Background: High motility of aggressive breast cancer cells is associated with high SLUG and low plakoglobin levels.
Results: SLUG binds to plakoglobin gene promoter and represses its expression.
Conclusion: SLUG-induced increase in breast cancer cell motility is due to repression of plakoglobin by SLUG.
Significance: Management of SLUG level should diminish the motility and thus aggressiveness in breast cancer cells.
Keywords: Breast Cancer, Cell Motility, Metastasis, Repressor Protein, Transcription Promoter, Plakoglobin, SLUG
Abstract
One of highly pathogenic breast cancer cell types are the triple negative (negative in the expression of estrogen, progesterone, and ERBB2 receptors) breast cancer cells. These cells are highly motile and metastatic and are characterized by high levels of the metastasis regulator protein SLUG. Using isogenic breast cancer cell systems we have shown here that high motility of these cells is directly correlated with the levels of the SLUG in these cells. Because epithelial/mesenchymal cell motility is known to be negatively regulated by the catenin protein plakoglobin, we postulated that the transcriptional repressor protein SLUG increases the motility of the aggressive breast cancer cells through the knockdown of the transcription of the plakoglobin gene. We found that SLUG inhibits the expression of plakoglobin gene directly in these cells. Overexpression of SLUG in the SLUG-deficient cancer cells significantly decreased the levels of mRNA and protein of plakoglobin. On the contrary, knockdown of SLUG in SLUG-high cancer cells elevated the levels of plakoglobin. Blocking of SLUG function with a double-stranded DNA decoy that competes with the E2-box binding of SLUG also increased the levels of plakoglobin mRNA, protein, and promoter activity in the SLUG-high triple negative breast cancer cells. Overexpression of SLUG in the SLUG-deficient cells elevated the motility of these cells. Knockdown of plakoglobin in these low motility non-invasive breast cancer cells rearranged the actin filaments and increased the motility of these cells. Forced expression of plakoglobin in SLUG-high cells had the reverse effects on cellular motility. This study thus implicates SLUG-induced repression of plakoglobin as a motility determinant in highly disseminating breast cancer.
Introduction
Breast cancer cells with the triple-negative phenotype (TNBC)2 lack estrogen, progesterone, and ERBB2 receptors and represent one of the most aggressive and difficult to treat subtypes of human breast cancer (1, 2). TNBC is highly overrepresented in the African American breast cancer patients (1, 2). The genetic and molecular basis for this incidence and the biological basis for the aggressiveness of TNBCs are largely unknown and of high priority (3–6). The transcription factor SLUG, which controls epithelial to mesenchymal transition, stem cell phenotypes, and therapeutic responsiveness (7–9), is highly expressed in the highly aggressive basal and mesenchymal subtypes of the TNBC cells (1, 10), making it a candidate master regulator of the TNBC phenotype. The uncanny ability of the TNBC cells to metastasize to other vital tissues and organs causes severe pain and mortality in the TNBC patients (3–6). The ability of these cells to breach the basement membrane of epithelial barriers and migrate distinguishes highly metastatic TNBC cells from the non-metastatic breast cancer cells (5, 6). We postulate that transcriptional repression of few key cell adhesion and migration regulatory protein genes by SLUG in the basal and mesenchymal subtypes of the TNBC cells contributes toward the metastatic behavior of these cells, and targeted inhibition of SLUG should reverse the process.
SLUG, also known as SNAI2, is a member of the SNAI superfamily of zinc finger transcriptional repressors (7–9). It is a C2H2-type zinc finger transcription factor that binds to E2-box motif (5′-CAGGTG-3′/5′-CACCTG-3′) and silences gene expression by chromatin remodeling (7–9). SLUG has essential SNAG and SLUG motifs in its repressor domain that mediate its repressor activity (7–9). SLUG is shown to repress many genes including E-cadherin, BRCA2, cytokeratins, UbE2D3, and vitamin D receptor (11–15). SLUG is reported recently to coordinate with another transcription factor SOX9 and thus to determine the stem cell-ness of aggressive breast cancer cells (16). Although coordinated repression of several cell-cell junction proteins and other associated molecules by SLUG is implicated as the cumulative cause for SLUG-induced invasiveness of breast cancer cells (7–9), it is not known how dysregulation in SLUG expression increases the motility of these cells.
Out of several epithelial cell molecules implicated in the direct or indirect regulation of cellular motility, the catenin molecule plakoglobin stands out as a major player in negatively regulating the motility of these cells (17–23). Plakoglobin (also known as junction plakoglobin (JUP)) is a member of the armadillo family of proteins and a close relative of β-catenin (24). Plakoglobin comprises 12 central armadillo repeats, which are flanked by N- and C-terminal domains (17–19). By interacting with both the desmosomal cadherins and the N terminus of desmoplakin, plakoglobin is positioned to play a role in linking intermediate filaments to the desmosomal plaque (17–19). Recent report indicates that plakoglobin not only inhibits motility of keratinocytes in contact but also inhibits Src-dependent single cell motility (17). These results indicate that plakoglobin is capable of regulating single cell motility through matrix deposition in concert with Rho GTPases independently of its role as a cell-cell adhesion molecule (17, 18).
Using isogenic breast cancer cell systems, we have shown here that the motility of human breast cancer cells is directly related to their SLUG levels. Evidence presented here suggests that SLUG regulates the motility of these cells by directly suppressing the transcription of the plakoglobin gene through chromatin remodeling. We also have shown that knockdown of plakoglobin in the SLUG-negative breast cancer cells leads to the rearrangement of the actin filaments and formation of actin-rich membrane ruffling resembling invadopodia (25–27). Forced expression of plakoglobin in the SLUG-high breast cancer cells has the similar effect on their motility as that with SLUG knockdown.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
All the cells used in this study were procured from American Type Culture Collection (ATCC, Manassas, VA) and were cultured in ATCC-recommended media (12). Briefly, MCF7, MDA-MB-453, M4A4, NM2C5, and BT20 cells were cultured in DMEM medium with 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA); MDA-MB-468 and MDA-MB-231 cells were grown in L15 medium with 10% FBS, whereas HCC1419, ZR7530, HCC1187, and BT549 cells were cultured in RPMI 1640 medium with 10% FBS. Authentications of the cell lines are routinely performed in our laboratory following the instructions provided in ATCC Bulletin 8. Mouse anti-FLAG M2 antibody was purchased from Sigma. Rabbit anti-SLUG (C19G7) and rabbit anti-JUP (2309S) antibodies were purchased from Cell Signaling Technology (Danvers, MA). Rabbit anti-SLUG (H-140) for chromatin immunoprecipitation assay and mouse anti SLUG (A-71) antibody for Western blotting and immunofluorescence microscopy were procured from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Rabbit anti-CtBP1 (C-terminal binding protein-1), anti-HDAC1 (histone deacetylase-1), histone H3 (Lys-9 and Lys-14), and acetylated histone H4 (Lys5, Lys8, Lys12, and Lys16) antibodies were purchased from Upstate Millipore (Burlington, MA). Rabbit monoclonal antibody (#3879, Cell Signaling Technology) against SNAIL (CD15D3) was used to detect SNAIL in the Western blots. Rabbit antibody against α-actinin 4 (ab108198) and mouse antibody against non-muscle myosin IIA (ab55456) were procured from AbCam (Cambridge, MA).
Expression of Recombinant Proteins in Breast Cancer Cells
Human SLUG coding sequence (open reading frame (ORF)) was amplified (12) from the RNA isolated from BT549 cells using SLUG-specific primers (supplemental Table S1). The amplified cDNA (831 bp) was sequence-verified, digested with ClaI/BamHI, and cloned at the ClaI/BamHI sites of p3XFLAG-CMV-14 plasmid (Sigma). MCF7 and MDA-MB-468 cells were transfected with the SLUG ORF construct using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. After 48 h, cells were plated in G418-containing medium to select a stable cell population expressing SLUG. The non-functional mutant of SLUG, in which the SLUG domain amino acid sequence PSDTSSK was changed to AAAAAAA, was generated from the wild-type SLUG construct as a template using recombination PCR and cloned into the BamHI/ClaI site of the multiple cloning site of p3XFLAG-CMV-14 vector (Sigma). Briefly, the wild-type SLUG construct was used as a template to amplify overlapping fragments template 1 (T1) and template 2 (T2) using specifically designed primers (supplemental Table S1). The mutated nucleotides are underlined. The gel-purified amplicons (T1 and T2) were used as templates to perform recombination PCR to generate the mutant. The gel-purified mutant amplicon was digested with BamHI and ClaI and column-purified. The amplicon was then cloned at the BamHI/ClaI sites of p3XFLAG-CMV14 plasmid (Sigma). Human plakoglobin ORF cloned in the plasmid pcDNA3.0 (plasmid number 16827) was procured from Addgene (28) and was used to transiently express plakoglobin in the BT549 cells. The efficiency of expression of the cloned mRNAs and proteins in the recombinant cells was evaluated by real-time RT-PCR and Western blot analysis, respectively (12).
Knockdown of SLUG and Plakoglobin Gene Expression
SLUG-specific stealth siRNAs and corresponding control siRNAs were designed using the Block-IT RNAi designer software (Invitrogen) and were procured from Invitrogen (supplemental Table S2). Plakoglobin siRNA (#6226) and control siRNA (#6568S) were procured from Cell Signaling Technology. We also designed stealth siRNAs against human plakoglobin (supplemental Table S2), and they had comparable efficacies to knockdown the plakoglobin gene (data not shown). Transfection of these siRNAs into the breast cancer cells was done by lipofection using Lipofectamine 2000 (Invitrogen) according to manufacturer's protocol (12). Briefly, cells were transfected at ∼50% confluency using 100 pmol of the siRNA in 6-well plates, and whole-cell lysates were prepared 48 h after transfection. RNA was isolated from these cells using TRIzol (Invitrogen). The efficiency of knockdown of the target genes was evaluated by real-time RT-PCR and immunoblot analysis (12, 29).
Real-time RT-PCR Analysis
Total RNA was isolated from the cultured cells using TRIzol reagent (Invitrogen). RNA was further treated with RNase-free DNase (Invitrogen) to get rid of any DNA contamination. The cDNA was synthesized from 1 μg of DNase-treated RNA using the iScript cDNA synthesis reagents (Bio-Rad). Real-time RT-PCR quantification was performed following standard protocols using SYBR Green dye (Bio-Rad). The sequences of the primers used for quantitative PCR are shown in the supplemental Table S2. RT-PCR was performed in the iCycler (Bio-Rad) as described (12, 29). The -fold change over control samples was calculated using Ct, ΔCt, and ΔΔCt values (12, 29). β-Actin RNA was used as an internal control.
Immunoblot Analysis
Whole cell extracts were obtained according to our standard protocol and probed with appropriate antibodies as described previously (12). Antibodies were used at a 1:1000 dilution. The antibody-protein complexes were visualized using horseradish peroxidase-conjugated goat anti-rabbit antibody following enhanced chemiluminescence method (12).
Dual Luciferase Reporter Assay
We PCR-amplified human plakoglobin gene promoter (-447 to +761, NM_021991; supplemental nucleotide sequences) from DNA isolated from BT549 cells with specific primers (supplemental Table S2). This promoter sequence has six E2 boxes. The amplified DNA was cloned into the pCR4.0/TOPO plasmid (Invitrogen) and subsequently subcloned into the EcoRI site of pRL-Null plasmid (Promega, Madison, WI). Colony PCR was performed to select forward and reverse orientation clones of the promoter DNA in pRL-Null. Cells were seeded on 24-well tissue culture plates in triplicate and allowed to grow overnight to reach 90–95% confluency. The following day cells were transfected with pGL3-Control plasmid (Promega) and pRL-JUP promoter construct plasmid using Lipofectamine 2000 transfection reagent (Invitrogen). Forty-eight hours later, luciferase activity was measured using the Dual Luciferase reporter assay reagents (Promega) (12). Renilla luciferase activity was normalized with firefly luciferase activity as described (12).
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay was performed as described previously (12). A chromatin pulldown assay was performed using antibodies against human SLUG (H140), CtBP1, HDAC1, and acetylated histones H3 and H4. For quantitative ChIP analysis, SLUG was knocked down with different stealth siRNAs (supplemental Table S1) in MDA-MB-231 and BT549 cells for 48 h (12). Knockdown of SLUG was evaluated by real-time RT-PCR, and Western blot analysis and subsequently ChIP assay was performed. Real-time PCR was performed using primers described in supplemental Table S2. Real-time RT-PCR data for antibody-bound fractions were compared with a 1:10 dilution of input DNA.
Decoy Treatment
The design, synthesis, and biochemical characterization of the double-stranded (ds)-DNA decoy against SLUG is described under “Results.” To form the dsDNA decoy, equimolar amounts of the complementary and antiparallel single-stranded oligonucleotides were dissolved in sterile TE buffer (10 mm Tris, 1 mm EDTA, pH 8.0, and annealed for 3 h during which time the temperature was reduced from 90 to 25 °C. Synthetic dsDNA decoy (25 nm) was used for the transfection of human SLUG-high breast cancer cells using Lipofectamine. Forty-eight hours after transfection, protein and RNA were isolated from the transfected cells, and the levels of SLUG, plakoglobin, and β-actin mRNAs and proteins were evaluated. For promoter activity assay, the cells were co-transfected with the promoter construct, the pGL3-Control plasmid, and the dsDNA decoy. A dual luciferase assay was performed with the cell extract from the transfected cells 48 h after incubation under growth conditions (12).
Streptavidin Pulldown Assay
SLUG-high breast cancer cells MDA-MB-231 and BT549 were used for this experiment. Nuclear and cytoplasmic fractions were isolated using NE-PER nuclear, and cytoplasmic protein extraction reagents were from Thermo Fisher (Pittsburg, PA) following their protocol. The identity of the fractions was determined by standard assays (29). The levels of SLUG and SNAIL in the nuclear extracts were evaluated by Western blotting (12). For the evaluation of the binding of the dsDNA decoy to SLUG or SNAIL in the nuclear extracts, 5 pmol of the annealed 5′-biotinylated wild-type or mutated decoy was incubated with gentle rocking with 20 μg of proteins of the nuclear fraction in TBST (50 mm Tris·HCl, pH 7.4, 150 mm NaCl, 0.1% Tween 20) overnight at 4 °C. To pull down the protein-bound biotinylated dsDNA decoy, 50 μl of streptavidin magnetic bead (Thermo Fisher) slurry in TBST was added to the reaction mixture and incubated for 1 h at room temperature. Beads were then collected by magnetic separation, and residual non-bead fraction was collected for the evaluation of SLUG or SNAIL. The beads were washed three times with TBST before elution of the bound proteins from the streptavidin-coated beads by boiling (10 min) with 1× Laemmli SDS sample buffer (5×; 60 mm Tris-Cl, pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.01% bromphenol blue).
Transwell Migration Assay
For the transwell migration assay to determine the motility of the breast cancer cells, 5 × 104 cells were plated in 100 μl of serum-free medium in the upper chamber of the transwell plates. Complete medium was used in the lower chamber of the plates. Cells were then incubated at 37 °C for 24 h to allow cell migration through the membrane. The migrated cells were then fixed with formaldehyde and stained with 1% crystal violet. Cells were counted under the microscope (20× magnification objective lens), and the average of the data from six independent experiments was computed.
Cell Culture Wound Healing Assay
Wounds were created in confluent cell cultures in 24-well plates using a pipette tip. The cells were then rinsed with PBS to remove any free-floating cells and debris. Serum-free medium was then added, and culture plates were incubated at 37 °C. The width of the wound was evaluated at 0, 24, 48, and 72 h within the scrap line, and representative scrape lines for each set were photographed. Duplicate wells of each condition were examined for each experiment, and each experiment was repeated 6 times.
Immunohistochemistry for Tissue Microarray Analysis
Immunohistochemistry was performed as described (12) using tissue microarray procured from US Biomax (BR1503a). Identification of the spots are according to the datasheet (see supplemental Fig. S1 and tissue microarray data). A tissue microarray slide was incubated in primary antibodies (rabbit SLUG and mouse plakoglobin; 1:100) overnight at 4 °C, and thereafter the slide was washed 3 times with PBS. It was further incubated with the corresponding Alexafluor-conjugated secondary antibodies (donkey anti-rabbit R488 and donkey anti-mouse R555; Invitrogen) for 1 h at room temperature. The slide was again washed 5 times with PBS, embedded in glycerol/PBS-based mounting medium, and examined for painted cells using a fluorescent microscope (Nikon TE2000-E).
Immunofluorescence Confocal Microscopy
Immunofluorescence staining and confocal analyses were performed as described (12). In brief, cells were cultured on coverslips in 24-well plates for 24 h, rinsed with ice-cold PBS, fixed with 3.7% formaldehyde for 30 min, washed three times with PBS, and fixed in ice-cold methanol for 10 min. Cells were permeabilized with 0.2% Triton X-100 in PBS for 10 min. Thereafter, the cells were washed 3 times with ice-cold PBS. The slides were blocked in PBS containing 5% goat serum and then were incubated with primary antibodies (SLUG 1:400, plakoglobin 1:400, β-actin 1:1000) overnight at 4 °C in the blocking buffer. Slides were then washed 5 times with PBS followed by incubation for 45 min with the respective Alexafluor-conjugated secondary antibody. The slides were again washed 3 times with PBS, nuclei were stained with TOPRO (1:1000 in PBS) for 5 min, and the slides were washed again with PBS, embedded in glycerol/PBS-based mounting medium, and examined for painted cells using a fluorescent microscope (Nikon TE2000-E). Confocal images were obtained with a Nikon TE2000-UC1 laser-scanning microscope (12). Immunofluorescence images were quantitated in a Nikon TE2000-E inverted wide-field microscope using NIS AR software (Nikon, Melville, NY).
Statistical Analysis
Each experiment was repeated 4–6 times. Results are expressed as the means ± S.E. Statistical analyses were performed using GraphPad Prism software. p values were calculated using the two-sided Student's t test (paired or unpaired, as appropriate) and analysis of variance test for significance. p values of <0.05 and <0.01 were considered as significant.
RESULTS
SLUG Level and in Vitro Motility Are Directly Correlated in Human Breast Cancer Cells
High motility is usually associated with oncogenic epithelial cells expressing high levels of the SLUG protein (8, 30–33). Here we present direct evidence with isogenic cell systems that the level of SLUG is indeed directly correlated with in vitro motility of human breast cancer cells. We selected three non-metastatic non-TNBC (MCF7, MDA-MB-468, and T47D) and two highly metastatic TNBC (MDA-MB-231 and BT549) cell lines and evaluated SLUG mRNA (supplemental Fig. S2A) and protein (supplemental Fig. S2, B and C) levels in them. As expected, the non-invasive cells are negative or low in SLUG mRNA and protein levels, whereas the invasive cells tested are high in SLUG (supplemental Fig. S2). Relative motility of the SLUG-high cells, as determined by transwell assay, was also significantly higher than that of the SLUG-low cells (Fig. 1A). Recently, high levels of SLUG have been reported in the basal and the mesenchymal subtypes of several human TNBC primary tissues (see supplemental Fig. S6 in Lehmann et al. (1)). Our data are thus in sync with the similar observation made by others.
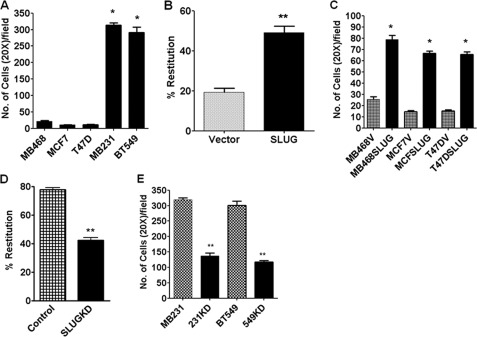
FIGURE 1.
SLUG level and in vitro motility are directly correlated in human breast cancer cells. A, shown is relative motility of different breast cancer cells as was determined by transwell assay. Results are the mean ± S.E. (n = 6). * indicates statistical significance p < 0.001. B, shown is evaluation of in vitro motility in the scratch assay of the control and the SLUG-expressing MDA-MB-468 cells. Results are the mean ± S.E. (n = 6). ** indicates statistical significance p < 0.01. C, shown is relative motility of human breast cancer cells ectopically expressing SLUG in the transwell migration assay. Results are the mean ± S.E. (n = 6). * indicates statistical significance p < 0.001. D, shown is evaluation of in vitro motility in the scratch assay of the control and the SLUG-knocked down MDA-MB-231 cells. Results are the mean ± S.E. (n = 6). ** indicates statistical significance, p < 0.01. E, shown is relative motility of human breast cancer cells with or without knockdown of SLUG in the transwell migration assay. Results are the mean ± S.E. (n = 6). ** indicates statistical significance, p < 0.01.
We then evaluated the effect of ectopic expression of SLUG in the SLUG-negative human breast cancer cells on their in vitro motility. We expressed C-terminal FLAG-tagged human SLUG in the SLUG-negative MCF7, MDA-MB-468, and T47D cells and evaluated the levels of SLUG in these cells (supplemental Fig. S3, A–C). Western blot analysis of the levels of SLUG in the vector-transfected and SLUG-overexpressing MDA-MB-468 cells is shown in supplemental Fig. S3, B and C. An increase in the SLUG levels was also noted in the MCF7 and T47D cells (data not shown). Two methods were employed to evaluate the in vitro motility of the vector transfected and the SLUG-expressing breast cancer cells. Data for the evaluation of in vitro motility in the “scratch” assay of the control and the SLUG-expressing MDA-MB-468 cells are shown in Fig. 1B. The motility of the SLUG-expressing cells was significantly higher than the vector-transfected cells (Fig. 1B). Similar results were obtained with the other cells tested. Transwell migration assay also revealed a significant increase in the relative motility of human breast cancer cells ectopically expressing SLUG as compared with the corresponding vector-transfected cells (Fig. 1C). These data support our hypothesis that breast cancer cell motility is increased by the dysregulation of SLUG expression in these cells.
We further verified this hypothesis by knocking down SLUG in the SLUG-high human breast cancer cells and evaluating their in vitro motility. We knocked down SLUG with two different stealth siRNAs as described (12) in the SLUG-high MDA-MB-231 and BT549 cells and evaluated the levels of SLUG in these cells. Data for the stealth siRNA#1 (supplemental Table S2) are shown (supplemental Fig. S4, A–C). Western blot analysis of the levels of SLUG in the control and SLUG siRNA-treated MDA-MB-231 cells are shown in supplemental Fig. S4, B and C. A decrease in the SLUG levels was also noted in the BT549 cells (data not shown). Again, we evaluated the in vitro motility of the control and the SLUG siRNA-treated breast cancer cells by two independent methods. Evaluation of in vitro motility by the scratch assay of the control and the SLUG-knocked down MDA-MB-231 cells revealed a significant decrease in the motility of these cells (Fig. 1D). Similar results were obtained with the other cells tested. A significant decrease in the relative motility of the SLUG-knocked down human breast cancer cells was also noted by the transwell migration assay (Fig. 1E). These data further strengthen our notion that breast cancer cell motility is regulated by SLUG in these cells.
Intracellular Levels of SLUG and Motility Regulatory Protein Plakoglobin Are Inversely Related in Breast Cancer Cells and Tissues
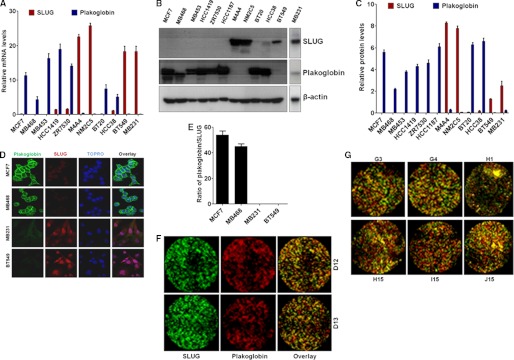
As mentioned above, the armadillo motif-containing protein plakoglobin is known to negatively regulate the motility of epithelial cells (17–23). Because SLUG in the breast cancer cells regulates the motility of these cells, we hypothesized that SLUG represses plakoglobin gene in these cells to increase their motility. Toward testing this hypothesis, our initial effort was to evaluate whether plakoglobin levels in the breast cancer cells are inversely related to the level of SLUG in these cells. We tested several breast cancer cell lines for their SLUG and plakoglobin mRNA and protein levels. We found that indeed, higher the levels of SLUG, the lower the levels of plakoglobin in these cells (Fig. 2, A–C). Immunofluorescence confocal microscopy also validated our notion that lower plakoglobin level is associated with high levels of SLUG in the highly motile MDA-MB-231 and BT549 cells (Fig. 2, D and E). Immunofluorescence imaging analysis of human malignant breast tissue microarray also revealed a large number of SLUG-high cells in the tumor samples that lack plakoglobin (green staining cells in the overlay images, Fig. 2, F and G, and supplemental Fig. S1). There are also cells in the tumor tissues that are rich in plakoglobin and lack SLUG (red staining cells in the overlay images (Fig. 2, F and G, and supplemental Fig. S1) as expected. In the breast tumor samples there are apparently cells that stained orange to yellow, possibly indicating cells that have both SLUG and plakoglobin (orange-yellow staining cells in the overlay images, Fig. 2, F and G, and supplemental Fig. S1). These cells may represent intermediate cells where SLUG expression occurs, but SLUG is not functional as a repressor because of the possibility of missing downstream regulators of SLUG. These preliminary analysis data thus indicate inverse relationship between SLUG and plakoglobin also in the human breast cancer tissues, suggesting physiological relevance of our finding.
FIGURE 2.
Inverse relationship in the levels of plakoglobin and SLUG in breast cancer cells and tissues. A, shown are relative levels of SLUG and plakoglobin mRNAs in different breast cancer cells as was determined by real-time RT-PCR analysis. β-Actin mRNA was used as a normalization control. Results are the mean ± S.E. (n = 6). B, shown is evaluation of the levels of SLUG and plakoglobin protein in different human breast cancer cells by immunoblotting. β-Actin was used as a loading control. C, shown is densitometric scanning of the Western blots to evaluate the relative levels of SLUG and plakoglobin in different breast cancer cells. Results are the mean ± S.E. (n = 4). D, immunofluorescence confocal microscopy shows the relative levels of SLUG (red) and plakoglobin (green) in four different human breast cancer cells. TOPRO was used to stain the nucleus. E, shown is a quantitative evaluation of the ratio of plakoglobin/SLUG in the immunofluorescence images as in D. F and G, immunofluorescence microscopic image shows distinct cells with high levels of SLUG (green) but no plakoglobin (red) and vice versa in human malignant breast cancer tissues. F, images show selected tissue microarray dots stained with SLUG (green), plakoglobin (red), and overlay. G, shown are examples of additional immunofluorescence images from the tissue microarray analysis. Refer to the supplemental Fig. S4 for the designations of the tissue dots and additional data.
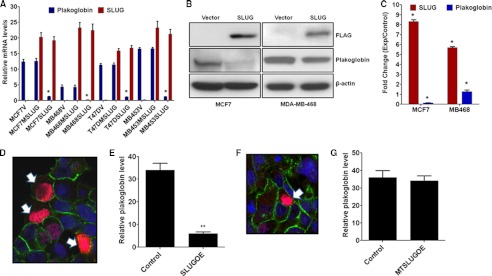
To test our hypothesis further we ectopically expressed functional and non-functional SLUG (see the supplemental amino acid sequences) in the SLUG-negative plakoglobin-high breast cancer cells and evaluated their plakoglobin levels. As expected, stable ectopic expression of functionally active wild-type SLUG but not that of the functionally inactive mutated SLUG in the SLUG-negative cells knocked down the plakoglobin mRNA levels in these cells (Fig. 3A). Unfortunately, these cells with stable expression of SLUG gradually lost the SLUG protein and became SLUG-negative after four-five generations. Although we found that proteasomal degradation of SLUG was responsible for the disappearance of recombinant SLUG in these cells, we do not know the exact mechanism of this degradation. Transient expression of SLUG in the SLUG-negative MDA-MB-468 and MCF7 cells also showed down-regulation of plakoglobin protein in these cells (Fig. 3, B and C). Immunofluorescence confocal microscopic analysis data showed that transient ectopic expression of SLUG in the MDA-MB-468 cells resulted in the down-regulation of plakoglobin only in those cells that have abundant expression of SLUG (red staining in the nucleus) but not in the cells that lack the recombinant SLUG (Fig. 3, D and E). Ectopic expression of the non-functional SLUG protein in the MCF7 cells did not down-regulate the plakoglobin level in these cells (Fig. 3, F and G) further suggesting the functional repression of plakoglobin by SLUG.
FIGURE 3.
Effect of ectopic expression of SLUG on the levels of plakoglobin in SLUG-negative breast cancer cells. A, shown is real-time RT-PCR analysis of the levels of SLUG and plakoglobin mRNAs in different SLUG-negative breast cancer cells transfected with empty vector (V), plasmid construct with mutated non-functional SLUG (MSLUG), or plasmid construct with wild-type functional SLUG (SLUG). Results are the mean ± S.E. (n = 6). * indicates statistical significance p < 0.001. B, shown is immunoblot analysis for SLUG and plakoglobin proteins in the control (vector-transfected) and SLUG-expressing (transfected with FLAG-SLUG construct) MCF7 and MDA-MB-468 cells. Recombinant SLUG protein was detected using FLAG antibody. β-Actin was used as a loading control. C, shown is a densitometric scan for SLUG and plakoglobin levels in six independent SLUG-transfected populations and the corresponding vector-transfected control cells. Results are the mean ± S.E. (n = 6). The * indicates that the -fold changes were statistically significant (p < 0.001). D, shown are immunofluorescence confocal microscopic analysis data showing down-regulation of plakoglobin in the MCF7 cells transfected with functional wild-type SLUG expressing plasmid. The cells were transiently transfected, and only those cells that have abundant expression of SLUG (red staining in the nucleus, shown by white arrows) lack plakoglobin on their cell membrane regions. Blue, TOPRO; green, plakoglobin; red, SLUG. E, quantitative evaluation of the relative levels of plakoglobin (with respect to TOPRO staining) in the immunofluorescence images was as in D. SLUG-overexpressing cells (SLUGOE) are those with a red stain in the nucleus. For adjacent cells with or without SLUG overexpression, only the boundaries of the cells not shared are quantitated. Results are the mean ± S.E. (n = 5). ** indicates statistical significance p < 0.01. F, immunofluorescence confocal microscopic analysis data show no detectable effect of non-functional mutated SLUG protein (red) on plakoglobin levels (green) in MCF7 cells transfected with mutated SLUG-expressing plasmid. The cell that expressed mutated SLUG is shown by a white arrow. G, quantitative evaluation of the relative levels of plakoglobin (with respect to TOPRO staining) in the immunofluorescence images was as in F. Results are the mean ± S.E. (n = 5). The changes between the control and the mutant SLUG-expressing (MTSLUGOE) cells were not statistically significant.
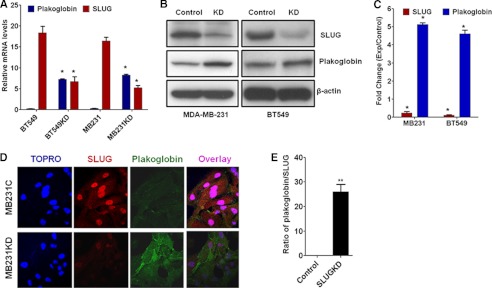
We knocked down SLUG in the SLUG-high MDA-MB-231 and BT549 cells to verify whether this action increased plakoglobin levels in these cells. SLUG knockdown in these cells significantly increased the levels of plakoglobin mRNA (Fig. 4A) and protein (Fig. 4, B and C) as expected. We also documented the increase of plakoglobin protein in the SLUG-knocked down MDA-MB-231 cells by immunofluorescence confocal microscopy (Fig. 4, D and E). We repeated our experiment with two different stealth siRNAs against SLUG with MDA-MB-231 and BT549 cells and obtained similar results. These data conclude that the levels of SLUG and plakoglobin in the breast cancer cells tested are inversely related and point toward the hypothesis that SLUG induces increased motility in the aggressive breast cancer cells through the repression of plakoglobin gene expression.
FIGURE 4.
Effect of knockdown of SLUG on plakoglobin levels in MDA-MB-231 and BT549 cells. A, shown is quantitative RT-PCR analysis for SLUG and plakoglobin mRNA levels in MDA-MB-231 and BT549 cells treated with control or SLUG siRNAs. β-Actin mRNA was used as a normalization control. Results are the mean ± S.E. (n = 6). * indicates statistical significance p < 0.001. B, shown is an immunoblot analysis of plakoglobin levels in MDA-MB-231 and BT549 cells by with (KD) or without (Control) knocking down SLUG. Control cells were transfected with control siRNA. C, shown is evaluation of SLUG and plakoglobin protein levels in MDA-MB-231 and BT549 cells with or without knockdown of SLUG. Six independent SLUG-knocked down cell populations and corresponding control siRNA-treated cells were used. Results are the mean ± S.E. (n = 6). * indicates that the -fold changes observed were statistically significant (p < 0.001). D, immunofluorescence confocal microscopic analysis data show the effect of SLUG (red) knockdown on the level of plakoglobin (green) in MDA-MB-231 cells. TOPRO was used to stain the nuclei of the cells. E, shown is a quantitative evaluation of the ratio of plakoglobin/SLUG in the immunofluorescence images as in D. Results are the mean ± S.E. (n = 10). ** indicates statistical significance p < 0.01.
SLUG Binds to and Inhibits Activity of Plakoglobin Gene Promoter in Human Breast Cancer Cells
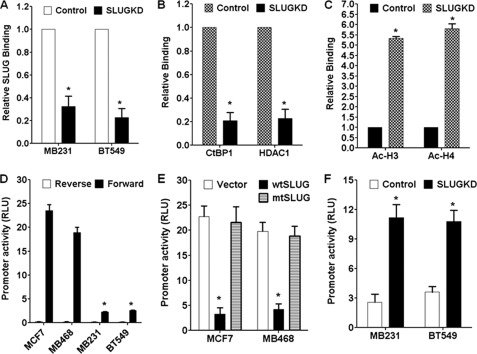
To evaluate the mode of action of SLUG in the repression of plakoglobin level in human breast cancer cells, we studied the binding to and inhibition of the promoter of plakoglobin gene in these cells. Our initial promoter array analysis experiment to identify gene promoters that bind to SLUG in human breast cancer cells revealed plakoglobin gene as a candidate (12). A quantitative chromatin immunoprecipitation assay further verified the binding of SLUG to the plakoglobin gene promoter in the nucleus of MDA-MB-231 and BT549 cells, which is inhibited significantly when SLUG is knocked down in these cells (Fig. 5A). As expected, the co-recruitment of the co-repressor CtBP1 and the effector HDAC1 at the plakoglobin gene promoter was also decreased significantly due to the knockdown of SLUG in the SLUG-high BT549 cells (Fig. 5B). The levels of acetylated histones at this promoter were increased in SLUG-knocked down BT549 cells (Fig. 5C). The deacetylation of histones at the plakoglobin gene promoter was also decreased by preincubation of the BT549 cells with the HDAC1 inhibitor trichostatin A (data not shown), further indicating the involvement of HDAC1 in the repression process. We have shown previously that SLUG inhibits the expression of its target genes by binding to its promoter and through the recruitment of the co-repressor CtBP1 and the effector HDAC1, which deacetylate the nucleosomal histones at the target gene promoter (12, 13). Our data presented here thus indicate that SLUG similarly represses plakoglobin gene expression by hetero-chromatinization of this gene promoter (supplemental Fig. S5).
FIGURE 5.
Effect of SLUG on the plakoglobin promoter activity in human breast cancer cells. A, quantitative chromatin immunoprecipitation analysis data show the effect of SLUG knockdown on the binding of SLUG to the promoter of plakoglobin gene in MDA-MB-231 and BT549 cells. Results are the mean ± S.E. (n = 6). * indicates that the changes observed in the SLUG-knockdown cells were statistically significant (p < 0.001). B, quantitative chromatin immunoprecipitation analysis data shows the effect of SLUG knockdown on the binding of CtBP1 and HDAC1 to the promoter of plakoglobin gene in BT549 cells. Results are the mean ± S.E. (n = 6). * indicates that the changes observed in the SLUG-knockdown cells were statistically significant (p < 0.001). C, quantitative chromatin immunoprecipitation analysis data shows the effect of SLUG knockdown on the abundance of acetylated histones H3 and H4 at the promoter of plakoglobin gene in BT549 cells. Results are the mean ± S.E. (n = 6). * indicates that the changes observed in the SLUG-knockdown cells were statistically significant (p < 0.001). D, shown is relative activity of plakoglobin gene promoter in different SLUG negative (MCF7 and MDA-MB-468) and SLUG-positive (MDA-MB-231 and BT549) human breast cancer cells. Results are the mean ± S.E. (n = 6). * indicates statistically significant changes in the promoter activity in the SLUG-positive cells as compared with that in the SLUG-negative MCF7 cells. RLU, relative light units. E, shown is plakoglobin promoter activity in the control (vector-transfected), wild-type (wtSLUG), and mutated non-functional (mtSLUG) SLUG-expressing MCF7 and MDA-MB-468 cells. Results are the mean ± S.E. (n = 6). * indicates statistically significant decrease in the promoter activity in the SLUG-expressing cells as compared with that in the vector-transfected cells (p < 0.001). F, shown is derepression of plakoglobin promoter activity in the SLUG-knocked down (SLUGKD) MDA-MB-231 and BT549 cells. Results are the mean ± S.E. (n = 6). * indicates a statistically significant increase in the promoter activity in the SLUG-knocked down cells as compared with that in the control cells (p < 0.001).
To further evaluate the inhibition of plakoglobin gene promoter activity by SLUG, we tested the activity of the cloned human plakoglobin gene promoter in SLUG-manipulated isogenic breast cancer cell systems. We cloned the 1208-bp plakoglobin promoter (−447 to +761 with respect to the transcription start site) in the forward or the reverse orientation with respect to the Renilla luciferase reporter gene in pRL-Null plasmid. We found that the orientation-specific activity of the cloned plakoglobin gene promoter is significantly higher in the SLUG-negative MCF7 and MDA-MB-468 cells as compared with that in the SLUG-high MDA-MB-231 and BT549 cells (Fig. 5D). Ectopic expression of wild-type SLUG but not the functionally inactive mutated SLUG in the SLUG-negative MCF7 and MDA-MB-468 cells inhibited the plakoglobin gene promoter activity significantly (Fig. 5E). In contrast, knockdown of SLUG in the SLUG-high MDA-MB-231 and BT549 cells increased the activity of the cloned plakoglobin gene promoter (Fig. 5F). The cloned plakoglobin gene promoter has six E2-box sequences that are potential SLUG binding sites. Site-directed mutagenesis of individual E2-box sequences to 5′-TAAATT-3′ did not affect the activity of the promoter significantly (data not shown), suggesting probable action of SLUG through multiple E2-box sequences at this promoter. These data indicate direct involvement of SLUG in the inhibition of plakoglobin gene promoter in the SLUG-expressing human breast cells.
Double-stranded DNA Decoy Developed against SLUG Alleviated SLUG Repression of Plakoglobin Gene in SLUG-high Breast Cancer Cells
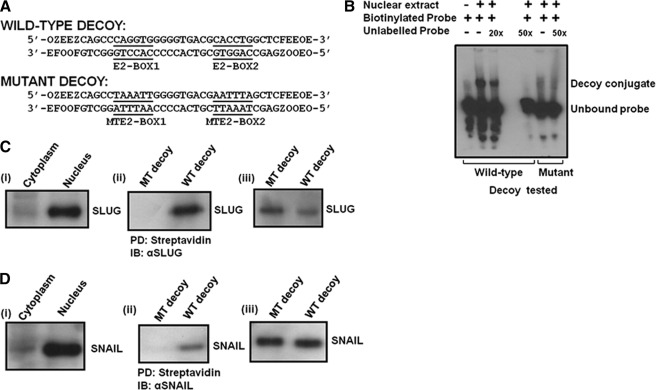
To validate the E2-box-dependent mode of action of SLUG to repress plakoglobin gene in human breast cells, we developed a SLUG binding double-stranded DNA molecular decoy. We have shown previously that SLUG inhibits vitamin D receptor gene promoter through its binding to the E2-box sequences of this promoter (13). We selected a 40-bp sequence from the human vitamin D3 receptor gene promoter that has two SLUG binding E2-box (5′-CACCTG-3′/3′-CAGGTG-5′) sequences (Fig. 6A). We custom synthesized individual oligonucleotides with a terminal five nucleotides having phosphorothioate groups to make these oligonucleotides resistant to the exonucleases (Fig. 6A). We also custom-synthesized the corresponding oligonucleotides with mutation at the two E2-box sequences to be used as a control (Fig. 6A). Using 5′-end biotin-tagged dsDNA decoys, we performed EMSA analysis with nuclear extract isolated from MDA-MB-231 cells. The probe binds tightly with proteins in the extract (Fig. 6B). This binding could be inhibited by unlabeled probe indicating specificity of binding (Fig. 6B). E-box-mutated dsDNA decoy failed to bind this protein in the EMSA assay (Fig. 6B). A supershift assay with the SLUG antibody indicated that the binding protein included SLUG (data not shown). We performed pulldown assay with streptavidin-coated paramagnetic particles to further evaluate the binding of SLUG to wild-type dsDNA decoy. Biotin-tagged dsDNA decoy was incubated with nuclear extract isolated from SLUG-high MDA-MB-231 cells. As expected, the transcriptional repressor protein SLUG was almost exclusively present in the nuclear fraction (Fig. 6Ci). The wild-type decoy but not the E2-box-mutated decoy could pulldown SLUG from the nuclear fraction (Fig. 6Cii). Evaluation of the supernatant after the pulldown for SLUG revealed little left in the supernatant from the wild-type dsDNA decoy experiment (Fig. 6Ciii). A similar experiment done for SNAIL binding to the decoy with the same nuclear extract revealed less binding of this isofunctional transcriptional repressor protein to the dsDNA decoy for SLUG (Fig. 6D). These experiments suggest relative specificity of the dsDNA decoy for SLUG binding.
FIGURE 6.
Characteristics of the molecular decoy developed against SLUG. A, shown are nucleotide sequences of the wild-type and the mutant molecular decoys used. F, O, E, and Z indicate phosphorothioate modifications of A, C, G, and T, respectively. B, EMSA analysis shows E2-box-dependent specific binding of nuclear proteins from MDA-MB-231 cell nuclear extract to the wild-type decoy but not to the mutated decoy. Biotin-tagged decoys were used, and the blots were developed with HRP-tagged streptavidin. C, an immuno pulldown assay followed by immunoblot analysis shows the binding of SLUG to the wild-type decoy but not to the mutated decoy. PD, pulldown. (i) the immunoblot shows exclusive location of SLUG in the nuclear fraction of MDA-MB-231 cells. (ii) shown is a pulldown assay for SLUG from the nuclear fraction of MDA-MB-231 cells using biotin-tagged decoys. (iii) the immunoblot shows leftover SLUG in the supernatant after the pulldown assay (one-fifth of the supernatant was loaded). D, an immuno pulldown assay followed by immunoblot (IB) analysis shows weak binding of SNAIL to the decoy. (i) the immunoblot shows exclusive location of SNAIL in the nuclear fraction of MDA-MB-231 cells. (ii) shown is a pulldown assay for SNAIL from the nuclear fraction of MDA-MB-231 cells using biotin-tagged decoys. (iii) the immunoblot shows leftover SNAIL in the supernatant after the pulldown assay (one-fifth of the supernatant was loaded).
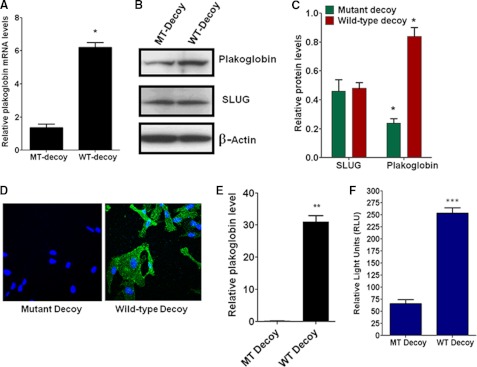
We tested the ability of the wild-type dsDNA decoy developed against SLUG to alleviate the repression of plakoglobin gene expression in the MDA-MB-231 cells. Treatment of the cells with the wild-type decoy but not the E2-box mutated decoy significantly increased the levels of plakoglobin mRNA (Fig. 7A) and protein without affecting SLUG protein levels (Fig. 7, B and C). Up-regulation of plakoglobin protein in the MDA-MB-231 cells treated with the wild-type dsDNA decoy was also revealed by immunofluorescence confocal microscopy (Fig. 7, D and E). The wild-type decoy, but not the mutant decoy, could significantly increase the activity of plakoglobin gene promoter in the MDA-MB-231 cells (Fig. 7F). These experiments were repeated with BT549 cells, and similar results were obtained. These data further support our conclusion that SLUG represses plakoglobin gene expression in the aggressive breast cancer cells.
FIGURE 7.
Alleviation of the repressor activity of SLUG against plakoglobin gene expression by molecular decoy against SLUG binding to E2-box. A, shown is the effect of the decoy on plakoglobin mRNA level in MDA-MB-231 cells as was determined by real-time RT-PCR analysis. β-Actin mRNA was used as a normalization control. Results are the mean ± S.E. (n = 6). * indicates statistical significance p < 0.001. MT-Decoy. decoy with mutation at the E2-box sequences; WT-Decoy, Decoy with wild-type E2-box sequences. B, Western blot analysis shows the effect of the decoy on plakoglobin protein level in MDA-MB-231 cells. SLUG levels did not significantly alter. β-Actin was used as a loading control. C, shown is densitometric scanning of the Western blots to evaluate the relative levels of SLUG and plakoglobin in MDA-MB-231 cells. Results are the mean ± S.E. (n = 4). * indicates statistical significance p < 0.001. D, shown is immunofluorescence confocal microscopy analysis of the effect of the SLUG decoy on plakoglobin protein (green) levels in MDA-MB-231 cells. TOPRO (blue) was used to stain the nucleus of the cells. E, quantitative evaluation of the relative levels of plakoglobin (with respect to TOPRO staining) in the immunofluorescence images was as in D. Results are the mean ± S.E. (n = 7). The difference in the relative (normalized with TOPRO level) plakoglobin levels between the cells treated with mutant decoy (MT Decoy) and wild-type Decoy (WT Decoy) was statistically significant. ** indicates statistical significance p < 0.01. F, shown is the effect of the decoy on the activity of plakoglobin gene promoter in MDA-MB-231 cells. Results are the mean ± S.E. (n = 6). *** indicates statistically significant changes between the control and the experimental sets (p < 0.05).
Forced Alteration of Plakoglobin Levels in Human Breast Cancer Cells with or without SLUG Altered Their in Vitro Motility
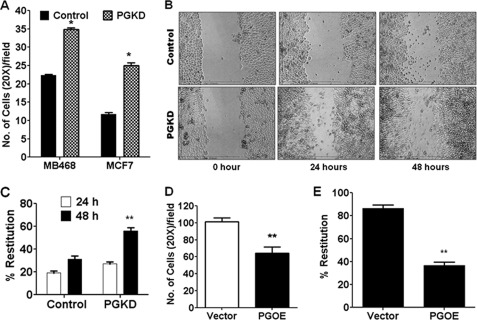
Our data presented above implicated higher levels of plakoglobin in the SLUG-negative human breast cancer cells as the cause of their lower in vitro motility. To verify the correlation of plakoglobin level and motility, we knocked down plakoglobin in SLUG-negative MCF7 and MDA-MB-468 cells with siRNA and evaluated their motility. The siRNA used successfully knocked down the levels of plakoglobin mRNA (supplemental Fig. S6A) and protein (supplemental Fig. S6, B and C) levels in these cells. Plakoglobin knockdown significantly increased the relative in vitro motility of these cells (Fig. 8, A–C). We also forced expressed plakoglobin in the SLUG-high BT549 cells (supplemental Fig. S7) and evaluated the effect of such expression on the motility of these cells. Ectopic expression of plakoglobin significantly decreased the relative in vitro motility of these cells (Fig. 8, D and E). These data thus support our conclusion that plakoglobin knockdown in the SLUG-high breast cancer cells is responsible for their higher motility.
FIGURE 8.
Effect of alteration of plakoglobin levels on the motility, actin filament reorganization, and invadopodia formation in breast cancer cells. A, shown is the effect of plakoglobin knockdown (PGKD) on the motility of MCF7 and MDA-MB-468 cells in transwell migration assay. Results are the mean ± S.E. (n = 4). * indicates statistical significance p < 0.001. B, shown is the effect of PGKD on the motility of MDA-MB-468 cells as was evaluated by scratch assay. C, shown is a quantitative evaluation of in vitro motility in the scratch assay (as in B) of the vector control and the PGKD MDA-MB-468 cells at 24 and 48 h. Results are the mean ± S.E. (n = 6). The difference of % restitution at 48 h was statistically significant. ** indicates statistical significance p < 0.01. D, shown is the effect of ectopic expression of plakoglobin (PGOE) on the motility of BT549 cells in the transwell migration assay. Results are the mean ± S.E. (n = 4). ** indicates statistical significance p < 0.01. E, shown is an evaluation of in vitro motility in the scratch assay of the vector control and the PGOE BT549 cells. Results are the mean ± S.E. (n = 6). ** indicates statistical significance p < 0.01.
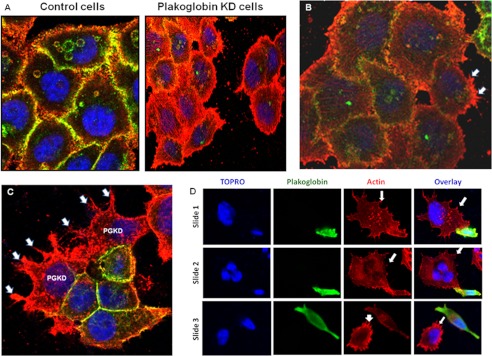
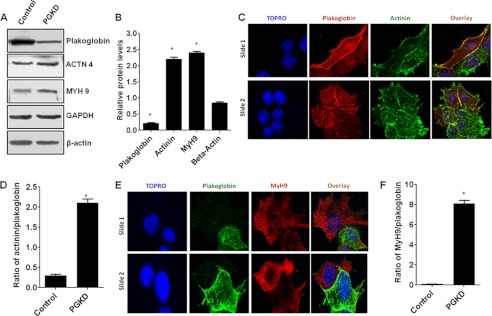
Interestingly, our immunofluorescence confocal microscopy data generated with the plakoglobin-knocked down MCF7 and MDA-MB-468 cells revealed reorganization of actin filaments in these cells, which is the hallmark of increased motility. These data suggest that the knockdown of plakoglobin reorganizes actin filaments in the MDA-MB-468 (Fig. 9, A and B) and MCF7 (Fig. 9C) cells showing invadopodia-like projections emanated from the plakoglobin-knocked down cells and not from the cells with plakoglobin expression (Fig. 9). Forced expression of plakoglobin in the SLUG-high BT549 cells showed significant changes in cell morphology and actin filament profile (Fig. 9D). Because actin filament reorganization in the highly motile metastatic cancer cells is also associated with higher levels of actinin and myosin heavy chain 9 (MyH9) proteins (30), we evaluated the relative levels of these two proteins in the plakoglobin-knocked down MCF7 cells. Western blot analysis showed significant increase in the levels of α-actinin 4 and MyH9 proteins in the plakoglobin-knocked down cells (Fig. 10, A and B). Immunofluorescence confocal microscopy also shows significant increases in the levels of these proteins in the plakoglobin-knocked down MCF7 cells (Fig. 10, C–F). These data further support our notion that SLUG-induced repression of plakoglobin gene in the SLUG-high breast cancer cells causes increased motility of these cells, contributing toward their aggressiveness.
FIGURE 9.
Effect of alteration of plakoglobin levels on actin filament reorganization and invadopodia formation in breast cancer cells. A, immunofluorescence confocal microscopy shows the effect of knockdown of plakoglobin (green) on actin filament (red) reorganization in MDA-MB-468 cells. Cell nuclei are painted blue with TOPRO. B, higher magnification of the plakoglobin-knocked down MDA-MB-468 cells shows actin filament projections (white arrows), suggesting invadopodia formation in these cells. C, immunofluorescence confocal microscopy shows the effect of transient knockdown of plakoglobin (green) on actin filament projections (red) from the MCF7 cells. Cell nuclei are painted blue with TOPRO. The field shows both plakoglobin-knocked down (PGKD) and control cells. The PGKD cells show actin filament projections (white arrows) characteristic of invadopodia. D, immunofluorescence confocal microscopy shows the effect of forced expression of plakoglobin (green) on actin filament (red) reorganization in BT549 cells. Cell nuclei are painted blue with TOPRO. Snapshots from three slides are shown. Each slide has at least one cell with overexpression of plakoglobin. The invadopodia-like structures are apparent (white arrows) on the cells lacking plakoglobin.
FIGURE 10.
Effect of knockdown of plakoglobin levels on the levels of actin filament-associated protein α-actinin 4 (ACTN4) and MyH9 in the MCF7 cells. A, a Western blot shows a significant increase in the levels of actinin and MyH9 proteins in the plakoglobin-knocked down cells. GAPDH was used as a loading control. B, shown are a densitometric scan for the protein levels in six independent plakoglobin-knocked down populations and the corresponding control siRNA-transfected control cells. Results are the mean ± S.E. (n = 6). * indicates that the -fold changes were statistically significant (p < 0.001). C, immunofluorescence confocal microscopy shows higher levels of actinin (green) in the plakoglobin (red) knocked down MCF7 cells. D, quantitative evaluation of the ratio of actinin/plakoglobin in the immunofluorescence images was in C. Results are the mean ± S.E. (n = 5). * indicates that the changes were statistically significant (p < 0.001). E, immunofluorescence confocal microscopy shows higher levels of MyH9 (red) in the plakoglobin (green) knocked down MCF7 cells. F, quantitative evaluation of the ratio of MyH9/plakoglobin in the immunofluorescence images was as in E. Results are the mean ± S.E. (n = 5). * indicates that the changes were statistically significant (p < 0.001). Snapshots from two slides are shown in C and E. Each slide shows at least one cell with significant knockdown of plakoglobin. Cell nuclei were stained with TOPRO.
DISCUSSION
The transcriptional repressor protein SLUG is implicated in several key aspects of metastasis in the epithelial cancer cells (7–9). It is shown to participate in epithelial to mesenchymal transition through the repression of the cell-cell adhesion molecule E-cadherin (11). It promotes cancer cell growth by direct inhibition of the tumor suppressor protein BRCA2 (15) and by indirect stimulation of cyclin D1 levels through the repression of cyclin D1 ubiquitinating enzyme UbE2D3 (12). It inhibits PUMA to block the induction of anoikis in the cancer cells (31–33). Although cancer cells with abnormally high levels of SLUG protein are highly motile (8, 35–38), direct association of SLUG with cellular motility is not established until our study. Using the isogenic breast cancer cell system, we have shown here that the motility of the SLUG-negative cells is increased significantly with the expression of SLUG in these cells. The increase in cellular motility is contingent upon the repressor function of SLUG, as a non-functional mutant of SLUG failed to increase the motility of these cells. These data suggest that transcriptional suppression of one or more genes by SLUG is responsible for the increase in the motility of the transfected breast cancer cells. We further verified that SLUG is responsible for the increased motility of the SLUG-high breast cancer cells by knocking down SLUG in these cells and showing the decrease in the motility of the SLUG-knocked down cells.
To be metastatic, the breast cancer cells must have a higher motility rate. Motility of the transformed epithelial cancer cells is increased through the trigger of formation of membrane ruffles known as invadopodia (25–27). Among the proteins that are attributed to control the motility of epithelial cells, the catenin protein plakoglobin is implicated as having the central role (17–23). A high plakoglobin level in the cells is translated to lower motility and vice versa (17–23). Our data show that SLUG-negative non-aggressive breast cancer cells express high levels of plakoglobin. Ectopic expression of SLUG in these cells suppressed the levels of plakoglobin, increasing their motility. On the other hand, knockdown of SLUG in the SLUG-high breast cancer cells increased the plakoglobin level and decreased the motility of these cells. This inverse relationship between SLUG and plakoglobin led to our hypothesis that negative regulation of plakoglobin by SLUG increases the motility of the SLUG-high breast cancer cells. Although causal relationship was not established, inverse relationship between SLUG and plakoglobin was also reported in other cancer cells including human pancreatic (39) and colon cancer (40).
To evaluate direct inhibition of plakoglobin gene expression by SLUG, we studied the binding and activity of SLUG to human plakoglobin gene promoter. SLUG is shown to bind to the E2-box sequences at its target gene promoters through its zinc finger domains and recruits co-repressor CtBP1 and the effector HDAC1 to heterochromatinize the promoter DNA, thus to silence the expression of the gene (12, 13, 15). In this report we have shown that SLUG binds to the plakoglobin gene promoter in vivo and inhibits plakoglobin promoter activity in SLUG-expressing human breast cells through chromatin remodeling. Our study also validated that the unique seven-amino acid SLUG motif located at the repressor domain of human SLUG protein is critical for its repressor function, as mutation of this motif abolished the ability of SLUG to inhibit plakoglobin gene promoter activity. The biochemical role of this motif of SLUG in its repressor activity is not known. We found that although this motif has similarity with the classical CtBP1 binding sequence (41, 42), this sequence of SLUG is not responsible for CtBP1 binding (43).3
Our study also reflects upon the importance of E2-box binding of SLUG and its repressor activity. We developed an exonuclease-resistant dsDNA decoy against SLUG that has two E2-box sequences. This decoy E-box dependently binds preferentially to SLUG but not to the similar E2-box-binding protein SNAIL in the breast cancer cells, suggesting potential role of the context sequences around the E2-boxes in determining the specificity. Through in vitro studies we have established the proof of concept that this decoy may be used as a potential drug to inhibit SLUG function in vivo. Such dsDNA decoys are developed against several transcription factors and are being used to successfully treat several experimental diseases (44–49).
To establish further that inhibition of plakoglobin gene alone in the SLUG-negative cells will increase the cellular motility, we knocked down plakoglobin mRNA in these cells by RNA interference. Our data indicated that knockdown of plakoglobin did indeed increase the motility of these cells. We also have shown that knockdown of plakoglobin changes the actin filaments of the cells, indicating the appearance of stress fibers within the cells and increasing the ruffled appearance on the periphery of the plakoglobin-knocked down cells. These phenotypes are indicative of induction of motile behavior of the cells (25–27) and supports our notion that knockdown of plakoglobin is directly responsible for the increase in the motility of these cells.
Several extracellular signals are known to trigger the induction of extracellular signal-regulated kinases (ERKs), which then induce the expression of the transcription factors FRA1 and cJUN (34). These transcription factors are shown to induce SLUG gene expression in human breast cancer cells (34). Induction of ERKs is implicated for increased breast cancer cell motility through the induction of SLUG (34). Our research established that SLUG induction knocks down plakoglobin gene expression and thus increases the motility of the cells, thus contributing toward the understanding of how extracellular signals influence the metastatic behavior of aggressive breast cancer cells. Because SLUG and plakoglobin levels are inversely related in other cancers including pancreatic and colorectal cancers as well (39, 40), our findings will also help in the mechanistic evaluation of metastatic progression of other cancers.
Supplementary Material
Acknowledgments
Confocal microscopy was performed through the use of the Meharry Medical College Morphology Core, which was supported in part by National Institutes of Health grants U01NS041071, U54RR026140, U54CA091408, and S10RR0254970. Other Core facility research was supported in part by Vanderbilt Clinical and Translational Science Awards Grant UL1 RR024975 from the Center for Research Resources, National Institutes of Health.
This work was supported, in whole or in part, by National Institutes of Health Grants 1U54RR026140-01 (to S. M.) and R25GM059993, T32HL007737, and T32HL007735 (graduate student fellowships to C. K. B.). This work was also supported in part by Department of Defense Congressionally Directed Medical Research Program Grants W81XWH-06-1-0466, BC103645, and W81XWH-09-1-0676 (to G. C.)

This article contains supplemental nucleotide sequences, amino acid sequences, Tables S1 and S2, Figs. S1–S7, and tissue microarray data.
C. K. Bailey, M. K. Mittal, and G. Chaudhuri, unpublished information.
- TNBC
- triple negative breast cancer
- CtBP1
- C-terminal binding protein-1
- HDAC1
- histone deacetylase-1
- JUP
- junction plakoglobin
- MyH9
- myosin heavy chain 9
- PGKD
- plakoglobin knockdown.
REFERENCES
- 1. Lehmann B. D., Bauer J. A., Chen X., Sanders M. E., Chakravarthy A. B., Shyr Y., Pietenpol J. A. (2011) Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 121, 2750–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huo D., Ikpatt F., Khramtsov A., Dangou J. M., Nanda R., Dignam J., Zhang B., Grushko T., Zhang C., Oluwasola O., Malaka D., Malami S., Odetunde A., Adeoye A. O., Iyare F., Falusi A., Perou C. M., Olopade O. I. (2009) Population differences in breast cancer. Survey in indigenous African women reveals over-representation of triple-negative breast cancer. J. Clin. Oncol. 27, 4515–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anders C. K., Carey L. A. (2009) Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clin. Breast Cancer. 9, S73–S81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. O'Brien K. M., Cole S. R., Tse C. K., Perou C. M., Carey L. A., Foulkes W. D., Dressler L. G., Geradts J., Millikan R. C. (2010) Intrinsic breast tumor subtypes, race, and long term survival in the Carolina Breast Cancer Study. Clin. Cancer Res. 16, 6100–6110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Amirikia K. C., Mills P., Bush J., Newman L. A. (2011) Higher population-based incidence rates of triple-negative breast cancer among young African-American women. Implications for breast cancer screening recommendations. Cancer 117, 2747–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tawfik O., Davis K., Kimler B. F., Davis M. K., Hull S., Fan F., Khan Q. J., O'Dea A. P., Thomas P. (2010) Clinicopathological characteristics of triple-negative invasive mammary carcinomas in African-American versus Caucasian women. Ann. Clin. Lab. Sci. 40, 315–323 [PubMed] [Google Scholar]
- 7. Shirley S. H., Hudson L. G., He J., Kusewitt D. F. (2010) The skinny on Slug. Mol. Carcinog. 49, 851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alves C. C., Carneiro F., Hoefler H., Becker K. F. (2009) Role of the epithelial-mesenchymal transition regulator Slug in primary human cancers. Front. Biosci. 14, 3035–3050 [DOI] [PubMed] [Google Scholar]
- 9. Nieto M. A. (2002) The snail superfamily of zinc finger transcription factors. Nat. Rev. Mol. Cell Biol. 3, 155–166 [DOI] [PubMed] [Google Scholar]
- 10. Storci G., Sansone P., Trere D., Tavolari S., Taffurelli M., Ceccarelli C., Guarnieri T., Paterini P., Pariali M., Montanaro L., Santini D., Chieco P., Bonafé M. (2008) The basal-like breast carcinoma phenotype is regulated by SLUG gene expression. J. Pathol. 214, 25–37 [DOI] [PubMed] [Google Scholar]
- 11. Hajra K. M., Chen D. Y., Fearon E. R. (2002) The SLUG zinc finger protein represses E-cadherin in breast cancer. Cancer Res. 62, 1613–1618 [PubMed] [Google Scholar]
- 12. Mittal M. K., Singh K., Misra S., Chaudhuri G. (2011) SLUG-induced elevation of D1 cyclin in breast cancer cells through the inhibition of its ubiquitination. J. Biol. Chem. 286, 469–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mittal M. K., Myers J. N., Misra S., Bailey C. K., Chaudhuri G. (2008) In vivo binding to and functional repression of the VDR gene promoter by SLUG in human breast cells. Biochem. Biophys. Res. Commun. 372, 30–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tripathi M. K., Misra S., Chaudhuri G. (2005) Negative regulation of the expressions of cytokeratins 8 and 19 by SLUG repressor protein in human breast cells. Biochem. Biophys. Res. Commun. 329, 508–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tripathi M. K., Misra S., Khedkar S. V., Hamilton N., Irvin-Wilson C., Sharan C., Sealy L., Chaudhuri G. (2005) Regulation of BRCA2 gene expression by the SLUG repressor protein in human breast cells. J. Biol. Chem. 280, 17163–17171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guo W., Keckesova Z., Donaher J. L., Shibue T., Tischler V., Reinhardt F., Itzkovitz S., Noske A., Zürrer-Härdi U., Bell G., Tam W. L., Mani S. A., van Oudenaarden A., Weinberg R. A. (2012) Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148, 1015–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Todorovi V., Desai B. V., Patterson M. J., Amargo E. V., Dubash A. D., Yin T., Jones J. C., Green K. J. (2010) Plakoglobin regulates cell motility through Rho- and fibronectin-dependent Src signaling. J. Cell Sci. 123, 3576–3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang H., Asimaki A., Lo D., McKenna W., Saffitz J. (2008) Disparate effects of different mutations in plakoglobin on cell mechanical behavior. Cell. Motil. Cytoskeleton 65, 964–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shair K. H., Schnegg C. I., Raab-Traub N. (2008) EBV latent membrane protein 1 effects on plakoglobin, cell growth, and migration. Cancer Res. 68, 6997–7005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pan H., Gao F., Papageorgis P., Abdolmaleky H. M., Faller D. V., Thiagalingam S. (2007) Aberrant activation of γ-catenin promotes genomic instability and oncogenic effects during tumor progression. Cancer Biol. Ther. 6, 1638–1643 [DOI] [PubMed] [Google Scholar]
- 21. Shafiei F., Rahnama F., Pawella L., Mitchell M. D., Gluckman P. D., Lobie P. E. (2008) DNMT3A and DNMT3B mediate autocrine hGH repression of plakoglobin gene transcription and consequent phenotypic conversion of mammary carcinoma cells. Oncogene 27, 2602–2612 [DOI] [PubMed] [Google Scholar]
- 22. Rieger-Christ K. M., Ng L., Hanley R. S., Durrani O., Ma H., Yee A. S., Libertino J. A., Summerhayes I. C. (2005) Restoration of plakoglobin expression in bladder carcinoma cell lines suppresses cell migration and tumorigenic potential. Br. J. Cancer. 92, 2153–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yin T., Getsios S., Caldelari R., Kowalczyk A. P., Müller E. J., Jones J. C., Green K. J. (2005) Plakoglobin suppresses keratinocyte motility through both cell-cell adhesion-dependent and -independent mechanisms. Proc. Natl. Acad. Sci. U.S.A. 102, 5420–5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Coates J. C. (2003) Armadillo repeat proteins. Beyond the animal kingdom. Trends Cell Biol. 13, 463–471 [DOI] [PubMed] [Google Scholar]
- 25. Ridley A. J. (2011) Life at the leading edge. Cell 145, 1012–1022 [DOI] [PubMed] [Google Scholar]
- 26. Murphy D. A., Courtneidge S. A. (2011) The “ins” and “outs” of podosomes and invadopodia. Characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12, 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Destaing O., Block M. R., Planus E., Albiges-Rizo C. (2011) Invadosome regulation by adhesion signaling. Curr. Opin. Cell Biol. 23, 597–606 [DOI] [PubMed] [Google Scholar]
- 28. Caca K., Kolligs F. T., Ji X., Hayes M., Qian J., Yahanda A., Rimm D. L., Costa J., Fearon E. R. (1999) β- and γ-catenin mutations, but not E-cadherin inactivation, underlie T-cell factor/lymphoid enhancer factor transcriptional deregulation in gastric and pancreatic cancer. Cell Growth Differ. 10, 369–376 [PubMed] [Google Scholar]
- 29. Misra S., Sharma S., Agarwal A., Khedkar S. V., Tripathi M. K., Mittal M. K., Chaudhuri G. (2010) Cell cycle-dependent regulation of the bidirectional overlapping promoter of human BRCA2/ZAR2 genes in breast cancer cells. Mol. Cancer 9, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wei S., Gao X., Du J., Su J., Xu Z. (2011) Angiogenin enhances cell migration by regulating stress fiber assembly and focal adhesion dynamics. PLoS One 6, e28797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu W. S., Heinrichs S., Xu D., Garrison S. P., Zambetti G. P., Adams J. M., Look A. T. (2005) Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell 123, 641–653 [DOI] [PubMed] [Google Scholar]
- 32. Zilfou J. T., Spector M. S., Lowe S. W. (2005) Slugging it out. Fine tuning the p53-PUMA death connection. Cell 123, 545–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leong K. G., Niessen K., Kulic I., Raouf A., Eaves C., Pollet I., Karsan A. (2007) Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J. Exp. Med. 204, 2935–2948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen H., Zhu G., Li Y., Padia R. N., Dong Z., Pan Z. K., Liu K., Huang S. (2009) Extracellular signal-regulated kinase signaling pathway regulates breast cancer cell migration by maintaining slug expression. Cancer Res. 69, 9228–9235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McConkey D. J., Choi W., Marquis L., Martin F., Williams M. B., Shah J., Svatek R., Das A., Adam L., Kamat A., Siefker-Radtke A., Dinney C. (2009) Role of epithelial-to-mesenchymal transition (EMT) in drug sensitivity and metastasis in bladder cancer. Cancer Metastasis Rev. 28, 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Anastassiou D., Rumjantseva V., Cheng W., Huang J., Canoll P. D., Yamashiro D. J., Kandel J. J. (2011) Human cancer cells express Slug-based epithelial-mesenchymal transition gene expression signature obtained in vivo. BMC Cancer 11, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Katafiasz D., Smith L. M., Wahl J. K., 3rd (2011) Slug (SNAI2) expression in oral SCC cells results in altered cell-cell adhesion and increased motility. Cell Adh. Migr. 5, 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wels C., Joshi S., Koefinger P., Bergler H., Schaider H. (2011) Transcriptional activation of ZEB1 by Slug leads to cooperative regulation of the epithelial-mesenchymal transition-like phenotype in melanoma. J. Invest. Dermatol. 131, 1877–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang A. D., Camp E. R., Fan F., Shen L., Gray M. J., Liu W., Somcio R., Bauer T. W., Wu Y., Hicklin D. J., Ellis L. M. (2006) Vascular endothelial growth factor receptor-1 activation mediates epithelial to mesenchymal transition in human pancreatic carcinoma cells. Cancer Res. 66, 46–51 [DOI] [PubMed] [Google Scholar]
- 40. Li Y., Zhu X., Zeng Y., Wang J., Zhang X., Ding Y. Q., Liang L. (2010) FMNL2 enhances invasion of colorectal carcinoma by inducing epithelial-mesenchymal transition. Mol. Cancer Res. 8, 1579–1590 [DOI] [PubMed] [Google Scholar]
- 41. Chinnadurai G. (2009) The transcriptional corepressor CtBP: a foe of multiple tumor suppressors. Cancer Res. 69, 731–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chinnadurai G. (2007) Transcriptional regulation by C-terminal binding proteins. Int. J. Biochem. Cell Biol. 39, 1593–1607 [DOI] [PubMed] [Google Scholar]
- 43. Bailey C. K., Misra S., Mittal M. K., Chaudhuri G. (2007) Human SLUG does not directly bind to CtBP1. Biochem. Biophys. Res. Commun. 353, 661–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cabrini G., Bezzerri V., Mancini I., Nicolis E., Dechecchi M. C., Tamanini A., Lampronti I., Piccagli L., Bianchi N., Borgatti M., Gambari R. (2010) Targeting transcription factor activity as a strategy to inhibit pro-inflammatory genes involved in cystic fibrosis. Decoy oligonucleotides and low molecular weight compounds. Curr. Med. Chem. 17, 4392–4404 [DOI] [PubMed] [Google Scholar]
- 45. Gambari R., Borgatti M., Bezzerri V., Nicolis E., Lampronti I., Dechecchi M. C., Mancini I., Tamanini A., Cabrini G. (2010) Decoy oligodeoxyribonucleotides and peptide nucleic acid-DNA chimeras targeting nuclear factor κB. Inhibition of IL-8 gene expression in cystic fibrosis cells infected with Pseudomonas aeruginosa. Biochem. Pharmacol. 80, 1887–1894 [DOI] [PubMed] [Google Scholar]
- 46. De Stefano D., De Rosa G., Carnuccio R. (2010) NFκB decoy oligonucleotides. Curr. Opin. Mol. Ther. 12, 203–213 [PubMed] [Google Scholar]
- 47. Piva R., Gambari R. (2002) Transcription factor decoy (TFD) in breast cancer research and treatment. Technol. Cancer Res. Treat. 1, 405–416 [DOI] [PubMed] [Google Scholar]
- 48. Piva R., Penolazzi L., Zennaro M., Bianchini E., Magri E., Borgatti M., Lampronti I., Lambertini E., Tavanti E., Gambari R. (2006) Induction of apoptosis of osteoclasts by targeting transcription factors with decoy molecules. Ann. N.Y. Acad. Sci. 1091, 509–516 [DOI] [PubMed] [Google Scholar]
- 49. Penolazzi L., Zennaro M., Lambertini E., Tavanti E., Torreggiani E., Gambari R., Piva R. (2007) Induction of estrogen receptor α expression with decoy oligonucleotide targeted to NFATc1 binding sites in osteoblasts. Mol. Pharmacol. 71, 1457–1462 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.