Background: The role of IKKβ in the production of type 1 interferons by plasmacytoid dendritic cells (pDCs) is unknown.
Results: Inhibition of IKKβ and its activator TAK1 prevents the production of IFNβ in pDCs, and hence the production of IFNα.
Conclusion: Toll-like receptor 7/9-stimulated production of interferons in pDCs requires the canonical IKKs and TAK1.
Significance: IKKβ inhibitors may have potential for the treatment of autoimmunity.
Keywords: Immunology, Innate Immunity, Interferon, Protein Phosphorylation, Toll-like Receptors (TLR), IKKβ, TAK1, Kinase Inhibitor, Plasmacytoid Dendritic Cells
Abstract
Plasmacytoid dendritic cells (pDCs) are characterized by their ability to produce high levels of type 1 interferons in response to ligands that activate TLR7 and TLR9, but the signaling pathways required for IFN production are incompletely understood. Here we exploit the human pDC cell line Gen2.2 and improved pharmacological inhibitors of protein kinases to address this issue. We demonstrate that ligands that activate TLR7 and TLR9 require the TAK1-IKKβ signaling pathway to induce the production of IFNβ via a pathway that is independent of the degradation of IκBα. We also show that IKKβ activity, as well as the subsequent IFNβ-stimulated activation of the JAK-STAT1/2 signaling pathway, are essential for the production of IFNα by TLR9 ligands. We further show that TLR7 ligands CL097 and R848 fail to produce significant amounts of IFNα because the activation of IKKβ is not sustained for a sufficient length of time. The TLR7/9-stimulated production of type 1 IFNs is inhibited by much lower concentrations of IKKβ inhibitors than those needed to suppress the production of NFκB-dependent proinflammatory cytokines, such as IL-6, suggesting that drugs that inhibit IKKβ may have a potential for the treatment of forms of lupus that are driven by self-RNA and self-DNA-induced activation of TLR7 and TLR9, respectively.
Introduction
Plasmacytoid dendritic cells (pDCs)2 are a subset of dendritic cells characterized by their capacity to produce high levels of type 1 IFNs in response to nucleic acids that are recognized by Toll-like receptors 7 and 9 (TLR7 and TLR9) (1). Single-stranded RNA of viral origin is detected by TLR7, whereas DNA of viral origin, and perhaps unmethylated CpG-containing sequences of bacterial origin, are detected by TLR9 (2). The type 1 IFNs produced by pDCs play a prominent role in triggering an antiviral state and in inducing innate and adaptive immune responses against pathogens (reviewed Ref. 3). However, pDCs are also thought to play an important role in autoimmunity, due to the fact that TLR7 and TLR9 have the potential to trigger immune responses if they recognize immune complexes consisting of autoantibodies bound to self-RNA and self-DNA (4). This is thought to be of particular importance in the pathogenesis of systemic lupus erythematosus, which presents features characteristic of deregulated type 1 interferon production (5). Similar observations have also been made for pDCs responding to autoantigens in inflammatory skin diseases (6, 7). These findings suggest that inhibitors of TLR7 and TLR9, and/or the signaling pathways that they activate, may have therapeutic potential for the treatment of autoimmune diseases.
A key molecular feature of pDCs is the basal expression of high levels of transcription factor interferon regulatory factor 7 (IRF7), which stimulates transcription of the genes encoding both IFNα and IFNβ and is believed to underlie the ability of pDCs to secrete large amounts of type 1 IFNs compared with other cells (8, 9). The translocation of IRF7 from the cytoplasm to the nucleus by ligands that activate TLR7 and TLR9 drives IFNα production, because pDCs from IRF7−/− mice are unable to produce IFNα (10). IRF3 is crucial for transcription of the gene encoding IFNβ, in response to ligands that activate TLR3 (double-stranded RNA) and TLR4 (lipopolysaccharide) and by cytoplasmic RNA sensors in response to viral infection (11), whereas IRF1 has been suggested to control IFNβ production in conventional dendritic cells (DCs) (12). However, these transcription factors are not required for the production of type 1 IFNs by ligands that activate TLR7 and TLR9, because the secretion of IFNβ is unimpaired in pDCs from IRF3−/− or IRF1−/− mice (10).
TLR7 and TLR9 are associated with the endosomal membranes of pDCs. When activated by their respective ligands, they recruit the adaptor MyD88, tumor necrosis factor receptor-associated factor 6, interleukin 1 receptor-associated kinase (IRAK1) and IRF7, as suggested by experiments in cells expressing tagged versions of these proteins. Moreover, IFNα production is impaired in pDCs from MyD88−/− or IRAK1Y/− mice (13–17). In other innate immune cells, “downstream” signaling from MyD88 is thought to be initiated when interleukin 1 receptor-associated kinase interacts with tumor necrosis factor receptor-associated factor 6, leading to the activation of this E3 ubiquitin ligase and the formation of Lys63-linked polyubiquitin chains. These polyubiquitin chains then bind to regulatory components of the TAK1 complex, inducing conformational changes that lead to the autoactivation of this protein kinase. Polyubiquitin chains also bind to the NEMO component of the canonical IKK complex (containing IKKα and IKKβ) leading to its activation by TAK1. The substrates of the canonical IKKs include IκBα, an inhibitor of transcription factor NFκB (18). The phosphorylated IκBα then interacts with SCFβTRCP allowing this E3 ubiquitin ligase to catalyze the Lys48-linked polyubiquitylation of IκBα, followed by its proteasomal degradation. The RelA and c-Rel components of NFκB can then translocate to the nucleus and stimulate the transcription of genes encoding inflammatory mediators.
Conventional (12) and plasmacytoid (19) dendritic cells from IKKα−/− mice are greatly impaired in their ability to produce type 1 IFNs, and it has been suggested that IKKα might be the protein kinase that phosphorylates IRF7 directly in response to ligands that activate TLR7 and TLR9 (19). In contrast, the role of IKKβ in the TLR7/9-induced production of type 1 IFNs by pDCs has not yet been addressed.
There are two other members of the IKK subfamily, TBK1 and IKKϵ, which are critical for the activation of IRF3 and hence for the production of type 1 IFNs by ligands that activate TLR3 and TLR4 and by cytoplasmic RNA sensors in response to viral infection (11). TBK1/IKKϵ have also been reported to phosphorylate IRF7 (20), but seems to be dispensable for TLR7/9-induced IFN production by pDCs derived from fetal liver cells of TBK1−/−IKKϵ−/− embryos (21).
Studies aimed at understanding how ligands that activate TLR7 and TLR9 stimulate the production of type 1 IFNs in pDCs have been hampered by the limited availability of these cells. A stable cell line providing enough material to analyze the TLR7/9 signaling pathways using chemical and genetic approaches would therefore be extremely valuable. Gen2.2 is a stable pDC line generated from the tumor blood cells of a patient (22). Gen2.2 cells express pDC markers, secrete IFNα in response to TLR7/9 stimulation, express maturation markers, and are able to present antigens in a similar manner to primary pDCs (22–24). These cells have recently been used to confirm aspects of pDC biology in relationship with DNA sensing (25) or HIV infection (26). Here we use these human cells, in conjunction with primary mouse pDCs, to show that IKKβ activity is required for type 1 IFN production in response to ligands that activate TLR7 and TLR9 by a mechanism that is largely independent of the IKKβ-mediated degradation of IκBα. We also demonstrate that IKKβ activity, together with the IFNβ-stimulated activation of the JAK-STAT signaling pathway is required for subsequent production of IFNα.
EXPERIMENTAL PROCEDURES
Compounds
BI605906 (27) and MLN4924 (28) were synthesized as described. The chemical synthesis of NG25 (supplemental Fig. S4C) will be described elsewhere. 5Z-7-Oxozeaenol was purchased from BioAustralis Fine Chemicals, INCB18424 from ChemieTek, and MG132 from Calbiochem. CpG type B for stimulation of Gen2.2 and mouse cells (ODN1826) and CpG type A for experiments with Gen2.2 cells (ODN2216) and murine cells (ODN1585), together with the TLR7 agonists CL097 and R848, were purchased from Invivogen. Human IFNβ was obtained from PBL Interferon Source.
DNA Constructs
Restriction digests and ligations were performed using standard protocols. All PCR were carried out using KOD Hot Start DNA polymerase (Novagen). The coding region for IRF7 (NCBI accession number NP_001563) was amplified from IMAGE consortium EST 5201404, cloned into vector pSC-b (Stratagene) and sequenced. The insert was subcloned into mammalian expression vector pCMV5HA-1 to create a DNA vector for expression of IRF7 with an N-terminal HA tag. DNA encoding IKKα (NCBI accession number O15111) was amplified from IMAGE consortium EST 5275799, and IKKβ (GenBankTM accession number AF080158) was cloned in a similar way by amplifying from IMAGE EST 5784717. Catalytically inactive mutants of IKKα and IKKβ were obtained by mutating Lys44 to Ala using the QuikChange mutagenesis method (Stratagene), but using KOD Hot Start DNA polymerase. The IFNβ promoter cloned into the pLuc-MCS vector was kindly provided by Katherine Fitzgerald (University of Massachusetts). pTK-RL was from Stratagene.
Mice
129SvJ × C57BL/6 mice on a mixed background were bred at Dundee under pathogen-free conditions according to European Union regulations. The work was approved by local ethical review and performed with a UK Home Office project license.
Cell Culture
HEK293 and 293T cells were maintained in DMEM with 10% FBS. Gen2.2 cells (generously provided by Drs. Joel Plumas and Laurence Chaperot, Research and Development Laboratory, French Blood Bank Rhône-Alpes, Grenoble, France) require murine stromal cells (MS5) as feeder cells to propagate (22). MS5 were obtained from DSMZ (Braunschweig, Germany) and cultured with Gen2.2 cells in RPMI supplemented with 10% FBS, sodium pyruvate (Lonza), nonessential amino acids (Lonza), penicillin (100 units/ml), and streptomycin (100 μg/ml). 24 h before passage, 1 × 106 MS5 cells were split into 25-cm flasks and then irradiated (60 gray). 24 h prior to stimulation, the Gen2.2 cells were harvested and incubated overnight without feeder cells.
Bone marrow cells extracted from the femur and tibia of mice were differentiated into Flt3 bone marrow-derived DCs by incubation for 7 days in RPMI supplemented with 100 ng/ml of recombinant Flt3-ligand (Peprotech), 10% FBS, 50 μm 2-mercaptoethanol, 10 mm HEPES, penicillin (100 units/ml), and streptomycin (100 μg/ml) (29).
RNA Interference
5 × 106 cells were transfected (25) with 0.4 nmol of siRNA in Amaxa Nucleofector (Lonza) with the Amaxa Cell Line Nucleofector V-kit (Lonza), using program A33. siRNAs were siGenome pools (Dharmacon), and resuspended according to the manufacturer's instructions. After transfection, the cells were left for 48 h in the absence of feeder cells, counted, and stimulated for the times indicated in the figure legends.
Measurement of Cytokines
3.5 × 105 Gen2.2 cells or Flt3-DCs were incubated for 1 h in 96-well plates without or with the indicated concentrations of inhibitor, then stimulated with 1 μm CpG (type A or B) or 1 μg/ml of CL097 or R848. After 5 or 12 h the cell culture supernatants were collected, clarified by centrifugation, and frozen at −80 °C until cytokine levels were analyzed. For cell viability assays, unstimulated cells were incubated for 12 h in the absence or presence of inhibitors. Cells were then fixed and the percentage of live cells analyzed by flow cytometry.
Immunoblotting
Gen2.2 cells were incubated for 1 h with or without inhibitors, and then treated with the indicated amounts of the ligand. At the times indicated, the cells were rinsed in ice-cold PBS and extracts were prepared and subjected to immunoblotting (27).
The antibodies used for immunoblotting were horseradish peroxidase-conjugated secondary antibodies (Pierce), anti-IKKβ (Millipore), anti-HA (Roche Applied Science), anti-tubulin (Sigma), anti-Stat1, anti-IκBα, and anti-IKKα (Cell Signaling Technology). Phosphospecific antibodies recognizing phospho-Tyr701 of Stat1, phospho-Ser933 of p105, phospho-Ser536 of p65/RelA, and another recognizing phospho-Ser177/Ser181 of IKKβ and phospho-Ser176/Ser180 of IKKα (catalog number 2697) were also from Cell Signaling Technology.
Quantitative RT-PCR
Total RNA extracted from cells using the RNeasy Micro kit (Ambion, Austin, TX) was reverse transcribed using random and oligo(dT) primers, qScript reverse transcriptase, and the accompanying reagents (Quanta Biosciences) according to the manufacturer's instructions. PCR were performed using the PerfeCT SYBR Green Fast mix (Quanta Biosciences) in the Bio-Rad iCycler or CFX96 (Bio-Rad). Human IFNB, IFNA1, and IFNA2 primers have been described (23). Primer sequences for murine transcripts were: Ifnb forward, 5′-GGAAAAGCAAGAGGAAAGATTGAC-3′; Ifnb reverse, 5′-CCACCATCCAGGCGTAGC-3′; Ifna4 forward, ACCCACAGCCCAGAGAGTGACC, Ifna4 reverse, AGGCCCTCTTGTTCCCGAGGT; Ifna6 forward, CAGGAGGTGGGGGTGCAGGA; Ifna6 reverse, TCACTCGTCCTCACTCAGTCT. Normalization and quantitation were performed using 18 S RNA and the ΔΔCt method
ELISA
The concentrations of IFNα, IFNβ, and IL-6 in the cell culture supernatant were measured by ELISA using the Verikine Human IFNα or IFNβ kit (PBL Interferon Source) and the Development IL-6 ELISA kit (Peprotech).
Luciferase Assays
1.8 × 105 HEK 293 cells were seeded on 24-well plates and transfected with 50 ng of the reporter plasmid encoding the firefly luciferase gene under control of the IFNβ promoter together with various expression plasmids (20 ng) using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Transfected DNA was maintained at 400 ng by adjusting the DNA concentration with empty vector. 10 ng of Renilla luciferase encoding plasmid (pTK-RL) was co-transfected as an internal control plasmid. 48 h later, cells were extracted in Passive lysis buffer (Promega). Luciferase activity was measured with a dual-luciferase assay system (Promega) according to the manufacturer's instructions.
Phosphatase Treatment
Lysates of 293T cells transfected with HA-IRF7 (0.5 mg) were subjected to overnight immunoprecipitation with anti-HA affinity matrix (Roche Applied Science). After extensive washing, beads were incubated for 30 min at 30 °C with 80 units of λ-phosphatase (New England Biolabs) in the presence or absence of phosphatase inhibitors (Calbiochem). Samples were subjected to SDS-PAGE on 6% acrylamide gels and immunoblotted with an HA antibody.
Proteins and Kinase Assays
IRF7 was expressed in Escherichia coli as a glutathione S-transferase (GST) fusion protein with a PreScission proteinase cleavage site between the GST and the IRF7. The GST-IRF7 was captured on glutathione-Sepharose and IRF7 released from GST and glutathione-Sepharose by digestion with PreScission proteinase. His6-tagged IKKβ and TBK1 were expressed in their active phosphorylated forms in insect Sf21 cells and purified by affinity chromatography on nickel nitrilotriacetate-agarose. Active GST-IKKα was purchased from Millipore and assayed as described previously (30).
RESULTS
A Specific Inhibitor of IKKβ Prevents Secretion of Type 1 Interferons in Human Plasmacytoid Dendritic Cell Line Gen2.2
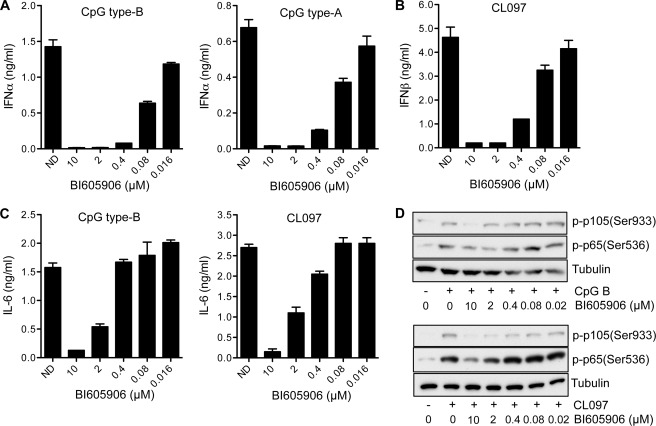
BI605906 is a potent and specific inhibitor of IKKβ (supplemental Fig. S1A) (27), which does not affect the closely related IKKα in vitro (supplemental Fig. S1B). Moreover, BI605906 potently inhibits the phosphorylation of p65/RelA and p105 in IKKα−/− mouse embryonic fibroblasts (MEF), but not in IKKβ−/− MEFs where their phosphorylation is catalyzed by IKKα rather than IKKβ (supplemental Fig. S1C). We therefore used this compound to investigate the potential importance of IKKβ in the signaling pathway by which TLR9 and TLR7 ligands stimulate the secretion of type 1 IFNs. TLR9 is activated by unmethylated CpG DNA sequences, which can be subdivided into two types. CpG type A is a potent stimulator of interferon production but a weak activator of proinflammatory cytokine production, whereas CpG type B stimulates both interferon and proinflammatory cytokine production. TLR7 is activated by single-stranded RNA and, in the present study, we used the synthetic agonist CL097, a water-soluble derivative of the imidazoquinoline compound R848, which is used as a vaccine adjuvant. The CpG A- and CpG B-stimulated secretion of IFNα (Fig. 1A) and IFNβ (results not shown) were potently inhibited by BI605906 with an IC50 of 0.1 μm, with complete inhibition occurring at 2 μm (Fig. 1A). Interestingly, CL097 and the structurally related TLR7 ligand R848 did not induce the synthesis of IFNα, but stimulated the secretion of IFNβ, which was also blocked by BI605906 (Fig. 1B). BI605906 also reduced the secretion of IL-6 by CpG B and CL097, although the IC50 was about 10-fold higher (Fig. 1C). The suppression of phosphorylation of IKKβ substrates p65/RelA and p105/NFκB1 correlated with the suppression of IL-6 production (Fig. 1D), indicating that less inhibition of IKKβ is needed to suppress IFN secretion than to suppress IL-6 secretion. CpG A did not stimulate the secretion of significant amounts of IL-6. Importantly, similar results were obtained with a structurally unrelated IKKβ inhibitor PS1145 (supplemental Fig. S2). Neither BI605906 (supplemental Fig. S3A) nor PS1145 (supplemental Fig. S3B) affected viability of the cells.
FIGURE 1.
A specific inhibitor of IKKβ blocks type 1 interferon production in Gen2.2 cells in response to ligands that activate TLR7 and TLR9. A, Gen2.2 cells were incubated for 1 h in the absence (ND, no drug) or presence of the indicated concentrations of BI605906 and then stimulated for 12 h with 1 μm CpG type B (left-hand panel) or CpG type A (right-hand panel). The levels of IFNα in the culture medium were quantified by ELISA. The experiment was performed in 96-well plates with two wells being used for each concentration of inhibitor studied and the results being averaged. Similar results were obtained in three independent experiments. Error bars show the variation in duplicate determinations. B, the experiment was carried out as in A, except that the cells were stimulated for 5 h with 1 μg/ml of the TLR7 agonist CL097 and IFNβ in the culture medium was measured by ELISA. Similar results were obtained in three independent experiments. Error bars show the variation in duplicate determinations. C, the experiment was carried out as in A (left-hand panel) or B (right-hand panel) except that the concentration of IL-6 in the culture medium was determined by ELISA. Similar results were obtained in three independent experiments. Error bars show the variation in duplicate determinations. D, Gen2.2 cells were stimulated for 5 h with 1 μm CpG B (top three panels) or for 30 min with 1 μg/ml of the TLR7 agonist CL097 (bottom panel) in the absence or presence of the indicated concentrations of BI605906. Following cell lysis, aliquots of cell extract (30 μg of protein (top three panels) or 12 μg of protein (bottom three panels) were subjected to SDS-PAGE followed by immunoblotting with the antibodies indicated. The immunoblot shown is representative of two independent experiments.
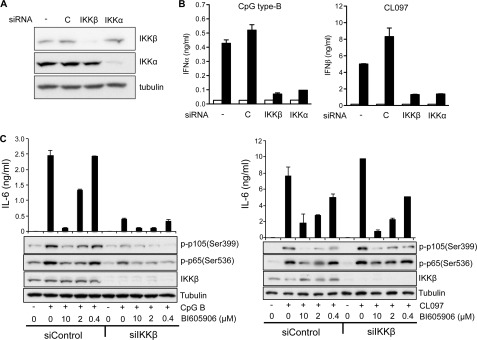
To check that BI605906 was exerting its effect by inhibiting IKKβ, and not via an “off-target” effect, we also reduced the expression of IKKβ using RNA interference. The knockdown of IKKβ (Fig. 2A) suppressed the CpG B-stimulated secretion of IFNα and the CL097-stimulated secretion of IFNβ (Fig. 2B). siRNA knockdown of IKKβ also suppressed CpG B-stimulated IL-6 secretion (Fig. 2C, left-hand panels) but did not reduce CL097-stimulated IL-6 secretion (Fig. 2C, right-hand panels). This is because despite the reduction in IKKβ expression, CL097-stimulated activation of the residual IKKβ was just as robust as in control cells expressing normal levels of IKKβ (Fig. 2C, right-hand panels). The CL097-stimulated IL-6 secretion could, however, be reduced by including BI605906 in the culture medium (Fig. 2C).
FIGURE 2.
RNA interference of IKKα or IKKβ blocks type 1 interferon production in Gen2.2 cells in response to ligands that activate TLR7 and TLR9. A, Gen2.2 cells were transfected in the absence of siRNA (−) or in the presence of 0.4 nmol of a non-targeting pool of siRNAs (C, control), siRNA blocking IKKβ (IKKβ), or siRNA blocking IKKα (IKKα). Cell extracts were prepared 48 h later and, after SDS-PAGE and transfer to PVDF, immunoblotting was carried out with the indicated antibodies. One experiment, which is representative of three that were carried out, is shown. B, the experiment was carried out as in A, except that after 48 h, the transfected cells were stimulated for 12 h with 1 μm CpG type B (left-hand panel) or for 5 h with 1 μg/ml of CL097 (right-hand panel). The concentrations of IFNα (left-hand panel) and IFNβ (right-hand panel) in the cell culture medium were then measured by ELISA. The experiment was performed in 96-well plates with two wells being used for each concentration of inhibitor studied and the results being averaged. Similar results were obtained in 3 independent experiments. Error bars show the variation in duplicate determinations. C, the experiment was carried out as in A, and cells transfected with a non-targeting pool of siRNAs (siControl) or siRNA blocking IKKβ (siIKKβ) were stimulated with 1 μm CpG type B (left panel) or 1 μg/ml of CL097 (right panel). Cells were lysed 5 h after CpG stimulation (left panel) or 30 min after CL097 stimulation and aliquots of the cell extracts were subjected to SDS-PAGE followed by immunoblotting with the antibodies indicated as described in the legend to Fig. 1. Supernatants were collected 12 h after stimulation with CpG B (left-hand panel) or 5 h after stimulation with CL097 (right-hand panel). IL-6 concentrations in the cell culture supernatant were measured by ELISA. A representative experiment of several performed is shown.
RNA interference of IKKα had a similar effect on IFN secretion to the knockdown of IKKβ (Fig. 2B). This is consistent with an earlier report showing that secretion of IFNα induced by TLR9 and TLR7 ligands is greatly reduced in pDCs from IKKα−/− mice (19).
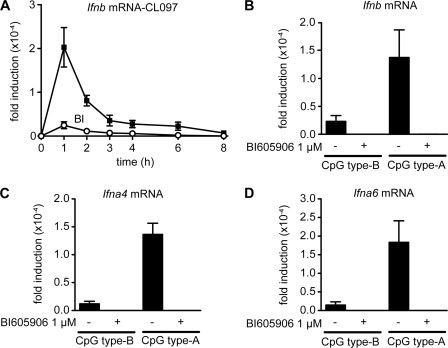
We also tested the effectiveness of BI605906 in primary mouse Flt3-derived DCs, which have been used as a model for studying type 1 IFN production by pDCs. These experiments showed that, as for human transformed Gen2.2 cells, the production of Ifnb mRNA by CL097 (Fig. 3A) or CpG (Fig. 3B), or the production of Ifna4 (Fig. 3C) and Ifna6 mRNA (Fig. 3D) by CpG was abolished by BI605906.
FIGURE 3.
Pharmacological inhibition of IKKβ blocks type 1 interferon secretion in primary murine Flt3-derived dendritic cells. Murine bone marrow cells were incubated for 7 days with Flt3-ligand to obtain Flt3-derived dendritic cells. Cells were pretreated with 1 μm BI605906 and stimulated for the times indicated with 1 μg/ml of the TLR7 ligand CL097 (A) or for 6 h with 0.05 μm CpG type B or 1 μm CpG type A (B–D). RNA was extracted from the cells and mRNA encoding Ifnb (A and B), Ifna4 (C), and Ifna6 (D) was determined by quantitative PCR. The results are plotted as the fold-increase in mRNA relative to the level determined in unstimulated cells. The experiments were performed in 96-well plates with three wells being used for each condition, each containing Flt3-derived dendritic cells from a different mouse. The results are presented as the mean ± S.E. for one representative experiment. Similar results were obtained in two independent experiments.
IKKβ Activity Is Required for Production of IFNβ mRNA
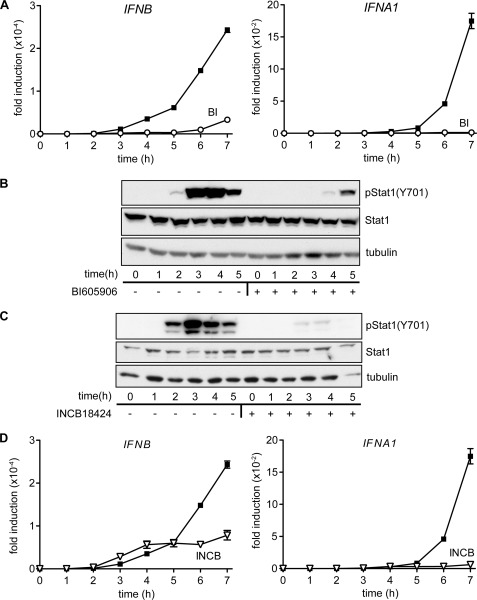
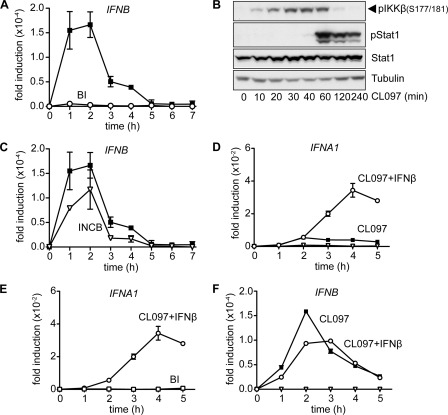
In MEFs, the production of Ifnb mRNA and IFNβ secretion induced by viral infection has been reported to precede the production of IFNα (8, 31). In the present study, we found that the production of IFNβ initiated after stimulation with ligands that activate TLR9 also occurred several hours before transcription of the IFNα gene (Fig. 4A). The inclusion of BI605906 in the cell culture medium at 1 μm greatly reduced the production of IFNB mRNA (Fig. 4A), and abolished subsequent transcription of the genes encoding IFNA1 (Fig. 4A) and IFNA2 (results not shown). IFNβ signals through the type 1 IFN receptor leading to activation of the JAK-STAT1/2 pathway. This explains why BI605906 suppresses the CpG B-stimulated phosphorylation of STAT1 at Tyr701 in GEN2.2 cells (Fig. 4B), reflecting suppression of the early secretion of IFNβ.
FIGURE 4.
Inhibition of IKKβ suppresses the CpG B-stimulated production of IFNβ, whereas inhibition of IKKβ or JAK kinases prevents the subsequent production of IFNα. A, Gen2.2 cells were stimulated for the times indicated with 1 μm CpG type B in the absence (black squares) or presence (open circles) of 1 μm BI605906. RNA was then extracted from the cells and IFNB mRNA (left-hand panel) or IFNA1 mRNA (right-hand panel) were measured by quantitative PCR. The results are plotted as the fold-increase in mRNA relative to the level determined in unstimulated cells. The results are shown for one representative experiment of three. B and C, Gen2.2 cells were stimulated for the times indicated with 1 μm CpG type B in the absence (−) or presence (+) of 10 μm BI605906 (B) or 1 μm INCB18424 (C) and cell lysates were subjected to SDS-PAGE followed by immunoblotting with the indicated antibodies. The blot shown is representative of two independent experiments. D, as in A, except that the cells were stimulated with 1 μm CpG type B in the absence (black squares) or presence (open triangles) of 1 μm INCB18424. The results are shown for one representative experiment of two.
To investigate further the importance of the JAK-STAT1/2 pathway in driving late-stage IFNα production, we examined the effects of the potent and specific JAK (Janus) inhibitor INCB18424 (also called Ruxolitinib) (supplemental Fig. S4A), which was recently approved for the treatment of the bone marrow disease myelofibrosis. This compound completely blocked the CpG B-stimulated phosphorylation of STAT1 (Fig. 4C). Incubation with INCB18424 did not inhibit the production of IFNB mRNA up to 5 h but blocked the further increase in IFNB mRNA production after this time (Fig. 4D), presumably by suppressing a JAK-STAT1/2-dependent positive autocrine loop required to sustain production of IFNB. INCB18424 also completely blocked the production of IFNA1 mRNA (Fig. 4D), establishing that it is dependent on the catalytic activity of the JAKs. Similar results were obtained when CpG A was used instead of CpG B, although, the production of IFNB was lower and more delayed with this ligand (results not shown). Taken together, these experiments showed that inhibition of IKKβ blocks the formation of IFNB mRNA and hence IFNβ secretion resulting in a failure of IFNβ to activate the JAK-STAT1/2 pathway and stimulate the transcription of IFNα genes.
IKKβ and IFNβ Are Both Required for Production of IFNα mRNA
When the Gen2.2 cells were stimulated with the TLR7 ligand CL097, there was a much more rapid induction of IFNB mRNA, which then declined to a very low level after 5 h (Fig. 5A). This induction was also inhibited by BI605906 (Fig. 5A). Consistent with the rapid kinetics of IFNB mRNA formation, there was also a rapid activation of IKKβ, and the phosphorylation of STAT1 also reached a maximum after 1 h (Fig. 5B). In contrast, the JAK inhibitor INCB 18424 had little effect on the CL097-stimulated formation of IFNB (Fig. 5C).
FIGURE 5.
IKKβ and IFNβ are both required for the production of IFNα mRNA by the TLR7 ligand CL097. A, Gen2.2 cells were stimulated with 1 μg/ml of the TLR7 agonist CL097 in the absence (black squares) or presence (open circles) of 1 μm BI605906 (BI) for the times indicated. RNA was then extracted from the cells and IFNB mRNA was measured by quantitative PCR. The results are plotted as the fold-increase in mRNA relative to the level determined in unstimulated cells. One representative experiment of four is shown. B, Gen2.2 cells were stimulated with 1 μg/ml of CL097 in the absence of inhibitors. The cell extracts were then subjected to SDS-PAGE followed by immunoblotting with the indicated antibodies. The phosphospecific antibody that recognizes IKKβ phosphorylated at Ser177 and Ser181 also recognizes IKKα phosphorylated at Ser176 and Ser180. However, the major band detected by the antibody corresponds to phosphorylated IKKβ only. Phosphorylated IKKα migrates more rapidly than IKKβ and is recognized very poorly by the antibody (supplemental Fig. S1A in Ref. 27, see also Fig. 6C), which may be explained by lower levels of expression and/or weaker activation of IKKα compared with IKKβ. One blot representative of two independent experiments is shown. C, similar to A, cells were stimulated with 1 μg/ml of CL097 after incubation without (black squares) or with INCB18424 (open triangles). Similar results were obtained in two other independent experiments. D, Gen2.2 cells were stimulated with 1 μg/ml of CL097 (black squares), 500 units/ml of human recombinant IFNβ (open triangles), or CL097 in combination with IFNβ (open circles) for the indicated times. IFNA1 mRNA was measured by quantitative PCR. Similar results were obtained in three other independent experiments. E, Gen2.2 cells were stimulated with a combination of 1 μg/ml of CL097 and 500 units/ml of human IFNβ in the absence (open circles) or presence (open squares) of 1 μm BI605906. The results are plotted as the fold-increase in IFNA1 mRNA relative to the level determined in unstimulated cells. Similar results were obtained in a second independent experiment. F, as in D but IFNB mRNA production was measured. Similar results were obtained in three other independent experiments.
The activation of IKKβ by CL097 was transient, reaching a maximum between 30 and 60 min and declining thereafter (Fig. 5B). The production of IFNB mRNA was also transient, peaking after 2 h and almost returning to basal levels after 5 h (Fig. 5A). The transient activation of IKKβ and IFNB mRNA was quite different from the sustained activation seen after stimulation with CpG Type B (Fig. 4A) and suggested that the failure of CL097 to induce the production of significant amounts of IFNα might be explained by its failure to activate IKKβ for a sufficient length of time. If this were true, we reasoned that if IFNβ was added to the cell culture medium together with CL097, then the production of IFNA mRNA might be induced before the activity of IKKβ had declined to basal levels. This hypothesis turned out to be correct. We found that there was a robust production of IFNA1 mRNA when Gen2.2 cells were incubated with CL097 plus IFNβ, but little induction with CL097 alone and no induction with IFNβ alone (Fig. 5D). The formation of IFNA1 mRNA induced by CL097 plus IFNβ was prevented by BI605906 (Fig. 5E), demonstrating that IKKβ is required for the production of IFNα as well as IFNβ. In contrast, the amount of IFNB mRNA formed when Gen2.2 cells were incubated with CL097 plus IFNβ was similar to those produced in response to CL097 alone (Fig. 5F).
Inhibition of TAK1 Suppresses Type 1 Interferon Production
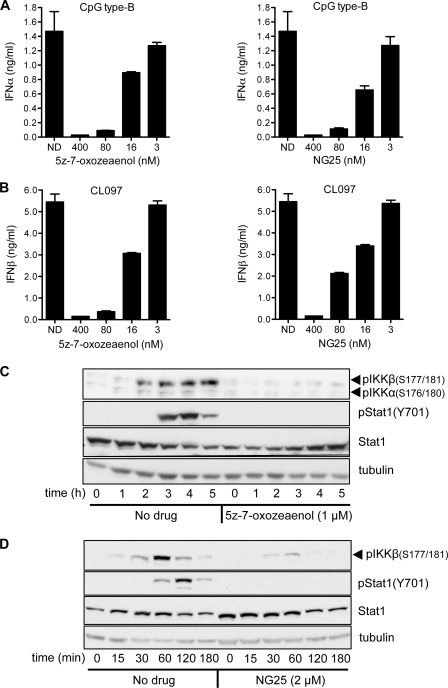
In other immune cells, the activation of IKKα and IKKβ by TLR ligands requires the protein kinase TAK1 (see Introduction). We therefore tested the effects of two structurally unrelated inhibitors of TAK1, 5Z-7-oxozeaenol (32) and NG25 on the secretion of type 1 interferons. NG25 has not been described or used previously and its specificity and structure are shown in supplemental Fig. S4, B and C. These experiments showed that both compounds were very potent suppressors of CpG B- (Fig. 6A) or CpG A-stimulated (supplemental Fig. S5A) secretion of IFNα and CL097-stimulated secretion of IFNβ (Fig. 6B), with complete inhibition by 400 nm. As expected, 5Z-7-oxozeanol (Fig. 6C) and NG25 (Fig. 6D) prevented the activation of IKKα/IKKβ and, due to the suppression of IFNβ secretion, the subsequent activation of STAT1 was also blocked (Fig. 6, C and D). The TAK1 inhibitors did not affect the viability of the cells at 400 nm (supplemental Fig. S5B).
FIGURE 6.
TAK1 inhibitors block type 1 interferon production in Gen2.2 cells. A and B, Gen2.2 cells were incubated for 1 h without (ND, no drug) or with the indicated concentrations of the TAK1 inhibitors 5Z-7-oxozeaenol (left-hand panels) or NG25 (right-hand panels) and then stimulated for 12 h with 1 μm CpG type B (A) or for 5 h with 1 μg/ml of CL097. B, the concentration of IFNα (A) or IFNβ (B) in the cell culture medium was then measured by ELISA. The experiment was performed in 96-well plates with two wells being used for each concentration of inhibitor studied and the results being averaged. Similar results were obtained in two other independent experiments. Error bars show the variation in duplicate determinations. C, Gen2.2 cells were stimulated for the times indicated with 1 μm CpG type B in the absence (no drug) or presence of 1 μm 5Z-7-oxozeaenol and the cell lysates were probed with the indicated antibodies. Similar results were obtained in a second independent experiment. D, as in C except that the cells were stimulated with 1 μg/ml of CL097 in the absence (no drug) or presence of 2 μm NG-25. Similar results were obtained in a second independent experiment.
IKKβ and IKKα Stimulate Type 1 Interferon Production by a Mechanism Independent of Degradation of IκBα
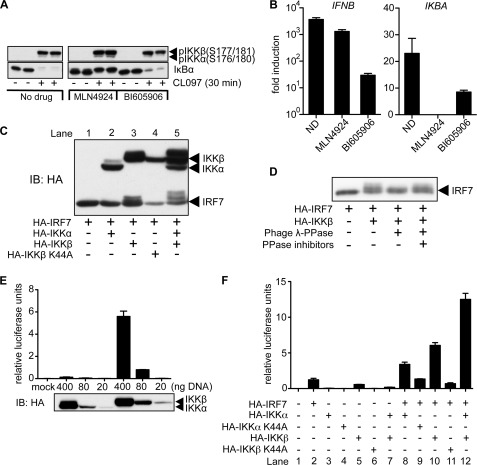
One of the major roles of the canonical IKKs (IKKα and IKKβ) is to phosphorylate IκBα, which leads to activation of the transcription factor NFκB. We therefore investigated whether inhibiting the degradation of IκBα would affect IFNβ gene transcription. The IKKs phosphorylate IκBα at Ser32 and Ser36, priming it for recognition by SCFβTRCP. This E3 ubiquitin ligase then catalyzes the Lys48-linked polyubiquitylation of IκBα, leading to its proteasomal destruction. We therefore tested the effect of MLN4924, a specific inhibitor of the E1 activating enzyme for NEDDylation. MLN4924 prevents the NEDDylation and hence the activation of SCFβTRCP (and other Cullin-based RING E3 ligases) (28) and blocks the polyubiquitylation of phosphorylated IκBα. We found that MLN4924 prevented the destruction of IκBα (Fig. 7A) and totally suppressed transcription of the NFκB-dependent gene IKBA (Fig. 7B), as expected, but the production of IFNB mRNA was still increased over 1000-fold. Conversely, the IKKβ inhibitor BI605906 suppressed IFNB mRNA production by 99% (Fig. 7B), but the degradation of IκBα observed after 30 min was only inhibited slightly (Fig. 7A). Similar results were obtained with the proteasomal inhibitor MG132, which also prevents the degradation of polyubiquitylated IκBα (results not shown). The failure of BI605906 to prevent degradation of IκBα is presumably explained by IKKα catalyzing the phosphorylation of IκBα if IKKβ is inhibited. Taken together, the results indicate that IKKβ stimulates IFNβ production by a pathway that is independent of the degradation of IκBα.
FIGURE 7.
The canonical IKKs act synergistically with IRF7 to stimulate IFNB transcription by an NFκB-independent mechanism. A, Gen2.2 cells were incubated for 1 h in the absence (no drug) or presence of 3 μm MLN4924 or 10 μm BI605906, then stimulated for 30 min with 1 μg/ml of CL097. Cell extracts were prepared and subjected to SDS-PAGE, followed by immunoblotting (IB) with the indicated antibodies. Similar results were obtained in two other independent experiments. B, same as A, except that the cells were stimulated for 1 h with 1 μg/ml of CL097 and the levels of IFNB and IKBA mRNA were measured by quantitative PCR. The results are plotted as the fold-increase in mRNA relative to the level determined in unstimulated cells. The experiment was performed in 24-well plates with two wells being used for each concentration of inhibitor studied and the results being averaged. Similar results were obtained in two other independent experiments. C, HEK293 cells were transfected with different combinations of plasmids encoding HA-tagged forms of IRF7, IKKα, IKKβ, or a catalytically inactive mutant of IKKβ. 48 h after transfection, cell extracts were prepared and subjected to SDS-PAGE, followed by immunoblotting with an anti-HA antibody. Similar results were obtained in several other independent experiments. D, lysates of HEK293 cells transfected with the indicated plasmids were subjected to HA immunoprecipitation and treated with phage λgt10-phosphatase in the absence or presence of phosphatase inhibitors. Samples were subjected to SDS-PAGE, transferred to PVDF membranes, and immunoblotted with an anti-HA antibody. Similar results were obtained in a second independent experiment. E, HEK293 cells were transfected with a reporter plasmid encoding a firefly luciferase gene under control of the IFNβ promoter, together with the indicated concentrations of plasmid DNA encoding HA-tagged forms of IKKα and IKKβ. 48 h after transfection, cell lysates were obtained and used to quantify the relative amounts of luciferase or subjected to SDS-PAGE followed by immunoblotting with anti-HA. Transfections and luciferase measurements were performed in triplicate and the results are presented as mean ± S.D. Similar results were obtained in a second independent experiment. As in E, except that different combinations of the indicated plasmids were transfected into HEK293 cells together with a luciferase IFNβ reporter and relative luciferase measurements were performed 48 h later.
The CL097-stimulated phosphorylation of the activation loop of IKKα and IKKβ was unaffected by BI605906 (Fig. 7A), indicating that binding of this compound to IKKβ does not prevent its phosphorylation by TAK1 in these cells. However, interestingly, we found that MLN4924 consistently increased the CL097-stimulated phosphorylation of the IKKs (Fig. 7A), suggesting that a Cullin-RING E3 ligase may be involved in restricting the extent of activation of the IKKs.
Canonical IKKs Synergize with IRF7 to Stimulate IFNβ Transcription
IRF7 is known to play a critical role in IFN gene transcription in pDCs (see Introduction). To understand further how IKKβ may stimulate IFNB gene transcription, we transfected DNA encoding this protein kinase into HEK293 cells with and without DNA encoding IRF7 and/or IKKα. We noticed that the electrophoretic mobility of IRF7 was decreased when it was transfected with IKKβ, but not when it was transfected with IKKα or with a catalytically inactive mutant of IKKβ (Fig. 7C). However, a more pronounced decrease in the mobility of IRF7 was observed when it was co-transfected with both IKKα and IKKβ (Fig. 7C, lane 5). The decrease in the electrophoretic mobility of IRF7 could be reversed by treatment with the phosphatase from bacteriophage λgt10 (Fig. 7D), demonstrating that it was the result of phosphorylation. The transcription of a luciferase reporter under the control of the IFNB promoter was increased by transfection with IKKβ DNA in a dose-dependent manner, whereas IKKα was a very poor inducer of IFNB gene transcription compared with IKKβ (Fig. 7E).
We next co-transfected IRF7 DNA with the lowest amounts of IKKβ and/or IKKα used in Fig. 7E (20 ng). As has been described previously (31), transfection with IRF7 alone stimulated IFNB gene transcription (Fig. 7F, lane 2) but this was increased by co-transfection with IKKβ (Fig. 7F, lane 10) and, to a lesser extent, by IKKα (Fig. 7F, lane 6). An even greater enhancement of IFNB gene transcription was observed when IRF7 DNA was co-transfected with both of these IKKs (Fig. 7F, lane 12).
Identification of Major Site on IRF7 Phosphorylated by IKKβ
IRF7 was phosphorylated with purified IKKβ or TBK1 (supplemental Fig. S6A) and the bands corresponding to IRF7 were excised, digested with trypsin, and subjected to chromatography on a Vydac C18 column. The same major 32P-labeled tryptic peptide T1 was generated after phosphorylation by either IKKβ (supplemental Fig. S6B) or TBK1 (results not shown). Mass spectrometry showed that this fraction contained two peptides, one corresponding to the unphosphorylated form of the peptide comprising amino acid residues 304 to 322 of IRF7, and the other to a monophosphorylated derivative of the peptide comprising residues 180 to 209 of IRF7. The phosphopeptide obtained after phosphorylation by either IKKβ (supplemental Fig. S6C) or TBK1 (results not shown) showed a burst of 32P radioactivity after the 21st cycle of Edman degradation following solid phase sequencing (33), corresponding to Ser200 (supplemental Fig. S6C).
It has been reported that IRF7 translocates from the cytosol to the nucleus when Gen2.2 cells are stimulated with CpG B (25), raising the possibility that IKKβ may exert its effect on IRF7 indirectly by promoting entry to the nucleus. We found that some of the endogenous IRF7 in Gen2.2 cells was already present in the nucleus of unstimulated cells, but were unable to observe any increased accumulation of IRF7 in the nucleus after stimulation with either CL097 or CpG B.3 This suggests that IKKβ does not exert its effect by stimulating the translocation of IRF7 to the nucleus.
DISCUSSION
We have shown in this study that the production of type 1 IFNs in the human pDC cell line Gen2.2 (Fig. 1) or in murine Ft3-derived dendritic cells (Fig. 3) can be prevented by inhibiting IKKβ activity with two structurally distinct inhibitors, BI605906 (Figs. 1 and 3) and PS1145 (Fig. S2), or by siRNA knockdown of IKKβ (Fig. 2). This effect appears to be independent of the role of IKKβ in degrading IκBα, because CL097 stimulated IFNB mRNA levels 1000-fold even when the degradation of IκBα was completely blocked by the Neddylation inhibitor MLN4924 (Fig. 7). These findings are consistent with a report that the CpG A-stimulated production of type 1 interferons was hardly impaired in pDCs from mice that do not express NFκB subunits RelA/p65 or p50 and c-Rel (34). Nevertheless, a proteasomal or Cullin-RING E3 ligase-dependent event, which might be the degradation of IκBα, may contribute to CL097-stimulated production of IFNB mRNA because it was reduced by 70% in the presence of MLN4924 (Fig. 7) or MG132 (results not shown).
The formation of type 1 IFNs by ligands that activate TLR7 and TLR9 is suppressed in pDCs and conventional DCs of IKKα−/− mice (12, 19) and, consistent with these reports, we found that siRNA knockdown of IKKα impaired the secretion of type 1 IFNs in human Gen2.2 cells (Fig. 2). IKKβ and IKKα are both components of the canonical IKK complex raising the question of whether inhibition and depletion of IKKβ prevents the activation of IKKα or vice versa. However, this is not the case, because the CL097-stimulated phosphorylation of p105, a physiological substrate of IKKβ (35) was not suppressed by siRNA knockdown of IKKα, and was prevented by the IKKβ-specific inhibitor BI605906 in IKKα-deficient Gen2.2 cells (results not shown). Moreover, ligands that activate TLR7 or TLR9 have been reported to stimulate transcription of NFκB-dependent genes il-6 and il-12 similarly in pDCs from IKKα−/− or wild type mice (19).
The activation of IKKβ does not occur in embryonic fibroblasts from mice that do not express TAK1 (36) or express a truncated, inactive form of TAK1 instead of the wild type protein (27, 37). The activation of IKKβ is also prevented in macrophages by the TAK1 inhibitor 5Z-7-oxozeaenol in response to LPS (see supplemental Fig. S2A in Ref. 27) or the TLR1/2 ligand Pam3Csk4 (38). We found that two structurally unrelated TAK1 inhibitors, NG25 and 5Z-7-oxozeaenol, both inhibited the activation of IKKβ by TLR7 and TLR9 agonists (Fig. 5C) and prevented the secretion of type 1 IFNs induced by these ligands in Gen2.2 cells. To our knowledge, these findings provide the first evidence that TAK1 may be required for IFN production by pDCs.
We also established that the production of IFNα in Gen2.2 cells depends on the prior production of IFNβ, which upon secretion into the culture medium activates the type 1 IFN receptor and switches on the JAK-STAT1/2 pathway that is needed for IFNα production. Thus the JAK inhibitor INCB18424 (Ruxolitinib) blocked the production of IFNA1 (Fig. 4D) and IFNA2 (results not shown). In contrast, INCB18424 did not suppress the CpG type B-stimulated formation of IFNB mRNA up to 4–5 h, and only prevented the further rise in IFNB mRNA that occurred after this time (Fig. 4D). This indicates that the JAK-STAT1/2 pathway drives a positive feedback loop that sustains the production of IFNβ. Such feedback activation had previously been suggested by the observation that the formation of Ifnb and Ifna mRNA by CpG type A was impaired in pDCs from mice that do not express the type 1 IFN receptor (10).
Interestingly, the TLR7 agonists CL097 and R848 induced a much more rapid increase in IFNB mRNA than the TLR9 ligand CpG type B, yet little IFNA mRNA was produced and no secretion of IFNα could be detected in response to these TLR7 ligands. However, we found that large amounts of IFNA mRNA could be produced rapidly in Gen2.2 cells if they were co-stimulated with both CL097 and IFNβ, whereas either agonist alone was ineffective (Fig. 5D). Moreover, under these conditions, IFNA1 mRNA production was prevented by pharmacological inhibition of IKKβ (Fig. 5E). Taken together, these experiments establish that IKKβ activity and signaling by IFNβ are both critical for the production of IFNA mRNA in Gen2.2 cells and that CL097 does not produce significant amounts of IFNA mRNA because it does not maintain the activation of IKKβ for a sufficient period of time (Fig. 5B).
A transcription factor that is crucial for type 1 IFN production in pDCs is IRF7 (10). In the present study we confirmed that transcription of a luciferase reporter under the control of the IFNB promoter could be increased by transfection with DNA encoding IRF7 and that this could be enhanced by co-transfection with IKKα DNA. However, IKKβ DNA was much more effective than IKKα DNA in enhancing IRF7 transcriptional activity (Fig. 7F) and, in contrast to IKKα, could also stimulate luciferase reporter gene expression in the absence of transfected IRF7 (Fig. 7E). The overexpression of components of signaling pathways can frequently cause the specificity of signaling to break down and lead to erroneous conclusions being made. Nevertheless, these experiments show that IKKβ is potentially capable of synergizing with IRF7 to promote IFNβ gene transcription.
When high concentrations of IKKβ DNA were co-transfected with IRF7 DNA, the expressed IRF7 became phosphorylated (Fig. 7, C and D), raising the possibility that IKKβ, or another protein kinase(s) activated by IKKβ, may exert its effects on this pathway by phosphorylating IRF7. Like TBK1, bacterially expressed IKKβ phosphorylated IRF7 at Ser200 in vitro, although phosphorylation by IKKβ was far weaker than with TBK1 (supplemental Fig. S6A). TBK1 is thought to be the protein kinase that phosphorylates IRF7 downstream of the viral double-stranded RNA receptors TLR3 and RIG-I/MDA5 but not in pDCs stimulated with TLR7 or TLR9 agonists (see Introduction). Ser200 was also identified as a site that became phosphorylated when DNA encoding IRF7 and IKKβ or TBK1 were co-transfected into HEK293 cells (results not shown). However, we found that the mutation of Ser200 to alanine had no effect on the ability of IRF7 to stimulate luciferase reporter expression under control of the IFNB promoter when it was transfected alone or together with IKKβ DNA (results not shown). Therefore, the physiological relevance of the phosphorylation of IRF7 at Ser200 that can be forced by transfection with high concentrations of IKKβ DNA is unclear.
Although we cannot exclude the possibility that IKKβ activates another protein kinase that then phosphorylates IRF7 at a distinct site, it is also possible that IRF7 is not the only transcription factor important for the activation of the IFNβ promoter in pDCs. Indeed, the combinatorial interaction of multiple transcription factors is commonly required for the transcription of many genes. IKKβ may therefore stimulate IFNβ gene transcription by activating another protein(s). IRF1 and IRF5 have previously been implicated in the regulation of type 1 IFN gene transcription, although studies with IRF1−/− (10) and IRF5−/− mice (39) may have excluded an essential role for these proteins in IFN production in pDCs. Therefore further work is still needed to understand how IKKβ stimulates IFNB transcription in pDCs.
It is unlikely that IKKα and IKKβ exert their effects on IFN gene transcription in pDCs by targeting the same phosphorylation site on the same protein, otherwise IKKα would be expected to be able to compensate for the loss of IKKβ activity or vice versa, which is not the case. A further hint that IKKα and IKKβ may be exerting their effects in distinct ways comes from our observation that IRF7-stimulated IFNB gene transcription was consistently higher when IRF7 was co-transfected with both IKKα and IKKβ than with either of these protein kinases alone (Fig. 7F).
Autoimmune diseases, such as systemic lupus erythematosus, are associated with the production of high levels of type 1 IFNs, which may be caused by self-RNA and self-DNA activating TLR7 and TLR9, respectively. The present study therefore raises the possibility that IKKβ inhibitors may have therapeutic potential for the treatment of autoimmunity. Particularly noteworthy is our finding that both of the IKKβ inhibitors we examined, BI605906 (Fig. 1) and PS1145 (supplemental Fig. S2), prevented the production of type 1 IFNs at much lower concentrations than those needed to suppress IL-6 secretion. To understand why IFN production is so sensitive to inhibition by BI605906 will require the identification of the IKKβ substrate that controls this process and the phosphatases that act on this site(s), as well as an understanding of how phosphatases are regulated. However, the sensitivity of IFN production to BI605906 is of great interest because it raises the possibility of suppressing the production of type 1 IFNs in vivo at concentrations of IKKβ inhibitors that are much lower than those required to block the activation of NFκB. This might help to avoid the side effects that may be caused by the inhibition of such a crucial transcription factor.
Supplementary Material
Acknowledgments
We thank our colleague Kristopher Clark for performing the experiments in IKKα-deficient and IKKβ-deficient MEFs (supplemental Fig. S1C) and Lorna Plater for the in vitro kinase assays (supplemental Fig. S1B). David Campbell and Robert Gourlay provided assistance with mass spectrometry and solid phase sequencing, whereas the Gen2.2 cells used in this study were generously provided by Joel Plumas and Laurence Chaperot. We are grateful to Inder Verma (Salk Institute, La Jolla, CA) for the immortalized MEFs from IKKα- and IKKβ-deficient mice. The compound NG25 was developed in the laboratory of Nathanael Gray, Dana Farber Cancer Institute, Harvard University, Cambridge, MA.
This work was supported by the United Kingdom Medical Research Council.

This article contains supplemental Figs. S1–S6.
E. Pauls, unpublished data.
- pDC
- plasmacytoid dendritic cell
- TLR
- Toll-like receptor
- IRF7
- interferon regulatory factor 7
- MEF
- mouse embryonic fibroblast.
REFERENCES
- 1. Gilliet M., Cao W., Liu Y. J. (2008) Plasmacytoid dendritic cells. Sensing nucleic acids in viral infection and autoimmune diseases. Nat. Rev. Immunol. 8, 594–606 [DOI] [PubMed] [Google Scholar]
- 2. Hemmi H., Takeuchi O., Kawai T., Kaisho T., Sato S., Sanjo H., Matsumoto M., Hoshino K., Wagner H., Takeda K., Akira S. (2000) A Toll-like receptor recognizes bacterial DNA. Nature 408, 740–745 [DOI] [PubMed] [Google Scholar]
- 3. Trinchieri G. (2010) Type I interferon. Friend or foe? J. Exp. Med. 207, 2053–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barrat F. J., Meeker T., Gregorio J., Chan J. H., Uematsu S., Akira S., Chang B., Duramad O., Coffman R. L. (2005) Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J. Exp. Med. 202, 1131–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Banchereau J., Pascual V. (2006) Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity 25, 383–392 [DOI] [PubMed] [Google Scholar]
- 6. Gregorio J., Meller S., Conrad C., Di Nardo A., Homey B., Lauerma A., Arai N., Gallo R. L., Digiovanni J., Gilliet M. (2010) Plasmacytoid dendritic cells sense skin injury and promote wound healing through type I interferons. J. Exp. Med. 207, 2921–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guiducci C., Tripodo C., Gong M., Sangaletti S., Colombo M. P., Coffman R. L., Barrat F. J. (2010) Autoimmune skin inflammation is dependent on plasmacytoid dendritic cell activation by nucleic acids via TLR7 and TLR9. J. Exp. Med. 207, 2931–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sato M., Suemori H., Hata N., Asagiri M., Ogasawara K., Nakao K., Nakaya T., Katsuki M., Noguchi S., Tanaka N., Taniguchi T. (2000) Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-α/β gene induction. Immunity 13, 539–548 [DOI] [PubMed] [Google Scholar]
- 9. Izaguirre A., Barnes B. J., Amrute S., Yeow W. S., Megjugorac N., Dai J., Feng D., Chung E., Pitha P. M., Fitzgerald-Bocarsly P. (2003) Comparative analysis of IRF and IFN-α expression in human plasmacytoid and monocyte-derived dendritic cells. J. Leukoc. Biol. 74, 1125–1138 [DOI] [PubMed] [Google Scholar]
- 10. Honda K., Yanai H., Negishi H., Asagiri M., Sato M., Mizutani T., Shimada N., Ohba Y., Takaoka A., Yoshida N., Taniguchi T. (2005) IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434, 772–777 [DOI] [PubMed] [Google Scholar]
- 11. Honda K., Takaoka A., Taniguchi T. (2006) Type I interferon α gene induction by the interferon regulatory factor family of transcription factors. Immunity 25, 349–360 [DOI] [PubMed] [Google Scholar]
- 12. Hoshino K., Sasaki I., Sugiyama T., Yano T., Yamazaki C., Yasui T., Kikutani H., Kaisho T. (2010) Critical role of IκB kinase α in TLR7/9-induced type I IFN production by conventional dendritic cells. J. Immunol. 184, 3341–3345 [DOI] [PubMed] [Google Scholar]
- 13. Hemmi H., Kaisho T., Takeda K., Akira S. (2003) The roles of Toll-like receptor 9, MyD88, and DNA-dependent protein kinase catalytic subunit in the effects of two distinct CpG DNAs on dendritic cell subsets. J. Immunol. 170, 3059–3064 [DOI] [PubMed] [Google Scholar]
- 14. Honda K., Ohba Y., Yanai H., Negishi H., Mizutani T., Takaoka A., Taya C., Taniguchi T. (2005) Spatiotemporal regulation of MyD88-IRF-7 signaling for robust type I interferon induction. Nature 434, 1035–1040 [DOI] [PubMed] [Google Scholar]
- 15. Honda K., Yanai H., Mizutani T., Negishi H., Shimada N., Suzuki N., Ohba Y., Takaoka A., Yeh W. C., Taniguchi T. (2004) Role of a transductional-transcriptional processor complex involving MyD88 and IRF-7 in Toll-like receptor signaling. Proc. Natl. Acad. Sci. U.S.A. 101, 15416–15421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kawai T., Sato S., Ishii K. J., Coban C., Hemmi H., Yamamoto M., Terai K., Matsuda M., Inoue J., Uematsu S., Takeuchi O., Akira S. (2004) Interferon-α induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 5, 1061–1068 [DOI] [PubMed] [Google Scholar]
- 17. Uematsu S., Sato S., Yamamoto M., Hirotani T., Kato H., Takeshita F., Matsuda M., Coban C., Ishii K. J., Kawai T., Takeuchi O., Akira S. (2005) Interleukin-1 receptor-associated kinase-1 plays an essential role for Toll-like receptor (TLR) 7- and TLR9-mediated interferon-α induction. J. Exp. Med. 201, 915–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Adhikari A., Xu M., Chen Z. J. (2007) Ubiquitin-mediated activation of TAK1 and IKK. Oncogene 26, 3214–3226 [DOI] [PubMed] [Google Scholar]
- 19. Hoshino K., Sugiyama T., Matsumoto M., Tanaka T., Saito M., Hemmi H., Ohara O., Akira S., Kaisho T. (2006) IκB kinase-α is critical for interferon-α production induced by Toll-like receptors 7 and 9. Nature 440, 949–953 [DOI] [PubMed] [Google Scholar]
- 20. Sharma S., tenOever B. R., Grandvaux N., Zhou G. P., Lin R., Hiscott J. (2003) Triggering the interferon antiviral response through an IKK-related pathway. Science 300, 1148–1151 [DOI] [PubMed] [Google Scholar]
- 21. Matsui K., Kumagai Y., Kato H., Sato S., Kawagoe T., Uematsu S., Takeuchi O., Akira S. (2006) Cutting edge. Role of TANK-binding kinase 1 and inducible IκB kinase in IFN responses against viruses in innate immune cells. J. Immunol. 177, 5785–5789 [DOI] [PubMed] [Google Scholar]
- 22. Chaperot L., Blum A., Manches O., Lui G., Angel J., Molens J. P., Plumas J. (2006) Virus or TLR agonists induce TRAIL-mediated cytotoxic activity of plasmacytoid dendritic cells. J. Immunol. 176, 248–255 [DOI] [PubMed] [Google Scholar]
- 23. Di Domizio J., Blum A., Gallagher-Gambarelli M., Molens J. P., Chaperot L., Plumas J. (2009) TLR7 stimulation in human plasmacytoid dendritic cells leads to the induction of early IFN-inducible genes in the absence of type I IFN. Blood 114, 1794–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lui G., Manches O., Angel J., Molens J. P., Chaperot L., Plumas J. (2009) Plasmacytoid dendritic cells capture and cross-present viral antigens from influenza virus-exposed cells. PLoS One 4, e7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim T., Pazhoor S., Bao M., Zhang Z., Hanabuchi S., Facchinetti V., Bover L., Plumas J., Chaperot L., Qin J., Liu Y. J. (2010) Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 107, 15181–15186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lepelley A., Louis S., Sourisseau M., Law H. K., Pothlichet J., Schilte C., Chaperot L., Plumas J., Randall R. E., Si-Tahar M., Mammano F., Albert M. L., Schwartz O. (2011) Innate sensing of HIV-infected cells. PLoS Pathog. 7, e1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Clark K., Peggie M., Plater L., Sorcek R. J., Young E. R., Madwed J. B., Hough J., McIver E. G., Cohen P. (2011) Novel cross-talk within the IKK family controls innate immunity. Biochem. J. 434, 93–104 [DOI] [PubMed] [Google Scholar]
- 28. Soucy T. A., Smith P. G., Milhollen M. A., Berger A. J., Gavin J. M., Adhikari S., Brownell J. E., Burke K. E., Cardin D. P., Critchley S., Cullis C. A., Doucette A., Garnsey J. J., Gaulin J. L., Gershman R. E., Lublinsky A. R., McDonald A., Mizutani H., Narayanan U., Olhava E. J., Peluso S., Rezaei M., Sintchak M. D., Talreja T., Thomas M. P., Traore T., Vyskocil S., Weatherhead G. S., Yu J., Zhang J., Dick L. R., Claiborne C. F., Rolfe M., Bolen J. B., Langston S. P. (2009) An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 [DOI] [PubMed] [Google Scholar]
- 29. Naik S. H., O'Keeffe M., Proietto A., Shortman H. H., Wu L. (2010) CD8+, CD8−, and plasmacytoid dendritic cell generation in vitro using flt3 ligand. Methods Mol. Biol. 595, 167–176 [DOI] [PubMed] [Google Scholar]
- 30. Hastie C. J., McLauchlan H. J., Cohen P. (2006) Assay of protein kinases using radiolabeled ATP. A protocol. Nat. Protoc. 1, 968–971 [DOI] [PubMed] [Google Scholar]
- 31. Marié I., Durbin J. E., Levy D. E. (1998) Differential viral induction of distinct interferon-α genes by positive feedback through interferon regulatory factor-7. EMBO J. 17, 6660–6669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ninomiya-Tsuji J., Kajino T., Ono K., Ohtomo T., Matsumoto M., Shiina M., Mihara M., Tsuchiya M., Matsumoto K. (2003) A resorcylic acid lactone, 5Z-7-oxozeaenol, prevents inflammation by inhibiting the catalytic activity of TAK1 MAPK kinase kinase. J. Biol. Chem. 278, 18485–18490 [DOI] [PubMed] [Google Scholar]
- 33. Campbell D. G., Morrice N. A. (2002) Identification of protein phosphorylation sites by a combination of mass spectrometry and solid phase Edman sequencing. J. Biomol. Tech. 13, 119–130 [PMC free article] [PubMed] [Google Scholar]
- 34. Wang X., Hussain S., Wang E. J., Li M. O., García-Sastre A., Beg A. A. (2007) Lack of essential role of NF-κB p50, RelA, and cRel subunits in virus-induced type 1 IFN expression. J. Immunol. 178, 6770–6776 [DOI] [PubMed] [Google Scholar]
- 35. Heissmeyer V., Krappmann D., Wulczyn F. G., Scheidereit C. (1999) NF-κB p105 is a target of IκB kinases and controls signal induction of Bcl-3-p50 complexes. EMBO J. 18, 4766–4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shim J. H., Xiao C., Paschal A. E., Bailey S. T., Rao P., Hayden M. S., Lee K. Y., Bussey C., Steckel M., Tanaka N., Yamada G., Akira S., Matsumoto K., Ghosh S. (2005) TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 19, 2668–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sato S., Sanjo H., Takeda K., Ninomiya-Tsuji J., Yamamoto M., Kawai T., Matsumoto K., Takeuchi O., Akira S. (2005) Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat. Immunol. 6, 1087–1095 [DOI] [PubMed] [Google Scholar]
- 38. Clark K., Takeuchi O., Akira S., Cohen P. (2011) The TRAF-associated protein TANK facilitates crosstalk within the IκB kinase family during Toll-like receptor signaling. Proc. Natl. Acad. Sci. U.S.A. 108, 17093–17098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Purtha W. E., Swiecki M., Colonna M., Diamond M. S., Bhattacharya D. (2012) Spontaneous mutation of the Dock2 gene in Irf5−/− mice complicates interpretation of type I interferon production and antibody responses. Proc. Natl. Acad. Sci. U.S.A. 109, E898–E904 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.