Abstract
Cell migration is involved in physiological processes such as wound healing, host defense, and cancer metastasis. The movement of various cell types can be directed by chemical gradients (i.e., chemotaxis). In addition to chemotaxis, many cell types can respond to direct current electric fields (dcEF) by migrating to either the cathode or the anode of the field (i.e., electrotaxis). In tissues, physiological chemical gradients and dcEF can potentially co-exist and the two guiding mechanisms may direct cell migration in a coordinated manner. Recently, microfluidic devices that can precisely configure chemical gradients or dcEF have been increasingly developed and used for chemotaxis and electrotaxis studies. However, a microfluidic device that can configure controlled co-existing chemical gradients and dcEF that would allow quantitative cell migration analysis in complex electrochemical guiding environments is not available. In this study, we developed a polydimethylsiloxane-based microfluidic device that can generate better controlled single or co-existing chemical gradients and dcEF. Using this device, we showed chemotactic migration of T cells toward a chemokine CCL19 gradient or electrotactic migration toward the cathode of an applied dcEF. Furthermore, T cells migrated more strongly toward the cathode of a dcEF in the presence of a competing CCL19 gradient, suggesting the higher electrotactic attraction. Taken together, the developed microfluidic device offers a new experimental tool for studying chemical and electrical guidance for cell migration, and our current results with T cells provide interesting new insights of immune cell migration in complex guiding environments.
INTRODUCTION
Cell migration plays an essential role in a broad range of physiological and pathological processes such as embryogenesis, inflammatory responses, wound healing, and cancer metastasis.1, 2, 3, 4 In particular, immune cells migrate in tissues in response to various cellular guiding signals, orchestrating both innate and adaptive immune responses.5, 6 The ability of immune cells to sense and migrate to concentration gradients of chemical factors in tissues, a process termed chemotaxis, together with the highly ordered tissue-specific chemoattractant network, provide a robust mechanism for coordinating different immune cell subsets to defined tissue sites.7, 8 In addition, physical parameters such as mechanical stimulation, magnetic fields, and electric fields can also mediate the migration of different cell types.3, 9, 10, 11, 12, 13 Particularly, various cell types such as epithelial cells, tumor cells, and leukocytes were found to respond to physiological or externally applied direct current electric fields (dcEF) by migrating toward either the cathode or the anode of the fields through a process commonly referred as electrotaxis or galvanotaxis.12, 14, 15, 16, 17, 18, 19 For some cell types such as epithelial cells, the roles of their electrotactic responses to the wound-produced dcEF for mediating wound recovery have been clearly demonstrated.3 Among immune cells, previous studies showed that most of human peripheral blood circulating leukocyte subsets are capable of undergoing electrotaxis invitro (Ref. 12) and the quantitative characteristics of several specific leukocyte subsets have been investigated in details using in-vitro electrotaxis assays.3, 11, 12 Moreover, a study has also demonstrated the possibility of manipulating T lymphocyte trafficking invivo by electrotactic guidance.12 However, compared to the relatively matured field of chemotaxis, the mechanisms of electrotaxis are not clearly defined.14, 20 A particular complication results from the co-existence of chemical fields and electric fields in tissues that can potentially cross-modify the chemical or electric field profiles.21 Studies toward dissecting chemotactic and electrotactic guidance for cell migration are hindered by the technical barrier of the existing electrotaxis assays that are incapable of generating well controlled co-existing chemical gradients and electric fields.
Conventional chemotaxis assays such as transwell assays produce chemical gradients by free diffusion of chemoattractant molecules so that the gradient profiles vary over time.22, 23, 24, 25, 26 The gradients in these assays can be further modified by electric field applications. The commonly used electrotaxis assays measure cell migration in electric fields inside a cell chamber built within a petri dish.3, 10, 17, 18, 19, 27, 28 The dish-based electrotaxis assays are not capable of creating chemical gradients. Microfluidic devices offer the advantages in miniaturization and cellular environmental control, and various gradient-generating devices have been increasingly developed and applied to cell migration and chemotaxis research over the last decade or so.21, 29 More recently, the potential of microfluidic devices for electrotaxis studies of different cell types started to be realized.11, 12, 15, 21, 30, 31, 32 The existing microfluidic devices basically generate controlled electric fields inside a straight microfluidic channel but do not allow superposition with chemical gradients.21 Integration of chemical gradient-generation and electric fields applications in microfluidic devices for studying interactions of these two guiding factors for cell migration is technically challenging because electric fields may modify chemical gradient profiles due to the electromigration mobility of chemoattractant molecules. On the other hand, an earlier study employing a stripe assay has demonstrated the altered chemotaxis of human blood granulocytes by an externally applied electric field.33 In addition, our recent study comparing human blood T cell chemotaxis and electrotaxis using separate microfluidic devices suggested the differential potency of chemokine gradients and dcEF for attracting T cells.11 Taken together, the importance of understanding cell migration in co-existing chemical gradients and electric fields, the demonstrated alteration of cell migration in combined chemical and electric fields, and the anticipated trend of microfluidics-based research for electrotaxis strongly motivated us to explore new microfluidics-based strategies of controlling single and co-existing chemical gradients and electric fields for cell migration research.
In the present study, we developed a polydimethylsiloxane (PDMS)-based microfluidic device that integrates a flow-based gradient-generating module with electric fields applications through multiple parallel side microchannels. Such a device would in principle allow superposition of stable chemical gradients and dcEF across the width of the main microfluidic channel because the chemical gradient formation is dominated by continuous laminar flow mixing with negligible influence by dcEF. We characterized single and co-existing chemical gradients and dcEF in this device by multiphysics modeling to verify the hypothesized strategy of generating better controlled co-existing chemical and electric fields. To validate the function of this developed microfluidic device and to explore its use for studying chemotaxis and electrotaxis, we tested the migration of activated human peripheral blood T cells in single or co-existing chemokine CCL19 gradients and externally applied dcEF. Our results showed chemotaxis of T cells to single CCL19 gradients and electrotaxis of T cells to the cathode of the applied dcEF. These results are consistent with our previous studies using separate microfluidic chemotaxis or electrotaxis devices,11 and thus serve as important controls for further testing of cell migration in co-existing chemical and dcEF in the new microfluidic device. Although we did not set specific anticipation for T cell migration in co-existing CCL19 gradients and dcEF in our experiments, we found that T cells show stronger migration toward the cathode of the dcEF in the presence of a competing CCL19 gradient that is interestingly consistent with our previous comparison between T cell chemotaxis and electrotaxis.11 This observed differential strength of chemical and electrical guidance for T cell migration suggests various possible interesting scenarios for cell migration and trafficking in complex tissue environments and thus opens up new possibilities of future research.
MATERIALS AND METHODS
Cell preparation
Preparation of activated T cells has been described in detail previously.11 Briefly, human peripheral blood mononuclear cells (PBMCs) were isolated from whole blood from healthy adult blood donors (in collaboration with the Victoria General Hospital in Winnipeg under an approved human ethics protocol) using the standard gradient centrifugation method. Isolated PBMCs were activated by anti-CD3/CD28 antibodies in a 37 °C incubator with 8% CO2 injection for 2 days. After the activation, cells in the culture medium (RPMI-1640 with 10% FBS) were transferred to a new flask and cultured in the presence of IL-2 for a few days before cell migration experiments.
Microfluidic device preparation
The device was designed in Freehand 9.0 (Macromedia) and the design was printed to a transparency mask by a high resolution printer with at least 2400 dpi resolution. The device masters were fabricated in The Nano Systems Fabrication Laboratory (NSFL) at the University of Manitoba. Briefly, the design was patterned on a silicon wafer by contact photolithography with SU-8 photoresist (MicroChem, MA), yielding ∼100 µm thickness. The PDMS replicas were then fabricated by molding PDMS (Dow Corning, MI) against the master.11 For the device in the current study (Fig. 1), one outlet well (4 mm diameter holes) at the end of a 350 µm (W) × 1 cm (L) channel (the main channel), two fluid inlets (1 mm diameter holes) at the other end of the main channel, and 2 electrode wells (4 mm diameter holes) connected by the thin side channels to the main channel were punched using sharpened needles. Then the PDMS replica was plasma bonded to a glass slide. Polyethylene tubing (PE-20, Becton Dickinson, MD) was inserted into the inlet holes to connect the microfluidic device to syringe pumps (KD Scientific, MA) with two 100 µl KD syringes containing medium or chemokine solution for fluid infusion. The main channel was connected by 20 thin channels (40 µm in width and 3 mm in length) on each side to the 2 electrode wells filled with medium, in where platinum electrodes (SPPL-010, Omega Engineering, Inc.) were inserted. The electrodes were buried in a 1% agarose gel block inside the well to prevent contact of possible toxic electrode byproducts with the cell culture. The electrodes were then wired to a DC power supply for electric field application. A new microfluidic device was used for each experiment.
Figure 1.
Schematic illustration of the microfluidic device. (a) 3D schematic drawing of the PDMS microfluidic device. Green indicates CCL19 solution and orange indicates medium solution. Platinum electrodes were buried in agarose gel blocks and inserted into the electrode wells. The two electrodes were then wired to the blue (cathode) and red (anode) wires, respectively, that were connected to a DC power supply to apply electric fields to the device. The drawing of the side channels was simplified with symmetric configurations. (b) Top view drawing of the microfluidic device with the channel dimensions indicated. (c) Illustration of the cell migration experiment setup. Microfluidic device was placed on a microscope stage; dcEF was applied to the device through a pair of electrodes; chemokine and medium solutions were infused into the device through tubing from syringe pumps for generating chemical gradients; cell migration in the device was then recorded by time-lapse microscopy. (d) Illustration of cell migration data analysis in the microfluidic device. Particularly, the O.I. is defined as the ratio of the displacement of cells (Δy) relative to the electric field or the chemical gradients to the total migration distance (d) using the equation O.I. = Δy/d.
Multiphysics modeling
Multiphysics modeling and simulations were performed using comsol Multiphysics (v4.2) with specialized modules for modeling chemical transport and electric fields, and the parameter values were adapted from the literatures (particularly, the diffusion coefficient is ∼1.7 × 10−6 cm2/s (Ref. 34) and net charge value is +7 (Refs. 35, 36) for chemokine CCL19).
Cell migration experiment setup
The microfluidic channel was coated with fibronectin (BD Biosciences, MA) for 1 h at room temperature and blocked with 0.4% BSA (bovine serum albumin) for another hour before the cell migration experiment. The experimental setup is illustrated in Fig. 1c. Briefly, for each experiment, a few thousand cells were loaded into the microfluidic channel and allowed to settle in the fibronectin-coated channel. The device was maintained at 37 °C by attaching a transparent heater to the back of the coverslide (Minco, MN). The heater was powered by a DC power supply (Harrison, Canada) and was controlled by a sensorless temperature controller (Minco, MN). The temperature was calibrated to 37 °C using a digital thermometer (VWR, Canada). After the cells were settled in the channel, ∼150 μl of medium (RPMI 1640 with 0.4% of BSA) was added to each well containing the agarose gel blocks and the electrodes, which are connected to a DC power supply (Central Scientific, NY) to complete the circuit. The assembled device was placed on a microscope stage (BX60, Olympus). To generate chemical concentration gradient, medium and chemokine solutions were infused into the device from the fluid inlets through tubing by syringe pumps at the total flow rate of 0.6 μl/min. The chemokine gradient was confirmed by measuring the fluorescence intensity profile of FITC-Dextran 10 kDa that has similar molecular weight of CCL19 and was added to the chemokine solution. Cell migration inside the main channel (in the region ∼7 mm downstream of the main channel) was recorded by time-lapse microscopy at 6 frames/min for at least 30 min using a CCD camera (Sensicam, Optikon). The image acquisition was controlled by NIH ImageJ (v.1.34 s).
Data analysis
Movement of individual cells was tracked using NIH ImageJ (v.1.34 s). The background noise of the image was removed using the “despeckle” function. Then the images were calibrated to distance. Only the cells that migrated within the relatively uniform dcEF region of the channel over the entire time-lapse were selected and tracked using the “Manual Tracking” plug-in in NIH ImageJ. The tracking data were then exported to Excel and Origin for analysis. A custom LabVIEW program was also used to analyze the tracking data. At least 40 cell tracks from multiple independent experiments for each condition were analyzed. The data analysis method is illustrated in Fig. 1d. The movement of cells was quantitatively evaluated by (1) the Orientation Index (O.I.), which is the ratio of the displacement of cells (Δy) toward the cathode of the electric field or the chemical gradients to the total migration distance (d) using the equation O.I. = Δy/d, presented as the average value ± standard error of the mean (SEM). In the current experimental configuration, positive O.I. indicates cell migration toward the cathode of the dcEF or away from the chemokine gradient; negative O.I. indicates cell migration toward the anode of the dcEF or toward the chemokine gradient; (2) the average speed (V), calculated as d/Δt and presented as the average value ± SEM of all cells; and (3) statistical analysis of migration angles, performed using Origin (v8.5, OriginLab Corporation, Northampton, MA) and the custom Labview program to examine the directionality of the cell movement. Specifically, migration angles (calculated from x-y coordinates at the beginning and the end of the cell tracks) were summarized in a direction plot, which is a rose diagram showing the distribution of angles grouped in 20° intervals, with the radius of each wedge indicating the cell number.
RESULTS
Generation of controlled chemical gradients and dcEF using the microfluidic device
As detailed in the microfluidic device preparation section and illustrated in Fig. 1, the developed PDMS microfluidic device is a “Y” type chemical gradient-generating module with electric field applied to the main channel from the side direction through multiple thin microchannels. The fundamental principle behind this design for generating better controlled single or co-existing chemical and electric fields is that gradient profiles will be controlled by the flows and thus minimizes the influence from the electric fields. Comparing to static assays, in which electromigration of chemokine molecules will significantly modify chemokine gradient over time, the introduction of the continuous flow in the current device limits the electromigration of chemokine molecules across the channel width as the chemokines flow through the channel length.
The use of multiple parallel channels for applying dcEF is expected to help increase the homogeneity of dcEF in the main channel. In addition, the alternating configuration of the small channels from each side of the main channel is found to help reduce small channel blocking by cells in our experiments and thus allow easier cell loading to the main channel. To verify the hypothesized functions of this design, we performed multiphysics modeling and simulations using comsol Multiphysics to characterize chemical gradients and electric fields in the device.
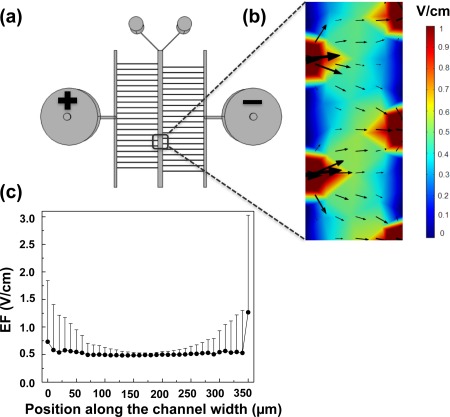
First, we modeled and simulated the dcEF in the main channel of the device. As shown in our later cell migration experiments, T cells showed clear electrotaxis toward the cathode of the applied dcEF in the region ∼7 mm downstream of the main channel (0.75 mm (L) × 0.35 mm (W)) when a 10 V of electrical potential difference was applied between the 2 electrode wells at the defined flow rate. Therefore, similar experimental parameters including the cell observation region, the flow rate, and the applied electrical potential difference were adapted in the simulations. Our results showed that the dcEF in the center region (i.e., away from the channel edges over a spans of ∼200 µm and cells migrated within this region were selected for cell tracking analysis in the later cell migration experiments) of the main channel is relatively uniform (Fig. 2) (the average dcEF strength is approximately 0.52 V/cm if a 10 V electrical potential difference is applied to the device). As we expected, the dcEF in the thin channels is much higher due to more significant electrical potential drop in those thin channels. In addition, the dcEF in the regions (in the main channel) where the thin channels connect the main channel is higher than the center region resulting from the boundary effect as we expected. Moreover, the dcEF varies along the 1 cm long main channel as expected. Superposition of dcEF with a 100 nM CCL19 gradient does not significantly change the dcEF or the current density in the main channel. Thus, the microfluidic device can produce a relatively uniform dcEF in the defined region of the main channel and the magnitude of the dcEF can be controlled to be within the physiological range (0.4–1.4 V/cm),12 allowing us to analyze cell migration in defined dcEF. Second, we modeled and simulated the chemokine gradient in the main channel of the device. Our results showed that the gradient profile in the same region of the main channel where we characterized the dcEF is maintained by the flows (total linear flow velocity = 2.9 × 10−4 m/s) and is not significantly affected by the dcEF induced electromigration of the chemokine molecules (Fig. 3) as shown by the F test (p = 0.82) to compare the gradient profiles with or without dcEF.
Figure 2.
Simulation of dcEF in the microfluidic device. (a) Top view of the microfluidic device. A 10 V of electrical potential difference was applied to the device from the two electrode wells. (b) The color map and the arrows indicate the magnitude and the direction of the dcEF in the main channel (0.75 mm (L) × 0.35 mm (W) at ∼7 mm downstream of the main channel). (c) Plot of simulated dcEF across the main channel width in the region as in (b). The dcEF is presented as the average value with the error bar as the standard deviation (SD). The simulation results show that dcEF is relatively uniform in the defined center region of the main channel and its magnitude can be configured to be within the physiological strength range.
Figure 3.
Simulation of chemical concentration gradients with dcEF application in the microfluidic device. (a) Simulated normalized CCL19 gradient in the main channel of the microfluidic device when a 10 V electrical potential difference is applied to the device through the 2 electrode wells. (b) Enlarged view of the CCL19 gradient in the boxed region in the main channel as indicated in (a) (∼7 mm downstream of the main channel). (c) Plot of simulated gradient across the main channel width in the region as in (b) with (grey) or without (black) the applied dcEF. The simulation results show that the gradient profile is not significantly affected by the applied dcEF. The p value of the F test comparing the gradient profiles is 0.82.
Taken together, modeling and simulation of the microfluidic device validated the hypothesized functions of the device for configuring better controlled single or co-existing chemical gradients and dcEF, and provided the quantitative characteristics of chemical and electric fields in the device that will guide the designs of cell migration experiments for studying chemotaxis, electrotaxis and their interactions.
T cell chemotaxis to single chemokine gradients in the microfluidic device
Using the developed microfluidic device, we experimentally analyzed the migration of activated human peripheral blood T cells in a single 100 nM CCL19 gradient. As we expected, T cells show chemotaxis to the CCL19 gradient (Fig. 4) that is consistent with our previous studies using the simple “Y” shape microfluidic device.11 By contrast, in the uniform medium control experiment, cells migrate randomly with reduced speed (Fig. 4). It is worth pointing out that the exact values of these cell migration parameters vary between the current data and our previous studies using the simple “Y” shape device,11 possibly due to the device design difference, changes of several experimental conditions (e.g., cell observation region), and blood donor variations. Thus, the chemotaxis experiments demonstrate the function of the developed microfluidic device for analyzing cell chemotaxis in single chemokine gradients.
Figure 4.
Chemotaxis of activated T cells in the microfluidic device. Angular histograms of cell migration angles in the control condition (medium only) or in a 100 nM CCL19 gradient are showed in (a) and (b), respectively. The rose diagrams show the distribution of migration angles of all cells analyzed from multiple independent experiments for each condition. The migration angles were calculated from x-y coordinates at the beginning and the end of the cell tracks and were grouped in 20° intervals, with the radius of each wedge indicating the cell number (i.e., the radius of each circle indicates the cell number with the increment of one). (c) O.I. and speed of cells in the control condition or in a 100 nM CCL19 gradient. The values are presented as the average ± SEM. The results show the effectiveness of the developed microfluidic device for analyzing cell chemotaxis in single chemokine gradients.
T cell electrotaxis to single dcEF in the microfluidic device
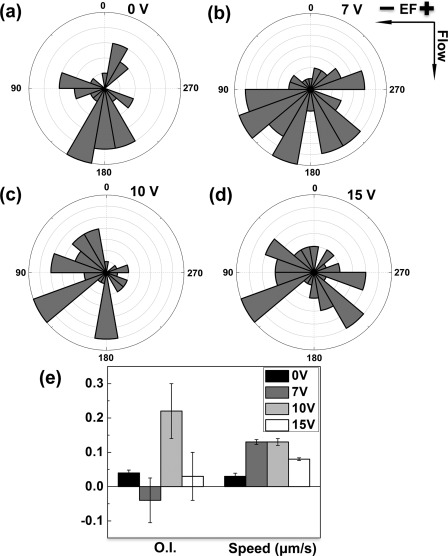
Next, we performed electrotaxis experiments to analyze activated human peripheral blood T cells in different applied dcEF using the microfluidic device. We applied a range of DC electrical potential difference to the device and our results identified 10 V as the most effective electrical potential difference for inducing electrotactic migration of T cells toward the cathode of the dcEF in this specific microfluidic device (Fig. 5). As described in Sec. 3A, this electrical potential difference results in a ∼0.5 V/cm dcEF in the center region of the main channel, confirming the ability of T cells to respond to the physiological strength of dcEF. The speed of the cells is higher for the intermediate electrical potential difference (7 V and 10 V) within the range of the electrical potential difference tested. Thus, the electrotaxis experiments confirm the function of the developed microfluidic device for analyzing T cell electrotaxis in single dcEF.
Figure 5.
Electrotaxis of activated T cells in the microfluidic device. (a)–(d) Angular histogram of cell migration angles in different dcEF (i.e., 0 V, 7 V, 10 V, and 15 V electrical potential difference between the two electrode wells). The rose diagrams show the distribution of migration angles of all cells analyzed from multiple independent experiments for each condition. The migration angles were calculated from x-y coordinates at the beginning and the end of the cell tracks and were grouped in 20° intervals, with the radius of each wedge indicating the cell number (i.e., the radius of each circle indicates the cell number with the increment of one). (e) O.I. and speed of cells in different applied dcEF. The values are presented as the average ± SEM. The results show the cathode-directing electrotaxis of cells when a 10 V electrical potential difference was applied to the device and thus demonstrate the effectiveness of the developed microfluidic device for analyzing cell electrotaxis in single dcEF.
T cell migration in competing chemokine gradient and dcEF in the microfluidic device
Based upon the separate chemotaxis and electrotaxis experiments as describe above, we further tested the migration of activated human peripheral blood T cells in co-existing CCL19 gradient and dcEF. In this experiment, we configured the CCL19 gradient (100 nM) and the dcEF (10 V across the device) along the opposite directions, creating a competing scenario between the chemokine gradient and the dcEF for directing T cell migration (Fig. 6). Our results show that the migration of T cells to the CCL19 gradient or to the dcEF is significantly altered compared to it in single CCL19 gradients or single dcEF. Interestingly, the net migration of T cells is toward the cathode of dcEF with reduced orientation index. The speed of T cells is not affected compared to it in single CCL19 gradient or single dcEF. These results demonstrate the function of the developed microfluidic device for analyzing T cell migration in co-existing chemical and electric fields and the altered cell migration suggests interesting interactions between chemotaxis and electrotaxis.
Figure 6.
T cell migration in competing CCL19 gradients and dcEF in the microfluidic device. (a) Migration tracks of 6 cells from a representative experiment with 2 cells (grey) migrating toward the 100 nM CCL19 gradient (left) and 4 cells (black) migrating toward the cathode of the applied dcEF (10 V across the device with the cathode on the right). (b) Angular histogram shows the distribution of migration angles of all cells analyzed from multiple independent experiments. The migration angles were calculated from x-y coordinates at the beginning and the end of the cell tracks and were grouped in 20° intervals, with the radius of each wedge indicating the cell number (i.e., the radius of each circle indicates the cell number with the increment of one). (c) O.I. and speed of T cells in single CCL19 gradient, single dcEF or competing CCL19 gradient and dcEF. The values are presented as the average ± SEM. The results show the stronger cell migration toward the cathode of the applied dcEF in the presence of a competing CCL19 gradient.
Altogether, our results characterized the developed microfluidic device for configuring better controlled single or co-existing chemical gradients and dcEF. The functions of the device for analyzing cell chemotaxis, electrotaxis, and cell migration in co-existing chemical gradients and electric fields were successfully demonstrated.
DISCUSSION
In the present study, we developed a PDMS-based microfluidic device that can generate separate chemical concentration gradients and electric fields or better controlled co-existing chemical gradients and electric fields. Such a device is critically required for analyzing cell migration in complex chemical and electrical guiding environments but it was not available previously.21 The T cell migration experiments using this developed device successfully demonstrated these unique functions of the device. Thus, the developed device presents advancement from the previous single function-based microfluidic chemotaxis and electrotaxis devices11 to an integrated device for configuring more complex but better controlled electrochemical guiding environments for cell migration analysis.
The current design of the device serves as a first step to explore the hypothesized principle for generating better controlled single or co-existing chemical gradients and dcEF. We successfully demonstrated the effectiveness of this concept in our modeling and cell migration studies. On the other hand, the design of the microfluidic device can be further improved for better performance. For example, the thickness of the connecting thin channels can be decreased using a multi-height fabrication procedure37 to further reduce flow disturbance and facilitate cell loading. Similarly, the connecting junctions between the thin channels and the main channel can be narrowed by increasing the fabrication resolution for the same purpose of reducing flow disturbance. Furthermore, the joining channel for the thin channels on each side of the main channel can be designed to bring them to equal electrical potential (possibly by making long and big electrode wells that connect all the thin channels or by patterning on-chip electrodes across the thin channels11). Our preliminary modeling and simulations suggest that such a design can improve the uniformity of the dcEF along the main channel. To exclude the possible effect of electrolysis (around the electrodes) induced pH change on gradient formation and cell migration, we measured the pH in our device before and after the dcEF application. The results showed that the pH in the anode electrode well decreased and the pH in the cathode electrode well increased, confirming the expected electrolysis of medium. However, the pH value remains unchanged in the medium well for the main channel. We have confirmed this unaffected pH in the main channel at the equilibrium state by multiphysics simulation. In the current study, we used FITC-Dextran 10 kDa that has similar molecular weight of CCL19 to indirectly verify the CCL19 gradient without the dcEF, and we also showed that the CCL19 gradient is similar with or without dcEF by multiphysics simulation. Ultimately, it will be important in the future to experimentally verify the chemokine gradients with or without dcEF by for instance using fluorescently tagged chemokines (but this will require the fluorescently tagged CCL19 without affecting its diffusivity and net charge).
A key interesting finding from the current T cell migration experiments using this developed device is that T cells show stronger migration toward the cathode of the applied dcEF in the presence of a competing CCL19 gradient (the dcEF strength and the chemokine gradient condition for inducing optimal electrotaxis or chemotaxis were chosen). This result is consistent with the comparison between chemotaxis and electrotaxis of T cells in single chemokine gradients or dcEF using this device or as shown in our previous studies using single function microfluidic devices,11 providing the initial experimental basis for discussing potential mechanisms behind the competition of chemotaxis and electrotaxis. For example, cells respond to chemical gradients through their specific surface receptors for the chemoattractant molecules and the activated receptors trigger complex downstream chemotactic signaling cascades.38, 39, 40 By contrast, such receptor specific signaling was not found for electrotaxis. On the other hand, it has been demonstrated that electrotaxis shares many downstream signaling processes for chemotaxis.3, 41 Therefore, assuming chemotaxis and electrotaxis employ separate non-interacting upstream signaling mechanisms but share the downstream signaling pathways, the competition of chemotaxis and electrotaxis can be possibly viewed as a simple algebraic integration between the chemical gradient activated and the dcEF activated downstream signaling events within the same downstream signaling molecule pool (assuming it is unlimited). This will explain the observed stronger migration of T cells toward the applied dcEF over the chemokine gradient due to the stronger electrotactic attraction. In the second scenario, previous studies and our recent modeling study have suggested that dcEF may polarize various cell surface receptors by electrophoresis allowing cells to sense the dcEF and moreover to overcome chemoattractant gradient by preferentially migrate toward the dcEF.14, 42, 43, 44 Such a unified chemoattractant receptor based picture is also consistent with our observed stronger electrotactic migration of T cells over a competing CCL19 gradient. Base on this theory, we would predict that CCR7, the receptor for CCL19, can be polarized toward the cathode of the dcEF (note that receptor polarization is a complex process that cannot be simply predicted by the receptor net charge and thus needs to be experimentally determined43). We would further predict that blocking CCR7 (using antibodies for example) will not only inhibit T cell chemotaxis to CCL19 or CCL21, but may also reduce T cell migration toward the dcEF in competing CCL19 gradient and dcEF. On the other hand, such a theory does not account for the observed T cell electrotaxis in single dcEF wherein no CCL19 is presented. Finally, the current cell migration data in competing CCL19 and dcEF with selected chemokine dose and dcEF strength does not exclude the possibility that the competition outcomes may vary for other combinations of chemokine gradients and dcEF and for other cell types, which will be interesting to investigate in future studies using this developed microfluidic device.
In tissues, multiple chemical concentration gradients as well as electric fields can potentially co-exist, and thus cells can be exposed to different spatiotemporal configurations of single or overlapping chemoattractant gradients and dcEF.3, 21 In the wound setting, both wound released chemical gradients and wound-generated dcEF can potentially attract the surrounding epithelial cells in a coordinated manner to facilitate wound healing.21 Similarly, combined chemical gradients and dcEF may enhance immune cell recruitment or cancer metastasis based on the reported chemotaxis and electrotaxis ability of various immune cell subsets and different metastatic cancer cells.11, 12, 15, 19 Equally importantly, there are likely situations that chemical gradients and dcEF direct cells along the opposite directions, and the competition outcomes will lead to different physiological consequences. The observed stronger electrotactic migration of cells in competing chemokine gradients and dcEF may support the possible scenario, in which cells from distant tissues are effectively recruited by dcEF to the surrounding regions of a defined target (e.g., the wound) without distraction by the surrounding tissue derived “irrelevant” chemoattractant gradients. Once the cells get closer to the final target, the target derived chemoattractant gradients and dcEF will collaborate to enhance cell recruitment to the target. In this regard, the more potent but non-specific dcEF may work more effectively for long-range recruitment of relevant cells to the surrounding regions of the target compared to the complete chemical based multi-step cell recruitment model through multiple sequentially arrayed chemoattractant sources as proposed previously.26 The flow conditions used in our device is not physiologically relevant to the real situations in tissues and therefore dcEF may indeed modify chemical gradients in tissues. On the other hand, in the absence of flows, one can imagine that dcEF may either weaken the competing gradient strength of chemoattractants with positive net charge (e.g., CCL19 and CCL21) or enhance these chemical gradients when they are along the dcEF direction. In those cases, cell migration toward dcEF may be further enhanced. The predictions will be different for chemoattractants with negative net change or for anode-electrotaxing cell types. Therefore, a better understanding of the relative potency, and more importantly, the competition of chemical gradients and dcEF for attracting different cell types, will help dissect the complex coordination of cell migration and trafficking in tissues in the context of different physiological processes. The developed microfluidic device will critically enable research toward this direction. Furthermore, characterizations of cell migration in competing chemoattractant gradients and dcEF may provide important scientific basis for developing new therapeutic strategies for cell trafficking mediated diseases or physiological processes such as autoimmune diseases, cancers, and wound healing, by electrically manipulating cell trafficking and positioning in tissues. The observed stronger cell migration toward dcEF over chemoattractant gradients in the current study argues the possibility of such clinical applications.
CONCLUSION
In conclusion, the present study developed a novel microfluidic device that offers the ability to quantitatively analyze cell migration in better controlled single or competing chemoattractant gradients and dcEF. The developed device can be useful for a broad range of cell migration and trafficking related research areas. Further development of the device and cell migration and trafficking studies enabled by the device will generate important insights into the complex biological mechanisms of electrotaxis and its interaction with chemotaxis with implications for physiological processes, disease pathologies, as well as clinical applications.
ACKNOWLEDGMENTS
This study is supported by grants from Natural Sciences and Engineering Research Council of Canada (NSERC), Manitoba Health Research Council (MHRC), and the University of Manitoba. We thank The Nano Systems Fabrication Laboratory (NSFL) at the University of Manitoba and The Victoria General Hospital in Winnipeg for research support. We also thank MHRC and the Faculty of Science at the University of Manitoba for fellowships to J.L. We thank Ouyang Lipan for helping with statistical analysis.
References
- Luster A., Alon R., and von Andrian U., Nat. Immunol. 6, 1182 (2005). 10.1038/ni1275 [DOI] [PubMed] [Google Scholar]
- Muller A., Homey B., Soto H., Ge N., Catron D., Buchanan M. E., McClanahan T., Murphy E., Yuan W., Wagner S. N., Barrera J. L., Mohar A., Verastegui E., and Zlotnik A., Nature 410, 50 (2001). 10.1038/35065016 [DOI] [PubMed] [Google Scholar]
- Zhao M., Song B., Pu J., Wada T., Reid B., Tai G., Wang F., Guo A., Walczysko P., Gu Y., Sasaki T., Suzuki A., Forrester J., Bourne H., Devreotes P., McCaig C., and Penninger J., Nature 442, 457 (2006). 10.1038/nature04925 [DOI] [PubMed] [Google Scholar]
- Behar T., Schaffner A., Colton C., Somogyi R., Olah Z., Lehel C., and Barker J., J. Neurosci. 14, 29 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. and Butcher E., Curr. Opin. Immunol. 12, 336 (2000). 10.1016/S0952-7915(00)00096-0 [DOI] [PubMed] [Google Scholar]
- Kubes P., Semin. Immunol. 14, 65 (2002). 10.1006/smim.2001.0343 [DOI] [PubMed] [Google Scholar]
- Kunkel E. and Butcher E., Immunity 16, 1 (2002). 10.1016/S1074-7613(01)00261-8 [DOI] [PubMed] [Google Scholar]
- Butcher E. C. and Picker L. J., Science 272, 60 (1996). 10.1126/science.272.5258.60 [DOI] [PubMed] [Google Scholar]
- Menon S. and Beningo K. A., PLoS ONE 6, e17277 (2011). 10.1371/journal.pone.0017277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B., Gu Y., Pu J., Reid B., Zhao Z., and Zhao M., Nat. Protoc. 2, 1479 (2007). 10.1038/nprot.2007.205 [DOI] [PubMed] [Google Scholar]
- Li J., Nandagopal S., Wu D., Romanuik S. F., Paul K., Thomson D. J., and Lin F., Lab Chip 11, 1298 (2011). 10.1039/c0lc00371a [DOI] [PubMed] [Google Scholar]
- Lin F., Baldessari F., Gyenge C., Sato T., Chambers R., Santiago J., and Butcher E., J. Immunol. 181, 2465 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel R. B. and Blakemore R. P., Bioelectromagnetics 10, 223 (1989). 10.1002/bem.2250100303 [DOI] [PubMed] [Google Scholar]
- McCaig C., Rajnicek A., Song B., and Zhao M., Physiol. Rev. 85, 943 (2005). 10.1152/physrev.00020.2004 [DOI] [PubMed] [Google Scholar]
- Huang C., Cheng J., Yen M., and Young T., Biosens. Bioelectron. 24, 3510 (2009). 10.1016/j.bios.2009.05.001 [DOI] [PubMed] [Google Scholar]
- Zhang J., Calafiore M., Zeng Q., Zhang X., Huang Y., Li R., Deng W., and Zhao M., Stem Cell Rev. Rep. 7, 987 (2011). 10.1007/s12015-011-9247-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Kuwayama H., van Egmond W., Takayama A., Takagi H., van Haastert P., Yanagida T., and Ueda M., Proc. Natl. Acad. Sci. U.S.A. 106, 6667 (2009). 10.1073/pnas.0809974106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M. J., Ueda M., Takagi H., Watanabe T. M., and Yanagida T., Biosystems 88, 261 (2007). 10.1016/j.biosystems.2006.06.008 [DOI] [PubMed] [Google Scholar]
- Djamgoz M. B. A., Mycielska M., Madeja Z., Fraser S., and Korohoda W., J. Cell Sci. 114, 2697 (2001). [DOI] [PubMed] [Google Scholar]
- Zhao M., Semin. Cell Dev. Biol. 20, 674 (2009). 10.1016/j.semcdb.2008.12.009 [DOI] [PubMed] [Google Scholar]
- Li J. and Lin F., Trends in Cell Biol. 21, 489 (2011). 10.1016/j.tcb.2011.05.002 [DOI] [PubMed] [Google Scholar]
- Nelson R. D., Quie P. G., and Simmons R. L., J. Immunol. 115, 1650 (1975). [PubMed] [Google Scholar]
- Boyden S., J. Exp. Med. 115, 453 (1962). 10.1084/jem.115.3.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohof A., Quillan M., Dan Y., and Poo M., J. Neurosci. 12, 1253 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond S., J. Cell Biol. 75, 606 (1977). 10.1083/jcb.75.2.606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxman E. F., Campbell J. J., and Butcher E. C., J. Cell Biol. 139, 1349 (1997). 10.1083/jcb.139.5.1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerick K. E., Longaker M. T., and Prinz F. B., Biochem. Biophys. Res. Commun. 397, 12 (2010). 10.1016/j.bbrc.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai G., Reid B., Cao L., and Zhao M., Methods Mol. Biol. 571, 77 (2009). 10.1007/978-1-60761-198-1 [DOI] [PubMed] [Google Scholar]
- Kim S., Kim H. J., and Jeon N. L., Integr. Biol. 2, 584 (2010). 10.1039/c0ib00055h [DOI] [PubMed] [Google Scholar]
- Rezai P., Siddiqui A., Selvaganapathy P., and Gupta B., Lab Chip 10, 220 (2010). 10.1039/b917486a [DOI] [PubMed] [Google Scholar]
- Minc N. and Chang F., Curr. Biol. 20, 710 (2010). 10.1016/j.cub.2010.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.-C., Kao Y.-C., Chi P.-Y., Huang C.-W., Lin J.-Y., Chou C.-F., Cheng J.-Y., and Lee C.-H., Lab Chip 11, 695 (2011). 10.1039/c0lc00155d [DOI] [PubMed] [Google Scholar]
- Aly A. A., Cheema M. I., Tambawala M., Laterza R., Zhou E., Rathnabharathi K., and Barnes F. S., IEEE Trans. Biomed. Eng. 55, 795 (2008). 10.1109/TBME.2007.912636 [DOI] [PubMed] [Google Scholar]
- Lin F. and Butcher E., Lab Chip 6, 1462 (2006). 10.1039/b607071j [DOI] [PubMed] [Google Scholar]
- Hori Y., Winans A. M., Huang C. C., Horrigan E. M., and Irvine D. J., Biomaterials 29, 3671 (2008). 10.1016/j.biomaterials.2008.05.033 [DOI] [PubMed] [Google Scholar]
- Christopherson K. W., Campbell J. J., Travers J. B., and Hromas R. A., J. Pharmacol. Exp. Ther. 302, 290 (2002). 10.1124/jpet.302.1.290 [DOI] [PubMed] [Google Scholar]
- Taylor A. M., Blurton-Jones M., Rhee S. W., Cribbs D. H., Cotman C. W., and Jeon N. L., Nat. Methods 2, 599 (2005). 10.1038/nmeth777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P. and Weigelin B., Nat. Immunol. 9, 960 (2008). 10.1038/ni.f.212 [DOI] [PubMed] [Google Scholar]
- Förster R., Davalos-Misslitz A., and Rot A., Nat. Rev. Immunol. 8, 362 (2008). 10.1038/nri2297 [DOI] [PubMed] [Google Scholar]
- Legler D. F., Krause P., Scandella E., Singer E., and Groettrup M., J. Immunol. 176, 966 (2006). [DOI] [PubMed] [Google Scholar]
- Zhao M., Br. J. Pharmacol. 152, 1141 (2007). 10.1038/sj.bjp.0707503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D. and Lin F., Biochem. Biophys. Res. Commun. 411, 695 (2011). 10.1016/j.bbrc.2011.07.004 [DOI] [PubMed] [Google Scholar]
- Poo M.-M. and Robinson K. R., Nature 265, 602 (1977). 10.1038/265602a0 [DOI] [PubMed] [Google Scholar]
- Zhao M., Bai H., Wang E., Forrester J. V., and McCaig C. D., J. Cell Sci. 117, 397 (2004). 10.1242/jcs.00868 [DOI] [PMC free article] [PubMed] [Google Scholar]