Background: Schwanniomyces occidentalis β-fructofuranosidase synthesizes 6-kestose and 1-kestose, prebiotics that stimulate beneficial bacteria.
Results: The β-sandwich domain is involved in substrate binding, and the Gln-228/Asn-254 pair plays a crucial role in acceptor/donor substrate binding and product specificity.
Conclusion: First evidence of β-sandwich domain functionality. Variants with new specificities, synthesizing neokestose, were obtained.
Significance: Understanding the molecular mechanism that regulates product specificity is crucial for designing improved new enzymes.
Keywords: Carbohydrate-binding Protein, Carbohydrate Chemistry, Carbohydrate Processing, Glycoside Hydrolases, Kinetics, Protein Complexes, Protein Engineering, Protein Structure, X-ray Crystallography
Abstract
Schwanniomyces occidentalis β-fructofuranosidase (Ffase) is a GH32 dimeric enzyme that releases fructose from the nonreducing end of various oligosaccharides and essential storage fructans such as inulin. It also catalyzes the transfer of a fructosyl unit to an acceptor producing 6-kestose and 1-kestose, prebiotics that stimulate the growth of bacteria beneficial for human health. We report here the crystal structure of inactivated Ffase complexed with fructosylnystose and inulin, which shows the intricate net of interactions keeping the substrate tightly bound at the active site. Up to five subsites were observed, the sugar unit located at subsite +3 being recognized by interaction with the β-sandwich domain of the adjacent subunit within the dimer. This explains the high activity observed against long substrates, giving the first experimental evidence of the direct role of a GH32 β-sandwich domain in substrate binding. Crucial residues were mutated and their hydrolase/transferase (H/T) activities were fully characterized, showing the involvement of the Gln-228/Asn-254 pair in modulating the H/T ratio and the type β(2–1)/β(2–6) linkage formation. We generated Ffase mutants with new transferase activity; among them, Q228V gives almost specifically 6-kestose, whereas N254T produces a broader spectrum product including also neokestose. A model for the mechanism of the Ffase transfructosylation reaction is proposed. The results contribute to an understanding of the molecular basis regulating specificity among GH-J clan members, which represent an interesting target for rational design of enzymes, showing redesigned activities to produce tailor-made fructooligosaccharides.
Introduction
Fructans and fructooligosaccharides (FOS)5 are β-d-fructose (Fru)-based carbohydrates, often linked to a sucrose skeleton, that are synthesized by 15–20% of the flowering plants and also by a number of bacteria and fungi. These compounds are related to carbohydrate storage and the processes of drought and freezing tolerance in plants (1). In addition, FOS are considered prebiotics because they selectively stimulate the growth/activity of Lactobacilli and Bifidobacteria from the animal digestive tract (1, 2) and exert a beneficial effect on human health, thus contributing to the prevention of cardiovascular diseases, colon cancer, and osteoporosis (3). Different types of FOS series can be distinguished depending on the linkage type between the monosaccharide residues: 1F-FOS, containing β-(2→1)-linked fructose units (with an inulin-type structure, e.g. 1-kestose or nystose); 6F-FOS, containing β-(2→6)-linked fructose units (with a levan-type structure, e.g. 6-kestose); and 6G-FOS, where the β-(2→6)-link connects fructose and the glucosyl moiety of sucrose (neo-FOS, e.g. neokestose or neonystose). Although the molecules included in the three series show prebiotic effect (4, 5), β-(2→6)-linked FOS may have enhanced prebiotic properties and chemical stability as compared with 1F-FOS (4). Enzymatic synthesis of these oligosaccharides has been reported using different enzymes from microorganisms. Thus, the β-fructofuranosidases from Saccharomyces cerevisiae (6), Schwanniomyces occidentalis (7, 8), and Rhodotorula dairenensis (9) produce mainly 6-kestose, whereas the enzymes from Xanthophyllomyces dendrorhous synthesize neokestose (10, 11).
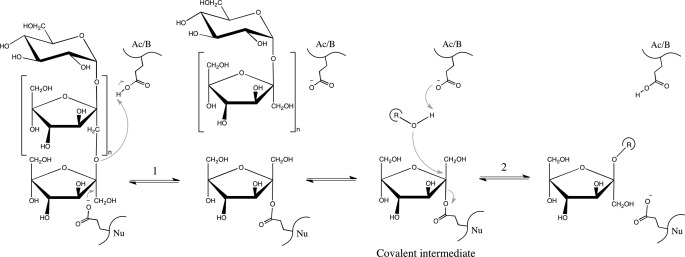
On the basis of their amino acid sequences, fructans processing enzymes are classified into the glycoside-hydrolase (GH) GH-J clan, in which two families are included: GH32 (invertases, β-fructofuranosidases, inulinases, and fructosyltransferases) and GH68 (inulosucrases and levansucrases) (12). GH-J clan enzymes share a five-blade β-propeller N-terminal domain, in which β-sheets are arranged around a central pocket that accommodates the active site (catalytic domain). Three key acidic residues located in the active site and surrounded by conserved sequences in the GH32 family, NDPNG (Asp (D) acting as nucleophile), RDP (Asp (D) acting as a stabilizer of the transient state) and EC (Glu (E) acting as the acid base catalyst), are implicated in substrate binding and hydrolysis (13). These enzymes display either hydrolysis or transfructosylation reactions (Fig. 1) depending on whether water or an oligosaccharide (the acceptor substrate) molecule accepts the fructose released by fructan hydrolysis (the donor substrate). The main differences between proteins included in GH32 and GH68 are the presence of an additional β-sandwich C-terminal domain in the former and the general transfructosylating character of the latter. The number of studies giving insight into the structural determinants involved in GH-J clan specificity has grown significantly in recent years (14–18). These studies show that the loops and turns connecting the different elements of the β-propeller domain, in the environs of the active site, seem to define the specificity unique to each enzyme. Thus, only a few substitutions in the vicinity of the residue acting as a nucleophile transform a plant cell wall invertase into a fructosyltransferase (19, 20), whereas a single substitution in a fructan 6-exohydrolase (F233D) turns it into a fructan 1-exohydrolase (21). Moreover, only three amino acids (Asn-340, Trp-343, and Ser-415) appear to be responsible for donor substrate specificity in 6-glucosyl-1-fructosyl-fructosyltransferase from Lolium perenne (22). Within this framework, the first three-dimensional structure of a GH32 fructosyltransferase (FT) in complex with nystose (a medium size substrate) has already been reported (16), and in agreement with previous works (21), it was postulated that residues in binding subsites +1 and +2 (nomenclature according to Davies et al. (23)) seem to be responsible for the specificity of the products formed in the transfructosylation reaction.
FIGURE 1.
General reaction catalyzed by GH-J clan enzymes through a double displacement mechanism that involves a covalent intermediate. The β-(2→1)-linked fructan represented (the donor substrate) would be hydrolyzed in the second step of the reaction when water is the acceptor substrate of the fructosyl (r = H). In the case of an oligosaccharide acting as acceptor, a transfructosylating product is obtained. The inulin-type scaffold is: sucrose (n = 0), 1-kestose (n = 1), nystose (n = 2), fructosylnystose (n = 3), inulin (n ≈ 30–50).
The β-fructofuranosidase (Ffase) from the yeast Sw. occidentalis hydrolyzes sucrose, 1-kestose, nystose, or inulin and efficiently produces the trisaccharides 6-kestose and 1-kestose (in the ratio of 3:1); both are prebiotic FOS (7). The Ffase three-dimensional structure has been solved as a homodimer, each modular subunit arranged like other GH32 enzymes including a N-terminal β-sandwich domain attached to the catalytic β-propeller. Interestingly, the active site cleft is shaped by both subunits within the dimer, which seem to be involved in substrate binding (15). The Asp-50 (NDPNG) and Glu-230 (EC) located at the center of the β-propeller are the catalytic residues implicated in substrate binding and hydrolysis, whereas Arg-178 and Asp-179 form the RDP motif. The structural analysis of Ffase active site, in combination with a mutational and kinetic analysis, has recently suggested the existence of a novel donor substrate binding site located in the EC motif environment (including residues such as Ser-196, Gln-176, Gln-228, Pro-232, and Asn-254) that presumably is responsible for the transferase specificity of the enzyme (8). In this article we report the crystal structure of two different Ffase inactivated mutants complexed with medium and long chain substrates, giving the first experimental evidence of the direct role of a GH32 supplementary β-sandwich domain in substrate binding. Moreover, potential oligosaccharide-binding residues were mutated and their hydrolase/transferase (H/T) activities characterized. Our results directly and unambiguously associate the specificity of the Ffase transferase activity with residues located in the environment of Glu that act as the acid/base catalyst in the GH32 catalytic mechanism.
EXPERIMENTAL PROCEDURES
Materials, Organisms, Plasmids, and Mutagenesis
Nystose (α-d-glucopyranosyl-[(1→2)-β-d-fructofuranosyl]2-(1→2)-β-d-fructofuranose), fructosylnystose (α-d-glucopyranosyl-[(1→2)-β-d-fructofuranosyl]3-(1→2)-β-d-fructofuranose), and inulin from chicory (α-d-glucopyranosyl-[(1→2)-β-d-fructofuranosyl]n-(1→2)-β-d-fructofuranose); n-36) were obtained from TCI-Europe, Megazyme, and Sigma-Aldrich, respectively. S. cerevisiae EUROSCARF Y02321 (BY4741; Mat a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YIL162w(SUC2)::kanMX4) was used as the expression host, and it was transformed by the standard lithium acetate method. Escherichia coli DH5α (λ−φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK−mK−) supE44 thi-1 gyrA relA1) was used for DNA manipulation and amplification by standard techniques.
The Ffase gene (CQ890277) from Sw. occidentalis ATCC 26077 used in this work was included in the construct Ffase-pYES and contained the only encoding CTG triplet (positions +586 to +588) mutated to TCA (L196S), which encodes serine in the standard code to obtain a heterologously expressed protein with the wild-type amino acid sequence (8). This construct was used as template to obtain all of the enzymatic mutants generated and analyzed in this work. Site-directed mutagenesis was carried out using specific primers (supplemental Table S1) and the method described previously (15). The full-length Ffase, including the substitution D50A or E230A fused to a C-terminal His6 tag, was obtained using the Ffase-pYES D50A or E230A construct as template and the oligonucleotides SwF-BamHI (5′-TAGGATCCAACATGGTACAAGTTTTAAGTGTATTAG-3′) and SwRHXbaI (5′-CCCTCTAGACTTATTTAGTTCCCTTATGATAAC-3′). Restriction sites for BamHI and XbaI (shown in bold) were included in these primers to clone the PCR products into the pYES2/CT (Invitrogen) vector, thereby generating the Ffase-pYESCT D50A and E230A constructs. DNA sequencing (SIDI, Universidad Autónoma de Madrid, Spain) was used to verify that only the desired mutations were present in all of the constructs obtained.
Protein Purification and Detection
β-Fructofuranosidase mutants from Sw. occidentalis expressed in S. cerevisiae were purified as described elsewhere (8). Basically, yeasts were grown in YPGal medium (1 liter), and the culture filtrates were concentrated and applied to a DEAE-Sephacel chromatography column and eluted using a 0–0.5 m NaCl gradient. Samples containing fructouranosidase activity eluted in 0.1 m NaCl and were analyzed by Coomassie-stained SDS-PAGE (8%). Only those containing apparently pure proteins were pooled, dialyzed, concentrated, and stored at −70 °C. Protein concentration was estimated photometrically at 280 nm (NanoDrop spectrophotometer ND-1000).
His-tagged proteins (Ffase including D50A or E230A substitution) were purified from yeast cellular fractions. Basically, around 30 g of cells (expressing each inactive variant) were lysed by the addition of YeastBuster reagent (Novagen) including the protease inhibitor mixture for fungal and yeast extracts (Sigma) as indicated by the manufacturers. The cellular debris was removed by centrifugation, and the supernatant was fitted to 40% glycerol, 0.5% Tween 20, 300 mm NaCl, and 20 mm sodium phosphate, pH 8.0. The samples were then incubated for 2 h at 4 °C with nickel-nitrilotriacetic acid-agarose (Qiagen) (1 ml of resin g−1 cells) using an orbital shaker and applied to a suitable column support as recommended by the manufacturer. No retained proteins were eluted using 8 mm imidazole. The protein of interest eluted in 50 mm sodium acetate, pH 4.5. All fractions were analyzed by Western blot as described below. Fractions containing Ffase protein were dialyzed in 20 mm HCl-Tris, pH 7.0, and concentrated by using the Microcon YM-10 (Amicon) system.
Western blot analysis was carried out using standard procedures and probed with anti-His6 antibody (Bio-Rad) as indicated by the manufacturer and/or the anti-S. cerevisiae invertase antibody, which was obtained and used as indicated previously (8).
Enzyme and Kinetic Analysis
Ffase hydrolytic activity was quantified by the dinitrosalicylic acid method adapted to a 96-well microplate as described previously (7). Enzymatic assays were performed in 100 mm sodium acetate buffer, pH 5.0, at 50 °C. One unit of activity was defined as that catalyzing the formation of 1 μmol of reducing sugar/min. For kinetic analysis, the initial velocity was measured in triplicate using 0.3–5.3 μg ml−1 of enzyme, sucrose, or nystose (2.5–100 mm) and inulin (1–10 mm). The reaction time was 20 min. Plotting and analysis of the curves were carried out using SigmaPlot software (version 7.101). Kinetic parameters were calculated by fitting the initial rate values to the Michaelis-Menten equation.
To study the effect of sucrose concentration on transfructosylation activity, the concentration of sucrose was varied between 250 and 1000 mm at a standard reaction time of 3 h and 47.5 °C. For the analysis of the selectivity of transfructosylation products (6-kestose, 1-kestose, and neokestose), 1 m sucrose and a 24-h reaction time were used. Reactions were stopped (5 min at 100 °C), and the carbohydrate concentration was determined by high-performance liquid chromatography (HPLC) as indicated previously (7).
Crystallization and Data Collection
As deglycosylation of wild-type Ffase increased the diffraction resolution limit from 2.9 to 1.9 Å (15), the recombinant proteins expressed by the Ffase-pYESCT D50A and E230A constructs were submitted to treatment with endoglycosidase H (New England Biolabs), which yielded a unique protein band below 66 kDa, as observed from the SDS-PAGE analysis performed for both mutant proteins (data not shown). The samples were concentrated, and screening of new crystallization conditions was performed with the PACT suite from Qiagen. Plate-shaped crystals appeared in solutions containing either PEG 3350 or PEG 6000 at pH 6 or 8. Further optimization was performed at room temperature by the vapor diffusion method by mixing 1 μl of protein solution with an equal volume of different reservoir solutions and using the sitting-drop procedure.
The best crystals of the E230A mutant concentrated to 10.5 mg ml−1 grew from 16% PEG 6000, 3% MPD, 0.2 m MgCl2, and 0.1 m MES, pH 6, and reached the maximum size in 10–15 days. To obtain the E230A-fructosylnystose complex, crystals were transferred to a soaking solution containing 19% PEG 6000, 3% MPD, 0.1 m MES, pH 6, and 100 mm fructosylnystose for 5 min, and subsequently to a fresh drop supplemented with 20% glycerol prior to cooling to 100 K in a nitrogen stream. A full data set was collected to 2.68 Å at the ESRF ID23-2 beamline.
Plate-like crystals of the D50A mutant concentrated to 11.2 mg ml−1 grew from 15% PEG 6000, 3% MPD, 0.2 m MgCl2, and 0.1 m HCl-Tris, pH 8. The D50A-inulin complex was obtained by soaking a crystal in 17% PEG 6000, 3% MPD, 0.1 m MES, pH 6, and 2.65% inulin for 4 h before being cryoprotected in a drop supplemented with 20% glycerol to be flash-frozen to 100 K. A full data set was collected at the ESRF ID14-1 beamline.
Analysis of the diffraction patterns was performed using MOSFLM (24) to yield integrated intensities, and data were merged with the CCP4 suite (25). A summary of data collection and data reduction statistics is shown in Table 1.
TABLE 1.
X-ray data collection and structure refinements statics
| Crystal | D50A-Inu | E230A-Fnys |
|---|---|---|
| Data collection | ||
| Unit cell dimensions (Å) | 67.1, 108.6, 96.1 (β = 91.4°) | 65.21, 92.66, 117.75 89.91, 82.96, 85.82 |
| Space group | P21 | P1 |
| Molecules in asymmetric unit | 2 | 4 |
| Solvent content (%) | 42 | 42 |
| Limiting resolution (Å) (outer shell) | 2.24 (2.36–2.24) | 2.68 (2.82–2.68) |
| Unique reflections | 65,603 | 74,146 |
| Rrima | 0.14 (0.51) | 0.22 (0.44) |
| Rpimb | 0.07 (0.24) | 0.11 (0.44) |
| Completeness (%) | 98.4 (98.2) | 96.4 (88.4) |
| Mean multiplicity | 5.2 (5.0) | 4.3 (2.0) |
| Mean I/σI | 5.3 (1.5) | 8.3 (2.9) |
| Final refinement parameters | ||
| Protein atoms (non-H) | 8,100 | 16,200 |
| Solvent molecules | 623 | 279 |
| Ligand molecules | 2 Fru6 | 4 Fructosylnystose |
| Glycosylation molecules | 8 N-acetylglucosamine | 20 N-acetylglucosamine |
| R-factorc | 0.19 | 0.23 |
| Rfree | 0.23 | 0.31 |
| r.m.s.d bonds (Å)/angles (°) | 0.012/1.8 | 0.014/1.8 |
| Averaged B-factors (Å2) | ||
| Main chain | 15.6 | 32.8 |
| Side chain | 16.5 | 33.1 |
| Ligand | 31.2 | 30.0 |
| Solvent | 23.2 | 45.6 |
| Ramachandran plot | ||
| Favored (%) | 98.8 | 96.7 |
| Outliers (%) | 1.2 | 3.3 |
| Protein Data Bank code | 3U14 | 3U75 |
a Rrim = Σ[N/(N − 1)]1/2Σ|I(h)I − 〈I(h)〉|]/ΣΣ〈I(h)〉.
b Rpim = Σ[1/(N − 1)]1/2Σ|I(h)i − 〈I(h)〉|/ΣΣ〈I(h)〉.
c R-factor = Σ(|Fobs − Fcalc|)/Σ|Fobs| (Rfree is equivalent to R-factor for a randomly selected 5% subset of reflections not used in structure refinement).
d r.m.s., root mean square.
Structure Solution and Refinement
The structure of the complexes was solved using the molecular replacement method. Initial phases were obtained with the MOLREP program (26) using the atomic coordinates of deglycosylated wild-type Ffase (PDB code: 3KF3) as the search model. A clear solution was obtained in both mutants, which was followed by crystallographic refinement using Refmac (27) in the CCP4 suite (25) with flat bulk-solvent correction, including low resolution data to 50 Å, and using maximum likelihood target features. A subset of 5% randomly selected structure-factor amplitudes was excluded from automated refinement to compute a free R-factor. Several rounds of refinement applying non-crystallographic symmetry restraints between the molecules in the asymmetric unit were alternated with rebuilding using the program O (28) after which the Fo − Fc maps showed unequivocally the positions of the substrates, well ordered at the active site. The inulin chain coordinates were retrieved from the crystal structure determined from electron diffraction data (29) and deposited in the Cambridge Structural DatabaseCSD; Refcode VIPRAN), whereas the fructosylnystose was modeled manually on the basis of the nystose coordinates retrieved from the crystal structure (30) deposited in the CSD (Refcode PEKHES). Both substrates were fitted manually into the electron density and further refined. At the last stage, water molecules and several N-acetylglucosamine were included, and further model completion was performed with the program Coot (31) combined with more rounds of positional and individual restrained B-factor refinement. This led to a final R-factor of 0.19 (Rfree = 0.23) for the D50A-inulin complex data set up to 2.24 Å resolution and 0.23 (Rfree = 0.31) for the E230A-fructosylnystose data set up to 2.68 Å. Final refinement parameters for both complexes are reported in Table 1. The stereochemistry of the model was checked by using ProCheck (32), and the figures were generated with PyMOL (33).
RESULTS
Ffase Inactive Mutants Trap the Substrates
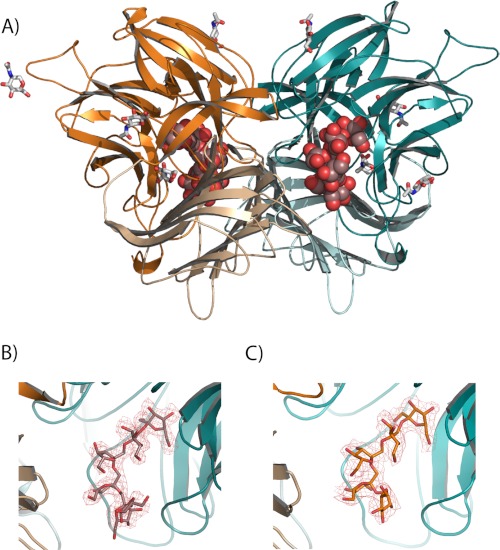
Prior to this work, the Ffase from Sw. occidentalis (also SoInv) fused to a His6 tag was actively expressed in S. cerevisiae, but initial attempts to obtain good quality crystals from this recombinant enzyme were unsuccessful (34). Therefore, the three-dimensional structure of the enzyme was solved from extracellular enzyme purified directly from Sw. occidentalis. To determine the crystal structure of the substrate complexes, the two catalytic residues, Asp-50 and Glu-230, were changed to Ala, a change that previously had been demonstrated to yield variants with undetectable activity against both short (sucrose) as well as medium and long size (nystose and inulin) substrates (15). These mutated variants of Ffase were submitted to deglycosylating digestion with Endo H, which proved to be crucial in getting good crystals. The best crystals from both mutants were used for soaking experiments, and finally the complexes D50A-inulin and E230A-fructosylnystose were solved. In accordance with the deglycosylated wild type, the recombinant Ffase D50A-inulin complex belongs to the P21 space group, with two molecules in the asymmetric unit, whereas the E230A-fructosylnystose complex loses symmetry, with the crystal showing the P1 space group and four molecules in the asymmetric unit. The structure of both complexes was solved by molecular replacement with the coordinates of native Ffase and refined to the final values given in Table 1. The initial difference maps revealed unequivocally the presence of the substrates in all of the active sites that were refined, showing apparently full occupancies. The final electron density of both substrates, together with the folding of the Ffase dimer, is shown in Fig. 2. Several N-acetylglucosamine units, also visible, were attached to Asn-72, Asn-126, Asn-219, Asn-334, and Asn-394.
FIGURE 2.
Structure of the Ffase complexes. A, overall structure of Ffase dimer is represented in ribbon diagram, with two inulin chains found in the D50A crystals represented as spheres and the glycosylation units represented as sticks. Each subunit (cyan and orange) is bimodular and consists of a catalytic β-propeller domain (dark) and a C-terminal β-sandwich domain (light). B and C, close-up view of D50A-inulin (B) and E230A-fructosylnystose (C) complex binding sites. The colored portion of the 2Fo − Fc electron density map at 2.2 Å covering the inulin (B) and 2.7 Å covering the oligosaccharide (C) is contoured at 1.0 σ level.
Overall, the crystal structure of both mutants (Fig. 2) is essentially identical, as revealed by the root-mean-square deviation of 0.321 Å between their equivalent Cα atom coordinates. They are also mostly similar with respect to the wild type, the D50A-inulin, and E230A-fructosylnystose complexes, showing root-mean-square deviation values of 0.303 and 0.279 Å, respectively, as superimposed on the same Cα atoms of the deglycosylated wild-type structure (PDB code: 3KF3). On the other hand, there were no significant alterations in the active site upon complexion, apart from a change in the conformation of the catalytic Glu-230 side chain and some minor differences in the orientation of the aromatic Trp-47, Tyr-293, Trp-310, and Trp-314 side chains, all of them located at the active site.
Ffase Dimer Induces a Fixed Conformation of the Substrate
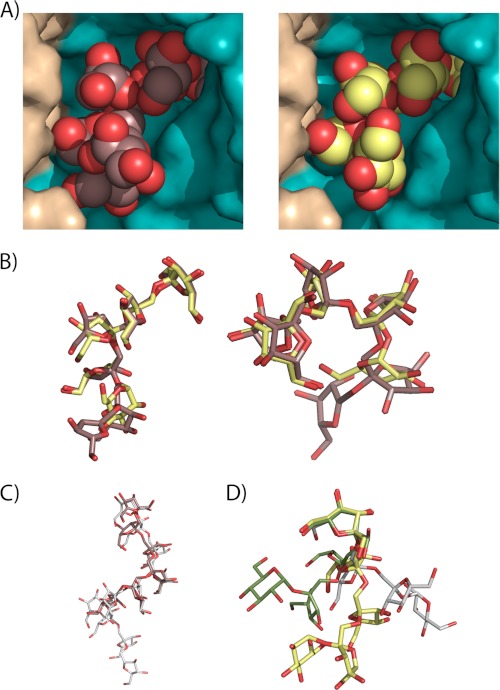
As shown previously, each active site of the Ffase dimer is made up from both subunits that contour a cavity 25 Å deep, accessible to the solvent by a slot 18 Å wide. Fig. 3A displays a close-up view of this cavity with the bound substrates observed in the complexes. We observed that the non-reducing end of the substrates was hidden deeply inside the catalytic pocket, as expected, but the remaining sugar units were also well inserted into the cavity and well adjusted to both subunits within the dimer. A subsequent fructose ring of inulin should be dangling out into the solvent with no atomic interactions to the protein and, consequently, not visible in the electron density maps. Remarkably, both oligosaccharides adopt a very similar conformation along the fructose-fructose bonds, as illustrated in Fig. 3B, the only differences being observed in the glucose moiety at the reducing end of fructosylnystose, which in any case is located in an equivalent position to the fifth fructose unit of inulin. This means that the substrates are accommodated into the Ffase cavity in a very precise conformation that is imposed by a highly specific catalytic active site, which is able to recognize oligosaccharide units distal from the scissile bond.
FIGURE 3.
The conformation of the substrates at the Ffase active site. A, close-up view of the Ffase active site showing the catalytic domain of one subunit colored in cyan and the C-terminal β-sandwich domain of the adjacent subunit in beige. The six units of inulin (left) and fructosylnystose (right) molecules found in the Ffase inactivated mutants are represented as spheres. B, inulin (brown) and fructosylnystose (lime green) moieties, in stick representation, are superimposed in two perpendicular views to highlight the fact that both substrates adopt the same conformation at the Ffase active site. C, inulin found at the D50A crystal (brown) is compared with inulin found in its free crystalline state (white). D, fructosylnystose found in the Glu-230 crystal (lime green) is superimposed on nystose found in the A. japonicus fructosyltransferase complex (green) and crystallized free nystose (white), showing that they adopt very different conformations.
On the other hand, the structure of bound inulin is very similar to the helical conformation observed in its free crystalline state (29), with only small differences observed at the last visible fructose unit (Fig. 3C). On the contrary, the conformation of fructosylnystose differs considerably from that found in crystallized free nystose (30) and nystose bound to Aspergillus japonicus FT (16) as shown in Fig. 3D. Therefore, it is apparent that the molecular surface of the dimer has evolved to accommodate the polymeric chain of inulin. The fact that dimerization of this enzyme is involved in the degradation of inulin and other long chain substrates has been proposed previously by us (15). Furthermore, and considering the Ffase catalytic efficiency on sucrose (S) and inulin (I) that gives an S/I ratio of 12, its catalytic behavior (Table 2) seems to be more typical of that of an inulinase (S/I ratio < 10) as compared with that expected from an invertase (S/I ratio > 1600) (35). Inulinases are generally monomeric enzymes with a molecular mass ranging from 50 to 70 kDa (36), although a few multimeric proteins such as that from the yeast Kluyveromyces fragilis (37) or the fungi Aspergillus fumigatus (38) have also been described. Unfortunately, no functional role for the oligomerization of these enzymes has been assigned, making the Ffase the unique enzyme described to date for which functionality and oligomerization have been correlated.
TABLE 2.
Kinetic analysis of Ffase mutants
The kcat values were calculated assuming a protein molecular mass of 180 kDa. The ± sign refers to standard errors based on curve fitting using SigmaPlot.
| Ffase mutant |
kcat |
Km |
kcat/Km |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Sucrose | Nystose | Inulin | Sucrose | Nystose | Inulin | Sucrose | Nystose | Inulin | |
| s−1 | mm | s−1mm−1 | |||||||
| WT | 105 ± 14 | 99 ± 6 | 19.9 ± 3 | 6.4 ± 0.9 | 2.1 ± 0.4 | 14.7 ± 2.4 | 16.4 | 47.1 | 1.3 |
| Q176S | 144 ± 28 | 208 ± 7 | 1220 ± 20 | 6.0 ± 0.8 | 30 ± 1.4 | 814 ± 8 | 24 | 6.9 | 1.5 |
| Q176N | 32 ± 3 | 50 ± 7 | 53.8 ± 6 | 6.7 ± 0.5 | 2.9 ± 0.2 | 102 ± 9 | 4.8 | 17.2 | 0.5 |
| Q176E | 82 ± 11 | 91 ± 2 | 18.5 ± 7 | 8.2 ± 1 | 2.5 ± 0.3 | 20.5 ± 2.7 | 10 | 36.4 | 0.9 |
| Q228V | 28 ± 5 | 1.8 ± 0.2 | 3.4 ± 0.2 | 10.7 ± 2 | 9.4 ± 0.9 | 52.5 ± 7.9 | 2.6 | 0.2 | 0.06 |
| Q228T | 29 ± 4 | 1.8 ± 1.9 | 5.0 ± 4.5 | 18.0 ± 8.4 | 11.3 ± 2 | 57.1 ± 6 | 1.6 | 0.2 | 0.087 |
| Q228N | 64 ± 11 | 30 ± 3.3 | 91 ± 8.1 | 16.1 ± 2.7 | 1.0 ± 0.2 | 23.9 ± 2.8 | 4 | 30 | 3.8 |
| Q228E | 38 ± 6 | 25 ± 3 | 409 ± 40 | 3.3 ± 0.5 | 1.2 ± 0.4 | 245 ± 32 | 11.5 | 20.8 | 1.7 |
| N254A | 80 ± 9 | 61 ± 3 | 11.3 ± 0.9 | 8.0 ± 0.9 | 9.2 ± 2 | 8.3 ± 0.8 | 10 | 6.6 | 1.3 |
| N254T | 10 ± 1 | 40 ± 8 | 2.1 ± 0.2 | 5.0 ± 5.5 | 9 ± 0.3 | 12.7 ± 1.1 | 2 | 4.4 | 0.2 |
| N254D | 31 ± 6 | 82 ± 3 | 4.4 ± 0.4 | 13.0 ± 2.4 | 4.8 ± 0.7 | 10.2 ± 1.2 | 2.4 | 17.1 | 0.4 |
| N254E | 16 ± 1 | 72 ± 2 | 1.3 ± 0.3 | 3.5 ± 0.4 | 19.5 ± 2 | 8.8 ± 1.3 | 4.6 | 3.7 | 0.1 |
Ffase Accommodates up to Five Sugar Units in Its Active Site
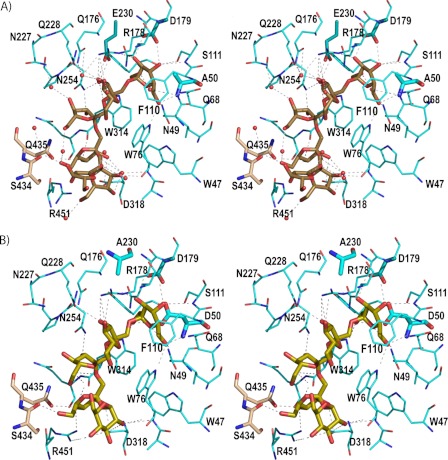
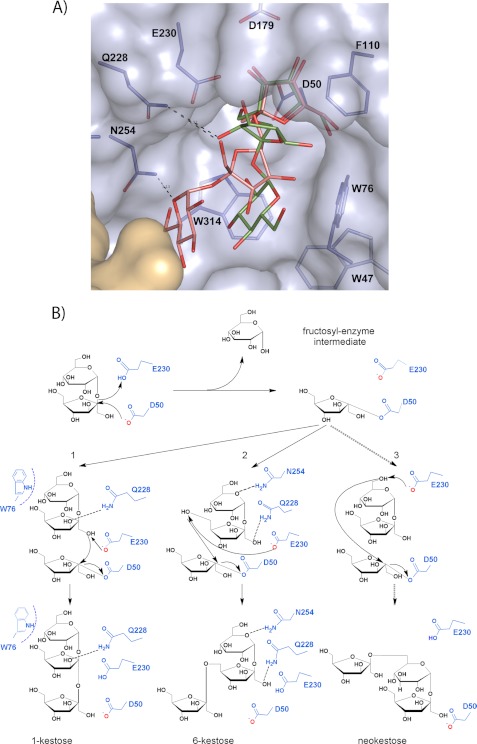
A detail of the inulin and fructosylnystose recognition mode by Ffase is displayed in Fig. 4. Inspection of the complexes at the active site reveals that the enzyme presents five binding subsites, i.e. it delineates subsites −1 to +4, following the nomenclature used for sugar-binding substrates (23) that defines cleavage as occurring between subsites −1 and +1. The subsites are made up from residues in all five blades of one subunit and also from the β-sandwich domain of the adjacent subunit. These residues establish a net of hydrogen bonding, some of them through well ordered water molecules, which keeps the substrates in a very fixed conformation. In addition, even when none of the stacking interaction typically found in protein-sugar interactions is found, several aromatic residues (Trp-47, Trp-76, Phe-110, and Trp-314) define a hydrophobic patch at one wall of the active site cavity (see Fig. 4) that directs the β-1,2 glycosidic bonds of the oligosaccharide.
FIGURE 4.
Binding mode of the substrates. A, stereo view of the D50A active site with important protein residues engaged in inulin recognition. The major interactions are indicated by a dotted line. Inulin is colored in brown and residues from the adjacent subunit B in beige. The catalytic triad is represented by thicker sticks. Water molecules are shown as red spheres. B, stereo view of the Glu-230 active site showing the fructosylnystose in lime green. Direct polar links between the substrate and the enzyme are shown as dotted lines.
The fructose unit bound at subsite −1 is tightly fixed by the hydrogen bonds of all of its hydroxyl groups to Asn-49, Gln-68, Trp-76, Arg-178, and Asp-179 side chains and to both the main and side chains of Ser-111. The only difference found between the two complexes is the fructosylnystose O1 that is hydrogen-bonded to Trp-314 and the nucleophile Asp-50 side chains, whereas in the D50A mutant a water molecule occupies a position close to the removed carboxylate and links the O1 to the Ala-50 main chain. Apart from this feature due to the removal of the Asp-50 side chain, the interaction pattern at subsite −1 is highly conserved along the GH32 complexes.
At subsite +1, the fructose O3 atom makes direct hydrogen bonds to the side chains of Glu-230, Gln-228, and Arg-178, with the last also interacting with O4. Furthermore, there are two highly ordered water molecules that link both hydroxyls to these residues, and also to other residues such as Gln-176 and Asn-254, in a net of polar interactions that keeps the fructose ring in a very fixed orientation. Also, the glycoside links by hydrogen bond to its fructose O2. In turn, the fructose unit located at subsite +2 makes hydrogen bonds at O3 to the Asn-254 side chain and at O4 to Asn-227, the latter through a water molecule. Moreover, there is an intramolecular link between the O6 of the fructose units, located at +2 and +3, through a water molecule in the case of the complex with inulin (Fig. 4).
Subsite +3 is defined mainly by subunit B through hydrogen bonds of the Ser-434 main chain and the side chain of Gln-435 to fructose O6 and O4 atoms, respectively. Additional links are made with subunit A through the main chain of Trp-314 to O4 and Asp-318 and Arg-451 side chains to O3 and O4, respectively. In contrast, subsite +4 is lined up only by residues from subunit A with the main chain of Trp-76, both through its N and O atoms, and with the Asp-318 side chain making hydrogen bonds to O3 and O4 of the corresponding fructose unit of inulin and to the glucose O2 in the fructosylnystose complex. Interestingly, the glucose O2 atom in this complex is located in a position equivalent to a well ordered water molecule found in the complex with inulin that has many interactions with this substrate. Thus, even when O2 is the only glucose hydroxyl making polar interactions with residues at the active site, its interaction keeps the glucose tightly bound to the enzyme. In turn, the corresponding fructose unit from inulin is fixed by further additional links to Arg-451 through its O6 atom. Finally, the last visible fructose unit of inulin does not make any direct interactions at the active site, which precludes the definition of a +5 subsite and explains why no further inulin chain is evident from the electron density maps.
The Catalytic Center of Ffase Differs from Other GH32 Members
To date, four GH32 enzymes have been crystallized in complex with substrates longer than sucrose, a β-fructofuranosidase from Thermotoga maritima with raffinose (39), a plant fructan exohydrolase (FEH) from Cichorium intybus with 1-kestose (40), a FT from A. japonicus with raffinose, 1-kestose, and nystose (16), and recently, a FT (6-SST/6-SFT) from Pachysandra terminalis in complex with its acceptor substrate, 6-kestose (41). With the exception of the A. japonicus FT, the complexes analyzed show that the sugar unit bound at subsite +2 is loosely bound, mostly through a few polar interactions mediated by water molecules, and that additional binding sites for longer substrates have been clearly discarded. On the contrary, and as it happens with Ffase, the FT from A. japonicus displays a deep active site able to recognize sugar units from the −1 to +3 subsites (16). However, an inspection of the active site of both enzymes reveals that only subsite −1 is conserved, whereas significant differences are found from subsite +1 toward the reducing end of the substrate.
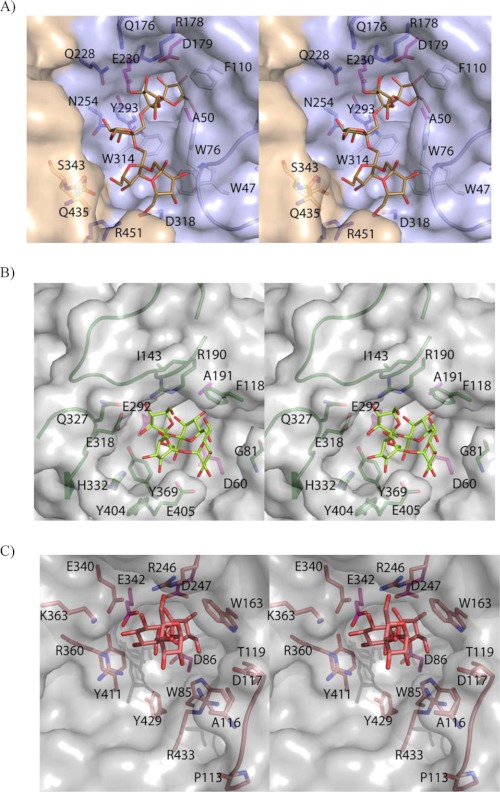
As seen in Fig. 5, A and B, among all of the aromatic residues that shape the hydrophobic wall of the cavity in Ffase, i.e. Trp-47, Trp-76, Phe-110, and Trp-314, only Phe-118 is present in A. japonicus FT. Furthermore, the terminal fructose unit bound at subsite +3 in the A. japonicus FT-nystose complex is located exactly in the position corresponding to the side chain of Trp-76 in Ffase. In fact this part of the A. japonicus FT cavity is wider because of its lack of loop 70–80 (Ffase numbering), which nevertheless is conserved in all the GH32 solved up to present from plants, fungi, and bacteria. On the contrary, the cavity at the opposite wall is wider in Ffase than in A. japonicus FT, where two loops linking β-strands B and C present two long insertions, residues 140–153 and 320–331, at blades II and IV, respectively, which is unique to the FT enzyme. Interestingly, these two insertions provide Ile-143 and Gln-327, two residues involved in binding raffinose and therefore proposed to provide a +2 subsite responsible for the formation of levan- or neo-type products in A. japonicus FT (16). An alternative +2 subsite for the formation of an inulin-type product is defined by the A. japonicus FT Tyr-404 and Glu-405, both also involved in subsite +3 and located near the position of Ffase Trp-314, with this last residue conserved in all the microbial enzymes. Therefore, it is apparent that none of determinants proposed as responsible for the transferase activity in A. japonicus FT are found in the Ffase active site. Moreover, it is worth noting that the subsites described for A. japonicus FT also diverge significantly from the sucrose-binding box motif (WMNDPN), including the nucleophile Asp, which had been identified as the binding site for the donor sucrose in plant enzymes and is not conserved in GH32 FTs.
FIGURE 5.
Ffase active site specificity. A, stereo pair of the D50A-inulin complex with relevant residues highlighted. Only five fructose units that interact with Ffase are shown for clarity. Loop 70–80, absent in the A. japonicus FT, is in ribbon representation. Aromatic residues defining the hydrophobic wall of the active site are colored in white, and the three catalytic residues are pink. B, A. japonicus FT complexed with nystose (PDB code: 3LYH). Relevant residues are represented as sticks. Phe-118 is the unique hydrophobic residue present in the A. japonicus FT active site, with the fructose unit located at subsite +3 at an equivalent position to Ffase Trp-76. Two long insertions, absent in the known GH32 hydrolases, residues 140–153 and 320–331, are highlighted and hold Ile-143 and Gln-327, two residues proposed to provide a +2 subsite responsible for the formation of levan- or neo-type products. C, levansucrase from B. subtilis complexed with raffinose (PDB code: 3BYN). The Glu-340/Arg-360 pair, participating in subsites +1 and +2, is topologically and structurally equivalent to the Ffase Gln-228/Asn-254 pair. The interactions observed with the sugar at subsites +1 and +2 are conserved with levansucrases, and the different product specificities are proposed to be modulated by a distant loop that is represented in the figure.
Finally, in contrast to the deep well found in A. japonicus FT, which probably precludes the approach of water molecules to the catalytic pocket favoring the transferase versus the hydrolytic activity, Ffase shows a rather wide cleft consistent with its hydrolytic activity. Furthermore, the polar side chains of Gln-176, Gln-228, and Asn-254 protrude noticeably at the opposite face of the hydrophobic wall within this cleft (see Fig. 5A), delineating a hydrophilic patch that might well be involved in binding the acceptor sucrose for transferring to fructose, as we have proposed previously (8). It is worth noting that the Ffase residues Gln-228/Asn-254 are structurally equivalent to the Asp-239/Lys-242 pair of Arabidopsis thaliana invertase and also to the Glu/Arg pair of levansucrases and inolusucrases (Fig. 5C) (17). This is a relevant issue taking into account that the presence of an acidic residue in this couple seems necessary for both binding and hydrolysis of sucrose in many clan GH-J enzymes (14). Therefore, the corresponding Gln/Asn Ffase pair might play a unique crucial role in its enzymatic activity that needs further investigation.
Kinetic Analysis of Mutated Ffase Shows Key Residues for Activity
Previous work on Sw. occidentalis Ffase points to the region in the vicinity of Glu-230, the residue acting as acid-base catalyst in the hydrolytic reaction, as responsible for substrate recognition and as a potential acceptor subsite for sucrose in the transfructosylation reaction (8, 15). In fact, the substitution Q228V caused a large reduction in the enzymatic catalytic efficiency (kcat/Km) on substrates of different size (short, sucrose; medium, nystose; long, inulin), whereas N254A showed a significant reduction on nystose. In addition the substitutions K181F, S196L, and P232V also reduced this parameter on sucrose, whereas the two last increased twice the transferase capacity of the enzyme. Moreover, Ffase crystal structure shows that residues in this region also participate in an intricate net of polar interactions with other residues located at the active site within the dimer interface, which may also be directly involved in substrate recognition. Therefore, substitutions on these residues may affect/distort the dimer interface and consequently the enzymatic activity even when they are not directly involved in substrate binding.
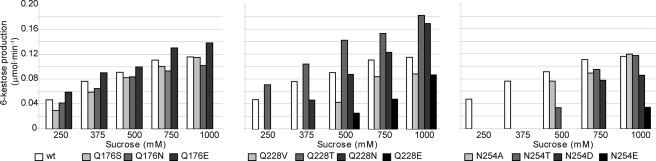
With the aim of determining the precise role of the Gln-176, Gln-228, and Asn-254 positions, we extended the previous mutational study and, in addition to the hydrolytic activity (Table 2), the role of these substitutions on the transferase activity (Table 3 and Fig. 6) was evaluated. Gln-176 is involved in substrate recognition at subsite +1 by interaction through a water molecule, but it also interacts with Arg-178 (in the RDP motif). Although the Q176S mutation significantly reduced (∼6-fold) the enzyme catalytic efficiency in nystose, and the Q176N replacement produced a moderately decreased efficiency (≤3-fold) in the three substrates, no clear influence of this position on the general hydrolytic behavior can be highlighted (Table 2). Therefore, we suggest that the contribution of Gln-176 to the enzymatic activity might be related to its interactions with other amino acids. Indeed, the transfructosylating activity of the enzyme was not affected by any of the three changes made in this position (Fig. 6 and Table 3), indicating that most likely Gln-176 does not determine the acceptor site for sucrose in transfructosylation reactions.
TABLE 3.
FOS produced by Ffase mutants after 24-h reactions
Enzymes (1 unit ml−1) and 1 m sucrose were incubated for 24 h. FOS is indicated as g liter−1. ND, not detected.
| Ffase mutant | Neokestose | 1-Kestose | 6-Kestose | Total FOS | 6-K/1-K ratio |
|---|---|---|---|---|---|
| WT | ND | 9.5 | 33.4 | 42.9 | 3.5 |
| Q176S | ND | 5.3 | 21.1 | 26.4 | 3.9 |
| Q176N | ND | 6.6 | 24.7 | 31.3 | 3.7 |
| Q176E | ND | 6.1 | 18.9 | 25.0 | 3.1 |
| Q228V | 0.5 | 0.8 | 28.7 | 30.0 | 35.9 |
| Q228T | 1.0 | 2.6 | 3.3 | 6.9 | 1.3 |
| Q228N | 0.8 | 2.1 | 24.6 | 27.6 | 11.7 |
| Q228E | ND | 2.1 | 23.2 | 25.3 | 11.0 |
| N254A | 1.7 | 1.3 | 13.5 | 16.5 | 10.4 |
| N254T | 11.8 | 8.6 | 28.4 | 48.8 | 3.3 |
| N254D | 1.9 | 3.6 | 12.5 | 18.0 | 3.5 |
| N2254E | ND | 4.1 | 7.3 | 11.4 | 1.8 |
FIGURE 6.
Effect of sucrose concentration on Ffase mutant transferase activity. Enzymes (1 unit ml−1) and sucrose (250–1000 mm) were incubated for 3 h. The amount of carbohydrates formed or hydrolyzed/min was quantified by HPLC in the 1–3 h interval (initial velocity conditions). 6-Kestose was the only transfructosylation product detected in all reaction mixtures, and the amount of sucrose hydrolyzed was lower than 10% (w/w) in all cases.
On the contrary, some Gln-228 substitutions had a greater effect on Ffase hydrolytic activity (Table 2). Thus, the presence of short side chain amino acids in this position (Thr and Val) clearly reduced the catalytic efficiency of the enzyme toward long (inulin, ∼10–20-fold) and medium size substrates (nystose, ∼102-fold), whereas replacement by similar size residues (Asn or Glu) had little effect on this parameter using medium (∼1.5–2-fold decreased) and long size substrates (∼1.3–3-fold increase); this suggests that the length of amino acid chain at position 228 is essential for its hydrolytic activity. Moreover, the transfructosylation assay proved that Gln-228 has to be involved in the transfructosylating reaction mechanism, as its replacement by Val or Glu considerably reduced the initial rate at sucrose concentrations in the 0.25–1 m range (Fig. 6). On the contrary, replacement by Thr and Asn increased (∼1.5-fold) the initial transfructosylating ability at 1 m sucrose, thereby suggesting that polar residues smaller than glutamine might allow a better recognition or positioning of the transferred sucrose. However, as seen in Table 3, none of the four substitutions analyzed was able to improve the total amount of FOS produced after 24 h of reactions. Interestingly, the specificity of the product is affected by the nature of the replacement, and thus the Q228V substitution had a more drastic effect in reducing the production of 1-kestose, whereas the Q228T change affected largely to that of 6-kestose.
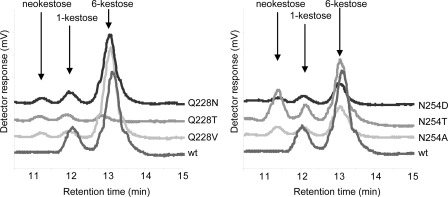
With regard to Asn-254, all of the substitutions confirmed the contribution of this residue in the recognition/hydrolysis of medium (catalytic efficiency reduced ∼3–11-fold) and long size substrates (catalytic efficiency reduced ∼3–13-fold for N254D/N254T/N254E). In particular, the N254E replacement produced a rearrangement of Trp-314 orientation and a subsequent change in subsite +3 (Fig. 4), which agrees with the 10-fold decreased efficiency of this mutant toward medium and long chain substrates. Furthermore, all mutations tested showed a decrease in the initial Ffase transferase activity at sucrose up to 0.75 m (Fig. 6). This effect is more evident in the N254E mutant, which shows about 20% of the transferase activity even at 1 m sucrose after 24 h of reaction (Table 3 and Fig. 6). With the exception of the N254E mutant, for which the active site might be putatively distorted as proposed above, the other variants produced, at high sucrose concentrations, an amount of FOS analogous to the wild-type enzyme. However, it is noteworthy that most substitutions at this position seemed to modify the product specificity, with Ffase mutants able to synthesize neokestose (Fig. 7). Moreover, short chain amino acids, such as Thr or Ala, produced neokestose to a higher extent than 1-kestose, which may well indicate the relevance of this position in product specificity.
FIGURE 7.
FOS profile produced by Ffase mutants. Frame of HPLC chromatograms (10.5–15 min) obtained using enzymatic variants with new product specificity. Enzymes (1 unit ml−1) and 1 m sucrose were incubated for 24 h.
In conclusion, it is clear that both, Gln-228 and Asn-254, configure the acceptor substrate binding site for transfructosylation reactions in Ffase and appear to be responsible for the inulin/levan-type FOS specificity of this enzyme.
Variants of Ffase Show Redesigned Activity
Our mutagenesis analysis gave valuable insights into the GH-J clan enzymes and delineated an interesting region for further studies in protein engineering to obtain tailor-made products. In fact, we found two variants of the enzyme with clearly redesigned functional features. First, as shown in Table 3 and Figs. 6 and 7, the Q228V mutant yields a 30% decrease in the total amount of FOS produced (∼30 versus 43 g/liter), but the product is almost specifically 6-kestose. On the other hand, the N254T mutant produces a similar amount of FOS as the native enzyme in these conditions, but these FOS are of a more varied type, with neokestose produced in similar quantity to 1-kestose. Therefore, this is an excellent example of an efficient way of manipulating the specificity of the recovered FOS, which may be adjusted to the requirements in the pursued prebiotic features, i.e. a specific versus a broader spectrum product.
Proposed Transfructosylating Mechanism
On the basis of the experimental complexes described here, and by superimposition with other reported complexes, we have modeled the putative 1-kestose and 6-kestose products into the Ffase active site (Fig. 8A). As observed, Gln-228 could be linked to the fructose accommodated at subsite +1 of both trisaccharides, whereas Asn-254 seems able to link the glucose of 6-kestose but is too far from the position inferred for glucose in 1-kestose, which in turn would stack against the Trp-76 side chain. These aspects are most relevant when taking into account that the binding mode of the product complexes at subsites +1 and +2 illustrates how the acceptor sucrose would be positioned in the catalytic site of a putative enzyme-fructose intermediate complex for the subsequent transfer of this sugar with the formation of β(2–1) or β(2–6) linkage to produce 1-kestose and 6-kestose, respectively (Fig. 8B). Therefore, it seems that the binding mode of the acceptor sucrose linked to Gln-228 and Asn-254 is preferred by Ffase, with the concomitant production of a 3-fold amount of 6-kestose/1-kestose. Furthermore, this model is consistent with the Q228V mutant having a decreased yield of FOS and an almost exclusive production of 6-kestose, as this mutant showed a reduced affinity against sucrose but still kept a polar link to Asn-254 only in this binding mode, consequently selecting 6-kestose as main product. On the other hand, the N254T substitution may conform to a larger subsite that allows a broader range of orientations to accommodate the acceptor sucrose and therefore also be able to generate neokestose efficiently (Fig. 8B).
FIGURE 8.
Proposed mechanism of Ffase transfructosylating activity. A, the inferred position of the product 1-kestose (green) at the Ffase active site was obtained by superimposition of the C. intybus FEH 1-kestose coordinates (PDB code: 2AEZ) onto the fructose at the −1 subsite of the experimental Ffase-substrate complexes. The position of the product 6-kestose (pink) is inferred from superposition of its coordinates extracted from the CSD (Refcode CELGIC) onto the fructose found at subsite −1. The position of the Fru-Glu portion in both complexes represents a potential binding mode of acceptor sucrose for the formation of β(2–1) or β(2–6) linkage. B, schematic illustration of the proposed mechanism. The donor substrate sucrose is hydrolyzed by nucleophilic attack forming the fructosyl-Ffase intermediate. A subsequent sucrose molecule (the acceptor substrate) can enter the active site pocket with binding mode 1 generating 1-kestose or, alternatively, with binding mode 2 generating 6-kestose. Additionally, the Q228V/Q228T/Q228N and the N254T/N254D/N254A Ffase mutants are able to produce neokestose.
DISCUSSION
We proposed previously that, unlike other known GH32 hydrolases, Sw. occidentalis Ffase presents a unique active site putatively able to recognize medium and long chain fructooligosaccharides. The structure of its complexes with fructosylnystose and inulin described here presents a detailed picture of the extended net of interactions that keeps the substrate tightly bound at the active site. Accordingly, the enzyme presents up to five subsites for substrate binding (−1 to +4), and what is more outstanding, the sugar unit located at subsite +3 is recognized by direct interaction with residue from the β-sandwich domain of the adjacent subunit within the dimer, not only through their side chain, as is the case of Gln-435, but also through the Ser-434 main chain. This explains the higher hydrolytic activity observed against nystose than sucrose and gives the first experimental evidence of the direct role of a GH32 supplementary β-sandwich domain in substrate binding. Furthermore, this restricted architecture of the active site seems to be adapted to the polymeric conformation of inulin and to induce a fixed conformation of bound fructooligosaccharides.
The complexes described here also assign a specific role to the previously identified key residues, Gln-228 and Asn-254, in shaping subsites +1 and +2 for donor substrate binding. Thus, Gln-228 is linked to fructose located at subsite +1 through its O3 atom, whereas Asn-254 is linked to the O3 of fructose located at +2, and both residues are involved in oligosaccharide binding of both fructose units through water molecules. Furthermore, on the basis of these experimental complexes, the binding mode of putative transfructosylating product complexes can be modeled. This allows us to infer how the acceptor sucrose would be positioned in subsites +1 and +2 of an enzyme-fructose intermediate complex for the subsequent transfer of this sugar with the formation of a β(2–1) or β(2–6) linkage to produce both 1-kestose and 6-kestose. This model illustrates how both residues may be involved in the transfructosylating activity of the enzyme, and certainly, their implied involvement in both H/T activity and specificity has been proved by mutagenesis analysis. According to the activity observed in the mutated proteins, the Gln-228 would be essentially responsible for conferring affinity for acceptor sucrose, whereas Asn-254 seems to be most responsible for product specificity. Furthermore, we have generated two variants of Ffase that show redesigned transfuctosylating ability; thus, the Q228V mutant gives almost specifically 6-kestose, whereas the N254T mutant yields a broad spectrum FOS product, adding neokestose to the 6-kestose/1-kestose mix produced by the wild-type enzyme.
As mentioned above, the Gln-228/Asn-254 Ffase residues are structurally equivalent to Asp-239/Lys-242 of A. thaliana invertase, which is reported to accomplish a crucial role in sucrose binding and stabilization. In fact, no structural equivalents are found in FEH, and this feature is proposed to confer invertase/FEH specificity in plant enzymes (14). In agreement with this role, the Aspergillus niger exoinulinase presents Val-239/Asn-265 at these positions (42). However, the mutagenesis analysis of Ffase shows an opposite trend, and the Q228V and Q228T mutations (Thr is found in C. intybus FEH) abolish the hydrolysis of long substrates while keeping moderate invertase activity. Moreover, this pair is equivalent to the A. japonicus FT Glu-318/His-332 (16), where it is involved in shaping subsite +1 (Fig. 5B), and also to the Asp-244/Gln-247 pair of the FT (6-SST/6-SFT) from P. terminalis that has been reported recently to be involved in both donor and acceptor substrate binding (41). In fact, the uniqueness of a Gln in this Asp/Gln pair has been related to the wide range of acceptor specificities and branching capacities of this plant FT. Consequently it is clear that all of the structural work carried out on GH32 enzymes assigns a crucial role to the corresponding equivalent couple in modulating the distinctive features particular to each enzyme, but Ffase shows a unique Gln substituting for the acidic residue within this pair, which may be related to its hydrolytic activity against both long and short substrates.
Accordingly, the structurally equivalent pair of the GH68 family (Fig. 5C) stabilizes the glucose part of sucrose in levansucrases (Glu-340/Arg-360 in Bacillus subtilis levansucrase), and it has been shown by mutagenesis that changing Arg altered the polymerization degree of the product and the transfructosylation/hydrolysis ratio but not the linkage type of the products, i.e. it does not modulate the product specificity (43). Moreover, it has been shown recently (17) that both residues are present in GH68 levansucrases and inulosacarases, the product linkage type being determined by non-conserved residues in the more distant +3 and +4 subsites. Interestingly, and unlike the other GH32 enzymes, the corresponding pair of Ffase Gln-228/Asn-254 is not only structurally but also topologically equivalent to that in levansucrases and inulosucrases. However, in this case the involvement of both residues in the activity is different, as both show a direct role in altering the ratio H/T, but Asn-254 seems also to be involved in modulating the specificity of the transfructosylating product.
In the case of Ffase, and relating to the chemical nature of the Gln/Asn pair, its particular activity and specificity pattern have to be explained in terms of its unique active site conformed by both subunits between the dimer as well as the participation of residues from the close β-sandwich domain in the enzymatic activity. Polymeric carbohydrate degradation or modification is usually facilitated and potentiated by carbohydrate-binding modules (44). However, only an inulin-binding module has been postulated for a cycloinulofructanotransferase that is not structurally related to the GH-J clan (45). On the other hand GH-F clan enzymes, which share the 5-fold β-propeller with GH-J enzymes but display an inverting mechanism, usually contain carbohydrate-binding modules (46). Further studies in fructan-related enzymes would be interesting to determine whether the architecture of Ffase active site is a unique feature of this yeast enzyme or a general strategy to cope with polymeric structures in a particular group of enzymes.
In summary, the structural information of the Ffase complexes with long substrates sheds light on the uniquely rigid architecture of its active site, which involves the β-sandwich domain in substrate binding, a role that has not been experimentally evidenced until now. On the other hand, extensive mutagenesis and biochemical analysis of the EC motif environment, combined with structural information, has enlightened us as to the crucial involvement of the Gln-228/Asn-254 pair in modulating the H/T ratio and type β(2–1)/β(2–6) linkage formation. This contributes to unraveling the molecular basis that regulates specificity within GH32 and GH68 enzymes, where the structurally equivalent pair of residues seems to be a potential target for the rational design of superior enzymes showing redesigned activities. This is an issue of utmost biotechnological interest for generating tailor-made FOS directed, ideally, to the manipulation of the gut microbial communities (47) with an associated benefit to human health (48).
Supplementary Material
Acknowledgments
We thank the staff of the European Synchrotron Radiation Facility for providing beam time and technical assistance at beamlines ID23-2 and ID14-1 and also the Fundación Ramon Areces for the institutional grant to the Centro de Biología Molecular Severo Ochoa. We also acknowledge Asunción Martin for excellent technical assistance.
This work was supported by Grants BIO2010-20508-C04-01, -03, and -04, as well as BIO2007-67708-C04-01, -03, and -04, from the Ministry of Education and Science, Spain.

This article contains supplemental Table S1.
The atomic coordinates and structure factors (codes 3U14 and 3U75) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- FOS
- fructooligosaccharide(s)
- FEH
- fructan exohydrolase
- Ffase
- β-fructofuranosidase from Schwanniomyces occidentalis
- FT
- fructosyltransferase
- GH
- glycosylhydrolase
- H/T
- hydrolase/transferase activity ratio
- PDB
- Protein Data Bank
- CSD
- Cambridge Structural Database
- MPD
- 2-methyl-2,4-pentanediol.
REFERENCES
- 1. Franck A., De Leenheer L. (2005) Inulin. In Biopolymers Online, DOI: 10.1002/3527600035.bpol6014, Wiley Online Library [DOI] [Google Scholar]
- 2. Swennen K., Courtin C. M., Delcour J. A. (2006) Non-digestible oligosaccharides with prebiotic properties. Crit. Rev. Food Sci. Nutr. 46, 459–471 [DOI] [PubMed] [Google Scholar]
- 3. Kaur N., Gupta A. K. (2002) Applications of inulin and oligofructose in health and nutrition. J. Biosci. 27, 703–714 [DOI] [PubMed] [Google Scholar]
- 4. Kilian S., Kritzinger S., Rycroft C., Gibson G., du Preez J. (2002) The effects of the novel bifidogeneic trisaccharide, neokestose, on the human colonic flora. World J. Microbiol. Biotechnol. 18, 637–644 [Google Scholar]
- 5. Sabater-Molina M., Larqué E., Torrella F., Zamora S. (2009) Dietary fructooligosaccharides and potential benefits on health. J. Physiol. Biochem. 65, 315–328 [DOI] [PubMed] [Google Scholar]
- 6. Farine S., Versluis C., Bonnici P. J., Heck A., L'homme C., Puigserver A., Biagini A. (2001) Application of high performance anion exchange chromatography to study invertase-catalysed hydrolysis of sucrose and formation of intermediate fructan products. Appl. Microbiol. Biotechnol. 55, 55–60 [DOI] [PubMed] [Google Scholar]
- 7. Alvaro-Benito M., de Abreu M., Fernández-Arrojo L., Plou F. J., Jiménez-Barbero J., Ballesteros A., Polaina J., Fernández-Lobato M. (2007) Characterization of a β-fructofuranosidase from Schwanniomyces occidentalis with transfructosylating activity yielding the prebiotic 6-kestose. J. Biotechnol. 132, 75–81 [DOI] [PubMed] [Google Scholar]
- 8. Alvaro-Benito M., de Abreu M., Portillo F., Sanz-Aparicio J., Fernández-Lobato M. (2010) New insights into the fructosyltransferase activity of Schwanniomyces occidentalis β-fructofuranosidase emerging from nonconventional codon usage and directed mutation. Appl. Environ. Microbiol. 76, 7491–7499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gutiérrez-Alonso P., Fernández-Arrojo L., Plou F. J., Fernández-Lobato M. (2009) Biochemical characterization of a β-fructofuranosidase from Rhodotorula dairenensis with transfructosylating activity. FEMS Yeast Res. 9, 768–773 [DOI] [PubMed] [Google Scholar]
- 10. Linde D., Macias I., Fernández-Arrojo L., Plou F. J., Jiménez A., Fernández-Lobato M. (2009) Molecular and biochemical characterization of a β-fructofuranosidase from Xanthophyllomyces dendrorhous. Appl. Environ. Microbiol. 75, 1065–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Linde D., Rodríguez-Colinas B., Estévez M., Poveda A., Plou F. J., Fernández Lobato M. (2012) Analysis of neofructooligosaccharides production mediated by the extracellular β-fructofuranosidase from Xanthophyllomyces dendrorhous. Bioresource Technol. 109, 123–130 [DOI] [PubMed] [Google Scholar]
- 12. Henrissat B., Sulzenbacher G., Bourne Y. (2008) Glycosyltransferases, glycoside hydrolases: surprise, surprise!. Curr. Opin. Struct. Biol. 18, 527–533 [DOI] [PubMed] [Google Scholar]
- 13. Pons T., Naumoff D. G., Martínez-Fleites C., Hernández L. (2004) Three acidic residues are at the active site of β-propeller architecture in glycoside hydrolase families 32, 43, 62 and 68. Proteins 54, 424–432 [DOI] [PubMed] [Google Scholar]
- 14. Lammens W., Le Roy K., Schroeven L., Van Laere A., Rabijns A., Van den Ende W. (2009) Structural insights into the glycoside hydrolase family 32 and 68 enzymes: functional implications. J. Exp. Bot. 60, 727–740 [DOI] [PubMed] [Google Scholar]
- 15. Alvaro-Benito M., Polo A., González B., Fernández-Lobato M., Sanz-Aparicio J. (2010) Structural and kinetic analysis of Schwanniomyces occidentalis invertase reveals a new oligomerization pattern and the role of its supplementary domain in substrate binding. J. Biol. Chem. 285, 13930–13941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chuankhayan P., Hsieh C. Y., Huang Y. C., Hsieh Y. Y., Guan H. H., Hsieh Y. C., Tien Y. C., Chen C. D., Chiang C. M., Chen C. J. (2010) Crystal structures of Aspergillus japonicus fructosyltransferase complex with donor/acceptor substrates reveal complete subsites in the active site for catalysis. J. Biol. Chem. 285, 23251–23264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pijning T., Anwar M. A., Böger M., Dobruchowska J. M., Leemhuis H., Kralj S., Dijkhuizen L., Dijkstra B. W. (2011) Crystal structure of inulosucrase from Lactobacillus: insights into the substrate specificity and product specificity of GH68 fructansucrases. J. Mol. Biol. 412, 80–93 [DOI] [PubMed] [Google Scholar]
- 18. Bujacz A., Jedrzejczak-Krzepkowska M., Bielecki S., Redzynia I., Bujacz G. (2011) Crystal structures of the apo form of β-fructofuranosidase from Bifidobacterium longum and its complex with fructose. FEBS J. 278, 1728–1744 [DOI] [PubMed] [Google Scholar]
- 19. Altenbach D., Rudiño-Pinera E., Olvera C., Boller T., Wiemken A., Ritsema T. (2009) An aceptor-substrate binding site determining glycosyl transfer emerges from mutant analysis of a plant vacuolar invertase and a fructosyltransferase. Plant Mol. Biol. 69, 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schroeven L., Lammens W., Van Laere A., Van den Ende W. (2008) Transforming wheat vacuolar invertase into a high affinity sucrose:sucrose 1-fructosyltransferase. New Phytol. 180, 822–831 [DOI] [PubMed] [Google Scholar]
- 21. Le Roy K., Lammens W., Van Laere A., Van den Ende W. (2008) Influencing the binding configuration of sucrose in the active sites of chicory fructan 1-exohydrolase and sugar beet fructan 6-exohydrolase. New Phytol. 178, 572–580 [DOI] [PubMed] [Google Scholar]
- 22. Lasseur B., Schroeven L., Lammens W., Le Roy K., Spangenberg G., Manduzio H., Vergauwen R., Lothier J., Prud'homme M. P., Van den Ende W. (2009) Transforming a fructan:fructan 6G-fructosyltransferase from perennial ryegrass into a sucrose 1-fructosyltransferase. Plant Physiol. 149, 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Davies G. J., Wilson K. S., Henrissat B. (1997) Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem. J. 321, 557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leslie A. G. W. (1992) Molecular data processing. In Crystallographic Computing 5: From Chemistry to Biology (Moras D., Podjarny A. D., Thierri J. C., eds) pp. 39–50, Oxford University Press, Oxford, UK [Google Scholar]
- 25. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 26. Vagin A., Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 27. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 28. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Improved methods for building protein methods in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 29. André I., Mazeau K., Putaux W., Winter W. T., Taravel F. R., Chanzy H. (1996) Molecular and crystal structures of inulin from electron diffraction data. Macromolecules 29, 4626–4635 [Google Scholar]
- 30. Okuyama K., Noguchi K., Saitoh M., Ohno S., Fujii S., Tsukada M., Takeda H., Hidano T. (1993) Crystal structure of nystose trihydrate. Bull. Chem. Soc. Japan 66, 374–379 [Google Scholar]
- 31. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 32. Laskowsky R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 33. DeLano W. L. (2002) The PyMOL Molecular Graphics System, Version 1.5.0.1, Scrödinger, LLC [Google Scholar]
- 34. Polo A., Alvaro-Benito M., Fernández-Lobato M., Sanz-Aparicio J. (2009) Crystallization and preliminary X-ray diffraction analysis of the fructofuranosidase from Schwanniomyces occidentalis. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 1162–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vandamme E. J., Derycke D. G. (1983) Microbial inulinases: fermentation process, properties and applications. Adv. Appl. Microbiol. 29, 139–176 [DOI] [PubMed] [Google Scholar]
- 36. Chi Z., Chi Z., Zhang T., Liu G., Yue L. (2009) Inulinase-expressing microorganisms and applications of inulinases. Appl. Microbiol. Biotechnol. 82, 211–220 [DOI] [PubMed] [Google Scholar]
- 37. Pandey A., Soccol C. R., Selvakumar P., Soccol V. T., Krieger N., Fontana J. D. (1999) Recent developments in microbial inulinases. Its production, properties, and industrial applications. Appl. Biochem. Biotechnol. 81, 35–52 [DOI] [PubMed] [Google Scholar]
- 38. Gill P. K., Manhas R. K., Singh J., Singh P. (2004) Purification and characterization of an exoinulinase from Aspergillus fumigatus. Appl. Biochem. Biotechnol. 117, 19–32 [DOI] [PubMed] [Google Scholar]
- 39. Alberto F., Jordi E., Henrissat B., Czjzek M. (2006) Crystal structure of inactivated Thermotoga maritima invertase in complex with the trisaccharide substrate raffinose. Biochem. J. 395, 457–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Verhaest M., Lammens W., Le Roy K., De Ranter C. J., Van Laere A., Rabijns A., Van den Ende W. (2007) Insights into the fine architecture of the active site of chicory fructan 1-exohydrolase: 1-kestose as substrate vs. sucrose as inhibitor. New Phytol. 174, 90–100 [DOI] [PubMed] [Google Scholar]
- 41. Lammens W., Le Roy K., Yuan S., Vergauwen R., Rabijns A., Van Laere A., Strelkov S. V., Van den Ende W. (2011) Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose. Plant J., in press [DOI] [PubMed] [Google Scholar]
- 42. Nagem R. A., Rojas A. L., Golubev A. M., Korneeva O. S., Eneyskaya E. V., Kulminskaya A. A., Neustroev K. N., Polikarpov I. (2004) Crystal structure of exoinulinase from Aspergillus awamori: the enzyme fold and structural determinants of substrate recognition. J. Mol. Biol. 344, 471–480 [DOI] [PubMed] [Google Scholar]
- 43. Homann A., Biedendieck R., Götze S., Jahn D., Seibel J. (2007) Insights into polymer versus oligosaccharide synthesis: mutagenesis and mechanistic studies of a novel levansucrases from Bacillus megaterium. Biochem. J. 407, 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. (2004) Carbohydrates-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee J. H., Kim K. N., Choi Y. J. (2004) Identification and characterization of a novel inulin binding module (IBM) from the CFTase of Bacillus macerans CFC1. FEMS Microbiol. Lett. 234, 105–110 [DOI] [PubMed] [Google Scholar]
- 46. Shoseyov O., Shani Z., Levy I. (2006) Carbohydrate-binding modules: biochemical properties and novel applications. Microbiol. Mol. Biol. Rev. 70, 283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D. R., Fernandes G. R., Tap J., Bruls T., Batto J. M., et al. (2011) Enterotypes of the human gut microbiome. Nature 473, 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sonnenburg E. D., Zheng H., Joglekar P., Higginbottom S. K., Firbank S. J., Bolam D. N., Sonnenburg J. L. (2010) Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141, 1241–1252 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.