Background: RNAs modulate immune responses. Neutrophils represent the major fraction of immune cells, but receptors by which neutrophils sense RNA are poorly characterized.
Results: Neutrophils and differentiated HL-60 cells express the RNA receptors RIG-I, MDA-5, and TLR8. RIG-I and MDA-5 are localized in secretory vesicles.
Conclusion: Neutrophils express a distinct pattern of RNA receptors.
Significance: RNA receptors on neutrophils could have implications for RNA-based therapeutics.
Keywords: Cell Surface Receptor, Inflammation, Innate Immunity, Neutrophil, RNA, MDA-5, RIG-I, Secretory Vesicles
Abstract
RNAs are capable of modulating immune responses by binding to specific receptors. Neutrophils represent the major fraction of circulating immune cells, but receptors and mechanisms by which neutrophils sense RNA are poorly defined. Here, we analyzed the mRNA and protein expression patterns and the subcellular localization of the RNA receptors RIG-I, MDA-5, TLR3, TLR7, and TLR8 in primary neutrophils and immortalized neutrophil-like differentiated HL-60 cells. Our results demonstrate that both neutrophils and differentiated HL-60 cells express RIG-I, MDA-5, and TLR8 at the mRNA and protein levels, whereas TLR3 and TLR7 are not expressed at the protein level. Subcellular fractionation, flow cytometry, confocal laser scanning microscopy, and immuno-transmission electron microscopy provided evidence that, besides the cytoplasm, RIG-I and MDA-5 are stored in secretory vesicles of neutrophils and showed that RIG-I and its ligand, 3p-RNA, co-localize at the cell surface without triggering neutrophil activation. In summary, this study demonstrates that neutrophils express a distinct pattern of RNA recognition receptors in a non-canonical way, which could have essential implications for future RNA-based therapeutics.
Introduction
In addition to their role in transcription, RNAs have been shown to demonstrate immunomodulatory potential (1). Immunostimulatory RNAs (isRNAs)2 are known to generate a potent immune response and are currently under intense investigation for new antiviral and anticancer treatment strategies (2–5). Recent evidence suggests that leukocytes recognize isRNA through cytosolic receptors (6–8). The relevance of cytosolic RNA receptors is highlighted by the fact that siRNAs engage similar pathways (3, 4). RIG-I (retinoic acid-inducible gene I; also known as DDX58) was identified as a key candidate receptor for cytoplasmic viral RNA detection (7–9). Together with MDA-5 (melanoma differentiation-associated gene 5) and LGP2 (laboratory of genetics and physiology 2), RIG-I forms the family of RIG-like helicases (RLHs) based on their high similarities among helicase domains (9). Activation of these receptors by RNA triggers production of type I IFNs (1, 10). For instance, RLHs interact with dsRNAs through their helicase domain, and dsRNA stimulation induces their ATP catalytic activity. The N-terminal caspase recruitment domains are then responsible for activating downstream signaling pathways that mediate, for example, dsRNA-induced type I IFN production (2). 5′-Triphosphorylated and uncapped viral RNAs are recognized by RIG-I (6, 7). MDA-5 is thought to respond to uncapped, 5′-unmodified dsRNA, such as poly(I:C) (6).
One promising therapeutic approach is to stimulate RNA receptors with modified siRNA to knock down cancer-related target genes and simultaneously boost the innate immune system (4). Poeck et al. (3) showed simultaneous knockdown of Bcl-2 and RIG-I-dependent IFN induction to be superior to either therapy alone utilizing an in vivo melanoma mouse model. Moreover, mRNAs have recently gained further attention as potential tools for gene therapy (11). However, evidence regarding the possible side effects of such RNA-based therapies is still scarce. Stimulating off-target cells in the circulation, such as neutrophils (polymorphonuclear leukocytes), may limit the therapeutic potential of RNA in vivo.
Neutrophils represent the major fraction of circulating immune cells and provide the first cellular line of antibacterial and antifungal host defense, whereas their potential contribution to viral infections is rather enigmatic. Neutrophils sense pathogens through recognition of pathogen-associated molecular patterns by innate pattern recognition receptors, such as the prototypical family of Toll-like receptors (TLRs). Previous studies showed that primary neutrophils express a broad range of bacterial recognition TLRs but lack expression of TLR3 and TLR7 (12, 13). Notably, TLR8 was found to be expressed and was functional in mediating neutrophil effector responses (13). Another study showed that neutrophils express RNA receptors of the RLH family and are capable of responding to intracellularly delivered poly(I:C), suggesting that neutrophils could recognize viral RNA through helicases and may play a role in antiviral immunity (21). Aside from these studies, the expression, subcellular localization, and functionality of RNA receptors in primary neutrophils and immortalized neutrophil-like differentiated HL-60 (dHL-60) cells are still poorly defined.
EXPERIMENTAL PROCEDURES
Isolation and Culture of Human Neutrophils
Peripheral blood was obtained from healthy volunteers in accordance with the Institutional Review Board and as approved by the Ethical Committee of the Ludwig Maximilians University of Munich and the University of Tübingen. After ammonium chloride erythrocyte lysis, human neutrophils and peripheral blood mononuclear cells (PBMCs) were prepared by Ficoll-Hypaque density gradient centrifugation of heparinized blood from healthy volunteers. Neutrophils were additionally positive-enriched using anti-CD16 MicroBeads (magnetic cell sorting, Miltenyi Biotec). Evaluation of the neutrophil population by FACS showed purities of 85% (S.E., 2.5%) for isolation with Ficoll-Hypaque density gradient centrifugation and 95% (S.E., 2%) for additional cell sorting with anti-CD16 MicroBeads. Cells were cultured in RPMI 1640 medium (Biochrom) supplemented with 10% FCS, 10 mm HEPES (Sigma-Aldrich), 1.5 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (all from PAA Laboratories) in 96-well round-bottom plates for stimulation experiments.
Isolation of Murine Neutrophils
To isolate murine neutrophils, bone marrow cells or splenic cells from healthy C57BL/6 mice were obtained, suspended in PBS, and washed. Neutrophils were then isolated as described above. The purity of the neutrophil populations analyzed by FACS was 93% (S.E., 3%). Animal studies were approved by the local regulatory agency (Regierung von Oberbayern, Munich, Germany).
HL-60 Cells
HL-60 cells were passaged at 3-day intervals to maintain exponential growth, and all experiments were performed with cells between passages 35 and 60. dHL-60 cells were obtained by treatment of 5 × 105 HL-60 cells/ml with 1.25% dimethyl sulfoxide (DMSO) for 7 days as described previously (14). The cells obtained were neutrophilic based on microscopic and flow cytometric criteria as described previously (15).
Subcellular Fractionation of Neutrophils
Subcellular fractionation of neutrophils was performed by nitrogen cavitation and sedimentation of the post-nuclear supernatant on a four-layer Percoll density gradient as described previously in detail by Clemmensen et al. (16). In brief, neutrophils isolated from peripheral blood were resuspended in disruption buffer (100 mm KCl, 3 mm NaCl, 1 mm Na2ATP, 3.5 mm MgCl2, and 10 mm PIPES, pH 7.2) with a protease inhibitor mixture (11836153001, Roche Applied Science) added as recommended by the manufacturer. Neutrophils were disrupted by nitrogen cavitation at 600 p.s.i. for 5 min and collected in 1.5 mm EGTA. Furthermore, the cavitate was centrifuged at 400 × g for 15 min to remove nuclei and unbroken cells, and the supernatant was added to a Percoll solution with a density of 1.11 g/ml at a ratio of 1:1, resulting in a final density of 1.055 g/ml. Next, 9 ml was layered on top of a two-layer Percoll gradient (9 ml with a density of 1.12 g/ml and with 9 ml with a density of 1.09 g/ml) to separate azurophilic, specific, and gelatinase granules. Then, 9 ml of Percoll solution with a density of 1.03 g/ml was layered on top to create a flotation medium for separation of plasma membranes/cytosol and secretory vesicles. The pH was adjusted to 7.0 with HCl. The four-layer gradient was centrifuged at 20,000 × g for 40 min, resulting in five major bands: the α-band enriched in primary/azurophilic granules (marker, myeloperoxidase), the β1-band enriched in secondary/specific granules (marker, neutrophil gelatinase-associated lipocalin), the β2-band enriched in tertiary/gelatinase granules (marker, gelatinase/MMP-9), the γ1-band enriched in secretory vesicles (marker, albumin), and the γ2-band containing plasma membranes (marker, human leukocyte antigen). Samples were subjected to ELISA analysis or to SDS-PAGE and Western blot analysis. Myeloperoxidase, neutrophil gelatinase-associated lipocalin, MMP-9, human serum albumin, and human leukocyte antigen were quantified in each fraction by ELISA and used as marker proteins for azurophilic granules, specific granules, gelatinase granules, secretory vesicles, and the plasma membrane, respectively (17). Where indicated, a three-layer Percoll gradient was used (17).
Western Blotting
Neutrophil fractions were separated on NuPAGE 4–12% Bis-Tris gels (Invitrogen), and immunoblotting was performed by standard procedures using XCell II blotting chambers (Invitrogen). After blocking, primary antibody against RIG-I or MDA-5 (both from Abcam) was incubated overnight. Blots were processed using alkaline phosphatase-conjugated secondary antibodies (Millipore) and 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium solution (Millipore). Semiquantitative analysis was performed with the Quantity One software system (Bio-Rad).
Stimulation of Cells
All TLR ligands were titrated to define optimal stimulating conditions (activation of cells without affecting cell survival/apoptosis). Neutrophils were stimulated with LPS from Escherichia coli (Sigma-Aldrich), phorbol 12-myristate 13-acetate (Sigma-Aldrich), 3M-003 (3M Pharmaceuticals), and poly(I:C) (Sigma-Aldrich). Poly(I:C) was subjected to 10 precipitations and washes in ethanol to remove endotoxin. The final concentrations of TLR ligands used for stimulation are given below. For stimulation of TLR7 and TLR8, we used isRNA9.2as (5′-UUGAAGGACAGGUUAAGCUdTdT-3′; synthesized by Eurogentec Deutschland GmbH). Upon stimulation, we complexed nucleic acid (200 ng) using the polycationic polypeptide poly-l-arginine (P7762, Sigma-Aldrich). As a control, we used non-stimulatory poly(A) RNA repeats (Eurogentec Deutschland GmbH). Details of the synthesized stimulants are given in Table 1.
TABLE 1.
Stimulants used in this study
| Stimulant | Target | Sequence |
|---|---|---|
| Stimulants synthesized by Eurogentec | ||
| Poly(A) (control) | 5′-AAAAAAAAAAAAAAAAAAA-3′ | |
| RNA9.2s, unmodified | TLR7/8 | 5′-AGCUUAACCUGUCCUUCA-3′ |
| Stimulants synthesized by Ambion | ||
| siRNA (OHBcl2_2) | RIG-I | 5′-GCAUGCGACCUCUGUUUGAUU-3′ (sense) |
| 5′-UCAAACAGAGGUCGCAUGCUU-3′ (antisense) | ||
| siRNA, mismatched (control) | 5′-UUCUCCGAACGUGUCACGUUU-3′ (sense) | |
| 5′-ACGUGACACGUUCGGAGAAUU-3′ (antisense) | ||
| In house-synthesized stimulants | ||
| 3p-RNA | RIG-I | 5′-pppGCAUGCGACCUCUGUUUGAUU-3′ (sense) |
| 5′-pppUCAAACAGAGGUCGCAUGCUU-3′ (antisense) | ||
| 3p-RNA (control) | 5′-pppUUCUCCGAACGUGUCACGUUU-3′ (sense) | |
| 5′-pppACGUGACACGUUCGGAGAAUU-3′ (antisense) |
Generation of 3p-RNA
For generation of in vitro transcribed double-stranded RNA, the DNA templates of the sense and antisense strands were transcribed for 6 h in separate reactions. An extra guanosine was added to the 5′-ends of both the sense and antisense strands to transcribe with T7 RNA polymerase. The reactions were then mixed and incubated overnight at 37 °C to anneal the transcribed RNA strands. The DNA template was digested using DNase I (Ambion), and RNAs were subsequently purified by phenol/chloroform extraction and alcohol precipitation. Excess salts and NTPs were removed by passing the RNAs through a mini Quick SpinTM Oligo column (Roche Applied Science). To test the immunostimulatory potential of 3p-RNA, T2 cells were incubated for 24 h, and MHC-I expression was compared with OH siRNA with the same nucleotide sequence.
RNA Isolation, cDNA Synthesis, and Quantitative Real-time PCR
For RNA preparation, cells were washed with 0.9% sodium chloride, and cell pellets were lysed in 300 μl of lysis buffer from the MagNA Pure LC mRNA isolation kit I (Roche Applied Science) supplemented with 1% DTT and frozen at 80 °C until further handling. Preparation of mRNA was performed with the MagNA Pure LC device using the mRNA I standard protocol. An aliquot of 8.2 μl of RNA was reverse-transcribed using avian myeloblastosis virus reverse transcriptase and oligo(dT) as the primer (first strand cDNA synthesis kit, Roche Applied Science) according to the manufacturer's protocol in a thermocycler. The reaction mixture was diluted to a final volume of 0.5 ml and stored at 20 °C until PCR analysis. Parameter-specific primer sets optimized for the LightCycler (Roche Applied Science) were developed and purchased from Search-LC GmbH (Table 2). PCR was performed with the LightCycler FastStart DNA SYBR Green I kit (Roche Applied Science) according to the protocol provided in the parameter-specific kits. The transcript numbers were normalized according to the expression of TATA box-binding protein/μl of cDNA.
TABLE 2.
Sequences
| Gene | Sequence |
|
|---|---|---|
| Forward | Reverse | |
| TLR3 | TCCCAAGCCTTCAACGACTG | TGGGTGAAGGAGAGCTATCCACA |
| TLR7 | TTACCTGGATGGAAACCAGCTAC | TCAAGGCTGAGAAGCTGTAAGCTA |
| TLR8 | GAGAGCCGAGACAAAAACGTTC | TGTCGATGATGGCCAATCC |
| MDA-5 | ATGTGGCAGCAAGAGCATCC | GGTAAGGCCTGAGCTGGAGTT |
Flow Cytometry
Flow cytometric data were obtained on a FACSCalibur flow cytometer and analyzed with CellQuest software (BD Biosciences). Staining was performed following standard procedures. The monoclonal antibodies used were FITC-conjugated anti-CD62L and allophycocyanin-conjugated anti-CD11b (BioLegend); anti-RIG-I and FITC-conjugated anti-TLR7 (Santa Cruz Biotechnology); anti-MDA-5 (Abcam); and phycoerythrin-conjugated anti-TLR8, FITC-conjugated goat anti-rabbit secondary Ab, and FITC-conjugated donkey anti-rat secondary Ab (Imgenex Corp.). Appropriate isotype controls were used for all applied detection antibodies. For intracellular cytokine detection, cells were preincubated with brefeldin A at 10 μg/ml. After surface staining, cells were fixed with 2% paraformaldehyde, subsequently permeabilized with 0.1% saponin, and then stained for intracellular proteins.
Confocal Microscopy
For visualization of TLR8, MDA-5, and RIG-I, the samples were incubated with rabbit anti-human TLR8 (clone 44C143; Imgenex), anti-MDA-5 polyclonal (Abcam), or anti-RIG-I polyclonal (Santa Cruz Biotechnology) antibody. The primary antibodies were detected by confocal laser scanning microscopy using Alexa Fluor 555-conjugated anti-rabbit secondary antibody (Invitrogen). DNA was stained with DAPI (Sigma-Aldrich), and concanavalin A-Alexa Fluor 488 conjugate (Invitrogen) was used for detecting glycoconjugates in the cytoplasm. The specimens were analyzed with an Olympus IX51 confocal laser scanning microscope.
Immuno-transmission Electron Microscopy
Isolated peripheral blood neutrophils were fixed, washed with PBS, permeabilized, and blocked (10% normal goat serum, 10 mm glycine, and 0.2% Tween 20 in diluent containing 0.5% bovine serum albumin and 0.5% Triton X-100 in PBS). For visualization of RIG-I, the grids were incubated with anti-RIG-I antibody and a gold-conjugated secondary antibody (gold sphere diameter, 5 nm; ab27237, Abcam). Finally, the grids were stained with 1% uranyl acetate (Sigma-Aldrich). Negative controls were obtained by omitting the primary antibody.
RNA-FITC Labeling
RNA was labeled using an N-hydroxysuccinimidyl-fluorescein labeling kit (Thermo Scientific) according to the manufacturer's instructions. Briefly, 3p-RNA was incubated with the N-hydroxysuccinimidyl-fluorescein dye for 2 h on ice. Excessive dye was removed by Zeba desalt spin columns (Thermo Scientific) according to the manufacturer's instructions. The degree of fluorescein labeling was determined by photometry.
ELISA
ELISA kits (Invitrogen) were used to quantify TNF-α levels in cell culture supernatants 40 min after stimulation. Protocols were performed according to the manufacturer's recommendations.
Statistical Analysis
Data are presented as means ± S.E. The statistical significance of differences was determined by Student's paired two-tailed t test. In all tests, differences were considered significant at p < 0.05.
RESULTS
TLR8, MDA-5, and RIG-I Are Expressed by Both Primary Neutrophils and Immortalized Neutrophil-like dHL-60 Cells
We analyzed TLR3, TLR7, TLR8, MDA-5, and RIG-I expression at the mRNA and protein levels in isolated primary neutrophils by RT-PCR, flow cytometry, and confocal microscopy (Fig. 1). Quantitative RT-PCR showed significant TLR8, MDA-5, and RIG-I expression levels, with the highest mRNA expression for TLR8, whereas TLR3 or TLR7 expression was below the detection limit (Fig. 1A, upper panel). In line with mRNA studies, we observed a robust protein expression of TLR8 but could not detect any expression of TLR3 and TLR7 by human neutrophils at the protein level (Fig. 1A, lower panel). In contrast to the low/moderate mRNA expression levels, protein expression of MDA-5 and RIG-I in neutrophils was higher compared with TLR8 expression. Comparison of the intracellular/cytoplasmic and cell surface/membranous expression sites showed that MDA-5 and RIG-I protein expression was detectable in both cytoplasmic and cell surface/membrane-associated compartments, whereas TLR8 seemed to be restricted mainly to the cytoplasm (Fig. 1A, lower panel, and Fig. 1C, merged images). Confocal microscopy showed a more pronounced intracellular staining pattern for MDA-5 and RIG-I compared with flow cytometry (Fig. 1A, lower panel, and Fig. 1C).
FIGURE 1.
Expression of RNA receptors by neutrophils and HL-60 cells. A, upper panel, isolated neutrophils were analyzed by quantitative real-time PCR for the expression profiles of TLR3, TLR7, TLR8, RIG-I, and MDA-5. TATA box-binding protein was used as a reference for expression. The mean ± S.E. of six different healthy donors is shown. Lower panel, intracellular (white bars) and cell surface (gray bars) expression of TLR3, TLR7, TLR8, MDA-5, and RIG-I in isolated human neutrophils as analyzed by flow cytometry. Hatched bars indicate experiments in which isolated neutrophils were stimulated for 10 min with 40 ng/ml phorbol 12-myristate 13-acetate at 37 °C. *, p < 0.05. The mean ± S.E. of six different healthy donors is shown. MFI, mean fluorescence intensity. B, upper panel, cultured HL-60 cells were either untreated or treated with DMSO for 5 days and analyzed by quantitative real-time PCR for the expression profiles of TLRs, RIG-I, and MDA-5. TATA box-binding protein was used as a reference for expression. *, p < 0.05. Lower panel, intracellular (white bars) and cell surface (gray bars) expression of TLR3, TLR7, TLR8, MDA-5, and RIG-I in cultured HL-60 cells untreated or treated with DMSO for 5 days as analyzed by flow cytometry. Hatched bars indicate experiments with DMSO treatment. *, p < 0.05. C, the subcellular localization of TLR8, MDA-5, and RIG-I (red); DAPI (DNA; blue); and cytosolic proteins (concanavalin A (ConA); green) is shown in isolated neutrophils using confocal microscopy.
Next, we tested whether neutrophil stimulation by PKC activation changes the expression pattern/cellular distribution of the analyzed RNA receptors. Stimulation of neutrophils with the PKC activator phorbol 12-myristate 13-acetate (40 ng/ml) led to an increase in intracellularly detectable MDA-5 receptor pools, whereas RIG-I surface expression levels decreased upon phorbol 12-myristate 13-acetate stimulation (Fig. 1A, lower panel), suggesting cell activation-induced redistribution/translocation mechanisms.
Because of the limited life span of primary neutrophils ex vivo/in vitro, dHL-60 cells, a leukemia cell line with characteristics and differentiation markers similar to those of human granulocytes, are frequently used to study neutrophil-like responses in cell culture conditions. Stimulation of HL-60 cells with DMSO results in differentiation to mature granulocytic cells. We compared the expression of RNA receptors in undifferentiated and DMSO-stimulated dHL-60 cells. Real-time PCR analysis showed that dHL-60 cells expressed a similar RNA receptor repertoire as primary human neutrophils did, with the exception of positive expression of TLR3 detectable in dHL-60 cells, which was completely absent in primary neutrophils (Fig. 1C). Further studies showed that differentiation of HL-60 cells with DMSO decreased the mRNA expression levels of TLR3 and TLR8, whereas those of MDA-5 and RIG-I increased. Protein expression analysis by flow cytometry demonstrated TLR8, MDA-5, and RIG-I expression in HL-60 cells. Similar to human neutrophils, TLR3 and TLR7 were completely absent at the protein level. Intracellular flow cytometric staining indicated that cytoplasmic TLR8, MDA-5, and RIG-I expression levels did not change upon DMSO differentiation. In contrast, surface MDA-5 and RIG-I expression levels were decreased or nearly lost, respectively, after DMSO differentiation. These studies demonstrate that both primary human neutrophils and neutrophil-like dHL-60 cells express a distinct set of RNA receptors, namely TLR8, MDA-5, and RIG-I.
In-depth Characterization and Subcellular Localization of RIG-I and MDA-5 in Neutrophils
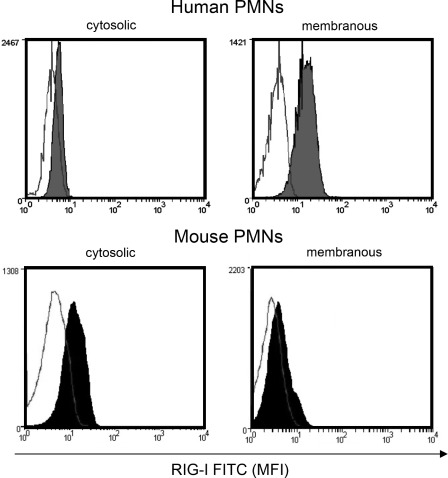
In contrast to the intracellular localization reported for other cell types (6, 9, 18, 19), we initially observed that RIG-I and MDA-5 protein expression in human neutrophils seemed not to be restricted to intracellular sites but was also detectable in the cell surface/membrane-associated compartments (Fig. 1, lower panels in A and C). As determined by flow cytometry, isotype control-corrected mean fluorescence intensities of MDA-5 and RIG-I were equal or higher, respectively, on the cell surface compared with intracellular pools, independent of the blood donor or cell fixation/permeabilization protocols used (Fig. 1A) (data not shown). We used two different specific antibodies with distinct RIG-I-binding sites to reduce the risk for unspecific binding. Both antibodies yielded similar staining results. We found a substantially higher RIG-I surface expression in human versus murine neutrophils, with RIG-I being expressed mainly in the cytosol of murine neutrophils (Fig. 2).
FIGURE 2.
Localization of RIG-I in human and murine neutrophils. Shown is the cytosolic and membranous RIG-I expression in human and murine neutrophils. For both experiments, a representative histogram of six independent experiments is shown. Intracellular flow cytometry was performed using cell permeabilization as described under “Experimental Procedures.” PMNs, polymorphonuclear leukocytes; MFI, mean fluorescence intensity.
However, because these observations were based mainly on flow cytometric detection assays, we decided to study the subcellular localization of these RLH proteins in neutrophils in more depth by comparing different protein detection and localization methods. For this purpose, previously established subcellular fractionation methods were used (16, 17). The subcellular components were stained for RIG-I or MDA-5 and analyzed separately by Western blotting (Fig. 3, A and B). As a positive control, autologous PBMCs were utilized, which are well known to express cytosolic RIG-I and MDA-5 (7–9). These experiments demonstrated that human neutrophils expressed both RIG-I and MDA-5 with a similar molecular mass compared with PBMCs (Fig. 3B). Remarkably, RIG-I and MDA-5 in neutrophils were detectable in fractions characteristic for secretory vesicles and plasma membranes, whereas the RLH proteins were absent in primary/azurophilic, secondary/specific, or tertiary/gelatinase granule fractions. Besides secretory vesicles, the proteins were detectable in cytosolic fractions. In immuno-transmission electron microscopy, RIG-I proteins were frequently associated with membranous/vesicle-like structures (Fig. 3C, red arrows). When viewed in combination, these experiments provide evidence that, besides the well known cytoplasmic expression sites reported for a variety of cell types, human neutrophils store the RLHs RIG-I and MDA-5 in secretory vesicles. Because secretory vesicles communicate and fuse with the plasma membrane, we speculate that the vesicular localization of the RLHs in neutrophils could explain their appearance on the cell surface, an issue necessitating further investigation.
FIGURE 3.
Subcellular localization of RIG-I in human neutrophils. A, resting neutrophils were gently disrupted using the nitrogen cavitation method, followed by four-layer gradient subcellular fractionation. Shown are the distribution profiles of myeloperoxidase (MPO; marker of azurophilic granules), neutrophil gelatinase-associated lipocalin (NGAL; marker of secondary/specific granules), MMP-9 (marker of tertiary granules), human serum albumin (marker of secretory vesicles), and MICA/human leukocyte antigen (marker of plasma membranes) as measured by ELISA. The y axis represents an arbitrary scale, where the highest measured value of each protein is normalized to 1. B, Western blot analysis of neutrophil subcellular fractions in autologous neutrophils (polymorphonuclear leukocytes (PMN)) and PBMCs. Upper panel, four-layer fractionation and MDA-5 immunoblotting. Middle panel, four-layer fractionation and RIG-I immunoblotting. PM, plasma membranes; SV, secretory vesicles; 3°, tertiary granules; 2°, tertiary granules; 1°, primary granules. Lower panel, three-layer fractionation and RIG-I immunoblotting. Cyt., cytosol. C, immuno-transmission electron microscopy. RIG-I was immunolabeled with 5-nm gold particles. Arrows indicate RIG-I staining. The red arrows mark RIG-I associated with a vesicle-like structure.
Finally, we investigated whether cell membrane-associated RIG-I binds to its ligand, 3p-RNA, and mediates downstream activation. We designed 3p-RNA containing a triphosphate chemical modification at the 5′-ends of both strands as described previously (2, 4). Confocal microscopy provided evidence that FITC-labeled 3p-RNA co-localized, at least partially, at the cell surface of neutrophils with RIG-I (Fig. 4A). Therefore, we asked whether binding of 3p-RNA to membrane-associated RIG-I triggers neutrophil activation. We used LPS (ligand for TLR4), R-848 (ligand for TLR7/TLR8), 3p-RNA, a double-stranded siRNA (with identical sequence but without the triphosphate modification of 3p-RNA), isRNA9.2s, and Lipofectamine or poly-l-arginine (RNA-based ligand for TLR7/TLR8) for complexation and as controls (see Table 1 for details). Neutrophil activation was analyzed by CD62L shedding (Fig. 4B), CD11b up-regulation (Fig. 4B), and TNF-α production (Fig. 4C). Both LPS and R-848 dose-dependently decreased CD62L surface expression, triggered CD11b up-regulation, and elicited TNF-α production. Whereas LPS activated neutrophils at a concentration of 0.1 μg/ml, cell activation elicited by R-848 required higher concentrations. Uncomplexed 3p-RNA or siRNA had no effects on neutrophil activation, whereas complexed RNAs had a slight, but non-significant effect at higher concentrations on CD62L shedding (Fig. 4B) (data not shown). In particular, isRNA9.2s complexed with poly-l-arginine triggered CD62L shedding, but this effect was also observed for poly-l-arginine alone (Fig. 4B, lower panel).
FIGURE 4.
Effect of RNAs on neutrophils. A, neutrophils were isolated and stained for co-localization of RIG-I (red), DAPI (DNA; blue), and FITC-labeled 3p-RNA (green). B, isolated neutrophils were stimulated at the indicated concentrations with LPS, R-848, complexed 3p-RNA (complexed with Lipofectamine), uncomplexed 3p-RNA (without Lipofectamine), siRNA (complexed with Lipofectamine), isRNA9.2s (complexed with poly-l-arginine), poly(A) RNA (complexed with poly-l-arginine), Lipofectamine only, or poly-l-arginine only. Forty minutes after stimulation, CD62L shedding and CD11b up-regulation were examined by flow cytometry. The mean ± S.E. of three donors is shown. MFI, mean fluorescence intensity. C, isolated neutrophils were stimulated for 40 min with the indicated reagents, and supernatants were analyzed for TNF-α production by ELISA. siRNA was complexed with poly-l-arginine, and 3p-RNA was complexed with Lipofectamine. p < 0.05.
DISCUSSION
Our results demonstrate that both primary human neutrophils and immortalized dHL-60 cells express the RNA receptors TLR8, RIG-I, and MDA-5 at the mRNA and protein levels. These studies further demonstrate that both RLHs RIG-I and MDA-5 are stored in secretory vesicles in neutrophils, suggesting that circulating granulocytes carry an intracellular reservoir of RNA receptors, reminiscent of other vesicle-stored receptors, such as CR1/CD35, Mac-1, CD13, and CD16. Further studies are required to understand the subcellular regulation and functional role of these receptors in the context of RNA recognition and their relevance for in vivo disease conditions.
Neutrophils are a substantial part of the innate immune system and recognize pathogens through pattern recognition receptors, such as TLRs. These professional phagocytes are usually the first cell population to be involved in an immune response and subsequently modulate the response not only due to their potential to directly eliminate pathogens by phagocytosis but also by the induction of modulatory chemokines/cytokines such as CXCL8 (IL-8) and others. Previous studies assessing TLRs in granulocytes (13) demonstrated that primary neutrophils express TLR7 (12) and TLR8 (12, 13) and respond to TLR8 ligands functionally (12, 13). Beyond TLRs, Ekman and Cardell (20) showed that neutrophils express functionally active non-TLR pattern recognition receptors, particularly NOD-like receptors. Despite these studies, our understanding of pattern recognition receptors mediating RNA recognition in these immune cells is still limited. In this study, we comprehensively characterized TLRs and non-TLR RNA receptors in primary neutrophils and immortalized neutrophil-like cells at the mRNA and protein levels. Using this approach, we detected no protein expression of TLR3 and TLR7 in human neutrophils, regardless of their state of activation. TLR3 mRNA was expressed in untreated HL-60 cells but was decreased upon DMSO differentiation. As for human neutrophils, TLR7 was not expressed in HL-60 cells. On the other hand, our results provide evidence for the substantial expression of TLR8 in human neutrophils, which is in line with a previous study by Janke et al. (13). In HL-60 cells, TLR8 mRNA expression decreased upon differentiation. We have currently no explanation for this phenomenon. When viewed in combination, our studies indicate that HL-60 cells express, with the exception of TLR3 mRNA, a similar RNA receptor expression pattern as primary human neutrophils do and may therefore represent a useful tool in studying RNA receptor expression and regulation in an immortalized cell line.
Canonically, RIG-I is regarded as an intracellular RNA receptor because cytosolic expression has been reported for dendritic cells, monocytes, mesothelial cells, monocytes/PBMCs, and other cell types (1, 9, 18, 22). In line with these previously reported cell types, we detected RIG-I protein expression in cytosolic neutrophil fractions at a similar molecular mass as in PBMCs, which are well known to express cytosolic RIG-I. Unexpectedly, we also observed that RIG-I was detectable at the cell surface of human (but not murine) neutrophils. We currently have no explanation for this interspecies discrepancy in RIG-I surface expression, but we found this RIG-I staining pattern consistently in both murine bone marrow- and peripheral blood-derived neutrophils independent of the isolation procedure. However, because these initial surface expression findings were based mainly on flow cytometric assays, which critically depend on antibody specificity and accessibility, we studied the expression and subcellular localization of the RLH proteins RIG-I and MDA-5 in greater detail using subcellular fractionation and immunoblotting. These studies demonstrated that both RIG-I and MDA-5 were localized in secretory vesicles, an easily mobilizable compartment, and in cytoplasmic fractions of human neutrophils. In both compartments, the RLH proteins were detected at a similar molecular mass as in PBMCs. Immuno-transmission electron microscopy analyses supported these findings by showing that RIG-I protein in neutrophils was associated, at least partially, with vesicle-like structures. Based on these studies, it is conceivable that RIG-I and MDA-5, in analogy to other secretory vesicle-associated receptors, such as CR1/CD35, and Mac-1, shuttle between intracellular pools and the plasma membrane. The surface expression of RIG-I and MDA-5 observed by flow cytometry could be due to plasma membrane reorganization events after fusion with secretory vesicles, a process occurring during neutrophil activation or after cell isolation procedures. To define the precise subcellular localization and translocation/redistribution mechanisms of RLHs in neutrophil vesicles in greater detail, methods to separate plasma membrane vesicles from secretory vesicles, such as free-flow electrophoresis, followed by proteomic approaches, will be required.
The effects of TLR7 and TLR8 ligands on granulocytes have recently been investigated by Janke et al. (13). TLR8 was found to be highly expressed by neutrophils, whereas it was absent in eosinophils. Isolated human neutrophils were stimulated with isRNA or R-848, and neutrophils (but not eosinophils) could be activated through the TLR8 downstream pathway. However, this effect was abolished when the RNA backbone of isRNA consisted not of nuclease-stable phosphothioate but of regular phosphodiester binding. Our findings support the data of Janke et al. because we were unable to observe neutrophil activation using unstable isRNA with a phosphodiester backbone but observed robust activation elicited by the TLR7/TLR8-ligand R-848. Taken together, these data highlight the notion that TLR8 ligands, including nuclease-stable RNA, could bear limitations for clinical applicability of RNA-based therapies due to potential off-target effects.
In a previous study by Hornung et al. (6), 5′-triphosphate RNA was reported to be a potent inducer of IFN-α in human monocytes and plasmacytoid dendritic cells. In vitro transcription was used to generate a dsRNA oligonucleotide with an overhang of one nucleotide at the 5′-position. The two single-stranded oligonucleotides and the double-stranded oligonucleotide induced comparable levels of IFN-α in monocytes but not in plasmacytoid dendritic cells. Cleavage of the 5′-overhang (including the 5′-triphosphate) of the dsRNA or dephosphorylation of the 5′-end completely abrogated the IFN-α response. The authors also observed that plasmacytoid dendritic cells showed no decrease in IFN-α production when oligonucleotides were dephosphorylated. When viewed in combination, these results of Hornung et al. suggest that the 5′-triphosphate is at least one well defined structural feature responsible for the IFN-α-inducing activity of in vitro transcribed RNA in monocytes and that a 5′-triphosphate confers IFN-α-inducing activity to both single-stranded RNA and dsRNA. In addition, Hornung et al. demonstrated a direct binding of RIG-I by 5′-triphosphate RNA in HEK293 cells. In light of these findings, the expression of RIG-I in human neutrophils and the functional response of RIG-I on human neutrophils were investigated in our study by treating isolated human neutrophils with 5′-triphosphate RNA and monitoring RIG-I co-localization and neutrophil activation. We obtained evidence for the co-localization of RIG-I and its labeled ligand, 3p-RNA, on the cell surface on neutrophils by confocal microscopy, thereby taking the possibility of early RNA degradation into account. However, the conclusions based on these findings are limited because they rely solely on confocal laser scanning microscopy, warranting future in-depth analyses to characterize in detail the interaction of RIG-I and 3p-RNA in primary human neutrophils. Despite the observed ligand-receptor co-localization at the cell surface, we were unable to detect specific activation of neutrophils by the RIG-I ligand 5′-triphosphate RNA under the experimental conditions used in this study. Of note, we did not study the effects of RIG-I ligands on gene expression, which might be more sensitive. A previous study clearly demonstrated that transfection with poly(I:C), a synthetic mimetic of viral dsRNA activating TLR3, MDA-5, and/or RIG-I in a cell-type and length-specific manner, elicited an orchestrated immunoregulatory and antiviral gene expression program in primary neutrophils (21). Although our study did not include poly(I:C) transfection or analysis of antiviral gene expression patterns, both studies support the notion that RLH ligands, in particular poly(I:C) and 3p-RNA, are unable to elicit the secretion of proinflammatory cytokines despite increased mRNA transcripts, as found by Tamassia et al. (21). The underlying reasons remain to be investigated and have been previously discussed in more detail (21).
On the basis of previous studies showing that neutrophils express non-canonical receptors at their cell surface, as demonstrated for the HIV co-receptor CCR5, and employ these receptors as ligand-binding/scavenging instead of signaling receptors (23), we are tempted to speculate that secretory vesicle-derived RIG-I in neutrophils could serve as an RNA-binding and inactivation receptor at sites of neutrophilic inflammation, a hypothesis that remains to be tested using in vitro and in vivo models. Overall, the pathophysiological relevance of RNA receptors expressed by neutrophils in the modulation of immune responses remains a subject for future investigation.
Acknowledgments
We thank Stefan Endres (Department of Clinical Pharmacology, Ludwig Maximilians University of Munich) for support with RNA synthesis and Dr. Samuel Wagner for proofreading the manuscript.
This work was supported by Deutsche Forschungsgemeinschaft (DFG) Emmy Noether Programme Grant HA 5274/3-1, PINA e.V., and the Novartis Foundation (to D. H.); the German Academic Exchange Service (DAAD) and the Friedrich-Baur-Institut (to M. B.); and the German Society of Pediatric Pneumology (to D. H. and V. M.).
- isRNA
- immunostimulatory RNA
- RLH
- RIG-like helicase
- TLR
- Toll-like receptor
- dHL-60
- differentiated HL-60
- PBMC
- peripheral blood mononuclear cell
- DMSO
- dimethyl sulfoxide
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Hornung V., Guenthner-Biller M., Bourquin C., Ablasser A., Schlee M., Uematsu S., Noronha A., Manoharan M., Akira S., de Fougerolles A., Endres S., Hartmann G. (2005) Sequence-specific potent induction of IFN-α by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 11, 263–270 [DOI] [PubMed] [Google Scholar]
- 2. Castanotto D., Rossi J. J. (2009) The promises and pitfalls of RNA-interference-based therapeutics. Nature 457, 426–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Poeck H., Besch R., Maihoefer C., Renn M., Tormo D., Morskaya S. S., Kirschnek S., Gaffal E., Landsberg J., Hellmuth J., Schmidt A., Anz D., Bscheider M., Schwerd T., Berking C., Bourquin C., Kalinke U., Kremmer E., Kato H., Akira S., Meyers R., Häcker G., Neuenhahn M., Busch D., Ruland J., Rothenfusser S., Prinz M., Hornung V., Endres S., Tüting T., Hartmann G. (2008) 5′-Triphosphate siRNA: turning gene silencing and RIG-I activation against melanoma. Nat. Med. 14, 1256–1263 [DOI] [PubMed] [Google Scholar]
- 4. Schlee M., Hornung V., Hartmann G. (2006) siRNA and isRNA: two edges of one sword. Mol. Ther. 14, 463–470 [DOI] [PubMed] [Google Scholar]
- 5. Hornung V., Barchet W., Schlee M., Hartmann G. (2008) RNA recognition via TLR7 and TLR8. Handb. Exp. Pharmacol. 183, 71–86 [DOI] [PubMed] [Google Scholar]
- 6. Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K. K., Schlee M., Endres S., Hartmann G. (2006) 5′-Triphosphate RNA is the ligand for RIG-I. Science 314, 994–997 [DOI] [PubMed] [Google Scholar]
- 7. Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K. J., Yamaguchi O., Otsu K., Tsujimura T., Koh C. S., Reis e Sousa C., Matsuura Y., Fujita T., Akira S. (2006) Differential roles of MDA-5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105 [DOI] [PubMed] [Google Scholar]
- 8. Kato H., Sato S., Yoneyama M., Yamamoto M., Uematsu S., Matsui K., Tsujimura T., Takeda K., Fujita T., Takeuchi O., Akira S. (2005) Cell type-specific involvement of RIG-I in antiviral response. Immunity 23, 19–28 [DOI] [PubMed] [Google Scholar]
- 9. Takeuchi O., Akira S. (2008) MDA-5/RIG-I and virus recognition. Curr. Opin. Immunol. 20, 17–22 [DOI] [PubMed] [Google Scholar]
- 10. Berger M., Ablasser A., Kim S., Bekeredjian-Ding I., Giese T., Endres S., Hornung V., Hartmann G. (2009) TLR8-driven IL-12-dependent reciprocal and synergistic activation of NK cells and monocytes by immunostimulatory RNA. J. Immunother. 32, 262–271 [DOI] [PubMed] [Google Scholar]
- 11. Kormann M. S., Hasenpusch G., Aneja M. K., Nica G., Flemmer A. W., Herber-Jonat S., Huppmann M., Mays L. E., Illenyi M., Schams A., Griese M., Bittmann I., Handgretinger R., Hartl D., Rosenecker J., Rudolph C. (2011) Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 29, 154–157 [DOI] [PubMed] [Google Scholar]
- 12. Hayashi F., Means T. K., Luster A. D. (2003) Toll-like receptors stimulate human neutrophil function. Blood 102, 2660–2669 [DOI] [PubMed] [Google Scholar]
- 13. Janke M., Poth J., Wimmenauer V., Giese T., Coch C., Barchet W., Schlee M., Hartmann G. (2009) Selective and direct activation of human neutrophils but not eosinophils by Toll-like receptor 8. J. Allergy Clin. Immunol. 123, 1026–1033 [DOI] [PubMed] [Google Scholar]
- 14. Novak-Hofer I., Bläuenstein P., Schubiger P. A. (1990) Regulation of the cell surface expression of a nonspecific cross-reacting antigen variant during differentiation of HL-60 cells. Cancer Res. 50, 7437–7443 [PubMed] [Google Scholar]
- 15. Blair O. C., Carbone R., Sartorelli A. C. (1986) Differentiation of HL-60 promyelocytic leukemia cells: simultaneous determination of phagocytic activity and cell cycle distribution by flow cytometry. Cytometry 7, 171–177 [DOI] [PubMed] [Google Scholar]
- 16. Clemmensen S. N., Jacobsen L. C., Rørvig S., Askaa B., Christenson K., Iversen M., Jørgensen M. H., Larsen M. T., van Deurs B., Ostergaard O., Heegaard N. H., Cowland J. B., Borregaard N. (2011) α1-Antitrypsin is produced by human neutrophil granulocytes and their precursors and liberated during granule exocytosis. Eur. J. Haematol. 86, 517–530 [DOI] [PubMed] [Google Scholar]
- 17. Udby L., Borregaard N. (2007) Subcellular fractionation of human neutrophils and analysis of subcellular markers. Methods Mol. Biol. 412, 35–56 [DOI] [PubMed] [Google Scholar]
- 18. Wörnle M., Sauter M., Kastenmüller K., Ribeiro A., Roeder M., Schmid H., Krötz F., Mussack T., Ladurner R., Sitter T. (2009) Novel role of Toll-like receptor 3, RIG-I, and MDA-5 in poly(I:C) RNA-induced mesothelial inflammation. Mol. Cell. Biochem. 322, 193–206 [DOI] [PubMed] [Google Scholar]
- 19. Jeong E., Lee J. Y. (2011) Intrinsic and extrinsic regulation of innate immune receptors. Yonsei Med. J. 52, 379–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ekman A. K., Cardell L. O. (2010) The expression and function of Nod-like receptors in neutrophils. Immunology 130, 55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tamassia N., Le Moigne V., Rossato M., Donini M., McCartney S., Calzetti F., Colonna M., Bazzoni F., Cassatella M. A. (2008) Activation of an immunoregulatory and antiviral gene expression program in poly(I:C)-transfected human neutrophils. J. Immunol. 181, 6563–6573 [DOI] [PubMed] [Google Scholar]
- 22. Kawai T., Akira S. (2011) Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34, 637–650 [DOI] [PubMed] [Google Scholar]
- 23. Ariel A., Fredman G., Sun Y. P., Kantarci A., Van Dyke T. E., Luster A. D., Serhan C. N. (2006) Apoptotic neutrophils and T cells sequester chemokines during immune response resolution through modulation of CCR5 expression. Nat. Immunol. 7, 1209–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]