Background: FKBP38 regulates the prosurvival activity of Bcl-2 in apoptosis signaling.
Results: The catalytic domain of FKBP38 interacts with helix 4 in Bcl-2.
Conclusion: An electrostatic interaction modulates transient complex formation between FKBP38 and Bcl-2.
Significance: The protein complex provides a molecular basis for FKBP38-mediated Bcl-2 regulation.
Keywords: Bcl-2, Cell Signaling, Immunophilin, Protein Structure, Site-directed Mutagenesis, FK506-binding Protein, NMR, Chemical Shift Perturbation, Peptide Library
Abstract
FKBP38 is a regulator of the prosurvival protein Bcl-2, but in the absence of detailed structural insights, the molecular mechanism of the underlying interaction has remained unknown. Here, we report the contact regions between Bcl-2 and the catalytic domain of FKBP38 derived by heteronuclear NMR spectroscopy. The data reveal that a previously identified charge-sensitive loop near the putative active site of FKBP38 is mainly responsible for Bcl-2 binding. The corresponding binding epitope of Bcl-2 could be identified via a peptide library-based membrane assay. Site-directed mutagenesis of the key residues verified the contact sites of this electrostatic protein/protein interaction. The derived structure model of the complex between Bcl-2 and the FKBP38 catalytic domain features both electrostatic and hydrophobic intermolecular contacts and provides a rationale for the regulation of the FKBP38/Bcl-2 interaction by Ca2+.
Introduction
FKBP38 is a peptidylprolyl cis/trans-isomerase (PPIase6; EC 5.2.1.8) that belongs to the family of FK506-binding proteins (FKBPs) and has been associated with a number of important cell regulatory processes (1). The multidomain protein consists of a Glu-rich segment at the N terminus, followed by the structurally well characterized PPIase domain (2, 3), a tetratricopeptide repeat (TPR) domain, a calmodulin (CaM)-binding site, and a putative C-terminal membrane anchor, which is unique among the human members of the FKBP family (4). Moreover, an exceptional cation-binding site was recently discovered in the PPIase domain close to the putative active site (3).
FKBP38 participates in various regulatory mechanisms such as (i) cell size regulation (5), (ii) sonic hedgehog signaling and thus the development of neuronal tissues in mice (6, 7), and (iii) apoptosis (8, 9). Interactions between FKBP38 and different proteins such as NS5A (i.e. a subunit of the hepatitis C virus replicase) (10), HIF1 prolyl 4-hydroxylase (11, 12), Hsp90 (13), and mTOR (14) and its activator Rheb (15) have been shown. The binding of FKBP38 to calmodulin for PPIase activation (9, 13, 16) and to Bcl-2 has been implicated in apoptosis regulation (8, 9, 13, 17–19).
The prosurvival Bcl-2 protein is a core regulator of apoptosis. Bcl-2 shares a common fold with other prosurvival Bcl-2 family members (Bcl-xL, Mcl-1, Bcl-w, and Bcl-B), containing four Bcl-2 homology (BH) domains and a C-terminal membrane anchor. Bcl-2 is localized in the membranes of mitochondria and the endoplasmic reticulum, just like FKBP38, and in the nucleus (1, 20). Bcl-2 is associated with many diseases that are related to deviations in the cell cycle and can lead to pathological regulation of mitochondrial apoptosis, including breast and prostate cancers, schizophrenia, and autoimmune disorders such as type 1 diabetes (21, 22).
Bcl-2 was found previously to interact with FKBP38 in a yeast two-hybrid system (8). In recent years, this interaction was further analyzed by several groups with partly contradictory results (1). Kang et al. (17) suggested that a long flexible loop between the BH4 and BH3 domains of Bcl-2 mediates the binding to FKBP38 and that this protein/protein interaction can prevent phosphorylation of the flexible loop, which would lead to apoptosis as a result of Bcl-2 degradation by caspase-3. Edlich et al. (9, 13) identified a CaM-dependent binding interaction between the FKBP38 catalytic domain and Bcl-2, which gave rise to a pro-apoptotic effect in cell experiments. Also on the basis of cell experiments, Choi et al. (19) reported that the PPIase and TPR domains of FKBP38 bind to Bcl-2 by affecting primarily its flexible loop between the BH4 and BH3 domains. Taken together, all of these results differ (i) in the localization of the contact regions, (ii) in the dependence on additional components, and (iii) in the pro- or anti-apoptotic nature of the FKBP38/Bcl-2 interaction.
In contrast with the above-described biological approaches, we present NMR-based data that aim to elucidate the structural features of the interaction between the FKBP38 catalytic domain (FKBP38(35–153)) and Bcl-2. On the basis of a peptide library derived from the Bcl-2 sequence, we identified a binding epitope that revealed specific binding to FKBP38(35–153). The corresponding Bcl-2(119–131) peptide induced significant chemical shift changes in the charge-sensitive β5-α1 loop of FKBP38. Further characterization of the respective binding interface in the transient FKBP38(35–153)·Bcl-2(1–211) complex revealed a predominantly electrostatic interaction. A structural model of the complex has been derived from docking calculations based on chemical shift perturbation (CSP) data.
EXPERIMENTAL PROCEDURES
Preparation of Peptides and Proteins
All peptides were generated by solid-phase synthesis as described (16). Wild-type FKBP38(35–153), the FKBP38(35–153) D92N/D94N variant, His6-CaM, and the His6-tagged variants of human Bcl-2(1–211) lacking the membrane anchor were expressed and purified as reported previously (2, 9, 16).
The His-tagged Bcl-2(1–211) variants were mutated by overlay extension PCR (23), expressed in Escherichia coli BL21 by adding isopropyl 1-thio-β-d-galactopyranoside at 30 °C to a final concentration of 1 mm, and purified with a nickel-nitrilotriacetic acid affinity column (Qiagen, Hilden, Germany). The cell lysate was solubilized in 70 mm Tris buffer containing 0.5 m NaCl and 20 mm imidazole (pH 8.7). After loading the lysate on the column, it was washed using 70 mm Tris buffer containing 0.5 m NaCl and 100 mm imidazole (pH 8.7). The imidazole was eluted with 50 mm EDTA in 50 mm HEPES buffer containing 1 m NaCl (pH 8.2), followed by elution with 120 mm EDTA in 50 mm HEPES containing 1 m NaCl (pH 8.2).
Peptide Library Assay
Two peptide-coupled cellulose membranes representing a peptide library array of overlapping decamers that span the entire Bcl-2 sequence with a one-residue shift were synthesized as described previously (24). Each membrane was incubated overnight at 4 °C with 50 ml of buffer (15 mm MES, 150 mm NaCl, 6.3 mm KCl, 5% (w/v) saccharose, and 0.05% Tween), 0.030 μm human FKBP38(1–336), 0.072 μm CaM, and 1.8 mm CaCl2 (1 m stock solution). In addition, 0.018 μm Bcl-2(1–211) was added to the second membrane for incubation. Both membranes were analyzed by Western blotting using anti-FKBP38 antibody.
For detailed analysis, intensities of the chemiluminescence signals were determined for individual spots using NIH ImageJ software. The intensities of spot signals in the presence of Bcl-2 were normalized using maximal signal intensity and background signal and subsequently subtracted from the corresponding normalized spot signals in the absence of Bcl-2. Analysis of the means ± S.D. was performed with SigmaPlot 12.
NMR Experiments
All NMR data were collected at 10 °C using a DRX500 spectrometer (500.13-MHz proton resonance frequency, 5-mm TXI probe with gradient capability) or an AVANCE 800 spectrometer (800.23-MHz proton resonance frequency, 5-mm TCI cryoprobe) (both from Bruker, Rheinstetten, Germany). Two-dimensional 1H/15N heteronuclear single-quantum correlation (HSQC) spectra were collected with the carrier placed on the water resonance, which was suppressed by a WATERGATE sequence. The NMR spectra were processed on a Silicon Graphics O2 workstation using the XWINNMR 3.5 software package with the DRX500 spectrometer or TopSpin 2.1 with the AVANCE 800 spectrometer. A 90° phase-shifted squared sine-bell function was used for apodization in all dimensions. After Fourier transform, a polynomial base-line correction was applied to the processed spectra in the directly detected 1H dimension. All chemical shifts were referenced to external 2,2-dimethyl-2-silapentane-5-sulfonate to ensure consistency among the spectra (25). Peak picking of backbone amide resonances was performed with the program FELIX 2000 (Accelrys Inc., San Diego, CA). Chemical shift differences in the amide proton (Δδ1HN) and nitrogen (Δδ15N) resonances of the free and complexed protein forms were combined for each residue using the expression ((Δδ1HN)2 + (Δδ15N/6.5)2)1/2 (26).
All experiments involving the Bcl-2(119–131) peptides listed in Table 1 were performed in 10 mm HEPES buffer (pH 7.5). For the titration experiments, the Bcl-2(119–131) peptides, FKBP38(35–153) (both non-labeled and 15N-labeled), and CaM were all dialyzed as highly concentrated stock solutions in the same buffer solution. Furthermore, CaCl2 was also solubilized in this buffer to prepare a Ca2+ stock solution, and, to avoid false-positive effects due to Ca2+ contamination, all protein samples had been pretreated with EDTA. Unless noted otherwise, all data points were derived from 1H/15N HSQC spectra obtained with the DRX500 spectrometer.
TABLE 1.
Sequences and notations of Bcl-2(119–131) peptides
| Peptide | Notation | Sequence |
|---|---|---|
| Non-labeled | ||
| Bcl-2(119–131) | LHLTPFTARGRFA-NH2 | |
| Bcl-2(119–131) R127Q | LHLTPFTAQGRFA-NH2 | |
| Bcl-2(119–131) R129Q | LHLTPFTARGQFA-NH2 | |
| Bcl-2(119–131) R127Q/R129Q | LHLTPFTAQGQFA-NH2 | |
| 15N-Labeleda | ||
| Bcl-2(119–131) | Npep1 | LHLTPFTARGRFA-NH2 |
| Bcl-2(119–131) (Arg-127/Arg-129) | Npep2 | LHLTPFTARGRFA-NH2 |
| Bcl-2(119–131) (Arg-127) | NpepR127 | LHLTPFTARGRFA-NH2 |
| Bcl-2(119–131) (Arg-129) | NpepR129 | LHLTPFTARGRFA-NH2 |
a 15N-Labeled residues are indicated in boldface.
The resonance assignment of non-labeled Bcl-2(119–131) was achieved using total correlation spectroscopy, rotating-frame Overhauser enhancement spectroscopy, and 1H/13C HSQC experiments performed with a 3 mm peptide sample. For the partly 15N-labeled peptides, 1H/15N HSQC spectra were additionally recorded. The complete resonance assignment is listed in supplemental Table S1. For secondary structure prediction of the peptide, the chemical shift index consensus of the Hα, Cα, and Cβ resonances was determined using the CSI Program of the University of Alberta (27).
The characterization of the interaction between the FKBP38 catalytic domain and the Bcl-2(119–131) peptide was started with a 0.6 mm 15N-labeled FKBP38(35–153) sample that was first titrated with Bcl-2(119–131) to a final concentration of 1.8 mm (i.e. 3-fold molar excess), next with apo-CaM to a final concentration of 1.2 mm (i.e. 2-fold molar excess), and finally with CaCl2 to a Ca2+ concentration of 4.6 mm (i.e. nearly 8-fold molar excess) to generate mostly Ca2+-saturated CaM without leaving any excess Ca2+ to interact with the charge-sensitive β5-α1 loop. In further CSP experiments with 15N-labeled FKBP38(35–153), a 0.4 mm sample was titrated with the non-labeled peptide variants Bcl-2(119–131) R127Q, Bcl-2(119–131) R129Q, and Bcl-2(119–131) R127Q/R129Q to a final concentration of 1.2 mm (i.e. 3-fold molar excess) each.
Furthermore, several Bcl-2(119–131) peptides enriched with 15N isotope at different positions (see Table 1) were analyzed with respect to their interactions with FKBP38(35–153) and FKBP38(35–153) D92N/D94N. Samples of 300 μm Npep1 or Npep2 with a 3-fold molar excess of non-labeled FKBP38(35–153) were measured and compared with the respective non-complexed peptide.
Additional experiments with the Bcl-2(119–131) peptides that included 15N-labeled arginines (i.e. Npep2, NpepR127, and NpepR129) were performed using the AVANCE 800 spectrometer with higher peptide (700 μm) and FKBP38(35–153) (2.1 mm) concentrations to assure detection of the arginine side chain amide signals.
For the study of the interaction between Bcl-2(1–211) and the catalytic domain of FKBP38, a different buffer system (50 mm HEPES, 100 mm NaCl, and 1 mm EDTA at pH 8.2) had to be used because of the stability and solubility requirements of Bcl-2 and to avoid false-positive results due to divalent cations. Because of the extremely low solubility of Bcl-2(1–211) (∼10 μm), the CSP experiments were performed at a 1:5 ratio between ligand and 15N-labeled FKBP38(35–153). The same experiment was repeated with the Bcl-2 variants Bcl-2(1–211) R127Q, Bcl-2(1–211) R129Q, and Bcl-2(1–211) R127Q/R129Q. For CSP experiments with CaM, a 2-fold molar excess of lyophilized apo-CaM was added to a sample of 50 μm 15N-labeled FKBP38(35–153) and 10 μm Bcl-2(1–211) in 50 mm HEPES buffer containing 100 mm NaCl (pH 8.2) prior to dialysis. To generate holo-CaM, the sample was titrated with a CaCl2 stock solution. All experiments involving Bcl-2(1–211) were measured using the AVANCE 800 spectrometer.
Docking Calculations
The three-dimensional model of the FKBP38(35–153)·Bcl-2 complex was obtained from docking calculations using the HADDOCK program (28). To drive the docking process, ambiguous interaction restraints (AIRs) were derived from the experimental NMR data. Based on CSP effects in combination with the results from the protein mutations, the residues mediating the electrostatic intermolecular interaction between FKBP38(35–153) and Bcl-2(1–211) were selected as “active AIR residues”, i.e. Asp-92 and Asp-94 in FKBP38 as well as Arg-129 in Bcl-2. A surface accessibility cutoff value of 25% was generally applied. In addition, surface-accessible residues located next to the active AIR residues were selected as “passive AIR residues”, i.e. Thr-89, Gly-91, and Cys-93 in FKBP38 as well as Gln-118, His-120, Thr-125, Gly-128, and Thr-132 in Bcl-2. Upper distance limit restraints were introduced between the residues responsible for the electrostatic interaction. The distance restraints from Arg-129 Cζ (Bcl-2) to Asp-92 Cγ and Asp-94 Cγ (both FKBP38) were set to 4.5 ± 0.2 and 7.0 ± 0.2 Å, respectively, according to a previously determined cation-binding model of FKBP38(35–153) (3).
Surface accessibility calculations with the program NACCESS (29) and subsequent docking calculations were carried out using the atomic coordinates of FKBP38(35–153) (Protein Data Bank code 3EY6; x-ray structure) and Bcl-2(3–207) (code 1GJH; minimized average NMR structure). For the calculation of the complex, 400 conformers were initially generated using the rigid-body docking protocol of HADDOCK. Next, the 20 lowest energy structures were submitted to semiflexible simulated annealing in torsion angle space, with the side chains of residues that define the protein/protein interface kept flexible. The resulting top 20 structures were subsequently refined in an 8-Å shell of explicit TIP3P water molecules (30).
RESULTS AND DISCUSSION
Bcl-2 Peptide Library Assay Reveals Specific FKBP38-binding Epitope
The interplay between Bcl-2 and its regulator FKBP38 has been shown in a number of experimental settings (3, 8, 9, 13, 17). Inside the cell, this interaction is apparently dependent on prior formation of the FKBP38·CaM·Ca2+ complex (9, 13). In this study, we identified the contact regions between the FKBP38 catalytic domain (FKBP38(35–153)) and the cytosolic segment of Bcl-2 (Bcl-2(1–211)).
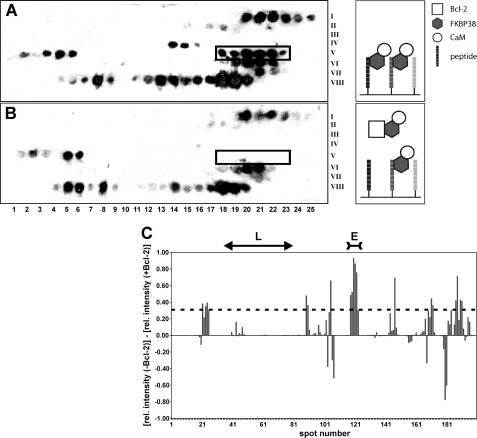
In a competitive membrane assay (Fig. 1), decapeptides derived from the Bcl-2 sequence were immobilized on two cellulose membranes. Each membrane was incubated with a mixture of FKBP38 and holo-CaM. The second membrane was incubated additionally with soluble Bcl-2(1–211) as FKBP38 competitor. Both membranes were analyzed by immunoblotting against FKBP38. Thereby, zones of consecutive peptide spots in the first but not the second membrane indicated specific FKBP38 binding to the membrane-bound Bcl-2 peptides, which was subsequently reduced due to competition when Bcl-2(1–211) was added to the solution.
FIGURE 1.
A and B, Western blot analysis of Bcl-2 peptide library assays incubated with FKBP38 and holo-CaM in the absence and presence of Bcl-2(1–211), respectively. The membrane is partitioned into a matrix of eight rows (Roman numerals on the right), each consisting of 25 spots (Arabic numbers at the bottom). Every spot features a different decapeptide derived from the Bcl-2 sequence with a one-residue shift to the adjacent peptide spot. The spot number thus represents the Bcl-2 residue number of the first amino acid in each decapeptide. Black spots in A reveal a binding interaction of the respective decapeptide with FKBP38. Weaker spot intensities occur in B when Bcl-2 competed with the immobilized decapeptide for FKBP38. The spot range 118–123 (boxed) shows several contiguous spots with very pronounced intensity differences between A and B. The corresponding Bcl-2 segment emerged in subsequent tests as the only specific FKBP38-binding epitope. C, intensity bar graph showing the differences in the relative (rel.) chemiluminescence intensity between corresponding spots on the membrane without Bcl-2 (i.e. A) and on the membrane with Bcl-2 added (i.e. B). The effects observed in the spot range defining the flexible loop (L) region of Bcl-2 are negligible. The spot range defining the primary FKBP38-binding epitope (E) is also indicated at the top. The dashed line represents the mean + 1 S.D.
Comparison of the resulting spot intensities in the absence and presence of Bcl-2 revealed one particular series of contiguous spots with pronounced intensity differences within the spot range 118–123, i.e. a region of the Bcl-2 sequence that comprises residues 118–132. In addition, minor binding interactions could be envisioned also in the spot ranges 21–25, 89–91, 169–172, and 187–191, even though none of these ranges featured more than two consecutive spots with above-average intensity changes (Fig. 1C), as would be expected for a potential binding epitope wherein several successive residues define the interaction site. Nevertheless, the corresponding peptides were all tested in a streptavidin matrix assay for FKBP38 binding and competition with Bcl-2(1–211). Although all peptides showed interaction with FKBP38, in analogy to the peptide array, Bcl-2 competed for FKBP38 binding only with the peptide that corresponded to spots 118–123 (supplemental Fig. S1).
Interestingly, the long flexible Bcl-2 loop, which had been reported earlier to be predominantly responsible for the FKBP38 interaction (17), showed no notable effects in the membrane assay (Fig. 1). Furthermore, the peptide Bcl-2(51–80), representing the core region of this supposedly non-structured loop, exhibited no FKBP38 binding in additional binding assays (supplemental Figs. S2 and S3).
Hence, only the peptide Bcl-2(119–131), derived from spots 118–123, showed specific binding to FKBP38 (supplemental Figs. S2 and S3), with a Kd value in the lower micromolar range as determined by isothermal titration calorimetry (supplemental Fig. S4). Moreover, this interaction between Bcl-2(119–131) and FKBP38 was hindered by the presence of FK506 (supplemental Fig. S3), which indicates an involvement of the putative FKBP38 active site in the binding of the identified epitope, corroborating prior results on the interaction between FKBP38 and Bcl-2 (1, 9). We therefore selected the Bcl-2 segment Leu-119–Ala-131 as the primary FKBP38-binding epitope to be employed in further investigation of the FKBP38/Bcl-2 interaction.
FKBP38(35–153) Interacts with Bcl-2(119–131) Peptide in Electrostatic Manner
NMR-based titration experiments were performed to study the binding of the Bcl-2(119–131) peptide to the catalytic domain of FKBP38. 1H/15N HSQC spectra of 15N-labeled FKBP38(35–153) were collected before and after the addition of Bcl-2(119–131) (supplemental Fig. S5). Furthermore, the influence of apo- and holo-CaM on the FKBP38(35–153)/Bcl-2(119–131) interaction was monitored.
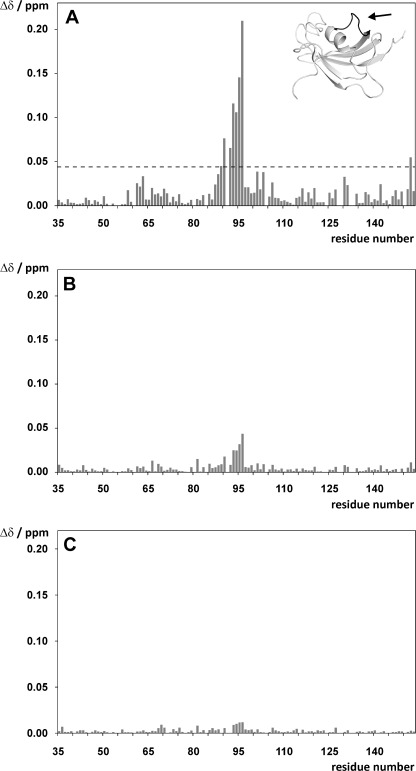
The addition of Bcl-2(119–131) to 15N-labeled FKBP38(35–153) revealed significant chemical shift changes in the Leu-90–Ile-96 segment (Fig. 2A), which corresponds to the charge-sensitive β5-α1 loop of FKBP38 (3). This loop has been shown to interact with positively charged moieties such as salt cations or guanidinium groups from arginine side chains via the aspartates Asp-92 and Asp-94 (3), thus suggesting a comparable electrostatic interaction with Bcl-2(119–131). Hence, one or both arginines in the Bcl-2(119–131) peptide (i.e. Arg-127 and/or Arg-129) may act as counterpart(s) for the negatively charged aspartates Asp-92 and Asp-94 in the β5-α1 loop of FKBP38.
FIGURE 2.
A and B, overview of the chemical shift changes (Δδ) observed in 15N-labeled FKBP38(35–153) upon addition of a 3-fold molar excess of Bcl-2(119–131) (A), followed by the addition of a 2-fold molar excess of apo-CaM (B). C, CSP changes (Δδ) observed in 15N-labeled FKBP38(35–153) upon addition of a 3-fold molar excess of Bcl-2(119–131) R127Q/R129Q. The arrow in A indicates the location of the charge-sensitive β5-α1 loop (black ribbon) in the catalytic domain of FKBP38. The dashed line in A represents the mean CSP value + 1 S.D.
The CSP effects caused by the Bcl-2(119–131) peptide in the β5-α1 loop region of FKBP38 decreased significantly when apo-CaM was added (Fig. 2B) but were essentially restored upon subsequent addition of Ca2+ to generate holo-CaM. These results suggest a reduction of the FKBP38(35–153)/Bcl-2(119–131) interaction in the presence of apo-CaM, whose negatively charged Ca2+-binding sites (31) may compete for the positive charges in Bcl-2(119–131) under these experimental conditions, which apparently is not the case in the presence of fully Ca2+-saturated holo-CaM.
To further explore the electrostatic interaction between FKBP38(35–153) and Bcl-2(119–131), analogous peptides lacking one or both of the positively charged arginine side chains were synthesized, and their CSP effects in 15N-labeled FKBP38(35–153) were determined (supplemental Fig. S6). The data revealed that binding of the peptide Bcl-2(119–131) R127Q/R129Q to the catalytic domain of FKBP38 was abolished by the two arginine-to-glutamine substitutions (Fig. 2C). Minor residual shifts in the β5-α1 loop region may be the result of hydrophobic contact(s), which are described below. However, this experiment identified the electrostatic attraction as the main driving force in the intermolecular interaction between Bcl-2(119–131) and FKBP38(35–153).
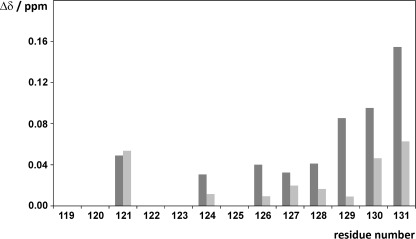
To analyze the effects of the FKBP38 catalytic domain on the Bcl-2 epitope, the reverse experiment was performed with two different partially 15N-labeled Bcl-2(119–131) peptides, Npep1 and Npep2 (Table 1). Upon addition of non-labeled FKBP38(35–153), the 15N-labeled residues of both Npep1 and Npep2 all showed significant chemical shift changes (supplemental Fig. S7), suggesting that the entire peptide sequence may be affected by the catalytic domain of FKBP38 (Fig. 3). The largest chemical shift changes occurred in the C-terminal half of the peptide, where the two arginines Arg-127 and Arg-129 are located. An additional indicator for the involvement of both arginines in the formation of the protein·peptide complex is the fact that the NϵH side chain signals of Arg-127 and Arg-129 were observable in the 1H/15N HSQC spectra only in the presence of FKBP38(35–153) (supplemental Fig. S8), implying slowed solvent exchange of the Nϵ protons due to contact with the FKBP38 catalytic domain. Moreover, CSP experiments of 15N-labeled FKBP38(35–153) with the peptide variants Bcl-2(119–131) R127Q and Bcl-2(119–131) R129Q, which both affect the residues in the β5-α1 loop (supplemental Fig. S9), demonstrated that either of these positively charged Bcl-2(119–131) side chains can participate in the interaction with FKBP38(35–153).
FIGURE 3.
Combined chemical shift changes (Δδ) observed in Bcl-2(119–131) peptides Npep1 and Npep2 upon addition of 3-fold molar excess of FKBP38(35–153) (dark gray bars) and FKBP38(35–153) D92N/D94N (light gray bars).
In contrast, upon addition of the FKBP38(35–153) D92N/D94N variant, which renders the charge-sensitive β5-α1 loop insensitive to cationic moieties (3), a reduction of the CSP effects in Npep1 and Npep2 was observed (Fig. 3), corroborating that aspartates Asp-92 and Asp-94 play a key role in this intermolecular interaction. Interestingly, however, most peptide residues still showed residual CSP effects, and Leu-121 even displayed the same shift as with wild-type FKBP38(35–153), suggesting a possible hydrophobic component to this intermolecular interaction.
According to the Bcl-2 structure (32), the positively charged guanidinium groups of Arg-127 and Arg-129, which are both integrated in α-helix 4, are ∼15 Å apart. Therefore, as long as the helical structure of α-helix 4 persists, it seems highly unlikely that both arginine side chains act simultaneously as counterparts to aspartates Asp-92 and Asp-94 in the β5-α1 loop of FKBP38. In case of the Bcl-2(119–131) peptide, the fact that both arginines reveal contacts to the charge-sensitive β5-α1 loop may thus be an artifact that is due to a lack of an organized structure in the peptide, as indicated also by the chemical shift index consensus of the Hα, Cα, and Cβ resonances (data not shown). Hence, the extensive data obtained from the above-described experiments with the Bcl-2(119–131) peptides could not unambiguously resolve which arginine residue is mainly responsible for the electrostatic protein/protein interaction between FKBP38 and Bcl-2.
Bcl-2(1–211) Binds to β5-α1 Loop of FKBP38 Catalytic Domain
At this point, we prepared Bcl-2(1–211), despite its rather low solubility even when the membrane anchor is removed (33), to determine whether the above-described interaction between the Bcl-2(119–131) segment and the charge-sensitive β5-α1 loop of FKBP38 occurs also in the Bcl-2 protein. Eventually, experimental conditions were identified to reach a maximal protein concentration of ∼10 μm with this Bcl-2 construct. This allowed the performance of NMR measurements at ultrahigh magnetic field using a cryoprobe for optimal sensitivity but applying merely substoichiometric ligand concentrations.
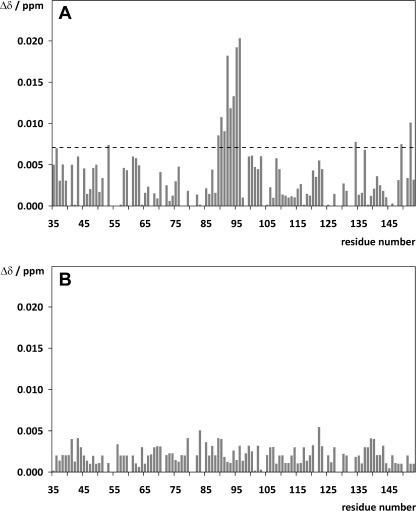
Analogous to the experiments with the Bcl-2(119–131) peptides, the interaction between 15N-labeled FKBP38(35–153) and Bcl-2(1–211) was analyzed based on CSP effects (supplemental Fig. S10) in both the absence and presence of CaM. Despite the low ligand concentration (FKBP38(35–153)/Bcl-2(1–211) ratio of 5:1), small effects were observed again in the β5-α1 loop of the FKBP38 domain (Fig. 4A), corroborating the electrostatic character of the FKBP38(35–153)/Bcl-2(1–211) interaction. However, the addition of either apo-CaM or holo-CaM revealed no influence on this interaction (data not shown). To elucidate which arginine residue in the Bcl-2 sequence is most crucial for the electrostatic protein/protein interaction, the Bcl-2 variants Bcl-2(1–211) R127Q, Bcl-2(1–211) R129Q, and Bcl-2(1–211) R127Q/R129Q were generated for further CSP experiments with 15N-labeled FKBP38(35–153). Analogous to the experiment with the peptide Bcl-2(119–131) R127Q/R129Q, the Bcl-2(1–211) double mutant also caused no significant CSP effects in FKBP38(35–153) (Fig. 4B), thus validating the hypothesis that either Arg-127 or Arg-129 in the Bcl-2 segment Leu-119–Ala-131 is responsible for the interaction with the FKBP38 catalytic domain. Remarkably, despite the large number of 16 arginine side chains that are highly solvent-accessible on the Bcl-2 surface (supplemental Fig. S11), the experiments with the Bcl-2(1–211) R127Q/R129Q variant clearly showed that only the Bcl-2 epitope identified in the peptide library-based assay is responsible for this protein/protein interaction.
FIGURE 4.
Chemical shift changes (Δδ) observed in 15N-labeled FKBP38(35–153) upon addition of 0.2 m ratio of Bcl-2(1–211) (A) and Bcl-2(1–211) R127Q/R129Q (B). Mainly the β5-α1 loop region of FKBP38 (i.e. Thr-89–Ile-96) was affected by the addition of Bcl-2(1–211), whereas the addition of Bcl-2(1–211) R127Q/R129Q caused no significant effects. The dashed line in A represents the mean CSP value + 1 S.D.
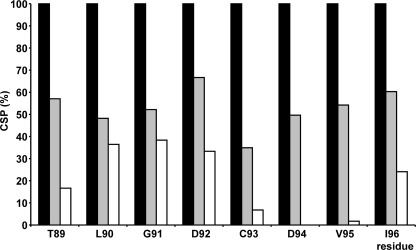
The single mutants of Bcl-2(1–211), with at least one of the two arginine residues intact, caused chemical shift changes in the β5-α1 loop of FKBP38(35–153) that were smaller than in case of the wild-type protein but larger than in case of the Bcl-2(1–211) R127Q/R129Q double mutant (Fig. 5). Compared with the double mutant, which precludes an electrostatic interaction with the β5-α1 loop but may feature hydrophobic interactions with FKBP38(35–153), the CSP effects were generally larger in Bcl-2(1–211) R127Q (with Arg-129 intact) than in Bcl-2(1–211) R129Q (with Arg-127 intact), suggesting that Arg-129 of Bcl-2 is the primary counterpart for the electrostatic interaction with the catalytic domain of FKBP38. However, Arg-127 nevertheless appears to play an additional role in the binding of FKBP38 because (i) the Bcl-2(1–211) R127Q variant caused a decrease in the CSP effects compared to the double mutant and (ii) the Bcl-2(1–211) R129Q variant showed no complete elimination of the CSP effects. Arg-127 may, for example, participate in the recognition process between the binding partners or affect other intermolecular interactions at the protein/protein interface.
FIGURE 5.
Overview of CSP effects observed in β5-α1 loop region (i.e. Thr-89–Ile-96) relative to Bcl-2(1–211) R127Q/R129Q double mutant upon addition of 0.2 m ratio of wild-type Bcl-2(1–211) (black bars), Bcl-2(1–211) R127Q (gray bars), or Bcl-2(1–211) R129Q (white bars). The CSP effects were normalized to the respective shifts observed in the presence of wild-type Bcl-2.
Model of FKBP38(35–153)·Bcl-2 Complex
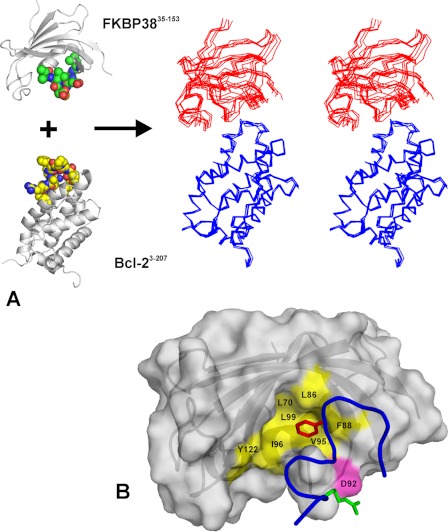
Based on the CSP data, docking calculations were performed to obtain a structural model of the FKBP38(35–153)·Bcl-2 complex. Distance restraints defining the electrostatic interaction between the β5-α1 loop of FKBP38(35–153) and Arg-129 at the surface of Bcl-2 were employed as additional structural input. Based on structural convergence (Table 2), a cluster of five conformers out of the top seven calculated structures was chosen as representative for the FKBP38(35–153)·Bcl-2 complex (Fig. 6A). Average backbone root mean square deviations of 2.70 ± 0.81 and 3.94 ± 1.15 Å were obtained when the Cα traces of Bcl-2 or FKBP38 were superposed, respectively. The values for the intermolecular energies reflect the predominantly electrostatic character of the protein/protein interaction, as the total energy of the complex consists mainly of the electrostatic energy contribution. The electrostatic character and the size of the buried surface area in the complex are typical for transient protein·protein complexes (34).
TABLE 2.
Structural statistics of the selected cluster consisting of HADDOCK conformers 2–5 and 7 (chosen as representative model of FKBP38(35–153)·Bcl-2(3–207) complex)
The root mean square deviation (r.m.s.d.) values correspond to the backbone deviations in FKBP38(35–153) when Bcl-2(3–207) is superposed (boldface) or in Bcl-2(3–207) when FKBP38(35–153) is superposed (italic). BSA, buried surface area; vdW, van der Waals; elec, electrostatic.
| Conformer | Backbone r.m.s.d. |
Intermolecular energies |
BSA | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 7 | Total | van der Waals | Electrostatic | AIRs | ||
| Å | kcal/mol | Å2 | ||||||||
| 2 | 2.73 | 2.86 | 1.74 | 2.24 | −298.2 | −31.0 | −267.1 | 0.031 | 922 | |
| 3 | 4.89 | 3.86 | 2.06 | 4.08 | −268.2 | −33.1 | −235.1 | 0.012 | 982 | |
| 4 | 3.21 | 3.74 | 3.03 | 1.66 | −237.3 | −34.1 | −203.3 | 0.040 | 894 | |
| 5 | 5.68 | 3.30 | 3.99 | 2.78 | −249.4 | −31.2 | −218.2 | 0.019 | 849 | |
| 7 | 2.62 | 5.15 | 2.13 | 4.71 | −251.5 | −29.8 | −221.7 | 0.015 | 810 | |
| Average | r.m.s.d. = 2.70 ± 0.81; r.m.s.d. = 3.94 ± 1.15 | Etotal = −260.9 ± 23.6; EvdW = −31.8 ± 1.7; Eelec = −229.1 ± 24.1; EAIRs = 0.023 ± 0.012; BSA = 897 ± 66 | ||||||||
FIGURE 6.
Structural model of FKBP38(35–153)·Bcl-2 complex. A, the spatial arrangement between the β5-α1 loop (green carbon spheres) of FKBP38 and the primary FKBP38-binding epitope (yellow carbon spheres) of Bcl-2 is shown schematically on the left. The stereo view plot on the right represents the selected cluster of five docking conformers with the Cα traces of Bcl-2 (blue lines) superposed at the bottom and the catalytic domain of FKBP38 (red lines) shown at the top. B, surface plot of FKBP38(35–153) with the segment Leu-119–Ala-131 of bound Bcl-2 shown as a blue ribbon. The hot spot of the electrostatic interaction is located between Asp-92 of FKBP38 (magenta surface) and Arg-129 of Bcl-2 (green sticks). In addition, the side chain of Bcl-2 residue Phe-124 (red sticks) extends into the hydrophobic pocket of the FKBP38 catalytic domain (yellow).
All selected conformers featured a hydrogen bond between the Arg-129 side chain of Bcl-2 and the Asp-92 side chain of FKBP38(35–153), according to the predefined distance restraints that define this hot spot of the complex. Other characteristic intermolecular interactions in the cluster occur between Gly-128 (Bcl-2) and Asp-94 (FKBP38), Thr-125 (Bcl-2) and Asp-94 (FKBP38), and the aromatic rings of Phe-124 (Bcl-2) and Phe-88 (FKBP38). In fact, the hydrophobic side chain of Phe-124 (Bcl-2) is immersed in a hydrophobic pocket of FKBP38(35–153) that consists of Leu-70, Leu-86, Phe-88, Val-95, Ile-96, Leu-99, and Tyr-122 and corresponds to the active site in the prototypic FKBP12. Aside from a possible conformational change in the β5-α1 loop upon complex formation, this structural binding arrangement could be yet another reason why the backbone amide signals of Val-95 and Ile-96 show significant CSP effects in the NMR titration experiments (Fig. 4A).
The interface of the protein·protein complex is thus characterized by a potential salt bridge between Asp-92 of FKBP38 and Arg-129 of Bcl-2 as well as by a non-polar contact of Bcl-2 residue Phe-124 with the putative active site of FKBP38 (Fig. 6B). The latter hydrophobic interaction might be too weak to produce significant CSP effects in the presence of the Bcl-2(1–211) R127Q/R129Q double mutant. However, a small CSP effect observed for the backbone amide signal of Tyr-122 (supplemental Fig. S10), whose side chain ring is also located in the putative active site, further supports such a hydrophobic binding contact.
Conclusions
Our results identify the molecular details of the interaction between Bcl-2 and the β5-α1 loop in the catalytic domain of FKBP38. The binding interface consists of a salt bridge formed between Asp-92 of FKBP38 and Arg-129 of Bcl-2 as well as additional hydrophobic contacts at the putative active site of FKBP38. The predominant electrostatic interaction involves two negative charges in the β5-α1 loop, which are unusual among the human FKBPs and mediate also the binding of divalent salt cations such as Ca2+ (3).
Remarkably, the experiment with the Bcl-2(1–211) R127Q/R129Q double mutant demonstrated that of 16 solvent-accessible arginine side chains in Bcl-2, only the identified epitope featuring Arg-127 and Arg-129 mediates the interaction with Asp-92 and Asp-94 in the β5-α1 loop of the FKBP38 catalytic domain. This specificity of interaction between the catalytic domain of FKBP38 and the identified epitope of Bcl-2 is apparently defined by additional hydrophobic contacts between nearby Bcl-2 residues and the putative active site of FKBP38.
A second interaction interface between the two protein domains can be excluded based on the globular shape of both molecules and the lack of additional CSP effects in the catalytic domain of FKBP38. Previous studies have reported, however, that Bcl-2 is stabilized by interactions with both the catalytic domain and the TPR motif of FKBP38 (17, 19). Moreover, the long flexible Bcl-2 loop, spanning 56 residues between the BH4 and BH3 domains from Asp-35 to Pro-90 (32), has been implicated to protect Bcl-2 from degradation by caspase-3. A close look at the Bcl-2 structure (supplemental Fig. S11) reveals that the here presented interaction site at Arg-129 and the long flexible loop are located at opposite ends of the Bcl-2 molecule, i.e. >25 Å apart. Hence, if this supposedly non-structured Bcl-2 loop should in fact interact with the TPR motif of FKBP38, thereby forming a “bidentate” protein·protein complex, this latter contact site may be extremely difficult to characterize on a structural level due to the very low solubility of Bcl-2 constructs featuring this flexible loop.
Electrostatic binding interactions, as observed here between Bcl-2 and the FKBP38 catalytic domain, are typical for transient complexes, which are often involved in signal transduction processes. Bcl-2 is a central regulator of mitochondrial apoptosis signaling, and the interaction of Bcl-2 with FKBP38 inhibits the prosurvival activity of Bcl-2 (9). Detailed structural insight into the binding process between both complex partners provides the molecular basis of FKBP38-mediated Bcl-2 regulation in human cells. The results presented here show that the formation of the FKBP38·Bcl-2 complex could be affected by the presence of other electrostatic factors such as Ca2+ and thus provide a rationale as to why high Ca2+ concentrations reduce the regulatory interaction between FKBP38 and Bcl-2 despite the requirement of CaM/Ca2+ in cells (3, 13).
Supplementary Material
Acknowledgments
We thank Monika Seidel for excellent technical assistance and Prof. Jochen Balbach (University of Halle, Halle, Germany) for providing access to the 800-MHz NMR spectrometer.

This article contains supplemental Figs. S1–S11 and Table S1.
- PPIase
- peptidylprolyl cis/trans-isomerase
- FKBP
- FK506-binding protein
- TPR
- tetratricopeptide repeat
- CaM
- calmodulin
- BH
- Bcl-2 homology
- CSP
- chemical shift perturbation
- HSQC
- heteronuclear single-quantum correlation
- AIR
- ambiguous interaction restraint.
REFERENCES
- 1. Edlich F., Lücke C. (2011) From cell death to viral replication: the diverse functions of the membrane-associated FKBP38. Curr. Opin. Pharmacol. 11, 348–353 [DOI] [PubMed] [Google Scholar]
- 2. Maestre-Martínez M., Edlich F., Jarczowski F., Weiwad M., Fischer G., Lücke C. (2006) Solution structure of the FK506-binding domain of human FKBP38. J. Biomol. NMR 34, 197–202 [DOI] [PubMed] [Google Scholar]
- 3. Maestre-Martínez M., Haupt K., Edlich F., Neumann P., Parthier C., Stubbs M. T., Fischer G., Lücke C. (2011) A charge-sensitive loop in the FKBP38 catalytic domain modulates Bcl-2 binding. J. Mol. Recognit. 24, 23–34 [DOI] [PubMed] [Google Scholar]
- 4. Lam E., Martin M., Wiederrecht G. (1995) Isolation of a cDNA encoding a novel human FK506-binding protein homolog containing leucine zipper and tetratricopeptide repeat motifs. Gene 160, 297–302 [DOI] [PubMed] [Google Scholar]
- 5. Rosner M., Hofer K., Kubista M., Hengstschläger M. (2003) Cell size regulation by the human TSC tumor suppressor proteins depends on PI3K and FKBP38. Oncogene 22, 4786–4798 [DOI] [PubMed] [Google Scholar]
- 6. Bulgakov O. V., Eggenschwiler J. T., Hong D. H., Anderson K. V., Li T. (2004) FKBP8 is a negative regulator of mouse sonic hedgehog signaling in neural tissues. Development 131, 2149–2159 [DOI] [PubMed] [Google Scholar]
- 7. Cho A., Ko H. W., Eggenschwiler J. T. (2008) FKBP8 cell-autonomously controls neural tube patterning through a Gli2- and Kif3a-dependent mechanism. Dev. Biol. 321, 27–39 [DOI] [PubMed] [Google Scholar]
- 8. Shirane M., Nakayama K. I. (2003) Inherent calcineurin inhibitor FKBP38 targets Bcl-2 to mitochondria and inhibits apoptosis. Nat. Cell Biol. 5, 28–37 [DOI] [PubMed] [Google Scholar]
- 9. Edlich F., Weiwad M., Erdmann F., Fanghänel J., Jarczowski F., Rahfeld J. U., Fischer G. (2005) Bcl-2 regulator FKBP38 is activated by Ca2+/calmodulin. EMBO J. 24, 2688–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Okamoto T., Nishimura Y., Ichimura T., Suzuki K., Miyamura T., Suzuki T., Moriishi K., Matsuura Y. (2006) Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 25, 5015–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barth S., Nesper J., Hasgall P. A., Wirthner R., Nytko K. J., Edlich F., Katschinski D. M., Stiehl D. P., Wenger R. H., Camenisch G. (2007) The peptidyl prolyl cis/trans isomerase FKBP38 determines hypoxia-inducible transcription factor prolyl-4-hydroxylase PHD2 protein stability. Mol. Cell. Biol. 27, 3758–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barth S., Edlich F., Berchner-Pfannschmidt U., Gneuss S., Jahreis G., Hasgall P. A., Fandrey J., Wenger R. H., Camenisch G. (2009) Hypoxia-inducible factor prolyl-4-hydroxylase PHD2 protein abundance depends on integral membrane anchoring of FKBP38. J. Biol. Chem. 284, 23046–23058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Edlich F., Maestre-Martínez M., Jarczowski F., Weiwad M., Moutty M. C., Malešević M., Jahreis G., Fischer G., Lücke C. (2007) A novel calmodulin-Ca2+ target recognition activates the Bcl-2 regulator FKBP38. J. Biol. Chem. 282, 36496–36504 [DOI] [PubMed] [Google Scholar]
- 14. Bai X., Ma D., Liu A., Shen X., Wang Q. J., Liu Y., Jiang Y. (2007) Rheb activates mTOR by antagonizing its endogenous inhibitor, FKBP38. Science 318, 977–980 [DOI] [PubMed] [Google Scholar]
- 15. Ma D., Bai X., Guo S., Jiang Y. (2008) The switch I region of Rheb is critical for its interaction with FKBP38. J. Biol. Chem. 283, 25963–25970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maestre-Martínez M., Haupt K., Edlich F., Jahreis G., Jarczowski F., Erdmann F., Fischer G., Lücke C. (2010) New structural aspects of FKBP38 activation. Biol. Chem. 391, 1157–1167 [DOI] [PubMed] [Google Scholar]
- 17. Kang C. B., Tai J., Chia J., Yoon H. S. (2005) The flexible loop of Bcl-2 is required for molecular interaction with immunosuppressant FK506-binding protein 38 (FKBP38). FEBS Lett. 579, 1469–1476 [DOI] [PubMed] [Google Scholar]
- 18. Wang R., Kovalchin J. T., Muhlenkamp P., Chandawarkar R. Y. (2006) Exogenous heat shock protein 70 binds macrophage lipid raft microdomain and stimulates phagocytosis, processing, and MHC-II presentation of antigens. Blood 107, 1636–1642 [DOI] [PubMed] [Google Scholar]
- 19. Choi B. H., Feng L., Yoon H. S. (2010) FKBP38 protects Bcl-2 from caspase-dependent degradation. J. Biol. Chem. 285, 9770–9779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Adams J. M., Cory S. (1998) The Bcl-2 protein family: arbiters of cell survival. Science 281, 1322–1326 [DOI] [PubMed] [Google Scholar]
- 21. Thompson C. B. (1995) Apoptosis in the pathogenesis and treatment of disease. Science 267, 1456–1462 [DOI] [PubMed] [Google Scholar]
- 22. Chao D. T., Korsmeyer S. J. (1998) Bcl-2 family: regulators of cell death. Annu. Rev. Immunol. 16, 395–419 [DOI] [PubMed] [Google Scholar]
- 23. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 24. Malešević M., Poehlmann A., Hernandez Alvarez B., Diessner A., Träger M., Rahfeld J. U., Jahreis G., Liebscher S., Bordusa F., Fischer G., Lücke C. (2010) The protein-free IANUS peptide array uncovers interaction sites between Escherichia coli parvulin 10 and alkyl hydroperoxide reductase. Biochemistry 49, 8626–8635 [DOI] [PubMed] [Google Scholar]
- 25. Wishart D. S., Bigam C. G., Yao J., Abildgaard F., Dyson H. J., Oldfield E., Markley J. L., Sykes B. D. (1995) 1H, 13C, and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR 6, 135–140 [DOI] [PubMed] [Google Scholar]
- 26. Mulder F. A., Schipper D., Bott R., Boelens R. (1999) Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J. Mol. Biol. 292, 111–123 [DOI] [PubMed] [Google Scholar]
- 27. Wishart D. S., Sykes B. D. (1994) The 13C chemical shift index: a simple method for the identification of protein secondary structure using 13C chemical shift data. J. Biomol. NMR 4, 171–180 [DOI] [PubMed] [Google Scholar]
- 28. Dominguez C., Boelens R., Bonvin A. M. (2003) HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 29. Hubbard S. J., Thornton J. M. (1993) NACCESS Computer Program, University College, London, United Kingdom [Google Scholar]
- 30. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 [Google Scholar]
- 31. Kuboniwa H., Tjandra N., Grzesiek S., Ren H., Klee C. B., Bax A. (1995) Solution structure of calcium-free calmodulin. Nat. Struct. Biol. 2, 768–776 [DOI] [PubMed] [Google Scholar]
- 32. Petros A. M., Medek A., Nettesheim D. G., Kim D. H., Yoon H. S., Swift K., Matayoshi E. D., Oltersdorf T., Fesik S. W. (2001) Solution structure of the anti-apoptotic protein Bcl-2. Proc. Natl. Acad. Sci. U.S.A. 98, 3012–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schendel S. L., Xie Z., Montal M. O., Matsuyama S., Montal M., Reed J. C. (1997) Channel formation by anti-apoptotic protein Bcl-2. Proc. Natl. Acad. Sci. U.S.A. 94, 5113–5118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jones S., Thornton J. M. (1996) Principles of protein-protein interactions. Proc. Natl. Acad. Sci. U.S.A. 93, 13–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.