Background: PTTG1 is an oncogene with its expression levels correlating with tumor development and metastasis.
Results: Modulation of PTTG1 expression levels revealed that PTTG1 promotes invasive and migratory properties and expansion of CD44high CD24low cell population via AKT activation in breast cancer cells.
Conclusion: PTTG1 induces EMT and promotes cancer stem cells via activation of AKT.
Significance: PTTG1 represents a potential target for therapeutic intervention against the spread of breast cancer.
Keywords: Akt, Breast Cancer, Cancer Stem Cells, Epithelial Mesenchymal Transition, Metastasis, PTTG1
Abstract
The prognosis of breast cancer patients is related to the degree of metastasis. However, the mechanisms by which epithelial tumor cells escape from the primary tumor and colonize at a distant site are not entirely understood. Here, we analyzed expression levels of pituitary tumor-transforming gene-1 (PTTG1), a relatively uncharacterized oncoprotein, in patient-derived breast cancer tissues with corresponding normal breast tissues. We found that PTTG1 is highly expressed in breast cancer patients, compared with normal tissues. Also, PTTG1 expression levels were correlated with the degree of malignancy in breast cancer cell lines; the more migratory and invasive cancer cell lines MDA-MB-231 and BT549 displayed the higher expression levels of PTTG1 than the less migratory and invasive MCF7 and SK-BR3 and normal MCF10A cell lines. By modulating PTTG1 expression levels, we found that PTTG1 enhances the migratory and invasive properties of breast cancer cells by inducing epithelial to mesenchymal transition, as evidenced by altered morphology and epithelial/mesenchymal cell marker expression patterns and up-regulation of the transcription factor Snail. Notably, down-regulation of PTTG1 also suppressed cancer stem cell population in BT549 cells by decreasing self-renewing ability and tumorigenic capacity, accompanying decreasing CD44high CD24low cells and Sox2 expression. Up-regulation of PTTG1 had the opposite effects, increasing sphere-forming ability and Sox2 expression. Importantly, PTTG1-mediated malignant tumor properties were due, at least in part, to activation of AKT, known to be a key regulator of both EMT and stemness in cancer cells. Collectively, these results suggest that PTTG1 may represent a new therapeutic target for malignant breast cancer.
Introduction
Breast cancer is the second-most lethal cancer in women. The main cause of death due to breast cancer is relapse and the resultant metastatic spread of malignant neoplasm to distant sites. Metastasis is a multistep process in which cancer cells are disseminated from the primary tumor and locally invade the surrounding tissue, where they enter blood vessels (intravasation) and subsequently exit the bloodstream (extravasation), colonizing the new microenvironment and forming secondary tumors (1–3). However, the mechanisms by which cancer cells are dissociated from primary tumors, survive in the bloodstream, and ultimately form secondary tumors remain largely unknown.
Recent studies have furthered our understanding of the contribution of epithelial to mesenchymal transition (EMT)4 to invasive and metastatic tumor growth (4, 5). EMT is a multistep process in which cells acquire molecular alterations that cause dysfunctional cell-cell adhesive interactions, loss of cell-cell junctions, and restructuring of the cytoskeleton, which collectively result in the loss of apical polarity and the acquisition of a more spindle-shaped morphology. This process is thought to ultimately culminate in cancer cell progression through the basement membrane and invasion into the surrounding microenvironment, including lymph and blood vascular systems (6).
Accumulating recent evidence suggests that a subpopulation of cancer cells is responsible for tumor formation (4, 7, 8), maintenance (9), and malignant progression (10–12). These rare tumor cells, termed cancer stem cells or cancer-initiating cells, are characterized by their strong tumorigenic properties and ability to self-renew (13, 14). Together with metastasis, cancer stem cell populations have been considered novel targets for cancer treatment. Thus, the development of novel therapeutic strategies that specifically target cancer stem cells may more effectively eradicate malignant tumors than conventional treatments, while reducing the risk of relapse and metastasis. However, the molecular features of cancer stem cells that orchestrate the enrichment and maintenance of stemness and tumorigenic properties remain to be elucidated.
The protein product of pituitary tumor-transforming gene-1 (PTTG1) was first isolated from GH4 rat pituitary tumor cells (15, 16) and shown to be transforming in vitro and tumorigenic in vivo (15–18). PTTG1 was also identified as human securin, a critical regulator of sister chromatid separation in late stage mitosis (19, 20). PTTG1 is expressed at very low or undetectable levels in most normal human tissues but is abundantly expressed in malignant cell lines and pituitary tumors (18, 21–23). However, the mechanisms by which PTTG1 contributes to tumor progression are not well understood.
In this study, we sought to determine the mechanisms and signaling pathway by which PTTG1 contributes to tumor malignancy in breast cancers. To this end, we modulated PTTG1 expression levels in breast cancer cell lines and normal breast cells by exogenously overexpressing PTTG1 or knocking down endogenous PTTG1 using small interfering RNA (siRNA). We found that PTTG1 expression is necessary and sufficient for acquisition of mesenchymal properties in both breast cancer cell lines and normal breast cells. In addition, we demonstrated that overexpression of PTTG1 leads to an expansion of the cancer stem cell population through activation of AKT, suggesting that PTTG1-mediated tumor malignancy occurs, at least in part, via the AKT signaling pathway.
EXPERIMENTAL PROCEDURES
Cell Culture
Human breast cancer cell lines, MCF-7, SK-BR3, MDA-MB-231, and BT549, and normal breast cell line, MCF10A, were established from the American Type Culture Collection (Manassas, VA). Cells were cultured in a humidified 5% CO2 atmosphere at 37 °C. The normal human breast epithelial cell line MCF10A was maintained in DMEM/F-12 medium supplemented with 5% heat-inactivated horse serum (Invitrogen), 10 μg/ml insulin, 20 ng/ml EGF, 0.1 μg/ml cholera toxin, 0.5 μg/ml hydrocortisone, penicillin (100 units/ml), and streptomycin (100 μg/ml). MCF7 cells were grown in minimum Eagle's medium supplemented with 10% fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 μg/ml). MDA-MB-231 and SK-BR3 cells were grown in DMEM supplemented with 10% fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 μg/ml). BT549 cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 μg/ml). For sphere formation, breast cancer cells were resuspended in DMEM/F-12 (Invitrogen) containing 20 ng/ml epidermal growth factor (EGF), basic fibroblast growth factor, and B27 (1:50) as sphere-forming conditions, a stem cell-permissive medium. Spheres were collected after 10 days, and protein was extracted for Western blotting and kinase assay or dissociated with Accutase (Innovative Cell Technologies, Inc.).
Chemical Reagents and Antibodies
Polyclonal antibodies to phospho-Akt (Ser-473), phospho-Akt (Thr-308), phospho-ERK1/2 (Thr-202/Tyr-204), phospho-p38 (Thr-180/Tyr-182), ERK1, p38, phospho-JNK1/2 (Thr-183/Tyr-185), and N-cadherin were obtained from Cell Signaling Technology (Beverly, MA). Polyclonal antibodies to Akt, JNK1, Zeb1, Snail, and Slug were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Polyclonal antibody CD44 was purchased from Abcam. The polyclonal antibody vimentin was obtained from Thermo Science. 4,6-Diamidino-2-phenylindole (DAPI), epidermal growth factor (EGF), and monoclonal antibodies to β-actin were obtained from Sigma. Basic fibroblast growth factor was purchased from R&D Systems. Anti-mouse Alexa Fluor 488, anti-rabbit Alexa Fluor 488, and B27 were purchased from Invitrogen. CD44 (directly conjugated with phycoerythrin), CD24 (directly conjugated with FITC), and mIgG2b-PE were purchased from Miltenyi Biotec Ltd. Inhibitors specific to JNK (SP600125), p38 MAPK (PD169316), MEK (U0126), and PI3K (LY294002) were obtained from Calbiochem.
Quantification of Cell Death
Cell death was measured by FACS analysis using propidium iodide and annexin-V double staining. Cells were harvested by trypsinization, washed in phosphate-buffered saline, and then incubated in propidium iodide (50 ng/ml) and annexin-V for 5 min at room temperature. Cells (10,000 per sample) were analyzed on a FACScan flow cytometer, using CellQuest software.
Cell Cycle
The cells were harvested and fixed with ice-cold 70% ethanol. The cells were washed in PBS and incubated with 0.1% Triton X-100 for 5 min at 4 °C. After washing in PBS, the cells were suspended in PBS containing 50 mg/ml of RNase A (Sigma), and incubated for 10 min at 37 °C. The cells were stained with 50 mg/ml of propidium iodide in PBS. The DNA content was then analyzed by FACScan flow cytometer, using CellQuest software.
Sphere Colony Counting
After treatment of sphere-cultured breast cancer cells with shRNA for 48 h, sixty spheres per group were randomly taken to measure sphere sizes by Motic Images Plus 2.0. For clonal analysis, spheres were dissociated to single cells and were plated into 96-well plates with a density of single cell per well. After 24 h, individual wells were visually checked for the presence of a single cell. The clones were grown and clone formation was monitored at 1, 5, 10, 15 days. The size of clones was measured under inverted microscope using Motic Images Plus 2.0.
Transfection
siRNA duplexes (40 nm) were introduced into cells using a Microporator-mini (Digital Bio Technology) by following the procedure recommended by the manufacturer. After 48 h, cells were harvested for additional experiments. All siRNAs targeted to PTTG1 (5′-GAGAAGACUGUUAAAGCAATT-3′), SNAIL (5′-CCAAUGCUCAUCUGGGACUTT-3′), slug (5′-CAUUAGUGAUGAAGAGGAATT-3′), AKT (5′-GCACCUUCAUUGGCUACAATT-3′), and negative control siRNA specific for green fluorescent protein (5′-CCACTACCTGAGCACCCAG-3′) were purchased from Samchully Pharmaceutical Co. Ltd. (Seoul, Korea). The full-length PTTG1 cDNA was cloned into the pcDNA3. The pcDNA3 PTTG1 was introduced by Lipofectamine Plus reagent (Invitrogen), following the procedure recommended by the manufacturer.
Transduction
Lentiviral construct encoding small hairpin RNA (shRNA) targeted to human PTTG1 (Clone NKI_p3205C032Q) was purchased from OriGene Technologies, Inc., Rockville, MD. Nontargeting shRNA was purchased from Sigma (SHC002). 293T cells were seeded (3 × 106 cells/ml) in a 60-mm dish, and viral particles were produced by transfection as described previously (24, 25). Viral supernatant was collected ∼65 h after transfection. The supernatant was used for infection. Cells were infected for 24 h and were then treated with 1 μg/ml puromycin to eliminate uninfected cells. Media were then replaced after 2 days, and cells were harvested for Western blot or additional experiments.
To clone PTTG1 into MSCV retroviral vector (Clontech), PTTG1 DNA fragment was amplified by using pSPORT-PTTG1 as templates and were cloned into MSCV retroviral vector derived from murine Moloney leukemia virus. For retrovirus production, the HD29D cell line was cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum, 2 mmol/liter GlutaMAX (Invitrogen), 50 units/ml penicillin/streptomycin, 1 μg/ml tetracycline, 2 μg/ml puromycin, and 0.6 mg/ml G418 sulfate (Calbiochem) and transfected with MSCV-PTTG1 using the Lipofectamine 2000 reagent (Invitrogen). 48 h after the transfection, virus supernatant was harvested every day by replenishing with fresh medium for 5 days and passed through a 0.45-μm filter, and the viral supernatant was frozen at −80 °C. The supernatant was used for infection after adding 4 μg/ml Polybrene (Sigma).
Invasion and Motility Assays
Cells (2 × 104 cells/well) were suspended in 0.2 ml of growth medium for invasion and motility assays. For invasion assay, the cells were loaded in the upper well of the Transwell chamber (8-mm pore size; Corning Glass) that was precoated with 10 mg/ml growth factor-reduced Matrigel (BD Biosciences) on an upper side of the chamber with the lower well filled with 0.8 ml of growth medium. After incubation for 48 h at 37 °C, noninvaded cells on the upper surface of the filter were removed with a cotton swab, and migrated cells on the lower surface of the filter were fixed and stained with a Diff-Quick kit (Fisher) and photographed (magnification ×20). Invasiveness was determined by counting cells in five microscopic fields per well, and the extent of invasion was expressed as an average number of cells per microscopic field. Cells were imaged by phase contrast microscopy (Leica Microsystems, Bannockburn, IL). For motility studies, we used invasion chambers with control inserts that contained the same type of membrane but without the Matrigel coating (one chamber per well of a 24-well plate). 2 × 104 cells in 0.2 ml of growth medium were added to the apical side of each insert, and 0.8 ml of growth medium was then added to the basal side of each insert. The inserts were processed as described above for the invasion assay.
Soft Agar Colony Formation Assay
To examine anchorage-independent growth, a cell suspension (2 × 104 cells) was suspended in 0.4% agar in growth medium and seeded in triplicate on 60-mm dishes precoated with 0.8% agar in growth medium and incubated at 37 °C, 5% CO2. After 12–30 days, colonies were photographed and counted in four randomly chosen fields and expressed as means of triplicates, representative of two independent experiments.
RNA Extraction and Reverse Transcription-PCR
RNA was extracted using the RNeasy kit (Qiagen, Valencia, CA). RNA was treated with DNase I (Ambion) to eliminate any contaminating DNA. Reverse transcription-PCRs were performed with SuperScript III (Invitrogen) according to the manufacturer's instructions. Primer sequences are described as follows: PTTG1 (forward, 5′-ATTCCCGCCACCATGGCATA-3′, and reverse, 5′-ATCCTTAAATATCTATG-3′); SNAIL (forward, 5′-TTTACCTTCCAGCAGCCCTA-3′, and reverse, 5′-CCAGGCTGAGGTTTCCTTG-3′); SLUG (forward, 5′-GAGCATACAGCCCCATCACT-3′, and reverse, 5′-CTCCCCCGTGTGAGTTCTAA-3′); TWIST (forward, 5′-GTCCGCAGTCTTACGAGGAG-3′, and reverse, 5′-CTAGTGGGACGCGGACAT-3′); ZEB1 (forward, 5′-GCACCTGAAGAGGACCAGAG-3′, and reverse, 5′-TGGTGATGCTGAAAGAGACG-3′); CD24 (forward, 5′-TGAAGAACATGTGAGAGGTTTGAC-3′, and reverse, 5′-GAAAACTGAATCTCCATTCCACAA-3′); CD44 (forward, 5′-CCCAGATGGAGAAAGCTCTG-3′, and reverse, 5′-CATCTACCCCAGCAACCCTA-3′); and GAPDH (forward, 5′-CCATGGAGAAGGCTGGGG-3′, and reverse, 5′-CAAAGTTGTCATGGATGACC-3′).
Western Blot Analysis
Cell lysates were prepared by extracting proteins with lysis buffer (40 mm Tris-HCl (pH 8.0), 120 mm NaCl, 0.1% Nonidet P-40) supplemented with protease inhibitors. Proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane (Amersham Biosciences). The membrane was blocked with 5% nonfat dry milk in Tris-buffered saline and incubated with primary antibodies overnight at 4 °C. Blots were developed with a peroxidase-conjugated secondary antibody, and proteins were visualized by enhanced chemiluminescence (ECL) procedures (Amersham Biosciences), using the manufacturer's protocol.
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 in PBS. Following cell fixation, cells were incubated with the appropriate primary antibodies in a solution of PBS with 1% bovine serum albumin and 0.1% Triton X-100 at 4 °C overnight. Antibodies used were as follows: human anti-E-cadherin (rabbit polyclonal antibody, 1:200), vimentin (rabbit polyclonal antibody, 1:200). Stainings were visualized using anti-rabbit or anti-mouse Alexa Fluor 488 (Molecular Probes). Nuclei were counterstained using 4,6-diamidino-2-phenylindole (DAPI; Sigma). Stained cells were visualized with a fluorescence-microscope (Olympus IX71).
Immunohistochemistry
Breast cancer tissue arrays with corresponding normal tissues (AccuMax Array, ISU ABXIS) were stained by using Vectastain Elite ABC kit (PK-6101 for PTTG-1 and PK-6102 for N-cadherin, Vector Laboratories, Burlingame, CA). The rabbit anti-PTTG-1 antibody (34-1500, Invitrogen) diluted 1:1000 and mouse anti-N-cadherin antibody (610920, BD Transduction Laboratories) diluted 1:50,000 in blocking solution were applied to the sections and incubated at 4 °C overnight. After washing in PBS, 1:200 dilution of biotinylated goat anti-rabbit IgG or biotinylated horse anti-mouse IgG antibodies in blocking solution were applied to the sections and incubated for 30 min. After washing in PBS, ABC reagent was applied to the sections and incubated for 30 min. After washing in PBS, color reaction was performed with 3,3′-diaminobenzidine (Vector Laboratories), and slides were washed with PBS. After counterstaining with hematoxylin and clearing with graded ethanol series and xylene, the sections were mounted with Canada balsam. Observation and photography were conducted using a IX71 microscope (Olympus, Tokyo, Japan) equipped with DP71 digital imaging system (Olympus).
PI3K Assay
Cell lysates (300 μg in 500 μl) were subjected to immunoprecipitation using anti-p85 antibodies (Upstate Biotechnology). The precipitates were washed twice with PBS, 1% Nonidet P-40, followed by PBS, 0.1 mol/liter Tris-HCl (pH 7.5), 0.5 mol/liter LiCl, and finally 25 mmol/liter HEPES (pH 7.5), 100 mmol/liter NaCl, 1 mmol/liter EDTA. The precipitates were then resuspended in 50 μl of presonicated phosphatidylinositol substrate and incubated for 10 min at room temperature. Each sample was labeled with 10 μCi of [32P]ATP for 10 min at room temperature. Reactions were stopped by adding 100 μl of chloroform/methanol/HCl (50:100:1). Lipids were extracted with 200 μl of chloroform, and after mixing and centrifugation, the lower “organic” phase was transferred to a new tube. The organic phase was washed once with 100 μl of methanol, 1 mol/liter HCl (1:1), and the upper phase was discarded. Finally, the lipid fraction was resuspended in 20 μl of chloroform and applied to a silica gel thin layer chromatography (TLC) plate pre-impregnated with 1% potassium oxalate. Phospholipids were resolved by TLC in freshly prepared buffer, chloroform/methanol/ammonia/water (43:38:5:7), for 45 min in a closed glass chamber at room temperature (26).
Tumor Xenografts on Nude Mice
After transfection with shRNA, BT549 cells (2 × 106 cells/ml) were subcutaneously inoculated into the right flank of athymic BALB/c female nude mice (5 weeks of age). Tumor size was measured with a caliper (calculated volume = shortest diameter2 × longest diameter/2) with 5-day intervals. This study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of Korea Institute of Radiological and Medical Sciences.
Statistical Analysis
All experimental data are reported as means, and the error bars represent the standard deviation. Statistical analysis was performed by the nonparametric Student's t test.
RESULTS
PTTG1 Is Expressed in Malignant Breast Cancer Cells
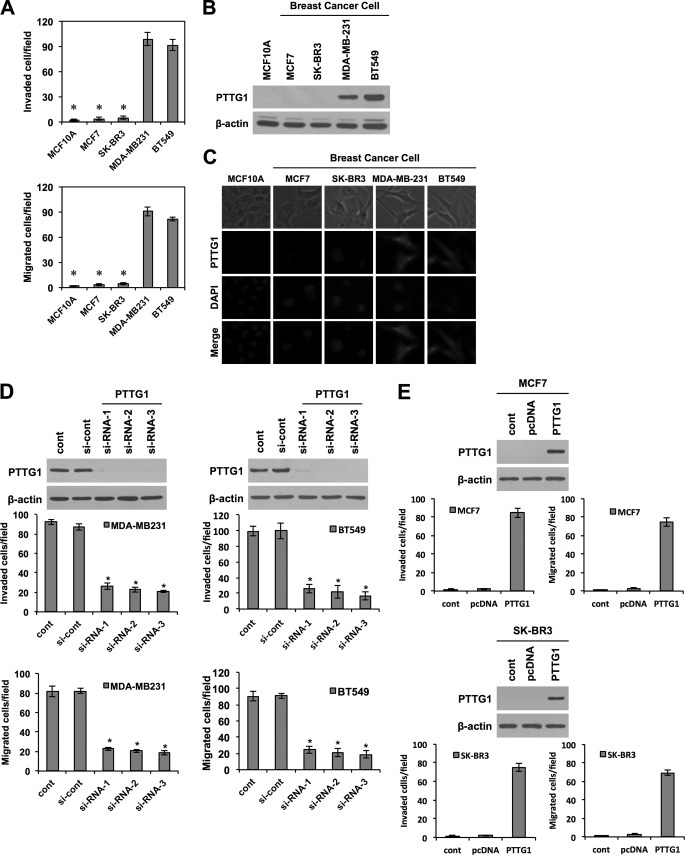
To assess whether PTTG1 expression levels have correlation with malignant breast cancer, we performed immunohistochemical staining on tissue arrays containing human breast cancer with corresponding normal tissues. Notably, PTTG1 was strongly expressed in breast tumor tissues in 9 of 11 breast cancer tissues, compared with normal breast tissues (Fig. 1A). In addition, we verified higher expression levels of PTTG1 in malignant breast cancer tissues than normal breast tissues by using publicly available microarray data base, Genevestigator (supplemental Fig. S1A). Because PTTG1 expression levels have correlation with malignant breast cancer, we next examined the correlation of PTTG1 with prognosis. Using an on-line survival analysis tool, we analyzed the correlation of PTTG1 expression levels with relapse free survival rate by the Kaplan-Meier plot (Fig. 1B). We found that breast cancer patients expressing higher levels of PTTG1 displayed a significantly lower relapse-free survival rate, compared with patients expressing relatively lower levels of PTTG1. We next examined the relationship between PTTG1 expression levels and tumor malignancy in breast cancer cell lines. Previous studies have shown that, among breast cancer cell lines, MDA-MB-231 and BT549 cells are more malignant than MCF7 and SK-BR3 cells (27). We also observed that MDA-MB-231 and BT549 cells have more migratory and invasive traits than MCF7 and SK-BR3 (Fig. 2A and supplemental Fig. S1B). In line with immunohistochemical staining on breast cancer tissues, PTTG1 expression levels were distinctly higher in MDA-MB-231 and BT549 cells compared with MCF7 and SK-BR3 cells (Fig. 2, B and C). Moreover, PTTG1 expression was not detected in normal MCF10A epithelial cells. Collectively, these results demonstrate a correlation between PTTG1 expression levels and malignancy in breast cancers.
FIGURE 1.
Correlation of PTTG1 expression with poor prognosis in breast cancer patients. A, tissue array containing breast cancer with normal counterpart tissues was immunostained with PTTG1 antibody. High expression levels of PTTG1 were shown in 9 of 11 cases of breast cancer tissues, compared with normal breast tissues. Representative images of three cases in normal breast tissues (upper) and breast cancer tissues (lower) are shown. Photomicrograph (×20 magnification) was taken by IX71 microscope (Olympus, Tokyo, Japan) equipped with DP71 digital imaging system (Olympus). B, Kaplan-Meier plot of human breast cancer patients using publicly available clinical breast cancer data. Breast cancer patients (2324) were grouped to high (1162) and low expression (1162) of PTTG1. Note that PTTG1 expression levels have strong correlation with poor prognosis and low survival rate.
FIGURE 2.
PTTG1 expression in malignant breast cancer cells. A, invasive (upper) and migratory (lower) properties of various breast cancer cell lines (MDA-MB-231, BT549, MCF7, and SK-BR3) and the normal breast cell line (MCF10A) were analyzed in trans-well by counting migrated cells in randomly selected five microscopic fields per well. Western blots (B) and immunocytochemistry (C) show that MDA-MB-231 and BT549 cells express higher levels of PTTG1 than MCF10A, MCF7, and SK-BR3. β-Actin was used as the loading control in Western blot. C, immunocytochemistry. MDA-MB-231 and BT549 cells express higher levels of PTTG1 expression than MCF10A, MCF7, and SK-BR3. D and E, transfection with three different PTTG1-targeting siRNAs (D) attenuated migratory and invasive properties of MDA-MB-231 and BT549 cells, compared with transfection with control siRNA (si-cont). Transfection with PTTG1 (E) promoted migratory and invasive properties of MCF7 and SK-BR2 cells, compared with transfection with control vector, pcDNA. Invasive and migratory properties were analyzed in trans-well by counting migrated cells in randomly selected five microscopic fields per well. All error bars represent mean ± S.D. of triplicate samples. *, p < 0.001.
To further investigate a possible causal connection between PTTG1 expression levels and malignancy, we modulated PTTG1 expression levels in these four human breast cancer cell lines. Three different siRNA-mediated knockdowns of PTTG1 in MDA-MB-231 and BT549 cells, which express higher levels of endogenous PTTG1, drastically attenuated the invasive and migratory properties of these cells compared with those of untreated cells or cells treated with scrambled siRNA (Fig. 2D and supplemental Fig. S1C). By contrast, transfection of MCF7 and SK-BR3 cells with the gene PTTG1 transformed MCF7 and SK-BR-3 cells, which normally express lower endogenous levels of PTTG1, into more migratory and invasive cells (Fig. 2E and supplemental Fig. S1D). Taken together, these results suggest that PTTG1 expression is necessary and sufficient for invasion and migration of breast cancer cells.
Ectopic Expression of PTTG1 Promotes Acquisition of Invasive and Migratory Properties by Normal Breast Cells
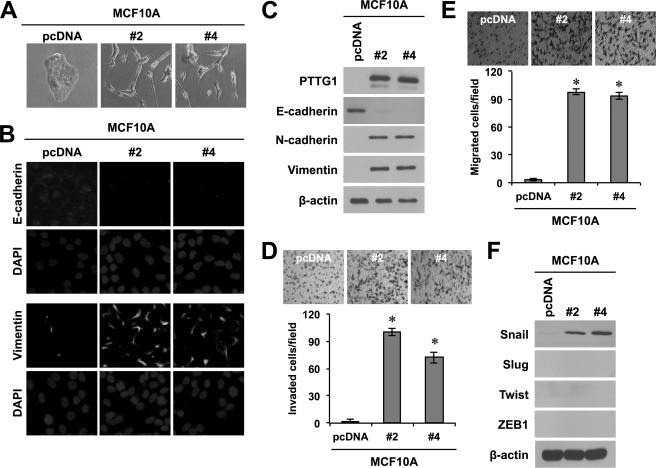
Tumor cell invasion involves the loss of cell-cell interaction together with acquisition of migratory properties and is often associated with EMT (6). Because PTTG1 expression levels were correlated with migratory and invasive properties of breast cancer cells, we further studied whether PTTG1 is involved in EMT in breast epithelial cells. To this end, we introduced PTTG1 into MCF10A normal breast epithelial cells by transfection and generated stable cell lines (supplemental Fig. S2A). Ectopic expression of PTTG1 changed the morphology of MCF10A cells to a more spindle shape and increased the spacing between cells, making monolayers less compact (Fig. 3A). Notably, PTTG1 expression led to a decrease in the levels of E-cadherin, which is expressed normally in epithelial cells (Fig. 3, B and C). In addition, PTTG1-expressing MCF10A cells expressed higher levels of N-cadherin and vimentin (Fig. 3, B and C), which are generally expressed in mesenchymal cells, compared with control. Consistent with these results, PTTG1-expressing MCF10A cells displayed more invasive and migratory properties compared with cells in the control group (Fig. 3, D and E). However, PTTG1 expression in MCF10A cells did not cause cell cycle arrest and death, compared with the control group, implicating that PTTG1 induces invasive and migratory traits, independently from the function of securin (supplemental Fig. S3). Next, we examined whether PTTG1 promotes EMT through regulation of the transcription factors, Snail, Slug, Twist, and ZEB1, which are known to be involved in EMT. We found that the transcription factor Snail was up-regulated at the level of both protein and mRNA by ectopic expression of PTTG1 in MCF10A normal epithelial cells (Fig. 3F and supplemental Fig. S2B). However, other transcription factors, Slug, Twist, and ZEB1, were not regulated by PTTG1 expression. To confirm the effect of PTTG1 expression on EMT, we also introduced PTTG1 into MCF10A normal breast epithelial cells by the retrovirus-mediated gene delivery system. Consistently, PTTG1-transduced MCF10A cells expressed lower levels of E-cadherin and higher levels of N-cadherin and vimentin, compared with control, and accompanied the acquisition of invasive and migratory properties (supplemental Fig. S4, A and B). We also performed immunohistochemistry with a breast cancer tissue array and observed that 1 of 8 cases displayed higher expression of N-cadherin, compared with normal counterpart tissues (supplemental Fig. S4C). In parallel with this, transduction with PTTG1 also led to the increase of Snail (supplemental Fig. S4D). Taken together, these results suggest that PTTG1 promotes EMT via regulation of the transcription factor Snail.
FIGURE 3.
Ectopic expression of PTTG1 promotes acquisition of invasive and migratory properties by normal breast cells. A, ectopic expression of PTTG1 in MCF10A cells induced morphological changes to a more spindle shape and increased the spacing between cells. Representative phase-contrast images of MCF10A-PTTG1 (clone #2 and #4) and control vector, pcDNA-transfected cells are shown. B, immunocytochemistry. PTTG1 overexpression results in loss of membrane E-cadherin and gain of membrane N-cadherin by immunofluorescence analysis (magnification, ×400). C, Western blots. Expression of PTTG1 results in loss of E-cadherin and gain of N-cadherin and vimentin. β-actin is used to show equal loading. D and E, MCF10A cells acquired invasive (D) and migratory properties (E) by ectopic expression of PTTG1. Invasive and migratory properties were analyzed in trans-well by counting migrated cells in randomly selected five microscopic fields per well. Error bars represent mean ± S.D. of triplicate samples. F, Western blots. Expression of PTTG1 induced transcription factor, Snail. However, expression levels of Slug, Twist, and ZEB1 were not changed by PTTG1 expression. β-Actin is used to show equal loading. *, p < 0.001.
PTTG1 Induces EMT via PI3K/AKT Signaling
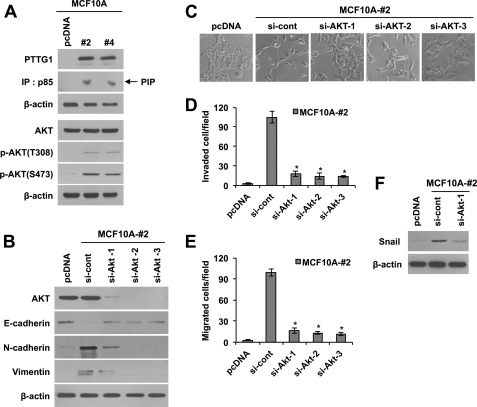
Next, we investigated the signaling mechanisms by which PTTG1 regulates EMT. Previous studies have shown that PTTG1 expression is promoted by growth factors such as EGF and FGF-2 (28, 29). We also observed that PTTG1 expression is promoted by treatment with EGF (data not shown). Because the growth factor signals are commonly integrated through mitogen-activated protein kinase (MAPK) or phosphoinositide 3-kinase (PI3K)/AKT pathways, we next examined whether these signaling pathways are involved in PTTG1-induced EMT. To this end, we analyzed the activity of MAPK, PI3K, and the phosphorylation status of AKT in PTTG1-transfected MCF10A cells. PI3K activity was determined by immunoprecipitating PI3K protein with an anti-p85 antibody and performing kinase assays with immunoprecipitates using PIP as a substrate. Separation of phospholipids by thin layer chromatography showed an increase in the lipid phosphorylation activity of the p85 subunit of PI3K in PTTG1-transfected MCF10A cells compared with control cells (Fig. 4A). In parallel with these data, both p-AKT (Thr-308) and p-AKT (Ser-473), the activated forms of AKT, were increased by PTTG1 expression. Consistent with these data, transduction of MCF10A cells with PTTG1 also showed the increase of phosphorylated AKT at Ser-473 and Thr-308 (supplemental Fig. S5A). Notably, treatment of PTTG1-expressing MCF10A cells with siRNA targeting AKT up-regulated E-cadherin and down-regulated N-cadherin and vimentin to the basal levels of MCF10A cells (Fig. 4B), suggesting that PTTG1 induces EMT through AKT activation. Moreover, down-regulation of AKT in PTTG1-expressing MCF10A cells restored cell morphology to MCF10A cells prior to overexpression of PTTG1, which are more round and stick together (Fig. 4C). In line with these results, down-regulation of AKT decreased migratory and invasive properties in PTTG1-expressing MCF10A cells, compared with control (Fig. 4, D and E, and supplemental Fig. S5C), verifying that PTTG1 promotes migratory and invasive properties through AKT activation. Because PTTG1-mediated EMT was triggered by induction of Snail, we determined whether AKT also regulates Snail. Importantly, treatment of PTTG1-expressing MCF10A cells with siRNA targeting AKT down-regulated the transcription factor Snail to the basal level observed in control groups (Fig. 4F). However, PTTG1 expression did not induce the activation of MAPK such as ERK1/2, JNK, and p38 in MCF10A cells (supplemental Fig. S5B). Taken together, these results suggest that PTTG1-mediated tumor malignancy, as reflected in migratory and invasive behavior, occurs via the AKT signaling pathway.
FIGURE 4.
PTTG1 induces EMT via PI3K/Akt signaling. A, Western blots. PTTG1 expression led to an increase of phosphorylated AKT at Thr-308 and Ser-473 in MCF10A cells. PI3K assay showed that PTTG1 expression increases the activity of p85, a component of PI3K. β-Actin was used as the loading control. IP, immunoprecipitation. B, Western blots. Transfection of PTTG1-expressing MCF10A (clone #2) with three different Akt-targeting siRNAs decreased epithelial marker, E-cadherin, and increased mesenchymal markers, N-cadherin and vimentin. β-Actin was used as the loading control. C, transfection of PTTG1-expressing MCF10A (clone #2) with three different Akt-targeting siRNAs recovered cell morphology to MCF10A cells. Representative phase-contrast images are shown. D and E, treatment with three different sRNA targeting AKT suppressed invasive (D) and migratory properties (E) of PTTG1-expressing MCF10A cells (clone #2). Invasive and migratory properties were analyzed in trans-well by counting migrated cells in randomly selected five microscopic fields per well. Error bars represent mean ± S.D. of triplicate samples. *, p < 0.001, versus control. F, Western blots. Treatment with siRNA targeting AKT attenuated PTTG1-induced expression of Snail in PTTG1-expressing MCF10A cells (clone #2). β-Actin was used as the loading control.
PTTG1 Induces EMT via AKT Signaling and Snail in Breast Cancer Cells
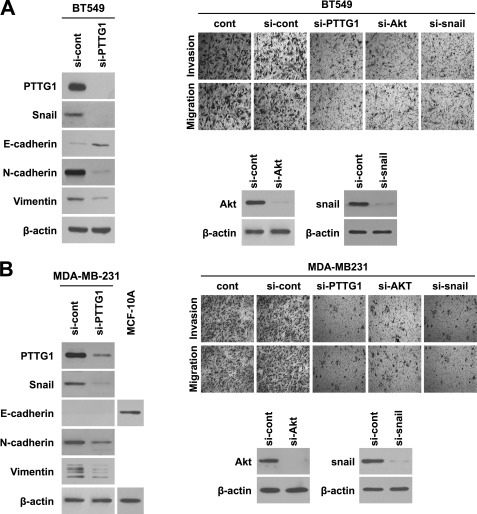
Because ectopic expression of PTTG1 in normal breast MCF10A cells led to AKT activation, induction of transcription factor Snail, and acquisition of migratory and invasive properties, we next examined whether down-regulation of PTTG1 attenuates mesenchymal properties and thereby suppresses migration and invasiveness in breast cancer cells BT549 and MDA-MB-231. As shown in Fig. 5, treatment with siRNA targeting PTTG1 caused an increase in epithelial cell marker E-cadherin and a decrease in mesenchymal cell markers such as N-cadherin and vimentin in both BT549 and MDA-MB-231 cells. Correspondingly, down-regulation of PTTG1 led to a decrease in expression of Snail, known as an EMT regulator (30). Because PTTG1-mediated EMT was induced via AKT activation and Snail in PTTG1-expressing breast MCF10A cells, we also attempted to down-regulate AKT and Snail in breast cancer cells BT549 and MDA-MB-231. In parallel with PTTG1-expressing MCF10A cells, treatment with siRNA targeting either AKT or Snail drastically suppressed migratory and invasive properties in breast cancer cells, BT549 and MDA-MB-231 (Fig. 5 and supplemental Fig. S6). Collectively, these data suggest that PTTG1 induces EMT via AKT signaling and Snail in breast cancer cells.
FIGURE 5.
PTTG1 induces EMT via AKT signaling and Snail in breast cancer cells. A and B, transfection with siRNA targeting PTTG1 increased epithelial marker, E-cadherin, and decreased transcription factor Snail and mesenchymal markers, N-cadherin and vimentin in BT549 (A) and MDA-MB-231 (B), that express high levels of PTTG1. Detection of E-cadherin failed in mesenchymal phenotype MDA-MB-231 cells. MCF10A cells lysate was loaded as a positive control of E-cadherin. Treatment of BT549 (A) and MDA-MB-231 cells (B) with either siRNA targeting PTTG1, AKT, or Snail suppressed invasive and migratory properties. Invasive and migratory properties were analyzed in trans-well by counting migrated cells in randomly selected five microscopic fields per well. Representative phase-contrast images are shown. β-Actin was used as the loading control. *, p < 0.001.
PTTG1 Plays a Pivotal Role in Maintenance of Self-renewal in Breast Cancer Stem Cells
Because EMT is often accompanied by an increase in the cancer stem cell population (4), we next examined whether the induction of EMT by PTTG1 expression was associated with an expansion of the cancer stem cell population. Previously, breast cancer stem cells were enriched by using culture conditions similar to that used to culture normal neural stem cells (31, 32). We also cultured BT549 breast cancer cells in serum-free medium supplemented with growth factors EGF and basic FGF (33). In these culture conditions, subsets of BT549 cells formed spheres and expressed higher levels of CD44 and lower levels of CD24, compared with adherent cultured cells (supplemental Fig. S7A). Moreover, sphere-cultured BT549 cells had higher expression of stemness-regulating transcription factors, CD44, Oct4, Sox2, and Nanog (34, 35), compared with cells in adherent culture conditions (supplemental Fig. S7A). Thus, to verify whether PTTG1 plays a role in breast cancer stem cells, we transduced sphere-cultured BT549 cells with shRNA targeting of PTTG1 and analyzed the sphere-forming ability. Notably, down-regulation of PTTG1 suppressed sphere formation in BT549 cells, compared with control (Fig. 6A). In line with this result, clone-forming ability at a single cell level was also markedly inhibited by transduction with shRNA targeting of PTTG1, suggesting that PTTG1 is required for self-renewal of breast cancer stem cells (Fig. 6B). Because breast cancer stem cells are enriched in CD44high CD24low cell population in previous studies (8), we next examined whether down-regulation of PTTG1 causes a decrease in the CD44high CD24low cell population in BT549 cells. Notably, FACS analysis showed that CD44high CD24low cell population is decreased by down-regulation of PTTG1 (Fig. 6C). Consistent with these findings, transduction with shRNA targeting of PTTG1 led to a decrease of stemness-regulating transcription factor Sox2, compared with scrambled shRNA (Fig. 6D). In parallel with these results, we also examined whether cancer stem cell population is increased by up-regulation of PTTG1 in MCF7 cells. In parallel with BT549 cells, up-regulation of PTTG1 promoted the sphere-forming ability in MCF7 (supplemental Fig. S7B). Also, transduction with PTTG1 induced Sox2 and CD44 expression (supplemental Fig. S7C). In addition, FACS analysis revealed that up-regulation of PTTG1 promotes CD44high CD24low population (supplemental Fig. S7D). Taken together, these data suggest that PTTG1 plays a pivotal role in maintaining breast cancer stem cell populations.
FIGURE 6.
PTTG1 regulates the breast cancer stem cell population. A, measurement of sphere sizes after treatment with shRNA targeted to PTTG1 for 72 h in BT549 cells. Sixty spheres per group were randomly taken to measure the size. B, clonal analysis of sphere-forming cells at single cell levels after treatment with shRNA targeting of PTTG1. C, quantification of breast cancer stem cell population by FACS. CD44high CD24low cell population was drastically increased in sphere-cultured conditions; however, treatment with shRNA targeting of PTTG1 suppressed CD44high CD24low cell population in sphere-cultured BT549 cells. D, treatment with shRNA targeting of PTTG1 decreased stemness-regulating transcription factor Sox2, but not Oct4 and Nanog. Error bars represent mean ± S.D. *, p < 0.001.
Targeting PTTG1 Suppresses Tumor Growth
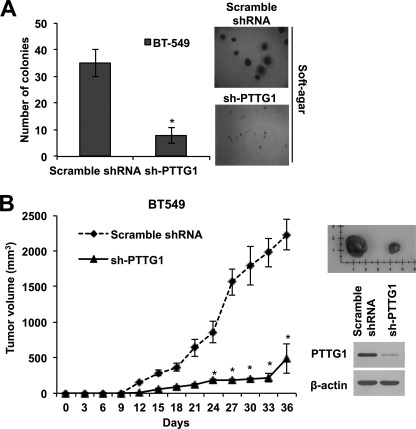
Cancer stem cells are characterized by high tumor initiating capacity (11, 33). To address the requirement for PTTG1 in maintaining tumorigenic capacity of breast cancer cells, we next examined anchorage-independent colony-forming ability of BT549 cells in soft agar after transduction with shRNA targeted to PTTG1. Contrary to normal cells that do not proliferate without anchorage, tumorigenic cancer cells can grow in a condition without anchorage such as semisolid medium. Thus, colony formation assay in soft agar is widely adapted to measure tumorigenic capacity in vitro, as a good predictor of the tumor-forming ability in vivo. As shown in Fig. 6A, BT549 breast cancer cells that are transduced by shRNA targeting PTTG1 displayed drastically lower colony-forming ability in soft agar, compared with control (Fig. 7A). To confirm the requirement of PTTG1 expression on the tumorigenic capacity of breast cancer stem cells in vivo, we transduced BT549 breast cancer cells with shRNA targeting PTTG1 and subcutaneously implanted 2 × 106 cells into female athymic BALB/c nude mice and monitored tumor growth every 3 days. Notably, tumor formation 2 weeks after implantation was dramatically attenuated in mice transplanted with BT549 cells treated with shRNA-PTTG1 compared with those transplanted with scrambled shRNA-treated cells (Fig. 7B). These results implicate that down-regulation of PTTG1 suppresses tumorigenic capacity of breast cancer cells through targeting of the breast cancer stem cell population. Taken together, these data demonstrates that PTTG1 expression is required in maintaining the breast cancer stem cell population.
FIGURE 7.
Down-regulation of PTTG1 decreases tumorigenic capacity. A, soft agar colony assay after transduction of breast cancer BT549 cells with shRNA targeting of PTTG1 (sh-PTTG1). Down-regulation of PTTG1 suppressed the colony-forming ability of BT549 cells. The number of colony was counted in randomly selected four microscopic fields per plate. Photomicrographs were taken at magnification ×200. Error bars represent mean ± S.D. of four different fields. *, p < 0.001, versus control. B, tumorigenic capacity in vivo. BT549 (2 × 106 cells/ml) cells were transfected with either control scrambled shRNA or shRNA targeting of PTTG1. Transfected cells were then subcutaneously inoculated to the right flank of athymic BALB/c female nude mice (5 weeks of age, n = 4 per group). Tumor formation was dramatically attenuated 2 weeks after implantation in mice transplanted with BT549 cells treated with shRNA-PTTG1, compared with those implanted with scrambled shRNA-treated cells. Representative photo shows differential volume of tumor at 36 days after implantation. Error bars represent mean ± S.D. *, p < 0.001, versus control.
DISCUSSION
The prognosis of breast cancer patients is closely correlated with the degree of cancer spread beyond the primary tumor. However, the mechanisms by which epithelial tumor cells escape from the primary tumor and colonize at a distant site are not entirely understood. In this study, we found that PTTG1 expression levels are higher in patient-derived breast cancer tissues than the normal counterpart. In addition, PTTG1 expression levels were correlated with poor prognosis, the relapse-free survival rate in publicly available clinical breast cancer data. This tendency was also observed in breast cancer cell lines; the more migratory and invasive basal phenotype cancer cell lines displayed higher expression levels of PTTG1. In parallel with these results, we found that down-regulation of PTTG1 expression in malignant breast cancer cell lines suppresses migratory and invasive properties, whereas overexpression of PTTG1 in less malignant breast cancer cells leads to the acquisition of migratory and invasive properties. Notably, normal breast cells also acquired migratory and invasive properties through ectopic expression of PTTG1. Our study suggests that PTTG1 expression is necessary and sufficient for acquisition of migratory and invasive behavior associated with tumor malignancy.
Recent studies in metastatic tumors have recently postulated EMT as a potential mechanism by which epithelial tumor cells acquire a more motile and invasive phenotype and escape from the primary tumor (2, 5, 24). In our study, PTTG1-driven acquisition of migratory and invasive properties was followed by suppression of E-cadherin, a marker of epithelial cells, and an increase in N-cadherin and vimentin, markers of mesenchymal cells. These changes in expression profile were accompanied by morphological changes to a more spindle-shape and a less compact growth pattern, implying that PTTG1 promotes EMT in breast cancer cells. This PTTG1-induced EMT was triggered by increases in Snail but not Slug, Twist, and Zeb1 transcription factors. These factors are thought to act collectively through dual repressor and activator transcriptional activities to induce EMT by targeting both genes associated with epithelial properties, such as E-cadherin, and mesenchymal properties, such as N-cadherin and vimentin (27, 38, 39). These transcription factors are known to induce normal EMT processes involved in complex body patterning and morphogenesis during embryonic development and wound healing. Although these transcription factors are suppressed in adult tissues, they are aberrantly expressed in various tumors, contributing to tumor progression through promotion of EMT. However, the mechanisms by which these factors are regulated in tumors are not understood.
Previously, several oncogenic pathways, including those involving peptide growth factors, Src, Ras, Ets, integrin, Wnt/β-catenin and Notch, were found to induce EMT. In particular, Ras-MAPK has been shown to activate the two related transcription factors, Snail and Slug, which, as noted above, transcriptionally repress E-cadherin and promote EMT. Activation of the PI3K/AKT axis has recently emerged as a central feature of EMT (27, 40). AKT is frequently activated in human epithelial cancer (41–43), and activation of AKT has been shown to induce Snail gene expression and thereby down-regulate the levels of E-cadherin mRNA and protein (27). Consistent with these previous studies, we observed that down-regulation of AKT caused a decrease in the migratory and invasive properties of breast cancer cells, accompanied by a decrease in Snail mRNA levels. Additionally, we found here that AKT activation is regulated by PTTG1. Specifically, we showed that overexpression of PTTG1 promoted phosphorylation of AKT at Thr-308 and Ser-473 in breast cells. Moreover, down-regulation of AKT in PTTG1-expressing cells blocked PTTG1-mediated promotion of migration and invasion, indicating that PTTG1 induces EMT in breast cancer cells, at least in part, via activation of AKT.
The PI3K-AKT pathway was previously found to be activated in cancer stem cells (36, 37). In our study, we found that the PI3K/AKT pathway is activated by PTTG1 expression. By activation of AKT signaling, PTTG1 expression contributed to expand the breast cancer stem cell population. Reversely, down-regulation of PTTG1 led to a decrease in self-renewing ability, CD44high CD24low cell population, and Sox2 expression. Also, targeting PTTG1 suppressed anchorage-independent colony-forming ability at clonal density and in vivo tumorigenic capacity, suggesting that PTTG1 is necessary for the maintenance of self-renewing and tumorigenic cancer stem cells.
In conclusion, our study suggests that PTTG1 expression levels are directly correlated with poor prognosis in breast cancer patients. PTTG1 promotes EMT and the maintenance of cancer stem cell population at least via activation of AKT. Taken together, these results suggest that PTTG1 may represent a new therapeutic target for malignant breast cancer.
Supplementary Material
This work was supported by National Nuclear Technology Program Grant 2008-2003935 and Converging Research Center Program Grant 2011K000877 funded by the Ministry of Education, Science, and Technology.

This article contains supplemental Figs. S1–S7.
- EMT
- epithelial to mesenchymal transition.
REFERENCES
- 1. Chaffer C. L., Weinberg R. A. (2011) A perspective on cancer cell metastasis. Science 331, 1559–1564 [DOI] [PubMed] [Google Scholar]
- 2. Liang X. (2011) EMT. New signals from the invasive front. Oral Oncol. 47, 686–687 [DOI] [PubMed] [Google Scholar]
- 3. Christofori G. (2006) New signals from the invasive front. Nature 441, 444–450 [DOI] [PubMed] [Google Scholar]
- 4. Singh A., Settleman J. (2010) EMT, cancer stem cells and drug resistance. An emerging axis of evil in the war on cancer. Oncogene 29, 4741–4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Karreth F., Tuveson D. A. (2004) Twist induces an epithelial-mesenchymal transition to facilitate tumor metastasis. Cancer Biol. Ther. 3, 1058–1059 [DOI] [PubMed] [Google Scholar]
- 6. Thiery J. P. (2002) Epithelial-mesenchymal transitions in tumor progression. Nat. Rev. Cancer 2, 442–454 [DOI] [PubMed] [Google Scholar]
- 7. Schatton T., Frank M. H. (2008) Cancer stem cells and human malignant melanoma. Pigment Cell Melanoma Res. 21, 39–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Al-Hajj M., Wicha M. S., Benito-Hernandez A., Morrison S. J., Clarke M. F. (2003) Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 100, 3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Singh S. K., Hawkins C., Clarke I. D., Squire J. A., Bayani J., Hide T., Henkelman R. M., Cusimano M. D., Dirks P. B. (2004) Identification of human brain tumor initiating cells. Nature 432, 396–401 [DOI] [PubMed] [Google Scholar]
- 10. Bao S., Wu Q., Sathornsumetee S., Hao Y., Li Z., Hjelmeland A. B., Shi Q., McLendon R. E., Bigner D. D., Rich J. N. (2006) Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 66, 7843–7848 [DOI] [PubMed] [Google Scholar]
- 11. Zhou J., Wulfkuhle J., Zhang H., Gu P., Yang Y., Deng J., Margolick J. B., Liotta L. A., Petricoin E., 3rd, Zhang Y. (2007) Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc. Natl. Acad. Sci. U.S.A. 104, 16158–16163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Inagaki A., Soeda A., Oka N., Kitajima H., Nakagawa J., Motohashi T., Kunisada T., Iwama T. (2007) Long term maintenance of brain tumor stem cell properties under nonadherent and adherent culture conditions. Biochem. Biophys. Res. Commun. 361, 586–592 [DOI] [PubMed] [Google Scholar]
- 13. Sullivan J. P., Minna J. D., Shay J. W. (2010) Evidence for self-renewing lung cancer stem cells and their implications in tumor initiation, progression, and targeted therapy. Cancer Metastasis Rev. 29, 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He S., Nakada D., Morrison S. J. (2009) Mechanisms of stem cell self-renewal. Annu. Rev. Cell Dev. Biol. 25, 377–406 [DOI] [PubMed] [Google Scholar]
- 15. Pei L., Melmed S. (1997) Isolation and characterization of a pituitary tumor-transforming gene (PTTG). Mol. Endocrinol. 11, 433–441 [DOI] [PubMed] [Google Scholar]
- 16. Zhang X., Horwitz G. A., Prezant T. R., Valentini A., Nakashima M., Bronstein M. D., Melmed S. (1999) Structure, expression, and function of human pituitary tumor-transforming gene (PTTG). Mol. Endocrinol. 13, 156–166 [DOI] [PubMed] [Google Scholar]
- 17. Zhang X., Horwitz G. A., Heaney A. P., Nakashima M., Prezant T. R., Bronstein M. D., Melmed S. (1999) Pituitary tumor transforming gene (PTTG) expression in pituitary adenomas. J. Clin. Endocrinol. Metab. 84, 761–767 [DOI] [PubMed] [Google Scholar]
- 18. Hamid T., Kakar S. S. (2003) PTTG and cancer. Histol. Histopathol. 18, 245–251 [DOI] [PubMed] [Google Scholar]
- 19. Zou H., McGarry T. J., Bernal T., Kirschner M. W. (1999) Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science 285, 418–422 [DOI] [PubMed] [Google Scholar]
- 20. Molina-Jiménez F., Benedicto I., Murata M., Martín-Vílchez S., Seki T., Antonio Pintor-Toro J., Tortolero M., Moreno-Otero R., Okazaki K., Koike K., Barbero J. L., Matsuzaki K., Majano P. L., López-Cabrera M. (2010) Expression of pituitary tumor-transforming gene 1 (PTTG1)/securin in hepatitis B virus (HBV)-associated liver diseases. Evidence for an HBV X protein-mediated inhibition of PTTG1 ubiquitination and degradation. Hepatology 51, 777–787 [DOI] [PubMed] [Google Scholar]
- 21. Domínguez A., Ramos-Morales F., Romero F., Rios R. M., Dreyfus F., Tortolero M., Pintor-Toro J. A. (1998) hpttg, a human homologue of rat pttg, is overexpressed in hematopoietic neoplasms. Evidence for a transcriptional activation function of hPTTG. Oncogene 17, 2187–2193 [DOI] [PubMed] [Google Scholar]
- 22. Filippella M., Galland F., Kujas M., Young J., Faggiano A., Lombardi G., Colao A., Meduri G., Chanson P. (2006) Pituitary tumor transforming gene (PTTG) expression correlates with the proliferative activity and recurrence status of pituitary adenomas. A clinical and immunohistochemical study. Clin. Endocrinol. 65, 536–543 [DOI] [PubMed] [Google Scholar]
- 23. Thompson A. D., 3rd, Kakar S. S. (2005) Insulin and IGF-1 regulate the expression of the pituitary tumor transforming gene (PTTG) in breast tumor cells. FEBS Lett. 579, 3195–3200 [DOI] [PubMed] [Google Scholar]
- 24. Singh A., Greninger P., Rhodes D., Koopman L., Violette S., Bardeesy N., Settleman J. (2009) A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell 15, 489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moffat J., Grueneberg D. A., Yang X., Kim S. Y., Kloepfer A. M., Hinkle G., Piqani B., Eisenhaure T. M., Luo B., Grenier J. K., Carpenter A. E., Foo S. Y., Stewart S. A., Stockwell B. R., Hacohen N., Hahn W. C., Lander E. S., Sabatini D. M., Root D. E. (2006) A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high content screen. Cell 124, 1283–1298 [DOI] [PubMed] [Google Scholar]
- 26. Hyun K. H., Yoon C. H., Kim R. K., Lim E. J., An S., Park M. J., Hyun J. W., Suh Y., Kim M. J., Lee S. J. (2011) Eckol suppresses maintenance of stemness and malignancies in glioma stem-like cells. Toxicol. Appl. Pharmacol. 254, 32–40 [DOI] [PubMed] [Google Scholar]
- 27. Grille S. J., Bellacosa A., Upson J., Klein-Szanto A. J., van Roy F., Lee-Kwon W., Donowitz M., Tsichlis P. N., Larue L. (2003) The protein kinase Akt induces epithelial mesenchymal transition and promotes enhanced motility and invasiveness of squamous cell carcinoma lines. Cancer Res. 63, 2172–2178 [PubMed] [Google Scholar]
- 28. Panguluri S. K., Yeakel C., Kakar S. S. (2008) PTTG. An important target gene for ovarian cancer therapy. J. Ovarian Res. 1, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tfelt-Hansen J., Yano S., Bandyopadhyay S., Carroll R., Brown E. M., Chattopadhyay N. (2004) Expression of pituitary tumor transforming gene (PTTG) and its binding protein in human astrocytes and astrocytoma cells. Function and regulation of PTTG in U87 astrocytoma cells. Endocrinology 145, 4222–4231 [DOI] [PubMed] [Google Scholar]
- 30. Shah P. P., Fong M. Y., Kakar S. S. (2011) PTTG induces EMT through integrin αVβ3-focal adhesion kinase signaling in lung cancer cells. Oncogene doi: 10.1038/onc.2011.488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Uchida N., Buck D. W., He D., Reitsma M. J., Masek M., Phan T. V., Tsukamoto A. S., Gage F. H., Weissman I. L. (2000) Direct isolation of human central nervous system stem cells. Proc. Natl. Acad. Sci. U.S.A. 97, 14720–14725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dontu G., Abdallah W. M., Foley J. M., Jackson K. W., Clarke M. F., Kawamura M. J., Wicha M. S. (2003) In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 17, 1253–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ponti D., Costa A., Zaffaroni N., Pratesi G., Petrangolini G., Coradini D., Pilotti S., Pierotti M. A., Daidone M. G. (2005) Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 65, 5506–5511 [DOI] [PubMed] [Google Scholar]
- 34. Wernig M., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B. E., Jaenisch R. (2007) In vitro reprogramming of fibroblasts into a pluripotent ES cell-like state. Nature 448, 318–324 [DOI] [PubMed] [Google Scholar]
- 35. Wang Z., Oron E., Nelson B., Razis S., Ivanova N. (2012) Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell 10, 440–454 [DOI] [PubMed] [Google Scholar]
- 36. Bleau A. M., Hambardzumyan D., Ozawa T., Fomchenko E. I., Huse J. T., Brennan C. W., Holland E. C. (2009) PTEN/PI3K/Akt pathway regulates the side population phenotype nd ABCG2 activity in glioma tumor stem-like cells. Cell Stem Cell 4, 226–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hambardzumyan D., Becher O. J., Rosenblum M. K., Pandolfi P. P., Manova-Todorova K., Holland E. C. (2008) PI3K pathway regulates survival of cancer stem cells residing in the perivascular niche following radiation in medulloblastoma in vivo. Genes Dev. 22, 436–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Batlle E., Sancho E., Francí C., Domínguez D., Monfar M., Baulida J., García De Herreros A. (2000) The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumorcells. Nat. Cell Biol. 2, 84–89 [DOI] [PubMed] [Google Scholar]
- 39. Cano A., Pérez-Moreno M. A., Rodrigo I., Locascio A., Blanco M. J., del Barrio M. G., Portillo F., Nieto M. A. (2000) The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2, 76–83 [DOI] [PubMed] [Google Scholar]
- 40. Larue L., Bellacosa A. (2005) Epithelial-mesenchymal transition in development and cancer. Role of phosphatidylinositol 3′-kinase/AKT pathways. Oncogene 24, 7443–7454 [DOI] [PubMed] [Google Scholar]
- 41. Brognard J., Clark A. S., Ni Y., Dennis P. A. (2001) Akt/protein kinase B is constitutively active in non-small cell lung cancer cells and promotes cellular survival and resistance to chemotherapy and radiation. Cancer Res. 61, 3986–3997 [PubMed] [Google Scholar]
- 42. Ringel M. D., Hayre N., Saito J., Saunier B., Schuppert F., Burch H., Bernet V., Burman K. D., Kohn L. D., Saji M. (2001) Overexpression and overactivation of Akt in thyroid carcinoma. Cancer Res. 61, 6105–6111 [PubMed] [Google Scholar]
- 43. Sun M., Wang G., Paciga J. E., Feldman R. I., Yuan Z. Q., Ma X. L., Shelley S. A., Jove R., Tsichlis P. N., Nicosia S. V., Cheng J. Q. (2001) AKT1/PKBα kinase is frequently elevated in human cancers and its constitutive activation is required for oncogenic transformation in NIH3T3 cells. Am. J. Pathol. 159, 431–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.