Abstract
This study presents a method for density-based separation of monodisperse encapsulated cells using a standing surface acoustic wave (SSAW) in a microchannel. Even though monodisperse polymer beads can be generated by the state-of-the-art technology in microfluidics, the quantity of encapsulated cells cannot be controlled precisely. In the present study, mono-disperse alginate beads in a laminar flow can be separated based on their density using acoustophoresis. A mixture of beads of equal sizes but dissimilar densities was hydrodynamically focused at the entrance and then actively driven toward the sidewalls by a SSAW. The lateral displacement of a bead is proportional to the density of the bead, i.e., the number of encapsulated cells in an alginate bead. Under optimized conditions, the recovery rate of a target bead group (large-cell-quantity alginate beads) reached up to 97% at a rate of 2300 beads per minute. A cell viability test also confirmed that the encapsulated cells were hardly damaged by the acoustic force. Moreover, cell-encapsulating beads that were cultured for 1 day were separated in a similar manner. In conclusion, this study demonstrated that a SSAW can successfully separate monodisperse particles by their density. With the present technique for separating cell-encapsulating beads, the current cell engineering technology can be significantly advanced.
INTRODUCTION
Cell encapsulation is a highly promising method for forming spherical cell bodies, which can mimic a multi-cellular in-vivo environment, for use as a three-dimensional cell culture model.1, 2, 3 A single cell or cell cluster encapsulated in a biocompatible polymer (alginate,4, 5, 6 poly ethylene glycol (PEG),7, 8, 9 hyaluronic acid (HA),10, 11, 12 etc.13, 14, 15, 16) is a promising tool for tissue engineering applications, such as drug screening,17, 18, 19 cell therapy,20, 21 transplantation,22, 23, 24 or organ bioprinting.25 These encapsulated cells can be used in a variety of applications because of their essential features: they are less vulnerable to both mechanical stress and the host’s immune system, and their properties are better controllable for the purposes of exchanging nutrients and therapeutic agents than normal cells without encapsulation.26, 27
Cell encapsulation technology has been required to produce mono-disperse polymer beads containing a large quantity of cells (such as embryonic stem cells13, 28 and embryonic carcinoma cells4, 5, 18). In particular, in cell therapy, including a large number of cells in a bead is advantageous for delivering a sufficient number of cells with a minimum loss in cell viability.5, 29, 30, 31 Recent microfluidic encapsulation studies have demonstrated successful production of monodisperse polymer beads containing cells using microfluidic techniques.4, 5, 6, 18, 32, 33, 34 These techniques produced highly uniformly sized beads33 with a complex, multi-layered structure4 using various biocompatible polymer materials.4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16 In fact, the dimension of the bead is an essential parameter for determining the biocompatibility and immune response of the encapsulated cells.35, 36 Despite the advancement of microfluidic encapsulation technology, it is difficult to produce polymer beads encapsulating a uniform quantity of cells because of the tendency of cells suspended in a suspending fluid to aggregate. In fact, in the past, random amounts of cells were encapsulated in each polymer bead, resulting in some beads having no cells (empty beads) and some having a small number of cells.26 Thus, an innovative sorting technique that sorts cell-encapsulating beads according to the cell quantity needs to be developed.
Microfluidic separation technology has advanced significantly. Passive separation techniques used to utilize the channel geometry37, 38, 39 and flow characteristics,40, 41 whereas active ones adopted external forces such as optical,42 electric,43, 44, 45, 46 pneumatic,47, 48 and acoustic49, 50, 51, 52 forces. These separation techniques can successfully separate various types of particles, including polymer beads. In particular, surface acoustic wave (SAW)-based acoustophoresis53 has shown a promising potential for various applications because of its exceptional features for handling particles and cells.49, 50, 51, 52, 53, 54 The SAW-based separation method is label-free and noninvasive and features relatively low electric power consumption and a smaller effect on cell viability.55, 56, 57, 58 Most previous studies adopting the SAW method have demonstrated excellent performance in particle separation according to particle size.50, 52, 57 However, density-based separation of particles using SAWs has not yet been reported, even though the theoretical equation apparently indicated the possibility of particle separation using solely the effect of the density difference among the particles.59
In this article, therefore, we describe a simple but novel approach to separation of cell-encapsulating beads using standing surface acoustic waves (SSAWs) to separate beads according to the bead density in a microfluidic channel. The essential feature of this method is that it separates uniformly sized polymer beads containing different quantities of cells according to the bead density, or, in other words, it performs the so-called density-dependent acoustophoresis of cell-encapsulating beads. In addition, another objective of this report is to demonstrate the cell viability after separation using the SSAW method. Considering these requirements, interdigitated transducers (IDTs) and microchannels have been carefully designed, and the hydrodynamic focusing technique has been adopted as well.
MATERIALS AND METHODS
Sample preparation
Alginate beads with P19 mouse embryonic carcinoma (EC) stem cells were produced as described in a previous report.5 Sodium alginate (A2158-250G, Sigma-Aldrich, St. Louis, MO) was dissolved in culture medium (α-MEM, Gibco, Grand Island, NY) to a 2% (w/v) concentration and filtered with a 0.2 μm syringe filter (Pall Life Science, Port Washington, NY). The P19 mouse EC stem cell line was obtained from American Type Culture Collection (CRL1825; Manassas, VA). The detailed culture method is described elsewhere.5 The P19 EC stem cells were prepared and suspended in the alginate solution. Alginate droplets including cells were gelified with calcified oleic acid using two-phase microfluidics. Next, 1.2 g of calcium chloride (C7902-500G, Sigma-Aldrich) was dissolved in 50 ml of 2-methyl-1-propanol (Junsei Chemical, Tokyo, Japan) using ultrasonication (5510-MT, Branson, Danbury, CT). After the calcium solution and oleic acid were mixed, 2-methyl-1-propanol was distilled overnight at 120 °C to prevent damage to the cells and filtered with a 0.2 μm syringe filter to remove any debris.4, 5 The average diameter of the alginate beads was approximately 150.7 ± 11.3 μm.5 The prepared alginate beads were suspended in a cell culture medium (Dulbecco’s Modified Eagle Medium, DMEM) for separation. For the separation experiment, the concentration of the sample was 50% (v/v).
Cell viability test
The viability of the encapsulated cells was examined using MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assays. Prior to the MTT assay, we adopted the Trypan blue stain, which is most commonly used to distinguish viable cells from nonviable cells. Viable cells exclude the dye, while nonviable cells absorb the dye and appear blue. The cells were dispersed in buffered saline and then counted. In result, the 0-day cells, which were selected as a control, yielded cell viability higher than 95%. Then, for the MTT assay, the optical density of the control sample was set to 100% cell viability as a reference value, and the relative optical densities of other cells were evaluated to determine their viability. The encapsulated cells were cultured in flat-bottomed, 96-well tissue culture plates. A volume of 20 μl of MTT solution was added to the wells, and the cells were incubated for 4 h at 37 °C. The MTT dye was reduced to insoluble purple formazan dye crystals in metabolically active cells. Detergent was then added to the wells, solubilizing the crystals so that the absorbance could be read using a spectrophotometer. The absorbance was measured at 570 nm with a plate reader (Wellscan MK3, Labsystems, Finland).
Working principle
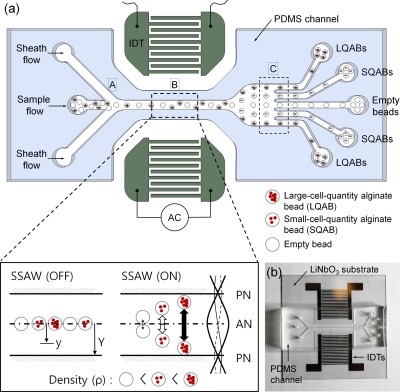
The fundamental principle of the SSAW method is similar to those of methods used for separating and patterning particles in prior studies.50, 52, 55, 57 However, the structures and the dimensions of the flow channel and the IDT electrodes were carefully examined and optimized for the separation of cell-encapsulating beads. As illustrated in Fig. 1a, three inlet channels converge to a single straight channel, and the main test section, in which the acoustic radiation force is applied, connects to five outlets. The inlet channels introduce the sample flow containing the alginate beads through a center channel and the sheath flow through two outer channels (“A” region in Fig. 1a). The alginate beads are focused hydrodynamically toward the center of the channel by the sheath flow. Notably, adopting a sheath flow in the outer inlets allows all beads of the sample fluid to be initially aligned along the centerline of the stream at the entrance of the straight microchannel, as shown in Fig. 1 (inlet). In fact, initializing the particle position using sheath fluid injection provides a starting line for particles to migrate toward the side walls.
Figure 1.
(a) Schematic illustration of the working principle of the SSAW for density-based alginate bead separation. (b) Photograph of the device consisting of a PDMS microchannel and patterned IDTs on a piezoelectric LiNbO3 wafer.
Two SAWs generated across the channel propagate and encounter each other in the channel flow. Two SAWs encountering each other superpose and form standing waves, which cause pressure fluctuations in the channel. Upon the superposition of these two pressure waves, pressure nodes (PNs, minimum pressure amplitude) and pressure antinodes (ANs, maximum pressure amplitude) are formed, and lateral acoustic radiation forces are generated between the nodes. Therefore, any particle in a suspending medium can be driven laterally to either the PNs (minimum pressure amplitude) or the ANs (maximum pressure amplitude), depending on the density and compressibility of the particle relative to those of the medium, as described in Eq. 1.59 In the present study, the lateral acoustic force drives alginate beads containing cells toward the PNs (“B” region in Fig. 1).
The acoustic force, which is a function of the bead properties, is described in Eq. 159
| (1) |
where Fr, p0, Vp, λ, k, x, ρ, and β correspond to the acoustic force, pressure amplitude, particle volume, wavelength, wave vector, distance from the AN to the PN, density, and compressibility, respectively. The p and l subscripts indicate particles and liquid, respectively. The pressure amplitude is also proportional to electrical voltage.50, 57 Since the acoustic radiation force is proportional to the particle volume (Vp ∼ r3), poly-disperse particles can easily be separated by size by the acoustic force.50, 52
In the present study, the alginate beads are almost uniform in size and monodisperse, but they contain different numbers of cells. Thus, the density of each cell-encapsulated bead is a unique physical parameter affecting the separation. Fortunately, the acoustic force is a function of φ, which includes the particle density (ρp), as described in Eq. 1. In fact, φ is a function of the densities and compressibilities of the medium and the particles, and the sign of φ indicates whether the particles are trapped at the PN (for positive φ) or the AN (for negative φ). The φ values for the present beads, which depend on the quantity of cells in each bead, are unfortunately unknown. For reference, φ is 0.32 for red blood cells (RBCs), 0.97 for white blood cells (WBCs), and 0.67 for platelets, and these particles with positive φ values tend to be trapped in the PN.57 Since the beads migrated to the PN, we can confirm that the beads have positive φ values. In the present results, the value of φ is directly proportional to the acoustic force, even though its contribution to the acoustic force (Fν ∝ φ) is smaller than the contribution of the particle size (Fr ∝ Vp ∝ r3).
Thus, cell-encapsulating alginate beads can be separated according to the bead density. Specifically, when subjected to the acoustic radiation force under optimized conditions, alginate beads containing different numbers of cells show different lateral displacements toward the PN. The more cells contained in a bead, the larger the lateral displacement of the bead is, as shown in the “B” region in Fig. 1a. Among the beads, empty beads yield the least lateral displacement. Although all the alginate beads pass through the SSAW working zone at the same time, the densities of the beads are different. Thus, the displacements of the alginate beads encapsulating different numbers of cells are different at the end of the channel.
Microsystem design and fabrication
Fig. 1b shows the device used in the present study. An Au/Cr (100 nm/10 nm) thin film was deposited on a piezoelectric LiNbO3 wafer using an electron beam evaporator (Ulvac, Japan), patterned using a conventional lithographic process and wet-etched to form IDTs (128° Y-cut, X-propagation, 500 μm thick; NEL Crystal Co, Fukushima). A polydimethylsiloxane (PDMS) microfluidic channel with two inlets and five outlets was cured on a silicon wafer, which was patterned using a SU-8 negative photoresist (MicroChem, MA) and bonded to the IDTs patterned on the piezoelectric substrate via oxygen plasma treatment (CUTE, Femto Science Co., Korea). The width and pitch of the IDTs were 250 μm and 500 μm, respectively, corresponding to a SAW working wavelength of 1000 μm. The main channel was 300 μm in width and 200 μm in depth (“B” region in Fig. 1a). Downstream, the main channel divided into outlet channels with 5 outlets to collect the separated alginate beads. The variations in the displacement of the alginate beads were evaluated in this region by normalizing the final displacement of each alginate bead in the direction from the center to the wall (“C” region in Fig. 1a).
Experiments were conducted using an inverted microscope (IX71, Olympus, Japan) equipped with a fast CCD camera (FASTCAM Ultima APX, Photron, Japan). A sample containing the alginate beads was injected through the front inlet and focused by the sheath flow, which was injected simultaneously through the rear inlet. All flows were driven by two syringe pumps (KDS101, KD Scientific) and Hamilton glass syringes (5.0 ml, 1005TTL SYR, Hamilton). The sample and the sheath flow rates were fixed at 8 μl/min and 16 μl/min, respectively. The IDTs were actuated using a signal generator (8657 A, HP), an amplifier (ZHL-1-2 W, Mini-Circuits), and a DC power supply (E3634A, Agilent). The working frequency was set to 3.94 MHz. The applied voltage was altered from 4 V to 22 V in 2 V steps to observe the variations in the normalized displacement due to voltage changes. The corresponding applied input power ranged from 55 mW to 1658 mW. For statistical analysis, we conducted five repeated tests, and the measured values are expressed as mean ± standard deviation (SD). The unpaired two-tailed Student’s t-test was used to determine the statistical significance.
RESULTS AND DISCUSSION
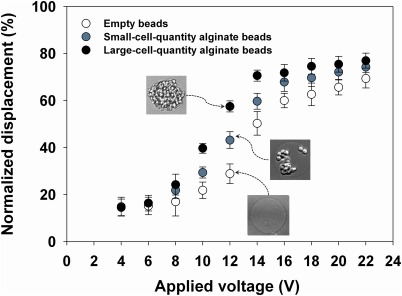
Prior to separating the cell-encapsulating beads according to the bead density, we examined the effect of the voltage applied to the SSAW on the displacement of the cell-encapsulating beads in a microfluidic system. Fig. 2 shows the normalized displacements of alginate beads with different numbers of cells for different applied voltages at a fixed flow rate of 18 µl/min. The normalized displacement was defined as the ratio of the lateral distance (y) of an alginate bead from the centerline of the main channel to half the channel width (Y), as defined in Fig. 1a. For displacement analysis, alginate beads containing various quantities of cells were classified into three groups: large-cell-quantity alginate beads (LQABs), small-cell-quantity alginate beads (SQABs), and empty beads. Through microscopic observation and image analyses, the ratio of cell-projected area to a bead area was determined. LQABs were determined when cell occupying area in a bead is greater than 80%. Since the cell area ratios for most SQABs are less than 50%, there was no difficulty in classifying bead groups between LQABs and SQABs.
Figure 2.
Normalized displacements of alginate beads as a function of applied voltage for different cell quantities.
Overall, the displacement of each bead was approximately proportional to the applied voltage between 8 V and 14 V as well as the bead density: the higher the applied voltage was, the more the alginate beads moved toward the channel wall. Under the optimized voltage of 12 V, the three groups of alginate beads showed an apparent difference in normalized displacement of more than 15%. These results imply that the LQABs, which may eventually be used to form a uniform embryonic body (EB) in a bead,18, 60, 61, 62, 63, 64, 65 could be separated from the other beads with high purity by tuning the applied voltage and controlling the flow rate. After the experiments, the separation/collection performance was evaluated.
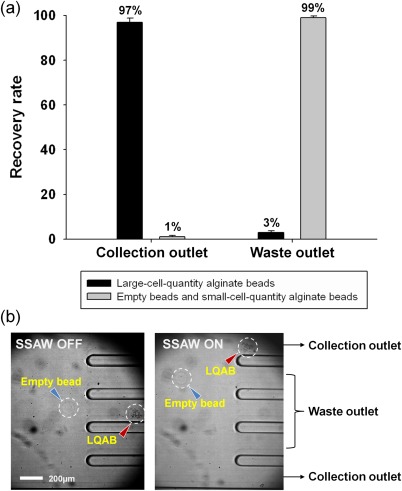
Fig. 3 shows the recovery rate and the captured images of the cell-encapsulating beads in the divided channels at a fixed applied voltage of 12 V. For selective collection of LQABs among the other beads, we defined the two outermost channels as the collection outlets and the three remaining channels in the middle as waste outlets [Fig. 3b]. The LQABs were observed to flow into the collection outlets, since they had the largest displacement induced by the acoustic radiation force in the straight channel flow. In other words, the target beads encapsulating a large quantity of cells (LQABs) were successfully separated according to their bead density difference with high purity by using SSAW-based microfluidics.
Figure 3.
(a) Recovery rate of alginate beads with large quantities of cells from the collection outlet and the waste outlet. (b) Captured images of the divided channel separating alginate beads based on cell quantity at 12 V.
The separation efficiency of the present experiment was evaluated through the recovery rate, which is defined as the ratio of the number of target alginate beads collected in a particular outlet to the number of target beads collected at the all outlets. Fig. 3a shows the recovery rate of target beads at each outlet. In the collection outlet, 99% of the unwanted beads (empty beads and SQABs) were removed through the waste outlets and 97% of the total number of LQABs was collected under a flow rate of 2300 beads per minute. In addition, when the purity was defined as the ratio of the number of collected target beads to the total number of all collected alginate beads in an outlet, the LQAB purity was over 98% in the collection outlet.
The recovery rate in Fig. 3a shows that only 1% of the unwanted beads (empty beads and SQABs) flowed into the collection outlet. The unwanted beads could flow into the wrong outlet because of the different sizes of the alginate beads, even though they were assumed to be monodisperse particles. In fact, a small difference in diameter among the beads would translate to a significantly large difference in volume, and the corresponding acoustic effect on the particles would be quite different. Relatively large-sized empty beads would be strongly affected by the surface acoustic force and collected along with the normal-sized beads containing cells. Thus, it is necessary to sort beads by their size in the first step, with adopting either pinched flow fractionation or acoustic method. It is of note that serial, repetitive applications of acoustic wave in the first and second stages would be a good option since acoustic separation is sensitive for particle size (FSAW ∼ r3) but not for density (FSAW ∼ ρ). Then, for mono-disperse beads, the present density-dependent separation using surface acoustic wave method would be useful.
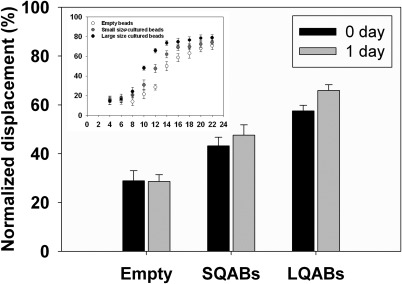
In addition, we examined the feasibility of separation of encapsulated cells cultured for 1 day in an alginate bead. Through a culture process, the cells in a bead grew and formed 1 or 2 EBs. In fact, the size of the EB formed in the alginate beads was dependent on the quantity, viability, and proliferation rate of the encapsulated cells.1, 3, 18, 63 If the last two parameters are constant, the EB size simply depends on the initial quantity of cells in a bead. Under the optimal conditions found as mentioned above (12 V, flow rate = 18 µl/min), the beads encapsulating EBs were successfully separated according to the size of the EB in each alginate bead using the same acoustophoresis-based microfluidic system, as shown in Fig. 4. As in the preceding results, three alginate bead groups were apparently separated based on the size of the EB as a result of a more than 10% difference in normalized displacement. The separated beads in the target outlets were analyzed, and the approximate size of the EBs in the beads was found to be 130 ± 10 μm, which would occupy most of a spherical bead with a 150.7 μm ± 11.3 diameter. In addition, the 1-day cultured beads collected in the target outlets showed slightly more displacement than the 0-day cultured beads, as shown in Fig. 4. These results demonstrate that encapsulated cells can be successfully cultured and became EBs with increasing densities in the alginate beads. In addition, EB-encapsulating beads were easily separated according to the density of the beads using the SSAW method.
Figure 4.
Normalized displacement of cultured beads as a function of the applied voltage for different cultured cell quantities.
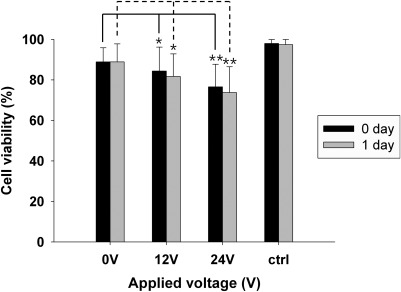
The viability of the encapsulated cells was also examined using MTT assays, as described in Sec. 2. We used freshly encapsulated cells as a control, and we used encapsulated cells that had passed through the microchip without an applied voltage as the 0 V reference sample. Figure 5 shows the effect of the acoustic radiation force on the viability of the cells in alginate beads. As shown in Fig. 5, applying a 12 V acoustic force yielded 85% cell viability, whereas the reference sample had 90% cell viability. Because the corresponding statistical p-value was greater than 0.05, the reduction in cell viability in the 12 V sample can be neglected. For the 24 V case, however, the cell viability was 75%, which is significantly decreased from the reference value (p = 0.015). Although the cell viability for such high voltages would vary with the flow rate and cell-exposure time, the application of high voltages should be carefully considered based on its effect on cell viability.
Figure 5.
Cell viability in beads separated under different applied voltages in 1 day. The p-value was calculated using the unpaired two-tailed Student’s t-test to determine whether the differences between the 0 V reference sample and the sample collected under high voltage conditions were significant (*p > 0.05; **p = 0.015).
The viability of cells would be affected by various factors including electrical, mechanical, and chemical ones. In the present study, the applying voltages range 0–24 V, but the flowing current is relatively low (0.013 A–0.075 A). Thus, there would be minimal effect of electricity on cell viability. In the present study, surface acoustic waves generated in fluid as a mechanical stress might affect the cells encapsulated in beads and the cell viability might be gradually decreased with increasing electric voltage, as shown in Fig. 5. In fact, as described in Eq. 1, the acoustic force is proportional to the pressure amplitude p0, which is also proportional to electric voltage.52, 55 However, in the present study, there was no noticeable change in cell viability for an applied voltage of 12 V, which was found to be the optimum applied voltage. In addition, we evaluated the viability of the cells 1 day after the separation. The separated cells encapsulated in the alginate beads were cultured for 1 day in α-MEM (Gibco) supplemented with 10% FBS (Gibco) and 1% penicillin and streptomycin (Gibco), and their viability was tested. The result indicated a less than 5% difference in cell viability between the 0-day and 1-day cultures (p > 0.05), and thus we confirmed that the alginate beads provide the encapsulated cells with good environments protecting them from mechanical and other stresses.
CONCLUSION
We have demonstrated a simple, novel method for separating cells encapsulated in alginate beads according to the bead density by adopting SSAWs and hydrodynamic focusing. The present study demonstrated that LQABs can be successfully collected with a recovery rate of over 97% and a purity of over 98% at a rate of 2300 beads per minute with acceptable cell viability. We also confirmed that beads could be separated according to the size of the EBs in the alginate beads after 1 day of culturing. The proposed SSAW-based microfluidic method was proven to separate cell-encapsulating beads according to the quantity and size of the cells encapsulated in monodisperse beads, which is the same as the bead density. Even though the operating principle of the SSAW is well known, its density-based separation capability has been reported for the first time in the present study. Since the proposed method allows easy tunability of the governing parameters, it can be further applied to separate various polymer beads, not just alginate beads, with minimum modification. Above all, the present separation method can be widely used for biomedical cell engineering as a result of the unique characteristics of the SSAW principle, such as local actuation and label-free sorting with high cell viability and high purity. In addition, the simple design of our device can be easily integrated with various droplet-generation systems, and thus it can be especially useful for the separation of alginate beads of uniform size containing different numbers of encapsulated cells.
ACKNOWLEDGMENTS
This research was supported by the Nano Material Technology Development Program (Green Nano Technology Development Program) through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (No. 2011-0020090).
References
- Dang S. M., Kyba M., Perlingeiro R., Daley G. Q., and Zandstra P. W., Biotechnol. Bioeng. 78, 442 (2002). 10.1002/bit.10220 [DOI] [PubMed] [Google Scholar]
- Griffith L. G. and Swartz M. A., Nat. Rev. Mol. Cell Biol. 7, 211 (2006). 10.1038/nrm1858 [DOI] [PubMed] [Google Scholar]
- Konno T., Akita K., Kurita K., and Ito Y. J., Biosci. Bioeng. 100, 88 (2005). 10.1263/jbb.100.88 [DOI] [PubMed] [Google Scholar]
- Kim C., Chung S., Kim Y. E., Lee K. S., Lee S. H., Oh K. W., and Kang J. Y., Lab Chip 11, 246 (2011). 10.1039/c0lc00036a [DOI] [PubMed] [Google Scholar]
- Kim C., Lee K. S., Kim Y. E., Lee K. J., Lee S. H., Kim T. S., and Kang J. Y., Lab Chip 9, 1294 (2009). 10.1039/b819044e [DOI] [PubMed] [Google Scholar]
- Tan W. H. and Takeuchi S., Adv. Mater. 19, 2696 (2007). 10.1002/adma.200700433 [DOI] [Google Scholar]
- Cruise G. M., Scharp D. S., and Hubbell J. A., Biomaterials 19, 1287 (1998). 10.1016/S0142-9612(98)00025-8 [DOI] [PubMed] [Google Scholar]
- Weber L. M., Hayda K. N., Haskins K., and Anseth K. S., Biomaterials 28, 3004 (2007). 10.1016/j.biomaterials.2007.03.005 [DOI] [PubMed] [Google Scholar]
- Weber L. M., He J., Bradley B., Haskins K., and Anseth K. S., Acta Biomater. 2, 1 (2006). 10.1016/j.actbio.2005.10.005 [DOI] [PubMed] [Google Scholar]
- Bae K. H., Yoon J. J., and Park T. G., Biotechnol. Prog. 22, 297 (2006). 10.1021/bp050312b [DOI] [PubMed] [Google Scholar]
- Chung C., Mesa J., Miller G. J., Randolph M. A., Gill T. J., and Burdick J. A., Tissue Eng. 12, 2665 (2006). 10.1089/ten.2006.12.2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khademhosseini A., Eng G., Yeh J., Fukuda J., J.BlumlingIII, Langer R., and Burdick J. A., J. Biomed. Mater. Res. Part A, 79, 522 (2006). 10.1002/jbm.a.30821 [DOI] [PubMed] [Google Scholar]
- Gerecht S., Townsend S. A., Pressler H., Zhu H., Nijst C. L. E., Bruggeman J. P., Nichol J. W., and Langer R., Biomaterials 28, 4826 (2007). 10.1016/j.biomaterials.2007.07.039 [DOI] [PubMed] [Google Scholar]
- Hoshikawa A., Nakayama Y., Matsuda T., Oda H., Nakamura K., and Mabuchi K., Tissue Eng. 12, 2333 (2006). 10.1089/ten.2006.12.2333 [DOI] [PubMed] [Google Scholar]
- Nicodemus G. D. and Bryant S. J., Tissue Eng. 14, 149 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoichet M. S., Li R. H., White M. L., and Winn S. R., Biotechnol. Bioeng. 50, 374 (1996). [DOI] [PubMed] [Google Scholar]
- Chen M. C. W., Gupta M., and Cheung K. C., Biomed. Microdevices 12, 647 (2010). 10.1007/s10544-010-9417-2 [DOI] [PubMed] [Google Scholar]
- Kim C., Lee K. S., Bang J. H., Kim Y. E., Kim M. C., Oh K. W., Lee S. H., and Kang J. Y., Lab Chip 11, 874 (2011). 10.1039/c0lc00516a [DOI] [PubMed] [Google Scholar]
- Lan S. F., Safiejko-Mroczka B., and Starly B., Toxicol. in Vitro 24, 1314 (2010). 10.1016/j.tiv.2010.02.015 [DOI] [PubMed] [Google Scholar]
- Gonzalez McQuire R., Green D. W., Partridge K., Oreffo R. O. C., Mann S., and Davis S. A., Adv. Mater. 19, 2236 (2007). 10.1002/adma.200602770 [DOI] [Google Scholar]
- Wendt D., Riboldi S. A., Cioffi M., and Martin I., Adv. Mater. 21, 3352 (2009). 10.1002/adma.200802748 [DOI] [PubMed] [Google Scholar]
- Declercq H. A., Gorski T. L., Tielens S. P., Schacht E. H., and Cornelissen M. J., Biomacromolecules 6, 1608 (2005). 10.1021/bm050031s [DOI] [PubMed] [Google Scholar]
- Hoffman D., Breakefield X. O., Short M. P., and Aebischer P., Exp. Neurol. 122, 100 (1993). 10.1006/exnr.1993.1111 [DOI] [PubMed] [Google Scholar]
- Shao S., Gao Y., Xie B., Xie F., Lim S. K., and Li G. D. J., Endocrinology 208, 245 (2011). [DOI] [PubMed] [Google Scholar]
- Norotte C., Marga F. S., Niklason L. E., and Forgacs G., Biomaterials 30, 5910 (2009). 10.1016/j.biomaterials.2009.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murua A., Portero A., Orive G., Hernandez R. M., De Castro M., and Pedraz J. L. J., J. Controlled Release 132, 76 (2008). 10.1016/j.jconrel.2008.08.010 [DOI] [PubMed] [Google Scholar]
- Orive G., Hernandez R. M., Gascon A. R., Calafiore R., Chang T. M. S., De Vos P., Hortelano D., Hunkeler G., Lacik I., and Shapiro A. M., J. Nat. Med. 9, 104 (2003). 10.1038/nm0103-104 [DOI] [PubMed] [Google Scholar]
- Bauwens C. L., Peerani R., Niebruegge S., Woodhouse K. A., Kumacheva E., Husain M., and Zandstra P. W., Stem Cells 26, 2300 (2008). 10.1634/stemcells.2008-0183 [DOI] [PubMed] [Google Scholar]
- Gage F. H., Nature 392, 18 (1998). [PubMed] [Google Scholar]
- Malda J. and Frondoza C. G., Trends Biotechnol. 24, 299 (2006). 10.1016/j.tibtech.2006.04.009 [DOI] [PubMed] [Google Scholar]
- Rogers P. R., Friend J. R., and Yeo L. Y., Lab Chip 10, 2979 (2010). 10.1039/c004822d [DOI] [PubMed] [Google Scholar]
- Chabert M. and Viovy J. L., Proc. Natl. Acad. Sci. U.S.A. 105, 3191 (2008). 10.1073/pnas.0708321105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C. H., Jung J. H., Rhee Y. W., Kim D. P., Shim S. E., and Lee C. S., Biomed. Microdevices 9, 855 (2007). 10.1007/s10544-007-9098-7 [DOI] [PubMed] [Google Scholar]
- Koster S., Angile F. E., Duan H., Agresti J. J., Wintner A., Schmitz C., Rowat A. C., Merten C. A., Pisignano D., and Griffiths A. D., Lab Chip 8, 1110 (2008). 10.1039/b802941e [DOI] [PubMed] [Google Scholar]
- Chicheportiche D. and Reach G., Diabetologia 31, 54 (1988). [DOI] [PubMed] [Google Scholar]
- Sakai S., Mu C., Kawabata K., Hashimoto I., and Kawakami K., J. Biomed. Mater. Res. Part A 78, 394 (2006). 10.1002/jbm.a.30676 [DOI] [PubMed] [Google Scholar]
- Tan Y. C., Fisher J. S., Lee A. I., Cristini V., and Lee A. P., Lab Chip 4, 292 (2004). 10.1039/b403280m [DOI] [PubMed] [Google Scholar]
- Tan Y. C., Ho Y. L. and Lee A. P., Microfluid. Nanofluid. 4, 343 (2008). 10.1007/s10404-007-0184-1 [DOI] [Google Scholar]
- Yang C. H., Lin Y. S., Huang K. S., Huang Y. C., Wang E. C., Jhong J. Y., and Kuo C. Y., Lab Chip 9, 145 (2009). 10.1039/b807454b [DOI] [PubMed] [Google Scholar]
- Maenaka H., Yamada M., Yasuda M., and Seki M., Langmuir 24, 4405 (2008). 10.1021/la703581j [DOI] [PubMed] [Google Scholar]
- Tan Y. C. and Lee A. P., Lab Chip 5, 1178 (2005). 10.1039/b504497a [DOI] [PubMed] [Google Scholar]
- Hung S. H., Lin Y. H., and Lee G. B., J. Micromech. Microeng. 20, 045026 (2010). 10.1088/0960-1317/20/4/045026 [DOI] [Google Scholar]
- Ahn B., Lee K., Louge R., and Oh K. W., Biomicrofluidics 3, 044102 (2009). 10.1063/1.3250303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn K., Agresti J., Chong H., Marquez M., and Weitz D., Appl. Phys. Lett. 88, 264105 (2006). 10.1063/1.2218058 [DOI] [Google Scholar]
- Fidalgo L. M., Whyte G., Bratton D., Kaminski C. F., Abell C., and Huck W. T. S., Angew. Chem. 120, 2072 (2008). 10.1002/ange.200704903 [DOI] [PubMed] [Google Scholar]
- Guo F., Ji X. H., Liu K., He R. X., Zhao L. B., Guo Z. X., Liu W., Guo S. S., and Zhao X. Z., Appl. Phys. Lett. 96, 193701 (2010). 10.1063/1.3360812 [DOI] [Google Scholar]
- Lee C. Y., Lin Y. H., and Lee G. B., Microfluid. Nanofluid. 6, 599 (2009). 10.1007/s10404-008-0340-2 [DOI] [Google Scholar]
- Wakui D., Takahashi S., Sekiguchi T., and Shoji S., in 2010 IEEE 23rd International Conference Micro Electro Mechanical Systems (MEMS) (IEEE, Wanchai, Hong Kong, 2010), pp. 144–147.
- Franke T., Abate A. R., Weitz D. A., and Wixforth A., Lab Chip 9, 2625 (2009). 10.1039/b906819h [DOI] [PubMed] [Google Scholar]
- Nam J., Lee Y., and Shin S., Microfluid. Nanofluid. 11, 317 (2011). 10.1007/s10404-011-0798-1 [DOI] [Google Scholar]
- Shemesh J., Bransky A., Khoury M., and Levenberg S., Biomed. Microdevices 12, 907 (2010). 10.1007/s10544-010-9445-y [DOI] [PubMed] [Google Scholar]
- Shi J., Huang H., Stratton Z., Huang Y., and Huang T. J., Lab Chip 9, 3354 (2009). 10.1039/b915113c [DOI] [PubMed] [Google Scholar]
- Orloff N. D., Dennis J. R., Cecchini M, Schonbrun Et., Rocas E., Wang Y., Novotny D., Simmonds R. W., Moreland J., Takeuchi I., and Booth J. C., Biomicrofluidics 5, 044107 (2011). 10.1063/1.3661129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Cai F., Zhang Z., Niu L., Jin Q., Yan F., Wu J., Wang Z., and Zheng H., Biomicrofluidics 5, 044104 (2011) 10.1063/1.3652872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Ahmed D., Mao X., Lin S. C. S., Lawit A., and Huang T. J., Lab Chip 9, 2890 (2009). 10.1039/b910595f [DOI] [PubMed] [Google Scholar]
- Yeo L. Y. and Friend J. R., Biomicrofluidics 3, 012002 (2009). 10.1063/1.3056040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J., Lim H., Kim D., and Shin S., Lab Chip 11, 3361 (2011). 10.1039/c1lc20346k [DOI] [PubMed] [Google Scholar]
- Li H., Friend J., Yeo L., Dasvarma A., and Traianedes K., Biomicrofluidics 3, 034102 (2009). 10.1063/1.3194282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosioka K. and Kawasima Y., Acustica 5, 167 (1955). [Google Scholar]
- Dang S. M., Gerecht Nir S., Chen J., Itskovitz Eldor J., and Zandstra P. W., Stem Cells 22, 275 (2004). 10.1634/stemcells.22-3-275 [DOI] [PubMed] [Google Scholar]
- Cameron C., Hu W. S., and Kaufman D. S., Biotechnol. Bioeng. 94, 938 (2006). 10.1002/bit.20919 [DOI] [PubMed] [Google Scholar]
- Khademhosseini A., Ferreira L., J.BlumlingIII, Yeh J., Karp J. M., Fukuda J., and Langer R., Biomaterials 27, 5968 (2006). 10.1016/j.biomaterials.2006.06.035 [DOI] [PubMed] [Google Scholar]
- Karp J. M., Yeh J., Eng G., Fukuda J., J.BlumlingIII, Suh K. Y., Cheng J., Mahdavi A., Borenstein J., Langer R., and Khademhosseini A., Lab Chip 7, 786 (2007). 10.1039/b705085m [DOI] [PubMed] [Google Scholar]
- Torisawa Y., Chueh B., Huh D., Ramamurthy P., Roth T. M., Barald K. F., and Takayama S., Lab Chip 7, 770 (2007). 10.1039/b618439a [DOI] [PubMed] [Google Scholar]
- Hwang Y. S., Chung B. G., Ortmann D., Hattori N., Moeller H. C., and Khademhosseini A., Proc. Natl. Acad. Sci. U.S.A. 106, 16978 (2009). 10.1073/pnas.0905550106 [DOI] [PMC free article] [PubMed] [Google Scholar]