Background: Are there differences in protein patterns relating to different cartilage properties?
Results: Quantitative proteomics of cartilage from articulating joints, trachea, rib and intervertebral disc revealed distinct differences.
Conclusion: Observed differences are pronounced between different types of cartilage, whereas less marked significant between subtypes of articular cartilages.
Significance: The data provides novel insights into tissue structure-function and tropism of disease.
Keywords: Cartilage, Cartilage Biology, Extracellular Matrix Proteins, Proteoglycan, Proteomics, Intervertabral Disc, Meniscus
Abstract
Human synovial joints display a characteristic anatomic distribution of arthritis, e.g. rheumatoid arthritis primarily affects the metacarpophalangeal and proximal finger joints, but rarely the distal finger joints, whereas osteoarthritis occurs in the distal and proximal finger joints. Pelvospondylitis has a selective localization to the spine and sacroiliac joints. Is this tropism due to differences between the cartilages at the molecular level? To substantiate this concept the present study provides a background detailed compositional analysis by relative quantification of extracellular matrix proteins in articular cartilages, meniscus, intervertebral disc, rib, and tracheal cartilages on samples from 5–6 different individuals using an optimized approach for proteomics. Tissue extraction followed by trypsin digestion and two-dimensional LC separations coupled to tandem mass spectrometry, relative quantification with isobaric labeling, iTRAQTM, was used to compare the relative abundance of about 150 proteins. There were clear differences in protein patterns between different kinds of cartilages. Matrilin-1 and epiphycan were specific for rib and trachea, whereas asporin was particularly abundant in the meniscus. Interestingly, lubricin was prominent in the intervertebral disc, especially in the nucleus pulposus. Fibromodulin and lumican showed distributions that were mirror images of one other. Analyses of the insoluble residues from guanidine extraction revealed that a fraction of several proteins remained unextracted, e.g. asporin, CILP, and COMP, indicating cross-linking. Distinct differences in protein patterns may relate to different tissue mechanical properties, and to the intriguing tropism in different patterns of joint pathology.
Introduction
Cartilage consists of a relatively sparse number of chondrocytes surrounded by an extensive extracellular matrix (ECM),2 where the prominent components are collagens, proteoglycans, and water. Collagens, mainly composed of type II together with type XI and type IX, form a fibrillar network that contributes tensile stiffness and strength to the tissue (1). The large aggregating proteoglycan, aggrecan, is a major highly negatively charged component attracting positively charged counterions and thereby water enabling the tissue to withstand load and compressive forces (2). There are also other noncollagenous proteins with different important roles, e.g. maintaining overall structure and modulating tissue formation (3, 4). Although forming only a small fraction of the total matrix, these proteins may be vital for cartilage function. The numbers of identified proteins in cartilage have been rather limited and therefore new means such as proteomics technology have been applied to obtain a deeper understanding of the molecular composition of the tissues. The progress in technology development during the last decade has made tissue proteomics available also for ECM-rich tissues such as cartilage. Some of the obvious targets for cartilage proteomics include surveillance and search for alterations in disease. Characterization of molecular differences between cartilage subtypes will provide a background for understanding different functionalities and disease patterns. There are two major concerns in cartilage proteomics, (i) the highly anionic macromolecules (mainly aggrecan) and (ii) the highly abundant collagens (mainly type II). A single aggrecan molecule may contain in the order of 10,000 fixed negative charges in the form of oligosaccharides and glycosaminoglycan chains. This extreme charge density can interfere with protein separations such as ion exchange chromatography. Upon SDS-PAGE, the heterogeneity of post-translational modifications causes streaking and smearing of bands. Several approaches circumventing this problem have been reported, e.g. the use of cetylpyridinium chloride to precipitate proteoglycans (5, 6), the use of CsCl gradient ultracentrifugation to remove aggrecan (7), and the use of chondroitinase ABC for digestion of glycosaminoglycan chains into disaccharides (8, 9). All these approaches have been used in combination with gel electrophoresis for the protein separation. However, the use of chondroitinase and N-glycosidase might change the proteome significantly due to incubations at physiological conditions with potential unknown enzymatic activity still present in the sample. Moreover, there are several challenging features of other ECM molecules including processing of procollagen, covalent cross-linking, and post-translational modifications such as glycosylation, sulfation, and phosphorylation. These variables in analyses can be circumvented by focusing on the unmodified peptides released upon well defined trypsin proteolysis in solution. These peptides can be analyzed using two-dimensional liquid chromatography separations in combination with mass spectrometry. The use of chemical isotope labels such as isobaric tags for relative and absolute quantitation (iTRAQ) enable multiplex proteomics (10). The 4-plex system used in the current study allowed three tissue samples to be analyzed in each sample set with an internal constant reference sample used for comparison. The amine-reactive isobaric label release signature ions upon MS-MS fragmentation that can be used for relative quantitation between tissue samples, in this case relative to a reference made up to represent all the tissues analyzed. The analysis revealed distinct differences in the composition of different cartilaginous tissues at the same time as many proteins were similarly represented.
EXPERIMENTAL PROCEDURES
Materials
Guanidine hydrochloride (GdnHCl) and anhydrous sodium acetate (NaAc) were purchased from Merck (Darmstadt, Germany). N-Ethylmaleimide, 6-aminocaproic acid, benzamidine hydrochloride hydrate, dithiothreitol (DTT), iodoacetamide, KCl, potassium phosphate, KH2PO4, ammonium bicarbonate, formic acid, and the mass calibrant erythromycin were purchased from Sigma. The HPLC grade acetonitrile was from Rathburn (Walkerburn, Scotland). Trypsin gold mass spectrometry grade was purchased from Promega (Madison, WI). Homemade stagetips (11) were made from 47-mm Empore C18 extraction discs (3M, Minneapolis, MN). ITRAQ kits were purchased from AB Sciex (Foster City, CA). The water used in the experiments was purified using a MilliQ apparatus (Millipore, Billerica, MA).
Cartilage Sample Preparation
Dissection, Pulverization, and Extraction
Samples, 4 males and one female with no previous disease history, were obtained from forensic medicine cases at the University of Oslo and approved by the local ethical committee. All samples were from ages within a range of 36–50 years. Approximately 1 × 1-cm cartilage pieces were dissected from each tissue. The macroscopically normal articular cartilage samples were taken to full-depth. The cartilage samples were taken from the following locations. Costal cartilage from rib 5 at the cartilage to bone transition; tracheal cartilage from the third tracheal ring over the bifurcation; meniscus from the medial part proximal to the articular surface of the medial tibial condyle; intervertebral disc with a wedge taken frontally from the disc between Th12 and L1 and then separating nucleus pulposus and annulus fibrosus leaving a significant part of the interface between the two structures; humerus and femoral head cartilage were isolated with a tangential section of the top of the joint and the full cartilage thickness sample taken perpendicularly at the center of the joint surface; knee cartilage from the medial tibial condyle were taken perpendicularly to the cartilage surface and representing the full thickness of the tissue.
The following tissues from each five individuals were extracted: articular cartilage (femoral head (F), humeral head (H), and knee medial tibial condyle (K)), intervertebral disc (annulus fibrosus (AF) and nucleus pulposus (NP)) and finally meniscus (M). Tracheal (T) and rib (R) cartilage samples (n = 6) were obtained from different individuals (24–36 years) than those described above. Clean dissected, frozen cartilages were pulverized in liquid nitrogen in a chilled metal tube with a mixer mill type 301 (Retsch, Haan, Germany). Visual inspections of particles formed were made in a light microscope to optimize settings (3 × 60 s at 25 Hz) with 30 min cooling between steps, resulting in a homogeneous powder. About 100 mg of powder was prepared per tissue. The frozen samples were weighed and extracted using 15 volumes (v/w) of chaotropic extraction buffer (4 m GdnHCl, 50 mm NaAc, 100 mm 6-aminocaproic acid, 5 mm benzamidine, 5 mm N-ethylmaleimide, pH 5.8) for 24 h on an orbital shaker at +4 °C. Extracts were collected after centrifugation at 13,200 × g and +4 °C for 30 min. The pellet (extraction residue) was further processed as described below.
Quantitative Sample Preparation
A volume of 200 μl of extract was transferred into a 2-ml tube reduced by 4 mm DTT (at +56 °C for 30 min shaking) and alkylated by 16 mm iodoacetamide (room temperature for 1 h in the dark). The extracts were precipitated with ethanol (9:1) overnight at +4 °C before centrifugation (13,200 × g at +4 °C for 30 min) followed by ethanol wash for 4 h at −20 °C to remove residual GdnHCl and other salts. Samples were dried in a SpeedVac and suspended in 100 μl of 0.1 m triethylammonium bicarbonate, pH 8.5, before trypsination with 2 μg of trypsin gold at +37 °C on a shaker for about 16 h. An aliquot of 50 μl was dried and used for the iTRAQ labeling.
The isobaric 4-plex iTRAQ label was used to enable mixing of up to 3 samples and a reference sample per sample set. The reference sample, 117, was a mixture of cartilage extracts originating from femoral head, rib, meniscus, and nucleus pulposus to have as many proteins as possible represented in the relative quantification. All samples were compared with this reference sample and ratios between sample and reference were calculated. The labeling step was carried out as follows: samples were redissolved in 30 μl of 0.5 m triethylammonium bicarbonate, pH 8.5. The iTRAQ reagents were brought to room temperature, redissolved in 70 μl of ethanol, vortexed, and centrifuged immediately before addition to the samples. Incubation was performed for 1 h at room temperature. The reaction was stopped by the addition of 100 μl of water. To check the labeling efficiency an aliquot (0.5%) of the labeled sample was injected onto the iontrap LC-MS. The generated data were checked for incomplete labeling using variable iTRAQ modification in the database search. Labeled digests were dried in a SpeedVac and washed twice with 500 μl of 50% acetonitrile in 0.1% formic acid for salt removal and then dried again. Labeled samples were redissolved in 100 μl of 2% formic acid. The samples were mixed in a 1:1:1 ratio together with reference sample and diluted up to 500 μl with cation exchange starting buffer before injection onto the ion exchange column. Because all analyses used the same reference, all sample data could be compared by normalization against this. We selected the articular cartilages (femoral head, humeral head, and knee medial tibial condyle) as one mixture (FHK) to analyze because they were expected to be most similar. The other mixture (MAN) was meniscus together with the disc structures annulus fibrosus and nucleus pulposus, whereas the last mixture was rib and trachea in various combinations. All sample sets were treated separately for the database search before combining them into one dataset. Only the average ratio (weighted) of all peptide quantities for each protein is presented.
Sample Preparation (Extraction Residue)
Extraction residues were washed in 4 steps each with 15 volumes of 4 m GdnHCl buffer first for 24 h, the second for overnight, the third for 2 h, and finally for 1 h. All washes were performed on a shaker at +4 °C. Samples were centrifuged between all washing steps to remove the supernatant. Pellets were re-suspended in 15 volumes of 4 m GdnHCl buffer and reduced with 10 mm DTT at +56 °C for 1 h, alkylated with 50 mm iodoacetamide at room temperature in the dark for 1 h, followed by ethanol precipitation as described above. Dried samples were re-suspended in 500 μl of 0.1 m triethylammonium bicarbonate, pH 8.5, and digested by trypsin, 25 μg were added in two steps, at +37 °C for 16 + 5 h on a shaker. Digests were dried and re-dissolved in 1500 μl of 0.1 m triethylammonium bicarbonate. An aliquot of the 100-μl sample was used for iTRAQ labeling, i.e. the same amount of sample, based on original tissue, as for the main guanidine extract.
Off-line Cat-Ion Exchange Chromatography
Initial ion exchange separations were performed on a micro-LC system (SMART, Pharmacia, Uppsala Sweden) using a strong cation exchange column (2.1 mm inner diameter × 100 mm, 5-μm polysulfoethyl aspartamide, pore size 300 Å, PolyLC, Columbia, MD). Elution was isocratic for 13 min with 99.5% solvent A (10 mm potassium phosphate, 20% acetonitrile, pH 2.8) and 0.5% solvent B (1 m KCl, 10 mm potassium phosphate, 20% acetonitrile, pH 2.8) followed by a 60-min linear gradient of increasing salt concentration to 130 mm. Subsequently the gradient increased over 15 min up to 385 mm KCl and finally for 6 min up to 1 m salt. Fractions of 300–500 μl were collected in 0.5-ml Eppendorf tubes (SafelockTM, Eppendorf AG, Hamburg, Germany).
Mass Spectrometry
Fractions from the off-line strong cation exchange were dried, redissolved in 60 μl of 0.2% formic acid, whereof 10 μl were purified and concentrated using homemade reversed phase tips, 4 discs thick (11, 12). Retained peptides were eluted using 10 μl of 50% acetonitrile in 0.1% formic acid into autosampler glass vials (Qsert, Waters). The organic solvent was evaporated using a SpeedVac and peptides were redissolved in 10 μl of 0.2% formic acid before injection onto the various LC-MS systems.
Iontrap LC-MS
The HCT iontrap MS (Bruker Daltonics, Bremen, Germany) was equipped with an Ultimate HPLC system (LC-Packings, Amsterdam, Netherlands) with a PepmapTM nano-precolumn (LC Packings, C18, 300 μm inner diameter and 5-mm long) and an AtlantisTM analytical column (Waters, C18, 3 μm particles, 150 μm inner diameter × 150-mm long). The analytical column was coupled to the MS instrument through a microflow nebulizer. The on-line reversed-phase separation was performed using a flow rate of about 1 μl/min and a linear gradient from 5% B (A = 3% acetonitrile in 0.1% formic acid and B = 80% acetonitrile in 0.1% formic acid) to 55% B in 50 min, followed by a wash for 3 min with 95% B, and reconditioning to initial conditions in 10 min. To avoid cross-contamination a blank run for 30 min was inserted between each sample fraction. The instrument was operating using data-dependent acquisition by selecting the three most intense ions, as long as they are multiply charged, for MS-MS. The equipment was controlled by HyStarTM software (Bruker Daltonics) and spectra generated were processed using DataAnalysisTM (Bruker Daltonics), and database (SwissProt 56.9) searches were performed using MASCOT (version 2.1) MS/MS Ions Search.
Database Searching
The protocol used in this study was optimized to study peptides lacking major post-translational modifications such as glycosylations, phosphorylations, and sulfation. However, due to high abundance of collagens in these samples, hydroxylation of proline residues were allowed in our database searches. MASCOT search parameters included: carbamidomethylation of cysteine as fixed modification, deamidation (Asn and Gln), and oxidation (Met and Pro) were considered as variable modifications. Other MASCOT search parameters were: monoisotopic masses, ±0.4Da peptide mass tolerance, ±0.4Da fragment mass tolerance, max miss cleavage of 2, ion score minimum 20, only highest ranked peptide matches, and taxonomy Homo sapiens.
Q-TOF LC-MS
The Q-TOF MS (Q-TOF Micro, Waters) was connected to a CapLC (Waters) with the same type of precolumn as described above and a Waters Symmetry, C18, 3.5-μm particles, 150 mm long × 75 μm inner diameter analytical column. The analytical column was coupled to a PicoTipTM needle (New Objective, Woburn, MA). The on-line reversed-phase separation was performed using a flow rate of about 250 nl/min and a linear gradient from 5% B (mobile phases as for the iontrap LC-MS) to 51% B in 60 min, followed by a wash for 6 min with 95% B, and reconditioning to initial conditions in 12 min. Blank runs were run as before to minimize cross-contamination between fractions. The instrument was operating using data-dependent acquisition by selecting the four most intense ions as long, as they are multiply charged, for MS-MS during 5 s.
Database Searching
The mass spectrometric raw data were processed using Protein Lynx 2.1 (Waters) and nano-lockspray calibration using the erythromycin peak at 716.45 Da as calibrant mass. The processed files were searched using MASCOT with the following search parameters: iTRAQ (N-term), iTRAQ (Lys) and carbamidomethylation (Cys) as fixed modifications. Mannosylation (Trp), iTRAQ (Tyr), deamidation (Asn and Gln), and oxidation (Met and Pro) were considered variable modifications. Other MASCOT search parameters were: monoisotopic masses, ±0.2 Da peptide mass tolerance, ±0.2 Da MS-MS fragment mass tolerance, max miss cleavage of 2, ion score of interest >20, only highest ranked peptide matches and taxonomy Homo sapiens.
Data Analysis
iTRAQ quantification parameters were: significant threshold p < 0.05, weighted ratios, no normalization, minimum number of peptides of 1, minimum precursor charges of 2, at least homology of 0.05, and software correction factors for each reporter were included. The individual sample lists were manually inspected and a few noncollagenous proteins that were identified based on hydroxylation of proline were removed. A final table was made with all tissue samples containing average ratios of all proteins (relative to the reference sample). This table, containing 340 identified proteins (supplemental Table S1), was further investigated by searching relevant information on the protein hits in the UniProt knowledge database. Proteins known to represent plasma, e.g. produced in liver, as well as intracellular, were excluded to obtain a filtrated list particularly relevant to the extracellular matrix. Thus, the data filtration with omitting proteins was based on the following criteria: (i) intracellular proteins, (ii) plasma proteins, and (iii) additional proteins only quantified in <50% of all tissues (see supplemental Table 2). The resulting filtrated list of 92 proteins is presented in Table 2.
TABLE 2.
Filtered data on relative quantitative proteomics data
Average ratios versus the reference sample are listed, n = 6 for rib and tracheal cartilages, whereas n = 5 for the other tissues. Accession numbers are from the SwissProt database. The complete list is available in supplemental Table S1. Single peptide quantifications are shown in italics.
| Protein name | Accession No. | Femur | Humerus | Tibia | Meniscus | AFa | NPb | Rib | Trachea |
|---|---|---|---|---|---|---|---|---|---|
| Aggrecan core protein | P16112 | 1.83 | 1.20 | 1.34 | 0.13 | 1.22 | 1.03 | 1.30 | 1.14 |
| α1-Antichymotrypsin | P01011 | 0.47 | 0.96 | 0.95 | 1.87 | 1.07 | 3.42 | 0.57 | 0.48 |
| α1-Antitrypsin | P01009 | 0.41 | 0.81 | 0.81 | 1.55 | 0.99 | 2.25 | 1.41 | 0.67 |
| Angiogenin | P03950 | 2.69 | 2.98 | 2.26 | 0.40 | 3.65 | 3.39 | 2.96 | 2.21 |
| Angiopoietin-related protein 2 | Q9UKU9 | 1.93 | 0.77 | 0.95 | 0.31 | 1.47 | 0.80 | 0.52 | 0.81 |
| Angiopoietin-related protein 7 | O43827 | NDc | ND | ND | 1.38 | 1.11 | 0.64 | 2.61 | 0.84 |
| Apolipoprotein A-I | P02647 | 0.76 | 2.06 | 1.93 | 2.60 | 0.60 | 1.33 | 0.57 | 1.36 |
| Apolipoprotein D | P05090 | ND | ND | ND | 0.62 | 2.21 | 6.99 | 3.53 | 1.89 |
| Asporin | Q9BXN1 | 0.15 | 0.24 | 0.54 | 3.08 | 0.36 | 0.38 | 0.13 | 0.16 |
| Augurin | Q9H1Z8 | 1.19 | 1.57 | 1.31 | 0.17 | 0.45 | 0.12 | 1.52 | 0.41 |
| Basement membrane-specific HS proteoglycan (Perlecan) | P98160 | 1.09 | 1.30 | 0.97 | 0.78 | 1.07 | 0.44 | 1.39 | 1.94 |
| Biglycan | P21810 | 1.21 | 1.75 | 1.14 | 0.73 | 0.52 | 0.33 | 1.44 | 1.29 |
| Cartilage intermediate layer protein 1 | O75339 | 1.16 | 0.38 | 0.68 | 0.39 | 1.20 | 0.70 | 0.55 | 0.13 |
| Cartilage intermediate layer protein 2 | Q8IUL8 | 1.86 | 1.52 | 1.69 | 0.84 | 1.27 | 1.13 | 0.29 | 0.26 |
| Cartilage matrix protein | P21941 | 0.10 | 0.14 | 0.12 | 0.08 | 0.10 | 0.07 | 4.29 | 5.48 |
| Cartilage oligomeric matrix protein | P49747 | 1.70 | 1.22 | 2.02 | 1.40 | 1.49 | 1.23 | 0.26 | 0.08 |
| C-C motif chemokine 21 | O00585 | 0.26 | 0.28 | 0.25 | ND | ND | ND | 2.83 | 2.29 |
| Chondroadherin | O15335 | 1.27 | 1.24 | 0.58 | 0.12 | 1.14 | 1.31 | 1.95 | 1.85 |
| Chondroitin sulfate proteoglycan 4 | Q6UVK1 | ND | ND | ND | ND | ND | ND | 1.69 | 1.65 |
| Chondromodulin-1 | O75829 | 0.29 | 0.12 | 0.09 | ND | ND | ND | 2.47 | 2.82 |
| Clusterin | P10909 | 1.59 | 0.81 | 1.29 | 0.93 | 1.67 | 1.85 | 0.63 | 0.49 |
| Coiled-coil domain-containing protein 80 | Q76M96 | 0.52 | 0.52 | 0.35 | 0.10 | 1.49 | 1.78 | 1.80 | 0.76 |
| Collagen α-1(I) | P02452 | 1.21 | 2.34 | 1.16 | 1.83 | 1.14 | 0.67 | 1.10 | 1.06 |
| Collagen α-1(II) | P02458 | 2.04 | 3.92 | 1.67 | 0.15 | 1.01 | 0.82 | 1.57 | 1.46 |
| Collagen α-1(III) | P02461 | 1.06 | 2.20 | 2.17 | 2.30 | 0.82 | 0.85 | 0.31 | 0.32 |
| Collagen α-1(IX) | P20849 | 3.96 | 6.76 | 4.21 | ND | ND | ND | 3.86 | 3.12 |
| Collagen α-1(V) | P20908 | 1.09 | 2.11 | 1.53 | 2.44 | 1.69 | 0.37 | ND | ND |
| Collagen α-1(VI) | P12109 | 0.47 | 0.98 | 0.88 | 2.15 | 0.62 | 0.49 | 0.49 | 0.94 |
| Collagen α-1(X) | Q03692 | 0.22 | 0.20 | 0.08 | 0.12 | 0.24 | 0.10 | 3.78 | 1.19 |
| Collagen α-1(XI) | P12107 | 0.88 | 1.76 | 1.14 | 1.55 | 1.58 | 1.03 | 1.14 | 1.47 |
| Collagen α-2(I) | P08123 | 0.24 | 0.49 | 0.46 | 2.47 | 1.32 | 0.55 | 0.40 | 0.69 |
| Collagen α-2(IX) | Q14055 | ND | ND | ND | 0.16 | 0.41 | 0.58 | 2.57 | 3.60 |
| Collagen α-2(V) | P05997 | 1.35 | 1.88 | 1.64 | 0.36 | 0.72 | 0.60 | 1.93 | 1.13 |
| Collagen α-2(VI) | P12110 | 0.56 | 1.19 | 1.07 | 2.17 | 0.72 | 0.55 | 0.60 | 1.10 |
| Collagen α-2(XI) | P13942 | 0.88 | 1.31 | 0.76 | 2.13 | 2.29 | 1.12 | 1.18 | 1.75 |
| Collagen α-3(VI) | P12111 | 0.48 | 0.97 | 0.82 | 2.17 | 0.67 | 0.51 | 0.55 | 1.06 |
| C-type lectin domain family 11A | Q9Y240 | 1.02 | 1.55 | 1.03 | 0.08 | 0.71 | 0.47 | 0.77 | 0.83 |
| C-type lectin domain family 3A | O75596 | 0.87 | 1.60 | 0.52 | 0.14 | 0.77 | 0.56 | 1.62 | 1.48 |
| C-X-C motif chemokine 14 | O95715 | ND | ND | ND | ND | ND | ND | 1.91 | 1.84 |
| Decorin | P07585 | 0.85 | 0.82 | 1.24 | 1.56 | 1.18 | 0.55 | 0.40 | 0.15 |
| Dermatopontin | Q07507 | ND | ND | ND | 2.30 | 1.00 | 1.51 | ND | ND |
| Ectonucleotide pyrophosphatase/phosphodiesterase 1 | P22413 | ND | ND | ND | ND | ND | ND | 3.15 | 4.04 |
| EGF-like repeat discoidin I-like domain-cont. protein 3 | O43854 | 0.90 | 0.73 | 0.55 | 0.35 | 1.24 | 0.93 | 1.01 | 0.38 |
| Epiphycan | Q99645 | ND | ND | ND | ND | ND | ND | 2.70 | 2.36 |
| Extracellular superoxide dismutase [Cu-Zn] | P08294 | 0.54 | 0.88 | 0.84 | 1.23 | 1.15 | 1.13 | 1.47 | 0.62 |
| FGF-binding protein 2 | Q9BYJ0 | 2.25 | 2.02 | 1.25 | 0.25 | 1.66 | 1.30 | 1.23 | 0.82 |
| Fibromodulin | Q06828 | 1.38 | 1.66 | 1.24 | 0.80 | 0.75 | 0.30 | 1.59 | 1.14 |
| Fibronectin | P02751 | 0.52 | 0.32 | 0.70 | 0.17 | 1.68 | 1.72 | 0.70 | 0.12 |
| Galectin-1 | P09382 | 0.74 | 1.42 | 1.45 | 2.15 | 0.61 | 0.37 | 1.23 | 2.12 |
| Galectin-3 | P17931 | ND | ND | ND | ND | ND | ND | 1.56 | 3.01 |
| Glia-derived nexin | P07093 | 0.29 | 0.22 | 0.14 | 0.29 | 1.72 | 2.17 | ND | ND |
| HHIP-like protein 2 | Q6UWX4 | 2.39 | 2.08 | 1.36 | 0.44 | 1.86 | 1.73 | ND | ND |
| Hyaluronan and proteoglycan link protein | P10915 | 2.28 | 1.26 | 0.93 | 0.06 | 0.53 | 0.33 | 1.72 | 1.45 |
| Immunoglobulin superfamily containing LRR protein | O14498 | ND | ND | ND | ND | ND | ND | 1.21 | 1.15 |
| Inter-α-trypsin inhibitor heavy chain H5-like protein | Q6UXX5 | 1.31 | 0.88 | 0.61 | ND | ND | ND | 1.52 | 5.70 |
| Lactadherin | Q08431 | 0.92 | 0.59 | 0.45 | 0.37 | 0.76 | 0.37 | 2.25 | 1.82 |
| Leukocyte cell-derived chemotaxin-2 | O14960 | 1.42 | 1.46 | 1.14 | 0.20 | 1.60 | 1.43 | 1.43 | 3.08 |
| Lubricin (Proteoglycan 4) | Q92954 | 0.51 | 0.33 | 0.36 | 0.32 | 1.89 | 4.59 | 0.08 | 0.09 |
| Lumican | P51884 | 0.32 | 0.51 | 1.01 | 1.47 | 1.61 | 1.30 | 0.18 | 0.12 |
| Lysozyme C | P61626 | 1.18 | 0.68 | 0.66 | 0.11 | 1.20 | 1.37 | 3.64 | 10.91 |
| Matrilin-3 | O15232 | 1.46 | 0.54 | 0.77 | 0.10 | 0.09 | 0.06 | 1.89 | 1.25 |
| Matrix Gla protein | P08493 | ND | ND | ND | ND | ND | ND | 1.22 | 2.25 |
| Microfibril-associated glycoprotein 4 | P55083 | ND | ND | ND | 1.53 | 1.83 | 1.01 | 0.95 | 1.55 |
| Mimecan | P20774 | 0.37 | 1.05 | 1.52 | 2.35 | 1.38 | 0.97 | 0.12 | 0.21 |
| Nidogen-2 | Q14112 | ND | ND | ND | ND | ND | ND | 1.65 | 4.45 |
| Osteoadherin | Q99983 | 1.62 | 1.53 | 1.67 | 0.36 | 1.62 | 0.97 | 1.62 | 0.55 |
| Phospholipase A2, membrane associated | P14555 | 6.82 | 2.61 | 4.97 | 0.14 | 1.14 | 0.98 | 1.20 | 1.63 |
| Pleckstrin homology domain-containing family A6 | Q9Y2H5 | 5.47 | 1.99 | 4.06 | 0.10 | 0.94 | 1.01 | 0.83 | 2.16 |
| Procollagen C-endopeptidase enhancer 1 | Q15113 | 1.09 | 2.00 | 1.16 | 4.02 | 1.56 | 2.15 | ND | ND |
| Procollagen C-endopeptidase enhancer 2 | Q9UKZ9 | 1.41 | 1.42 | 1.56 | 1.84 | 1.44 | 1.75 | 1.10 | 1.45 |
| PRELP | P51888 | 1.01 | 1.27 | 1.21 | 1.24 | 0.88 | 0.43 | 1.27 | 0.91 |
| Protein S100-A1 | P23297 | 3.56 | 3.70 | 2.55 | 1.11 | 0.75 | 1.18 | 5.99 | 4.53 |
| Protein S100-A9 | P06702 | 0.68 | 1.00 | 0.48 | 0.61 | 3.95 | 5.37 | 0.37 | 2.13 |
| Protein S100-B | P04271 | 1.59 | 2.31 | 1.15 | 0.16 | 0.27 | 0.33 | 2.85 | 4.36 |
| Retinoic acid receptor responder protein 2 | Q99969 | 2.69 | 2.79 | 2.06 | 0.99 | 1.10 | 0.75 | 1.40 | 1.81 |
| Secreted frizzled-related protein 3 | Q92765 | ND | ND | ND | 0.15 | 0.26 | 0.35 | 1.83 | 1.47 |
| Serine protease HTRA1 | Q92743 | 0.74 | 0.36 | 0.73 | 0.43 | 2.13 | 1.92 | 0.20 | 0.24 |
| SPARC | P09486 | 0.23 | 0.86 | 0.34 | 0.16 | 0.36 | 0.19 | 1.81 | 0.56 |
| SPARC-related modular calcium-binding protein 2 | Q9H3U7 | 2.14 | 0.87 | 1.08 | ND | ND | ND | ND | ND |
| Sushi repeat-containing protein SRPX2 | O60687 | 0.70 | 1.25 | 0.92 | 0.27 | 0.49 | 0.42 | 2.10 | 1.64 |
| Target of Nesh-SH3 | Q7Z7G0 | 0.15 | 0.24 | 0.33 | 0.97 | 2.00 | 2.55 | 0.06 | 0.10 |
| Tenascin | P24821 | 0.73 | 16.22 | 2.86 | 3.22 | 1.77 | 1.07 | ND | ND |
| Tenascin-X | P22105 | 0.48 | 1.10 | 0.84 | 2.88 | 0.45 | 0.42 | ND | ND |
| Tetranectin | P05452 | 1.19 | 2.41 | 1.81 | 2.40 | 0.22 | 0.10 | 0.76 | 2.29 |
| TSP type-1 domain-containing protein 4 | Q6ZMP0 | ND | ND | ND | ND | ND | ND | 13.50 | 23.96 |
| Thrombospondin-1 | P07996 | 0.99 | 1.10 | 0.98 | 1.96 | 0.92 | 0.34 | 1.11 | 0.96 |
| Thrombospondin-3 | P49476 | 1.01 | 0.96 | 1.29 | 1.30 | 1.18 | 0.82 | 0.39 | 0.10 |
| TGF-β-induced protein ig-h3 | Q15582 | 0.52 | 2.84 | 1.39 | 2.49 | 1.22 | 1.20 | 1.13 | 1.38 |
| TNF receptor superfamily member 11B | O00300 | 0.14 | 0.09 | 0.03 | 0.21 | 2.38 | 1.80 | ND | ND |
| Versican | P13611 | 0.24 | 0.20 | 0.32 | 1.49 | 2.27 | 3.01 | 0.30 | 0.31 |
| Vitrin | Q6UXI7 | 3.22 | 11.35 | 1.77 | ND | ND | ND | ND | ND |
| Vitronectin | P04004 | 1.05 | 1.55 | 2.19 | 4.07 | 0.69 | 0.89 | 0.73 | 1.41 |
a AF, annulus fibros.
b NP, nucleus pulposus.
c ND, not determined.
Western Blot Analysis
Western blot was used on cartilage extracts from one individual to provide information on the quality of the protein and corroborate the quantitative data obtained by mass spectrometry. Aliquots (5 μl) of cartilage extracts were ethanol precipitated and then separated by gradient 8–16% SDS-polyacrylamide gel electrophoresis under reducing conditions at 70 V for 4 h. Proteins were transferred electrophoretically to nitrocellulose membranes (Amersham Biosciences) blocked with 1% bovine serum albumin (Serva electrophoresis, Germany) for asporin and with 5% fat-free milk in 10 mm Tris, 0.15 m NaCl, 0.2% Tween, pH 7.4, for chondroadherin. Blots were incubated for 1 h at room temperature with primary antibodies (1:1000) from our laboratory raised in rabbits against chondroadherin and asporin, respectively. This was followed by incubation for 1 h at room temperature with the secondary antibodies: polyclonal swine anti-rabbit HRP conjugate (DAKO A/S Glostrup, Denmark) at 1:1000 for chondroadherin and due to background problems an AffiniPure donkey anti-rabbit IgG peroxidase conjugate (Jackson Immunoresearch, West Grove, PA) used at 1:5000 dilution for asporin. Protein bands were visualized using the ECL chemiluminescence system (GE Healthcare).
Multivariate Analysis
The filtrated list of 92 proteins in Table 2 was used for multivariate analysis. The analysis was performed at the Lund University SCIBLU genomics center (Dr. Srinivas Veerla). Unsupervised hierarchical clustering analysis using Pearson correlation (×500 randomized data) as the distance matrix was performed (supplemental Fig. 1). t test analyses were performed to detect significantly different protein expressions in separate “pairwise” tissue comparisons (p ≤ 0.05) (supplemental Table S3).
RESULTS
Optimization of Procedures
As mentioned above the strongly anionic macromolecules mainly represented by aggrecan, and the highly abundant collagen, make the protein extraction and identification of other proteins challenging. In the non-gel approach where the separations are performed at the peptide level, the anionic pool of modified peptides (and protein fragments) should not interact with the strong cation-exchange column in the first dimension of separation. Indeed in pilot experiments,3 comparing the normal extract with extract depleted in aggrecan and larger glycosaminoglycan-containing aggrecan fragments using CsCl gradient ultracentrifugation, neither showed any significant differences in the number of proteins identified nor the appearance of cationic separation. Therefore the ultracentrifugation step was omitted, enabling a less complex sample protocol compatible with very small amounts of tissue samples.
Initially, we tried a pre-extraction with phosphate-buffered saline to wash out noncartilage proteins such as plasma proteins from the tissue samples. However, this buffer wash also contained typical ECM proteins as in GdnHCl extracts resulting in an apparent risk for loss of relevant sample components. Therefore, this step was omitted. Direct tissue extraction by trypsin digestion yielded lower numbers of identified proteins. In the initial experiments, we compared guanidine extraction for 24 and 48 h and found no significant quantitative changes (average top 40 proteins, p = 0.05) by doubling the extraction time. Thus, 24 h was used throughout this work.
Method Performance
The generated data are based on the weighted average ratio for each protein and the number of measurements vary from n = 1 (in the few instances listed in italics) to several hundreds. There is only one set of runs for each sample set because the instrumental variation was found to be small with coefficients of variation (CVs) in the range of <10%. One trypsin-digested sample was diluted at three levels 1:2:5, and we obtained a measured ratio of 1:1.82(±0.05):5.21(±0.17) as an average for the top 25 proteins in the identification list and the relative S.D. was <10% (p = 0.05). The errors increase with a lower number of peptides used for quantification as well as for larger differences with ratios far from 1. We also estimated the experimental error for the entire procedure by preparing 4 equal samples from extract to iTRAQ quantification using the same articular cartilage powder (femoral head) and the observed ratios with one sample as reference were calculated (p = 0.05) to: 1:0.97(±0.02):1.02(±0.04):0.98(±0.02) as the average for the top 100 proteins. The biological variation is notably much larger.
Qualitative Proteomics
We investigated the feasibility of a quantitative proteomics study by performing the preliminary screening of eight cartilage subtypes using a qualitative proteomics approach including off-line two-dimensional LC-MS in combination with an Ion Trap mass spectrometer. This set up is faster than the Q-TOF MS allowing more MS-MS experiments to be performed but is lacking the capability of quantification, obtained by the iTRAQ technology, due to its low mass cut-off limit. This qualitative proteomics approach identified 121–200 proteins in each tissue extract and ∼2000–4000 peptides (see Table 1). All tested tissues included typical proteins of the extracellular matrix but significant amounts of plasma originating proteins were detected in some samples. However, the typical extracellular cartilage proteins were dominating the protein lists. Collagens were not overwhelming, in line with a poor extractability due to cross-linking. If a single tissue had created particular problems, these would have been identified before pursuing mixing the selected tissue samples. For data comparisons between various tissues, Proteincenter (Proxeon A/S, Odense, Denmark) software was used. This software enabled us to make simple comparisons, e.g. what proteins are present in rib or tracheal cartilages although not present in articular cartilage. The matching proteins were matrilin 1, type X collagen, and epiphycan, being unique to rib and tracheal cartilages. This type of comparison (n = 1) illustrated the feasibility of performing quantitative tissue comparisons using relative quantifications with iTRAQ labeling.
TABLE 1.
Screening of cartilage subtypes using qualitative proteomics for 8 tissue samples
Data were merged from 28 LC-MS runs in off-line two-dimensional LC-MS (iontrap).
| Tissue extractions | No. proteins/peptides identified |
|---|---|
| Femoral head | 123/2959 |
| Humeral head | 197/3345 |
| Tibial plateu (knee) | 182/2806 |
| Meniscus | 198/2705 |
| Disc annulus fibrosus | 120/2507 |
| Disc nucleus pulposus | 141/4158 |
| Rib | 160/2421 |
| Trachea | 164/854 |
Quantitative Proteomics
All tissues tested in the qualitative proteomics experiment were selected for quantitative comparison. The sample preparation for quantitative proteomics included dissection, pulverization, extraction, reduction, alkylation, precipitation, and digestion by trypsin into peptide mixtures and their derivatization by a given iTRAQ reagent for each tissue and individual. All samples were weighed after pulverization and this wet weight was the reference for subsequent calculations. The multiplex strategy involved 3 tissue extract peptide preparations being mixed together with a constant reference sample of a representative cartilage tissue mixture treated in the same way. The isobaric labels react to give the same mass adduct to all peptides, regardless of their origin. All peptides of a given identity will have identical properties in chromatography and mass screening. However, upon fragmentation in MS-MS the individual peptide origin will be revealed from the release of a reporter ion, a fragment from the iTRAQ label, and quantification relative to the reference sample peptide signal can be performed, see also Fig. 1. In this example the MS-MS experiment identified an asporin peptide and the relative signal clearly shows that the level of this peptide is far greater in meniscus compared with the intervertebral disc structures (see also protein levels in Fig. 4a). The resulting protein list, including the weighted average protein ratio (versus reference sample) for each individual sample, is presented in supplemental Table S1 (totally 340 identified proteins). To simplify the analysis a biased list of 92 proteins considered to be particularly relevant for the extracellular matrix was generated (Table 2). In this attempt to filter the data, plasma and intracellular proteins were omitted. The data obtained in this work were further analyzed using two approaches. The relative presence of individual proteins between tissues was determined or all proteins in all tissues were investigated using multivariate analysis (similar to the established approach for analysis of DNA microarray data).
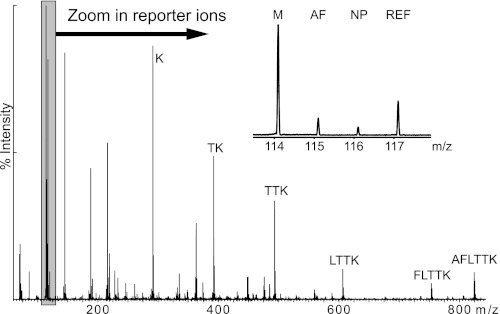
FIGURE 1.
MS-MS spectrum of the peptide ion m/z = 484.74Da (2+) illustrating the multiplex method. Samples from the meniscus, M (label 114), annulus fibrosus, AF (115), and nucleus pulposus, NP (116) were mixed with a reference sample (117) and the individual low mass reporter signals (114–117) are shown in the inset for the relative quantification. The other fragment ions are used for the identification using a database search in this case identifying the asporin peptide AFLTTK.
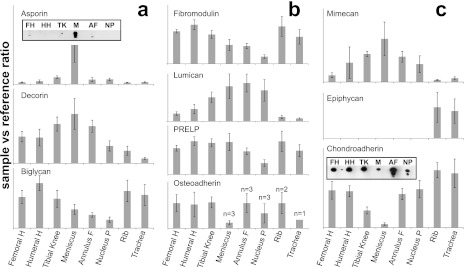
FIGURE 4.
Relative protein abundance pattern, in cartilaginous tissues, of SLRPs family. The SLRPs of class I are shown in a, class II in b, and class III and IV in c. Error bars are calculated using the confidence interval at p = 0.05 and n = 5 (n = 6 for rib and tracheal cartilages). The Western blot insets for asporin and chondroadherin show bands with intensities that correlate with the iTRAQ data.
Protein Extractability
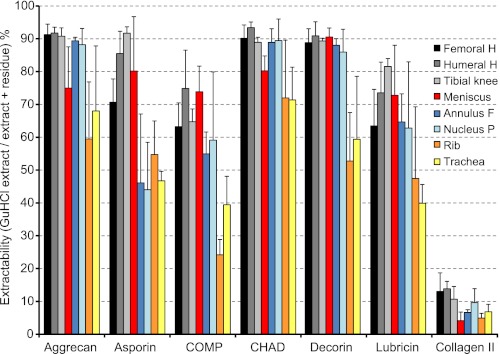
Most work on extraction yields of cartilage has been focused on aggrecan being efficiently extracted and collagen retained due to cross-linking. Interestingly with age an increasing proportion of matrilin-1 (cartilage matrix protein) in bovine tracheal cartilage resists such extraction possibly due to cross-linking (13). A small fraction of aggrecan that resists extraction from bovine nasal cartilage has also been reported (14). This un-extracted pool becomes larger if the tissue is not cut into sufficiently small pieces shown for articular cartilage (15). In view of potential and variable cross-linking and thus extractability we decided to quantify proteins in GdnHCl extraction residues from tissue samples. These were digested with trypsin after reduction and alkylation to release peptides that were not covalently bound. The iTRAQ labeling was performed for the peptides released from residues for relative quantification as described previously. All extraction residues showed predominant identifications of collagens (mainly I, II, and III). However, also certain noncollagenous matrix proteins such as members of the leucine-rich repeat protein family, cartilage oligomeric matrix protein, COMP (articular cartilage), and high levels of matrilin-1 (rib/tracheal cartilages) were found in the residues. It should be noted that there was still an insoluble residue even after trypsin digestion. We believe that this material primarily represents insoluble cross-linked collagen.
The data were used to calculate protein extractability, i.e. the amount that was extracted by 4 m GdnHCl (the extract) versus the sum of the extract plus the amount released upon trypsin digestion of the residue (residual extract). Most ECM proteins were extracted with yields of 80–100% from most tissues. However, the overall extractability was generally lower from rib and tracheal cartilages. The results for representative proteins are presented in Fig. 2, where aggrecan, chondroadherin (CHAD), and decorin show high yields in the extract, whereas asporin, lubricin, and COMP have lower yields from rib and tracheal cartilages as well as from the intervertebral disc structures. The level of collagen II extracted with GdnHCl was close to 10% of that extracted with enzyme treatment but the bulk of the protein is probably still present in the final residues because this triple helical collagen is quite insensitive to trypsin digestion. The collagen VI chains, which form beaded filamentous networks (16), are not covalently cross-linked and are consequently extracted efficiently more like the noncollagenous proteins (data not shown). An additional protein, fibrillin-1, was detected in the extraction residues only. This protein supposedly associated with matrix microfibrils that are suggested to be cross-linked (17–19), as corroborated by our findings. Notably, we found significant quantities of only one of the fibrillins.
FIGURE 2.
Extractability of selected proteins. Extractability is defined as the percentage recovered in extracts using 4 m GdnHCl compared with the sum of amounts in extract + residue, the latter released upon trypsin digestion of the extraction residue, error bars indicate the S.D. at p = 0.05. Selected tissues include femoral head, humeral head, tibial knee articular cartilages, meniscus, annulus fibrosus, nucleus pulposus, rib, and tracheal cartilages.
Individual Protein Distributions
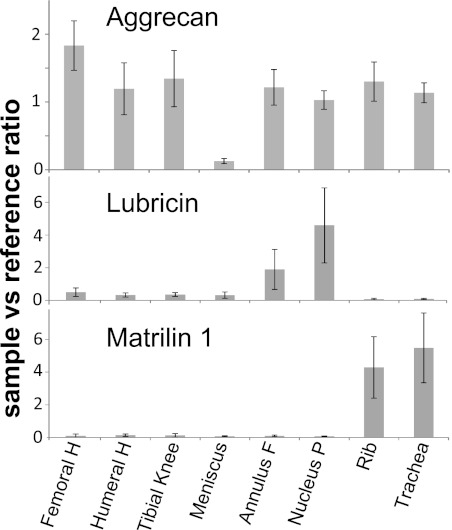
The distributions of individual proteins across various tissues were studied in detail and some of the more striking findings are presented in Fig. 3 where aggrecan, matrilin-1, and lubricin are presented. Overall the average levels of aggrecan were similar in tissues with the meniscus as a striking exception, showing pronouncedly lower amounts than present in all other tissues. Matrilin-1 is only detected in rib and tracheal cartilages, confirming previous findings where the protein was neither detected in articular cartilage nor in the intervertebral disc (20). In mice, mRNA expression was neither found in the intervertebral disc nor in the articular cartilage (21). Lubricin, also called superficial zone protein, is highly expressed in the superficial zone of the articular cartilage (22). We have measured the average level of lubricin protein in full-depth cartilage where a small contribution from the superficial parts would be diluted and partially obscured. We found markedly higher levels of lubricin in the intervertebral disc structures than in other cartilage subtypes, with the highest amount detected in the nucleus pulposus. We detected significant amounts of lubricin in the meniscus, whereas the levels in rib and tracheal cartilages were barely detectable. Lubricin has previously been reported to be present in the bovine meniscus (23).
FIGURE 3.
Average ratios of the individual proteins aggrecan, lubricin and matrilin-1 in various tissues. Tissues include articular cartilage from femoral head, humeral head, and tibial knee condyle as well as samples from meniscus, annulus fibrosus, nucleus pulposus, rib, and tracheal cartilages. Error bars are calculated using the confidence interval at p = 0.05 and n = 5 (n = 6 for rib and trachea).
The small leucine-rich repeat proteins (SLRPs) are commonly expressed in cartilage tissues. Protein levels of members of the four subfamilies are presented in Fig. 4, a (class I), b (class II), and c (class III and IV). In class I, the asporin content in articular cartilage seems to be the highest in the tibial condyle, but the level in the meniscus was even higher, i.e. 6- and 20-fold compared with medial tibial condyle and femoral head cartilage, respectively. The distribution between tissues of the related proteins decorin and asporin shows similarities with the highest levels in the meniscus of both (Fig. 4a). The biglycan distribution is quite different with higher levels in tissues where the other two members of the class I are lower and vice versa. Interestingly, as shown in Fig. 4b the relative ratios of fibromodulin and lumican differ between the tissues such that when one of the proteins is high the other is low. For the other class II group members in Fig. 4b, proline and arginine-rich end leucine-rich repeat protein (PRELP) showed a similar distribution as fibromodulin. Osteoadherin was detected in samples from articular cartilages but was variably present in the other tissues. From class III (Fig. 4c) mimecan and epiphycan presented completely different protein patterns. Mimecan is easily quantified in all cartilages except from the rib and trachea, where the levels are very low. Epiphycan is on the other hand only detected in rib and tracheal cartilages. The integrin and collagen binding CHAD, class IV, showed uniquely low levels in the meniscus compared with other tissues investigated in a kind of mirror image to asporin.
Validation of Proteomics Data with Corresponding Western Blots
The quantitative data from asporin and CHAD, two proteins with opposite patterns, uncovered using the described mass spectrometric approach was further evaluated by Western blots of a selected range of tissues (Fig. 4, a and c). The blot was performed on samples from one individual, whereas the MS data shown are an average of five samples. The asporin blot (Fig. 4a, inset), corroborating the MS data, detected a strong signal in meniscus, whereas only very weak bands were found in cartilage samples from femoral head, tibial knee, and annulus fibrosus. Also the blot indicated that the asporin in the meniscus was fragmented and that this may also be the case for the protein in the disc. The CHAD Western blot (Fig. 4c, inset) shows good correlation with the MS data, detecting the lowest amount in the meniscus. Additionally, the blot indicates that there are protein fragments primarily in the intervertebral disc.
Multivariate and Pairwise Tissue Comparisons
The multivariate analysis, based on the data in Table 2, demonstrated a sample clustering according to supplemental Fig. S1. The individual samples from the same tissue types were clustered together allowing them to be defined based on the data set. The meniscus tissue is the prime tissue to diverge and therefore it is most different from the other tissues. The disc structures were next to separate from the other tissues. The related structures of the annulus fibrosus and nucleus pulposus were somewhat intermingled, probably due to smaller differences in their protein patterns. The other cartilages were separated into three distinct groups, knee-hip, shoulder, and trachea-rib, respectively. Because overall clustering was promising it was reasonable to make pairwise comparisons of different tissues. An example is comparison of the individual samples from femoral head versus the medial tibial cartilage graphically presented in Fig. 5. Significant distinctions between the tissues are observed with high probability (p = 0.01). The details of pairwise comparisons are presented in supplemental Table S3, a–k and include femoral head as a representative of an articular cartilage being compared with all the other tissues except meniscus, which was compared with tibial condyle. The disc structures of the annulus fibrosus versus nucleus pulposus, the annulus fibrosus versus meniscus, as well as rib versus tracheal cartilage were also compared. There were significant differences (t test with p < 0.05) even between different articular cartilages such as femoral head versus tibial condyle (supplemental Table S3a) where a relative ratio above 2 includes proteins such as hyaluronan and proteoglycan link protein 1 (LINK) and CHAD, whereas proteins below a ratio of 0.5 include proteins such as lumican, asporin, and mimecan. By comparing femoral versus humeral head cartilages (supplemental Table S3b), the most significantly high ratio proteins were observed for cartilage intermediate layer protein 1 (CILP-1) (3.0) and matrilin-3 (2.7), whereas mimecan showed a low ratio (0.3). The comparison of humeral head versus tibial knee (supplemental Fig. S3c) revealed that these tissues were more similar in composition with mainly two proteins differing: CHAD (2.1) and asporin (0.4). In the comparison of articular cartilage to meniscus (supplemental Fig. S3d), we selected the knee tibial condyle as the representative because they represent structures of the same joint. The relative abundance of aggrecan, LINK, and extracted collagen II were more than 10-fold and CHAD was ∼5-fold higher in the knee tibial condyle. On the other hand both asporin and versican were around 5-fold higher in the meniscus. Additionally, apolipoprotein D was only found in meniscus, whereas SPARC-related modular calcium-binding protein 2 (SMOC-2) was present only in tibial condylar cartilage.
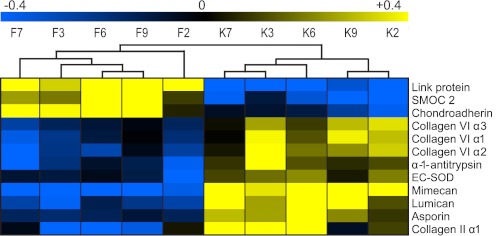
FIGURE 5.
Proteins significantly different (p = 0.01) in abundance between femoral head (F) and tibial knee (K) articular cartilage using paired t tests. High protein abundances are indicated in yellow, whereas low abundances are in blue. Samples with the same number, e.g. F7 and K7, correspond to the same individual.
The most striking differences in the other cartilages versus the intervertebral disc annulus fibrosus and nucleus pulposus (supplemental Table S3, e and f) were observed for matrilin-3 levels (some 20-fold higher in femoral head cartilage), whereas versican and the rather unknown “target of Nesh-SH3” protein were about 10-fold higher in the disc. The distribution patterns of these two latter proteins were very similar, possibly indicating some type of unknown correlation. In general the disc structures were very similar to each other with the exception of lubricin being 9-fold higher in nucleus pulposus, although less than 4-fold higher in the annulus fibrosus. However, because the articular cartilage was obtained at full thickness, we compared the average lubricin value in this case and not only the superficial layer. The significant differences between annulus fibrosus and nucleus pulposus (supplemental Table S3i) were showing a trend of more ECM proteins present in the annulus fibrosus compared with femoral head cartilage. However, lubricin had a tendency (p = 0.09) of being higher in the nucleus pulposus. The tensile loading in meniscus and annulus fibrosus with an ordered collagen network made it interesting to compare these tissues as well. The comparison is presented in supplemental Table S3j and proteins such as aggrecan, LINK, and CHAD were significantly more abundant (almost 10-fold) in annulus fibrosus, whereas asporin (0.12), collagen VI (0.3), and tenascin-X (0.16) were more abundant in the meniscus.
The femoral head cartilage was also compared with rib and tracheal cartilages (supplemental Tables S3, g and h) where higher levels were found for proteins such as COMP (6- and 22-fold compared with rib and tracheal cartilages, respectively) and CILP-2 (6- and 7-fold, respectively). In contrast chondromodulin was almost 10-fold higher in both rib and trachea. Matrilin-1 stood out as highly specific for both rib and tracheal cartilages with at least 50-fold higher levels detected. Another specific protein was epiphycan being detected in rib and tracheal cartilages only. The comparison of rib versus tracheal cartilages (supplemental Table S3k) showed small differences and only COMP (3-fold) and lysozyme C (0.33) were significantly different at p = 0.01.
DISCUSSION
The group of articular cartilages, meniscus, and intervertebral disc samples were from one set of individuals 36–50 years, whereas the tracheal and rib cartilages were from another set of individuals 24–36 years old. Although the overall age range represent mature cartilages prior to a major disease becoming evident, we made use of the large data set to investigate for trends in protein contents over age. We found no such trends in either group, whereas the differences between, e.g. the articular cartilages on one hand and the tracheal and rib cartilages, are very clear (data can be found in the supplemental data). Thus, these differences are not a result of the different ages.
The concentration of collagen (mainly type II) in cartilage is estimated to be in the order of 10–15% of the tissue wet weight, whereas that of proteoglycans is 5–10% (24). The major part of the collagen network is insoluble in 4 m GdnHCl due to cross-links and therefore is not extracted. The number of cross-links increase with age but should be high for the selected individuals in this study, 36–50 years. This group is below the age when major joint pathology occurs and should represent normal tissue. The same areas on the tissues of the various individuals were selected for dissection and analysis. It should be emphasized that most of our studies reflect proteins extracted using 4 m GdnHCl and not necessarily total protein content.
In studies of proteins extracted with strongly chaotropic agents we have demonstrated major differences in selected proteins between cartilaginous tissues. The overall data show that articular cartilages were rather similar although some distinct and significant differences were identified. Similarly, patterns of disc structures for the annulus fibrosus and nucleus pulposus were quite similar. The same was true for the rib and tracheal cartilages. However, the meniscus was rather different from all other tissues investigated with proteins like asporin being very high, whereas aggrecan was indeed low. We detected most of the proteins expected to be present in cartilage extracellular matrix. However, some low abundant proteins such as the fibulins were not detected. This could be due to the data-dependent approach where medium to high abundant proteins are favored over low abundant proteins. The highest signals are selected for MS-MS and therefore low abundant proteins are rarely selected. To address the low abundant proteins, targeted proteomics will become an alternative for low abundance proteins of interest.
The work presented herein also address the often overlooked, insoluble fraction after GdnHCl extraction. Surprisingly, several molecular entities were only partially extracted by 4 m GdnHCl despite indications of efficient extraction in general, in the form of almost other completely extracted proteins. Significant amounts could be recovered in the residue by protease digestion. These proteins include matrilin-1, COMP, and CILP. At the same time aggrecan was mainly present in the extract and collagens in the residue to verify that extraction was not generally poor but at the levels previously reported.
The data presented in this work are based on normal human tissue and will serve as an important reference material for future studies of pathological cartilage. Although not investigated in this study of normal tissues it has been shown that some proteins such as asporin, CILP, and COMP are elevated in pathological tissue in both early and late osteoarthritic cartilage (25). The D14 allele of asporin has been associated with an increased risk of osteoarthritis in Chinese (26), Japanese (27), and Korean populations (28). Additionally, the same D14 allele was found to be associated with lumbar disc degeneration in Chinese and Japanese populations (29). Recently, asporin has been reported to be more abundant in the more degenerate human discs and more frequently in the annulus fibrosus (30). The data in our study demonstrate the presence of asporin in all selected tissues but the levels are in general low except for the meniscus. Any specific functions within this tissue are largely unexplored. The increased levels of asporin in diseases such as osteoarthritis and disc degeneration might be a result of a cellular repair response within the affected tissue. The protein is suggested to interact with TGF-β and modulate its signaling pathway (31). However, decorin and biglycan also binds TGF-β (32, 33). The similar distribution pattern of asporin and decorin is especially interesting in view of the overlapping binding of collagen between the two (34). Biglycan shows a different distribution pattern with particularly high levels in the hyaline cartilages and lower levels in the intervertebral disc. Biglycan primarily binds to collagen type VI rather than the fibril forming collagens. The molecule therefore appears to have a different role despite the fact that decorin and biglycan both bind the same N-terminal site of collagen VI albeit affecting beaded filament formation differently (35).
Most SLRPs are known to interact with collagen (for references, see Kalamajski and Oldberg (3)). Fibromodulin and lumican, both class II members, show complementary distribution patterns. These proteins seem to compete for the same binding site in collagen type I although with different affinity (36, 37). Fibromodulin-deficient mice have weaker tendons with a larger proportion of thin and abnormally shaped collagen fibrils and show 4-fold increased levels of lumican protein (36), whereas lumican-deficient mice have reduced corneal transparency and skin fragility but an unaltered fibromodulin level (38). It has been suggested that lumican promotes formation of thinner collagen fibrils, whereas fibromodulin appears to be necessary for thick fibril formation (3). The fact that lumican protein concentration is increased in the fibromodulin null mouse despite a lower expression (mRNA level) implies that there is a sequence of events, such that lumican is expressed early to be replaced by fibromodulin and that this replacement induces fibril lateral growth. Whether this change resulted in thicker collagen fibrils observed in articular, rib, and tracheal cartilages compared with thinner fibrils in the meniscus and the intervertebral disc remains to be shown.
The similar distribution pattern of PRELP and fibromodulin is interesting in view of the demonstrated binding of the N-terminal tyrosine sulfate domain of fibromodulin to the basic cluster heparin-binding domain of PRELP. It is possible that this interaction can link the two molecules bound at the surface of collagen fibers and enhance tissue mechanical stability. Representing SLRPs class III, we detected epiphycan in rib and tracheal cartilages only. Epiphycan has previously been detected in epiphyseal cartilage (39, 40) and in the articular cartilage of young animals (41) but not in adults. Consequently, to our knowledge, this is the first time that epiphycan was detected in rib and tracheal cartilages. Its presence may reflect some chondrocyte hypertrophy and tissue mineralization often seen in these two tissues. CHAD distinguishes itself from other SLRPs (with the exception of osteoadherin) by binding cells mediated via the α2β1-integrin. In addition, it contains a heparin-binding domain but with different binding specificity than that of PRELP. It is interesting to speculate why the protein levels in the meniscus are uniquely low. It may relate to a low abundance of cells with a chondrocyte phenotype.
We found markedly higher levels of lubricin in the intervertebral disc structures compared with other cartilage subtypes, with the highest levels detected in the nucleus pulposus. Recently, the distribution of lubricin in the intervertebral disc was reported and positive immune histochemical staining of lubricin was shown in all parts of the intervertebral disc, with the highest abundance in the nucleus pulposus (42). The high levels of lubricin in the intervertebral disc could have a functional role as a lubricant to decrease friction during movement of the spine. Lubricin or proteoglycan 4, a mucin-like glycoprotein, was originally identified as a lubricating protein present in synovial fluid (43). Mutations result in severe joint dysfunction, but also in problems at other sites such as the pericardium (44). In the joint, the protein is produced by superficial chondrocytes, but appears not to be efficiently retained in cartilage (22). The protein is also produced in the synovial membrane (45). The lower levels found in articular cartilage could be explained by the fact that full thickness cartilage was analyzed obscuring the higher levels in the superficial zone layer. Recent studies using animal models of osteoarthritis have reported a protective effect against cartilage degeneration by intra-articular injections of lubricin (46) and the protein is considered promising as a putative therapeutic agent (47).
We have chosen not to show all the data in graphic form to focus on particularly prominent and interesting differences. However, all the data on the individual proteins detected are provided in supplemental Fig. S1 and Tables S1–S3.
Taken together the data provides novel insights into different structure-function relationships, where at the molecular level even articular cartilages can be divided into different groups, but with overall very similar compositions. The different compositions provide some insight into why disease shows tropism to different cartilages, where notable selectivity ranges from arthritis affecting articular cartilages while neither the other cartilages nor the intervertebral disc are engaged. On the other hand, relapsing polychondritis in its typical form does affect cartilage in trachea, nose, and ear, although does not affect the articular cartilage. The observation of differences between articular cartilages may reflect that knee and hip osteoarthritis primarily affect different individuals.
Future work to further our understanding needs to take into account differences in composition over the joint, exemplified by the medial and lateral compartments of the knee, which are differently affected by osteoarthritis. Also, a detailed analysis of variability in molecular composition with distance from the surface of the articular cartilage will provide important information that when coupled to an understanding of, e.g. mechanical properties, will increase our understanding of the biology of the joint and lay groundwork for comprehending its pathology. The technology presented here lays the groundwork for such studies and applying information to targeted approaches by mass spectrometry will allow more broad-reaching studies of the tissue detailed organization.
Supplementary Material
Acknowledgments
Dr. Kari Ormstad is gratefully acknowledged for scientific input and providing the tissue samples. The mass spectrometers were financed by the Inga-Britt and Arne Lundberg foundation.
This work was supported, in whole or in part, by National Institutes of Health Grant U01 AR050926 from the NIAMS, Swedish Research Council Grant 52X-05668, grants from the European Union FP7-ICT project NanoDiaRA under contract FP7-NMP-2008-LARGE-2, Lund University (to ALF and TIM), Alfred Österlund Foundation, Magnus Bergvall Foundation, King Gustaf V 80-year fund, the Crafoord Foundation, Anamar Medical, and the Greta and Johan Kock Foundation.

This article contains supplemental Fig. S1 and Tables S1–S3.
P. Önnerfjord, A. Khabut, F. P. Reinholt, O. Svensson, and D. Heinegård, unpublished data.
- ECM
- extracellular matrix
- iTRAQ
- isobaric tags for relative and absolute quantitation
- GdnHCl
- guanidine HCl
- SLRP
- small leucine-rich repeat protein
- PRELP
- proline and arginine-rich end leucine-rich repeat protein.
REFERENCES
- 1. Eyre D. R., Weis M. A., Wu J. J. (2006) Articular cartilage collagen. An irreplaceable framework? Eur. Cell Mater. 12, 57–63 [DOI] [PubMed] [Google Scholar]
- 2. Heinegård D. (2009) Proteoglycans and more. From molecules to biology. Int. J. Exp. Pathol. 90, 575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kalamajski S., Oldberg A. (2010) The role of small leucine-rich proteoglycans in collagen fibrillogenesis. Matrix Biol. 29, 248–253 [DOI] [PubMed] [Google Scholar]
- 4. Merline R., Schaefer R. M., Schaefer L. (2009) The matricellular functions of small leucine-rich proteoglycans (SLRPs). J. Cell Commun. Signal. 3, 323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hermansson M., Sawaji Y., Bolton M., Alexander S., Wallace A., Begum S., Wait R., Saklatvala J. (2004) Proteomic analysis of articular cartilage shows increased type II collagen synthesis in osteoarthritis and expression of inhibin betaA (activin A), a regulatory molecule for chondrocytes. J. Biol. Chem. 279, 43514–43521 [DOI] [PubMed] [Google Scholar]
- 6. Vincourt J. B., Lionneton F., Kratassiouk G., Guillemin F., Netter P., Mainard D., Magdalou J. (2006) Establishment of a reliable method for direct proteome characterization of human articular cartilage. Mol. Cell Proteomics 5, 1984–1995 [DOI] [PubMed] [Google Scholar]
- 7. Wu J., Liu W., Bemis A., Wang E., Qiu Y., Morris E. A., Flannery C. R., Yang Z. (2007) Comparative proteomic characterization of articular cartilage tissue from normal donors and patients with osteoarthritis. Arthritis Rheum. 56, 3675–3684 [DOI] [PubMed] [Google Scholar]
- 8. Belluoccio D., Wilson R., Thornton D. J., Wallis T. P., Gorman J. J., Bateman J. F. (2006) Proteomic analysis of mouse growth plate cartilage. Proteomics 6, 6549–6553 [DOI] [PubMed] [Google Scholar]
- 9. Wilson R., Belluoccio D., Bateman J. F. (2008) Proteomic analysis of cartilage proteins. Methods 45, 22–31 [DOI] [PubMed] [Google Scholar]
- 10. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 11. Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 12. Rappsilber J., Mann M., Ishihama Y. (2007) Protocol for micropurification, enrichment, prefractionation, and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 13. Paulsson M., Inerot S., Heinegård D. (1984) Variation in quantity and extractability of the 148-kDa cartilage protein with age. Biochem. J. 221, 623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Christner J. E., Baker J. R., Caterson B. (1983) Studies on the properties of the inextractable proteoglycans from bovine nasal cartilage. J. Biol. Chem. 258, 14335–14341 [PubMed] [Google Scholar]
- 15. Bayliss M. T., Venn M., Maroudas A., Ali S. Y. (1983) Structure of proteoglycans from different layers of human articular cartilage. Biochem. J. 209, 387–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wiberg C., Heinegård D., Wenglén C., Timpl R., Mörgelin M. (2002) Biglycan organizes collagen VI into hexagonal-like networks resembling tissue structures. J. Biol. Chem. 277, 49120–49126 [DOI] [PubMed] [Google Scholar]
- 17. Keene D. R., Jordan C. D., Reinhardt D. P., Ridgway C. C., Ono R. N., Corson G. M., Fairhurst M., Sussman M. D., Memoli V. A., Sakai L. Y. (1997) Fibrillin-1 in human cartilage. Developmental expression and formation of special banded fibers. J. Histochem. Cytochem. 45, 1069–1082 [DOI] [PubMed] [Google Scholar]
- 18. Sakai L. Y., Keene D. R., Engvall E. (1986) Fibrillin, a new 350-kDa glycoprotein, is a component of extracellular microfibrils. J. Cell Biol. 103, 2499–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu J., Urban J. P. (2010) The elastic network of articular cartilage. An immunohistochemical study of elastin fibers and microfibrils. J. Anat. 216, 533–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Paulsson M., Heinegård D. (1981) Purification and structural characterization of a cartilage matrix protein. Biochem. J. 197, 367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aszódi A., Hauser N., Studer D., Paulsson M., Hiripi L., Bösze Z. (1996) Cloning, sequencing, and expression analysis of mouse cartilage matrix protein cDNA. Eur. J. Biochem. 236, 970–977 [DOI] [PubMed] [Google Scholar]
- 22. Schumacher B. L., Block J. A., Schmid T. M., Aydelotte M. B., Kuettner K. E. (1994) A novel proteoglycan synthesized and secreted by chondrocytes of the superficial zone of articular cartilage. Arch. Biochem. Biophys. 311, 144–152 [DOI] [PubMed] [Google Scholar]
- 23. Schumacher B. L., Schmidt T. A., Voegtline M. S., Chen A. C., Sah R. L. (2005) Proteoglycan 4 (PRG4) synthesis and immunolocalization in bovine meniscus. J. Orthop. Res. 23, 562–568 [DOI] [PubMed] [Google Scholar]
- 24. Martel-Pelletier J., Boileau C., Pelletier J. P., Roughley P. J. (2008) Cartilage in normal and osteoarthritis conditions. Best Pract. Res. Clin. Rheumatol. 22, 351–384 [DOI] [PubMed] [Google Scholar]
- 25. Lorenzo P., Aspberg A., Onnerfjord P., Bayliss M. T., Neame P. J., Heinegard D. (2001) Identification and characterization of asporin. A novel member of the leucine-rich repeat protein family closely related to decorin and biglycan. J. Biol. Chem. 276, 12201–12211 [DOI] [PubMed] [Google Scholar]
- 26. Kizawa H., Kou I., Iida A., Sudo A., Miyamoto Y., Fukuda A., Mabuchi A., Kotani A., Kawakami A., Yamamoto S., Uchida A., Nakamura K., Notoya K., Nakamura Y., Ikegawa S. (2005) An aspartic acid repeat polymorphism in asporin inhibits chondrogenesis and increases susceptibility to osteoarthritis. Nat. Genet. 37, 138–144 [DOI] [PubMed] [Google Scholar]
- 27. Jiang Q., Shi D., Yi L., Ikegawa S., Wang Y., Nakamura T., Qiao D., Liu C., Dai J. (2006) Replication of the association of the aspartic acid repeat polymorphism in the asporin gene with knee-osteoarthritis susceptibility in Han Chinese. J. Hum. Genet. 51, 1068–1072 [DOI] [PubMed] [Google Scholar]
- 28. Song J. H., Lee H. S., Kim C. J., Cho Y. G., Park Y. G., Nam S. W., Lee J. Y., Park W. S. (2008) Aspartic acid repeat polymorphism of the asporin gene with susceptibility to osteoarthritis of the knee in a Korean population. Knee. 15, 191–195 [DOI] [PubMed] [Google Scholar]
- 29. Song Y. Q., Cheung K. M., Ho D. W., Poon S. C., Chiba K., Kawaguchi Y., Hirose Y., Alini M., Grad S., Yee A. F., Leong J. C., Luk K. D., Yip S. P., Karppinen J., Cheah K. S., Sham P., Ikegawa S., Chan D. (2008) Association of the asporin D14 allele with lumbar-disc degeneration in Asians. Am. J. Hum. Genet. 82, 744–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gruber H. E., Ingram J. A., Hoelscher G. L., Zinchenko N., Hanley E. N., Jr., Sun Y. (2009) Asporin, a susceptibility gene in osteoarthritis, is expressed at higher levels in the more degenerate human intervertebral disc. Arthritis Res. Ther. 11, R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kou I., Nakajima M., Ikegawa S. (2010) Binding characteristics of the osteoarthritis-associated protein asporin. J. Bone Miner Metab. 28, 395–402 [DOI] [PubMed] [Google Scholar]
- 32. Hildebrand A., Romarís M., Rasmussen L. M., Heinegård D., Twardzik D. R., Border W. A., Ruoslahti E. (1994) Interaction of the small interstitial proteoglycans biglycan, decorin, and fibromodulin with transforming growth factor β. Biochem. J. 302, 527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yamaguchi Y., Mann D. M., Ruoslahti E. (1990) Negative regulation of transforming growth factor-β by the proteoglycan decorin. Nature 346, 281–284 [DOI] [PubMed] [Google Scholar]
- 34. Kalamajski S., Aspberg A., Lindblom K., Heinegård D., Oldberg A. (2009) Asporin competes with decorin for collagen binding, binds calcium, and promotes osteoblast collagen mineralization. Biochem. J. 423, 53–59 [DOI] [PubMed] [Google Scholar]
- 35. Wiberg C., Hedbom E., Khairullina A., Lamandé S. R., Oldberg A., Timpl R., Mörgelin M., Heinegård D. (2001) Biglycan and decorin bind close to the N-terminal region of the collagen VI triple helix. J. Biol. Chem. 276, 18947–18952 [DOI] [PubMed] [Google Scholar]
- 36. Svensson L., Aszódi A., Reinholt F. P., Fässler R., Heinegård D., Oldberg A. (1999) Fibromodulin-null mice have abnormal collagen fibrils, tissue organization, and altered lumican deposition in tendon. J. Biol. Chem. 274, 9636–9647 [DOI] [PubMed] [Google Scholar]
- 37. Kalamajski S., Oldberg A. (2009) Homologous sequence in lumican and fibromodulin leucine-rich repeat 5–7 competes for collagen binding. J. Biol. Chem. 284, 534–539 [DOI] [PubMed] [Google Scholar]
- 38. Chakravarti S., Magnuson T., Lass J. H., Jepsen K. J., LaMantia C., Carroll H. (1998) Lumican regulates collagen fibril assembly. Skin fragility and corneal opacity in the absence of lumican. J. Cell Biol. 141, 1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Johnson H. J., Rosenberg L., Choi H. U., Garza S., Höök M., Neame P. J. (1997) Characterization of epiphycan, a small proteoglycan with a leucine-rich repeat core protein. J. Biol. Chem. 272, 18709–18717 [DOI] [PubMed] [Google Scholar]
- 40. Kurita K., Shinomura T., Ujita M., Zako M., Kida D., Iwata H., Kimata K. (1996) Occurrence of PG-Lb, a leucine-rich small chondroitin/dermatan sulfate proteoglycan in mammalian epiphyseal cartilage. Molecular cloning and sequence analysis of the mouse cDNA. Biochem. J. 318, 909–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nuka S., Zhou W., Henry S. P., Gendron C. M., Schultz J. B., Shinomura T., Johnson J., Wang Y., Keene D. R., Ramírez-Solis R., Behringer R. R., Young M. F., Höök M. (2010) Phenotypic characterization of epiphycan-deficient and epiphycan/biglycan double-deficient mice. Osteoarthritis Cartilage 18, 88–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shine K. M., Simson J. A., Spector M. (2009) Lubricin distribution in the human intervertebral disc. J. Bone Joint Surg. Am. 91, 2205–2212 [DOI] [PubMed] [Google Scholar]
- 43. Swann D. A., Slayter H. S., Silver F. H. (1981) The molecular structure of lubricating glycoprotein-I, the boundary lubricant for articular cartilage. J. Biol. Chem. 256, 5921–5925 [PubMed] [Google Scholar]
- 44. Marcelino J., Carpten J. D., Suwairi W. M., Gutierrez O. M., Schwartz S., Robbins C., Sood R., Makalowska I., Baxevanis A., Johnstone B., Laxer R. M., Zemel L., Kim C. A., Herd J. K., Ihle J., Williams C., Johnson M., Raman V., Alonso L. G., Brunoni D., Gerstein A., Papadopoulos N., Bahabri S. A., Trent J. M., Warman M. L. (1999) CACP, encoding a secreted proteoglycan, is mutated in camptodactyly arthropathy-coxa vara-pericarditis syndrome. Nat. Genet. 23, 319–322 [DOI] [PubMed] [Google Scholar]
- 45. Jay G. D., Britt D. E., Cha C. J. (2000) Lubricin is a product of megakaryocyte stimulating factor gene expression by human synovial fibroblasts. J. Rheumatol. 27, 594–600 [PubMed] [Google Scholar]
- 46. Flannery C. R., Zollner R., Corcoran C., Jones A. R., Root A., Rivera-Bermúdez M. A., Blanchet T., Gleghorn J. P., Bonassar L. J., Bendele A. M., Morris E. A., Glasson S. S. (2009) Prevention of cartilage degeneration in a rat model of osteoarthritis by intraarticular treatment with recombinant lubricin. Arthritis Rheum. 60, 840–847 [DOI] [PubMed] [Google Scholar]
- 47. Bao J. P., Chen W. P., Wu L. D. (2011) Lubricin, a novel potential biotherapeutic approaches for the treatment of osteoarthritis. Mol. Biol. Rep. 38, 2879–2885 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.