Abstract
RNA exosomes are multi-subunit complexes conserved throughout evolution1 and emerging as the major cellular machinery for processing, surveillance, and turnover of a diverse spectrum of coding and non-coding RNA substrates essential for viability2. By exome sequencing, we discovered recessive mutations in exosome component 3 (EXOSC3) in four siblings with infantile spinal motor neuron disease, cerebellar atrophy, progressive microcephaly, and profound global developmental delay, consistent with pontocerebellar hypoplasia type 1 [PCH1; OMIM 607596]3–6. We identified mutations in EXOSC3 in an additional 8 of 12 families with PCH1. Morpholino knockdown of exosc3 in zebrafish embryos caused embryonic maldevelopment with small brain and poor motility, reminiscent of human clinical features and largely rescued by coinjected wildtype but not mutant exosc3 mRNA. These findings represent the first example of an RNA exosome gene responsible for a human disease and further implicate dysregulation of RNA processing in cerebellar and spinal motor neuron maldevelopment and degeneration.
Pontocerebellar hypoplasia (PCH) is a clinically and genetically heterogeneous group of autosomal recessive disorders characterized by cerebellar hypoplasia or atrophy, variable pontine atrophy, and progressive microcephaly with global developmental delay7. PCH1 is a distinctive subtype of PCH, characterized by diffuse muscle wasting secondary to spinal cord anterior horn cell loss and cerebellar hypoplasia.3–6 The diagnosis of PCH1 is often delayed or never made because the combination of cerebellar and spinal motor neuron degeneration is not commonly recognized, and the presentation of diffuse weakness and devastating brain involvement is atypical of classical proximal spinal muscular atrophy (SMA)8. The literature contains only a handful of case series9–12 and reports on PCH113–19. The responsible gene has not been identified in the majority of PCH1 patients. Recessive mutations have been found in vaccinia-related kinase 1 (VRK1)20, mitochondrial arginyl-transfer RNA synthetase (RARS2)21, and tRNA splicing endonuclease homolog 54 (TSEN54)22 in single individuals with PCH1. In PCH without SMA, TSEN54 mutations account for most cases of PCH2 and PCH423,21, while RARS2 mutations have been found in two families with PCH624,25.
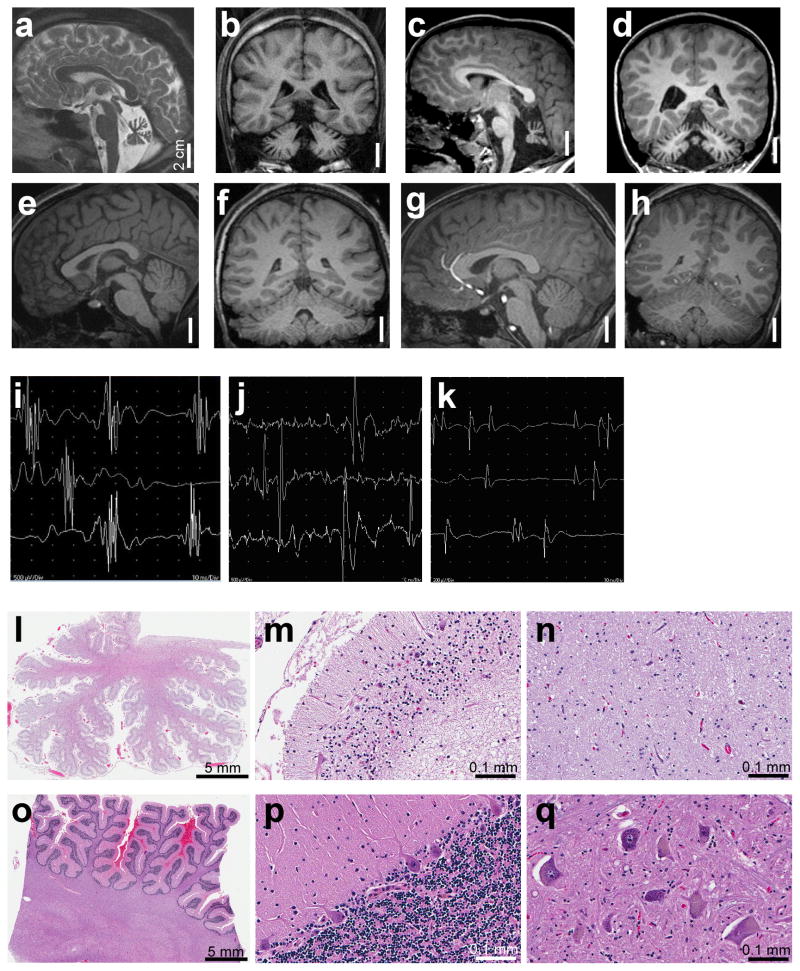
We identified Family 1 in which four children were floppy at birth, with ocular motor apraxia, progressive muscle wasting, distal contractures, progressive microcephaly, growth retardation, global developmental delay, and never reached any motor milestone or spoke. Although normal in size at birth, in all four the head circumference, height, and weight dropped to below the 5th percentile by age 7–10 months. Magnetic resonance imaging demonstrated marked cerebellar atrophy with prominent sulci and decreased volume of folia (Fig. 1a–d) compared to age- and gender-matched normal individuals (Fig. 1e–h). In the patients, the brainstem and the cerebral cortex appear normal in configuration but are small. Electromyography showed in one patient neurogenic motor changes (Fig. 1i) exemplified by a single fast-firing (25 Hz) wave complex that was polyphasic (crossing the baseline multiple times) and unstable. The high frequency firing in the absence of other complexes suggests a loss of axons, while unstable polyphasic units are manifestations of reinnervation in response to denervation. In a younger patient we observed borderline neurogenic motor changes (Fig. 1j) with a normal recruitment pattern but occasional large-amplitude motor unit action potentials (~4.5 mV) suggestive of reinnervation, compared to normal (Fig. 1k), which demonstrates multiple distinct wave-complexes of normal amplitudes (200–400 μV) that represent preserved motor axons without injury. Nerve conduction studies showed motor response with severely reduced amplitudes but normal sensory responses (Supplementary Table 1). Furthermore, when the oldest child died at age 18 years after a respiratory infection, the autopsy revealed a severe loss of cerebellar (Fig. 1l–m) and spinal motor neurons (Fig. 1n) compared to control (Fig. 1o–q). These clinical features are most consistent with PCH1.
Figure 1.
Neuroimaging, neuromuscular, and pathological features in Family 1. a. Sagittal T2- and b. coronal T1-weighted images from the oldest surviving sibling who is now 18 years old demonstrate the presence of all cerebellar lobules yet with marked atrophy compared to T1-weighted e. sagittal and f. coronal images from an age-matched normal male. c. Sagittal and d. coronal T1-weighted images from the youngest surviving sibling who is now 9 years old demonstrate cerebellar volume loss comparable to a.-b. in contrast to g. sagittal and h. coronal T1-weighted images from an age-matched normal male. Needle EMG tracings in the i. left triceps muscle of the 18-year-old sibling and j. right vastus lateralis muscle of the 9-year-old sibling showed neurogenic changes compared to control in k. l. Brain autopsy of the patient who died at age 18 years shows profound cerebellar atrophy (compared to control o), with dysmorphic Purkyně (also known as Purkinje) cells and loss of granule cells seen at higher magnification in m. compared to control p. n. Diffuse loss of motor neurons in the anterior horn of the spinal cord, compared to q. control with normal appearing spinal motor neurons.
No PCH1 genes were known when this study began. We performed a genome scan, which narrowed the candidate regions to four subchromosomal loci with more than 100 candidate genes (Supplementary Fig. 1). To identify the responsible gene, we captured the exome using SureSelect Human All Exon kit (Agilent G3362) and sequenced on a Genome Analyzer IIx (Illumina Inc.). This analysis yielded one candidate variant fulfilling the requirement of rare biallelic variants within the intervals identical by descent in all the affected individuals: EXOSC3:g.9:37783990T>G (ENST00000327304.4:c.395A>C, ENSP00000323046.4:p.Asp132Ala). We did not observe variants in VRK1, RARS2, or TSEN54 previously reported in PCH1.
There are multiple alternative splice forms of EXOSC3, with the longest reading frame spanning 4 exons over 5,119 bases (NM_016042.2) and encoding a 275-amino acid protein, human exosome component 3 (EXOSC3), also known as the ribosomal RNA-processing protein 40 (RRP40) (NP_057126.2). EXOSC3 is a core component of the human RNA exosome complex (distinct from exosome vesicles) present in the cytoplasm and the nucleus, especially enriched in the nucleolus26. The N-terminal (NT) domain and putative RNA binding S1 and KH domains are evolutionarily conserved (Fig. 2).
Figure 2.
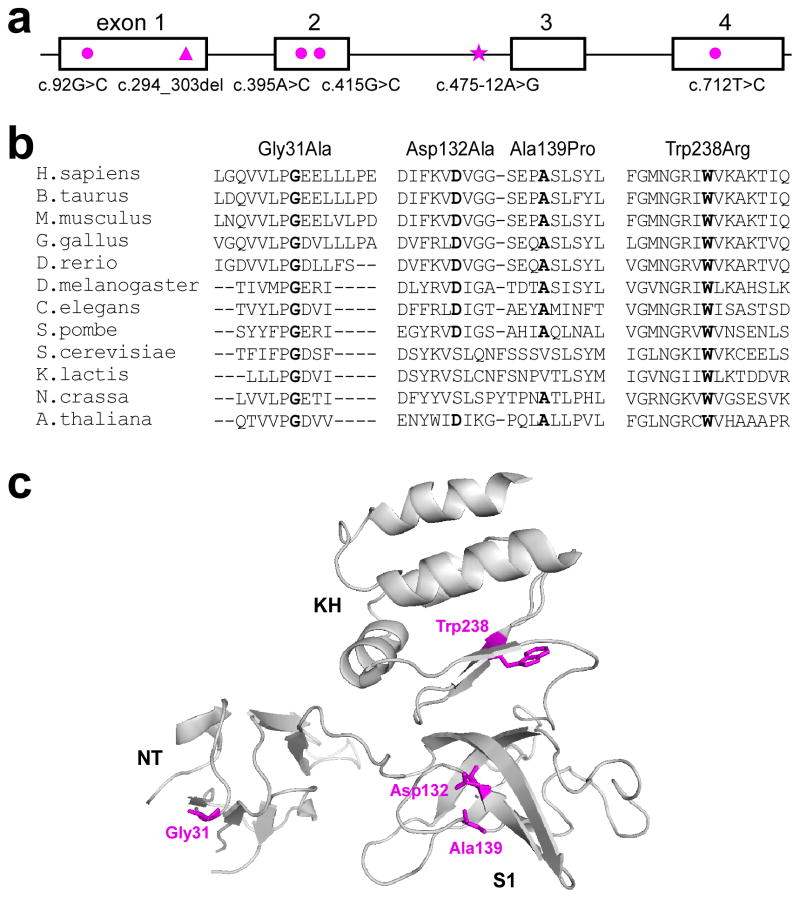
EXOSC3 mutations in PCH1. a. Genomic structure of EXOSC3, with four exons in open boxes and mutations highlighted in magenta. circle- missense mutation; triangle- deletion mutation; star- splice site mutation. b. Alignment of orthologous sequences in human and other eukaryotic organisms, including vertebrate, insect, plant, and yeast, demonstrating that the mutated residues are conserved (highlighted in bold). c. Graphic representation of the locations of the mutated residues (highlighted in magenta) in EXOSC3, with conserved domains NT, S1, and KH (PDB code 2NN6).
We confirmed genotype-phenotype cosegregation in this family by Sanger sequencing. To validate the association between EXOSC3 mutations and PCH1, we sequenced all exons and flanking introns of EXOSC3 (Supplementary Table 2) in the index patients from 12 additional PCH1 families. Eight probands had recessive mutations in the gene (Fig. 2; Table 1). All available parent samples were heterozygous. None of the mutations were found in Turkish (n=94), Czech (n=96), or North American (n=189) control individuals. A more recent review of databases, including the NHLBI Exome Sequencing Project, showed that Asp132Ala has been observed in 6 of 4870 exomes, with an estimated allele frequency of 0.0012. None of the other mutations has been previously reported.
Table 1.
Ethnic origins and EXOSC3 mutations in subjects with PCH1. Mutations in EXOSC3 were identified by exome sequencing in affected subjects in Family 1 and further investigated in DNA samples from Families 2–13 by targeted sequencing.
| Family | Subjects | Ethnicity | Age (death) | Nucleotide change | Amino acid change | Ref. |
|---|---|---|---|---|---|---|
| 1 | 4M | American/European | (18 yrs) 18 yrs 16 yrs 9 yrs |
c.395A>C, homozygous | Asp132Ala | - |
| 2 | 1F | Canadian/Cuban | (40mos) | c.395A>C, homozygous | Asp132Ala | 14 |
| 3* | 1F/1M | German/Turkish | 20 yrs 16 yrs |
c.395A>C, homozygous | Asp132Ala | - |
| 4 | 1 M/1F | Czech | (8 mos) (8 mos) |
c.92G>C c.712T>C |
Gly31Ala Trp238Arg |
- |
| 5 | 1M | New Caledonian |
(seen at 3 mos) | c.294_303del c.395A>C |
99fsX11 Asp132Ala |
11 |
| 6 | 1F | Australian | (26 mos) | c.395A>C c.475-12A>G |
Asp132Ala exon 3 skipping/ aberrant splicing | - |
| 7* | 1M | Australian/ Turkish | (3 yrs) | c.395A>C, homozygous | Asp132Ala | - |
| 8 | 1M | Australian | (11 mos ) | c.395A>C c.415G>C |
Asp132Ala Ala139Pro |
11 |
| 9 | 1M | Czech | (17 mo) | c.92G>C homozygous | Gly31Ala | - |
| 10 | 1M | Spanish | 10 yrs | - | - | 15 |
| 11 | 1M | Japanese | (15 yrs) | - | - | 19 |
| 12 | 1M/1F | Australian | (9 mos ) | - | - | - |
| 13 | 1F | Australian | (20 mos ) | - | - | - |
parental consanguinity.
The Asp132Ala mutation was present in seven of the nine mutation-positive families (Fig. 2; Table 1). This mutation altered a highly conserved amino acid residue in the putative RNA binding S1 domain; the crystal structure suggests that Asp132 may be important for inter-subunit interaction within the exosome complex27. We genotyped the probands of Families 1–3 homozygous for Asp132Ala to find identical haplotypes in a 1cM region flanking the mutation locus, suggesting an ancestral origin (Supplementary Table 3).
We found three additional missense mutations. Two mutations, Gly31Ala and Trp238Arg, were present in Family 4, with parents as carriers. Gly31Ala was homozygous in the patient in Family 9. Strictly conserved from yeast to human, Gly31 in the NT domain appears to be involved in inter-subunit interaction27, while Trp238 is in the putative RNA-binding KH domain27. In Family 8, the patient harbored Asp132Ala in trans with another missense mutation in the S1 domain: c.415G>C; p.Ala139Pro. (Fig. 2; Supplementary Note)
We identified one frameshift mutation: a 10-nucleotide deletion in Family 5, predicted to prematurely terminate the protein; the faulty transcript may be subject to nonsense-mediated mRNA degradation. In silico analysis of the intronic mutation c.475-12A>G in Family 6 suggested that it may introduce a new splice site just upstream of the normal splice acceptor for exon 3. RT-PCR in expression studies demonstrated mainly skipping of exon 3 (shifting the reading frame) and evidence of aberrant splicing (which incorporated 11 nucleotides upstream of the normal splice site), with a minority of transcripts having normal splicing (Supplementary Fig. 2 & Supplementary Table 2).
That biallelic missense, frameshift, and splice site mutations all led to the same clinical manifestations suggests that they may be null or hypomorphic alleles. Since all components of the exosome are essential for viability1, it is unlikely that PCH1 patients harbor biallelic null mutations; it is more likely that the missense mutations are hypomorphic while the frameshift mutations could be null. In silico analyses predicted detrimental consequences from the missense mutations (Supplementary Table 4). The standard marker for impaired exosome function has long been an abnormal accumulation of unprocessed rRNA1, which we did not observe in fibroblasts from the patients in Family 1 (Supplementary Fig. 3), suggesting that the impact of the homozygous Asp132Ala mutations in EXOSC3 may be more nuanced and subtle than a complete elimination of exosome function.
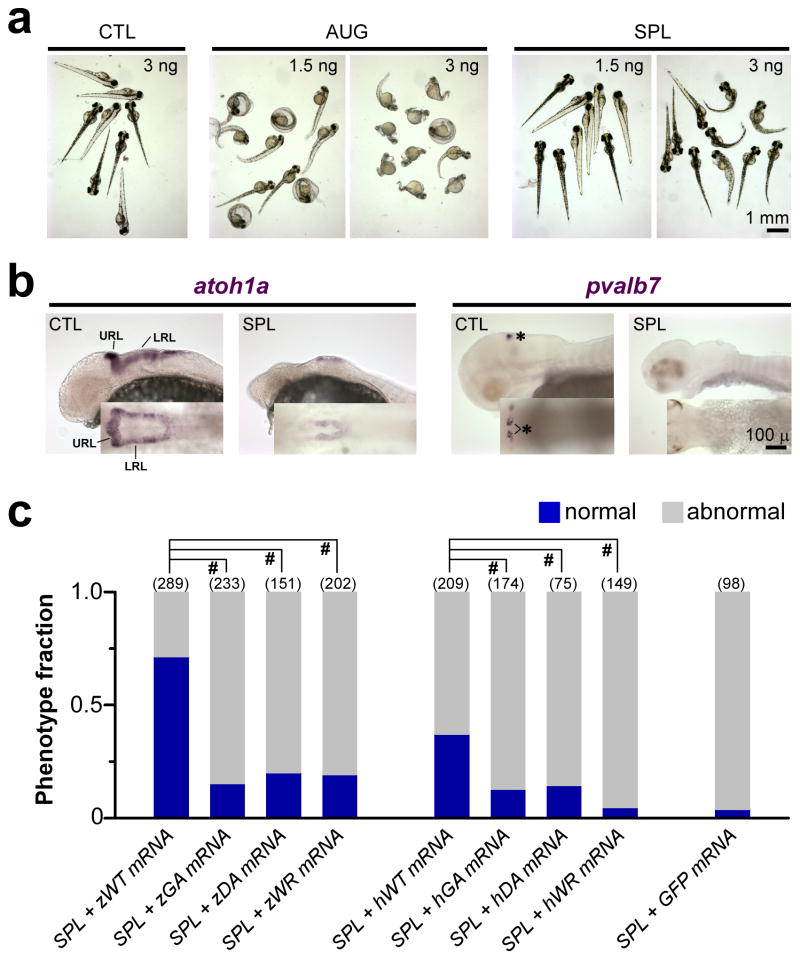
To further examine the functional effects of the mutations, we knocked down the endogenous exosc3 expression in zebrafish embryos by exosc3-specific antisense morpholino injection (Figure 3; Supplementary Table 2; Supplementary Fig. 4). Zebrafish embryos injected with antisense morpholinos directed against the start codon (AUG) or the splice donor site of exon 2 (SPL) of exosc3 led to a dose-dependent phenotype with short curved spine and small brain with poor motility and even death by 3 days post fertilization (dpf), compared to embryos injected with nonspecific control morpholinos (CTL) (Fig. 3a).
Figure 3.
Knockdown of exosc3 in zebrafish embryos disrupts normal development. a. Zebrafish embryos injected with exosc3-specific antisense morpholinos AUG (directed against the start codon) or SPL (directed against the splice donor site for exon 2), compared to nonspecific CTL. b. Whole-mount in situ hybridization in SPL morpholino-injected embryos in lateral view (inset- dorsal view, with rostral to the left) demonstrated diminished expression of dorsal hindbrain progenitor-specific marker atoh1a and cerebellar-specific marker pvalb7 compared to control. URL- upper rhombic lip; LRL-lower rhombic lip. *-distinct clusters of differentiated Purkinje cells in embryos 3dpf. c. Survival data from embryos 3dpf co-injected with 3 ng SPL and 240 pg of human EXOSC3 or zebrafish exosc3 mRNA vs. GFP mRNA as control, from three separate experiments. z-zebrafish; h-human; WT- wild type EXOSC3/exosc3; GA- mutant Gly31Ala in human or Gly20Ala in zebrafish; DA-mutant Asp132Ala in human or Asp102Ala in zebrafish; WR-mutant Trp238Arg in human or Trp208Arg in zebrafish. Embryos were classified as normal (blue) or abnormal (grey, which includes embryos that are mildly abnormal, severely abnormal, or dead). #, p<0.0001, two-tailed Pearson’s Chi-squared test.
The observation of shrunken or collapsed hindbrain in SPL-injected embryos prompted us to further investigate hindbrain-specific cells. Whole-mount in situ hybridization demonstrated decreased expression of atoh1a (marker specific for dorsal hindbrain progenitors28) by 1 dpf in the upper rhombic lip (URL) and lower rhomic lip (LRL) in embryos injected with SPL, compared to the normal pattern of robust expression of atoh1a in CTL-injected embryos of hindbrain progenitors in the URL and the LRL bilaterally28 (Fig. 3b). Whole-mount in situ hybridization further showed a lack of expression of pvalb7, which is specific for differentiated cerebellar Purkinje neurons28, by 3 dpf in embryos injected with SPL, compared to the normal expression in distinct clusters (highlighted by *) of differentiated Purkinje cells in embryos injected with CTL (Fig. 3b).
The abnormal phenotype from exosc3-specific morpholino injections was largely rescued by co-injection with wildtype zebrafish exosc3 mRNA (zWT, Fig. 3c; Supplementary Table 5), suggesting that the detrimental effects of the antisense morpholinos were specific to exosc3 knockdown. Co-injection with wildtype human EXOSC3 mRNA (hWT), which shares 67% identity with the zebrafish ortholog, was less effective in the rescue. Co-injection with zebrafish or human mRNA containing the mutations was ineffective, suggesting that the mutations disrupted the normal function of EXOSC3 (Fig. 3c; Supplementary Table 5). Survival data of embryos 1–3 dpf are stratified and summarized in Supplementary Table 5.
We have discovered disease-causing mutations in a gene encoding the exosome component EXOSC3 leading to PCH1 with combined cerebellar and spinal motor neuron degeneration of infantile onset. There is clinical heterogeneity. Affected individuals in families 1 and 3 do not present with primary hypoventilation and have survived beyond infancy and early childhood, which is exceptionally unusual for ‘classical’ PCH123,7. Furthermore, in families 1 and 2, autopsy showed profound cerebellar atrophy and variable involvement of the pons and inferior olives, suggesting a degenerative process in addition to a developmental disorder. Additional studies will facilitate endophenotype stratification of PCH1. There is clear genetic heterogeneity in PCH1, as some patients do not harbor mutations in any known PCH1 genes.
RNA exosomes are the principal enzymes that process and degrade RNA. The bulk of the human genome is transcribed to produce an extraordinary diversity of RNAs.29 The versatility and specificity of the exosome regulate the activity and maintain the fidelity of gene expression. Although exosomes are immunogenic in some patients with polymyositis-scleroderma 30,31 or chronic myelogenous leukemia 32,33, the findings in this report are the first to establish a pathogenic role for exosome mutations in human disease. Despite a growing effort to examine exosome function and subunit contribution, its substrates have not been fully characterized in human or lower animals, and the specific contribution of each component is incompletely understood. The discovery of naturally occurring mutations in exosome components provides a valuable opportunity to define subunit contribution to exosome function. Our findings suggest that normal function of the EXOSC3 component is essential to the survival of cerebellar and spinal motor neurons. Intriguingly, RNA dysregulation is emerging as important in the etiology of motor and cerebellar degeneration. RNA processing defects are implicated in SMN1 deficiency in SMA8. Mutations in RNA/DNA-binding proteins34–37 and pathogenic repeat expansions generating likely toxic RNA38,39 cause amyotrophic lateral sclerosis (ALS), an adult-onset motor neuron disease. RNA gain of function from noncoding repeat expansions was recently proposed to cause combined spinocerebellar and brainstem motoneuron degeneration of late onset in SCA3640. Dysregulation of tRNA processing underlies other subtypes of PCH24,23,21. The elucidation of the pathomechanism underlying PCH1 may lead to new insights regarding RNA processing in the development and survival of cerebellar and spinal motor neurons.
METHODS
Clinical characterization
PCH1 was diagnosed in 13 unrelated families from around the world, with documented congenital combined cerebellar and spinal motor neuron disease. Almost all subjects were hypotonic from birth. All had spontaneous breathing. Neurogenic muscle atrophy with spinal motor neuron disease was confirmed by EMG, muscle biopsy, or autopsy. Many subjects developed progressive microcephaly, with prominent cerebellar atrophy and variable involvement of the brainstem by MRI or autopsy.
Genetic analysis
DNA was extracted from peripheral blood from patients with parental consent and consenting subjects using standard methods. The study was approved by the UCLA Institutional Review Board. For exome sequencing, each library produced approximately 28 million single-end 76-base reads. The mean coverage of bases in the target exomes was 23X. Raw reads that passed Illumina’s quality filters were aligned to the reference human genome build 37 with Novoalign from Novocraft. The GATK UnifiedGenotyper was used to call single nucleotide variants and indels. Each case had ~15,000 non-reference variants, amounting to 19,098 total variants in the four cases. We limited the search to variants within the coding region and flanking intronic essential splice site of protein coding genes in the Ensembl dataset. Under the hypothesis that the disorder was rare and, therefore the causative allele(s) would not be common, we filtered out variants that were in dbSNP13241,42 and the 1000 Genomes Project43, leaving ~400 variants in each case, and a total of 699 variants. Under a recessive model we searched for homozygous variants and compound heterozygous variants (defined as two variants in the same transcript), shared by all four cases (n=15 and n=10, respectively). To compensate for bias in our own analytical system, we then filtered out variants we had identified in 25 exomes from unrelated unaffected cases. Sanger sequencing for further validation was performed using standard protocols (Supplementary Table 2).
Zebrafish morpholino injection & in situ hybridization
Cloning, mutagenesis, in vitro mRNA synthesis were performed using standard protocols (Supplementary Table 2). Zebrafish embryos were provided by the UCLA Zebrafish Core Facility and procedures were approved by UCLA Animal Research Committee. Fish were maintained at 28°C using a 14/10h light/dark cycle and bred to obtain embryos. Morpholino oligonucleotides (Gene Tools LLC) were designed to block translation initiation or the splice donor site of exon 2 of zebrafish exosc3 pre-mRNA (Supplementary Table 2). We obtained a standard control from Gene Tools. We used fine glass needles and a microinjector to perform injections on embryos at 1-cell stage. The injection volume ranged from 0.5 to 2.0 nL at a concentration of 3 ng/nL. Embryos were incubated in E3 medium at 28°C.
Digoxigenin-linked riboprobes were generated by using mMESSAGE mMACHINE SP6 KIT or mMESSAGE mMACHINE T7 Ultra KIT substituing the DIG RNA labeling Mix (Roche Applied Science). Whole-mount in situ hybridization were performed and the expression of specific genes was detected using an alkaline phosphatase-conjugated antibody against digoxigenin and a chromogenic substrate, as described previously.44
Riboprobe generation
To generate the anti-sense probe, full length zebrafish exosc3 cDNA was digested with EcoR I and Not I, then ligated into pCR-Blunt II-TOPO (Invitrogen) with T4 DNA ligase. The sense probe was transcribed using full length zebrafish exosc3 in pcDNA3.1/Zeo(+) (Invitrogen).
Zebrafish full length pvalb7 clone ID 7087368 (Thermo Scientific Open Biosystems) digested with EcoR I and Not I was ligated into pCR-Blunt II-TOPO (Invitrogen) with T4 DNA ligase. The sense probe was transcribed in vitro with the SP6 promoter and the anti-sense with the T7 promoter.
Full length zebrafish atoh1a clone ID 7428977 was obtained from Open Biosystems (Thermo Scientific), digested with EcoR I and Not I, then ligated into pcDNA3.1/Zeo(+) to generate the sense probe and pcDNA3.1/Zeo(−) (Invitrogen) for the anti-sense probe.
RNA probes for in situ hybridization were generated with mMESSAGE mMACHINE SP6 KIT or mMESSAGE mMACHINE T7 Ultra KIT (Ambion) substituting the DIG RNA labeling Mix (Roche Applied Science) for the NTP mix supplied in the kit. All probes were analyzed by denaturing agarose gel electrophoresis and quantified by NanoDrop.
Assessing the functional impact on EXOSC3 splicing of an intronic variant c.475-12A>G
We investigated the functional impact of the intronic variant c.475-12A>G on splicing using semi-quantitative RT-PCR in transfected cells. We obtained full-length EXOSC3 constructs (all exons and intervening introns) by PCR amplification using Phusion High-Fidelity DNA Polymerase (NEB; Supplementary Table 2) from genomic DNA of a normal subject and the proband of Family 6 (harboring mutation c.475-12A>G). The gel extracted amplicons were cloned into pcDNA3.1/Zeo(+) (linearized with BamH I and Xho I; Invitrogen) by using the In-Fusion® HD Cloning System (Clonetech). The full-length clones were confirmed by bi-directional Sanger sequencing.
The wildtype or mutant full-length constructs were introduced into HeLa cells by Lipofectamine 2000 (Invitrogen). Two days after transfection, RNA was extracted by using TRIzol Reagent (Invitrogen). cDNA was generated using Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Science) and BGH reverse primer located upstream of the BGH polyadenylation signal on pcDNA3.1/Zeo(+) (Invitrogen; Supplementary Figure 2). Reverse transcription with BGHr ensures that cDNA is synthesized exclusively from exogenous mRNA and eliminates transcription from endogenous EXOSC3. 1 μl of cDNA was PCR amplified with Phusion High-Fidelity DNA Polymerase (NEB) and primers hEXOSC3-c.423f and hEXOSC3-c.670r (Supplementary Table 2; Supplementary Figure 2). RT-PCR products were resolved on a 12% polyacrylamide gel visualized by GelRed (Biotium). We observed multiple products for both wild type and mutant constructs. To ascertain the identity of each product, amplicons were further cloned into pCR-Blunt II-TOPO (Invitrogen) and directly sequenced.
Cloning, mutagenesis, in vitro transcription
The human full length EXOSC3 cDNA ID 3346075 was obtained from Open Biosystems (Thermo Scientific), digested with EcoR I and Xho I, then ligated into pcDNA3.1/Zeo(+) (Invitrogen) with T4 DNA ligase. The zebrafish full length exosc3 clone ID 7282897 was obtained from Open Biosystems (Thermo Scientific), digested with EcoR I and Not I, then ligated into pcDNA3.1/Zeo(+) (Invitrogen) with T4 DNA ligase. Specific missense mutations were introduced into the wild type cDNA by QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies; Supplementary Table 2). All clones were fully sequenced bidirectionally by Sanger sequencing. In vitro transcription was performed with mMESSAGE mMACHINE T7 Ultra Kit (Ambion). The resulting mRNA was analyzed by denaturing agarose gel electrophoresis and quantified by NanoDrop.
Cell culture, siRNA Transfection, and RNA Extraction
HeLa cells were culutured in DMEM supplemented with 10% fetal bovine serum at 37 C and 5% CO2. Cells were transfected every 48 hours over 6 days with 60nM siRNA duplexes (sense: 5′-cacgcacaguacuaggucatt-3′) with Lipofectamine 2000 (Invitrogen) or left untreated, as detailed by Preker and colleagues45. After the cells were lysed in TRIzol Reagent (Invitrogen), RNA was extracted from the aqueous phase and protein from the remaining organic phase.
Fibroblast were grown in DMEM supplemented with 20% fetal bovine serum, 100nM MEM Non-Essential Amino Acids, 1 mM GlutaMAX, 100 U penicillin, and 100 μg streptomycin at 37 C and 5% CO2. Total RNA was extracted with TRIzol Reagent (Invitrogen).
Northern blotting
Northern Blotting was performed following standard protocols. The integrity of the RNA was analyzed by denaturing agarose gel electrophoresis and quantified by NanoDrop. 4 μg total RNA was resolved on a 6% denaturing polyacrylamide gel and transferred to positively charged nylon membranes (Roche). After crosslinking the RNA to membranes, blots were hybridized with Digoxigenin (DIG)-labled LNA (locked nucleic acid)-modified antisense oligonucleotide probe to 5.8s rRNA (5′-CGAAGTGTCGATGATCAAT-3xDig-3′; LNA in bold). Bound probes were detected with an alkaline phosphatase-congugated anti-DIG antibody (Roche Applied Science; 1:10000) and CSPD chemiluminescence (Roche Applied Science).
Western blotting
The protein concentration was determined by using the Micro BCA protein assay kit (Pierce). 10 μg lysates were separated by SDS-PAGE, then transferred onto a nitrocellulose membrane. Western blotting was performed following standard protocols with mouse monoclonal anti-EXOSC3 antibody (Santa Cruz Biotechnology; 1:400) followed by a secondary antibody horseradish peroxidase-conjugated horse anti-mouse IgG (Vector Laboratories; 1:5000). The blot was subsequently stripped and reprobed with chicken anti-GAPDH (Millipore; 1:1000) and secondary antibody horseradish peroxidase-conjugated goat anti-Chicken IgY (Abcam; 1:5000). Bound antibodies were visualized with Amersham ECL Plus™ Western Blotting Detection Reagents (Amersham).
Supplementary Material
Acknowledgments
The authors are grateful for the generosity of the families who have participated in this study. The sequencing and analytical work was supported by the Bioinformatics and Genomics Core of the UCLA Muscular Dystrophy Core Center (NIH/NIAMS P30AR057230) within the Center for Duchenne Muscular Dystrophy. We acknowledge the contribution of clinical data by P. Carpenter, University of California, Irvine, CA, USA. The work was supported by NIH/NEI R01 EY015311 & NINDS R01 NS064183 (J.C.J.), Deutsche Forschungsgemeinschaft Ru-746/1-1 (S.R.-S. & K.Z.), IZKF Aachen N5-4 (S.R.-S. & J.S.), Australian NHMRC Centre for Research Excellence (M.M.R.), Internal Grant Agency of Ministry of Health of the Czech Republic NS 10552-3 (P.S.), and Xunta de Galicia-INCITE 10PXIB9101280PR (M.-J.S.).
Footnotes
URLs. NHLBI Exome Sequencing Project http://snp.gs.washington.edu/EVS/; Novocraft (http://www.novocraft.com); Ensembl http://uswest.ensembl.org; dbSNP132 http://www.ncbi.nlm.nih.gov/projects/SNP/; 1000 Genome Project http://www.1000genomes.org/
Accession numbers. human EXOSC3 NM_016042.2; EXOSC3 NP_057126.2; zebrafish exosc3 NM_001029961.1, exosc3 NP_001025132.1
AUTHOR CONTRIBUTIONS
S.F.N. and J.C.J. designed the study. M.Y. Z.C., and S.F.N. analyzed data from SNP genotyping and exome sequencing. S. R.-S., M.P.M., J.S., M.S.S., D.C., P.S., A.v.M., L.G.-N, A.J.K., M.C.-G., M.-J.S., M.S., P.B.S., N.S., R.C.K., H.V.V., K.Z., M.M.R. provided and analyzed clinical material from subjects. H.M. J.H., and D.W.L. performed and analyzed data from Sanger sequencing of subjects and controls. H.M. and J.H. generated constructs and performed molecular genetic studies. J.W. performed and analyzed the functional studies in zebrafish. All authors contributed to the manuscript written by J.C.J.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-->5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 2.Jensen THe. RNA Exosome. Adv Exp Med Biol. 2010;702 [PubMed] [Google Scholar]
- 3.Norman RM. Cerebellar hypoplasia in Werdnig-Hoffmann disease. Arch Dis Child. 1961;36:96–101. doi: 10.1136/adc.36.185.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goutieres F, Aicardi J, Farkas E. Anterior horn cell disease associated with pontocerebellar hypoplasia in infants. J Neurol Neurosurg Psychiatry. 1977;40:370–378. doi: 10.1136/jnnp.40.4.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Leon GA, Grover WD, D’Cruz CA. Amyotrophic cerebellar hypoplasia: a specific form of infantile spinal atrophy. Acta Neuropathol. 1984;63:282–286. doi: 10.1007/BF00687334. [DOI] [PubMed] [Google Scholar]
- 6.Barth PG. Pontocerebellar hypoplasias. An overview of a group of inherited neurodegenerative disorders with fetal onset. Brain Dev. 1993;15:411–422. doi: 10.1016/0387-7604(93)90080-r. [DOI] [PubMed] [Google Scholar]
- 7.Namavar Y, Barth PG, Poll-The BT, Baas F. Classification, diagnosis and potential mechanisms in pontocerebellar hypoplasia. Orphanet J Rare Dis. 2011;6:50. doi: 10.1186/1750-1172-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melki J, et al. De novo and inherited deletions of the 5q13 region in spinal muscular atrophies. Science. 1994;264:1474–1477. doi: 10.1126/science.7910982. [DOI] [PubMed] [Google Scholar]
- 9.Gorgen-Pauly U, Sperner J, Reiss I, Gehl HB, Reusche E. Familial pontocerebellar hypoplasia type I with anterior horn cell disease. Eur J Paediatr Neurol. 1999;3:33–38. doi: 10.1053/ejpn.1999.0177. [DOI] [PubMed] [Google Scholar]
- 10.Muntoni F, et al. Clinical spectrum and diagnostic difficulties of infantile pontocerebellar hypoplasia type 1. Neuropediatrics. 1999;30:243–248. doi: 10.1055/s-2007-973498. [DOI] [PubMed] [Google Scholar]
- 11.Ryan MM, Cooke-Yarborough CM, Procopis PG, Ouvrier RA. Anterior horn cell disease and olivopontocerebellar hypoplasia. Pediatr Neurol. 2000;23:180–184. doi: 10.1016/s0887-8994(00)00166-1. [DOI] [PubMed] [Google Scholar]
- 12.Rudnik-Schoneborn S, et al. Extended phenotype of pontocerebellar hypoplasia with infantile spinal muscular atrophy. Am J Med Genet A. 2003;117A:10–17. doi: 10.1002/ajmg.a.10863. [DOI] [PubMed] [Google Scholar]
- 13.Chou SM, et al. Infantile olivopontocerebellar atrophy with spinal muscular atrophy (infantile OPCA + SMA) Clin Neuropathol. 1990;9:21–32. [PubMed] [Google Scholar]
- 14.Salman MS, et al. Pontocerebellar hypoplasia type 1: new leads for an earlier diagnosis. J Child Neurol. 2003;18:220–225. doi: 10.1177/08830738030180031201. [DOI] [PubMed] [Google Scholar]
- 15.Gomez-Lado C, Eiris-Punal J, Vazquez-Lopez ME, Castro-Gago M. Pontocerebellar hypoplasia type I and mitochondrial pathology. Rev Neurol. 2007;45:639–640. [PubMed] [Google Scholar]
- 16.Lev D, et al. Infantile onset progressive cerebellar atrophy and anterior horn cell degeneration--a late onset variant of PCH-1? Eur J Paediatr Neurol. 2008;12:97–101. doi: 10.1016/j.ejpn.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Szabo N, Szabo H, Hortobagyi T, Turi S, Sztriha L. Pontocerebellar hypoplasia type 1. Pediatr Neurol. 2008;39:286–288. doi: 10.1016/j.pediatrneurol.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 18.Tsao CY, Mendell J, Sahenk Z, Rusin J, Boue D. Hypotonia, weakness, and pontocerebellar hypoplasia in siblings. Semin Pediatr Neurol. 2008;15:151–153. doi: 10.1016/j.spen.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Sanefuji M, et al. Autopsy case of later-onset pontocerebellar hypoplasia type 1: pontine atrophy and pyramidal tract involvement. J Child Neurol. 2010;25:1429–1434. doi: 10.1177/0883073810372991. [DOI] [PubMed] [Google Scholar]
- 20.Renbaum P, et al. Spinal muscular atrophy with pontocerebellar hypoplasia is caused by a mutation in the VRK1 gene. Am J Hum Genet. 2009;85:281–289. doi: 10.1016/j.ajhg.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Namavar Y, et al. Clinical, neuroradiological and genetic findings in pontocerebellar hypoplasia. Brain. 2011;134:143–156. doi: 10.1093/brain/awq287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simonati A, Cassandrini D, Bazan D, Santorelli FM. TSEN54 mutation in a child with pontocerebellar hypoplasia type 1. Acta Neuropathol. 2011;121:671–673. doi: 10.1007/s00401-011-0823-1. [DOI] [PubMed] [Google Scholar]
- 23.Budde BS, et al. tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia. Nat Genet. 2008;40:1113–1118. doi: 10.1038/ng.204. [DOI] [PubMed] [Google Scholar]
- 24.Edvardson S, et al. Deleterious mutation in the mitochondrial arginyl-transfer RNA synthetase gene is associated with pontocerebellar hypoplasia. Am J Hum Genet. 2007;81:857–862. doi: 10.1086/521227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rankin J, et al. Pontocerebellar hypoplasia type 6: A British case with PEHO-like features. Am J Med Genet A. 2010;152A:2079–2084. doi: 10.1002/ajmg.a.33531. [DOI] [PubMed] [Google Scholar]
- 26.Brouwer R, et al. Three novel components of the human exosome. J Biol Chem. 2001;276:6177–6184. doi: 10.1074/jbc.M007603200. [DOI] [PubMed] [Google Scholar]
- 27.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 28.Kani S, et al. Proneural gene-linked neurogenesis in zebrafish cerebellum. Dev Biol. 2010;343:1–17. doi: 10.1016/j.ydbio.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 29.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolfe JF, Adelstein E, Sharp GC. Antinuclear antibody with distinct specificity for polymyositis. J Clin Invest. 1977;59:176–178. doi: 10.1172/JCI108616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allmang C, et al. The yeast exosome and human PM-Scl are related complexes of 3′ --> 5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang XF, et al. CML28 is a broadly immunogenic antigen, which is overexpressed in tumor cells. Cancer Res. 2002;62:5517–5522. [PMC free article] [PubMed] [Google Scholar]
- 33.Xie LH, et al. Activation of cytotoxic T lymphocytes against CML28-bearing tumors by dendritic cells transduced with a recombinant adeno-associated virus encoding the CML28 gene. Cancer Immunol Immunother. 2008;57:1029–1038. doi: 10.1007/s00262-007-0434-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kabashi E, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 35.Sreedharan J, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwiatkowski TJ, Jr, et al. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 37.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dejesus-Hernandez M, et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renton AE, et al. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobayashi H, et al. Expansion of intronic GGCCTG hexanucleotide repeat in NOP56 causes SCA36, a type of spinocerebellar ataxia accompanied by motor neuron involvement. Am J Hum Genet. 2011;89:121–130. doi: 10.1016/j.ajhg.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biesecker LG, et al. The ClinSeq Project: piloting large-scale genome sequencing for research in genomic medicine. Genome Res. 2009;19:1665–1674. doi: 10.1101/gr.092841.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhagwat M. Searching NCBI’s dbSNP database. Curr Protoc Bioinformatics. 2010;Chapter 1(Unit 1):19. doi: 10.1002/0471250953.bi0119s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 45.Preker P, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.