Background: Sclerites of alcyonarian species are biominerals formed of organic matrix molecules trapped inside a mineral inorganic fraction.
Results: Scleritin is a small basic organic matrix protein, synthetized by scleroblasts and incorporated into sclerites.
Conclusion: Scleritin is the first organic matrix protein fully characterized and localized in the sclerites of C. rubrum.
Significance: Scleritin provides information on the biomineralization pathway in C. rubrum.
Keywords: Biomineralization, Calcification, Genomics, Invertebrates, Proteomics, Organic Matrix Proteins, Red Coral, Sclerites
Abstract
We report here for the first time the isolation and characterization of a protein from the organic matrix (OM) of the sclerites of the alcyonarian, Corallium rubrum. This protein named scleritin is one of the predominant proteins extracted from the EDTA-soluble fraction of the OM. The entire open reading frame (ORF) was obtained by comparing amino acid sequences from de novo mass spectrometry and Edman degradation with an expressed sequence tag library dataset of C. rubrum. Scleritin is a secreted basic phosphorylated protein which exhibits a short amino acid sequence of 135 amino acids and a signal peptide of 20 amino acids. From specific antibodies raised against peptide sequences of scleritin, we obtained immunolabeling of scleroblasts and OM of the sclerites which provides information on the biomineralization pathway in C. rubrum.
Introduction
Biomineralization is defined as both the study of biogically produced mineral materials (or biominerals) and the processes that lead to their formation (1). Calcium carbonate skeletal structures are the most abundant biominerals encountered in the metazoan world (2). Much of what is known about biomineralization is to date inferred from marine calcifying model organisms such as mollusks and echinoderms where data are available from the ultrastructural to the biochemical and molecular levels (3–5). For other organisms, data are far less abundant despite their importance for a better basic understanding of the biomineralization process. This is the case for cnidarians among which species from the subclass Hexacorallia are typically known for their role in producing tropical coral reefs.
This is also the case for various other taxa such as alcyonarian species (6). Among Alcyonaria many families typically contain species with skeletal structures called sclerites (or spicules) which are considered to form a fragmented skeleton located in the soft tissues (coenenchyme). Sclerites are suggested to provide structural support (7) and may have a potential role of physical defense against predation (8, 9). Sclerites are also a main source of information for systematic studies because their morphology and morphometry are species-specific (10, 11).
Among alcyonarian, the precious coral Corallium rubrum is a well known sclerites-producer species. This Mediterranean species has a cultural and commercial importance, and its axial skeleton has been used for jewelry and art objects for centuries (12–14). The sclerites are distributed within the animal tissues, and they have been described in detail by means of light and scanning electron microscopy (15, 16). Their microstructure never reaches more than a few micrometers in axial or lateral directions (60–90 μm) (17). Their density can reach about 106/mg of tissue proteins (18), and they are initially formed in intracellular vesicles within cells, named scleroblasts, present in the mesoglea (17). Sclerites are also incorporated in the axial skeleton, but the pathway of incorporation remains debated (19, 13, 20).
Sclerites of C. rubrum are, as for other biominerals, a composite material formed of an organic fraction called organic matrix (OM)3 trapped inside a mineral inorganic fraction. This inorganic fraction of calcium carbonate is crystallized under the form of Mg-calcite, and the OM represents <2% of the dry weight of the skeletal structure (21). From biomimetic experiments, it is suggested that the OM plays a role of assembler between inorganic building blocks forming mesocrystals (22). These blocks have been recently evidenced in the sclerites of C. rubrum (23).
Previous biochemical works on the axial skeleton and sclerites of C. rubrum have revealed that their OM is composed of proteins, glycosaminoglycans, and proteoglycans (20, 21, 24, 25), as well as pigments such as carotenoids (26, 27) or trans-polyacetylene molecules (28). However, the most abundant literature on the biochemistry of OM in Alcyonaria concerns: (i) the Alcyoniidae: Lobophytum crassum and Synularia polydactyla (29–34); (ii) the Gorgoniidae: Leptogorgia virgulata (35–40) and Pseudoplexaura flagellosa (41). Nevertheless, to date only partial sequences of OM proteins have been obtained for alcyonarian sclerites (32, 42), and even when widening to the Anthozoa, only one OM protein has been fully characterized in a scleractinian coral (43) (for review, see also Refs. 44–46).
Based on the methodology of OM protein extraction and separation by gel electrophoresis that we previously set up (20, 47) and in combination with transcriptomic data from an EST library,4 we have isolated and fully characterized one protein of OM in the sclerites of C. rubrum. To our knowledge, this is the first report of a complete sequence of an organic matrix protein in the Alcyonaria subclass.
EXPERIMENTAL PROCEDURES
Organic Matrix Extraction
Sclerites from colonies of C. rubrum collected at 30-m depth in Marseille, Riou Island (outside cave, Gulf of Lion; Mediterranean coast of France) were prepared as described by Debreuil et al. (20). Extraction of the organic fraction from the mineral fraction was adapted from Puverel et al. (48) and Debreuil et al. (47). Briefly, after demineralization of sclerites powder with 0.25 m EDTA (pH 7.8, 23 h, 4 °C; Sigma), the solution was prefiltered (0.2 μm of polyethersulfone) and centrifuged (10,000 × g, 15 min, 4 °C). The supernatant was filtered, concentrated, and rinsed thoroughly (8 times) with ultrapure water (3,500 × g, Centricon Plus 70, cut-off 5 kDa). Protein content was determined using a Quant-iTTM protein assay kit (QubitTM; Molecular Probes) with bovine serum albumin (BSA) as a standard and Qubit fluorometer apparatus (Q32857; Invitrogen).
Electrophoresis and Immunolabeling
One- and two-dimensional gel electrophoresis and Western blotting were performed as described by Debreuil et al. (20, 47). BisTris 12% polyacrylamide gels (Criterion, Bio-Rad) were used for wide range molecular weight protein; and Tris-Tricine 16.5% polyacrylamide gels (Criterion, Bio-Rad) for small molecular weight proteins. The protein markers used were: Silver Stain Molecular Weight Marker (M6539; Sigma) for the BisTris silver-stained electrophoresis gel, Kaleidoscope Polypeptide Standards (Bio-Rad 161-0325) for the Tris-Tricine silver-stained electrophoresis gels, and Precision Plus Protein WesternCTM standard (161-0376 Bio-Rad) both for the BisTris and Tris-Tricine gels of Western blots.
The primary antibodies used for Western blotting were anti-phosphoserine (9332; Abcam), anti-phosphothreonine (9337; Abcam), anti-phosphotyrosine (9319; Abcam), and polyclonal antibodies raised against two amino acid sequences of the OM protein identified in the present study, i.e. scleritin: NH2-FIELSKRMQRESSNFC-COOH and NH2-CNTRPVQPISRQLDDL-COOH. For these last antibodies, both peptides were mixed with Freund's complete adjuvant and injected intraperitoneally into a rabbit (Oryctolagus cuniculus; Eurogentec). Rabbit was immunoboosted by several injections of peptides mixed with incomplete Freund's adjuvant (at 7, 10, and 18 days after the first injection). The final bleeding was conducted 28 days after the last injection. Preimmune (from 0 day bleeding) and final sera were drawn off and stored at −20 °C.
Electroelution Method
After one-dimensional SDS-PAGE (16.5% polyacrylamide gel; Criterion, Bio-Rad) of the whole extract of OM of sclerites and a staining step overnight with ImperialTM Protein Stain (Thermo Scientific), the electroelution of the 18-kDa protein band was performed with an Electro-Eluter (model 422; Bio-Rad) as used by Rahman et al. (42) for the purification of OM proteins in alcyanorian sclerites. Electroelution was performed for 5 h at 60 mA, using dialyze caps (cutoff 3.5 kDa) in an elution buffer (25 mm Tris, 192 mm glycine, 0.1% SDS). The samples were then concentrated, rinsed thoroughly (eight times) by ultrafiltration using Amicon-Ultra (cutoff 5 kDa; Millipore), freeze-dried, and then used for the two-dimensional electrophoresis experiment.
Sequencing Methods
The amino acid sequence determination based on Edman degradation was performed using an Applied Biosystems gas-phase sequencer (model 492; s/n: 9510287J; Institut de Biologie Structurale, CEA/CNRS/UJF, Grenoble, France). Phenylthiohydantoin amino acid derivatives generated at each sequence cycle were identified and quantified on-line with an HPLC system (Applied Biosystems 140C and Applied Biosystems 610A data analysis software 2.1) with PTH-amino acid standard kit (PerkinElmer Life Sciences P/N 4340968; according to instructions 900776 Rev D).
Mass spectrometry analysis was performed on protein spot from the two-dimensional electrophoresis gel. Proteins were digested with trypsin, blocked with 5% formic acid, and analyzed by capillary-LC/ESI/MS/MS (LTQ/FT-Orbitrap mass spectrometer; Thermo Fisher) coupled with pumps and autosampler under standard conditions (275 °C; 4500 V). Helium was used as the collision gas, and experiments were performed in parallel mode (30,000 survey resolution and 5 data-dependent ion trap MS/MS scans). De novo analysis was operated on PEAKS Studio 4.5 (Bioinformatics Solutions; Waterloo, ON, Canada). Then putative amino acid sequences obtained were analyzed against nucleotide sequences data base dynamically translated in all reading frames (tBLASTn) (49) from an EST library of C. rubrum.4
Cloning Protein Gene
Total RNA extraction and cDNA synthesis from three colonies of C. rubrum (Riou Island) were, respectively performed as described by Zoccola et al. (50) and Moya et al. (51). PCR amplification was carried out using platinum Taq DNA polymerase (Invitrogen) and primers designed using nucleotide sequences of the contig corru5764135 from the EST library of C. rubrum.4 Nucleotides sequences were chosen from the deduced consensus between amino acid sequence from Edman degradation, mass spectrometry analyses, and the translated contig corru5764135: 5′-AGGAAGGTTGGTCAGGGCGT-3′ for the forward primer and 5′-AGCGAACTTCGGAGTCCTTA-3′ for reverse primer. PCR amplification product was ligated into a plasmid vector (pGEM-T easy vector system II; Promega), and DNA sequencing of three independent clones was carried out on both strands with SP6 and T7 primer sequences (Macrogen).
After the sequence confirmation, RACE experiments were performed using the 5′/3′-RACE kit (Roche Applied Science) with the primers 5′-CATTAAGCTCTGAAGCAGTGG-3′ and 5′-AGGAAGGTTGGTCAGGGCGT-3′ for 5′-RACE; and the primers 5′-AAACGTATGCAACGAGAGTCA-3′ and 5′-AGCGAACTTCGGAGTCCTTA-3′ for 3′-RACE. Both ends were cloned and sequenced as mentioned above. All sequences were assembled using the Lasergene software (SEQMAN LASERGENE 9.0.4; DNASTAR) to obtain the whole open reading frame (ORF).
Nucleotide and Amino Acid Sequences Analysis
The theoretical mass, isoelectric point, and amino acid composition of the protein identified in the present study were obtained with Editseq 9.0.4 (DNASTAR Inc.). The complete translated ORF was interpreted with Bioworks 3.3 (Thermo Fisher) with both b and y ion series for charge 2, 3, and 4 and valuate Xcorr for recovery confirmation (xc (± 1, 2, 3) = 1.50, 2.00, 2.50); supplemental Fig. S1a). BLASTp and tBLASTn analysis (49) were carried out to identify homologous sequences in UniProt (52) and GenBank database on the NCBI server (53). From the EXPASY online server, the putative glycosylation sites were identified using DictyOGlyc1.1, NetCGlyc 1.0, NetNGlyc 1.0, YinOYang1.2; phosphorylation sites with NetPhos2.0; and the signal peptide with SignalP 4.0. The subcellular localization were predicted using TargetP 1.1 (54) and PsortII prediction tool (55).
Histology and Immunolabeling on Tissue Cross-sections
To obtain a histological overview of a demineralized branch of C. rubrum, a longitudinal section was incubated with hematoxylin/eosin/aniline blue to stain, respectively, nuclei and cytoplasmic and connective regions. Then, the polyclonal anti-scleritin antibodies used for Western blotting were used for immunohistochemistry. Briefly as described by Debreuil et al. (20), pieces of C. rubrum colonies (2–3-cm length) were fixed, demineralized, dehydrated, and embedded in paraffin wax. Deparaffinized cross-sections were mounted on a silane-coated glass slide and incubated in saturating solution. Sections were then incubated with anti-scleritin antibodies. Controls were performed with preimmune serum as primary antibodies. All samples were then incubated with biotinylated anti-rabbit antibodies (Sigma) as secondary antibodies, stained with streptavidin-Alexa Fluor 568 (Molecular Probes, Invitrogen). 0.002% DAPI (4′6-diamidino-2-phenylindole, Sigma) was used to stain the nuclei. Samples were embedded in Pro-Long antifade solution (Molecular Probes, Invitrogen), and observed with a confocal laser scanning microscope (Leica SP5) equipped with UV and visible laser lines.
RESULTS
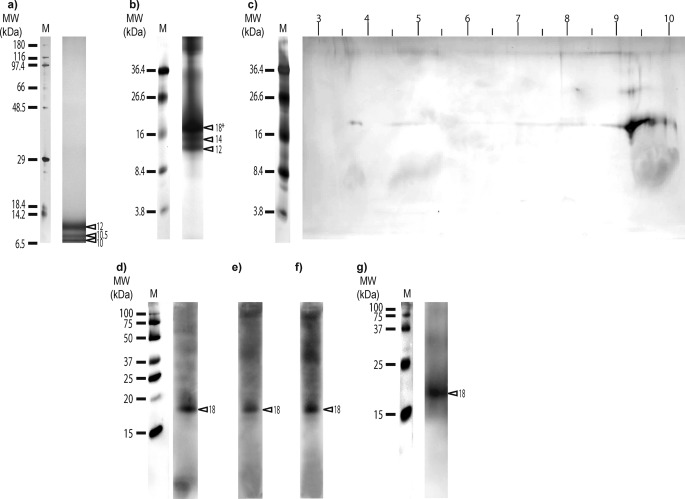
Separation of OM proteins of sclerites from C. rubrum by one-dimensional electrophoresis (BisTris 12% polyacrylamide gel; Fig. 1a), shows a set of protein bands distributed mainly in the range of low apparent molecular mass (≤12 kDa) as in Debreuil et al. (20). To obtain a better resolution of small molecular mass proteins, we performed electrophoresis with Tris-Tricine 16.5% polyacrylamide gels (Fig. 1b) which are the preferred electrophoretic systems for the resolution of proteins smaller than 30 kDa (56). The bands previously obtained at 12, 10.5, and 10 kDa in BisTris 12% polyacrylamide gel (Fig. 1a) and in our previous work (20), respectively, give three major bands at 18, 14, and 12 kDa in Tris-Tricine 16.5% polyacrylamide gel (Fig. 1b). This difference in apparent molecular mass is common when using different percentage concentration of acrylamide in electrophoresis gels (56). We chose to study the predominant 18-kDa protein band. This band was excised, electroeluted, and used for two-dimensional gel electrophoresis (Fig. 1c). The protein spot obtained with a high isoelectric point (pI) at nearly 9.4 was excised, destained, digested, and analyzed by capillary-LC/ESI/MS/MS. Reliable peptide sequences were obtained in the MS/MS spectra with identical y and b series ions: PLSSEAVALFFNK from 711.89 m/z and KWNTFIELSK from 633.35 m/z (supplemental Fig. S1, b and c). These amino acid sequences and the one obtained from Edman degradation (RKVGQGVIN) matched only with one translated nucleotide contig from an EST library of C. rubrum (81,497 sequences).4 This contig (corru5764135) of 368 nucleotides long encodes for a sequence of 122 amino acids but not a complete ORF. From this contig, primers were designed and by 3′-RACE and 5-′RACE, a complete ORF was obtained (Fig. 2).
FIGURE 1.
Silver staining of electrophoresis gels and Western blots on organic matrix extracted from sclerites of C. rubrum. a, one-dimensional SDS-PAGE (10 μg protein/lane; BisTris 12% polyacrylamide gel, silver stain molecular mass marker (M6539; Sigma) as protein marker). b, one-dimensional SDS-PAGE (10 μg protein/lane, Tris-Tricine 16.5% polyacrylamide gel, kaleidoscope polypeptide standards (161-0325; Bio-Rad) as protein marker); c, two-dimensional electrophoresis gel (Tris-Tricine 16.5% polyacrylamide gel, kaleidoscope polypeptide standards (161-0325) as protein marker) of the major protein band (18*) excised from one dimensional SDS-PAGE. d, Western blot (5 μg of proteins/lane; BisTris 12% polyacrylamide gel, Precision Plus Protein WesternCTM standard (161-0376; Bio-Rad) as protein marker) with antibodies against phosphorylated serine. e, Western blot with antibodies against the phosphorylated threonine (5 μg of proteins/lane; BisTris 12% polyacrylamide gel, Precision Plus Protein WesternCTM standard (161-0376) as protein marker). f, Western blot with antibodies against phosphorylated tyrosine (5 μg of proteins/lane; BisTris 12% polyacrylamide gel, Precision Plus Protein WesternCTM standard (161-0376) as protein marker). g, Western blot with anti-scleritin antibodies (Tris-Tricine 16.5% polyacrylamide gel, Precision Plus Protein WesternCTM standard (161-0376) as protein marker). MW, molecular mass; M, protein marker; and triangles show apparent molecular mass of proteins (in kDa).
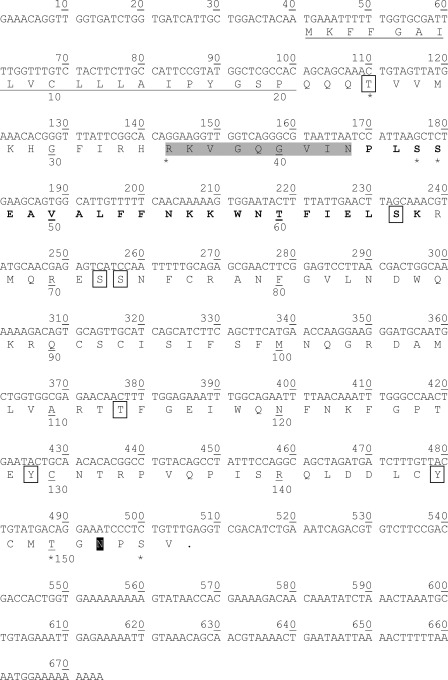
FIGURE 2.
Nucleotide gene and amino acid sequences of scleritin. Nucleotide sequence above (GenBank accession number JQ652458) and deduced translation of the ORF (nucleotides 40–507) below. The putative signal peptide is underlined, the peptide sequence obtained after Edman degradation is highlighted in gray, the two consecutive peptides obtained by MS/MS are indicated in bold letters, the potential N-glycosylation site is highlighted in black (Asp-152), the five potential O-glycosylation sites (Thr-24, Ser-56, Ser-57, Thr-150, and Ser-154) are indicated by asterisks, and the seven potential phosphorylation sites are boxed (Thr-24, Ser-65, Ser-72, Ser-73, Thr-113, Tyr-129, and Tyr-147).
The full-length cDNA sequence obtained is 665 bp long and contains, at 40 bp from the 5′-end, a methionine in a Kozak (57) cnidarian (58) context followed by an ORF of 468 bp (GenBank accession number JQ652458). This ORF encodes a protein of 155 amino acids composed of an N-terminal signal peptide of 20 amino acids and a C-terminal sequence of 135 amino acids (Fig. 2). This protein has a calculated molecular mass of 17.78 kDa and a calculated pI of 9.35 (DNASTAR Editseq 9.0.4). This protein is a secreted protein, its secretion is confirmed by the Target 1.1 (54) and PsortII (55) tools with, respectively, 86.9% and 55.6% probability. Indeed, the signal peptide is no more present once the protein is incorporated in the OM (confirmed by MS/MS recovery; supplemental Fig. S1, a and d). This secreted protein is named scleritin. Scleritin is a basic small protein (calculated molecular mass of 15.65; calculated pI of 9.39; DNASTAR Editseq 9.0.4) composed mainly of polar, hydrophobic, and amine forms of amino acids (Phe, Asn, and Gln in majority).
The analysis of the amino acid sequence of scleritin reveals several potential sites of post-translational modifications (Fig. 2) such as seven phosphorylations on Thr-24, Ser-65, Ser-72, Ser-73, Thr-113, Tyr-129, and Tyr-147 which are confirmed by the Western blotting results (Fig. 1, d–f); five O-glycosylations on Thr-24, Ser-56, Ser-57, Thr-150, and Ser-154; and one N-glycosylation on Asp-152.
We looked for homologies between scleritin and other sequences from the available public sequence databases, GenBank nonredundant proteins, and nucleotide databases (NCBI-NRdb, NCBI-dbEST, UniProt). No significant hits were obtained by BLASTp and tBLASTn (e-value cutoff: 0.01) (49).
We then looked at the localization of scleritin in the tissues of C. rubrum. First, we tested the polyclonal antibodies raised against scleritin by Western blotting on OM extract from the sclerites. The Western blotting shows that only one band at 18 kDa is labeled (Fig. 1g), confirming that the antibodies are specific for scleritin (18-kDa protein band from the OM extract; Fig. 1b).
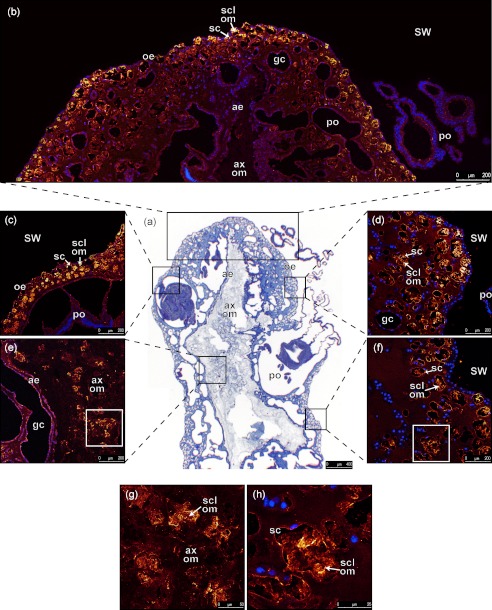
These antibodies were then used to label the tissues after skeleton demineralization (Fig. 3). A longitudinal section of tissues stained with hematoxylin/eosin/aniline blue shows the typical histology of a colony of C. rubrum with oral epithelium, polyps, gastrodermic canals, and aboral epithelium surrounding the OM of the axial skeleton (Fig. 3a). The labeling of these tissues (Fig. 3, b–h) with the anti-scleritin antibodies revealed by Alexa Fluor 568 appears in orange to yellow with increasing the intensity of labeling; and nuclei in cells stained with DAPI appear in blue. Z-stack reconstructions of 10 tissue sections show that: (i) the OM of the sclerites is labeled (Fig. 3, b–h); (ii) the scleroblasts but no other cell types are labeled (Fig. 3, b–f and h); (iii) labeling occurs from the apex to the base of the colony (Fig. 3, b–f); (iv) the OM of the axial skeleton is not labeled whereas labeling occurs in the OM of sclerites present in the medullar part of the axial skeleton (Fig. 3, b, e, and g). It must also be noted that most of the immunolabeling signal is observed at the periphery of the colony (i.e. near the oral epithelium) where sclerites are more numerous.
FIGURE 3.
Histology and immunohistochemistry of a longitudinal section of a demineralized branch of C. rubrum. a, overview of a section stained with hemalun/eosin/aniline blue which, respectively, stain nuclei and cytoplamic and connective regions observed with a bright field light microscope. b–h, labeling with antibodies against scleritin (orange to yellow) is merged with blue labeling of cell nuclei with DAPI: b, immunolabeling of apical region of the colony; c, immunolabeling of tissues close to a polyp; d, immunolabeling of oral epithelium; e, immunolabeling at the interface between aboral epithelium and organic matrix of axial skeleton; f, immunolabeling at the base of a branch; g, magnification of e showing labeling of organic matrix of sclerites inside the organic matrix of axial skeleton. h, magnification of f showing labeling of organic matrix of a sclerite inside a scleroblast. Observations from b to h were performed with a confocal microscope. SW, sea water; po, polyp; oe, oral epithelium; sc, scleroblast; gc, gastrodermic canal; ae, aboral epithelium; ax om, organic matrix of axial skeleton; scl om, organic matrix of sclerite. Each image was obtained by Z-stack reconstruction of 10 tissue sections, with 1-μm step.
DISCUSSION
Molecular and Biochemical Characterization of Scleritin
By combining transcriptomic and proteomic approaches, we identified a coral OM protein in the sclerites of C. rubrum which we named scleritin. Scleritin is a basic small molecular mass protein composed mainly of polar, hydrophobic, and amine forms of amino acids (Phe, Asn, and Gln in majority). This result is not trivial because OM proteins are usually acidic (i.e. low range of isoelectric point) (59), and in C. rubrum the OM of sclerites is composed mainly of acidic and polar amino acids (Asp and Gly) (21). However, basic small molecular mass OM proteins have already been evidenced in other calcifying organisms (for review, see Ref. 60); for mollusks, see Ref. 3; for echinoderms, see Ref. 61. In corals the only OM protein fully characterized, i.e. galaxin from the tropical hexacoral Galaxea fascicularis, does not show any acidic domain (43).
The analysis of the amino acid sequence of scleritin reveals several potential sites of post-translational modifications such as phosphorylations and glycosylations which could explain the difference between the calculated molecular mass of scleritin (15.65 kDa) and that of the secreted form observed on the electrophoresis gels (18 kDa; Fig. 1, b and c). OM proteins of several calcifying models are known to be post-translationally modified (3, 60, 62–64). Such modifications can participate in the binding of calcium ions as proposed for marine calcifying invertebrates (see Refs. 65–67 for phosphorylations in mollusks and crustaceans and Ref. 68 for glycosylations in mollusks). These post-translational modifications could even be a prerequisite step in the calcification process of corals. Indeed, Allemand et al. (69) have shown that the inhibition of N-glycosylations by tunicamycin reduces both the rate of calcification and the incorporation of macromolecules (OM proteins) into the skeleton of the tropical coral Stylophora pistillata. Moreover, glycosylated proteins (or glycoproteins) were also reported in the OM of (i) the sclerites of L. virgulata (37, 38, 70), (ii) the skeletal structures of other alcyonarian species (3, 29, 42, 71), and (iii) the axial skeleton of C. rubrum (25).
Concerning scleritin, the potential phosphorylation sites at both extremities of the amino acid sequence (i.e. Thr-24 at the N terminus and Tyr-147 at the C terminus; Fig. 2) are certainly highly accessible and could be crucial for proper folding, calcium binding, and other functions in the calcification process (3, 60, 63). Scleritin also possesses six predicted sites of glycosylation (N-glycosylation on Asp-152; and O-glycosylations on Thr-24, Ser-56, Ser-57, Thr-150 and Ser-154; Fig. 2). This high ratio of predicted glycosylation sites relative to the shortness of the amino acid sequence suggests a potential activity of scleritin in playing different roles such as stabilization of the protein (as in echinoderms) (61) and/or scaffold for the control of mineral deposition as in mollusks (72–76, 4, 3, 63, 64).
The fact that we did not find any homology (insignificant e-values) between scleritin and other sequences from all sequence databases available (GenBank nonredundant proteins and nucleotide databases: NCBI-NRdb, NCBI-dbEST, UniProt) could indicate that scleritin is a species-specific OM protein of C. rubrum. However, this could also be because even for phylogenetically neighbor species, currently there are very few data available (i.e. approximately 1,000 EST sequences and nearly 2,600 proteins sequences for Alcyonaria).
Insights in Biomineralization Process: from Scleritin Synthesis to Incorporation
Even if we cannot precisely determine the role of scleritin, from the sequence analysis and the existing literature on alcyonarian sclerites formation (77), we can propose a pathway from synthesis to incorporation within the crystalline structure of the sclerites.
Scleroblasts are the only cells that produce scleritin because only these cells in the tissues are labeled by the anti-scleritin antibodies. Because scleritin is a secreted protein, this shows a trafficking pathway where scleritin is targeted to the endoplamic reticulum. As for other proteins, in the endoplasmic reticulum the peptide signal is lost, and the potential N-glycosylations together with conformational changes in an active form (secondary and tertiary configurations) (78) can occur. Then as shown for mammal cells (79, 80), phosphorylations (Fig. 3) and potential O-glycosylations can occur in the Golgi apparatus. From the Golgi apparatus, scleritin is secreted into vesicles. From ultrastructural observations such a process has also been proposed for the formation of sclerites in L. virgulata (37) where the first step of formation of the sclerites occurs within “intracellular vesicles probably originating from the Golgi apparatus.”
From immunolabeling experiments, some other important piece of information concerns the growth mechanism of the axial skeleton. In the literature two possibilities have been proposed: (i) the axial skeleton only results from the migration and the fusion of sclerites (i.e. theory of Lacaze-Duthiers) (19) and (ii) the axial skeleton results from the incorporation of sclerites at the apex allowing mainly for vertical growth (medullar part) and then a concentric secretion (annular part) by the skeletogenic epithelium allowing mainly for centrifugal growth (i.e. theory of Allemand) (13). This last hypothesis was supported by kinetic experiments of biocalcification using 45Ca as a tracer (18, 81). In addition, Grillo et al. (17) showed that microprotuberances on the axial skeleton are smaller, more spinose, and less numerous per surface unit than sclerites; and later Marschal et al. (82) observed a growth ring pattern of OM in the annular part of the axial skeleton but not in the medullar part.
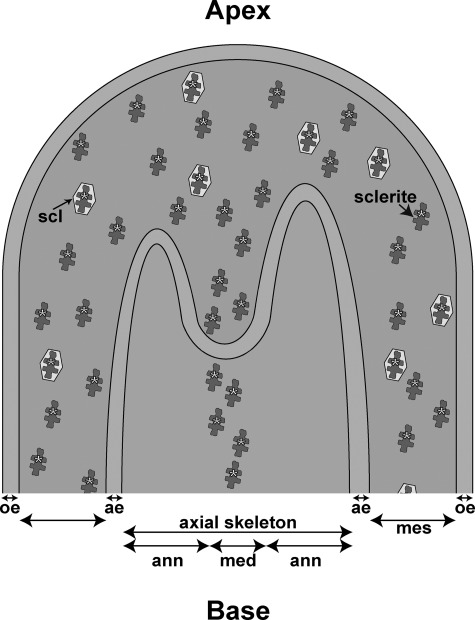
Considering the two hypotheses for the axial skeleton mechanism of growth of C. rubrum, in a previous paper (20) we used antibodies raised against the whole OM extract of sclerites and axial skeleton. However, the antibodies cross-reacted with the two organic matrices, and we could not discriminate between labeling from sclerites or axial skeleton. In the present study we raised antibodies against scleritin which is, as described above, a specific protein of the OM of sclerites. When we used these antibodies against the demineralized axial skeleton, only the scleritin from sclerites located in the medullar part of the axial skeleton was labeled (Fig. 3g). On the opposite side, the OM surrounding the medullar part of the axial skeleton (i.e. the annular part) was not labeled. This result thus confirms that the annular part results from another process than the fusion of sclerites and that the incorporation of sclerites only occurs in the medullar part of the axial skeleton (Fig. 4).
FIGURE 4.
Diagram of sclerite incorporation in medullar part of axial skeleton at apex of a branch of C. rubrum. From the inside to the outside: medullar part of the axial skeleton (med); annular part of the axial skeleton (ann); aboral epithelium (ae); mesoglea (mes); scleroblast (scl); and oral epithelium (oe). Scleritin from the organic matrix of sclerites is indicated by yellow asterisks.
Scleritin is the first organic matrix protein fully characterized in an alcyonarian species. With a low molecular mass and a high isoelectric point, it presents no homology with any other sequence available in data bases. Our study illustrates the efficiency of combining transcriptomic and proteomic approaches for the identification of coral organic matrix proteins, opens gates for further protein characterization among alcyonarian species, and provides information on sclerites and axial skeleton formation in C. rubrum.
Supplementary Material
Acknowledgments
We thank F. Zuberer for assistance during coral sampling and the RAMOGE Agreement for the Alain Vatrican Award; A. Bertucci, N. Techer, and N. Segonds for technical help and fruitful discussions; A. Venn for proofreading the English in the paper; and two anonymous reviewers for comments that helped to improve the paper.
This work was conducted as part of the Centre Scientifique de Monaco research program, supported by the Government of the Principality of Monaco.

This article contains supplemental Fig. S1.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) JQ652458.
Centre Scientifique de Monaco, unpublished data.
- OM
- organic matrix
- BisTris
- bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane
- EST
- expressed sequence tag
- MS/MS
- tandem MS
- RACE
- rapid amplification of cDNA ends
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Estroff L. A. (2008) Introduction: biomineralization. Chem. Rev. 108, 4329–4331 [DOI] [PubMed] [Google Scholar]
- 2. Lowenstam H. A., Weiner S. (1989) On Biomineralization, pp. 7–16, Oxford University Press, New York [Google Scholar]
- 3. Marin F., Luquet G., Marie B., Medakovic D. (2008) Molluscan shell proteins: primary structure, origin, and evolution. Curr. Top. Dev. Biol. 80, 209–276 [DOI] [PubMed] [Google Scholar]
- 4. Mann S. (2001) Biomineralization: Principles and Concepts in Bioinorganic Materials Chemistry (Compton R. G., Davies S. G., Evans J., eds) pp. 6–23, Oxford University Press, New York [Google Scholar]
- 5. Dubois P., Chen C. P. (1989) in Echinoderm Studies (Jangoux M., Lawrence J. M., eds) pp. 109–178, Rotterdam [Google Scholar]
- 6. Ehrlich H., Koutsoukos P. G., Demadis K. D., Pokrovsky O. S. (2009) Principles of demineralization: modern strategies for the isolation of organic frameworks. Part II. Decalcification. Micron 40, 169–193 [DOI] [PubMed] [Google Scholar]
- 7. Wainwright S. A., Biggs W. D., Currey J. D., Gosline J. M. (1982) Mechanical Design in Organisms, Princeton University Press, Princeton, NJ [Google Scholar]
- 8. Harvell C. D., Fenical W. (1989) Chemical and structural defenses of Caribbean gorgonians (Pseudopterogorgia spp.): intracolony localization of defense. Limnol. Oceanogr. 34, 382–389 [Google Scholar]
- 9. Lewis J. C., Wallis E. V. (1991) The function of surface sclerites in gorgonians (Coelenterata, Octocorallia). Biol. Bull. 181, 275–288 [DOI] [PubMed] [Google Scholar]
- 10. Bayer F. M. (1961) The Shallow-water Octocorallia of the West Indian Region, Martinus Nijhoff, The Hague [Google Scholar]
- 11. Vargas S., Breedy O., Siles F., Guzman H. M. (2010) How many kinds of sclerite? Towards a morphometric classification of gorgoniid microskeletal components. Micron 41, 158–164 [DOI] [PubMed] [Google Scholar]
- 12. Tescione G. (1973) The Italians and Their Coral Fishing, Fausto Fiorentino, Napoli [Google Scholar]
- 13. Allemand D. (1993) The biology and skeletogenesis of the Mediterranean red coral: a review. Precious Corals Octocoral Res. 2, 19–39 [Google Scholar]
- 14. Morel J.-P., Rondi-Costanzo C., Ugolini D. (2000) Corallo di Ieri, Corallo di Oggi (Bari, ed), Centro Universitario Europeo per i beni culturali, Ravello Edipuglia [Google Scholar]
- 15. Weinberg S. (1976) Revision of the common octocorallia of the Mediterranean circalittoral. I. Gorgonacea. Beaufortia 24, 63–103 [Google Scholar]
- 16. Mateu G., Traveria A., Fontarnau R., Masso C. (1986) Biodiagénesis mineralògica del Corallium rubrum (L.). Bol. Inst. Esp. Oceanogr. 3, 1–12 [Google Scholar]
- 17. Grillo M. C., Goldberg W. M., Allemand D. (1993) Skeleton and sclerite formation in the precious red coral Corallium rubrum. Mar. Biol. 117, 119–128 [Google Scholar]
- 18. Allemand D., Grillo M.-C. (1992) Biocalcification mechanism in gorgonians: 45Ca uptake and deposition by the Mediterranean red coral Corallium rubrum. J. Exp. Zool. 262, 237–246 [Google Scholar]
- 19. Lacaze-Duthiers H. (1864) Histoire Naturelle du Corail, pp. 110–124, J. B. Baillière et Fils, Paris [Google Scholar]
- 20. Debreuil J., Tambutté S., Zoccola D., Segonds N., Techer N., Allemand D., Tambutté E. (2011) Comparative analysis of the soluble organic matrix of axial skeleton and sclerites of Corallium rubrum: insights for biomineralization. Comp. Biochem. Physiol. B 159, 40–48 [DOI] [PubMed] [Google Scholar]
- 21. Allemand D., Cuif J.-P., Watabe N., Oishi M., Kawaguchi T. (1994) Bulletin Institut Océanographique Monaco 14, 129–139 [Google Scholar]
- 22. Cölfen H., Antonietti M. (2005) Mesocrystals: inorganic superstructures made by highly parallel crystallization and controlled alignment. Angew. Chem. Int. Ed. 44, 5576–5591 [DOI] [PubMed] [Google Scholar]
- 23. Floquet N., Vielzeuf D. (2011) Mesoscale twinning and crystallographic registers in biominerals. Am. Mineralogist 96, 1228–1237 [Google Scholar]
- 24. Borelli G., Mayer-Gostan N., Merle P. L., De Pontual H., Boeuf G., Allemand D., Payan P. (2003) Composition of biomineral organic matrices with special emphasis on turbot (Psetta maxima) otolith and endolymph. Calcif. Tissue Int. 72, 717–725 [DOI] [PubMed] [Google Scholar]
- 25. Dauphin Y. (2006) Mineralizing matrices in the skeletal axes of two Corallium species (Alcyonacea). Comp. Biochem. Physiol. A 145, 54–64 [DOI] [PubMed] [Google Scholar]
- 26. Merlin J. C., Dele M. L. (1983) Etude par spectroscopie Raman de résonance de la pigmentation des squelettes calcaires de certains coraux. Bull. Soc. Zool. Fr. 108, 289–301 [Google Scholar]
- 27. Cvejic J., Tambutté S., Lotto S., Mikov M., Slacanin I., Allemand D. (2007) Determination of canthaxanthin in the red coral (Corallium rubrum) from Marseilles by HPLC combined with UV and MS detection. Mar. Biol. 152, 855–862 [Google Scholar]
- 28. Fritsch E., Karampelas S. (2008) Comment on Determination of canthaxanthin in the red coral (Corallium rubrum) from Marseille by HPLC combined with UV and MS detection. Mar. Biol. 154, 929–930 [Google Scholar]
- 29. Rahman M. A., Isa Y. (2005) Characterization of proteins from the matrix of spicules from the alcyonarian, Lobophytum crassum. J. Exp. Mar. Biol. Ecol. 321, 71–82 [Google Scholar]
- 30. Rahman M. A., Isa Y., Uehara T. (2005) Blood-based proteomics for personalized medicine: examples from neurodegenerative disease. Proteomics 5, 1–9 [DOI] [PubMed] [Google Scholar]
- 31. Rahman M. A., Isa Y., Uehara T. (2006) Studies on two closely related species of octocorallians: biochemical and molecular characteristics of the organic matrices of endoskeletal sclerites. Mar. Biotechnol. 8, 415–424 [DOI] [PubMed] [Google Scholar]
- 32. Rahman M. A., Isa Y., Takemura A., Uehara T. (2006) Analysis of proteinaceous components of the organic matrix of endoskeletal sclerites from the alcyonarian Lobophytum crassum. Calcif. Tissue Int. 78, 178–185 [DOI] [PubMed] [Google Scholar]
- 33. Rahman M. A., Oomori T., Uehara T. (2008) Carbonic anhydrase in calcified endoskeleton: novel activity in biocalcification in alcyonarian. Mar. Biotechnol. 10, 31–38 [DOI] [PubMed] [Google Scholar]
- 34. Rahman M. A., Oomori T. (2008) Structure, crystallization, and mineral composition of sclerites in the alcyonarian coral. J. Crystal. Growth 310, 3528–3534 [Google Scholar]
- 35. Kingsley R. J., Tsuzaki M., Watabe N., Mechanic G. L. (1990) Collagen in the spicule organic matrix of the gorgonian Leptogorgia virgulata. Biol. Bull. 179, 207–213 [DOI] [PubMed] [Google Scholar]
- 36. Kingsley R. J., Melaro E. W., Coe K. E., Flory M. R., Skorupa A. M., Harclerode K. A. (1996) Mechanisms of the annual cycling of organic-matrix collagen from spicules of the gorgonian Leptogorgia virgulata. Invert. Biol. 115, 89–98 [Google Scholar]
- 37. Kingsley R. J., Watabe N. (1982) Ultrastructural investigation of spicule formation in the gorgonian Leptogorgia virgulata (Lamarck) (Coelenterata: Gorgonacea). Cell Tissue Res. 223, 325–334 [DOI] [PubMed] [Google Scholar]
- 38. Kingsley R. J., Watabe N. (1983) Analysis of proteinaceous components of the organic matrices of spicules from the gorgonian Leptogorgia virgulata. Comp. Biochem. Physiol. B 76, 443–447 [Google Scholar]
- 39. Kingsley R. J., Watabe N. (1989) The dynamics of spicule calcification in whole colonies of the gorgonian Leptogorgia virgulata Lam. (Coelenterata, Gorgonacea). J. Exp. Mar. Biol. Ecol. 133, 57–65 [Google Scholar]
- 40. Samata T., Kingsley R. J., Watabe N. (1989) Ca-binding glycoprotein from the spicules of the octocoral Leptogorgia virgulata. Comp. Biochem. Physiol. B 94, 651–654 [Google Scholar]
- 41. Goldberg W. M. (1988) Chemistry, histochemistry, and microscopy of the organic matrix of spicules from a gorgonian coral. Histochem. Cell Biol. 89, 163–170 [DOI] [PubMed] [Google Scholar]
- 42. Rahman A., Oomori T., Wörheide G. (2011) Calcite formation in soft coral sclerites is determined by a single reactive extracellular protein. J. Biol. Chem. 286, 31638–31649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fukuda I., Ooki S., Fujita T., Murayama E., Nagasawa H., Isa Y., Watanabe T. (2003) Molecular cloning of a cDNA encoding a soluble protein in the coral exoskeleton. Biochem. Biophys. Res. Commun. 304, 11–17 [DOI] [PubMed] [Google Scholar]
- 44. Tambutté S., Tambutté É., Zoccola D., Allemand D. (2007) in Handbook of Biomineralization: The Biology of Biominerals Structure Formation (Baeuerlein E., ed) pp. 243–259, Wiley-VCH, New York [Google Scholar]
- 45. Allemand D., Tambutté É., Zoccola D., Tambutté S. (2011) in Coral Reefs: An Ecosystem in Transition (Dubinsky Z., Stambler N., eds) pp. 119–150, Springer, The Netherlands [Google Scholar]
- 46. Tambutté S., Holcomb M., Ferrier-Pagès C., Reynaud S., Tambutté É., Zoccola D., Allemand D. (2011) Coral biomineralization: from the gene to the environment. J. Exp. Mar. Biol. Ecol. 408, 58–78 [Google Scholar]
- 47. Debreuil J., Tambutté S., Zoccola D., Segonds N., Techer N., Marschal C., Allemand D., Kosuge S., Tambutté É. (2011) Specific organic matrix characteristics in skeletons of Corallium species. Mar. Biol. 158, 2765–2774 [Google Scholar]
- 48. Puverel S., Tambutté E., Pereira-Mouriès L., Zoccola D., Allemand D., Tambutté S. (2005) Soluble organic matrix of two scleractinian corals: partial and comparative analysis. Comp. Biochem. Physiol. B 141, 480–487 [DOI] [PubMed] [Google Scholar]
- 49. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zoccola D., Tambutté E., Sénégas-Balas F., Michiels J. F., Failla J. P., Jaubert J., Allemand D. (1999) Cloning of a calcium channel α1 subunit from the reef-building coral, Stylophora pistillata. Gene 227, 157–167 [DOI] [PubMed] [Google Scholar]
- 51. Moya A., Tambutté S., Bertucci A., Tambutté E., Lotto S., Vullo D., Supuran C. T., Allemand D., Zoccola D. (2008) Carbonic anhydrase in the scleractinian coral Stylophora pistillata: characterization, localization, and role in biomineralization. J. Biol. Chem. 283, 25475–25484 [DOI] [PubMed] [Google Scholar]
- 52. Jain E., Bairoch A., Duvaud S., Phan I., Redaschi N., Suzek B. E, Martin M. J, McGarvey P., Gasteiger E. (2009) Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics 10, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Geer L. Y., Marchler-Bauer A., Geer R. C., Han L., He J., He S., Liu C., Shi W., Bryant S. H. (2010) The NCBI BioSystems database. Nucleic Acids Res. 38, D492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP, and related tools. Nat. Protocols 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 55. Horton P., Park K. J., Obayashi T., Fujita N., Harada H., Adams-Collier C. J., Nakai K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585-W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schägger H. (2006) Tricine-SDS-PAGE. Nat. Protocols 1, 16–22 [DOI] [PubMed] [Google Scholar]
- 57. Kozak M. (1984) Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 12, 857–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mankad R. V., Gimelbrant A. A., McClintock T. S. (1998) Consensus translational initiation sites of marine invertebrate phyla. Biol. Bull. 195, 251–254 [DOI] [PubMed] [Google Scholar]
- 59. Weiner S. (1986) Organization of extracellularly mineralized tissues: a comparative study of biological crystal growth. CRC Crit. Rev. Biochem. 20, 365–408 [DOI] [PubMed] [Google Scholar]
- 60. Treccani L., Khoshnavaz S., Blank S., von Roden K., Schulz U., Weiss I. M., Mann K., Radmacher M., Fritz H. M. (2005) in Biopolymers Online (Fahnestock S. R., Steinbüchel A., eds) pp. 289–298, Wiley-VCH, New York [Google Scholar]
- 61. Ameye L., De Becker G., Killian C., Wilt F., Kemps R., Kuypers S., Dubois P. (2001) Proteins and saccharides of the sea urchin organic matrix of mineralization: characterization and localization in the spine skeleton. J. Struct. Biol. 134, 56–66 [DOI] [PubMed] [Google Scholar]
- 62. Luquet G., Marin F. (2004) Biomineralisations in crustaceans: storage strategies. Comptes Rendus Palevol. 3, 515–534 [Google Scholar]
- 63. Mann K., Poustka A. J., Mann M. (2010) Phosphoproteomes of Strongylocentrotus purpuratus shell and tooth matrix: identification of a major acidic sea urchin tooth phosphoprotein, phosphodontin. Proteome Sci. 8, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Furuhashi T., Schwarzinger C., Miksik I., Smrz M., Beran A. (2009) Molluscan shell evolution with review of shell calcification hypothesis. Comp. Biochem. Physiol. B 154, 351–371 [DOI] [PubMed] [Google Scholar]
- 65. Borbas J. E., Wheeler A. P., Sikes C. S. (1991) Molluscan shell matrix phosphoproteins: correlation of degrees of phosphorylation to shell mineral microstructure and to in vitro regulation of mineralization. J. Exp. Zool. 258, 1–13 [Google Scholar]
- 66. Hecker A., Testenière O., Marin F., Luquet G. (2003) Phosphorylation of serine residues is fundamental for the calcium-binding ability of Orchestin, a soluble matrix protein from crustacean calcium storage structures. FEBS Lett. 535, 49–54 [DOI] [PubMed] [Google Scholar]
- 67. Testenière O., Hecker A., Le Gurun S., Quennedey B., Graf F., Luquet G. (2002) Characterization and spatiotemporal expression of orchestin, a gene encoding an ecdysone-inducible protein from a crustacean organic matrix. Biochem. J. 361, 327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sarashina I., Endo K. (2001) The complete primary structure of molluscan shell protein 1 (MSP-1), an acidic glycoprotein in the shell matrix of the scallop Patinopecten yessoensis. Mar. Biotechnol. 3, 362–369 [DOI] [PubMed] [Google Scholar]
- 69. Allemand D., Tambutté E., Girard J. P., Jaubert J. (1998) Organic matrix synthesis in the scleractinian coral Stylophora pistillata: role in biomineralization and potential target of the organotin tributyltin J. Exp. Biol. 201, 2001–2009 [DOI] [PubMed] [Google Scholar]
- 70. Weiner S., Traub W., Lowenstam M. A. (1983) in Biomineralization and Biological Metal Accumulation (Westbrock P., De Jong E. W., eds) pp. 205–224, Reidel Publishing Company, Dordrecht, The Netherlands [Google Scholar]
- 71. Kingsley R. J., Watabe N. (1982) Ultrastructure of the axial region in Leptogorgia virgulata (Cnidaria: Gorgonaceae). Trans. Am. Microsc. Soc. 101, 325–339 [Google Scholar]
- 72. Weiner S., Addadi L. (1997) Design strategies in mineralized biological materials. J. Mater. Chem. 5, 689–702 [Google Scholar]
- 73. Marin F., Luquet G. (2004) Molluscan shell proteins. Comptes Rendus Palevol. 3, 469–492 [Google Scholar]
- 74. Marie B., Le Roy N., Zanella-Cléon I., Becchi M., Marin F. (2011) Molecular evolution of mollusc shell proteins: insights from proteomic analysis of the edible mussel Mytilus. J. Mol. Evol. 72, 531–546 [DOI] [PubMed] [Google Scholar]
- 75. Marie B., Trinkler N., Zanella-Cleon I., Guichard N., Becchi M., Paillard C., Marin F. (2011) Proteomic identification of novel proteins from the calcifying shell matrix of the Manila clam Venerupis philippinarum. Mar. Biotechnol. 13, 955–962 [DOI] [PubMed] [Google Scholar]
- 76. Marie B., Zanella-Cléon I., Guichard N., Becchi M., Marin F. (2011) Novel proteins from the calcifying shell matrix of the Pacific oyster Crassostrea gigas. Mar. Biotechnol. 13, 1159–1168 [DOI] [PubMed] [Google Scholar]
- 77. Sethmann I., Helbig U., Worheide G. (2007) Octocoral sclerite ultrastructures and experimental approach to underlying biomineralisation principles. Cryst. Eng. Comm. 9, 1262–1268 [Google Scholar]
- 78. Segev N., Tokarev A. A., Alfonso A. (2009) in Trafficking Inside Cells Pathways, Mechanisms, and Regulation (Segev N., ed) pp. 1–12, Landes Bioscience and Springer Science+Business Media, New York [Google Scholar]
- 79. Capasso J. M., Keenan T. W., Abeijon C., Hirschberg C. B. (1989) Mechanism of phosphorylation in the lumen of the Golgi apparatus: translocation of adenosine 5′-triphosphate into Golgi vesicles from rat liver and mammary gland. J. Biol. Chem. 264, 5233–5240 [PubMed] [Google Scholar]
- 80. Anitei M., Hoflack B. (2011) Exit from the trans-Golgi network: from molecules to mechanisms. Curr. Opin. Cell Biol. 23, 443–451 [DOI] [PubMed] [Google Scholar]
- 81. Allemand D., Bénazet-Tambutté S. (1996) Dynamics of calcification in the Mediterranean red coral, Corallium rubrum (Linnaeus) (Cnidaria, Octocorallia). J. Exp. Zool. 276, 270–278 [Google Scholar]
- 82. Marschal C., Garrabou J., Harmelin J. G., Pichon M. (2004) A new method for measuring growth and age in the precious red coral Corallium rubrum (L.). Coral Reefs 23, 423–432 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.