Background: Trypanosoma cruzi regulates gene expression by means of post-transcriptional mechanisms.
Results: A 43-nt U-rich element was found in the 3′-UTR of a large number of mRNAs that are more abundant in intracellular amastigotes.
Conclusion: The 43-nt U-rich element might be involved in the modulation of abundance and/or translation of transcripts in amastigotes.
Significance: Results suggest the existence of stage-specific RNA regulons in T. cruzi.
Keywords: Gene Expression, Parasite, RNA Abundance, RNA-binding Protein, Trypanosoma brucei, Trypanosome
Abstract
Trypanosoma cruzi, the agent of Chagas disease, does not seem to control gene expression through regulation of transcription initiation and makes use of post-transcriptional mechanisms. We report here a 43-nt U-rich RNA element located in the 3′-untranslated region (3′-UTR) of a large number of T. cruzi mRNAs that is important for mRNA abundance in the intracellular amastigote stage of the parasite. Whole genome scan analysis, differential display RT-PCR, Northern blot, and RT-PCR analyses were used to determine the transcript levels of more than 900 U-rich-containing mRNAs of large gene families as well as single and low copy number genes. Our results indicate that the 43-nt U-rich mRNA element is preferentially present in amastigotes. The cis-element of a protein kinase 3′-UTR but not its mutated version promoted the expression of the green fluorescent protein reporter gene in amastigotes. The regulatory cis-element, but not its mutated version, was also shown to interact with the trypanosome-specific RNA-binding protein (RBP) TcUBP1 but not with other related RBPs. Co-immunoprecipitation experiments of TcUBP1-containing ribonucleoprotein complexes formed in vivo validated the interaction with representative endogenous RNAs having the element. These results suggest that this 43-nt U-rich element together with other yet unidentified sequences might be involved in the modulation of abundance and/or translation of subsets of transcripts in the amastigote stage.
Introduction
Trypanosoma cruzi, the agent of Chagas disease, possesses a complex life cycle involving several morphological and functionally different stages that are adapted to diverse environments in the insect vector and the mammalian host. These adaptations require up-regulation and down-regulation of multiple genes (1). Current knowledge suggests that trypanosomatids lack precise transcriptional control because no classical promoters have been identified (2–4). Given this peculiarity, transcription is polycistronic, and genes coding for proteins with unrelated functions are transcribed in large polycistrons (2–4). These polycistronic units are co-transcriptionally processed by trans-splicing and polyadenylation to produce mature monocistronic transcripts. Regulation of gene expression is mainly at the post-transcriptional level, and in T. cruzi it has been proposed to occur through pre-mRNA processing, RNA degradation (5, 6), or translational repression (7–9).
Both 5′- and 3′-untranslated regions (UTRs) can be involved in stabilization/destabilization mechanisms, up-regulating and down-regulating mRNA levels in a developmentally regulated manner. Some cis-elements and trans-acting factors controlling mRNA stability have been characterized in trypanosomatids (10). In most cases the association of RNA-binding proteins (RBPs)2 with cis-elements located in the 3′-UTRs of regulated transcripts leads to changes in stability and/or translation of target mRNA (for review, see Ref. 11). A case in point is the AU-rich (ARE) and G-rich elements identified in the 3′-UTR of the small mucin gene (SMUG) mRNA in T. cruzi. These elements were shown to confer stage-specific transcript stability or instability, respectively. Particularly, AREs are recognized by specific trans-acting factors such as the RBP that binds U-rich sequences 1 (TcUBP1) and -2 (TcUBP2). These two RBPs together with poly (A)-binding protein (TcPABP) form a ribonucleoprotein complex that is developmentally regulated and plays a stabilizing effect on SMUG mRNA in epimastigotes (for review, see Ref. 12).
It has been reported that multiple mRNAs can be co-regulated by one or more sequence-specific RBPs that orchestrate their splicing, export, stability, localization, and translation (13–16). The existence of these post-transcriptional regulons would be a very appropriate mechanism for the developmental regulation of gene expression in trypanosomatids. However, searches for shared motifs in clusters of co-regulated genes in trypanosomatids have met with limited success (17). In this regard, a common mechanism of stage-regulated expression of at least 85 genes mediated by a conserved 3′-UTR 450-nt cis-element has been proposed in Leishmania major (18).
The most clinically relevant stages of T. cruzi, the intracellular and replicative amastigotes, have radical changes in their protein expression pattern, but how this is achieved is completely unknown at present. These stages are difficult to study because of their intracellular habitat and the need to genetically manipulate the parasites in their epimastigote form, differentiate them into invasive trypomastigotes, and then infect tissue culture cells to obtain amastigotes.
We report here the identification of a 43-nt cis-element in the 3′-UTR of numerous mRNAs that appear to be up-regulated in intracellular amastigotes, as demonstrated by differential display RT-PCR, Northern blot, and RT-PCR analyses. The list of developmentally regulated transcripts includes more than 900 mRNAs of large gene families (trans-sialidase (TS), mucin-associated protein (MASP), mucin, surface protease GP63, and protein kinase) as well as single or low copy number genes. This cis-element, but not its mutated version, was shown to interact with the trypanosome-specific factor TcUBP1. These results suggest that messenger ribonucleoprotein (mRNP) complexes containing this 43-nt RNA element could act as a post-transcriptional regulon in the amastigote stages of the parasite.
EXPERIMENTAL PROCEDURES
Cell Culture
T. cruzi amastigotes and trypomastigotes, Y strain, were obtained from the culture medium of L6E9 myoblasts by a modification of the method of Schmatz and Murray (19) as we have described before (20). The contamination of trypomastigotes with amastigotes and intermediate forms or of amastigotes with trypomastigotes or intermediate forms was always less than 5%. T. cruzi epimastigotes (Y strain) were grown at 28 °C in liver infusion tryptose medium (LIT) (21) supplemented with 5% newborn calf serum unless indicated. The epimastigotes transformed with pTREX constructs were maintained in LIT medium supplemented with 5% heat-inactivated fetal bovine serum and 250 μg/ml Geneticin (G418). Epimastigotes were differentiated into intermediate forms or metacyclic trypomastigotes and isolated using a complement selection procedure (22). Trypomastigotes and amastigotes were later obtained from infected cell cultures (20).
Chemicals
Fetal bovine serum, newborn calf serum, Dulbecco's phosphate-buffered saline (PBS), 4′, 6-diamidino-2-phenylindole (DAPI), paraformaldehyde, bovine serum albumin, actinomycin D, adipic acid dihydrazide-agarose beads, and TRI® reagent were purchased from Sigma. Restriction enzymes, T4 DNA ligase, and goat serum were from New England BioLabs. pCR2.1-TOPO cloning kit, superscript reverse transcriptase, antibodies against GFP, Alexa Fluor 488-conjugated goat anti-mouse secondary antibodies, and 1Kb plus DNA ladder were from Invitrogen. RNeasy kit and PCR clean up columns were from Qiagen. Hybond-N nylon membrane and [32P]dCTP (3000 mCi/mmol) were obtained from PerkinElmer Life Sciences. Taq polymerase was purchased from Denville Scientific Inc. RQ1 RNase-free DNase I and T7 RNA polymerase were from Promega. All other reagents were analytical grade. The oligonucleotides were ordered from Sigma or IDT (Coralville, IA). The oligonucleotides used in this study are listed in supplemental Table S1.
Bioinformatic Analysis
Use of the motif discovery program MEME (Multiple Em for Motif Elicitation) (23) revealed the presence of a 43-nt element in ∼60 T. cruzi genomic sequences that had been discovered via BLASTN analyses to share regions of similarity. The parameters used in the MEME analysis were “-dna -mod zoops -nmotifs 1 -minw 3 -maxw 50 -evt 1e-5.” Subsequent motif discovery was implemented on the entire T. cruzi genome (CL Brener) using the MAST (Motif Alignment and Search Tool) program (24). The parameters for MAST were “-mt 1e−10 -comp -text.” The T. cruzi genome sequence was from TriTrypDB v2.1 (52).
Total RNA Extraction, Differential Display RT-PCR (DD-RT-PCR), and RT-PCR
Total RNA was isolated from different stages of T. cruzi using the TRI® reagent by following the manufacturer's instructions. The extracted total RNAs were further treated with RQ1 RNase-free DNase I for 30 min at 37 °C to remove genomic DNA contamination. The purified total RNAs were cleaned up by RNeasy kit and used as reverse transcription template in the presence of trace amount of [32P]dCTP. The cDNA was further purified by PCR clean-up columns and quantified by liquid scintillation counting. Equal amount of cDNAs from the three stages were used for differential display RT-PCR. The DD-RT-PCR was performed in a 50-μl volume with 1× PCR buffer, 1.2 mm dNTPs, 0.5 μm spliced leader (SL) RNA primer, 0.5 μm reverse primer and 2 units of Taq polymerase. The differential display PCR was first carried out at low annealing temperature (94 °C for 30 s, 38 °C for 30 s, and 72 °C for 1 min) for 3 cycles and then changed to high stringent PCR conditions (94 °C for 30 s, 56 °C for 30 s, and 72 °C for 1 min) for another 20 cycles. Trace amounts of [32P]dCTP were added so the PCR products could be separated and visualized by radioautography. The DD-RT-PCR products from amastigote stages were gel-purified and cloned into the TA-cloning pCR 2.1-TOPO vector. All other RT-PCR were performed in a PTC-100 Programmable Thermal Controller (MJ Research, Inc., Atertown, MA) at 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 1 min/cycle (25 cycles) using Taq polymerase. Sequencing of the cloned product was done by Yale DNA analysis facility.
Construction of Reporter Gene with Wild Type and Mutant Cis-element
The plasmid pTREX-1-eGFP-tTS-3′-UTR in which the 3′-UTR of a trypomastigote TS (tTS) gene was cloned downstream the eGFP reporter gene was a gift from Oscar Campetella (National University of General San Martin, Buenos Aires, Argentina) and was used as control (25). The tTS 3′-UTR was removed by HindIII and XhoI digestion. The entire 3′-UTR of a protein kinase gene (Tc00.1047053507881.60) plus a fragment of the 5′-end-coding sequence of the contiguous gene Tc00.10475035007881.30 was amplified by PCR (supplemental Table S1, primers 7 and 8) and cloned into HindIII and XhoI sites of pTREX-GFP to generate pTREX-GFP-WT-PK-3′-UTR. The 3′-UTR has the wild type U-rich element (5′-TTATTTTGATTATTGTTTAAATTTATTTTTATTTTTATTTT-3′). A 3′-UTR containing a mutated cis-element (5′-TTATGGTGCTTGTTGCAGCGGCCGCTCTTGCTTCC-3′) by removing most T residues from the element was generated by overlapping PCR (see supplemental Table S1 for overlapping primers 9 and 10) and was also cloned into the HindIII and XhoI sites of pTREX-GFP to generate pTREX-GFP-MT-PK-3′-UTR.
Northern Blot Analyses
For the Northern blot analyses total RNA was isolated from different stages of T. cruzi using the TRI® reagent. RNA samples were subjected to electrophoreses in 1% agarose gels containing 2.2 m formaldehyde, 20 mm Mops (pH 7.0), 1 mm EDTA, and 8 mm sodium acetate, transferred to nylon membranes, and hybridized with radiolabeled probes. The PCR primers used to generate the probes are listed in supplemental Table S1. DNA probes were labeled with [α-32P]dCTP using random hexanucleotide primers and the Klenow fragment of DNA polymerase I (Prime-a-Gene Labeling System). The rRNA gene was used as a loading control. The results of northern blots were quantified by using a phosphorimaging system.
Actinomycin D Treatment to Determine mRNA Half-life in Epimastigotes and Amastigotes
A total of 108 parasites in culture medium (LIT with 5% newborn calf serum for epimastigotes and DMEM supplemented with 20% fresh FBS for amastigotes) was incubated with 10-μg/ml actinomycin D for different time periods. After incubation for the appropriate time, cells were pelleted. Total RNA was harvested using the TRI® reagent and subjected to electrophoresis in 1% denaturing agarose. The gel was blotted to nylon and hybridized to probes described above. The membranes were washed and rehybridized with an rRNA probe as a loading control. The results of northern blots were quantified by using a phosphorimaging system.
Transfection of T. cruzi
T. cruzi epimastigotes growing at a density of 5 × 106–10 × 106 parasites/ml in LIT medium plus 5% newborn calf serum were harvested, washed once with PBS, and resuspended at a density of 108 parasites/ml in electroporation buffer (120 mm KCl, 0.15 mm CaCl2, 10 mm K2HPO4, 25 mm Hepes, 2 mm EDTA, 5 mm MgCl2, pH 7.6). Aliquots (0.4 ml) of cell suspensions were mixed with 50 μg of DNA on ice-cold 0.2-cm cuvettes and electroporated using a Bio-Rad gene pulser set at 0.3 kV and 500 microfarads, with 2 pulses (10 s between pulses). Parasites were recovered in 5 ml of LIT supplemented with 5% fetal bovine serum at 28 °C, and after 48 h in culture, Geneticin was added to a final concentration of 250 μg/ml.
Fluorescence Microscopy
T. cruzi epimastigotes (Y strain) were harvested by centrifugation and washed with PBS. The parasites were then resuspended in PBS, and the fluorescence of eGFP was directly recorded with an Olympus IX-71 inverted fluorescence microscope with a Photometrix-cooled CCD camera (CoolSnapHQ) driven by DeltaVision software from Applied Precision (Seattle, WA). Intracellular amastigotes were grown inside host cells plated on coverslips. The coverslips were washed with PBS and fixed with freshly prepared 4% paraformaldehyde (Electron Microscopy Sciences) in PBS for 1 h at room temperature. The trypomastigotes and extracellular amastigotes were collected and fixed with freshly prepared 4% paraformaldehyde in PBS for 1 h at room temperature, then adhered to poly-l-lysine-coated coverslips. For indirect immunofluorescence microscopy analysis, the cells were permeabilized with 0.3% Triton X-100 in PBS for 5 min. The cells were blocked with 3% bovine serum albumin (BSA) in PBS, 1% cold fish gelatin, 2% normal goat serum, and 50 mm ammonium chloride for 1 h at room temperature. After washing with PBS, the slides were incubated with a monoclonal antibody against GFP (1:300 dilution) in 1% BSA-PBS for 1 h at room temperature. After washing 5 times with PBS, the slides were incubated with Alexa Fluor 488-conjugated goat anti-mouse secondary antibody (1:400) diluted in 1% BSA-PBS plus 1 μg/ml of DAPI for 45 min in the dark. Differential interference contrast and fluorescence images were collected using the Olympus IX-71 inverted fluorescence microscope described above. DeltaVision software (sofWoRx) (Applied Precision, Seattle, WA) was used to deconvolve the images.
Dihydrazide-agarose RNA Cross-linking
pGEM-T Easy plasmid containing the 43-nt cis-element of the TS1 gene (Tc00.1047053509217.40) 5′-CTTTTACTTTTTTTGCGTATTTTAAATTTATTTTACTGTTTGC-3′ and a pGEM-T polylinker transcript without any insert (used as a negative control) were digested with SpeI for in vitro transcription with T7 RNA polymerase. The integrity of each RNA was verified in 1.5% agarose gels. RNAs were oxidized with NaIO4 and cross-linked to adipic acid dihydrazide-agarose beads as previously indicated (26). Trypanosome protein extracts from amastigote and epimastigote stages (5 × 108 cells) were incubated with RNA-cross-linked beads for 1 h at room temperature in binding buffer (80 mm KCl, 0.1 mm EDTA, 0.1 mm PMSF, 1 mm DTT, 10% glycerol, 5 mm MgCl2, 20 mm Tris-HCl, pH 7.6, and 10 units of RNase inhibitor) and washed. Elution was done with 1.5 m KCl, and samples were resolved by electrophoresis in SDS-PAGE gels. Proteins were detected by Western blot analysis using specific antibodies.
In Vitro RNA Binding Assay
The general procedure has been described previously (27). The 43-nt cis-element or the mutated version were in vitro transcribed from PCR products obtained with the following complementary primers: T7-WTPK4 and R-WTPK4 (for the 43-nt wild type) and T7-MTPK4 and R-MTPK4 (for the 43-nt mutant) (see the primer sequences in supplemental Table S1). The integrity of each RNA was verified in 2% agarose gels. Purified GST-tagged TcUBP1, TcRBP3, and GST alone were evaluated in this assay. cDNAs were PCR-amplified with F-Anchor oligonucleotide and reverse specific primers.
RNA Extraction from mRNP Complexes
The in vivo formaldehyde fixation of epimastigotes and immunoprecipitation (IP) assays were carried out as described (28). After the washing steps, RNA was extracted from IP material using TRI® reagent. RNA samples were resuspended in water and then used in RT-PCRs. PCRs were performed with the following primers: C1 and C2 covering a portion of the intercistronic region of TcUBP RNA, Amastin, protein kinase 4, putative (PK4), nuclear transport factor 2 (NTF2), short basic transcription factor, putative (BTF-3a), nucleolar RNA-binding protein, putative, kinetoplast DNA-associated protein, putative, GTP-binding protein, putative (GTP), fatty acid desaturase, putative (FAD), MASP, and RNA-binding protein, putative (RBP) (see supplemental Table S1). Immunoprecipitations using rabbit preimmune serum or TcPTB2 antibodies were performed as experimental controls.
RESULTS
Conserved Motif Is Widely Distributed in Genome of T. cruzi
When analyzing transcriptional changes in epimastigotes submitted to osmotic stress (29), we found a short 65-nt fragment downstream a TS open reading frame (ORF) (Tc00.1047053506909.100) that was not present in other TS genes analyzed: 5′-GGGAAAGAATGTTTTTGATACTTTTACTTTTTTGTGTGTATTTTAAATTTATTTTAATGTTTGCT-3′.
This motif is only ∼60 nt downstream of the annotated stop codon. Further BLASTN analyses indicated that a subset of TS genes has this short fragment downstream of their coding regions. Surprisingly, subsets of large gene families such as MASP, GP63, and mucin genes seem to have similar conserved sequences downstream of their ORF. Initial analysis of the BLASTN hits indicated that 43 nucleotides of the short fragment were well conserved: 5′-CCATCCTTTTTTGTGCGTATTTTAAATTTATTTTCCTGTTTGC-3′.
A comparison of the sequences obtained revealed that they all have a high degree of conservation. On average, the fragments share about 75% identity (supplemental Fig. S1). Sequence logo graphics show that all the sequences bear an AT-rich core sequence composed by 5′-TATTTTAAATTTATTTT (Fig. 1A). A whole genome scan on all available contigs was then performed to search for sequences similar to the 43-nt element. The scan results revealed that there were more than 900 copies of this sequence in the genome of T. cruzi when the cut off was set at e−10. The detailed results of this search can be found in supplemental Table S2. The element is widely distributed in the genome as shown in supplemental Fig. S2. When we randomly selected ∼300 of 900 hits (we were not able to use all sequences due to program constraints) and analyzed them by linear MEME, we identified a common conserved AT-rich core sequence. This core sequence is very rich in U residues if it is transcribed into RNA (Fig. 1B), and we named it U-rich element.
FIGURE 1.
Sequence logo representation of the cis-element. A, 43-nt motif present in some large gene family members. B, U-rich element.
Conserved U-rich Element Is in 3′-UTR of mRNAs
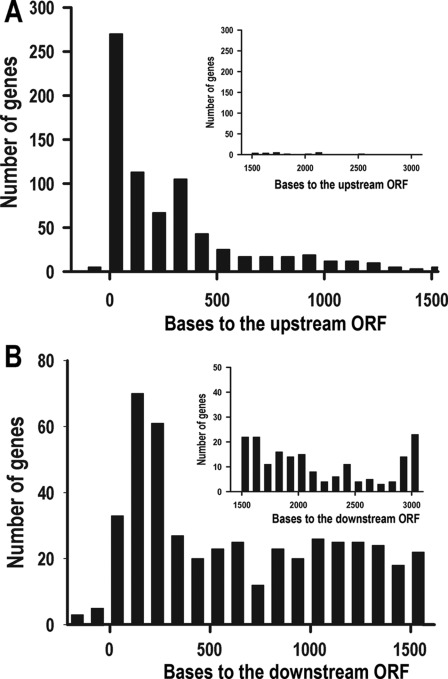
The whole genome scan indicated that the short element is predominantly localized in non-coding sequences (supplemental Table S2). The relative distance of this element to its upstream gene fits well with the typical 3′-UTR length (Fig. 2A), whereas its relative distance to its downstream gene (Fig. 2B) does not fit with the typical 5′-UTR length in T. cruzi (30), suggesting that the element is located in the 3′-UTR of a large number of genes. Other evidence supporting the 3′-UTR localization of the 43-nt cis-element is provided by the position of the element in MASP genes and the EST information available in the databases. It has been determined that the length of the majority of MASP 3′-UTRs is ∼400–500 nucleotides (31), whereas the distance of the cis-element to the MASP coding region of 62 of the 67 genes that have the cis-element is also within this range (TriTrypDB). Furthermore, although few ESTs are available of genes possessing the cis-element, those that have been identified (for example Tc00.1047053503543.20 and Tc00.1047053509965.260) have the cis-element in the 3′-UTR (TritrypDB).
FIGURE 2.
The cis-element is located in the 3′-UTR of a subset of genes. A, distance to the upstream gene is shown. Only 4.6% of the cis-elements are more than 1500 nucleotides away from their upstream genes (shown in the inset). B, shown is distance to the downstream gene. About 44% of the motifs are more than 1500 nucleotides away from their downstream genes (shown in the inset).
The cis-element is widely distributed in all chromosomes, but its distribution is not random. Only 169 of 1430 TS and 67 of 1377 MASP genes in the genome of T. cruzi (32) have this cis-element, and we detected it in only two genes belonging to the retrotransposon hot spot (RHS) protein family (752 genes in T. cruzi genome) and none in the dispersed gene family protein 1 (DGF-1) (565 genes) (32). More than 90% of the cis-elements found have the same orientation as the coding sequences. For the few cis-elements found with the opposite orientation, there is often another cis-element with the same orientation. Most of the genes have only one cis-element in their 3′-UTR, although we found a few genes harboring multiple copies (supplemental Table S2).
Cis-element Is Preferentially Present in 3′-UTR of mRNAs of Amastigote Stage
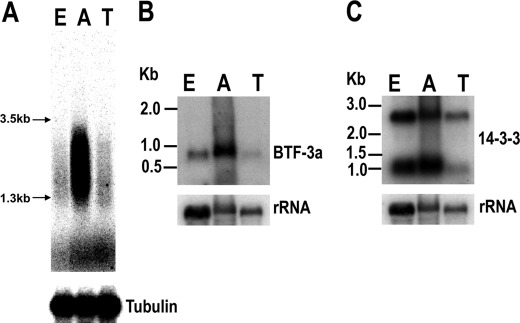
mRNA degradation/stabilization is one of the main mechanisms that controls gene expression in trypanosomatids, and cis-elements are known to regulate mRNA abundance in these parasites (10). We hypothesized that the cis-element that we identified could be involved in developmental regulation of gene expression in T. cruzi. To test this hypothesis, we took advantage of a strategy similar to differential display PCR (33) to examine whether this 43-nt U-rich element is preferentially expressed in certain developmental stages. Total RNA from different stages was reverse-transcribed, and DD-RT-PCR was performed. Because every mRNA in T. cruzi has the conserved 5′-end SL RNA sequence that constitutes the 5′-terminal exon of the mature mRNA, a specific primer against the SL RNA was used as the forward primer for DD-RT-PCR. The reverse primer had 14 nucleotides of the most conserved sequence in the cis-element plus 6 nucleotides in the 5′-end used for sequence amplification of the 43-nt element-containing mRNAs. PCR was first done for 3 cycles at low annealing temperature, then changed to stringent annealing conditions (56 °C) for the remaining 20 cycles. The results are shown in Fig. 3A. Clearly, there is more RT-PCR signal in the amastigote stages, suggesting that these U-rich element-containing mRNAs are more abundant in amastigotes than in epimastigotes and trypomastigotes (Fig. 3A). We sequenced five clones of the DD-RT-PCR products from amastigotes. Four clones were found in the whole genome scan list (two clones encoding for ribosomal protein S12, one clone encoding for a protein kinase, and one clone encoding for a 14-3-3 protein; supplemental Table S2). Northern blot analyses confirmed that a short basic transcription factor (BTF-3a, Tc00.1047053504017.10) (which was the one not present in the scan list) was preferentially expressed in amastigotes (Fig. 3B). Northern blot analysis of the 14-3-3 protein gene (Tc00.1047053508851.180) revealed 2 distinct mRNA bands (Fig. 3C). The bigger mRNA seems uniformly expressed in all stages, whereas the smaller mRNA (with stronger signal) is also preferentially expressed in amastigotes. These results suggest that paralogs may have completely different stage-specific patterns of expression.
FIGURE 3.
U-rich element-containing mRNAs are more abundant in amastigotes. A, differential display RT-PCR products were separated by 1% agarose gel electrophoresis and visualized by phosphorimaging scanning. RT-PCR of tubulin was also performed using the same sample as for differential display RT-PCR. B and C, Northern blot analyses of the mRNA levels of BTF-3a (B) and 14-3-3 protein (C) genes in three different stages of T cruzi are shown. The membranes hybridized to BTF-3a and 14-3-3 were stripped and hybridized to the rRNA probe as a loading control. E, epimastigotes; A, amastigotes; T, trypomastigotes.
Bioinformatic Analysis Suggests That Genes with U-rich Element Are Preferentially Expressed in Amastigotes
We performed a bioinformatics analysis of previous microarray (1) data available in TriTrypDB to investigate whether there was a stage-specific pattern of expression of genes encoding the U-rich element. It is important to note, however, that those microarray studies (1) were done using the Brazil strain of T. cruzi and that the amastigotes were obtained after in vitro differentiation from tissue culture trypomastigotes, whereas in our bioinformatics studies we used the CL strain (the strain used for the genome project), and the amastigotes of the Y strain were obtained from tissue cultures.
Because microarrays are based on the coding sequences of genes and most members of gene families (e.g. TS, MASP, etc) have similar coding sequences, we selected 294 genes that do not belong to these gene families, encode for the U-rich element, and for which there is transcriptome information available in TriTrypDB to study. A plot of the total scores of the 294 genes in the 4 developmental stages is shown in Fig. 4A. The plot shows that the genes encoding the U-rich element tend to have higher mRNA levels in the amastigote stage. We also randomly chose six single-copy or low-copy number genes from the whole genome scan list and did Northern blot analyses to determine the steady state mRNA levels in epimastigotes and tissue culture amastigotes and trypomastigotes (Fig. 4B). The relative steady state mRNA level of each gene was plotted in Fig. 4C. Fig. 4D shows the higher transcript abundance of 7 genes in amastigotes, as analyzed by Northern blot. Both the previous microarray study of the 294 genes (despite the limitations described above) and our Northern blot analyses suggest that the cis-element-containing genes tend to have higher transcript levels in amastigotes.
FIGURE 4.
Predominant expression of U-rich element-containing genes in amastigotes. A, a summary of microarray analysis of 294 genes with the U-rich element suggests the U-rich element-containing genes have relative higher mRNA levels in the amastigote stage. The relative mRNA levels in 4 developmental stages of these genes were obtained from TriTrypDB. We assigned the stage with the highest mRNA level with score 2, the 2nd highest mRNA stage with score 1, then scores −1 and −2 to the 2 stages with lower mRNA abundance. The total score of each stage were then plotted. The gene families with multiple members were filtered out for this study because it is not possible to assign a microarray spot to a specific gene ID. B, shown is a Northern blot analysis to detect steady state transcript levels of some single copy or low copy number genes. Each membrane was washed and reprobed with rRNA. Only one representative membrane probed with rRNA is shown here. C, the mRNA/rRNA ratios in the three stages were determined by phosphorimaging scan/quantification. The genes studied here are: NTF-2 (nuclear transport factor 2, putative (Tc00.1047053509567.40)); β-AP3 (β subunit of adaptor protein 3, putative (Tc00.1047053506673.60)); NRBP (nucleolar RNA-binding protein, putative (Tc00.1047053508277.230)); KDNA (kinetoplast DNA-associated protein, putative (Tc00.1047053511529.80)); GTP (GTP-binding protein, putative (Tc00.1047053509099.10)); CYC2 (CYC2-like cyclin, putative (Tc00.1047053507089.260)); D, a Northern blot analysis indicated the average steady state mRNA levels of seven single copy genes with the cis-element (BSF-3a and genes in C) in amastigote stage are significantly higher than that in epimastigote (p = 0.00276) and trypomastigote stages (p = 0.00369). The mean ± S.E. of the relative mRNA levels in three different stages are shown. ** = p < 0.01. E, epimastigotes; A, amastigotes; T, trypomastigotes; M, metacyclics. AU, arbitrary units.
Paralogs of Large Gene Families Containing 43-nt Cis-element in Their 3′-UTRs Are Preferentially Detected in Amastigotes
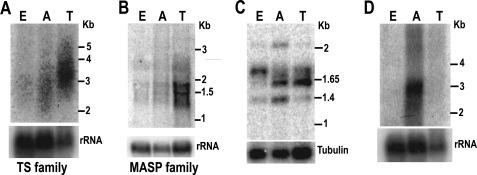
The TS gene family has ∼1500 members (32). Northern blot analyses indicated that the TS mRNAs are more abundant in the trypomastigote stage (Fig. 5A and Ref. 34). The MASP gene family has ∼1400 genes (32), and a recent study (31) showed that the highest MASP mRNA levels were also found in trypomastigotes. Fig. 5B shows similar results. We used two primers to amplify a subset of MASP genes with the cis-element in their 3′-UTR by RT-PCR. The forward primer was against SL RNA. The cis-element specific reverse primer (oligo 14 in supplemental Table S1) is 100% conserved in 7 MASP genes. 5 of the 7 MASP genes (Tc00.1047053507237.170, Tc00.1047053511609.30, Tc00.1047053511611.20, Tc00.104705350 6965.100, Tc00.1047053507237.100) have ORFs (969–978 bp) of similar size. The motif location is also very similar (∼340 nucleotides downstream the stop codon). The theoretical RT-PCR product sizes for these 5 genes should be around 1.38 kb (SL RNA + 5′-UTR + ORF + 3′-UTR = 30 + 50 + 970 + 340 = 1.38 kb). The PCR products were separated by agarose gel electrophoresis and hybridized to a radiolabeled MASP probe. As shown in Fig. 5C, there is a band with size close to 1.4 kb that hybridizes to the MASP probe. This band had the strongest intensity when the amastigote total RNA was used for RT-PCR. We purified the band and cloned it for sequencing. Sequencing results confirmed that it is indeed the RT-PCR product of the MASP genes Tc00.1047053511609.30 and Tc00.1047053511611.20. The RT-PCR was done under stringent PCR conditions for 20, 24, 28, and 32 cycles. Under all PCR conditions the 1.4 kb Southern blot analysis band was always stronger in amastigote stages than in the other 2 stages. There was no signal when the total RNA without reverse transcription was used as template (data not shown). There is also a strong 1.6-kb band mainly found in the trypomastigote stage. Cloning and sequencing results suggest that it is the MASP gene Tc00.1047053509613.80 with the ORF of 1287 bp. It was amplified because its 3′-UTR has 80% similarity with the reverse primer (but it is not in the scan list). The PCR/Southern blot analyses indicate that this MASP gene has the highest transcript level in trypomastigotes. Our results indicate that the MASP paralogs have different expression patterns. The subset of MASP paralogs with the U-rich element behaves differently than the majority of its family members. Their transcripts seem predominantly present in amastigote stages (Fig. 5C).
FIGURE 5.
Gene family transcripts containing the cis-element are more abundant in amastigotes. Northern blot analysis detects transcripts of TS gene family (A) and MASP gene family (B) in the three stages. The 5′-end of the coding sequence of a TS gene and a full-length MASP gene were used as probes. The membrane was also hybridized with a probe for the rRNA gene as a loading control. C, shown is RT-PCR amplification of a small subset of MASP mRNAs with the most conserved U-rich sequence. PCR was done under stringent conditions. The PCR products were then separated by agarose gel and hybridized with a MASP probe. RT-PCR of tubulin was done as a control for the amount of cDNA used in this assay. D, Northern blot analysis detects the steady state level of a protein kinase subfamily. The probe is for the 3′-UTR of 9 protein kinase genes. E, epimastigotes; A, amastigotes; T, trypomastigotes.
By whole genome scanning (supplemental Table S2) we found 21 members of the protein kinase gene family (of a total of 156 protein kinase genes annotated) (32) that have this U-rich element. At least nine of them seem to have well conserved 3′-UTRs, and therefore, it is possible to detect the mRNA levels of this subset of protein kinase genes by Northern blot analyses. A ∼300-bp-long probe derived from one of the protein kinase (Tc00.1047053507881.60) 3′-UTR was labeled with 32P and used for Northern blot analysis. The results confirmed that this subset of protein kinase genes also has the highest steady state mRNA levels in amastigote stages (Fig. 5D).
Deletion of Cis-element Results in Loss of Gene Expression in Amastigote Stages
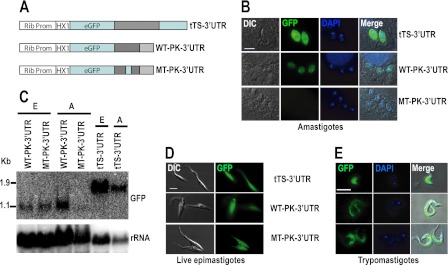
We asked whether the higher mRNA abundance of the cis-element-containing transcripts in amastigotes was due to the presence of the element in the 3′-UTRs. To test this possibility, we investigated the expression of the protein kinase 4 (Tc00.1047053507881.60) of known expression profile (Fig. 5D) and with a small 3′-UTR easy to manipulate in vitro. We fused the 3′-UTRs of this protein kinase mRNA downstream of a GFP reporter gene (Fig. 6A). The control plasmid pTREX-1-eGFP-tTS-3′-UTR, pTREX-1-eGFP-PK-WT-3′-UTR (with the wild type cis-element), and pTREX-1-eGFP-PK-MT-3′-UTR (with a mutated cis-element) were transfected into Y strain epimastigotes by electroporation. G418 was added 48 h later to select stable transfectants. The epimastigotes were differentiated in vitro and used to infect myoblasts. Northern blot analysis was used to detect the eGFP mRNA in these transfected cell lines. As shown in Fig. 6C, a Northern blot detected a similar level of the eGFP signal in epimastigotes transfected with both constructs, whereas in amastigotes, the eGFP signal is much stronger in parasites transfected with construct containing wild type PK4 3′-UTR than containing the mutant PK4 3′-UTR. Both wild type and mutant PK4 3′-UTR produce mature mRNAs with the same size, indicating that the mutation of this cis-element has no effect in mRNA maturation. This result suggests that this cis-element plays an important role in maintaining the steady state eGFP mRNA level in amastigote stages. We were unable to obtain a relatively pure population of trypomastigotes expressing either pTREX-1-eGFP-PK-WT-3′-UTR or pTREX-1-eGFP-PK-MT-3′-UTR as they rapidly differentiated into extracellular amastigotes and epimastigotes. The expression of the eGFP protein was further analyzed by live cell imaging or by imunofluorescence analyses. Expression of eGFP was detected in the cytosol of live epimastigotes (Fig. 6D). Immunofluorescence was performed to detect eGFP expression in amastigotes and trypomastigotes using an antibody against eGFP. As shown in Fig. 6B, mutation of the element resulted in loss of eGFP signal in amastigotes. The expression of eGFP in epimastigotes and trypomastigotes was not affected by the mutation of the cis-element (Fig. 6, D and E).
FIGURE 6.
Loss of reporter gene expression in amastigote stage after mutation of the regulatory cis-element. A, constructs of reporter genes with TS 3′-UTR, and WT and mutated (MT) protein kinase 3′-UTR are shown. B, immunofluorescence detects eGFP expression in the cytosol of amastigotes. DIC, differential interference contrast. C, Northern blot analysis detects steady state eGFP levels in epimastigote and amastigote stages transfected with different constructs. The probe was stripped and hybridized to rRNA probe as a loading control. D, live cell imaging examines eGFP expression in the cytosol of epimastigotes (left) and E, immunofluorescence to examine eGFP expression in the cytosol of trypomastigote stages. Bars in B, D, and E = 5 μm. The HX1 sequence is used in the expression vector pTREX to obtain a highly efficient pre-mRNA processing. E, epimastigotes; A, amastigotes.
Differential Stabilities of Genes Containing 3′-UTR Cis-element in Epimastigotes and Amastigotes
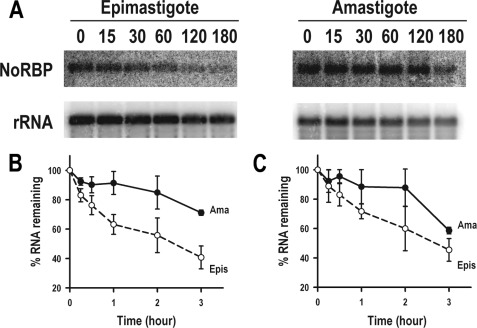
To investigate whether mRNA stability is responsible for the higher steady state level of the genes containing the 3′-UTR cis-element, the half-lives of NoRBP and NTF-2 mRNA derived from these two life cycle stages were determined by actinomycin D treatment. Epimastigotes and amastigotes were incubated in the presence of 10 μg/ml actinomycin D, and total RNA was harvested at various time points. The amount of NoRBP and NTF-2 mRNA was then determined by Northern blot analysis and quantitated by phosphorimaging analysis (Fig. 7A). The mRNA half-life in amastigotes was found to be ∼8 ± 1 h (n = 3) for NoRBP transcript (Fig. 7B) and ∼6.5 ± 1.5 h (n = 2) for NTF-2 mRNA (Fig. 7C). The decay rate in epimastigotes was much faster, with a half-life of only ∼2 ± 0.2 h and ∼2.5 ± 0.1 h for NoRBP and NTF-2 mRNA, respectively (Fig. 7, B and C). These results suggest that NoRBP and NTF-2 mRNA are either stabilized in amastigotes and/or destabilized in epimastigotes.
FIGURE 7.
NoRBP and NTF-2 decay in epimastigotes and amastigotes. A, total epimastigote (left panel) or amastigote RNA (right panel) was harvested at the indicated times (in minutes) after actinomycin D addition and separated as described under “Experimental Procedures.” The membrane was first hybridized to a probe against NoRBP coding region and then washed and reprobed with rRNA as a loading control. One representative experiment of three independent experiments is shown. B and C, shown is a graphic representation of NoRBP (B) and NTF-2 (C) mRNA half-life quantitation in amastigotes (Ama) and epimastigotes (Epis) from three and two independent experiments, respectively, as determined by phosphorimaging analysis. The percentage of mRNA remaining is plotted. NoRBP, nucleolar RNA-binding protein, putative (Tc00.1047053508277.230 (B)); NTF-2, nuclear transport factor 2, putative (Tc00.1047053509567.40 (C)).
Element Recognition by Trypanosome-specific RBP
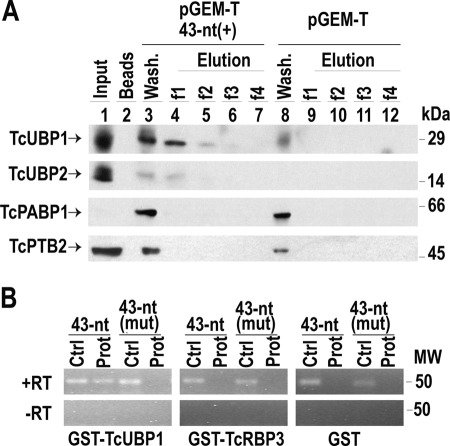
TcUBP1 is a small RNA recognition motif (RRM)-containing protein that has been reported to control mRNA levels of some genes (27). Given that the element under study has a U-rich composition and TcUBP1 preferentially recognize U-rich sequences in RNA-binding assays (27), we decided to examine how many of the previously identified mRNA targets for this RRM-type protein (28) harbor the 43-nt cis-element. To this end, we searched for the best motif for TcUBP1 binding, termed UBP1m, in the genes listed in supplemental Table S2. The data obtained revealed that the UBP1-motif was enriched in the group of 43-nt cis-element-containing genes, compared with the entire T. cruzi genome (χ2 test, p < 0.001). Therefore, a significant number of genes (218 of 905) have a co-occurrence of both elements in their 3′-UTRs (supplemental Table S3). Because binding of TcUBP1 can occur at distinct sites of a given 3′-UTR target sequence (28), our observations led us to investigate if this RRM protein can interact directly with the 43-nt RNA element. We therefore performed dihydrazide-agarose RNA cross-linking assays with this and other RRM-type proteins described in T. cruzi (35). For this purpose we transcribed a 43-nt element sequence inserted into a pGEM-T polylinker transcript (pGEM-T 43-nt(+)) and a pGEM-T polylinker transcript without any insert (used as a negative control). Parasite extracts were incubated separately with both probes, and the presence of proteins was tested by Western blot analysis. The results showed that TcUBP1 interacted with the transcript pGEM-T 43-nt(+) but failed to recognize the control transcript pGEM-T. Other RRM-containing proteins such as TcUBP2, polypyrimidine-tract binding protein 2 (TcPTB2), (Tc00.1047053511727.160) (36), or TcPABP1 (Tc001047053506885.70) failed to show binding to any mRNAs (Fig. 8A).
FIGURE 8.
RNA binding assay using an amastigote stage protein extract and purified recombinant RBPs. A, cytosolic cell-free extracts of amastigotes were incubated with dihydrazide-agarose beads cross-linked to the following transcripts: pGEM-T polylinker (pGEM-T) or the 43-nt cis-element (pGEM-T 43-nt(+)). After washing and elution with KCl, samples were resolved by electrophoresis in SDS-PAGE gels and analyzed by Western blot. Lane 1, input. Lane 2, beads alone. Lanes 3 and 8, washing step. Lanes 4–7, RNA pGEM-T 43-nt(+); lanes 9–12, negative control pGEM-T polylinker. RRM-type proteins are indicated by arrows. Molecular mass protein standards Dalton Mark VII-L are indicated at the right. B, in vitro transcripts encoding the 43-nt motif (43-nt) or the mutated version (43-nt (mut)) were incubated with recombinant GST-tagged TcUBP1, TcRBP3, or GST alone. After binding and washing, a RT-PCR was made with the primers indicated above the panels. PCR products were electrophoresed through a 3% agarose gel and stained with ethidium bromide. A reverse transcription experiment was performed with (+RT) or without (−RT) SuperScript II enzyme. Ctrl, positive PCR control; Prot, RNA-binding reaction. The systematic gene name for the RBPs are: TcUBP1, Tc00.1047053507093.220; TcUBP2, Tc00.1047053507093.229; TcRBP3, Tc00.1047053507093.250; TcPABP1, Tc001047053506885.70; TcPTB2, Tc00.1047053511727.160.
To further analyze the specificity of the interaction between the 43-nt element and TcUBP1, the following transcripts were in vitro transcribed and tested for binding with purified recombinant proteins; 1) RNA encoding the wild type cis-element (43-nt) and 2) the mutated version previously described in Fig. 6 (43-nt (mut)). Each RNA was incubated with recombinant GST-tagged TcUBP1 immobilized on a glutathione-agarose matrix. After washing, the eluted RNAs were reverse-transcribed, and the cDNA obtained was tested in PCRs with specific primers. As shown in Fig. 8B, TcUBP1 effectively bound to the 43-nt element but failed to show binding when the motif was mutated or substituted by a random sequence (Fig. 8B and data not shown). GST and GST-TcRBP3 were used as experimental controls and did not recognize any of the transcripts tested.
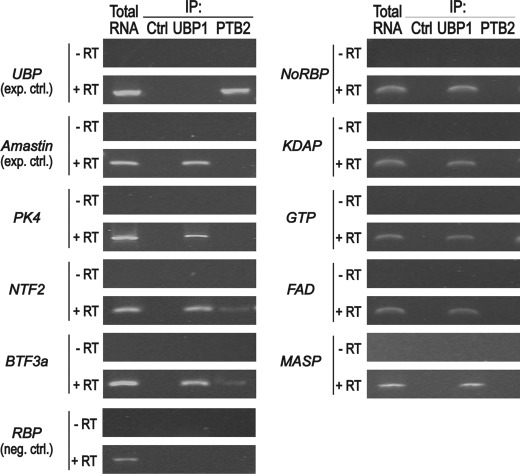
To test whether the 43-nt cis-element-containing mRNAs might indeed be able to interact with TcUBP1, we performed in vivo IP of the mRNP complexes with anti-TcUBP1 antibody. The association of targets with the RRM protein was then investigated by RNA extraction from purified mRNP complexes followed by RT-PCR. A preimmune serum was used as a negative control and did not immunoprecipitate visible amount of mRNAs. Additionally, a specificity control was done using polyclonal antibodies against the related RRM protein TcPTB2. The IP samples were analyzed for the presence of different transcripts: the abundant dicistronic TcUBP RNA (as a positive control of TcPTB2 IP (5, 6)), Amastin (a known target of TcUBP1 (27)), and the 43-nt-containing transcripts previously analyzed in this work. All the transcripts tested harboring the 43-nt cis-element were found in the pool of UBP1-bound RNAs, whereas no product was amplified in IP experiments performed with antibodies against PTB2 or control serum (Fig. 9). These findings suggest that TcUBP1 interacts with cellular mRNAs having the 43-nt cis-elements. Taken together these results demonstrate that TcUBP1 specifically recognizes the short cis-element identified.
FIGURE 9.
43-nt cis-element-containing cellular transcripts interact in vivo with TcUBP1. Agarose gels show the RT-PCR products of the indicated transcripts from total RNA coimmunoprecipitated in vivo with control rabbit serum (immunoprecipitation control (ctrl.)), anti-UBP1, or anti-PTB2 antibodies. For TcUBP cDNA synthesis, we used the internal specific primer NH2/AS-ubp1. For the remaining transcripts tested, the oligo(dT)18 was used (see supplemental Table S1). RT was performed with (+) or without (−) SuperScript II enzyme. RT, reverse transcriptase enzyme; UBP, dicistronic TcUBP RNA; PK4, protein kinase mRNA; NTF-2, nuclear transport factor 2, putative (Tc00.1047053509567.40); BTF-3a, basic transcription factor 3a (Tc00.10470535–4017.10); NoRBP, nucleolar RNA-binding protein, putative (Tc00.1047053508277.230); KDAP, kinetoplast DNA-associated protein, putative (Tc00.1047053511529.80); GTP, GTP-binding protein, putative (Tc00.1047053509099.10); FAD, fatty acid desaturase, putative (Tc00.1047053429257.20); MASP, mucin-associated protein (Tc00.104705351160 9.30); RBP, RNA-binding protein (Tc00.1047053506649.80) (negative control).
DISCUSSION
In this study we identified a 43-nt U-rich element in the 3′-UTR of more than 900 mRNAs of large gene families (TS, MASP, mucin, surface protease GP63, and protein kinase) as well as in single and low copy number genes that are predominantly detected in the intracellular amastigote stages of T. cruzi. There are potentially more genes containing this element in their 3′-UTR but not identified by this search because we set a cut-off value of e−10. For example, the nucleolar RNA-binding protein in Fig. 4B was not identified in the latest scan, but it was originally identified when we first searched the available T. cruzi contigs (we used a cutoff of e−8). Accordingly, a protein kinase 3′-UTR containing the 43-nt cis-element, but not its mutated version, promoted the expression of the GFP reporter gene in amastigotes. These results would indicate that in the absence of the 3′-UTR 43nt cis-element, this protein kinase mRNA is specifically destabilized in amastigotes but not in trypomastigotes or epimastigotes. The 43-nt RNA element, but not its mutated form, was shown to specifically interact in vitro with the RBP TcUBP1. However, this element was not recognized by other RRM-related RBPs such as TcUBP2, TcPABP1, TcPTB2, and TcRBP3.
Results shown in Fig. 9 suggest that cellular transcripts having the 43-nt cis-element can be identified in TcUBP1-containing mRNP complexes, suggesting its interaction in living cells. Because TcUBP1 binds the 43-nt cis-element-containing transcripts in both amastigote (Fig. 8) and epimastigote (data not shown) forms, it might be postulated that it plays a destabilizing role in this last stage.
The sequence motifs that form AREs were first identified in higher eukaryotes within the 3′-UTR of mRNA encoding several cytokines and lymphokines (37, 38), and since then other mRNAs encoding for proteins important for a variety of functions have been found to have this motif (38, 39). It has been estimated that 5–8% of human genes code for ARE-containing mRNAs (38, 40). This percentage would be considerably higher in T. cruzi taking into account the large number of mRNAs containing the 43-nt cis-element that we have described here. These results are consistent with the lack of transcriptional control in trypanosomes. Unlike mammalian cells that can coordinate expression of multiple related genes by having specific promoters or enhancers, trypanosomes have to rely almost exclusively on post-transcriptional control of mRNA abundance. A general characteristic of AREs is the presence of 5′-AUUUA pentamers, and based on the number and distribution of these pentamers, AREs have been grouped in three classes (39, 41). Because the T. cruzi 43-nt cis-element has only one 5′-AUUUA pentamer, it would correspond to Class I AREs, which contain 1–5 dispersed 5′-AUUUA motifs in a U-rich context. The T. cruzi 43-nt U-rich element does not contain the 5′-UUAUUUA(U/A)(U/A) nonamers found in class II AREs, whereas class III AREs do not contain the 5′-AUUUA pentamer (40). Other AREs have also been identified in the 3′-UTR of the small mucin gene (SMUG) in T. cruzi (42, 43). Although both sequences are similar and share a U-rich core, they have different flanking regions and stage-specific expression. These elements conferred selective mRNA destabilization in stage-specific manner, and they were recognized by trans-acting factors (12). Interestingly, these AREs contain the 5′-UUAUUUA(U/A)(U/A) nonamer and could be classified as class II AREs.
Although AREs can be different in sequence, it is clear that most are able to bind more than one ARE-binding protein (ARE-BP) and that some ARE-BPs can bind multiple mRNAs (40). Our results indicate the interaction of the RNA element described with TcUBP1 present in amastigote cell-free extracts. In this regard TcUBP1 has been shown to interact with AU- and GU-rich elements present in a great variety of trypanosome transcripts in vitro (12). It was previously demonstrated that TcUBP1 could act in the formation of distinct developmentally regulated complexes, playing stabilizing or destabilizing roles in these stages (12). It is likely that the binding of more than one RBP is necessary to stabilize/destabilize the mRNAs in vivo or that these interactions occur preferentially in a particular developmental stage according to transcripts levels.
Previous studies have reported that T. cruzi displays significant stage-dependent regulation of relative transcript abundances for thousands of genes (1). For many gene families relative transcript abundance correlates to known protein expression profiles (1). For example, proteomic analyses indicated that most TS gene expression occurs in trypomastigotes, fewer TS proteins are detected in amastigotes and metacyclics, and none in epimastigotes (44). Similar correlations were found with ribosomal protein transcripts (down-regulated in metacyclics), genes in the histidine-to-glutamate pathway (up-regulated in epimastigotes), mucin genes (up-regulated in trypomastigotes), and flagellum-associated genes (down-regulated in amastigotes) (1). These results suggest that in some cases mRNA relative abundances could be useful indicators of the protein expression levels in T. cruzi (1).
Another interesting observation related to our findings is that genes in paralog clusters could have divergent mRNA relative abundance patterns (1). These results were proposed to be due to differences in the 3′-UTR of paralogs with divergent expression patterns (1). In agreement with this hypothesis, we found that only 169 of 1430 TS and 67 of 1377 MASP genes have the 43-nt cis-element, and their expression is up-regulated in amastigote stages in contrast with the other family members that are up-regulated in trypomastigotes. The presence of this element in some but not all members of large gene families suggests that these protein-coding sequences were co-selected with these elements because they are developmentally relevant genes. These results also suggest that the identification of the presence of the 43-nt element in these genes could help to identify and study in more detail amastigote-specific paralogs of large gene families.
Coevolution of the ORFs and UTRs probably made some of paralogs more appropriate for a particular stage. Further studies are needed to investigate differential expression of members of the same gene family.
Keene (13–15) initially suggested that RBPs organize nascent RNA transcripts into groups to facilitate their splicing, nuclear export, stability, and translation so that proteins are produced to meet the needs of the cell. The trans-acting factors (primarily RBPs, but also non-coding RNAs or metabolites) would interact with regulatory elements within the mRNAs, which were termed USERs (untranslated sequence elements for regulation) (15). The presence of the 43-nt U-rich element in mRNAs that are up-regulated in the amastigote stages of T. cruzi provides new evidence in support of a post-transcriptional regulon to coordinate the stage-specific expression of a large number of genes. The existence of such regulons could be extremely important for trypomastigote to amastigote differentiation in the mammalian host, a process that occurs in only a few hours but with drastic morphological and metabolic changes. For example, it has been reported that incubation of trypomastigotes in medium at pH 5.0 for 2 h is sufficient to trigger their transformation into amastigotes (45). Differentiation of epimastigotes to metacyclic trypomastigotes also results in drastic changes in mRNA and protein expression, and these changes are accompanied by the formation of stress granules that contain proteins orthologous to those present in P bodies and stress granules from metazoan organisms (8). Among the proteins localized in these granules are TcUBP1 and TcUBP2 (8). It is possible that the pH stress during differentiation of trypomastigotes to amastigotes could also result in the formation of stress granules where RBPs could store the 43-nt cis-element-containing mRNAs until translation occurs. The results suggest that a subset of stage-specific transcripts that harbor the 43-nt cis-element could be organized by one (or more) sequence-specific RBP, such as TcUBP1, into a post-transcriptional regulon.
Although the notion of post-transcriptional regulons in Trypanosoma brucei and Leishmania spp. has been discussed previously (17, 46–51), it has been pointed out that searches for shared motifs in clusters of co-regulated genes in T. brucei met with limited success (17). Our work shows that these shared motifs exist in T. cruzi and suggests the presence of functional post-transcriptional regulons in these parasites.
Supplementary Material
Acknowledgments
We thank Oscar Campetella (National University of General San Martin, Argentina) for construct pTREX-GFP-tTS-3′-UTR, Todd A. Minning for useful discussions, and Melina Galizzi for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01AI-068647, D43TW-07888, and D43TW-07820 (to R. D.). Work at the Frasch laboratory was supported in part by an international scholarship from the Howard Hughes Medical Institute, Agencia Nacional de Promoción Científica y Tecnológica, Argentina (to A. C. F. and J. G. D. G.) and Consejo Nacional de Investigaciones Clentíficas y Técnicas (to J. G. D. G.).

This article contains supplemental Tables S1-S3 and Figs. S1 and S2.
- RBP
- RNA-binding protein
- ARE
- AU-rich element
- DD-RT-PCR
- differential display RT-PCR
- MASP
- mucin-associated protein
- PABP1
- poly(A)-binding protein 1
- RRM
- RNA recognition motif
- TS
- trans-sialidase
- tTS
- trypomastigote TS
- UBP1
- RNA-binding protein that binds U-rich sequences 1
- UBP2
- RNA-binding protein that binds U-rich sequences 2
- mRNP
- messenger ribonucleoprotein
- SL
- spliced leader
- IP
- immunoprecipitation
- BTF
- basic transcription factor
- PK4
- protein kinase 4
- nt
- nucleotide(s).
REFERENCES
- 1. Minning T. A., Weatherly D. B., Atwood J., 3rd, Orlando R., Tarleton R. L. (2009) The steady state transcriptome of the four major life-cycle stages of Trypanosoma cruzi. BMC Genomics 10, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clayton C. E. (2002) Life without transcriptional control? From fly to man and back again. EMBO J. 21, 1881–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Haile S., Papadopoulou B. (2007) Developmental regulation of gene expression in trypanosomatid parasitic protozoa. Curr. Opin. Microbiol. 10, 569–577 [DOI] [PubMed] [Google Scholar]
- 4. Palenchar J. B., Bellofatto V. (2006) Gene transcription in trypanosomes. Mol. Biochem. Parasitol. 146, 135–141 [DOI] [PubMed] [Google Scholar]
- 5. D'Orso I., De Gaudenzi J. G., Frasch A. C. (2003) RNA-binding proteins and mRNA turnover in trypanosomes. Trends Parasitol. 19, 151–155 [DOI] [PubMed] [Google Scholar]
- 6. Jäger A. V., De Gaudenzi J. G., Cassola A., D'Orso I., Frasch A. C. (2007) mRNA maturation by two-step trans-splicing/polyadenylation processing in trypanosomes. Proc. Natl. Acad. Sci. U.S.A. 104, 2035–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nardelli S. C., Avila A. R., Freund A., Motta M. C., Manhães L., de Jesus T. C., Schenkman S., Fragoso S. P., Krieger M. A., Goldenberg S., Dallagiovanna B. (2007) Small-subunit rRNA processome proteins are translationally regulated during differentiation of Trypanosoma cruzi. Eukaryot. Cell 6, 337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cassola A., De Gaudenzi J. G., Frasch A. C. (2007) Recruitment of mRNAs to cytoplasmic ribonucleoprotein granules in trypanosomes. Mol. Microbiol. 65, 655–670 [DOI] [PubMed] [Google Scholar]
- 9. Holetz F. B., Correa A., Avila A. R., Nakamura C. V., Krieger M. A., Goldenberg S. (2007) Evidence of P-body-like structures in Trypanosoma cruzi. Biochem. Biophys. Res. Commun. 356, 1062–1067 [DOI] [PubMed] [Google Scholar]
- 10. Kramer S., Carrington M. (2011) Trans-acting proteins regulating mRNA maturation, stability, and translation in trypanosomatids. Trends Parasitol. 27, 23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hendriks E. F., Matthews K. R. (2007) in Trypanosomes: After The Genome (Barry D., McCulloch R., Mottram J., Acosta-Serrano A., eds) pp. 209–238, Horizon Bioscience, Norfolk, UK [Google Scholar]
- 12. De Gaudenzi J. G., Noé G., Campo V. A., Frasch A. C., Cassola A. (2011) Gene expression regulation in trypanosomatids. Essays Biochem. 51, 31–46 [DOI] [PubMed] [Google Scholar]
- 13. Keene J. D., Tenenbaum S. A. (2002) Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell 9, 1161–1167 [DOI] [PubMed] [Google Scholar]
- 14. Keene J. D. (2001) Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc. Natl. Acad. Sci. U.S.A. 98, 7018–7024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Keene J. D. (2007) RNA regulons. Coordination of post-transcriptional events. Nat. Rev. Genet 8, 533–543 [DOI] [PubMed] [Google Scholar]
- 16. Mansfield K. D., Keene J. D. (2009) The ribonome. A dominant force in co-ordinating gene expression. Biol. Cell 101, 169–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ouellette M., Papadopoulou B. (2009) Coordinated gene expression by post-transcriptional regulons in African trypanosomes. J. Biol. 8, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boucher N., Wu Y., Dumas C., Dube M., Sereno D., Breton M., Papadopoulou B. (2002) A common mechanism of stage-regulated gene expression in Leishmania mediated by a conserved 3'-untranslated region element. J. Biol. Chem. 277, 19511–19520 [DOI] [PubMed] [Google Scholar]
- 19. Schmatz D. M., Murray P. K. (1982) Cultivation of Trypanosoma cruzi in irradiated muscle cells. Improved synchronization and enhanced trypomastigote production. Parasitology 85, 115–125 [DOI] [PubMed] [Google Scholar]
- 20. Moreno S. N., Silva J., Vercesi A. E., Docampo R. (1994) Cytosolic-free calcium elevation in Trypanosoma cruzi is required for cell invasion. J. Exp. Med. 180, 1535–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bone G. J., Steinert M. (1956) Isotopes incorporated in the nucleic acids of Trypanosoma mega. Nature 178, 308–309 [DOI] [PubMed] [Google Scholar]
- 22. Caler E. V., Vaena de Avalos S., Haynes P. A., Andrews N. W., Burleigh B. A. (1998) Oligopeptidase B-dependent signaling mediates host cell invasion by Trypanosoma cruzi. EMBO J. 17, 4975–4986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bailey T. L., Elkan C. (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 [PubMed] [Google Scholar]
- 24. Bailey T. L., Gribskov M. (1998) Combining evidence using p values. Application to sequence homology searches. Bioinformatics 14, 48–54 [DOI] [PubMed] [Google Scholar]
- 25. Jäger A. V., Muiá R. P., Campetella O. (2008) Stage-specific expression of Trypanosoma cruzi trans-sialidase involves highly conserved 3'-untranslated regions. FEMS Microbiol. Lett. 283, 182–188 [DOI] [PubMed] [Google Scholar]
- 26. de Diego J. L., Katz J. M., Marshall P., Gutiérrez B., Manning J. E., Nussenzweig V., González J. (2001) The ubiquitin-proteasome pathway plays an essential role in proteolysis during Trypanosoma cruzi remodeling. Biochemistry 40, 1053–1062 [DOI] [PubMed] [Google Scholar]
- 27. De Gaudenzi J. G., D'Orso I., Frasch A. C. (2003) RNA recognition motif-type RNA-binding proteins in Trypanosoma cruzi form a family involved in the interaction with specific transcripts in vivo. J. Biol. Chem. 278, 18884–18894 [DOI] [PubMed] [Google Scholar]
- 28. Noé G., De Gaudenzi J. G., Frasch A. C. (2008) Functionally related transcripts have common RNA motifs for specific RNA-binding proteins in trypanosomes. BMC Mol. Biol. 9, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Z. H., Alvarez V. E., De Gaudenzi J. G., Sant'Anna C., Frasch A. C., Cazzulo J. J., Docampo R. (2011) Hyperosmotic stress induces aquaporin-dependent cell shrinkage, polyphosphate synthesis, amino acid accumulation, and global gene expression changes in Trypanosoma cruzi. J. Biol. Chem. 286, 43959–43971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Campos P. C., Bartholomeu D. C., DaRocha W. D., Cerqueira G. C., Teixeira S. M. (2008) Sequences involved in mRNA processing in Trypanosoma cruzi. Int. J. Parasitol. 38, 1383–1389 [DOI] [PubMed] [Google Scholar]
- 31. Bartholomeu D. C., Cerqueira G. C., Leão A. C., daRocha W. D., Pais F. S., Macedo C., Djikeng A., Teixeira S. M., El-Sayed N. M. (2009) Genomic organization and expression profile of the mucin-associated surface protein (masp) family of the human pathogen Trypanosoma cruzi. Nucleic Acids Res. 37, 3407–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. El-Sayed N. M., Myler P. J., Bartholomeu D. C., Nilsson D., Aggarwal G., Tran A. N., Ghedin E., Worthey E. A., Delcher A. L., Blandin G., Westenberger S. J., Caler E., Cerqueira G. C., Branche C., Haas B., Anupama A., Arner E., Aslund L., Attipoe P., Bontempi E., Bringaud F., Burton P., Cadag E., Campbell D. A., Carrington M., Crabtree J., Darban H., da Silveira J. F., de Jong P., Edwards K., Englund P. T., Fazelina G., Feldblyum T., Ferella M., Frasch A. C., Gull K., Horn D., Hou L., Huang Y., Kindlund E., Klingbeil M., Kluge S., Koo H., Lacerda D., Levin M. J., Lorenzi H., Louie T., Machado C. R., McCulloch R., McKenna A., Mizuno Y., Mottram J. C., Nelson S., Ochaya S., Osoegawa K., Pai G., Parsons M., Pentony M., Pettersson U., Pop M., Ramirez J. L., Rinta J., Robertson L., Salzberg S. L., Sanchez D. O., Seyler A., Sharma R., Shetty J., Simpson A. J., Sisk E., Tammi M. T., Tarleton R., Teixeira S., Van Aken S., Vogt C., Ward P. N., Wickstead B., Wortman J., White O., Fraser C. M., Stuart K. D., Andersson B. (2005) The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309, 409–415 [DOI] [PubMed] [Google Scholar]
- 33. Liang P., Pardee A. B. (1992) Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257, 967–971 [DOI] [PubMed] [Google Scholar]
- 34. Frasch A. C. (2000) Functional diversity in the trans-sialidase and mucin families in Trypanosoma cruzi. Parasitol. Today 16, 282–286 [DOI] [PubMed] [Google Scholar]
- 35. De Gaudenzi J., Frasch A. C., Clayton C. (2005) RNA-binding domain proteins in Kinetoplastids. A comparative analysis. Eukaryot. Cell 4, 2106–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stern M. Z., Gupta S. K., Salmon-Divon M., Haham T., Barda O., Levi S., Wachtel C., Nilsen T. W., Michaeli S. (2009) Multiple roles for polypyrimidine tract binding (PTB) proteins in trypanosome RNA metabolism. RNA 15, 648–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Caput D., Beutler B., Hartog K., Thayer R., Brown-Shimer S., Cerami A. (1986) Identification of a common nucleotide sequence in the 3'-untranslated region of mRNA molecules specifying inflammatory mediators. Proc. Natl. Acad. Sci. U.S.A. 83, 1670–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Khabar K. S. (2005) The AU-rich transcriptome. More than interferons and cytokines, and its role in disease. J. Interferon Cytokine Res. 25, 1–10 [DOI] [PubMed] [Google Scholar]
- 39. Chen C. Y., Shyu A. B. (1995) AU-rich elements. Characterization and importance in mRNA degradation. Trends Biochem. Sci. 20, 465–470 [DOI] [PubMed] [Google Scholar]
- 40. Barreau C., Paillard L., Osborne H. B. (2005) AU-rich elements and associated factors. Are there unifying principles? Nucleic Acids Res. 33, 7138–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bakheet T., Frevel M., Williams B. R., Greer W., Khabar K. S. (2001) ARED. Human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 29, 246–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Di Noia J. M., D'Orso I., Sánchez D. O., Frasch A. C. (2000) AU-rich elements in the 3'-untranslated region of a new mucin-type gene family of Trypanosoma cruzi confers mRNA instability and modulates translation efficiency. J. Biol. Chem. 275, 10218–10227 [DOI] [PubMed] [Google Scholar]
- 43. D'Orso I., Frasch A. C. (2001) Functionally different AU- and G-rich cis-elements confer developmentally regulated mRNA stability in Trypanosoma cruzi by interaction with specific RNA-binding proteins. J. Biol. Chem. 276, 15783–15793 [DOI] [PubMed] [Google Scholar]
- 44. Atwood J. A., 3rd, Weatherly D. B., Minning T. A., Bundy B., Cavola C., Opperdoes F. R., Orlando R., Tarleton R. L. (2005) The Trypanosoma cruzi proteome. Science 309, 473–476 [DOI] [PubMed] [Google Scholar]
- 45. Tomlinson S., Vandekerckhove F., Frevert U., Nussenzweig V. (1995) The induction of Trypanosoma cruzi trypomastigote to amastigote transformation by low pH. Parasitology 110, 547–554 [DOI] [PubMed] [Google Scholar]
- 46. Mayho M., Fenn K., Craddy P., Crosthwaite S., Matthews K. (2006) Post-transcriptional control of nuclear-encoded cytochrome oxidase subunits in Trypanosoma brucei. Evidence for genome-wide conservation of life-cycle stage-specific regulatory elements. Nucleic Acids Res. 34, 5312–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Queiroz R., Benz C., Fellenberg K., Hoheisel J. D., Clayton C. (2009) Transcriptome analysis of differentiating trypanosomes reveals the existence of multiple post-transcriptional regulons. BMC Genomics 10, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Archer S. K., Luu V. D., de Queiroz R. A., Brems S., Clayton C. (2009) Trypanosoma brucei PUF9 regulates mRNAs for proteins involved in replicative processes over the cell cycle. PLoS Pathog 5, e1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mao Y., Najafabadi H. S., Salavati R. (2009) Genome-wide computational identification of functional RNA elements in Trypanosoma brucei. BMC Genomics 10, 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Archer S. K., Inchaustegui D., Queiroz R., Clayton C. (2011) The cell cycle regulated transcriptome of Trypanosoma brucei. PLoS One 6, e18425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bringaud F., Müller M., Cerqueira G. C., Smith M., Rochette A., El-Sayed N. M., Papadopoulou B., Ghedin E. (2007) Members of a large retroposon family are determinants of post-transcriptional gene expression in Leishmania. PLoS Pathog. 3, 1291–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Aslett M., Aurrecoechea C., Berriman M., Brestelli J., Brunk B. P., Carrington M., Depledge D. P., Fischer S., Gajria B., Gao X., Gardner M. J., Gingle A., Grant G., Harb O. S., Heiges M., Hertz-Fowler C., Houston R., Innamorato F., Iodice J., Kissinger J. C., Kraemer E., Li W., Logan F. J., Miller J. A., Mitra S., Myler P. J., Nayak V., Pennington C., Phan I., Pinney D. F., Ramasamy G., Rogers M. B., Roos D. S., Ross C., Sivam D., Smith D. F., Srinivasamoorthy G., Stoeckert C. J., Jr., Subramanian S., Thibodeau R., Tivey A., Treatman C., Velarde G., Wang H. (2010) TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 38, D457–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.