Background: Bacterial plasminogen (Pg) activators like SUPA subvert mammalian Pg activation to facilitate infection in a species-restricted manner.
Results: Fibrin blood clots permit SUPA to activate human Pg, thereby overriding species restriction.
Conclusion: Fibrin provides a source of enzyme cofactor and enhances the catalytic efficiency of SUPA.
Significance: Molecular interactions that override species-restricted Pg activation may be used to modulate infections or create new human therapeutics.
Keywords: Bacteria, Fibrin, Fibrinolysis, Plasminogen, Streptococcus, Pau A, α2-Antiplasmin, Plasminogen Activation, Serpin, Streptococcus uberis
Abstract
Bacterial plasminogen (Pg) activators generate plasmin to degrade fibrin blood clots and other proteins that modulate the pathogenesis of infection, yet despite strong homology between mammalian Pgs, the activity of bacterial Pg activators is thought to be restricted to the Pg of their host mammalian species. Thus, we found that Streptococcus uberis Pg activator (SUPA), isolated from a Streptococcus species that infects cows but not humans, robustly activated bovine but not human Pg in purified systems and in plasma. Consistent with this, SUPA formed a higher avidity complex (118-fold) with bovine Pg than with human Pg and non-proteolytically activated bovine but not human Pg. Surprisingly, however, the presence of human fibrin overrides the species-restricted action of SUPA. First, human fibrin enhanced the binding avidity of SUPA for human Pg by 4–8-fold in the presence and absence of chloride ion (a negative regulator). Second, although SUPA did not protect plasmin from inactivation by α2-antiplasmin, fibrin did protect human plasmin, which formed a 31-fold higher avidity complex with SUPA than Pg. Third, fibrin significantly enhanced Pg activation by reducing the Km (4-fold) and improving the catalytic efficiency of the SUPA complex (6-fold). Taken together, these data suggest that indirect molecular interactions may override the species-restricted activity of bacterial Pg activators; this may affect the pathogenesis of infections or may be exploited to facilitate the design of new blood clot-dissolving drugs.
Introduction
The plasminogen (Pg)2 and plasmin system play key roles in physiologic processes such as fibrin blood clot dissolution, wound healing, cell migration, and the pathogenesis of certain types of bacterial infections. Pg activators convert Pg into plasmin, a proteolytic enzyme that cleaves fibrin polymers into soluble degradation products (1). Both mammals and bacteria produce Pg activators, but they are markedly different in structure and mechanisms of action. The mammalian Pg activators, tissue Pg activator and urinary-type Pg activator, are serine proteases that directly cleave a broad spectrum of Pgs from different animal species (2–4). In contrast, the bacterial Pg activators lack inherent protease activity and form complexes to cleave a restricted spectrum of mammalian Pgs (5–7).
The mechanisms responsible for the species-restricted action of bacterial Pg activators are not fully understood, but they affect the consideration of bacterial Pg activators as potential therapies for human clotting or thrombotic diseases such as heart attacks and strokes. For example, the prototypical bacterial Pg activator streptokinase (SK), which was isolated from bacteria that infect humans, preferentially activates human but not bovine Pg. In a similar fashion, SUPA (PauA), a Pg activator purified from Streptococcus uberis, a bacterium that infects cows, has been shown to activate bovine Pg but not human Pg (8–11). These differences are remarkable considering that both Pg activators function through similar mechanisms. Both SK and SUPA non-proteolytically activate their cognate Pgs, form activator complexes with plasmins, reduce the susceptibility of plasmin to inhibition by α2-antiplasmin, and alter the substrate specificity of plasmin (10, 12).
The species-restricted activation of Pg by bacterial Pg activators is due in part to key sequence differences that alter the intermolecular complementarity between the Pg and the Pg activator (6, 13, 14). In this study, we show that the species-restricted activation of human Pg activation by SUPA is profoundly affected by fibrin and other molecules that interact with Pg. These findings provide new insights into the regulation of the activity of the bacterial Pg activators in different mammalian hosts that can be used to modulate blood clot-dissolving activity or to regulate their function during bacterial infection.
EXPERIMENTAL PROCEDURES
Protein and Reagents
Reagents were purchased from the indicated sources: human Glu-Pg, bovine Pg, human Pg-depleted fibrinogen, and CNBr-digested fibrinogen (American Diagnostica Inc., Stamford, CT); α2-antiplasmin (Calbiochem); citrated frozen human and bovine plasma (Lampire Biological Laboratories, Pipersville, PA); tissue Pg activator (Baxter Biotechnology, Hayward, CA); 125I-fibrinogen (PerkinElmer Life Sciences); and all other reagents if not specified (Sigma).
Cloning, Expression, and Purification of Recombinant Protein
SK, staphylokinase, and SUPA were cloned, expressed, and purified as described (10, 12, 15). Human micro-Pg and the cleavage-resistant mutant micro-Pg(R561A) were prepared as described previously (16). Protein concentrations were determined using a BCA protein kit (Thermo Fisher Scientific, Rockford, IL), and the active concentration of each protein was determined by active site titration as described below.
Active Site Titration
Non-proteolytic active site generation by a Glu-Pg and SK complex or a bovine Pg and SUPA complex was determined at 25 °C in a multi-detection microplate reader (BioTek Instruments, Winooski, VT) by active site titration with the fluorogenic substrate 4-methylumbelliferyl p-guanidinobenzoate (15, 17, 18). Briefly, 400 nm Pg or micro-Pg was added to a microplate well containing 1 μm 4-methylumbelliferyl p-guanidinobenzoate in filtered HEPES assay buffer (10 mm HEPES, pH 7.2) at 25 °C. After 10 min, SK or SUPA (200–800 nm) or buffer alone (control) was added to test wells. In certain runs, a complex of Pg and Pg activator, which was preformed on ice by mixing both proteins for 10–20 min, was added directly to the assay buffer. The development of fluorescence was monitored continuously with excitation at 360 nm and emission at 460 nm. The fluorescence increase was used to calculate the active site generation using 4-methylumbelliferone as a control.
Binding Assay
SUPA and Pg or plasmin binding assays were performed in microtiter plates coated with 100 nm SUPA for 1 h at room temperature. Wells were washed, and nonspecific binding sites were blocked with 1% bovine serum albumin. After that, varying concentrations of Pg or aprotinin-inactivated plasmin were added for 1 h. After washing, mouse anti-plasmin(ogen) monoclonal antibody was added for 1 h, followed by washing and the addition of horseradish peroxidase-conjugated anti-mouse IgG for 1 h. The bound antibody was detected by 3,3′,5,5′-tetramethylbenzidine substrate, and the reaction was monitored at A370 nm within the dynamic range of the microplate reader. Binding constants were calculated using GraphPad Prism software.
Pg Activation
The activation of human Glu-Pg/bovine Pg/human micro-Pg by various amounts of Pg activator (SK, SUPA, and staphylokinase) was examined at 37 °C in either HEPES buffer (10 mm HEPES with or without 150 mm NaOAc or NaCl, pH 7.4) or Tris/NaCl buffer (50 mm Tris-HCl and 100 mm NaCl, pH 7.4). Pgs were pretreated with aprotinin-agarose beads for 4 h at 4 °C to remove contaminating plasmin. Pg was added to the assay buffer in the presence of 500 μm S-2251 (Chromogenix-Instrumentation Laboratory, Lexington, MA) (19), followed by Pg activator (final volume of 100 μl). The change in absorbance at 405 nm was continuously recorded in a Thermomax microtiter plate reader. To examine the effect of fibrinogen or CNBr-fibrin fragments on Pg activation, Pg was preincubated with each reagent in the assay buffer for 10 min before the addition of Pg activator. Pg activation parameters were calculated using amidolytic parameters for plasmin as described by Wohl et al. (20).
Plasma Fibrinogenolysis
Human or bovine plasma fibrinogenolysis was determined by incubating 180 μl of plasma with 20 μl assay buffer containing varying concentrations of SUPA or no SUPA (control) at 37 °C. Samples were collected after 2 h and mixed with 200 kallikrein inhibitor units of aprotinin/ml of plasma to quench plasmin-mediated lysis. Residual fibrinogen concentrations were determined by the sodium sulfite method (21).
Plasma Fibrinolysis
Human or bovine plasma fibrinolysis was determined by simultaneous mixing 180 μl of plasma with various amounts of SUPA in the presence of 1 mm Ca2+, 1 IU/ml thrombin, and trace amounts of 125I-fibrinogen (final volume of 200 μl). The total radioactivity of the sample was monitored using a Cobra II γ-counter (PerkinElmer Life Sciences). After incubation of the samples at 37 °C for 2 h, 200 kallikrein inhibitor units of aprotinin was added to each milliliter of plasma clot to inhibit further lysis. The radioactivity of the clot and supernatant plasma was monitored separately. The percentage of fibrinolysis was defined as the difference between the initial and residual clot radioactivity divided by the initial clot radioactivity.
SDS-PAGE and Immunoblotting
The generation of plasmin by SUPA was detected using SDS-PAGE assay under denaturing conditions. The effect of SUPA or SK on the inhibition of plasmin by α2-antiplasmin was examined. Plasmin (0.25 μm) was mixed with varying concentrations of SK or SUPA (0.25–2.5 μm) in HEPES/NaOAc buffer, pH 7.4, on ice for 20 min, followed by the addition of 0.25 μm α2-antiplasmin. After 10 min, samples were analyzed by nonreducing SDS-PAGE, followed by immunoblotting with a monoclonal antibody against α2-antiplasmin (RWR) (22).
RESULTS
Species-restricted Pg Activation
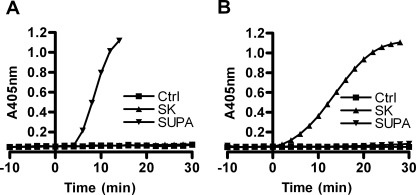
To confirm that SUPA is restricted in its ability to activate different mammalian Pgs, we examined its action on human and bovine Pgs. SUPA rapidly and efficiently activated purified bovine Pg (Fig. 1A) but did not efficiently activate purified human Pg (Fig. 1B). In contrast, as a control, SK had no activity against bovine Pg (Fig. 1A), but it rapidly and efficiently activated human Pg (Fig. 1B).
FIGURE 1.
Species-restricted Pg activation by SUPA. Bovine Pg (A) or human Glu-Pg (B) (100 nm) was activated by 40 nm SK or SUPA in Tris/NaCl buffer at 37 °C. The production of plasmin was monitored using S-2251 substrate (500 μm) as described under “Experimental Procedures.” The results shown are representative of at least three experiments. Ctrl, control.
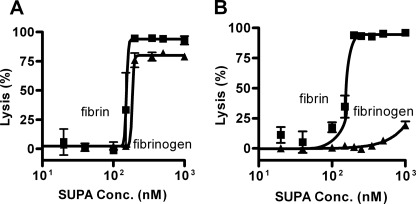
Pg activation with purified reagents in vitro is quite different from Pg activation in complex biologic solutions such as plasma and serum that contain molecules (e.g. fibrinogen, fibrin, α2-antiplasmin, etc.) that modify the activity of both Pg and plasmin. In plasma or serum, the plasmin generated by Pg activators degrades target substrates such as fibrinogen and fibrin. When SUPA was added to bovine plasma (Fig. 2A), it triggered the degradation of fibrinogen in a potent dose-related process with an EC50 of 185 nm. In a similar fashion, when added to bovine serum containing fibrin clots, there was a potent degradation of fibrin with a comparable EC50 of 153 nm. When SUPA was added to human plasma (Fig. 2B), it triggered fibrinogen degradation with a potency that was markedly lower (estimated EC50 of ∼1000 nm) than in bovine plasma. In contrast, when SUPA was added to human serum containing fibrin, it initiated fibrin degradation with a similar potency (EC50 = 154 nm) as that seen in bovine plasma and serum. These results indicate that fibrin and, to a much lesser extent, fibrinogen significantly alter the SUPA species-restricted mechanism of human Pg activation.
FIGURE 2.
Fibrin-targeted activation of human but not bovine Pg by SUPA. The effect of SUPA-mediated Pg activation on fibrinolysis and fibrinogenolysis was examined in bovine (A) and human (B) plasma. The results shown are representative of two independent experiments. Means ± S.D. are shown.
SUPA-Pg Complexes
Pg activation and subsequent degradation of fibrinogen and fibrin by SUPA require the initial formation of SUPA-Pg complexes. Consistent with this notion, in the presence of physiologic levels of chloride ion, bovine Pg bound well to SUPA (KD = 6.9 ± 0.5 nm). However, the relative binding avidity of SUPA for human Pg was 118-fold less (KD = 811 ± 356 nm) in the same chloride-containing buffer (Table 1). Chloride ion is known to regulate the conformation of Pg and its interactions with other molecules. Indeed, in the absence of chloride ion, there was enhanced binding (4-fold, KD = 194 ± 22 nm) of SUPA to human Pg. In the presence of chloride ion, SUPA binding was also enhanced by fibrin (8-fold, KD = 99 ± 50 nm).
TABLE 1.
Binding constants for SUPA-Pg or SUPA-plasmin complexes
|
KD |
|||
|---|---|---|---|
| +Chloride (150 mm) | −Chloride | +Chloride + fibrin (0.1 mg/ml) | |
| ×10−9m | |||
| SUPA-human Pg | 811 ± 356 | 194 ± 22 | 99 ± 50 |
| SUPA-human plasmin | 26 ± 4 | 33 ± 3 | 26 ± 1 |
SUPA-Plasmin Complexes
By comparison with human Pg, human plasmin bound with greater avidity to SUPA. In the presence of chloride ion, the KD for SUPA-plasmin binding was 31-fold lower (KD = 26 ± 4 nm) (Table 1). In contrast to Pg, the binding of human plasmin was minimally affected, if at all, by the absence of chloride ion (KD = 33 ± 3 nm) or the presence of fibrin (KD = 26 ± 1 nm).
Fibrin Enhances Activation of Human Pg by SUPA
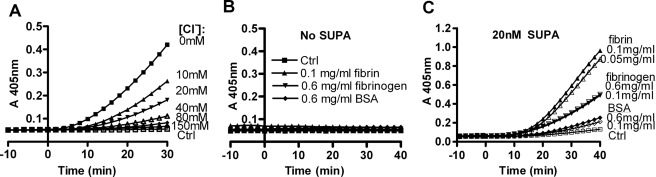
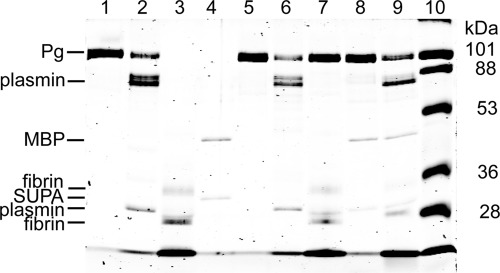
After formation, the SUPA-activator complex interacts with Pg substrate to catalyze the production of plasmin. In fluids containing physiologic concentrations of chloride ion, human Glu-Pg adopts a folded conformation that protects it from activation (24, 25). Fibrin opposes the effects of chloride ion and unfolds the Glu-Pg substrate, which may modulate the ability of the SUPA-activator complex to activate human Glu-Pg substrate (26, 27). Although chloride did not significantly affect the formation of the SUPA-plasmin activator complex (see below and Table 1), increasing chloride concentrations inhibited SUPA-induced Pg activation in a concentration-dependent manner (Fig. 3A). In the presence of chloride ion, fibrinogen modestly enhanced Pg activation by SUPA compared with BSA alone (as a nonspecific protein control) (Fig. 3C). However, Pg activation by SUPA (Fig. 3C) was enhanced more markedly by cyanogen bromide-digested fibrin fragments, a widely used soluble form of fibrin (28, 29). Direct analysis of these reactions by SDS-PAGE (Fig. 4) confirmed that fibrin fragments enhanced human Pg activation by SUPA compared with Pg activation in the absence of fibrin. Kinetic analysis of Pg activation by SUPA in HEPES buffer containing 150 mm chloride ion indicated that the presence of 0.1 mg/ml CNBr-fibrin decreased KPg by 4.2-fold, increased kPg by 1.4-fold, and enhanced the catalytic efficiency (kPg/KPg) by 6.1-fold (Table 2).
FIGURE 3.
Fibrin and chloride ions modulate human Glu-Pg activation by SUPA. A, effect of chloride on Pg activation. Human Glu-Pg (100 nm) was mixed with HEPES buffer containing various amounts of NaCl (0–150 mm) and 500 μm S-2251. The total ionic strength of the buffers was maintained at 150 mm by the addition of NaOAc to buffers with chloride concentrations <150 mm. After incubation at 37 °C for 10 min, SUPA (40 nm) was added at t = 0, and the absorbance at 405 nm was monitored to detect the generation of plasmin. Results are representative of two experiments. B and C, fibrin fragments enhance Glu-Pg activation by SUPA. Human Glu-Pg (100 nm) was added to Tris/NaCl buffer containing 500 μm S-2251 at 37 °C in the absence (control (Ctrl)) or presence of varying concentrations of CNBr-fibrin fragments, fibrinogen, or BSA. After 10 min, Pg activation was initiated by the addition of 0 (B) or 20 (C) nm SUPA, and the absorbance at 405 nm was monitored. The results shown are representative of two independent experiments.
FIGURE 4.
Analysis of human Glu-Pg activation by SUPA in presence of fibrin. Human Glu-Pg (500 nm) was mixed with 40 nm SUPA in Tris/NaCl buffer in the absence or presence of 0.1 mg/ml CNBr-fibrin fragments for 0.5 h at 37 °C. Samples were collected, reduced, and analyzed by SDS-PAGE. Lanes 1–4, reagents alone at t = 0: Glu-Pg (lane 1), human plasmin (lane 2), fibrin (lane 3), and SUPA cut from the fusion partner maltose-binding protein (MBP; lane 4). Lanes 5–9, reagents after incubation at 37 °C for 0.5 h: Glu-Pg (lane 5), human plasmin (lane 6), Glu-Pg in the presence of fibrin (lane 7), Glu-Pg activated by SUPA (lane 8), and Glu-Pg activated by SUPA in the presence of fibrin (lane 9). Lane 10, protein standards and relative molecular mass (shown in kilodaltons).
TABLE 2.
Effects of chloride and fibrin on activation of human Pg by SUPA
Activation experiments were carried out at 37 °C in a total volume of 100 μl with HEPES/NaCl or HEPES/NaOAc buffer in the absence or presence of 0.1 mg/ml fibrin (CNBr-fibrin fragments). The kinetic analysis of activation parameters is described under “Experimental Procedures.” Values represent the mean ± S.D.
| Buffer | Plasminogen activation parameters |
||
|---|---|---|---|
| KPg | kPg | kPg/KPg | |
| nm | min−1 | μm−1min−1 | |
| HEPES/NaCl | 202 ± 61 | 0.98 ± 0.02 | 4.8 |
| HEPES/NaOAc | 96 ± 15 | 3.52 ± 0.01 | 36.6 |
| HEPES/NaCl + fibrin | 48 ± 7 | 1.41 ± 0.01 | 29.6 |
| HEPES/NaOAc + fibrin | 30 ± 2 | 3.48 ± 0.01 | 114.3 |
SUPA Is Unable to Activate Pg through Non-proteolytic Mechanisms
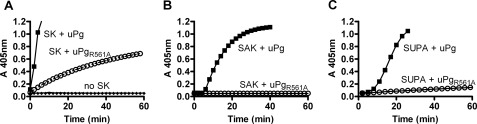
In part, SK efficiently activates human Pg in plasma through a non-proteolytic mechanism that does not require cleavage of the Arg561–Val bond to form plasmin. In a similar fashion, SUPA can non-proteolytically activate bovine Pg (10, 11). Because plasmin is a trace contaminant of Pg preparations, we examined whether SUPA can non-proteolytically activate human Pg using micro-Pg(R561A); this contains an activation loop mutation of Arg561 to alanine that prevents it from forming plasmin (16, 23). Consistent with its ability to non-proteolytically activate Pg, SK generated active complexes with both micro-Pg and mutant micro-Pg(R561A) (Fig. 5A). In contrast, staphylokinase, which cannot non-proteolytically activate human Pg, activated micro-Pg but not micro-Pg(R561A) (Fig. 5B). Similar to staphylokinase, SUPA activated micro-Pg but not micro-Pg(R561A) (Fig. 5C). These observations confirm that human Pg activation by staphylokinase or SUPA requires the presence of microplasmin, which is a trace contaminant of micro-Pg but not micro-Pg(R561A) (16, 23). Thus, SUPA appears to activate human Pg in plasma through a mechanism akin to staphylokinase: by forming an activator complex with plasmin.
FIGURE 5.
Activation of micro-Pg or micro-Pg(R561A) by different Pg activators. Human micro-Pg (uPg) or the cleavage-resistant mutant micro-Pg(R561A) (uPgR561A) was added to HEPES buffer containing 500 μm S-2251. After incubation for 10 min, 200 nm SK (A), 200 nm staphylokinase (B), 1000 nm SUPA (C), or no Pg activators were added to micro-Pg or micro-Pg(R561A) at an equimolar concentration. Controls consisted of micro-Pg (200 nm) or the cleavage-resistant mutant micro-Pg(R561A) (200 nm) in HEPES buffer containing 500 μm S-2251 without Pg activator. Results are representative of two experiments.
SUPA Does Not Prevent Formation of Plasmin-α2-Antiplasmin Complexes
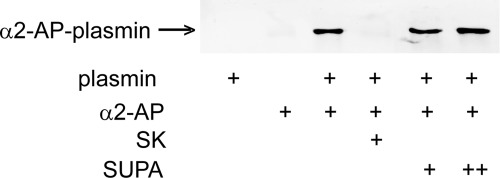
By forming covalent plasmin-α2-antiplasmin complexes, α2-antiplasmin rapidly inactivates plasmin in plasma. The binding of plasmin to fibrin partially protects from inactivation by α2-antiplasmin (30, 31). Thus, as expected, when mixed with plasmin, α2-antiplasmin formed inactive plasmin-α2-antiplasmin complexes (Fig. 6). SK protected against the formation of inactive human plasmin-α2-antiplasmin complexes. However, SUPA did not prevent the formation of human plasmin-α2-antiplasmin complexes (Fig. 6). Thus, in contrast to its interactions with bovine plasmin, SUPA is unable to protect human plasmin from inactivation by α2-antiplasmin.
FIGURE 6.
SK-plasmin and SUPA-plasmin resistance to α2-antiplasmin inhibition. Plasmin (0.25 μm) was mixed with SK (0.25 μm) or varying concentrations of SUPA (0.25 or 2.5 μm) in HEPES/NaOAc buffer and incubated on ice for 20 min, followed by the addition of 0.25 μm α2-antiplasmin (α2AP) and incubation for an additional 10 min. Samples were analyzed by SDS-PAGE under nonreducing conditions using Western blotting. The formation of the plasmin-α2-antiplasmin complex was detected by mouse anti-human α2-antiplasmin monoclonal antibody at the appropriate molecular size as indicated. First lane, plasmin alone; second lane, α2-antiplasmin alone; third lane, plasmin and α2-antiplasmin; fourth lane, plasmin, 0.25 μm SK, and α2-antiplasmin; fifth and sixth lanes, plasmin and 0.25 or 2.5 μm SUPA (respectively) mixed with α2-antiplasmin.
DISCUSSION
The restricted ability of bacterial Pg activators to activate different mammalian Pgs has long been considered a fundamental distinction between mammalian and bacterial Pg activators (6). Thus, in plasma or in purified systems with physiologic concentrations of chloride, SUPA efficiently activates bovine but not human Pg. However, in the presence of fibrin, SUPA acquires the ability to efficiently activate human Pg. This fibrin-targeted activity of SUPA with human Pg is the result of several different mechanisms.
First, fibrin modifies the avidity of binding interactions between SUPA and human Pg. In plasma and fluids that contain physiologic concentrations of chloride ion (e.g. ∼120–150 mm), the binding interactions between SUPA and human Pg are of much lower avidity (118-fold higher KD) than the binding interactions between SUPA and bovine Pg. Fibrin significantly enhances the binding interactions between SUPA and human Pg (8-fold lower KD), as does the absence of chloride ion (4-fold lower KD). Fibrin has a minimal effect on the binding interactions between SUPA and human plasmin, which are already of much higher avidity (31-fold lower KD) than the binding interactions with human Pg.
Second, fibrin provides a protective environment for the existence of plasmin, which permits the formation of the SUPA-plasmin complex. This is critical because SUPA cannot non-proteolytically activate human Pg as it does bovine Pg. Similar to staphylokinase, SUPA does not protect human plasmin from inhibition by α2-antiplasmin; fibrin does (30, 31). This effectively limits Pg activation by the SUPA-plasmin complex to the fibrin surface, where it is partially protected from inhibitors.
Third, fibrin enhances the catalytic efficiency of the SUPA-plasmin complex with human Pg substrate by 6-fold. Because fibrin does not significantly affect the formation of the SUPA-plasmin complex (Table 1), this enhanced catalytic efficiency is likely to reflect the known effects of fibrin on unfolding the Pg substrate conformation (26, 27). In a similar fashion, the negative effects of chloride ion on the catalytic efficiency of the SUPA-plasmin complex may well be attributed to the effect that chloride ion has on promoting a folded, less readily activated Pg substrate.
Species-restricted activation of Pg by bacterial Pg activators has been noted for decades, but the mechanisms responsible for it are still poorly understood. This species restriction occurs despite homology between Pgs from different species and between different SK-like bacterial Pg activators. For example, the α- and β-domains of SUPA show considerable sequence identity (30.9 and 27.4%, respectively) and structural similarity to SK and significant but less sequence identity (11.9 and 15.6%, respectively) and structural similarity to staphylokinase (10, 11). Species restriction also occurs despite evidence of binding interactions between different SK-like molecules and Pgs from different species (13, 32). One key determinant of species restriction is the requirement of intermolecular complementarity at sites necessary for function (6, 14). Alteration of key loop residues in human Pg makes it susceptible to activation by SUPA (6). In a complementary fashion, replacing the SK β-domain with the SUPA β-domain creates an SK chimera that activates horse Pg, a non-cognate substrate of native SK (6). Still, it has not been appreciated that Pg-interacting molecules may also regulate species-specific Pg activation by altering the interactions, conformation, etc., of Pg. In this study, we have shown that human fibrin, which binds and alters the conformation of Pg, improves the substrate binding and processing of human Pg by SUPA.
The Pg-plasmin system has been exploited by bacteria to facilitate infection (6). For example, group A streptococci producing SK (which does not activate mouse Pg) cannot infect normal mice, but they can infect mice carrying a transgene for human Pg (33). In a similar fashion, vaccines generated against SUPA (PauA) have been shown to reduce infection by S. uberis (34). Both Pg and fibrinogen are key modulators of the pathogenesis and inflammatory response to bacteria producing Pg activators such as Yersinia pestis (35). The action of bacterial Pg activators on the host Pg system can degrade the extracellular matrix to facilitate the spread and invasion of bacteria (36). In addition to Pg activators, bacteria also produce plasmin-binding molecules that have been implicated in the pathogenesis of infection because 1) they protect plasmin from inhibition by serpins like α2-antiplasmin (37) and 2) they modify the activity of the Pg activator complexes. It has also recently been shown that bacteria that lack intrinsic fibrinolytic activity produce a molecule (Skizzle) that can enhance Pg activation by either urinary-type or tissue-type Pg activator (32).
Our results suggest that a number of different bacterial Pg activators, once thought to be specific for cognate mammalian Pgs, can be induced to become activators of human Pg. Further insights into the mechanisms of action of Pg activators may result in new therapeutic agents targeted to human clots as well as novel approaches for ablating the contribution of bacterial Pg activators to the pathogenesis of infections.
Acknowledgment
We thank Dr. Irina Sazonova (Georgia Health Sciences University) for discussions regarding this work.
The work was supported, in whole or in part, by National Institutes of Health Grants HL58496 and HL78562 (to G. L. R.). This work was also supported by American Heart Association Grant SDG 0835376N (to I. P. G.).
- Pg
- plasminogen
- SK
- streptokinase
- SUPA
- S. uberis plasminogen activator.
REFERENCES
- 1. Castellino F. J., Ploplis V. A. (2005) Structure and function of the plasminogen/plasmin system. Thromb. Haemost. 93, 647–654 [DOI] [PubMed] [Google Scholar]
- 2. Bode W., Renatus M. (1997) Tissue-type plasminogen activator: variants and crystal/solution structures demarcate structural determinants of function. Curr. Opin. Struct. Biol. 7, 865–872 [DOI] [PubMed] [Google Scholar]
- 3. Blasi F., Vassalli J. D., Danø K. (1987) Urokinase-type plasminogen activator: proenzyme, receptor, and inhibitors. J. Cell Biol. 104, 801–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Collen D., Lijnen H. R. (2004) Tissue-type plasminogen activator: a historical perspective and personal account. J. Thromb. Haemost. 2, 541–546 [DOI] [PubMed] [Google Scholar]
- 5. Cliffton E. E., Cannamela D. A. (1953) Proteolytic and fibrinolytic activity of serum: activation by streptokinase and staphylokinase indicating dissimilarity of enzymes. Blood 8, 554–562 [PubMed] [Google Scholar]
- 6. Gladysheva I. P., Turner R. B., Sazonova I. Y., Liu L., Reed G. L. (2003) Coevolutionary patterns in plasminogen activation. Proc. Natl. Acad. Sci. U.S.A. 100, 9168–9172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lähteenmäki K., Kuusela P., Korhonen T. K. (2001) Bacterial plasminogen activators and receptors. FEMS Microbiol. Rev. 25, 531–552 [DOI] [PubMed] [Google Scholar]
- 8. Johnsen L. B., Rasmussen L. K., Petersen T. E., Etzerodt M., Fedosov S. N. (2000) Kinetic and structural characterization of a two-domain streptokinase: dissection of domain functionality. Biochemistry 39, 6440–6448 [DOI] [PubMed] [Google Scholar]
- 9. Leigh J. A. (1994) Purification of a plasminogen activator from Streptococcus uberis. FEMS Microbiol. Lett. 118, 153–158 [DOI] [PubMed] [Google Scholar]
- 10. Sazonova I. Y., Houng A. K., Chowdhry S. A., Robinson B. R., Hedstrom L., Reed G. L. (2001) The mechanism of a bacterial plasminogen activator intermediate between streptokinase and staphylokinase. J. Biol. Chem. 276, 12609–12613 [DOI] [PubMed] [Google Scholar]
- 11. Ward P. N., Field T. R., Rosey E. L., Abu-Median A. B., Lincoln R. A., Leigh J. A. (2004) Complex interactions between bovine plasminogen and streptococcal plasminogen activator PauA. J. Mol. Biol. 342, 1101–1114 [DOI] [PubMed] [Google Scholar]
- 12. Sazonova I. Y., Robinson B. R., Gladysheva I. P., Castellino F. J., Reed G. L. (2004) α-Domain deletion converts streptokinase into a fibrin-dependent plasminogen activator through mechanisms akin to staphylokinase and tissue plasminogen activator. J. Biol. Chem. 279, 24994–25001 [DOI] [PubMed] [Google Scholar]
- 13. Gladysheva I. P., Sazonova I. Y., Chowdhry S. A., Liu L., Turner R. B., Reed G. L. (2002) Chimerism reveals a role for the streptokinase β-domain in non-proteolytic active site formation, substrate, and inhibitor interactions. J. Biol. Chem. 277, 26846–26851 [DOI] [PubMed] [Google Scholar]
- 14. Wang X., Lin X., Loy J. A., Tang J., Zhang X. C. (1998) Crystal structure of the catalytic domain of human plasmin complexed with streptokinase. Science 281, 1662–1665 [DOI] [PubMed] [Google Scholar]
- 15. Reed G. L., Lin L. F., Parhami-Seren B., Kussie P. (1995) Identification of a plasminogen-binding region in streptokinase that is necessary for the creation of a functional streptokinase-plasminogen activator complex. Biochemistry 34, 10266–10271 [DOI] [PubMed] [Google Scholar]
- 16. Wang S., Reed G. L., Hedstrom L. (1999) Deletion of Ile1 changes the mechanism of streptokinase: evidence for the molecular sexuality hypothesis. Biochemistry 38, 5232–5240 [DOI] [PubMed] [Google Scholar]
- 17. Chase T., Jr., Shaw E. (1969) Comparison of the esterase activities of trypsin, plasmin, and thrombin on guanidinobenzoate esters. Titration of the enzymes. Biochemistry 8, 2212–2224 [DOI] [PubMed] [Google Scholar]
- 18. Jameson G. W., Roberts D. V., Adams R. W., Kyle W. S., Elmore D. T. (1973) Determination of the operational molarity of solutions of bovine α-chymotrypsin, trypsin, thrombin, and factor Xa by spectrofluorometric titration. Biochem. J. 131, 107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Friberger P., Knös M., Gustavsson S., Aurell L., Claeson G. (1978) Methods for determination of plasmin, antiplasmin, and plasminogen by means of substrate S-2251. Haemostasis 7, 138–145 [DOI] [PubMed] [Google Scholar]
- 20. Wohl R. C., Summaria L., Robbins K. C. (1980) Kinetics of activation of human plasminogen by different activator species at pH 7.4 and 37 °C. J. Biol. Chem. 255, 2005–2013 [PubMed] [Google Scholar]
- 21. Rampling M. W., Gaffney P. J. (1976) The sulfite precipitation method for fibrinogen measurement: its use on small samples in the presence of fibrinogen degradation products. Clin. Chim. Acta 67, 43–52 [DOI] [PubMed] [Google Scholar]
- 22. Reed G. L., 3rd, Matsueda G. R., Haber E. (1990) Synergistic fibrinolysis: combined effects of plasminogen activators and an antibody that inhibits α2-antiplasmin. Proc. Natl. Acad. Sci. U.S.A. 87, 1114–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang S., Reed G. L., Hedstrom L. (2000) Zymogen activation in the streptokinase-plasminogen complex. Ile1 is required for the formation of a functional active site. Eur. J. Biochem. 267, 3994–4001 [DOI] [PubMed] [Google Scholar]
- 24. Chibber B. A., Castellino F. J. (1986) Regulation of the streptokinase-mediated activation of human plasminogen by fibrinogen and chloride ions. J. Biol. Chem. 261, 5289–5295 [PubMed] [Google Scholar]
- 25. Boxrud P. D., Bock P. E. (2000) Streptokinase binds preferentially to the extended conformation of plasminogen through lysine-binding site and catalytic domain interactions. Biochemistry 39, 13974–13981 [DOI] [PubMed] [Google Scholar]
- 26. Bok R. A., Mangel W. F. (1985) Quantitative characterization of the binding of plasminogen to intact fibrin clots, lysine-Sepharose, and fibrin cleaved by plasmin. Biochemistry 24, 3279–3286 [DOI] [PubMed] [Google Scholar]
- 27. Christensen U., Mølgaard L. (1992) Positive cooperative binding at two weak lysine-binding sites governs the Glu-plasminogen conformational change. Biochem. J. 285, 419–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee P. P., Wohl R. C., Boreisha I. G., Robbins K. C. (1988) Kinetic analysis of covalent hybrid plasminogen activators: effect of CNBr-degraded fibrinogen on kinetic parameters of Glu1-plasminogen activation. Biochemistry 27, 7506–7513 [DOI] [PubMed] [Google Scholar]
- 29. Lijnen H. R., Zamarron C., Blaber M., Winkler M. E., Collen D. (1986) Activation of plasminogen by pro-urokinase. I. Mechanism. J. Biol. Chem. 261, 1253–1258 [PubMed] [Google Scholar]
- 30. Kolev K., Léránt I., Tenekejiev K., Machovich R. (1994) Regulation of fibrinolytic activity of neutrophil leukocyte elastase, plasmin, and miniplasmin by plasma protease inhibitors. J. Biol. Chem. 269, 17030–17034 [PubMed] [Google Scholar]
- 31. Schneider M., Nesheim M. (2004) A study of the protection of plasmin from antiplasmin inhibition within an intact fibrin clot during the course of clot lysis. J. Biol. Chem. 279, 13333–13339 [DOI] [PubMed] [Google Scholar]
- 32. Wiles K. G., Panizzi P., Kroh H. K., Bock P. E. (2010) Skizzle is a novel plasminogen- and plasmin-binding protein from Streptococcus agalactiae that targets proteins of human fibrinolysis to promote plasmin generation. J. Biol. Chem. 285, 21153–21164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sun H., Ringdahl U., Homeister J. W., Fay W. P., Engleberg N. C., Yang A. Y., Rozek L. S., Wang X., Sjöbring U., Ginsburg D. (2004) Plasminogen is a critical host pathogenicity factor for group A streptococcal infection. Science 305, 1283–1286 [DOI] [PubMed] [Google Scholar]
- 34. Leigh J. A., Finch J. M., Field T. R., Real N. C., Winter A., Walton A. W., Hodgkinson S. M. (1999) Vaccination with the plasminogen activator from Streptococcus uberis induces an inhibitory response and protects against experimental infection in the dairy cow. Vaccine 17, 851–857 [DOI] [PubMed] [Google Scholar]
- 35. Sifringer M., Stefovska V., Zentner I., Hansen B., Stepulak A., Knaute C., Marzahn J., Ikonomidou C. (2007) The role of matrix metalloproteinases in infant traumatic brain injury. Neurobiol. Dis. 25, 526–535 [DOI] [PubMed] [Google Scholar]
- 36. Lähteenmäki K., Edelman S., Korhonen T. K. (2005) Bacterial metastasis: the host plasminogen system in bacterial invasion. Trends Microbiol. 13, 79–85 [DOI] [PubMed] [Google Scholar]
- 37. Boyle M. D., Lottenberg R. (1997) Plasminogen activation by invasive human pathogens. Thromb. Haemost. 77, 1–10 [PubMed] [Google Scholar]