Background: The C5b-6 complex triggers assembly of the Membrane Attack Complex.
Results: The structure of C5b-6 at 4.2 Å resolution allowed an atomic model to be built.
Conclusion: C5b is stabilized by an interdomain linker of C6 and N-terminal elements that simultaneously engage N- and C-terminal elements.
Significance: In stabilizing C5b, C6 must change its conformation so that it becomes “primed” for initiating MAC assembly.
Keywords: Complement System, Crystal Structure, Host Defense, Innate Immunity, Membrane Proteins
Abstract
The complement membrane attack complex (MAC) forms transmembrane pores in pathogen membranes. The first step in MAC assembly is cleavage of C5 to generate metastable C5b, which forms a stable complex with C6, termed C5b-6. C5b-6 initiates pore formation via the sequential recruitment of homologous proteins: C7, C8, and 12–18 copies of C9, each of which comprises a central MAC-perforin domain flanked by auxiliary domains. We recently proposed a model of pore assembly, in which the auxiliary domains play key roles, both in stabilizing the closed conformation of the protomers and in driving the sequential opening of the MAC-perforin β-sheet of each new recruit to the growing pore. Here, we describe an atomic model of C5b-6 at 4.2 Å resolution. We show that C5b provides four interfaces for the auxiliary domains of C6. The largest interface is created by the insertion of an interdomain linker from C6 into a hydrophobic groove created by a major reorganization of the α-helical domain of C5b. In combination with the rigid body docking of N-terminal elements of both proteins, C5b becomes locked into a stable conformation. Both C6 auxiliary domains flanking the linker pack tightly against C5b. The net effect is to induce the clockwise rigid body rotation of four auxiliary domains, as well as the opening/twisting of the central β-sheet of C6, in the directions predicted by our model to activate or prime C6 for the subsequent steps in MAC assembly. The complex also suggests novel small molecule strategies for modulating pathological MAC assembly.
Introduction
Complement is an immunoeffector system, consisting of ∼30 blood plasma proteins and 10 cell surface receptors, that plays a major role in host defense against microorganisms (1). The ultimate outcome of complement activation on target phospholipid membranes is the formation of the membrane attack complex (MAC).4 The first step in MAC assembly is the specific cleavage of C5 (Mr = 196,000) to form the major product, C5b (Mr = 185,000), and the proinflammatory “anaphylatoxin,” C5a (Mr = 11,000) (2–5). Newly formed C5b is metastable and must bind rapidly to C6 to form the first stable intermediate, C5b-6, on the assembly pathway (6–8).
The MAC appears as a transmembrane tubule (∼100 Å inner diameter) in electron micrographs. Single copies of C6, C7, and C8 together with 12–18 copies of C9 (9–13) form the circular pore, whereas C5b binds to the upper segments of C6 and C7 and projects upwards from the pore.
We recently determined the crystal structure of complement C6 and proposed a mechanism for MAC assembly, including the structural basis for sequential and unidirectional assembly (14). Based on comparisons with C8 and perforin, we proposed that the auxiliary domains play key roles in regulating conformation and assembly of the MAC. Specifically, we suggested how the rotation of auxiliary modules at the leading edge of the nascent MAC could mediate optimal packing interactions with the new recruit and trigger the opening of its β-sheet, leading to the release of clusters of helices (CH), which would ultimately form the membrane attachment/spanning elements (14).
In the absence of direct structural data, our model remained speculative, particularly in the early steps of initiation by C5b. To address this, we have crystallized C5b-6 and solved its structure to a resolution of 4.2 Å. The map is of excellent quality, allowing an almost complete atomic model of the biomolecular ∼300-kDa complex to be built. The complex structure provides a wealth of new insights into how C5b primes C6 for MAC assembly, allowing us to refine and extend our model (14). In particular, it shows how C5b engages the auxiliary domains of C6 in an intimate embrace that “primes” C6 for initiating MAC assembly.
EXPERIMENTAL PROCEDURES
Purification and Crystallization of C5b-6
C5 and C6 were purified from a single batch of human plasma as described previously (14, 15). C5 (6 mg/ml) and C6 (3 mg/ml) in 5 mm imidazole HCl, pH 7.6, and 80 mm Li2SO4 were incubated with the fluid phase C5 convertase, CVF-Bb (EC 3.4.21.47), a complex of cobra venom factor and the Bb subunit of complement factor B, as described (16), except that 30 μm Ni2+ was substituted for 3 mm Mg2+. The reactants were incubated in 300-μl polypropylene tubes at room temperature for 2–4 weeks. During this time, orthorhombic crystals of C5b-6 formed spontaneously as long thin plates.
C5b-6 hemolytic activity was evaluated using sheep erythrocytes (Colorado Serum Co.) pretreated for 1 h with 10 mm dithiothreitol at 37 °C in 20 mm Tris, pH 9.0, 0.15 m NaCl, 10 mm EDTA. After washing several times, cells were suspended in 0.14 m dextrose, 0.1% gelatin, 2.5 mm sodium barbital pH 8.0, 70 mm NaCl, and 5 mm MgCl2. Samples of C5b-6 (0–0.2 μg) were preincubated for 1 min with 107 erythrocytes, followed by the addition of C8 (0.05 μg) and C9 (0.4 μg). Finally, C7 (0.05 μg) was added, and the cells were incubated at 37 °C for 30 min. One ml of buffer was then added to each tube, and the samples were centrifuged. Hemolysis was measured by optical density measurement of the supernatants at 413 nm, and complement hemolysis CH50 activity units were determined as previously described (17).
X-ray Data Collection and Structure Solution
Crystals with a typical size ∼0.1 × 0.06 × 0.4 mm were suspended in mother liquor and mounted in glass capillaries for room temperature data collection. The best crystals diffracted to ∼4 Å resolution but were radiation-sensitive. The diffraction data were collected at SSRL, Beamline 11-1, equipped with a Pilatus 6M detector. The data were collected at a rate of 1 frame/s in a beam collimated to 0.15 × 0.1 mm, without attenuation. One or two batches of data were collected from distinct volumes of each crystal. Each batch contained ∼15 frames with 0.5° rotation per frame. Each crystal volume typically survived for ∼10 frames (5°) before radiation damage became severe. Because most crystals adopted a similar orientation when mounted in the capillary, we were able to manually align them for data collection, so that reasonable completeness was obtained using 17 batches from 10 crystals. The data were processed using HKL2000, and radiation damage was assessed by changes in scale and B factor, as well as Rmerge. Frames with Rmerge > 25–30% were rejected.
Table 1 summarizes data collection and refinement statistics. The structure was solved initially by placing the major domains, using a molecular replacement method implemented in the program PHASER from CCP4 (18). Search models were based on C3b (Protein Data Bank code 3IDH) (19), C5 (Protein Data Bank code 3PRX) (20), and C6 (Protein Data Bank code 3T5O) (14). Template 1 contained the first seven domains of C5 overlaid onto the equivalent domains of C3b; template 2 comprised the MAC-perforin (MACPF) and selected auxiliary domains of C6; and templates 3–5 comprised C5d, the C1r/C1s, Uegf, and BMP-1 (bone morphogenetic protein-1) (CUB) domain, and C345C from the structure of C5. Template 6 comprised the C-terminal auxiliary domains (complement control proteins (CCPs) and factor I modules (FIMs) of C6). The missing parts of both proteins, including the linker regions, were built manually into difference Fourier maps.
TABLE 1.
Data collection and refinement statistics
| Data collection | |
| Number of crystals/segments | 10/17 |
| Temperature (K) | 298 |
| Resolution (Å) | 30–4.2 |
| Space group | I212121 |
| Cell dimensions (Å) | 159, 228, 278 |
| Rmerge | 0.16 (0.56)a |
| I/σI | 5.7 (1.6)a |
| Completeness (%) | 82 (67)a |
| Redundancy | 3.0 (2.6)a |
| Refinement statistics | |
| Number of reflections (work/free) | 28,605/1529 |
| Completeness (%) | 82 |
| Rwork | 0.221 (0.33)a |
| Rfree | 0.278 (0.37)a |
| Mean B (Å2) | 167 |
| Wilson B, Å2 | 120 |
| Number of protein atoms | 19,517 |
| Root mean square deviations from ideality | |
| Bond lengths (Å) | 0.01 |
| Angle (°) | 1.7 |
| Ramachandran plot (from PROCHECK) | |
| Favored (%) | 82.6 |
| Allowed (%) | 16.2 |
| Generally allowed (%) | 1.0 |
| Disallowed (%) | 0.3 |
a The value for the outer resolution shell, 4.4–4.2 A.
The model was refined using REFMAC5 (version 6), which has a number of tools that enable reliable atomic models to be built at relatively low (∼4 Å) resolution, including anisotropic scaling and water components modeled by Babinet's principle (21). In addition, “jelly body” restraints (σ = 0.012) enabled atomic coordinates and B-factors to be refined. Reciprocal space refinement was iterated with manual model building and real space refinement in COOT (22). The final cycles of refinement were conducted with TLS (translation, libration, screw) in five groups, with B-factor restraints increased by 1.5 from the default values. The quality of the structure was validated with CCP4. The final R factors are 0.21 (Rwork) and 0.27 (Rfree). Portions of the final electron density map are shown in supplemental Fig. S1.
The final model comprises 2450 residues (of a total of 2495), 15 saccharide units at eight attachments sites, and one divalent metal ion (modeled as Ca2+) bound at the C6 low density lipoprotein receptor (LR) element. The only missing elements are four loops within C5. The entire C6 structure is defined, with the exception of part of the CH1 domain (which was also disordered in crystals of C6) and the CCP2-FIM1 linker.
RESULTS
Structure Determination of C5b-6 Complex
C5b-6 was previously shown to form paracrystals at concentrations ∼1 mg/ml (23). By using higher protein concentrations (>5 mg/ml), we obtained orthorhombic crystals of C5b-6 that grew spontaneously from the reaction mixture, comprising C5, C6, and a soluble C5 convertase (CVF-Bb). The integrity of the complex was confirmed by dissolving crystals in 30% glycerol followed by the dialysis and assaying for specific hemolytic activity. The measured value (∼2 × 104 CH50 units/mg was comparable with that of the initially prepared soluble C5b-6 (see “Experimental Procedures”).
Crystals diffracted to ∼4 Å resolution at room temperature but were highly radiation-sensitive. We were unable to freeze crystals without substantial loss of diffraction quality; we therefore collected room temperature data from multiple capillary-mounted crystals. The data from 10 crystals merged satisfactorily to a resolution of 4.2 Å. The structure was solved by a combination of molecular replacement and ab initio model building of novel elements, notably a key linker region that was invisible in the structure of C6 alone. Refinement utilized jelly body restraints to stabilize convergence and minimize overfitting (21). The quality of the final electron density and derived model are exceptionally good for this resolution (Fig. 1, Table 1, and supplemental Fig. S1).
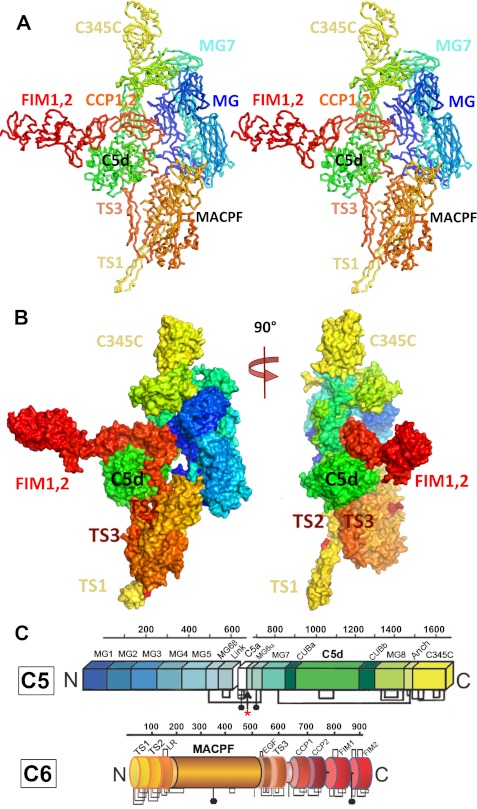
FIGURE 1.
Structure of the C5b-6 complex. A, stereo view of the C5b-6 complex shown as Cα chain trace. C5b is depicted in blue, green, and yellow; C6 is depicted in yellow, orange, and red. B, C5b-6 shown as molecular surface representation, colored as in A. The left view is the same as in A, and the right view is rotated by 90° about a vertical axis. C, domain organizations of C5 and C6, colored as in A and B. N-Linked carbohydrates are indicated by black hexagons, and disulfide linkages are indicated by brackets. The C5 convertase cleavage site in C6, which leads to the loss of C5a and formation of C5b, is indicated by an arrow.
Structure of C5b
C5b (Mr = ∼185,000) is the major proteolytic product of C5 after cleavage by C5 convertase, which excises a 74-residue helical domain, the C5a anaphylatoxin, from the center of the molecule. The two chains of C5b remain covalently linked via a disulfide bond. Although the structure of C5 (20, 24, 26) is known, as well as the homologous C3 and its proteolytic product, C3b, the structure of C5b has not previously been reported.
The conversion of C5 to C5b is accompanied by large conformational changes that are similar in nature to those observed in the homologous C3b, but with several important differences. C5b contains 12 domains. The first seven comprise ∼100-residue α-macroglobulin-like (MG) domains, each of which folds as a small β-sandwich. MG1–MG7 assemble into a large “MG scaffold” that supports the more flexible C-terminal domains. The anaphylatoxin C5a was an insert into the MG6 domain, connected via long loops to the opposite side of the protein. The link domain (residues 608–673) retains a similar conformation in C5b and includes two helical elements that form part of the MG scaffold.
The chain continues as a CUB domain, which is another β-sandwich into which a large α-helical domain, C5d (Mr∼35,000), has been inserted. After returning to complete the CUB domain, the chain continues as another MG domain (MG8) and finally a C-terminal α/β-domain, related to members of the netrin family, called C345C (27) (Mr = ∼17,000) that sits at the top of the molecule (Figs. 1 and 2).
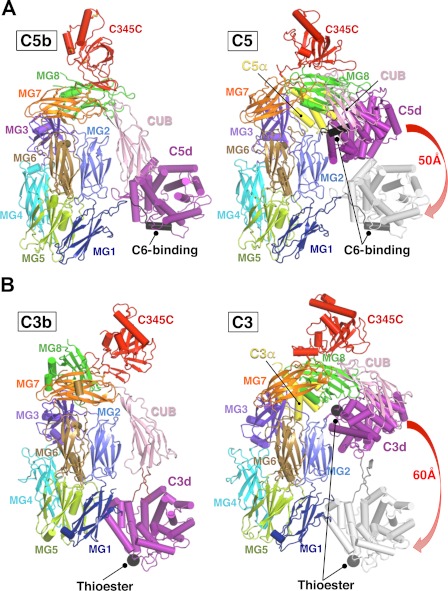
FIGURE 2.
Structural differences between C5b and C5 versus C3b and C3. A, side by side comparison of C5b and C5, overlaid on the MG scaffold. The large (50 Å) motion of the C5d domain is illustrated by overlaying its C5d location onto the C5 structure (right panel). The major binding site for C6 (the TS3-CCP1 linker) is located in C5d and is indicated by a black helix in both C5 and C5b, illustrating how it is cryptic in C5 and becomes exposed in C5b. B, a similar comparison of C3b and C3, illustrating the larger movement of the C3d domain compared with C5d; the extension of the CUB-C3d linker; and the distinct final location of C5d, which packs against the body of MG1 domain. The thioester site is shown as a black ball, illustrating its analogous location and exposure compared with the C6 binding site in C5d (black helix).
In the transition from C5 to C5b, the subset of the MG scaffold comprising MG1, MG2, MG5, and MG6 is quite rigid (superposing with an root mean square difference of only 1.2 Å (Cα)), providing a convenient reference frame. The other domains rotate about their centers of mass, especially MG3 and MG7, which directly contact the C-terminal elements.
In C5, the CUB, C5d, MG8, and C5a domains form a compact bundle that packs against the MG scaffold (24). Following the excision of C5a, MG8 moves ∼20 Å laterally to fill the cavity created by the loss of C5a. This releases constraints on the CUB-C5d unit, and C5d “unfurls” from the CUB, rotating ∼120° and shifting (downward and outward (away from the MG scaffold) by ∼40 Å (Fig. 2A). The CUB domain accommodates this movement by rotating ∼40° and by making a new interface with MG2. C345C is perched at the top of the MG scaffold in a loose association and is displaced in the complex partly in response to the shift of MG8.
A related conformational change in the transition from C3 to C3b has been described (19) (Fig. 2B). The most obvious difference is the final position of C3d vis-à-vis C5d. In C3b and all of its complexes determined thus far (19, 28–30), the linker between the CUB and C3d is extended, and the C3d domain unfurls but further downward (∼60 Å) to pack against the base of the MG scaffold (MG1) (19, 31, 32).
In the C5b-6 complex, the CUB-C5d linkage is much more compact, and although C5b swings downward, it does not move as far (∼40Å); its direction is also different, such that it remains 50 Å from the base of MG1. This distinct location for C5d appears to be stabilized by the unique packing of C6 against C5b (see below).
Structure of C6
We recently reported the crystal structure of C6 in its uncomplexed form (14). It comprises a central α/β globular MACPF domain, with a highly bent four-stranded β-sheet at its center; in addition, nine “auxiliary” domains (Fig. 1C) either wrap around or extend from the body of the MACPF. The last four domains (two CCP modules and two FIMs) extend upward from the MACPF body, attached via a flexible linker. In the C5b-6 complex, the linker and its flanking domains are sequestered by C5b, such that the C-terminal domains adopt a very different conformation (supplemental Fig. S2).
FIM2 was poorly defined in crystals of C6, but in the C5b-6 complex, both FIMs are clearly seen to fold as a single module that is distinct from the structure of the C7 FIM pair (33) because of the insertion of a helix and disulfide-linked loop. More subtle C5b-induced changes in C6, which we propose to be linked to activation, are described below.
Overview of C5b-6 Complex
C5b-6 is a bimolecular complex of Mr∼300,000. In the view shown in Fig. 1, C5b sits on top of C6, and because both domains stand “upright,” the complex is very tall (>200 Å). This packing mode is distinct from any known C3b-ligand interaction.
In the complex, C5b grabs the top of C6 like a pair of pincers. On one side, the rigid MG scaffold of C5b locks onto the TS2 and LR domains of C6. On the other, the C5d and CUB domains of C5b engage the top of the third thrombospondin module (TS3), the TS3-CCP1 linker, and the first CCP domain in an extensive and intimate interface. Table 2 lists the buried interfaces for all C5b-C6 interactions in the crystal lattice and estimates and energy for each of them.
TABLE 2.
Interacting surface areas and estimated energetic contributions
| C5b | C6 | Area | ΔG |
|---|---|---|---|
| Å2 | kcal/mol | ||
| C5d | Linker + TS3 | 1480 | −17.8 |
| CCP1 | 350 | −1.6 | |
| CCP2 | 410 | +0.2 | |
| Total | 2320 | −19.2 | |
| MG1–6 | CCP1 | 180 | −0.6 |
| LR + TS2 | 540 | − 1.0 | |
| Total | 720 | −1.6 | |
| CUB | CCP1 | 430 | −1.2 |
| C345C | FIM2 | 560 | −7.2 |
| C6 | C5b | Area | ΔG |
|---|---|---|---|
| Å2 | kcal/mol | ||
| CCP1 | C5d | 350 | −1.6 |
| CUB | 430 | −1.2 | |
| MG1–6 | 180 | −0.6 | |
| Total | 960 | −3.4 | |
An additional contact is made by the C-terminal FIMs, which form an extensive contact with the C345C domain of C5b across a 2-fold symmetry axis in the crystal; although the domains belong to different molecules in the crystal, there is reason to believe that a similar intramolecular contact exists in solution (i.e., this may be an example of domain swapping; see below). We will now describe each interface in detail.
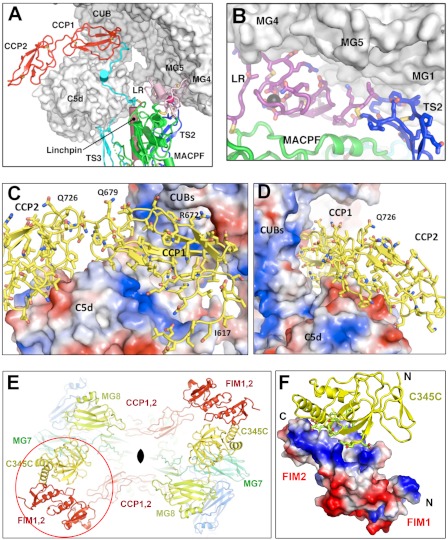
Base of C5b MG Scaffold Engages TS2 and LR Modules of C6
Two auxiliary domains of C6 (TS2, the LR module and the linker between them) form a continuous ridge at the top of the MACPF that packs against the base of the MG scaffold (contacting MG1, MG4, and MG5), burying ∼550 Å2 of protein surface. The interface has reasonable charge and shape complementarity, and docking involves only minor conformational changes on either side of the interface; however, the predicted binding energy is relatively small (Fig. 3B and Table 2).
FIGURE 3.
Structure of the C5b-C6 interface. A, overview of the binding site of C6 with C5b, illustrating the major elements of the interface (with the exception of the domain-exchanged FIM-C345C interaction, shown in Fig. 3E). The major interaction between the TS3-CCP1 linker is only partly visible in this view and is shown in detail in Fig. 4. B, close-up of the interface between the LR (magenta cartoon; side chains colored by atom-type; disulfide bonds in yellow; Ca2+ ion is shown as a black ball) and TS2 domains (in blue) of C6 with the base of the MG scaffold of C5b (shown as molecular surface). The top of the MACPF domain is shown as a green ribbon. C, close-up of the interface between the CCP modules (yellow cartoon with atom-colored side chain sticks and selected residues labeled) of C6 with C5b, shown as a molecular surface colored by surface potential. Red, negative; blue, positive; white, neutral. CCP1 packs into a large groove formed by the juxtaposition of the C5d, CUB, and MG2 domains. CCP2 makes limited contacts, exclusively with C5d. D, same as C, but the view is orthogonal (about vertical axis), revealing the tight packing at the base and loose packing at the top of the pocket. E, a three-dimensional slice through the crystal of C5b-6 showing the interchange of C-terminal arms across a 2-fold axis of symmetry (black lens at center) that enables binding of FIMs from one C5b-6 complex with the C345C domain from the symmetry-related complex. F, close-up view of the FIM2-C345C interaction. C345C is shown as colored ribbons (colored as in E), the FIMs as electrostatic potential surfaces.
Top of TS3 and TS3-CCP1 Linker Make Intimate Contacts with C5d Domain
The long linker segment (from TS3 to CCP1) emanating from the last β-strand of TS3 (residues 590–623), together with the adjacent β-hairpin at the top of TS3 (residues 556CDATY560), bury a total of ∼1500 Å2 of protein surface (Fig. 4). This is by far the largest interface in the complex, and energy calculations suggest that it dominates the overall binding (ΔG = −18 kCal/mol).
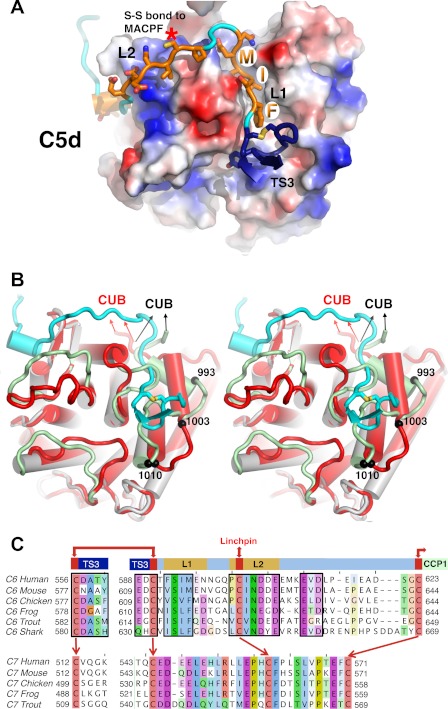
FIGURE 4.
The TS3-CCP1 of C6 linker interface with the C5d domain of C5. A, structure of the linker region of C6 (ribbon main chain and side chains as sticks, with atomic coloring) inserted into a groove on the surface of C5d (colored by electrostatic potential). The regions (TS3, L1, and L2) are labeled as in C. B, stereo superposition of the linker region of C6 (cyan) onto a cartoon (cylindrical helices and main chain loops) overlay of C5d (red) and C5 (gray helices, green loops), showing the multiple changes in helical elements and loops, as well as a shift in helical register between residues 993 and 1010, that together create the major groove observed in A. Arrows point to the connections with the CUB domain. C, sequence alignments of C6 (upper sequence) and C7 (lower sequence) TS3-CCP1 linker regions among vertebrate orthologs, including the cartilaginous fish, the shark. In C6, the first loop at the top of TS3 is discontinuous with the adjacent linker sequence, but joined by a disulfide bond (Cys556–Cys590 in human). The major interaction sites are boxed, revealing strong conservation of sequence, as well as length between the delimiting cysteines. Homologous cysteines in C6 and C7 are linked by blue arrows. C7 also has conserved motifs, but they are distinct from those in C6. Notably, the hydrophobic motif (FSIM in human C6) that inserts into C5d is absent in C7. Furthermore, the lengths of the intracysteine segments are different: S1 is longer in C7, whereas S2 is much shorter. The numbering of human C6 sequence does not include the leader peptide.
The linker is delimited by three disulfides (Cys590, which bonds to Cys556 within TS3; Cys602, which bonds to Cys478 within the MACPF domain (the top of the Linchpin helix); and Cys623–Cys665, which forms the first disulfide of the CCP1 domain. Clear and continuous electron density exists for the entire segment, with most side chains visible (this entire region was largely disordered in crystals of C6).
The interaction begins with the hairpin loop of the TS3 module and the first disulfide bond, which pack firmly into a crevice at the base of C5d. This is followed by a motif, 592FSIM595, which packs its three hydrophobic residues (Phe, Ile, and Met) into a deep hydrophobic crevice on the C5d domain. The crevice is created by the reorganization of several helices and connecting loops of C5d (Fig. 4B).
Interestingly, the thioester “warhead” of C3b is located in the analogous region of the C5d domain that makes contact with the TS3 domain of C6. The thioester is cryptic in native C3 by virtue of being buried and shielded from water, but it is exposed on the transition to C3b. The analogous region of C5d is similarly cryptic in C5 and becomes exposed in C5b; remarkably, it is this newly exposed analogous surface that forms a major interaction site with C6.
The linker continues as a hydrophilic loop that extends away from the interface before returning to the surface of C5b by forming a disulfide (Cys602–Cys478) with the top of the Linchpin helix of MACPF. The disulfide is flanked by two hydrophobic residues (Pro601 and Ile603) that pack against C5b. This is followed by four consecutive acidic residues that interact with a basic surface on C5b. The remaining 15 residues meander across the surface of C5d before connecting to the first disulfide bond of the CCP1 domain.
This linker is highly conserved evolutionarily in both sequence and length from human through shark and other cartilaginous fish. Moreover, it contrasts sharply with the sequence of C7, which, although also conserved among species, has very different properties and lengths (Fig. 4). For example, the FSIM motif of C6 is replaced by an acidic motif in C7, and the second half of the linker (“L2”) is too short in C7 to span the distance from the Linchpin disulfide to the beginning of CCP1. We propose that the differences between these linker segments provide a basis for strong discrimination between C6 and C7 for binding to this site on C5b.
CCP1 Domain Lies in Cradle Created by C5d, CUB, and MG2 Domains
A binding pocket for the CCP1 domain is created by the juxtaposition of the CUB and C5d domains, as well as the MG2 domain (Fig. 3, C and D). The major interactions involve hydrogen bonding and salt bridges, burying a total of ∼1000 Å2. Notably, CCP1 does not completely fill the pocket formed at the CUB-C5d interface, and many interactions with C5d are long range. This may be reflected in the low estimated energy for an interface of this size (ΔG = −3.4 kCal/mol). The organization suggests that modified packing arrangements are possible; for example, a closing of the hinge angle between the CUB and C5d would bring CCP1 into closer apposition with C5d.
We also note that one of the β-strands of the CUB domain contains a β-bulge in the structure of C5 (at position Tyr939), whereas in the C5b-C6 complex, the bulge is eliminated by shifting the register of residues 936–939 with respect to the β-sheet. This change creates a binding pocket for Tyr660 of C6 and enhances charge complementarity between the CUB domain and MG2. Perhaps the new interaction with MG2, which was not present in C5, explains the need for two conformations.
The CCP2 domain makes much more limited contacts with C5; it wraps around one helix of C5d, burying a small interface (∼400 Å2), and its contribution to overall binding is predicted to be minimal or unfavorable (ΔG = +0.2 kCal/mol).
FIMs Interact with C345C Domain
In the crystals, the FIM pair makes no contacts with its “own” C5b protein, but intriguingly, FIM2, as well as the unique insertion between FIM1 and FIM2, forms an interface with the C345C domain of a second molecule in the crystal lattice (Fig. 3, E and F). The interface, which includes one end of the inserted helix and loop, buries nearly 600 Å2 of protein surface and has good shape complementarity, and its predicted energy (−7.2 kCal/mol) is the second largest of all C5b-C6 interactions in the crystal lattice. The interaction occurs around a 2-fold symmetry axis within the crystals (it involves a “pairwise” exchange of arms), and we hypothesize that the interaction represents a genuine C5b-6 interface that has been “domain-swapped” (34–36). Indeed, simple modeling suggests that in solution, a C5b-C6 intracomplex can be made, requiring only a 45° rotation of the flexibly attached C345C domain about a vertical axis, together with a distinct bend at the FIM1-FIM2 boundary (supplemental Fig. S3). We further note that the C6 FIM interaction involves only one face of the C345C. The opposite face presents an extensive surface that could conceivably engage the C7 FIMs (37).
Conformational Changes in C6
In addition to the gross conformational changes observed in the C-terminal CCPs and FIMs (supplemental Fig. S2), more subtle changes can be discerned that we propose to be linked to the activation or “priming” of C6 to initiate MAC assembly. Overlay of the uncomplexed C6 structure onto the C5b-6 complex (Fig. 5) clearly reveals C5b-induced conformational changes in C6 in the directions that we predicted (14). For example, the intermolecular packing results in the TS3 domain being pushed downward by ∼5 Å with respect to the body of MACPF. We previously showed that the Y-shaped unit comprising TS1, TS2, TS3, and EGF domains behaved as a rigid body (the “regulatory” segment (Fig. 5A), and indeed the shift of TS3 is clearly linked to a large concerted clockwise rotation of TS1, the lower part of TS2, and the central EGF elements (Fig. 5, B and C). However, the expected “upward” motion of TS2 does not occur in this complex, because it is attached to the rigid MG scaffold, and it moves horizontally instead.
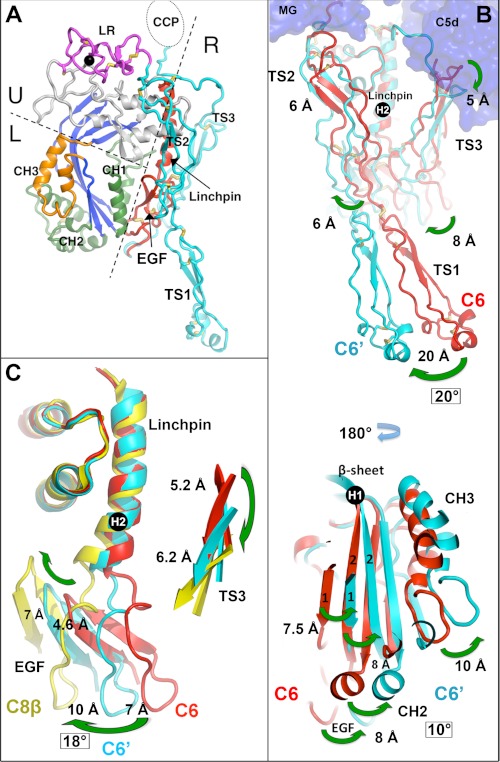
FIGURE 5.
Conformational changes in C6 induced by complex formation with C5b. A, ribbon diagram of the C6 (with the CCP modules and FIMs removed for clarity), indicating the three segments: lower (L), upper (U), and regulatory (R) that we previously defined as subdomains that rotate as rigid bodies about two hinge points (one at the bend in the β-sheet, the second at the end of the Linchpin helix), based on comparisons of C6 and C8 (14). B, unliganded C6 (red) and C5b-bound C6 (cyan) overlaid on their upper segments, which are identical within experimental error (root mean square deviation for 125 Cα = 0.51 Å). The upper panel, which is viewed from the “outside” of the nascent pore, reveals a downward shift of TS3 by ∼5 Å, and a concerted rotation of TS1, TS2, and TS3, three elements of the regulatory segment (the fourth element, the EGF domain, cannot be seen in this view. The largest rotation (∼20°) occurs in TS1, leading to a shift of ∼20 Å at its tip. Smaller rotations occur nearer to the top of the TS2, which is constrained by its contact with the MG scaffold. The lower panel is an “inside” view of C6 (i.e., looking from what will become the lumen of the pore. The counterclockwise twist of the β-sheet (∼10°) occurs about hinge point 1, at the bend in the β-sheet (labeled H1), and is similar but smaller to that observed in C8α (see Ref. 14). The CH1–3 elements rotate in concert with the sheet. C6 is red, and C5b-6 C6 is cyan. C, comparison between C6 (in red), C6 in the C5b-6 complex (C6′ in cyan), and C8β (in yellow), aligned on the upper segment (the root mean square difference for the overlaid upper segments of C6 and C8β is 0.8 Å for 92 on Cαs). The view is similar to that in B (upper panel) but cut away to reveal the rotation of the EGF domains in concert with the downward motion of the TS3 domains. The rotation is about the second hinge point (H2). The total rotation of the EGF domain is ∼18°, in the direction of the incoming recruit to the nascent pore. Note that liganded C6 (C6′) lies about halfway between unliganded C6 and C8β (∼8° rotation); note also the intermediate bend at the base of the Linchpin helix. C8β provides one model of the “open” conformation (see Ref. 14). Thus, C5b appears to prime C6 by inducing conformational changes toward the open conformation.
There is also a distinct twisting (and partial opening) of the β-sheet, in concert with the motion of the CH2 and CH3 elements that constitute the “lower” segment (see also Fig. 5, B and C). The twisting motion is similar to that seen in C8α. Finally, in support of our model, we note that the conformation of C6 in the C5b-6 complex is in many respects midway between its conformation in C6 and C8β. Fig. 5C shows a comparison between these three structures, illustrating the concerted downward motion of TS3 and the rotation of the EGF domain about the hinge point (“H”) at the end of the Linchpin helix.
DISCUSSION
C5b (like C3b) is metastable in solution in the absence of a binding partner. When viewed by EM, for example, it exhibits multiple conformations, presumably because the CUB and C5d domains are not firmly attached to the rest of the molecule (38, 39). Our crystal structure of the C5b-6 complex shows how the binding of C6 stabilizes C5b: by “tying together” the N- and C-terminal parts of the molecule; i.e., by simultaneously engaging the rigid MG scaffold on one side and by forming strong multifaceted interactions with the CUB and C5d domains on the other (Figs. 1, 3, and 4). Essentially all of the interactions involve the auxiliary domains at or near the upper surface of C6. This mode of interaction has no known counterpart in the ligand complexes of C3b.
In forming this complex, C6 also changes its conformation, and the directions of movement are consistent with the model that we recently proposed for propagation of pore assembly. The major elements are a clockwise rotation (∼20°) of the regulatory segment, with a concerted motion of the EGF domain of ∼10–20 Å toward the presumed location of the incoming recruit for pore formation. Within C6, there is also a linked twisting of the central β-sheet of the MACPF and its associated elements CH1–CH3, which we have proposed to be the first step in releasing the CH1 and CH2 elements prior to reorganization into membrane-binding β-hairpins.
The major surprise of our study was the role of the linker region between TS3 and CCP1. The linker was largely invisible in crystals of C6, but in the C5b-6 complex it appears to create a groove in the C5d domain into which it inserts a series of hydrophobic residues. This hydrophobic segment is conserved among vertebrates (Fig. 4). The buried surface area and estimated free energy change for this interface dominate the entire C5b-6 interaction (Table 2). The analogous linker in C7 has distinct motifs and lengths, and we propose that these differences provide the major discriminatory elements for binding of C6 versus C7 at this site on C5b.
The second major interaction is between the FIMs of C6 and the C345C domain at the C terminus of C5b. Such an interaction was expected, because binding was reported between recombinant fragments of C6 and the C345C domain (7, 37). The actual interaction we observe is between two complexes in the crystal lattice that exchange C-terminal arms (CCP2 and both FIMS) across a 2-fold symmetry axis (Fig. 3, E and F). We believe this to be an example of domain swapping, a phenomenon that is fairly common in crystal lattices. Simple model building (supplemental Fig. S3) suggests that in solution, the same intracomplex FIM-C345C interaction could be generated by rotations about two hinge points: the linker between CCP1 and CCP2 and a 45° rotation of the C345C domain about a vertical axis. These rotations are not predicted to be energetically costly, because CCP2 is loosely attached to C5d (its calculated binding free energy is actually positive), whereas C345C is flexibly attached to the top of C5b (Fig. 3E).
C345C likely provides an initial, reversible, attachment point for C6; the site is available in C5 (7, 40), whereas we have shown that the major interaction site lies on the C5d domain, which is cryptic in C5, and only becomes available following the cleavage step that creates C5b. Notably, C7 was also shown to engage the C345C domain through its FIMs (40, 41), and binding was competitive with C6.
The next step in MAC assembly is the recruitment of C7 to the C5b-6 complex, the final outcome of which is an amphiphilic transition that enables the complex to tether to the outer leaflet of a phospholipid membrane (42) presumably by elongation of β hairpins formed out of the CH elements. An atomic structure of this complex is likely to provide the next major insights into MAC assembly.
How might our mechanistic studies benefit human health? Remarkably, the absence of MAC is not seriously detrimental to human health, but its dysregulation can be very harmful. MAC formation is regulated by CD59, but localized hyperactivation of complement can overcome its protective effects. For example, in Alzheimer disease, opsozination of β-amyloid activates MAC, leading to damage of nearby neurites (43). The only drug that currently targets MAC formation is an antibody, eculizumab, that is directed against C5. It is used to treat the inherited disease, paroxysmal nocturnal hemoglobinuria, in which CD59 fails to attach to erythrocyte membranes (44). Targeting complement activation at the early stages of activation (45, 46) is likely to produce significant side effects, because C3 convertase is required for opsonization and phagocytosis of pathogens (47, 48). A large number of other complement-mediated disorders are known, and in many situations indications were found for the importance of the MAC. They include inflammation caused by trauma (49), rheumatoid arthritis (50), macular degeneration (51, 52), hemolytic anemias (53, 54), nephritis (55, 56), and demyelinating neuropathies such as multiple sclerosis and Guillain-Barré syndrome (57, 58). In rodent models, MAC formation increased the severity of rheumatoid arthritis, whereas a CD59 derivative decreased it (59, 60). Similarly, in a mouse model of renal ischemia reperfusion injury, mice deficient in CD59 suffered more tubular injury than wild type littermates (61).
Very recently, small molecule inhibitors (aurin carboxylic acids) have been shown to inhibit C9 recruitment to the MAC without interfering with other complement functions (25). Our new structure offers the possibility of rational design of inhibitors, for example based on peptides derived from the TS3-CCP1 linker region.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants AI055789 and AI055860 (to R. C. L.). This work was also supported by United States Army Medical Research and Materiel Command Grant DAMD17-03-2-0038 (to R. C. L.) and Multiple Sclerosis National Research Institute Grant 4061 (to R. G. D.).

This article contains supplemental Figs. S1–S3.
The atomic coordinates and structure factors (code 4E0S) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- MAC
- membrane attack complex
- CCP
- complement control protein
- CH
- clusters of helices
- CUB
- C1r/C1s, Uegf, and BMP-1 (bone morphogenetic protein-1)
- FIM
- factor I module
- LR
- low density lipoprotein receptor class A repeat
- MACPF
- membrane attack complex-perforin (domain)
- MG
- macroglobulin (domain)
- TS
- thrombospondin.
REFERENCES
- 1. Daha M. R. (2010) Role of complement in innate immunity and infections. Crit. Rev. Immunol. 30, 47–52 [DOI] [PubMed] [Google Scholar]
- 2. Fernandez H. N., Hugli T. E. (1978) Primary structural analysis of the polypeptide portion of human C5a anaphylatoxin. J. Biol. Chem. 253, 6955–6964 [PubMed] [Google Scholar]
- 3. Ehrengruber M. U., Geiser T., Deranleau D. A. (1994) Activation of human neutrophils by C3a and C5A. Comparison of the effects on shape changes, chemotaxis, secretion, and respiratory burst. FEBS Lett. 346, 181–184 [DOI] [PubMed] [Google Scholar]
- 4. Guo R. F., Ward P. A. (2005) Role of C5a in inflammatory responses. Annu. Rev. Immunol. 23, 821–852 [DOI] [PubMed] [Google Scholar]
- 5. Manthey H. D., Woodruff T. M., Taylor S. M., Monk P. N. (2009) Complement component 5a (C5a). Int. J. Biochem. Cell Biol. 41, 2114–2117 [DOI] [PubMed] [Google Scholar]
- 6. Cooper N. R., Müller-Eberhard H. J. (1970) The reaction mechanism of human C5 in immune hemolysis. J. Exp. Med. 132, 775–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. DiScipio R. G., Linton S. M., Rushmere N. K. (1999) Function of the factor I modules (FIMS) of human complement component C6. J. Biol. Chem. 274, 31811–31818 [DOI] [PubMed] [Google Scholar]
- 8. DiScipio R. G. (1981) The conversion of human complement component C5 into fragment C5b by the alternative-pathway C5 convertase. Biochem. J. 199, 497–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Müller-Eberhard H. J. (1986) The membrane attack complex of complement. Annu. Rev. Immunol. 4, 503–528 [DOI] [PubMed] [Google Scholar]
- 10. Kolb W. P., Haxby J. A., Arroyave C. M., Müller-Eberhard H. J. (1972) Molecular analysis of the membrane attack mechanism of complement. J. Exp. Med. 135, 549–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tschopp J., Müller-Eberhard H. J., Podack E. R. (1982) Formation of transmembrane tubules by spontaneous polymerization of the hydrophilic complement protein C9. Nature 298, 534–538 [DOI] [PubMed] [Google Scholar]
- 12. Tschopp J., Engel A., Podack E. R. (1984) Molecular weight of poly(C9). 12 to 18 C9 molecules form the transmembrane channel of complement. J. Biol. Chem. 259, 1922–1928 [PubMed] [Google Scholar]
- 13. DiScipio R. G. (1998) Late Components. in The Complement System (Rother K., Till G. O., Hansch G. M., eds) 2nd Ed., pp. 50–68, Springer-Verlag, New York [Google Scholar]
- 14. Aleshin A. E., Schraufstatter I. U., Stec B., Bankston L. A., Liddington R. C., Discipio R. G. (2012) Structure of complement C6 suggests a mechanism for initiation and unidirectional, sequential assembly of the membrane attack complex (MAC). J. Biol. Chem. 287, 10210–10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DiScipio R. G., Sweeney S. P. (1994) The fractionation of human plasma proteins. II. The purification of human complement proteins C3, C3u, and C5 by application of affinity chromatography. Protein Expr. Purif. 5, 170–177 [DOI] [PubMed] [Google Scholar]
- 16. DiScipio R. G., Smith C. A., Muller-Eberhard H. J., Hugli T. E. (1983) The activation of human complement component C5 by a fluid phase C5 convertase. J. Biol. Chem. 258, 10629–10636 [PubMed] [Google Scholar]
- 17. Whaley K., North J. (1997) Haemolytic assays for whole complement activity and individual components, in Complement: A Practical Approach (Dodds A. W., Sim R. B. eds) pp. 19–47, IRL Press, Oxford [Google Scholar]
- 18. McCoy A. J. (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Janssen B. J., Christodoulidou A., McCarthy A., Lambris J. D., Gros P. (2006) Structure of C3b reveals conformational changes that underlie complement activity. Nature 444, 213–216 [DOI] [PubMed] [Google Scholar]
- 20. Laursen N. S., Gordon N., Hermans S., Lorenz N., Jackson N., Wines B., Spillner E., Christensen J. B., Jensen M., Fredslund F., Bjerre M., Sottrup-Jensen L., Fraser J. D., Andersen G. (2010) Structural basis for inhibition of complement C5 by the SSL7 protein from Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 107, 3681–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Podack E. R., Esser A. F., Biesecker G., Müller-Eberhard H. J. (1980) Membrane attack complex of complement. A structural analysis of its assembly. J. Exp. Med. 151, 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fredslund F., Laursen N. S., Roversi P., Jenner L., Oliveira C. L., Pedersen J. S., Nunn M. A., Lea S. M., Discipio R., Sottrup-Jensen L., Andersen G. R. (2008) Structure of and influence of tick complement inhibitor on complement component 5. Nat. Immunol. 9, 753–760 [DOI] [PubMed] [Google Scholar]
- 25. Lee M., Guo J. P., Schwab C., McGeer E. G., McGeer P. L. (2012) Selective inhibition of the membrane attack complex of complement by low molecular weight components of the aurin tricarboxylic acid synthetic complex. Neurobiol. Aging, http://dx.doi.org/10.1016/j.neurobiolaging.2011.12.005 [DOI] [PubMed]
- 26. Laursen N. S., Andersen K. R., Braren I., Spillner E., Sottrup-Jensen L., Andersen G. R. (2011) Substrate recognition by complement convertases revealed in the C5-cobra venom factor complex. EMBO J. 30, 606–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bányai L., Patthy L. (1999) The NTR module. Domains of netrins, secreted frizzled related proteins, and typ I procollagen C-proteinase enhancer protein are homologous with tissue inhibitors of metalloproteases. Protein Sci. 8, 1636–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Garcia B. L., Ramyar K. X., Tzekou A., Ricklin D., McWhorter W. J., Lambris J. D., Geisbrecht B. V. (2010) Molecular basis for complement recognition and inhibition determined by crystallographic studies of the staphylococcal complement inhibitor (SCIN) bound to C3c and C3b. J. Mol. Biol. 402, 17–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rooijakkers S. H., Wu J., Ruyken M., van Domselaar R., Planken K. L., Tzekou A., Ricklin D., Lambris J. D., Janssen B. J., van Strijp J. A., Gros P. (2009) Structural and functional implications of the alternative complement pathway C3 convertase stabilized by a staphylococcal inhibitor. Nat. Immunol. 10, 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wiesmann C., Katschke K. J., Yin J., Helmy K. Y., Steffek M., Fairbrother W. J., McCallum S. A., Embuscado L., DeForge L., Hass P. E., van Lookeren Campagne M. (2006) Structure of C3b in complex with CRIg gives insights into regulation of complement activation. Nature 444, 217–220 [DOI] [PubMed] [Google Scholar]
- 31. Wu J., Wu Y. Q., Ricklin D., Janssen B. J., Lambris J. D., Gros P. (2009) Structure of complement fragment C3b-factor H and implications for host protection by complement regulators. Nat. Immunol. 10, 728–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Forneris F., Ricklin D., Wu J., Tzekou A., Wallace R. S., Lambris J. D., Gros P. (2010) Structures of C3b in complex with factors B and D give insight into complement convertase formation. Science 330, 1816–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Phelan M. M., Thai C. T., Soares D. C., Ogata R. T., Barlow P. N., Bramham J. (2009) Solution structure of factor I-like modules from complement C7 reveals a pair of follistatin domains in compact pseudosymmetric arrangement. J. Biol. Chem. 284, 19637–19649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu Y., Eisenberg D. (2002) 3D domain swapping. As domains continue to swap. Protein Sci. 11, 1285–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gronenborn A. M. (2009) Protein acrobatics in pairs: Dimerization via domain swapping. Curr. Opin. Struct. Biol. 19, 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bennett M. J., Choe S., Eisenberg D. (1994) Domain swapping. Entangling alliances between proteins. Proc. Natl. Acad. Sci. U.S.A. 91, 3127–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thai C. T., Ogata R. T. (2004) Complement components C5 and C7. Recombinant factor I modules of C7 bind to the C345C domain of C5. J. Immunol. 173, 4547–4552 [DOI] [PubMed] [Google Scholar]
- 38. Nishida N., Walz T., Springer T. A. (2006) Structural transitions of complement component C3 and its activation products. Proc. Natl. Acad. Sci. U.S.A. 103, 19737–19742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. DiScipio R. G. (1992) Formation and structure of the C5b-7 complex of the lytic pathway of complement. J. Biol. Chem. 267, 17087–17094 [PubMed] [Google Scholar]
- 40. Thai C. T., Ogata R. T. (2005) Recombinant C345C and factor I modules of complement components C5 and C7 inhibit C7 incorporation into the complement membrane attack complex. J. Immunol. 174, 6227–6232 [DOI] [PubMed] [Google Scholar]
- 41. Thai C. T., Ogata R. T. (2003) Expression and characterization of the C345/NTR domains of complement components C3 and C5. J. Immunol. 171, 6565–6573 [DOI] [PubMed] [Google Scholar]
- 42. Podack E. R., Biesecker G., Müller-Eberhard H. J. (1979) Membrane attack complex of complement. Generation of high affinity phospholipid binding sites by fusion of five hydrophilic plasma proteins. Proc. Natl. Acad. Sci. U.S.A. 76, 897–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McGeer P. L., Akiyama H., Itagaki S., McGeer E. G. (1989) Activation of the classical complement pathway in brain tissue of Alzheimer patients. Neurosci. Lett. 107, 341–346 [DOI] [PubMed] [Google Scholar]
- 44. McKeage K. (2011) Eculizumab. A review of its use in paroxysmal nocturnal haemoglobinuria. Drugs 71, 2327–2345 [DOI] [PubMed] [Google Scholar]
- 45. Katschke K. J., Jr., Stawicki S., Yin J., Steffek M., Xi H., Sturgeon L., Hass P. E., Loyet K. M., Deforge L., Wu Y., van Lookeren Campagne M., Wiesmann C. (2009) Structural and functional analysis of a C3b-specific antibody that selectively inhibits the alternative pathway of complement. J. Biol. Chem. 284, 10473–10479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ricklin D., Lambris J. D. (2007) Complement-targeted therapeutics. Nat. Biotechnol. 25, 1265–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ross G. D., Newman S. L., Lambris J. D., Devery-Pocius J. E., Cain J. A., Lachmann P. J. (1983) Generation of three different fragments of bound C3 with purified factor I or serum. II. Location of binding sites in the C3 fragments for factors B and H, complement receptors, and bovine conglutinin. J. Exp. Med. 158, 334–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ross G. D., Yount W. J., Walport M. J., Winfield J. B., Parker C. J., Fuller C. R., Taylor R. P., Myones B. L., Lachmann P. J. (1985) Disease-associated loss of erythrocyte complement receptors (CR1, C3b receptors) in patients with systemic lupus erythematosus and other diseases involving autoantibodies and/or complement activation. J. Immunol. 135, 2005–2014 [PubMed] [Google Scholar]
- 49. Qiao F., Atkinson C., Song H., Pannu R., Singh I., Tomlinson S. (2006) Complement plays an important role in spinal cord injury and represents a therapeutic target for improving recovery following trauma. Am. J. Pathol. 169, 1039–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Okroj M., Heinegård D., Holmdahl R., Blom A. M. (2007) Rheumatoid arthritis and the complement system. Ann. Med. 39, 517–530 [DOI] [PubMed] [Google Scholar]
- 51. Dinu V., Miller P. L., Zhao H. (2007) Evidence for association between multiple complement pathway genes and AMD. Genet. Epidemiol. 31, 224–237 [DOI] [PubMed] [Google Scholar]
- 52. Nozaki M., Raisler B. J., Sakurai E., Sarma J. V., Barnum S. R., Lambris J. D., Chen Y., Zhang K., Ambati B. K., Baffi J. Z., Ambati J. (2006) Drusen complement components C3a and C5a promote choroidal neovascularization. Proc. Natl. Acad. Sci. U.S.A. 103, 2328–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ståhl A. L., Kristoffersson A., Olin A. I., Olsson M. L., Roodhooft A. M., Proesmans W., Karpman D. (2009) A novel mutation in the complement regulator clusterin in recurrent hemolytic uremic syndrome. Mol. Immunol. 46, 2236–2243 [DOI] [PubMed] [Google Scholar]
- 54. Rother R. P., Rollins S. A., Mojcik C. F., Brodsky R. A., Bell L. (2007) Discovery and development of the complement inhibitor eculizumab for the treatment of paroxysmal nocturnal hemoglobinuria. Nat. Biotechnol. 25, 1256–1264 [DOI] [PubMed] [Google Scholar]
- 55. Camussi G., Brentjens J. R., Noble B., Kerjaschki D., Malavasi F., Roholt O. A., Farquhar M. G., Andres G. (1985) Antibody-induced redistribution of Heymann antigen on the surface of cultured glomerular visceral epithelial cells. Possible role in the pathogenesis of Heymann glomerulonephritis. J. Immunol. 135, 2409–2416 [PubMed] [Google Scholar]
- 56. Welch T. R., Frenzke M. (2001) Glomerulonephritis associated with deficiencies and polymorphisms of complement components encoded in the class III region of the MHC. Front. Biosci. 6, D898–D903 [DOI] [PubMed] [Google Scholar]
- 57. Ingram G., Hakobyan S., Robertson N. P., Morgan B. P. (2009) Complement in multiple sclerosis. Its role in disease and potential as a biomarker. Clin. Exp. Immunol. 155, 128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Koski C. L., Sanders M. E., Swoveland P. T., Lawley T. J., Shin M. L., Frank M. M., Joiner K. A. (1987) Activation of terminal components of complement in patients with Guillain-Barré syndrome and other demyelinating neuropathies. J. Clin. Invest. 80, 1492–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Williams A. S., Mizuno M., Richards P. J., Holt D. S., Morgan B. P. (2004) Deletion of the gene encoding CD59a in mice increases disease severity in a murine model of rheumatoid arthritis. Arthritis Rheum. 50, 3035–3044 [DOI] [PubMed] [Google Scholar]
- 60. Fraser D. A., Harris C. L., Williams A. S., Mizuno M., Gallagher S., Smith R. A., Morgan B. P. (2003) Generation of a recombinant, membrane-targeted form of the complement regulator CD59. Activity in vitro and in vivo. J. Biol. Chem. 278, 48921–48927 [DOI] [PubMed] [Google Scholar]
- 61. Turnberg D., Botto M., Lewis M., Zhou W., Sacks S. H., Morgan B. P., Walport M. J., Cook H. T. (2004) CD59a deficiency exacerbates ischemia-reperfusion injury in mice. Am. J. Pathol. 165, 825–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.