Abstract
A vast number of cellular processes and signaling pathways are regulated by various receptors, ranging from transmembrane to nuclear receptors. These receptor-mediated processes are modulated by a diverse set of regulatory proteins. TNFα-induced protein 3-interacting protein 1 is such a protein that inhibits both transduction by transmembrane receptors, such as TNFα-receptor, EGF-R, and TLR, and nuclear receptors’ PPAR and RAR activity. These receptors play key roles in regulating inflammation and inflammatory diseases. A growing number of references have implicated TNIP1 through GWAS and expression studies in chronic inflammatory diseases such as psoriasis and rheumatoid arthritis, although TNIP1’s exact role has yet been determined. In this review, we aim to integrate the current knowledge of TNIP1’s functions with the diseases in which it has been associated to potentially elucidate the role this regulator has in promoting or alleviating these inflammatory diseases.
Keywords: TNIP1, ABIN-1, TNFα receptor, nuclear receptor, inflammation
1. Introduction
Receptor-mediated signals and the changes in gene expression they instigate are regulated by the availability of receptor ligands as well as supernumerary checks on post-receptor pathways thus providing a more rheostat-type control rather than an all-on or all-off switch. The TNFα-induced protein 3 (TNFAIP3)-interacting protein 1 (TNIP1, also known as ABIN-1, Naf1 and VAN) appears to be one of these pathway-modulating factors. Known as a human cellular protein capable of interaction with the HIV proteins nef and matrix [1,2], it was also discovered to interact with the cytoplasmic protein A20 (also known as TNFAIP3) and to dampen subsequent NF-κB signaling following TNFα receptor (TNFα–R) activation [3]. Even with this latter function well established, TNIP1’s interaction portfolio continues to expand [3–7]. For instance, we have found it acts as a corepressor of ligand-bound retinoic acid receptors (RAR) [8] and peroxisome proliferator activated receptors (PPAR) [9], while other researchers discovered it prevents EGF-R-induced ERK2 nuclear translocation [10]. Additionally, genetic ablation of TNIP1 in mice [11] has suggested the phenotype due to its loss may not simply be the inverse of over-expression studies. Unexpectedly, TNIP1 null cells had levels of NF-κB-dependent gene expression similar to wild-type cells. Nevertheless, in these cells, another post-TNFα receptor pathway was affected, leading to increased apoptosis [11]. These roles for TNIP1 in model systems raise the questions of what do we know of TNIP1 in human disease, and, how might one correlate TNIP1 reduction of TNFα signaling with other findings of altered TNIP1 expression levels in inflamed tissues. The aim of this review is to provide some perspective by examining the association of TNIP1 and disease states in human genetic studies along with candidate signaling pathways based on cellular and molecular studies.
2. TNIP1 and associated diseases and phenotypes
2.1 Implications from GWAS and array studies
Current connections between TNIP1 and human pathologies cross several tissues including skin, connective tissue, possibly blood vessels and some internal organs. These associations derive from high throughput approaches such as genome-wide association studies (GWAS) and expression microarrays [12–18] (Table 1). Whether through sequence variations or expression levels, these approaches have linked TNIP1 with psoriasis, systemic lupus erythematosus (SLE), systemic sclerosis (SSc), rheumatoid arthritis (RA) and leukemia/lymphoma. Although family genealogies and twin studies have documented hereditary components or shared genetic loci for these pathologies [19–24], no one gene such as TNIP1 has been identified as definitively causative for any one of them, unlike the variant coding sequence for a “classic” single-gene disease, viz. hemophilia and mutations in genes for clotting factors VIII or IX. Further compounding the study of psoriasis, SLE, and RA is the participation of often multiple cell types, broadly grouped as non-immune and immune, suggesting TNIP1 expression changes specific to any one of them might confer some disease pathology to the affected organ. Although no human disease correlate has yet been identified with the pathology seen in the TNIP1 mouse knockout [11] (see section 2.3), a lupus-like autoimmune disease was seen in a TNIP1 mutant mouse knock-in [25], which is consistent with current genome wide association scans (GWAS). Additionally, the inflammation-associated defects seen using both in vitro and in vivo experimental systems are consistent with current reports of TNIP1 alterations associated with human auto-immune and chronic inflammatory diseases. TNIP1’s wide tissue distribution [2,9,26] and involvement in a number of receptor-mediated signaling pathways would likely extend impact of its altered function to non-immune cells. For instance, we found TNIP1 antibody staining in both stratified epithelial cells and germinal centers of human tonsil [26]. More clearly defined roles for TNIP1 in normo- and patho-physiology and will benefit from organ- and cell-specific knockout systems.
Table 1.
TNIP1 and associated diseases
| Disease | TNIP1 Association | Experimental Approach | Reference |
|---|---|---|---|
| Psoriasis | Intronic SNP; Increased expression | GWAS; Gene expression microarray | Nair et al. [13]; Psoriasis Consortium [33]; Ellinghaus et al. [30] |
| Psoriatic Arthritis | Intronic SNP | GWAS | Bowes et al.[29] |
| Systemic Lupus Erythematosus | Intronic SNP | GWAS | Kawasaki et al. [17] |
| Systemic Sclerosis | Intronic SNP | GWAS | Allanore et al. [18] |
| Leukemia-Lymphoma | Splice Variants | RT-PCR and sequencing | Shiote et al. [32] |
| Leukemia-Lymphoma | Point or frameshift mutations | PCR and sequencing | Dong et al. [41] |
| Rheumatoid Arthritis | Increased expression | Gene Expression microarray | Gallagher et al. [12] |
The TNIP1 gene has been implicated in psoriasis, SLE and SSc through at least three independent GWAS reports. In each case however, the strongest disease-associated single nucleotide polymorphisms (SNP) were in non-coding regions. In the psoriasis study [13], despite strong association with the disease (P-value 1 × 10−20) and ~1.5 fold increase in TNIP1 expression between lesioned and uninvolved skin (i.e., tissues from the same individual), the SNP was several kilobases upstream from the TNIP1 locus. Psoriasis is classically recognized as epidermal keratinocyte hyperproliferation with incomplete differentiation, incomplete barrier formation, and immune cell infiltration [27]. Notably, there is often the comorbidity of psoriatic arthritis, a chronic inflammatory disease where immune cells target the patient’s joints promoting cartilage breakdown and bone damage [28]. It is not unexpected then that SNP alleles were also confirmed for psoriatic arthritis [29,30].
Similar to psoriasis, SNPs in non-coding regions were also disease associated with SSc. Three different TNIP1 SNPs were identified in European populations in the second GWAS report for SSc [18]. Intriguingly, when TNIP1 mRNA and protein levels were assessed from cultured dermal fibroblasts of SSc patients, a ~1.7-fold decrease was observed. A separate GWAS study also identified SNPs in SLE. Two TNIP1 intronic SNP variants were found in SLE patients from Chinese Han, Caucasian, and Japanese populations, with the latter two groups having the same SNP [14,15,17]. Unlike the altered expression of TNIP1 in psoriasis and SSc, there was no TNIP1 mRNA change associated with this SLE SNP [17]. However, Kawasaki and colleagues suggested the SNP location in intron 1 could impact TNIP1 splicing possibly affecting the use of alternative exons 1A and B with exon 2 and thereby contributing to the numerous splice variants of TNIP1 [31,32] with as yet unrecognized consequences. Perhaps reflecting the polygenic nature of these pathologies, it is interesting to note that a protein-protein interaction partner for TNIP1, TNFAIP3 (also known as A20), is also a susceptibility locus for psoriasis [13,33], SLE [14,34], and RA [35,36]. Most challenging in understanding these results will be to appreciate how SNP variants in non-coding regions can go from association with the disease to at least contributory if not causative. Some context for that comes from a recent report that about 88% of trait or disease-associated SNPs are located in gene introns or intergenic regions [37]. Far from being innocuous spacers between coding regions of genes, introns are now recognized as possible sites of transcription-regulating factors at the DNA level and/or potential effectors of splicing at the RNA level [38,39]. Likewise, proximal or intergenic regions, especially those covering the disease-associated gene’s promoter/enhancer region, may affect expression levels or tissue-specific expression [37]. Most recently, copy number variations were reported for TNFAIP3 and TNIP1 suggesting other forms of genome-wide analyses could prove productive in relating these genes to the disease states [40].
In addition to gene analysis, TNIP1 mRNA expression has been analyzed from several human cell lines and tissues. Several splice variants having either 5′ truncated ends or lacking specific exons were detected in samples derived from patients with acute myeloid leukemia (AML) [32]. Although variant 5′ ends have been mapped to the use of alternative first exons, the 3′ truncations described in these samples are the first of their kind to be reported. Most of the splice variants did not confer changes in amino acid sequence. However, one variant lacking exons 16 and 17 was less effective at reducing NF-κB activity. Decreased TNIP1 mRNA levels, for with full-length or splice variants, were observed in AML patient samples post chemotherapy treatments. Separately, several TNIP1 mutations have been detected in gastrointestinal diffuse large B cell lymphomas [41]. These sequence alterations are either point or frame-shift mutations, the latter resulting in a protein truncation. One mutant in particular, causing a glutamic acid to lysine change (E476K), lost its NF-κB inhibitory properties; other missense mutations did not alter this TNIP1 property. Thus, sequence variations, either at the mRNA level possibly affecting message stability, exon content, or amino acid sequence could impact ultimate TNIP1 protein function. Additionally, we should consider that there could be functional consequence to even wild-type TNIP1 protein if its levels or post-translation processing, e.g. phosphorylation (see sections 2.3 and 5.2) were altered.
In contrast to other TNIP1 associated diseases, the connection between TNIP1 and RA appears strictly at the expression level, not at a susceptibility locus or nucleotide mutation. Three SNP type GWAS reports [17,36,42] concluded loci-disease association(s) did not meet the cut-offs used for the analyses. However, when compared to knee synovial membrane biopsies from osteoarthritis patients, similar samples from patients with RA showed a 2.5–3.5 fold TNIP1 mRNA increase. Osteoarthritis and RA are referred to as non-inflammatory versus inflammatory forms of the disease, respectively. Consistent with this inflammatory association, TNIP1 was one of the genes with increased expression following TNFα treatment of cultured synovial fibroblasts [12]. Nevertheless, TNFα-increased TNIP1 expression may be tissue specific by following one of multiple post-TNFα-receptor signaling pathways. For instance, retrovirus-mediated increases in NF-κB signaling, one of several post-TNFα-receptor consequences, did not increase TNIP1 expression in dermal fibroblasts but did in epidermal keratinocytes [43]. TNIP1 up-regulation in response to signaling from inflammatory mediators coupled with dampening of NF-κB activity, at least in experimental systems, suggests its disregulation may be contributory and/or consequential to cytokine signaling.
2.2 Non-coding changes in TNIP1 and possible connections to disease
The quandary of how TNIP1 non-coding region SNPs affect psoriasis, SLE and SSc is much the same as for any other extra-exonic sequence changes associated with disease. Sequence alterations in promoter regions, even those distant to transcription start sites may affect transcription factor binding and, in turn, mRNA production. Likewise, SNPs in non-coding regions may alter transcript conformation resulting in changes in its stability, translational efficiency, or interaction with RNA regulatory factors [44]. Thus even the wild-type TNIP1 protein sequence at altered levels could impact the associated disease states given the ability of TNIP1 to modulate post-receptor signaling as detailed below. In the case of RA, experiments using fibroblast-like synoviocytes show wild-type TNIP1 increases pro-inflammatory cytokines, potentially advancing the disease [45]. In this vein, as psoriasis, SLE, and RA are at least in part regulated [46] by receptor pathways (TNFα–R and EGF-R) modulated by TNIP1, TNIP1 itself could be a focal point for clinical intervention. This possibility is again echoed by TNIP1 corepression of nuclear receptors currently used as therapeutic targets (RAR) [47] or suggested for such use (PPAR) [48,49] for treatment of psoriasis or other inflammatory diseases [50]. However, a discrepancy arises in TNIP1’s inhibitory effect on TNFα–R signaling and increasing pro-inflammatory cytokines in RA. The authors hypothesize that TNIP1 could regulate these molecules through a separate pathway distinct from TNFR [45]. Therefore, while several targets have been elucidated, it is plausible that other TNIP1-mediated pathways are yet to be discovered.
2.3 Experimentally altered TNIP1 levels and the resulting phenotypes
Building on the cell culture findings of NF-κB inhibition by TNIP1 overexpression, Beyaert and colleagues used adenoviral gene delivery of TNIP1 to test its effect on allergic airway inflammation [51]. Constructs expressing either TNIP1, the NF-κB inhibitor IκB (as a positive control), or a LacZ (as a negative control), were generated and delivered intratracheally to mice prior to irritant exposure. Comparing tissue histology, protein markers, and mRNA production, they demonstrated increased TNIP1 levels could, like the NF-κB inhibitor IkB, decrease sensitivity to the experimental irritant compared to animals receiving the negative control construct. These finding confirmed and extended the role of TNIP1 negatively regulating NF-κB-dependent gene expression associated with inflammation.
In addition to an anti-inflammatory effect, the protective role for TNIP1 has been extended to inhibition of apoptosis. TNIP1 overexpression from adenoviral constructs protected mice from TNFα/galactosamine (TNF/GalN) induced acute liver failure [52]. TNF/GalN-treated animals receiving control constructs showed massive sinusoidal hemorrhage, significant infiltration of neutrophils and macrophages, and increases in apoptotic markers such as caspase. However, these changes were significantly restricted or undetectable in mice over expressing TNIP1. This data suggests that TNIP1 has an anti-apoptotic function in addition to its regulation of inflammation. Importantly, NF-κB suppresses TNF-mediated hepatocyte apoptosis. This makes the hepato-protective effects of increased levels of TNIP1, with their expected inhibition of NF-κB, particularly intriguing. Thus, as Beyaert and colleagues point out, the hepato-protective effect of TNIP1 in this murine model of liver failure highlights an additional, anti-apoptotic function of TNIP1, possibly distinct from its NF-κB inhibition.
Using a TNIP1 knockout mouse line, Oshima and colleagues confirmed Beyaert’s results showing TNIP1 control over programmed cell death (PCD). TNIP1 deficient mice are smaller and present with anemia and hypocellular livers showing markers of apoptosis but not inflammation [11]. This phenotype is surprising in light of TNIP1’s suppression of NF-κB and presumably inflammation [5,6,53]. Additionally, although TNIP1- null mice developed in normal Mendelian ratios up to embryonic day 18.5, only ~ 1 in 40 mice were live-born suggesting TNIP1 is essential in the development of normal embryos. Interbreeding heterozygote TNIP1 mice with heterozygote TNF knockout mice resulted in animals born in normal ratios [11]. This suggests TNIP1 is involved in embryonic development, and has anti-apoptotic effects through regulation of TNFα signaling cascade in a similar but distinct way compared to TNFα-induced NF-κB activation.
Subsequent to Oshima et al., Zhou and colleagues also generated a TNIP1 knockout mouse line via the gene trap method [11]. Similar to prior observations, a decrease in the number of live-born homozygous null mice was noted (4.3% vs. expected 25%). However, new born live pup survival was increased to 10.3% when the C57BL/6 strain mice were backcrossed with the 129S2 strain, suggesting that the mouse genetic background influences TNIP1’s role in embryonic development. Interestingly, the live-born TNIP1 deficient mice die within 4 months. These mice present with enlarged lymph nodes and spleen, and develop an inflammatory disease similar to SLE. Deletion of the TNFα-R in addition to TNIP1 prevented the embryonic lethality. These mice, however still retained their inflammatory disease state, suggesting that the inflammation could be caused in a TNFα-R-independent pathway. Both these murine experiments indicate TNIP1 plays key roles in regulating TNFα-R dependent and TNFα-R independent pathways.
To contrast animal phenotypes caused by a lack of TNIP1 from those expressing a TNIP1 variant, Nanda and colleagues generated a TNIP1 knockin mouse where a single amino acid in TNIP1’s UBAN (ubiquitin binding in ABIN and NEMO) domain was mutated [25]. The inability of TNIP1 to bind ubiquitin chains led to the development of a lupus-like autoimmune disease in these knock-in mice. Different from the knockout mice, these knockin mice were the born in normal Mendelian ratios; examination of the spleen and lymph nodes, however, showed an enlargement of these organs. Within 5 months, all recombinant mice developed the autoimmune disease and were sacrificed by 6 months. Analysis of immunological mediators showed a TLR ligand-dependent increase in levels of serum immunoglobulins and autoantibodies. This SLE-like phenotype derives from an experimentally induced change in a conserved amino acid of the core UBAN sequence. Intriguingly, the mutation associated with a human large B cell lymphoma (see section 2.1 above) also mapped to the UBAN domain, although it occurred at a different amino acid in the core sequence [41].
Recently, TNIP1’s cytoprotective role was also reported using cultured Saos-2 cells [54]. Over-expression of TNIP1 provided cells with more resistance to treatment with the histone deacetylase inhibitor trichostatin-A. Zhang and colleagues observed that during the cell cycle, there was an M phase-dependent TNIP1 phosphorylation. Taking a cue from when in the cell cycle this modification arose and that the phosphorylated TNIP1 protein was degraded with the progression from M to G1 phase, the investigators hypothesized that TNIP1 phosphorylation may be an important event as a cell cycle protective factor. Presumably, the effects trichostatin A induces from accumulated acetylation impaired chromosome condensation that would be incompatible with cell cycle progression. The protection from chemically-induced apoptosis by TNIP1 over-expression may be the reciprocal of the increased sensitivity to cytokine- (TNFα) induced apoptosis in cells derived from TNIP1 null mice [11].
Taken together, these in vivo and in vitro models strongly suggest TNIP1 plays a crucial role in regulating several cellular pathways involved in inflammation and immune-related disorders (Table 2). The phenotypes arising from experimentally altering TNIP1 levels or ubiquitin binding are consistent with diseases correlated with TNIP1 from GWAS and expression arrays. Since expression array studies found increased TNIP1 levels in psoriasis and RA, these elevated levels could play a crucial role in reducing the pathogenesis of these diseases. We speculate that if TNIP1 protein amounts could be further increased, the inflammatory phenotype may be lessened, providing a potential treatment for these diseases.
Table 2.
Experimentally altered TNIP1 and the resulting phenotypes
| Experimental Model | Phenotype | Reference |
|---|---|---|
| Overexpression of WT TNIP1 in vivo via adenoviral tail vein delivery | Protection from TNFα/Galactosamine induced acute liver failure | El Bakkouri et al. [51] |
| Overexpression of WT TNIP1 in vivo via adenoviral intratracheal delivery | Protection from allergen induced airway inflammation | Wullaert et al. [52] |
| Mouse knockout by BAC recombineering and Cre mediated excision to delete sequences including exons 12–15 | 1 in 40 mice were live-born; Embryonic lethal at day 18.5; Anemic; Hypocellular livers; Increased apoptosis in embryonic livers | Oshima et al. [11] |
| Gene trap mutation mouse model | 1 in 40 mice were live-born; Embryonic lethal at day 18.5; Live- born mice die within 40 days post- birth; Enlarged lymph nodes and spleen | Zhou et al. [80] |
| Mouse knock-in model mutating TNIP1’s UBAN domain | Development of lupus-like autoimmune disease within 5 months; Enlarged lymph nodes and spleen; | Nanda et al. [25] |
| Overexpression of WT TNIP1 in vitro in Saos-2 osteosarcoma cells | Protection from trichostatin A induced apoptosis | Zhang et al. [54] |
2.4 TNIP1 modulation of HIV proteins and viral life cycle
While this review focuses on the role of TNIP1 via its interactions with other endogenous proteins, we would be remiss not to note the possible significant implications of its interactions with the HIV encoded proteins nef and matrix. Beyond these physical associations, early work demonstrated higher levels of TNIP1 increased levels of cell surface CD4; these CD4 amounts were decreased by co-expression of HIV Nef [1]. Additionally, TNIP1 interaction with HIV matrix suggested it might regulate the nuclear localization of that viral protein [2]. Better understanding of the opportunistic use of TNIP1 by these viral proteins awaits more in-depth investigation in such systems as has accrued for TNIP1 and TNFα, EGF-Rand nuclear receptor signaling. Until then, we note three potentially important intersections between TNIP1, its target proteins or signal pathways, and players in HIV pathogenesis or treatment: i)the HIV Nef increase in ERK enzymatic activity [55] and the physical association of ERK and TNIP1 [10], ii)the activation of NF-κB by Nef ([56] for review) and the dampening of NF-κB signaling by TNIP1, and iii)the shortcomings of PPARγ agonists in the treatment of HIV antiretroviral therapy-related lipoatrophy (highlighted in [57], see also [58]) and corepression of agonist-bound PPAR activity by TNIP1 [9]. Together, these overlaps suggest TNIP1’s functions regulating the activity of post-TNFα receptor signaling, NF-κB activation, ERK activity, and PPARγ function could have significant impact on cellular consequences of HIV infection and patient therapy.
3. TNIP1 functional motifs
3.1 Ubiquitin binding regions
It was within the post- TNFα-R pathway that TNIP1 was recognized as a protein-protein interaction partner of A20 (also named TNFAIP3) and, like A20 [59], an inhibitor of NF-κB signaling [3]. A later report [5] dissected this relationship and suggested that interaction of A20 and TNIP1 was not required for either’s inhibition of TNFα-activated NF-κB signaling. Nevertheless, a TNIP1-A20 functional cooperation may exist to form an A20-ubiquitin editing complex ([60] for review) or other functional interactions since A20 knockdown does decrease recombinant TNIP1’s ability to inhibit NF-κB [6]. Some constituents of the complex may be redundant in this important control step of NF-κB signaling and/or vary in relative expression levels dependent on cell type thereby accounting for differences in interpretation of protein function. These constituents include proteins controlling protein ubiquitination and/or recognizing ubiquitinated proteins. For the latter case, ubiquitin binding motifs, called UBAN (ubiquitin binding in ABIN and NEMO) domains, are present in TNIP1 [61] (Fig. 1). The UBAN domain, previously named ABIN homology domain 2 (AHD2), is present in all ABIN proteins [62] and NEMO. TNIP1’s similarities with the other ABIN family members, ABIN-2 and ABIN-3, extend past its homologous amino acid sequences. All three ABINs contain the UBAN domain, capable of binding ubiquitin chains to inhibit NF-κB activation.
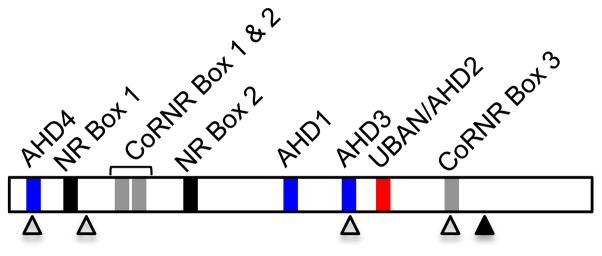
Figure 1.
Linear representation of TNIP1. Specific functional domains of TNIP1 are depicted. ABIN homology domains (AHD) 1, 3 and 4 are shown in blue. TNIP1’s ubiquitin binding domain, UBAN/AHD2, is shown in red and contains the DFXXER core sequence motif. The LXXLL nuclear receptor (NR) and LXXXI/LXXXI/L corepressor/nuclear receptor (CoRNR) boxes are shown in black and gray, respectively. The putative nuclear export sequences (NES) and nuclear localization sequences (NLS) are shown in open and filled triangles, respectively.
In addition to the presence of these motifs, TNIP1’s ability to bind ubiquitin proteins was shown through protein-protein interaction experiments [7,11,25]. It is this direct binding that facilitates TNIP1’s inhibitory effects on transmembrane receptor-mediated pathways. UBAN motifs, found between amino acids 461 to 483, are essential to the NF-κB inhibitory function of TNIP1. A core sequence within the domain, DFXXER (X = any amino acid), appears to be crucial for binding ubiquitin as mutations of these residues abolish TNIP1’s repressive effect [5,25,41].
3.2 Nuclear Receptor Interaction Motifs
Research in our laboratory has identified TNIP1 as a nuclear receptor (NR) coregulator. Coregulators physically associate with NRs and are key determinants of NR-mediated transcriptional activity. They are broadly subdivided into two functional classes – coactivators and corepressors – which either facilitate or inhibit the transcription of NR-targeted genes. To examine what coregulators may be present in human skin cells, we [63] conducted a yeast two-hybrid screen of a keratinocyte cDNA library using the carboxyl two-thirds of PPARκ which contain the contiguous ligand-binding domain and ligand-dependent activation function (LBD and AF-2, respectively). TNIP1 was one of the proteins isolated in that screen.
The TNIP1 amino acid sequence contains two types of potential interaction motifs for association with NRs, LXXLL (L = leucine, X = any amino acid), referred to as an NR box, and LXXXI/LXXXI/L (I = isoleucine), referred to as a corepressor (CoRNR) box (Fig. 1). Typically, NR boxes are associated with coactivators and CoRNR boxes with corepressors. Agonist binding causes a repositioning of the NR AF-2 domain resulting in the shedding of a corepressor and the recruitment of a coactivator [64]. Although TNIP1 contains both types of motifs, further experiments have characterized it as a corepressor of agonist-bound PPARs and RARs [8,9].
4. TNIP1 inhibits TNFα–R -induced signaling cascades
4.1 Overview of the TNFα–R-induced signaling cascades
TNFα binding to its receptor can trigger intracellular signaling consequences including NF-κB-dependent transcription and programmed cell death ([65,66] for reviews). For both TNFα-R induced outcomes, the activated receptor recruits and binds numerous intracellular proteins, including the kinase enzyme RIP1, forming a group of proteins named “Complex I”. To initiate the cascade for NF-κB activation, RIP1 is ubiquitinated thus activating its kinase activity. RIP1 activation facilitates the phosphorylation which results in the subsequent ubiquitination of the NF-κB essential modulator (NEMO, also named IKKγ), which is a subunit in the trimeric protein complex, the inhibitor NF-κB (IκB) kinase (IKK). The IKK complex is comprised of two additional kinase enzymes, IKKα and IKKβ. Ubiquitination of NEMO activates the associated kinase enzymes in the IKK complex, which results in the phosphorylation and eventual degradation of IκB, a protein which physically retains the homo-or hetero-dimeric NF-κB in the cytoplasm. Degradation of IκB releases NF-κB, exposing its nuclear localization sequences allowing the transcription factor to translocate to the nucleus and to bind NF-κB sites in responsive gene promoters (see Fig. 2).
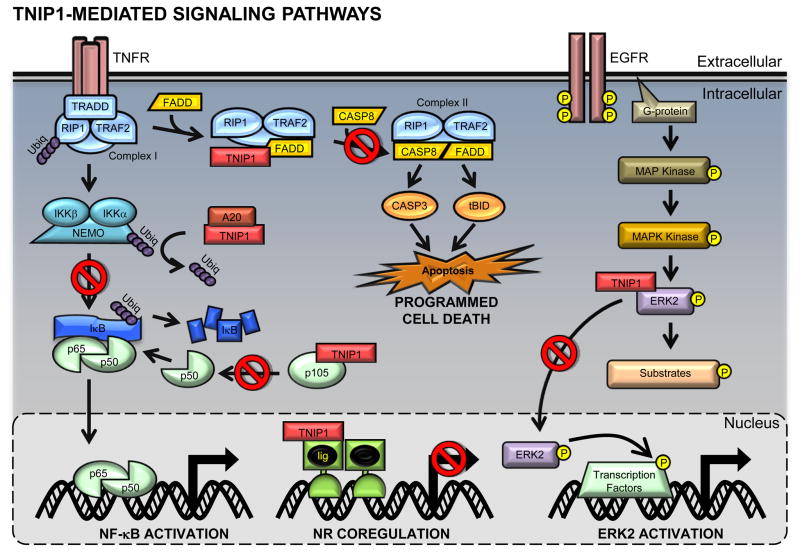
Figure 2.
TNIP1-mediated signaling pathways. The canonical NF-κB activation (left) and cell death (middle top) pathways are mediated by the TNFα-R. (Left) In the NF-κB pathway, intracellular proteins are recruited to the TNFα-R to form complex I, which facilitates the phosphorylation and subsequent activation and polyubiquitination of NEMO. IκB is then targeted for degradation, allowing the p65/p50 NF-κB heterodimer to translocate into the nucleus. TNIP1 inhibits NF-κB activation by preventing NEMO’s polyubiquitination. Additionally, TNIP1 blocks the processing of p105 to the NF-κB subunit p50, therefore decreasing the available pool of NF-κB. (Middle top) In the programmed cell death pathway, complex I dissociates from the TNFα-R and recruits FADD and caspase 8 to form complex II. Complex II mediates the activation of caspase 3 and tBid, leading to apoptosis. TNIP1 prevents the recruitment of caspase 8 to complex I which prevents the signaling cascade. (Right) EFGR initiates a MAP kinase pathway leading to the phosphorylation and activation of ERK2, a MAPKK kinase. ERK2 can phosphorylate cytosolic and nuclear substrates including various transcription factors. ERK2 nuclear translocation is inhibited by TNIP1. (Middle nucleus) TNIP1 represses nuclear receptors transcriptional activity. Upon ligand-NR binding, TNIP1 exerts its inhibitory effects on either PPAR or RAR. Red (□) denotes TNIP1’s inhibitory functions.
TNFα-R mediated programmed cell death begins with TNFα-R binding to its transmembrane receptor followed by the intracellular recruitment of various proteins to the receptor, forming Complex I. Deviating from the NF-κB activating pathway, this group of proteins dissociates from the TNFα-R. Complex I then recruits the fas associated death domain (FADD), which facilitates the binding and activation of procaspase 8 to caspase 8, forming the aptly named Complex II. Complex II further mediates the cleavage and activation of pro-apoptotic enzymes, such as caspase 3 and tBid. tBid interacts with the mitochondria, disrupting its plasma membrane, allowing the release of more pro-apoptotic mitochondrial proteins such as cytochrome c and Smac/DIABLO into the cytoplasm further leading to the activation of caspases, DNA cleavage, and eventual cell death [67] (see Fig. 2). The ultimate fate of this signaling pathway depends on the intracellular proteins available during TNF-R activation.
4.2 TNIP1 protein-protein interactions and NF-κB activation (NEMO, p105 & TNFα-R)
TNFα-R-mediated NF-κB activation is largely dependent on a series of phosphorylation and ubiquitination steps on various proteins. Typically, ubiquitination is part of the proteasome-mediated protein degradation pathway; however, there are proteins, including NEMO, whose ubiquitination results not only in protein breakdown but also in protein-protein interaction and enzyme activation [60]. Although not an enzyme itself, ubiquitinated NEMO interacts with IKKα and IKKβ in the IKK complex and facilitates activation of these kinases. The ubiquitination of NEMO is essential in this cascade because without it, the subsequent degradation of IκB and release of NF-κB will be prevented. TNIP1 over-expression inhibits NF-κB signaling downstream of TRAF2, apparently at the level of IKK (see Fig. 2), specifically NEMO. There is a direct physical interaction [6] between TNIP1 and NEMO (in addition to TNIP1 and A20) and with TNIP1 levels experimentally increased, A20-mediated removal of ubiquitin from NEMO is likely facilitated, decreasing the activity of the IKK complex, blocking NF-κB gene regulation [6,7].
In addition to the interaction with NEMO, TNIP1 can also prevent NF-κB activation through decreasing the pool of one of the NF-κB subunits -- p50. NF-κB is a homo- or hetero-dimeric transcription factor consisting of proteins in the Rel family. The p50 and p65 complex is the most common NF-κB dimer, with the p50 subunit derived from proteolytic processing of the precursor, and IκB protein, p105. Endogenous [68] and overexpressed [53] TNIP1 was found to bind and inhibit the processing of p105 resulting in a reduction of active p50. While the two proteins can physically interact, this is not an absolute requirement for the effect on p105 (see Fig. 2). Interestingly, for any effect TNIP1 may have on intracellular signaling, increases in p105 expression significantly increased TNIP1 half-life. This protein-protein interaction could prevent NF-κB activation in two ways: (1)decreasing available p50 to form an active NF-κB dimer and (2)increasing TNIP1 expression to prevent IKK activation.
Further upstream of NEMO or Complex II, TNIP1 was found to interact with the TNFα–R. Haas and colleagues identified the various intracellular proteins recruited post TNFα–R ligand binding, including the IKK trimeric complex and TNIP1. Although the specific details of how TNIP1 associates with the complex were not elucidated, mechanisms of NEMO’s association in this complex were discussed. NEMO’s ubiquitin binding domain, UBAN, facilitates the recruitment of the complex to other ubuquitinated TNFα–R bound proteins, such as RIP1 and TNFα–R associated factor 2 (TRAF2). Given that TNIP1 also has the same UBAN domain, it is likely that its presence in the TNFα–R complex is mediated through TNIP1’s ability to bind ubiquitin chains.
Through physical association with NEMO, p105 and the TNFα–R, the molecular mechanisms of TNIP1’s function to inhibit NF-κB-dependent gene transcription may explain its potential role in inflammatory and immune-related diseases. Deregulation of this pathway can result in a myriad of diseases and disorders, including but not limited to the progression of arthritis and psoriasis, and yet, controlled TNFα–R signaling can lead to differentiation and immunomodulation in equally diverse cell types [65,66]. As previously mentioned for leukemia-lymphoma [32,41] and psoriasis [13,33], TNIP1 association with disease states need not be limited to variants in its protein sequence. Wild-type TNIP1 could still play a key role in pathologies or as a pharmacologic target if its levels were altered, as in this case subsequent to increases in p105.
4.3 Control over programmed cell death post-TNFα–R signaling
Additional in vitro studies using TNIP1 knockout- or wild type-derived mouse embryonic fibroblasts (MEF) and three TNIP1 deficient human cell lines were performed to further characterize the specific molecular interactions in which TNIP1 is involved. These cell culture experiments mirrored the results observed from whole animal studies [11]. As observed by Oshima et al., mice lacking both TNIP1 and TNF exhibited hypocellular livers due to increased apoptosis; human and mouse cell lines deficient in TNIP1 were prone to TNFα-induced PCD. Conversely, exogenous TNIP1 expression into these cells recovered the normal phenotype. Specifically, the presence of TNIP1 prevented the activation of apoptosis regulating enzymes tBid, caspase 3 and caspase 8, which are downstream targets of the TNFα cascade. Similar to TNFα-mediated NF-κB activation, initiation of PCD by TNFα involves the recruitment of numerous intracellular proteins to the TNFα-R. Deviating from the prior NF-κB activating cascade, these intracellular proteins dissociate from the receptor to recruit the Fas associated death domain (FADD), which facilitates recruiting and activating caspase 8 from pro-caspase 8. This secondary protein complex mediates the cleavage and activation of additional pro-apoptotic intracellular proteins such as Bid to tBid and procaspase 3 to caspase 3. tBid interacts with the mitochondria, disrupting its plasma membrane, allowing the release of more pro-apoptotic mitochondrial proteins such as cytochrome c and Smac/DIABLO into the cytoplasm further leading to the activation of caspases, DNA cleavage, and eventual cell death [67] (see Fig. 2). Protein-protein interaction analysis show TNIP1 inhibits secondary protein complex formation by blocking the FADD and caspase 8 interactions (see Fig. 2). These experiments revealed TNIP1 is involved in inhibiting apoptosis early in the TNF signaling pathway [11].
4.4 TNIP1 anti-apoptotic roles are unexpected from prior TNIP1-NF-κB studies
A discord in determining TNIP1’s function arises comparing the results observed from Oshima et al. and previous published data. Prior in vitro data concluded TNIP1 inhibits TNFα-mediated NF-κB activation [3,6], whereas the new in vivo and in vitro research suggest TNIP1’s inhibitory effect is seen in TNFα-mediated programmed cell death [11]. It is possible that TNIP1 has the capability of blocking both pathways, but favors inhibition of PCD depending on which intracellular proteins are available to regulate the cascade. The presence of intracellular proteins could be a result of the different cell types used. Earlier NF-κB related research used HEK293 cells and later experiments used Jurkat T cells, HepG2 hepatoma cells and HT1080 fibrosarcoma cells, in addition to MEF’s. The intracellular proteins available in each cell type may play a factor in determining TNIP1’s regulatory function.
Apoptosis plays a key role in the physiology of many tissues and cells, both in normal and diseased states. Keratinocyte differentiation, for example, depends on cell death to make the necessary epidermal layers to produce the outermost barrier layer, the stratum corneum, which protects our bodies from vast physical or chemical insults and injuries [69]. Aberrant keratinocyte cell death leads to hyperproliferation of the skin, which could contribute to the progression of psoriasis [70]. Since keratinocyte hyperproliferation is observed in psoriatic lesions, the increased TNIP1 levels in lesioned psoriatic skin [13] may be inhibiting the TNFα-induced PCD therefore contributing to the progression of the disease.
5. TNIP1 control over the epidermal growth factor receptor pathway
5.1 TNIP1 blocks NF-κB activation
Although the canonical NF-κB activation occurs through the TNFα-R pathway, the epidermal growth factor receptor (EFGR) pathway can also initiate NF-κB translocation through an as yet uncharacterized mechanism. However, it is known that the induction of NF-κB by EGF-R is independent of IκB degradation, thus a different pathway involving different regulatory proteins is mediating its activation. Similar to the TNFα-R cascade, TNIP1 prevents NF-κB dependent gene regulation in cells stimulated with EGF [71]. Because the induction of NF-κB by EGF-R is independent of IκB degradation, TNIP1 is likely interacting with different, yet determined protein(s) in the EGF-R pathway. Interestingly, TNIP1’s UBAN domain plays a functional role in this inhibition. Mutations in the DFXXER sequence abolish TNIP1inhibitory effects, suggesting that ubiquitin binding is necessary for this function. These results, along with data involving TNFα-R, demonstrate that TNIP1 may regulate inflammation through more than one mechanism. Furthermore, there appears to be a redundancy in TNIP1 function to prevent NF-κB activation regardless of which post-receptor signaling initiates its nuclear translocation.
5.2 TNIP1 blocks ERK2 signaling
TNIP1’s regulation of the EGF-R pathway was extended to its inhibition of extracellular signal-regulated kinase (ERK) 2 nuclear translocation. ERK [72] is one cytoplasmic stepping stone in a signaling pathway initiated by activation of cell membrane receptors such as G-protein coupled receptors, EGF-R, and integrins. These receptors translate extracellular signals through the cytoplasm via the sequential phosphorylation and therefore activation of Raf, MEK and eventually ERK. Phosphorylated (i.e., enzymatically activated) ERK can be found both in the cytoplasm and nucleus, increasing opportunity for it to in turn phosphorylate more than 100 target proteins, thus regulating a multitude of physiological processes including cell proliferation, migration and death [72]. Over-expression of TNIP1 in osteosarcoma (Saos-2) cells [10] inhibited the nuclear translocation of EGF-activated ERK2 recalling the suggestion from Gupta and colleagues [2] that increased levels of TNIP1 may titrate out cellular factors necessary for its ability to shuttle into the nucleus. This TNIP1-induced stalling of ERK in the cytoplasm reduced transactivation of ERK-dependent reporter constructs. Coincident and perhaps as a direct result of the TNIP1-ERK2 interaction, over-expressed TNIP1 itself was phosphorylated subsequent to EGF stimulation [10] (Fig. 2).
TNIP1 reduction of ERK2 signaling and translocation to the nucleus might also have a collateral effect on TNFα signaling. ERK2 activation promotes the translocation of TNFα mRNA from the nucleus into the cytoplasm [73,74]. With ERK2 signaling lessened by TNIP1, it will be important to formally test if TNFα production is decreased and possibly its promotion of inflammatory disease states e.g., psoriasis. It is possible the increase in TNIP1 expression seen in psoriasis [13] and the experimental increase in TNIP1 brought about by TNFα exposure [75,76] or recombinant expression [43] of NF-κB may be a compensatory mechanism to negatively regulate inflammatory signals.
6. TNIP1 represses agonist-bound PPARs and RARs
6.1 TNIP1 is a PPAR and RAR Corepressor
The isolation and characterization of TNIP1 under the alias VAN (virion-associated matrix-interacting protein), a human protein possibly involved with the HIV replication cycle, and on its own, possessing a nuclear-cytoplasmic shuttling ability [2] raised the question of a nuclear function for TNIP1 in absence of HIV infection. Since TNIP1 localization was determined in both cytoplasmic and nuclear compartments [26], we became interested in determining TNIP1’s possible role in the nucleus.
In a series of mammalian two-hybrid studies, we observed that TNIP1 interacted with nuclear receptors PPARs and RARs only in the presence of agonist ligand [8,9], suggesting TNIP1 functions as a coactivator. PPAR and RAR interaction with TNIP1 is strictly dependent on the integrity of the NR AF-2 as conservative amino acid substitutions within this domain completely abolished TNIP1 interaction, a coregulator characteristic again suggestive of TNIP1 as a coactivator. However, mutagenesis of the NR boxes in TNIP1 complicated its coactivator candidacy. Conservative amino acid substitutions in either of the TNIP1 NR boxes significantly reduced its interaction with PPARs, with an additive effect observed when both were mutated equaling about 50% reduction [9]. For its interaction with RARs, only the mutation of both TNIP1 NR boxes had a significant effect leading to about a 30% decrease in mammalian two-hybrid determined interaction [8]. Beyond NR boxes, we observed that for both PPAR and RAR interaction the central portion of TNIP1 between amino acids 206 and 418 played an important role despite the absence of recognizable NR box motifs. For the NRs tested to date, the interaction of TNIP1 with them is neither universal nor equal. TNIP1 exhibits a strong subtype preference amongst PPARs (γ > δ ≫> α) and RARs (α ≫ γ) [8,9]. There is no interaction between TNIP1 and retinoid X receptor (RXR) α, the common obligate heterodimer partner of both PPARs and RARs, regardless of presence of RXR agonist.
Characteristics of TNIP1-NR interaction – requirement for ligand presence, requirement for the integrity of receptor AF-2 domain, the partial dependence on TNIP1 NR boxes – all suggest TNIP1 as a NR coactivator. We were somewhat surprised then to see that in the presence of the respective NR agonists, TNIP1 attenuated the activity of both PPARs and RARs [8,9]. TNIP1, however, had no effect on the activities of estrogen receptors α and β, androgen receptor, and progesterone receptor (Encarnacao and Aneskievich, unpublished). Although increased expression of TNIP1 decreases PPAR and RAR activity, the protein levels of these receptors does not decrease [8,9,26] supporting the interpretation that the repressive effect was due to the alteration in NR transcriptional activity by TNIP1, i.e., that TNIP1 is a bona fide corepressor of agonist-bound PPAR and RAR.
6.2 Towards resolving the role(s) of a corepressor of agonist-bound PPAR and RAR
As a NR coregulator, TNIP1 is in a still relatively small class of corepressors of agonist-bound NRs exemplified by this group’s archetype, receptor interacting protein 140 (RIP140) [77]. This group is distinct from the better-known typical corepressors e.g., nuclear receptor corepressor (NCoR) and silencing mediator of retinoid and thyroid receptors (SMRT) which associate with NRs in the absence of ligand and abolish the receptor activity entirely [64]. Rather, these corepressors of agonist-bound NRs attenuate receptor activity acting as something of an emergency brake in cases of excessive receptor activation either by toxic ligand levels or exposure to the ligand at inappropriate times. Additionally, even under normal ligand conditions, they may contribute to a combinatorial approach to NR regulation, providing for a finer level of control over receptor activity instead of the all-on or all-off effect of typical coactivators or corepressors.
Many corepressors of agonist-bound NRs contain both NR and CoRNR boxes [78]; TNIP1 fits this pattern. However, their mechanisms of NR repression vary. Unlike what has been reported for RIP140 [79], we found no association between TNIP1 and chromatin remodeling histone deacetylase (HDAC) enzymes. TNIP1 repression is partially relieved by over-expression of the NR coactivator steroid receptor coactivator 1 (SRC1) suggesting that interference with coactivator recruitment by liganded NRs is a mechanism of TNIP1 repression [8]. We found the transcriptional repression effect of TNIP1 was mostly localized to its carboxyl-terminus and surprisingly, that its amino terminus could activate transcription [9]. With such traits, TNIP1 may be a coregulator of mixed function akin to other such proteins including NR binding SET domain containing protein 1 (NSD1) and comodulator of PPAR and RXRα 1(COPR1) which act as a coactivator and corepressor, respectively, yet possess separable activation and repression domains [63,71,78].
7. TNIP1 and TLR signaling
Additional receptor-mediated signaling pathways regulating inflammation may also be controlled by TNIP1. Most recently, these include a role for TNIP1 in toll-like receptor (TLR) signaling [25] likely via a complex with MyD88 [80], an essential signal transducer in the TLR pathway. Using TNIP1 knockout mouse bone marrow-derived macrophages, Zhou and colleagues determined that absence of TNIP1 had the functional consequence of increased mRNA expression and DNA binding of the transcription factor CCAAT/enhancer-binding protein β (C/EBPβ) post TLR stimulation, resulting in the increased expression of C/EBPβ target genes. Given that TLR signaling has been implicated to play a role in inflammatory diseases, including psoriasis [81], SSc [82], SLE and RA [83], TNIP1’s regulation of the TLR pathway could likely contribute to the pathogenesis of these diseases.
8. Conclusions & perspectives
TNIP1 appears to play important roles in regulating multiple receptor mediated signal pathways— from the membrane bound TNFα-R [3,6,11], EGF-R [10], and TLR [25,80] signaling cascades to modulating the transcriptional activity of nuclear PPAR [9] and RAR [8]. Additionally, TNIP1 has been implicated in several disease states through changes in protein expression [12,13,18] and/or gene SNPs [13–15,17,18], which could alter its transcription or TNIP1 mRNA splicing [31,32]. Though TNIP1’s specific role has not been identified in these diseases, we postulated mechanisms of TNIP1’s potential roles in either the progression or regression of these disorders. In psoriasis, increased TNIP1 protein expression could be abrogating the TNFα–induced PCD pathway instead of the pro-inflammatory NF-κB signaling, consequently leading to hyperproliferation and increased inflammation typical of psoriatic lesions. This hypothesis could account for the discord in increased levels of NF-κB and TNIP1, an NF-κB inhibitor, as seen in psoriatic inflammation.
However, another potential explanation for increased levels of both TNIP1 and NF-κB could be that both are involved in a negative feedback regulation loop. In transcriptional analysis studies, several NF-κB binding sites have been elucidated in TNIP1’s promoter [75] and [84]. Additionally, Tian and colleagues concluded that increased TNIP1 expression is delayed post-TNFα stimulation. We suggest that NF-κB activation primarily activates pro-inflammatory molecules, thus progressing these diseases. TNIP1 upregulation could be a secondary event arising from this transcription factor’s activation, and TNIP1 could then partially alleviate inflammation.
Furthermore, TNIP1 could be involved in regulating inflammation through regulation of other receptors, namely PPAR [9], RAR [8], and TLR [25,80]. Both PPAR and RARs are pharmacologic targets used or proposed to treat inflammation [47–50], whereas TLRs have gained attention as potential targets for immunotherapeutics [85]. TNIP1 may then regulate inflammation as a NR corepressor and/or inhibitor of C/EBPβ activation, preventing the transcription of these inflammatory proteins. Depending on the function of these target genes, the pathogenesis of these inflammatory diseases could be greatly affected by TNIP1.
In conclusion, while this review summarizes the various experiments defining TNIP1’s roles regulating receptor mediated events and presents GWAS studies implicating TNIP1 in psoriasis, SLE and RA, the compiled research to date has yet to determine the exact role this regulator plays in normal physiology or the progression of these diseases. Much work remains to determine whether the identified or yet potential undiscovered TNIP1-modulated pathways are responsible for regulating inflammation. It is likely that further work regarding TNIP1 will establish it as a significant regulator of these pathways and diseases.
Acknowledgments
We thank Dr. C. Giardina for critical review of the manuscript. We appreciate the efforts of the journal reviewers and editor for helpful suggestions in improving this review. Research from the authors’ laboratory was supported by the National Institutes of Health (NIAMS Grant No. AR048860, BJA); pre-doctoral fellowships from Boehringer–Ingelheim Pharmaceuticals (IG) and the American Foundation for Pharmaceutical Education (IG) provided partial stipend support. The University of Connecticut Center for Regenerative Biology Excellence in Graduate Research Award (VPR) and the Edward A. Khairallah Graduate Fellowship (IG and VPR) provided partial summer support.
Biographies

Vincent P. Ramirez is a Ph.D. candidate in the Department of Pharmaceutical Sciences at the University of Connecticut. He received his B.S. in Biochemistry & Molecular Biology at the University of California, Davis. Prior to starting his graduate studies, he worked at a pharmaceutical company as a Research Associate in the DMPK department. For his Ph.D thesis, his main focus is determining the molecular mechanisms of TNIP1 influence over keratinocyte homeostasis and differentiation. He is a recipient of a Khairallah Graduate Fellowship, a 2012 Graduate Student Travel Support Award from the Society of Toxicology, and a 2012 Batelle Research Award for graduate students in dermal toxicology.

Igor Gurevich, Ph.D, completed his undergraduate degree at University of Massachusetts Amherst and then worked for four years as a Research Technician in the drug metabolism and pharmacokinetics group at a contract research organization. During this time he became interested in the drug discovery process and decided to pursue a doctoral degree. He completed his Ph.D. in Pharmaceutical Sciences with a specialty in toxicology under the guidance of Dr. Brian J. Aneskievich at the University of Connecticut School of Pharmacy. His Ph.D. research focused on characterizing TNIP1 as a coregulator of retinoid receptors and the transcriptional regulation of TNIP1 gene expression. He is now a postdoctoral fellow with Dr. Theodore P. Rasmussen studying prevention of spontaneous epigenetic drift in human ES and iPS cells.

Brian J. Aneskievich, Ph.D., received his doctorate from the State University of New York at Stony Brook in cell and developmental biology under the mentorship of Lorne Taichman and then completed a post-doctoral fellowship with Elaine Fuchs at the University of Chicago. He is now Associate Professor of Pharmacology at the University of Connecticut School of Pharmacy and member of the University Stem Cell Institute. His research covers molecular control of keratinocyte replication and differentiation, particularly by nuclear receptors, their ligands, and their coregulatory proteins. As Principal Investigator, his laboratory has been funded by the National Institutes of Health, including a FIRST Award from the NIAMS, the Skin Cancer Foundation Henry Shotmeyer Research Award, the American Institute for Cancer Research, and the Connecticut Stem Cell Initiative through CT Innovations.
Footnotes
Conflict of Interest
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fukushi M, Dixon J, Kimura T, Tsurutani N, Dixon MJ, Yamamoto N. Identification and cloning of a novel cellular protein Naf1, Nef-associated factor 1, that increases cell surface CD4 expression. FEBS Lett. 1999;442:83–8. doi: 10.1016/s0014-5793(98)01631-7. [DOI] [PubMed] [Google Scholar]
- 2.Gupta K, Ott D, Hope TJ, Siliciano RF, Boeke JD. A human nuclear shuttling protein that interacts with human immunodeficiency virus type 1 matrix is packaged into virions. J Virol. 2000;74:11811–24. doi: 10.1128/jvi.74.24.11811-11824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heyninck K, De Valck D, Vanden Berghe W, Van Criekinge W, Contreras R, Fiers W, et al. The zinc finger protein A20 inhibits TNF-induced NF-kappaB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-kappaB-inhibiting protein ABIN. J Cell Biol. 1999;145:1471–82. doi: 10.1083/jcb.145.7.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beyaert R, Heyninck K, Van Huffel S. A20 and A20-binding proteins as cellular inhibitors of nuclear factor-kappa B-dependent gene expression and apoptosis. Biochem Pharmacol. 2000;60:1143–51. doi: 10.1016/s0006-2952(00)00404-4. [DOI] [PubMed] [Google Scholar]
- 5.Heyninck K, Kreike MM, Beyaert R. Structure-function analysis of the A20-binding inhibitor of NF-kappa B activation, ABIN-1. FEBS Lett. 2003;536:135–40. doi: 10.1016/s0014-5793(03)00041-3. [DOI] [PubMed] [Google Scholar]
- 6.Mauro C, Pacifico F, Lavorgna A, Mellone S, Iannetti A, Acquaviva R, et al. ABIN-1 binds to NEMO/IKKgamma and co-operates with A20 in inhibiting NF-kappaB. J Biol Chem. 2006;281:18482–8. doi: 10.1074/jbc.M601502200. [DOI] [PubMed] [Google Scholar]
- 7.Wagner S, Carpentier I, Rogov V, Kreike M, Ikeda F, Lohr F, et al. Ubiquitin binding mediates the NF-kappaB inhibitory potential of ABIN proteins. Oncogene. 2008;27:3739–45. doi: 10.1038/sj.onc.1211042. [DOI] [PubMed] [Google Scholar]
- 8.Gurevich I, Aneskievich BJ. Liganded RARalpha and RARgamma interact with but are repressed by TNIP1. Biochem Biophys Res Commun. 2009;389:409–14. doi: 10.1016/j.bbrc.2009.08.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flores AM, Gurevich I, Zhang C, Ramirez VP, Devens TR, Aneskievich BJ. TNIP1 is a corepressor of agonist-bound PPARs. Arch Biochem Biophys. 2011 doi: 10.1016/j.abb.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang S, Fukushi M, Hashimoto S, Gao C, Huang L, Fukuyo Y, et al. A new ERK2 binding protein, Naf1, attenuates the EGF/ERK2 nuclear signaling. Biochem Biophys Res Commun. 2002;297:17–23. doi: 10.1016/s0006-291x(02)02086-7. [DOI] [PubMed] [Google Scholar]
- 11.Oshima S, Turer EE, Callahan JA, Chai S, Advincula R, Barrera J, et al. ABIN-1 is a ubiquitin sensor that restricts cell death and sustains embryonic development. Nature. 2009;457:906–9. doi: 10.1038/nature07575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallagher J, Howlin J, McCarthy C, Murphy EP, Bresnihan B, FitzGerald O, et al. Identification of Naf1/ABIN-1 among TNF-alpha-induced expressed genes in human synoviocytes using oligonucleotide microarrays. FEBS Lett. 2003;551:8–12. doi: 10.1016/s0014-5793(03)00823-8. [DOI] [PubMed] [Google Scholar]
- 13.Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, Goldgar D, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41:1234–7. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 15.Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41:1228–33. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He CF, Liu YS, Cheng YL, Gao JP, Pan TM, Han JW, et al. TNIP1, SLC15A4, ETS1, RasGRP3 and IKZF1 are associated with clinical features of systemic lupus erythematosus in a Chinese Han population. Lupus. 2010;19:1181–6. doi: 10.1177/0961203310367918. [DOI] [PubMed] [Google Scholar]
- 17.Kawasaki A, Ito S, Furukawa H, Hayashi T, Goto D, Matsumoto I, et al. Association of TNFAIP3 interacting protein 1, TNIP1 with systemic lupus erythematosus in a Japanese population: a case-control association study. Arthritis Res Ther. 2010;12:R174. doi: 10.1186/ar3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allanore Y, Saad M, Dieude P, Avouac J, Distler JH, Amouyel P, et al. Genome-Wide Scan Identifies TNIP1, PSORS1C1, and RHOB as Novel Risk Loci for Systemic Sclerosis. PLoS Genet. 2011;7:e1002091. doi: 10.1371/journal.pgen.1002091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farber EM, Nall ML, Watson W. Natural history of psoriasis in 61 twin pairs. Arch Dermatol. 1974;109:207–11. [PubMed] [Google Scholar]
- 20.Deapen D, Escalante A, Weinrib L, Horwitz D, Bachman B, Roy-Burman P, et al. A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis Rheum. 1992;35:311–8. doi: 10.1002/art.1780350310. [DOI] [PubMed] [Google Scholar]
- 21.Gudjonsson JE, Elder JT. Psoriasis: epidemiology. Clin Dermatol. 2007;25:535–46. doi: 10.1016/j.clindermatol.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 22.Rhodes B, Vyse TJ. The genetics of SLE: an update in the light of genome-wide association studies. Rheumatology (Oxford) 2008;47:1603–11. doi: 10.1093/rheumatology/ken247. [DOI] [PubMed] [Google Scholar]
- 23.Perdriger A, Werner-Leyval S, Rollot-Elamrani K. The genetic basis for systemic lupus erythematosus. Joint Bone Spine. 2003;70:103–8. doi: 10.1016/s1297-319x(03)00007-1. [DOI] [PubMed] [Google Scholar]
- 24.Feghali-Bostwick CA. Genetics and proteomics in scleroderma. Curr Rheumatol Rep. 2005;7:129–34. doi: 10.1007/s11926-005-0065-0. [DOI] [PubMed] [Google Scholar]
- 25.Nanda SK, Venigalla RK, Ordureau A, Patterson-Kane JC, Powell DW, Toth R, et al. Polyubiquitin binding to ABIN1 is required to prevent autoimmunity. J Exp Med. 2011;208:1215–28. doi: 10.1084/jem.20102177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gurevich I, Zhang C, Francis N, Aneskievich BJ. TNIP1, a Retinoic Acid Receptor Corepressor and A20-binding Inhibitor of NF-kappaB, Distributes to Both Nuclear and Cytoplasmic Locations. J Histochem Cytochem. 2011;59:1101–12. doi: 10.1369/0022155411427728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nestle FO, Kaplan DH, Barker J. Psoriasis. N Engl J Med. 2009;361:496–509. doi: 10.1056/NEJMra0804595. [DOI] [PubMed] [Google Scholar]
- 28.Mease PJ. Psoriatic arthritis: update on pathophysiology, assessment and management. Ann Rheum Dis. 2011;70 (Suppl 1):i77–84. doi: 10.1136/ard.2010.140582. [DOI] [PubMed] [Google Scholar]
- 29.Bowes J, Orozco G, Flynn E, Ho P, Brier R, Marzo-Ortega H, et al. Confirmation of TNIP1 and IL23A as susceptibility loci for psoriatic arthritis. Ann Rheum Dis. 2011 doi: 10.1136/ard.2011.150102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellinghaus E, Stuart PE, Ellinghaus D, Nair RP, Debrus S, Raelson JV, et al. Genome-Wide Meta-Analysis of Psoriatic Arthritis Identifies Susceptibility Locus at REL. J Invest Dermatol. 2012;132:1133–40. doi: 10.1038/jid.2011.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Favre M, Butticaz C, Stevenson B, Jongeneel CV, Telenti A. High frequency of alternative splicing of human genes participating in the HIV-1 life cycle: a model using TSG101, betaTrCP, PPIA, INI1, NAF1, and PML. J Acquir Immune Defic Syndr. 2003;34:127–33. doi: 10.1097/00126334-200310010-00002. [DOI] [PubMed] [Google Scholar]
- 32.Shiote Y, Ouchida M, Jitsumori Y, Ogama Y, Matsuo Y, Ishimaru F, et al. Multiple splicing variants of Naf1/ABIN-1 transcripts and their alterations in hematopoietic tumors. Int J Mol Med. 2006;18:917–23. [PubMed] [Google Scholar]
- 33.Strange A, Capon F, Spencer CC, Knight J, Weale ME, et al. Genetic Analysis of Psoriasis Consortium & the Wellcome Trust Case Control Consortium 2. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42:985–90. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graham RR, Cotsapas C, Davies L, Hackett R, Lessard CJ, Leon JM, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:1059–61. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kochi Y, Okada Y, Suzuki A, Ikari K, Terao C, Takahashi A, et al. A regulatory variant in CCR6 is associated with rheumatoid arthritis susceptibility. Nat Genet. 2010;42:515–9. doi: 10.1038/ng.583. [DOI] [PubMed] [Google Scholar]
- 36.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–14. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tung YC, Yeo GS. From GWAS to biology: lessons from FTO. Ann N Y Acad Sci. 2011;1220:162–71. doi: 10.1111/j.1749-6632.2010.05903.x. [DOI] [PubMed] [Google Scholar]
- 39.Berulava T, Horsthemke B. The obesity-associated SNPs in intron 1 of the FTO gene affect primary transcript levels. Eur J Hum Genet. 2010 doi: 10.1038/ejhg.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uddin M, Sturge M, Rahman P, Woods MO. Autosome-wide Copy Number Variation Association Analysis for Rheumatoid Arthritis Using the WTCCC High-density SNP Genotype Data. J Rheumatol. 2011;38:797–801. doi: 10.3899/jrheum.100758. [DOI] [PubMed] [Google Scholar]
- 41.Dong G, Chanudet E, Zeng N, Appert A, Chen YW, Au WY, et al. A20, ABIN-1/2, and CARD11 mutations and their prognostic value in gastrointestinal diffuse large B-cell lymphoma. Clin Cancer Res. 2011;17:1440–51. doi: 10.1158/1078-0432.CCR-10-1859. [DOI] [PubMed] [Google Scholar]
- 42.Orozco G, Eyre S, Hinks A, Bowes J, Morgan AW, Wilson AG, et al. Study of the common genetic background for rheumatoid arthritis and systemic lupus erythematosus. Ann Rheum Dis. 2011;70:463–8. doi: 10.1136/ard.2010.137174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hinata K, Gervin AM, Jennifer Zhang Y, Khavari PA. Divergent gene regulation and growth effects by NF-kappa B in epithelial and mesenchymal cells of human skin. Oncogene. 2003;22:1955–64. doi: 10.1038/sj.onc.1206198. [DOI] [PubMed] [Google Scholar]
- 44.Halvorsen M, Martin JS, Broadaway S, Laederach A. Disease-associated mutations that alter the RNA structural ensemble. PLoS Genet. 2010;6:e1001074. doi: 10.1371/journal.pgen.1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Igarashi H, Yahagi A, Saika T, Hashimoto J, Tomita T, Yoshikawa H, et al. A pro-inflammatory role for A20 and ABIN family proteins in human fibroblast-like synoviocytes in rheumatoid arthritis. Immunol Lett. 2011 doi: 10.1016/j.imlet.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 46.Dinarello CA. Anti-inflammatory Agents: Present and Future. Cell. 2010;140:935–50. doi: 10.1016/j.cell.2010.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sbidian E, Maza A, Montaudie H, Gallini A, Aractingi S, Aubin F, et al. Efficacy and safety of oral retinoids in different psoriasis subtypes: a systematic literature review. J Eur Acad Dermatol Venereol. 2011;25 (Suppl 2):28–33. doi: 10.1111/j.1468-3083.2011.03993.x. [DOI] [PubMed] [Google Scholar]
- 48.Michalik L, Wahli W. Peroxisome proliferator-activated receptors (PPARs) in skin health, repair and disease. Biochim Biophys Acta. 2007;1771:991–8. doi: 10.1016/j.bbalip.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 49.Schmuth M, Jiang YJ, Dubrac S, Elias PM, Feingold KR. Thematic review series: skin lipids. Peroxisome proliferator-activated receptors and liver X receptors in epidermal biology J Lipid Res. 2008;49:499–509. doi: 10.1194/jlr.R800001-JLR200. [DOI] [PubMed] [Google Scholar]
- 50.Huang W, Glass CK. Nuclear receptors and inflammation control: molecular mechanisms and pathophysiological relevance. Arterioscler Thromb Vasc Biol. 2010;30:1542–9. doi: 10.1161/ATVBAHA.109.191189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El Bakkouri K, Wullaert A, Haegman M, Heyninck K, Beyaert R. Adenoviral gene transfer of the NF-kappa B inhibitory protein ABIN-1 decreases allergic airway inflammation in a murine asthma model. J Biol Chem. 2005;280:17938–44. doi: 10.1074/jbc.M413588200. [DOI] [PubMed] [Google Scholar]
- 52.Wullaert A, Wielockx B, Van Huffel S, Bogaert V, De Geest B, Papeleu P, et al. Adenoviral gene transfer of ABIN-1 protects mice from TNF/galactosamine-induced acute liver failure and lethality. Hepatology. 2005;42:381–9. doi: 10.1002/hep.20785. [DOI] [PubMed] [Google Scholar]
- 53.Cohen S, Ciechanover A, Kravtsova-Ivantsiv Y, Lapid D, Lahav-Baratz S. ABIN-1 negatively regulates NF-kappaB by inhibiting processing of the p105 precursor. Biochem Biophys Res Commun. 2009;389:205–10. doi: 10.1016/j.bbrc.2009.08.074. [DOI] [PubMed] [Google Scholar]
- 54.Zhang S, Mahalingam M, Tsuchida N. Naf1alpha is phosphorylated in mitotic phase and required to protect cells against apoptosis. Biochem Biophys Res Commun. 2008;367:364–9. doi: 10.1016/j.bbrc.2007.12.141. [DOI] [PubMed] [Google Scholar]
- 55.Schrager JA, Der Minassian V, Marsh JW. HIV Nef increases T cell ERK MAP kinase activity. J Biol Chem. 2002;277:6137–42. doi: 10.1074/jbc.M107322200. [DOI] [PubMed] [Google Scholar]
- 56.Herbein G, Gras G, Khan KA, Abbas W. Macrophage signaling in HIV-1 infection. Retrovirology. 2010;7:34. doi: 10.1186/1742-4690-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hadigan C. Peroxisome proliferator-activated receptor gamma agonists and the treatment of HIV-associated lipoatrophy: unraveling the molecular mechanism of their shortcomings. J Infect Dis. 2008;198:1729–31. doi: 10.1086/593180. [DOI] [PubMed] [Google Scholar]
- 58.Mallon PW, Sedwell R, Rogers G, Nolan D, Unemori P, Hoy J, et al. Effect of rosiglitazone on peroxisome proliferator-activated receptor gamma gene expression in human adipose tissue is limited by antiretroviral drug-induced mitochondrial dysfunction. J Infect Dis. 2008;198:1794–803. doi: 10.1086/593179. [DOI] [PubMed] [Google Scholar]
- 59.Jaattela M, Mouritzen H, Elling F, Bastholm L. A20 zinc finger protein inhibits TNF and IL-1 signaling. J Immunol. 1996;156:1166–73. [PubMed] [Google Scholar]
- 60.Harhaj EW, Dixit VM. Deubiquitinases in the regulation of NF-kappaB signaling. Cell Res. 2011;21:22–39. doi: 10.1038/cr.2010.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iwai K, Tokunaga F. Linear polyubiquitination: a new regulator of NF-kappaB activation. EMBO Rep. 2009;10:706–13. doi: 10.1038/embor.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verstrepen L, Carpentier I, Verhelst K, Beyaert R. ABINs: A20 binding inhibitors of NF-kappa B and apoptosis signaling. Biochem Pharmacol. 2009;78:105–14. doi: 10.1016/j.bcp.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 63.Flores AM, Li L, Aneskievich BJ. Isolation and functional analysis of a keratinocyte-derived, ligand-regulated nuclear receptor comodulator. J Invest Dermatol. 2004;123:1092–101. doi: 10.1111/j.0022-202X.2004.23424.x. [DOI] [PubMed] [Google Scholar]
- 64.Privalsky ML. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu Rev Physiol. 2004;66:315–60. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- 65.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–62. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 66.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 67.Guicciardi ME, Gores GJ. Life and death by death receptors. FASEB J. 2009;23:1625–37. doi: 10.1096/fj.08-111005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 69.Polakowska RR, Piacentini M, Bartlett R, Goldsmith LA, Haake AR. Apoptosis in human skin development: morphogenesis, periderm, and stem cells. Dev Dyn. 1994;199:176–88. doi: 10.1002/aja.1001990303. [DOI] [PubMed] [Google Scholar]
- 70.Raj D, Brash DE, Grossman D. Keratinocyte apoptosis in epidermal development and disease. J Invest Dermatol. 2006;126:243–57. doi: 10.1038/sj.jid.5700008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang L, Verstrepen L, Heyninck K, Wullaert A, Revets H, De Baetselier P, et al. ABINs inhibit EGF receptor-mediated NF-kappaB activation and growth of EGF receptor-overexpressing tumour cells. Oncogene. 2008;27:6131–40. doi: 10.1038/onc.2008.208. [DOI] [PubMed] [Google Scholar]
- 72.Ramos JW. The regulation of extracellular signal-regulated kinase (ERK) in mammalian cells. Int J Biochem Cell Biol. 2008;40:2707–19. doi: 10.1016/j.biocel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 73.Zheng ZM, Specter S. Dynamic production of tumour necrosis factor-alpha (TNF-alpha) messenger RNA, intracellular and extracellular TNF-alpha by murine macrophages and possible association with protein tyrosine phosphorylation of STAT1 alpha and ERK2 as an early signal. Immunology. 1996;87:544–50. doi: 10.1046/j.1365-2567.1996.513591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dumitru CD, Ceci JD, Tsatsanis C, Kontoyiannis D, Stamatakis K, Lin JH, et al. TNF-alpha induction by LPS is regulated posttranscriptionally via a Tpl2/ERK-dependent pathway. Cell. 2000;103:1071–83. doi: 10.1016/s0092-8674(00)00210-5. [DOI] [PubMed] [Google Scholar]
- 75.Tian B, Nowak DE, Brasier AR. A TNF-induced gene expression program under oscillatory NF-kappaB control. BMC Genomics. 2005;6:137. doi: 10.1186/1471-2164-6-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tian B, Nowak DE, Jamaluddin M, Wang S, Brasier AR. Identification of direct genomic targets downstream of the nuclear factor-kappaB transcription factor mediating tumor necrosis factor signaling. J Biol Chem. 2005;280:17435–48. doi: 10.1074/jbc.M500437200. [DOI] [PubMed] [Google Scholar]
- 77.Fritah A, Christian M, Parker MG. The metabolic coregulator RIP140: an update. Am J Physiol Endocrinol Metab. 2010;299:E335–40. doi: 10.1152/ajpendo.00243.2010. [DOI] [PubMed] [Google Scholar]
- 78.Gurevich I, Flores AM, Aneskievich BJ. Corepressors of agonist-bound nuclear receptors. Toxicol Appl Pharmacol. 2007;223:288–98. doi: 10.1016/j.taap.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wei LN, Hu X, Chandra D, Seto E, Farooqui M. Receptor-interacting protein 140 directly recruits histone deacetylases for gene silencing. J Biol Chem. 2000;275:40782–7. doi: 10.1074/jbc.M004821200. [DOI] [PubMed] [Google Scholar]
- 80.Zhou J, Wu R, High AA, Slaughter CA, Finkelstein D, Rehg JE, et al. A20-binding inhibitor of NF-kappaB (ABIN1) controls Toll-like receptor-mediated CCAAT/enhancer-binding protein beta activation and protects from inflammatory disease. Proc Natl Acad Sci U S A. 2011;108:E998–E1006. doi: 10.1073/pnas.1106232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McInturff JE, Modlin RL, Kim J. The role of toll-like receptors in the pathogenesis and treatment of dermatological disease. J Invest Dermatol. 2005;125:1–8. doi: 10.1111/j.0022-202X.2004.23459.x. [DOI] [PubMed] [Google Scholar]
- 82.Lafyatis R, York M. Innate immunity and inflammation in systemic sclerosis. Curr Opin Rheumatol. 2009;21:617–22. doi: 10.1097/BOR.0b013e32832fd69e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Drexler SK, Foxwell BM. The role of toll-like receptors in chronic inflammation. Int J Biochem Cell Biol. 2010;42:506–18. doi: 10.1016/j.biocel.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 84.Gurevich I, Zhang C, Encarnacao PC, Struzynski CP, Livings SE, Aneskievich BJ. PPARgamma and NF-kappaB regulate the gene promoter activity of their shared repressor. TNIP1 Biochim Biophys Acta. 2011;1819:1–15. doi: 10.1016/j.bbagrm.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gomariz RP, Gutierrez-Canas I, Arranz A, Carrion M, Juarranz Y, Leceta J, et al. Peptides targeting Toll-like receptor signalling pathways for novel immune therapeutics. Curr Pharm Des. 2010;16:1063–80. doi: 10.2174/138161210790963841. [DOI] [PubMed] [Google Scholar]