Background: Living cells utilize several mechanisms to commit suicide when stressed.
Results: We find that dying yeast cells utilize the V-ATPase to release toxic materials into the cytoplasm.
Conclusion: Stressed yeast cells regulate cell death in a manner similar to stressed neurons.
Significance: Yeast may be a powerful new model system for understanding disease-related necrosis-like cell death in humans.
Keywords: Calcineurin, Cell Death, Lysosomal Acidification, Necrosis (Necrotic Death), Yeast
Abstract
Stress in the endoplasmic reticulum caused by tunicamycin, dithiothreitol, and azole-class antifungal drugs can induce nonapoptotic cell death in yeasts that can be blocked by the action of calcineurin (Cn), a Ca2+-dependent serine/threonine protein phosphatase. To identify additional factors that regulate nonapoptotic cell death in yeast, a collection of gene knock-out mutants was screened for mutants exhibiting altered survival rates. The screen revealed an endocytic protein (Ede1) that can function upstream of Ca2+/calmodulin-dependent protein kinase 2 (Cmk2) to suppress cell death in parallel to Cn. The screen also revealed the vacuolar H+-ATPase (V-ATPase), which acidifies the lysosome-like vacuole. The V-ATPase performed its death-promoting functions very soon after imposition of the stress and was not required for later stages of the cell death program. Cn did not inhibit V-ATPase activities but did block vacuole membrane permeabilization (VMP), which occurred at late stages of the cell death program. All of the other nondying mutants identified in the screens blocked steps before VMP. These findings suggest that VMP is the lethal event in dying yeast cells and that fungi may employ a mechanism of cell death similar to the necrosis-like cell death of degenerating neurons.
Introduction
Programmed cell death (PCD)2 occurs in metazoans as a means of removing infected, damaged, mutant, misplaced, or superfluous cells from the bodies of individuals. Three main types of PCD have been classified (1). Apoptosis (PCD-I) involves a core set of proteases from the caspase family and usually members of the Bcl-2 family of apoptosis regulators that link caspase activation to a wide array of cellular signals. Autophagy (PCD-II) involves uptake of organelles and cytoplasm into the lysosome for degradation, which is usually beneficial and cytoprotective in its early stages but self-destructive when taken to extremes. These types of PCD have been thoroughly characterized at the molecular and cytological levels in a wide array of cell types in metazoan species. On the other hand, necrosis (PCD-III) seems much more heterogeneous and morphologically variable from one cell type to another, probably as a consequence of distinct molecular mechanisms at work in the different circumstances (2).
One form of necrosis-like cell death has been studied genetically in degenerating neurons of the nematode Caenorhabditis elegans. Mutant animals that express hyperactive ion channels in their neurons exhibit very high frequencies of necrosis-like cell death that resembles neurodegenerative diseases of humans (3). Genetic screens have revealed lysosomal membrane permeabilization (LMP) and release of lysosomal proteases and acids as the lethal event in this system (4–6). LMP seemed to be triggered by elevation of cytosolic free Ca2+ concentrations ([Ca2+]i) and activation of calpain proteases in the cytoplasm (4, 7, 8). LMP also required functions of the proton-pumping V-ATPase that normally acidifies lysosomes (5) and is associated with accumulation of reactive oxygen species (3). Autophagy seemed to accelerate the progression toward LMP but was not absolutely required for the necrotic cell death to proceed (9). Other aspects of membrane trafficking also have been implicated (10). Other kinds of necrosis-like cell death have been linked to LMP in a variety of mammalian cells (11), leading to the hypothesis that LMP-dependent PCD processes may be broadly conserved among metazoans. There is growing evidence that PCD in plants also depends on the rupture or leakage of the vacuole, a lysosome-like organelle (12).
PCD has been observed in a variety of fungi at different stages of development. One example is mating incompatibility of filamentous fungi such as Neurospora crassa, where fusion of genetically incompatible hyphal cells results in rapid death of the fused cells (13). This PCD benefits the species by preventing the transmission of viruses, prions, and plasmids. Something similar may occur in the mating of haploid Saccharomyces cerevisiae (yeast) cells, which form diploid cells in a process that utilizes secreted mating pheromones as cues for guidance and differentiation (14). Interestingly, when exposed to high concentrations of mating pheromones in the absence of mating partners, rapid cell death can occur in a significant portion of the population (15, 16). This manner of pheromone-induced cell death depends on the expression of pheromone-inducible Fig1 protein of the plasma membrane and seems to involve the inappropriate removal of cell wall material, which is normally removed only when a mating partner is properly positioned (16). Mating pheromones induce a second manner of cell death in yeast that is slower than Fig1-dependent cell death and independent of Fig1 and cell wall remodeling (16). In wild-type cells, this “slow” form of pheromone-induced cell death is normally blocked by the activation of a high-affinity Ca2+ influx system (HACS) and the calcium signaling pathway downstream of HACS (17–24). The genetic disruption of HACS, calmodulin, or calcineurin (Cn) or the pharmacological inhibition of Cn with either FK506 or cyclosporine was not harmful to yeast growth or mating in ordinary circumstances. However, deficiencies in this calcium signaling pathway were completely lethal during prolonged exposures to mating pheromones. The findings suggest that HACS, [Ca2+]i elevation, calmodulin, and Cn constitute a signaling pathway that actively suppresses a pheromone-inducible cell death program through regulation of protein phosphorylation. The pathogenic yeast Candida albicans employs the homologous pathway for similar purposes (25), suggesting broad conservation of the pheromone-induced cell death program in fungi.
HACS, calmodulin, and Cn also actively suppress cell death in a variety of yeast species during exposure to azole-class antifungal drugs (26), which selectively inhibit enzymes in the endoplasmic reticulum (ER) involved in sterol biosynthesis. Similarly, the calcium signaling pathway suppresses death of yeast cells exposed to tunicamycin, a natural antifungal compound that interferes with N-glycosylation of secretory proteins in the ER (27, 28). Thus, azoles and tunicamycin are fungistatic to wild-type yeasts but fungicidal to yeasts that have lost functionality of their calcium signaling pathways. FK506 and cyclosporine convert the fungistats to fungicides by blocking Cn, thereby increasing the efficacy of the antifungals and minimizing the possibility of acquired drug resistance. Unfortunately, most fungal infections cannot be treated with combinations of azoles and Cn inhibitors because FK506 and cyclosporine suppress immunity functions in humans.
The targets of Cn that mediate these fungal cell death phenomena have not yet been identified. In Cn-deficient mutants of yeast, reactive oxygen species (ROS) accumulate ∼70 min before cell death, as indicated by staining with the fluorescent probe dihydro-DCFDA (28), but little else is known about the sequence of events that lead to death of Cn-deficient yeasts. The death of Cn-deficient yeast cells in these circumstances was independent of metacaspase (Mca1) and other factors that had been implicated in apoptosis-like death of yeasts (28, 29). Therefore, the manner of cell death in Cn-deficient yeasts and the possible existence of “pro-death” signaling pathways that are responsive to stimuli remain completely undefined.
In this study, we present evidence that the vacuolar membranes of yeast cells can become permeabilized in response to tunicamycin, and we employ a forward genetic screen to reveal networks of cellular factors that control vacuole membrane permeabilization (VMP) and cell death. Autophagy was not essential for these phenomena. Instead, we find V-ATPase activity to be necessary for death of yeast cells similar to the requirement for V-ATPase activity in the necrosis-like death of degenerating neurons (5) and the chloroquine-induced death of cerebellar granule neurons (30).
EXPERIMENTAL PROCEDURES
Yeast Strains, Culture Media, and Reagents
The yeast strains used in this study were obtained from collections of viable gene knock-out mutants in the BY4741 and BY4742 strain backgrounds (31). The ede1::NatR knock-out mutation was introduced into BY4741 and BY4741-cmk2::G418r strains using standard PCR-based methods (32) to yield strains HK081 and HK082. The vma1::NatR knock-out mutation was made similarly in BY4741 to yield HK083. The cmk2::G418r cnb1::G418r double knock-out mutant strain HK006 was constructed from a cross between BY4741-cmk2::G418r and BY4742-cnb1::G418r. Yeast strains were cultured in rich YPD medium or synthetic SC medium (33). Stocks of tunicamycin (Sigma-Aldrich), concanamycin C (Santa Cruz Biotechnology), and FK506 (Astellas Pharma) were dissolved in DMSO and stored at −20 °C. Aqueous 45CaCl2 was purchased from MP Biosciences. Propidium iodide (Sigma-Aldrich) was dissolved in PBS, and carboxy-DCFDA (Invitrogen), dihydro-DCFDA (Invitrogen), and FM4-64 (Invitrogen) were dissolved in DMSO.
Genetic Screen for Death-inhibiting and Death-promoting Factors
A collection of all viable gene knock-out mutants of yeast strain BY4741 (31) was grown overnight at 30 °C in synthetic complete (SC) medium containing all 20 amino acids plus adenine and uracil. The saturated cultures were diluted 7-fold into 90 μl of fresh SC medium containing 2.5 μg/ml tunicamycin with and without 1 μg/ml FK506. After 24 h of incubation at room temperature, 100 μl of 1 μm propidium iodide (PI) in phosphate-buffered saline was added to each culture. After mixing, 5,000 cells in each culture were immediately counted as live (PI-negative) or dead (PI-positive) using a 96-well flow cytometer (BD FACSArray). The entire collection was analyzed in 11 nonoverlapping batches. Although the batch-to-batch variation was small, it was further minimized by converting the raw cell death frequencies to Z-scores using the average and standard deviation of each batch after log transformation of the raw data. To identify mutants with poor responses to FK506, the differences between the +FK506 and the −FK506 cell death frequencies were scaled to the proportion of the culture that survived and converted to Z-scores as above except without log transformation. All mutants in the collection that exhibited a Z-score of 2 or greater were identified, rearrayed, and rescreened for cell death as before except using 4 mm dithiothreitol (Sigma-Aldrich) instead of tunicamycin as the death-inducing stimulus. Z-Scores for the dithiothreitol experiment were calculated as before using averages and standard deviations derived from a randomly chosen control set of 94 mutants. The raw data and the statistical transformations are recorded in supplemental Table 1. Some data were represented as heat maps using Java TreeView (34).
45Ca2+ Accumulation Measurements
Total cellular accumulation of Ca2+ was measured as described previously (35). Briefly, cells were grown to log phase in YPD medium overnight, harvested, and resuspended in fresh YPD medium supplemented with tracer quantities (10 μCi/ml) of 45CaCl2 (MP Biomedicals) and various drugs. After a 2-h incubation in a 96-well filtration plate (Millipore) at 30 °C, cells were harvested, washed with ice-cold buffer W (10 mm CaCl2, 5 mm HEPES-NaOH, pH 6.5) by using a vacuum filtration unit (Millipore), and dried at room temperature overnight. MicroScint-20 scintillation mixture (PerkinElmer Life Sciences) was added to each well, and the radioactivity was counted using a TopCount NXT instrument (Packard).
Cell Staining Methods
Log-phase yeast cells growing in SC medium were exposed to tunicamycin (2.5 μg/ml), dithiothreitol (4 mm), concanamycin C (3 μm), FK506 (1 μg/ml), or combinations thereof for varying lengths of time, pelleted, and stained for 15 min at room temperature in the same medium containing freshly prepared PI (0.25 μg/ml), carboxy-DCFDA (1 μg/ml), and/or dihydro-DCFDA (1 μg/ml). Stained cells were imaged using a LSM 510 META confocal microscope (Zeiss) with appropriate filter sets, photographed, and counted manually after appropriate binning. Live/dead counting was also performed automatically using the FACSArray instrument described earlier (28). In some experiments, log-phase cells were prestained with FM4-64 (10 μg/ml) for 15 min at 30 °C, washed, and incubated for an additional 15 min at 30 °C before loading onto ONIX microfluidic slides (CellASIC) and the addition of stressors and inhibitors.
RESULTS
Genome-wide Screen for Factors Involved in Tunicamycin-induced Cell Death
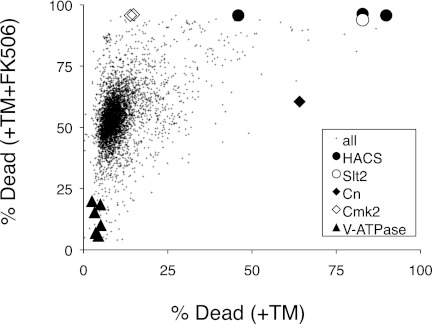
The direct targets of Cn that regulate nonapoptotic cell death in yeasts are presently unknown. To search for these targets and for other death-inhibiting or death-promoting factors, a flow cytometer was used to count the numbers of live and dead cells in individual cultures of 4,847 different nonessential gene knock-out mutants of yeast after exposures to tunicamycin and to tunicamycin plus FK506. The experimental conditions were carefully chosen such that the average frequencies of cell death for all the mutant strains were 10.6 and 54.5% in the absence and presence of FK506, respectively. As can be seen in Fig. 1, most mutants were distributed close to these averages, and relatively few mutants appeared as outliers. As expected, cultures of the Cn-deficient cnb1 mutant exhibited a very high frequency of cell death in the absence of FK506 (64% death), and there was no additional effect of FK506. The Cmk2-deficient cmk2 mutant was indistinguishable from the mean in the absence of FK506 (14.6% death) but was significantly higher than the mean in the presence of FK506 (96% death), confirming a mild death-inhibiting activity of Cmk2 in the absence of Cn (28). The HACS-deficient mutants cch1, mid1, and ecm7 exhibited higher frequencies of cell death in both conditions (83 and 96%, 90 and 96%, and 46 and 96% death, respectively), which was closely mimicked by cultures of cnb1 cmk2 double mutants that simultaneously lack both targets of HACS (data not shown). These findings show that the primary genetic screen recovered all the known components of the death-inhibiting calcium signaling pathway.
FIGURE 1.
Genome-wide screen for mutants with altered rates of cell death. Individual mutants from the yeast gene knock-out collection were cultured in the presence of tunicamycin (+TM) and tunicamycin plus FK506 (+TM+FK506), stained with PI, analyzed by flow cytometry, and plotted as a single point in the chart. Each point represents the percentage of dead cells in the culture containing tunicamycin versus that in the culture containing tunicamycin plus FK506. Selected mutants have been highlighted with large symbols.
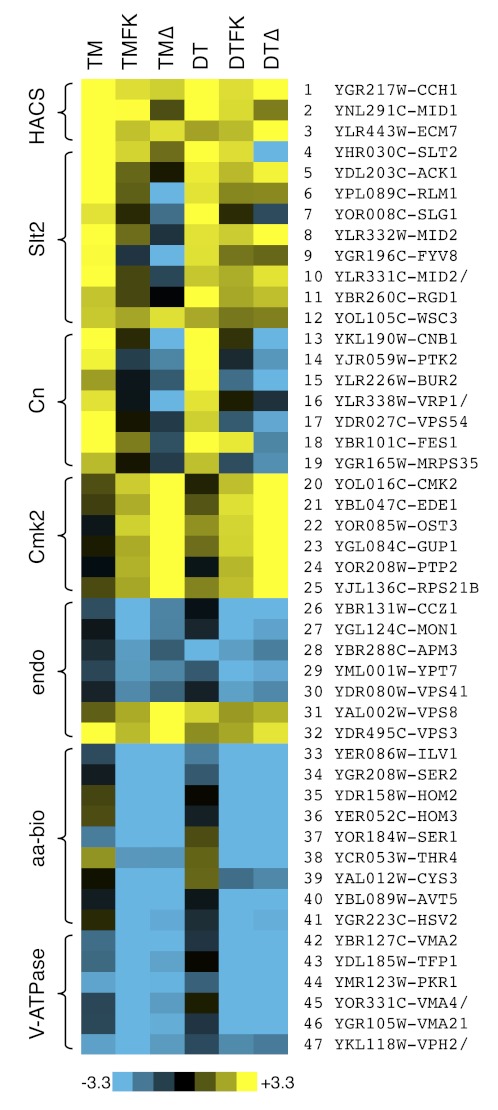
A total of 461 mutants exhibited significantly higher or lower frequencies of cell death in the primary screen. Another 92 mutants exhibited significantly higher or lower FK506 responsiveness, defined as the scaled difference between the two screening conditions. To increase confidence in the results, all these outliers and many control strains were rearrayed and screened for cell death a second time after exposures to dithiothreitol and dithiothreitol plus FK506. Like tunicamycin, dithiothreitol inhibits essential enzymes in the ER involved in secretory protein biogenesis, triggering ER stress, growth inhibition, and calcineurin-suppressible cell death (28). A total of 111 mutants exhibited significantly higher frequencies of cell death in both screens lacking FK506 (column HD-FK in supplemental Table 1). As expected, the HACS-deficient mutants (cch1, mid1, ecm7) were among this group (Fig. 2, lanes 1–3). An additional nine mutants of this group lacked the stress-activated MAP kinase Slt2 or one of its upstream regulators in the cell wall integrity signaling pathway (Fig. 2, lanes 4–12). Cell wall integrity signaling was shown previously to be important for HACS activation in these conditions (36). The 98 remaining mutants were deficient in a wide array of cellular processes, and the set was enriched in the Gene Ontology process “vesicle-mediated transport” (19 mutants; p value < 0.001). This same process is targeted by tunicamycin and dithiothreitol, so many factors in this group may inhibit cell death by compensating for the effects of ER stress.
FIGURE 2.
Heat maps of cell death for selected mutants. Raw data from Fig. 1 and a secondary screen involving dithiothreitol (DT) instead of tunicamycin (TM) were converted to Z-scores as described under “Experimental Procedures,” and mutants with patterns of cell death similar to or opposite from those of HACS-, Cn-, and Cmk2-deficient cells are illustrated. The scale bar illustrates the number of standard deviations below (blue) or above (yellow) the population averages. TMΔ and DTΔ represent Z-scores of the FK506 responsiveness of each mutant strain.
To identify cellular factors that may mediate the death-inhibiting effects of Cn, we ranked the mutants of this set by their unresponsiveness to FK506. Only one mutant (ptk2) exhibited significantly poor responsiveness to FK506 in both screens similar to the cnb1 mutant, and just five other mutants exhibited significant unresponsiveness in one or the other condition (Fig. 2, lanes 13–19). The ptk2 mutant lacks a serine/threonine protein kinase that regulates general ion homeostasis in yeast, in part through the phosphorylation of the electrogenic H+ ATPase (Pma1) in the plasma membrane (37). Although Ptk2 and Pma1 lack recognizable PXIXIT motifs that are often found in Cn substrates (38), these proteins are candidate effectors of Cn signaling.
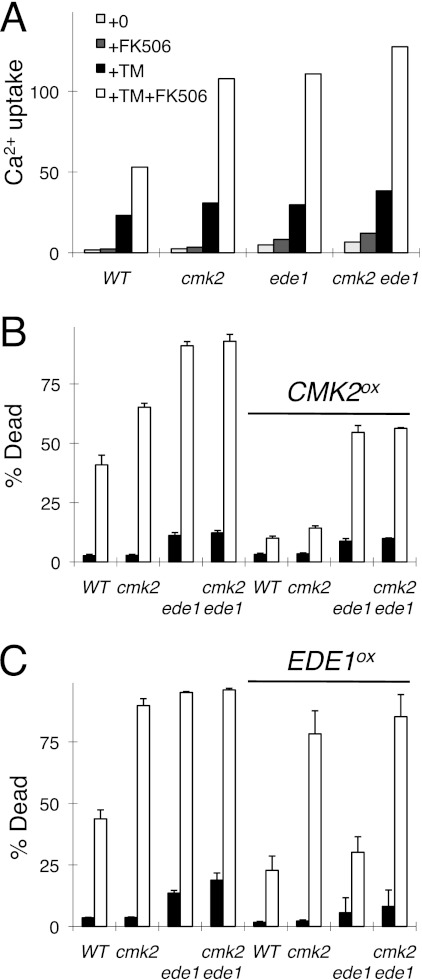
The genetic screens may also reveal components of the Cmk2-dependent branch of the death-inhibiting mechanism. The cmk2 mutants exhibit wild-type cell death in the absence of FK506 and very high cell death in the presence of FK506. After filtering the data from both genetic screens and ranking the mutants by their high FK506 responsiveness, only ede1 and four other mutants consistently behaved in a manner similar to cmk2 mutants (Fig. 2, lanes 20–25). The ede1 mutant also behaved like the cmk2 mutant in another genetic screen that measured HACS activity during exposure to tunicamycin plus FK506 (39), and a high-throughput study demonstrated a physical association between Ede1 and Cmk2 (40). Ede1 is a well known regulator of endocytosis in yeast (41, 42). To explore the interactions between Cmk2 and Ede1, the knock-out mutants and the double knock-out mutant were recreated and retested in the presence and absence of CMK2 and EDE1 overexpression plasmids. The cmk2 ede1 double mutant exhibited the same rates of cell death and 45Ca2+ uptake in response to tunicamycin plus FK506 as the single mutants (Fig. 3A), suggesting that Cmk2 and Ede1 may function in a common pathway that regulates both HACS and cell death. Overexpressed Cmk2 partially suppressed both phenotypes of ede1 mutants (Fig. 3B), whereas overexpressed Ede1 failed to suppress either phenotype of cmk2 mutants (Fig. 3C). Altogether, these findings suggest that Cmk2 may function downstream of Ede1 in a pathway that weakly inhibits both HACS and cell death in the absence of Cn. Cn strongly inhibits HACS and cell death in the absence of Cmk2. Thus, Cn and Cmk2 function independently in these processes despite an ability of Cn to induce expression of Cmk2 (28).
FIGURE 3.
Ede1 stimulates Cmk2 functions. A–C, mutants lacking Ede1, Cmk2, or both proteins were compared with wild-type yeast in assays of 45Ca2+ uptake (A) and cell death (B and C) in response to tunicamycin (TM) and FK506. The strains bearing control plasmids (left half of B and C) or Cmk2 and Ede1 overexpression plasmids (right half of B and C) were analyzed in parallel. Error bars indicate ±S.D.
Death-promoting Factors in Yeast
Cn and Cmk2 may antagonize enzymes or pathways that become toxic in cells exposed to tunicamycin, dithiothreitol, or other death-inducing stimuli. To identify such death-promoting factors, the data from the genetic screens were analyzed for mutants that exhibit significantly lower rates of cell death in both of the death-inducing conditions (tunicamycin plus FK506 and dithiothreitol plus FK506). A total of 96 mutants exhibited cell death frequencies that were at least 1.5 standard deviations below the population average (column LD+FK in supplemental Table 1). All of these mutants remained sensitive to tunicamycin or dithiothreitol as judged by growth assays (data not shown). Therefore, this set of nondying mutants appears to be enriched for genes that encode death-promoting factors rather than stress-promoting factors.
The set of 96 nondying mutants was not enriched for any Gene Ontology “process” or “function” terms. However, many of these mutants could be grouped into a few functional categories based on prior knowledge of their functions. One class of 15 mutants (ada2, cdc73, elf1, hos4, leo1, rtt103, ydr290w, sif2, sir3, yjl175w, snf5, snf6, swc3, vps71, vps72, yaf9) was defective in several aspects of chromatin remodeling and transcription. A second class of five nondying mutants (ccz1, mon1, ypt7, vps41, apm3) was defective in maturation and fusion of late endosomes with the vacuole, a lysosome-like organelle of yeast (43). Interestingly, mutants that block an earlier step of endosome maturation (vps3, vps8) exhibited elevated frequencies of cell death in all the conditions (Fig. 2, endo). Thus, endosome maturation and fusion complexes exhibit strong death-promoting and death-inhibiting effects.
A third class of seven nondying mutants (cys3, hom2, hom3, ilv1, ser1, ser2, thr4) was defective for biosynthesis of several amino acids (Fig. 2, aa-bio). All 20 amino acids were present in excess in the culture media during the screening conditions, so the enhanced survival of these auxotrophic mutants could not be attributed to slower rates of protein synthesis. One additional nondying mutant (avt5) lacked an SLC36 family protein that probably functions as a vacuolar H+/amino acid symporter or antiporter (44). Although Avt5 has not been carefully studied, avt5 knock-out mutants exhibited a spectrum of genetic interactions that were very closely correlated with those of hsv2 mutants (45), which also failed to die in the genetic screens (Fig. 2, lanes 40–41). Hsv2 binds phosphatidylinositol 3,5-bisphosphate that is generated on the surface of vacuolar membranes (46), but its cellular functions have not yet been determined.
As mentioned earlier, a genetic screen in C. elegans revealed subunits of the V-ATPase as death-promoting factors in stressed and degenerating neurons (5). The V-ATPase is a multisubunit proton pump responsible for acidification of vacuoles, lysosomes, endosomes, and secretory organelles in eukaryotes (47). Six V-ATPase-deficient mutants of yeast (tfp1, vma2, vma4, vma21, pkr1, vph2) were deficient in cell death in our screening conditions (Fig. 2, lanes 42–47). Eight other V-ATPase-deficient mutants all exhibited strong nondying phenotypes after a careful rescreen (supplemental Table 2). The vph1 and stv1 single mutants that lack partially redundant subunits of the V0-sector of the V-ATPase exhibited weaker nondying phenotypes (supplemental Table 2). The findings suggest that the assembled functional V-ATPase is crucial for the death of stressed yeast cells in these conditions.
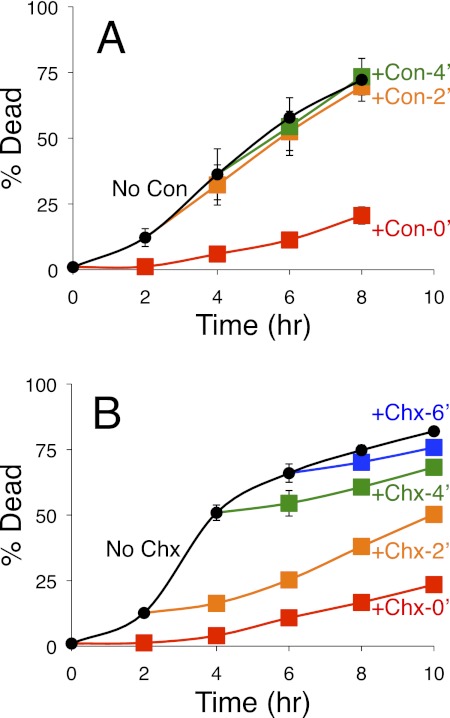
To define more precisely the death-promoting functions of the V-ATPase, the inhibitor concanamycin C was added to cultures of cnb1 mutants at various times after exposure to tunicamycin. When added at the same time as tunicamycin, concanamycin strongly delayed the onset of cell death (Fig. 4A). Surprisingly, concanamycin failed to block cell death when added 2 h or more after initial exposure to tunicamycin (Fig. 4A), a time at which almost all of the cnb1 mutant cells were still alive. A protein synthesis inhibitor (cycloheximide) strongly delayed cell death when added at all times following tunicamycin exposure (Fig. 4B). These findings suggest that ongoing protein synthesis was required for cell death even in the few surviving cells that remained alive after 6 h of tunicamycin exposure. The findings also suggest that cell death in these circumstances is an active process and that the V-ATPase can accomplish all its death-promoting functions in cnb1 mutant cells within 2 h of tunicamycin exposure.
FIGURE 4.
Inhibition of cell death by inhibitors of V-ATPase and translation. A and B, cultures of the Cn-deficient cnb1 mutants were exposed to tunicamycin at 0 h with additions of either the V-ATPase inhibitor concanamycin (Con) (A) or the translation inhibitor cycloheximide (Chx) (B) at 0 h (red), 2 h (orange), 4 h (green), 6 h (blue), or never (black). The frequencies of cell death in triplicate cultures were measured at the indicated times by staining with PI and flow cytometry. Error bars indicate ±S.D.
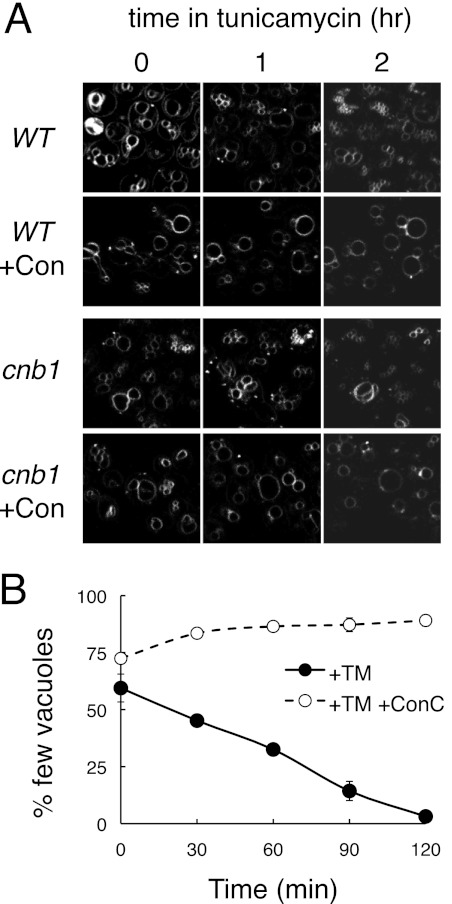
Several experiments were designed to test the hypothesis that Cn inhibits V-ATPase activity. First, acidity of the vacuoles was measured in tunicamycin-treated cnb1 mutants and wild-type cells. Using a fluorescent pH probe and an established in vivo method (48), we observed complete neutralization of the vacuoles of both strains treated with concanamycin but no detectable neutralization in response to tunicamycin and no significant differences between the two strains within the first 2 h of exposure (data not shown). Second, the ability of Cn to suppress V-ATPase-dependent changes in vacuole morphology was examined. Vacuoles are dynamic organelles that undergo continuous fission and fusion reactions, with V-ATPase activity being required for vacuole fission (49). In cnb1 mutants, tunicamycin exposure caused the slow disappearance of cells with large vacuoles and appearance of cells with many small vacuoles (Fig. 5). A similar effect of tunicamycin was observed in Cn-replete wild-type cells (Fig. 5). In both cases, the V-ATPase inhibitor concanamycin completely blocked the tunicamycin-induced vacuole fragmentation in both cell types (Fig. 5). Thus, V-ATPase-dependent vacuole fragmentation appeared insensitive to the presence or absence of Cn.
FIGURE 5.
V-ATPase-dependent vacuole fragmentation in response to tunicamycin. A, wild-type and Cn-deficient cnb1 mutants were stained with the fluorescent dye FM4-64 to reveal vacuole membranes and imaged in a microfluidic device at various times of exposure to tunicamycin or tunicamycin plus concanamycin (+Con) as indicated. B, wild-type cells prestained with FM4-64 were imaged by fluorescence microscopy at various time points after exposure to tunicamycin (+TM) or tunicamycin plus FK506 (+TM+Con) and counted manually as having few vacuoles (two or fewer) or several vacuoles (three or more). The averages of three parallel experiments (±S.D.) were charted.
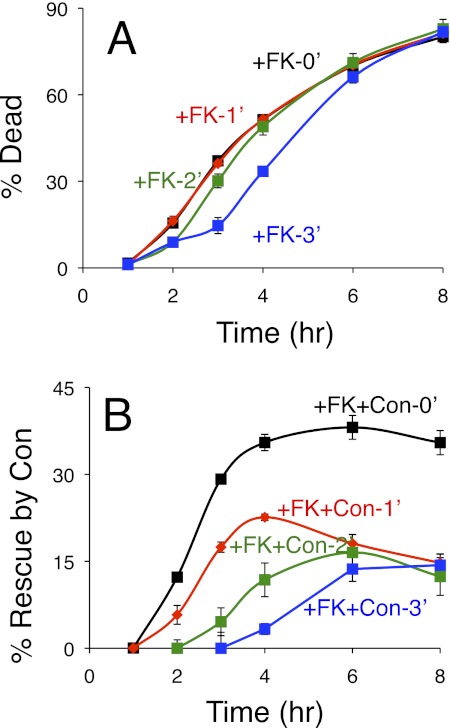
In a third experiment, we asked whether the presence of functional Cn in wild-type cells could delay the death-promoting function of the V-ATPase. For this experiment, wild-type cells were exposed to tunicamycin for varying amounts of time before the addition of FK506 or FK506 plus concanamycin and then assayed at later times for the extent of cell death. Starting 2 h after initial exposure to tunicamycin, the delayed additions of FK506 resulted in delayed waves of cell death in the populations (Fig. 6A and data not shown). The ability of concanamycin to prevent cell death was very strong when added with FK506 at the same time as tunicamycin, and it remained significant when added with FK506 after 3 h of tunicamycin pre-exposure (Fig. 6B). Thus, in Cn-proficient cells, the V-ATPase required more than 3 h to complete its death-promoting functions. In Cn-deficient cells, the V-ATPase completed its death-promoting functions within 2 h of tunicamycin exposure (Fig. 4A). The simplest model consistent with all these findings is that Cn inhibits a toxic enzyme or process positioned downstream of the V-ATPase in a process that controls nonapoptotic cell death.
FIGURE 6.
Cn delays V-ATPase-dependent cell death. Cultures of wild-type yeast cells were exposed to tunicamycin at 0 h with additions of FK506 (FK) or FK506 plus concanamycin (FK+Con) at 0 h (black), 1 h (red), 2 h (green), or 3 h (blue). A and B, the frequencies of cell death in triplicate cultures were measured at the indicated times by staining with PI and flow cytometry and plotted directly as percentage of dead (A) or plotted after determining the percentage of the population that was rescued by the addition of concanamycin (B), which was calculated by subtraction of the values obtained in the FK506 plus concanamycin cultures from those shown in A. Error bars indicate ±S.D.
Cn Inhibits VMP and ROS Production in Stressed Yeast Cells
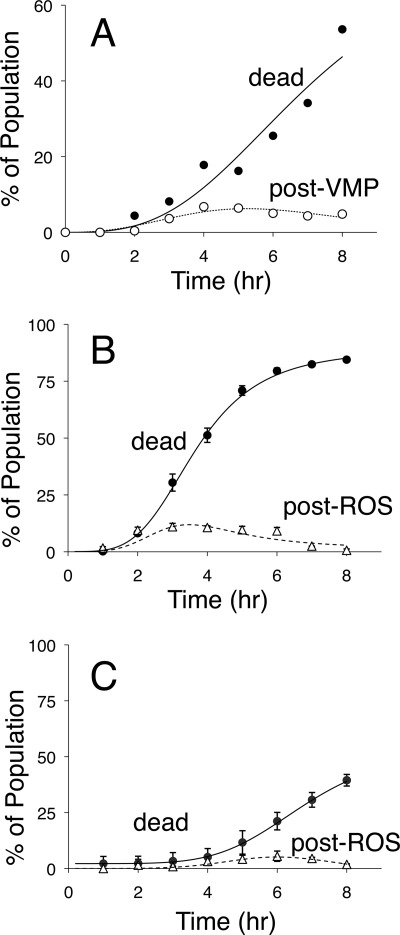
Necrosis-like cell death in degenerating neurons has been shown to require V-ATPase activity, which is necessary for changes in morphology and permeability of lysosomal membranes (5, 6). To determine whether the permeability of the vacuolar membrane changes in dying yeast cells, we employed the fluorogenic compound carboxy-DCFDA, which becomes fluorescent and membrane impermeant upon hydrolysis of the acetate groups by esterases in the vacuole lumen (50). Vacuoles of wild-type and Cn-deficient cnb1 mutants fluoresced brightly after staining with carboxy-DCFDA, existing mainly as one or two large spheres in standard growth conditions. Tunicamycin induced changes in the morphology of vacuoles in both cell types, as expected for the increased vacuole fragmentation described earlier. Fluorescence was never observed in the cytoplasms of wild-type cells at any time after exposure to tunicamycin. In contrast, cytoplasmic fluorescence was observed in significant numbers of cnb1 mutant cells that had not yet died beginning around 2 h after tunicamycin exposure. The frequency of nondead cells in the population with fluorescent cytoplasms rose, peaked, and declined over time (Fig. 7A), suggesting that dying cells pass through a brief intermediate stage before cell death where either the vacuolar esterases or the vacuole-generated fluorophore (carboxy-DCF) leak into the cytoplasm. When sigmoid curves are fit to these data using nonlinear regression (28), the leakage of vacuolar esterases or fluorophore could be estimated at roughly 0.5 h prior to cell death.
FIGURE 7.
Vacuole membrane permeabilization and ROS accumulation in dying populations of yeast cells. A, the Cn-deficient cnb1 mutant was exposed to tunicamycin, incubated at room temperature, stained at the indicated times with either PI or carboxy-DCFDA, and immediately observed by epifluorescence microscopy. The frequency of dead cells in the population that stained with PI was plotted (black circles), and the frequency of cells in the population that failed to stain with PI and exhibited cytoplasmic staining with carboxy-DCFDA was plotted (white circles). B and C, wild-type (B) and V-ATPase-deficient vma1 mutant cells (C) were exposed to tunicamycin plus FK506, stained with PI at the indicated times, counted by flow cytometry, and plotted as the average (±S.D.) from the three replicate experiments (black circles). The same cultures were also sampled, stained with dihydro-DCFDA at the indicated times, counted manually in the fluorescence microscope, and plotted (white triangles). The PI staining data were fit by nonlinear regression to the standard sigmoid equation (solid lines), and the other staining data were similarly to fit to the difference between a standard sigmoid equation and the solid lines (dashed lines).
Previous studies of tunicamycin-induced cell death in yeast have employed dihydro-DCFDA, an analog of carboxy-DCFDA that requires oxidation by ROS in addition to maturation by vacuolar esterases to become fluorescent (28). Wild-type cells stained with dihydro-DCFDA remained alive and nonfluorescent at all times of tunicamycin exposure, whereas Cn-deficient cells of the W303-1A strain background became fluorescent at ∼1.2 h prior to cell death and nonfluorescent after cell death (28). A similar pattern was observed in wild-type cells of the BY4741 strain background after exposure to tunicamycin plus FK506 (Fig. 7B). The peak of staining with dihydro-DCFDA was somewhat lower and broader than that of the W303-1A background because the latter strain died faster and more synchronously. Nevertheless, the kinetics of staining with dihydro-DCFDA closely paralleled the kinetics of staining with carboxy-DCFDA, with the dying cells staining positive at ∼0.8 h prior to cell death (Fig. 7B). A similar experiment using V-ATPase-deficient vma1 mutants exposed to tunicamycin plus FK506 revealed much lower rates of cell death and staining with dihydro-DCFDA (Fig. 7C). When taken together, these results suggest that dying yeast cells release vacuolar esterases or fluorophores and generate ROS roughly an hour before cell death.
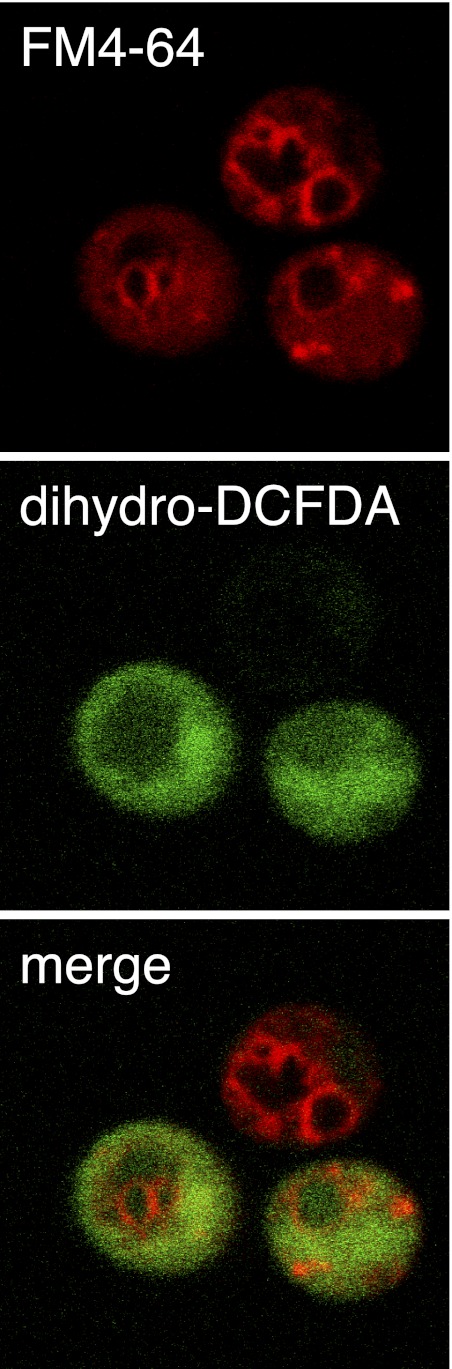
Microscopic examination of dying cnb1 mutants stained with dihydro-DCFDA revealed very little fluorescence in the vacuole and most fluorescence in the cytoplasmic regions (Fig. 8). Unlike carboxy-DCFDA, dihydro-DCFDA never stained vacuoles in the cell without also staining the cytoplasm. The absence of fluorescent vacuoles after staining with dihydro-DCFDA suggests that ROS had not yet accumulated in the cytoplasm or culture medium prior to VMP. Therefore, ROS accumulation is likely to be a consequence of VMP in these conditions.
FIGURE 8.
Nonvacuolar localization of ROS. Wild-type yeast cells were stained with FM4-64 and subsequently exposed to tunicamycin plus FK506. After 4 h of incubation, cells were stained with dihydro-DCFDA and imaged by confocal microscopy for vacuole membrane architecture (FM4-64) and ROS accumulation (dihydro-DCFDA). The image shows three live cells, two of which are representative of cells that stain positive for ROS. Fluorescence products of dihydro-DCFDA were almost always excluded from the region of the cell containing vacuoles.
Although the V-ATPase appears to function upstream of VMP and ROS accumulation, other nondying mutants may be necessary for downstream processes. Such mutants would exhibit higher than wild-type frequencies of staining with dihydro-DCFDA after exposure to tunicamycin plus FK506. To test this possibility, all 96 mutants of the nondying set were exposed to tunicamycin plus FK506 for 4 h, stained with dihydro-DCFDA, and examined microscopically. None of the nondying mutants exhibited higher frequencies of staining than the wild-type cells, and the great majority of these mutants exhibited lower frequencies of staining (data not shown). Our inability to find mutants with increased survival after ROS accumulation suggests that VMP is the lethal event in the response of yeast cells to ER stressors.
DISCUSSION
The findings presented above demonstrate important roles for the V-ATPase and vacuole membrane permeabilization in the death of Cn-deficient yeast cells responding to ER stressors. In wild-type cells exposed to tunicamycin, V-ATPase activity was necessary for vacuole fragmentation, and in Cn-deficient cells the V-ATPase was necessary for VMP, ROS accumulation, and subsequent cell death. Because Cn did not inhibit the V-ATPase-dependent fragmentation of vacuoles or the V-ATPase-dependent acidification of vacuoles, Cn probably does not inhibit the V-ATPase directly and seemingly inhibits some process that depends on V-ATPase activity. Indeed, experiments with inhibitors suggest that Cn can delay the V-ATPase-dependent process of cell death most likely at the step upstream of VMP.
Another new addition to the working model of cell death regulation is Ede1, which we find to function upstream of Cmk2 in a branch of the death-inhibiting regulatory loop. High-throughput studies also identified a physical interaction between Ede1 and Cmk2 (40). Ede1 is a scaffold protein that recruits several different proteins and promotes the process of endocytosis (42). Mutants lacking the endocytic partners of Ede1 (ent1, ent2, syp1, yap1801, yap1802) did not exhibit the same phenotypes of the ede1 and cmk2 mutants, so the regulation of Cmk2 by Ede1 may be independent of endocytosis. Although Cmk2 expression can be strongly induced by Cn activation and overexpressed Cmk2 can partially suppress the death of Cn-deficient mutants (28), the roles of Cmk2 in the regulation of cell death seem weaker and less well defined than those of Cn. Because tunicamycin-treated cmk2 mutants and ede1 mutants do not exhibit more death than wild-type when Cn is functioning, Cmk2 may inhibit some earlier step of the pathway leading to cell death. The only step earlier than VMP observed to date is vacuole fragmentation. Purified Cmk2 was able to phosphorylate the Vma13 or associated proteins in a protein microarray experiment (51), potentially indicating a direct effect of Cmk2 on the V-ATPase and its role in vacuole fragmentation. More detailed studies will be required to determine precisely how Cmk2 modulates cell death in yeast and how a closely related Ca2+/calmodulin-dependent protein kinase (Cmk1) does not.
The role of vacuole fragmentation in tunicamycin-induced cell death was unexpected and remains of unknown significance. Vacuole fragmentation results from an imbalance between fission and fusion processes. Fission requires V-ATPase activity and all the subunits of the V0 and V1 sectors, whereas fusion requires subunits of the V0 sector but not the V1 sector or V-ATPase activity (49). Because cell death required all subunits of the V-ATPase and is sensitive to V-ATPase inhibitors, it is tempting to suggest that vacuole fission promotes cell death in these circumstances. Several other observations complicate this simple picture. First, other fission-defective mutants (vps15, vps34, fab1, vps1) exhibited very high cell death when exposed to tunicamycin or dithiothreitol alone, as well as very robust increases in response to FK506. Vps34 and Fab1 sequentially phosphorylate phosphatidylinositol at the 3 and 5 positions to produce phosphatidylinositol 3-phosphate and phosphatidylinositol 3,5-bisphosphate on the cytoplasmic surface of vacuole membranes (52, 53), which recruit specific binding proteins such as Pib2 and Hsv2 with unknown roles in vacuole fission (46, 54). The pib2 and hsv2 mutants each exhibited striking defects in cell death, which was opposite to the behaviors of vps34 and fab1 mutants. Second, several mutants defective in vacuole fusion (ccz1, mon1, ypt7, vps41) were also strikingly deficient in cell death during the response to tunicamycin plus FK506, whereas others (vps3, vps8) exhibited elevated rates of cell death (Fig. 2). Finally, all these mutants are known to have defects in other processes such as vesicle-mediated delivery of newly synthesized and endocytosed proteins to the vacuole (43). No simple model emerges from the seemingly contradictory behaviors of these fusion- and fission-deficient mutants. Except for V-ATPase subunits and regulators, the vast majority of the nondying mutants identified here underwent normal vacuole fragmentation in response to tunicamycin plus FK506 as indicated by staining with FM4-64 (data not shown). Vacuole fragmentation therefore does not represent a commitment to cell death but perhaps is necessary for subsequent steps that lead to cell death.
Data presented here suggest that leakage of vacuolar materials into the cytoplasm may represent the “point of no return” that commits yeast cells to death. This conclusion is based on the observed staining patterns with carboxy-DCFDA and dihydro-DCFDA, membrane-permeable molecules that become membrane-impermeable after hydrolysis by vacuolar esterases (50). In both cases, cytoplasmic fluorescence was only observed in dying yeast cells and never observed in nondying cells with either functional Cn or nonfunctional death-promoting factors. The dying cells released either vacuolar esterases or the matured probes into the cytoplasm well before they died, as indicated by their ability to exclude PI. In the case of dihydro-DCFDA, which becomes fluorescent only after oxidation by ROS and cleavage by vacuolar esterases (55), vacuolar fluorescence was not observed in either nondying or dying cells. The absence of vacuolar staining suggests that ROS never accumulate in the vacuole and only accumulate in the cytoplasm after permeabilization of the vacuolar membrane. This conclusion rests on the assumption that oxidized dihydro-DCFDA is as permeable to the vacuolar membrane as carboxy-DCFDA. Because all the nondying mutants studied here failed to stain with dihydro-DCFDA, vacuole membrane permeabilization and subsequent cellular damage and ROS accumulation represent the lethal events in stressed yeast cells.
LMP is thought to be the lethal event in necrosis-like death of neurons that are stressed by expressing hyperactive variants of ion channels (4–6). Similar to the situation in yeast, LMP and subsequent cell death depended on V-ATPase activity. However, unlike the situation in yeast, death of degenerating neurons was not detectably altered by mutations or inhibitors of Cn (7), and it was significantly delayed if the neurons lacked proteases of the calpain or cathepsin families, which function upstream and downstream of LMP, respectively (4). The mutants of yeast lacking the sole calpain-like protease (Rim13) or one of the vacuolar proteases (Pep4, Prb1, Prc1, Cps1, Ybr139w, Lap4) or their cytoplasmic inhibitors (Pai3, Pbi2, Tfs1) all behaved like wild-type cells in our screening conditions. Previous studies showed the GFP-tagged Pep4 protein was released into the cytoplasm after exposure of yeast cells to hydrogen peroxide or acetic acid, although little or no lifespan extension was observed in pep4 knock-out mutants (56–58). Functional redundancy among the proteases and other hydrolases of the vacuole may have obscured their executioner roles in yeast cell death in all these circumstances. LMP contributes to necrotic cell death in other cell types as well, with variable utilization of autophagic and apoptotic processes depending on the extent and the types of hydrolytic enzymes released in each case (reviewed in Refs. 11, 59, and 60).
We found little or no role for autophagy in the rate or extent of cell death in yeast cells stressed with tunicamycin. Likewise, we confirm on a genome-wide level earlier studies that argue against a role for apoptosis in the death of tunicamycin-stressed cells (28). Earlier genome-wide studies of tunicamycin susceptibility in yeast have relied on growth assays at limiting drug concentrations (61, 62) or viability assays after brief drug exposures (63). Those screens revealed unfolded protein response-deficient mutants (ire1, hac1) as having extreme phenotypes with HACS- and Cn-deficient mutants having little or no phenotype. Conversely, HACS- and Cn-deficient mutants exhibited high rates of cell death following tunicamycin exposure in our screens, whereas the unfolded protein response-deficient mutants behaved like wild-type cells (Fig. 2). The distinct outcomes of these screens highlights the fact that growth assays, cell viability assays, and cell death assays correlate poorly with one another. Unfortunately, these studies and many others frequently conflate poor growth and decreased viability with cell death, thereby confounding the field of research on mechanisms cell death in yeast. The simplest interpretation of the findings is that unfolded protein response signaling promotes repair and adaptation processes to cope with the damage caused by limited tunicamycin exposure, whereas having little or no role in regulating cell death in yeast. Conversely, HACS and Cn can be viewed simply as factors that prevent cell death during unlimited tunicamycin exposure.
The present studies also demonstrate that cycloheximide can suppress the death of Cn-deficient cells even in the small population of survivors that remain after several hours of tunicamycin exposures. The inhibition of new protein synthesis may eliminate the stress by diminishing consumption of N-glycan precursors whose synthesis is blocked by tunicamycin (64). Alternatively, cycloheximide may block the production of specific enzymes or molecules that end up permeabilizing the vacuolar membrane and killing the cell. Precisely how Cn prevents the death of yeast cells with particular types of membrane stress remains an open question. Similarly, it is not yet clear which protein kinase(s) oppose the effects of Cn and whether their activities become stimulated by these membrane stresses. At present, there has been no demonstration that any of the effective death-promoting factors (V-ATPase, translation, amino acid biosynthetic enzymes, Hsp90 (28), etc.) respond to the death-inducing stimuli or to Cn, and thus, we cannot yet confirm the existence of a pro-death signaling pathway in these conditions.
HACS, Cn, and Cmk2 become activated and suppress the death of yeast cells responding to their own natural mating pheromones (16, 22), and this branched death-inhibiting pathway is at least partially conserved in C. albicans (25). As VMP seems to precede the death of these cells (16), the molecular mechanisms that promote cell death in these conditions may be similar to those that promote the death of tunicamycin-exposed yeast cells. In the hyphae-forming filamentous fungi, a vacuole-related cell death can occur when genetically dissimilar individuals of the same species fuse their hyphal cells in a mating-like process (13). This phenomenon of self-incompatibility is thought to limit the spread of viruses, prions, and other parasitic elements and therefore meets most criteria of a pro-death signaling pathway. It is not clear whether calcineurin or other components of the calcium signaling pathway suppress the death of self-compatible hyphal fusions or whether self-incompatibility relates in any mechanistic way to the pheromone- or tunicamycin-induced cell deaths in yeast. Nevertheless, changes in vacuolar membrane permeability in fungi and the necrosis-like death of degenerating neurons may be modern manifestations of a mechanism that arose in a common ancestor to help control cell death and survival in populations.
Supplementary Material
Acknowledgment
We thank Drew Dudgeon for help with optimization and implementation of the genetic screens.
This work was supported, in whole or in part, by National Institutes of Health Research Awards GM053082, NS057023, and NS074072 (to K. W. C.).

This article contains supplemental Tables 1 and 2.
- PCD
- programmed cell death
- Cn
- calcineurin
- LMP
- lysosomal membrane permeabilization
- VMP
- vacuolar membrane permeabilization
- V-ATPase
- vacuolar H+-ATPase
- HACS
- high-affinity calcium influx system
- ROS
- reactive oxygen species
- ER
- endoplasmic reticulum
- DCFDA
- dichlorofluorescein diacetate
- PI
- propidium iodide
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1. Kroemer G., Galluzzi L., Vandenabeele P., Abrams J., Alnemri E. S., Baehrecke E. H., Blagosklonny M. V., El-Deiry W. S., Golstein P., Green D. R., Hengartner M., Knight R. A., Kumar S., Lipton S. A., Malorni W., Nuñez G., Peter M. E., Tschopp J., Yuan J., Piacentini M., Zhivotovsky B., Melino G. (2009) Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 16, 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Golstein P., Kroemer G. (2007) Cell death by necrosis: toward a molecular definition. Trends Biochem. Sci. 32, 37–43 [DOI] [PubMed] [Google Scholar]
- 3. Driscoll M., Gerstbrein B. (2003) Dying for a cause: invertebrate genetics takes on human neurodegeneration. Nat. Rev. Genet. 4, 181–194 [DOI] [PubMed] [Google Scholar]
- 4. Syntichaki P., Xu K., Driscoll M., Tavernarakis N. (2002) Specific aspartyl and calpain proteases are required for neurodegeneration in C. elegans. Nature 419, 939–944 [DOI] [PubMed] [Google Scholar]
- 5. Syntichaki P., Samara C., Tavernarakis N. (2005) The vacuolar H+-ATPase mediates intracellular acidification required for neurodegeneration in C. elegans. Curr. Biol. 15, 1249–1254 [DOI] [PubMed] [Google Scholar]
- 6. Artal-Sanz M., Samara C., Syntichaki P., Tavernarakis N. (2006) Lysosomal biogenesis and function are critical for necrotic cell death in Caenorhabditis elegans. J. Cell Biol. 173, 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xu K., Tavernarakis N., Driscoll M. (2001) Necrotic cell death in C. elegans requires the function of calreticulin and regulators of Ca2+ release from the endoplasmic reticulum. Neuron 31, 957–971 [DOI] [PubMed] [Google Scholar]
- 8. Bianchi L., Gerstbrein B., Frøkjaer-Jensen C., Royal D. C., Mukherjee G., Royal M. A., Xue J., Schafer W. R., Driscoll M. (2004) The neurotoxic MEC-4(d) DEG/ENaC sodium channel conducts calcium: implications for necrosis initiation. Nat. Neurosci. 7, 1337–1344 [DOI] [PubMed] [Google Scholar]
- 9. Samara C., Syntichaki P., Tavernarakis N. (2008) Autophagy is required for necrotic cell death in Caenorhabditis elegans. Cell Death Differ. 15, 105–112 [DOI] [PubMed] [Google Scholar]
- 10. Troulinaki K., Tavernarakis N. (2012) Endocytosis and intracellular trafficking contribute to necrotic neurodegeneration in C. elegans. EMBO J. 31, 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boya P., Kroemer G. (2008) Lysosomal membrane permeabilization in cell death. Oncogene 27, 6434–6451 [DOI] [PubMed] [Google Scholar]
- 12. van Doorn W. G. (2011) Classes of programmed cell death in plants, compared with those in animals. J. Exp. Bot. 62, 4749–4761 [DOI] [PubMed] [Google Scholar]
- 13. Glass N. L., Dementhon K. (2006) Non-self-recognition and programmed cell death in filamentous fungi. Curr. Opin Microbiol. 9, 553–558 [DOI] [PubMed] [Google Scholar]
- 14. Dohlman H. G., Thorner J. W. (2001) Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70, 703–754 [DOI] [PubMed] [Google Scholar]
- 15. Severin F. F., Hyman A. A. (2002) Pheromone induces programmed cell death in S. cerevisiae. Curr. Biol. 12, R233–235 [DOI] [PubMed] [Google Scholar]
- 16. Zhang N. N., Dudgeon D. D., Paliwal S., Levchenko A., Grote E., Cunningham K. W. (2006) Multiple signaling pathways regulate yeast cell death during the response to mating pheromones. Mol. Biol. Cell 17, 3409–3422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iida H., Yagawa Y., Anraku Y. (1990) Essential role for induced Ca2+ influx followed by [Ca2+]i rise in maintaining viability of yeast cells late in the mating pheromone response pathway: a study of [Ca2+]i in single Saccharomyces cerevisiae cells with imaging of fura-2. J. Biol. Chem. 265, 13391–13399 [PubMed] [Google Scholar]
- 18. Cyert M. S., Kunisawa R., Kaim D., Thorner J. (1991) Yeast has homologs (CNA1 and CNA2 gene products) of mammalian calcineurin, a calmodulin-regulated phosphoprotein phosphatase. Proc. Natl. Acad. Sci. U.S.A. 88, 7376–7380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cyert M. S., Thorner J. (1992) Regulatory subunit (CNB1 gene product) of yeast Ca2+/calmodulin-dependent phosphoprotein phosphatases is required for adaptation to pheromone. Mol. Cell. Biol. 12, 3460–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Foor F., Parent S. A., Morin N., Dahl A. M., Ramadan N., Chrebet G., Bostian K. A., Nielsen J. B. (1992) Calcineurin mediates inhibition by FK506 and cyclosporin of recovery from α-factor arrest in yeast. Nature 360, 682–684 [DOI] [PubMed] [Google Scholar]
- 21. Iida H., Nakamura H., Ono T., Okumura M. S., Anraku Y. (1994) MID1, a novel Saccharomyces cerevisiae gene encoding a plasma membrane protein, is required for Ca2+ influx and mating. Mol. Cell. Biol. 14, 8259–8271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moser M. J., Geiser J. R., Davis T. N. (1996) Ca2+-calmodulin promotes survival of pheromone-induced growth arrest by activation of calcineurin and Ca2+-calmodulin-dependent protein kinase. Mol. Cell. Biol. 16, 4824–4831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fischer M., Schnell N., Chattaway J., Davies P., Dixon G., Sanders D. (1997) The Saccharomyces cerevisiae CCH1 gene is involved in calcium influx and mating. FEBS Lett. 419, 259–262 [DOI] [PubMed] [Google Scholar]
- 24. Paidhungat M., Garrett S. (1997) A homolog of mammalian, voltage-gated calcium channels mediates yeast pheromone-stimulated Ca2+ uptake and exacerbates the cdc1(Ts) growth defect. Mol. Cell. Biol. 17, 6339–6347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alby K., Schaefer D., Sherwood R. K., Jones S. K., Jr., Bennett R. J. (2010) Identification of a cell death pathway in Candida albicans during the response to pheromone. Eukaryot Cell 9, 1690–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Steinbach W. J., Reedy J. L., Cramer R. A., Jr., Perfect J. R., Heitman J. (2007) Harnessing calcineurin as a novel anti-infective agent against invasive fungal infections. Nat. Rev. Microbiol. 5, 418–430 [DOI] [PubMed] [Google Scholar]
- 27. Bonilla M., Nastase K. K., Cunningham K. W. (2002) Essential role of calcineurin in response to endoplasmic reticulum stress. EMBO J. 21, 2343–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dudgeon D. D., Zhang N., Ositelu O. O., Kim H., Cunningham K. W. (2008) Nonapoptotic death of Saccharomyces cerevisiae cells that is stimulated by Hsp90 and inhibited by calcineurin and Cmk2 in response to endoplasmic reticulum stresses. Eukaryot. Cell 7, 2037–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hauptmann P., Riel C., Kunz-Schughart L. A., Fröhlich K. U., Madeo F., Lehle L. (2006) Defects in N-glycosylation induce apoptosis in yeast. Mol. Microbiol. 59, 765–778 [DOI] [PubMed] [Google Scholar]
- 30. Shacka J. J., Klocke B. J., Shibata M., Uchiyama Y., Datta G., Schmidt R. E., Roth K. A. (2006) Bafilomycin A1 inhibits chloroquine-induced death of cerebellar granule neurons. Mol. Pharmacol. 69, 1125–1136 [DOI] [PubMed] [Google Scholar]
- 31. Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., Arkin A. P., Astromoff A., El-Bakkoury M., Bangham R., Benito R., Brachat S., Campanaro S., Curtiss M., Davis K., Deutschbauer A., Entian K. D., Flaherty P., Foury F., Garfinkel D. J., Gerstein M., Gotte D., Güldener U., Hegemann J. H., Hempel S., Herman Z., Jaramillo D. F., Kelly D. E., Kelly S. L., Kötter P., LaBonte D., Lamb D. C., Lan N., Liang H., Liao H., Liu L., Luo C., Lussier M., Mao R., Menard P., Ooi S. L., Revuelta J. L., Roberts C. J., Rose M., Ross-Macdonald P., Scherens B., Schimmack G., Shafer B., Shoemaker D. D., Sookhai-Mahadeo S., Storms R. K., Strathern J. N., Valle G., Voet M., Volckaert G., Wang C. Y., Ward T. R., Wilhelmy J., Winzeler E. A., Yang Y., Yen G., Youngman E., Yu K., Bussey H., Boeke J. D., Snyder M., Philippsen P., Davis R. W., Johnston M. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 [DOI] [PubMed] [Google Scholar]
- 32. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 33. Sherman F., Hicks J. B., Fink G. R. (1986) Methods in Yeast Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 34. Saldanha A. J. (2004) Java Treeview: extensible visualization of microarray data. Bioinformatics 20, 3246–3248 [DOI] [PubMed] [Google Scholar]
- 35. Cunningham K. W., Fink G. R. (1996) Calcineurin inhibits VCX1-dependent H+/Ca2+ exchange and induces Ca2+ ATPases in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 2226–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bonilla M., Cunningham K. W. (2003) Mitogen-activated protein kinase stimulation of Ca2+ signaling is required for survival of endoplasmic reticulum stress in yeast. Mol. Biol. Cell 14, 4296–4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eraso P., Mazón M. J., Portillo F. (2006) Yeast protein kinase Ptk2 localizes at the plasma membrane and phosphorylates in vitro the C-terminal peptide of the H+-ATPase. Biochim. Biophys. Acta 1758, 164–170 [DOI] [PubMed] [Google Scholar]
- 38. Hogan P. G., Li H. (2005) Calcineurin. Curr. Biol. 15, R442–443 [DOI] [PubMed] [Google Scholar]
- 39. Martin D. C., Kim H., Mackin N. A., Maldonado-Báez L., Evangelista C. C., Jr., Beaudry V. G., Dudgeon D. D., Naiman D. Q., Erdman S. E., Cunningham K. W. (2011) New regulators of a high-affinity Ca2+ influx system revealed through a genome-wide screen in yeast. J. Biol. Chem. 286, 10744–10754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Breitkreutz A., Choi H., Sharom J. R., Boucher L., Neduva V., Larsen B., Lin Z. Y., Breitkreutz B. J., Stark C., Liu G., Ahn J., Dewar-Darch D., Reguly T., Tang X., Almeida R., Qin Z. S., Pawson T., Gingras A. C., Nesvizhskii A. I., Tyers M. (2010) A global protein kinase and phosphatase interaction network in yeast. Science 328, 1043–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gagny B., Wiederkehr A., Dumoulin P., Winsor B., Riezman H., Haguenauer-Tsapis R. (2000) A novel EH domain protein of Saccharomyces cerevisiae, Ede1p, involved in endocytosis. J. Cell Sci. 113, 3309–3319 [DOI] [PubMed] [Google Scholar]
- 42. Reider A., Wendland B. (2011) Endocytic adaptors: social networking at the plasma membrane. J. Cell Sci. 124, 1613–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nordmann M., Cabrera M., Perz A., Bröcker C., Ostrowicz C., Engelbrecht-Vandré S., Ungermann C. (2010) The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr. Biol. 20, 1654–1659 [DOI] [PubMed] [Google Scholar]
- 44. Russnak R., Konczal D., McIntire S. L. (2001) A family of yeast proteins mediating bidirectional vacuolar amino acid transport. J. Biol. Chem. 276, 23849–23857 [DOI] [PubMed] [Google Scholar]
- 45. Hillenmeyer M. E., Fung E., Wildenhain J., Pierce S. E., Hoon S., Lee W., Proctor M., St Onge R. P., Tyers M., Koller D., Altman R. B., Davis R. W., Nislow C., Giaever G. (2008) The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320, 362–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Burd C. G., Emr S. D. (1998) Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol. Cell 2, 157–162 [DOI] [PubMed] [Google Scholar]
- 47. Kane P. M. (2005) Close-up and genomic views of the yeast vacuolar H+-ATPase. J. Bioenerg. Biomembr. 37, 399–403 [DOI] [PubMed] [Google Scholar]
- 48. Plant P. J., Manolson M. F., Grinstein S., Demaurex N. (1999) Alternative mechanisms of vacuolar acidification in H+-ATPase-deficient yeast. J. Biol. Chem. 274, 37270–37279 [DOI] [PubMed] [Google Scholar]
- 49. Baars T. L., Petri S., Peters C., Mayer A. (2007) Role of the V-ATPase in regulation of the vacuolar fission-fusion equilibrium. Mol. Biol. Cell 18, 3873–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Preston R. A., Murphy R. F., Jones E. W. (1989) Assay of vacuolar pH in yeast and identification of acidification-defective mutants. Proc. Natl. Acad. Sci. U.S.A. 86, 7027–7031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fasolo J., Sboner A., Sun M. G., Yu H., Chen R., Sharon D., Kim P. M., Gerstein M., Snyder M. (2011) Diverse protein kinase interactions identified by protein microarrays reveal novel connections between cellular processes. Genes Dev. 25, 767–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schu P. V., Takegawa K., Fry M. J., Stack J. H., Waterfield M. D., Emr S. D. (1993) Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science 260, 88–91 [DOI] [PubMed] [Google Scholar]
- 53. Gary J. D., Wurmser A. E., Bonangelino C. J., Weisman L. S., Emr S. D. (1998) Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J. Cell Biol. 143, 65–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dove S. K., Piper R. C., McEwen R. K., Yu J. W., King M. C., Hughes D. C., Thuring J., Holmes A. B., Cooke F. T., Michell R. H., Parker P. J., Lemmon M. A. (2004) Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 23, 1922–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jakubowski W., Bartosz G. (2000) 2,7-Dichlorofluorescin oxidation and reactive oxygen species: what does it measure? Cell Biol. Int. 24, 757–760 [DOI] [PubMed] [Google Scholar]
- 56. Mason D. A., Shulga N., Undavai S., Ferrando-May E., Rexach M. F., Goldfarb D. S. (2005) Increased nuclear envelope permeability and Pep4p-dependent degradation of nucleoporins during hydrogen peroxide-induced cell death. FEMS Yeast Res. 5, 1237–1251 [DOI] [PubMed] [Google Scholar]
- 57. Gourlay C. W., Ayscough K. R. (2006) Actin-induced hyperactivation of the Ras signaling pathway leads to apoptosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 26, 6487–6501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pereira C., Chaves S., Alves S., Salin B., Camougrand N., Manon S., Sousa M. J., Côrte-Real M. (2010) Mitochondrial degradation in acetic acid-induced yeast apoptosis: the role of Pep4 and the ADP/ATP carrier. Mol. Microbiol. 76, 1398–1410 [DOI] [PubMed] [Google Scholar]
- 59. Blum E. S., Driscoll M., Shaham S. (2008) Noncanonical cell death programs in the nematode Caenorhabditis elegans. Cell Death Differ. 15, 1124–1131 [DOI] [PubMed] [Google Scholar]
- 60. Kirkegaard T., Jäättelä M. (2009) Lysosomal involvement in cell death and cancer. Biochim. Biophys. Acta 1793, 746–754 [DOI] [PubMed] [Google Scholar]
- 61. Parsons A. B., Brost R. L., Ding H., Li Z., Zhang C., Sheikh B., Brown G. W., Kane P. M., Hughes T. R., Boone C. (2004) Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 22, 62–69 [DOI] [PubMed] [Google Scholar]
- 62. Tan S. X., Teo M., Lam Y. T., Dawes I. W., Perrone G. G. (2009) Cu,Zn superoxide dismutase and NADP(H) homeostasis are required for tolerance of endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol. Biol. Cell 20, 1493–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen Y., Feldman D. E., Deng C., Brown J. A., De Giacomo A. F., Gaw A. F., Shi G., Le Q. T., Brown J. M., Koong A. C. (2005) Identification of mitogen-activated protein kinase signaling pathways that confer resistance to endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol. Cancer Res. 3, 669–677 [DOI] [PubMed] [Google Scholar]
- 64. Lehle L., Tanner W. (1976) The specific site of tunicamycin inhibition in the formation of dolichol-bound N-acetylglucosamine derivatives. FEBS Lett. 72, 167–170 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.