Abstract
Rapid, volumetric imaging of hyperpolarized 13C compounds allows the real time measurement of metabolic activity and can be useful in distinguishing between normal and diseased tissues. This work extends a fast 2D under-sampled spiral magnetic resonance spectroscopic imaging (MRSI) sequence to provide volumetric coverage, acquiring a 16×16×12 matrix with a nominal 5 mm isotropic resolution in 4.5 s. The rapid acquisition enables a high temporal resolution for dynamic imaging. This dynamic 3D MRSI method was used to investigate hyperpolarized [1-13C]-pyruvate metabolism modulated by the administration of ethanol in rat liver. A significant increase in the pyruvate to lactate conversion was observed in the liver due to the greater availability of NADH from ethanol metabolism.
Keywords: hyperpolarized 13C, dynamic metabolic imaging, fast volumetric imaging, liver ethanol, spiral MRSI
Introduction
Hyperpolarized 13C magnetic resonance spectroscopic imaging (MRSI) is a powerful tool for non-invasive in vivo measurement of metabolism in real time. Several studies have demonstrated metabolic imaging of hyperpolarized [1-13C]-pyruvate (Pyr) and its downstream products lactate (Lac) and alanine (Ala) for a variety of applications, including cancer diagnosis, monitoring therapeutic response, and cardiovascular pathologies [1–7]. One of the challenges of imaging hyperpolarized 13C labeled compounds is the rapid decay of the hyperpolarized magnetization. The non-equilibrium magnetization decays to thermal polarization with relaxation time constant T1, which is typically on the order of 20–40 s in vivo. Several fast spectroscopic imaging techniques have been developed to capture the metabolic information within the limited lifetime of the hyperpolarized signal [8–17]. Rapid acquisition methods are especially needed in order to capture the metabolic kinetics with adequate temporal resolution. One such fast sequence is the 2D spectrally under-sampled spiral MRSI [9–10], which allows imaging of a slice in less than one second. However, 2D multi-slice approaches suffer from the problems of gaps between slices and chemical shift displacement. For example, chemical shift mis-registration between slice locations of Pyr and Lac, which are 390 Hz apart at 3T, using an RF excitation bandwidth of 2.3 kHz is 1.7 mm for a 10-mm thick slice. Though chemical shift displacement is also present for the Pyr and Lac slab profiles in a 3D acquisition, the phase encoding provides contiguous slices without mis-registration at the expense of some potential slice aliasing at the edge of the field of view (FOV). The 3D approach also allows phase modulating in the slice encode direction during reconstruction to shift the slice locations by a fraction of the slice thickness, as well as retrospectively reformatting the image plane.
The conversion of Pyr to Lac takes place via the enzyme lactate dehydrogenase (LDH) and the concomitant oxidation of NADH to nicotinamide adenine dinucleotide (NAD+). At a sufficiently high dose of Pyr, this reaction is saturated, with both LDH activity and NADH availability being rate-limiting steps. The metabolism of ethanol in the liver takes place through the breakdown of ethanol to acetaldehyde and acetaldehyde to acetate. These reactions are catalyzed by the enzymes alcohol dehydrogenase and acetaldehyde dehydrogenase respectively, and both of these reactions reduce the coenzyme NAD+ to NADH. Thus, ethanol consumption leads to accumulation of NADH in the liver [18–20], and provides a mechanism by which NADH availability can be altered and used to modulate the Pyr-to-Lac metabolic pathway [21].
Spielman et al. [21] have previously reported MRS measurements of hyperpolarized [1-13C]-Pyr to study rat liver metabolism modulated by ethanol. Dynamic 13C MRS was used to measure the conversion rates of Pyr to Lac and Ala in the absence and presence of ethanol. However, the MRS data were not spatially localized within a slice, which contained vascular and other structures in addition to the targeted organs, and may underestimate the conversion rates due to the large Pyr signal contribution from the vasculature. Dynamic 3D 13C MRSI can provide improved rate-constant estimates by allowing the analysis of spectra from organ-specific regions-of-interest (ROIs). This work extends the fast 2D spiral MRSI sequence to provide volumetric coverage and applies it to investigate Pyr metabolism in rat liver modulated by ethanol.
Method
The polarized sample consisted of 25 μL of a mixture of 14-M [1-13C] pyruvic acid and 15-mM Ox063 trityl radical, to which 3 μL of a 1:50 dilution of Dotarem (Guerbet, France) was added prior to polarization. The sample was polarized via dynamic nuclear polarization [22] using a HyperSense system (Oxford Instruments Molecular Biotools, Oxford, UK) to achieve approximately 24% liquid-state polarization at dissolution. The polarized sample was dissolved with a solution of 80 mM NaOH mixed with 40 mM Tris buffer and 0.1 g/L EDTA-Na2, leading to an 80-mM solution of hyperpolarized pyruvate with a pH of 7.4–7.5. Healthy male Wistar rats (n=5, 275–352 g body weight) were injected with 3 mL of the hyperpolarized pyruvate solution through a tail vein catheter at a rate of approximately 0.2 mL/s. All animal procedures were approved by the Institutional Animal Care and Use Committee.
All experiments were performed on a clinical 3T Signa MR scanner (GE Healthcare, Waukesha, WI) equipped with self-shielded gradients (40 mT/m, 150 mT/m/ms). A custom-built dual-tuned (1H/13C) quadrature rat coil (inner diameter=80 mm, length=90 mm), operating at 127.7 MHz and 32.1 MHz, respectively, was used for both RF excitation and signal reception. The transmit 13C RF power was calibrated using a reference phantom containing an 8-M solution of 13C-urea, which was placed on top of the animal. Single-shot fast spin-echo 1H MR images with nominal in-plane resolution of 0.47 mm and 2-mm slice thickness were acquired in the axial, sagittal, and coronal planes throughout the scan session as anatomical references for prescribing the 13C-MRSI experiments. Additional 1H 3D spoiled gradient echo (SPGR) images (0.625 mm in-plane resolution, 1.25 mm slice thickness, 96 slices) were also acquired matching the 13C MRSI prescription for overlay of the metabolic images.
The 2D under-sampled spiral spectroscopic imaging sequence described in [9–10] was extended for volumetric coverage. A spiral readout trajectory encoded the spectral and two spatial (x–y) dimensions using 32 echoes and 3 spiral interleaves. The slice (z) direction used 12 phase encoding steps in a centric order. Imaging parameters were: FOV=80×80×60 mm3, 5×5×5 mm3 nominal resolution, spectral width=280 Hz, 42-mm slab excitation, 36 excitations/volume, flip angle=5.625o, TE=2.3 ms, readout duration=114.3 ms, nominal spectral resolution=8.7 Hz, total acquisition time=4.5 s. The cumulative effect of 36 excitations of 5.625° each leads to an effective flip angle of 32.8° for the volume acquisition. The effective flip angle was estimated as αeff = cos−1(cosN αk), where αk = 5.625° and N=36. This calculation assumes instantaneous successive excitations, ignoring any changes in pyruvate magnetization during the time between excitations. Dynamic 13C MRSI data were acquired from a 3D volume that included both liver and kidney every 5 s for 80 s, starting at the same time as the Pyr injection. The time from dissolution to start of injection was kept constant at 24 s.
After acquiring a baseline 13C MRSI measurement, 1.0 g/kg of a 20% ethanol solution was injected into the tail vein at the rate of 1 mL/min. Another 13C MRSI measurement was acquired approximately 45 min after the ethanol injection to achieve a targeted steady-state blood alcohol level (BAL) of approximately 100 mg/dL at the time of the second acquisition. At the end of the exam, blood was collected for BAL analysis.
The data were reconstructed similarly as the 2D case described in [9–10] with an additional apodization and Fourier transform step in the slab-select direction. The data were zero-filled by a factor of 2 in each spatial direction. Due to spectral undersampling and aliasing of the metabolites, a separate reconstruction is required for each metabolite. With the center frequency approximately at alanine, pyruvate and lactate were aliased once in opposite directions. In each reconstruction, one of the spectral resonances is corrected for the chemical shift phase accrual and reconstructed ‘in-focus’ while the aliased resonances are blurred. A zero-order phase correction is then applied to put the ‘in-focus’ peak into absorption mode. The details of the reconstruction are given in [9–10].
Metabolic maps for Pyr, Lac, and Ala were calculated by integrating the signal within ±20 Hz around each peak in absorption mode. The metabolic images were corrected for differences in the solid-state polarization level of each sample. ROIs were drawn manually for liver (mean ROI size was 1.22 cm3), kidney (0.94 cm3) and vasculature (0.28 cm3) in both the liver and kidney slices to calculate the mean time-resolved signal intensities for Pyr, Lac, and Ala. The liver and kidney ROIs were also used to calculate signal-to-noise ratios (SNR) in the time-averaged metabolic images. The noise was measured as the standard deviation in a region of the FOV outside the animal in a slice outside the excited slab. Additionally, a large ROI encompassing the entire animal (17.8 cm3) was used to generate whole-slice time-courses for the liver and kidney slices. The metabolite time-courses were then fit using a three-site exchange model to estimate the apparent Pyr-to-Lac (kpl) and Pyr-to-Ala (kpa) rate constants. The model used here was the same as described in [21], except that the measured Pyr time-curve was used as the bolus input function into the model. It used linear gray-box modeling (idgrey, MatLab, MathWorks, Natick, MA) to estimate the parameters by minimizing the sum of squares of the residual error at each time point. The parameters fit by this model were apparent kpl and kpa exchange rate constants, and apparent T1 decay time constants for Pyr (T1pyr), Lac (T1lac) and Ala (T1ala). In order to simplify the model, the backward exchange rates, klp and kap, were set to zero. The apparent exchange rate constants include contributions from metabolic flux, isotopic exchange, and transport kinetics. Similarly, the apparent T1 decay constant represents polarization loss over time from T1 relaxation as well as from RF excitation and inflow/outflow of spins.
A statistical analysis of the estimated metabolic rate constants pre- and post-ethanol was performed to identify significant changes. Due to the small sample size and non-normal distribution, a non-parametric Wilcoxon signed rank test was used for hypothesis testing.
Results and Discussion
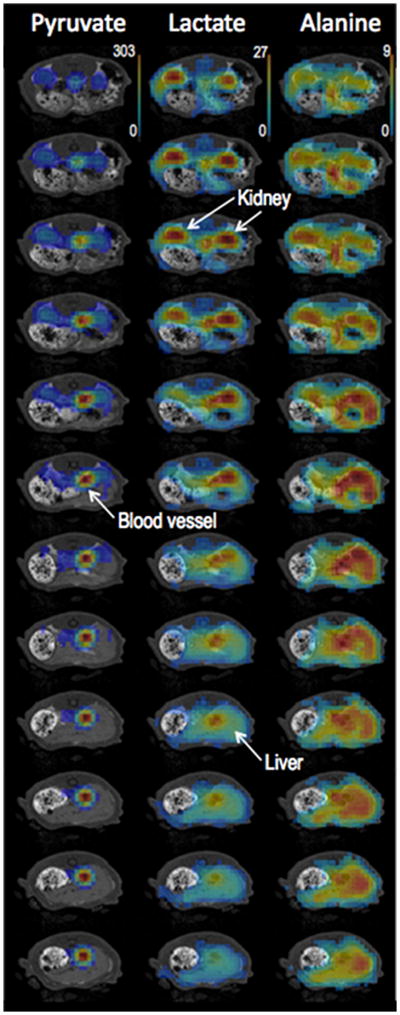
Figure 1 shows representative post-ethanol 13C metabolic maps of Pyr, Lac, and Ala from the 3D volume covering kidney and liver superimposed onto corresponding 1H 3D SPGR slices. The central 12 of the 24 reconstructed slices are shown here. The 13C images were averaged over all of the 16 time-points acquired during the 80-s scan. The time series of 13C-Lac maps from the kidney and liver slices shown in Fig. 2 illustrate the temporal dynamics and spatial distribution of this metabolite. The fast acquisition achieved by spiral MRSI enabled a temporal resolution of 5 s for dynamic imaging. These figures present an example of time-resolved volumetric imaging, which allows the simultaneous characterization of metabolic profiles from different tissues.
Figure 1.
Representative post-ethanol 13C metabolic maps of pyruvate, lactate and alanine superimposed on corresponding 1H MRI images from the 3D volume covering kidneys and liver. The 13C images are averaged over all 16 time-points acquired. The central 12 of the 24 reconstructed slices are shown here. The thresholds for the display of 13C maps were 9% of the maximum intensity for pyruvate, and 20% for lactate and alanine.
Figure 2.
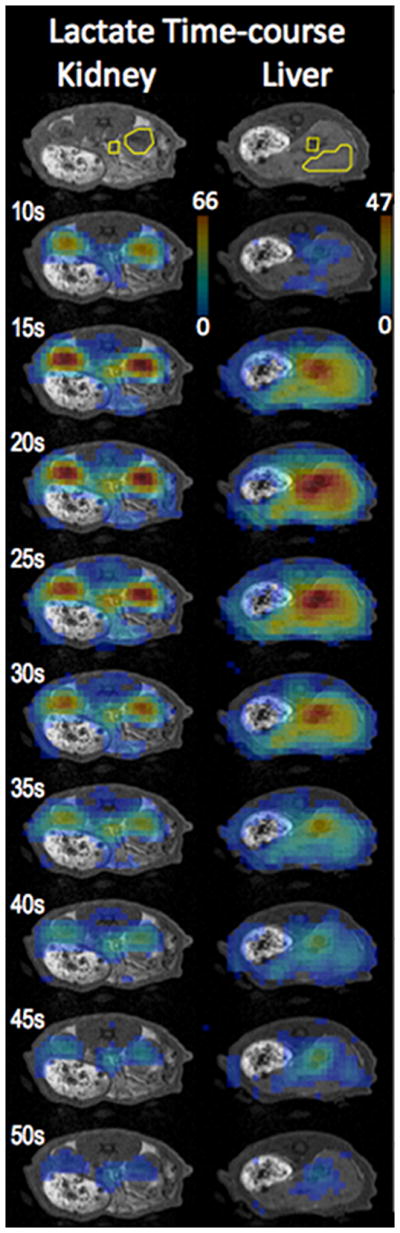
Time series of lactate maps from kidney and liver slices illustrate the metabolite dynamics and distribution. The labeled time indicates time from beginning of the pyruvate injection. The threshold for the display of 13C images was 15% of the maximum intensity. The ROIs used to obtain the metabolite time-courses are also shown.
The baseline (pre-ethanol) SNR values were calculated for the time-averaged Pyr, Lac and Ala in liver and kidney. For liver, the SNR of Pyr, Lac, and Ala were 63.8±10.7 (mean±standard deviation, n=5), 37.5±6.6, and 29.3±3.2 respectively. For kidney, the SNR of Pyr, Lac, and Ala were 304.7±79.1, 70.4±8.6, and 30.8±3.4 respectively. The kidney SNR was measured in the right kidney, which lies relatively close to the aorta and vena cava. The pyruvate level in vasculature is much higher than that in the kidney, and at the 5 mm spatial resolution used here, there can be some pyruvate signal contribution from the vasculature into the kidney ROI. This may be the cause for the high inter-subject variability seen in the pyruvate SNR in kidney. This signal contamination from vasculature is much lower for lactate and alanine, and hence could also lead to underestimation of apparent kpl and kpa. Imaging with higher spatial resolution would help alleviate this problem.
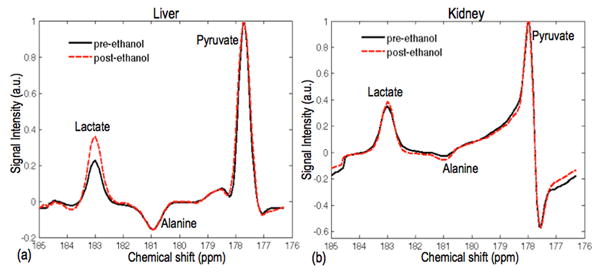
Figure 3 shows the time-averaged lactate spectra, normalized to pyruvate, pre- and post-ethanol in liver and in kidney. The spectra from the liver show higher lactate signal post-ethanol compared to pre-ethanol, as expected from the NADH accumulation due to ethanol metabolism. The alanine signal was unchanged. The kidney lactate signal pre- and post-ethanol was also similar. The spectra shown here are from the lactate reconstruction where lactate is corrected for the chemical shift phase accrual and reconstructed ‘in-focus’ while the aliased resonances are blurred
Figure 3.
Representative time-averaged 13C lactate spectra for liver (a) and kidney (b). The liver spectra show higher lactate signal post-ethanol compared to pre-ethanol while the kidney spectra showed similar lactate signal pre- and post-ethanol.
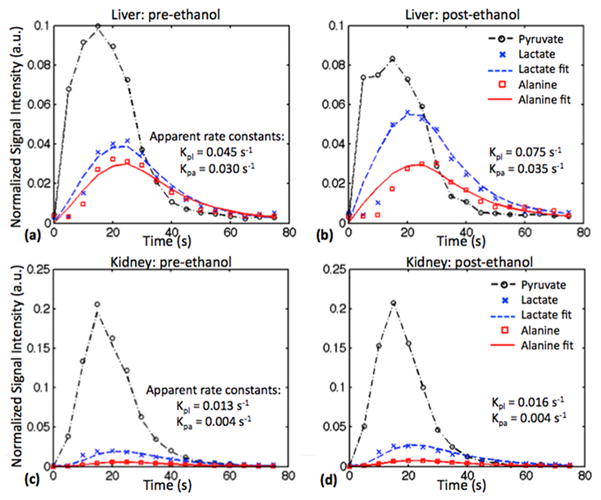
Figure 4 plots the time-courses of the mean Pyr, Lac, and Ala signals from ROIs in the liver and the kidney, measured pre- and post-ethanol, from one animal. The corresponding fits with the apparent rate constants are also shown. The liver plots clearly show increased lactate signal post-ethanol compared to pre-ethanol, while there was little change in the kidney lactate signal. Alanine signal also was similar before and after ethanol in both organs. The estimated apparent kpl and kpa rate constants for all 5 rats are summarized in Fig. 5 and in Table 1. The BALs for the 5 animals ranged from 81.1 to 101.6 mg/dL. The average apparent liver kpl increased from 0.049 ± 0.007 s−1 (mean ± standard deviation) pre-ethanol to 0.064 ± 0.012 s−1 post-ethanol. This increase in kpl was found to be statistically significant (P<0.044), and is consistent with the hypothesis that NADH levels are rate limiting for liver Pyr-to-Lac conversion at the given Pyr dose [21]. As a control, no significant changes were observed for the labeled lactate production in kidney or vasculature ROIs or for alanine production in any of the tissues. The inter-subject variability leads to high standard deviations in Table 1, but the intra-subject comparison between pre- and post-ethanol in Fig. 5 clearly shows the change in rate constants.
Figure 4.
Representative time course data, curve fits, and calculated apparent rate constants, from the liver acquired pre-ethanol (a) and 45 min post-ethanol (b), and from the kidney pre-ethanol (c) and post-ethanol (d). Higher lactate was measured in the liver after ethanol administration, whereas there was little change in kidney. The conversion of pyruvate to alanine was similar before and after ethanol in both organs.
Figure 5.
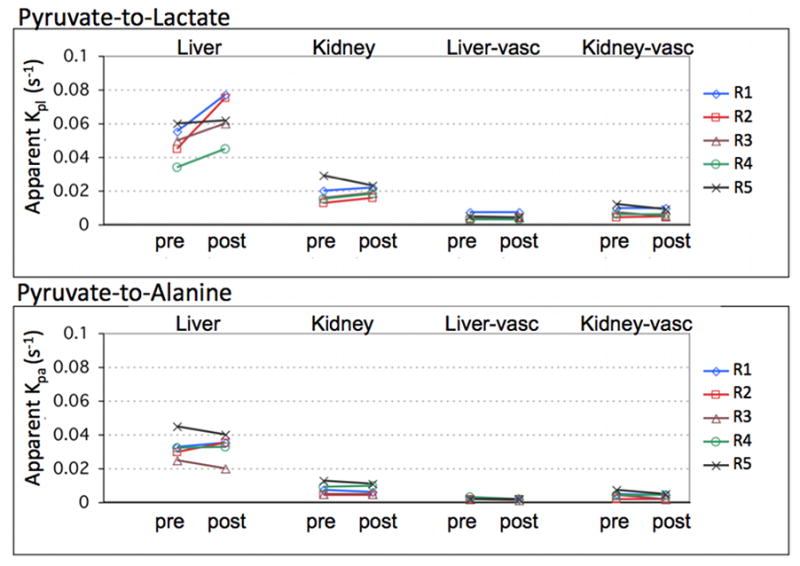
Estimated apparent kpl and kpa rate constants in liver, kidney, and vasculature in the respective slices. Conversion of labeled pyruvate to lactate increased in the liver after ethanol administration as seen by the increase in kpl values. There was no significant change in kpl in kidney or in vasculature or in kpa.
Table 1.
Average apparent kpl and kpa rate constants with corresponding standard deviations in liver, kidney and vasculature ROIs from 5 animals. Conversion of labeled pyruvate to lactate increased in the liver after ethanol administration as seen by the increase in kpl. There was no significant change in kpl for kidney or vasculature, or in kpa in any of the tissues. Though the inter-subject variability leads to relatively high standard deviations, the intra-subject comparison between pre-and post-ethanol in Fig. 5 clearly shows the change in rate constants.
| Liver kpl (s−1) | Liver kpa (s−1) | Kidney kpl (s−1) | Kidney kpa (s−1) | Liver vasc. kpl (s−1) | Liver vasc. kpa (s−1) | Kidney vasc. kpl (s−1) | Kidney vasc. kpa (s−1) | |
|---|---|---|---|---|---|---|---|---|
| Pre-ethanol | 0.049 ± 0.009 | 0.033 ± 0.007 | 0.018 ± 0.004 | 0.008 ± 0.003 | 0.004 ± 0.002 | 0.002 ± 0.001 | 0.008 ± 0.003 | 0.004 ± 0.002 |
| Post-ethanol | 0.064 ± 0.012 | 0.032 ± 0.007 | 0.019 ± 0.003 | 0.007 ± 0.003 | 0.004 ± 0.002 | 0.002 ± 0.001 | 0.007 ± 0.002 | 0.003 ± 0.001 |
| Wilcoxon p-value | 0.0431 | 0.8927 | 0.4982 | 0.5930 | 1.0000 | 0.1573 | 0.2850 | 0.1025 |
Compared to the slice-selective MRS measurements in [21], the organ specific apparent kpl and kpa values obtained here at baseline were considerably higher in the liver but similar in the kidney. For example, the average apparent kpl at baseline in liver was 0.049 ± 0.007 s−1 for the 3D MRSI and 0.014 ± 0.003 s−1 for the slice-selective MRS in ref. 21. The corresponding values in kidney were 0.018 ± 0.004 s−1 for the 3D MRSI and 0.012 ± 0.002 s−1 for the slice-selective MRS. The greater difference in liver kpl is presumably because the fraction of Pyr signal contribution from the vasculature to the total Pyr signal in the slice is higher for the liver slice than for the kidney slice. This effect is also seen in the Pyr images in Fig. 1. The Pyr signal in the liver is much lower compared to that in the blood vessel in the same slice, and is below the threshold used for display of the 13C maps. However, the Pyr signal in the kidney relative to the vessel in the kidney slice is easily visible. Due to the large contribution of the vascular Pyr signal, the whole-slice ROI is a better comparison to the slice-selective estimates. For the whole-slice time-courses generated from the 3D MRSI for liver and kidney slices, the apparent kpl were 0.015 ± 0.003 s−1 for liver and 0.016 ± 0.006 s−1 for kidney, and the apparent kpa were 0.010 ± 0.002 s−1 for liver and 0.011 ± 0.003 s−1 for kidney, all of which are similar to the slice-selective MRS estimates from [18]. These values demonstrate how the organ-specific rate constants are underestimated by slice-selective dynamic data due to the large vascular Pyr component.
The three-site exchange model used for fitting does not account for the RF excitation scheme used for data acquisition. The slice-selective measurements in [21] used a single RF excitation with a 5° flip angle for each time-point, while the 3D imaging here used 36 excitations of 5.6° each, resulting in a much higher effective flip angle per time-point. Also, the 3D sequence had an acquisition duration of 4.5 s and a TR of 5 s with inflow occurring throughout, as compared to the short 409.6 ms readout and TR of 3 s in [21]. These differences in the RF sampling schemes and the inflow effects could lead to differences in the absolute values of the apparent rate constant and T1 decay estimates. However, the impact of the RF sampling is the same for both pre- and post-ethanol measurements, and should have only a small effect on the change in apparent kpl and kpa from pre- to post-ethanol infusion.
A key difference from the fitting in [21] was that instead of modeling the bolus input function by a trapezoid, this study used the measured Pyr time-curve directly as the bolus input into the model. As the measured Pyr time-curve includes the T1 decay information, it obviates the need to separately fit for Pyr T1, and so apparent T1pyr was fixed to 1 s. Good fits were obtained with apparent T1lac = 12 s and T1ala = 15 s for liver and kidney, and apparent T1lac = T1ala = 20 s for vasculature. These values were chosen empirically and kept the same for all animals.
The increase in apparent liver kpl after infusion of ethanol (post-ethanol/pre-ethanol) observed here was 1.33 ± 0.11 SEM compared to the approximately 2-fold (2.0 ± 0.5 SEM) increase reported in [21]. This smaller effect may be due to the use of 80-mM Pyr instead of 100-mM Pyr as in that study, whereby the LDH enzyme activity may not be completely saturated. Though the total dose was comparable in both studies (240 μmol vs. 250 μmol), the lower concentration Pyr solution may be just at the boundary between the linear and saturated regimes of LDH enzyme activity, especially given the time-varying Pyr concentration during the ~15 s bolus injection.
The analysis in [21] used separate control measurements with saline injection instead of ethanol injection to confirm that the increased lactate production in the liver was an effect of ethanol. Based on those results, additional saline control measurements were not repeated for this study. The alanine kinetics as well as the lactate kinetics in kidney and in vasculature serve as self-control within each animal.
Conclusion
This work demonstrates fast volumetric imaging of hyperpolarized 13C Pyr and its metabolic products in vivo using spectrally under-sampled spiral MRSI. The method was applied to measure the changes in Pyr-to-Lac conversion after infusion of ethanol. Spiral MRSI is an efficient acquisition method, achieving 80×80×60 mm3 volume coverage with a nominal 5-mm isotropic resolution in 4.5 s on a clinical 3T scanner. This implementation using clinical gradients makes the method suitable for translation to human use, e.g., the imaging parameters listed above would be suitable for prostate cancer imaging which is the target for the first clinical trial currently underway [23]. Combining this method with parallel imaging techniques would allow greater volume coverage, higher resolution, or even faster acquisition times, enabling other applications. Alternatively, a high performance insert gradient [11] can be used to further speed up the acquisition for small animal imaging, achieving a 2.7×2.7×5-mm3 resolution for the same 16×16×12 matrix size in 1.5 s. The short acquisition times enable a high temporal resolution for monitoring dynamic processes. The dynamic 3D acquisition allows the analysis of spectra from organ-specific ROIs, thus providing improved rate-constant estimates and permitting quantitative evaluation of metabolic kinetics.
Acknowledgments
Funded by:
NIH RR09784, AA018681, AA05965, AA13521-INIA, EB009070
Abbreviations used
- MRSI
magnetic resonance spectroscopic imaging
- FOV
field of view
- ROI
region of interest
- LDH
lactate dehydrogenase
- NAD+
nicotinamide adenine dinucleotide
- SPGR
spoiled gradient echo
- BAL
blood alcohol level
- SNR
signal-to-noise ratio
References
- 1.Golman K, Zandt R, Thaning M. Real-time metabolic imaging. Proc Natl Acad Sci USA. 2006;103:11270–11275. doi: 10.1073/pnas.0601319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kohler SJ, Yen Y, Wolber J, Chen AP, Albers MJ, Bok R, Zhang J, Tropp J, Nelson S, Vigneron SB, Kurhanewicz J, Hurd RE. In vivo 13carbon metabolic imaging at 3T with hyperpolarized 13C-1-pyruvate. Magn Reson Med. 2007;58:65–69. doi: 10.1002/mrm.21253. [DOI] [PubMed] [Google Scholar]
- 3.Larson PEZ, Bok R, Kerr AB, Lustig M, Hu S, Chen AP, Nelson SJ, Pauly JM, Kurhanewicz J, Vigneron DB. Investigation of tumor hyperpolarized [1-13C] pyruvate dynamics using time resolved multiband RF excitation echo-planar MRSI. Magn Reson Med. 2010;63:582–591. doi: 10.1002/mrm.22264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darpolor MM, Yen Y-F, Chua M-S, Xing L, Clarke-Katzenberg RH, Shi W, Mayer D, Josan S, Hurd R, Pfefferbaum A, Senadheera L, So S, Hoffman L, Glazer G, Spielman D. In vivo MRSI of hyperpolarized [1-13C]pyruvate metabolism in rat hepatocellular carcinoma. NMR in Biomedicine. 2011;24:n/a. doi: 10.1002/nbm.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park I, Larson P, Zierhut ML, Vigneron DB, Nelson SJ. Hyperpolarized 13C magnetic resonance metabolic imaging: application to brain tumors. Neuro-oncology. 2010;12:133–144. doi: 10.1093/neuonc/nop043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Day SE, Kettunen MI, Cherukuri MK, Mitchell JB, Lizak MJ, Morris D, Matsumoto S, Koretsky AP, Brindle M. Detecting response of rat C6 glioma tumors to radiotherapy using hyperpolarized [1-13C]pyruvate and 13C magnetic resonance spectroscopic imaging. Magn Reson Med. 2011;65:557–563. doi: 10.1002/mrm.22698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Golman K, Petersson JS, Magnusson P, Johansson E, Akeson P, Chai C-M, Hansson G, Mansson S. Cardiac metabolism measured noninvasively by hyperpolarized 13C MRI. Magn Reson Med. 2008;59:1005–1013. doi: 10.1002/mrm.21460. [DOI] [PubMed] [Google Scholar]
- 8.Lau AZ, Chen AP, Ghugre NR, Ramanan V, Lam WW, Connelly KA, Wright GA, Cunnigham CH. Rapid multislice imaging of hyperpolarized 13C pyruvate and bicarbonate in the heart. Magn Reson Med. 2010;64:1323–31. doi: 10.1002/mrm.22525. [DOI] [PubMed] [Google Scholar]
- 9.Mayer D, Yen Y, Tropp J, Pfefferbaum A, Hurd RE, Spielman DM. Application of subsecond spiral chemical shift imaging to real-time multislice imaging of rat in vivo after injection of hyperpolarized 13C1 pyruvate. Magn Reson Med. 2009;62:557–564. doi: 10.1002/mrm.22041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mayer D, Yen Y, Levin Y, Tropp J, Pfefferbaum A, Hurd RE, Spielman DM. In vivo application of subsecond spiral chemical shift imaging to hyperpolarized 13C1 metabolic imaging: Comparison with phase encoded CSI. J Magn Reson. 2010;204:340–345. doi: 10.1016/j.jmr.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayer D, Yen Y, Takahashi A, Josan S, Tropp J, Rutt BK, Hurd RE, Spielman DM, Pfefferbaum A. Dynamic and High-Resolution Metabolic Imaging of Hyperpolarized [1-13C]-Pyruvate in the Rat Brain Using a High-Performance Gradient Insert. Magn Reson Med. 2011;65:1228–1233. doi: 10.1002/mrm.22707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larson PEZ, Hu S, Lustig M, Kerr AB, Nelson SJ, Kurhanewicz J, Pauly JM, Vigneron DB. Fast dynamic 3D MR spectroscopic imaging with compressed sensing and multiband excitation pulses for hyperpolarized 13C studies. Magn Reson Med. 2011;65:610–619. doi: 10.1002/mrm.22650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cunnigham CH, Chen AP, Lustig M, Hargreaves BA, Lupo J, Xu D, Kurhanewicz J, Hurd RE, Yen Y, Pauly JM, Nelson SJ, Vigneron DB. Pulse sequence for dynamic volumetric imaging of hyperpolarized metabolic products. J Magn Reson. 2008;193:139–146. doi: 10.1016/j.jmr.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurd RE, Yen Y-F, Tropp J, Pfefferbaum A, Spielman D, Mayer D. Cerebral dynamics and metabolism of hyperpolarized [1-13C]pyruvate using time-resolved MR spectroscopic imaging. Journal of Cerebral blood flow & metabolism. 2010:1–8. doi: 10.1038/jcbfm.2010.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arunachalam A, Whitt D, Fish K, Gianquinto R, Piel J, Watkins R, Hancu I. Accelerated spectroscopic imaging of hyperpolarized C-13 pyruvate using SENSE parallel imaging. NMR in Biomedicine. 2009;22(8):867–73. doi: 10.1002/nbm.1401. [DOI] [PubMed] [Google Scholar]
- 16.Tropp J, Lupo J, Chen A, Calderon P, McCune D, Grafendorfer T, Ozturk-Isik E, Larson PEZ, Hu S, Yen Y-F, Rob F, Bok R, Schulte R, Xu D, Hurd R, Vigneron D, Nelson S. Multi-channel metabolic imaging, with SENSE reconstruction, of hyperpolarized [1-13C] pyruvate in a live rat at 3. 0tesla on a clinical MR scanner. Journal of Magnetic Resonance. 2011;208:171–177. doi: 10.1016/j.jmr.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perman WH, Bhattacharya P, Leupold J, Lin AP, Harris KC, Norton VA, Hoverner J-B, Ross B. Fast volumetric spatial-spectral MR imaging of hyperpolarized 13C-labeled compounds using multiple echo 3D bSSFP. Magn Reson Imaging. 2010;28:459–465. doi: 10.1016/j.mri.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jue T, Arias-Mendoza F, Gonnella NC, Shulman GI, Shulman RG. 1H NMR technique for observing metabolite signals in the spectrum of perfused liver. Proc Natl Acad Sci USA. 1985;82:5246–5249. doi: 10.1073/pnas.82.16.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forsander OA. Influence of the metabolism of ethanol on the lactate/pyruvate ratio of rat liver slices. J Biochem. 1966;98:244–247. doi: 10.1042/bj0980244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith ME, Newman HW. The rate of ethanol metabolism in fed and fasting animals. J Biol Chem. 1959:1544–1549. [PubMed] [Google Scholar]
- 21.Spielman DM, Mayer D, Yen Y-F, Tropp J, Hurd RE, Pfefferbaum A. In vivo measurement of ethanol metaboism in the rat liver using magnetic resonance spectroscopy of hyperpolarized [1-13C] pyruvate. Magn Reson Med. 2009;62:307–313. doi: 10.1002/mrm.21998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K. Increase in signal-to-noise ratio of >10,000 times in liquid-state NMR. Proc Natl Acad Sci USA. 2003;100:10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurhanewicz J, Vigneron DB, Brindle K, Chekmenev EY, Comment A, Cunningham CH, Deberardinis RJ, Green GG, Leach MO, Rajan SS, Rizi RR, Ross BD, Warren WS, Malloy CR. Analysis of cancer metabolism by imaging hyperpolarized nuclei: prospects for translation to clinical research. Neoplasia. 2011;13:81–97. doi: 10.1593/neo.101102. [DOI] [PMC free article] [PubMed] [Google Scholar]