Background: N-Glycan structures of the human pathogenic yeast C. neoformans have not yet been elucidated.
Results: Cryptococcal N-glycans were composed of mostly mannoses with addition of xylose and xylose phosphate residues.
Conclusion: Cryptococcal N-glycans show serotype-specific structures with variation in the length and in the presence of xylose.
Significance: This is the first survey on the structure and biosynthesis pathway of C. neoformans N-glycans.
Keywords: Glycomics, Glycoprotein, Glycosylation, Pathogenesis, Yeast, Cryptococcus neoformans, N-Linked Glycosylation, Cell Wall Mannoproteins, Glycan Structure
Abstract
The encapsulated fungal pathogen Cryptococcus neoformans causes cryptococcosis in immunocompromised individuals. Although cell surface mannoproteins have been implicated in C. neoformans pathogenicity, the structure of N-linked glycans assembled on mannoproteins has not yet been elucidated. By analyzing oligosaccharide profiles combined with exoglycosidase treatment, we report here that C. neoformans has serotype-specific high mannose-type N-glycans with or without a β1,2-xylose residue, which is attached to the trimannosyl core of N-glycans. Interestingly, the neutral N-glycans of serotypes A and D were shown to contain a xylose residue, whereas those of serotype B appeared to be much shorter and devoid of a xylose residue. Moreover, analysis of the C. neoformans uxs1Δ mutant demonstrated that UDP-xylose is utilized as a donor sugar in N-glycan biosynthesis. We also constructed and analyzed a set of C. neoformans mutant strains lacking genes putatively assigned to the reconstructed N-glycan biosynthesis pathway. It was shown that the outer chain of N-glycan is initiated by CnOch1p with addition of an α1,6-mannose residue and then subsequently extended by CnMnn2p with multiple additions of α1,2-mannose residues. Finally, comparative analysis of acidic N-glycans from wild-type, Cnoch1Δ, Cnmnn2Δ, and Cnuxs1Δ strains strongly indicated the presence of xylose phosphate attached to mannose residues in the core and outer region of N-glycans. Our data present the first report on the unique structure and biosynthesis pathway of N-glycans in C. neoformans.
Introduction
The encapsulated basidiomycetous Cryptococcus neoformans species complex is an opportunistic fungal pathogen causing fatal cryptococcal meningoencephalitis in immunocompromised populations, such as AIDS patients, if left untreated (1). The capsule of C. neoformans, a major immunomodulatory and antiphagocytic cellular structure, is mainly composed of two polysaccharides, glucuronoxylomannan (GXM)3 and glucuronoxylomannogalactan. C. neoformans species are categorized into serotypes A–D based on the number of xylose residues on the mannose backbone in GXM (2). Recently, serotypes B and C have been classified as an independent species, named Cryptococcus gattii, because they cause a fatal cryptococcosis even in immunocompetent persons (3). The pathogenicity of the Cryptococcus species complex has long been linked to its polysaccharide capsule as a key virulence factor, but it was recently also associated with cell-bound mannoproteins (4). Several studies showed that cryptococcal mannoproteins secreted or localized on the cell surface stimulate host T-cell responses (1, 5, 6).
Glycans attached to proteins serve various functions, including correct protein conformation, stabilization of proteins against denaturation and proteolysis, and mediation of host-pathogen protein interactions (7). In yeast and fungi belonging to Ascomycota and Basidiomycota, N-linked oligosaccharides are mostly high mannose types with some modifications such as addition of N-acetylglucosamine, galactose, galactofuranose, fucose, pyruvate, or phosphate (8–10). In the ascomycetous yeast Saccharomyces cerevisiae, hypermannosylation of the Man8GlcNAc2 core glycan is initiated with the addition of an α1,6-linked mannose residue by ScOch1p in the Golgi. Subsequently, the α1,6 backbone of the outer chain is extended by mannan polymerase (M-pol) I and II and further elaborated with α1,2- and 1,3-linked mannoses by various mannosyltransferases such as ScMnn2p/ScMnn5p, ScKtr protein family, and ScMnn1p (11–13). The outer chains and core N-glycans of S. cerevisiae are modified by the addition of phosphomannan by ScMnn4p/ScMnn6p, generating acidic glycans with negative charge (14). S. cerevisiae mutants lacking the OCH1 gene (Scoch1Δ) exhibit hypersensitivity to high temperature and cell wall perturbation reagents (15). Candida albicans, an ascomycetous yeast and opportunistic human pathogen, has an N-glycan structure different from that of S. cerevisiae, because Candida N-glycans are modified with both α- and β-linked mannoses (16, 17). Deletion of Candida OCH1 results in hypersensitivity to cell wall-perturbing agents and an attenuation of virulence in a murine model of systemic candidiasis (18). The OCH1 deletion mutants of other ascomycetous yeast species, such as Kluyveromyces lactis, Hansenula polymorpha, Yarrowia lipolytica, Schizosaccharomyces pombe, and Pichia pastoris, and an ascomycetous filamentous fungus Neurospora crassa also exhibit altered morphological phenotypes under stress conditions, suggesting that outer chain N-glycans are important for cell wall integrity in yeast and fungal species (19–24). The only exception was observed in the ascomycetous filamentous fungus and opportunistic human pathogen Aspergillus fumigatus, where deletion of OCH1 does not affect normal growth even under various stress conditions (25).
Based on the Cryptococcus genome database, C. neoformans is predicted to have more than 50 putative mannoproteins, which typically contain potential Asn (N)-glycosylation sites, putative Ser/Thr (S/T)-rich regions for O-glycosylation, and a glycosylphosphatidylinositol anchor (6, 26). However, only a small number of C. neoformans glycoproteins have been characterized, and detailed investigation on oligosaccharide structures has not been carried out except for carbohydrate composition (27–29). Previous work involving bioinformatics and radioactive N-glycan analyses indicated that the structure of the dolichol-linked N-glycosylation precursor in the ER of C. neoformans is Man9GlcNAc2-PP-Dol with no addition of glucose residues due to a lack of Alg glucosyltransferases (Alg6p, Alg8p, and Alg10p), implying different processing of cryptococcal N-glycans in the ER (30). In addition, C. neoformans was predicted to lack most of the genes encoding M-pol I and II subunits, suggesting N-glycans have less elongated α1,6-mannan chains compared with those of S. cerevisiae (31). However, only limited information is available on the structural characteristics of the N-linked glycans in Cryptococcus species. In this study, we performed a comparative N-glycan profile analysis of C. neoformans species complex using MALDI-TOF mass spectrometry and HPLC, and we present the first report on the serotype-specific presence of a β1,2-xylose residue in high mannose type N-glycans in C. neoformans. In addition, we showed that the OCH1 and MNN2 genes play major roles in cryptococcal N-glycan processing in the Golgi. Furthermore, we provide a line of data supporting the presence of xylose phosphate residues on the core and outer regions of N-glycans in Cryptococcus.
EXPERIMENTAL PROCEDURES
Strains and Culture Conditions
The yeast strains used in this study are listed in supplemental Table 1 and were generally cultured in YPD broth medium (1% w/v yeast extract, 2% w/v peptone, 2% w/v glucose) with shaking (200 rpm) at 30 °C. YPDNAT (YPD solid medium containing 100 μg/ml nourseothricin, Werner BioAgents; where NAT is nourseothricin acetyltransferase) or YPDG418 (YPD solid medium containing 200 μg/ml geneticin, Sigma) was used for selection of C. neoformans transformants.
DNA Manipulation
The primer sets used for disruption and reintegration of genes in this study are listed in supplemental Table 2. Each gene was disrupted in the C. neoformans serotype A H99 (MATα) strain background using overlap PCR or double joint-PCR strategies followed by biolistic transformation as described previously (32). Genomic DNA of C. neoformans transformants grown on YPDNAT was isolated from cell lysate using acid-washed glass beads (425–600 μm, Sigma), and deletion of CnOCH1, CnMNN2, and CnKTR3 was confirmed by PCR (supplemental Fig. 1). For reintegration of wild-type genes into the corresponding mutant strains, genomic DNA fragments of CnOCH1 and CnMNN2 were obtained and introduced into the original genomic locus using pJAF1 (33) with the G418 resistance marker (supplemental Fig. 1).
Preparation of Glycoproteins
To obtain cell wall mannoproteins (cwMPs), C. neoformans cells freshly grown on YPD plates for 2 days were inoculated in 200 ml of YPD and incubated at 30 °C for 24 h with shaking at 200 rpm. The cells at the stationary phase (A600 = 40–50) were harvested and washed with water. To reduce the possibility of capsular polysaccharide contamination, the washed cells were incubated twice for 30 min at room temperature in an equal volume of DMSO as described previously (34). After centrifugation, the supernatants were decanted, and the cell pellets were washed with water. The washed cells were then resuspended in 0.1 m citrate buffer, pH 7.0, autoclaved at 121 °C for 120 min, and centrifuged at 4,000 × g for 10 min at 4 °C (35). The supernatants (mixture of crude cell wall proteins and capsular polysaccharides) were recovered, and capsular polysaccharides were precipitated from the supernatants by slowly adding an equal volume of cold ethanol to the supernatants and removed by filtration with a syringe filter (0.45-μm, Sartorius Stedim Biotech). After addition of 3 volumes of ethanol to the filtrate and incubation at 4 °C overnight, crude cell wall proteins were collected by centrifugation at 4,000 × g for 30 min at 4 °C. The dried cell wall proteins were then dissolved with 10 ml of concanavalin A (ConA) binding buffer (20 mm Tris-HCl, pH 7.4, 0.5 m NaCl, 1 mm CaCl2, 1 mm MnCl2) and incubated with 1 ml of ConA-Sepharose beads (GE Healthcare) in a column for 2 h with slow rotation. The beads were then washed with 10 ml of ConA binding buffer, and cwMPs were eluted by addition of 5 ml of 1 m methyl-α-d-mannopyranoside. For isolation of secretory mannoproteins (sMPs), supernatants from 500 ml of YPD cultures incubated for 48 h at 30 °C were obtained by centrifugation and filtration using a 0.45-μm filter membrane, concentrated, and then exchanged with PBS buffer by tangential flow filtration using a 30-kDa cassette (PXC030C50, Millipore). The supernatants were further concentrated by using an Amicon-15 (30,000 molecular weight cutoff, Millipore). sMPs were purified from the concentrated culture supernatants using ConA, as described for the purification of cwMPs, except that PBS buffer was used instead of Tris-HCl buffer. The eluted glycoproteins were dialyzed with water for 2 days and dried using a vacuum evaporator (Hanil Scientific). The dried glycoproteins were dissolved in water and quantified by spectrophotometer (NanoDrop, Thermoscientific).
Profiling of N-Linked Oligosaccharides by MALDI-TOF
From purified cwMPs or sMPs (100–200 μg), N-linked glycans were isolated using 3 μl of peptide:N-glycanase F (500 units/μl, New England Biolabs) and then purified by Carbograph Extract-CleanTM column (150 mg, Alltech). For MALDI-TOF analysis, a matrix solution consisting of 6-aza-2-thiothymine and 2,5-dihydroxybenzoic acid (Bruker Daltonics Inc.) (v/v, 1:1) in 0.25% acetonitrile (Burdick & Jackson) and 0.075% trifluoroacetic acid was mixed with samples of equal volume. The glycan samples were dried and then analyzed using MicroflexTM mass spectrometer (Bruker Daltonics Inc.) operated in the reflective positive mode for neutral glycan analysis or in the linear negative mode for acidic glycan analysis. Alternatively, isolated N-glycans were dried, resuspended in 50 μl of fresh 1% (w/v) sodium acetate·3H2O, and then labeled with 100 μl of 2-aminobenzoic acid (2-AA) solution (30 mg of 2-AA and 30 mg of NaBH3CN in 1 ml of 4% sodium acetate·3H2O, 2% boric acid, in methanol) at 80 °C for 45 min. Labeled N-glycans were purified using a SampliQ Cyano cartridge (100 mg, Agilent) to remove excess 2-AA.
Exoglycosidase Treatment
Purified N-glycans were reacted with 1 μl of α1,2-mannosidase (α1,2-MNS, 0.1 milliunit/μl, Prozyme) in 20 mm ammonium acetate buffer, pH 5.0, for 12 h at 37 °C, and half of the mixture with an additional 1 μl of α1,2-MNS was further incubated for 12 h. The other half of the α1,2-MNS-treated mixture was subsequently reacted with 1 μl of α1,6-mannosidase (α1,6-MNS, 40 units/μl, New England Biolabs) for 12 h at 37 °C. Enzymes were removed using a 10K Microcon (Millipore), dried in a vacuum evaporator (Hanil Scientific), and reconstituted with 3–5 μl of water for mass analysis. To determine the presence of xylose residue, 2-AA labeled N-glycans in 20 mm ammonium acetate buffer, pH 5.0, were mixed with 2 μl of jack bean α-mannosidase (JBM, 150 milliunits/μl, Prozyme), incubated for 48 h at 25 °C, and then successively treated with 1 μl of β1,2-xylosidase (from Xanthomonas sp., 20 microunits/μl, Calbiochem) for 24 h at 37 °C. After removal of exoglycosidases by filtration through a 10K Microcon, glycan profiles were analyzed by normal phase HPLC. Two peaks generated after JBM treatment were fractionated and further analyzed by MALDI-TOF.
α1,6-Mannosyltransferase Activity Assay
Membrane fractions were obtained as described previously with slight modification (21). Pre-cultured C. neoformans in YPD was inoculated in 500 ml of YPD medium at initial optical density (A600 = 0.5), and cultured to mid-log phase (A600 = 5). The cells were harvested by centrifugation (3,000 × g, 10 min), washed with water, and resuspended in 5 ml of PMS buffer (50 mm Tris-HCl, pH 7.5, 5% glycerol, and 2 μl/ml protease inhibitor mixture (Sigma)). Aliquots (500 μl) were transferred to 1.5-ml microcentrifuge tubes; glass beads were added at half the volume of the cell suspension, and cell lysis was then carried out through 5–10 cycles of alternating 1-min vortexing and 1-min cooling on ice. After ∼50% of the cells were disrupted (as assessed by microscopy), lysates were centrifuged at 10,000 × g for 20 min. The supernatant was separated, and an equal volume of PMS buffer was added to the pellets. After a second round of cell lysis, the supernatant was removed and added to the first round supernatant. Total supernatant (S1) was further centrifuged at 100,000 × g for 1 h. High speed pellets were collected, resuspended in 100 μl of 50 mm Tris-HCl buffer, pH 7.5, plus 5% glycerol, and stored at 4 °C. Protein concentrations of high speed pellets were determined using the protein assay reagent (Bio-Rad). α1,6-Mannosyltransferase activity was assayed as described previously with slight modification (21). High speed pellets (500 μg) were incubated in 100 μl of 50 mm Tris-HCl, pH 7.5, buffer containing 2 mm MnCl2, 1 mm GDP-mannose, 0.5 mm 1-deoxymannojirimycin, and 0.1 μg of Man8GlcNAc2-AA (Prozyme) as an acceptor at 30 °C overnight. The reaction mixture was filtered through a Microcon (YM-10, Millipore), and the filtrate was analyzed by HPLC. To identify linkage, the Golgi reaction mixture was treated with α1,2-MNS.
N-Glycan Analysis by HPLC
Normal phase HPLC was conducted using an Asahipak NH2P-50 4E column (0.46 × 25 cm, 5 μm, Shodex) at a rate of 1.0 ml/min with Solvent A (100% acetonitrile) and Solvent B (50 mm ammonium formate in water, pH 4.4). The column was equilibrated with a solution containing 50% Solvent A and 50% Solvent B. After sample injection, the proportion of Solvent B was increased in a linear fashion to 68% for 50 min. 2-AA-oligosaccharides were analyzed with a Waters HPLC system composed of 515 dual pumps, a 717 plus autosampler, and a 2475 fluorescence detector with excitation and emission wavelengths of 360 and 425 nm, respectively. Data were collected using EmpowerTM 2 chromatography data software (Waters).
For acidic glycan analysis, the column was equilibrated with a solution containing 90% Solvent C (2% acetic acid, 1% tetrahydrofuran in acetonitrile) and 10% Solvent D (5% acetic acid, 3% triethylamine, 1% tetrahydrofuran in water). After sample injection (50 μl), the proportion of Solvent D was increased in a linear fashion up to 90% for 60 min. 2-AA-oligosaccharides were detected using the same Waters HPLC system described above. After fractionation, each sample was dried and dissolved in water. The peaks separated on HPLC were further identified by MALDI-TOF mass spectrometry using the linear negative mode. All chemicals were purchased from Sigma unless mentioned otherwise.
RESULTS
Neutral N-Glycan Structures of C. neoformans
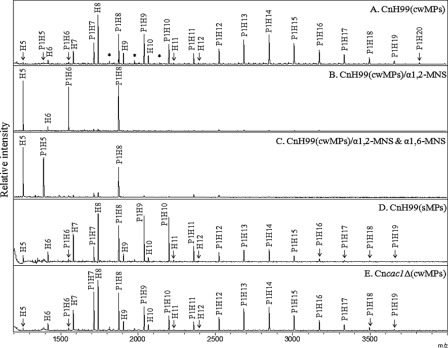
To obtain general information on the structure of neutral N-glycans of the Cryptococcus species complex, MALDI-TOF analysis was carried out in the positive mode with N-linked oligosaccharides assembled on cwMPs derived from serotype A strain H99 (α-mating type) (Fig. 1A). A series of major peaks observed at m/z of 1258.1, 1420.1, 1582.1, 1744.1, 1906.0, 2067.9, 2230.1, and 2392.1 were matched well to high mannose type N-glycans ranging from Hex5HexNAc2 to Hex12HexNAc2 (H5–12), originating from the ascomycetous yeast and fungal species (glycan structure database of the Consortium for Functional Glycomics). In contrast, other major peaks with a mass value 30.0 m/z smaller than those of HexnHexNAc2 could not be assigned to any reported glycans. Considering that the mass difference of each peak corresponds to the molecular weight of C1H2O1, it was speculated that the unassigned glycan structures might be oligosaccharides containing a pentose residue (P1H5–20, Pen1Hex5–20HexNAc2).
FIGURE 1.
N-Linked oligosaccharide profiles of C. neoformans serotype A. N-Glycans of cwMPs and sMPs from the serotype A H99 strain, cultivated up to the stationary phase, were analyzed by MALDI-TOF mass spectrometry in the positive mode. A, no mannosidase treatment. B, α1,2-mannosidase (α1,2-MNS) treatment. C, α1,6-mannosidase (α1,6-MNS) treatment of α1,2-MNS-treated N-glycans. D and E, sMPs and cwMPs from the capsule-defective strain YSB42 (Cncac1Δ), respectively. The mass difference between peaks in each type of glycan is about 162 m/z, which corresponds to the mass of a single hexose residue. P, pentose; H, hexose; *, unidentified peak.
Given that most fungal N-glycans are of high mannose type, we predicted that N-linked oligosaccharides of C. neoformans would also be mostly constituted of mannose residues. As expected, digestion of N-glycans from serotype A strain H99 with α1,2-MNS resulted in the convergence of most peaks to H5–6 and P1H6–8, indicating that the outer chains of cryptococcal N-glycans were mostly extended by α1,2-mannose residues without a terminal α1,3-mannose cap (Fig. 1B). Subsequent digestion with α1,6-MNS resulted in the shift of H6 to H5 and P1H6 to P1H5, although the peak of Pen1Hex8GlcNAc2 (P1H8) was not shifted (Fig. 1C). The P1H8 glycan seems to be an incompletely digested product that was derived from a fraction of glycans lacking α1,6-mannose extension, because it was converted to P1H5 by additional prolonged digestion with α1,2-mannosidase (data not shown). These results indicated that the outer chains contained a single α1,6-mannose extension. The profiles of N-linked oligosaccharides obtained from sMPs of the H99 strain also showed the same pattern of H5–12 glycans containing one residue of pentose as observed in those from cwMPs (Fig. 1D). We also analyzed glycan profiles of the C. neoformans cac1Δ mutant, which has a deletion of the adenylyl cyclase gene and is deficient in capsule production (36), and confirmed the same profile pattern (Fig. 1E) thus excluding the possibility of capsule oligosaccharide contamination in the preparation of N-linked glycans. Exoglycosidase treatment of glycan samples from the Cncac1Δ mutant also showed the same result as the wild type (data not shown). The findings strongly suggested that the N-glycans of serotype A strain were extended with mostly α1,2-linked mannose residues and a single α1,6-linked mannose residue and were further modified with a pentose residue bound to the inner region of the N-glycan.
Comparison of Neutral N-Glycan Profiles among Different Serotype Strains
Next, we examined whether the neutral N-glycan profiles of C. neoformans vary depending on mating type or serotypes. It was previously reported that phenotypic variants of one C. neoformans strain differ with respect to virulence and the arrangement of xylose moieties within the GXM of the capsule (2). Mating type has also been implicated as a virulence factor in C. neoformans. Epidemiological studies have shown that clinical isolates mostly have the α-mating type (96% average) (37). In addition, α strains were reported as more virulent than congenic a strains in serotype D but not in serotype A (38).
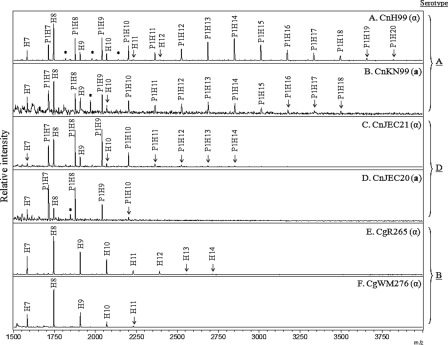
There were no apparent differences found when the N-glycan profiles of strain H99 were compared with mating type α or when congenic strain KN99 was compared with mating type a (Fig. 2, A and B), indicating that glycan profiles were not affected by mating type. The N-glycans of serotype D strains, JEC21 (mating type α) and JEC20 (mating type a), were shown to be shorter than those of serotype A; however, they also contained a pentose residue (H5–10 and P1H5–14) (Fig. 2, C and D). In contrast, the N-glycans of C. gattii serotype B strains, R265 and WM276 (both mating type α but different molecular types), consisted of H5–14 without a pentose residue, similar to those of other yeast species in general (Fig. 2, E and F). The N-glycans of JEC21 and CgR265 were shifted to P1H5 and H5, respectively, by serial treatment with α1,2- and 1,6-MNSs, indicating their core N-glycans were also extended by addition of α1,2- and α1,6-linked mannose residues even though the extension is quite limited in serotypes B and D strains (supplemental Fig. 2, A and B). These results strongly suggested that cryptococcal N-glycans displayed serotype-specific differences not only in length but also in the presence of pentose.
FIGURE 2.
Comparison of N-glycan profiles between different serotypes and mating types. N-Glycans of cell wall mannoproteins from various serotypes of C. neoformans, cultivated up to the stationary phase, were analyzed by MALDI-TOF spectrometry in the positive mode. A and B, serotype A strains H99 (MATα) and KN99 (MATa), respectively; C and D, serotype D strains JEC21 (MATα) and JEC20 (MATa), respectively; and E and F, serotype B strains R265 and WM276 (both MATα), respectively. P, pentose; H, hexose; *, unidentified peak.
Identification of a Pentose Residue as Xylose
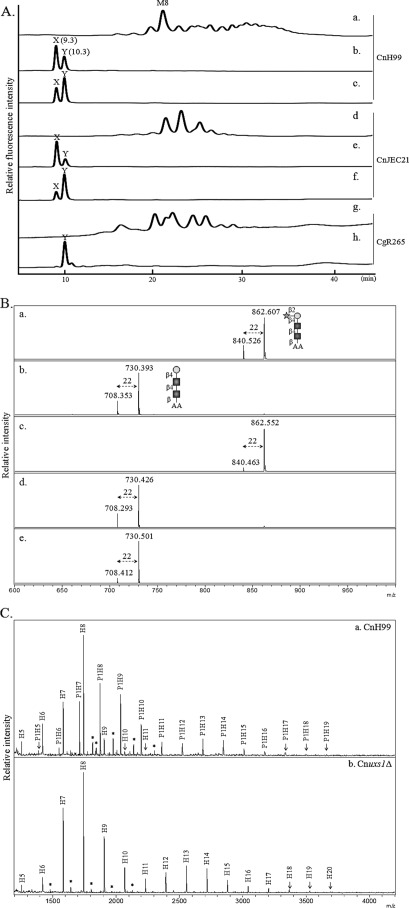
In Cryptococcus, xylose is a sole pentose residue that is incorporated as a component of GXM, glucuronoxylomannogalactan, glucosylinositol phosphorylceramides, and O-glycans (39). The possible presence of xylose in N-glycans of C. neoformans has been implicated by a previous report showing that the cryptococcal laccase 1 protein (Lac1) from serotype D strain contains 4 mol of glucosamine and 22 mol of mannose/xylose/mol of protein (27). To determine whether the pentose attached to the inner region of the N-glycans was a xylose residue, 2-AA-labeled N-glycans fractionated in HPLC using an amine column were digested by treatment with JBM, which cleaves α1,2/3/6-linked mannoses. After JBM treatment, the N-glycans of serotypes A and D (Fig. 3A, panels a and d) converged to two peaks (X and Y, Fig. 3A, panels b and e). Then the subsequent β1,2-xylosidase treatment of the JBM-digested N-glycans resulted in an increased intensity of peak Y (Fig. 3A, panels c and f). In the case of the serotype B, N-glycans were already shifted to a single peak (Y, Fig. 3A, panels g and h) by treatment with JBM only. Peaks X and Y from JMB-treated N-glycans of three serotype strains were fractionated and analyzed for mass by MALDI-TOF. As shown in Fig. 3B, peaks X and Y corresponding to m/z of 862.6 and 730.3, respectively, were assigned as sodium adducts of P1H1-AA and H1-AA, respectively. Based on the data showing that a pentose residue was still retained even after removal of α1,2/3-mannoses from the trimannosyl core of N-glycans and that the pentose residue was removed by β1,2-xylosidase treatment, we speculated that P1H1 and H1 correspond to Xyl1Man1GlcNAc2 and Man1GlcNAc2, respectively. Thus, it is highly likely that a single xylose residue is attached to the first mannose residue of the trimannosyl core of N-glycans.
FIGURE 3.
Structural analysis of xylose-containing neutral N-glycans. A, HPLC analysis. N-Glycans of C. neoformans H99 (panel a) and JEC21 (panel d) were treated serially with jack bean α-mannosidase (panels b and e) and β1,2-xylosidase (panels c and f). In the case of C. gattii R265 (panel g), the N-glycans were treated only with JBM (panel h). B, MALDI-TOF analysis. Two peaks (X and Y) from JBM-treated N-glycans of H99 (A, panel b) and JEC21 (A, panel e) as well as a single peak (Y) from JBM-treated N-glycans in R265 (A, panel h) were fractionated and analyzed by MALDI-TOF. Sodium adducts (+22) of the mass spectra (m/z) were identified as major mass peaks. Square, circle, and star symbolize N-acetylglucosamine, mannose, and xylose residues, respectively. C, N-glycan profile of Cnuxs1Δ strain. N-Glycans of cell wall mannoproteins from the C. neoformans H99 wild-type (panel a) and Cnuxs1Δ mutant (panel b) strains were analyzed by MALDI-TOF spectrometry in the positive mode. P, pentose; H, hexose; *, unidentified peak.
Moreover, we constructed a Cnuxs1Δ mutant strain with a defect in the synthesis of UPD-xylose (UDP-Xyl) and compared its N-glycan profile to that of the wild-type strain. It was previously reported that the addition of xylose to the capsular polysaccharides GXM and glucosylinositol phosphorylceramides is defective in the Cnuxs1Δ mutant (40). As seen in Fig. 3C, the N-glycans from cwMPs of the Cnuxs1Δ mutant showed only high mannose-type N-glycans without pentose residues, strongly supporting the idea that the pentose residue was xylose. These results also indicated that UDP-Xyl was utilized as a sugar donor not only for capsule biosynthesis but also for N-glycan biosynthesis of C. neoformans through the classical protein secretory pathway.
In Silico Analysis of C. neoformans N-Glycan Biosynthesis Pathway
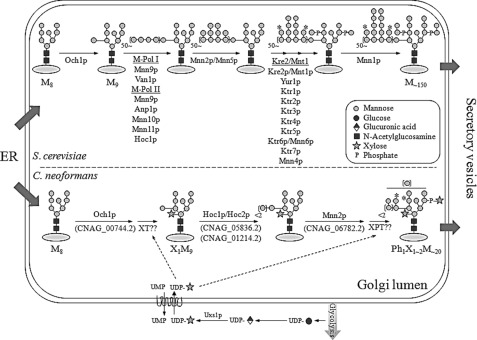
To reconstruct the N-glycan outer chain biosynthesis pathway of C. neoformans, we aimed to identify C. neoformans genes orthologous to S. cerevisiae genes participating in the N-glycan biosynthesis pathway (Table 1). The in silico analysis based on the genome database of C. neoformans serotype A (H99) and serotype D (JEC21) suggested that several Golgi glycosyltransferase genes involved in processing the outer chain of N-glycans in S. cerevisiae were missing in C. neoformans. For example, C. neoformans does not have orthologs for M-pol I and II complex formation, such as VAN1, ANP1, MNN10, or MNN11, which are responsible for elongation of the α1,6-mannose backbone of outer chains. Furthermore, C. neoformans did not appear to contain orthologs of MNN4 responsible for mannosyl phosphorylation and MNN1 for addition of the terminal α1,3-mannose residue in S. cerevisiae glycans, indicating a different structure of the cryptococcal N-glycan outer chains.
TABLE 1.
S. cerevisiae genes involved in the outer chain N-linked glycosylation pathway and homologous genes identified in the C. neoformans genome
| Gene family | Related member |
E value | Function in S. cerevisiae | |
|---|---|---|---|---|
| S. cerevisiae | Cn_serotype A (H99) | |||
| OCH1 | α1,6-Mannosyltransferase | |||
| Och1 | CNAG_00744.2 | 8 e-37 | α1,6-Initiating mannosyltransferase | |
| Hoc1 | CNAG_05836.2, CNAG_01214.2 | 4 e-23, 2 e-15 | α1,6-Mannosyltransferase | |
| MNN9 | Subunit of Golgi mannosyltransferase complex | |||
| Mnn9 | Subunit of Golgi mannosyltransferase complex | |||
| Anp1 | Subunit of the α1,6-mannosyltransferase complex | |||
| Van1 | Component of the mannan polymerase I | |||
| MNN10 | Subunit of a Golgi mannosyltransferase complex | |||
| Mnn10 | Subunit of a Golgi mannosyltransferase complex | |||
| Mnn11 | Subunit of a Golgi mannosyltransferase complex | |||
| MNN2/MNN5 | α1,2-Mannosyltransferase | |||
| Mnn2 | CNAG_06782.2 | 2 e-15 | α1,2-Mannosyltransferase | |
| Mnn5 | α1,2-Mannosyltransferase | |||
| KRE2/MNT1 | α1,2-Mannosyltransferase | |||
| Kre2/Mnt1 | α1,2-Mannosyltransferase | |||
| Ktr1 | α1,2-Mannosyltransferase | |||
| Ktr2 | α1,2-Mannosyltransferase | |||
| Ktr3 | CNAG_03832.2 | 2 e-98 | α1,2-Mannosyltransferase | |
| Ktr4 | Putative mannosyltransferase | |||
| Ktr5 | Putative mannosyltransferase | |||
| Ktr6/Mnn6 | Probable mannosylphosphate transferase | |||
| Ktr7 | Putative mannosyltransferase | |||
| Yur1 | Mannosyltransferase of the KTR1 family | |||
| MNN4 | Putative positive regulator of mannosylphosphate transferase (Ktr6p) | |||
| Mnn4 | Putative positive regulator of mannosylphosphate transferase (Ktr6p) | |||
| MNN1 | α1,3-Mannosyltransferase | |||
| Mnn1 | α1,3-Mannosyltransferase | |||
Yeast and fungus-specific outer chain biosyntheses are initiated by Och1p having an α1,6-mannosyltransferase activity in the Golgi. In S. cerevisiae, Hoc1p (homologous to Och1p) resides in the M-pol II complex, although its function has not been defined (11). We identified three Cryptococcus genes encoding proteins homologous to yeast Och1p (CNAG_00744.2, CNAG_05836.2, and CNAG_01214.2) from the H99 genome database, designated CnOCH1, CnHOC1, and CnHOC2, respectively. We also found a single gene (CNAG_06782.2, designated CnMNN2) homologous to yeast MNN2/MNN5 genes encoding α1,2-mannosyltransferase and a single gene (CNAG_03832.2, designated CnKTR3) homologous to the S. cerevisiae KRE2/MNT1 family containing nine members of Golgi mannosyltransferases involved in both N- and O-linked glycan synthesis (13). The bioinformatics analysis indicated that the glycosylation pathway of C. neoformans might be simpler with a fewer number of components than those of other yeast species, which have larger protein families playing functionally redundant or inactive roles in glycosylation.
Functional Characterization of C. neoformans Och1p Homologs
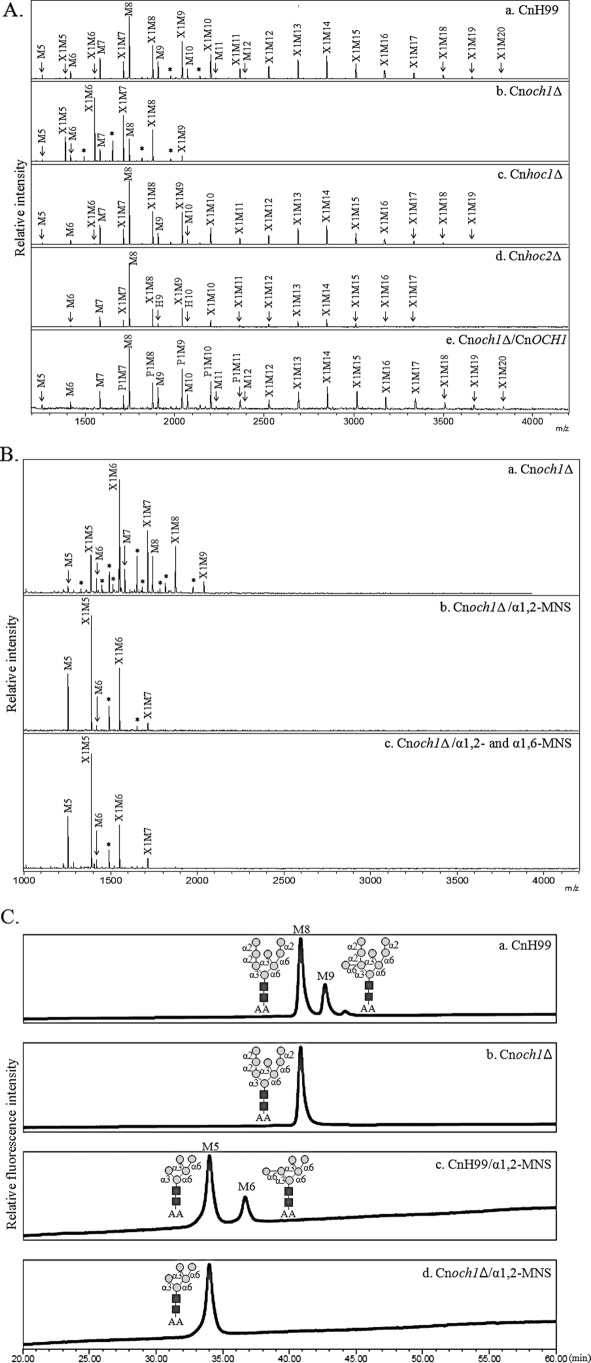
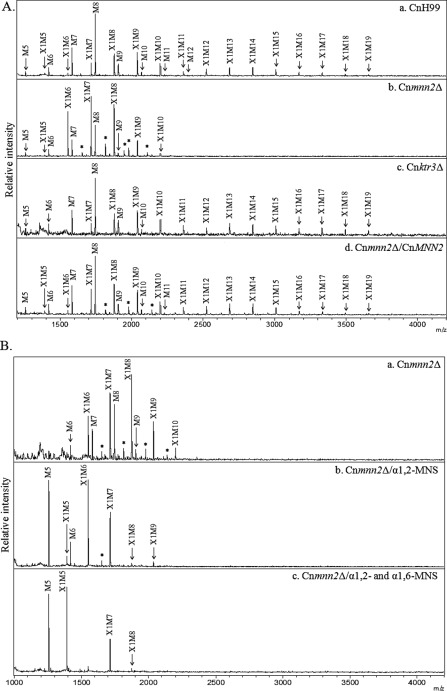
To analyze the function of the C. neoformans Och1p homologs, we constructed gene deletion mutants for each homolog (Cnoch1Δ, Cnhoc1Δ, and Cnhoc2Δ) and examined their neutral N-glycans structures by MALDI-TOF mass spectrometry. As seen Fig. 4A, N-glycans from the Cnoch1Δ mutant (Fig. 4A, panel b) were noticeably much shorter than those of the H99 wild-type strain (Fig. 4A, panel a), although no apparent changes were detected in N-glycan profiles from the mutant Cnhoc1Δ and Cnhoc2Δ strains (Fig. 4A, panels c and d). In the Cnoch1Δ mutant, N-glycans larger than Xyl1Man10GlcNAc2 (X1M10) were hardly detected, but reintroduction of the wild-type CnOCH1 gene into the Cnoch1Δ strain restored its N-glycan profile to that of the H99 strain (Fig. 4A, panel e), thus verifying the involvement of CnOch1p in the N-glycan processing of C. neoformans.
FIGURE 4.
Analysis of neutral N-glycan structures and in vitro mannosyltransferase activity of the Cnoch1Δ mutant. A, N-glycan profiles of C. neoformans H99 wild-type (CnH99, panel a), Cnoch1Δ (panel b), Cnhoc1Δ (panel c), Cnhoc2Δ (panel d), and Cnoch1Δ/CnOCH1 (panel e) strains by MALDI-TOF analysis in the positive mode. B, linkage analysis of the outer region in N-glycans from the Cnoch1Δ strain without (panel a) and with treatment by α1,2- (panel b) and α1,6-MNS (panel c) treatment. X, xylose; M, mannose; *, unidentified peak. C, analysis of α1,6-mannosyltransferase activity in CnH99 and Cnoch1Δ strains. The enriched Golgi membrane fraction of CnH99 or Cnoch1Δ strain was used to analyze α1,6-mannosyltransferase activity using Man8GlcNAc2-AA as an acceptor glycan. The reaction products were then treated with α1,2-MNS. Reaction products of the membrane fractions of CnH99 and Cnoch1Δ strains (panels a and b, respectively) and the reaction products of α1,2-MNS treatment (panels c and d, respectively) were analyzed by HPLC. Squares and circles with linkage information symbolize N-acetylglucosamine and mannose, respectively.
To examine which step of the outer chain biosynthesis pathway was affected by disruption of the CnOCH1 gene, N-glycans obtained from the Cnoch1Δ mutant strain were sequentially treated with α1,2- and α1,6-MNSs (Fig. 4B). After treatment with α1,2-MNS, most glycan peaks were converted to Man5–6GlcNAc2 (M5–6) or Xyl1Man5–6GlcNAc2 (X1M5–6) with M5 and X1M5 as major peaks (Fig. 4B, panel b). However, subsequent treatment with α1,6-MNS in the Cnoch1Δ mutant did not generate any further changes in the N-glycan profiles (Fig. 4B, panel c), whereas the treatment with α1,6-MNS in the wild-type strain did result in further shifts of X1M6 and M6 to X1M5 and M5, respectively (Fig. 1C). The exoglycosidase α1,6-MNS can remove only linear α1,6-mannose residues at the nonreducing end. Because the presence of a branched α1,3-mannose in the core N-glycans inhibits the removal of the 1,6-mannose, the α1,6-MNS treatment of N-glycans from the wild-type strain would convert X1M6 and M6 to X1M5 and M5, respectively. Thus, no shift of any N-glycan peak after the α1,6-MNS treatment of N-glycans from the Cnoch1Δ mutant strain strongly supported the idea that the outer chains of N-glycans in the Cnoch1Δ mutant do not possess α1,6-linked mannose residues because of the loss of CnOch1p function. We further confirmed that Cryptococcus Och1p is a functional homolog of S. cerevisiae Och1p by analyzing its complementation capacity to recover the defective phenotype of the S. cerevisiae och1Δ mutant (supplemental Fig. 3A). Heterologous expression of CnOCH1, but not of CnHOC1 or CnHOC2, recovered the N-glycosylation defect of secreted invertase as well as the decreased resistance to hygromycin B in the Scoch1Δ mutant (supplemental Fig. 3B). These results strongly suggest that CnHoc1p and CnHoc2p are not functional homologs of ScOch1p, despite sequence similarities between them. Further investigation will be necessary to determine whether they are functional orthologs of ScHoc1p or play Cryptococcus-specific functions. Moreover, we also observed decreased additional activity of an α1,6-mannose residue to the M8 core form oligosaccharide in the och1Δ mutant of C. neoformans (Fig. 4C). These results further support that the Cryptococcus OCH1 gene is a functional ortholog of the S. cerevisiae OCH1 gene encoding α1,6-mannosyltransferase, which initiates outer chain branching of N-glycans in the Golgi by addition of a single α1,6-linked mannose residue to the Man8GlcNAc2 core.
Glycan Profile Analysis of Cnmnn2Δ and Cnktr3Δ Strains
To examine the function of the CnMNN2 and CnKTR3 genes in cryptococcal N-glycan biosynthesis, N-glycan profiles of the Cnmnn2Δ and Cnktr3Δ mutants were analyzed (Fig. 5A, panels b and c). N-Glycans of the Cnmnn2Δ strain were much shorter than those of the wild-type strain, consisting mostly of M6–9 and X1M6–10, but were slightly longer than those of the Cnoch1Δ mutant (Fig. 5A, panels a and b). Reintroduction of the CnMNN2 gene into the Cnmnn2Δ strain recovered its glycan profile to that of the wild-type CnH99 strain (Fig. 5A, panel d). When treated with α1,2-MNS, N-glycans of the Cnmnn2Δ mutant were shifted to M5–6 and X1M5–7 (Fig. 5B, panel b), and subsequent treatment with α1,6-MNS generated peaks of M5 and X1M5 as major forms (Fig. 5B, panel c). Compared with the α1,2-MNS-treated N-glycans from the Cnoch1Δ mutant, those from the Cnmnn2Δ mutant were larger by a single hexose residue, which was removed by subsequent α1,6-MNS treatment. These results suggest that CnMnn2p mediates the addition of α1,2-mannose residues after the first step of α1,6-mannose addition to the core N-glycan by CnOch1p in C. neoformans. In contrast to the notable defects in the elongation of outer chains in Cnoch1Δ and Cnmnn2Δ mutants, the N-glycan profile of the Cnktr3Δ mutant was almost identical to that of the wild-type strain (Fig. 5A, panel c), indicating that CnKtr3p is not involved in the processing of N-glycans in C. neoformans. Interestingly, however, we observed a significant defect in O-glycan biosynthesis in the Cnktr3Δ mutant (supplemental Fig. 4), suggesting a possible role of CnKTR3 in O-glycan elongation but not in N-glycan processing in C. neoformans.
FIGURE 5.
Analysis of neutral N-glycan structures of Cnmnn2Δ and Cnktr3Δ mutants. A, MALDI-TOF analysis in the positive mode of N-glycans from CnH99 (panel a), Cnmnn2Δ (panel b), Cnktr3Δ (panel c), and Cnoch1Δ/CnOCH1 complemented (panel d) strains. B, linkage analysis of the outer region of N-glycans from the Cnmnn2Δ strain without (panel a) and with treatment by α1,2- (panel b) and α1,6-MNS (panel c) treatment. X, xylose; M, mannose; *, unidentified peak.
Presence of Acidic N-Linked Oligosaccharides Containing Xylose Phosphate
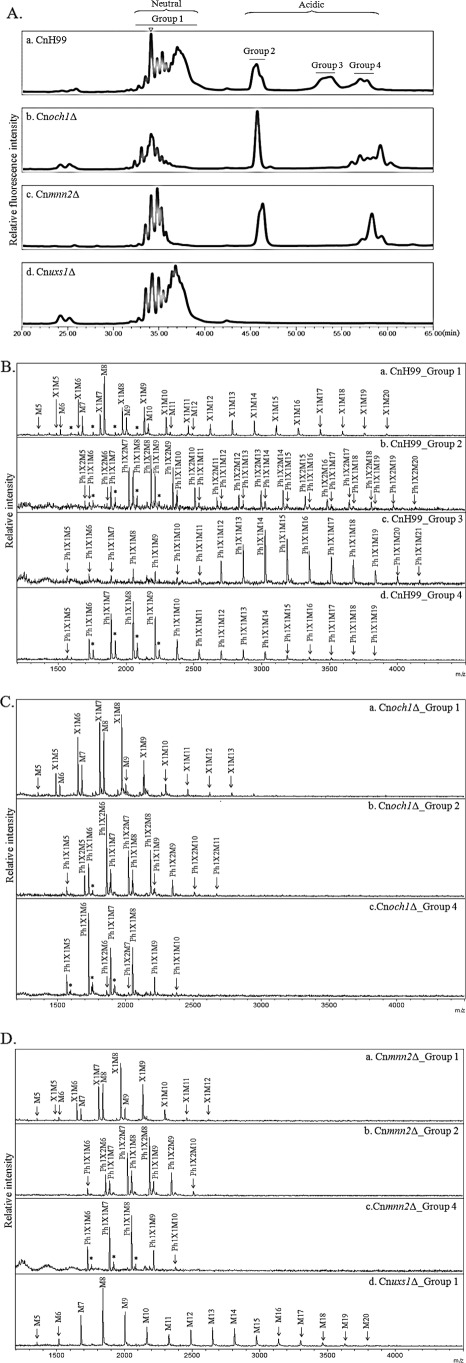
A recent study on xylosylphosphotransferase 1 (Xpt1p) of C. neoformans reported that this enzyme catalyzes O-linked glycosylation of proteins by adding xylose phosphate to O-glycans using UDP-Xyl as a reaction donor (39). To investigate the possible presence of xylose phosphate residues in cryptococcal N-glycans, total N-glycan profiles, including both neutral and acidic glycans, were analyzed by HPLC using an amine column at an acidic pH, which can separate oligosaccharides based on their charges and sizes. Interestingly, all N-glycans of the wild-type H99 strain (serotype A) could be separated into four major peaks (groups 1–4) (Fig. 6A, panel a). Notably, Cnoch1Δ, Cnmnn2Δ, and Cnuxs1Δ mutants showed distinctive profiles compared with the wild-type strain. The peak assigned to group 3 was missing in Cnoch1Δ and Cnmnn2Δ mutants (Fig. 6A, panels b and c), and peaks corresponding to groups 2–4 were not detected in Cnuxs1Δ mutant (Fig. 6A, panel d). Each group exhibited a similar HPLC profile with an amide column separating oligosaccharides based mainly on sizes (supplemental Fig. 5).
FIGURE 6.
Acidic N-glycan analysis of C. neoformans mutant strains. A, total N-glycan profiles of C. neoformans H99 wild-type (panel a), Cnoch1Δ (panel b), Cnmnn2Δ (panel c), and Cnuxs1Δ (panel d) strains by HPLC analysis using an amine column. Triangle indicates the retention time for Man8. B, MALDI-TOF analysis in the negative reflector mode for the detection of acidic N-glycans, group 1 (panel a), group 2 (panel b), group 3 (panel c), and group 4 (panel d), released from CnH99. C, MALDI-TOF analysis in the negative reflector mode for the detection of acidic N-glycans, group 1 (panel a), group 2 (panel b), and group 4 (panel c), released from Cnoch1Δ. D, MALDI-TOF analysis in the negative reflector mode for the detection of acidic N-glycans, group 1 (panel a), group 2 (panel b), and group 4 (panel c) from Cnmnn2Δ and group 1 from Cnuxs1Δ (panel d) strains, respectively. Ph, phosphate; X, xylose; M, mannose; *, unidentified peak.
Further analysis using MALDI-TOF in the negative reflector mode revealed that group 1 and groups 2–4 in H99 were composed of neutral and acidic N-glycans, respectively (Fig. 6B). Group 1 consisted of neutral N-glycans with or without a xylose residue. In contrast, N-glycans from group 2 in the wild-type strain were composed of Ph1P1–2H5–20 glycans containing an additional pentose phosphate residue with or without a xylose residue (Fig. 6B, panel b). Group 3 consisted of large glycans with a pentose phosphate residue (Ph1P1H12–20) and group 4 mostly consisted of a core form of N-glycans containing a pentose phosphate residue (Ph1P1H5–10) as the major portion (Fig. 6B, panels c and d). In particular, the disappearance of only group 3 in both Cnoch1Δ and Cnmnn2Δ mutants indicated that the glycans in group 3 were large N-glycan species containing a single pentose phosphate on the outer chains extended by addition of α1,6- and α1,2-mannoses. Therefore, the absence of all peaks of groups 2–4 corresponding to acidic glycans in the Cnuxs1Δ mutant (Fig. 6D, panel d) strongly supported that the pentose phosphate residues found in the acidic glycans were xylose phosphate, which appeared to be added to both the core and outer regions of N-glycans in C. neoformans. The absence of xylose phosphate in N-glycans from the Cnuxs1Δ mutant also suggested that UDP-Xyl is also used as a donor to add xylose phosphate to N-glycans assembled on proteins.
We observed very small peaks corresponding to groups 2 and 4, and no peak for group 3, in the total N-glycan profiles of serotypes B and D (supplemental Fig. 6A), implying the presence of acidic N-glycans in the core form of N-glycans of serotypes B and D. However, the proportion of acidic glycans in serotypes B and D was much smaller (less than 10%) compared with those of serotype A, which contained more than 50%. The lack of a group 3 peak was consistent with the much shorter length of outer chains of N-glycans from serotypes B and D. Acidic N-glycans from serotypes B and D were also shown to consist of Ph1P1H5–11 containing an additional pentose phosphate residue (supplemental Fig. 6, B and C), suggesting that C. neoformans N-glycans are modified by addition of a xylose phosphate regardless of the serotypes. Notably, an interesting observation is the presence of acidic glycans carrying a phosphate moiety without xylose, particularly in N-glycans from serotype B.
Growth Phenotypes of C. neoformans Mutant Strains Defective in Glycan Biosynthesis
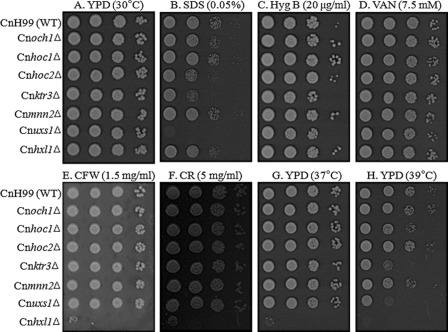
In general, yeast mutant strains with defects in glycosylation show alteration in cell wall integrity and thus exhibit increased sensitivity to cell wall-disturbing reagents and high temperatures (41). We assessed the growth phenotypes of the C. neoformans mutants Cnoch1Δ, Cnhoc1Δ, Cnhoc2Δ, Cnmnn2Δ, Cnktr3Δ, and Cnuxs1Δ in the presence of diverse cell wall- and membrane-disturbing reagents, including SDS, Calcofluor white, Congo red, sodium orthovanadate, and hygromycin B, and high temperature. All mutants grew as well as the wild-type strain under normal unstressed conditions on YPD at 30 °C (Fig. 7). Unexpectedly, however, despite a significant alteration in the structure of outer chain N-glycans, the Cnoch1Δ and Cnmnn2Δ mutants showed no apparent change. In contrast, the Cnktr3Δ mutant, which appeared to have a defect in O-glycosylation but not in N-glycosylation, and the Cnuxs1Δ mutant with a defect in UDP-Xyl synthesis, displayed increased sensitivity to SDS and high temperature at 39 °C, indicating that CnKtr3p and CnUxs1p are required for maintenance of cell wall stability.
FIGURE 7.
Phenotype analysis of C. neoformans H99 wild-type, Cnoch1Δ, Cnhoc1Δ, Cnhoc2Δ, Cnmnn2Δ, Cnktr3Δ, and Cnuxs1Δ mutant strains. Yeast cells were spotted on YPD plates only or YPD plates containing different cell wall-disturbing reagents. A, YPD at 30 °C. B, YPD with 0.05% SDS. C, YPD with 20 μg/ml hygromycin B (Hyg B). D, YPD with 7.5 mm vanadate (VAN). E, YPD with 1.5 mg/ml Calcofluor white (CFW). F, YPD with 5 mg/ml Congo red (CR). G, YPD at 37 °C. H, YPD at 39 °C. The Cnhxl1Δ mutant strain with a defect in unfolded protein response (42) was included as a positive control for a defect in cell wall integrity.
The capsule of C. neoformans is intimately associated with the cell wall, which underlies the capsule and provides yeast mechanical strength under stressful conditions. However, we did not observe any defects in capsule formation in Cnoch1Δ, Cnhoc1Δ, Cnhoc2Δ, Cnmnn2Δ, or Cnktr3Δ mutant strains (data not shown). These results imply that the truncated outer chain structure in N-glycans might be tolerable for maintaining the cell wall integrity of C. neoformans, different from other yeast species such as S. cerevisiae and C. albicans with hypermannosylated outer chains (8). Similarly, it was also reported that the outer chain structure of N-glycans is not important for maintenance of cell wall integrity in the filamentous fungi A. fumigatus (25).
DISCUSSION
N-Glycosylation, the most common type of eukaryotic protein glycosylation, involves the linkage of an oligosaccharide core to Asn residues in the ER, which is well conserved from yeast to mammals. However, subsequent processing of N-glycans is significantly different among different organisms and even in yeast species, generating diversity in N-glycan structures. Although N-glycans in yeasts are generally extended by addition of mannose to the core oligosaccharide in the Golgi, additional monosaccharide units such as galactose and N-acetylglucosamine (GlcNAc) are added in some species (11). N-Glycans of several yeast species also contain acidic sugars, which are composed of mannosyl phosphorylated sugars in most yeast species such as S. cerevisiae, C. albicans, P. pastoris, and Y. lipolytica. However, in S. pombe, the addition of pyruvate generates acidic glycans (11, 43).
Historically, C. neoformans and its related species, such as C. gattii, have been further categorized by serotypes based on a defined set of capsular-reactive immune sera (44). C. gattii includes serotypes B and C, whereas strains classified as serotypes A, D, or AD hybrids make up C. neoformans. Serotypes A and D have been also classified as varieties of C. neoformans, var. grubii and var. neoformans, respectively (45). Serotypes A and D generally associated with diseases in immunocompromised individuals are distributed worldwide, whereas serotypes B and C known to infect immunocompetent persons are typically found in, but not limited to, tropical and subtropical regions. The number of xylose residues on the major repeat unit of capsular polysaccharides exhibits a serotype-specific pattern: 2:3:4:1 = serotype A, serotype B, serotype C, and serotype D (46).
In this study, we performed comparative N-glycan profile analysis of several C. neoformans serotype strains, including C. neoformans var. grubii H99 and KN99 (serotype A), C. neoformans var. neoformans JEC21 and JEC20 (serotype D), and C. gattii R265 and WM276 (serotype B). We report, for the first time, the serotype-specific presence of a β1,2-xylose residue in high mannose type N-glycans of cryptococcal glycoproteins. Unexpectedly, serotype B strains containing the higher number of xylose residues in their capsule compared with serotypes A and D showed only high mannose-type N-glycans without xylose (Figs. 2 and 3). It is speculated that the differences in N-glycan structures among the serotypes tested in this study might reflect evolutionary divergence.
Interestingly, we also present data strongly indicating that C. neoformans N-glycans also contain xylose phosphate residues as acidic sugars (Fig. 6 and supplemental Fig. 6). The relative proportion of acidic sugars among total N-glycans was more than 50% in serotype A, much higher than the reported proportion of less than 1% of total acidic O-glycans (39). It is noticeable that the portion of acidic sugars was much lower in serotypes B and D, compared with that of serotype A. N-Glycans from cwMPs of the Cnuxs1Δ mutant contained high mannose-type oligosaccharides without any xylose and xylose phosphate residues. Based on our data, it is conceivable that UDP-xylose is used as a substrate not only for xylosylation but also for xylosyl phosphorylation of various glycoconjugates, including N-glycans. The physiological role of acidic glycans in yeast species has been implicated in the stress response to environmental changes. However, it has been reported the mannosyl phosphorylation is not required for macrophage interactions or for virulence in C. albicans, despite significant loss of β1,2-mannose oligosaccharides (47).
The in silico analysis indicated that C. neoformans might have a simpler N-glycan outer chain biosynthetic pathway. As expected from the lack of genes encoding M-pol I and M-pol II subunits, our data confirmed that the outer chains of N-glycans from C. neoformans have a very short α1,6-mannose extension, consisting mostly of a single α1,6-mannose residue, and are extended mainly by α1,2-mannose residues. Based on the structural information of N-glycans in C. neoformans mutants constructed in this study, we propose the N-linked outer chain biosynthetic pathway in C. neoformans as shown in Fig. 8. Similar to S. cerevisiae, the Man8GlcNAc2 core glycan attached to the cryptococcal proteins in the ER is also elongated via the addition of an α1,6-mannose unit by CnOch1p, an initiating α1,6-mannosyltransferase in the Golgi. However, the α1,6 outer chain backbone is not further elongated in C. neoformans. Instead, the outer chain and core N-glycans are further elongated mainly via α1,2-mannose addition mediated by CnMnn2p. Moreover, although the exact order is not yet clear, a single β1,2-xylose residue is added to the first mannose at the trimannosyl core of N-glycan during the early processing stages in the Golgi. As in the case of the xylose addition to C. neoformans N-glycans, the modification of N-glycans by bisecting GlcNAc at the β-mannose of the N-glycan core was previously reported, and the corresponding transferase has been characterized in the basidiomycete Coprinopsis cinerea (10). Then, similar to the addition of mannosyl phosphate in S. cerevisiae, the xylose phosphate might be added to some N-glycans at the core and outer chain regions, generating acidic N-glycans with negative charges during the late processing stages. Considering that the addition of a xylose phosphate residue occurs at the late stage of N-glycan processing in C. neoformans, it is highly likely that a transporter for UDP-Xyl would be present at the Golgi membrane. There has been a report on the Golgi localization of a human transporter for UPD-Xyl, which can transport UPD-Xyl over the Golgi membrane (48). However, we could not exclude the possibility that a transporter for UPD-xylose could be also present in the ER. As indicated by the absence of a MNN1 ortholog in C. neoformans, our structural analysis data supported the idea that C. neoformans N-glycans do not undergo the final modification step to add terminal α1,3-linked mannoses to the outer chains.
FIGURE 8.
Proposed pathway for cryptococcal N-linked glycan biosynthesis in the Golgi. Upper and lower panels depict Golgi N-glycan biosynthetic pathways in S. cerevisiae and C. neoformans, respectively. Uncharacterized glycosyltransferases such as xylosyltransferase (XT) and xylosylphosphotransferase (XPT) are marked in the N-glycan biosynthesis pathway in C. neoformans. *, a putative site for mannose or xylose phosphorylation. Mn is a MannGlcNAc2 (n = number of mannose residue).
In Cryptococcus, Cxt1p, a β1,2 xylosyltransferase, uses UDP-Xyl as a donor for the incorporation of xylose as a component of capsular polysaccharides and glucosylinositol phosphorylceramides (49). However, we observed that the Cncxt1Δ mutant displayed the same N-glycan profile as that of the wild-type strain (data not shown), excluding the possibility that Cxt1p may act solely to transfer xylose to mannose residues during N-glycan biosynthesis. In fact, we identified several Cxt1 homologs from the C. neoformans genome database. Moreover, in our preliminary analysis of N-glycan profiles in the xpt1Δ strain, which is defective in adding xylose phosphate to O-glycans due to lack of Xpt1p, we could still detect the presence of acidic glycans.4 This indicates the presence of unknown xylosyltransferase(s) and xylosylphosphotransferase(s) specific for N-glycan biosynthesis or the involvement of other redundant enzymes in the addition of xylose and xylose phosphate residues to N-glycans in C. neoformans. In this study, we report the comprehensive information on the structure and biosynthesis pathway of C. neoformans N-glycome. However, our experimental approach based on the enrichment of the mannose-containing structures by using ConA might miss a fraction of glycans that are not bound by ConA. Also, the release of N-glycans using peptide:N-glycanase F from cryptococcal mannoproteins might impose a bias on the structures analyzed, because the modification of the core GlcNAc by α1,3-linked fucose prevents the action of peptide:N-glycanase F. Thus, the possibility still remains that cryptococcal N-glycans with yet uncharacterized structures might be identified by further systematic analysis.
Cell surface mannoproteins contain both O- and N-linked oligosaccharides. Most information about glycosylation of proteins has been generated through studies of the model yeast S. cerevisiae. Very recently, information assessing the relevance of glycosylation for virulence has emerged for medically important fungal pathogens, including C. albicans, A. fumigatus, and C. neoformans. Previous studies have demonstrated that O-glycosylation is required for host-fungus interactions and virulence in all strains of C. albicans, A. fumigatus, and C. neoformans (50). However, the roles of N-glycan outer chains for host-fungus interactions and virulence were shown to be quite different between C. albicans and A. fumigatus. The Caoch1 null mutant with loss of the α1,6-linked polymannose backbone was shown be attenuated in virulence. However, similar infection experiments revealed no difference between the afoch1 null mutant and control strains, suggesting that N-glycan outer chains of A. fumigatus are not associated with virulence (25). To address whether the modification of N-glycan outer chains affects the virulence of C. neoformans, we tested the virulence of the Cnoch1Δ mutant, which is defective in initiating extension of N-glycan outer chains, using a murine model of systemic cryptococcosis. Although the Cnoch1Δ mutant showed slightly attenuated virulence compared with the wild-type strain, the Cnoch1Δ/CnOCH1 complemented strain was as virulent as the Cnoch1Δ mutant (data not shown), suggesting that the function of CnOch1p is not critical for virulence in C. neoformans. Our data also strongly suggest that perturbation of outer chain processing of N-glycans in C. neoformans will not significantly affect virulence. For example, the N-glycans of serotype B appeared to be mostly core forms without extended outer chains or the addition of xylose. Moreover, they were shown to be much less modified with the addition of xylose phosphate compared with those of serotype A. Despite such noticeable differences in the outer chain N-glycans, serotype B can infect even immunocompetent persons.
In conclusion, we present evidence supporting the idea that C. neoformans N-glycans are high mannose type modified with addition of a β1,2-xylose in a serotype-specific way. Moreover, our data strongly indicate the presence of differential modification by addition of xylose phosphate. It remains a challenge to identify the glycosyltransferases responsible for these unique modifications of N-glycans with xylose and xylose phosphate in C. neoformans. It would be intriguing to elucidate regulatory mechanisms underlying the serotype-specific processing of cryptococcal N-glycans. Furthermore, we expect that the glycosylation-defective mutant strains developed in this study will be useful for systematic investigation on how structural alterations of N-/O-glycans affect the intensity of virulence and the extent of host immunological interactions in C. neoformans.
Supplementary Material
Acknowledgments
We are grateful to Dr. Yin-Won Lee for inspiring comments and encouragement. We thank Dr. Hong-Jin Kim for help with virulence analysis.
This work was supported in part by the National Research Foundation of Korea Grants 2010-0029117 and 2012-0001150 from the Korean Ministry of Education, Science, and Technology.

This article contains supplemental “Materials and Methods,” Figs. 1–6, Tables 1 and 2, and additional references.
J.-N. Park, D.-J. Lee, O. Kwon, D.-B. Oh, Y.-S. Bahn, and H. A. Kang, unpublished data.
- GXM
- glucuronoxylomannan
- ER
- endoplasmic reticulum
- cwMPs
- cell wall mannoproteins
- sMPs
- secretory mannoproteins
- α1,2-MNS
- α1,2-mannosidase
- α1,6-MNS
- α1,6-mannosidase
- JBM
- jack bean α-mannosidase
- UDP-Xyl
- UPD-xylose
- ConA
- concanavalin A
- 2AA
- 2-aminobenzoic acid
- M-pol
- mannan polymerase.
REFERENCES
- 1. Perfect J. R. (2005) Cryptococcus neoformans. A sugar-coated killer with designer genes. FEMS Immunol. Med. Microbiol. 45, 395–404 [DOI] [PubMed] [Google Scholar]
- 2. Doering T. L. (2009) How sweet it is! Cell wall biogenesis and polysaccharide capsule formation in Cryptococcus neoformans. Annu. Rev. Microbiol. 63, 223–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kwon-Chung K. J., Boekhout T., Fell J. W., Díaz M. (2002) Proposal to conserve the name Cryptococcus gattii against C. hondurianus and C. bacillisporus (Basidiomycota, Hymenomycetes, Tremellomycetidae). Taxon 51, 804–806 [Google Scholar]
- 4. Wozniak K. L., Levitz S. M. (2009) Isolation and purification of antigenic components of Cryptococcus. Methods Mol. Biol. 470, 71–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mansour M. K., Latz E., Levitz S. M. (2006) Cryptococcus neoformans glycoantigens are captured by multiple lectin receptors and presented by dendritic cells. J. Immunol. 176, 3053–3061 [DOI] [PubMed] [Google Scholar]
- 6. Levitz S. M., Specht C. A. (2006) The molecular basis for the immunogenicity of Cryptococcus neoformans mannoproteins. FEMS Yeast Res. 6, 513–524 [DOI] [PubMed] [Google Scholar]
- 7. Helenius A., Aebi M. (2004) Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019–1049 [DOI] [PubMed] [Google Scholar]
- 8. Gemmill T. R., Trimble R. B. (1999) Overview of N- and O-linked oligosaccharide structures found in various yeast species. Biochim. Biophys. Acta 1426, 227–237 [DOI] [PubMed] [Google Scholar]
- 9. Deshpande N., Wilkins M. R., Packer N., Nevalainen H. (2008) Protein glycosylation pathways in filamentous fungi. Glycobiology 18, 626–637 [DOI] [PubMed] [Google Scholar]
- 10. Buser R., Lazar Z., Kaser S., Kunzler M., Aebi M. (2010) Identification, characterization, and biosynthesis of a novel N-glycan modification in the fruiting body of the basidiomycete Coprinopsis cinerea. J. Biol. Chem. 285, 10715–10723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dean N. (1999) Asparagine-linked glycosylation in the yeast Golgi. Biochim. Biophys. Acta 1426, 309–322 [DOI] [PubMed] [Google Scholar]
- 12. Nakayama K., Nakanishi-Shindo Y., Tanaka A., Haga-Toda Y., Jigami Y. (1997) Substrate specificity of α-1,6-mannosyltransferase that initiates N-linked mannose outer chain elongation in Saccharomyces cerevisiae. FEBS Lett. 412, 547–550 [DOI] [PubMed] [Google Scholar]
- 13. Lussier M., Sdicu A. M., Bussey H. (1999) The KTR and MNN1 mannosyltransferase families of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1426, 323–334 [DOI] [PubMed] [Google Scholar]
- 14. Jigami Y., Odani T. (1999) Mannosyl phosphate transfer to yeast mannan. Biochim. Biophys. Acta 1426, 335–345 [DOI] [PubMed] [Google Scholar]
- 15. Nakayama K., Nagasu T., Shimma Y., Kuromitsu J., Jigami Y. (1992) OCH1 encodes a novel membrane-bound mannosyltransferase: outer chain elongation of asparagine-linked oligosaccharides. EMBO J. 11, 2511–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mille C., Bobrowicz P., Trinel P. A., Li H., Maes E., Guerardel Y., Fradin C., Martínez-Esparza M., Davidson R. C., Janbon G., Poulain D., Wildt S. (2008) Identification of a new family of genes involved in β1,2-mannosylation of glycans in Pichia pastoris and Candida albicans. J. Biol. Chem. 283, 9724–9736 [DOI] [PubMed] [Google Scholar]
- 17. Mora-Montes H. M., Bates S., Netea M. G., Castillo L., Brand A., Buurman E. T., Díaz-Jiménez D. F., Jan Kullberg B., Brown A. J., Odds F. C., Gow N. A. (2010) A multifunctional mannosyltransferase family in Candida albicans determines cell wall mannan structure and host-fungus interactions. J. Biol. Chem. 285, 12087–12095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bates S., Hughes H. B., Munro C. A., Thomas W. P., MacCallum D. M., Bertram G., Atrih A., Ferguson M. A., Brown A. J., Odds F. C., Gow N. A. (2006) Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J. Biol. Chem. 281, 90–98 [DOI] [PubMed] [Google Scholar]
- 19. Song Y., Choi M. H., Park J. N., Kim M. W., Kim E. J., Kang H. A., Kim J. Y. (2007) Engineering of the yeast Yarrowia lipolytica for the production of glycoproteins lacking the outer chain mannose residues of N-glycans. Appl. Environ. Microbiol. 73, 4446–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Uccelletti D., Farina F., Rufini S., Magnelli P., Abeijon C., Palleschi C. (2006) The Kluyveromyces lactis α1,6-mannosyltransferase KlOch1p is required for cell wall organization and proper functioning of the secretory pathway. FEMS Yeast Res. 6, 449–457 [DOI] [PubMed] [Google Scholar]
- 21. Kim M. W., Kim E. J., Kim J. Y., Park J. S., Oh D. B., Shimma Y., Chiba Y., Jigami Y., Rhee S. K., Kang H. A. (2006) Functional characterization of the Hansenula polymorpha HOC1, OCH1, and OCR1 genes as members of the yeast OCH1 mannosyltransferase family involved in protein glycosylation. J. Biol. Chem. 281, 6261–6272 [DOI] [PubMed] [Google Scholar]
- 22. Yoko-o T., Tsukahara K., Watanabe T., Hata-Sugi N., Yoshimatsu K., Nagasu T., Jigami Y. (2001) Schizosaccharomyces pombe och1(+) encodes α1,6-mannosyltransferase that is involved in outer chain elongation of N-linked oligosaccharides. FEBS Lett. 489, 75–80 [DOI] [PubMed] [Google Scholar]
- 23. Choi B. K., Bobrowicz P., Davidson R. C., Hamilton S. R., Kung D. H., Li H., Miele R. G., Nett J. H., Wildt S., Gerngross T. U. (2003) Use of combinatorial genetic libraries to humanize N-linked glycosylation in the yeast Pichia pastoris. Proc. Natl. Acad. Sci. U.S.A. 100, 5022–5027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maddi A., Free S. J. (2010) α-1,6-Mannosylation of N-linked oligosaccharide present on cell wall proteins is required for their incorporation into the cell wall in the filamentous fungus Neurospora crassa. Eukaryot. Cell 9, 1766–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kotz A., Wagener J., Engel J., Routier F. H., Echtenacher B., Jacobsen I., Heesemann J., Ebel F. (2010) Approaching the secrets of N-glycosylation in Aspergillus fumigatus: characterization of the AfOch1 protein. PLoS One 5, e15729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Biondo C., Mancuso G., Midiri A., Bombaci M., Messina L., Beninati C., Teti G. (2006) Identification of major proteins secreted by Cryptococcus neoformans. FEMS Yeast Res. 6, 645–651 [DOI] [PubMed] [Google Scholar]
- 27. Williamson P. R. (1994) Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J. Bacteriol. 176, 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Biondo C., Messina L., Bombaci M., Mancuso G., Midiri A., Beninati C., Cusumano V., Gerace E., Papasergi S., Teti G. (2005) Characterization of two novel cryptococcal mannoproteins recognized by immune sera. Infect. Immun. 73, 7348–7355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Turner K. M., Wright L. C., Sorrell T. C., Djordjevic J. T. (2006) N-Linked glycosylation sites affect secretion of cryptococcal phospholipase B1, irrespective of glycosylphosphatidylinositol anchoring. Biochim. Biophys. Acta 1760, 1569–1579 [DOI] [PubMed] [Google Scholar]
- 30. Samuelson J., Banerjee S., Magnelli P., Cui J., Kelleher D. J., Gilmore R., Robbins P. W. (2005) The diversity of dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc. Natl. Acad. Sci. U.S.A. 102, 1548–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Klutts J. S., Yoneda A., Reilly M. C., Bose I., Doering T. L. (2006) Glycosyltransferases and their products. Cryptococcal variations on fungal themes. FEMS Yeast Res. 6, 499–512 [DOI] [PubMed] [Google Scholar]
- 32. Kim M. S., Kim S. Y., Yoon J. K., Lee Y. W., Bahn Y. S. (2009) An efficient gene-disruption method in Cryptococcus neoformans by double-joint PCR with NAT-split markers. Biochem. Biophys. Res. Commun. 390, 983–988 [DOI] [PubMed] [Google Scholar]
- 33. Fraser J. A., Subaran R. L., Nichols C. B., Heitman J. (2003) Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot. Cell 2, 1036–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bryan R. A., Zaragoza O., Zhang T., Ortiz G., Casadevall A., Dadachova E. (2005) Radiological studies reveal radial differences in the architecture of the polysaccharide capsule of Cryptococcus neoformans. Eukaryot. Cell 4, 465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chiba Y., Suzuki M., Yoshida S., Yoshida A., Ikenaga H., Takeuchi M., Jigami Y., Ichishima E. (1998) Production of human compatible high mannose-type (Man5GlcNAc2) sugar chains in Saccharomyces cerevisiae. J. Biol. Chem. 273, 26298–26304 [DOI] [PubMed] [Google Scholar]
- 36. Bahn Y. S., Hicks J. K., Giles S. S., Cox G. M., Heitman J. (2004) Adenylyl cyclase-associated protein Aca1 regulates virulence and differentiation of Cryptococcus neoformans via the cyclic AMP-protein kinase A cascade. Eukaryot. Cell 3, 1476–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kwon-Chung K. J., Bennett J. E. (1978) Distribution of α and α-mating types of Cryptococcus neoformans among natural and clinical isolates. Am. J. Epidemiol. 108, 337–340 [DOI] [PubMed] [Google Scholar]
- 38. Kwon-Chung K. J., Edman J. C., Wickes B. L. (1992) Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60, 602–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reilly M. C., Aoki K., Wang Z. A., Skowyra M. L., Williams M., Tiemeyer M., Doering T. L. (2011) A xylosylphosphotransferase of Cryptococcus neoformans acts in protein O-glycan synthesis. J. Biol. Chem. 286, 26888–26899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moyrand F., Klaproth B., Himmelreich U., Dromer F., Janbon G. (2002) Isolation and characterization of capsule structure mutant strains of Cryptococcus neoformans. Mol. Microbiol. 45, 837–849 [DOI] [PubMed] [Google Scholar]
- 41. Ram A. F., Klis F. M. (2006) Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat. Protoc. 1, 2253–2256 [DOI] [PubMed] [Google Scholar]
- 42. Cheon S. A., Jung K. W., Chen Y. L., Heitman J., Bahn Y. S., Kang H. A. (2011) Unique evolution of the UPR pathway with a novel bZIP transcription factor, Hxl1, for controlling pathogenicity of Cryptococcus neoformans. PLoS Pathog. 7, e1002177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Park J. N., Song Y., Cheon S. A., Kwon O., Oh D. B., Jigami Y., Kim J. Y., Kang H. A. (2011) Essential role of YlMPO1, a novel Yarrowia lipolytica homolog of Saccharomyces cerevisiae MNN4, in mannosylphosphorylation of N- and O-linked glycans. Appl. Environ. Microbiol. 77, 1187–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bovers M., Hagen F., Boekhout T. (2008) Diversity of the Cryptococcus neoformans-Cryptococcus gattii species complex. Rev. Iberoam. Micol. 25, S4–S12 [DOI] [PubMed] [Google Scholar]
- 45. Franzot S. P., Salkin I. F., Casadevall A. (1999) Cryptococcus neoformans var. grubii: separate varietal status for Cryptococcus neoformans serotype A isolates. J. Clin. Microbiol. 37, 838–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cherniak R., Sundstrom J. B. (1994) Polysaccharide antigens of the capsule of Cryptococcus neoformans. Infect. Immun. 62, 1507–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hobson R. P., Munro C. A., Bates S., MacCallum D. M., Cutler J. E., Heinsbroek S. E., Brown G. D., Odds F. C., Gow N. A. (2004) Loss of cell wall mannosyl phosphate in Candida albicans does not influence macrophage recognition. J. Biol. Chem. 279, 39628–39635 [DOI] [PubMed] [Google Scholar]
- 48. Ashikov A., Routier F., Fuhlrott J., Helmus Y., Wild M., Gerardy-Schahn R., Bakker H. (2005) The human solute carrier gene SLC35B4 encodes a bifunctional nucleotide sugar transporter with specificity for UDP-xylose and UDP-N-acetylglucosamine. J. Biol. Chem. 280, 27230–27235 [DOI] [PubMed] [Google Scholar]
- 49. Castle S. A., Owuor E. A., Thompson S. H., Garnsey M. R., Klutts J. S., Doering T. L., Levery S. B. (2008) β1,2-Xylosyltransferase Cxt1p is solely responsible for xylose incorporation into Cryptococcus neoformans glycosphingolipids. Eukaryot. Cell 7, 1611–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leach M. D., Brown A. J. (2012) Post-translational modifications of proteins in the pathobiology of medically relevant fungi. Eukaryot. Cell 11, 98–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.