Abstract
In previous linkage and genome-wide association studies we identified 17 susceptibility loci for generalized vitiligo. By a second genome-wide association study, meta-analysis, and independent replication study, we have now identified 13 additional vitiligo-associated loci, including OCA2-HERC2, a region of 16q24.3 containing MC1R, a region of chromosome 11q21 near TYR, several immunoregulatory loci including IFIH1, CD80, CLNK, BACH2, SLA, CASP7, CD44, IKZF4, SH2B3, and a region of 22q13.2 where the causal gene remains uncertain. Functional pathway analysis shows that most vitiligo susceptibility loci encode immunoregulatory proteins or melanocyte components that likely mediate immune targeting and genetic relationships among vitiligo, malignant melanoma, and normal variation of eye, skin, and hair color.
In generalized vitiligo patches of depigmented skin and hair result from autoimmune destruction of epidermal melanocytes1, epidemiologically associated with other autoimmune diseases2. In previous linkage analyses and a genome-wide association study (GWAS1; Supplementary Table 1), we identified 14 confirmed and 3 suggestive vitiligo susceptibility loci3–5 in persons of non-Hispanic European (EUR) ancestry. Most encode immunoregulatory proteins, and several are associated with other autoimmune diseases. However, one, TYR, encodes tyrosinase, a key enzyme of melanin biosynthesis that likely mediates immune targeting of melanocytes. Causal variation at TYR is inversely associated with vitiligo and malignant melanoma6, suggesting vitiligo may be related to immune surveillance of melanoma7.
To identify additional vitiligo susceptibility loci, we performed a second GWAS (GWAS2; Supplementary Table 1), with meta-analysis of GWAS1 and GWAS2 (GWAS-MA) to enhance statistical power. GWAS2 included 450 EUR generalized vitiligo cases and genotype data from 3,182 EUR controls from the database of Genotypes and Phenotypes (dbGaP). The GWAS-MA demonstrated improved significance of almost all significant loci from GWAS1 (Supplementary Table 2) and suggestive association (P < 10−4 for multiple SNPs across a region) of 24 novel loci (Supplementary Figure 1 and Supplementary Table 3), of which six achieved genome-wide significance (P < 5 × 10−8) (Table 1). At all 24 novel loci, we imputed genotypes using 1000 Genomes Project data, performed logistic regression to identify independent association signals, and genotyped the most significant SNPs at each locus (Supplementary Table 3) in an independent replication cohort of 1440 EUR cases and 1316 EUR controls. We then performed overall meta-analysis of GWAS1, GWAS2, and the replication study, using conservative criteria for confirming association: (i) nominal association in the replication study (P < 0.05), (ii) consistent high-risk alleles in GWAS1, GWAS2, and the replication study, (iii) non-significant heterogeneity across all three studies (P > 1.09 × 10−3), and (iv) overall combined genome-wide significance (P < 5 × 10−8).
Table 1.
GWAS meta-analysis, replication, and overall meta-analysis of novel generalized vitiligo susceptibility loci
| GWAS-MA (GWAS 1 + GWAS 2) | Replication study | Overall Meta-Analysis (GWAS 1 + GWAS 2+ replication study) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | SNP | Positional Candidate gene | RA | EA | EAF | P | OR | TREND P | OR | P | OR | Breslow-Day P |
| 2q24.2 | rs2111485 | IFIH1 | G | A | 0.38 | 1.67 × 10−10 | 0.77 | 7.26 × 10−6 | 0.78 | 4.91 × 10−15 | 0.77 | 1.14 × 10−2 |
| rs1990760 | T | C | 0.38 | 2.00 × 10−9 | 0.78 | 1.18 × 10−5 | 0.78 | 9.33 × 10−14 | 0.78 | 2.83 × 10−2 | ||
| 3q13.33 | rs59374417 | CD80 | A | C | 0.13 | 3.97 × 10−7 | 1.33* | 3.02 × 10−4 | 1.33 | 3.78 × 10−10* | 1.34* | 5.41 × 10−1 |
| rs4330287 | C | A | 0.13 | 3.97 × 10−7 | 1.33 | 5.90 × 10−4 | 1.31 | 7.80 × 10−10 | 1.33 | 5.80 × 10−1 | ||
| 4p16.1 | rs11940117 | CLNK | C | T | 0.47 | 9.00 × 10−8 | 1.24* | 3.03 × 10−2 | 1.13 | 1.96 × 10−8* | 1.20* | 3.99 × 10−1 |
| rs16872571 | T | C | 0.56 | 2.50 × 10−7 | 1.23 | 1.54 × 10−2 | 1.16 | 1.56 × 10−8 | 1.21 | 7.31 × 10−1 | ||
| 6q15 | rs3757247 | BACH2 | G | A | 0.47 | 2.14 × 10−5 | 1.18 | 2.91 × 10−4 | 1.22 | 2.53 × 10−8 | 1.20 | 5.16 × 10−1 |
| 8q24.22 | rs853308 | SLA | A | G | 0.48 | 1.14 × 10−6 | 1.21 | 3.24 × 10−3 | 1.18 | 1.58 × 10−8 | 1.20 | 8.33 × 10−1 |
| 10q25.3 | rs3814231 | CASP7 | G | A | 0.25 | 1.20 × 10−5 | 0.81 | 8.80 × 10−4 | 0.81 | 3.56 × 10−8 | 0.81 | 9.77 × 10−1 |
| rs4353229 | T | C | 0.25 | 1.32 × 10−* | 0.84 | 9.10 × 10−4 | 0.81 | 4.35 × 10−7 | 0.83 | 9.18 × 10−1 | ||
| 11p13 | rs736374 | CD44 | G | A | 0.38 | 3.06 × 10−8 | 1.25 | 1.45 × 10−2 | 1.15 | 3.41 × 10−9 | 1.22 | 1.51 × 10−1 |
| rs10768122 | A | G | 0.41 | 6.13 × 10−8 | 1.24 | 5.74 × 10−3 | 1.17 | 1.78 × 10−9 | 1.22 | 1.66 × 10−1 | ||
| 11q21 | rs4409785 | TYR regulation | T | C | 0.19 | 2.26 × 10−10 | 1.36 | 1.58 × 10−4 | 1.30 | 1.57 × 10−13 | 1.34 | 4.63 × 10−1 |
| rs11021232 | T | C | 0.20 | 9.20 × 10−10 | 1.35 | 3.23 × 10−5 | 1.32 | 1.91 × 10−13* | 1.34* | 6.07 × 10−1 | ||
| 12q13.2 | rs1701704 | IKZF4 | A | C | 0.35 | 1.53 × 10−9 | 1.28 | 4.44 × 10−6 | 1.30 | 3.19 × 10−14 | 1.29 | 8.09 × 10−1 |
| rs2456973 | A | C | 0.35 | 1.22 × 10−9 | 1.28 | 4.97 × 10−6 | 1.30 | 2.75 × 10−14* | 1.29* | 8.16 × 10−1 | ||
| 12q24.12 | rs3184504 | SH2B3 | T | C | 0.49 | 1.32 × 10−11 | 0.76 | 2.31 × 10−7 | 0.75 | 2.46 × 10−17 | 0.76 | 1.17 × 10−1 |
| rs4766578 | T | A | 0.48 | 9.10 × 10−12 | 0.76 | 4.07 × 10−8 | 0.73 | 3.54 × 10−18* | 0.76* | 1.01 × 10−1 | ||
| 15q12-13.1 | rs1129038 | OCA2-HERC2 | T | C | 0.27 | 3.23 × 10−7 | 1.26 | 2.06 × 10−2 | 1.14 | 3.91 × 10−8* | 1.22* | 4.66 × 10−1 |
| rs12913832 | G | A | 0.27 | 3.29 × 10−7 | 1.26 | 2.01 × 10−2 | 1.14 | 3.81 × 10−8 | 1.22 | 4.71 × 10−1 | ||
| 16q24.3 | rs4785587 | MC1R | G | A | 0.49 | 1.08 × 10−8 | 0.80 | 1.44 × 10−4 | 0.81 | 6.35 × 10−12* | 0.80* | 7.98 × 10−1 |
| rs9926296 | A | G | 0.46 | 4.34 × 10−11 | 0.77 | 5.16 × 10−4 | 0.82 | 1.82 × 10−13* | 0.79* | 1.35 × 10−1 | ||
| 19p13.3† | rs6510827 | TICAM1 | C | T | 0.41 | 6.98 × 10−6 | 1.20 | 3.87 × 10−3 | 1.17 | 8.80 × 10−8 | 1.19 | 6.37 × 10−1 |
| 22q13.2 | rs4822024 | TOB2 | G | A | 0.21 | 1.02 × 10−7 | 0.76 | 1.54 × 10−3 | 0.81 | 6.81 × 10−10* | 0.78* | 2.00 × 10−1 |
| rs79008 | G | A | 0.19 | 1.44 × 10−6 | 0.78 | 1.13 × 10−3 | 0.79 | 6.19 × 10−9 | 0.78 | 7.95 × 10−1 | ||
This table reports data for newly identified vitiligo susceptibility loci that had P < 1 × 10−4 in the GWAS meta-analysis (GWAS-MA; GWAS1 + GWAS2), P < 0.05 in the replication study, P < 5 × 10−8 in the overall meta-analysis (GWAS1 + GWAS2 + replication study), with consistent effect allele and non-significant Breslow-Day heterogeneity statistics for the ORs across all three studies (P > 1.09 × 10−3). RA, reference allele; EA, effect allele; EAF, effect allele frequency (among all cases and controls).
Imputed from 1000 Genomes Project data.
The association signal at 19p13.3 does not quite achieve the criterion for genome-wide significance but is included for completeness
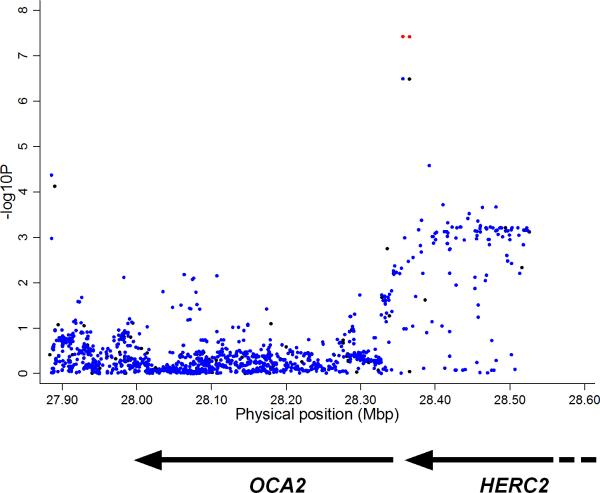
As shown in Table 1, we confirmed association of vitiligo with 13 novel loci. Among the most interesting, at chromosome 15q12–q13.1 the GWAS-MA showed suggestive association of SNPs (nt 27886016–28392261) spanning OCA2 upstream to within HERC2 (Fig. 1), especially rs12913832 (P = 3.29 × 10−7) and imputed SNP rs1129038 (P = 3.23 × 10−7) (r2 = 0.99). OCA2 is causal for oculocutaneous albinism type 2, encodes a melanosomal membrane transporter8, and plays a major role in determining skin, hair, and eye color. The replication study and overall meta-analysis confirmed association of both rs1129038 (P = 3.91 × 10−8, OR 1.22) and rs12913832 (P = 3.81 × 10−8, OR 1.22) (Table 1). Furthermore, the SNP alleles that are low-risk for vitiligo are strongly associated with gray/blue eye color9–11 and with elevated risk of malignant melanoma12,13, tagging a founder variant within HERC2 that down-regulates transcription of the OCA2 allele in cis11,14. OCA2 is thus analogous to TYR: both encode melanocyte antigens presented by HLA-A*0215,16, for both vitiligo protection is associated with reduced functional protein, and for both susceptibility to vitiligo and melanoma constitute genetic opposites7, perhaps modulating immune surveillance for melanoma (Supplementary Fig. 2). Furthermore, we predict that gray/blue eye color should be under-represented, and tan/brown eye color over-represented, among vitiligo patients. To test this, we surveyed 1206 EUR vitiligo patients, confirming both predictions: among vitiligo patients the prevalence of gray/blue eye color (26.8%) was greatly reduced and tan/brown eye color (43.2%) greatly elevated compared to both USA17 (P < 0.0001) and Australian18 (P < 0.0001) EUR individuals; Table 2). Compared to persons with gray/blue eye color, the OR for vitiligo was 2.98 in persons with tan/brown eye color and 2.25 in persons with green/hazel eye color, indicating additional eye color genes besides OCA2 constitute risk loci for vitiligo, and indeed TYR is associated both with vitiligo3 and with green/hazel eye color19.
Figure 1.
Association of generalized vitiligo with SNPs in the OCA2-HERC2 region of chromosome 15q12–q13.1. Results of Cochran-Mantel-Haenszel meta-analysis of GWAS1 and GWAS2 data (GWAS-MA) for genotyped (black) and imputed (blue) SNPs on the y axis versus chromosomal nucleotide position (GRCh37/hg19) on the x axis. Red circles indicate the Cochran-Mantel-Haenszel P values from the GWAS1, GWAS2, and replication studies for rs12913832 and rs1129038 (see Table 1). Arrows indicate gene positions and transcriptional orientation.
Table 2.
Eye color among Non-Hispanic/Latino European-derived vitiligo patients versus normal individuals
| Eye color | USA/Canada/UK vitiligo patients (%) | USA European-derived normals17 (%) | Australian European-derived normals18 (%) |
|---|---|---|---|
| Blue/gray | 323 (26.8) | 1856 (51.6) | 1314 (46.1) |
| Green/hazel | 362 (30.0) | 788 (21.7) | 789 (27.7) |
| Tan/brown | 521 (43.2) | 968 (26.7) | 749 (26.3) |
| P-value | - | <0.0001 | <0.0001 |
P values were obtained by chi-square distribution comparison of the number of individuals with tan/brown, green/hazel, and blue/gray eyes between the vitiligo patient and indicated normal groups.
At chromosome 16q24.3, the GWAS-MA showed complex association of SNPs spanning nt 89647951–90078022, particularly rs8049897 (P = 2.03 × 10−7) and imputed SNPs rs9926296 (P = 4.34 × 10−11) and rs4785587 (P = 1.08 × 10−8) (Supplementary Fig. 3a), confirmed by the replication study and overall meta-analysis (rs9926296 P = 1.82 × 10−13, OR 0.79). The associated region contains 20 genes, notably including MC1R, encoding the melanocortin receptor, a regulator of melanogenesis and minor vitiligo autoantigen, associated with malignant melanoma and with skin and hair color20.
At 11q21, the GWAS-MA showed association with rs4409785 (nt 95311422) (P = 2.26 × 10−10) and imputed SNP rs11021232 (P = 9.20 × 10−10) (Supplementary Fig. 3b), confirmed by the replication study and overall meta-analysis (rs4409785 P = 1.57 × 10−13, OR 1.34). These SNPs are located in a 559 kb region containing no known genes, approximately 6.28 Mb distal to TYR. These SNPs are not in linkage disequilibrium with TYR SNPs (r2=0), and remain highly significant when conditioned on common causal TYR SNPs rs1042602 and rs1126809. We speculate this region might harbor a regulatory element affecting TYR transcription in cis.
Besides OCA2, MC1R, and the chromosome 11q21 locus, most vitiligo-associated loci encode immunoregulatory proteins. At chromosome 2q24.2, the GWAS-MA showed association with SNPs spanning nt 163076146–163154363, between IFIH1 and FAP, particularly rs2111485 (P = 1.67 × 10−10) (Supplementary Fig. 3c), confirmed by the replication study and overall meta-analysis (rs2111485 P = 4.91 × 10−15, OR 0.77). IFIH1 encodes an interferon-induced RNA helicase involved in antiviral innate immune responses21, associated with type 1 diabetes22, Graves' disease23, multiple sclerosis24, psoriasis25, and perhaps lupus26.
At 3q13.33, the GWAS-MA showed suggestive association of SNPs (nt 119276377–119197379) spanning and upstream of CD80, particularly rs4330287 and imputed SNP rs59374417 (both P = 3.97 × 10−7; r2 = 1.0) (Supplementary Fig. 3d), confirmed by the replication study and overall meta-analysis (rs59374417 P = 3.78 × 10−10, OR 1.34). CD80 is a surface protein on activated B-cells, monocytes, and dendritic cells that co-stimulates T cell priming27,28.
At 4p16.1, the GWAS-MA showed suggestive association of SNPs (nt 10702156–10729386) upstream of CLNK, including rs16872571 (P = 2.50 × 10−7) and several imputed SNPs, particularly rs11940117 (P = 9.00 × 10−8) (Supplementary Fig. 3e), confirmed by the replication study and overall meta-analysis (rs16872572 P = 1.56 × 10−8, OR 1.21). CLNK encodes mast cell immunoreceptor signal transducer, a positive regulator of immunoreceptor signaling29.
At 6q15, the GWAS-MA showed suggestive association of SNPs (nt 90941239-91915693) spanning BACH2, particularly rs3757247 (P = 2.14 × 10−5) (Supplementary Fig. 3f), confirmed by the replication study and overall meta-analysis (P = 2.53 × 10−8, OR 1.20). BACH2 encodes a transcriptional repressor of B cells30, and is associated with type 1 diabetes31,32, celiac disease33, and Crohn's disease34.
At 8q24.22, the GWAS-MA showed suggestive association of SNPs (nt 133929917-133979872) spanning the TG/SLA locus, at which the two genes are interdigitated and encoded on the opposite strands. Most significant were rs853308 (P = 1.14 × 10−6) and several imputed SNPs (Supplementary Fig. 3g), confirmed by the replication study and overall meta-analysis (rs853308 P = 1.58 × 10−8, OR 1.20). TG encodes thyroglobulin, while SLA encodes Src-like adaptor protein, a regulator of antigen receptor signaling35. TG is associated with autoimmune thyroid disease36, which affects approximately 17 percent of vitiligo patients2, suggesting association of rs853308 with vitiligo might derive from patients with concomitant autoimmune thyroid disease. However, stratification showed association of rs853308 with vitiligo derives both from patients with (P = 2.43 × 10−3) and without (P = 3.98 × 10−7) autoimmune thyroid disease, with virtually identical ORs in the two subgroups (1.20 and 1.19, respectively). Moreover, directly comparing rs853308 in vitiligo patients with (N = 608) versus without (N = 2579) autoimmune thyroid disease showed no difference (P = 0.94, OR 1.01). It is not apparent what role thyroglobulin might play in vitiligo pathogenesis, suggesting association of vitiligo with the TG/SLA locus may derive from SLA, rather than TG. SLA might likewise account for reported association of TG with autoimmune thyroid disease36.
At 10q25.3, the GWAS-MA showed suggestive association of SNPs (nt 115439530-115492092) spanning CASP7, particularly rs3814231 (P = 1.20 × 10−5) (Supplementary Fig. 3h), confirmed by the replication study and overall meta-analysis (P = 3.56 × 10−8, OR 0.81). CASP7 encodes caspase 7, an executioner protein of apoptosis and inflammation37, associated with rheumatoid arthritis38, and suggested as a candidate gene for IDDM17 in type 1 diabetes39.
At 11p13, the GWAS-MA showed association of SNPs (nt 35242907-35375280) spanning portions of CD44 and SLC1A2, particularly rs736374 (P = 3.06 × 10−8) and rs10768122 (P = 6.13 × 10−8) (Supplementary Fig. 3i), confirmed by the replication study and overall meta-analysis (rs10768122 P = 1.78 × 10−9, OR 1.21). CD44 encodes a cell surface glycoprotein with various functions, including a role in T cell development40, and is associated with lupus41.
At 12q13.2, the GWAS-MA showed association with SNPs (nt 56369506-56535251) in a region including IKZF4 (Supplementary Fig. 3j), particularly rs1701704 (P = 1.53 × 10−9) and imputed SNP rs2456973 (P = 1.22 × 10−9), confirmed by the replication study and overall meta-analysis (rs2456983 P = 2.75 × 10−14, OR 1.29). IKZF4 encodes a regulator of T cell activation42, and is associated with type 1 diabetes43 and alopecia areata44.
At 12q24.12, the GWAS-MA showed association with SNPs (nt 111708458-112906415) within and near SH2B3, particularly rs3184504 (P = 1.32 × 10−11) and imputed SNP rs4766578 (P = 9.10 × 10−12), located downstream, within ATXN2 (Supplementary Fig. 3k). Many SNPs in this region achieved genome-wide significance, but logistic regression analysis indicated all reflect a single association signal. The replication study and overall meta-analysis confirmed association of both rs3184504 (P = 2.46 × 10−17, OR 0.76) and rs4766578 (P = 3.54 × 10−18, OR 0.76). ATXN encodes Ataxin-2, and is causal for spinocerebellar ataxia type 2. SH2B3 encodes adaptor protein LNK, regulating development of both B and T cells45, and associated with type 1 diabetes46, celiac disease47, rheumatoid arthritis48, multiple sclerosis49, and perhaps lupus25. SH2B3 thus seems more likely relevant to vitiligo susceptibility than ATXN2.
At 22q13.2, the GWAS-MA showed suggestive association with SNPs in a broad region (nt 41707054-42062822), particularly rs79008 (P = 1.44 × 10−6), upstream of TOB2, and several imputed SNPs, including rs4822024 (P = 1.02 × 10−7), between ZC3H7B and TEF (Supplementary Fig. 3l). The replication study and overall meta-analysis confirmed association of both SNPs, greatest with rs4822024 (P = 6.81 × 10−10, OR 0.78). TOB2 encodes a regulator of cell cycle progression involved in T cell tolerance50. However, the locus contains 12 genes, and assignment of TOB2 as causal remains uncertain.
Besides these 13 confirmed vitiligo-associated loci, an additional locus at 19p13.3 did not quite achieve genome-wide significance. The GWAS-MA showed suggestive association of SNPs (nt 4830628-4837557) spanning TICAM1, particularly rs6510827 (P = 6.98 × 10−6) and imputed SNP rs811383825 (P = 1.49 × 10−5) (Supplementary Fig. 3m), with association with rs6510827 confirmed by the replication study and near-significance in the overall meta-analysis (P = 8.80 × 10−8, OR 1.19). TICAM1 encodes toll-like receptor adaptor molecule 1, which mediates innate immune responses to viral pathogens51. SNPs at ten additional loci that appeared suggestive in the GWAS-MA were not confirmed by the replication study (Supplementary Table 3).
Altogether, the vitiligo susceptibility loci we identified account for approximately 10% of vitiligo risk in EUR individuals, and about 18% of vitiligo heritability (h2 ~ 0.75). Most encode immunoregulatory proteins or melanocyte proteins that likely mediate antigenic triggering and immune targeting of melanocytes, and bioinformatic network analysis indicated at least 26 comprise a functional network spanning from the melanocyte to the immune system (Supplementary Fig. 4). Many vitiligo susceptibility loci are shared with other autoimmune diseases, most sharing the same high-risk alleles, consistent with epidemiological associations among these diseases (Supplementary Fig. 5). Functional prediction of all genotyped and imputed missense and splice junction variants in all confirmed non-MHC vitiligo loci (Supplementary Table 4) identified predicted deleterious variants at PTPN22, IFIH1, SLA, CD44, TYR, OCA2, MC1R, UBASH3A, and C1QTNF6, and for TYR two common variants, S192Y and R402Q, confer protection from vitiligo, whereas HLA-A *02:01 confers risk6. Additional vitiligo susceptibility loci undoubtedly remain undiscovered; nevertheless, the genes and pathways already identified provide insights into vitiligo pathobiology, optimal use of existing treatments, and even novel therapeutics.
METHODS
Subjects
GWAS1 has been described previously3. GWAS2 included 450 unrelated generalized vitiligo patients (cases) of non-Hispanic/Latino European ancestry (EUR) from North America and Europe, who met strict clinical criteria for generalized vitiligo52. Controls for GWAS2 were 3182 EUR individuals not specifically known to have any autoimmune disease or malignant melanoma, for whom genome-wide genotypes were obtained from the database of Genotypes and Phenotypes (dbGaP; phs000092v1, phs000125v1, phs000138v2, phs000168v1, and phs000206v3), or from the Illumina iControlDB. The replication study included 1440 unrelated EUR generalized vitiligo cases and 1316 unrelated EUR controls from North America and Europe, principally spouses of vitiligo patients from the GWAS2 study and the replication study itself. There was no overlap of cases and controls between the GWAS1, GWAS2, and replication cohorts. Cases and controls provided clinical history regarding vitiligo and other autoimmune diseases as described previously for GWAS13, and controls having known relatives with vitiligo or reporting any known autoimmune diseases or melanoma were excluded. Eye color was by self-report. Written informed consent was obtained from all study subjects. This study was approved by each institutional review board and was conducted according to Declaration of Helsinki principles.
Genotyping and quality control
Genomic DNA was prepared from saliva specimens using a DNA self-collection kit per the manufacturer's instructions (Oragene, DNA Genotek). For genome-wide genotyping, DNA concentrations were assayed by both fluorescence staining (PicoGreen method, Invitrogen) and ultraviolet A260 spectrophotometry (Nanodrop, Thermo Scientific). For genotyping specific SNPs in the replication study, DNA concentrations were assayed by Nanodrop.
Genome-wide genotyping for GWAS2 cases was performed for 657,366 SNPs using Illumina Human660W-Quad BeadChips per the manufacturer's instructions. GWAS2 controls obtained from dbGaP and Illumina iControlDB had been genotyped using Illumina Human 610-Quad and Human 1Mv1 BeadChips. Genotyping of 46 SNPs in the replication study was performed using a custom Illumina GoldenGate assay per the manufacturer's instructions.
Quality control filtering of genome-wide genotype data in GWAS2 was carried out as described for GWAS13 using Illumina GenomeStudio, version 3 and PLINK53, version 1.07. Cases were excluded on the basis of SNP call rates < 98.5% (N = 16), discordance between reported and observed sex (N = 0), and/or inadvertent subject duplication (N = 0). Beyond prior quality control procedures, controls were excluded on the basis of SNP call rates < 95% (N = 0). Additional cases (N = 6) and controls (N = 356) were excluded on the basis of cryptic relatedness based on pair-wise identity-by-descent estimation (Pi-hat > 0.05) of the entire (GWAS1 + GWAS2) summary dataset, in which case the individual with lower SNP call rate was excluded. SNPs were excluded on the basis of genotype missing rate ≥ 2% overall (N = 97,301) in cases plus controls, observed minor allele frequency < 0.01 (N = 23,064), significant deviation (P < 10−5) from Hardy-Weinberg equilibrium in the control dataset (N = 5,241), and/or significant difference (P < 10−5) in genotype missing rate in cases versus controls (N = 35,939). Data for SNPs with P values < 10−15 were reviewed with respect to genotype clusters and allele calls in cases, and minor allele frequency in controls compared to data in public data sources, and were excluded if there were apparent data quality problems (N = 2). To control for population stratification, we performed principle components analysis using EIGENSOFT54, version 4.2, and excluded as outliers cases (N = 10) and controls (N = 16) whose ancestry was ≥ 6 standard deviations from the mean on one of the top ten eigenvectors.
In the replication study we successfully determined genotypes for 46 SNPs that showed suggestive (P < 10−4) or significant P values in the genome-wide meta-analysis of GWAS1 and GWAS2, using a custom Illumina GoldenGate assay. SNP genotypes were subjected to quality control filters similar to those in GWAS1 and GWAS2, as appropriate.
Statistical analyses
For GWAS2, after quality control filtering of subjects and SNP data and removal of genetic outliers, we compared allele frequencies of the remaining 495,821 SNPs in the final 418 cases and 2810 controls using the unadjusted Cochran-Armitage trend test implemented in PLINK53 and the adjusted Cochran-Armitage trend test implemented in EIGENSOFT54, in which both phenotypes and genotypes of subjects were adjusted for ancestry using the top ten eigenvectors. The unadjusted Cochran-Armitage trend tests and the adjusted Cochran-Armitage trend tests yielded genomic inflation factors of 1.054 and 1.050, respectively, indicating that residual population stratification was negligible. Odds ratios and 95% confidence limits were calculated by logistic regression analysis by use of PLINK53.
To fully assess association at the 24 novel suggestive loci, we imputed genotypes for each locus using MaCH55, ver.1.0 based on patterns of haplotype variation in the 1000 Genomes Project European ancestry samples (Release Aug 4, 2010). We retained imputed SNPs with r2 > 0.3 and minor allele frequency > 0.01 for further analyses.
For the replication study, after quality control filtering, we compared allele frequencies for genotyped SNPs in the remaining 1377 patients and 1284 controls using the Cochran-Armitage trend test. Odds ratios and 95% confidence limits were calculated by logistic regression analysis.
To obtain combined ORs and P values for GWAS1 and GWAS2, and for the combined GWAS1, GWAS2, and replication studies, we performed meta-analysis using a Cochran-Mantel-Haenszel test. A Breslow-Day test was used to test for heterogeneity of ORs of the same SNP in different study cohorts.
Calculation of linkage disequilibrium between SNPs in regions of association was carried out using Haploview56, version 4.2. As a test of the independent effect of a given locus conditioned on the effect of another locus, we compared the fit of a model containing both loci to a model containing only the conditioning locus, assuming a multiplicative genotypic effect for the high-risk allele of each locus. Analyses were performed using STATA, version 10.0.
We estimated the contribution of known vitiligo susceptibility loci to the total variance in vitiligo liability as the difference between the variance accounted for by all SNPs genome-wide and the variance remaining after removing SNPs in known vitiligo susceptibility loci, using GCTA57 to estimate the two components of variance. We estimated heritability of vitiligo liability58,59 assuming a vitiligo prevalence in the EUR population of 0.003860 and sibling risk 0.062.
Functional network analysis
To gain insights into potential functional relationships among proteins encoded by vitiligo risk loci, we carried out functional interaction network analysis using the Search Tool for the Retrieval of Interacting Genes (STRING) 9.061. Input data were all known vitiligo susceptibility loci (including TICAM1, and selecting BTNL2 to represent the MHC class II gene region3). In addition, we iteratively tested inclusion of all proteins encoded by genes in the 16q24.3 and 22q13.2 association regions as well as FAM76B as potentially representing the 11q21 association region as a means of possible gene identification.
Supplementary Material
ACKNOWLEDGMENTS
We thank the many vitiligo patients and normal control individuals around the world who participated in this study. We thank the University of Colorado Cancer Center Genomics and Microarray Core for genomewide genotyping, and BodySync (Aurora, CO) for replication genotyping. Supported by grants R01AR045584, R01AR056292, and P30AR057212 from the U.S. National Institutes of Health.
Footnotes
URLs. 1000 Genomes Project, http://www.1000genomes.org/; 1000 Genomes Project data, http://www.sph.umich.edu/csg/abecasis/MACH/download/1000G-2010-08.html; CCDS database, http://www.ncbi.nlm.nih.gov/CCDS/CcdsBrowse.cgi; Functional Genome Server, http://gvs.gs.washington.edu/GVS/; Genomic Evolutionary Rate Profiling http://mendel.stanford.edu/SidowLab/downloads/gerp/index.html; Illumina iControlDB, http://www.illumina.com/science/icontroldb.ilmn; MACH, http://www.sph.umich.edu/csg/abecasis/MACH/download/; NIH Database of Genotypes and Phenotypes (dbGaP), http://www.ncbi.nlm.nih.gov/gap; PHylogenetic Analysis with Space/Time models Conservation, http://genome.ucsc.edu/goldenPath/help/phastCons.html; PLINK, http://pngu.mgh.harvard.edu/purcell/plink/; STATA, http://www.stata.com; STRING database, http://string-db.org
AUTHOR CONTRIBUTIONS Y.J. performed statistical analyses. K.G. managed computer databases and genotype data. T.M.F., S.B., S.A.B., S.L.R., and J.B.C. managed DNA samples and contributed to experimental procedures. P.J.H. managed subject coordination. S.A.B., D.C.B., R.L., A.W., J.P.v.d.V., M.R.W., W.T.M., E.H.K., D.J.G., A.P.W., M.P., G.L., A.T., T.J., K.E., N.v.G., J.L., A.O., A.H., R.M.H., and N.B.S. provided subject samples and phenotype information. P.R.F. and R.A.S. oversaw and managed all aspects of the study. R.A.S. wrote the first draft of the manuscript. All authors contributed to the final paper.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
References
- 1.Picardo M, Taïeb A, editors. Vitiligo. Springer; Heidelberg & New York: 2010. [Google Scholar]
- 2.Alkhateeb A, et al. Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res. 2003;16:208–214. doi: 10.1034/j.1600-0749.2003.00032.x. [DOI] [PubMed] [Google Scholar]
- 3.Jin Y, et al. Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. New Engl. J. Med. 2010;362:1686–1697. doi: 10.1056/NEJMoa0908547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin Y, et al. Common variants in FOXP1 are associated with generalized vitiligo. Nat. Genet. 2010;42:576–578. doi: 10.1038/ng.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birlea SA, et al. Comprehensive association analysis of candidate genes for generalized vitiligo supports XBP1, FOXP3, and TSLP. J. Invest. Dermatol. 2011;131:371–381. doi: 10.1038/jid.2010.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin Y, et al. Next-generation DNA re-sequencing identifies common variants of TYR and HLA-A that modulate the risk of generalized vitiligo via antigen presentation. J. Investig. Dermatol. 2012 doi: 10.1038/jid.2012.37. advance online publication 8 March 2012; doi: 10.1038/jid.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spritz RA. The genetics of generalized vitiligo: autoimmune pathways and an inverse relationship with malignant melanoma. Genome Med. 2010;19(2(10)):78. doi: 10.1186/gm199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rinchik EM, et al. A gene for the mouse pink-eyed dilution locus and for human type II oculocutaneous albinism. Nature. 1993;361:72–76. doi: 10.1038/361072a0. [DOI] [PubMed] [Google Scholar]
- 9.Kayser M, et al. Three genome-wide association studies and a linkage analysis identify HERC2 as a human iris color gene. Am J. Hum. Genet. 2008;82:411–423. doi: 10.1016/j.ajhg.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sturm RA, et al. A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am. J. Hum. Genet. 2008;82:424–431. doi: 10.1016/j.ajhg.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eiberg H, et al. Blue eye color in humans may be caused by a perfectly associated founder mutation in a regulatory element located within the HERC2 gene inhibiting OCA2 expression. Hum. Genet. 2008;123:177–187. doi: 10.1007/s00439-007-0460-x. [DOI] [PubMed] [Google Scholar]
- 12.Jannot A-S, et al. Allele variations in the OCA2 gene (pink-eyed-dilution locus) are associated with genetic susceptibility to melanoma. Eur. J. Hum. Genet. 2005;13:913–920. doi: 10.1038/sj.ejhg.5201415. [DOI] [PubMed] [Google Scholar]
- 13.Amos CI, et al. Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum. Molec. Genet. 2011;20:5012–5023. doi: 10.1093/hmg/ddr415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook AL, et al. Analysis of cultured human melanocytes based on polymorphisms within the SLC45A2/MATP,SLC24A5/NCKX5, and OCA2/P loci. J. Investig. Dermatol. 2009;129:392–405. doi: 10.1038/jid.2008.211. [DOI] [PubMed] [Google Scholar]
- 15.Skipper JC. An HLA-A2-restricted tyrosinase antigen on melanoma cells results from posttranslational modification and suggests a novel pathway for processing of membrane proteins. J. Exp. Med. 1996;183:527–534. doi: 10.1084/jem.183.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Touloukian CE, Leitner WW, Robbins PF, Rosenberg S, Restifo NP. Mining the melanosome for tumor vaccine targets: P.polypeptide is a novel tumor-associated antigen. Canc. Res. 2001;61:8100–8104. [PMC free article] [PubMed] [Google Scholar]
- 17.Tomany SC, Klein R, Klein BEK. The relationship between iris color, hair color, and skin sun sensitivity and the 10-year incidence of age-related maculopathy: the Beaver Dam Eye Study. Ophthalmology. 2003;110:1526–1533. doi: 10.1016/s0161-6420(03)00539-6. [DOI] [PubMed] [Google Scholar]
- 18.Duffy DL, et al. A three-single-nucleotide polymorphism haplotype in intron 1 of OCA2 explains most human eye-color variation. Am. J. Hum. Genet. 2007;80:241–252. doi: 10.1086/510885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sulem P. Genetic determinants of hair, eye, and skin pigmentation in Europeans. Nat. Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 20.Dessinioti C, Antoniou C, Katsambas A, Stratigos AJ. Melanocortin 1 receptor variants: functional role and pigmentary associations. Photochem. Photobiol. 2011;87:978–987. doi: 10.1111/j.1751-1097.2011.00970.x. [DOI] [PubMed] [Google Scholar]
- 21.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 22.Smyth DJ, et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat. Genet. 2006;38:617–619. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 23.Sutherland A, et al. Genomic polymorphism at the interferon-induced helicase (IFIH1) locus contributes to Graves' disease susceptibility. J. Clin. Endocrinol. Metab. 2007;92:3338–3341. doi: 10.1210/jc.2007-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martínez A, et al. IFIH1-GCA-KCNH7 locus: influence on multiple sclerosis risk. Eur. J. Hum. Genet. 2008;16:861–864. doi: 10.1038/ejhg.2008.16. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, et al. Carriers of rare missense variants in IFIH1 are protected from psoriasis. J. Invest. Dermatol. 2010;130:2768–2772. doi: 10.1038/jid.2010.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gateva V, et al. A large-scale replication study identifies TNIP1,PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat. Genet. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peach RJ, et al. Both extracellular immunoglobin-like domains of CD80 contain residues critical for binding T cell surface receptors CTLA-4 and CD28. J. Biol. Chem. 1995;270:21181–21187. doi: 10.1074/jbc.270.36.21181. [DOI] [PubMed] [Google Scholar]
- 28.Stamper CC, et al. Crystal structure of the B7-1/CTLA-4 complex that inhibits human immune responses. Nature. 2001;410:608–611. doi: 10.1038/35069118. [DOI] [PubMed] [Google Scholar]
- 29.Wu JN, Koretzky GA. The SLP-76 family of adapter proteins. Semin. Immunol. 2004;16:379–393. doi: 10.1016/j.smim.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 30.Sasaki S, et al. Cloning and expression of human B cell-specific transcription factor BACH2 mapped to chromosome 6q15”. Oncogene. 2000;19:3739–3749. doi: 10.1038/sj.onc.1203716. [DOI] [PubMed] [Google Scholar]
- 31.Cooper JD, et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat. Genet. 2008;40:1399–1401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grant SF, et al. Follow-up analysis of genome-wide association data identifies novel loci for type 1 diabetes. Diabetes. 2009;58:290–295. doi: 10.2337/db08-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubois PC, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dragone LL, Shaw LA, Myers MD, Weiss A. SLAP, a regulator of immunoreceptor ubiquitination, signaling, and trafficking. Immunol. Rev. 2009;232:218–228. doi: 10.1111/j.1600-065X.2009.00827.x. [DOI] [PubMed] [Google Scholar]
- 36.Tomer Y, Greenberg D. The thyroglobulin gene as the first thyroid-specific susceptibility gene for autoimmune thyroid disease. Trends Mol. Med. 2004;10:306–308. doi: 10.1016/j.molmed.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 37.Lamkanfi M, Kanneganti TD. Caspase-7: a protease involved in apoptosis and inflammation. Int. J. Biochem. Cell. Biol. 2010;42:21–24. doi: 10.1016/j.biocel.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.García-Lozano JR, et al. Caspase 7 influences susceptibility to rheumatoid arthritis. Rheumatology (Oxford) 2007;46:1243–1247. doi: 10.1093/rheumatology/kem096. [DOI] [PubMed] [Google Scholar]
- 39.Babu SR, et al. Caspase 7 is a positional candidate gene for IDDM 17 in a Bedouin Arab family. Ann. N.Y. Acad. Sci. 2003;1005:340–343. doi: 10.1196/annals.1288.054. [DOI] [PubMed] [Google Scholar]
- 40.Baaten BJ, Li CR, Bradley LM. Multifaceted regulation of T cells by CD44. Commun. Integr. Biol. 2010;3:508–512. doi: 10.4161/cib.3.6.13495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramos PS, et al. Genetic analyses of interferon pathway-related genes reveal multiple new loci associated with systemic lupus erythematosus. Arthritis Rheum. 2011;63:2049–2057. doi: 10.1002/art.30356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan F, et al. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science. 2009;325:1142–1146. doi: 10.1126/science.1176077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hakonarson H, et al. A novel susceptibility locus for type 1 diabetes on Chr12q13 identified by a genome-wide association study. Diabetes. 2008;57:1143–6. doi: 10.2337/db07-1305. [DOI] [PubMed] [Google Scholar]
- 44.Petukhova L, et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature. 2010;466:113–117. doi: 10.1038/nature09114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Devallière J, Charreau B. The adaptor Lnk (SH2B3): an emerging regulator in vascular cells and a link between immune and inflammatory signaling. Biochem. Pharmacol. 2011;82:1391–402. doi: 10.1016/j.bcp.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 46.Smyth DJ, et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N. Engl. J. Med. 2008;359:2767–77. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hunt KA, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat. Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coenen MJ, et al. Common and different genetic background for rheumatoid arthritis and coeliac disease. Hum. Mol. Genet. 2009;18:4195–4203. doi: 10.1093/hmg/ddp365. [DOI] [PubMed] [Google Scholar]
- 49.Alcina A, et al. The autoimmune disease-associated KIF5A, CD226 and SH2B3 gene variants confer susceptibility for multiple sclerosis. Genes Immun. 2010;11:439–445. doi: 10.1038/gene.2010.30. [DOI] [PubMed] [Google Scholar]
- 50.Jia S, Meng A. Tob genes in development and homeostasis. Dev. Dyn. 2007;236:913–921. doi: 10.1002/dvdy.21092. [DOI] [PubMed] [Google Scholar]
- 51.Seya T, Matsumoto M, Ebihara T, Oshiumi H. Functional evolution of the TICAM-1 pathway for extrinsic RNA sensing. Immunol. Rev. 2009;227:44–53. doi: 10.1111/j.1600-065X.2008.00723.x. [DOI] [PubMed] [Google Scholar]
- 52.Taïeb A, Picardo M. The definition and assessment of vitiligo: a consensus report of the Vitiligo European Task Force. Pigment Cell Res. 2007;20:27–35. doi: 10.1111/j.1600-0749.2006.00355.x. [DOI] [PubMed] [Google Scholar]
- 53.Purcell S, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 55.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 57.Yang J, et al. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Falconer DS. The inheritance of liability to certain diseases, estimated from the incidence among relatives. Ann. Hum. Genet. 1965;29:51–76. [Google Scholar]
- 59.Risch N. Assessing the role of HLA-linked and unlinked determinants of disease. Am. J. Hum. Genet. 1987;40:1–14. [PMC free article] [PubMed] [Google Scholar]
- 60.Howitz J, Brodthagen H, Schwartz M, Thompsen K. Arch. Dermatol. 1977;113:47–52. doi: 10.1001/archderm.113.1.47. [DOI] [PubMed] [Google Scholar]
- 61.Szklarczyk D, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucl. Acids Res. 2011;39(Database Issue):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.