Abstract
In extensive liver resection secondary to primary or metastatic liver tumors, or in living donor liver transplantation, strategies to quell deleterious inflammatory responses and facilitate regeneration are essential. The receptor for advanced glycation endproducts (RAGE) and myeloid differentiating factor 88 (Myd88) are implicated in the inflammatory response. To establish the contributions of RAGE vs. Myd88 signaling in extensive liver resection, we probed the effect of RAGE and/or Myd88, the latter primarily a key transducer of major toll-like receptors and also implicated in interleukin-1 (Il1) signaling, in a murine model of extensive (85%) hepatectomy. We report that, although Myd88 is thoroughly essential for survival via regulation of NF-κB and TNF-α, deletion of RAGE significantly improved survival compared to wild-type, Myd88-null, or RAGE-null/Myd88-null mice. RAGE opposes Myd88 signaling at multiple levels: by suppression of p65 levels, thereby reducing activation of NF-κB and consequent production of cyclin D1, and by suppression of Il6-mediated phosphorylation of Stat3, thereby down-regulating Pim1 and suppressing the hyperplastic response. Further, RAGE-dependent suppression of glyoxalase1, a detoxification pathway for pre-AGEs, enhances AGE levels and suppresses Il6 action. We conclude that blockade of RAGE may rescue liver remnants from the multiple signals that preclude adaptive proliferation triggered primarily by Myd88 signaling pathways.—Zeng, S., Zhang, Q. Y., Huang, J., Vedantham, S., Rosario, R., Ananthakrishnan, R., Yan, S. F., Ramasamy, R., DeMatteo, R. P., Emond, J. C., Friedman, R. A., Schmidt, A. M. Opposing roles of RAGE and Myd88 signaling in extensive liver resection.
Keywords: nuclear factor-κB, Pim1, inflammation, proliferation, stat 3 phosphorylation
Although the liver is capable of extensive regeneration, overwhelming damage triggers mechanisms that preclude repair and lead to rapid demise. In extensive liver resection secondary to primary or metastatic liver tumors, or in living donor liver transplantation, wherein the donor must provide sufficient liver tissue to both successfully restore function to the recipient yet retain adequate potential for its own regeneration, strategies to quell these damage responses are essential (1, 2). In animals, although partial (70%) resection of the liver is met with robust repair in which proliferating hepatic cells repopulate the remnant to full potential, 85% liver resection is met with significant mortality (3).
We showed previously that pharmacological blockade of the receptor for advanced glycation endproducts (RAGE, produced by the AGER gene) increased the probability of survival of mice subjected to extensive liver resection (4). A central question arose from this observation: Did the RAGE axis act in concert with or in opposition to fundamental innate immune response mechanisms? As establishing the answers to this question is essential for the design of optimal liver-sparing strategies in extensive resection or injury, we studied mice deficient in RAGE and/or myeloid differentiating factor 88 (Myd88), the latter a central signaling pathway transducing innate immune signals evoked by major toll receptors, such as 2 and 4 (5), or several non-toll-like receptors, such as interleukin (Il)1 pathways (6). These groups of mice were subjected to extensive liver resection, and the effect on survival, gene expression programs, and regeneration were assessed.
MATERIALS AND METHODS
Animals and procedures
Specific pathogen-free male C57BL/6 mice (Jackson Laboratories, Bar Harbor, ME, USA) at 10–12 wk of age were used. Homozygous RAGE-null mice and/or Myd88-null mice were backcrossed >10 generations into C57BL/6 before studies prior to interbreeding. Myd88-null mice were a generous gift of Dr. Ruslan Medzhitov (Howard Hughes Medical Institute, Yale University School of Medicine, New Haven, CT, USA). Mice were maintained at all times on 12-h light-dark cycles in a pathogen-free environment with free access to normal rodent chow and water. All experiments were approved by the Institutional Animal Care and Use Committee of Columbia University. Mice underwent 85% hepatectomy as described previously (4).
Immunoprecipitation and Western blotting
Whole protein was extracted from frozen liver tissues using lysis buffer (10 mM Tris HCl, pH 7.5; 150 mM NaCl; 2 mM EDTA; 1% Triton X-100; 10% glycerol; 2 mM sodium orthovanadate; and complete protease inhibitor cocktail, Roche Diagnostics, Indianapolis, IN, USA). For immunoprecipitation, samples were incubated with rabbit anti-mouse RAGE IgG (provided by A.M.S.) or nonimmune rabbit IgG (Sigma-Aldrich, St. Louis, MO, USA) at 4°C overnight (4). Protein A/G (Pierce Chemical Co., Rockford, IL, USA) was added for 1 h at room temperature, followed by washing and centrifugation. The pellet was solubilized in reduced SDS sample buffer and subjected to SDS-PAGE. Immunoblotting was performed as described previously (4). Antibodies to cleaved caspase 3, Erk, phosphorylated STAT3, and Gapdh were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). Other antibodies used were to Pim1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), Tlr4 (Imgenex Corp., San Diego, CA, USA), and Glo1 (Novus, Cambridge, MA, USA).
Electrophoretic mobility shift assay (EMSA)
EMSAs were performed as described previously (4).
Immunohistochemistry
Consecutive sections from paraffin-embedded liver were prepared for proliferating cell nuclear antigen (Pcna) immunohistochemistry. Staining for detection of Pcna was performed using mouse anti-Pcna IgG (Dako, Carpinteria, CA, USA; ref. 4).
Real-time PCR and GeneChip analysis
Total RNA was extracted from liver tissues or liver cells, reverse transcribed, and used with PCR TaqMan probes for mouse RAGE, Tlr4, Bax, cyclin D1, Pim1, Glo1, Il6, p65, TNF-α and Gapdh (Applied Biosystems, Foster City, CA, USA). Quantitative PCR was performed using the ABI 7900 system (Applied Biosystems). Data are calculated by the 2−ΔΔCt method as described by the manufacturer and are expressed as the fold increase over the indicated controls (set to 1) in each figure. For GeneChip analysis, cDNA was in vitro transcribed into biotin-labeled antisense cRNA using an Affymetrix kit (Affymetrix, Santa Clara, CA, USA) according to the standard kit protocol. cRNA (1 μg) from each sample was hybridized to Affymetrix Mouse Genome 430 2.0 GeneChips. Arrays were performed in the Core Facility of Columbia University according to the protocol described in the Affymetrix GeneChip expression analysis.
ELISA for detection of AGE epitopes
Liver lysates were prepared as described above. Protein (300 μg/well) was coated overnight onto an ELISA 96-well plate using carbonate-bicarbonate buffer (Sigma). AGE ELISA was performed using chicken anti-AGE (Pierce) as the primary antibody for 3 h at room temperature, followed by washing and adding secondary antibody (rabbit anti-chicken IgY; Sigma) for 1 h at room temperature. The wells were then washed and developed with 0.1 ml of peroxidase substrates (o-phenylenediamine tablets; Sigma) in the dark at room temperature.
Measurements of 3-methylglyoxal (MG) and Glo1 activity
MG, a key pre-AGE species, was measured in the neutralized perchloric acid extracts of tissue after derivatizing with ortho-phenyl diamine (o-PDA, 1%) at room temperature. The derivatives were quantified by high-performance liquid chromatography (HPLC) methods according to previously published procedures (7). The activity of Glo1 in liver remnants was measured using the hemithioacetal of methylglyoxal as substrate, as described previously (8).
Hepatocyte isolation and stimulation
Liver hepatocytes were isolated from mice by a 2-step collagenase perfusion method. Briefly, the liver was perfused with Ca2+-Mg2+-free HBSS (Invitrogen, Carlsbad, CA, USA) via the portal vein, followed by 0.03% collagenase (Worthington, Lakewood, NJ, USA) perfusion. Liver homogenates were filtered by 100-μm cell strainer, mixed gently with equal volume percoll solution, and spun down at 600 rpm for 8 min. The cell pellets were washed twice with serum-free DMEM for 2 min at 600 rpm and were resuspended in complete DMEM medium supplemented with 5% FCS and 1% penicillin/streptomycin antibiotics. Aliquots (2 ml, 1×106/ml) were dispensed onto 6-well collagen I-coated plates. Hepatocytes were stimulated by AGEs (20 μg/ml; provided by Dr. Eric Boulanger, Lille2 Medical School, Lille, France) or Il6 (50 ng/ml; R&D Systems, Inc., Minneapolis, MN, USA) or AGEs plus Il6. Hepatocytes isolated from RAGE-null mice were stimulated with AGEs (20 μg/ml) plus Il6 (50 ng/ml) or AGEs plus Il6 in the presence of an Il6 receptor antagonist (anti-Il6r, 50 ng/ml; R&D Systems).
Chromatin immunoprecipitation (ChIP) assay
ChIP was performed using the ChIP-IT kit (Active Motif, Carlsbad, CA, USA) according to the manufacturer's protocol. Briefly, 100 mg tissue was cut into small pieces and cross-linked with 1% formaldehyde in PBS for 15 min at room temperature. The cross-linking was stopped by addition of 0.125 M glycine for 5 min at room temperature. The tissue was washed briefly with cold PBS, and the tissue pieces were minced using a tissue disintegrator for ≥3 min. Once homogeneous, the cell suspension was centrifuged at 2500 rpm at 4°C for 10 min. The cells were lysed in 1× lysis buffer, provided by the manufacturer. The nuclear pellet was obtained by centrifuging the lysed cells at 5000 rpm for 10 min at 4°C. The nucleus was sonicated for 20 pulses, with each pulse consisting of 20 s shearing and 20 s placement in ice. The sonicated DNA was checked for sonication efficiency (200–1000 bp) by running the chromatin on an agarose gel. The chromatin was initially precleared using protein-G beads and subsequently treated with p-65 NF-κB antibody (Santa Cruz Biotechnology) at a concentration of 1:200 for immunoprecipitation overnight. The immunoprecipitated ChIP product was washed, reverse cross-linked, treated with proteinase K, and eluted to obtain the ChIP-enriched DNA. ChIP-enriched DNA (50 ng) was used to perform PCR for the Glo1 promoter using the following primers: forward primer, 5′-GGGAAGACCACTGAGCTGAT-3′; reverse primer, 5′-ATCCTTAGTGGTCCACAGGG-3′. The PCR products were run on a 2% agarose gel, and the amplicons were visualized using UV light.
Microarray studies
Microarray analysis is given in Supplemental Data.
Statistical analysis
All data are expressed as means ± se. Differences were detected by analysis of variance (ANOVA) and, as indicated, subjected to post hoc comparisons using a 2-tailed Student's t test or ANOVA followed by post hoc analysis using Tukey's procedure for comparison of individual groups. For survival studies, the Kaplan-Meier method was used to analyze survival. The log-rank test was used to compare survival curves. Differences were considered significant at values of P < 0.05.
RESULTS
Gene deletion of RAGE significantly improves survival after extensive hepatectomy
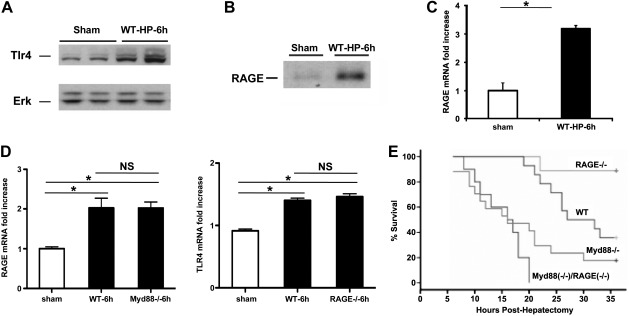
To discern the specific effect of Myd88- and RAGE-dependent signaling in massive liver resection, we probed hepatic remnants after 85% resection for the expression of RAGE and toll-like receptor 4 (Tlr4), as Myd88 is a key adapter for this molecule. Remnants retrieved 6 h after 85% hepatectomy revealed significant up-regulation of both Tlr4 and RAGE expression compared to sham-operated livers (Fig. 1A–C). To establish the functions of Myd88 and RAGE pathways in extensive liver resection, we subjected the following groups of animals to 85% hepatectomy: wild-type (WT), RAGE-null, and Myd88-null mice. We first established that RAGE mRNA transcripts did not differ significantly in the remnants of Myd88-null mice at 6 h after 85% hepatectomy compared to WT mice (Fig. 1D, left panel) and that Tlr4 mRNA transcripts did not significantly differ in the remnants of RAGE-null mice at 6 h after 85% hepatectomy compared to WT mice subjected to massive resection (Fig. 1D, right panel). Notably, compared to sham-operated WT livers, livers of both WT and Myd88-null mice displayed significantly higher RAGE mRNA transcripts at 6 h after 85% hepatectomy (Fig. 1D, left panel), and compared to sham-operated WT livers, livers of both WT and RAGE-null mice displayed significantly higher levels of Tlr4 transcripts at 6 h after massive resection (Fig. 1D, right panel). Animals of each of these four groups were subjected to 85% liver resection; Kaplan Meier product-limit estimates revealed that compared to WT mice, RAGE-null mice displayed significantly higher probability of survival in the first 36 h, ≈38 vs. 89%, respectively (P=0.01), whereas mice deficient in Myd88 displayed significantly reduced survival, ≈18% vs. WT or RAGE-null mice (P<0.05; Fig. 1E). In mice deficient in both Myd88 and RAGE subjected to 85% liver resection, the probability of survival was significantly less than that of either WT mice or RAGE-null mice and not significantly different from the probability of survival observed in Myd88-null mice (Fig. 1E). These results suggest that Myd88 signaling is essential for survival and that RAGE signaling is deleterious in extensive liver resection.
Figure 1.
Up-regulation of Tlr4 and RAGE in hepatic remnants and the effect of RAGE or/and Myd88 gene deletion on survival after extensive (85%) hepatectomy: Male C57BL/6 mice were subjected to 85% hepatectomy or sham surgery. A, B) At the indicated times, hepatic remnants were subjected to Western blotting with anti-Tlr4 IgG (A); immunoprecipitation for RAGE was performed with rabbit anti-RAGE IgG or nonimmune rabbit IgG as described in Materials and Methods and then subjected to Western blotting with same anti-RAGE IgG (B). Illustrated bands are representative of n = 3–4 mice/condition. C) mRNA transcript levels for RAGE were determined in the liver remnants by real-time PCR. Data are means ± se; n = 4. *P < 0.05. D) mRNA transcript levels for RAGE and Tlr4 were determined in the liver remnants of sham-operated WT mice or WT, Myd88-null, or RAGE-null mice at 6 h after massive resection by real-time PCR. NS, not significant. *P < 0.05. E) Kaplan-Meier curves for WT, RAGE-null, Myd88-null, or Myd88-null/RAGE-null mice were plotted, and the statistical significance of the probability of survival of the mutants vs. WT mice was calculated. n = 10 mice/group. Statistical considerations: RAGE−/− vs. WT, P = 0.01; RAGE−/− vs. Myd88−/−, P < 0.001; Myd88−/− vs. WT, P = 0.013; Myd88−/− vs. Myd88−/−/RAGE−/−, P = 0.144.
RAGE and Myd88 distinctly modulate apoptosis and proliferation after extensive hepatectomy
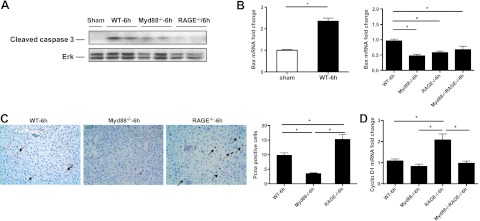
To determine the mechanisms by which Myd88 and RAGE mediated distinct responses to extensive liver resection, we examined cellular fate in the remnants. Levels of cleaved caspase 3, a marker of apoptosis, were detectable in the remnants of WT mice and, to lesser degrees, in Myd88-null remnants but were nearly undetectable in RAGE-null remnants by Western blotting at 6 h postinjury (Fig. 2A). Although levels of proapoptotic Bax mRNA were significantly higher in WT vs. sham remnants at 6 h after extensive hepatectomy (9), levels of this factor were significantly lower in the remnants of Myd88-null, RAGE-null, and Myd88-null/RAGE-null mice compared to WT mice (Fig. 2B). These findings suggested that apoptosis was not likely to be the pivotal mechanism distinguishing RAGE vs. Myd88 action in the survival response after 85% resection.
Figure 2.
Gene deletion of RAGE or Myd88 affects proliferation after extensive hepatectomy. Male C57BL/6 mice were subjected to 85% hepatectomy or sham surgery. A) At the indicated times, hepatic remnants were subjected to immunoblotting with anti-cleaved caspase 3 IgG. Illustrated bands are representative of n = 3–4 mice/condition. B) mRNA transcript levels for Bax were determined in the indicated mice by real time PCR. C) Pcna staining. Hepatic remnants were retrieved and subjected to immunohistochemistry with anti-Pcna IgG (×20 view). Arrows indicate Pcna+ cells. Data are representative of n = 4 mice/condition. D) mRNA transcript levels for cyclin D1 were determined in the liver remnants of the indicated mice by real-time PCR. Data are means ± se; n = 4. *P < 0.05.
Based on these data, we thus reasoned that changes in proliferation consequent to extensive resection were more likely to contribute to the significant survival effect of RAGE vs. Myd88 deletion. Pcna staining of liver remnants 6 h after injury revealed that compared to WT mice, Myd88-null remnants demonstrated only scant Pcna+ cells, but significantly more Pcna+ cells were noted in RAGE-null remnants after 85% resection vs. the other groups (Fig. 2C). Consistent with these findings, at 6 h after extensive hepatectomy, RAGE-null remnants displayed the highest levels of cyclin D1 mRNA transcripts compared to WT, Myd88-null, or Myd88-null/RAGE-null mice (Fig. 2D).
Gene deletion of RAGE modulates AGE production and NF-κB activation after extensive hepatectomy
To probe in detail the mechanisms by which the actions of RAGE and Myd88 diverged in 85% resection, we performed Affymetrix gene arrays on hepatic remnants at 2 h postinjury. Based on the survival time course, we reasoned that by 2 h after 85% hepatectomy, fundamental differences in the biological response to massive resection would be evident between WT, RAGE-null vs. Myd88-null remnants. Table 1 reveals the number of differentially expressed probesets with Benjamini-Hochberg false discovery rate (PBH) < 0.05 among the various groups of mice and comparisons, and Tables 2–4 indicate the number of unique Entrez genes with PBH < 0.05 between RAGE-null vs. WT remnants (Table 2), Myd88-null vs. WT remnants (Table 3), and RAGE-null/Myd88-null vs. WT remnants (Table 4).
Table 1.
Number of differentially expressed genes for each comparison
| Table | Comparison | Probesets, PBH < 0.05 | Unique Entrez genes, PBH < 0.05 |
|---|---|---|---|
| 2 | RAGE null vs. WT | 49 | 32 |
| 3 | Myd88 null vs.WT | 24 | 20 |
| 4 | RAGE null/Myd88 null vs.WT | 36 | 22 |
| Not shown | RAGE null vs. Myd88 null | 119 | 89 |
| Not shown | RAGE null/Myd88 null vs. RAGE null | 117 | 87 |
| Not shown | RAGE null/Myd88 null vs. Myd88 null | 20 | 10 |
Table 2.
RAGE-null vs. WT genes with PBH < 0.05
| Rank | ID | Symbol | Description | log2FC | PBH |
|---|---|---|---|---|---|
| 1 | 1458719_at | NA | NA | 3.37 | 3.E-06 |
| 2 | 1424108_at | Glo1 | Glyoxalase 1 | 1.30 | 3.E-06 |
| 3 | 1450531_at | H2-Bl | Histocompatibility 2, blastocyst | 2.13 | 1.E-04 |
| 4 | 1424109_a_at | Glo1 | Glyoxalase 1 | 0.83 | 1.E-04 |
| 5 | 1451240_a_at | Glo1 | Glyoxalase 1 | 0.86 | 1.E-04 |
| 6 | 1425496_at | Abca3 | ATP-binding cassette, subfamily A (ABC1), member 3 | 2.89 | 3.E-04 |
| 7 | 1447260_at | H2-Bl | Histocompatibility 2, blastocyst | 1.59 | 4.E-04 |
| 8 | 1451731_at | Abca3 | ATP-binding cassette, subfamily A (ABC1), member 3 | 2.85 | 5.E-04 |
| 9 | 1426995_a_at | Gfer | Growth factor, erv1 (Saccharomyces cerevisiae)-like (augmenter of liver regeneration) | 2.09 | 0.001 |
| 10 | 1425623_a_at | Cbs | Cystathionine β-synthase | 1.17 | 0.002 |
| 11 | 1436684_a_at | Riok2 | RIO kinase 2 (yeast) | −2.05 | 0.002 |
| 12 | 1426663_s_at | Slc45a3 | Solute carrier family 45, member 3 | 2.43 | 0.003 |
| 13 | 1435872_at | NA | NA | −2.57 | 0.004 |
| 14 | 1426664_x_at | Slc45a3 | Solute carrier family 45, member 3 | 2.17 | 0.006 |
| 15 | 1437911_at | 6330416L07Rik | RIKEN cDNA 6330416L07 gene | −1.25 | 0.008 |
| 16 | 1423844_s_at | Cbs | Cystathionine β-synthase | 0.78 | 0.008 |
| 17 | 1455648_at | NA | NA | −1.26 | 0.008 |
| 18 | 1455439_a_at | Lgals1 | Lectin, galactose binding, soluble 1 | −1.40 | 0.009 |
| 19 | 1419573_a_at | Lgals1 | Lectin, galactose binding, soluble 1 | −1.62 | 0.009 |
| 20 | 1439012_a_at | Dck | Deoxycytidine kinase | −0.78 | 0.010 |
| 21 | 1460425_at | 1700001C19Rik | RIKEN cDNA 1700001C19 gene | −1.49 | 0.012 |
| 22 | 1423006_at | Pim1 | Proviral integration site 1 | 1.24 | 0.014 |
| 23 | 1423481_at | Riok2 | RIO kinase 2 (yeast) | −1.29 | 0.015 |
| 24 | 1435385_at | Tshz2 | Teashirt zinc finger family member 2 | 0.93 | 0.015 |
| 25 | 1422888_at | Rnf5 | Ring finger protein 5 | 0.58 | 0.015 |
| 26 | 1427711_a_at | Ceacam1 | Carcinoembryonic antigen-related cell adhesion molecule 1 | 1.60 | 0.016 |
| 27 | 1428191_s_at | Mett11d1 | Methyltransferase 11 domain containing 1 | 0.76 | 0.020 |
| 28 | 1416256_a_at | Tubb5 | Tubulin, β 5 | 0.69 | 0.021 |
| 29 | 1436070_at | Glo1 | Glyoxalase 1 | 1.06 | 0.021 |
| 30 | 1431166_at | Chd1 | Chromodomain helicase DNA binding protein 1 | 1.51 | 0.024 |
| 31 | 1438289_a_at | Sumo1 | SMT3 suppressor of mif two 3 homolog 1 (yeast) | −0.55 | 0.024 |
| 32 | 1452272_a_at | Gfer | Growth factor, erv1 (S. cerevisiae)-like (augmenter of liver regeneration) | −0.78 | 0.028 |
| 33 | 1419766_at | Sik1 | Salt-inducible kinase 1 | −0.80 | 0.028 |
| 34 | 1452754_at | Creld2 | Cysteine-rich with EGF-like domains 2 | 0.97 | 0.030 |
| 35 | 1417843_s_at | Eps8l2 | EPS8-like 2 | 1.14 | 0.030 |
| 36 | 1449342_at | Ptplb | Protein tyrosine phosphatase-like (proline instead of catalytic arginine), member b | −0.49 | 0.032 |
| 37 | 1419866_s_at | Atxn2 | Ataxin 2 | 1.15 | 0.034 |
| 38 | 1435140_at | Ide | Insulin degrading enzyme | 0.96 | 0.035 |
| 39 | 1418442_at | Xpo1 | exportin 1, CRM1 homolog (yeast) | 0.81 | 0.035 |
| 40 | 1434625_at | 4930432O21Rik | RIKEN cDNA 4930432O21 gene | −0.98 | 0.037 |
| 41 | 1433859_at | Ino80c | INO80 complex subunit C | −0.93 | 0.037 |
| 42 | 1435650_at | Hapln4 | Hyaluronan and proteoglycan link protein 4 | 0.88 | 0.037 |
| 43 | 1417274_at | Snrpa | Small nuclear ribonucleoprotein polypeptide A | 1.05 | 0.037 |
| 44 | 1444138_at | Cyp2r1 | Cytochrome P450, family 2, subfamily r, polypeptide 1 | −0.62 | 0.037 |
| 45 | 1452224_at | Morc3 | Microrchidia 3 | −1.14 | 0.039 |
| 46 | 1417618_at | Itih2 | Inter-α trypsin inhibitor, heavy chain 2 | 0.47 | 0.039 |
| 47 | 1435458_at | Pim1 | Proviral integration site 1 | 1.62 | 0.042 |
| 48 | 1434188_at | Slc16a12 | Solute carrier family 16 (monocarboxylic acid transporters), member 12 | −0.70 | 0.044 |
| 49 | 1435072_at | Zfyve1 | Zinc finger, FYVE domain containing 1 | 1.17 | 0.049 |
Table 3.
Myd88-null vs. WT genes with PBH < 0.05
| Rank | ID | Symbol | Description | log2FC | PBH |
|---|---|---|---|---|---|
| 1 | 1435682_at | Lars2 | Leucyl-tRNA synthetase, mitochondrial | −2.23 | 1.E-06 |
| 2 | 1438238_at | 2010315B03Rik | RIKEN cDNA 2010315B03 gene | −3.02 | 8.E-06 |
| 3 | 1460285_at | Itga9 | Integrin α 9 | 1.55 | 5.E-05 |
| 4 | 1456182_x_at | Mela | Melanoma antigen | 5.63 | 3.E-04 |
| 5 | 1422731_at | Limd1 | LIM domains containing 1 | −2.24 | 7.E-04 |
| 6 | 1419272_at | Myd88 | Myeloid differentiation primary response gene 88 | −2.06 | 7.E-04 |
| 7 | 1433438_x_at | Mela | Melanoma antigen | 3.33 | 0.002 |
| 8 | 1428604_at | 2610305D13Rik | RIKEN cDNA 2610305D13 gene | 2.05 | 0.004 |
| 9 | 1425243_at | Cd207 | CD207 antigen | −1.59 | 0.009 |
| 10 | 1421653_a_at | Igh | Immunoglobulin heavy chain complex | 2.04 | 0.009 |
| 11 | 1423606_at | Postn | Periostin, osteoblast specific factor | 1.34 | 0.009 |
| 12 | 1433685_a_at | 6430706D22Rik | RIKEN cDNA 6430706D22 gene | 1.26 | 0.013 |
| 13 | 1437834_s_at | Pacsin3 | Protein kinase C and casein kinase substrate in neurons 3 | 1.30 | 0.017 |
| 14 | 1427764_a_at | Tcfe2a | Transcription factor E2a | 0.44 | 0.025 |
| 15 | 1426951_at | Crim1 | Cysteine rich transmembrane BMP regulator 1 (chordin like) | 1.26 | 0.034 |
| 16 | 1434825_at | Tnrc18 | Trinucleotide repeat containing 18 | 0.91 | 0.037 |
| 17 | 1455051_at | Rnf31 | Ring finger protein 31 | 0.58 | 0.037 |
| 18 | 1448779_at | Ciz1 | CDKN1A interacting zinc finger protein 1 | 0.86 | 0.037 |
| 19 | 1427051_at | Tnks1bp1 | Tankyrase 1 binding protein 1 | 0.77 | 0.037 |
| 20 | 1460559_at | Kank2 | KN motif and ankyrin repeat domains 2 | 0.83 | 0.040 |
| 21 | 1427177_at | Fyco1 | FYVE and coiled-coil domain containing 1 | 0.90 | 0.044 |
| 22 | 1418168_at | Zcchc14 | Zinc finger, CCHC domain containing 14 | 1.20 | 0.048 |
| 23 | 1456133_x_at | Itgb5 | Integrin β 5 | 0.76 | 0.050 |
| 24 | 1450622_at | Bcar1 | Breast cancer antiestrogen resistance 1 | 0.89 | 0.050 |
Table 4.
RAGE-null/Myd88-null vs. WT genes with PBH < 0.05
| Rank | ID | Symbol | Description | log2FC | PBH |
|---|---|---|---|---|---|
| 1 | 1435682_at | Lars2 | Leucyl-tRNA synthetase, mitochondrial | −2.10 | 2.E-06 |
| 2 | 1458719_at | NA | NA | 3.73 | 2.E-06 |
| 3 | 1438238_at | 2010315B03Rik | RIKEN cDNA 2010315B03 gene | −3.25 | 2.E-06 |
| 4 | 1424108_at | Glo1 | Glyoxalase 1 | 1.28 | 9.E-06 |
| 5 | 1460285_at | Itga9 | Integrin α 9 | 1.44 | 9.E-05 |
| 6 | 1422731_at | Limd1 | LIM domains containing 1 | −2.51 | 9.E-05 |
| 7 | 1450531_at | H2-Bl | Histocompatibility 2, blastocyst | 2.22 | 9.E-05 |
| 8 | 1426995_a_at | Gfer | Growth factor, erv1 (Saccharomyces cerevisiae)-like (augmenter of liver regeneration) | 2.66 | 9.E-05 |
| 9 | 1451240_a_at | Glo1 | Glyoxalase 1 | 0.93 | 9.E-05 |
| 10 | 1435872_at | NA | NA | −3.64 | 2.E-04 |
| 11 | 1419272_at | Myd88 | Myeloid differentiation primary response gene 88 | −2.10 | 3.E-04 |
| 12 | 1447260_at | H2-Bl | Histocompatibility 2, blastocyst | 1.71 | 3.E-04 |
| 13 | 1425496_at | Abca3 | ATP-binding cassette, sub-family A (ABC1), member 3 | 2.99 | 3.E-04 |
| 14 | 1424109_a_at | Glo1 | Glyoxalase 1 | 0.77 | 5.E-04 |
| 15 | 1451731_at | Abca3 | ATP-binding cassette, sub-family A (ABC1), member 3 | 2.87 | 0.001 |
| 16 | 1421653_a_at | Igh | Immunoglobulin heavy chain complex | 2.17 | 0.003 |
| 17 | 1434141_at | Gucy1a3 | Guanylate cyclase 1, soluble, α 3 | 0.84 | 0.003 |
| 18 | 1456182_x_at | Mela | Melanoma antigen | 4.40 | 0.003 |
| 19 | 1436684_a_at | Riok2 | RIO kinase 2 (yeast) | −2.10 | 0.003 |
| 20 | 1437911_at | 6330416L07Rik | RIKEN cDNA 6330416L07 gene | −1.44 | 0.004 |
| 21 | 1416256_a_at | Tubb5 | Tubulin, beta 5 | 0.83 | 0.011 |
| 22 | 1431648_at | 4930528F23Rik | RIKEN cDNA 4930528F23 gene | 1.77 | 0.013 |
| 23 | 1429385_at | Wdr68 | WD repeat domain 68 | 0.70 | 0.014 |
| 24 | 1452272_a_at | Gfer | Growth factor, erv1 (S. cerevisiae)-like (augmenter of liver regeneration) | −0.93 | 0.014 |
| 25 | 1434188_at | Slc16a12 | Solute carrier family 16 (monocarboxylic acid transporters), member 12 | −0.89 | 0.016 |
| 26 | 1428832_at | 1600002H07Rik | RIKEN cDNA 1600002H07 gene | −2.20 | 0.019 |
| 27 | 1423848_at | Mphosph6 | M-phase phosphoprotein 6 | 0.75 | 0.019 |
| 28 | 1418142_at | Kcnj8 | Potassium inwardly-rectifying channel, subfamily J, member 8 | 0.89 | 0.019 |
| 29 | 1423606_at | Postn | Periostin, osteoblast specific factor | 1.17 | 0.021 |
| 30 | 1419073_at | Tmeff2 | Transmembrane protein with EGF-like and two follistatin-like domains 2 | 1.33 | 0.025 |
| 31 | 1453198_at | 100043468 | Predicted gene, 100043468 | 1.18 | 0.025 |
| 32 | 1423659_a_at | Tbc1d17 | TBC1 domain family, member 17 | 0.78 | 0.030 |
| 33 | 1423844_s_at | Cbs | cystathionine beta-synthase | 0.72 | 0.039 |
| 34 | 1415750_at | Tbl3 | Transducin β-like 3 | 1.02 | 0.039 |
| 35 | 1425763_x_at | Igh | Immunoglobulin heavy chain complex | 1.52 | 0.044 |
| 36 | 1455648_at | NA | NA | −1.13 | 0.049 |
Overrepresented KEGG pathways (10), based on the comparisons between RAGE-null vs. WT and Myd88-null vs. WT pathways (hypergeometic P<0.20) identified by Pathway Express (11), are given in Tables 5 and 6. We particularly noted that RAGE-null vs. WT comparisons included the Jak/STAT pathway. The Jak/STAT pathway is known to play key roles in liver regeneration and thus was a logical direction for pursuit of unique RAGE-dependent mechanisms. From these data, we identified two genes whose expression casts light on the observed genotype-phenotype relationships in extensive liver resection: Pim1 (proviral integration site of the Moloney murine leukemia virus) and Glo1. Pim1 was highlighted, as the Pathway Express analysis (12) showed that it was influenced by RAGE as part of the Jak/STAT pathway (13, 14). The second gene, Glo1, deactivates a precursor of RAGE ligand AGEs, MG, by catalyzing a step in the transformation of MG to lactate (15). Thus, Glo1 slows the rate of AGE formation by reducing the concentration of MG.
Table 5.
RAGE-null vs. WT pathways with P < 0.05
| Rank | Pathway | Input genes in pathway | Pathway genes on chip | P |
|---|---|---|---|---|
| 1 | Biosynthesis of unsaturated fatty acids | 1 | 27 | 0.04 |
| 2 | Graft-versus-host disease | 1 | 39 | 0.06 |
| 3 | Allograft rejection | 1 | 40 | 0.06 |
| 4 | ABC transporters | 1 | 42 | 0.06 |
| 5 | Type I diabetes mellitus | 1 | 45 | 0.07 |
| 6 | Autoimmune thyroid disease | 1 | 50 | 0.08 |
| 7 | Acute myeloid leukemia | 1 | 56 | 0.08 |
| 8 | Antigen processing and presentation | 1 | 70 | 0.10 |
| 9 | Gap junction | 1 | 84 | 0.12 |
| 10 | Cell adhesion molecules (CAMs) | 1 | 131 | 0.19 |
| 11 | Jak-STAT signaling pathway | 1 | 137 | 0.19 |
Table 6.
Myd88-null vs. WT pathways with P < 0.05
| Rank | Pathway | Input genes in pathway | Pathway genes on chip | P |
|---|---|---|---|---|
| 1 | Focal adhesion | 3 | 190 | 8.E-04 |
| 2 | Regulation of actin cytoskeleton | 3 | 201 | 9.E-04 |
| 3 | ECM-receptor interaction | 2 | 77 | 3.E-03 |
| 4 | Asthma | 1 | 26 | 0.03 |
| 5 | Primary immunodeficiency | 1 | 35 | 0.03 |
| 6 | Allograft rejection | 1 | 40 | 0.04 |
| 7 | Autoimmune thyroid disease | 1 | 50 | 0.05 |
| 8 | B-cell receptor signaling pathway | 1 | 67 | 0.06 |
| 9 | Fc ε RI signaling pathway | 1 | 78 | 0.07 |
| 10 | Systemic lupus erythematosus | 1 | 79 | 0.07 |
| 11 | Hematopoietic cell lineage | 1 | 81 | 0.08 |
| 12 | Apoptosis | 1 | 85 | 0.08 |
| 13 | Toll-like receptor signaling pathway | 1 | 94 | 0.09 |
| 14 | Leukocyte transendothelial migration | 1 | 113 | 0.10 |
| 15 | Cell adhesion molecules (CAMs) | 1 | 131 | 0.12 |
| 16 | Calcium signaling pathway | 1 | 171 | 0.15 |
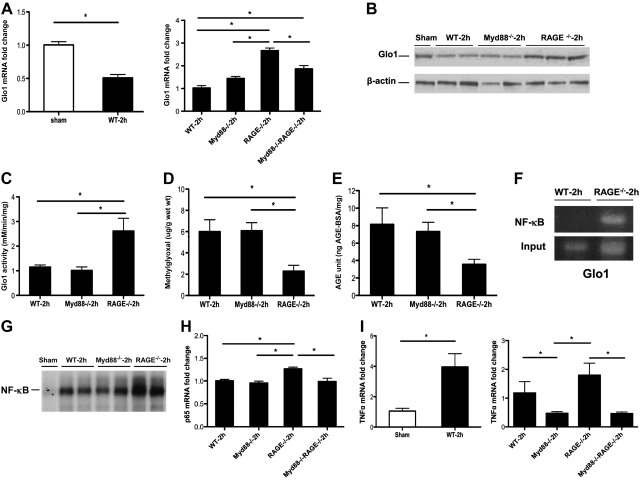
Based on these considerations, we sought to verify the Affymetrix gene arrays and to determine the potential effect of Glo1 and Pim1 in extensive liver resection. First, we analyzed Glo1 and report that in WT mice, compared to sham-treated mice, extensive hepatectomy resulted in 2-fold lower levels of Glo1 mRNA transcript levels at 2 h postresection. Quantitative real-time PCR experiments confirmed that, compared to all other groups, RAGE-null remnants displayed the highest levels of Glo1 mRNA transcripts at this time point (Fig. 3A). Similar patterns of Glo1 protein expression and activity in the remnant were observed (Fig. 3B, C). In parallel, levels of MG and AGEs were much lower in RAGE-null remnants vs. WT or Myd88-null remnants at 2 h postresection (Fig. 3D, E).
Figure 3.
Gene deletion of RAGE modulates Glo1, MG, and AGE production, and NF-κB activation. A) mRNA transcript levels for Glo1 in the liver remnants were determined in the indicated mice by real-time PCR. Data are means ± se; n = 5. B) Equal amounts of protein extracts from hepatic remnants were subjected to immunoblotting with anti-Glo1 IgG. Illustrated bands are representative of n = 4 mice/condition. C) Glo1 activity was assessed as described in the indicated groups; n = 4 mice/condition. D, E) At the indicated times, MG (D) and AGEs (E) were determined in the liver remnants as described; n = 4 mice/condition. F) WT and RAGE-null remnants were retrieved 2 h after extensive hepatectomy. ChIP was performed using anti-NF-κB p65 IgG, and the ChIP-enriched DNA was subjected to PCR for Glo1; n = 3 mice/condition. G) Nuclear extracts were prepared from the remnants of the indicated mice, and EMSA for NF-κB was performed; n = 4 mice/condition. H) mRNA transcript levels for p65 in the liver remnants were determined in the indicated mice by real-time PCR; n = 4–5. I) mRNA transcript levels for TNF-α in the liver remnants were determined in the indicated mice by real-time PCR; n = 4–5. Data are means ± se. *P < 0.05.
To identify the specific mechanisms by which RAGE deficiency was linked to up-regulation of Glo1 mRNA, protein, and activity, we examined the Glo1 promoter and identified potential binding sites for NF-κB, a key factor in liver regeneration. To test whether RAGE was linked to Glo1 regulation via NF-κB, we performed ChIP assays. WT and RAGE-null remnants were retrieved 2 h after 85% hepatectomy. ChIP was performed using anti-NF-κB p65 IgG, and the ChIP-enriched DNA was subjected to PCR for Glo1. PCR products revealed NF-κB bound to Glo1 promoters in RAGE-null remnants that were not detected in the WT mice (Fig. 3F).
As these data suggested that RAGE modulated NF-κB expression and/or activity, we performed EMSAs. At 2 h posthepatectomy, compared to WT and Myd88-null remnants, nuclear extracts retrieved from RAGE-null mice displayed the highest degree of nuclear NF-κB DNA binding (Fig. 3G). To assess whether RAGE regulated expression of NF-κB subunit p65, we performed quantitative real-time PCR on liver remnants at 2 h after resection and found that the highest levels of p65 mRNA transcripts were found in RAGE-null remnants vs. all other groups (Fig. 3H). Furthermore, as TNF-α has been linked to liver regeneration (16, 17) and is a key target of NF-κB, we examined its levels after 85% resection. Up-regulation of TNF-α occurred after extensive resection vs. sham treatment in the remnants of WT mice (Fig. 3I). Compared to WT mice after 85% hepatectomy, Myd88-null remnants displayed significantly lower levels of TNF-α transcripts (Fig. 3I). However, levels of TNF-α transcripts were not significantly different between WT vs. RAGE-null remnants, thereby suggesting that additional factors contributed to RAGE/NF-κB-dependent mechanisms in regulation of adaptive responses to extensive liver resection.
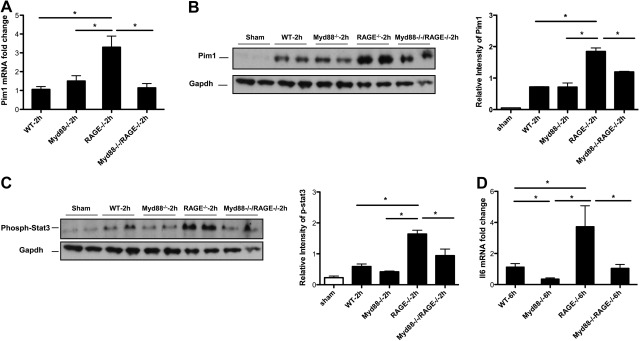
Gene deletion of RAGE up-regulates Pim1 and increases phospho-Stat3 after extensive hepatectomy
To explore this possibility further, we noted that the Affymetrix gene array and Pathway Express analyses suggested potential roles for Pim1 in extensive liver injury. Pim1 inhibits apoptosis and stimulates cellular proliferation (18), a pattern consistent with findings in RAGE-null remnants. Real-time quantitative PCR confirmed that levels of Pim1 mRNA were significantly higher in RAGE-null remnants at 2 h vs. WT, Myd88-null, or Myd88-null/RAGE-null remnants after 85% resection (Fig. 4A). Protein levels of Pim1 protein were significantly higher in the RAGE-null remnants vs. WT remnants at 2 h after extensive hepatectomy, but no significant difference was found between WT, Myd88-null, and Myd88-null/RAGE-null remnants (Fig. 4B). Notably, these data revealed that deletion of Myd88 had no significant effect on Pim1 levels compared to WT mice, suggesting that regulation of Pim1 was RAGE but not Myd88 dependent.
Figure 4.
Up-regulation of Pim1 and increased phospho-Stat3 by RAGE gene deletion after extensive hepatectomy. A) mRNA transcript levels for Pim1 in the liver remnants were determined in the indicated mice by real-time PCR. Data are means ± se; n = 4–5. B, C) Equal amounts of protein extracts from hepatic remnants in the indicated mice were subjected to immunoblotting with anti-Pim1 IgG (B), or with anti-phospho-Stat3 IgG (C), followed by anti-Gapdh IgG. Illustrated bands are representative of n = 4 mice/condition. D) At the indicated times, mRNA transcript levels for Il6 in the liver remnants were determined by real-time PCR. Data are means ± se; n = 4–5. *P < 0.05.
A prominent regulatory mechanism linked to transcription of Pim1 is the phosphorylation of Stat3 (19). Western blotting of remnants at 2 h after extensive injury revealed very low levels of phospho-Stat3 in WT, Myd88-null, and Myd88-null/RAGE-null remnants, but much greater degrees of Stat3 phosphorylation in RAGE-null remnants, which was significantly higher than that observed in all other groups (Fig. 4C). As phosphorylation of Stat3 is regulated indirectly by Il6, an NF-κB-dependent cytokine that contributes to hepatocyte proliferation and survival after extensive resection (20, 21), we examined Il6 levels in the remnants. Compared to WT, Myd88-null, or Myd88-null/RAGE-null mice, RAGE-null mouse remnants displayed the highest levels of Il6 mRNA transcripts (Fig. 4D). Thus, when Myd88 signaling was absent, Il6 was profoundly reduced, revealing the primacy of the Myd88 pathway in regulation of this molecule. However, when Myd88 signaling was active, deletion of RAGE resulted in much higher Il6 mRNA transcripts beyond those observed in all other groups, suggesting that RAGE action suppresses Il6 generation, even in Myd88-expressing mice.
AGE-RAGE suppresses Il6-induced phosphorylation of Stat3 and up-regulation of Pim1 in hepatocytes
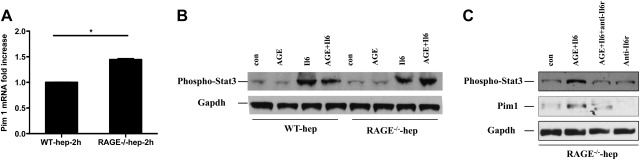
To account for these findings, we hypothesized that RAGE-mediated down-regulation of Glo1 increased production of RAGE ligand AGEs in the remnant, and that AGEs via RAGE antagonized Il6, thereby reducing phosphorylation of Stat3 and blocking up-regulation of Pim1. To test this hypothesis, we retrieved hepatocytes directly from WT and RAGE-null remnants at 2 h after extensive resection and found that the levels of Pim1 mRNA were indeed significantly higher in RAGE-null vs. WT hepatocytes (Fig. 5A).
Figure 5.
AGE-RAGE suppresses Il6 mediated-phosphorylation of Stat3 and up-regulation of Pim1 in hepatocytes. A) Ex vivo, hepatocytes were isolated from WT or RAGE-null mice 2 h after 85% resection, and the mRNA transcripts levels of Pim1 were determined by real-time PCR. Data are means ± se; n = 3. *P < 0.05. B) Hepatocytes were isolated from WT or RAGE-null mice and stimulated with AGE, Il6, or AGE plus Il6 as described. Immunoblotting was performed with anti-phospho-Stat3 IgG, followed by anti-Gapdh IgG; n = 3 mice/condition. C) Hepatocytes were isolated from RAGE-null mice and stimulated by AGE and Il6, and an Il6 receptor antagonist (anti-Il6r). Immunoblotting was performed with anti-phospho-Stat3 IgG or anti-Pim1 IgG, followed by anti-Gapdh IgG. Illustrated bands are representative of n = 3 mice/condition.
Accordingly, to trace the mechanisms linked to RAGE ligands and their effect on phosphorylation of Stat3 and Pim1 expression, we incubated WT and RAGE-null hepatocytes retrieved from uninjured mice with Il6 alone, or in the presence of RAGE ligand AGE. In WT hepatocytes, incubation with both Il6 and AGE suppressed phosphorylation of Stat3 compared to Il6 alone, but in RAGE-null hepatocytes, phosphorylation of Stat3 was not suppressed in the presence of AGE and Il6 (Fig. 5B). To specifically link Il6 as the key factor responsible for phosphorylation of Stat3 and up-regulation of Pim1, we incubated RAGE-null hepatocytes with AGE, Il6, and an Il6 receptor antagonist (anti-Il6r; ref. 22). We found markedly lower phosphorylation of Stat3 and production of Pim1 in the presence of Il6 receptor antagonist, even in RAGE-deficient hepatocytes, thereby confirming that Il6 was a key factor linked to Stat3 phosphorylation and regulation of Pim1 (Fig. 5C). An integrated model of the role of Myd88 and RAGE in the response to extensive liver resection is illustrated in Fig. 6.
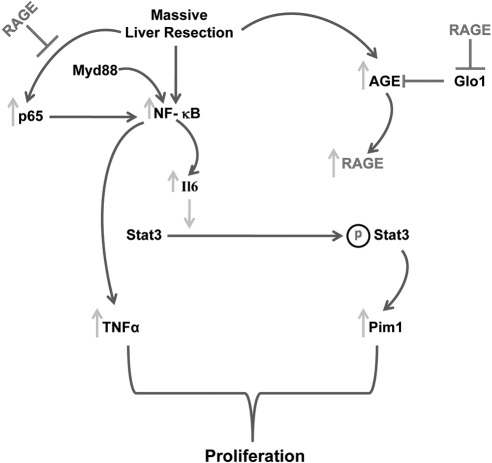
Figure 6.
Opposing roles of Myd88 and RAGE in extensive liver resection. In this study, we addressed the roles of Myd88 and RAGE in the response to massive liver resection. Our findings reveal that the Myd88 pathway is essential for survival consequent to 85% resection of the liver. Myd88 action stimulates activation of NF-κB and subsequent up-regulation of key genes involved in liver regeneration responses, including TNF-α and Il6. In contrast, RAGE opposes the actions of Myd88 signaling by suppression of p65 levels, thereby reducing activation of NF-κB and consequent production of cyclin D1, and by suppression of Il6-mediated phosphorylation of Stat3, thereby down-regulating Pim1 and reducing the hyperplastic response. Further, RAGE-dependent suppression of Glo1, a detoxification pathway for pre-AGEs, enhances AGE levels and fuels a mechanism that suppresses Il6 action.
DISCUSSION
Our findings demonstrate unequivocal roles for the Myd88 signaling pathway in extensive liver damage in countering the effects of the increased density of LPS and other toxins filtered through the exceptionally small remnant after 85% resection, at least in part through NF-κB (23). NF-κB regulates generation of Il6, which stimulates phosphorylation of Stat3 and up-regulation of key target,s such as Pim1 (24, 25); the role of Pim1 in survival mechanisms consequent to extensive liver resection is illustrated here for the first time. Furthermore, cell cycle progression is augmented by NF-κB-driven up-regulation of TNF-α (16) and cyclin D1 (26), both key factors in hepatocyte replication. Taken together, Myd88 signaling stimulates multiple pathways linked to proliferation in the remnant after extensive resection.
In striking contrast, in RAGE-deficient mice, activation of NF-κB is even higher than that observed in the remnants of WT mice, at least in part due to RAGE-dependent regulation of p65. Although binding of ligands to RAGE has been shown to induce NF-κB signaling in other cell types, in the liver, the specific action of RAGE to down-regulate p65 results in the opposite effect, that is, suppression of NF-κB activation. Indeed, this is not surprising, since p65 imparts potent antiapoptotic effects that suppress liver degeneration (27), consistent with the actions of RAGE to reduce survival after massive resection, thereby highlighting the exquisite cellular and tissue-specific roles for NF-κB in homeostasis or exaggerated cellular stress. Thus, RAGE action tempers activation of NF-κB in the liver, leading to antagonism of the beneficial roles of this factor in survival programs.
Key targets of RAGE-dependent suboptimal NF-κB activation in the liver were revealed by the Affymetrix array data. We found that the higher survival rate of RAGE-deficient mice was associated with an NF-κB-dependent up-regulation of Glo1 in the remnants. Using ChIP assays with anti-p65 IgG, we showed that transcripts for Glo1 were detectable selectively in RAGE-deficient but not WT remnants. We conclude that RAGE-driven suppression of Glo1 leads to increased production of RAGE ligand AGEs. Furthermore, consistent with this premise, in in vitro studies, treatment of WT hepatocytes with AGEs blocked the effects of Il6 on phosphorylation of Stat3 and up-regulation of Pim1. In contrast, AGEs had no deleterious effect on Il6 action in hepatocytes devoid of RAGE. These data add to the growing body of evidence that AGEs may be produced even in the absence of high levels of glucose, such as by primary inflammatory and prooxidative mechanisms (28, 29).
Notably, our findings link Pim1 to extensive liver resection and the role of RAGE in its down-regulation in this setting for the first time. We found that RAGE deletion increases levels of cyclin D1, Pim1, and Pcna+ cells in the hepatic remnant after extensive resection, suggesting that RAGE has a potent effect on suppression of key mechanisms that regulate proliferation. The Pim1 gene encodes for a serine/threonine kinase, which is a member of the group of calcium/calmodulin regulated kinases (CAMK; ref. 25). Major regulatory nodes for transcriptional regulation of Pim1 have been identified for STATs, such as Stat3 and Stat5 (30–32), and NF-κB (33, 34). Feedback loops within Pim1 actions sustain activation of NF-κB, as Pim1 has been reported to phosphorylate Ser276 in RelA/p65, thereby controlling NF-κB signaling (35). Among the roles of Pim1 is activation of proproliferation mechanisms, at least in part via regulation of discrete nodes within the cell cycle. For example, Pim1 binds and phosphorylates Cdc25a, a G1-specific cell cycle regulator (36); phosphorylates and activates the G2/M phosphatase Cdc25c (37); and, via Pim1 phosphorylation, inactivates the cyclin kinase inhibitor p21Waf, thereby blocking inhibition of G1/S progression (38). Together with increased levels of cyclin D1 mRNA transcripts in RAGE-null vs. WT or Myd88-null remnants, the deletion of RAGE exerts critical effect on mechanisms that regulate proliferation.
Intriguingly, Meloche et al. (39) recently reported that in vascular smooth muscle cells (VSMCs), proliferation was mediated via a RAGE-dependent increase in Stat3 activation, which, in turn, stimulated Pim1 expression and NFATc1 activation. These researchers showed that in VSMCs, RAGE-dependent activation of Stat3 and Pim1 increased VSMC proliferation, as small interfering RNAs that reduced RAGE expression blocked this effect. Our present findings reveal essentially opposite roles for RAGE—that is, RAGE signaling in hepatocytes suppresses optimal Il6-mediated phosphorylation of Stat3 and consequent up-regulation of Pim1. In this context, it is noteworthy that although Loppnow and Libby (40) found that Il6 may be produced by proliferating SMCs, they found no evidence that Il6 itself modulated SMC proliferation (40). Hence, we speculate that RAGE-dependent control of Pim1 may be unique in different cell types and under distinct forms of stress. Therefore, although Il6 is known to exert potent benefit in hepatocyte proliferation after injury, its direct role in SMC proliferation is less clear.
Finally, our data reveal that Myd88 is essential for NF-κB/TNF-α survival mechanisms in the remnant. These conclusions are affirmed by experiments in mice doubly deficient in both Myd88 and RAGE and establish the primacy of Myd88 signaling pathways in regulating survival after 85% liver resection. Toll-like receptor pathways have been shown to be significant among pathways in which Myd88 plays a role in the liver (41). Myd88 also plays a role in Il1 signaling, which, in turn, plays a role in liver regeneration (42). However, as we have not observed significant changes in Il1 expression (results not shown) in our Affymetrix array data between genotypes, we suggest that toll-like receptor signaling plays the predominant role by which Myd88 acts in liver regeneration, although we cannot discount the possibility that other Myd88-mediated pathways also contribute.
In contrast to the actions of Myd88 signaling, RAGE action antagonizes prosurvival mechanisms, at least in part through reduced Glo1-mediated increased AGEs and consequent signaling via this receptor. Among the consequences of RAGE action is reduction in transcription (p65) and activation of NF-κB, which results in decreased production of Il6. Reduction in Il6 levels suppresses Stat3 phosphorylation and, thereby, expression of its key target protein, Pim1. Of note, it is plausible that reduced Pim1 fuels further cycles of reduced NF-κB action, thereby preventing sustainable Il6 up-regulation and action. We conclude that in concert with effective innate immune signaling, antagonism of RAGE may maximize survival programs in the severely damaged liver by rescuing the remnant from RAGE-dependent suppression of adaptive proliferation. Understanding the distinct roles of Myd88 vs. RAGE signaling in the biological response to massive liver resection will help to establish optimal means to harness and/or inhibit these pathways, respectively.
Supplementary Material
Acknowledgments
The authors thank members of the laboratory of A.M.S. for technical assistance and helpful discussions. The authors gratefully acknowledge the assistance of Ms. Latoya Woods in the preparation of this manuscript.
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1. Samstein B., Emond J. (2001) Liver transplants from living related donors. Ann. Rev. Med. 52, 147–160 [DOI] [PubMed] [Google Scholar]
- 2. Koniaris L. G., McKillop I. H., Schwartz S. I., Zimmers T. A. (2003) Liver regeneration. J. Am. Coll. Surg. 197, 634–659 [DOI] [PubMed] [Google Scholar]
- 3. Panis Y., McMullan D. M., Emond J. C. (1997) Progressive necrosis after hepatectomy and the pathophysiology of liver failure after massive resection. Surgery 121, 142–149 [DOI] [PubMed] [Google Scholar]
- 4. Cataldegirmen G., Zeng S., Ippagunta N., Dun H., Qu W., Lu Y., Rong L. L., Hofmann M. A., Kislinger T., Pachydaki S. I., Jenkins D. G., Weinberg A., Lefkowitch J., Rogiers X., Yan S. F., Schmidt A. M., Emond J. C. (2005) RAGE limits regeneration after massive liver injury by coordinated suppression of TNF-alpha and NF-kappaB. J. Exp. Med. 201, 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kawai T., Akira S. (2007) TLR signaling. Sem. Immunol. 19, 24–32 [DOI] [PubMed] [Google Scholar]
- 6. Burns K., Martinon F., Esslinger C., Pahl H., Schneider P., Bodmer J. L., Di Marco F., French L., Tschopp J. (1998) MyD88, an adapter protein involved in interleukin-1 signaling. J. Biol. Chem. 273, 12203–12209 [DOI] [PubMed] [Google Scholar]
- 7. Ohmori S., Mori M., Kawase M., Tusboi S. (1987) Determination of methylglyoxal as 2-methylquinoxaline by high-performance liquid chromatography and its application to biological samples. J. Chromatogr. 414, 149–155 [DOI] [PubMed] [Google Scholar]
- 8. Bair W. B., III, Cabello C. M., Uchida K., Bause A. S., Wondrak G. T. (2010) Glo1 overexpression in human malignant melanoma. Melanoma Res. 20, 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oltvai Z. N., Milliman C. L., Korsmeyer S. J. (1993) Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell 74, 609–619 [DOI] [PubMed] [Google Scholar]
- 10. Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, D277–D280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Draghici S., Khatri P., Tarca A. L., Amin K., Done A., Voichita C., Georgescu C., Romero R. (2007) A systems biology approach for pathway level analysis. Genome Res. 17, 1537–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cuypers H. T., Selten G., Quint W., Zijlstra M., Maandag E. R., Boelens W., Van Wezenbeek P., Melief C., Berns A. (1984) Murine leukemia virus-induced T cell lymphomagenesis: integration of provirus in a distinct chromosomal region. Cell 37, 141–150 [DOI] [PubMed] [Google Scholar]
- 13. Chen X., Xu C., Zhang F., Ma J. (2010) Microarray approach reveals the relevance of interferon signaling pathways with rat liver restoration post 2/3 hepatectomy at the cellular level. J. Interferon Cytokine Res. 30, 525–539 [DOI] [PubMed] [Google Scholar]
- 14. Sowa J. P., Benko T., Bockhorn M., Gu Y., Niehues E. M., Bucchi A., Benedetto-Castro E. M., Gerken G., Rauen U., Schlaak J. F. (2008) Extent of liver resection modulates the activation of transcription factors and the production of cytokines involved in liver regeneration. World J. Gastroenterol. 14, 7093–7100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thornalley P. J. (1993) The glyoxalase system in health and disease. Mol. Aspects Med. 14, 287–371 [DOI] [PubMed] [Google Scholar]
- 16. Yamada Y., Fausto F. (1998) Deficient liver regeneration after carbon Tetrachloride injury in mice lacking type 1 but not type 2 tumor necrosis factor receptor. Am. J. Pathol. 152, 1577–1589 [PMC free article] [PubMed] [Google Scholar]
- 17. Akerman P., Cote P., Yang S. Q., McClain C., Nelson S., Bagby G. J., Diehl A. M. (1992) Antibodies to tumor necrosis factor-alpha inhibit liver regeneration after partial hepatectomy. Am. J. Physiol. 263, G579–G585 [DOI] [PubMed] [Google Scholar]
- 18. Wang Z., Bhattacharya N., Weaver M., Petersen K., Meyer M., Gapter L., Magnuson N. S. (2001) Pim-1: a serine/threonine kinase with a role in cell survival, proliferation, differentiation and tumorigenesis. J. Vet. Sci. 2, 167–179 [PubMed] [Google Scholar]
- 19. Zemskova M., Sahakian E., Bashkirova S., Lilly M. (2008) The PIM1 kinase is a critical component of a survival pathway activated by docetaxel and promotes survival of docetaxel-treated prostate cancer cells. J. Biol. Chem. 283, 20635–20644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jin X., Zhang Z., Beer-Stoiz D., Zimmers T. A., Koniaris L. G. (2007) Interleukin-6 oxidative injury and necrosis after extreme liver resection. Hepatology 46, 802–812 [DOI] [PubMed] [Google Scholar]
- 21. Cressman D. E., Greeenbaum L. E., DeAngelis R. A., Ciliberto G., Furth E. E., Poli V., Taub R. (1996) Liver failure and defective hepatocyte regeneration in interleukin-6 deficient mice. Science 274, 1379–1383 [DOI] [PubMed] [Google Scholar]
- 22. Brakenhoff J. P., de Hon F. D., Fontaine V., ten Boekel E., Schooltink. H., Rose-John S., Heinrich P. C., Content J., Aarden L. A. (1994) Development of a human interleukin-6 receptor antagonist. J. Biol. Chem. 269, 86–93 [PubMed] [Google Scholar]
- 23. Kawai T., Adachi O., Ogawa T., Takeda K., Akira S. (1999) Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity 11, 115–122 [DOI] [PubMed] [Google Scholar]
- 24. Libermann T. A., Baltimore D. (1990) Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol. Cell. Biol. 10, 2327–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bachmann M., Möröy T. (2005) The serine/threonine kinase Pim-1. Int. J. Biochem. Cell Biol. 37, 726–730 [DOI] [PubMed] [Google Scholar]
- 26. Hanse E. A., Nelsen C. J., Goggin M. M., Anttila C. K., Mullany L. K., Berthet C., Kaldis P., Crary G. S., Kuriyama R., Albrecht J. H. (2009) Cdk2 plays a critical role in hepatocytes cell cycle progression and survival in the setting of cycline D1 expression in vivo. Cell Cycle 8, 2802–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beg A. A., Sha W. C., Bronson R. T., Ghosh S., Baltimore D. (1995) Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature 376, 167–170 [DOI] [PubMed] [Google Scholar]
- 28. Anderson M. M., Requena J. R., Crowley J. R., Thorpe S. R., Heinecke J. W. (1999) The myeloperoxidase system of human phagocytes generates Nepsilon-(carboxymethyl)lysine on proteins: a mechanism for producing advanced glycation end products at sites of inflammation. J. Clin. Invest. 104, 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ramasamy R., Yan S. F., Schmidt A. M. (2009) RAGE: therapeutic target and biomarker of the inflammatory response—the evidence mounts. J. Leukoc. Biol. 86, 505–512 [DOI] [PubMed] [Google Scholar]
- 30. Matikainen S., Sareneva T., Ronni T., Lehtonen A., Koskinen P. J., Julkunen I. (1999) Interferon-alpha activates multiple STAT proteins and upregulates proliferation-associated IL-2R alpha, c-myc, and pim-1 genes in human T cells. Blood 93, 1980–1991 [PubMed] [Google Scholar]
- 31. Buckley A. R. (2000) Transcriptional regulation of pim-1 by prolactin: independence of a requirement for Jak2/Stat signaling. J. Neuroimmunol. 109, 40–46 [DOI] [PubMed] [Google Scholar]
- 32. Stout B. A., Bates M. E., Liu L. Y., Farrington N. N., Bertics P. J. (2004) IL-5 and granulocyte-macrophage colony-stimulating factor activate STAT3 and STAT5 to promote Pim-1 and cyclin D3 protein expression in human eosinophils. J. Immunol. 173, 6409–6417 [DOI] [PubMed] [Google Scholar]
- 33. Zhu N., Ramirez L. M., Lee R. L., Magnuson N. S., Bishop G. A., Gold M. R. (2002) CD40 signaling in B cells regulates the expression of the Pim-1 kinase via the NF-kappa B pathway. J. Immunol. 168, 744–754 [DOI] [PubMed] [Google Scholar]
- 34. Endo Y., Marusawa H., Kinoshita K., Morisawa T., Sakurai T., Okazaki I. M., Watashi K., Shimotohno K., Honjo T., Chiba T. (2007) Expression of activation-induced cytidine deaminase in human hepatocytes via NF-kappaB signaling. Oncogene 26, 5587–5595 [DOI] [PubMed] [Google Scholar]
- 35. Nihira K., Ando Y., Yamaguchi T., Kagami Y., Miki Y., Yoshida K. (2010) Pim-1 controls NF-kB signaling by stabilizing RelA/p65. Cell Death Differ. 17, 689–698 [DOI] [PubMed] [Google Scholar]
- 36. Mochizuki T., Kitanaka C., Noguchi K., Muramatsu T., Asai A., Kuchino Y. (1999) Physical and functional interactions between Pim1 kinase and Cdc25A phosphatase. Implications for the Pim-1 mediated activation of the c-myc signaling pathway. J. Biol. Chem. 274, 18659–18666 [DOI] [PubMed] [Google Scholar]
- 37. Bachmann M., Kosan C., Xing P. X., Montenarh M., Hoffmann I., Moroy T. (2006) The oncogenic serine/threonine kinase Pim-1 directly phosphorylates and activates the G2/M specific phosphatase Cdc25C. Int. J. Biochem. Cell Biol. 38, 430–443 [DOI] [PubMed] [Google Scholar]
- 38. Wang Z., Bhattacharya N., Mixter P. F., Wei W., Sedivy J., Magnuson N. S. (2002) Phosphorylation of the cell cycle inhibitor p21Cip1/WAF1 by Pim-1 kinase. Biochim. Biophys. Acta 1593, 45–55 [DOI] [PubMed] [Google Scholar]
- 39. Meloche J., Paulin R., Courboulin A., Lambert C., Barrier M., Bonnet P., Bisserier M., Roy M., Sussman M. A., Agharazii M., Bonnet S. (2011) RAGE-dependent activation of the oncoprotein Pim1 plays a critical role in systemic vascular remodeling process. Arterioscler. Thromb. Vasc. Biol. 31, 2114–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Loppnow H., Libby P. (1990) Proliferating or interleukin-1 activated human vascular smooth muscle cells secrete copious interleukin 6. J. Clin. Invest. 85, 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kluwe J., Mencin A., Schwabe R. F. (2009) Toll-like receptors, wound healing, and carcinogenesis. J. Mol. Med. (Berl.) 87, 125–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Taub R. (2004) Liver regeneration: from myth to mechanism. Nat. Rev. Mol. Cell. Biol. 5, 836–847 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.