Background: The H2A variant H2A.Z is an important regulatory component of chromatin.
Results: A novel form of human H2A.Z and mutants of yeast H2A.Z that share truncated C termini have altered properties.
Conclusion: The C-terminal tail of histone H2A.Z is important for its stable association with nucleosomes.
Significance: The strength of H2A.Z's association with nucleosomes is key to the biological functions of this histone variant.
Keywords: Chromatin, Chromatin Immunoprecipitation (ChiP), Chromatin Regulation, Chromatin Structure, Histones, Histone Variant
Abstract
Histone H2A variants generate diversity in chromatin structure and functions, as nucleosomes containing variant H2A histones have altered physical, chemical, and biological properties. H2A.Z is an evolutionarily ancient and highly conserved H2A variant that regulates processes ranging from gene expression to the DNA damage response. Here we find that the unstructured portion of the C-terminal tail of H2A.Z is required for the normal functions of this histone variant in budding yeast. We have also identified a novel splice isoform of the human H2A.Z-2 gene that encodes a C-terminally truncated H2A.Z protein that is similar to the truncation mutants we identified in yeast. The short forms of H2A.Z in both yeast and human cells are more loosely associated with chromatin than the full-length proteins, indicating a conserved function for the H2A.Z C-terminal tail in regulating the association of H2A.Z with nucleosomes.
Introduction
A common theme in biology is the combinatorial use of a core set of components to generate diverse products. For example, a single genome generates tissue-specific patterns of RNAs through selective transcription of subsets of genes, while at the level of a single gene a variety of protein isoforms can be produced via alternative splicing. These principles of combinatorial use are employed to great effect in the regulation of nucleosomes in chromatin, where individual histone proteins can be substituted by variant forms and/or chemically altered by post-translational modifications. Such diversification allows the specification of certain nucleosomes for particular functions, e.g. nucleosomes adjacent to telomeric heterochromatin in budding yeast contain the H2A variant H2A.Z and are acetylated at H4K16, and these features are critical for the maintenance of boundaries between heterochromatin and euchromatin (1–5). This is because the chemical constituents of nucleosomes affect their intrinsic physical properties such as stability, alter their nucleosome-extrinsic contacts that mediate higher order folding, and act as signals for protein recruitment or dissociation (6).
The H2A family is distinguished from the other core histones (H2B, H3, and H4) by two features. The first of these is their diversity, with a larger number of H2A-family histones than any other core histone. These include lineage-specific H2A family proteins that have evolved to cope with specific environmental conditions, such as the variants found specifically in Bdelloid rotifers (7) as well as more generally distributed variants such as macroH2A, H2A.Bbd, H2A.X, and H2A.Z that are found in mammalian species. Of the H2A variants, H2A.Z is the most evolutionarily ancient and widely distributed, being found in all eukaryotic lineages (8).
The second distinguishing feature of H2A family proteins is the presence of an extended C-terminal tail, part of which interacts with H3 and H4 within the nucleosome while the most C-terminal portion exits the core particle (9). Like N-terminal histone tails, the C-terminal tails of H2A proteins are targets for post-translational modifications, particularly bulky modifications like sumo and ubiquitin (10–15). Interestingly, the sequence divergence between H2A family histones is most pronounced in their C-terminal tails (16) indicating that this region may be key for imparting distinct functions to H2A variants. This variability is due to length alterations as well as sequence changes, with macroH2A and H2A.Bbd displaying particularly long and short C-terminal tails, respectively.
The length of the major H2A C terminus can be regulated by proteolytic and chemical cleavage (17, 18) and expression of artificially truncated H2A alters gene expression in human cells (19). Processes that regulate the length of other H2A family C termini have not been described to date. Here we show that alternative splicing of the H2A.Z-2 gene in human tissues produces a transcript that encodes a truncated protein with a distinct C terminus. In parallel we have isolated C-terminal truncation mutants that abrogate the normal functions of H2A.Z in budding yeast. The H2A.Z C terminus regulates the stable association of H2A.Z in nucleosomes in both yeast and human cells. The C-terminal tail of H2A.Z therefore has a conserved function in regulating nucleosomal association.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, and Molecular Cloning
Lists of yeast strains and plasmids used in this work are provided in supplemental Tables S1 and S2. Gene disruption and tagging in Saccharomyces cerevisiae were performed using standard techniques (20). Site-directed mutagenesis was carried out using Pfu Turbo polymerase (Agilent) followed by DpnI digestion of template plasmids. Htz1-M7-H2A and Htz1–120-H2A fusions were synthesized in vitro by Genscript. Residue numbering of Htz1 does not include the initiator methionine, which is cleaved from the mature protein. Human ESTs derived from the H2A.Z-2 (H2A.V) gene were obtained from Open Biosystems, verified by sequencing and subcloned into a pcDNA5/FRT/TO/Myc vector containing a single N-terminal Myc tag (21).
Western Blotting
Proteins were separated by electrophoresis through SDS-PAGE gels and transferred to nitrocellulose membranes. Antibodies to HA (12CA5; Roche) and Myc (4A6; Millipore) tags; histones H3 and H4 (Abcam); RNA polymerase II (8WG16; Covance); and tubulin (gift from Andy Sharrocks) were detected using HRP- (GE Healthcare) or IRDye- (LiCor) conjugated secondary antibodies. Antibody concentrations were titrated to ensure that measurements were within the linear range of the detection system.
Mammalian Cell Culture and Transfections
DLD-1 cells (gift from Stephen Taylor) were maintained in DMEM containing 10% fetal bovine serum, 2 mm l-glutamine, 100 units/ml penicillin, and 0.1 mg/ml streptomycin. Flp-InTM T-RExTM stable cell lines expressing H2A.Z were generated by co-transfection of pcDNA5/FRT/TO/H2A.Z-2.1/2/3/4/5 and pOG44 into a DLD-1 Flp-InTM T-RExTM host cell line (21) using LipofectamineTM 2000 (Invitrogen). Following transfection, site-specific integrant clones were selected and maintained in growth medium containing hygromycin B (400 μg/ml) and blasticidin (8 μg/ml).
Chromatin Stability Assay
Tests of H2A.Z resistance to buffers of increasing salt concentration were carried out with S. cerevisiae and human DLD-1 cells. Yeast cells were treated with Zymolyase 100T (Seikagaku) to remove the cell wall prior to lysis of the outer membrane followed by separation of the nuclear and cytoplasmic material by centrifugation through a sucrose cushion. Nuclei were lysed in 1% Triton in a Tris buffer containing 10 mm NaCl. Fractions were either analyzed directly or the chromatin pellet was divided into aliquots and washed with buffers of various salt concentrations. The proteins remaining in the insoluble pellet were analyzed by Western blotting. Mammalian cells were treated with 1 μg/ml tetracycline for 48 h to induce transgene expression prior to cell harvesting. Cells were first lysed with Triton extraction buffer containing 10 mm NaCl to extract cytoplasmic and soluble nuclear proteins. Permeablized nuclei were then divided into aliquots and washed with buffers of various salt concentrations (0.25, 0.5, 0.75, 1, and 1.5 m NaCl). Total cell protein and proteins soluble at each salt concentration were analyzed by Western blotting.
Nucleosome Isolation and Co-immunoprecipitation
Permeabilized nuclei from yeast or mammalian cells were treated with micrococcal nuclease (MNase,2 Worthington) in a buffer containing 1 mm CaCl2. DNA was purified from an aliquot of the MNase-solubilized fraction and assayed by gel electrophoresis. The relative intensity of the mononucleosomal band compared with other detectable bands was quantified using Image Lab software (Bio-Rad). The remaining material in MNase-solubilized fractions was immunoprecipitated with anti-HA/anti-Myc agarose or IgG Sepharose, washed and analyzed by Western blotting.
Chromatin Immunoprecipitation
Htz1-bound loci were immunoprecipitated from cross-linked extracts as described (22), but using anti-HA (12CA5) antibodies. Recovered DNA was analyzed by Q-PCR and/or amplified for hybridization to Affymetrix S. cerevisiae Tiling 1.0 arrays. Amplification, hybridization, and washing were performed according to the manufacturer's protocols, and ChIP data were analyzed by TAS (Affymetrix), using a two-sample comparison of immunoprecipitated to input DNA. Q-PCR reactions were performed as described below.
Q-PCR and RT-Q-PCR
Total RNA from a panel of human tissues (FirstChoice® Total RNA Human Normal Tissue from Applied Biosystems) was reverse transcribed into cDNA using SuperScript® III Reverse Transcriptase (Invitrogen). All Q-PCR reactions were carried out in triplicate using QuantiTect SYBR Green reagents (Qiagen). Standard curves based on serial dilutions of plasmid standards (RT-Q-PCR) or sonicated genomic DNA (ChIP) were performed for all primer sets to test for linear amplification. For RT-Q-PCR, mock reverse transcribed controls were included to check for amplification of products specifically from cDNA template and primer sets were designed to span exon junctions. All primer sequences are available upon request.
Computational Analysis
Raw microarray data (23) was downloaded and processed with Partek GS (v6.5, Partek Inc., St. Charles, MO). Gene expression analysis was by RMA (24), and data were normalized by Z-transformation for each probeset. Hierarchical clustering analysis was performed using Euclidian distance for both probesets and tissues.
Immunofluorescence
Cells were treated with 1 μg/ml tetracycline for 48 h to induce transgene expression prior to fixation in 1% (w/v) paraformaldehyde-PBS. Anti-Myc antibodies (4A6; Millipore) and anti-mouse Alexa Fluor® 594 secondary antibodies (Invitrogen) were used for H2A.Z detection; DNA was counterstained with DAPI (Sigma).
RESULTS
The C Terminus Is Required for Normal Function of H2A.Z in Yeast
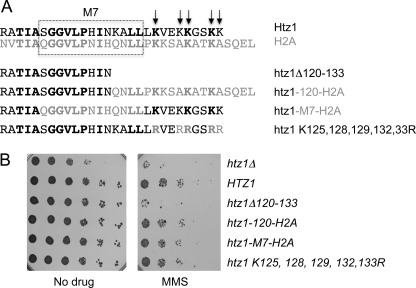
To identify regions of H2A.Z (called Htz1 in S. cerevisiae (25)) that are important for its function, we undertook an unbiased forward genetic screen to isolate mutants lacking wild-type Htz1 function in S. cerevisiae. This screen will be described in detail elsewhere.3 Briefly, randomly mutated HTZ1 alleles were screened for synthetic growth defects with a deletion of the gene encoding the chromatin assembly factor Asf1. Strains lacking both HTZ1 and ASF1 grow poorly or not at all (26) and we therefore selected clones that showed poor growth in two rounds of screening. One class of mutant alleles recovered in this way had altered the length of the C terminus of the Htz1 protein, which we defined as all residues C-terminal to the α-C helix, i.e. residues R106-K133. Several clones with premature stop codons were identified as well as a clone encoding a protein extended by 23 residues due to a frameshift caused by a single base deletion in the penultimate codon. The mutant clones are depicted in Fig. 1A, along with the aligned sequences of the proteins' C termini.
FIGURE 1.
An intact C terminus is required for Htz1 function. Mutations altering the C terminus of Htz1 were identified by screening for synthetic genetic interactions with asf1Δ. A, structural organization of the Htz1 protein is depicted, with missing or additional sequences present in the recovered mutants indicated above the full-length protein (black bars) and an alignment of the C-terminal residues shown below. B, rescreening of deletion clones in an independent asf1Δhtz1Δ strain carrying wild-type HTZ1 on a URA-marked plasmid and a wild-type or mutant HTZ1 allele on a LEU plasmid. The identity of the Htz1 allele is (clockwise from arrow): HTZ1 (WT); vector (htz1Δ); HTZ1 (WT); htz1Δ120–133; htz1Δ128–133; Htz1 + 23; Htz1Δ125–133; vector (htz1Δ). C, C-terminally truncated Htz1 proteins are stable and present at similar levels to full-length Htz1. The panels show a Western blot of total cell protein from the indicated strains probed with anti-HA antibodies (top) to detect Htz1 proteins and anti-H3 antibodies (bottom) as a loading control. D, phenotypes of Htz1 C-terminal truncation mutants in an ASF1+ strain. Serial dilutions of the indicated strains were spotted on plates containing 0.005% MMS and control drug-free plates and incubated for 48–72 H at 30 °C. E, summary of mutant phenotypes. The synthetic genetic interaction data represent the initial screen and the FOA assays (separated by /), with + indicating a genetic interaction. MMS sensitivity is denoted by +, no sensitivity by −.
To ensure that the synthetic phenotypes observed in the primary screen were reproducible and could not be the result of another mutation, a second set of plasmids containing the C-terminal mutants was created and transformed into an independent asf1Δhtz1Δ strain carrying a wild-type HTZ1 allele on a URA-marked plasmid. The dependence of these strains on the wild-type HTZ1 allele was tested by counter-selection of the URA plasmid with 5-FOA (Fig. 1B). The mutant forms of Htz1 identified in the primary screen also showed synthetic lethal/sick interactions with asf1Δ in this plasmid shuffle assay, with the exception of the Δ128–133 mutant, which could support growth in the absence of HTZ1 and ASF1 in this strain.
One explanation for the loss of function of Htz1 truncations is that the truncated proteins are present at a reduced level. To test this, N-terminally 3HA-tagged forms of the C-terminal deletion constructs were created and the cognate proteins were examined by Western blotting of total cell extracts (Fig. 1C). When compared with histone H3, it is clear that truncating the C terminus does not substantially alter the levels of Htz1 protein.
While the asf1Δ strain was useful for selection in our initial screen, we next wanted to ask whether the truncated HTZ1 alleles were functional in an otherwise wild-type (ASF1+) background. To do this, we took advantage of the fact that htz1Δ cells are sensitive to the DNA-damaging agent methyl methanesulfonate (MMS) (27, 28). htz1Δ cells were transformed with plasmids encoding full-length or truncated HTZ1 alleles and tested for growth on plates containing MMS (Fig. 1D). The two mutants with the largest deletions (Δ111–133 and Δ120–133) could not restore growth on MMS-containing plates, while the shorter deletions and the 23 amino acid extension (not shown) were indistinguishable from wild-type. The MMS sensitivity of the Δ111–133 mutant was reproducibly more severe than that of the Δ120–133 strain. These data collectively reveal a requirement for an intact C terminus for normal Htz1 functions, with residues up to 128 required for survival in the absence of Asf1 and residues up to 125 required for resistance to MMS (summarized in Fig. 1E).
The Length of the H2A.Z C-terminal Tail Rather than Specific Residues Is Important for Function
One question arising from the progressively worse MMS sensitivity phenotype of deletions within the Htz1 C-terminal tail is whether this is due to specific removal of certain residues or a general effect of reducing the length of the tail. To test this, we made constructs with altered groups of residues and tested their sensitivity to MMS. The C-terminal tail of H2A.Z contains a domain called M7 that is essential for viability in Drosophila melanogaster (29) as well as a number of lysine residues that are sites of sumoylation (12) (Fig. 2A). It seemed unlikely that the sumoylation sites were responsible for the MMS sensitivity as they are removed by the Δ125–133 deletion that does not render cells sensitive to MMS. Nonetheless, we directly tested the requirement for Htz1 sumoylation as Htz1su has been implicated in the response to DNA damage, which may be related to MMS sensitivity. When the 5 potential sumo acceptor lysines in the C terminus were changed to arginine, cells had a wild-type level of resistance to MMS, indicating that MMS sensitivity of C-terminal truncations is not a result of a loss of Htz1 sumoylation (Fig. 2B).
FIGURE 2.
The role of the Htz1 C terminus in MMS resistance can be provided by the H2A C-terminal tail. A, 28 residues at the C terminus of Htz1 (R106 onwards) are aligned with the corresponding region of H2A (top), with residues that are identical in Htz1 and H2A in bold type. The Htz1-H2A hybrid proteins and point mutants are depicted below relative to the Δ120–133 truncation mutant. The M7 region (boxed) or the C-terminal region distal to residue 119 from Htz1 were replaced with the corresponding residues from H2A, which results in 5 or 9 Htz1 residues being substituted with a different amino acid. Arrows indicate the 5 C-terminal lysines that were mutated to arginines. B, serial dilutions of the indicated strains were spotted on plates containing 0.005% MMS or drug-free control plates and incubated for 48–72 H at 30 °C.
To examine the role of the M7 domain, a hybrid protein where the M7 domain from yeast Htz1 was swapped with that from H2A (Htz1-M7-H2A; Fig. 2A) was generated. In parallel, all C-terminal residues from lysine 120 onwards were replaced with the corresponding residues from H2A (Htz1–120-H2A) to test whether Htz1 residues 120–133 are specifically required for MMS resistance. Both constructs behaved like full-length Htz1, meaning that the C terminus from H2A can fully compensate for the Htz1 C terminus in this assay (Fig. 2B). Taking all the constructs tested collectively, two C-terminal residues (leucines 122 and 123) remained unchanged due to conservation in H2A and it is formally possible that these two residues are key for the function of the Htz1 C-terminal tail. However, because of the increasing phenotypic severity of larger deletions and the complete rescue by the H2A C terminus, it is more likely that the length of the Htz1 C terminus, rather than specific amino acid sequences and post-translational modification sites, is critical for wild-type function of the protein.
C-terminally Truncated Htz1 Protein Is Normally Assembled into Nucleosomes
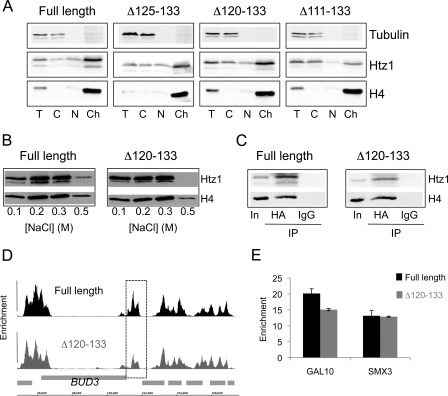
To biochemically test whether truncated forms of Htz1 are associated with chromatin in vivo, cell fractionation experiments were performed to assess the proportions of Htz1 resident in the cytoplasmic, nuclear and chromatin fractions. The Δ111–133 protein had a relatively reduced abundance in the nuclear and chromatin fractions, and a corresponding increased abundance in the cytoplasm, which indicates that the phenotypes observed in strains with this truncation are likely to be due to its reduced occupancy in chromatin (Fig. 3A). The Δ120–133 and Δ125–133 mutants, on the other hand, were present at normal levels in the chromatin fraction. The MMS sensitivity of the Δ120–133 mutant consequently cannot be attributed to a general defect in chromatin incorporation.
FIGURE 3.
The C terminus is required for stable retention of Htz1 in chromatin but not for normal deposition. A, proteins from various sub-cellular fractions (T, total; C, cytoplasm; n, nuclear; Ch, chromatin) derived from the indicated strains were analyzed by Western blotting. Membranes were probed with anti-HA antibodies to detect Htz1 and its truncated forms; anti-H4 and anti-tubulin were included to verify expected fractionation of proteins. B, chromatin fraction from wild-type (full-length) or Htz1Δ120–133 cells was washed with buffers containing various salt concentrations (indicated below), and the proteins remaining in the pellet were assayed by Western blotting. C, immunoprecipitation of HA-Htz1 proteins from the soluble, primarily mononucleosomal, fraction produced by MNase digestion (input; In) results in co-precipitation of H4. Immunoprecipitation with an unrelated antibody (IgG) does not precipitate H4. D, view of a region of chromosome III (coordinates indicated at bottom) displaying the enrichments of HA-Htz1 (top; black) and HA-htz1Δ120–133 (bottom; dark gray) relative to S. cerevisiae open reading frames (light gray boxes). This region around the BUD3 gene is representative of the pattern across the entire genome and was chosen for ease of comparison to previously published data (37). The dashed box surrounds a region with lower Htz1 occupancy in the Δ120–133 mutant. E, Q-PCR analysis of ChIP enrichments of full-length Htz1 and Htz1Δ120–133 at the GAL10 and SMX3 genes relative to a non-enriched control region at HMR.
Histones can be disassociated from nucleosomes by washing chromatin preparations with buffers containing salt ions and different histones are more or less resistant to salt washing, e.g. H3 and H4 are dissociated at higher salt concentrations than H2A and H2B. To test whether the stability of Δ120–133 was altered relative to full-length Htz1, chromatin preparations from strains expressing each protein were subjected to washing with buffers containing increasing amounts of salt. Proteins remaining in the insoluble chromatin pellet were assayed by Western blotting (Fig. 3B) to allow precise quantification of Htz1. We found that the Δ120–133 truncation protein is dissociated to a greater extent than full-length Htz1 from the chromatin pellet by washing with 0.5 m NaCl. Washing with 0.5 m NaCl removed up to 55% (average 54.4%; S.E. 1.56) of full-length Htz1 but up to 70% (average 67.3%; S.E. 4.5) of Htz1Δ120–133 was removed by the same treatment. The relative level of H4 was also reproducibly lower in the 0.5 m pellet from the Htz1Δ120–133 strain, indicating that the dissociation of the Δ120–133 protein was destabilizing some nucleosomes. The Δ125–133 mutant, on the other hand, is indistinguishable from the full-length protein in these assays (not shown). These results show that although the Δ120–133 protein is present in the chromatin fraction, it can be more easily dissociated than full-length Htz1, indicating that the last 14 residues of H2A.Z are important to stabilize the protein in yeast chromatin.
To ensure that the Htz1Δ120–133 protein present in the chromatin fraction is complexed with other core histones in nucleosomes, yeast chromatin was digested with micrococcal nuclease and the association of Htz1Δ120–133 with H4 within the resulting soluble fraction (average 88% mononucleosomal, supplemental Fig. S1A) was verified by co-immunoprecipitation (Fig. 3C). Next, ChIP was performed as an independent means to compare the in vivo nucleosomal occupancies of Htz1 and Htz1Δ120–133. Hybridization of Htz1-associated DNA fragments to high-resolution whole-genome tiling microarrays revealed that the genome-wide pattern of C-terminally truncated Htz1 is virtually indistinguishable from the full-length protein (Fig. 3D). The pattern of Htz1 deposition is therefore not affected by the Δ120–133 deletion. However, at some highly enriched regions, the abundance of Δ120–133 is slightly reduced compared with the full-length protein (e.g. boxed region, Fig. 3D). Q-PCR analysis of two loci confirms that enrichment of Δ120–133 is similar to (SMX3) or slightly lower (GAL10) than that of the wild-type protein (Fig. 3E). The slight decrease in the steady-state level of Htz1Δ120–133 in some highly-enriched nucleosomes is consistent with its increased susceptibility to removal from chromatin.
A Novel Splice Isoform of H2A.Z Encoding a Truncated Protein Is Present in Some Human Tissues
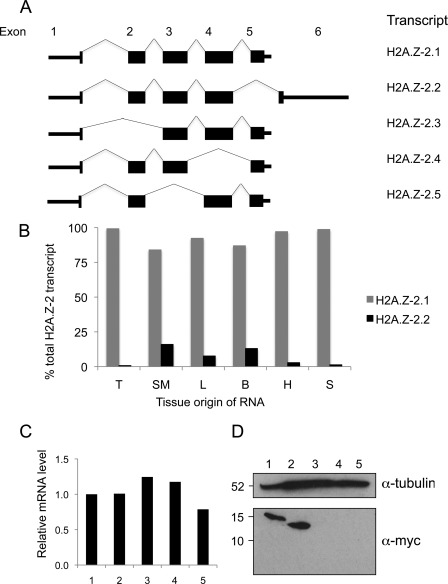
H2A.Z is a highly conserved protein and in vertebrate genomes there are two non-allelic genes that differ significantly in DNA sequence and give rise to very similar proteins termed H2A.Z-1 and H2A.Z-2 (30). Interestingly, our analysis of cDNAs and ESTs derived from the H2A.Z-2 locus (H2A.V) indicated that multiple alternatively spliced transcripts are produced from this gene in humans (Fig. 4A) including one (H2A.Z-2.2) coding for a 113-residue protein with a shorter C terminus much like the truncation mutants that we identified in yeast (supplemental Fig. S2A). Sequencing of human cDNAs derived from H2A.V confirmed the existence of the full-length correctly spliced transcript H2A.Z-2.2.
FIGURE 4.
The human H2A.Z-2 gene encodes a protein with a truncated C terminus. A, schematic diagram showing splice isoforms produced by the human H2A.Z-2 (H2A.V) gene. For convenience they are named 1–5 and correspond to H2A.V isoform 1 (NM_012412.4), isoform 2 (NM_138635.3), isoform 3 (NM_201436.2), isoform 4 (NM_201516.2), and isoform 5 (NM_201517.2). B, endogenous H2A.Z-2 transcript is detected in cDNA generated from normal human tissues. RT-Q-PCR measurements of the relative amounts of H2A.Z-2.1 and H2A.Z-2.2 transcripts in RNA from normal human tissues. T, thymus, SM, skeletal muscle; L, liver; B, brain; H, heart; S, spleen. C, mRNA levels produced from human cell lines engineered to express transcript 1, 2, 3, 4, or 5 in response to tetracycline were measured by RT-Q-PCR. Levels are normalized to splice isoform 1. (1–5, splice isoforms 1–5). D, Western blotting of whole-cell extracts generated from the same engineered cell lines upon transgene induction. Transcripts 3–5 do not produce detectable Myc-tagged protein in this assay, even upon over-exposure. Tubulin is included as a loading control.
H2A.Z-2 splice isoform 2 contains exon 6, which is not present in any of the other splice isoforms (Fig. 4A and supplemental Fig. S2B). This allowed us to examine existing expression datasets for evidence of transcripts containing exon 6. We identified two Affymetrix probes specific for exon 6 as well as a probe that is complementary to exons 1–5. Using a comprehensive microarray study of RNA levels derived from various human tissues (23), we compared the relative expression levels reported for these probes. Clustering of these data revealed that exon 6-specific RNA was relatively high in certain tissues, particularly those derived from brain (supplemental Fig. S2C; boxed). To validate our in silico analysis, we obtained RNA samples from a range of normal human tissues and compared the levels of H2A.Z-2 splice isoform 1 and splice isoform 2 RNAs by reverse transcription followed by quantitative PCR (RT-Q-PCR). The primers used to detect splice isoform 1 would also detect splice isoforms 3 and 5 but for simplicity we refer only to isoform 1 as the other isoforms are unlikely to produce protein in vivo (see below). Our analysis revealed that splice isoform 1 is the predominant form of H2A.Z-2 in all tissues tested but that levels of splice isoform 2 are higher in certain tissues, especially brain, liver, and skeletal muscle (Fig. 4B). These data show that certain human tissues are likely to contain a pool of H2A.Z protein with a truncated C terminus.
Lack of suitable specific reagents meant that we could not use antibodies to detect the endogenous proteins encoded by the H2A.Z-2 splice isoforms. Instead, we tested whether the proteins produced from different H2A.Z-2 splice isoforms could produce stable protein in the karyotypically stable human colon carcinoma cell line DLD-1. Site-directed recombination was used to make stable sister cell lines in which H2A.Z-2 cDNAs were targeted to precisely the same genomic locus, driven by a tetracycline-regulatable promoter, flanked by the same 5′- and 3′-UTRs, and fused to a small epitope tag. This approach rules out genome position effects on expression, allows regulated expression and permits detection and comparison of protein isoforms using a single antibody. In addition to splice isoforms 1 and 2, we also generated cell lines to express the three other transcripts identified by our in silico analysis. All of the cell lines produced transgene mRNA, as measured by RT-Q-PCR, only in the presence of tetracycline and the levels of mRNA for each splice isoform were similar upon induction (Fig. 4C). However, analysis of the cognate proteins by Western blotting of total cell extracts revealed that expression of only splice isoforms 1 and 2, encoding full-length and C-terminally truncated H2A.Z-2, resulted in detectable protein (Fig. 4D). This implies that the alternatively spliced transcripts 3–5 are unlikely to produce stable protein in vivo, but that proteins derived from splice isoforms 1 and 2 are stable in human cells.
The Short Form of H2A.Z-2 Is Less Stably Associated with Chromatin than Full-length H2A.Z-2
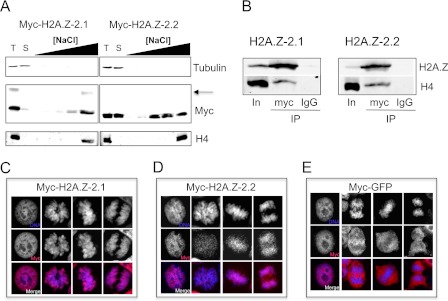
Given the similar shortening of the C-terminal tail in H2A.Z-2.2 and the truncation mutants that we identified in yeast, we tested whether the C-terminal tail similarly modulates stability of H2A.Z in human cells. To this end, we extracted soluble protein from cells expressing H2A.Z-2.1 or H2A.Z-2.2 and then exposed the resulting permeablized nuclei in parallel to buffers of various salt concentrations and assayed the proteins that became solubilized (Fig. 5A). Surprisingly, a large proportion of isoform 2 is soluble in a buffer containing 10 mm salt, while only a small proportion of isoform 1 is soluble under these conditions. Additionally, H2A.Z-2.2 is released from chromatin at lower salt concentrations than H2A.Z-2.1, indicating that it is less stably bound than the full-length protein. Nonetheless, H2A.Z-2.2, like H2A.Z-2.1 co-immunopreciptates H4 from the primarily (average 95%) mononucleosomal material released by MNase treatment of chromatin, thus verifying its occupancy in nucleosomes (Fig. 5B and supplemental Fig. S1B).
FIGURE 5.
The short form of human H2A.Z-2 protein is less stably bound to chromatin than the full-length protein. A, DLD-1 cells induced to express Myc-tagged H2A.Z-2.1 or 2.2 were treated to extract soluble protein, then the permeabilized nuclei were washed with buffers of various salt concentrations. Fractions corresponding to total (T) protein, soluble (S) protein and protein extracted at each salt concentration (0.25, 0.5, 0.75, 1.0, and 1.5 m NaCl) were analyzed by Western blotting. Membranes were probed with the antibodies indicated at the right-hand side. The arrow indicates mono-ubiquitylated H2A.Z-2.1 (data not shown). B, Western blotting reveals that H4 is co-immunoprecipitated (IP) by anti-myc antibodies that pull down H2A.Z-2.1/2 from the primarily mononucleosomal MNase-solubilized fraction (input; In) but not by an unrelated antibody (IgG). C–E, immunofluorescence staining of tetracycline-treated DLD-1 cells carrying a transgene encoding Myc-tagged H2A.Z-2.1 (C), H2A.Z-2.2 (D), or GFP (E). Images of cells in interphase and during various stages of mitosis are shown, with the DAPI and Myc channels shown separately above each merged image.
As an independent means to compare isoforms 1 and 2, the subcellular localization of the proteins was examined by immunofluorescence microscopy of asynchronously growing cells induced to express the Myc-tagged proteins. Both proteins are predominantly present in the nucleus but the extent of their chromatin association differs. This is best visualized in cells undergoing mitosis where H2A.Z-2.1 is mainly associated with mitotic chromosomes, as indicated by the co-localization of DAPI and anti-Myc signals, while the H2A.Z-2.2 protein is only partially co-localized (Fig. 5, C and D). Nonetheless, isoform 2 behaves entirely differently from a non-histone protein (GFP), which is apparently excluded from the condensed chromosomes (Fig. 5E). The anti-Myc signal is specific to H2A.Z-2, as no signal is seen without tetracycline induction (supplemental Fig. S3). Expression of H2A.Z-2.1 and H2A.Z-2.2 proteins to similar levels in human cells therefore reveals that the full-length H2A.Z-2.1 is predominantly associated with chromatin but the shorter H2A.Z-2.2 protein has a larger soluble pool and is more easily dissociated from chromatin by salt washing. This parallels the behavior of the C-terminal truncation mutant Htz1Δ120–133 that we isolated in yeast and indicates that the H2A.Z C-terminal tail regulates the stability and extent of H2A.Z's association with nucleosomes in both yeast and human cells.
DISCUSSION
Variant forms of H2A have heterogeneous C termini that appear to be central to the functional identity of these proteins. Post-translational modifications of the H2A.Z C terminus have previously been implicated in the DNA damage response in yeast and in heterochromatic silencing in mammals (11, 12). The experiments presented here demonstrate that in addition to modification at specific residues, the length of the H2A.Z C-terminal tail is also important for the normal functions of the protein.
Truncated Htz1 proteins are unable to perform the functions of full-length Htz1 in yeast, resulting in synthetic genetic interactions with asf1Δ and sensitivity of cells to MMS. While the shorter truncations Δ128–133 and Δ125–133 are not MMS sensitive, they were isolated as having growth defects in our original screen. They may therefore have subtle phenotypes that are not as severe as larger deletions and that do not affect the response to DNA damage. Known sites of sumoylation in the Htz1 C terminus that regulate the response to an irreparable DNA double-strand break (12) are not required for MMS resistance, indicating that the function of this region is not dependent on modification of these residues. Indeed, portions of the C-terminal tail of major H2A with markedly different amino acid sequences can fully compensate for the Htz1 C terminus in mediating resistance to MMS.
Removal of amino acids 111 onwards reduces the occupancy of H2A.Z in yeast chromatin, explaining the loss of function phenotypes observed for this mutant. For this reason, we have not characterized this mutant further, as chromatin localization is upstream of most H2A.Z functions. It is interesting to note that this mutant should have normal chromatin occupancy as it contains the M6 region that is required for association with the Htz1 deposition complex SWR-C (31, 32). This finding agrees with the work of Wang et al. (33), who recently showed that H2A.Z lacking the last 20 amino acids (Δ114–133) is not functional in yeast cells despite interacting with SWR-C components. As Htz1 can be enzymatically removed from nucleosomes by the INO80-C (34), one possibility is that the C-terminal tail regulates this process. Alternatively, the removal of amino acids may simply reduce the contact surfaces between Htz1 and other nucleosomal components, making the protein less able to stably associate. The C-terminal tail of major H2A is also required for stable association with nucleosomes and chromatin (35, 36), indicating that regulation of association with nucleosomes may be a general function of the C termini of H2A family proteins.
The mutant that we have focused on is the Δ120–133 truncation, which is not functional despite being associated with the chromatin fraction and having a normal localization pattern across the yeast genome. The slight but reproducible destabilization of this mutant relative to full-length Htz1, as measured by salt washes and ChIP, is the only detectable difference between the functional full-length protein and the non-functional Δ120–133 mutant. These data link the dynamics of the nucleosomal association of Htz1 to function in yeast cells.
Although the yeast C-terminal truncation mutants reveal a function for the H2A.Z C terminus and will be useful in further studies of the effects of modulating H2A.Z nucleosomal stability they do not, to our knowledge, represent physiological proteins. The novel splice isoform of human H2A.Z-2 containing exon 6, on the other hand, is a naturally occurring example of truncated H2A.Z. This splice isoform is expressed predominantly in human brain, skeletal muscle and liver, suggesting that this transcript may have tissue-specific roles. Although other transcripts derived from the H2A.Z-2 gene may exist in vivo, they encode proteins lacking sequences within the histone fold domain, and they do not produce stable protein in human cells.
A direct comparison of the characteristics of H2A.Z-2.1 and H2A.Z-2.2 in human cells showed that H2A.Z-2.2 is markedly less stably associated with chromatin than H2A.Z-2.1, as revealed by washing with buffers containing various salt concentrations. Additionally, isoform 1 is almost entirely chromatin bound while there is a large pool of soluble isoform 2. It is possible that the soluble pool of H2A.Z-2.2 is a result of the protein level in our system being above normal physiological levels, as isoforms 1 and 2 were induced to the same levels to allow direct comparison between them while RT-Q-PCR analysis indicates that the endogenous transcript encoding isoform 2 is less abundant than that encoding isoform 1. Nonetheless, the different behaviors of the proteins can only be attributed to the differences between their C termini. It will be interesting to discover whether the novel sequence in H2A.Z-2.2 provides a unique docking site for proteins regulating H2A.Z chromatin occupancy or if the shortening of the C terminus directly regulates stability.
Why might the nucleosomal stability of H2A.Z be important for its functions? H2A.Z is found in nucleosomes close to the position of transcription initiation in all organisms studied to date (37–43). These nucleosomes represent key control points as they can sterically occlude transcription factors and other proteins of the transcription machinery but may need to be easily disrupted in response to induction signals. Correspondingly, H2A.Z is required for normal kinetics of gene activation, and its chromatin occupancy is dynamic during gene activation and repression in yeast and mammalian cells (44–47). Yeast H2A.Z is dissociated from nucleosomes at lower salt concentrations than H2A (45) and nucleosomes containing H2A.Z are more susceptible to nuclease digestion than their canonical counterparts (48), indicating that H2A.Z nucleosomes in yeast are less stable than those containing H2A. In mammalian cells, nucleosomes containing H2A.Z at promoters and other regulatory regions are also highly salt-labile (49). These characteristics have been interpreted as important for H2A.Z functions but the effects of altering the stability of H2A.Z have not previously been tested. Our findings that a C-terminal truncation mutant with phenotypic defects is present with a normal distribution in chromatin but is nonetheless unable to function like wild-type Htz1 indicates that the altered stability of this mutant in nucleosomes interferes with its ability to function normally in yeast. In human cells, the naturally occurring shorter H2A.Z-2 isoform also displays altered affinity for chromatin relative to the full-length protein. The tissue-specific distribution of this isoform indicates that the relative instability of H2A.Z.2–2 may be harnessed as a novel regulatory mechanism in certain cell types. As the importance of the dynamic behavior of nucleosomes becomes clearer, the contribution of the H2A.Z C-terminal tail to nucleosome stability will help to reconcile the complex and diverse functional roles of H2A.Z.
Supplementary Material
Acknowledgments
We thank Stephen Taylor and Anthony Tighe for the DLD-1 host cell line and advice on cell culture, Enrique Amaya for access to his Q-PCR machine, and Tom Wood for generation of pCM556. We also thank Dean Jackson and Andy Sharrocks for comments on the manuscript and Sandra Hake for communication of results prior to publication.
This work was supported by a Wellcome Trust Career Development Fellowship (to C. B. M.) and a BBSRC Doctoral Training Studentship (to D. W.).

This article contains supplemental Tables S1 and S2 and Figs. S1–S3.
A. Thistlewaite, M. Harris, and C. B. Millar, manuscript in preparation.
- MNase
- micrococcal nuclease
- 5-FOA
- 5-fluoroorotic acid
- WT
- wild-type
- SWR-C
- Swr1 complex
- INO80-C
- Ino80 complex
- TAS
- tiling analysis software.
REFERENCES
- 1. Meneghini M. D., Wu M., Madhani H. D. (2003) Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112, 725–736 [DOI] [PubMed] [Google Scholar]
- 2. Guillemette B., Bataille A. R., Gévry N., Adam M., Blanchette M., Robert F., Gaudreau L. (2005) Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3, e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shia W. J., Li B., Workman J. L. (2006) SAS-mediated acetylation of histone H4 Lys 16 is required for H2A.Z incorporation at subtelomeric regions in Saccharomyces cerevisiae. Genes Dev. 20, 2507–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kimura A., Umehara T., Horikoshi M. (2002) Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 32, 370–377 [DOI] [PubMed] [Google Scholar]
- 5. Suka N., Luo K., Grunstein M. (2002) Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 32, 378–383 [DOI] [PubMed] [Google Scholar]
- 6. Ruthenburg A. J., Li H., Patel D. J., Allis C. D. (2007) Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Van Doninck K., Mandigo M. L., Hur J. H., Wang P., Guglielmini J., Milinkovitch M. C., Lane W. S., Meselson M. (2009) Phylogenomics of unusual histone H2A Variants in Bdelloid rotifers. PLoS Genet. 5, e1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Talbert P. B., Henikoff S. (2010) Histone variants–ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 11, 264–275 [DOI] [PubMed] [Google Scholar]
- 9. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 10. Vissers J. H., Nicassio F., van Lohuizen M., Di Fiore P. P., Citterio E. (2008) The many faces of ubiquitinated histone H2A: insights from the DUBs. Cell Div. 3, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sarcinella E., Zuzarte P. C., Lau P. N., Draker R., Cheung P. (2007) Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Mol. Cell. Biol. 27, 6457–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kalocsay M., Hiller N. J., Jentsch S. (2009) Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol. Cell 33, 335–343 [DOI] [PubMed] [Google Scholar]
- 13. Ikura T., Tashiro S., Kakino A., Shima H., Jacob N., Amunugama R., Yoder K., Izumi S., Kuraoka I., Tanaka K., Kimura H., Ikura M., Nishikubo S., Ito T., Muto A., Miyagawa K., Takeda S., Fishel R., Igarashi K., Kamiya K. (2007) DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol. Cell. Biol. 27, 7028–7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ogawa Y., Ono T., Wakata Y., Okawa K., Tagami H., Shibahara K. I. (2005) Histone variant macroH2A1.2 is mono-ubiquitinated at its histone domain. Biochem. Biophys. Res. Commun. 336, 204–209 [DOI] [PubMed] [Google Scholar]
- 15. Thambirajah A. A., Li A., Ishibashi T., Ausió J. (2009) New developments in post-translational modifications and functions of histone H2A variants. Biochem. Cell Biol. 87, 7–17 [DOI] [PubMed] [Google Scholar]
- 16. Ausió J., Abbott D. W. (2002) The many tales of a tail: carboxyl-terminal tail heterogeneity specializes histone H2A variants for defined chromatin function. Biochemistry 41, 5945–5949 [DOI] [PubMed] [Google Scholar]
- 17. Karaczyn A. A., Bal W., North S. L., Bare R. M., Hoang V. M., Fisher R. J., Kasprzak K. S. (2003) The octapeptidic end of the C-terminal tail of histone H2A is cleaved off in cells exposed to carcinogenic nickel(II). Chem. Res. Toxicol. 16, 1555–1559 [DOI] [PubMed] [Google Scholar]
- 18. Eickbush T. H., Godfrey J. E., Elia M. C., Moudrianakis E. N. (1988) H2a-specific proteolysis as a unique probe in the analysis of the histone octamer. J. Biol. Chem. 263, 18972–18978 [PubMed] [Google Scholar]
- 19. Karaczyn A. A., Cheng R. Y., Buzard G. S., Hartley J., Esposito D., Kasprzak K. S. (2009) Truncation of histone H2A's C-terminal tail, as is typical for Ni(II)-assisted specific peptide bond hydrolysis, has gene expression altering effects. Ann. Clin. Lab. Sci. 39, 251–262 [PMC free article] [PubMed] [Google Scholar]
- 20. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 21. Tighe A., Johnson V. L., Taylor S. S. (2004) Truncating APC mutations have dominant effects on proliferation, spindle checkpoint control, survival, and chromosome stability. J. Cell Sci. 117, 6339–6353 [DOI] [PubMed] [Google Scholar]
- 22. Millar C. B., Xu F., Zhang K., Grunstein M. (2006) Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 20, 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Su A. I., Wiltshire T., Batalov S., Lapp H., Ching K. A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Cooke M. P., Walker J. R., Hogenesch J. B. (2004) A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U.S.A. 101, 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 [DOI] [PubMed] [Google Scholar]
- 25. Jackson J. D., Gorovsky M. A. (2000) Histone H2A.Z has a conserved function that is distinct from that of the major H2A sequence variants. Nucleic Acids Res. 28, 3811–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Collins S. R., Miller K. M., Maas N. L., Roguev A., Fillingham J., Chu C. S., Schuldiner M., Gebbia M., Recht J., Shales M., Ding H., Xu H., Han J., Ingvarsdottir K., Cheng B., Andrews B., Boone C., Berger S. L., Hieter P., Zhang Z., Brown G. W., Ingles C. J., Emili A., Allis C. D., Toczyski D. P., Weissman J. S., Greenblatt J. F., Krogan N. J. (2007) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446, 806–810 [DOI] [PubMed] [Google Scholar]
- 27. Mizuguchi G., Shen X., Landry J., Wu W. H., Sen S., Wu C. (2004) ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348 [DOI] [PubMed] [Google Scholar]
- 28. Kobor M. S., Venkatasubrahmanyam S., Meneghini M. D., Gin J. W., Jennings J. L., Link A. J., Madhani H. D., Rine J. (2004) A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2, E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Clarkson M. J., Wells J. R., Gibson F., Saint R., Tremethick D. J. (1999) Regions of variant histone His2AvD required for Drosophila development. Nature 399, 694–697 [DOI] [PubMed] [Google Scholar]
- 30. Eirín-López J. M., González-Romero R., Dryhurst D., Ishibashi T., Ausió J. (2009) The evolutionary differentiation of two histone H2A.Z variants in chordates (H2A.Z-1 and H2A.Z-2) is mediated by a stepwise mutation process that affects three amino acid residues. BMC Evol. Biol. 9, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu W. H., Alami S., Luk E., Wu C. H., Sen S., Mizuguchi G., Wei D., Wu C. (2005) Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat. Struct. Mol. Biol. 12, 1064–1071 [DOI] [PubMed] [Google Scholar]
- 32. Jensen K., Santisteban M. S., Urekar C., Smith M. M. (2011) Mol. Genet. Genomics 285, 287–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang A. Y., Aristizabal M. J., Ryan C., Krogan N. J., Kobor M. S. (2011) Key functional regions in the histone variant H2A.Z C-terminal docking domain. Mol. Cell. Biol. 31, 3871–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Papamichos-Chronakis M., Watanabe S., Rando O. J., Peterson C. L. (2011) Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 144, 200–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shukla M. S., Syed S. H., Goutte-Gattat D., Richard J. L., Montel F., Hamiche A., Travers A., Faivre-Moskalenko C., Bednar J., Hayes J. J., Angelov D., Dimitrov S. (2011) The docking domain of histone H2A is required for H1 binding and RSC-mediated nucleosome remodeling. Nucleic Acids Res. 39, 2559–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vogler C., Huber C., Waldmann T., Ettig R., Braun L., Izzo A., Daujat S., Chassignet I., Lopez-Contreras A. J., Fernandez-Capetillo O., Dundr M., Rippe K., Längst G., Schneider R. (2010) Histone H2A C terminus regulates chromatin dynamics, remodeling, and histone H1 binding. PLoS Genet. 6, e1001234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Raisner R. M., Hartley P. D., Meneghini M. D., Bao M. Z., Liu C. L., Schreiber S. L., Rando O. J., Madhani H. D. (2005) Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123, 233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Albert I., Mavrich T. N., Tomsho L. P., Qi J., Zanton S. J., Schuster S. C., Pugh B. F. (2007) Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446, 572–576 [DOI] [PubMed] [Google Scholar]
- 39. Mavrich T. N., Jiang C., Ioshikhes I. P., Li X., Venters B. J., Zanton S. J., Tomsho L. P., Qi J., Glaser R. L., Schuster S. C., Gilmour D. S., Albert I., Pugh B. F. (2008) Nucleosome organization in the Drosophila genome. Nature 453, 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Whittle C. M., McClinic K. N., Ercan S., Zhang X., Green R. D., Kelly W. G., Lieb J. D. (2008) The genomic distribution and function of histone variant HTZ-1 during C. elegans embryogenesis. PLoS Genet. 4, e1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 42. Creyghton M. P., Markoulaki S., Levine S. S., Hanna J., Lodato M. A., Sha K., Young R. A., Jaenisch R., Boyer L. A. (2008) H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell 135, 649–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zilberman D., Coleman-Derr D., Ballinger T., Henikoff S. (2008) Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456, 125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Santisteban M. S., Kalashnikova T., Smith M. M. (2000) Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell 103, 411–422 [DOI] [PubMed] [Google Scholar]
- 45. Zhang H., Roberts D. N., Cairns B. R. (2005) Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123, 219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gévry N., Chan H. M., Laflamme L., Livingston D. M., Gaudreau L. (2007) p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 21, 1869–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gévry N., Hardy S., Jacques P. E., Laflamme L., Svotelis A., Robert F., Gaudreau L. (2009) Histone H2A.Z is essential for estrogen receptor signaling. Genes Dev. 23, 1522–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xi Y., Yao J., Chen R., Li W., He X. (2011) Nucleosome fragility reveals novel functional states of chromatin and poises genes for activation. Genome Res. 21, 718–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jin C., Zang C., Wei G., Cui K., Peng W., Zhao K., Felsenfeld G. (2009) H3.3/H2A.Z double variant-containing nucleosomes mark nucleosome-free regions of active promoters and other regulatory regions. Nat. Genet. 41, 941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.