vs. resistant comparison in the first study, coexisting DM in the second).
Studies of individuals with NASH provide additional opportunities to identify potential interactions between hepatic lipid accumulation, insulin resistance, and mitochondrial function in humans. Indeed, enzymatic activity of complexes I-V is reduced in liver extracts from patients with NASH and is inversely correlated with BMI and HOMA-IR (233,234). Moreover, NASH is characterized by prominent abnormalities in mitochondrial ultrastructure, with increased size, loss of cristae, and paracrystalline inclusion bodies similar to those observed in some mitochondrial myopathies (235). Although these data cannot address whether such changes are indeed pathogenic, it is interesting that reduced OXPHOS activity in this setting is accompanied by increased tissue long-chain acylcarnitines and reduced short-chain acylcarnitines, despite normal CPT1 activity and increased expression of β-oxidation genes (230,236). Similarly, circulating β-hydroxybutyrate levels are increased in NASH (235). Together, these data suggest excessive, but incomplete, fatty acid oxidation, potentially limited by reduced availability of NAD+ and FAD. Byproducts of incomplete fatty acid oxidation could act in concert with adipose tissue-derived inflammatory signals (e.g., TNFα), and altered expression and activation of proinflammatory (e.g., IL-1R family) and profibrotic genes (e.g., TGFB1, FGFR2), to increase production of ROS and ultimately contribute to the development of NASH and cirrhosis (235).
In summary, available data indicate that hepatic lipid accumulation and insulin resistance are intimately linked with mitochondrial oxidative dysfunction. We hypothesize that modest obesity may be associated with compensatory up-regulation of OXPHOS gene expression in response to sustained lipid load and/or functional defects in complete fatty acid oxidation. Up-regulation of PGC-1β in this context may contribute to increased gluconeogenesis and hyperlipidemia, in part via coactivation of sterol regulatory element binding transcription factor 1, as observed in high-fat diet-fed mice (223). With aging, chronic ROS exposure, and/or the development of insulin resistance related to obesity or sustained lipid accumulation, OXPHOS expression may fall. Although this may be an appropriate response, limiting oxidative stress, it may also contribute to a vicious cycle of further impairments in oxidative capacity, increased lipid accumulation, and progressive insulin resistance. To test this hypothesis, longitudinal measurements of gene expression, oxidative function, and lipid accumulation in humans with progressive obesity and evolution of insulin resistance would be required—but are unlikely to be performed due to the invasive nature of serial liver biopsies in humans.
D. Pancreatic β-cells
1. Roles of mitochondria in β-cells
Mitochondrial capacity is central to the key function of the pancreatic β-cell—regulated insulin secretion. Both rapid (first phase) and more prolonged (second phase) insulin secretion (237) are dependent on glucose metabolism and mitochondrial oxidative capacity; glucose oxidation increases the ATP/ADP ratio, inhibiting plasma membrane K-ATP channels and allowing voltage-gated calcium channels to open. Increased cytoplasmic calcium then triggers exocytosis of plasma-membrane docked insulin granules (first phase). Subsequent recruitment of granules to the plasma membrane (second phase) appears to depend on mitochondrial metabolites produced by anaplerosis (238). Mitochondrial metabolism is also required for the transient, controlled production of ROS, which is required for the mitochondrial signaling pathways that trigger granule exocytosis (239,240).
2. Evidence for reduced β-cell mitochondrial capacity in DM
Given the crucial role of mitochondrial ATP generation, anaplerosis, and ROS production in insulin secretion, mitochondrial dysfunction in β-cells would be expected to reduce insulin secretion and thus promote the development of DM. Consistent with this possibility, β-cell specific deletion of Tfam reduces insulin secretory capacity and β-cell mass, yielding so-called mitochondrial DM (241). Moreover, Tfam has recently been shown to be directly downstream of PDX1, a key transcription factor for β-cell development (242).
In humans, the key role of β-cell mitochondria is exemplified by the development of diabetes in families harboring mutations in mtDNA. Of these, the best studied is the 3243A>G mutation in the mtDNA-encoded tRNALeu, UUR gene, which is associated with maternally inherited diabetes and deafness (MIDD) (243,244). Another example is mutation 14577 T>C, a missense substitution in the NADH dehydrogenase 6 gene (245). In this case, mitochondrial respiratory chain complex I activity and O2 consumption rates are decreased by 65 and 62%, respectively, in hybrid cell lines derived from probands.
Interestingly, mitochondrial diabetes only develops upon aging, with an average age of onset between 35 and 40 yr for MIDD and 48 yr for 14577 T>C. This contrasts with the early childhood onset of diabetes in syndromes such as maturity-onset diabetes of the young 2 (MODY2), in which a mutation in glucokinase, the first step of glycolysis, results in attenuated glucose-stimulated ATP generation and insulin secretion. These data suggest that mitochondrial diabetes is more likely to result from a gradual deterioration of β-cell function, rather than from an acute functional impairment due to insufficient ATP production (246).
One of the mechanisms by which mtDNA mutations might lead to a gradual deterioration in β-cell function, and not to an acute failure of insulin secretion due to decreased ATP levels, could be the stress imposed by an increase in metabolic flux to compensate for inefficiencies in the ETC. Consistent with this view, clonal cytosolic hybrid cells harboring mitochondria derived from MIDD patients exhibit impaired calcium handling and elevated ROS under metabolic stress (247,248). Chronically increased ROS production could also induce β-cell death and result in gradual onset of diabetes (249,250,251,252,253).
3. Factors affecting mitochondrial function in β-cells
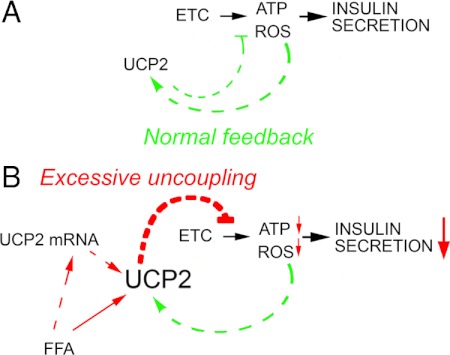
Mitochondrial function in β-cells is highly regulated by the levels and activities of UCPs, in turn regulated by ROS produced by the activity of the ETC. Low levels of ROS are necessary for insulin secretion, but chronic, high mitochondrial ROS production can have a deleterious effect on β-cell function (254,255,256). Thus, the activation of UCP2 protects the β-cell from the deleterious effects of excess ROS (257) by dissipating the proton gradient and decreasing ROS production in a controlled negative feedback manner (Fig. 4). However, it also leads to decreased ATP production, which impairs insulin secretion. Thus, UCPs must uncouple respiration sufficiently to mitigate toxic levels of ROS, but not enough to decrease ATP and ROS below the levels necessary for insulin secretion. This delicate balance in which UCP2 is desirable for β-cell protection, but undesirable for glycemic control, probably underlies the discrepancy in results between two reports on the phenotype of UCP2 knockout mice. In a mixed background, UCP2 knockout improves glycemic control in ob/ob mice (258), whereas in a pure C57BL6/J background, UCP2 knockout accelerates β-cell failure and diabetes (259).
Figure 4.
Hypothesized mechanism by which free fatty acid (FFA) excess impairs insulin secretion. A, As described above, the activity of the ETC leads to the synthesis of ATP and the generation of a small amount of ROS. In the β-cell, both ATP and ROS are signals that trigger insulin secretion. Excessive accumulation of ROS is mitigated normally by the activation of UCP2, which dissipates the proton gradient, decreasing both ATP and ROS production. The presence of this normal negative feedback loop suggests that the control of excessive ROS generation is imperative in the β-cell, even if it occurs at the expense of decreasing ATP synthesis. B, In the presence of excess FFA, this normal feedback loop is compromised by a direct activation of UCP2 by FFA, as well as an effect of FFA to increase the amount of UCP2. Thus, uncoupling occurs to an excessive degree, compromising ATP synthesis enough to impair insulin secretion and β-cell fitness.
The levels and activity of UCP2 and the rate of ROS production are both increased by high-fat diet and hyperphagia, possibly through the actions of nonesterified fatty acids and their ceramide derivatives (260). It is likely that decreased ATP production due to unbalanced activation of UCPs by direct actions of fatty acids and their derivatives, in addition to excessive ROS, could underlie the accumulation of β-cell damage that precedes type 2 DM (Fig. 4).
Although the tissues reviewed above are considered central to the pathophysiology of DM, other tissues such as gut, brain, kidney, neuronal tissues, and endothelium are also likely to be implicated in a primary or secondary manner in the pathophysiology of DM and /or its complications. The aspects of mitochondrial function unique to each of these tissues and the consequences of their potential dysfunction in relation to DM pathophysiology are relatively less explored areas and are thus outside the scope of this review.
IV. Experimental Strategies to Explore the Relationship between Mitochondrial Function and DM
Although available data demonstrate links between mitochondrial oxidative function and phenotypes linked to insulin resistance and diabetes, it remains unclear whether these are simply associations or whether oxidative dysfunction can contribute to insulin resistance and diabetes risk. To address this question, we will examine available data from experimental models in which OXPHOS function has been altered. Such studies have shed light on the basic mechanisms underlying mitochondrial biogenesis and on the consequences of disruption of normal mitochondrial homeostatic mechanisms on cell and whole-body oxidative metabolism. A summary of these studies is presented in Tables 1 and 2 and is discussed in Section IV.A and B.
Table 1.
Studies of PGC-1α transgenic expression models
| First author, year (Ref.) | Model | Mito DNA | Mito density (EM) | OXPHOS mRNA/protein | Non-OXPHOS mRNA/protein | Mito energetics | Exercise capacity | Insulin sensitivity | Other |
|---|---|---|---|---|---|---|---|---|---|
| Benton, 2008 (261) | PGC-1α electroporation | ↑ 113% | 180% ↑ COXIV protein | ↑ muscle | ↑ AMP kinase activity | ||||
| Ward, 2009 (262) | PCG-1α whole-body own promoter | 130% ↑ mRNA | ↑ muscle | Hepatic insulin resistance | |||||
| Miura, 2003, 2006 (263,264) | PGC-1α whole-body α-actin promoter | ↑ 200–300% | ↑ number | 150–200% ↑ mRNA | 300% ↑ in UCP2 mRNA | 60% ↓ ATP levels in homogenates | ↓ voluntary | ↓ whole-body | ↑ AMP kinase activity |
| Russell, 2004 (266) | PGC-1α inducible heart | ↑ 350% | Myofibrillar disorganization cardiac failure | ||||||
| Wende, 2007 (265) | PGC-1α inducible skeletal muscle | ↑ | 150–250% ↑ mRNA | 150% ↑ FAO gene mRNA | No change in low intensity, ↓ performance at high intensity | ↑ glucose uptake, glycogen deposition, decreased glycolysis | |||
| Lin, 2002 (268); Sandri, 2006 (269); Calvo, 2008 (267); Choi, 2008 (270) | PGC-1α MCK promoter | ↑ 166–250% | ↑ 250% in EDL | 170–300% ↑ mRNA | 200–400% ↑ in FAO gene mRNA | 50–60% ↑ ATP synthesis by NMR | ↑ exercise performance; ↓ fatigue in vitro, protection from denervation-induced atrophy | ↓ muscle and whole-body only in high-fat diet | No change in AMP kinase activity |
| Arany, 2007 (271) | PGC-1β MCK promoter | ↑ | 200–500% ↑ mRNA and protein | 200–500% ↑ FAO gene mRNA | 120–130% ↑ endurance | ||||
| Kamei, 2003 (272) | PGC1-β whole-body β-actin promoter | ↑ whole-body |
Mito, Mitochondrial; EM, electron microscopy; MCK, muscle creatine kinase; ↓, decrease; ↑, increase; EDL, extensor digitorum longus; FAO, fatty acid oxidation.
Table 2.
Studies of PGC-1α or PGC-1β knockout models
| First author, year (Ref.) | Model | Mito DNA | Mito density (EM) | OXPHOS mRNA/protein | Non-OXPHOS mRNA/protein | Mito energetics | Exercise capacity | Insulin sensitivity | Other |
|---|---|---|---|---|---|---|---|---|---|
| Lin, 2004 (69); Arany, 2005 (68) | PGC-1α whole-body KO | Normal | 30–60% ↓ mRNA; 50% ↓ CytC protein | ATP levels ↓ 20% in heart | 10–50% ↓ cardiac contractile performance | ↑ whole-body | ↓ body weight, increased AMP kinase activity | ||
| Leone, 2005 (67) | PGC-1α whole-body KO | 30% ↓ | 40–60% ↓ mRNA | 10% ↓ in state 3 respiration | 50% ↓ fatigue resistance, abnormal cardiac response to stress | ↑ whole-body | Hepatic steatosis | ||
| Lehman, 2008 (61) | PGC-1α whole-body KO | ↓ cristae density in heart | Slight ↓ mRNA | 60% ↓ in metabolic efficiency | ↓ cardiac power | ||||
| Handschin, 2007 (63,64) | PGC-1α muscle-specific KO | Normal | 30–40% ↓ mRNA | 50% ↓ ALAS1 | 60% ↓ grip strength, endurance, muscle damage basal and exercise | ↑ muscle | ↓ food consumption, ↓ body weight, muscle inflammation, β-cell dysfunction | ||
| Lai, 2008 (62) | PGC-1β whole-body KO | Not changed (heart) | Modest ↓ in state 3 respiration | ↓ running duration | |||||
| Lelliot, 2006 (66) | PGC-1β whole-body KO | ↓ 20% | 20–40% ↓ mRNA | ↓ state 3 and 4 respiration, ↓ ATP synthesis | ↓ chronotropic response to dobutamine | Normal | ↓ body weight, hepatic steatosis on high-fat diet | ||
| Vianna, 2006 (65) | PGC-1β whole-body hypomorph | ↓ 30% | 20–30% ↓ mRNA | Normal | ↑ hepatic lipid levels | ||||
| Sonoda, 2007 (59) | PGC-1β whole-body hypomorph | Normal | 30–40% ↓ mRNA | Normal | Normal | Hepatic steatosis on high-fat diet | |||
| Lai, 2008 (62) | PGC-1α and PGC-1β muscle-specific KO | ↓ 60%, heart | Perinatal lethality due to cardiac failure | Perinatal lethality due to cardiac failure |
↓, Decrease; ↑, increase; KO, knockout; CytC, cytochrome C; ALAS1, 5-aminolevulinate synthase 1.
A. PGC-1 α and β overexpression
PGC-1α and related coactivators are critical for the regulation of mitochondrial oxidative capacity, as demonstrated by the approximately 2-fold increases in mtDNA and oxygen consumption and a 50% increase in mitochondrial density in myotubes overexpressing PGC-1α (56). To address whether this family plays the same functional role in vivo, several different models of PGC-1α transgenic expression have been generated, each of which differs in tissue selectivity, levels of overexpression achieved, and resulting metabolic phenotype (Table 1). The lowest level of PGC-1α overexpression was achieved in rat muscle by means of electroporation (261). This resulted in modest up-regulation of mitochondrial proteins, increased palmitate oxidation, and increased insulin-stimulated glucose uptake. Similarly, transgenic mice expressing human PGC-1α driven by its own promoter displayed a modest (30% higher than basal) increase in mRNA expression of several OXPHOS genes, fiber type switching, and enhanced muscle insulin sensitivity (262). Importantly, this modest PGC-1α overexpression also was accompanied by decreased levels of ROS and inflammatory signaling. However, these same animals displayed increased liver gluconeogenic enzyme levels and impaired insulin suppression of hepatic glucose production, thus nullifying the potentially beneficial effect of modest muscle PGC-1α overexpression on whole-body glucose homeostasis.
Higher levels of overexpression of PGC-1α achieved through actin promoter-driven expression in transgenic animals display a strikingly different phenotype (263,264). In these animals, mitochondrial density and OXPHOS gene expression are more than double basal levels, and large increases in UCP2 gene expression are also observed. Mitochondrial energetics are impaired, with 60% decreases in ATP levels in muscle homogenates and concomitant increases in AMP kinase activation, probably as a compensatory response to decreased mitochondrial functionality. Muscle function appears compromised, as evidenced by decreased voluntary exercise, muscle atrophy, and decreased insulin sensitivity.
Inducible overexpression of PGC-1α in skeletal muscle (265) results in increased mitochondrial density and a robust increase in expression of OXPHOS genes and genes necessary for fatty acid oxidation. In these animals, both muscle glucose uptake and glycogen deposition were increased. Although low-intensity exercise performance did not differ in this model, high-performance exercise was impaired, in parallel with failure to mobilize stored glycogen. Similarly, inducible expression in cardiac muscle can have deleterious effects. When higher levels of PGC-1α overexpression are restricted to heart during early life (266), increased neonatal mitochondrial proliferation is observed, but it is accompanied by myofibrillar displacement. In adults, PGC-1α induction led to a more modest mitochondrial proliferation, which was nevertheless surprisingly accompanied by cardiomyopathy. Thus, whole-body and inducible skeletal muscle- or cardiac-specific overexpression of PGC-1α can produce deleterious effects on muscle structure and function.
When PGC-1α overexpression is restricted to skeletal muscle but overexpressed throughout development by the use of the creatine kinase gene promoter, very large increases in mitochondrial mass, OXPHOS, and fatty acid oxidation genes are observed (267,268,269). In this model, ATP synthesis rate and exercise performance are increased, and fatty acid oxidation is also enhanced. Despite these effects, insulin sensitivity is normal in PGC-1α transgenic mice fed a chow diet and, surprisingly, is reduced during high-fat feeding. Insulin resistance was paralleled by accumulation of triglycerides and long-chain acyl CoA (270).
Although PGC-1β-mediated gene expression and function appear to overlap considerably with that of PGC-1α, PGC-1β has distinct expression profiles in skeletal muscle, being expressed in parallel with myosin IIx rather than type I fibers (271). In addition, PGC-1α and PGC-1β coactivate different nuclear receptors (272). Muscle-specific transgenic overexpression of PGC-1β results in increased oxidative fiber content and increased expression of OXPHOS and multiple other mitochondrial genes. Mice displayed enhanced endurance and oxidative work (271). Consistent with these findings, whole-body transgenic overexpression of PGC-1β resulted in increased oxidative metabolism, protection from obesity induced by high-fat diet or hyperphagia, and enhanced insulin sensitivity (272).
Taken together, these studies suggest that modest overexpression of PGC-1α (261,262) and transgenic overexpression of PGC-1β (271,272) increase oxidative metabolism and insulin sensitivity. By contrast, high-level overexpression of PGC-1α may actually reduce insulin sensitivity via reduced glycolysis and/or glucose oxidation (265) or myofibrillar disruption/myopathy, potentially limiting exercise tolerance. Moreover, high-level expression of PGC-1α can induce not only increases in lipid oxidation genes but also parallel increases in expression of genes promoting lipid uptake and synthesis (270). The net balance of these effects, and thus the net accumulation of pathogenic lipid species, is likely to be determined by the metabolic and hormonal milieu. For example, in cultured myotubes overexpressing PGC-1α (273), lipid content is decreased in the serum-starved condition, reflecting increased oxidation of lipids; by contrast, in the serum-replete state, intracellular fatty acid levels are increased, likely due to both increased de novo fatty acid synthesis from glucose and fatty acid synthesis by chain elongation (273). Thus, high-level expression of PGC-1 may actually cause accumulation of lipids and/or incomplete oxidation products, leading to reduced insulin sensitivity, as observed in PGC-1α transgenic mice (267,268,269). These results also suggest that inappropriate, nonphysiological regulation of PGC-1α expression may prevent appropriate modulation of oxidative metabolism with feeding/fasting, or exercise/recovery—the hallmarks of impaired metabolic flexibility observed in human insulin resistance.
B. PGC-1 knockout models
Because enhanced insulin sensitivity is seen in some models of PGC-1 transgenic overexpression and human diabetes is associated with reduced PGC-1α and PGC-1β expression, an important question is whether PGC-1α or PBC-1β deficiencies per se could result in insulin resistance. The results from studies of PGC-1α or PGC-1β knockout models shed some light on this question (Table 2). Two independent lines of whole-body PGC-1α-null mice have been generated (61,67,68,69). These mice consistently display 30–60% reductions in OXPHOS gene expression in muscle, normal to decreased mitochondrial density in skeletal muscle, and decreased cristae density in heart. Functionally, decreases in ATP levels, state 3 respiration, and metabolic efficiency are seen, and cardiac and skeletal muscle performance is reduced in response to stress. Many of these phenotypes are recapitulated in a muscle-specific PGC-1α knockout line (63,64) (Table 2). In addition, both lines of whole-body PGC-1α knockout animals display cold sensitivity, likely related to impaired mitochondrial function in brown adipose tissue (67,69) (Table 2).
Interestingly, muscle is significantly more insulin sensitive in both whole-body and muscle-specific PGC-1α null mice. This apparently paradoxical finding—where decreased oxidative function is not paralleled by insulin resistance—may be explained by the leanness and hyperactivity noted in one of the whole-body knockout lines (68,69). In the muscle-specific knockout lines, reduction in food intake, body weight, and adiposity (despite decreased physical activity) may also contribute to insulin sensitivity (63,64). Thus, lack of PGC-1α per se does not lead to insulin resistance, despite clear alterations in muscle and metabolic phenotypes. It is possible that the compensatory mechanisms elicited by the complete lack of PGC-1α, which result in a leaner phenotype, have a paradoxical insulin-sensitizing effect. For example, decreased metabolic efficiency (61), defined as decreased ATP produced per unit of oxidized fuel and/or oxygen, may lead to enhanced whole-body fuel utilization and thus a leaner, more insulin-sensitive phenotype. Activation of AMP kinase in this setting of reduced ATP production may also contribute to enhanced insulin sensitivity (274).
Despite pronounced muscle and brown adipose tissue functional impairments seen in models of PGC-1α deficiency, mitochondrial density in muscle and brown adipose tissue is not notably decreased, indicating the possible compensatory role of PGC-1β in mitochondrial biogenesis. PGC-1β-null mice (59,62,65,66) display phenotypes similar to, but not identical to, those of PGC-1α knockouts, including reduced expression of OXPHOS genes, reduced ATP synthesis, reduced exercise capacity, and impaired thermogenesis (59,62). Insulin sensitivity in these animals is reported to be normal. PGC-1β-null mice have more striking hepatic phenotypes, with hepatic insulin resistance, even on normal chow, and steatosis with high-fat feeding (59,65,66). These data raise the possibility that reduced expression of PGC1 family genes and impaired oxidative function may play a more important, and potentially pathogenic role in liver than in muscle.
The existence of compensation for PGC-1β deficiency by PGC-1α and vice versa is evidenced by the phenotype of double PGC-1α and PGC-1β muscle-specific knockout mice (62). These animals display normal mitochondrial biogenesis during development but die early after birth due to failure of perinatal mitochondrial proliferation and consequent cardiac failure (62).
Together, data from knockout mice have provided important insights into the complex and multifaceted roles of PGC-1α and PGC-1β as mediators of basal and adaptive energy homeostasis. PGC-1α and PGC-1β are both required for the full complement of OXPHOS gene expression, the establishment of correct fuel partitioning, and optimal metabolic efficiency because whole-body and tissue-specific knockout of either of these genes leads to deranged multiorgan metabolic phenotypes that are more evident during energetic stress. In light of these phenotypes, it is interesting to consider whether the 40–50% reduction in both PGC-1α and PGC-1β expression observed in humans with obesity and type 2 DM in both liver (232) and muscle (122), potentially mediated by lipid excess, may contribute to further impairment in oxidative capacity and a vicious cycle of maladaptive responses during energetic stress.
C. Other mitochondrial function defects
PGC-1 family members interact with the family of estrogen-related nuclear receptors, or ERRs, to transactivate transcription of oxidative genes (275) and mitofusins (276). Interestingly, ERRα-null mice have reduced body weight and are normoglycemic, in parallel with up-regulation of medium-chain acyl-CoA dehydrogenase and reduced lipogenesis, both of which may contribute to resistance to diet-induced obesity (277). Although the physiological effects of the related gene ERRγ remain unclear, it interacts with PGC-1 to regulate expression of ERRα (278), and polymorphisms at this locus were found to be associated with increased glucose area under the curve in a genetic analysis of Old Order Amish subjects (279).
Pospisilik et al. (274) used an alternative strategy to test whether experimentally induced respiratory chain complex deficiency would induce metabolic phenotypes. Tissue-specific ablation of the nuclear-encoded gene, mitochondrial apoptosis-inducing factor (AIF), produced modest defects in respiratory chain gene expression and function, as expected; however, both muscle- and liver-specific null mice had increased insulin sensitivity, reduced fat mass, and improved glucose tolerance. Although defects in AIF may not precisely replicate respiratory chain defects mediated by other genes, these data mirror to a large extent those in PGC-1-α null mice and again suggest that modest defects in mitochondrial oxidative capacity may enhance insulin action, potentially via ATP deficiency and/or secondary activation of AMP kinase (280).
To address whether reductions in OXPHOS content or function may contribute to diabetes-related metabolic phenotypes, we can also consider phenotypes related to genes regulating transcription of mtDNA. The nuclear-encoded mitochondrial transcription factor Tfam is essential for transcription of mtDNA and thus mitochondrial development, as indicated by the embryonic lethality of Tfam-null mice (281). Similarly, skeletal and cardiac muscle-specific deletion of Tfam causes severe OXPHOS deficits, dilated cardiomyopathy, and death during early postnatal life (282). Skeletal muscle-specific Tfam deficiency is accompanied by increased AMP kinase activation and increased glycolysis, indicating that compensatory mechanisms to maintain ATP levels are activated in response to impaired mitochondrial energetics (282). In these mice, insulin sensitivity is normal and glucose tolerance is enhanced, consistent with a need for enhanced fuel utilization in response to inefficient mitochondrial ATP production. Thus, in these experimental models in which mitochondrial oxidative capacity is severely restricted, insulin action is enhanced.
DNA polymerase γ is a mtDNA polymerase critical for maintenance of mtDNA. POLG-null mice die during embryogenesis with severe mtDNA depletion (283), whereas knock-in mutant mice have accelerated aging potentially linked to impaired repair of mtDNA mutations (284). Mice null for the mitochondrial helicase Twinkle have impaired respiratory chain activity and muscle atrophy, but no defect in exercise capacity (285). Thus, these data indicate that primary defects in mtDNA, even when produced experimentally by ablation of nuclear-encoded genes, do not produce overt metabolic phenotypes. These observations are consistent with those in humans harboring mutations in mtDNA, where mitochondrial diabetes typically manifests as a gradual deterioration of pancreatic β-cell function (see Section III.D).
V. Conclusions
Mitochondrial function in tissues involved in the pathogenesis of DM (liver, muscle, adipose tissue, and pancreatic β-cells) is critical for multiple aspects of cellular metabolism. In each of these tissues, mitochondrial oxidative activity must be appropriate to fully oxidize nutrient loads, particularly fatty acids. Failure of complete oxidation can lead to accumulation of lipid intermediates, incomplete fatty acid oxidation products, and ROS, inducing both insulin resistance (muscle, liver, adipose) and altered secretion (β-cells).
The sufficiency to fully oxidize fatty acids resides in the balance between: 1) net mitochondrial oxidative activity, in turn determined by the need to generate energy to meet cellular demands, e.g., contraction and ion transport; and 2) fuel availability (determined by food intake, adiposity, and adipose storage capacity) (Fig. 5A). Balance is achieved when oxidative activity equals or exceeds fuel loads.
Under normal homeostatic conditions, both oxidative activity and cellular fuel availability could in principle be altered to ensure that mitochondrial function is appropriate for the ambient metabolic environment. For example, cellular demand for energy can be increased through exercise, and fuel availability can be reduced through weight loss and/or reduced food intake.
In this context, interindividual variation in oxidative capacity and/or activity, fuel load, or ability to modulate mitochondrial activity (acute response), increase mitochondrial capacity (chronic response), or resolve oxidative stress could determine the set point of metabolic balance. Such differences could become prominent particularly in an obesogenic environment. Thus, individuals with a high oxidative capacity or adaptive responses would have high tolerance to large fuel loads. Conversely, individuals with low oxidative capacity and/or suboptimal adaptive responses would be intolerant to moderate-high fuel loads, leading to lipid accumulation, incomplete oxidation, production of ROS, and acute insulin resistance (Fig. 5B). In turn, such alterations in mitochondrial activity could be mediated by genetic factors (family history, ethnicity), epigenetic mechanisms, developmental exposures, and aging. With time, insufficient oxidative capacity could be resolved by compensatory mechanisms that increase oxidative capacity (e.g., exercise, mitochondrial biogenesis) (Fig. 5C, top right) or decrease fuel load (weight loss) (Fig. 5C, middle right). Insufficient compensation could result in chronic insulin resistance (Fig. 5C, bottom right).
Two key questions arise from this model: 1) does interindividual variation in either baseline or adaptive mitochondrial oxidative responses alter risk for insulin resistance; and 2) does chronic imbalance itself resulting from chronic overnutrition (e.g., ROS damage to mtDNA) impair mitochondrial capacity, leading to further metabolic imbalance? With regard to the first question, mild deficiencies in mitochondrial activity, and/or an inability to increase activity and capacity in response to cellular energy demand, could explain the reduced exercise ability seen in individuals with a family history of DM (106). Over time, this phenotype could contribute to reduced voluntary exercise and increase the likelihood of an imbalance between mitochondrial activity and fatty acid load.
Secondly, chronic imbalance in energy metabolism due to overnutrition, obesity, and inactivity could directly contribute to increased cellular and mitochondrial ROS production. In turn, excessive ROS can induce both insulin resistance and mitochondrial dysfunction. For example, a high-fat, high-sucrose diet in the diabetes-prone C57BL6 mouse causes mitochondrial alterations in parallel with enhanced ROS production and impaired insulin sensitivity. Similarly, exposure of muscle cells in vitro to saturated fatty acids or high-fat feeding in mice results in alterations in mitochondrial structure and insulin resistance, both of which are reversed by antioxidants (286,287,288). Thus, oxidative stress can induce mitochondrial dysfunction in parallel with insulin resistance—perhaps an adaptive response aimed at limiting further oxidative damage. More importantly, resolution of oxidative stress can reverse insulin resistance. These data also suggest that defects in resolution of oxidative stress may be another mechanism conferring increased risk for both mitochondrial dysfunction and insulin resistance.
The effects of these variables on disease progression can be hypothesized to occur in three stages (Table 3). In the first stage, mitochondrial activity is adequate relative to fuel intake, and normal insulin sensitivity and secretion are observed. In a second stage, mitochondrial activity is inadequate relative to oxidative load, leading to the development of oxidative stress, insulin resistance, impaired insulin secretion, or both. Insulin resistance per se could result in further reductions in mitochondrial capacity (229,289), which in the absence of compensation (e.g., decreased food intake) would lead to a third stage, characterized by further imbalance, β-cell failure, and progression to DM.
Table 3.
Stages of disease progression
| Stage I: healthy insulin sensitive | Stage II: physiological stress with appropriate compensation | Stage III: failure of compensation | |
|---|---|---|---|
| Oxidative capacity and activity | Normal exercise capacity and activity | Inappropriately low relative to load, due to: | Further ⇊ |
| • Inactivity | |||
| • Reduced fitness | |||
| • Genetic susceptibility | |||
| • Aging | |||
| Oxidative load | Normal | Increased, due to: | Sustained ⇈ |
| • Cellular overnutrition | |||
| • Obesity | |||
| Cellular response patterns | Normal | • Initial ↑ mitochondrial biogenesis, inadequate to meet metabolic demand | • ⇊ Nuclear-encoded OXPHOS gene expression |
| • ↑ Incomplete oxidation | • Vicious cycle of suboptimal oxidative capacity | ||
| • ↑ ROS generation | |||
| • ↑ Tissue lipid accumulation | |||
| Physiological consequences | Normal insulin sensitivity and secretion | Insulin resistance | • Progressive insulin resistance |
| • Insulin secretory dysfunction | |||
| • Type 2 DM |
↓, Decrease; ↑, increase.
We return to the key question—do variations in oxidative activity underlie the risk for and development of DM in humans? Unfortunately, the current human data are insufficient to answer this question with certainty or to determine whether impaired oxidative capacity can cause insulin resistance, or result from insulin resistance. Animal data are inherently limited by interspecies comparisons. Moreover, it is also difficult to fully recapitulate in an animal model the subtle changes in mitochondrial activities and capacity and the interactions between mitochondrial function and relative fuel load that, in humans, can enhance diabetes risk. Nevertheless, results from multiple mouse models now indicate that absolute deficiency of the PGC-1 family of coactivators or mitochondrial OXPHOS per se does not directly produce insulin resistance. Thus, we interpret the current data to indicate that an individual’s intrinsic mitochondrial oxidative activity may determine the magnitude of chronic cellular dysfunction and influence adaptive responses to chronic fuel excess, as with obesity or inactivity. Moreover, experimental induction of insulin resistance and/or components of the insulin-resistant/diabetes milieu may contribute to reductions in mitochondrial oxidative capacity, further fueling a vicious cycle of diabetes risk (Fig. 6).
Figure 6.
Vicious cycle leading to progressively increased risk of DM. An individual’s intrinsic mitochondrial oxidative capacity is determined by numerous factors including genetic and ethnic background, intrauterine exposures, and age. Chronic fuel excess, in the setting of suboptimal oxidative activity, results in fatty acid accumulation, incomplete oxidation, increased oxidative stress, and ROS generation leading to impairment in oxidative capacity, compounding the deleterious effects of fuel excess. With time, mitochondrial damage ensues, further exacerbating the process. Together, these factors contribute to progressively impaired insulin sensitivity, insulin secretion, and heightened risk of DM.
To fully dissect these possibilities, better animal models, together with prospective longitudinal studies in humans analyzing OXPHOS capacity in different tissues, their variation with aging, weight gain, and fuel load, and their correlation with the development of metabolic disease are necessary. The recent emergence of metabolomic approaches will likely facilitate further assessment of mitochondrial function in humans, potentially allowing use of clinically accessible blood samples for such longitudinal studies (290,291,292,293). A still-unanswered but intriguing question is whether altered energetics during developmentally sensitive periods (e.g., intrauterine or early postnatal life) could ultimately increase susceptibility to insulin resistance.
In summary, we hypothesize that mitochondrial oxidative activity can be considered as a key determinant underlying diabetes risk. In isolation, reductions in mitochondrial activity mediated by genetic factors (family history, ethnicity), epigenetic mechanisms, developmental exposures, and aging may not be sufficient to induce insulin resistance. However, when sustained fuel excess (e.g., resulting from overnutrition or impaired fat storage) exceeds energetic demands and/or oxidative capacity, and/or appropriate compensatory mechanisms are insufficient (e.g., due to inactivity or failure of mitochondria to adapt to higher cellular oxidative demands), a vicious cycle of insulin resistance and impaired insulin secretion can be initiated (Fig. 6). Resulting lipid accumulation and oxidative stress can alter transcriptional responses and damage mitochondria, further reducing OXPHOS capacity, compounding the deleterious effects of fuel excess and increasing diabetes risk. Although the importance of the PGC-1 family of coactivators has been stressed thus far, it is likely there are many additional transcriptional and metabolic mechanisms by which mitochondrial function adapts to environmental demands. Critical studies of these gene-environment interactions is likely to be a fruitful area of investigation in the years to come.
Footnotes
The authors gratefully acknowledge research support from National Institutes of Health Grants DK062948 (to M.-E.P.), DK080366 (to S.C.), the Graetz Foundation (to M.-E.P.), LM008748 (to M.-E.P.), DK060837 (Diabetes Genome Anatomy Project, to M.-E.P., and S.C.), M01 RR001032 (General Clinical Research Center), DK36836 (Diabetes and Endocrinology Research Center, Joslin Diabetes Center), and DK32520 (Diabetes Endocrinology Research Center, University of Massachusetts Medical School).
Disclosure Summary: The authors have nothing to disclose.
Patti ME, Liu M, Zin W, Lerin C, Dreyfuss J, Vokes M, Schroeder J, Tatro E, Park P, Kohane I, Kasif S, Goldfine AB, submitted. Transcriptome analysis reveals parallel dysregulation of oxidative metabolism and inflammation in muscle and adipose tissue with progression of insulin resistance in humans.
Stender-Petersen KL, Poulsen P, Butte A, Jensen CB, Yee J, Leykin I, Vaag A, Pedersen O, Patti ME, manuscript under review. Gene expression analysis in monozygotic twins reveals heritable contributions to PGC-1/ERR pathways.
First Published Online February 15, 2010
Abbreviations: BMI, Body mass index; CoA, coenzyme A; COX, cytochrome oxidase; CPT1, carnitine palmitoylotransferase 1; DM, diabetes mellitus; ERR, estrogen-related receptor; ETC, electron transport chain; FADH2, reduced flavin adenine dinucleotide; MIDD, maternally inherited diabetes and deafness; mtDNA, mitochondrial DNA; NADH, reduced nicotinamide adenine dinucleotide; NASH, nonalcoholic steatohepatitis; NMR, nuclear magnetic resonance; NRF, nuclear respiratory factor; OXPHOS, oxidative phosphorylation; PGC, PPARγ coactivator; PPAR, peroxisome proliferator-activated receptor; ROS, reactive oxygen species; RQ, respiratory quotient; TCA, tricarboxylic acid; UCP, uncoupling protein.
References
- American Diabetes Association 2008 Diabetes statistics. http://www.diabetes.org/diabetes-basics/diabetes-statistics/ [Google Scholar]
- Centers for Disease Control and Prevention, US Department of Health and Human Services 2007 National diabetes fact sheet. http://www.cdc.gov/diabetes/pubs/factsheet07.htm [Google Scholar]
- Holloszy JO 2009 Skeletal muscle “mitochondrial deficiency” does not mediate insulin resistance. Am J Clin Nutr 89:463S–466S [DOI] [PubMed] [Google Scholar]
- Boyle JP, Honeycutt AA, Narayan KM, Hoerger TJ, Geiss LS, Chen H, Thompson TJ 2001 Projection of diabetes burden through 2050: impact of changing demography and disease prevalence in the U.S. Diabetes Care 24:1936–1940 [DOI] [PubMed] [Google Scholar]
- Roglic G, Unwin N, Bennett PH, Mathers C, Tuomilehto J, Nag S, Connolly V, King H 2005 The burden of mortality attributable to diabetes: realistic estimates for the year 2000. Diabetes Care 28:2130–2135 [DOI] [PubMed] [Google Scholar]
- Ogden CL, Carroll MD, Curtin LR, McDowell MA, Tabak CJ, Flegal KM 2006 Prevalence of overweight and obesity in the United States, 1999–2004. JAMA 295:1549–1555 [DOI] [PubMed] [Google Scholar]
- Hu FB, Manson JE, Stampfer MJ, Colditz G, Liu S, Solomon CG, Willett WC 2001 Diet, lifestyle, and the risk of type 2 diabetes mellitus in women. N Engl J Med 345:790–797 [DOI] [PubMed] [Google Scholar]
- Knowler WC, Barrett-Connor E, Fowler SE, Hamman RF, Lachin JM, Walker EA, Nathan DM 2002 Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med 346:393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomilehto J, Lindström J, Eriksson JG, Valle TT, Hämäläinen H, Ilanne-Parikka P, Keinänen-Kiukaanniemi S, Laakso M, Louheranta A, Rastas M, Salminen V, Uusitupa M 2001 Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N Engl J Med 344:1343–1350 [DOI] [PubMed] [Google Scholar]
- Barroso I, Gurnell M, Crowley VE, Agostini M, Schwabe JW, Soos MA, Maslen GL, Williams TD, Lewis H, Schafer AJ, Chatterjee VK, O'Rahilly S 1999 Dominant negative mutations in human PPARγ associated with severe insulin resistance, diabetes mellitus and hypertension. Nature 402:880–883 [DOI] [PubMed] [Google Scholar]
- Fajans SS, Bell GI, Polonsky KS 2001 Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N Engl J Med 345:971–980 [DOI] [PubMed] [Google Scholar]
- Florez JC, Jablonski KA, Bayley N, Pollin TI, de Bakker PI, Shuldiner AR, Knowler WC, Nathan DM, Altshuler D 2006 TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med 355:241–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, Magnusson KP, Walters GB, Palsdottir E, Jonsdottir T, Gudmundsdottir T, Gylfason A, Saemundsdottir J, Wilensky RL, Reilly MP, Rader DJ, Bagger Y, Christiansen C, Gudnason V, Sigurdsson G, Thorsteinsdottir U, Gulcher JR, Kong A, Stefansson K 2006 Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 38:320–323 [DOI] [PubMed] [Google Scholar]
- Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, Hinokio Y, Lindner TH, Mashima H, Schwarz PE, del Bosque-Plata L, Horikawa Y, Oda Y, Yoshiuchi I, Colilla S, Polonsky KS, Wei S, Concannon P, Iwasaki N, Schulze J, Baier LJ, Bogardus C, Groop L, Boerwinkle E, Hanis CL, Bell GI 2000 Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet 26:163–175 [DOI] [PubMed] [Google Scholar]
- O'Rahilly S, Barroso I, Wareham NJ 2005 Genetic factors in type 2 diabetes: the end of the beginning? Science 307:370–373 [DOI] [PubMed] [Google Scholar]
- Owen KR, McCarthy MI 2007 Genetics of type 2 diabetes. Curr Opin Genet Dev 17:239–244 [DOI] [PubMed] [Google Scholar]
- Knowler WC, Pettitt DJ, Saad MF, Bennett PH 1990 Diabetes mellitus in the Pima Indians: incidence, risk factors and pathogenesis. Diabetes Metab Rev 6:1–27 [DOI] [PubMed] [Google Scholar]
- Poulsen P, Kyvik KO, Vaag A, Beck-Nielsen H 1999 Heritability of type II (non-insulin-dependent) diabetes mellitus and abnormal glucose tolerance—a population-based twin study. Diabetologia 42:139–145 [DOI] [PubMed] [Google Scholar]
- Barker DJ, Hales CN, Fall CH, Osmond C, Phipps K, Clark PM 1993 Type 2 (non-insulin-dependent) diabetes mellitus, hypertension and hyperlipidaemia (syndrome X): relation to reduced fetal growth. Diabetologia 36:62–67 [DOI] [PubMed] [Google Scholar]
- Dabelea D, Hanson RL, Lindsay RS, Pettitt DJ, Imperatore G, Gabir MM, Roumain J, Bennett PH, Knowler WC 2000 Intrauterine exposure to diabetes conveys risks for type 2 diabetes and obesity: a study of discordant sibships. Diabetes 49:2208–2211 [DOI] [PubMed] [Google Scholar]
- Rich-Edwards JW, Colditz GA, Stampfer MJ, Willett WC, Gillman MW, Hennekens CH, Speizer FE, Manson JE 1999 Birthweight and the risk for type 2 diabetes mellitus in adult women. Ann Intern Med 130:278–284 [DOI] [PubMed] [Google Scholar]
- Després JP 1993 Abdominal obesity as important component of insulin-resistance syndrome. Nutrition 9:452–459 [PubMed] [Google Scholar]
- Lazar MA 2005 How obesity causes diabetes: not a tall tale. Science 307:373–375 [DOI] [PubMed] [Google Scholar]
- Manson JE, Rimm EB, Stampfer MJ, Colditz GA, Willett WC, Krolewski AS, Rosner B, Hennekens CH, Speizer FE 1991 Physical activity and incidence of non-insulin-dependent diabetes mellitus in women. Lancet 338:774–778 [DOI] [PubMed] [Google Scholar]
- Buchanan TA, Xiang AH 2005 Gestational diabetes mellitus. J Clin Invest 115:485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lönnroth P, Smith U 1986 Aging enhances the insulin resistance in obesity through both receptor and postreceptor alterations. J Clin Endocrinol Metab 62:433–437 [DOI] [PubMed] [Google Scholar]
- Eriksson J, Franssila-Kallunki A, Ekstrand A, Saloranta C, Widén E, Schalin C, Groop L 1989 Early metabolic defects in persons at increased risk for non-insulin-dependent diabetes mellitus. N Engl J Med 321:337–343 [DOI] [PubMed] [Google Scholar]
- Martin BC, Warram JH, Krolewski AS, Bergman RN, Soeldner JS, Kahn CR 1992 Role of glucose and insulin resistance in development of type 2 diabetes mellitus: results of a 25-year follow-up study. Lancet 340:925–929 [DOI] [PubMed] [Google Scholar]
- Matthews DR, Levy JC 2009 Impending type 2 diabetes. Lancet 373:2178–2179 [DOI] [PubMed] [Google Scholar]
- Goldfine AB, Bouche C, Parker RA, Kim C, Kerivan A, Soeldner JS, Martin BC, Warram JH, Kahn CR 2003 Insulin resistance is a poor predictor of type 2 diabetes in individuals with no family history of disease. Proc Natl Acad Sci USA 100:2724–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK 2008 A mitochondrial protein compendium elucidates complex I disease biology. Cell 134:112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, Patterson N, Lander ES, Mann M 2003 Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115:629–640 [DOI] [PubMed] [Google Scholar]
- Starkov AA 2008 The role of mitochondria in reactive oxygen species metabolism and signaling. Ann NY Acad Sci 1147:37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MP 2009 How mitochondria produce reactive oxygen species. Biochem J 417:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgia M, Giorgi C, Pinton P, Rizzuto R 2009 Controlling metabolism and cell death: at the heart of mitochondrial calcium signalling. J Mol Cell Cardiol 46:781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimessi A, Giorgi C, Pinton P, Rizzuto R 2008 The versatility of mitochondrial calcium signals: from stimulation of cell metabolism to induction of cell death. Biochim Biophys Acta 1777:808–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R, Mühlenhoff U 2008 Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu Rev Biochem 77:669–700 [DOI] [PubMed] [Google Scholar]
- Miller WL 2008 Steroidogenic enzymes. Endocr Dev 13:1–18 [DOI] [PubMed] [Google Scholar]
- Miller WL 2007 Steroidogenic acute regulatory protein (StAR), a novel mitochondrial cholesterol transporter. Biochim Biophys Acta 1771:663–676 [DOI] [PubMed] [Google Scholar]
- Miller WL 2005 Minireview: regulation of steroidogenesis by electron transfer. Endocrinology 146:2544–2550 [DOI] [PubMed] [Google Scholar]
- Gonzalez IL 2005 Barth syndrome: TAZ gene mutations, mRNAs, and evolution. Am J Med Genet A 134:409–414 [DOI] [PubMed] [Google Scholar]
- Muravchick S 2008 Clinical implications of mitochondrial disease. Adv Drug Deliv Rev 60:1553–1560 [DOI] [PubMed] [Google Scholar]
- Gargus JJ 2009 Genetic calcium signaling abnormalities in the central nervous system: seizures, migraine, and autism. Ann NY Acad Sci 1151:133–156 [DOI] [PubMed] [Google Scholar]
- Gandhi S, Wood-Kaczmar A, Yao Z, Plun-Favreau H, Deas E, Klupsch K, Downward J, Latchman DS, Tabrizi SJ, Wood NW, Duchen MR, Abramov AY 2009 PINK1-associated Parkinson’s disease is caused by neuronal vulnerability to calcium-induced cell death. Mol Cell 33:627–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard G, Rossignol R 2008 Ultrastructure of the mitochondrion and its bearing on function and bioenergetics. Antioxid Redox Signal 10:1313–1342 [DOI] [PubMed] [Google Scholar]
- Stuart RA 2008 Supercomplex organization of the oxidative phosphorylation enzymes in yeast mitochondria. J Bioenerg Biomembr 40:411–417 [DOI] [PubMed] [Google Scholar]
- Vonck J, Schäfer E 2009 Supramolecular organization of protein complexes in the mitochondrial inner membrane. Biochim Biophys Acta 1793:117–124 [DOI] [PubMed] [Google Scholar]
- Shaw JM, Nunnari J 2002 Mitochondrial dynamics and division in budding yeast. Trends Cell Biol 12:178–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chan DC 2005 Emerging functions of mammalian mitochondrial fusion and fission. Hum Mol Genet 14 Spec No. 2:R283–R289 [DOI] [PubMed] [Google Scholar]
- Parone PA, Da Cruz S, Tondera D, Mattenberger Y, James DI, Maechler P, Barja F, Martinou JC 2008 Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS ONE 3:e3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubannier V, McBride HM 2009 Positioning mitochondrial plasticity within cellular signaling cascades. Biochim Biophys Acta 1793:154–170 [DOI] [PubMed] [Google Scholar]
- Bach D, Pich S, Soriano FX, Vega N, Baumgartner B, Oriola J, Daugaard JR, Lloberas J, Camps M, Zierath JR, Rabasa-Lhoret R, Wallberg-Henriksson H, Laville M, Palacín M, Vidal H, Rivera F, Brand M, Zorzano A 2003 Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism. A novel regulatory mechanism altered in obesity. J Biol Chem 278:17190–17197 [DOI] [PubMed] [Google Scholar]
- Scarpulla RC 1997 Nuclear control of respiratory chain expression in mammalian cells. J Bioenerg Biomembr 29:109–119 [DOI] [PubMed] [Google Scholar]
- Virbasius JV, Scarpulla RC 1994 Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc Natl Acad Sci USA 91:1309–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC 2008 Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev 88:611–638 [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM 1999 Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98:115–124 [DOI] [PubMed] [Google Scholar]
- Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman BM 2002 Peroxisome proliferator-activated receptor γ coactivator 1β (PGC-1β), a novel PGC-1-related transcription coactivator associated with host cell factor. J Biol Chem 277:1645–1648 [DOI] [PubMed] [Google Scholar]
- Andersson U, Scarpulla RC 2001 Pgc-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Mol Cell Biol 21:3738–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Mehl IR, Chong LW, Nofsinger RR, Evans RM 2007 PGC-1β controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proc Natl Acad Sci USA 104:5223–5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leick L, Wojtaszewski JF, Johansen ST, Kiilerich K, Comes G, Hellsten Y, Hidalgo J, Pilegaard H 2008 PGC-1α is not mandatory for exercise- and training-induced adaptive gene responses in mouse skeletal muscle. Am J Physiol Endocrinol Metab 294:E463–E474 [DOI] [PubMed] [Google Scholar]
- Lehman JJ, Boudina S, Banke NH, Sambandam N, Han X, Young DM, Leone TC, Gross RW, Lewandowski ED, Abel ED, Kelly DP 2008 The transcriptional coactivator PGC-1α is essential for maximal and efficient cardiac mitochondrial fatty acid oxidation and lipid homeostasis. Am J Physiol Heart Circ Physiol 295:H185–H196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai L, Leone TC, Zechner C, Schaeffer PJ, Kelly SM, Flanagan DP, Medeiros DM, Kovacs A, Kelly DP 2008 Transcriptional coactivators PGC-1α and PGC-lβ control overlapping programs required for perinatal maturation of the heart. Genes Dev 22:1948–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C, Chin S, Li P, Liu F, Maratos-Flier E, Lebrasseur NK, Yan Z, Spiegelman BM 2007 Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1α muscle-specific knock-out animals. J Biol Chem 282:30014–30021 [DOI] [PubMed] [Google Scholar]
- Handschin C, Choi CS, Chin S, Kim S, Kawamori D, Kurpad AJ, Neubauer N, Hu J, Mootha VK, Kim YB, Kulkarni RN, Shulman GI, Spiegelman BM 2007 Abnormal glucose homeostasis in skeletal muscle-specific PGC-1α knockout mice reveals skeletal muscle-pancreatic β-cell crosstalk. J Clin Invest 117:3463–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vianna CR, Huntgeburth M, Coppari R, Choi CS, Lin J, Krauss S, Barbatelli G, Tzameli I, Kim YB, Cinti S, Shulman GI, Spiegelman BM, Lowell BB 2006 Hypomorphic mutation of PGC-1β causes mitochondrial dysfunction and liver insulin resistance. Cell Metab 4:453–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelliott CJ, Medina-Gomez G, Petrovic N, Kis A, Feldmann HM, Bjursell M, Parker N, Curtis K, Campbell M, Hu P, Zhang D, Litwin SE, Zaha VG, Fountain KT, Boudina S, Jimenez-Linan M, Blount M, Lopez M, Meirhaeghe A, Bohlooly-Y M, Storlien L, Strömstedt M, Snaith M, Oresic M, Abel ED, Cannon B, Vidal-Puig A 2006 Ablation of PGC-1β results in defective mitochondrial activity, thermogenesis, hepatic function, and cardiac performance. PLoS Biol 4:e369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, Chen Z, Holloszy JO, Medeiros DM, Schmidt RE, Saffitz JE, Abel ED, Semenkovich CF, Kelly DP 2005 PGC-1α deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol 3:e101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arany Z, He H, Lin J, Hoyer K, Handschin C, Toka O, Ahmad F, Matsui T, Chin S, Wu PH, Rybkin II, Shelton JM, Manieri M, Cinti S, Schoen FJ, Bassel-Duby R, Rosenzweig A, Ingwall JS, Spiegelman BM 2005 Transcriptional coactivator PGC-1 α controls the energy state and contractile function of cardiac muscle. Cell Metab 1:259–271 [DOI] [PubMed] [Google Scholar]
- Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jäger S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan MC, Rohas LM, Zavacki AM, Cinti S, Shulman GI, Lowell BB, Krainc D, Spiegelman BM 2004 Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell 119:121–135 [DOI] [PubMed] [Google Scholar]
- Vercauteren K, Gleyzer N, Scarpulla RC 2009 Short hairpin RNA-mediated silencing of PRC (PGC-1-related coactivator) results in a severe respiratory chain deficiency associated with the proliferation of aberrant mitochondria. J Biol Chem 284:2307–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, White R, Parker MG 2006 Metabolic regulation by the nuclear receptor corepressor RIP140. Trends Endocrinol Metab 17:243–250 [DOI] [PubMed] [Google Scholar]
- Powelka AM, Seth A, Virbasius JV, Kiskinis E, Nicoloro SM, Guilherme A, Tang X, Straubhaar J, Cherniack AD, Parker MG, Czech MP 2006 Suppression of oxidative metabolism and mitochondrial biogenesis by the transcriptional corepressor RIP140 in mouse adipocytes. J Clin Invest 116:125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debevec D, Christian M, Morganstein D, Seth A, Herzog B, Parker M, White R 2007 Receptor interacting protein 140 regulates expression of uncoupling protein 1 in adipocytes through specific peroxisome proliferator activated receptor isoforms and estrogen-related receptor α. Mol Endocrinol 21:1581–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardsson G, Steel JH, Christian M, Pocock V, Milligan S, Bell J, So PW, Medina-Gomez G, Vidal-Puig A, White R, Parker MG 2004 Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc Natl Acad Sci USA 101:8437–8442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MG, Christian M, White R 2006 The nuclear receptor co-repressor RIP140 controls the expression of metabolic gene networks. Biochem Soc Trans 34:1103–1106 [DOI] [PubMed] [Google Scholar]
- Seth A, Steel JH, Nichol D, Pocock V, Kumaran MK, Fritah A, Mobberley M, Ryder TA, Rowlerson A, Scott J, Poutanen M, White R, Parker M 2007 The transcriptional corepressor RIP140 regulates oxidative metabolism in skeletal muscle. Cell Metab 6:236–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MJ, Frazier AE, Gulbis JM, Ryan MT 2007 Mitochondrial protein-import machinery: correlating structure with function. Trends Cell Biol 17:456–464 [DOI] [PubMed] [Google Scholar]
- MacKenzie JA, Payne RM 2007 Mitochondrial protein import and human health and disease. Biochim Biophys Acta 1772:509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D, Neupert W 2008 Energetics of protein translocation into mitochondria. Biochim Biophys Acta 1777:758–762 [DOI] [PubMed] [Google Scholar]
- Walther DM, Rapaport D 2009 Biogenesis of mitochondrial outer membrane proteins. Biochim Biophys Acta 1793:42–51 [DOI] [PubMed] [Google Scholar]
- Hatch GM 2004 Cell biology of cardiac mitochondrial phospholipids. Biochem Cell Biol 82:99–112 [DOI] [PubMed] [Google Scholar]
- Mileykovskaya E, Zhang M, Dowhan W 2005 Cardiolipin in energy transducing membranes. Biochemistry (Mosc) 70:154–158 [DOI] [PubMed] [Google Scholar]
- Schlame M, Rua D, Greenberg ML 2000 The biosynthesis and functional role of cardiolipin. Prog Lipid Res 39:257–288 [DOI] [PubMed] [Google Scholar]
- Osman C, Haag M, Potting C, Rodenfels J, Dip PV, Wieland FT, Brügger B, Westermann B, Langer T 2009 The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J Cell Biol 184:583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutik S, Rissler M, Guan XL, Guiard B, Shui G, Gebert N, Heacock PN, Rehling P, Dowhan W, Wenk MR, Pfanner N, Wiedemann N 2008 The translocator maintenance protein Tam41 is required for mitochondrial cardiolipin biosynthesis. J Cell Biol 183:1213–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Condell M, Plesken H, Edelman-Novemsky I, Ma J, Ren M, Schlame M 2006 A Drosophila model of Barth syndrome. Proc Natl Acad Sci USA 103:11584–11588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Pfannschmidt S, Guiard B, Stojanovski D, Milenkovic D, Kutik S, Pfanner N, Meisinger C, Wiedemann N 2008 Biogenesis of the mitochondrial TOM complex: Mim1 promotes insertion and assembly of signal-anchored receptors. J Biol Chem 283:120–127 [DOI] [PubMed] [Google Scholar]
- Claypool SM, Oktay Y, Boontheung P, Loo JA, Koehler CM 2008 Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J Cell Biol 182:937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Sutachan JJ, Plesken H, Kelley RI, Schlame M 2005 Characterization of lymphoblast mitochondria from patients with Barth syndrome. Lab Invest 85:823–830 [DOI] [PubMed] [Google Scholar]
- Luft R, Ikkos D, Palmieri G, Ernster L, Afzelius B 1962 A case of severe hypermetabolism of nonthyroid origin with a defect in the maintenance of mitochondrial respiratory control: a correlated clinical, biochemical, and morphological study. J Clin Invest 41:1776–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducluzeau PH, Perretti N, Laville M, Andreelli F, Vega N, Riou JP, Vidal H 2001 Regulation by insulin of gene expression in human skeletal muscle and adipose tissue. Evidence for specific defects in type 2 diabetes. Diabetes 50:1134–1142 [DOI] [PubMed] [Google Scholar]
- Vaag A, Henriksen JE, Beck-Nielsen H 1992 Decreased insulin activation of glycogen synthase in skeletal muscles in young nonobese Caucasian first-degree relatives of patients with non-insulin-dependent diabetes mellitus. J Clin Invest 89:782–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratipanawatr W, Pratipanawatr T, Cusi K, Berria R, Adams JM, Jenkinson CP, Maezono K, DeFronzo RA, Mandarino LJ 2001 Skeletal muscle insulin resistance in normoglycemic subjects with a strong family history of type 2 diabetes is associated with decreased insulin-stimulated insulin receptor substrate-1 tyrosine phosphorylation. Diabetes 50:2572–2578 [DOI] [PubMed] [Google Scholar]
- Jacob S, Machann J, Rett K, Brechtel K, Volk A, Renn W, Maerker E, Matthaei S, Schick F, Claussen CD, Häring HU 1999 Association of increased intramyocellular lipid content with insulin resistance in lean nondiabetic offspring of type 2 diabetic subjects. Diabetes 48:1113–1119 [DOI] [PubMed] [Google Scholar]
- Malenfant P, Joanisse DR, Thériault R, Goodpaster BH, Kelley DE, Simoneau JA 2001 Fat content in individual muscle fibers of lean and obese subjects. Int J Obes Relat Metab Disord 25:1316–1321 [DOI] [PubMed] [Google Scholar]
- Dresner A, Laurent D, Marcucci M, Griffin ME, Dufour S, Cline GW, Slezak LA, Andersen DK, Hundal RS, Rothman DL, Petersen KF, Shulman GI 1999 Effects of free fatty acids on glucose transport and IRS-1-associated phosphatidylinositol 3-kinase activity. J Clin Invest 103:253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DB, Petersen KF, Shulman GI 2007 Disordered lipid metabolism and the pathogenesis of insulin resistance. Physiol Rev 87:507–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, Bain J, Stevens R, Dyck JR, Newgard CB, Lopaschuk GD, Muoio DM 2008 Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab 7:45–56 [DOI] [PubMed] [Google Scholar]
- Holland WL, Brozinick JT, Wang LP, Hawkins ED, Sargent KM, Liu Y, Narra K, Hoehn KL, Knotts TA, Siesky A, Nelson DH, Karathanasis SK, Fontenot GK, Birnbaum MJ, Summers SA 2007 Inhibition of ceramide synthesis ameliorates glucocorticoid-, saturated-fat-, and obesity-induced insulin resistance. Cell Metab 5:167–179 [DOI] [PubMed] [Google Scholar]
- Yu C, Chen Y, Cline GW, Zhang D, Zong H, Wang Y, Bergeron R, Kim JK, Cushman SW, Cooney GJ, Atcheson B, White MF, Kraegen EW, Shulman GI 2002 Mechanism by which fatty acids inhibit insulin activation of insulin receptor substrate-1 (IRS-1)-associated phosphatidylinositol 3-kinase activity in muscle. J Biol Chem 277:50230–50236 [DOI] [PubMed] [Google Scholar]
- Chibalin AV, Leng Y, Vieira E, Krook A, Björnholm M, Long YC, Kotova O, Zhong Z, Sakane F, Steiler T, Nylén C, Wang J, Laakso M, Topham MK, Gilbert M, Wallberg-Henriksson H, Zierath JR 2008 Downregulation of diacylglycerol kinase δ contributes to hyperglycemia-induced insulin resistance. Cell 132:375–386 [DOI] [PubMed] [Google Scholar]
- Houstis N, Rosen ED, Lander ES 2006 Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature 440:944–948 [DOI] [PubMed] [Google Scholar]
- Lanner JT, Bruton JD, Katz A, Westerblad H 2008 Ca(2+) and insulin-mediated glucose uptake. Curr Opin Pharmacol 8:339–345 [DOI] [PubMed] [Google Scholar]
- Lebeche D, Davidoff AJ, Hajjar RJ 2008 Interplay between impaired calcium regulation and insulin signaling abnormalities in diabetic cardiomyopathy. Nat Clin Pract Cardiovasc Med 5:715–724 [DOI] [PubMed] [Google Scholar]
- Park S, Scheffler TL, Gunawan AM, Shi H, Zeng C, Hannon KM, Grant AL, Gerrard DE 2009 Chronic elevated calcium blocks AMPK-induced GLUT-4 expression in skeletal muscle. Am J Physiol Cell Physiol 296:C106–C115 [DOI] [PubMed] [Google Scholar]
- Kacerovsky-Bielesz G, Chmelik M, Ling C, Pokan R, Szendroedi J, Farukuoye M, Kacerovsky M, Schmid AI, Gruber S, Wolzt M, Moser E, Pacini G, Smekal G, Groop L, Roden M 2009 Short-term exercise training does not stimulate skeletal muscle ATP synthesis in relatives of humans with type 2 diabetes. Diabetes 58:1333–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoneau JA, Colberg SR, Thaete FL, Kelley DE 1995 Skeletal muscle glycolytic and oxidative enzyme capacities are determinants of insulin sensitivity and muscle composition in obese women. FASEB J 9:273–278 [PubMed] [Google Scholar]
- Simoneau JA, Bouchard C 1995 Skeletal muscle metabolism and body fat content in men and women. Obes Res 3:23–29 [DOI] [PubMed] [Google Scholar]
- Simoneau JA, Kelley DE 1997 Altered glycolytic and oxidative capacities of skeletal muscle contribute to insulin resistance in NIDDM. J Appl Physiol 83:166–171 [DOI] [PubMed] [Google Scholar]
- Bruce CR, Anderson MJ, Carey AL, Newman DG, Bonen A, Kriketos AD, Cooney GJ, Hawley JA 2003 Muscle oxidative capacity is a better predictor of insulin sensitivity than lipid status. J Clin Endocrinol Metab 88:5444–5451 [DOI] [PubMed] [Google Scholar]
- Kelley DE, Goodpaster B, Wing RR, Simoneau JA 1999 Skeletal muscle fatty acid metabolism in association with insulin resistance, obesity, and weight loss. Am J Physiol 277:E1130–E1141 [DOI] [PubMed] [Google Scholar]
- Kelley DE, Mandarino LJ 2000 Fuel selection in human skeletal muscle in insulin resistance: a reexamination. Diabetes 49:677–683 [DOI] [PubMed] [Google Scholar]
- Zurlo F, Lillioja S, Esposito-Del Puente A, Nyomba BL, Raz I, Saad MF, Swinburn BA, Knowler WC, Bogardus C, Ravussin E 1990 Low ratio of fat to carbohydrate oxidation as predictor of weight gain: study of 24-h RQ. Am J Physiol 259:E650–E657 [DOI] [PubMed] [Google Scholar]
- Ritov VB, Menshikova EV, He J, Ferrell RE, Goodpaster BH, Kelley DE 2005 Deficiency of subsarcolemmal mitochondria in obesity and type 2 diabetes. Diabetes 54:8–14 [DOI] [PubMed] [Google Scholar]
- Boushel R, Gnaiger E, Schjerling P, Skovbro M, Kraunsøe R, Dela F 2007 Patients with type 2 diabetes have normal mitochondrial function in skeletal muscle. Diabetologia 50:790–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DE, He J, Menshikova EV, Ritov VB 2002 Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 51:2944–2950 [DOI] [PubMed] [Google Scholar]
- Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI 2004 Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med 350:664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Befroy DE, Petersen KF, Dufour S, Mason GF, de Graaf RA, Rothman DL, Shulman GI 2007 Impaired mitochondrial substrate oxidation in muscle of insulin-resistant offspring of type 2 diabetic patients. Diabetes 56:1376–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen M, Sahlin K, Fernström M, Glintborg D, Vind BF, Beck-Nielsen H, Højlund K 2007 Mitochondrial respiration is decreased in skeletal muscle of patients with type 2 diabetes. Diabetes 56:1592–1599 [DOI] [PubMed] [Google Scholar]
- Petersen KF, Dufour S, Shulman GI 2005 Decreased insulin-stimulated ATP synthesis and phosphate transport in muscle of insulin-resistant offspring of type 2 diabetic parents. PLoS Med 2:e233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC 2003 PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34:267–273 [DOI] [PubMed] [Google Scholar]
- Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, Landaker EJ, Goldfine AB, Mun E, DeFronzo R, Finlayson J, Kahn CR, Mandarino LJ 2003 Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc Natl Acad Sci USA 100:8466–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekumar R, Halvatsiotis P, Schimke JC, Nair KS 2002 Gene expression profile in skeletal muscle of type 2 diabetes and the effect of insulin treatment. Diabetes 51:1913–1920 [DOI] [PubMed] [Google Scholar]
- Morino K, Petersen KF, Dufour S, Befroy D, Frattini J, Shatzkes N, Neschen S, White MF, Bilz S, Sono S, Pypaert M, Shulman GI 2005 Reduced mitochondrial density and increased IRS-1 serine phosphorylation in muscle of insulin-resistant offspring of type 2 diabetic parents. J Clin Invest 115:3587–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair KS, Bigelow ML, Asmann YW, Chow LS, Coenen-Schimke JM, Klaus KA, Guo ZK, Sreekumar R, Irving BA 2008 Asian Indians have enhanced skeletal muscle mitochondrial capacity to produce ATP in association with severe insulin resistance. Diabetes 57:1166–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederiksen CM, Højlund K, Hansen L, Oakeley EJ, Hemmings B, Abdallah BM, Brusgaard K, Beck-Nielsen H, Gaster M 2008 Transcriptional profiling of myotubes from patients with type 2 diabetes: no evidence for a primary defect in oxidative phosphorylation genes. Diabetologia 51:2068–2077 [DOI] [PubMed] [Google Scholar]
- Crunkhorn S, Dearie F, Mantzoros C, Gami H, da Silva WS, Espinoza D, Faucette R, Barry K, Bianco AC, Patti ME 2007 Peroxisome proliferator activator receptor γ coactivator-1 expression is reduced in obesity: potential pathogenic role of saturated fatty acids and p38 mitogen-activated protein kinase activation. J Biol Chem 282:15439–15450 [DOI] [PubMed] [Google Scholar]
- Sparks LM, Xie H, Koza RA, Mynatt R, Hulver MW, Bray GA, Smith SR 2005 A high-fat diet coordinately downregulates genes required for mitochondrial oxidative phosphorylation in skeletal muscle. Diabetes 54:1926–1933 [DOI] [PubMed] [Google Scholar]
- Richardson DK, Kashyap S, Bajaj M, Cusi K, Mandarino SJ, Finlayson J, DeFronzo RA, Jenkinson CP, Mandarino LJ 2005 Lipid infusion decreases the expression of nuclear encoded mitochondrial genes and increases the expression of extracellular matrix genes in human skeletal muscle. J Biol Chem 280:10290–10297 [DOI] [PubMed] [Google Scholar]
- Garcia-Roves P, Huss JM, Han DH, Hancock CR, Iglesias-Gutierrez E, Chen M, Holloszy JO 2007 Raising plasma fatty acid concentration induces increased biogenesis of mitochondria in skeletal muscle. Proc Natl Acad Sci USA 104:10709–10713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock CR, Han DH, Chen M, Terada S, Yasuda T, Wright DC, Holloszy JO 2008 High-fat diets cause insulin resistance despite an increase in muscle mitochondria. Proc Natl Acad Sci USA 105:7815–7820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekumar R, Unnikrishnan J, Fu A, Nygren J, Short KR, Schimke J, Barazzoni R, Nair KS 2002 Effects of caloric restriction on mitochondrial function and gene transcripts in rat muscle. Am J Physiol Endocrinol Metab 283:E38–E43 [DOI] [PubMed] [Google Scholar]
- Parikh H, Nilsson E, Ling C, Poulsen P, Almgren P, Nittby H, Eriksson KF, Vaag A, Groop LC 2008 Molecular correlates for maximal oxygen uptake and type 1 fibers. Am J Physiol Endocrinol Metab 294:E1152–E1159 [DOI] [PubMed] [Google Scholar]
- Wisløff U, Najjar SM, Ellingsen O, Haram PM, Swoap S, Al-Share Q, Fernström M, Rezaei K, Lee SJ, Koch LG, Britton SL 2005 Cardiovascular risk factors emerge after artificial selection for low aerobic capacity. Science 307:418–420 [DOI] [PubMed] [Google Scholar]
- Wright DC, Han DH, Garcia-Roves PM, Geiger PC, Jones TE, Holloszy JO 2007 Exercise-induced mitochondrial biogenesis begins before the increase in muscle PGC-1α expression. J Biol Chem 282:194–199 [DOI] [PubMed] [Google Scholar]
- Rönn T, Poulsen P, Hansson O, Holmkvist J, Almgren P, Nilsson P, Tuomi T, Isomaa B, Groop L, Vaag A, Ling C 2008 Age influences DNA methylation and gene expression of COX7A1 in human skeletal muscle. Diabetologia 51:1159–1168 [DOI] [PubMed] [Google Scholar]
- Ling C, Poulsen P, Carlsson E, Ridderstråle M, Almgren P, Wojtaszewski J, Beck-Nielsen H, Groop L, Vaag A 2004 Multiple environmental and genetic factors influence skeletal muscle PGC-1α and PGC-1β gene expression in twins. J Clin Invest 114:1518–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönn T, Poulsen P, Tuomi T, Isomaa B, Groop L, Vaag A, Ling C 2009 Genetic variation in ATP5O is associated with skeletal muscle ATP50 mRNA expression and glucose uptake in young twins. PLoS ONE 4:e4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C, Groop L 2009 Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes 58:2718–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen KF, Befroy D, Dufour S, Dziura J, Ariyan C, Rothman DL, DiPietro L, Cline GW, Shulman GI 2003 Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science 300:1140–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yechoor VK, Patti ME, Saccone R, Kahn CR 2002 Coordinated patterns of gene expression for substrate and energy metabolism in skeletal muscle of diabetic mice. Proc Natl Acad Sci USA 99:10587–10592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakelides H, Asmann YW, Bigelow ML, Short KR, Dhatariya K, Coenen-Schimke J, Kahl J, Mukhopadhyay D, Nair KS 2007 Effect of insulin deprivation on muscle mitochondrial ATP production and gene transcript levels in type 1 diabetic subjects. Diabetes 56:2683–2689 [DOI] [PubMed] [Google Scholar]
- Meugnier E, Faraj M, Rome S, Beauregard G, Michaut A, Pelloux V, Chiasson JL, Laville M, Clement K, Vidal H, Rabasa-Lhoret R 2007 Acute hyperglycemia induces a global downregulation of gene expression in adipose tissue and skeletal muscle of healthy subjects. Diabetes 56:992–999 [DOI] [PubMed] [Google Scholar]
- Rabøl R, Højberg PM, Almdal T, Boushel R, Haugaard SB, Madsbad S, Dela F 2009 Effect of hyperglycemia on mitochondrial respiration in type 2 diabetes. J Clin Endocrinol Metab 94:1372–1378 [DOI] [PubMed] [Google Scholar]
- Southgate RJ, Bruce CR, Carey AL, Steinberg GR, Walder K, Monks R, Watt MJ, Hawley JA, Birnbaum MJ, Febbraio MA 2005 PGC-1α gene expression is down-regulated by Akt-mediated phosphorylation and nuclear exclusion of FoxO1 in insulin-stimulated skeletal muscle. FASEB J 19:2072–2074 [DOI] [PubMed] [Google Scholar]
- Barrès R, Osler ME, Yan J, Rune A, Fritz T, Caidahl K, Krook A, Zierath JR 2009 Non-CpG methylation of the PGC-1α promoter through DNMT3B controls mitochondrial density. Cell Metab 10:189–198 [DOI] [PubMed] [Google Scholar]
- Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P 2006 GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1α. Cell Metab 3:429–438 [DOI] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J 2006 Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell 127:1109–1122 [DOI] [PubMed] [Google Scholar]
- Astrup A 1986 Thermogenesis in human brown adipose tissue and skeletal muscle induced by sympathomimetic stimulation. Acta Endocrinol Suppl (Copenh) 278:1–32 [PubMed] [Google Scholar]
- Cannon B, Nedergaard J 1985 The biochemistry of an inefficient tissue: brown adipose tissue. Essays Biochem 20:110–164 [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, Kolodny GM, Kahn CR 2009 Identification and importance of brown adipose tissue in adult humans. N Engl J Med 360:1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber WA 2004 Brown adipose tissue and nuclear medicine imaging. J Nucl Med 45:1101–1103 [PubMed] [Google Scholar]
- Bouillaud F, Ricquier D, Thibault J, Weissenbach J 1985 Molecular approach to thermogenesis in brown adipose tissue: cDNA cloning of the mitochondrial uncoupling protein. Proc Natl Acad Sci USA 82:445–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsson A, Stadler U, Glotzer MA, Kozak LP 1985 Mitochondrial uncoupling protein from mouse brown fat. Molecular cloning, genetic mapping, and mRNA expression. J Biol Chem 260:16250–16254 [PubMed] [Google Scholar]
- Ricquier D, Bouillaud F 1997 The mitochondrial uncoupling protein: structural and genetic studies. Prog Nucleic Acid Res Mol Biol 56:83–108 [DOI] [PubMed] [Google Scholar]
- Luo GF, Yu TY, Wen XH, Li Y, Yang GS 2008 Alteration of mitochondrial oxidative capacity during porcine preadipocyte differentiation and in response to leptin. Mol Cell Biochem 307:83–91 [DOI] [PubMed] [Google Scholar]
- Kim BW, Choo HJ, Lee JW, Kim JH, Ko YG 2004 Extracellular ATP is generated by ATP synthase complex in adipocyte lipid rafts. Exp Mol Med 36:476–485 [DOI] [PubMed] [Google Scholar]
- Wilson-Fritch L, Burkart A, Bell G, Mendelson K, Leszyk J, Nicoloro S, Czech M, Corvera S 2003 Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol Cell Biol 23:1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen OE, Kalhan SC, Hanson RW 2002 The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem 277:30409–30412 [DOI] [PubMed] [Google Scholar]
- Koh EH, Park JY, Park HS, Jeon MJ, Ryu JW, Kim M, Kim SY, Kim MS, Kim SW, Park IS, Youn JH, Lee KU 2007 Essential role of mitochondrial function in adiponectin synthesis in adipocytes. Diabetes 56:2973–2981 [DOI] [PubMed] [Google Scholar]
- Shi X, Burkart A, Nicoloro SM, Czech MP, Straubhaar J, Corvera S 2008 Paradoxical effect of mitochondrial respiratory chain impairment on insulin signaling and glucose transport in adipose cells. J Biol Chem 283:30658–30667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marette A, Tulp OL, Bukowiecki LJ 1991 Mechanism linking insulin resistance to defective thermogenesis in brown adipose tissue of obese diabetic SHR/N-cp rats. Int J Obes 15:823–831 [PubMed] [Google Scholar]
- Nedergaard J, Bengtsson T, Cannon B 2007 Unexpected evidence for active brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab 293:E444–E452 [DOI] [PubMed] [Google Scholar]
- Saito M, Okamatsu-Ogura Y, Matsushita M, Watanabe K, Yoneshiro T, Nio-Kobayashi J, Iwanaga T, Miyagawa M, Kameya T, Nakada K, Kawai Y, Tsujisaki M 2009 High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes 58:1526–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ 2009 Cold-activated brown adipose tissue in healthy men. N Engl J Med 360:1500–1508 [DOI] [PubMed] [Google Scholar]
- Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerbäck S, Nuutila P 2009 Functional brown adipose tissue in healthy adults. N Engl J Med 360:1518–1525 [DOI] [PubMed] [Google Scholar]
- Zingaretti MC, Crosta F, Vitali A, Guerrieri M, Frontini A, Cannon B, Nedergaard J, Cinti S 2009 The presence of UCP1 demonstrates that metabolically active adipose tissue in the neck of adult humans truly represents brown adipose tissue. FASEB J 23:3113–3120 [DOI] [PubMed] [Google Scholar]
- Cinti S 2002 Adipocyte differentiation and transdifferentiation: plasticity of the adipose organ. J Endocrinol Invest 25:823–835 [DOI] [PubMed] [Google Scholar]
- Deveaud C, Beauvoit B, Salin B, Schaeffer J, Rigoulet M 2004 Regional differences in oxidative capacity of rat white adipose tissue are linked to the mitochondrial content of mature adipocytes. Mol Cell Biochem 267:157–166 [DOI] [PubMed] [Google Scholar]
- Bogacka I, Xie H, Bray GA, Smith SR 2005 Pioglitazone induces mitochondrial biogenesis in human subcutaneous adipose tissue in vivo. Diabetes 54:1392–1399 [DOI] [PubMed] [Google Scholar]
- Himms-Hagen J, Melnyk A, Zingaretti MC, Ceresi E, Barbatelli G, Cinti S 2000 Multilocular fat cells in WAT of CL-316243-treated rats derive directly from white adipocytes. Am J Physiol Cell Physiol 279:C670–C681 [DOI] [PubMed] [Google Scholar]
- Loncar D 1991 Convertible adipose tissue in mice. Cell Tissue Res 266:149–161 [DOI] [PubMed] [Google Scholar]
- Orci L, Cook WS, Ravazzola M, Wang MY, Park BH, Montesano R, Unger RH 2004 Rapid transformation of white adipocytes into fat-oxidizing machines. Proc Natl Acad Sci USA 101:2058–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ström K, Hansson O, Lucas S, Nevsten P, Fernandez C, Klint C, Movérare-Skrtic S, Sundler F, Ohlsson C, Holm C 2008 Attainment of brown adipocyte features in white adipocytes of hormone-sensitive lipase null mice. PLoS ONE 3:e1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiraby C, Tavernier G, Lefort C, Larrouy D, Bouillaud F, Ricquier D, Langin D 2003 Acquirement of brown fat cell features by human white adipocytes. J Biol Chem 278:33370–33376 [DOI] [PubMed] [Google Scholar]
- Toh SY, Gong J, Du G, Li JZ, Yang S, Ye J, Yao H, Zhang Y, Xue B, Li Q, Yang H, Wen Z, Li P 2008 Up-regulation of mitochondrial activity and acquirement of brown adipose tissue-like property in the white adipose tissue of fsp27 deficient mice. PLoS ONE 3:e2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Fritch L, Nicoloro S, Chouinard M, Lazar MA, Chui PC, Leszyk J, Straubhaar J, Czech MP, Corvera S 2004 Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J Clin Invest 114:1281–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maassen JA, 't Hart LM, Janssen GM, Reiling E, Romijn JA, Lemkes HH 2006 Mitochondrial diabetes and its lessons for common type 2 diabetes. Biochem Soc Trans 34:819–823 [DOI] [PubMed] [Google Scholar]
- Maasen JA 2008 Mitochondria, body fat and type 2 diabetes: what is the connection? Minerva Med 99:241–251 [PubMed] [Google Scholar]
- Heilbronn L, Smith SR, Ravussin E 2004 Failure of fat cell proliferation, mitochondrial function and fat oxidation results in ectopic fat storage, insulin resistance and type II diabetes mellitus. Int J Obes Relat Metab Disord 28(Suppl 4):S12–S21 [DOI] [PubMed] [Google Scholar]
- Frayn KN, Langin D, Karpe F 2008 Fatty acid-induced mitochondrial uncoupling in adipocytes is not a promising target for treatment of insulin resistance unless adipocyte oxidative capacity is increased. Diabetologia 51:394–397 [DOI] [PubMed] [Google Scholar]
- de Ferranti S, Mozaffarian D 2008 The perfect storm: obesity, adipocyte dysfunction, and metabolic consequences. Clin Chem 54:945–955 [DOI] [PubMed] [Google Scholar]
- Rossmeisl M, Flachs P, Brauner P, Sponarova J, Matejkova O, Prazak T, Ruzickova J, Bardova K, Kuda O, Kopecky J 2004 Role of energy charge and AMP-activated protein kinase in adipocytes in the control of body fat stores. Int J Obes Relat Metab Disord 28(Suppl 4):S38–S44 [DOI] [PubMed] [Google Scholar]
- Harper JA, Dickinson K, Brand MD 2001 Mitochondrial uncoupling as a target for drug development for the treatment of obesity. Obes Rev 2:255–265 [DOI] [PubMed] [Google Scholar]
- Stefl B, Janovská A, Hodný Z, Rossmeisl M, Horáková M, Syrový I, Bémová J, Bendlová B, Kopecký J 1998 Brown fat is essential for cold-induced thermogenesis but not for obesity resistance in aP2-Ucp mice. Am J Physiol 274:E527–E533 [DOI] [PubMed] [Google Scholar]
- Rossmeisl M, Syrový I, Baumruk F, Flachs P, Janovská P, Kopecký J 2000 Decreased fatty acid synthesis due to mitochondrial uncoupling in adipose tissue. FASEB J 14: 1793–1800 [DOI] [PubMed] [Google Scholar]
- Si Y, Palani S, Jayaraman A, Lee K 2007 Effects of forced uncoupling protein 1 expression in 3T3–L1 cells on mitochondrial function and lipid metabolism. J Lipid Res 48:826–836 [DOI] [PubMed] [Google Scholar]
- Choo HJ, Kim JH, Kwon OB, Lee CS, Mun JY, Han SS, Yoon YS, Yoon G, Choi KM, Ko YG 2006 Mitochondria are impaired in the adipocytes of type 2 diabetic mice. Diabetologia 49:784–791 [DOI] [PubMed] [Google Scholar]
- Okamoto Y, Higashiyama H, Rong JX, McVey MJ, Kinoshita M, Asano S, Hansen MK 2007 Comparison of mitochondrial and macrophage content between subcutaneous and visceral fat in db/db mice. Exp Mol Pathol 83:73–83 [DOI] [PubMed] [Google Scholar]
- Rong JX, Qiu Y, Hansen MK, Zhu L, Zhang V, Xie M, Okamoto Y, Mattie MD, Higashiyama H, Asano S, Strum JC, Ryan TE 2007 Adipose mitochondrial biogenesis is suppressed in db/db and high-fat diet-fed mice and improved by rosiglitazone. Diabetes 56:1751–1760 [DOI] [PubMed] [Google Scholar]
- Sutherland LN, Capozzi LC, Turchinsky NJ, Bell RC, Wright DC 2008 Time course of high-fat diet-induced reductions in adipose tissue mitochondrial proteins: potential mechanisms and the relationship to glucose intolerance. Am J Physiol Endocrinol Metab 295:E1076–E1083 [DOI] [PubMed] [Google Scholar]
- Kaaman M, Sparks LM, van Harmelen V, Smith SR, Sjölin E, Dahlman I, Arner P 2007 Strong association between mitochondrial DNA copy number and lipogenesis in human white adipose tissue. Diabetologia 50:2526–2533 [DOI] [PubMed] [Google Scholar]
- Dahlman I, Forsgren M, Sjögren A, Nordström EA, Kaaman M, Näslund E, Attersand A, Arner P 2006 Downregulation of electron transport chain genes in visceral adipose tissue in type 2 diabetes independent of obesity and possibly involving tumor necrosis factor-α. Diabetes 55:1792–1799 [DOI] [PubMed] [Google Scholar]
- Boden G, Homko C, Mozzoli M, Showe LC, Nichols C, Cheung P 2005 Thiazolidinediones upregulate fatty acid uptake and oxidation in adipose tissue of diabetic patients. Diabetes 54:880–885 [DOI] [PubMed] [Google Scholar]
- Bogacka I, Ukropcova B, McNeil M, Gimble JM, Smith SR 2005 Structural and functional consequences of mitochondrial biogenesis in human adipocytes in vitro. J Clin Endocrinol Metab 90:6650–6656 [DOI] [PubMed] [Google Scholar]
- Digby JE, Montague CT, Sewter CP, Sanders L, Wilkison WO, O'Rahilly S, Prins JB 1998 Thiazolidinedione exposure increases the expression of uncoupling protein 1 in cultured human preadipocytes. Diabetes 47:138–141 [DOI] [PubMed] [Google Scholar]
- Goetzman ES 2009 The regulation of acyl-CoA dehydrogenases in adipose tissue by rosiglitazone. Obesity (Silver Spring) 17:196–198 [DOI] [PubMed] [Google Scholar]
- Laplante M, Festuccia WT, Soucy G, Gélinas Y, Lalonde J, Berger JP, Deshaies Y 2006 Mechanisms of the depot specificity of peroxisome proliferator-activated receptor γ action on adipose tissue metabolism. Diabetes 55:2771–2778 [DOI] [PubMed] [Google Scholar]
- Granneman JG, Li P, Zhu Z, Lu Y 2005 Metabolic and cellular plasticity in white adipose tissue I: effects of β3-adrenergic receptor activation. Am J Physiol Endocrinol Metab 289:E608–E616 [DOI] [PubMed] [Google Scholar]
- Tedesco L, Valerio A, Cervino C, Cardile A, Pagano C, Vettor R, Pasquali R, Carruba MO, Marsicano G, Lutz B, Pagotto U, Nisoli E 2008 Cannabinoid type 1 receptor blockade promotes mitochondrial biogenesis through endothelial nitric oxide synthase expression in white adipocytes. Diabetes 57:2028–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katic M, Kennedy AR, Leykin I, Norris A, McGettrick A, Gesta S, Russell SJ, Bluher M, Maratos-Flier E, Kahn CR 2007 Mitochondrial gene expression and increased oxidative metabolism: role in increased lifespan of fat-specific insulin receptor knock-out mice. Aging Cell 6:827–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM 2007 Transcriptional control of brown fat determination by PRDM16. Cell Metab 6:38–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scimè A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, Spiegelman BM 2008 PRDM16 controls a brown fat/skeletal muscle switch. Nature 454:961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons JA, Wennmalm K, Larsson O, Walden TB, Lassmann T, Petrovic N, Hamilton DL, Gimeno RE, Wahlestedt C, Baar K, Nedergaard J, Cannon B 2007 Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc Natl Acad Sci USA 104:4401–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino N, Tamori Y, Tateya S, Kawaguchi T, Shibakusa T, Mizunoya W, Inoue K, Kitazawa R, Kitazawa S, Matsuki Y, Hiramatsu R, Masubuchi S, Omachi A, Kimura K, Saito M, Amo T, Ohta S, Yamaguchi T, Osumi T, Cheng J, Fujimoto T, Nakao H, Aiba A, Okamura H, Fushiki T, Kasuga M 2008 FSP27 contributes to efficient energy storage in murine white adipocytes by promoting the formation of unilocular lipid droplets. J Clin Invest 118:2808–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple RK, Crowley VC, Sewter CP, Laudes M, Christodoulides C, Considine RV, Vidal-Puig A, O'Rahilly S 2004 Expression of the thermogenic nuclear hormone receptor coactivator PGC-1α is reduced in the adipose tissue of morbidly obese subjects. Int J Obes Relat Metab Disord 28:176–179 [DOI] [PubMed] [Google Scholar]
- Kotronen A, Seppälä-Lindroos A, Bergholm R, Yki-Järvinen H 2008 Tissue specificity of insulin resistance in humans: fat in the liver rather than muscle is associated with features of the metabolic syndrome. Diabetologia 51:130–138 [DOI] [PubMed] [Google Scholar]
- Michael MD, Kulkarni RN, Postic C, Previs SF, Shulman GI, Magnuson MA, Kahn CR 2000 Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol Cell 6:87–97 [PubMed] [Google Scholar]
- Biddinger SB, Hernandez-Ono A, Rask-Madsen C, Haas JT, Alemán JO, Suzuki R, Scapa EF, Agarwal C, Carey MC, Stephanopoulos G, Cohen DE, King GL, Ginsberg HN, Kahn CR 2008 Hepatic insulin resistance is sufficient to produce dyslipidemia and susceptibility to atherosclerosis. Cell Metab 7:125–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltri KL, Espiritu M, Singh G 1990 Distinct genomic copy number in mitochondria of different mammalian organs. J Cell Physiol 143:160–164 [DOI] [PubMed] [Google Scholar]
- Benard G, Faustin B, Passerieux E, Galinier A, Rocher C, Bellance N, Delage JP, Casteilla L, Letellier T, Rossignol R 2006 Physiological diversity of mitochondrial oxidative phosphorylation. Am J Physiol Cell Physiol 291:C1172–C1182 [DOI] [PubMed] [Google Scholar]
- Schmid AI, Chmelík M, Szendroedi J, Krssák M, Brehm A, Moser E, Roden M 2008 Quantitative ATP synthesis in human liver measured by localized 31P spectroscopy using the magnetization transfer experiment. NMR Biomed 21:437–443 [DOI] [PubMed] [Google Scholar]
- Agarwal AK, Garg A 2006 Genetic disorders of adipose tissue development, differentiation, and death. Annu Rev Genomics Hum Genet 7:175–199 [DOI] [PubMed] [Google Scholar]
- Kotronen A, Juurinen L, Tiikkainen M, Vehkavaara S, Yki-Järvinen H 2008 Increased liver fat, impaired insulin clearance, and hepatic and adipose tissue insulin resistance in type 2 diabetes. Gastroenterology 135:122–130 [DOI] [PubMed] [Google Scholar]
- Petersen KF, Dufour S, Befroy D, Lehrke M, Hendler RE, Shulman GI 2005 Reversal of nonalcoholic hepatic steatosis, hepatic insulin resistance, and hyperglycemia by moderate weight reduction in patients with type 2 diabetes. Diabetes 54:603–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koska J, Stefan N, Permana PA, Weyer C, Sonoda M, Bogardus C, Smith SR, Joanisse DR, Funahashi T, Krakoff J, Bunt JC 2008 Increased fat accumulation in liver may link insulin resistance with subcutaneous abdominal adipocyte enlargement, visceral adiposity, and hypoadiponectinemia in obese individuals. Am J Clin Nutr 87:295–302 [DOI] [PubMed] [Google Scholar]
- Hwang JH, Stein DT, Barzilai N, Cui MH, Tonelli J, Kishore P, Hawkins M 2007 Increased intrahepatic triglyceride is associated with peripheral insulin resistance: in vivo MR imaging and spectroscopy studies. Am J Physiol Endocrinol Metab 293:E1663–E1669 [DOI] [PubMed] [Google Scholar]
- Korenblat KM, Fabbrini E, Mohammed BS, Klein S 2008 Liver, muscle, and adipose tissue insulin action is directly related to intrahepatic triglyceride content in obese subjects. Gastroenterology 134:1369–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Fillmore JJ, Chen Y, Yu C, Moore IK, Pypaert M, Lutz EP, Kako Y, Velez-Carrasco W, Goldberg IJ, Breslow JL, Shulman GI 2001 Tissue-specific overexpression of lipoprotein lipase causes tissue-specific insulin resistance. Proc Natl Acad Sci USA 98:7522–7527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Takamura T, Matsuzawa-Nagata N, Takayama H, Misu H, Noda H, Nabemoto S, Kurita S, Ota T, Ando H, Miyamoto K, Kaneko S 2009 Palmitate induces insulin resistance in H4IIEC3 hepatocytes through reactive oxygen species produced by mitochondria. J Biol Chem 284:14809–14818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruddock MW, Stein A, Landaker E, Park J, Cooksey RC, McClain D, Patti ME 2008 Saturated fatty acids inhibit hepatic insulin action by modulating insulin receptor expression and post-receptor signalling. J Biochem 144:599–607 [DOI] [PubMed] [Google Scholar]
- Hertz R, Magenheim J, Berman I, Bar-Tana J 1998 Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4α. Nature 392:512–516 [DOI] [PubMed] [Google Scholar]
- Lin J, Tarr PT, Yang R, Rhee J, Puigserver P, Newgard CB, Spiegelman BM 2003 PGC-1β in the regulation of hepatic glucose and energy metabolism. J Biol Chem 278:30843–30848 [DOI] [PubMed] [Google Scholar]
- Xu HE, Lambert MH, Montana VG, Parks DJ, Blanchard SG, Brown PJ, Sternbach DD, Lehmann JM, Wisely GB, Willson TM, Kliewer SA, Milburn MV 1999 Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Mol Cell 3:397–403 [DOI] [PubMed] [Google Scholar]
- Duplus E, Glorian M, Forest C 2000 Fatty acid regulation of gene transcription. J Biol Chem 275:30749–30752 [DOI] [PubMed] [Google Scholar]
- Seppälä-Lindroos A, Vehkavaara S, Häkkinen AM, Goto T, Westerbacka J, Sovijärvi A, Halavaara J, Yki-Järvinen H 2002 Fat accumulation in the liver is associated with defects in insulin suppression of glucose production and serum free fatty acids independent of obesity in normal men. J Clin Endocrinol Metab 87:3023–3028 [DOI] [PubMed] [Google Scholar]
- Park SY, Cho YR, Kim HJ, Hong EG, Higashimori T, Lee SJ, Goldberg IJ, Shulman GI, Najjar SM, Kim JK 2006 Mechanism of glucose intolerance in mice with dominant negative mutation of CEACAM1. Am J Physiol Endocrinol Metab 291:E517–E524 [DOI] [PubMed] [Google Scholar]
- Liu HY, Yehuda-Shnaidman E, Hong T, Han J, Pi J, Liu Z, Cao W 2009 Prolonged exposure to insulin suppresses mitochondrial production in primary hepatocytes. J Biol Chem 284:14087–14095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Guo S, Copps K, Dong X, Kollipara R, Rodgers JT, Depinho RA, Puigserver P, White MF 2009 Foxo1 integrates insulin signaling with mitochondrial function in the liver. Nat Med 15:1307–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misu H, Takamura T, Matsuzawa N, Shimizu A, Ota T, Sakurai M, Ando H, Arai K, Yamashita T, Honda M, Yamashita T, Kaneko S 2007 Genes involved in oxidative phosphorylation are coordinately upregulated with fasting hyperglycaemia in livers of patients with type 2 diabetes. Diabetologia 50:268–277 [DOI] [PubMed] [Google Scholar]
- Takamura T, Misu H, Matsuzawa-Nagata N, Sakurai M, Ota T, Shimizu A, Kurita S, Takeshita Y, Ando H, Honda M, Kaneko S 2008 Obesity upregulates genes involved in oxidative phosphorylation in livers of diabetic patients. Obesity (Silver Spring) 16:2601–2609 [DOI] [PubMed] [Google Scholar]
- Pihlajamäki J, Boes T, Kim EY, Dearie F, Kim BW, Schroeder J, Mun E, Nasser I, Park PJ, Bianco AC, Goldfine AB, Patti ME 2009 Thyroid hormone-related regulation of gene expression in human fatty liver. J Clin Endocrinol Metab 94:3521–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Carreras M, Del Hoyo P, Martín MA, Rubio JC, Martín A, Castellano G, Colina F, Arenas J, Solis-Herruzo JA 2003 Defective hepatic mitochondrial respiratory chain in patients with nonalcoholic steatohepatitis. Hepatology 38:999–1007 [DOI] [PubMed] [Google Scholar]
- Greenfield V, Cheung O, Sanyal AJ 2008 Recent advances in nonalcoholic fatty liver disease. Curr Opin Gastroenterol 24:320–327 [DOI] [PubMed] [Google Scholar]
- Sanyal AJ, Campbell-Sargent C, Mirshahi F, Rizzo WB, Contos MJ, Sterling RK, Luketic VA, Shiffman ML, Clore JN 2001 Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology 120:1183–1192 [DOI] [PubMed] [Google Scholar]
- Kohjima M, Enjoji M, Higuchi N, Kato M, Kotoh K, Yoshimoto T, Fujino T, Yada M, Yada R, Harada N, Takayanagi R, Nakamuta M 2007 Re-evaluation of fatty acid metabolism-related gene expression in nonalcoholic fatty liver disease. Int J Mol Med 20:351–358 [PubMed] [Google Scholar]
- O'Connor MD, Landahl H, Grodsky GM 1980 Comparison of storage- and signal-limited models of pancreatic insulin secretion. Am J Physiol 238:R378–R389 [DOI] [PubMed] [Google Scholar]
- Hasan NM, Longacre MJ, Stoker SW, Boonsaen T, Jitrapakdee S, Kendrick MA, Wallace JC, MacDonald MJ 2008 Impaired anaplerosis and insulin secretion in insulinoma cells caused by small interfering RNA-mediated suppression of pyruvate carboxylase. J Biol Chem 283:28048–28059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JL, Goldfine ID, Maddux BA, Grodsky GM 2003 Are oxidative stress-activated signaling pathways mediators of insulin resistance and β-cell dysfunction? Diabetes 52:1–8 [DOI] [PubMed] [Google Scholar]
- Leloup C, Tourrel-Cuzin C, Magnan C, Karaca M, Castel J, Carneiro L, Colombani AL, Ktorza A, Casteilla L, Pénicaud L 2009 Mitochondrial reactive oxygen species are obligatory signals for glucose-induced insulin secretion. Diabetes 58:673–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JP, Köhler M, Graff C, Oldfors A, Magnuson MA, Berggren PO, Larsson NG 2000 Impaired insulin secretion and β-cell loss in tissue-specific knockout mice with mitochondrial diabetes. Nat Genet 26:336–340 [DOI] [PubMed] [Google Scholar]
- Gauthier BR, Wiederkehr A, Baquié M, Dai C, Powers AC, Kerr-Conte J, Pattou F, MacDonald RJ, Ferrer J, Wollheim CB 2009 PDX1 deficiency causes mitochondrial dysfunction and defective insulin secretion through TFAM suppression. Cell Metab 10:110–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Kadowaki H, Mori Y, Tobe K, Sakuta R, Suzuki Y, Tanabe Y, Sakura H, Awata T, Goto Y, Hayakawa T, Matsuoka K, Kawamori R, Kamada T, Horai S, Nonaka I, Hagura R, Akanuma Y, Yazaki Y 1994 A subtype of diabetes mellitus associated with a mutation of mitochondrial DNA. N Engl J Med 330:962–968 [DOI] [PubMed] [Google Scholar]
- Maassen JA, Jahangir Tafrechi RS, Janssen GM, Raap AK, Lemkes HH, 't Hart LM 2006 New insights in the molecular pathogenesis of the maternally inherited diabetes and deafness syndrome. Endocrinol Metab Clin North Am 35:385–396, x–xi [DOI] [PubMed] [Google Scholar]
- Tawata M, Hayashi JI, Isobe K, Ohkubo E, Ohtaka M, Chen J, Aida K, Onaya T 2000 A new mitochondrial DNA mutation at 14577 T/C is probably a major pathogenic mutation for maternally inherited type 2 diabetes. Diabetes 49:1269–1272 [DOI] [PubMed] [Google Scholar]
- Maassen JA, ‘T Hart LM, Van Essen E, Heine RJ, Nijpels G, Jahangir Tafrechi RS, Raap AK, Janssen GM, Lemkes HH 2004 Mitochondrial diabetes: molecular mechanisms and clinical presentation. Diabetes 53(Suppl 1):S103–S109 [DOI] [PubMed] [Google Scholar]
- Maechler P, Carobbio S, Rubi B 2006 In β-cells, mitochondria integrate and generate metabolic signals controlling insulin secretion. Int J Biochem Cell Biol 38:696–709 [DOI] [PubMed] [Google Scholar]
- de Andrade PB, Rubi B, Frigerio F, van den Ouweland JM, Maassen JA, Maechler P 2006 Diabetes-associated mitochondrial DNA mutation A3243G impairs cellular metabolic pathways necessary for β-cell function. Diabetologia 49:1816–1826 [DOI] [PubMed] [Google Scholar]
- Hou N, Torii S, Saito N, Hosaka M, Takeuchi T 2008 Reactive oxygen species-mediated pancreatic β-cell death is regulated by interactions between stress-activated protein kinases, p38 and c-Jun N-terminal kinase, and mitogen-activated protein kinase phosphatases. Endocrinology 149:1654–1665 [DOI] [PubMed] [Google Scholar]
- Lim M, Park L, Shin G, Hong H, Kang I, Park Y 2008 Induction of apoptosis of β-cells of the pancreas by advanced glycation end-products, important mediators of chronic complications of diabetes mellitus. Ann NY Acad Sci 1150:311–315 [DOI] [PubMed] [Google Scholar]
- Newsholme P, Haber EP, Hirabara SM, Rebelato EL, Procopio J, Morgan D, Oliveira-Emilio HC, Carpinelli AR, Curi R 2007 Diabetes associated cell stress and dysfunction: role of mitochondrial and non-mitochondrial ROS production and activity. J Physiol 583:9–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa T, Araki E 2007 Impact of mitochondrial ROS production in the pathogenesis of diabetes mellitus and its complications. Antioxid Redox Signal 9:343–353 [DOI] [PubMed] [Google Scholar]
- Robertson RP 2009 β-Cell deterioration during diabetes: what’s in the gun? Trends Endocrinol Metab 20:388–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Frigerio F, Maechler P 2008 The sensitivity of pancreatic β-cells to mitochondrial injuries triggered by lipotoxicity and oxidative stress. Biochem Soc Trans 36:930–934 [DOI] [PubMed] [Google Scholar]
- Rachek LI, Thornley NP, Grishko VI, LeDoux SP, Wilson GL 2006 Protection of INS-1 cells from free fatty acid-induced apoptosis by targeting hOGG1 to mitochondria. Diabetes 55:1022–1028 [DOI] [PubMed] [Google Scholar]
- Simmons RA, Suponitsky-Kroyter I, Selak MA 2005 Progressive accumulation of mitochondrial DNA mutations and decline in mitochondrial function lead to β-cell failure. J Biol Chem 280:28785–28791 [DOI] [PubMed] [Google Scholar]
- Brand MD 2005 The efficiency and plasticity of mitochondrial energy transduction. Biochem Soc Trans 33:897–904 [DOI] [PubMed] [Google Scholar]
- Zhang CY, Baffy G, Perret P, Krauss S, Peroni O, Grujic D, Hagen T, Vidal-Puig AJ, Boss O, Kim YB, Zheng XX, Wheeler MB, Shulman GI, Chan CB, Lowell BB 2001 Uncoupling protein-2 negatively regulates insulin secretion and is a major link between obesity, β-cell dysfunction, and type 2 diabetes. Cell 105:745–755 [DOI] [PubMed] [Google Scholar]
- Pi J, Bai Y, Daniel KW, Liu D, Lyght O, Edelstein D, Brownlee M, Corkey BE, Collins S 2009 Persistent oxidative stress due to absence of uncoupling protein 2 associated with impaired pancreatic β-cell function. Endocrinology 150:3040–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshkin V, Wang X, Scherer PE, Chan CB, Wheeler MB 2003 Mitochondrial functional state in clonal pancreatic β-cells exposed to free fatty acids. J Biol Chem 278:19709–19715 [DOI] [PubMed] [Google Scholar]
- Benton CR, Nickerson JG, Lally J, Han XX, Holloway GP, Glatz JF, Luiken JJ, Graham TE, Heikkila JJ, Bonen A 2008 Modest PGC-1α overexpression in muscle in vivo is sufficient to increase insulin sensitivity and palmitate oxidation in subsarcolemmal, not intermyofibrillar, mitochondria. J Biol Chem 283:4228–4240 [DOI] [PubMed] [Google Scholar]
- Liang H, Balas B, Tantiwong P, Dube J, Goodpaster BH, O'Doherty RM, DeFronzo RA, Richardson A, Musi N, Ward WF 2009 Whole body overexpression of PGC-1α has opposite effects on hepatic and muscle insulin sensitivity. Am J Physiol Endocrinol Metab 296:E945–E954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura S, Kai Y, Ono M, Ezaki O 2003 Overexpression of peroxisome proliferator-activated receptor γ coactivator-1α down-regulates GLUT4 mRNA in skeletal muscles. J Biol Chem 278:31385–31390 [DOI] [PubMed] [Google Scholar]
- Miura S, Tomitsuka E, Kamei Y, Yamazaki T, Kai Y, Tamura M, Kita K, Nishino I, Ezaki O 2006 Overexpression of peroxisome proliferator-activated receptor γ co-activator-1α leads to muscle atrophy with depletion of ATP. Am J Pathol 169:1129–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wende AR, Schaeffer PJ, Parker GJ, Zechner C, Han DH, Chen MM, Hancock CR, Lehman JJ, Huss JM, McClain DA, Holloszy JO, Kelly DP 2007 A role for the transcriptional coactivator PGC-1α in muscle refueling. J Biol Chem 282:36642–36651 [DOI] [PubMed] [Google Scholar]
- Russell LK, Mansfield CM, Lehman JJ, Kovacs A, Courtois M, Saffitz JE, Medeiros DM, Valencik ML, McDonald JA, Kelly DP 2004 Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-activated receptor γ coactivator-1α promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circ Res 94:525–533 [DOI] [PubMed] [Google Scholar]
- Calvo JA, Daniels TG, Wang X, Paul A, Lin J, Spiegelman BM, Stevenson SC, Rangwala SM 2008 Muscle-specific expression of PPARγ coactivator-1α improves exercise performance and increases peak oxygen uptake. J Appl Physiol 104:1304–1312 [DOI] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM 2002 Transcriptional co-activator PGC-1 α drives the formation of slow-twitch muscle fibers. Nature 418:797–801 [DOI] [PubMed] [Google Scholar]
- Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, Goldberg AL, Spiegelman BM 2006 PGC-1α protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci USA 103:16260–16265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi CS, Befroy DE, Codella R, Kim S, Reznick RM, Hwang YJ, Liu ZX, Lee HY, Distefano A, Samuel VT, Zhang D, Cline GW, Handschin C, Lin J, Petersen KF, Spiegelman BM, Shulman GI 2008 Paradoxical effects of increased expression of PGC-1α on muscle mitochondrial function and insulin-stimulated muscle glucose metabolism. Proc Natl Acad Sci USA 105:19926–19931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arany Z, Lebrasseur N, Morris C, Smith E, Yang W, Ma Y, Chin S, Spiegelman BM 2007 The transcriptional coactivator PGC-1β drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab 5:35–46 [DOI] [PubMed] [Google Scholar]
- Kamei Y, Ohizumi H, Fujitani Y, Nemoto T, Tanaka T, Takahashi N, Kawada T, Miyoshi M, Ezaki O, Kakizuka A 2003 PPARγ coactivator 1β/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc Natl Acad Sci USA 100:12378–12383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza DO, Boros LG, Crunkhorn S, Gami H, Patti ME 11 November 2009 Dual modulation of both lipid oxidation and synthesis by peroxisome proliferator-activated receptor-γ coactivator-1α and -1β in cultured myotubes. FASEB J doi: 10.1096/fj.09-133728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospisilik JA, Knauf C, Joza N, Benit P, Orthofer M, Cani PD, Ebersberger I, Nakashima T, Sarao R, Neely G, Esterbauer H, Kozlov A, Kahn CR, Kroemer G, Rustin P, Burcelin R, Penninger JM 2007 Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell 131:476–491 [DOI] [PubMed] [Google Scholar]
- Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N, Willy PJ, Schulman IG, Heyman RA, Lander ES, Spiegelman BM 2004 ERRα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci USA 101:6570–6575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano FX, Liesa M, Bach D, Chan DC, Palacín M, Zorzano A 2006 Evidence for a mitochondrial regulatory pathway defined by peroxisome proliferator-activated receptor-γ coactivator-1 α, estrogen-related receptor-α, and mitofusin 2. Diabetes 55:1783–1791 [DOI] [PubMed] [Google Scholar]
- Luo J, Sladek R, Carrier J, Bader JA, Richard D, Giguère V 2003 Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor α. Mol Cell Biol 23:7947–7956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Zhang Z, Teng CT 2005 Estrogen-related receptor-γ and peroxisome proliferator-activated receptor-γ coactivator-1α regulate estrogen-related receptor-α gene expression via a conserved multi-hormone response element. J Mol Endocrinol 34:473–487 [DOI] [PubMed] [Google Scholar]
- Rampersaud E, Damcott CM, Fu M, Shen H, McArdle P, Shi X, Shelton J, Yin J, Chang YP, Ott SH, Zhang L, Zhao Y, Mitchell BD, O'Connell J, Shuldiner AR 2007 Identification of novel candidate genes for type 2 diabetes from a genome-wide association scan in the Old Order Amish: evidence for replication from diabetes-related quantitative traits and from independent populations. Diabetes 56:3053–3062 [DOI] [PubMed] [Google Scholar]
- Freyer C, Larsson NG 2007 Is energy deficiency good in moderation? Cell 131:448–450 [DOI] [PubMed] [Google Scholar]
- Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA 1998 Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet 18:231–236 [DOI] [PubMed] [Google Scholar]
- Wang J, Wilhelmsson H, Graff C, Li H, Oldfors A, Rustin P, Brüning JC, Kahn CR, Clayton DA, Barsh GS, Thorén P, Larsson NG 1999 Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat Genet 21:133–137 [DOI] [PubMed] [Google Scholar]
- Hance N, Ekstrand MI, Trifunovic A 2005 Mitochondrial DNA polymerase γ is essential for mammalian embryogenesis. Hum Mol Genet 14:1775–1783 [DOI] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly-Y M, Gidlöf S, Oldfors A, Wibom R, Törnell J, Jacobs HT, Larsson NG 2004 Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429:417–423 [DOI] [PubMed] [Google Scholar]
- Tyynismaa H, Sembongi H, Bokori-Brown M, Granycome C, Ashley N, Poulton J, Jalanko A, Spelbrink JN, Holt IJ, Suomalainen A 2004 Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum Mol Genet 13:3219–3227 [DOI] [PubMed] [Google Scholar]
- Stump CS, Short KR, Bigelow ML, Schimke JM, Nair KS 2003 Effect of insulin on human skeletal muscle mitochondrial ATP production, protein synthesis, and mRNA transcripts. Proc Natl Acad Sci USA 100:7996–8001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson EJ, Kypson AP, Rodriguez E, Anderson CA, Lehr EJ, Neufer PD 2009 Substrate-specific derangements in mitochondrial metabolism and redox balance in the atrium of the type 2 diabetic human heart. J Am Coll Cardiol 54:1891–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnard C, Durand A, Peyrol S, Chanseaume E, Chauvin MA, Morio B, Vidal H, Rieusset J 2008 Mitochondrial dysfunction results from oxidative stress in the skeletal muscle of diet-induced insulin-resistant mice. J Clin Invest 118:789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T, Kinugawa S, Hirabayashi K, Matsushima S, Inoue N, Ohta Y, Hamaguchi S, Sobirin MA, Ono T, Suga T, Kuroda S, Tanaka S, Terasaki F, Okita K, Tsutsui H 2009 Oxidative stress in skeletal muscle impairs mitochondrial respiration and limits exercise capacity in type 2 diabetic mice. Am J Physiol Heart Circ Physiol 297:H1069–H1077 [DOI] [PubMed] [Google Scholar]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, Rochon J, Gallup D, Ilkayeva O, Wenner BR, Yancy Jr WS, Eisenson H, Musante G, Surwit RS, Millington DS, Butler MD, Svetkey LP 2009 A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 9:311–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SH, Hauser ER, Bain JR, Muehlbauer MJ, Haynes C, Stevens RD, Wenner BR, Dowdy ZE, Granger CB, Ginsburg GS, Newgard CB, Kraus WE 2009 High heritability of metabolomic profiles in families burdened with premature cardiovascular disease. Mol Syst Biol 5:258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerin C, Goldfine AB, Tatro E, Boes T, Schroeder J, Patti ME 2009 Reductions in branched chain amino acid oxidation gene expression is a signature of insulin resistance in humans. Diabetes 58:A298 (Supplement) [Google Scholar]
- Shaham O, Wei R, Wang TJ, Ricciardi C, Lewis GD, Vasan RS, Carr SA, Thadhani R, Gerszten RE, Mootha VK 2008 Metabolic profiling of the human response to a glucose challenge reveals distinct axes of insulin sensitivity. Mol Syst Biol 4:214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj M, Medina-Navarro R, Suraamornkul S, Meyer C, DeFronzo RA, Mandarino LJ 2007 Paradoxical changes in muscle gene expression in insulin-resistant subjects after sustained reduction in plasma free fatty acid concentration. Diabetes 56:743–752 [DOI] [PubMed] [Google Scholar]