Background: Bnip3, a protein involved in initiation of cardiovascular disease and cancer, induces autophagy of damaged organelles by a poorly understood mechanism.
Results: Bnip3 homodimerization and binding to LC3 induces selective autophagy of mitochondria and ER.
Conclusion: Bnip3 plays a direct and selective role in autophagy.
Significance: Our studies will advance future studies investigating the role of Bnip3 in human disease.
Keywords: Autophagy, Bcl-2 Family Proteins, Endoplasmic Reticulum (ER), Mitochondria, Mitochondrial Apoptosis, Bnip3, LC3
Abstract
Autophagy plays an important role in cellular quality control and is responsible for removing protein aggregates and dysfunctional organelles. Bnip3 is an atypical BH3-only protein that is known to cause mitochondrial dysfunction and cell death. Interestingly, Bnip3 can also protect against cell death by inducing mitochondrial autophagy. The mechanism for this process, however, remains poorly understood. Bnip3 contains a C-terminal transmembrane domain that is essential for homodimerization and proapoptotic function. In this study, we show that homodimerization of Bnip3 is also a requirement for induction of autophagy. Several Bnip3 mutants that do not interfere with its mitochondrial localization but disrupt homodimerization failed to induce autophagy in cells. In addition, we discovered that endogenous Bnip3 is localized to both mitochondria and the endoplasmic reticulum (ER). To investigate the effects of Bnip3 at mitochondria or the ER on autophagy, Bnip3 was targeted specifically to each organelle by substituting the Bnip3 transmembrane domain with that of Acta or cytochrome b5. We found that Bnip3 enhanced autophagy in cells from both sites. We also discovered that Bnip3 induced removal of both ER (ERphagy) and mitochondria (mitophagy) via autophagy. The clearance of these organelles was mediated in part via binding of Bnip3 to LC3 on the autophagosome. Although ablation of the Bnip3-LC3 interaction by mutating the LC3 binding site did not impair the prodeath activity of Bnip3, it significantly reduced both mitophagy and ERphagy. Our data indicate that Bnip3 regulates the apoptotic balance as an autophagy receptor that induces removal of both mitochondria and ER.
Introduction
Autophagy is an evolutionary conserved catabolic process that occurs constitutively in most cells (1). It is also rapidly increased when there is a change in the cellular environment such as a reduction in nutrients or increased oxidative stress. Up-regulation of autophagy allows cells to reuse their own constituents for amino acids and fatty acids during the starvation period. Autophagy also plays an important role in cellular quality control where it degrades protein aggregates and dysfunctional organelles that can be harmful to the cell. Although autophagy is generally considered to be nonspecific, it has become clear that autophagy can be selective for organelles such as mitochondria (2, 3), ER2 (4, 5), and peroxisomes (6) under certain conditions. When autophagy is initiated, a double-membrane structure sequesters cytoplasmic material that is subsequently delivered to the lysosome for degradation via fusion (1). Exactly how specific organelles are removed by autophagosomes is still unclear. It has been suggested that there is an interaction between proteins on the target organelle and the Atg8-family proteins (e.g. LC3 and GABARAP) on the forming autophagosome. The Atg8 family proteins bind to proteins that contain a WXXL motif called the LC3-interacting region (LIR) (7–9).
The Bcl-2/adenovirus E1B 19-kDa interacting protein 3 (Bnip3) is a BH3-only protein primarily localized to the mitochondria. Bnip3 induces cell death by perturbing mitochondrial function (10–12). In addition, specific targeting of Bnip3 to the ER interferes with Ca2+ homeostasis and promotes cell death (13, 14). Bnip3 is also a potent inducer of autophagy in many different cell types (2, 15–18). Recent studies have demonstrated that mitophagy is specifically activated by Bnip3 and its homologue Nix/Bnip3L and that this process is separate from activation of cell death (2, 19, 20). Currently it is unclear how Bnip3 targets mitochondria for removal by the autophagosomes. Bnip3 is anchored in the outer mitochondrial membrane via its C-terminal transmembrane domain (TMD), whereas the N terminus faces the cytosol. The C-terminal TMD is essential for targeting Bnip3 to the mitochondria, homodimerization, and proapoptotic activity (13, 21, 22). Interestingly, the N terminus of Bnip3 contains a WXXL-like motif that might be important in binding to Atg8 family proteins. Binding of Bnip3 to Atg8 proteins such as LC3 might serve to dock mitochondria to the autophagosomes, thereby ensuring their removal. The Bnip3 homologue Nix/Bnip3L was reported recently to contain a similar motif that was important for its interaction with LC3A and GABARAP (23, 24). However, it is currently unknown whether Bnip3 also interacts with GABARAP and LC3 via its WXXL motif and whether this interaction is important for selective removal of mitochondria.
In this study, we investigated the mechanism by which Bnip3 promotes selective autophagy in cells. We report that endogenous Bnip3 is localized to both mitochondria and ER in cells and that Bnip3 induces autophagy from both locations. Moreover, our findings suggest that LC3 binds to the LIR in the Bnip3 homodimer and that this interaction is important for removal of mitochondria and ER.
EXPERIMENTAL PROCEDURES
Cell Culture
HeLa cells were maintained in DMEM plus 10% (v/v) FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. HeLa cells were infected with adenoviruses encoding GFP-LC3, β-galactosidase (β-gal), Bnip3, or the different Bnip3 mutants in DMEM + 2% heat-inactivated serum. After 3 h, the cells were rinsed in PBS and incubated in growth medium. Western blot analysis and cell death experiments were performed 24 h after the adenoviral infection. For fluorescence microscopy and immunoprecipitation experiments, HeLa cells were transiently transfected with empty Myc vector, Bnip3, or Bnip3 mutants using FuGENE 6 (Roche) according to the instructions of the manufacturer. For immunoprecipitation experiments, the cells were transfected for 24 h. For fluorescence microscopy experiments, cells were transfected for 48 h. The Bnip3Acta and Bnip3cb5 constructs were generously provided by Dr. Gary Isom, Purdue University (14). Bnip3W18A and Bnip3cb5W18A were prepared using site-directed mutagenesis by PCR with Bnip3 wild-type and Bnip3cb5 constructs as templates, respectively. The Bnip3 adenoviruses were generated using the pENTR directional TOPO cloning kit (Invitrogen) followed by recombination into the pAd/CMV/V5-DEST gateway vector (Invitrogen). All autophagy experiments were done in the presence of 50 μm zVAD-fmk (carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone) (EMD Biosciences).
Western Blotting and Coimmunoprecipitation
HeLa cells were lysed in ice-cold buffer (50 mm Tris/HCl (pH 7.4), 150 mm NaCl, 1 mm EGTA, 1 mm EDTA, 1% Triton X-100 and Completetm protease inhibitor cocktail (Roche)) and then cleared by centrifugation at 20,000 × g for 20 min at 4 °C. Proteins in the supernatants were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and immunoblotted with antibodies. The protein concentrations were determined by the Coomassie Blue binding assay (Pierce) using BSA standards. In the immunoprecipitation experiment, cell lysates were precleared with protein G PLUS-agarose (Santa Cruz Biotechnology) for 1 h and then incubated with anti-GFP or anti-GABARAP overnight. Immune complexes were captured with protein G PLUS-agarose beads, eluted in 2× SDS sample buffer, and analyzed by Western blotting. Blots were quantified and analyzed using Quantity One software (Bio-Rad).
Hypoxia and Subcellular Fractionation
HeLa cells in growth medium were placed in hypoxic pouches (GasPak EZ, BD Biosciences) at 37 °C. After 24 h of hypoxia, cells were scraped in ice-cold PBS, resuspended in ice-cold isolation buffer (200 mm sucrose, 1 mm EGTA, and 10 mm MOPS (pH7.4)) and subsequently lysed with a glass dounce homogenizer. The cellular fractions were separated by differential centrifugation: mitochondria, 8000 × g for 10 min and ER, 100,000 × g for 90 min. Rat hearts were homogenized in buffer containing 250 mm sucrose, 5 mm KH2PO4, 2 mm MgCl2, 10 mm MOPS, 1 mm EGTA, 0.1% fatty acid-free BSA, and protease inhibitors (Roche) and then centrifuged at 600 × g for 5 min. The pellet was discarded, and the supernatant was centrifuged at 3000 × g for 10 min to obtain the mitochondrial fraction. The supernatant was centrifuged at 100,000 × g for 90 min to obtain cytosol and ER/sarcoplasmic reticulum (SR)-rich fractions.
Isolation of Autophagosomes
GFP-LC3-positive autophagosomes were isolated from HeLa cells using a protocol adapted from Gao et al. (25). HeLa cells were homogenized in ice-cold buffer (250 mm Sucrose, 5 mm KH2PO4, 2 mm MgCl2, 10 mm MOPS (pH 7.4), 1 mm EGTA, 0.1% fatty acid-free BSA supplemented with protease inhibitors (Roche)) and then centrifuged at 4 °C for 10 min at 10,000 × g. The supernatant was incubated with a monoclonal mouse GFP antibody coupled to MACS magnetic beads (Miltenyi Biotech) for 1 h before being applied to a MACS MS separation column. Proteins were eluted in buffer containing 50 mm Tris-HCl (pH 6.8), 50 mm DTT, 1% SDS, 1 mm EDTA, and 10% glycerol and subsequently analyzed by Western blot analysis.
Immunofluorescence Analysis
Cells overexpressing vector or Bnip3 constructs plus GFP-LC3 or GABARAP-GFP for 48 h were fixed with 4% (w/v) paraformaldehyde for 15 min, permeabilized with 0.2% Triton X-100 in PBS, and then blocked in 5% goat serum. The cells were incubated with anti-TOM20 or anti-calnexin and then with goat anti-rabbit Alexa Fluor 594 secondary antibody. Cells were examined using a Carl Zeiss AxioObserver Z1 equipped with a motorized z-stage and ApoTome for optical sectioning. To assess autophagy, cells overexpressing GFP-LC3 were examined at ×60 magnification and classified as cells with diffuse GFP-LC3 fluorescence or as cells with > 20 GFP-LC3 puncta/cell. For quantitation of autophagosomes in single cells, z-stacks of at least 50 representative cells per condition in three separate experiments were acquired through the ×60 oil immersion lens. Maximum projection images of the z-stacks were produced using the AxioVision software, and the number of autophagosomes in each cell was counted. Mitophagy was assessed by analyzing colocalization between GFP-LC3 positive autophagosomes and Tom20-labeled mitochondria. ERphagy was assessed by measuring colocalization between GFP-LC3-positive autophagosomes and calnexin-labeled ER. The pseudo-line scan analysis was performed using ImageJ software. To assess cell death, HeLa cells infected with β-gal, Bnip3, or Bnip3W18A were stained with 100 nm tetramethylrhodamine methyl ester (TMRM) (Invitrogen) and 50 μm Hoechst 33342 (Invitrogen) for 10 min. Cells were examined at ×10 magnification using a fluorescence microscope. Cells stained with TMRM (red) and Hoechst 33342 (blue) were counted as live cells, whereas cells with Hoechst 33342 alone were considered dead.
Statistical Analysis
All values are expressed as mean ± S.E. Statistical analyses were performed using Student's t test to identify statistical significance between groups. p < 0.05 was considered significant.
RESULTS
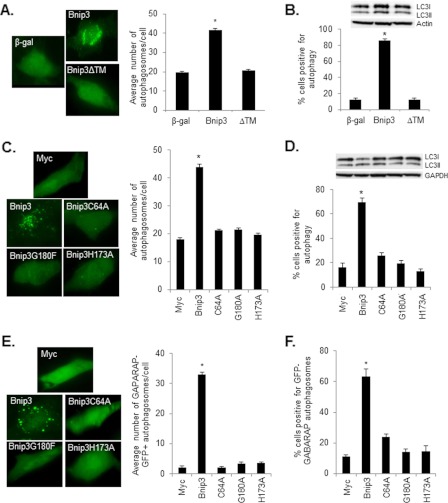
Bnip3 contains a C-terminal TMD that is important for its homodimerization and proapoptotic function (13, 21, 22). To investigate whether an intact TMD is also required for activation of autophagy, we overexpressed wild-type Bnip3 or the carboxyl terminal TMD deletion mutant of Bnip3 (Bnip3ΔTM) plus GFP-LC3 to monitor formation of autophagosomes in HeLa cells. We found that deletion of the TMD completely abrogated induction of autophagy as measured by fluorescence microscopy for GFP-LC3-positive autophagosomes (Fig. 1A). The average number of autophagosomes per cell (Fig. 1A) and the number of cells positive for induction of autophagy (Fig. 1B) were increased significantly in cells overexpressing Bnip3 but not Bnip3ΔTM. The conversion of LC3I to LC3II is also indicative of increased autophagic activity (25), and Western blot analysis of endogenous LC3 levels confirmed an increase in LC3II in cells overexpressing Bnip3 but not Bnip3ΔTM (Fig. 1B).
FIGURE 1.
Homodimerization of Bnip3 is essential for induction of autophagy. A, HeLa cells overexpressing Bnip3 or Bnip3 mutants were scored for the presence of GFP-LC3-positive autophagosomes. Bnip3, but not Bnip3ΔTM, significantly increased the number of autophagosomes/cell compared with β-gal-infected cells. *, p < 0.05. n = 3. B, Bnip3, but not Bnip3ΔTM, significantly increased the number of cells positive for autophagy compared with β-gal-infected cells. *, p < 0.05. n = 3. Shown is a representative Western blot analysis for endogenous LC3I/II. C, Bnip3, but not the homodimerization-deficient mutants, significantly increased the number of GFP-LC3 positive autophagosomes/cell compared with Myc-transfected control cells. *, p < 0.05. n = 3. D, Bnip3, but not the homodimerization-deficient mutants, induced significant autophagy compared with Myc-transfected control cells. *, p < 0.05. n = 4. Representative Western blot analysis for endogenous LC3I/II. E, the presence of GABARAP-GFP-positive autophagosomes in HeLa cells overexpressing Bnip3. Bnip3, but not the mutants, significantly increased the number of GABARAP-GFP-positive autophagosomes/cell compared with Myc-transfected control cells. *, p < 0.05. n = 3. Representative images are shown. F, Bnip3, but not the mutants, induced autophagy in a significant number of cell compared with Myc-transfected control cells. *, p < 0.05. n = 4.
Because the TMD is also important for targeting Bnip3 to membranes, it was still unclear whether the failure to induce autophagy was due to lack of homodimerization or simply a failure to localize to the mitochondria. Therefore, we investigated whether mutations in the TMD affected induction of autophagy. Previous studies by us and others found that TMD mutants Bnip3G180F or Bnip3H173A are unable to form homodimers but still localize to the mitochondria (22, 26, 27). The mutations in the TMD did not affect their stability in cells (22, 26). Interestingly, both Bnip3G180F and Bnip3H173A were unable to induce autophagy in HeLa cells (Fig. 1, C and D). We also examined the importance of the conserved cysteine residue in the Bnip3 N-terminal domain in induction of autophagy. Mutation of this residue to an alanine reduces Bnip3 homodimer formation and its proapoptotic activity (22). In this study, we found that Bnip3C64A had a reduced capability to induce autophagy compared with Bnip3 (Fig. 1, C and D).
Similar to LC3, GABARAP is a mammalian homologue of yeast Atg8 and has been reported to associate with the autophagosomal membrane (28). We found that GABARAP-GFP showed a predominantly diffuse cytoplasmic distribution of green fluorescence in control cells, whereas overexpression of Bnip3, but not the TMD or N-terminal domain Bnip3 mutants, resulted in increased punctate patterns, indicating enhanced formation of GABARAP-positive autophagosomes (Fig. 1, E and F). We confirmed that GABARAP-GFP colocalized with mCherry-LC3-positive autophagosomes in HeLa cells overexpressing Bnip3 (supplemental Fig. S1). Altogether, the data highlight the importance of Bnip3 homodimerization for its ability to induce autophagy in cells.
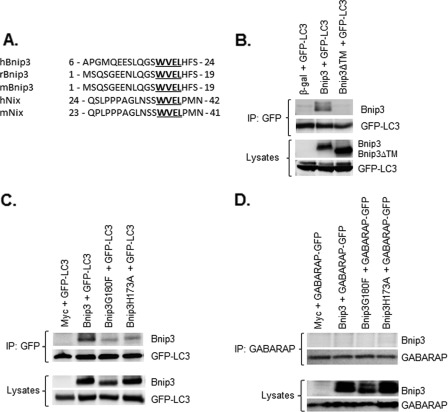
Bnip3 specifically activates mitophagy in cells (2, 29), but it is still unclear how Bnip3 targets mitochondria to the autophagosomes. The Atg8 family proteins bind to proteins that contain a WXXL motif called the LIR (8, 9). Interestingly, Bnip3 and its homologue Nix contain a LIR in the N-terminal region that is conserved between species (Fig. 2A). We have found previously that Bnip3 interacts with LC3 (19), but it is still unknown if Bnip3 must form a homodimer to interact with LC3 and whether Bnip3 preferentially interacts with LC3 or GABARAP. Immunoprecipitation of GFP-LC3 from cell lysates followed by Western blot analysis with an antibody to Bnip3 showed that Bnip3 but not Bnip3ΔTM coimmunoprecipitated with LC3 (Fig. 2B). In addition, we found that TMD mutants Bnip3G180F and Bnip3H173A had severely reduced interaction with LC3 compared with Bnip3 (Fig. 2C). This suggests that the Bnip3 homodimer has the strongest interaction with LC3. Because Bnip3 promoted translocation of the LC3 homologue GABARAP to autophagosomes, we also examined whether Bnip3 interacted with GABARAP. Interestingly, Bnip3 failed to coimmunoprecipitate with GABARAP (Fig. 2D). Previous studies have reported that Nix preferentially interacts with GABARAP but not LC3 (23, 24). We confirmed that Nix interacts with GABARAP but not with LC3 in our study (supplemental Fig. S2).
FIGURE 2.
Bnip3 interacts with LC3 but not GABARAP. A, the presence of a conserved LIR in Bnip3 and Nix. B, Bnip3, but not Bnip3ΔTM, coimmunoprecipitates (IP) with GFP-LC3 in HeLa cells. C, Bnip3G180F and Bnip3H173A have reduced interaction with LC3 compared with Bnip3. D, Bnip3 does not interact with GABARAP-GFP.
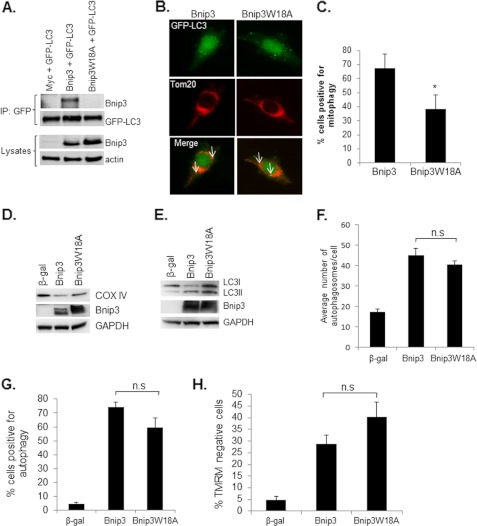
Next, we investigated if the interaction between Bnip3 and LC3 was dependent on the putative LIR motif. Mutation of the conserved tryptophan at position 18 in Bnip3 to an alanine completely abrogated the interaction with LC3 (Fig. 3A). The fact that Bnip3 interacts with LC3 on the autophagosome suggests that Bnip3 might function as an autophagy receptor that specifically targets mitochondria for removal by the autophagosomes. We therefore investigated whether disrupting the interaction between Bnip3 and LC3 had an effect on mitophagy. As shown in Figs. 3, B–D, mitophagy was reduced in cells overexpressing the LIR mutant Bnip3W18A. In contrast, disrupting the interaction between LC3 and Bnip3 had little effect on induction of general autophagy (Fig. 3, E–G). We found that the average number of autophagosomes per cell (Fig. 3F) and the number of cells positive for induction of autophagy (Fig. 3G) were not significantly different between Bnip3 and Bnip3W18A. The increase in the number of autophagosomes could reflect either increased autophagosome formation because of enhanced autophagic activity or an accumulation of autophagosomes due to an impairment in the downstream degradation pathway. The proton ATPase inhibitor Bafilomycin A1 (Baf A1) prevents lysosomal acidification and causes an accumulation of autophagosomes because of a block in the fusion between autophagosomes and lysosomes (30). We found that the presence of Baf A1 significantly increased the percentage of cells exhibiting GFP-LC3-positive autophagosomes in both Bnip3 and Bnip3W18A overexpressing HeLa cells (supplemental Fig. S3), confirming that these constructs increased autophagic activity. Moreover, analysis of cell viability revealed that Bnip3W18A-mediated cell death was similar to Bnip3 (Fig. 3H). The fact that mitophagy was not completely abrogated in response to Bnip3W18A suggests that LC3 can bind to other proteins on the mitochondria to ensure their removal. It also suggests that the function of the LIR in Bnip3 is separate from its prodeath activity.
FIGURE 3.
Disrupting the interaction between Bnip3 and LC3 reduces mitophagy. A, a point mutation in the conserved LIR disrupts the interaction between Bnip3 and LC3. Bnip3W18A failed to coimmunoprecipitate (IP) with LC3 in HeLa cells. Equal expression levels of Bnip3 and Bnip3W18A were confirmed by Western blot analysis. Actin was used as a loading control. B, representative images of HeLa cells infected with Bnip3 or Bnip3W18A plus GFP-LC3. Mitophagy was determined by assessing colocalization (white arrow) between GFP-LC3-positive autophagosomes and Tom20-labeled mitochondria using fluorescence microscopy. C, quantitation of the number of cells positive for mitophagy. Bnip3W18A induced significantly less mitophagy compared with Bnip3. *, p < 0.05. n = 4. D, mitophagy was also confirmed by Western blotting for cytochrome c oxidase subunit IV (COX IV), a mitochondrial inner membrane protein, in HeLa cells infected with β-gal, Bnip3, or Bnip3W18A. GAPDH was used as a loading control. E, Western blot analysis for endogenous LC3I/II. F, quantitation of the number of GFP-LC3-positive autophagosomes per cell. n = 4. n.s., not significant. G, HeLa cells overexpressing β-gal, Bnip3, or Bnip3W18A were scored for induction of autophagy (n = 3). H, HeLa cells overexpressing β-gal, Bnip3, or Bnip3W18A stained with tetramethylrhodamine methyl ester (TMRM) to measure mitochondrial membrane potential (n = 4).
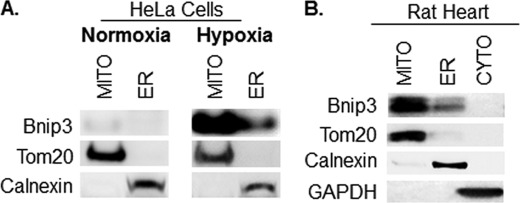
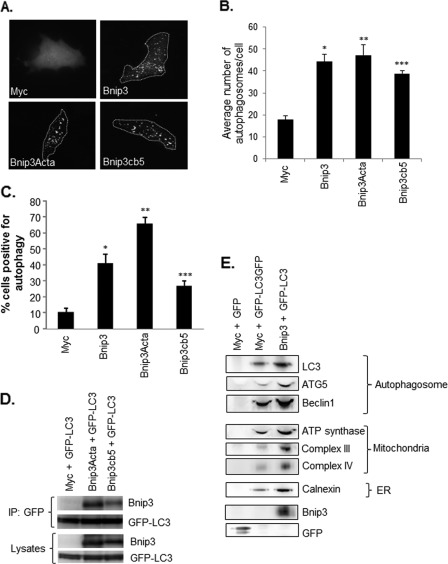
Several of the Bcl-2 proteins, including the Bnip3 homologue Nix, are also localized to the ER where they perturb Ca2+ homeostasis (31–33). Overexpression of a Bnip3 construct specifically targeted to the ER interferes with Ca2+ storage (13, 14). Because it is not known whether endogenous Bnip3 localizes to the ER, we investigated the cellular localization of Bnip3 in HeLa cells. Bnip3 was barely detectable in HeLa cells under normoxic conditions, but exposure to 24 h of hypoxia led to up-regulation of Bnip3. Subcellular fractionation experiments revealed that Bnip3 was found mostly in the mitochondrial fraction but also that some Bnip3 was found in the enriched ER fraction (Fig. 4A). Bnip3 is expressed in the adult heart under normal conditions (15), and, consistent with our findings in HeLa cells, we found that a portion of Bnip3 was located in the ER/SR-rich fraction (Fig. 4B). To investigate whether Bnip3 induced autophagy when localized to mitochondria or ER, we overexpressed Bnip3 mutants engineered to target mitochondria or ER by swapping the TMD with Acta or cytochrome b5 (cb5), respectively (14). Because only the region that spans the mitochondrial membrane is replaced, these mutants are still anchored in the outer membrane with their N-terminal domain facing the cytosol (13, 14). We also confirmed that Bnip3Acta and Bnip3cb5 were still able to form homodimers despite replacement of their TMDs (data not shown). Interestingly, we found that both Bnip3Acta and Bnip3cb5 induced autophagy in HeLa cells (Fig. 5, A–C). We also investigated whether these mutants had an effect on autophagic flux. We found that Baf A1 significantly increased the percentage of cells exhibiting GFP-LC3-positive autophagosomes in Bnip3-, Bnip3Acta-, and Bnip3cb5-transfected HeLa cells (supplemental Fig. S4), confirming that these constructs increased autophagic activity. We also verified that Bnip3Acta and Bnip3cb5 localized to the mitochondria and ER, respectively, when overexpressed in HeLa cells (supplemental Fig. S5).
FIGURE 4.
Endogenous Bnip3 localizes to mitochondria and ER in HeLa cells and in heart tissue. A, after 24 h of normoxia or hypoxia, HeLa cells were subjected to subcellular fractionation into cytosol, mitochondria (MITO), and ER-rich fractions. The fractions were separated by SDS-PAGE and immunoblotted with Bnip3, Tom20, or calnexin antibodies. B, hearts from adult rats were fractionated into cytosol (CYTO), mitochondria, and ER/SR-enriched fractions. The fractions were separated by SDS-PAGE and immunoblotted with Bnip3, Tom20, calnexin, or GAPDH antibodies.
FIGURE 5.
Bnip3 induces autophagy from both the mitochondria and the ER. A, representative images of HeLa cells transfected with vector, Bnip3, Bnip3Acta, or Bnip3cb5 plus GFP-LC3. B, quantitation of the number of GFP-LC3-positive autophagosomes per cell. n = 5. *, **, and ***, p < 0.05 versus Myc. C, quantitation of cells with activated autophagy. Bnip3, Bnip3Acta, and Bnip3cb5 significantly activated autophagy in HeLa cells compared with Myc cells. Data are mean ± S.E. n = 3. *, **, and ***, p < 0.05 versus Myc. D, Bnip3Acta and Bnip3cb5 coimmunoprecipitate (IP) with GFP-LC3 in HeLa cells. E, isolation of autophagosomes from HeLa cells using anti-GFP-linked to magnetic beads. Western blot analysis of isolated autophagosomes showed an increase in the number of autophagosomes and the presence of mitochondria and ER proteins in the autophagosomes in cells overexpressing Bnip3. Results are representative of three independent experiments.
Next, we investigated whether mitochondria and ER-targeted Bnip3 interacted with LC3. We found that both Bnip3Acta and Bnip3cb5 coimmunoprecipitated with LC3 (Fig. 5D). Bnip3 is known to perturb mitochondrial function in cells, which promotes the removal of the damaged mitochondria via autophagy (15). Because Zhang et al. (14) found that Bnip3cb5 perturbs ER Ca2+ stores and ER stress has been reported to induce autophagy (34), we examined whether Bnip3 also induced removal of ER in cells. Isolation of autophagosomes using a GFP antibody bound to magnetic beads confirmed an increased number of LC3-positive autophagosomes in cells overexpressing Bnip3 (Fig. 5E). Western blot analysis for Beclin1 and Atg5 confirmed that the GFP antibody also pulled down autophagosomes at different maturation steps, as reported previously (25). Early autophagic structures contain Beclin1, Atg5, and LC3, whereas only LC3 is retained on the mature autophagosome (35, 36). Further analysis of the isolated autophagosomes by Western blotting revealed that the autophagosomes contained both mitochondrial inner membrane and ER lumen proteins (Fig. 5E). Importantly, no autophagy, mitochondria, or ER proteins were pulled down with anti-GFP magnetic beads in GFP-transfected control cells. Moreover, we did not detect mitochondrial or ER proteins in autophagosomes isolated from HeLa cells after treatment with rapamycin (supplemental Fig. S6), confirming that the presence of these proteins was specifically due to elevated Bnip3.
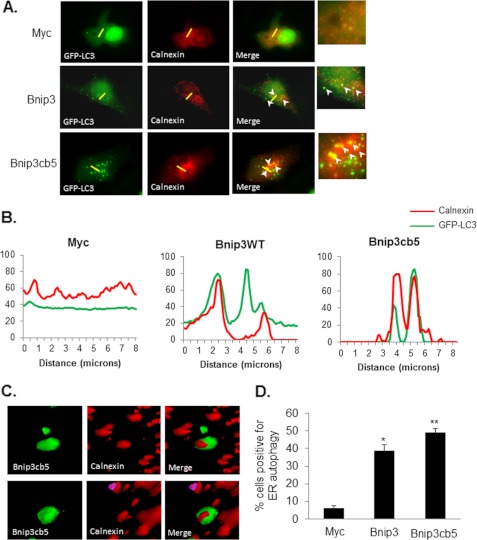
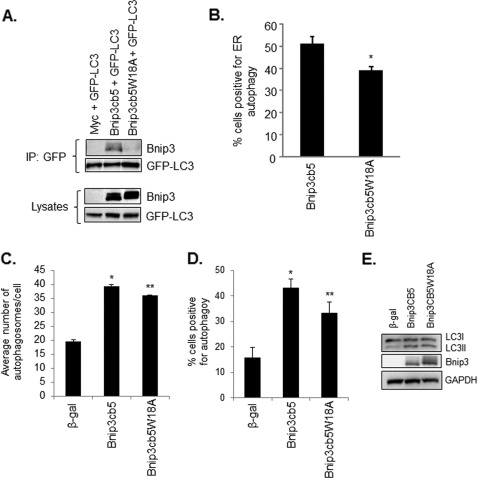
Next, we further investigated if Bnip3 promoted autophagy of ER. HeLa cells overexpressing vector, Bnip3, or Bnip3cb5 plus GFP-LC3 were fixed and stained with an antibody against calnexin to label the ER. Analysis of cells by high-resolution imaging revealed extensive colocalization between autophagosomes and ER in both Bnip3- and Bnip3cb5-overexpressing cells (Fig. 6A). A pseudo-line scan analysis confirmed that GFP-LC3-positive autophagosomes colocalized with ER in cells overexpressing Bnip3 and Bnip3cb5 (Fig. 6B). Three-dimensional surface rendering of z-stacks revealed ER inside the autophagosomes (Fig. 6C). Quantitation of cells positive for colocalization between autophagosomes and ER showed that both Bnip3 and Bnip3cb5 significantly induced autophagy of ER (Fig. 6D). Finally, we investigated the role of the LC3-Bnip3 complex in mediating removal of ER by mutating the tryptophan in the LIR of Bnip3cb5 to generate Bnip3cb5W18A. Coimmunoprecipitation experiments showed that Bnip3cb5 interacted with LC3, whereas Bnip3cb5W18A did not (Fig. 7A). Moreover, Bnip3cb5W18A induced significantly less ERphagy compared with Bnip3cb5 (Fig. 7B). Mutation of the tryptophan residue in the LIR of Bnip3cb5 did not reduce the number of autophagosomes per cell compared with Bnip3cb5 (Fig. 7C) or reduce the overall induction of autophagy in cells (Fig. 7, D and E). This suggests that Bnip3 promotes removal of ER via its interaction with LC3 on the autophagosomes. Disruption of this interaction did not completely abrogate autophagy of ER, indicating the presence of other LC3 receptors on the ER that ensure its removal.
FIGURE 6.
Bnip3 promotes autophagy of ER. HeLa cells overexpressing vector, Bnip3, or Bnip3cb5 plus GFP-LC3 were fixed and stained with anti-calnexin. ER autophagy was determined by assessing colocalization between GFP-LC3-positive autophagosomes and calnexin-labeled ER using fluorescence microscopy. A, representative images of HeLa cells. The yellow line indicates the segment used for line scan analysis. B, colocalization of GFP-LC3 and calnexin was quantified with line scan analysis using ImageJ software. The line scan shows colocalization between autophagosomes (GFP-LC3, green line) and the ER (calnexin, red line) in Bnip3 and Bnip3cb5 but not Myc-transfected cells. C, surface volume rendering of autophagosomes revealed ER inside autophagosomes in Bnip3cb5-overexpressing cells. D, quantitation of ERphagy. Bnip3 and Bnip3cb5 induced significant autophagy of ER compared with Myc-transfected control cells. * and **, p < 0.05. n = 3.
FIGURE 7.
Disrupting the interaction between Bnip3cb5 and LC3 reduces ERphagy. A, a point mutation in the conserved LIR disrupts the interaction between Bnip3cb5 and LC3. Bnip3cb5W18A failed to coimmunoprecipitate (IP) with LC3 in HeLa cells. Equal expression levels of Bnip3cb5 and Bnip3cb5W18A were confirmed by Western blot analysis. B, ERphagy was determined by assessing colocalization between GFP-LC3-positive autophagosomes and calnexin-labeled ER using fluorescence microscopy. Bnip3cb5W18A induced significantly less ERphagy compared with Bnip3cb5. *, p < 0.05. n = 3. C, quantitation of the number of autophagosomes/cell in cells infected with β-gal, Bnip3, or Bnip3cb5W18A. * and **, p < 0.05 versus β-gal. n = 3. D, quantitation of cells positive for activation of autophagy after infection with β-gal, Bnip3, or Bnip3cb5W18A. Bnip3 and Bnip3cb5 induced autophagy in a significant number of cells compared with β-gal. * and **, p < 0.05 versus β-gal. n = 3. E, Western blot analysis for endogenous LC3I/II.
DISCUSSION
In this study, we report several new and important findings. First, endogenous Bnip3 localizes to the ER where it induces autophagy of the ER. Second, the autophagy protein LC3 binds to the LIR motif in the Bnip3 homodimer to induce removal of mitochondria and ER. Third, disrupting the interaction between Bnip3 and LC3 significantly reduces the removal of mitochondria and ER but does not significantly alter the prodeath activity of Bnip3. Our data, therefore, reveal previously unknown mechanisms by which Bnip3 mediates autophagic removal of mitochondria and ER.
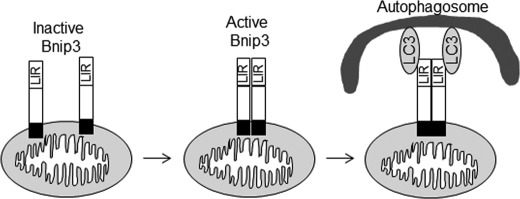
Severe consequences can result from the inability of a cell to remove damaged and dysfunctional organelles through the highly regulated process of autophagy (37, 38). However, it is still unclear how an autophagosome knows to remove a particular mitochondrion. The Bnip3 homologue Nix has been reported to interact with Atg8 family proteins to promote removal of damaged mitochondria (23, 24). Schwartzen et al. (24) initially identified Nix as a GABARAP but not a LC3B binding protein. Similarly, Novak et al. (23) discovered that Nix interacted with Atg8-family proteins, Atg-8, GABARAP, GABARAP-L1/L2, and LC3A but not with LC3B. This study also discovered that mitochondria clearance in Nix deficient reticulocytes was restored by overexpressing Nix but not a Nix LIR mutant (NixW35A). We confirmed that Nix interacts preferentially with GABARAP and not the LC3B isoform in our study. We reported previously that Bnip3 interacts with LC3B in cells (19). This finding was recently confirmed by Ma et al. (39). This study also found that Bnip3ΔTM did not interact with LC3. Interestingly, we have found that Bnip3ΔTM is unable to induce autophagy in cells (15). Here, we report that the TMD is primarily involved in membrane targeting and homodimerization and that replacing it with Acta or cb5 does not affect its ability to induce autophagy or interact with LC3. Instead, our results suggest that homodimerization of Bnip3 via the TMD facilitates the interaction between Bnip3 and LC3 (Fig. 8).
FIGURE 8.
A schematic illustrating the mechanism by which Bnip3 targets mitochondria for removal by autophagosomes. Inactive Bnip3 exists as a monomer in the mitochondria. Upon activation, Bnip3 undergoes homodimerization via the transmembrane domain. The conformational change in the homodimer facilitates the interaction between the LIR in Bnip3 and LC3.
Furthermore, we found that the interaction between LC3 and Bnip3 was important in removing both mitochondria and ER by autophagosomes. Although GABARAP was recruited to autophagosomes, we could not detect any interaction between Bnip3 and GABARAP. This is not surprising because GABARAP exhibits only 30% sequence homology with LC3. However, it is interesting that the LIR is completely conserved among Bnip3 and Nix and that a mutation of the conserved tryptophan residue (at position 18 in hBnip3 and 35 in hNix) abrogates the interaction. This suggests that the domain is needed to specify binding to Atg8 family proteins but that the non-homologous sequences in Nix and Bnip3 are involved in determining the binding specificity for GABARAP or LC3B. Although the LIR is conserved between Nix and Bnip3, they are only 56% homologous.
In this study, we report for the first time that Bnip3 induces autophagy of the ER. Interestingly, our data showed that both ERphagy and mitophagy were significantly reduced, but not completely abrogated, when the interaction between Bnip3 and LC3 was disrupted. This suggests that Bnip3 is not the only autophagy receptor on the mitochondria and the ER. Similar results were obtained in the study by Novak et al. (23) that reported that disrupting the interaction between Nix and LC3A or GABARAP caused only a partial reduction in mitochondrial clearance. It is very likely that several different proteins can act as receptors for autophagy proteins to ensure the removal of damaged mitochondria and ER. For instance, p62 is an autophagy adaptor protein that binds to proteins such as voltage-dependent anion channel and mitofusin1 on the mitochondria and also interacts with LC3 via its LIR (8). We have reported previously that induction of mitophagy protects against Bnip3-mediated cell death (29). Therefore, it was surprising that overexpression of Bnip3W18A did not result in increased cell death compared with Bnip3. However, a redundancy in autophagy receptors would ensure that damaged mitochondria and ER are still being cleared but most likely at a slower rate. Most studies suggest that autophagy is a protective response up-regulated by the cell in response to stress. It is unclear why Bnip3 activates two opposing pathways in cells. It is possible that autophagy functions as a repair mechanism to prevent accidental cell death upon Bnip3 activation. However, if the damage is too great and the cell is beyond rescue, apoptosis will become the dominant pathway, and the cell will die.
One limitation of our study is that we had to utilize overexpression of Bnip3 and the mutants in our experiments to determine their functional differences. There may be a concern that overexpression of a protein in cells may not induce a physiologically relevant response because the levels of the overexpressed protein are higher than endogenous levels. However, Bnip3 is barely detectable in many cells under normal conditions but is up-regulated approximately 8- to 10-fold in response to stress, such as hypoxia (40–43). Similarly, we found that Bnip3 is substantially up-regulated after hypoxia in HeLa cells, which correlated with a reduction in the mitochondrial protein Tom20 (supplemental Fig. S7). Bnip3 is also up-regulated in mouse hearts after a myocardial infarction (supplemental Fig. S7). Thus, overexpression of Bnip3 in HeLa cells achieves protein levels that are comparable with endogenous protein levels after hypoxia. Because hypoxia is associated with activation of multiple pathways, overexpression of Bnip3 in the absence of hypoxia allows us to isolate and study the pathways that are specifically activated by Bnip3. We reported previously that there is a dose-dependent increase in autophagy in response to Bnip3 overexpression, and we found that autophagy is induced even at a very low levels of Bnip3 overexpression (2). In a previous study from our laboratory, we confirmed that silencing Bnip3 using siRNA or inhibition using a dominant negative Bnip3 reduces autophagy in response to simulated ischemia/reperfusion (15), suggesting that Bnip3 is responsible for the induction of autophagy. In addition, down-regulation of Bnip3 using siRNA reduces hypoxia-mediated autophagy (40). Therefore, we are confident that the autophagy response that we are studying by overexpressing Bnip3 mimics the response of the cells when Bnip3 is up-regulated in response to hypoxia.
Activation of Bnip3 has been shown to play an important role in the initiation and progression of heart failure (12). Moreover, inactivation of Bnip3 is also implicated in disease progression and treatment resistance of multiple cancers such as pancreatic, colorectal, and gastric cancer (44–46). Our identification of how Bnip3 functions as a receptor for autophagy of mitochondria and the ER, therefore, has myriad implications toward human disease and highlights the importance of further understanding the role of Bnip3 in mitochondrial function and autophagy.
Supplementary Material
Acknowledgments
We thank Dr. Gary E. Isom (Purdue University, West Lafayette, IN) for the Bnip3Acta and Bnip3cb5 constructs.
This work was supported by National Institutes of Health Grants R01HL087023 and R01HL101217. This work was also supported by a scientist development grant from the American Heart Association.

This article contains supplemental Figs. S1–S6.
- ER
- endoplasmic reticulum
- LC3
- microtubule-associated protein 1 light chain 3
- GABARAP
- γ-aminobutyric acid type A receptor-associated protein
- LIR
- LC3-interacting region
- TMD
- transmembrane domain
- β-gal
- β-galactosidase
- ERphagy
- ER autophagy.
REFERENCES
- 1. Levine B., Klionsky D. J. (2004) Development by self-digestion. Molecular mechanisms and biological functions of autophagy. Dev. Cell 6, 463–477 [DOI] [PubMed] [Google Scholar]
- 2. Quinsay M. N., Thomas R. L., Lee Y., Gustafsson A. B. (2010) Bnip3-mediated mitochondrial autophagy is independent of the mitochondrial permeability transition pore. Autophagy 6, 855–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim I., Rodriguez-Enriquez S., Lemasters J. J. (2007) Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 462, 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hamasaki M., Noda T., Baba M., Ohsumi Y. (2005) Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic 6, 56–65 [DOI] [PubMed] [Google Scholar]
- 5. Bernales S., Schuck S., Walter P. (2007) ER-phagy. Selective autophagy of the endoplasmic reticulum. Autophagy 3, 285–287 [DOI] [PubMed] [Google Scholar]
- 6. Monastyrska I., Klionsky D. J. (2006) Autophagy in organelle homeostasis. Peroxisome turnover. Mol. Aspects Med. 27, 483–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. (2007) Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 8. Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 9. Kirkin V., Lamark T., Sou Y. S., Bjørkøy G., Nunn J. L., Bruun J. A., Shvets E., McEwan D. G., Clausen T. H., Wild P., Bilusic I., Theurillat J. P., Øvervatn A., Ishii T., Elazar Z., Komatsu M., Dikic I., Johansen T. (2009) A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516 [DOI] [PubMed] [Google Scholar]
- 10. Kubli D. A., Ycaza J. E., Gustafsson A. B. (2007) Bnip3 mediates mitochondrial dysfunction and cell death through Bax and Bak. Biochem. J. 405, 407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vande Velde C., Cizeau J., Dubik D., Alimonti J., Brown T., Israels S., Hakem R., Greenberg A. H. (2000) BNIP3 and genetic control of necrosis-like cell death through the mitochondrial permeability transition pore. Mol. Cell. Biol. 20, 5454–5468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Regula K. M., Ens K., Kirshenbaum L. A. (2002) Inducible expression of BNIP3 provokes mitochondrial defects and hypoxia-mediated cell death of ventricular myocytes. Circ. Res. 91, 226–231 [DOI] [PubMed] [Google Scholar]
- 13. Ray R., Chen G., Vande Velde C., Cizeau J., Park J. H., Reed J. C., Gietz R. D., Greenberg A. H. (2000) BNIP3 heterodimerizes with Bcl-2/Bcl-X(L) and induces cell death independent of a Bcl-2 homology 3 (BH3) domain at both mitochondrial and nonmitochondrial sites. J. Biol. Chem. 275, 1439–1448 [DOI] [PubMed] [Google Scholar]
- 14. Zhang L., Li L., Liu H., Borowitz J. L., Isom G. E. (2009) BNIP3 mediates cell death by different pathways following localization to endoplasmic reticulum and mitochondrion. FASEB J. 23, 3405–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hamacher-Brady A., Brady N. R., Logue S. E., Sayen M. R., Jinno M., Kirshenbaum L. A., Gottlieb R. A., Gustafsson A. B. (2007) Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ. 14, 146–157 [DOI] [PubMed] [Google Scholar]
- 16. Bellot G., Garcia-Medina R., Gounon P., Chiche J., Roux D., Pouysségur J., Mazure N. M. (2009) Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol. Cell. Biol. 29, 2570–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Azad M. B., Chen Y., Henson E. S., Cizeau J., McMillan-Ward E., Israels S. J., Gibson S. B. (2008) Hypoxia induces autophagic cell death in apoptosis-competent cells through a mechanism involving BNIP3. Autophagy 4, 195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kanzawa T., Zhang L., Xiao L., Germano I. M., Kondo Y., Kondo S. (2005) Arsenic trioxide induces autophagic cell death in malignant glioma cells by up-regulation of mitochondrial cell death protein BNIP3. Oncogene 24, 980–991 [DOI] [PubMed] [Google Scholar]
- 19. Rikka S., Quinsay M. N., Thomas R. L., Kubli D. A., Zhang X., Murphy A. N., Gustafsson Å. B. (2011) Bnip3 impairs mitochondrial bioenergetics and stimulates mitochondrial turnover. Cell Death Differ. 18, 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schweers R. L., Zhang J., Randall M. S., Loyd M. R., Li W., Dorsey F. C., Kundu M., Opferman J. T., Cleveland J. L., Miller J. L., Ney P. A. (2007) NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. U.S.A. 104, 19500–19505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen G., Ray R., Dubik D., Shi L., Cizeau J., Bleackley R. C., Saxena S., Gietz R. D., Greenberg A. H. (1997) The E1B 19K/Bcl-2-binding protein Nip3 is a dimeric mitochondrial protein that activates apoptosis. J. Exp. Med. 186, 1975–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kubli D. A., Quinsay M. N., Huang C., Lee Y., Gustafsson A. B. (2008) Bnip3 functions as a mitochondrial sensor of oxidative stress during myocardial ischemia and reperfusion. Am. J. Physiol. Heart Circ. Physiol. 295, H2025-H2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Novak I., Kirkin V., McEwan D. G., Zhang J., Wild P., Rozenknop A., Rogov V., Löhr F., Popovic D., Occhipinti A., Reichert A. S., Terzic J., Dötsch V., Ney P. A., Dikic I. (2010) Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11, 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schwarten M., Mohrlüder J., Ma P., Stoldt M., Thielmann Y., Stangler T., Hersch N., Hoffmann B., Merkel R., Willbold D. (2009) Nix directly binds to GABARAP. A possible cross-talk between apoptosis and autophagy. Autophagy 5, 690–698 [DOI] [PubMed] [Google Scholar]
- 25. Gao W., Kang J. H., Liao Y., Ding W. X., Gambotto A. A., Watkins S. C., Liu Y. J., Stolz D. B., Yin X. M. (2010) Biochemical isolation and characterization of the tubulovesicular LC3-positive autophagosomal compartment. J. Biol. Chem. 285, 1371–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sulistijo E. S., Jaszewski T. M., MacKenzie K. R. (2003) Sequence-specific dimerization of the transmembrane domain of the “BH3-only” protein BNIP3 in membranes and detergent. J. Biol. Chem. 278, 51950–51956 [DOI] [PubMed] [Google Scholar]
- 27. Bocharov E. V., Pustovalova Y. E., Pavlov K. V., Volynsky P. E., Goncharuk M. V., Ermolyuk Y. S., Karpunin D. V., Schulga A. A., Kirpichnikov M. P., Efremov R. G., Maslennikov I. V., Arseniev A. S. (2007) Unique dimeric structure of BNip3 transmembrane domain suggests membrane permeabilization as a cell death trigger. J. Biol. Chem. 282, 16256–16266 [DOI] [PubMed] [Google Scholar]
- 28. Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., Yoshimori T. (2004) LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117, 2805–2812 [DOI] [PubMed] [Google Scholar]
- 29. Lee Y., Lee H. Y., Hanna R. A., Gustafsson A. B. (2011) Mitochondrial autophagy by Bnip3 involves Drp1-Mediated mitochondrial fission and recruitment of parkin in cardiac myocytes. Am. J. Physiol. Heart Circ. Physiol. 301, 1924–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yoshimori T., Yamamoto A., Moriyama Y., Futai M., Tashiro Y. (1991) Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem. 266, 17707–17712 [PubMed] [Google Scholar]
- 31. Diwan A., Matkovich S. J., Yuan Q., Zhao W., Yatani A., Brown J. H., Molkentin J. D., Kranias E. G., Dorn G. W., 2nd (2009) Endoplasmic reticulum-mitochondria cross-talk in NIX-mediated murine cell death. J. Clin. Invest. 119, 203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scorrano L., Oakes S. A., Opferman J. T., Cheng E. H., Sorcinelli M. D., Pozzan T., Korsmeyer S. J. (2003) BAX and BAK regulation of endoplasmic reticulum Ca2+. A control point for apoptosis. Science 300, 135–139 [DOI] [PubMed] [Google Scholar]
- 33. Foyouzi-Youssefi R., Arnaudeau S., Borner C., Kelley W. L., Tschopp J., Lew D. P., Demaurex N., Krause K. H. (2000) Bcl-2 decreases the free Ca2+ concentration within the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 97, 5723–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Criollo A., Maiuri M. C., Tasdemir E., Vitale I., Fiebig A. A., Andrews D., Molgó J., Díaz J., Lavandero S., Harper F., Pierron G., di Stefano D., Rizzuto R., Szabadkai G., Kroemer G. (2007) Regulation of autophagy by the inositol trisphosphate receptor. Cell Death Differ. 14, 1029–1039 [DOI] [PubMed] [Google Scholar]
- 35. Fan W., Nassiri A., Zhong Q. (2011) Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) Proc. Natl. Acad. Sci. U.S.A. 108, 7769–7774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kang R., Zeh H. J., Lotze M. T., Tang D. (2011) The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 18, 571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nakai A., Yamaguchi O., Takeda T., Higuchi Y., Hikoso S., Taniike M., Omiya S., Mizote I., Matsumura Y., Asahi M., Nishida K., Hori M., Mizushima N., Otsu K. (2007) The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat. Med. 13, 619–624 [DOI] [PubMed] [Google Scholar]
- 38. Tanaka Y., Guhde G., Suter A., Eskelinen E. L., Hartmann D., Lüllmann-Rauch R., Janssen P. M., Blanz J., von Figura K., Saftig P. (2000) Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 406, 902–906 [DOI] [PubMed] [Google Scholar]
- 39. Ma X., Godar R. J., Liu H., Diwan A. (2012) Enhancing lysosome biogenesis attenuates BNIP3-induced cardiomyocyte death. Autophagy 8, 297–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Z., Yang X., Zhang S., Ma X., Kong J. (2007) BNIP3 up-regulation and EndoG translocation in delayed neuronal death in stroke and in hypoxia. Stroke 38, 1606–1613 [DOI] [PubMed] [Google Scholar]
- 41. Zhang H., Bosch-Marce M., Shimoda L. A., Tan Y. S., Baek J. H., Wesley J. B., Gonzalez F. J., Semenza G. L. (2008) Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J. Biol. Chem. 283, 10892–10903 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42. Webster K. A., Graham R. M., Bishopric N. H. (2005) BNip3 and signal-specific programmed death in the heart. J. Mol. Cell Cardiol. 38, 35–45 [DOI] [PubMed] [Google Scholar]
- 43. Jian B., Yang S., Chen D., Zou L., Chatham J. C., Chaudry I., Raju R. (2011) Aging influences cardiac mitochondrial gene expression and cardiovascular function following hemorrhage injury. Mol. Med. 17, 542–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Erkan M., Kleeff J., Esposito I., Giese T., Ketterer K., Büchler M. W., Giese N. A., Friess H. (2005) Loss of BNIP3 expression is a late event in pancreatic cancer contributing to chemoresistance and worsened prognosis. Oncogene 24, 4421–4432 [DOI] [PubMed] [Google Scholar]
- 45. Okami J., Simeone D. M., Logsdon C. D. (2004) Silencing of the hypoxia-inducible cell death protein BNIP3 in pancreatic cancer. Cancer Res. 64, 5338–5346 [DOI] [PubMed] [Google Scholar]
- 46. Murai M., Toyota M., Suzuki H., Satoh A., Sasaki Y., Akino K., Ueno M., Takahashi F., Kusano M., Mita H., Yanagihara K., Endo T., Hinoda Y., Tokino T., Imai K. (2005) Aberrant methylation and silencing of the BNIP3 gene in colorectal and gastric cancer. Clin. Cancer Res. 11, 1021–1027 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.