Background: The conservation of the only invariant noncysteine residue in the α-defensins, Gly17, is poorly understood.
Results: The G17A mutation impaired dimerization of the human α-defensin HNP1 and its ability to self-associate, inhibit anthrax lethal factor, and kill bacteria.
Conclusion: Gly17 is totally conserved for both structural and functional reasons.
Significance: The molecular determinants of human α-defensin function are better understood.
Keywords: Antimicrobial Peptides, Defensins, Host Defense, Peptide Chemical Synthesis, X-ray Crystallography
Abstract
The human α-defensins (HNP) are synthesized in vivo as inactive prodefensins, and contain a conserved glycine, Gly17, which is part of a β-bulge structure. It had previously been shown that the glycine main chain torsion angles are in a d-configuration, and that d-amino acids but not l-alanine could be substituted at that position to yield correctly folded peptides without the help of a prodomain. In this study, the glycine to l-alanine mutant defensin was synthesized in the form of a prodefensin using native chemical ligation. The ligation product folded correctly and yielded an active peptide upon CNBr cleavage. The l-Ala17-HNP1 crystal structure depicted a β-bulge identical to wild-type HNP1. However, dimerization was perturbed, causing one monomer to tilt with respect to the other in a dimerization model. Inhibitory activity against the anthrax lethal factor showed a 2-fold reduction relative to wild-type HNP1 as measured by the inhibitory concentration IC50. Self-association was slightly reduced, as detected by surface plasmon resonance measurements. According to the results of the virtual colony count assay, the antibacterial activity against Escherichia coli, Staphylococcus aureus, and Bacillus cereus exhibited a less than 2-fold reduction in virtual lethal dose values. Prodefensins with two other l-amino acid substitutions, Arg and Phe, at the same position did not fold, indicating that only small side chains are tolerable. These results further elucidate the factors governing the region of the β-bulge structure that includes Gly17, illuminating why glycine is conserved in all mammalian α-defensins.
Introduction
Antimicrobial peptides constitute a commonly deployed defensive strategy in the war against disease. They are found throughout the tree of life (1, 2). Taking advantage of the protein scaffold and the cellular equipment of transcription and translation, over evolutionary time scales antimicrobial peptides are mutable and often quite versatile, acquiring new functions as they strive to keep the host free from invaders (3). The same peptide might bind and disrupt a wide variety of target molecules, including lipids, carbohydrates, and proteins. An example of this type of multifunctional antimicrobial peptide is the defensins (4–7), which are active against a multitude of potentially dangerous microbes. For example, defensins can provide a measure of protection against Bacillus anthracis by binding and inhibiting anthrax lethal factor (LF),5 an enzyme required to form anthrax toxin (8). Defensins are also bactericidal against both Gram-positive and Gram-negative bacteria (9).
Defensins are small (2–5 kDa) cationic peptides, stabilized by three disulfide bonds, and there are three classes of defensins based on disulfide connectivity: α-, β-, and θ-defensins (10, 11). In humans there are six α-defensins, which share the same structural fold and connectivity of the disulfide pairs. Four of the six are expressed predominantly in human neutrophils and are termed human neutrophil peptides HNP1–HNP4 (12–15). The other two are expressed primarily in the Paneth cells of the intestinal crypts and are called human defensins HD5 and HD6 (16, 17). In addition to disulfide connectivity, all six have in common a Gly, and the reason for its strict conservation is only partially understood. A previous study (18) discovered that its flexibility allows the adoption of main chain torsion angles reserved for d-amino acids as part of a classical β-bulge (19, 20). In the structures of HNP1 (21, 22) and HNP3 (23), it is oriented so that its R-group hydrogen would be situated away from the protein and facing solvent. These characteristics led to the hypothesis that any d-amino acid could be substituted in place of Gly17. Seven d-amino acids were tried, Ala, Arg, Glu, Phe, Thr, Tyr, and Val, and all seven resulted in correctly folded, functionally active peptides with antibacterial activities proportional to hydrophobicity and net charge (18). The crystal structure of the d-Ala variant depicted its side chain methyl group pointed away from the interior of the protein toward solvent, and the β-bulge structure was maintained.
Not only were d-amino acids tolerated at that position, it appeared that the d-configuration was required. The l-Ala17-HNP2 mutant (HNP1 numbering) did not fold properly using the established oxidative folding method without the prodomain (24), resulting in massive precipitation and a highly heterogeneous liquid chromatographic trace. However, folding in the presence of the prodomain was not attempted at that time. The α-defensins are translated in vivo as inactive prodefensins. The prodomain is required for correct intracellular trafficking and for inhibition of defensin activity until it is proteolytically cleaved in the correct compartment, such as the azurophilic granules of the neutrophil in the case of the four HNPs (25–27). It also serves as a chaperone to ensure correct folding and disulfide pairing (28, 29). Here we report that l-Ala17-HNP1 folds productively as a prodefensin, and it is functional after cyanogen bromide (CNBr) cleavage to remove the prodomain. The synthetic access to l-Ala17-HNP1 enabled us for the first time to interrogate structural and functional properties of a “natural mutant” of HNP1 to answer the question of why the Gly17 residue is strictly conserved in all mammlian α-defensins. The crystal structure of l-Ala17-HNP1 was determined at 1.9-Å resolution. Its function was quantified with respect to self-association as measured by surface plasmon resonance, LF inhibition as measured by an enzyme kinetic assay (30, 31), and antibacterial activity against Escherichia coli, Staphylococcus aureus, and Bacillus cereus as measured using the “virtual colony count” assay (9).
MATERIALS AND METHODS
Synthesis and Folding of Defensins
HNP1 was synthesized as described previously (24). l-Ala62-pro-HNP1, l-Arg62-pro-HNP1, and l-Phe62-pro-HNP1 were synthesized as HNP2 mutants following native chemical ligation with H-(1–46)pro-HNP1-αCOSR as described (28, 29). Oxidative folding was performed at a concentration of 2 mg/ml in 2 m urea, 3 mm reduced and 0.3 mm oxidized glutathione (pH 8.1). Cleavage of the prodefensin was done with 25 mg/ml of CNBr in 2.5% TFA. All peptides were purified to homogeneity by C18 reversed-phase high-performance liquid chromatography (RP-HPLC), and their molecular masses were verified by electrospray ionization mass spectrometry (ESI-MS). Folding reactions were monitored by RP-HPLC. The quantification of defensins was done by UV measurements at 280 nm using molar extinction coefficients calculated from a published algorithm (32). Recombinant LF was purchased from List Biological Laboratories, Inc. A sequence-optimized chromogenic LF substrate, Ac-NleKKKKVLP-p-nitroaniline, was synthesized as previously described (22).
Structural Studies of l-Ala17-HNP1
l-Ala17-HNP1 at 20 mg/ml in water was mixed at a volume ratio of 1:1 with 0.2 m magnesium chloride, 0.1 m HEPES sodium salt (pH 7.5), and 30% isopropyl alcohol and left to equilibrate in the hanging drop by vapor diffusion at room temperature. Orthorhombic crystals appeared after 3 weeks of incubation and grew to ∼0.2 × 0.1 × 0.2 mm in about 5 weeks. Crystals were briefly soaked in 0.2 m magnesium chloride, 0.1 m HEPES sodium salt (pH 7.5), 30% glycerol and flash-frozen by plunging directly into liquid nitrogen. Diffraction data were collected at the Stanford Synchrotron Radiation Light source (SSRL) BL7-1 beamline on an ADSC QUANTUM 315 area detector and processed and reduced with HKL2000 (33). The structure was solved by molecular replacement with Phaser from the CCP4 suite (34) based on the coordinates of the HNP1 monomer (Protein Data Bank (PDB) code 3GNY (22)). Refinement was carried out with Refmac (35) and coupled with manual refitting and rebuilding with COOT (36). The data collection and refinement statistics are shown in Table 1. Ramachandran statistics were calculated with MolProbility (37) and all illustrations were prepared with the PyMol Molecular Graphic suite (DeLano Scientific, San Carlos, CA). Quaternary structure was modeled using PISA (38) (Protein Interfaces, Surfaces and Assemblies service at European Bioinformatics Institute). The coordinates and structure factors have been deposited in the PDB with accession code 4DU0.
TABLE 1.
Crystallographic data collection and refinement statistics
Values in parentheses are for the highest-resolution shell.
| Data collection | |
|---|---|
| Wavelength (Å) | 0.98 |
| Space group | P212121 |
| Cell parameters | |
| a, b, c (Å) | 38.5, 50.1, 78.9 |
| Molecules/a.u. | 4 |
| Resolution (Å) | 50-1.90 (1.93-1.90) |
| Number of reflections | |
| Total | 23,240 |
| Unique | 12,600 |
| Rmerga (%) | 14.4 (85.9) |
| I/σ | 18.1 (2.5) |
| Completeness (%) | 100 (100) |
| Redundancy | 6.5 (6.5) |
| Data refinement | |
| Resolution (Å) | 42-1.90 |
| Rb (%) | 17.9 |
| Rfreec (%) | 21.3 |
| Number of atoms | |
| Protein | 962 |
| Water | 106 |
| Ligand/ion | 26 |
| Root mean square deviation | |
| Bond lengths (Å) | 0.017 |
| Bond angles (°) | 1.8 |
| Ramachandrand | |
| Favored (%) | 96.4 |
| Allowed (%) | 3.6 |
| Outliers (%) | 0.0 |
a Rmerge = Σ I − 〈I〉|/ΣI, where I is the observed intensity and 〈I〉 is the average intensity obtained from multiple observations of symmetry-related reflections after rejections.
b r = Σ|Fo| − |Fc|/Σ|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively.
c Rfree = defined by Brünger (58).
d Calculated with MolProbity (37).
Functional Assays
The inhibition of LF by l-Ala17-HNP1 was quantified using an enzyme kinetic assay (30, 31). Briefly, freshly prepared LF at a final concentration of 1 μg/ml (∼10 nm) was incubated at 37 °C for 30 min with a 2-fold dilution series of defensin in 20 mm HEPES buffer containing 1 mm CaCl2 and 0.5% Nonidet P-40 (pH 7.2). 20 μl of LF substrate (1 mm in the buffer) was added to each well to a final concentration of 100 μm in a total volume of 200 μl. The enzyme activity, characterized as a time-dependent absorbance increase at 405 nm due to the release of p-nitroaniline, was monitored at 37 °C over a period of 15 min on a 96-well Tecan Infinite M1000 microplate reader (Tecan Group Ltd., Switzerland). Data are presented in a plot showing percent inhibition versus defensin concentration, from which IC50 values (the concentration of defensin that reduced the enzymatic activity of LF by 50%) were derived by a nonlinear regression analysis.
Virtual colony count (9) was employed to quantify the dose-dependent defensin killing of E. coli ATCC 25922, S. aureus ATCC 29213, and B. cereus ATCC 10876. Strains were obtained from the American Type Culture Collection (ATCC). A Tecan Infinite M1000 plate reader preset at 37 °C was used, and each assay was conducted in triplicate using seed cultures originating from single colonies grown on three separate days. The incubation buffer was 10 mm sodium phosphate (pH 7.4) containing 1% tryptic soy broth, which was also used previously (22). The low concentration of TSB ensures that the cells are actively metabolizing, while keeping the salt concentration low, both of which are important for defensin activity (13). Data analysis utilized a Visual Basic script to calculate the time necessary for each growth curve to reach a threshold change in optical density at 650 nm of 0.05. Rather than using the published calibration slope and y intercept values (9), recalibration was necessary to account for the tryptic soy broth addition and a different plate reader than was used previously. Calibration curves were generated to relate minutes to reach threshold with log(C′0) according to Fig. 2 and Table 1 of Ref. 9, yielding a slope of −68.3 and y intercept of 553.6 for E. coli (average of two calibration experiments whereby the slope differed by 1.0%), a slope of −65.3 and y intercept of 535.6 (a single calibration experiment) for S. aureus, and a slope of −51.8 and a y intercept of 437.8 (a single calibration experiment) for B. cereus. Cells were incubated on ice for 5 min to arrest growth before the defensin incubation step. During the 2-h incubation step, the 96-well plate was monitored in the plate reader under the same conditions as during the outgrowth step: 37 °C and shaking for 5 s every 5 min. A systematic decrease in absorbance was observed between the t = 0 and t = 5 min kinetic time points that was also noticed when a room temperature blank plate was placed in the 37 °C incubator. In all cases the optical density was reset to zero at 5 min after Mueller Hinton broth addition to start the outgrowth phase of the experiment.
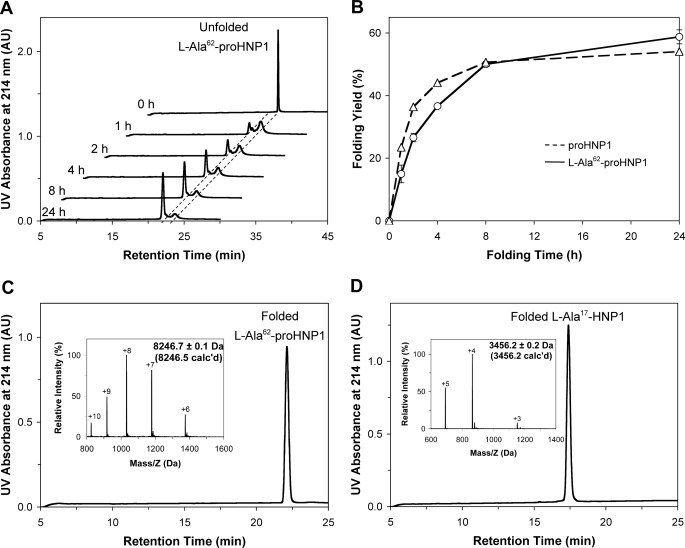
FIGURE 2.
A, HPLC-monitored folding of l-Ala62-pro-HNP1 at 2 mg/ml. B, time-dependent folding yield for l-Ala62-pro-HNP1. The data were obtained from two separate experiments. The published result of time-dependent folding yield for pro-HNP1 was cited here to compare the folding kinetics and rate. C, HPLC and ESI-MS analysis of oxidative-folded l-Ala62-pro-HNP1 after purification. D, HPLC and ESI-MS analysis of folded l-Ala17-HNP1 purified after cyanogen bromide (CNBr) cleavage.
l-Ala17-HNP1 interacting with HNP1 and itself was evaluated by surface plasmon resonance on a BIAcore T100 instrument (BIAcore, Inc., Piscataway, NJ). Synthetic HNP1 and l-Ala17-HNP1 in 10 mm acetate buffer (pH 5.0) were covalently coupled to CM5 carboxylated dextran chips using N-hydroxysuccinimide and N-ethyl-N′-(3-diethylaminopropyl)carbodiimide chemistry, respectively. Uncoupled N-hydroxysuccinimide ester groups were blocked with 1 m ethanolamine, and 233 resonance units of wild-type HNP1 and 244 resonance units of l-Ala17-HNP1 were coupled to the sensor chip by this procedure, respectively. Kinetic analysis was carried out at 25 °C in 10 mm HEPES, 150 mm NaCl, 3 mm EDTA, 0.05% surfactant P20 (pH 7.4). 2-Fold serial dilutions of HNP1 and l-Ala17-HNP1 were injected at a flow rate of 30 μl/min over each flow cell. Association and dissociation were assessed for 300 and 600 s, respectively. Resonance signals were corrected for nonspecific binding by subtracting the background of the control flow cell. After each analysis, the sensor chip surfaces were regenerated with 30 mm HCl and equilibrated with the buffer before the next injection. Binding kinetics was analyzed with BIAevaluation software.
RESULTS
Total Chemical Synthesis of l-Ala62-Pro-HNP1, l-Arg62-Pro-HNP1, and l-Phe62-Pro-HNP1 via Native Chemical Ligation
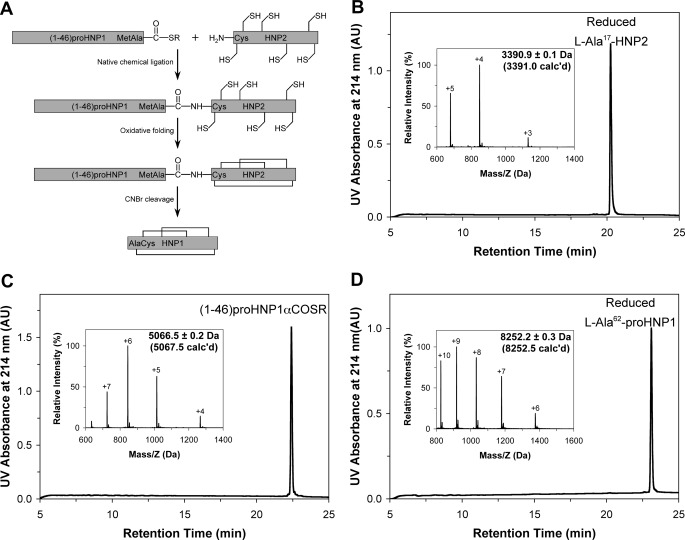
The 75-residue pro-HNP1 comprises a 45-residue N-terminal prodomain and the 30-residue C-terminal mature defensin HNP1. The presence of the only Met residue at position 45 enables convenient release of HNP1 from a folded pro-HNP1 by CNBr cleavage. Illustrated in Fig. 1A is the strategy for the synthesis of HNP1 or its analogs from a prodefensin, which is synthesized via native chemical ligation (39, 40) of two peptide fragments: residues 1–46 and 47–75 or HNP2. For this project, three variants of HNP2 with Ala, Arg, or Phe at position 17 (HNP1 numbering) were created.
FIGURE 1.
A, strategy for the preparation of HNP1 from pro-HNP1 synthesized via native chemical ligation. HNP1 differs from HNP2 by an extra Ala residue at the N terminus. Because native chemical ligation requires an N-terminal Cys in the downstream peptide, HNP2, which begins with an N-terminal Cys residue, was used for the reaction. To generate pro-HNP1, however, an Ala residue was C-terminal appended to Met45 of the upstream prodomain of 45 amino acid residues. CNBr cleavage of the only Met-45-Xxx bond in oxidatively folded pro-HNP1 generates HNP1. B–D, reversed phase high performance liquid chromatography (HPLC) and ESI-MS analysis of synthetic and purified l-Ala17-HNP2 (B), (1–46)-pro-HNP1-αCOSR (C), and fully reduced l-Ala62-pro-HNP1 (D). The chromatographic data were collected at 40 °C on a Waters XBridge C18 column using a linear gradient of 5–65% B at a flow rate of 1 ml/min over 30 min. ESI-MS data were obtained by suspending the sample in methanol/water (1:1) plus 2% acetic acid and infusing by a syringe pump at a flow rate of 10 μl/min. De-convolution of the data were performed using the Micromass MaxEnt software. The values in parentheses were calculated based on the average isotope compositions of the peptides.
We first synthesized l-Ala17-HNP2 and the N-terminal thioester peptide (1–46)pro-HNP1-αCOSR (r = CH2CH2CO-Leu-OH) as previously described (28). Both peptides were purified by RP-HPLC to homogeneity and their molecular masses were ascertained by ESI-MS (Fig. 1, B and C). The native chemical ligation reaction between (1–46)-pro-HNP1-αCOSR and l-Ala17-HNP2 proceeded to completion overnight, and the resultant ligation product l-Ala62-pro-HNP1 gave rise to a molecular mass of 8252.2 ± 0.3 Da, in good agreement with the theoretical value of 8252.5 Da calculated on the basis of the average isotopic compositions of reduced l-Ala62-pro-HNP1 (Fig. 1D). Highly purified l-Arg62-pro-HNP1 and l-Phe62-pro-HNP1 were also synthesized essentially as described (data not shown).
l-Ala62-pro-HNP1 but Not l-Arg62-pro-HNP1 or l-Phe62-pro-HNP1 Could Be Folded
Each of the three oxidative folding reactions began with pure prodefensin. As the reactions proceeded overnight, l-Ala62-pro-HNP1 produced a peak with mass corresponding to the prodefensin minus the mass of the six hydrogen atoms lost when the disulfide bonds were formed (Fig. 2, A and C). By contrast, the folding reactions with the Arg and Phe substitutions did not result in a single peak, and both prodefensin analogs were massively precipitated and misfolded during the folding reaction (data not shown). Importantly, the folding rate and yield of wild-type pro-HNP1 were similar to those of l-Ala62-pro-HNP1 (Fig. 2B), suggesting that the prodomain was capable of fully rescuing misfolding of l-Ala17-HNPs. After purification, l-Ala62-pro-HNP1 was subjected to CNBr cleavage, yielding folded l-Ala17-HNP1 with the correct molecular mass (3456.2 ± 0.2 Da found, 3456.2 Da calculated) (Fig. 2D).
l-Ala17-HNP1 Is Correctly Folded
We determined the crystal structure of l-Ala17-HNP1 at 1.9 Å resolution. The high-quality diffraction data resulted in readily discernible experimental electron density maps, which confirmed the identity of the Gly17 to l-Ala mutation in the synthetic defensin by showing a well defined conformation of l-Ala17 (Fig. 3A). In the refined structure the average B-factors of l-Ala17 atoms (17.1 and 16.2 Å2 for the backbone and side chain atoms, respectively) were significantly below the corresponding values calculated for all atoms in the asymmetric unit (19.2 and 24.8 Å2, respectively). The orthorhombic crystals contained four l-Ala17-HNP1 molecules in the asymmetric unit. These structurally independent monomers are almost identical and can be superimposed in pairs with root mean square deviations between 0.5 and 0.9 Å as calculated for the 30 Cα atoms (Fig. 3B). Some differences existed among the monomers in the backbone conformation of the loop connecting the β2 and β3 strands, a commonly observed variation typical for the β2-hairpin region in many known structures of α-defensins (22, 41, 42). Importantly, l-Ala17-HNP1 is correctly folded as shown by the native disulfide bonding (Cys2-Cys30, Cys4-Cys19, Cys9-Cys29) present in the synthetic defensin structure (Fig. 3B). The overall structure of l-Ala17-HNP1 contains all of the previously identified secondary structure elements, closely resembling wild-type HNP1 (Fig. 3C). Pairwise superimposition of l-Ala17-HNP1 and wild-type HNP1 monomers yields root mean square deviation values ranging from 0.3 to 0.9 Å as calculated for the 30 Cα atoms. These structural findings unequivocally demonstrate that the prodomain of HNP1 is capable of rescuing defensin misfolding as previously suggested (29).
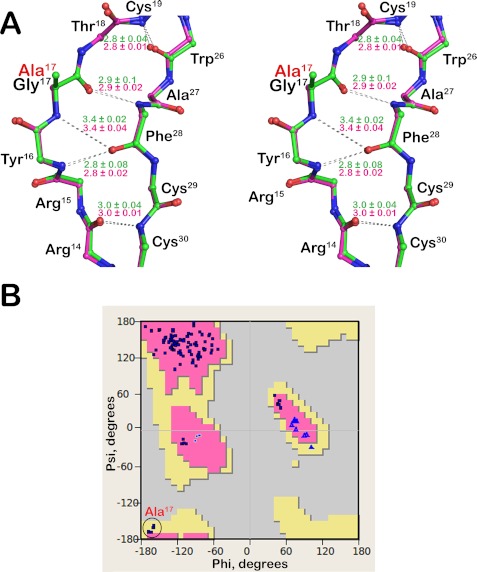
FIGURE 3.
Stereoviews of the l-Ala17-HNP1 structure. A, the 2Fo − Fc electron density map around Ala17 contoured at 1.5 σ. Ala17 and selected residues are labeled. B, stereo view of four superimposed monomers present in the asymmetric unit of orthorhombic crystal. Side chains are shown as balls and sticks, whereas the Cα-traces are shown as ribbons. C, structural alignment of crystallographically independent monomers of l-Ala17-HNP1 (green) and wild-type HNP1 (pink). Side chains are shown as lines, whereas backbone atoms are shown as balls and sticks.
β-Bulge Is Well Maintained in l-Ala17-HNP1
β-Bulges typically occur in the anti-parallel β-sheet region of a protein, where two residues on one β-strand form backbone H-bonds with a single residue on the opposite strand (19, 20). The extra residue creates a “bulge” that disrupts the normal pattern of alternating side chain direction, impacts the directionality of β-strands, and accentuates the typical right-handed twist of β-sheets (19, 20). In HNP1, the bulged strand comprises residues 15–18 (Arg15-Tyr16-Gly17-Thr18), of which Tyr16 and Gly17 participate in backbone H-bonding interactions with Phe28 of the opposing strand. Structural analysis of wild-type HNP1 and l-Ala17-HNP1 indicates that the β-bulge present in the wild-type defensin is fully maintained in the mutant. In fact, the H-bond networks that stabilize the β-bulges in both defensins are identical as judged by the H-bonding pattern and individual H-bond distances (Fig. 4A).
FIGURE 4.
β-Bulge of l-Ala17-HNP1. A, stereo view of superimposed β-bulge regions of l-Ala17-HNP1 (green) and wild-type HNP1 (pink). Only the backbone atoms are shown as balls and sticks. Main chain hydrogen bonds stabilizing the β-bulge are represented by gray dashes and their individual distances are shown in green (l-Ala17-HNP1) and pink (HNP1). B, the backbone dihedral angles (φ, ψ) of all amino acids of l-Ala17-HNP1 plotted on the Ramachandran diagram.
It was shown previously that the invariant Gly17 residue in α-defensins typically adopts the backbone dihedral angles (φ, ψ) centered at φ = 170° and ψ = −150° in the Ramachandran diagram, a region disallowed for l-amino acids but permissible for d-enantiomers (18, 43). A torsion angle analysis of l-Ala17 in the four structurally independent monomers indicates that it adopts the (φ, ψ) angles centered at φ = −163° ± 3.4, ψ = −163° ± 5.1 in an allowed but much less common β-sheet region for l-amino acid residues (Fig. 4B). The preservation of the β-bulge required for correct α-defensin folding likely forced l-Ala17 to adopt the energetically less favorable backbone dihedral angles, thereby explaining why l-Ala17-HNP2 alone, without the prodomain, was unable to fold into the native conformation (18). Given that small residues such as Ala and Ser are strictly required in place of Gly in classic β-bulges (19, 20), it is perhaps not surprising that both l-Arg62-pro-HNP1 and l-Phe62-pro-HNP1 misfolded even in the presence of the prodomain.
l-Ala17-HNP1 Monomers Associate into a Strained Canonical Dimer
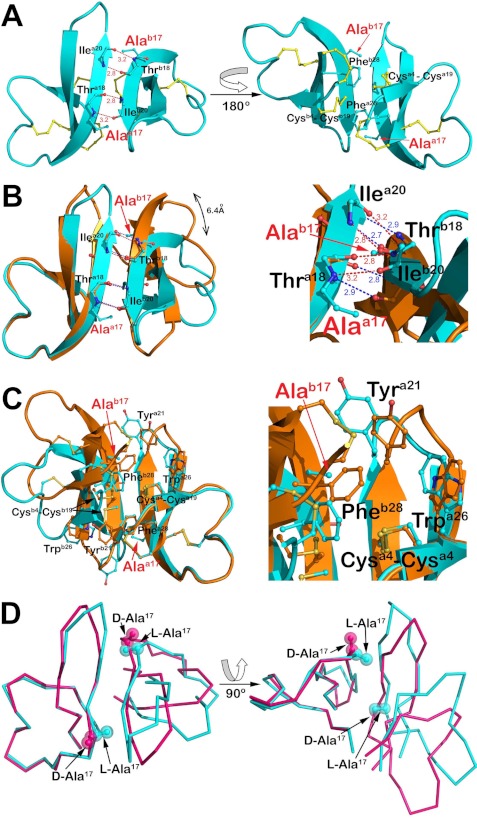
Although the asymmetric unit of the l-Ala17-HNP1 crystal contains four monomers, they are not arranged into dimers. However, further analysis of probable quaternary structures by PISA indicates that one of these structurally independent monomers could form a homodimer with a symmetry-related copy (Fig. 5A). This dimer is formed between the two β2-strands and stabilized by four backbone-backbone H-bonds donated reciprocally by the main chain nitrogen atoms of Thr18 and Ile20 of one monomer to the main chain oxygen atoms of Ile20 and Thr18 of another, a “canonical” mode of dimerization characteristic of α-defensins (44). Hydrophobic contacts involving the side chains of Ala17 and Phe28 from one monomer and the Cys4–Cys19 disulfide from the opposing counterpart also contribute to l-Ala17-HNP1 dimerization. The molecular surface buried within the l-Ala17-HNP1 dimer is 381 Å2 per monomer, which compares to 391 Å2 for the HNP1 dimer (PDB code 3GNY) (22) and 410 Å2 for the average of three independent d-Ala17-HNP2 dimers (PDB codes 1ZMK and 1ZMI) (18).
FIGURE 5.
Quaternary structure of l-Ala17-HNP1. A, putative dimeric assembly of l-Ala17-HNP1. Residues involved in dimer formation are shown as balls and sticks and reciprocal main chain hydrogen bonds and their distances are shown in red. B and C, comparison of l-Ala17-HNP1 and wild-type HNP1 dimers. Dimers were aligned based on monomer and colored cyan (Ala17-HNP1) and orange (HNP1). B, H-bond networks stabilizing monomer-monomer association (left) and close-up view of the dimer interface (rotated 30°, right). H-bonds (dashes) and their distances are shown colored in red (l-Ala17-HNP1) and blue (HNP1). C, the residues involved in hydrophobic packing (left) and the close-up view of the hydrophobic core (right). Displacement of Tyr21 and Trp26 of l-Ala17-HNP1 due to the relocation of the monomers enables its involvement in dimer stabilization. D, structural comparison of l-Ala17-HNP1 and d-Ala17-HNP2 (PDB code 1ZMK). Note that HNP2 is shorter than HNP1 by one amino acid residue at the N terminus; therefore, the d-Ala residue in HNP2 was numbered 16 in the original crystal structure.
Despite the canonical mode of dimerization, the dimer interface of l-Ala17-HNP1 is noticeably different from that of wild-type HNP1 as a result of the Gly17 to l-Ala mutation (Fig. 5B). The methyl side chain of l-Ala17 of one monomer points directly toward the β2-strand of the opposite monomer, tilting the molecules to avoid steric clashes between them. Both ends of the β2 strand were displaced, moving the first Cα of the β2 strand, Arg14, 6.4 Å, and the last Cα of the β2 strand, Tyr21, 3.5 Å. Consequently, the two reciprocal inter-monomer H-bonds between Thr-18N and Ile-20O are significantly lengthened from 2.9 to 3.2 Å with an energetically less favorable geometry as compared with those in the wild-type defensin, effectively destabilizing the l-Ala17-HNP1 dimer. The Gly to l-Ala17 mutation also causes fewer nonpolar contacts within the hydrophobic core of the dimer. In wild-type HNP1, dimerization is stabilized by extensive hydrophobic packing of the Cys4–Cys19 disulfide and its C-terminal aromatic residues, in that Tyr21 of one monomer, supported by the Trp26 residue, stacks against Phe28 of the opposing monomer (42, 44). However, in the Ala17-HNP1 dimer the hydrophobic core is reduced to only the side chains of Ala17, Phe28, and the Cys4–Cys19 disulfide as a result of the tilting of the two monomers with respect to each other (Fig. 5C). Of note, structural comparison of l-Ala17-HNP1 with d-Ala17-HNP2 shows that unlike l-Ala17, the methyl side chain of d-Ala17 points away from the dimer interface, posing no steric hindrance to d-Ala17-HNP2 dimerization (Fig. 5D).
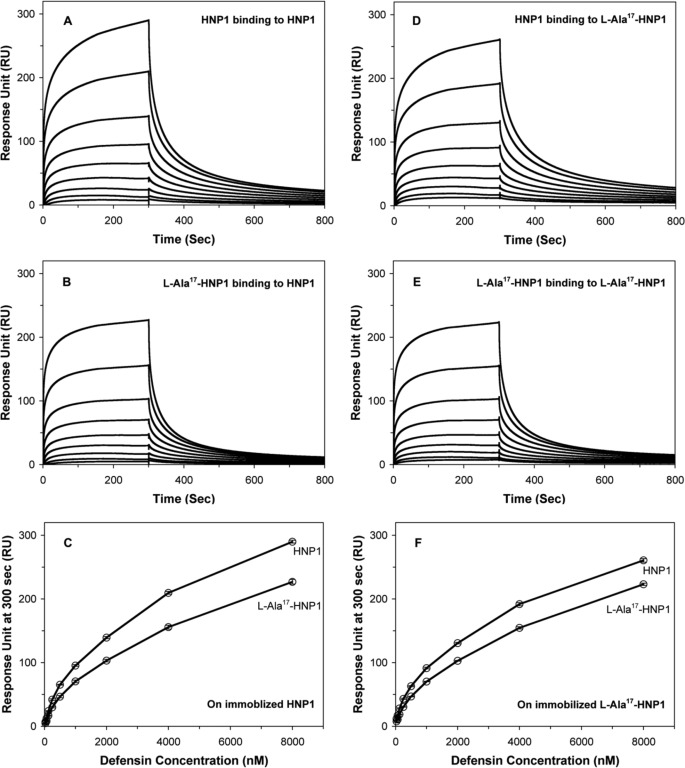
Self-association of l-Ala17-HNP1 Slightly Decreased Relative to HNP1 on Both HNP1 and l-Ala17-HNP1 Surfaces
HNP1 dimerizes and is capable of forming higher-ordered oligomers (10). It has recently been shown that HNP1 dimerization is important for many of its functions such as S. aureus killing, LF inhibition, and HIV-1 gp120 binding (44). To further examine the functional role of l-Ala17-HNP1, the binding kinetics of HNP1 or l-Ala17-HNP1 in solution on immobilized HNP1 or l-Ala17-HNP1 was measured by surface plasmon resonance. Representative sensorgrams are shown in Fig. 6. Self-association decreased due to the l-Ala17 mutation, such that HNP1 on HNP1 > HNP1 on l-Ala17-HNP1 > l-Ala17-HNP1 on HNP1 ≈ l-Ala17-HNP1 on l-Ala17-HNP1.
FIGURE 6.
Binding characteristics of l-Ala17-HNP1 to immobilized wt-HNP1 (233 resonance units) and l-Ala17-HNP1 (244 resonance units) as determined by surface plasmon resonance. Wild-type HNP1 is included for comparison. For both peptides, a concentration of 8000 nm was used. The curves are obtained from the averages of three measurements.
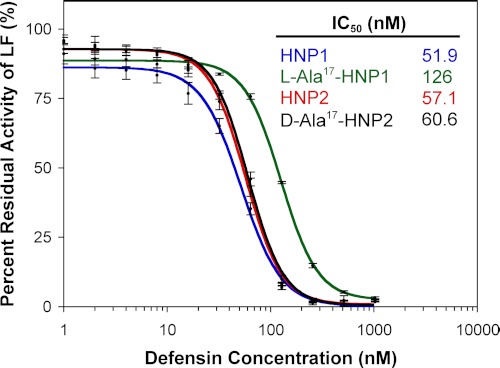
LF Activity Inhibition by l-Ala17-HNP1 Was Less Than Wild-type HNP1
The enzyme activity of LF in the presence of HNP1 or l-Ala17-HNP1 was assessed using a method adapted from Kaufmann and colleagues (30). Fig. 7 shows that the inhibitory activity of l-Ala17-HNP1 as measured by the concentration that gave 50% inhibition (IC50) differed slightly more than 2-fold compared with HNP1. By contrast, d-Ala17-HNP2 (HNP1 numbering) was equally active compared with HNP2. The d-Ala17 mutation was incorporated into HNP2 instead of HNP1 to conform to the previous study (18), but because HNP1 and HNP2 vary only by the deletion of a single Ala residue at the N terminus, they should be comparable with one another with respect to the internal Gly17 residue. LF inhibition is impaired only by the Gly17 to l-Ala17 mutation but not the Gly17 to d-Ala17 mutation, pointing to a local structural defect as the culprit.
FIGURE 7.
Inhibition of LF activity by different concentrations of l-Ala17-HNP1, wild-type HNP1, d-Ala17-HNP2, and wild-type HNP2. Each inhibition curve is the mean of three independent enzyme kinetic measurements.
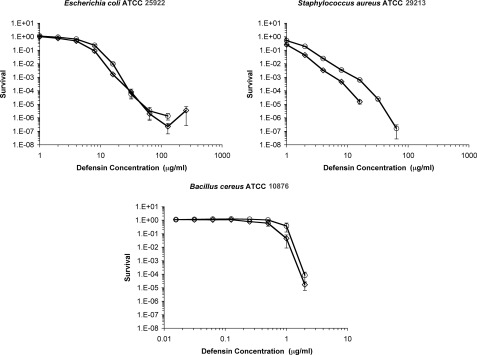
Antibacterial Activity of l-Ala17-HNP1 against S. aureus and E. coli Was Diminished Compared with Wild-type HNP1
To assess antibacterial activity, E. coli ATCC 25922, S. aureus ATCC 29213, and B. cereus ATCC 10876 were tested using the convenient virtual colony count assay (9). Activity is reported as the survival curves shown in Fig. 8 and virtual lethal dose values in Table 2. The antibacterial activity against E. coli differed slightly in the region of the virtual lethal dose values reported in Table 2. It was nearly identical at higher concentrations, except the highest (256 μg/ml) concentration of l-Ala17-HNP1 gave complete killing in two of the three assays and the calculated survival was 8.6 × 10−10 in the third replicate. Apparent survival of less than the order of 10−6 indicates that, in addition to measuring the time necessary for the surviving cell(s) to reach the threshold optical density of 0.05, there was a lag time after the twice-concentrated Mueller-Hinton broth was added to the defensin incubation mixture. This phenomenon has previously been reported to a lesser extent with both E. coli and S. aureus (9, 22). Virtual lethal dose values for S. aureus were almost 2-fold less for l-Ala17-HNP1 than wild-type HNP1, and this trend continued at each concentration throughout the survival curves. HNP1, but not l-Ala17-HNP1, gave complete killing at 32 and 64 μg/ml, and both gave complete killing at 128 and 256 μg/ml. Against B. cereus, both defensins gave complete killing in two of the three replicates at the highest concentration tested, and low survival rates in the third, once again indicative of a lag time after the addition of Mueller-Hinton broth. Error was higher with B. cereus than the other two strains.
FIGURE 8.
Virtual colony count survival curves of E. coli ATCC 25922, S. aureus ATCC 29213, and B. cereus ATCC 10876 exposed to HNP1 (♢) and l-Ala17-HNP1 (○). Strains were exposed a 2-fold dilution series of defensins at concentrations varying from 1 to 256 μg/ml (E. coli and S. aureus) or 0.016 to 4 (B. cereus). Each curve is the mean of triplicate experiments. Points equivalent to 0 survival cannot be plotted on a logarithmic scale, such that there was complete killing above 16 μg/ml for HNP1 and 64 μg/ml for l-Ala17-HNP1 against S. aureus. At 256 μg/ml against E. coli, l-Ala17-HNP1 gave complete killing in two replicates and a survival rate of 8.6 × 10−10 in the third (indicating a lag time before outgrowth). Similarly, at 4 μg/ml against B. cereus, HNP1 gave complete killing in two replicates and a survival rate of 8.5 × 10−13 in the third. Also, at 4 μg/ml against B. cereus, l-Ala17-HNP1 gave complete killing in two replicates and a survival rate of 2.3 × 10−7 in the third.
TABLE 2.
Virtual lethal doses obtained from the virtual colony count assay
The vLD50, vLD90, vLD99, and vLD99.9 are shown as the defensin concentrations in μm that resulted in a survival rate of 0.5, 0.1, 0.01, and 0.001, respectively. Mean ± S.E. is reported for n = three measurements.
| vLD50 |
vLD90 |
vLD99 |
vLD99.9 |
|||||
|---|---|---|---|---|---|---|---|---|
| HNP1 | l-Ala-17-HNP1 | HNP1 | l-Ala-17-HNP1 | HNP1 | l-Ala-17-HNP1 | HNP1 | l-Ala-17-HNP1 | |
| E. coli | 3.90 ± 0.53 | 5.19 ± 0.45 | 8.14 ± 0.73 | 11.2 ± 0.70 | 14.8 ± 0.35 | 16.8 ± 0.88 | 21.1 ± 1.26a | 29.8 ± 0.38a |
| S. aureus | <1 | 1.12 ± 0.01 | 1.67 ± 0.38a | 2.83 ± 0.13a | 3.55 ± 0.05a | 6.43 ± 0.16a | 6.99 ± 0.13a | 14.4 ± 0.65a |
| B. cereus | 0.55 ± 0.13a | 1.00 ± 0.21a | 0.96 ± 0.12 | 1.48 ± 0.28 | 1.40 ± 0.27 | 1.76 ± 0.22 | 1.88 ± 0.08 | 1.97 ± 0.03 |
a vLD values of wild-type HNP1 versus l-Ala17-HNP1 that are significantly different statistically (p < 0.05).
DISCUSSION
Mammalian α-defensins are capable of acting on a diverse array of bacterial, viral, and molecular targets, playing important roles in host innate immunity against microbial infection (10). These functionally versatile antimicrobial peptides are structurally conserved yet vary considerably in amino acid sequence and composition. There are a total of seven invariant residues in all known mammalian α-defensins: six cysteines that form the three intramolecular disulfides and Gly17 (HNP1 numbering) that constitutes part of the β-bulge structure. In addition, a salt bridge between Arg5 and Glu13 (HNP1 numbering) exists in most mammalian α-defensins, contributing a great deal to α-defensin folding and in vivo stability (45–48), but little to α-defensin functionality (10). Although disulfide bonding is critical for α-defensin biosynthesis (49) and the vast majority of reported α-defensin functions (10), the structural and functional basis for the strict conservation of Gly17 largely remains elusive. Of note, Gly17 also constitutes part of a larger structural motif (C-X7-GXC) known as the γ-core (50, 51), which is present across many classes of antimicrobial peptides. Elucidating the role of Gly17 in HNP1 will help identify the structural determinants that impart functional versatility to α-defensins, and afford new insights as well into how antimicrobial peptides function in general at the molecular level.
Previous studies showed that Gly17 lies within a “classical” type β-bulge stabilized by three hydrogen bonds (19, 20), but is atypical in the sense that it contains a Gly with backbone torsion angles permissible only to unnatural d-amino acids but not natural l-enantiomers (18). At the time, we commented that it appeared that only Gly could be tolerated at position 17. Our current findings reveal that this is not strictly the case. Replacement of Gly17 in HNP2 by l-Ala resulted in quantitative misfolding of the α-defensin peptide, whereas substitutions of multiple d-amino acids at the same position yielded correctly and productively folded functional analogs of HNP2. These results suggest that introducing an l-amino acid at position 17 disrupts the β-bulge, thus causing disulfide mispairing and defensin misfolding. However, these studies on HNP2 were done in the absence of its prodomain. Because HNPs are synthesized in vivo as prodefensins and the prodomain is capable of catalyzing correct folding of defensin mutants unable to fold on their own (29, 42, 45, 46), the question of whether or not a natural evolutionary substitution at position 17 of α-defensins is structurally allowed and functionally viable remained unanswered.
We have demonstrated in this work that a correctly folded l-Ala17-HNP1 could be efficiently produced in vitro from its corresponding prodefensin mutant l-Ala62-pro-HNP1. This finding confirms the catalytic role of the prodomain in α-defensin folding and suggests that natural selection is possible in vivo at the Gly17 position. In fact, l-Ala17 was tolerable to the β-bulge structure as evidenced by a well maintained H-bonding network identical to that of wild-type HNP1 (Fig. 4A). Despite the tolerance of l-Ala, natural selection at position 17 would be restricted to a few small amino acids as the prodomain failed to rescue misfolding of HNP1 mutants with bulky residues such as Phe and Arg in place of Gly17. Side chains other than Ala, Phe, and Arg were not tested in this capacity; therefore, we cannot rule out that other small side chains such as Ser and Val would behave like Ala. Nevertheless, replacement of Gly17 by the smallest l-amino acid, namely alanine, while having limited impact on the tertiary structure of the α-defensin HNP1, is detrimental to its quaternary structure and function. Overall, our findings strongly suggest that the conservation of Gly17 in mammalian α-defensins endows them with the ability to fold correctly into a functional quaternary structure.
Recent studies on HNP1 have shown that dimerization is functionally important for the α-defensin to kill S. aureus, inhibit LF, and bind HIV-1 gp120 (44). Substitution of l-Ala for Gly17 obviously disfavored defensin dimerization as evidenced structurally by lengthened intermolecular backbone H-bonds and less hydrophobic packing at the putative dimer interface, contributing to the impaired activity of l-Ala17-HNP1. However, the functional effects of the G17A mutation on LF inhibition, self-association, and antibacterial activity were rather modest (2-fold or less), contrasting with a more pronounced change in the strength of dimerization as indicated by structural analysis of l-Ala17-HNP1. Far greater differences were observed in a study of a monomeric form of HNP1 generated by methylation of the main chain NH of Ile20 (44). Monomeric HNP1 bound to surface-associated wild-type HNP1 with a 7-fold decrease in affinity compared with wild-type HNP1 bound to itself, and an 11-fold increase in LF IC50 was observed from the monomeric mutant. Also in contrast to the monomeric HNP1, which had dramatically decreased activity against S. aureus but not E. coli, l-Ala17-HNP1 had only mildly decreased activity against both strains although in all cases the effect against S. aureus was greater than the effect against E. coli. Taken together, the evidence points to perturbation, but not abrogation, of dimerization. Given the degree of conservation of Gly17, it may be expected that even the slight decreases in activity that we observed would be enough to prevent the l-Ala mutation from being fixed in natural populations, indicating that defensins are apparently under strong stabilizing selective pressures in the region that includes the β-bulge.
Investigators wishing to explore minimally perturbed structures at a glycine position often study glycine to alanine substitutions. One reason for the popularity of this particular mutation is that the Ala methyl side chain is the smallest save for the hydrogen atom in glycine, thus minimizing steric hindrance. Another reason to conduct Gly to Ala mutations is to probe the role of backbone flexibility on protein structure, because the φ and ψ torsion angles of the glycine are less restricted than those of Ala. The achiral Gly can adopt angles representative of either an l- or d-amino acid. Finally, each Gly to Ala mutation replaces an amino acid that is a likely secondary structure breaker into one that readily occupies regular α helical conformations. Gly tends to favor turns, kinks, and bulges due to the entropic penalty for participation in conventional backbone hydrogen bonding patterns. In this respect, the insertion of an Ala within a β-sheet can have an unpredictable effect. Ala is known to be one of the least likely amino acids to be found in a naturally occurring β-sheet, as was found in the case of staphylococcal IgG-binding protein G, for example (52). Meanwhile, it does not introduce a bulkier hydrophobic side chain more likely to create steric clashes, a polar side chain that might introduce hydrogen bonds, or a charged side chain that might invite electrostatic attraction or repulsion. Despite the fact that l-Ala17 is structurally tolerable to the β-bulge as well as to the tertiary structure of l-Ala17-HNP1, it is obviously constrained energetically as evidenced by quantitative misfolding of l-Ala17-HNP2 in the absence of its prodomain (18). The prodomain chaperones defensin folding by forming an intramolecular inhibitory complex with the defensin domain, thus reducing the free energy barrier to the transition state of folding intermediates (29). Two possible reasons may explain the misfolding of l-Arg62-pro-HNP1 and l-Phe62-pro-HNP1. First, the energy barrier to the formation of the β-bulge is too high to overcome; second, the bulky residues introduced in place of Gly may disrupt intramolecular interactions between the prodomain and C-terminal defensin domain, thus negating a folding catalysis.
In the region of the β-bulge there is a hydrophobic patch that includes the side chains of Trp26 and Phe28. Alanine scanning mutations identified hydrophobicity at those two positions as crucial for the structure and function of both HNP1 (42) and HD5.6 Both d-Arg17-HNP2 and d-Phe17-HNP2 folded correctly and productively without the prodomain and displayed improved activity against both E. coli and S. aureus compared with wild-type (18), underscoring the importance of the interplay between cationicity and hydrophobicity in the action of α-defensins against microbes. In general, net charge correlates with defensin antibacterial activity, as has been demonstrated previously (53–57), although the relationship is a complex one with other residues such as the hydrophobic residues near the C terminus playing a major role (42). The motifs of the β-bulge structure, dimer interface, cationicity, and hydrophobicity produce an intricate defensin architecture capable of many activities concentrated in a small package, which is gradually being elucidated through biochemical and microbiological analysis.
In summary, we have shown that although in principle it is possible to have a natural selection of small l-amino acids to replace the invariant Gly17 residue in mammalian of α-defensins, the resultant mutant(s) will likely be defective at the structural level. A defective quaternary structure of the mutant defensin bears the consequences of sufficiently impaired biological function such that its selection will become evolutionarily disfavored.
Acknowledgments
We thank the X-ray Crystallography Core Facility of the University of Maryland at Baltimore for providing crystallographic equipment and resources. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the United States Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by National Institutes of Health, National Center for Research Resources, Biomedical Technology Program Grant P41RR001209, and the National Institute of General Medical Sciences.
This work was supported, in whole or in part, by National Institutes of Health Grants AI072732 and AI061482 (to W. L.).
The atomic coordinates and structure factors (code 4DUO) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
M. Rajabi, B. Ericksen, X. Wu, E. de Leeuw, L. Zhao, M. Pazgier, and W. Lu, unpublished results.
- LF
- anthrax lethal factor
- RP-HPLC
- reversed phase high-performance liquid chromatography
- PDB
- Protein Data Bank
- HNP1
- human α-defensin 1.
REFERENCES
- 1. Wiesner J., Vilcinskas A. (2010) Antimicrobial peptides. The ancient arm of the human immune system. Virulence 1, 440–464 [DOI] [PubMed] [Google Scholar]
- 2. Zasloff M. (2002) Antimicrobial peptides of multicellular organisms. Nature 415, 389–395 [DOI] [PubMed] [Google Scholar]
- 3. Bernard J. J., Gallo R. L. (2011) Protecting the boundary. The sentinel role of host defense peptides in the skin. Cell Mol. Life Sci. 68, 2189–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ganz T. (2003) Defensins, antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3, 710–720 [DOI] [PubMed] [Google Scholar]
- 5. Bevins C. (2006) Paneth cell defensins. Key effector molecules of innate immunity. Biochem. Soc. Trans. 34, 263–266 [DOI] [PubMed] [Google Scholar]
- 6. Selsted M. E, Ouellette A. J. (2005) Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 6, 551–557 [DOI] [PubMed] [Google Scholar]
- 7. Lehrer R. I. (2004) Primate defensins. Nat. Rev. Microbiol. 2, 727–738 [DOI] [PubMed] [Google Scholar]
- 8. Abrami L., Reig N., van der Goot F. G. (2005) Anthrax toxin. The long and winding road that leads to the kill. Trends Microbiol. 13, 72–78 [DOI] [PubMed] [Google Scholar]
- 9. Ericksen B., Wu Z., Lu W., Lehrer R. I. (2005) Antibacterial activity and specificity of the six human α-defensins. Antimicrob. Agents Chemother. 49, 269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lehrer R. I., Lu W. (2012) α-Defensins in human innate immunity. Immunol. Rev. 245, 84–112 [DOI] [PubMed] [Google Scholar]
- 11. Pazgier M., Hoover D. M., Yang D., Lu W., Lubkowski J. (2006) Human β-defensins. Cell Mol. Life Sci. 63, 1294–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Selsted M. E., Harwig S. S., Ganz T., Schilling J. W., Lehrer R. I. (1985) Primary structures of three human neutrophil defensins. J. Clin. Invest. 76, 1436–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ganz T., Selsted M. E., Szklarek D., Harwig S. S., Daher K., Bainton D. F., Lehrer R. I. (1985) Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Invest. 76, 1427–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wilde C. G., Griffith J. E., Marra M. N., Snable J. L., Scott R. W. (1989) Purification and characterization of human neutrophil peptide 4, a novel member of the defensin family. J. Biol. Chem. 264, 11200–11203 [PubMed] [Google Scholar]
- 15. Gabay J. E., Scott R. W., Campanelli D., Griffith J., Wilde C., Marra M. N., Seeger M., Nathan C. F. (1989) Antibiotic proteins of human polymorphonuclear leukocytes. Proc. Natl. Acad. Sci. U.S.A. 86, 5610–5614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jones D. E., Bevins C. L. (1993) Defensin-6 mRNA in human paneth cells. Implications for antimicrobial peptides in host defense of the human bowel. FEBS Lett. 315, 187–192 [DOI] [PubMed] [Google Scholar]
- 17. Jones D. E., Bevins C. L. (1992) Paneth cells of the human small intestine express an antimicrobial peptide gene. J. Biol. Chem. 267, 23216–23225 [PubMed] [Google Scholar]
- 18. Xie C., Prahl A., Ericksen B., Wu Z., Zeng P., Li X., Lu W. Y., Lubkowski J., Lu W. (2005) Reconstruction of the conserved β-bulge in mammalian defensins using d-amino acids. J. Biol. Chem. 280, 32921–32929 [DOI] [PubMed] [Google Scholar]
- 19. Richardson J. S., Getzoff E. D., Richardson D. C. (1978) The β-bulge. A common small unit of nonrepetitive protein structure. Proc. Natl. Acad. Sci. U.S.A. 75, 2574–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chan A. W., Hutchinson E. G., Harris D., Thornton J. M. (1993) Identification, classification, and analysis of β-bulges in proteins. Protein Sci. 2, 1574–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pardi A., Zhang X. L., Selsted M. E., Skalicky J. J., Yip P. F. (1992) NMR studies of defensin antimicrobial peptides. 2. Three-dimensional structures of rabbit NP-2 and human HNP-1. Biochemistry 31, 11357–11364 [DOI] [PubMed] [Google Scholar]
- 22. Wei G., de Leeuw E., Pazgier M., Yuan W., Zou G., Wang J., Ericksen B., Lu W. Y., Lehrer R. I., Lu W. (2009) Through the looking glass, mechanistic insights from enantiomeric human defensins. J. Biol. Chem. 284, 29180–29192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hill C. P., Yee J., Selsted M. E., Eisenberg D. (1991) Crystal structure of defensin HNP-3, an amphiphilic dimer. Mechanisms of membrane permeabilization. Science 251, 1481–1485 [DOI] [PubMed] [Google Scholar]
- 24. Wu Z., Powell R., Lu W. (2003) Productive folding of human neutrophil α-defensins in vitro without the propeptide. J. Am. Chem. Soc. 125, 2402–2403 [DOI] [PubMed] [Google Scholar]
- 25. Michaelson D., Rayner J., Couto M., Ganz T. (1992) Cationic defensins arise from charge-neutralized propeptides. A mechanism for avoiding leukocyte autocytotoxicity? J. Leukoc. Biol. 51, 634–639 [DOI] [PubMed] [Google Scholar]
- 26. Liu L., Ganz T. (1995) The proregion of human neutrophil defensin contains a motif that is essential for normal subcellular sorting. Blood 85, 1095–1103 [PubMed] [Google Scholar]
- 27. Valore E. V., Martin E., Harwig S. S., Ganz T. (1996) Intramolecular inhibition of human defensin HNP-1 by its propiece. J. Clin. Invest. 97, 1624–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu Z., Prahl A., Powell R., Ericksen B., Lubkowski J., Lu W. (2003) From pro defensins to defensins. Synthesis and characterization of human neutrophil pro-α-defensin-1 and its mature domain. J. Pept. Res. 62, 53–62 [DOI] [PubMed] [Google Scholar]
- 29. Wu Z., Li X., Ericksen B., de Leeuw E., Zou G., Zeng P., Xie C., Li C., Lubkowski J., Lu W. Y., Lu W. (2007) Impact of prosegments on the folding and function of human neutrophil α-defensins. J. Mol. Biol. 368, 537–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim C., Gajendran N., Mittrücker H. W., Weiwad M., Song Y. H., Hurwitz R., Wilmanns M., Fischer G., Kaufmann S. H. (2005) Human α-defensins neutralize anthrax lethal toxin and protect against its fatal consequences. Proc. Natl. Acad. Sci. U.S.A. 102, 4830–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Min D. H., Tang W. J., Mrksich M. (2004) Chemical screening by mass spectrometry to identify inhibitors of anthrax lethal factor. Nat. Biotechnol. 22, 717–723 [DOI] [PubMed] [Google Scholar]
- 32. Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. (1995) How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4, 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 34. Collaborative Computational Project, Number 4 (1994) The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 35. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 36. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 37. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity. All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 39. Dawson P. E., Kent S. B. (2000) Synthesis of native proteins by chemical ligation. Annu. Rev. Biochem. 69, 923–960 [DOI] [PubMed] [Google Scholar]
- 40. Dawson P. E., Muir T. W., Clark-Lewis I., Kent S. B. (1994) Synthesis of proteins by native chemical ligation. Science 266, 776–779 [DOI] [PubMed] [Google Scholar]
- 41. Szyk A., Wu Z., Tucker K., Yang D., Lu W., Lubkowski J. (2006) Crystal structures of human α-defensins HNP4, HD5, and HD6. Protein Sci. 15, 2749–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wei G., Pazgier M., de Leeuw E., Rajabi M., Li J., Zou G., Jung G., Yuan W., Lu W. Y., Lehrer R. I., Lu W. (2010) Trp-26 imparts functional versatility to human α-defensin HNP1. J. Biol. Chem. 285, 16275–16285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mitchell J. B., Smith J. (2003) d-Amino acid residues in peptides and proteins. Proteins 50, 563–571 [DOI] [PubMed] [Google Scholar]
- 44. Pazgier M., Wei G., Ericksen B., Jung G., Wu Z., de Leeuw E., Yuan W., Szmacinski H., Lu W. Y., Lubkowski J., Lehrer R. I., Lu W. (2012) Sometimes it takes two to tango. Contributions of dimerization to functions of human α-DEFENSIN HNP1 PEPTIDE. J. Biol. Chem. 287, 8944–8953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rajabi M., de Leeuw E., Pazgier M., Li J., Lubkowski J., Lu W. (2008) The conserved salt bridge in human α-defensin 5 is required for its precursor processing and proteolytic stability. J. Biol. Chem. 283, 21509–21518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wu Z., Li X., de Leeuw E., Ericksen B., Lu W. (2005) Why is the Arg5–Glu13 salt bridge conserved in mammalian α-defensins? J. Biol. Chem. 280, 43039–43047 [DOI] [PubMed] [Google Scholar]
- 47. Rosengren K. J., Daly N. L., Fornander L. M., Jönsson L. M., Shirafuji Y., Qu X., Vogel H. J., Ouellette A. J., Craik D. J. (2006) Structural and functional characterization of the conserved salt bridge in mammalian paneth cell α-defensins. Solution structures of mouse Cryptdin-4 and (E15D)-Cryptdin-4. J. Biol. Chem. 281, 28068–28078 [DOI] [PubMed] [Google Scholar]
- 48. Andersson H. S., Figueredo S. M., Haugaard-Kedström L. M., Bengtsson E., Daly N. L., Qu X., Craik D. J., Ouellette A. J., Rosengren K. J. (2012) (January 29, 2012) The α-defensin salt-bridge induces backbone stability to facilitate folding and confer proteolytic resistance. Amino Acids [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Maemoto A., Qu X., Rosengren K. J., Tanabe H., Henschen-Edman A., Craik D. J., Ouellette A. J. (2004) Functional analysis of the α-defensin disulfide array in mouse cryptdin-4. J. Biol. Chem. 279, 44188–44196 [DOI] [PubMed] [Google Scholar]
- 50. Yount N. Y., Yeaman M. R. (2004) Multidimensional signatures in antimicrobial peptides. Proc. Natl. Acad. Sci. U.S.A. 101, 7363–7368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yeaman M. R., Yount N. Y. (2007) Unifying themes in host defense effector polypeptides. Nat. Rev. Microbiol. 5, 727–740 [DOI] [PubMed] [Google Scholar]
- 52. Smith C. K., Withka J. M., Regan L. (1994) A thermodynamic scale for the β-sheet forming tendencies of the amino acids. Biochemistry 33, 5510–5517 [DOI] [PubMed] [Google Scholar]
- 53. Xie C., Zeng P., Ericksen B., Wu Z., Lu W. Y., Lu W. (2005) Effects of the terminal charges in human neutrophil α-defensin 2 on its bactericidal and membrane activity. Peptides 26, 2377–2383 [DOI] [PubMed] [Google Scholar]
- 54. Zou G., de Leeuw E., Li C., Pazgier M., Li C., Zeng P., Lu W. Y., Lubkowski J., Lu W. (2007) Toward understanding the cationicity of defensins. Arg and Lys versus their noncoded analogs. J. Biol. Chem. 282, 19653–19665 [DOI] [PubMed] [Google Scholar]
- 55. Tanabe H., Qu X., Weeks C. S., Cummings J. E., Kolusheva S., Walsh K. B., Jelinek R., Vanderlick T. K., Selsted M. E., Ouellette A. J. (2004) Structure-activity determinants in paneth cell α-defensins. Loss-of-function in mouse cryptdin-4 by charge-reversal at arginine residue positions. J. Biol. Chem. 279, 11976–11983 [DOI] [PubMed] [Google Scholar]
- 56. Figueredo S. M., Weeks C. S., Young S. K., Ouellette A. J. (2009) Anionic amino acids near the pro-α-defensin N terminus mediate inhibition of bactericidal activity in mouse procryptdin-4. J. Biol. Chem. 284, 6826–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Llenado R. A., Weeks C. S., Cocco M. J., Ouellette A. J. (2009) Electropositive charge in α-defensin bactericidal activity. Functional effects of Lys for Arg substitutions vary with the peptide primary structure. Infect. Immun. 77, 5035–5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Brünger A. T. (1992) Free R value. A novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475 [DOI] [PubMed] [Google Scholar]