Background: Gp59 is required to correctly load DNA helicase during T4 recombination-dependent DNA replication and repair transactions.
Results: Gp59 forms helicase loading complexes with clusters of Gp32 (SSB) bound to ssDNA.
Conclusion: Gp59 follows Gp32 to single-stranded regions of recombination intermediates to promote strand-specific helicase loading.
Significance: Mediator proteins such as Gp59 play critical roles in the regulation of DNA recombination, repair, and replication.
Keywords: Bacteriophage, DNA Helicase, DNA Recombination, DNA Repair, DNA Replication, DNA-Protein Interaction, Fluorescence, Helicase Loading Protein, Mediator, ssDNA-binding Protein
Abstract
The Gp59 protein of bacteriophage T4 plays critical roles in recombination-dependent DNA replication and repair by correctly loading the replicative helicase, Gp41, onto recombination intermediates. Previous work demonstrated that Gp59 is required to load helicase onto single-stranded DNA that is saturated with Gp32, the T4 single-stranded DNA (ssDNA)-binding protein. Gp59 and Gp32 bind simultaneously to ssDNA, forming a Gp59-Gp32-ssDNA complex that is a key intermediate in helicase loading. Here we characterize the assembly and dynamics of this helicase loading complex (HLC) through changes in the fluorescent states of Gp32F, a fluorescein-Gp32 conjugate. Results show that HLC formation requires a minimum Gp32-ssDNA cluster size and that Gp59 co-localizes with Gp32-ssDNA clusters in the presence of excess free ssDNA. These and other results indicate that Gp59 targets helicase assembly onto Gp32-ssDNA clusters that form on the displaced strand of D-loops, which suggests a mechanism for the rapid initiation of recombination-dependent DNA replication. Helicase loading at the HLC requires ATP binding (not hydrolysis) by Gp41 and results in local remodeling of Gp32 within the HLC. Subsequent ATPase-driven translocation of Gp41 progressively disrupts Gp32-ssDNA interactions. Evidence suggests that Gp59 from the HLC is recycled to promote multiple rounds of helicase assembly on Gp32-ssDNA, a capability that could be important for the restart of stalled replication forks.
Introduction
Recombination-dependent DNA replication (RDR)3 is a central component of homology-directed DNA repair processes including DNA double strand break repair and replication restart (1). RDR occurs when a homologous DNA strand invasion event, catalyzed by a recombinase such as RecA or Rad51, is used to initiate DNA synthesis. The critical biochemical step in RDR is the conversion of a D-loop recombination intermediate into a replication fork. RDR has been studied most thoroughly in the bacteriophage T4, which uses RDR as its major mode of chromosomal replication and for which purified RDR in vitro systems have been established (Refs. 2–4; for reviews, see Refs. 5 and 6).
In T4 RDR, the UvsX recombinase in concert with recombination mediator protein (UvsY) and ssDNA-binding protein (Gp32) catalyzes the invasion of a 3′ ssDNA tail from one phage chromosome into the homologous duplex region of another chromosome, forming a D-loop (5). The invading 3′-end then serves as a primer for leading strand DNA synthesis catalyzed by DNA polymerase holoenzyme. Conversion of the D-loop into a replication fork also requires the loading of a DNA helicase onto the displaced ssDNA. The major helicase used for this conversion is Gp41. Gp41 is the essential, replicative helicase encoded by bacteriophage T4. A member of the ring hexamer helicase family, Gp41 stimulates DNA replication by translocating processively in the 5′ → 3′ direction on lagging strand ssDNA while catalyzing the unwinding of the parental duplex (7). During lagging strand synthesis, Gp41 serves as an essential component of the T4 primosome (helicase-primase complex). The loading of Gp41 and therefore of primosome onto D-loops requires the activity of a helicase loading protein, Gp59. Once loaded by Gp59, Gp41 can promote either the conservative or semiconservative mode of RDR (2, 5, 6). The conservative mode includes “bubble migration” (D-loop translocation) steps that are used for the synthesis-dependent strand annealing pathway of double strand break repair; the semiconservative mode, involving both leading and lagging strand synthesis machinery, is identical to break-induced replication (1, 6).
The helicase/primosome loading activity of Gp59 protein has emerged as a critical factor in the initiation of RDR (1, 5, 8, 9). Gp59 (26 kDa) consists of two α-helical domains. The N-terminal domain contains a duplex DNA binding motif that is structurally homologous to the high mobility group B family (10, 11). Gp59 exhibits intrinsic, structure-selective DNA binding activity with relative affinity for fork DNA > ssDNA > dsDNA (12, 13). Gp59 also exhibits high affinity interactions with both Gp32 and Gp41, and it can act as an adapter between these proteins (14–18). In DNA replication and RDR, efficient helicase or primosome assembly depends on Gp59 activity because Gp41 has an intrinsically low affinity for ssDNA saturated with Gp32. Being abundant and highly cooperative, Gp32 competes with Gp41 for ssDNA binding sites. Gp59 in contrast has been shown to readily form interactions with Gp32 either bound to ssDNA or free in solution (15, 16, 19, 20). This interaction plays a major role in the recruitment of Gp41 onto D-loops and replication forks (1, 5).
Some aspects of the Gp32-Gp59 interaction have been revealed from cross-linking studies and truncation mutagenesis (15, 16, 21). Gp32 interacts with Gp59 primarily through the C-terminal “A” domain of Gp32, whereas the N-terminal “B” domain of Gp32 is required for cooperativity. Central to the Gp32 structure is the core domain, which contains an oligonucleotide/oligosaccharide binding fold as well as bound Zn2+ and is the primary site of Gp32-ssDNA interactions (22). Previous studies demonstrated that the interaction of Gp59 with the Gp32 occurs with a stoichiometry of 1:1 and that both proteins can co-saturate ssDNA (19, 20). Gp32 and Gp59 are present on ssDNA in large, compact protein-DNA complexes that represent a major portion of the protein mass of the replication fork (19, 23). These results suggest that the co-localization of ssDNA with Gp59 and Gp32 is an important structural feature of T4 helicase/primosome assembly, forming an intermediate that we refer to as the helicase loading complex (HLC).
Although the tripartite Gp59-Gp32-ssDNA helicase loading complex has been observed in a number of previous studies, the specific conditions required for its formation and turnover have not been fully addressed. Unresolved issues include 1) whether Gp59 preferentially recognizes DNA versus Gp32-DNA targets, 2) the minimum ssDNA length and Gp32 cluster size for HLC formation, and 3) the fate of the HLC after helicase loading. To address some of the remaining questions, we developed ensemble fluorescence methods to quantify Gp59 interactions with various DNA structures and to monitor the formation and turnover of the HLC. The latter studies make use of a fluorescein-Gp32 conjugate, Gp32F (46), which was described previously as a real time probe for Gp32-ssDNA interactions (24). The unique fluorescent states of Gp32F allow for the distinction between its different binding states during HLC assembly and turnover: unbound, ssDNA-bound, and Gp59-bound on ssDNA. Results show that HLC formation requires a minimum cluster size of Gp32 on ssDNA and that Gp59 preferentially co-localizes with Gp32-ssDNA clusters even in the presence of excess free ssDNA. These and related findings support a model in which Gp59-Gp32 interactions promote helicase loading onto the displaced strand of D-loops, leading to efficient initiation of RDR and homology-directed repair. We further observed that helicase loading by the HLC requires ATP binding (not hydrolysis) by Gp41 and results in remodeling of the HLC. Subsequent ATPase-driven helicase translocation disrupts Gp32-ssDNA interactions; however, Gp59 appears to be recycled to promote multiple rounds of helicase loading, a capability that may be important for replication fork restart. The results of this study provide new insights into genome stability mechanisms that are used by all organisms to repair DNA damage and avoid disease.
EXPERIMENTAL PROCEDURES
Reagents and Resins
All chemicals were of analytical grade. All reagents were purchased from Sigma unless otherwise indicated. Deionized water was used to prepare all aqueous solutions and buffers. ssDNA cellulose resin was prepared by a previously published protocol (25). Hydroxyapatite resin was purchased from Bio-Rad. 6-Iodoacetamidofluorescein was purchased from Invitrogen.
Nucleic Acids
All DNA concentrations are expressed in moles/liter nucleotide residues. Circular M13mp18 ssDNA was purchased either from New England Biolabs or from USB. Concentrations were provided by the manufacturer and checked by measuring the absorbance at 260 nm using a conversion factor of 33 μg/ml/A260. HPLC-purified mixed sequence oligonucleotides were purchased either from Operon Biotechnologies, Inc. (Oligos 1 and 4), Bio-Synthesis, Inc. (Oligo 2), or Integrated DNA Technologies, Inc. (Oligo 3) (Table 1). HPLC-purified homopolymers dT12, dT20, dT40, and dT70 were purchased from Operon Biotechnologies, Inc. The concentration of each oligo was determined by the absorbance at 260 nm using extinction coefficients provided by the manufacturer.
TABLE 1.
Mixed sequence oligonucleotides used in experiments
(C2dT) denotes the position of the C2dT amino linker used for fluorescent labeling.
| Sequence | Length | |
|---|---|---|
| bases | ||
| Oligo 1 | 5′-TAACGTATTCAAGATACCTCGTACTCTGTATAACGC-AGGTTGCGATCCGACTGTCCTGCATCAGGTTGCG-3′ | 70 |
| Oligo 2 | 5′-GCTTCTGAAGTCTAATTCTATC(C2dT)CT-3′ | 25 |
| Oligo 3 | 5′-AGAGATAGAATTAGACTTCAGAAGC-3′ | 25 |
| Oligo 4 | 5′-TCCCTATCTTAATGACTTCAGAAGC-3′ | 25 |
| Oligo 5 | 5′-TAACGTATTCAAGATACCTCGTACTCTG TACAGGTTGCGATCCGACTGTCCTGCAT-3′ | 56 |
| Oligo 6 | 5′-GATCATGCAGGACAGTCGGATCGCAACC TGATTTACTGTGTCATATAGTACGTGATTCAG-3′ | 60 |
Preparation of Alexa Fluor 546-labeled DNA Molecules
Oligo 2 contained a C2dT amino linker at position 23. This oligo was labeled with Alexa Fluor 546 carboxylic acid succinimidyl ester, an amine-reactive probe, according to the manufacturer's instructions (Invitrogen). Labeling efficiency was determined spectrophotometrically by measuring the absorbance at wavelengths of 260 and 556 nm, using extinction coefficients of 235,200 and 104,000 m−1 cm−1, respectively. The labeling efficiency of Oligo 2 was calculated to be 69%. The A260 reading was corrected for contributions from the Alexa Fluor 546 by a correction factor provided by the manufacturer. The corrected A260 was used to calculate the concentration of Oligo 2. Alexa Fluor 546-labeled duplex and fork 25-mers were assembled by annealing Oligo 2 to Oligo 3 or to Oligo 4, respectively. The annealing process was carried out using a modification of a published procedure (10). A stoichiometric mixture of the two oligos was prepared in water and then heated to 95 °C. The water bath was then turned off and allowed to slowly cool to room temperature (∼2 h). The integrity of annealed DNA structures was confirmed by polyacrylamide gel electrophoresis as described (10).
Bacteriophage T4 Proteins
Gp32 and Gp41 proteins were purified using previously published protocols (16) with the following modifications. For growth and induction in Escherichia coli, the NZCYM medium (10 g/liter NZ-Amine A, 1 g/liter Casamino Acids, 5 g/liter yeast extract, 5 g/liter NaCl, 2 g/liter MgSo4) was replaced with LB. Also, both protocols were modified by replacing the Whatman P11 chromatography step with a Mono Q (GE Healthcare) chromatography step. Additionally, the storage conditions of both Gp41 and Gp32 were modified to include the following: 20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 0.1 mm EDTA, 1 mm β-mercaptoethanol, and 10% glycerol. Concentrations of protein stock solutions were calculated from extinction coefficients for Gp32 and Gp41 at 280 nm of 41,306 and 76,000 m−1 cm−1, respectively (26).
Gp59 was purified by either of two published methods (16, 27). The latter method was modified to include an additional column chromatography step to remove a contaminating nuclease activity. Nuclease-contaminated fractions were applied to a Bio-Rad Biogel-HTP hydroxyapatite column and eluted using a 50–500 mm potassium phosphate gradient, pH 6.8 in buffer containing 100 mm NaCl, 2 mm EDTA, 1 mm β-mercaptoethanol, and 10% glycerol. Purified fractions from either method were dialyzed into one of two Gp59 storage buffers: 1) 20 mm Tris-HCl, pH 7.4, 50 mm NaCl, 1 mm EDTA, 1 mm β-mercaptoethanol, and 60% glycerol and stored at −20 °C or 2) 10 mm Tris acetate, pH 7.8, 25 mm potassium acetate, 5 mm magnesium acetate, 2 mm DTT, and 20% glycerol and stored at −80 °C. Gp59 derived from either purification protocol and stored under either condition behaved identically in biochemical experiments. The concentration of Gp59 was calculated using an extinction coefficient of 37,800 m−1 cm−1 (26).
All stock solutions of Gp59, Gp32, and Gp41 proteins used in this study were nuclease-free according to published criteria (14). All proteins were determined to be >95% pure according to SDS-PAGE analysis.
Labeling of Gp32 with 6-Iodoacetamidofluorescein
The fluorescein-Gp32 conjugate Gp32F was generated by labeling Gp32 with 6-iodoacetamidofluorescein as described (24). Labeling efficiency was calculated according to the manufacturer's instructions using extinction coefficients of 41,300 and 68,500 m−1 cm−1 at 280 and 494 nm, respectively.
DNA Binding Assays
Alexa Fluor 546 fluorescence quenching assays for Gp59-DNA interactions were carried out on a Quantamaster QM-6 spectrofluorometer (Photon Technology International, South Brunswick, NJ). The slit widths were 3 nm for both emission and excitation monochromators. All fluorescence assays were carried out at room temperature (∼21 °C). The excitation wavelength was 540 nm, and the emission was measured at 569 nm. The following reaction buffer was used for the binding assays: 25 mm Tris acetate, pH 7.8, 90 mm potassium acetate, 6 mm magnesium acetate, and 1 mm DTT. To collect the data for these assays, the fluorescence emission at 569 nm was measured for 2 s at one data point per second. First, the buffer alone was measured as background. Then, 2 μm Oligo 2 (either single-stranded or annealed) was added to a cuvette in a total volume of 80 μl, and the fluorescence emission was measured. Finally, the Gp59 protein was added at the indicated concentration and incubated at room temperature for 2 min, and the fluorescence emission was measured again. This was repeated for each data point in the titration, each time adding an equal volume of increasing concentrations of Gp59. Each data point was corrected for background and then for dilution by subtracting the signal change caused by the addition of a storage buffer blank from the signal change caused by the addition of protein. All dilutions were <12% of the total volume. Contributions from photobleaching, inner filter effects, and fluorescence of protein and/or storage buffer were found to be negligible under the experimental conditions. Binding curves were generated by converting the differences in fluorescence emission between the unbound and bound oligo to the fraction of substrate bound and plotting it versus Gp59 concentration. Binding curves were each fit to Equation 1 in GraphPad Prism,
in which Y is the fraction of DNA bound, Bmax is the maximum DNA bound (assumed to equal 1.0 at plateau), X is the ligand (Gp59) concentration, Kd is the ligand (Gp59) concentration at half-saturation, and h is the Hill coefficient, an indicator of binding cooperativity. Error bars shown in the data represent the S.D. of the results of a minimum of two experiments.
Fluorescence Assays for Helicase Loading Complex Assembly
HLC assembly was monitored by changes in Gp32F fluorescence. Fluorescence emission spectra were recorded over an emission range of 480–600 nm while exciting at the absorbance maximum, 460 nm. A 495-nm longpass filter was used for the emission scans. The slit widths were set at 3 nm for both excitation and emission. The basic method for measuring HLC complex formation was as follows. The emission spectrum of Gp32F alone was measured first followed by the addition of the ssDNA (oligonucleotide or circular M13mp18 ssDNA) and incubation for 5 min at room temperature. The emission spectrum of the Gp32F-ssDNA complex was then measured followed by the addition of Gp59 and a 5-min incubation at room temperature. A final emission spectrum was then measured for the Gp32F-Gp59-ssDNA tripartite complex. This procedure was used to detect the HLC on oligonucleotides of different lengths. To do this, 100 nm Gp32F was bound to a 700 nm concentration of each of the following oligonucleotides: dT12, dT20, dT40, or dT70. This was followed by the addition of 100 nm Gp59. Again, the fluorescence emission was measured after the addition of each component of the HLC. All experiments were performed in HLC reaction buffer containing 25 mm Tris acetate, pH 7.4, 90 mm potassium acetate, 10 mm magnesium acetate, and 1 mm DTT. All Gp32F fluorescence data were corrected for background and for dilution. Photobleaching, inner filter effects, and signal contributions from proteins and storage buffers were found to be negligible under the experimental conditions. All Gp32F fluorescence intensities shown in the figures are normalized with respect to the signal from free Gp32F at its emission maximum, 519 nm.
To measure HLC formation at low binding density of proteins on ssDNA, the fluorescence of the following mixtures in HLC reaction buffer were measured: 30 nm Gp32F alone, 30 nm Gp32F + 3 μm M13mp18 ssDNA, and 30 nm Gp32F + 3 μm M13mp18 ssDNA + 30 nm Gp59. To test the interaction of Gp32F and Gp59 in the absence of ssDNA, the following fluorescence emission scans were compared: 100 nm Gp32F and 100 nm Gp32F + 100 nm Gp59.
Helicase Loading and Translocation Assays
Interactions of Gp41 helicase with the HLC were monitored by changes in Gp32F fluorescence. Assays were performed under two sets of conditions, those favoring detection of Gp41 loading versus detection of Gp41 translocation. The basic procedure for the assays was as follows. HLC components were added in the following order: Gp32F → ssDNA → Gp59. The fluorescence emission of this complex was followed as a function of time (excitation, 460 nm; emission, 519 nm). When the fluorescence intensity had stabilized, data acquisition was paused while Gp41 and ATP were added simultaneously. The scan was then resumed, and the fluorescence intensity was monitored for up to 1000 s after Gp41/ATP addition.
To study the effects of helicase loading in the absence of extensive translocation, the ssDNA substrate used was Oligo 1 (mixed sequence 70-mer; Table 1). The HLC was preformed using 100 nm Gp32F, 700 nm Oligo 1, and 100 nm Gp59. 100 nm Gp41 and 1 mm ATP were then added simultaneously. The time courses shown in the figures are the normalized Gp32F intensities versus time after Gp41/ATP addition. This procedure was repeated under conditions where ATP was replaced with ATPγS. Controls were done by individually replacing either Gp41 or Gp59 with their respective storage buffers or by replacing the oligonucleotide or ATP individually with water. Under these conditions, the following buffer was used: 25 mm Tris acetate, pH 7.4, 25 mm potassium acetate, 10 mm magnesium acetate, and 1 mm DTT. A lower potassium acetate concentration (25 versus 90 mm in experiments with homopolymeric oligos; as described above) was used to optimize saturation of the mixed sequence oligo with Gp59 and Gp32.
To study the effects of helicase translocation, the following complex was preformed: 100 nm Gp32F, 700 nm M13mp18 ssDNA, and 10 nm Gp59. When the fluorescence emission of this complex had stabilized, 100 nm Gp41 and 1 mm ATP were added simultaneously. The time courses shown in the figures are the normalized Gp32F intensities versus time after Gp41/ATP addition. Controls were done in which M13mp18 ssDNA or ATP was individually replaced with water. To determine the dependence of Gp32F remodeling on Gp41 concentration, the Gp32F concentration was increased to 200 nm to ensure complete saturation of the ssDNA while the Gp59 concentration remained at 10 nm and the Gp41 concentration was varied from 0 to 200 nm. All helicase translocation experiments were carried out in 25 mm Tris acetate, pH 7.8, 90 mm potassium acetate, 10 mm magnesium acetate, and 1 mm DTT.
Maximum rates of Gp32F fluorescence change in helicase translocation experiments were determined by fitting regression lines to the portion of each trace showing the steepest signal decrease. At most Gp41 concentrations, the steepest decrease occurred from 10 to 30 s after the addition of Gp41 + ATP. However, at one Gp41 concentration (25 nm), the steepest decrease was observed 110–190 s after the addition of Gp41 + ATP. The slopes of lines fit to two independent data sets at each Gp41 concentration were averaged. These slopes represent the maximum rate of Gp32F fluorescence change. The absolute values of these slopes were plotted as a function of Gp41 concentration and fit to Equation 1 to determine the Gp41 concentration at half-saturation.
Fluorescence Assays for Helicase Loading Complex Assembly and Helicase Loading on Fork DNA
A fork DNA oligonucleotide was prepared by annealing Oligos 5 and 6 (Table 1) in a 1:1 ratio. This generated a fork with a 26-base pair duplex region and two 30-nucleotide single-stranded arms. This substrate was used to monitor the formation of the HLC similarly to those experiments carried out on ssDNA oligonucleotides as described above. Buffer contained the following components: 20 mm Tris acetate, pH 7.8, 90 mm potassium acetate, 10 mm magnesium acetate, and 1 mm DTT. For the detection of helicase loading complex formation, the reaction components were added in the following order: 100 nm Gp32F, 1.66 μm (860 nm ssDNA on fork arms) fork DNA, and 100 nm Gp59. Fluorescence emission scans were taken after each addition as described above for experiments with ssDNA oligos. The fluorescence emission of this complex at 519 nm was then monitored over time. When the signal had stabilized, the scan was paused, and the following reaction components were added simultaneously: 100 nm Gp41 and 500 μm ATPγS. The fluorescence emission was monitored for ∼500 s after Gp41/ATPγS addition.
RESULTS
Detection of HLC Formation and Dependence on ssDNA Length
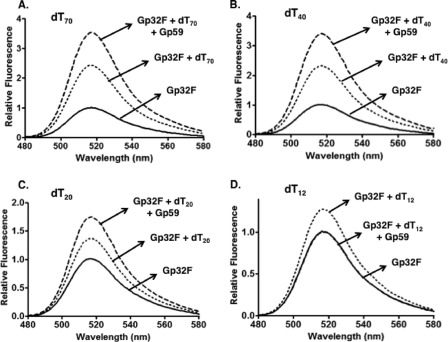
The fluorescein-Gp32 conjugate Gp32F was previously shown to have ssDNA binding properties similar to those of unlabeled Gp32 and was used as a real time probe for Gp32-ssDNA association/dissociation events (24). Gp32F was prepared as described under “Experimental Procedures,” and the labeling efficiency was determined to be 83%. As shown in Fig. 1, the addition of ssDNA enhanced Gp32F fluorescence by up to 2.4-fold, depending on ssDNA length. Homopolymeric oligos dT70 and dT40 contain 10 and five to six binding sites for Gp32F, respectively, assuming a binding site size of n = 7 nucleotide residues (24, 28). Both dT70 and dT40 support the formation of stable, cooperative clusters of Gp32F as reflected by the 2.4-fold increase in fluorescence upon addition of a stoichiometric amount of ssDNA to protein (Fig. 1, A and B). This level of Gp32F fluorescence enhancement is consistent with results obtained using long, mixed sequence M13mp18 ssDNA (24). Gp32F binding to shorter oligos is less efficient due to its high cooperativity parameter (ω ≥ 1000), and this is reflected by the decrease in fluorescence enhancement (1.3-fold) seen with dT20 and dT12, respectively (Fig. 1, C and D). These oligos have binding sites for only two to three and one to two protomers, respectively, which is insufficient for stable cluster formation (29).
FIGURE 1.
Effects of oligonucleotide length on Gp59-dependent fluorescence changes of Gp32F. Fluorescence measurements were performed as described under “Experimental Procedures.” Buffer contained the following components: 20 mm Tris acetate, pH 7.4, 10 mm magnesium acetate, 90 mm potassium acetate, and 1 mm DTT. Fluorescence emission scans monitored helicase loading complex formation at high binding density on each of the following homopolymeric oligonucleotides: dT70 (A), dT40 (B), dT20 (C), and dT12 (D). Solid line, 100 nm Gp32F alone; dotted line, 100 nm Gp32F + 700 nm oligonucleotide; dashed line, 100 nm Gp32F + 700 nm oligonucleotide + 100 nm Gp59.
The addition of Gp59 (equimolar with respect to Gp32F) to Gp32F-ssDNA complexes altered Gp32F fluorescence in a manner that depended on ssDNA length (Fig. 1). The fluorescence of Gp32F-dT70 and -dT40 complexes was enhanced an additional 1.5-fold upon Gp59 addition (Fig. 1, A and B). We interpret this signal change as a signature for the formation of the tripartite Gp59-Gp32-ssDNA helicase loading complex. The additional enhancement by Gp59 was slightly diminished (1.3-fold) in the complex with Gp32F-dT20 (Fig. 1C). In contrast, upon addition of Gp59 to Gp32F-dT12, the fluorescence intensity returned to approximately that of the unbound state of Gp32F (Fig. 1D), which we interpret as Gp59 competing with dT12 for binding to Gp32F. The dependence of HLC formation on oligo length is especially evident by noting the differences in scale of relative fluorescence between the panels of Fig. 1. These results indicate that HLC assembly requires ssDNA long enough for stable Gp32 cluster formation.
Gp59 Co-localizes with Gp32 at Low Binding Density on ssDNA
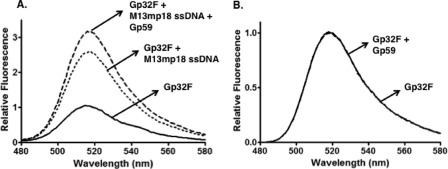
Gp32F was incubated with M13mp18 ssDNA circles at a low binding density of protein on ssDNA (ν = 0.07 assuming a Gp32 binding site size of 7 nucleotide residues). This means that given the high cooperativity of Gp32-ssDNA interactions (ω ≥ 1000), which is unaltered in Gp32F (24), 7% of the ssDNA is occupied by Gp32F clusters under the experimental conditions, whereas the remaining 93% exists as free ssDNA. Under these conditions, Gp32F fluorescence increased by a factor of 2.7 upon Gp32F binding to ssDNA (Fig. 2A), consistent with previous results (24). Under the same conditions of low binding density, Gp59 was added to Gp32F-ssDNA complexes at a stoichiometry of 1:1 Gp59:Gp32F, and fluorescence changes were monitored. Gp59 addition resulted in an additional 1.3-fold enhancement of Gp32F fluorescence above that caused by ssDNA alone (Fig. 2A), indicating an interaction between Gp59 and the sparsely distributed Gp32F clusters under these conditions. This result indicates that Gp59 preferentially co-localizes with Gp32 clusters on ssDNA even in the presence of a large excess of free ssDNA. In the absence of ssDNA, Gp59 did not affect the Gp32F fluorescence signal (Fig. 2B). The fact that Gp59 only affected Gp32F fluorescence when it was ssDNA-bound suggests that the environment of the fluorescein tag in Gp32F and possibly the conformation of Gp32 itself are quite different in the ssDNA-bound versus ssDNA-free forms of the Gp59-Gp32F complex.
FIGURE 2.
Effects of M13mp18 ssDNA and Gp59 on fluorescence of Gp32F at low binding density. Fluorescence measurements were performed as described under “Experimental Procedures.” Buffer contained the following components: 20 mm Tris acetate, pH 7.4, 10 mm magnesium acetate, 90 mm potassium acetate, and 1 mm DTT. A, solid line, 30 nm Gp32F alone; dotted line, 30 nm Gp32F + 3 μm M13mp18 ssDNA; dashed line, 30 nm Gp32F + 3 μm M13mp18 ssDNA + 30 nm Gp59. B, solid line, 100 nm Gp32F alone; dashed line,100 nm Gp32F + 100 nm Gp59.
Remodeling of HLC during Helicase Loading and Translocation
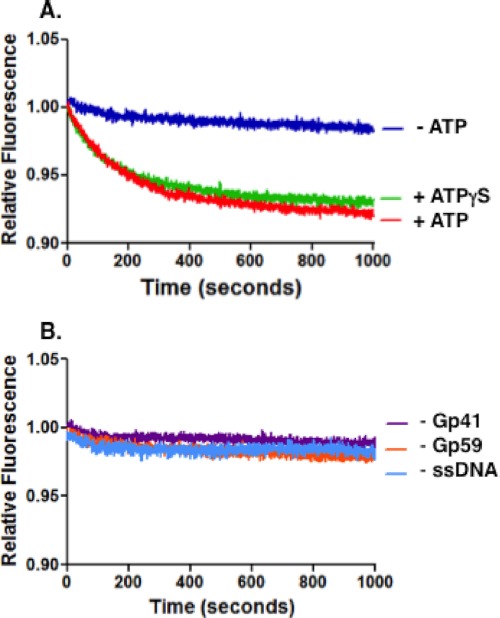
To monitor the fate of the HLC during helicase assembly, the helicase loading complex was preassembled using saturating amounts of Gp59 and Gp32F on a mixed sequence 70-mer (Oligo 1). The simultaneous addition of Gp41 helicase and ATP to the preassembled HLC resulted in a time-dependent decrease in Gp32F fluorescence (Fig. 3A), consistent with remodeling of the HLC. An identical result occurred when ATPγS was substituted for ATP (Fig. 3A); therefore, the reaction does not appear to be dependent on ATP hydrolysis by Gp41. The Gp32F fluorescence decrease did not occur if any component was individually removed: ATP or ATPγS (Fig. 3A), Gp59, Gp41, or ssDNA (Fig. 3B). This indicates that the change in signal is the result of active remodeling of the HLC that depends on ATP binding by Gp41 and Gp59-mediated helicase loading. We conclude that the event under observation is helicase loading rather than translocation as it did not depend on ATP hydrolysis. Also, the drop in fluorescence does not seem extensive enough to be the result of complete Gp59 or Gp32 displacement from the HLC. Instead, the cause of the signal change may be a conformational change of the HLC that repositions Gp32 and Gp59 to accommodate the presence of Gp41 on the ssDNA.
FIGURE 3.
Gp32F fluorescence effects under conditions of helicase loading. Helicase loading assays were performed as described under “Experimental Procedures.” Buffer contained the following: 20 mm Tris acetate, pH 7.4, 10 mm magnesium acetate, 25 mm potassium acetate, and 1 mm DTT. The components of the helicase loading complex were added in the following order and preincubated: 100 nm Gp32F, 700 nm Oligo 1 (mixed sequence 70-mer), and 100 nm Gp59. Data show normalized fluorescence of this complex versus time after addition of 100 nm Gp41 and 1 mm ATP simultaneously. A, Gp32F fluorescence changes in the presence of ATP (red) or ATPγS (green) or in the absence of nucleotide cofactor (dark blue). B, controls using the same procedure as in A except in the absence of Gp41 (purple), Gp59 (orange), or ssDNA (light blue).
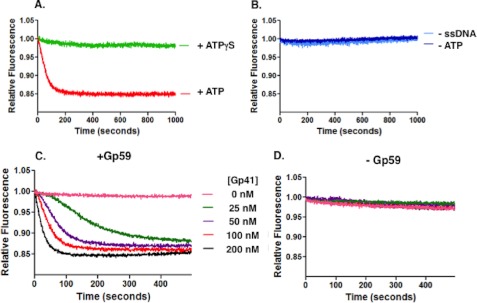
Similar assays were performed using circular M13mp18 ssDNA in place of the mixed sequence 70-mer (Fig. 4). In these experiments, the ssDNA was saturated with Gp32F, but the Gp59 concentration was limited to ∼10% of potential binding sites, leading to widely dispersed HLC sites. Under these conditions, the results in the presence of ATP versus ATPγS were dramatically different. The presence of ATP resulted in a faster and more extensive drop in Gp32F fluorescence, whereas no significant drop in fluorescence occurred in the presence of ATPγS (Fig. 4A). This suggests that Gp41 is translocating on the long ssDNA molecule, a process that is known to require ATP hydrolysis, and disrupting HLC and/or displacing Gp32F from the ssDNA as it goes. Controls indicate that the initial rapid signal decrease requires both ATP hydrolysis and ssDNA (Fig. 4B). The Gp41 helicase is known to be a processive enzyme, translocating ∼400 base pairs per second (30). The most likely explanation of the data in Fig. 4A appears to be that the rapidly decreasing phase represents the loading of Gp41 and initiation of translocation from discrete HLC sites dispersed along the Gp32F-covered ssDNA. Meanwhile, the final, diminished signal represents the establishment of a steady state in which Gp32F displaced by helicase translocation rebinds to the ssDNA at some distance behind the moving helicase.
FIGURE 4.
Gp32F fluorescence effects under conditions of helicase translocation. Helicase translocation assays were performed as described under “Experimental Procedures.” Buffer contained the following: 20 mm Tris acetate, pH 7.4, 10 mm magnesium acetate, 90 mm potassium acetate, and 1 mm DTT. A, the components of the helicase loading complex were added in the following order: 100 nm Gp32F, 700 nm M13mp18 ssDNA, and 10 nm Gp59. The data show the normalized fluorescence of Gp32F in this complex versus time after addition of 100 nm Gp41 and 1 mm ATP simultaneously. Red, Gp32F displacement in the presence of 1 mm ATP; light green, Gp32F displacement in the presence of 1 mm ATPγS. B, controls using the same procedure as A except in the absence of ATP (dark blue) or in the absence of ssDNA (light blue). C, Gp32F displacement resulting from the addition of 1 mm ATP and increasing concentrations of Gp41. The Gp32F concentration was increased to 200 nm to ensure no free ssDNA. The data show the normalized fluorescence of this complex after simultaneously adding 1 mm ATP and the following concentrations of Gp41: 0 (pink), 25 (green), 50 (purple), 100 (red), and 200 nm (black). D, reactions were repeated using the same conditions and the same range of concentrations as in C except that the Gp59 was removed from the reaction.
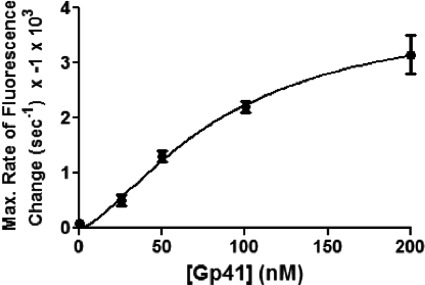
Other experiments were performed under helicase translocation conditions while varying Gp41 concentration with all other components held constant (Fig. 4C). The concentration of Gp59 (10 nm) was limiting in each of the reactions containing Gp41 (25–200 nm). The data show that the rate of the fluorescence change increased with increasing Gp41 concentration. In the absence of Gp59, however, no fluorescence change was observed at any Gp41 concentration (Fig. 4D). Therefore, reactions at all Gp41 concentrations are Gp59-dependent. As shown in Fig. 5, the curve describing the relationship between the maximum rate of fluorescence change (from Fig. 4C) and Gp41 concentration resembles that of a cooperative enzyme: saturable at high helicase concentrations and slightly sigmoidal at low helicase concentrations. We assume that the rate of Gp32F fluorescence decrease is related to the number of helicase hexamers loaded onto ssDNA by the HLC. If so, then the data suggest that Gp59 present in HLC complexes can act “catalytically” to promote multiple rounds of helicase loading. Analysis of the curve in Fig. 5 yielded an “apparent Km” value of 86 ± 30 nm Gp41 for this activity with an apparent Hill coefficient of 1.5.
FIGURE 5.
Rate of Gp59-dependent Gp32F fluorescence change as function of helicase concentration. Rates were derived from the maximum slope of each trace in Fig. 4C as described under “Experimental Procedures.” The Gp41 concentration for half-maximal activity was calculated to be 86 ± 30 nm. Error bars represent the S.D. of the results of a minimum of two experiments.
Another notable feature of the data in Fig. 4C is that all of the reactions eventually approach the same steady-state level of diminished Gp32F fluorescence, suggesting that it is independent of Gp41 concentration. This could reflect a limit to the number of helicase hexamers that can be loaded and/or translocate on each M13mp18 ssDNA circle. Alternatively, it could indicate that beyond a certain density of helicase hexamers on ssDNA the Gp32F fluorescence signal becomes insensitive to further increases in helicase density.
Quantitative Analysis of Gp59 Binding to Different DNA Structures
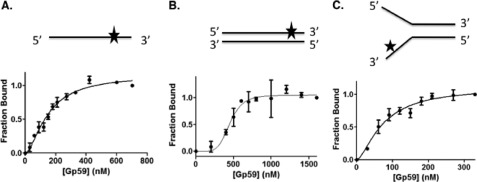
An important unresolved issue is how the relative affinities of Gp59 for Gp32 versus various DNA structures influence the location and timing of HLC assembly. Gp59 has been observed to bind with very high affinity (Kd = 2–3 nm) to certain Gp32 derivatives in the absence of ssDNA (20) and form complexes with full-length Gp32 that are stable at very high salt concentrations (14, 15, 31, 32). Estimates for the apparent Kd of Gp59-ssDNA interactions range from 50 to 100 nm under moderate salt conditions based on analysis of etheno-DNA binding data (13).4 Gp59 is reported to bind with higher affinity to specific DNA structures such as fork DNA (12); therefore, a quantitative assessment of structure-specific DNA binding is warranted. To this end, single-stranded, double-stranded, and fork oligonucleotides of the same length (25 bases or base pairs) and related sequence (Fig. 6 and Table 1) were labeled with Alexa Fluor 546 as described under “Experimental Procedures.” The binding of Gp59 to the ssDNA, dsDNA, and fork DNA ligands quenched Alexa Fluor 546 fluorescence at an emission wavelength of 569 nm by 27, 20, and 31%, respectively, at saturation (data not shown). Binding curves were generated by monitoring fluorescence quenching as a function of Gp59 concentration (Fig. 6). Based on the results of fitting to Equation 1, the order of binding affinities from strongest to weakest is fork DNA > ssDNA > dsDNA with all Kd values in the low to mid-nanomolar range (Table 2). The relative binding affinities are consistent with what was observed in electrophoretic mobility shift assays (12). Gp59 binds to all DNA types cooperatively, but its cooperativity for dsDNA is substantially higher than for ssDNA or fork DNA based on the different Hill coefficients (Fig. 6 and Table 2). Previous studies described cooperative binding of Gp59 to etheno-modified ssDNA and 2-aminopurine-labeled fork DNAs as well as hexamer formation on fork DNA (13, 21, 33). The presence of the Alexa Fluor 546 label significantly stabilizes Gp59-DNA interactions because fluorescence quenching of labeled fork, ssDNA, or dsDNA was not reduced by the addition of equimolar unlabeled DNA (data not shown). Thus, the data in Table 2 represent lower limits for the apparent Kd values for unmodified DNA. These results argue that Gp59 has higher affinity for Gp32 than it does for either fork DNA or single-stranded DNA.
FIGURE 6.
Binding curves from Alexa Fluor-labeled DNA substrates titrated with Gp59. DNA binding assays were performed as described under “Experimental Procedures.” Buffer contained the following components: 20 mm Tris acetate, pH 7.4, 10 mm magnesium acetate, 90 mm potassium acetate, and 1 mm DTT. All DNA substrates were 25-mers either as single-stranded DNA (A) or annealed to form double-stranded DNA (B) or fork DNA structures (C) as depicted with the relative position of the Alexa Fluor 546 probe indicated by the star. In each experiment, the concentration of the labeled Oligo 2 was 2 μm nucleotides. When present, unlabeled Oligo 3 or Oligo 4 was added in the same concentration for annealing as described under “Experimental Procedures.” Error bars represent the S.D. of the results of a minimum of two experiments.
TABLE 2.
Binding parameters for Gp59 binding to Alexa Fluor 546-labeled DNA substrates
Apparent Kd values (nm) and Hill coefficients were generated by fitting the data in Fig. 6 to Equation 1.
| Fork 25-mer | ssDNA 25-mer | dsDNA 25-mer | |
|---|---|---|---|
| Apparent Kd (nm) | 73 ± 12 | 149 ± 12 | 450 ± 29 |
| Hill coefficient | 1.6 ± 0.4 | 1.7 ± 0.2 | 5.3 ± 1.9 |
HLC Formation and Remodeling on Fork Oligonucleotide Substrate
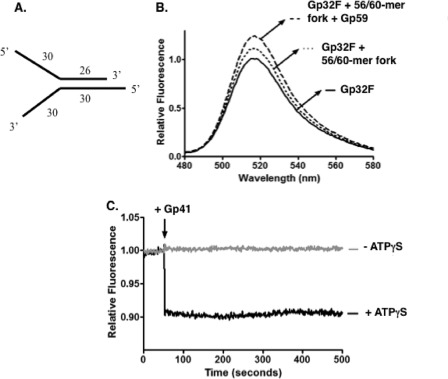
Fluorescence assays for detecting HLC formation and remodeling were repeated on a fork oligonucleotide structure (Fig. 7). The substrate used for these experiments is represented in Fig. 7A. The formation of the HLC occurred similarly on the fork oligonucleotide as seen with single-stranded oligonucleotides (Fig. 7B). However, the magnitudes of the fluorescence changes were somewhat smaller. Fig. 7C shows the results of the Gp32F displacement assays under conditions of helicase loading both in the presence and absence of ATPγS. In the presence of both Gp41 and ATPγS, there was an initial drop in Gp32F fluorescence followed by a rapid stabilization. This intensity drop did not occur when ATPγS was omitted, indicating that the fluorescence change occurs as a result of nucleotide-dependent helicase loading. This suggests that a helicase loading complex rearrangement may occur faster on fork structures than on ssDNA oligonucleotides.
FIGURE 7.
HLC formation and remodeling on fork oligonucleotide substrate. Fluorescence measurements were performed as described under “Experimental Procedures.” Buffer contained the following components: 20 mm Tris acetate, pH 7.8, 90 mm potassium acetate, 10 mm magnesium acetate, and 1 mm DTT. A, representation of the fork substrate used in the experiment; a 56-mer (Oligo 5) annealed to a 60-mer (Oligo 6). B, fluorescence emission scans monitored the helicase loading complex formation on the fork DNA. Solid line, 100 nm Gp32F alone; dotted line, 100 nm Gp32F + 1.66 μm (860 nm on ssDNA arms) 56-mer/60-mer fork; dashed line, 100 nm Gp32F + 1.66 μm (860 nm on ssDNA arms) 56-mer/60-mer fork + 100 nm Gp59. C, Gp32F fluorescence effects under conditions of helicase loading on fork DNA. The components of the helicase loading complex were added in the following order: 100 nm Gp32F, 1.66 μm (860 nm on ssDNA arms) 56-mer/60-mer fork, and 100 nm Gp59. Data show normalized fluorescence of this complex versus time. The arrow indicates the time of Gp41 addition. Black, 500 μm ATPγS was added simultaneously with 100 nm Gp41. Gray, 100 nm Gp41 was added in the absence of any nucleotide.
DISCUSSION
Across many conserved systems, the rate-limiting step of homology-directed repair is the conversion of a recombination intermediate into a replication fork (1, 5, 6). This step requires mediator proteins that recruit and properly load the appropriate helicase. In bacteriophage T4, this step is the loading of Gp41 onto the displaced strand of a D-loop, a task that is carried out by Gp59 protein. The initiation of RDR has been shown to depend on the presence of Gp59 (1). Furthermore, Gp59 is known to play a central role in the assembly of the T4 primosome, forming interactions with all other protein components of the primosome as well as with the replisome components Gp32 and Gp43 (DNA polymerase) (15, 16, 19, 34–36). The ternary Gp32-Gp59-ssDNA HLC is particularly important for the helicase loading function of Gp59. Our fluorescence assays have provided new insight into the formation of this complex in T4 RDR.
DNA Length Requirement of HLC Formation
Gp59 bound readily to Gp32F-covered 70- and 40-mers, oligonucleotides that support cooperative binding of Gp32 (Fig. 1, A and B). The high observed affinity of Gp32 for ssDNA is a consequence of its high cooperativity (37). On ssDNA molecules shorter than about 25 bases, Gp32 is unable to bind in a cooperative manner to form clusters, resulting in weaker observed binding. This correlates with decreased HLC formation on 20-mer and no HLC formation on 12-mer ssDNA in our experimental system (Fig. 1, C and D). Therefore, the formation of the HLC is only stable on ssDNA long enough to support Gp32 clusters. In fact, Gp59 selectively co-localized with clusters of Gp32 bound to ssDNA even when these clusters were present at low binding density and a large excess of free ssDNA was available (Fig. 2).
Further evidence that Gp59-mediated helicase assembly targets Gp32 clusters was presented in a published study of the effects of Gp32 mutations on the T4 DNA replication system (21). Gp32-B is an N-terminally truncated form of Gp32 that lacks cooperativity (38). Gp32 missense mutants R4G and R4T have greatly reduced cooperativity compared with wild type (39). All three mutants are defective in Gp59-mediated loading of Gp41 helicase at nascent replication forks, resulting in no (Gp32-B) or greatly reduced (R4G and R4T) stimulation of the strand displacement DNA synthesis activity of T4 DNA polymerase holoenzyme (21). Therefore, the inability to form stable Gp32 clusters curtails Gp59-mediated helicase assembly on nascent lagging strand ssDNA.
The requirement for a minimal ssDNA length and Gp32 cluster size suggests that Gp32 and Gp59 cannot co-localize on very short segments of ssDNA that may occur between Okazaki fragments during the maturation phase of lagging strand DNA synthesis, preventing improper helicase loading at such sites. Similarly, Gp59 and Gp32 co-localization is restricted from short ssDNA segments that may arise within presynaptic filaments during homologous recombination, preventing inappropriate antirecombination activity by Gp41 helicase (40).
Localized Rearrangement of HLC during Helicase Loading
In our Gp32F fluorescence assays, we defined conditions that distinguish between loading and translocation of Gp41 helicase. Under loading conditions (70-mer saturated with Gp32F and Gp59), the time-dependent decrease in Gp32F fluorescence required Gp41 and either ATP or ATPγS (Fig. 3). The requirement for both nucleotide cofactor and Gp59 indicates that the fluorescence change is dependent on active helicase loading by Gp59. The results in the presence of ATP and ATPγS were similar, which suggests that the conformational change of Gp59 and Gp32 upon Gp41 loading is dependent only on binding of Gp41-ATP to the complex rather than on ATP hydrolysis. An interesting question is whether Gp59 and Gp32 are still associated with the ssDNA at this point or whether they become displaced. In a previous study, it was shown that Gp59 and Gp32 remain bound to DNA in the presence of Gp41 and ATPγS but not in the presence of Gp41 and ATP (41). Therefore, it is likely that all three proteins are present on the ssDNA initially, and then both Gp59 and Gp32 are displaced at the start of ATP hydrolysis and translocation by Gp41. The high affinity of Gp59-Gp32 interactions (20) suggests that these proteins remain bound to each other after being displaced.
Gp32 Displacement during Gp41 Translocation
Gp41 displaces Gp32 as it translocates along ssDNA. This was seen in our Gp32F fluorescence experiments in which a limiting concentration of Gp59 was used to load helicase at dispersed sites on Gp32F-saturated M13 mp18 ssDNA circles (Fig. 4). Under these conditions, there is a relatively fast decrease in Gp32F fluorescence that depends on ATP hydrolysis by Gp41 because only a very slow signal decrease occurred in the presence of ATPγS. These results are consistent with the rapid displacement of Gp32F as Gp41 translocates along the circular ssDNA. Presumably, Gp32F rebinds to the ssDNA at some distance behind the moving helicase, establishing the intermediate level of signal decrease that we observed at steady state (Fig. 4).
Gp59 Promotes Multiple Rounds of Helicase Loading
Although Gp59 was present at a limiting concentration (10 nm) in the translocation assays, the amount of translocating helicase that can be placed on Gp32F-saturated ssDNA was not limited by Gp59 concentration in a stoichiometric sense. The rate of Gp32F displacement increased markedly with increasing Gp41 concentration from 25 to 200 nm (Figs. 4 and 5), the latter representing a 20-fold excess of Gp41 over Gp59. This suggests that Gp59 and/or the entire HLC can be recycled to promote multiple rounds of helicase loading onto the same or different ssDNA molecules. The saturable, sigmoidal plot of Gp32F displacement rate versus [Gp41] (Fig. 5) supports the notion of an enzyme-like helicase loading activity for the HLC. Turnover in this case may involve dissociation of Gp59-Gp32 from the ssDNA upon ATP hydrolysis by Gp41 as suggested above (Fig. 4) (41). As Gp41 translocates along ssDNA, it displaces more Gp32F and generates additional free ssDNA. This ssDNA is a target for rebinding by Gp59-Gp32, a notion supported by the finding that Gp32F prebound to Gp59 can rapidly bind to M13mp18 ssDNA.4 The two proteins could then form a new HLC and undergo another round of Gp41 loading at a nearby or distant site, depending on the relative rates of helicase translocation, HLC reassembly, and protein exchange.
Helicase Loading Complex Formation and Rearrangement on Fork DNA
The helicase loading complex forms on a fork oligonucleotide as indicated by changes in Gp32F fluorescence (Fig. 7). The signal changes were smaller than those observed with homopolymeric oligos and long M13mp18 ssDNA, which likely is due to the short length (30 bases) and mixed sequence of the ssDNA arms of the fork (Fig. 7A). Nevertheless, the addition of Gp59 strongly enhanced the fluorescence of the Gp32F-fork DNA complex (Fig. 7B), which is the signature of HLC formation. We conclude that the HLC forms on at least one of the ssDNA arms of fork DNA. We hypothesize that HLC formation occurs preferentially on the 5′ ssDNA arm, consistent with results suggesting that Gp32 binds preferentially to this strand (42). Our results from Fig. 7C show that the HLC is remodeled on fork DNA upon Gp41 loading in the presence of ATPγS. The relative extent of Gp32F signal change upon Gp41/ATPγS addition was similar in experiments with fork DNA (Fig. 7C) versus ssDNA (Fig. 3A), but the rate of signal change was much faster with fork DNA. This finding suggests that Gp32 and fork DNA recognition could act synergistically to load helicase onto Gp32 clusters that form on the lagging strand side of a fork junction or of a D-loop.
Interactions with Gp32 Direct Helicase Loading Activity of Gp59
Bacteriophage T4 appears to express a small amount of Gp59 during its infection cycle in E. coli, and this Gp59 exists in large deficit with respect to the concentration of expressed Gp32 (31). Gp59-Gp32 protein-protein interactions are stable in high salt (14, 15, 31, 32). Previous studies point to a Kd in the low nanomolar range for Gp59-Gp32 interactions in both the absence and presence of DNA (20, 41). Therefore, the affinity of Gp59 for Gp32 appears to exceed its affinity for fork or single-stranded DNA structures (Fig. 6 and Table 2). In addition, previous studies determined that Gp59 and Gp32 reduce each other's affinity for DNA, which may serve to prime HLC complexes for protein exchange during helicase loading (19, 21). Together, these observations indicate that Gp59 rarely exists as a free protein under physiological conditions; rather it is likely to exist in complexes with Gp32 and to interact at least initially with DNA and other proteins in this form.
Helicase Loading in Recombination-dependent Replication and Repair
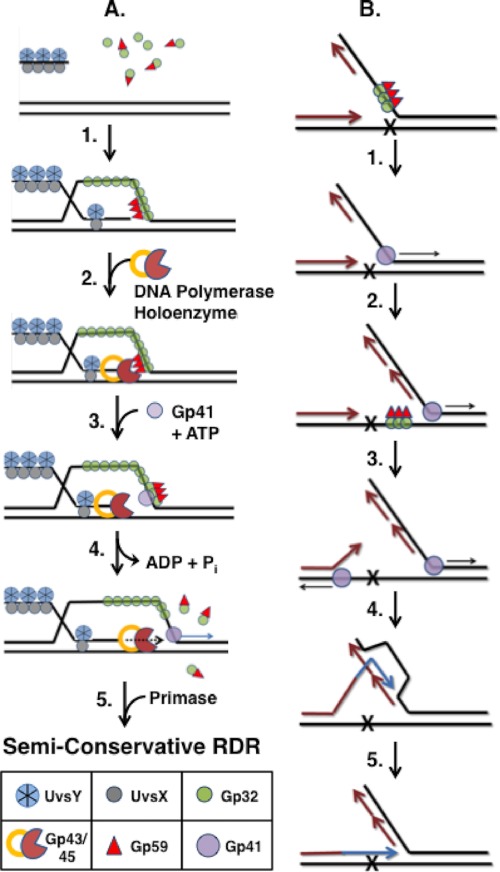
In the bacteriophage T4 recombination-dependent replication pathway (Fig. 8A), DNA strand exchange catalyzed by UvsX recombinase generates D-loop intermediates that are converted into replication forks, a handoff that requires the helicase loading activity of Gp59 (1, 5, 6). To successfully initiate RDR, Gp41 helicase must be loaded specifically onto the displaced strand of the D-loop, which will become the lagging strand of the nascent replication fork (Fig. 8A). If Gp41 is loaded onto the invading strand instead, its 5′ → 3′ helicase activity would result in dissolution of the D-loop, terminating RDR. For this reason, the structure-specific DNA binding activity of Gp59, which targets forks and cruciforms, is insufficient to enforce strand-specific helicase loading during RDR initiation. Instead, it appears that Gp59 has evolved to follow Gp32 to the correct strand of the D-loop to efficiently couple recombination to DNA synthesis. Kodadek (43) demonstrated that Gp32 rapidly sequesters the displaced strand of D-loops during UvsX-catalyzed DNA strand exchange (Fig. 8A, Step 1). In contrast, Gp32 is excluded from the invading strand, which is saturated with recombination proteins (40). Therefore, Gp59 would co-localize with Gp32 clusters on the displaced strand. In principle, HLC could form anywhere on this strand; however, the added affinity of Gp59 for fork DNA might lead preferentially to HLC assembly at the fork junction as suggested by the synergistic effect observed in Fig. 7 and as depicted in Fig. 8A, Step 1. There, Gp59 could perform its two known replication functions: inhibiting the leading strand polymerase (Step 2) and subsequently loading helicase while releasing polymerase (Steps 3 and 4) (Refs. 9 and 34–36; for reviews, see Refs. 1 and 5). Gp59-Gp32 released from the HLC (Step 4) may be recycled for additional rounds of helicase loading. The model in Fig. 8A depicts Gp59-Gp32 leaving the HLC as a free complex (Step 4); however, it is possible that the complex remains tethered to the replication fork via other interactions as suggested by electron microscopy studies (23).
FIGURE 8.
Models for Gp59-dependent helicase loading at Gp32-ssDNA clusters during recombination-dependent DNA replication and repair. A, strand-specific loading of Gp41 helicase during RDR (Step 1). Following DNA strand invasion promoted by UvsX recombinase, UvsY mediator, and Gp32, Gp59 co-localizes with Gp32 clusters on the displaced strand of the D-loop intermediate, forming a helicase loading complex. The structure-specific DNA binding activity of Gp59 may help target the HLC to the leading edge (fork junction) of the D-loop (Step 2). As the protein components of the T4 replisome assemble, the presence of Gp59 at the fork inhibits Gp43 (DNA polymerase) from replicating the leading strand (Step 3). Recruitment of Gp41-ATP rearranges the HLC so that it no longer blocks the leading strand polymerase (Step 4). ATP hydrolysis by Gp41 drives unwinding of the duplex ahead of Gp43. Gp32 and Gp59 both are ejected from the helicase assembly site but may be recycled for additional rounds of helicase loading (see text for details) (Step 5). Primase recruitment reconstitutes the Gp41/Gp61 primosome leading to semiconservative RDR. B, hypothetical model for multiple rounds of helicase loading during error-free lesion bypass. This process involves the uncoupling of leading/lagging strand DNA synthesis followed by a round of RDR using a newly synthesized Okazaki fragment as template. Newly synthesized DNA is shown as either magenta (semiconservative synthesis) or blue (bubble migration synthesis). Replication and recombination proteins other than Gp59, Gp32, and Gp41 are omitted for clarity (Step 1). Leading and lagging strand syntheses are initially coupled until the leading strand polymerase encounters a lesion and becomes stalled. Uncoupling of leading/lagging strand synthesis could require a new round of helicase loading by Gp59 targeting Gp32 clusters on the lagging strand ssDNA (Step 2). Continued helicase translocation allows for the synthesis of a new Okazaki fragment overlapping the site of damage. Helicase activity also exposes ssDNA on the damaged strand that is sufficiently long for HLC assembly (Step 3). An additional round of helicase loading and translocation from this site leads to the unwinding of the stalled daughter strand from the damaged template (Step 4). The displaced 3′-end of the daughter strand primes a round of RDR using the overlapping Okazaki fragment as a template (Step 5). Bubble migration RDR extends the daughter strand until it can reanneal to the original template and continue semiconservative synthesis, having bypassed the lesion. See text for details.
The ability of Gp59-Gp32 to perform multiple rounds of helicase loading could be important for genome stability mechanisms including replication fork restart and lesion bypass. It was previously shown that Gp59 is required to maintain leading strand synthesis by Gp43-A737V, a DNA polymerase mutant exhibiting low processivity and frequent pausing (27). Continuous reloading of helicase by Gp59 was proposed as a mechanism to explain this dependence. Another genome stability mechanism that is likely to involve multiple rounds of helicase loading is error-free lesion bypass (Fig. 8B) (1, 44, 45). Here, in a replisome encountering a lesion on the leading strand template, leading and lagging strand synthesis must be uncoupled (Steps 1 and 2). Gp59-Gp32 could promote this uncoupling by reloading Gp41 onto the lagging strand ssDNA. This helicase could form a primosome that primes synthesis of a new Okazaki fragment that spans the site of damage (Step 2). This Okazaki fragment would serve as the template for a round of recombination-dependent replication that bypasses the lesion (Steps 3–5). This would entail unwinding the stalled daughter strand from its template to make it accessible to recombination proteins. Gp59-Gp32 could facilitate this by loading helicase onto the exposed ssDNA of the leading strand template (Step 3). This mechanism has elements in common with Gp41- and Gp32-dependent models of template switching proposed by Kadyrov and Drake (44, 45) but may differ in detail from alternative lesion bypass mechanisms promoted by T4 Dda and/or UvsW helicase.
Our results provide further understanding of the interactions between Gp32 and Gp59 in the process of helicase loading in the coupled T4 replication and recombination systems. Our studies have significant implications for cancer and helicase-related diseases as improper helicase loading can lead to genome instability via the failure of double strand break repair and related pathways. Our findings should be useful for the design of future experiments to further investigate helicase loading in T4 and other systems.
Acknowledgment
We thank Dr. Stephen Benkovic for providing overexpression vectors for Gp59 and Gp32.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM48847 (to S. W. M.) and Training Grant T32ES07122 (to A. M. B.).
A. Branagan and S. Morrical, unpublished observations.
- RDR
- recombination-dependent DNA replication
- ssDNA
- single-stranded DNA
- ATPγS
- adenosine-5′-(3-thio)-triphosphate
- HLC
- helicase loading complex.
REFERENCES
- 1. Maher R. L., Branagan A. M., Morrical S. W. (2011) Coordination of DNA replication and recombination activities in the maintenance of genome stability. J. Cell. Biochem. 112, 2672–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Formosa T., Alberts B. M. (1986) DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell 47, 793–806 [DOI] [PubMed] [Google Scholar]
- 3. Morrical S. W., Alberts B. M. (1990) The UvsY protein of bacteriophage T4 modulates recombination-dependent DNA synthesis in vitro. J. Biol. Chem. 265, 15096–15103 [PubMed] [Google Scholar]
- 4. Morrical S. W., Wong M. L., Alberts B. M. (1991) Amplification of snap-back DNA synthesis reactions by the uvsX recombinase of bacteriophage T4. J. Biol. Chem. 266, 14031–14038 [PubMed] [Google Scholar]
- 5. Liu J., Morrical S. W. (2010) Assembly and dynamics of the bacteriophage T4 homologous recombination machinery. Virol. J. 7, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kreuzer K. N., Brister J. R. (2010) Initiation of bacteriophage T4 DNA replication and replication fork dynamics: a review in the Virology Journal series on bacteriophage T4 and its relatives. Virol. J. 7, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alberts B. M. (1987) Prokaryotic DNA replication mechanisms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 317, 395–420 [DOI] [PubMed] [Google Scholar]
- 8. Jones J. M., Nakai H. (2000) PriA and phage T4 gp59: factors that promote DNA replication on forked DNA substrates microreview. Mol. Microbiol. 36, 519–527 [DOI] [PubMed] [Google Scholar]
- 9. Jones C. E., Mueser T. C., Dudas K. C., Kreuzer K. N., Nossal N. G. (2001) Bacteriophage T4 gene 41 helicase and gene 59 helicase-loading protein: a versatile couple with roles in replication and recombination. Proc. Natl. Acad. Sci. U.S.A. 98, 8312–8318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mueser T. C., Jones C. E., Nossal N. G., Hyde C. C. (2000) Bacteriophage T4 gene 59 helicase assembly protein binds replication fork DNA. The 1.45 Å resolution crystal structure reveals a novel α-helical two-domain fold. J. Mol. Biol. 296, 597–612 [DOI] [PubMed] [Google Scholar]
- 11. Mueser T. C., Hinerman J. M., Devos J. M., Boyer R. A., Williams K. J. (2010) Structural analysis of bacteriophage T4 DNA replication: a review in the Virology Journal series on bacteriophage T4 and its relatives. Virol. J. 7, 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jones C. E., Mueser T. C., Nossal N. G. (2000) Interaction of the bacteriophage T4 gene 59 helicase loading protein and gene 41 helicase with each other and with fork, flap, and cruciform DNA. J. Biol. Chem. 275, 27145–27154 [DOI] [PubMed] [Google Scholar]
- 13. Lefebvre S. D., Morrical S. W. (1997) Interactions of the bacteriophage T4 gene 59 protein with single-stranded polynucleotides: binding parameters and ion effects. J. Mol. Biol. 272, 312–326 [DOI] [PubMed] [Google Scholar]
- 14. Morrical S. W., Hempstead K., Morrical M. D. (1994) The gene 59 protein of bacteriophage T4 modulates the intrinsic and single-stranded DNA-stimulated ATPase activities of gene 41 protein, the T4 replicative DNA helicase. J. Biol. Chem. 269, 33069–33081 [PubMed] [Google Scholar]
- 15. Morrical S. W., Beernink H. T., Dash A., Hempstead K. (1996) The gene 59 protein of bacteriophage T4. Characterization of protein-protein interactions with gene 32 protein, the T4 single-stranded DNA binding protein. J. Biol. Chem. 271, 20198–20207 [DOI] [PubMed] [Google Scholar]
- 16. Ishmael F. T., Alley S. C., Benkovic S. J. (2001) Identification and mapping of protein-protein interactions between gp32 and gp59 by cross-linking. J. Biol. Chem. 276, 25236–25242 [DOI] [PubMed] [Google Scholar]
- 17. Ishmael F. T., Alley S. C., Benkovic S. J. (2002) Assembly of the bacteriophage T4 helicase: architecture and stoichiometry of the gp41-gp59 complex. J. Biol. Chem. 277, 20555–20562 [DOI] [PubMed] [Google Scholar]
- 18. Delagoutte E., von Hippel P. H. (2005) Mechanistic studies of the T4 DNA (gp41) replication helicase: functional interactions of the C-terminal tails of the helicase subunits with the T4 (gp59) helicase loader protein. J. Mol. Biol. 347, 257–275 [DOI] [PubMed] [Google Scholar]
- 19. Lefebvre S. D., Wong M. L., Morrical S. W. (1999) Simultaneous interactions of bacteriophage T4 DNA replication proteins gp59 and gp32 with single-stranded (ss) DNA. Co-modulation of ssDNA binding activities in a DNA helicase assembly intermediate. J. Biol. Chem. 274, 22830–22838 [DOI] [PubMed] [Google Scholar]
- 20. Xu H., Wang Y., Bleuit J. S., Morrical S. W. (2001) Helicase assembly protein Gp59 of bacteriophage T4: fluorescence anisotropy and sedimentation studies of complexes formed with derivatives of Gp32, the phage ssDNA binding protein. Biochemistry 40, 7651–7661 [DOI] [PubMed] [Google Scholar]
- 21. Ma Y., Wang T., Villemain J. L., Giedroc D. P., Morrical S. W. (2004) Dual functions of single-stranded DNA-binding protein in helicase loading at the bacteriophage T4 DNA replication fork. J. Biol. Chem. 279, 19035–19045 [DOI] [PubMed] [Google Scholar]
- 22. Pant K., Karpel R. L., Rouzina I., Williams M. C. (2005) Salt dependent binding of T4 gene 32 protein to single and double-stranded DNA: single molecule force spectroscopy measurements. J. Mol. Biol. 349, 317–330 [DOI] [PubMed] [Google Scholar]
- 23. Chastain P. D., 2nd, Makhov A. M., Nossal N. G., Griffith J. (2003) Architecture of the replication complex and DNA loops at the fork generated by the bacteriophage t4 proteins. J. Biol. Chem. 278, 21276–21285 [DOI] [PubMed] [Google Scholar]
- 24. Liu J., Qian N., Morrical S. W. (2006) Dynamics of bacteriophage T4 presynaptic filament assembly from extrinsic fluorescence measurements of Gp32-single-stranded DNA interactions. J. Biol. Chem. 281, 26308–26319 [DOI] [PubMed] [Google Scholar]
- 25. Alberts B. M., Herrick G. (1971) DNA-cellulose chromatography. Methods Enzymol. 21, 198–217 [Google Scholar]
- 26. Gill S. C., von Hippel P. H. (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182, 319–326 [DOI] [PubMed] [Google Scholar]
- 27. Spacciapoli P., Nossal N. G. (1994) Interaction of DNA polymerase and DNA helicase within the bacteriophage T4 DNA replication complex. Leading strand synthesis by the T4 DNA polymerase mutant A737V (tsL141) requires the T4 gene 59 helicase assembly protein. J. Biol. Chem. 269, 447–455 [PubMed] [Google Scholar]
- 28. Kowalczykowski S. C. (1990) in Landolt-Börnstein: Numerical Data and Functional Relationships in Science and Technology (Saenger W., ed) pp. 244–263, Springer-Verlag, Berlin [Google Scholar]
- 29. Kowalczykowski S. C., Lonberg N., Newport J. W., von Hippel P. H. (1981) Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. I. Characterization of the binding interactions. J. Mol. Biol. 145, 75–104 [DOI] [PubMed] [Google Scholar]
- 30. Schrock R. D., Alberts B. (1996) Processivity of the gene 41 DNA helicase at the bacteriophage T4 DNA replication fork. J. Biol. Chem. 271, 16678–16682 [DOI] [PubMed] [Google Scholar]
- 31. Barry J., Alberts B. (1994) Purification and characterization of bacteriophage T4 gene 59 protein. A DNA helicase assembly protein involved in DNA replication. J. Biol. Chem. 269, 33049–33062 [PubMed] [Google Scholar]
- 32. Yonesaki T. (1994) The purification and characterization of gene 59 protein from bacteriophage T4. J. Biol. Chem. 269, 1284–1289 [PubMed] [Google Scholar]
- 33. Arumugam S. R., Lee T. H., Benkovic S. J. (2009) Investigation of stoichiometry of T4 bacteriophage helicase loader protein (gp59). J. Biol. Chem. 284, 29283–29289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xi J., Zhang Z., Zhuang Z., Yang J., Spiering M. M., Hammes G. G., Benkovic S. J. (2005) Interaction between the T4 helicase loading protein (gp59) and the DNA polymerase (gp43): unlocking of the gp59-gp43-DNA complex to initiate assembly of a fully functional replisome. Biochemistry 44, 7747–7756 [DOI] [PubMed] [Google Scholar]
- 35. Xi J., Zhuang Z., Zhang Z., Selzer T., Spiering M. M., Hammes G. G., Benkovic S. J. (2005) Interaction between the T4 helicase-loading protein (gp59) and the DNA polymerase (gp43): a locking mechanism to delay replication during replisome assembly. Biochemistry 44, 2305–2318 [DOI] [PubMed] [Google Scholar]
- 36. Nelson S. W., Yang J., Benkovic S. J. (2006) Site-directed mutations of T4 helicase loading protein (gp59) reveal multiple modes of DNA polymerase inhibition and the mechanism of unlocking by gp41 helicase. J. Biol. Chem. 281, 8697–8706 [DOI] [PubMed] [Google Scholar]
- 37. Lonberg N., Kowalczykowski S. C., Paul L. S., von Hippel P. H. (1981) Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. III. Binding properties of two specific proteolytic digestion products of the protein (G32P*I and G32P*III). J. Mol. Biol. 145, 123–138 [DOI] [PubMed] [Google Scholar]
- 38. Giedroc D. P., Khan R., Barnhart K. (1990) Overexpression, purification, and characterization of recombinant T4 gene 32 protein22–301 (g32P-B). J. Biol. Chem. 265, 11444–11455 [PubMed] [Google Scholar]
- 39. Villemain J. L., Giedroc D. P. (1993) Energetics of arginine-4 substitution mutants in the N-terminal cooperativity domain of T4 gene 32 protein. Biochemistry 32, 11235–11246 [DOI] [PubMed] [Google Scholar]
- 40. Bleuit J. S., Xu H., Ma Y., Wang T., Liu J., Morrical S. W. (2001) Mediator proteins orchestrate enzyme-ssDNA assembly during T4 recombination-dependent DNA replication and repair. Proc. Natl. Acad. Sci. U.S.A. 98, 8298–8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang Z., Spiering M. M., Trakselis M. A., Ishmael F. T., Xi J., Benkovic S. J., Hammes G. G. (2005) Assembly of the bacteriophage T4 primosome: single-molecule and ensemble studies. Proc. Natl. Acad. Sci. U.S.A. 102, 3254–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jones C. E., Mueser T. C., Nossal N. G. (2004) Bacteriophage T4 32 protein is required for helicase-dependent leading strand synthesis when the helicase is loaded by the T4 59 helicase-loading protein. J. Biol. Chem. 279, 12067–12075 [DOI] [PubMed] [Google Scholar]
- 43. Kodadek T. (1990) The role of the bacteriophage T4 gene 32 protein in homologous pairing. J. Biol. Chem. 265, 20966–20969 [PubMed] [Google Scholar]
- 44. Kadyrov F. A., Drake J. W. (2003) Properties of bacteriophage T4 proteins deficient in replication repair. J. Biol. Chem. 278, 25247–25255 [DOI] [PubMed] [Google Scholar]
- 45. Kadyrov F. A., Drake J. W. (2004) UvsX recombinase and Dda helicase rescue stalled bacteriophage T4 DNA replication forks in vitro. J. Biol. Chem. 279, 35735–35740 [DOI] [PubMed] [Google Scholar]
- 46. Morrical S., Qian N. (June 9, 2009) U. S. Patent 7,544,469 B2