Abstract
MRI is used for tracking of superparamagnetic iron oxide (SPIO)-labeled neural stem cells (NSCs). Studies have shown that long-term MR tracking of rapidly dividing cells underestimates their migration distance. Time-lapse microscopy of random cellular motility and cell division was performed to evaluate the effects of SPIO-labeling on NSC migration. Labeled cells divided symmetrically, and exhibited no changes in cell viability, proliferation, or apoptosis. However, SPIO-labeling resulted in decreased motility of NSCs as compared to unlabeled controls. When SPIO-labeled NSCs and human induced pluripotent stem cells (iPSCs) were transplanted into mouse brain, rapid exocytosis of SPIO by live cells was observed as early as 48 hours post-engraftment, with SPIO-depleted cells showing the farthest migration distance. As label dilution is negligible at this early time point, we conclude that MRI underestimation of cell migration can also occur as a result of reduced cell motility, which appears to be mitigated following SPIO exocytosis.
Keywords: superparamagnetic iron oxide, cell tracking, neural stem cell, exocytosis
Introduction
Successful repair of the damaged central nervous system (CNS) is difficult due to a limited regenerative capacity of the host. Introducing exogenous neural stem cells (NSCs) by transplantation is a promising new therapeutic paradigm but significant challenges remain. With phase I/II clinical trials underway to evaluate the safety and efficacy of stem cell-based therapy, there is a need for suitable imaging techniques that can ideally monitor the function, survival, migration, and host tissue integration of transplanted cells (1). Indeed, clinical MRI cell tracking has recently emerged as a predominant imaging modality (2), in particular for stem cell tracking in the CNS (3–5). In these studies, cells are made MR-visible by labeling them with superparamagnetic iron oxide (SPIO) particles (6).
The primary clinical application of MR tracking of NSCs is likely to be monitoring delivery of cells in real-time, using MR-compatible catheters, with visualization of their initial engraftment and homing shortly after administration. Long-term tracking of NSCs will be challenging, as it is difficult to discriminate live from dead cells, and rapidly dividing NSCs may exhibit a pronounced mismatch between the MRI and histological distribution pattern soon after their engraftment (7). It has been suggested that a primary reason for this MRI mismatch, i.e., cells becoming undetectable, is their cell division and associated dilution of label. However, to the best of our knowledge, there have been no studies on a possible direct effect of SPIO-labeling on NSC motility and migration.
In this study, C17.2 murine NSCs were labeled with rhodamine-SPIO and assessed in vitro for cell proliferation, random motility, and cell division. In addition, labeled cells were transplanted into immunodeficient mice and followed for 4–72 hours. We show here that SPIO-labeling can reduce the overall motility of transplanted cells, both in vitro and in vivo, and that exocytosis of SPIO mitigates this adverse effect making this only a temporary phenomenon.
Materials and Methods
Cell culture and labeling
The immortalized LacZ-transfected NSC line C17.2, derived from neonatal mouse cerebellum (8), was cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, NY, USA), supplemented with 10% fetal bovine serum (Gibco), 5% horse serum (Gibco), 2 mM L-glutamine (Gibco), 1% penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO, USA), and 1% amphotericin B (Sigma-Aldrich). For constitutive firefly luciferase expression, 2.5×106 cells were transduced for 6 hours with the lentiviral vector pLenti-FUGW-fLuc2, with a multiplicity of infection (MOI) of 10, using 0.1% polybrene (Chemicon, MA, USA). Cells were cultured on 75 cm2 polystyrene tissue culture dishes (Corning, NY, USA) at 37oC and 5% CO2.
Luc-expressing C17.2 cells were labeled with the fluorescently tagged SPIO formulation Molday ION Rhodamine B (BioPAL, Worcester, MA, USA), having a colloidal size of 50 nm. 25 μg/mL Fe of Molday ION Rhodamine B was mixed with culture medium and added to cells for 24 hours under normal culture conditions. Transfection agents were not utilized; the iron oxide particles were mixed with culture medium and applied to the cells.
Human iPS cells derived from skin fibroblasts (courtesy of Dr. James Thompson) were differentiated into oligodendrocytes by adding B-27, insulin, N1, triiodthyronine (T3), bFGF, EGF, pDGF, and IGF to the medium according to the protocol by Nistor et al (9). These cells expressed Olig1 and Olig2 at the time of transplantation, at day 57 into the differentiation protocol.
Time-lapse microscopy of cell motility
C17.2 cells (3,500 cells/well) were plated on borosilicate coverglass Lab-Tek 4-well plates (Nunc, Rochester, NY, USA), coated with 50 μg/ml poly-D-lysine (P1024, Sigma, St. Louis, MO, USA) for 30 minutes and 20 μg/ml laminin (23017, Invitrogen, Carlsbad, CA, USA) for 2 hours. Cells were cultured and observed in normal culture medium that was phenol red free (Gibco, NY, USA) at 37°C and in the presence of a humidified 5% CO2/air atmosphere. Microscopy was conducted using a Zeiss Axiovert 200M inverted epifluorescence microscope with a 10x objective (NA = 0.4). Acquisition was performed using Slidebook 4.2 software (Intelligent Innovations Inc., Denver, CO, USA). Phase-contrast images of live cells were taken using a cascade 512BII CCD camera (Photometrics, Tucson, AZ, USA) attached to the microscope with a 10X objective (NA = 0.4). Images were taken every 5 minutes for 12 hours for each experiment. In all experiments, cell images were visually examined before analysis; cells displaying morphologic changes characteristic of apoptosis were excluded.
Quantification of cellular motility
The Time Lapse Analyzer cell tracker analysis program described by Huth et al. (10), available from www.informatik.uni-ulm.de, was used to quantitatively track cell motility from live cell microscopy images. Tracked cells were manually evaluated to ensure that the track was recorded for healthy, live cells. One hundred unlabeled cells and 100 SPIO-labeled cells were analyzed every 5 minutes for 12 hours.
The following parameters were evaluated and statistically compared: 1) the average, 2) the median, and 3) the maximum speed for each cell cohort, with data plotted in a histogram. Statistical analysis was performed using a Student’s t- and ANalysis Of VAriance between groups (ANOVA) test.
Time-lapse microscopy of cell division
C17.2 NSCs were labeled with Molday ION Rhodamine B for 24 hours prior to microscopic imaging. Cells were additionally labeled with a cell membrane tracer (CellTracker Green CMFDA, Invitrogen, Carlsbad, CA, USA). For evaluation of cell division in cells undergoing differentiation, cells were cultured in regular maintenance medium for 2 days, then switched to a differentiation medium containing DMEM/F12 (Gibco, Carlsbad, CA, USA) supplemented with N-2 (2.5 mL of 100x stock into 250 mL of medium; 17502, Gibco), B-27 (5 mL of 50x stock into 250 mL of medium; 17504, Gibco), 1% penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO, USA), and 1% amphotericin B (Sigma-Aldrich). Differentiation into neurons was confirmed by anti-β3-tubilin immunohistochemistry (1:500, Covance, Princeton, NJ, USA).
MatTek glass bottom microwell dishes (P35G-1.5-14-C, Ashland, MA, USA were coated with 50 μg/ml poly-D-lysine (P1024, Sigma, St. Louis, MO, USA) for 30 minutes and then with 20 μg/ml laminin (23017, Invitrogen, Carlsbad, CA, USA) for 2 hours. C17.2 cells (40,000 cells/plate) were seeded on the coated MatTek dishes. Cells were cultured and observed in either normal culture medium or differentiation medium (at 2, 4, 6, or 8 days of differentiation prior to starting microscopy) at 37°C and in a humidified 5% CO2/air atmosphere using a Zeiss Axiovert 200M inverted epifluorescence microscope with a 40x objective (NA = 1.3). Acquisition was performed using Slidebook 4.2 software (Intelligent Innovations Inc., Denver, CO, USA). Three separate experiments for non-differentiating conditions and four experiments for differentiating conditions (at day 2, 4, 6, and 8) were performed. Phase-contrast and fluorescent (green and red channels) images of live cells were taken using a cascade 512BII CCD camera (Photometrics, Tucson, AZ, USA) attached to the microscope and using a 10X objective (NA = 0.4). Images were taken every 30 minutes for 24 hours for each experiment.
Quantification of cellular SPIO content during cell division
Live cell microscopy experiments were analyzed to locate cellular division events; over 150 division events were monitored. Using a custom-designed program in Matlab 7.9 (MathWorks, Natick, MA, USA), the region of interest (ROI) was manually drawn around cells presented in a graphical interface using either the green (CellTracker Green) fluorescent image or the phase-contrast image. The ROI was projected onto the corresponding red (Molday ION Rhodamine B) fluorescent image and the red fluorescence in each parent cell and daughter cells during a cell division event was quantified using Matlab 7.9.
Cell transplantation
All animal procedures were approved and conducted in accordance with the institutional guidelines for the care of laboratory animals. For transplantation, the cells were harvested, washed, and suspended in normal culture medium at a density of 5×104 cells/μL. Immunodeficient Rag2 mice (n=3 per time point, 8 weeks old, 129S6/SvEvTac-Rag2tm1Fwa, Taconic, Hudson, NY, USA) were placed in a stereotaxic device (Stoelting, Wood Dale, IL, USA) under 2% isoflurane general anesthesia. Cells (2×105/2μL) were injected using a Hamilton 31G microinjection needle (Hamilton, Reno, NV, USA) into the right corpus callosum. The coordinates were medial-lateral (ML) = +2.5 mm and dorso-ventral (DV) = −1.2 mm relative to bregma =0.0.
Bioluminescence Imaging
Bioluminescence imaging (BLI) was performed immediately following cell transplantation and every 24 hours for 3 days using an IVIS 200 optical imaging device (Caliper Life Sciences, Hopkinton, MA, USA) equipped with a high sensitivity, cryogenically-cooled CCD detection system. Prior to imaging, each mouse was intraperitoneally injected with 150 mg/kg of the substrate luciferin (Caliper Life Sciences, Hopkinton, MA, USA) in order to detect firefly luciferase activity. Mice were anesthetized with 1–2% isoflurane and imaged within 15 minutes after luciferin injection. BLI signal was processed using Xenogen Living Image 2.50 software. A region of interest (ROI) was selected around each brain and the total flux (photons/sec) was calculated. The size of the ROI was kept constant to compare the mice over time. The BLI signal was normalized for each mouse individually with the first imaging time point’s total flux (photos/sec) being 100% and the signal from each subsequent time point relative to the initial total flux.
Immunofluorescent histology
Mice were transcardially perfused with 10 mM phosphate buffered saline (PBS) followed by 4% paraformaldehyde (PFA) fixation at 4 hours (n=3), 1 day (n=3), 2 days (n=3), and 3 (n=3) days after transplantation. Brains were removed and fixed in 4% PFA overnight, cryopreserved in 30% sucrose, snap frozen on powdered dry ice, and cryo-sectioned at 30 μm slices.
For immunohistochemistry, the following primary antibodies were used: anti-firefly luciferase (ab21176, 1:1000, Abcam, Cambridge, MA, USA), anti-PCNA (sc-56, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-Caspase 3 (9661S, Cell Signaling Technology, Danvers, MA, USA), TUNEL (S7111, Millipore, Billerica, MA, USA), and anti- β3-tubulin (PRB-435P, Covance, Princeton, NJ, USA). Goat anti-rat 488 (A11042, 1:200) and goat anti-rabbit 488 (A11008, 1:200), both from Molecular Probes (Eugene, OR, USA), were used as secondary antibodies. Tissue samples were blocked for two hours at room temperature with PBS containing 5% bovine serum albumin (BSA) and 0.01% Triton, incubated overnight at 4°C with primary antibody diluted in PBS, 5% bovine serum albumin and 0.1% Triton, and then incubated for two hours at room temperature with secondary antibody in PBS containing 5% BSA and 0.1% Triton. Nuclei were stained using 1 μg/mL DAPI (Invitrogen, Carlsbad, CA, USA). Intracellular SPIO was detected by rhodamine fluorescence or Prussian blue staining. Slides were embedded with Vectashield mounting medium (Vector, Burlingame, CA, USA) to preserve the fluorescence signal. Microscopy was performed using an Olympus BX51 fluorescence microscope equipped with an Olympus DP-70 digital acquisition system.
Results
In vitro motility of SPIO-labeled cells
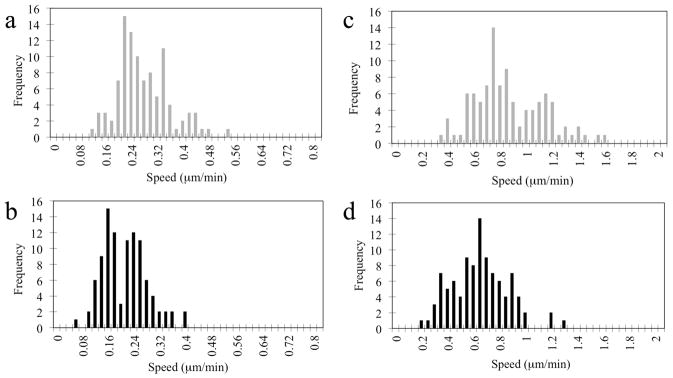
Over 99% of C17.2 NSCs were labeled with rhodamine-SPIO after 24 hours of incubation. The motility of C17.2 SPIO-labeled and unlabeled NSCs was evaluated in vitro using time-lapse bright field microscopy. The average speed and the maximum speed were calculated from recorded movies for labeled cells (Supplementary Movie 1) and unlabeled cells (Supplementary Movie 2). In Fig. 1, histograms are shown representing the frequency of speed distribution. The average speed for unlabeled and SPIO-labeled cells was 0.268 ± 0.040 and 0.202 ± 0.027 μm/min, respectively (95% confidence interval). The average of the maximum speed was 0.842 μm/min for unlabeled cells and 0.625 μm/min for Molday labeled cells. Clearly, unlabeled cells outperform the labeled cells in terms of motility for all in vitro measured parameters by a ratio of 1.3:1. Statistical analysis by Student’s t-test and ANOVA non-parametric test both had a p-value of 10−9. Thus, unlabeled cells are significantly more motile than SPIO-labeled cells as tested using a Student’s t- test and ANOVA non-parametric test (p<10−9).
Figure 1.
Cellular motility. Histograms of the distribution of (a) the average speed of unlabeled C17.2 cells, (b) the average speed of SPIO-labeled cells, (c) the maximum speed of unlabeled C17.2 cells, and (d) the maximum speed of SPIO-labeled cells. Recordings were obtained over a period of 12 hours.
Cell division studies
C17.2 cells were labeled with fluorescent SPIO, and live cell division events were recorded using fluorescent video microscopy to directly compare the relative distribution of label between parent and daughter cells (Supplementary Movie 3). Two different growth conditions were evaluated: one that maintains stem cells in an immature, undifferentiated state (three independent measurements) and one that promotes their differentiation towards neurons (four independent measurements).
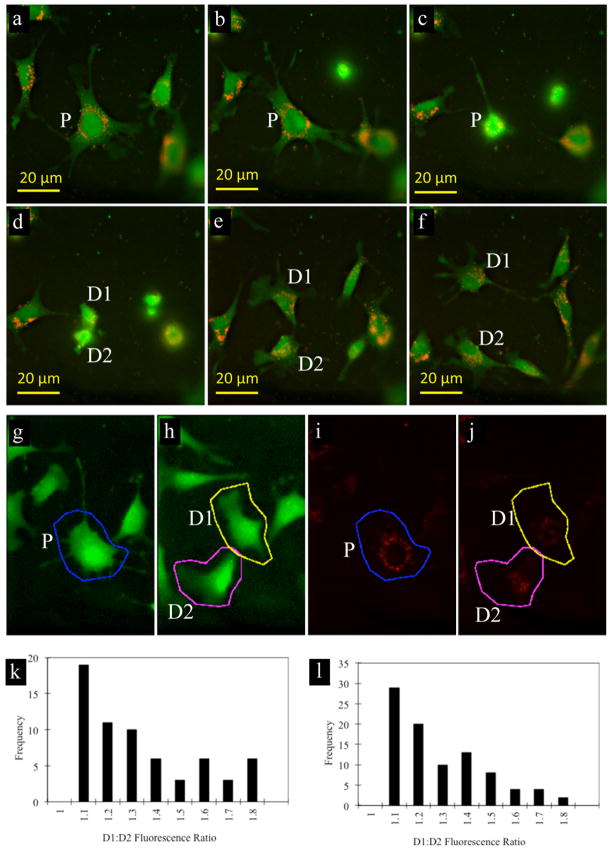
Over 150 cellular division events were analyzed for the amount of red fluorescence in the two daughter cells (Fig. 2a–g). The average D1:D2 ratio for cells under undifferentiating and differentiating conditions was 1.28 ± 0.06 and 1.24 ± 0.04, respectively (Fig. 2 h,i, 95% confidence interval). Thus, on average, 55% of the SPIO label went into one daughter cell and 45% into the other daughter cell. Based on these values, no significant asymmetric cellular division occurred either for differentiating or undifferentiated NSCs.
Figure 2.
Quantification of SPIO dilution due to cell division. (a–f) Fluorescent microscopy images of proliferating, SPIO-labeled C17.2 cells (red: SPIO; green: C17.2 cells). Each image (labeled sequentially a–f) represents a time point of 30 minutes. (g–j) Representative images demonstrating ROI selection for calculation of the amount of red fluorescence from SPIO in parent cell (g, i; blue) and two daughter cells D1, D2 (h, j; pink and yellow), following their division from the parent cell (P). (k,l) Histogram showing distribution of fluorescence (rhodamine-SPIO) between two daughter cells for undifferentiated (k) and differentiated (l) NSCs.
In vivo transplantation studies
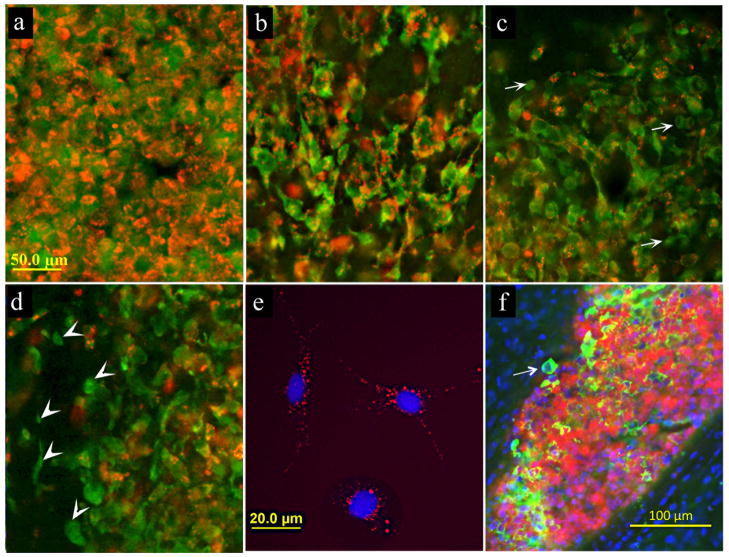
SPIO-labeled C17.2 NSCs were transplanted into the brains of immunodeficient mice, with sampling of tissue at 4, 24, 48, and 72 hours after transplantation. At 4 and 24 hours post-engrafting, cells were found to be densely clustered in the corpus callosum at the site of transplantation, with a good co-localization between the cells and the red fluorescent SPIO label (Fig. 3a, b). Starting at 48 hours after transplantation, a portion of transplanted C17.2 cells appeared to no longer contain iron. Among the cells that were found migrating 100μm or more from the core of the graft deposit 8.0±1.9% were unlabeled. Fig. 3 c–d is showing border region of the graft with migrating cells. At 48 and 72 hours after transplantation cells were found migrating away from the transplantation site with the distance of migration from the border of graft deposit of up to 300μm. Even though complete loss of iron was at 48 hours postgrafting infrequent, interestingly, cells devoid of the contrast agent were typically those that were found migrating away from the site of transplantation (less then 1% of unlabeled cells in the core and 8% in the periphery of the graft). This is in sharp contrast to SPIO-labeled NSCs in vitro in culture at 72 hours following labeling, where the SPIO label is retained in all cells (Fig. 3e).
Figure 3.
(a–d) Histological detection of SPIO-labeled transplanted C17.2 cells (red: SPIO; green: C17.2 cells) at 4 hours (a), 24 hours (b), 48 hours (c), and 72 hours (d) after transplantation. Anti-luciferase immunohistochemical staining for C17.2 cells (green) shows that at as early as 48 hours post-engrafting cells start to lose SPIO label (c, arrows indicate cells without SPIO). Majority of cells that migrated away from the site of transplantation typically do not co-localize with iron oxide label (d, arrowheads). In vitro, SPIO-labeled NSCs in culture retain label at 72 hours post-labeling (e). Histological staining for iPS-derived neural precursor cells (green: anti-luciferase staining) also demonstrates the occurrence of cells without label at 48 hours after transplantation (f, arrow). Scale bar for (a–d)=50μm.
In order to ensure that this observation was not cell type-specific, we repeated the experiment with neural progenitors derived from human iPS cells. Similar to that observed for C17.2 NSCs, a fraction of iPS-derived progenitors that were migrating away from the site of transplantation lost the iron label already at 48 hours after transplantation (Fig. 3f).
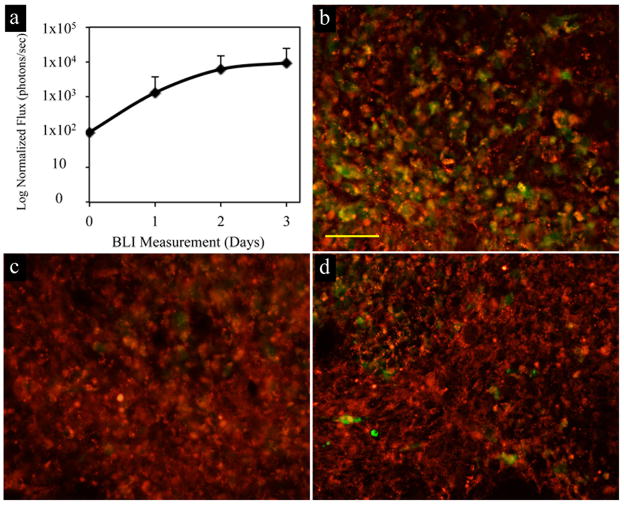
In order to determine the survival and proliferation of cells after transplantation, bioluminescence imaging (BLI) was performed on all mice. Most cells survived and started proliferating within the first 24 hours after transplantation as the BLI signal increased (Fig. 4a). From the BLI data, transplanted C17.2 NSCs were calculated to have an approximate doubling time of 19 hours, which is consistent with literature reports ranging from 16 to 24 hours (11–13). In vivo survival and proliferation was further confirmed by anti-proliferating cell nuclear antigen (PCNA) immunohistochemical staining, with several PCNA-positive cells co-localizing with the (rhodamine) SPIO label (Fig. 4b). In addition, terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining and anti-caspase 3 immunohistochemistry was performed to further determine the survival status of transplanted cells. TUNEL is used to identify the end stage of the apoptotic pathway while caspase 3 is indicated in the early or middle stages of apoptosis. At 48 hours post grafting TUNEL staining (Fig. 4c) did not detect positive cells while anti-caspase 3 (Fig. 4d) revealed only a few positive apoptotic cells, consistent with the BLI findings indicating negligible cell death in vivo following transplantation.
Figure 4.
Monitoring the survival and proliferation of C17.2 cells after transplantation. (a) Quantification of normalized bioluminescence imaging revealed signal increase at every time point after transplantation. (b) Anti-PCNA staining for proliferating cells at 48 hours post transplantation showed some cells proliferating (green) and were also positive for iron oxide nanoparticles (red). TUNEL staining (c, green) did not detect any positive cells and anti-caspase 3 staining for apoptotic cells (d, green) revealed only small amount of apoptotic cells. Scale bar (b–d) =50μm.
Discussion
This study was undertaken to look into the possible causes and mechanisms of the rapid loss of MRI signal in immortalized NSCs after their transplantation in vivo. We previously showed that, during in vitro differentiation of SPIO-labeled C17.2 NSCs, there can be significant heterogeneity of the intracellular contrast content starting at day 6 after labeling, with some cells heavily labeled and other cells containing just trace amounts of iron (7). It was hypothesized that rapidly dividing, differentiating C17.2 NSCs can undergo asymmetric cell division and therefore rapidly lose their SPIO label. This, in turn, could explain the sharp dividing line between labeled and unlabeled NSCs in vivo (7), that has also been observed for glial-restricted precursor cells (14). Another explanation to previously reported observation is that cells retaining the contrast are not dividing and are less migratory, while those that migrate have some way of clearing the iron either through proliferation or exocytosis. In this study, we performed detailed characterization of contrast dilution in vitro including time-lapse microscopy of live dividing cells and determined that assymetric cell division did not occur. Here we focused our analysis on an early time interval just after grafting and found that, SPIO-labeled cells became devoid of iron as early as 48 hours after transplantation. With nearly 100% of cells labeled and a doubling time of approximately 20 hours, the complete clearance of SPIO-label within 48 hours after labeling cannot be explained by label dilution alone, as on average only 2.5 cell divisions are occurring. Rather, an explanation for this rapid loss could be early cell death and extracellular, passive release of SPIO. However, BLI demonstrated that cell death in vivo following transplantation was negligible. In addition, TUNEL, anti-PCNA, and anti-caspase-3 immunostaining demonstrated that labeled cells did not undergo extensive apoptosis. Based on these observations, the only explanation for the rapid loss of SPIO is active exocytosis. Interestingly, we found that cells that migrated away from the injection site were those devoid of label (Fig. 3d).
In order to investigate the exocytosis and effects of SPIO label on cell migration further in detail, we performed in vitro experiments in a well-controlled environment. Live cell imaging showed that in vitro C17.2 stem cells divided symmetrically for both an immature, undifferentiated state and a differentiation state toward the neural lineage (Fig. 2). In this scenario, the SPIO label is approximately divided equally between daughter cells, which was verified by quantitative fluorescence measurements. Therefore, loss of iron in vivo due to asymmetric cell division is not supported by our data.
In order to assess whether the SPIO label can directly affect the migration of labeled cells, we performed an in vitro random motility assay. We found that SPIO-labeling significantly inhibits the motility of NSCs as compared to unlabeled cells (Fig. 1). In vivo, inhibitory effects of SPIO-labeling on cell migration may be mitigated by exocytosis, reducing or completely depleting the iron load, explaining why cells that migrated the greatest distance contained little or no iron (Fig 3d). C17.2 cells typically migrate extensively in the brain (7,8,15), where signaling cues are present to which these cells effectively respond, driving in vivo migration. These signals, which are only present in the brain and not in culture conditions, activate the microtubule network and may induce SPIO exocytosis in favor of maintaining a high rate of migration.
Overall, little or no adverse effects of SPIO labeling have been reported when labeled cells are compared to unlabeled cells, both in vitro and in vivo (16). Gene expression analysis has shown that, following SPIO-labeling of C17.2 cells, less than 1% of all genes exhibit greater than a 2-fold difference in expression, returning to pre-labeling levels after 7 days in culture (17). However, recent reports have shown that SPIO-labeling can affect the actin cytoskeleton and microtubule architecture of labeled cells in a dose-dependent manner, possibly due to sterical hindrance of SPIO-containing endosomes with the actin fiber network (18,19). Interestingly, the cytoskeleton morphology returned back to normal after several days of culture, when the cellular SPIO content diminished as a result of cell division (19). A proper microtubular configuration is essential for normal cell motility and migration. Slightly reduced migration has also been observed for SPIO-dendritic cells, although only at higher iron loads (30 pg Fe/cell) (20). Although unlikely unrelated to microtubule deconformation, other inhibitory effects of SPIO-labeling have been reported for mesenchymal stem cells, where chondrogenesis was inhibited in a dose-dependent manner as well (21–24).
It was reported that the impact of SPIO-labeling on cell function highly depends on the particle surface coating. Dextran coating was found to significantly inhibit in vitro cell movements (25). Wu et al. found that cells labeled with even very low concentrations (0.1 mM) of SPIO particles coated with dextran significantly inhibited cell migration and chemotaxis of human umbilical vein endothelial cells (26). The Molday SPIO preparation used in this study is dextran-coated. The composition and coating of the particles, the type of cell, and the site of transplantation are all independent factors that may contribute to whether or not cells inhibit reduced migration and exocytosis. Clearly, much needs to be learned about the cell fate in vivo under specific conditions. Most studies on the effect of SPIO-labeling have hitherto been performed in vitro, however, we were unable to reproduce SPIO exocytosis in vitro at least up to 72 hours (Fig. 3e). It is not clear why NSCs exocytose SPIO label in vivo but not in vitro. One possibility for this discrepancy is that activation of the microtubule network may only occur in vivo when cells respond to migratory cues present within host tissue. This may then simultaneously induce the active process of exocytosis, which uses the cytoskeleton (microtubuli) framework.
In summary, we have shown here that SPIO-labeled NSCs exhibit symmetric cell division and reduced migration in vitro. In vivo, cells rapidly loose their iron through exocytosis, upon which they initiate migration. The present results imply that MR tracking of SPIO-labeled NSCs for long-term studies may not always be reliable, and therefore should only be applied to monitor (real-time) cell delivery and initial dispersion of transplanted cells within the host tissue.
Supplementary Material
Bright field time-lapse microscopy of Molday labeled C17.2 cells for in vitro motility analysis. The images were acquired at 10X magnification; an image was taken every 5 minutes for 12 hours. The track (red) is marking one cell and the last image shows the entire track taken for that cell over 12 hours.
Bright field time-lapse microscopy 10X of unlabeled C17.2 cells for in vitro motility analysis. The images were acquired at 10x magnification; an image was taken every 5 minutes for 12 hours. The track (blue) is on one cell and the last image shows the entire track taken for that cell over 12 hours.
Fluorescent microscopic time-lapse video of SPIO-labeled C17.2 cell division (red: SPIO; green: C17.2 cells). Images were acquired at 40X magnification and taken every 30 minutes.
Acknowledgments
This work was supported by MSCRFII-0193, MSCRFII-0052, RO1DA026299, and RO1NS045062. We are grateful to Nidhi Rumpal, Ph.D. and Chulani Galpoththawela for culturing iPSCs.
References
- 1.Fink DW., Jr FDA regulation of stem cell-based products. Science. 2009;324(5935):1662–1663. doi: 10.1126/science.1173712. [DOI] [PubMed] [Google Scholar]
- 2.Bulte JW. In vivo MRI cell tracking: clinical studies. AJR American journal of roentgenology. 2009;193(2):314–325. doi: 10.2214/AJR.09.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karussis D, Karageorgiou C, Vaknin-Dembinsky A, Gowda-Kurkalli B, Gomori JM, Kassis I, Bulte JW, Petrou P, Ben-Hur T, Abramsky O, Slavin S. Safety and immunological effects of mesenchymal stem cell transplantation in patients with multiple sclerosis and amyotrophic lateral sclerosis. Archives of neurology. 2010;67(10):1187–1194. doi: 10.1001/archneurol.2010.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu J, Zhou L, XingWu F. Tracking neural stem cells in patients with brain trauma. N Engl J Med. 2006;355(22):2376–2378. doi: 10.1056/NEJMc055304. [DOI] [PubMed] [Google Scholar]
- 5.Jozwiak S, Habich A, Kotulska K, Sarnowska A, Kropiwnicki T, Janowski M, Jurkiewicz E, Lukomska B, Kmiec T, Walecki J, Roszkowski M, Litwin M, Oldak T, Boruczkowski D, Domanska-Janik K. Intracerebroventricular transplantation of cord blood-derived neural progenitors in a child with severe global brain ischemic injury. Cell Med Part B Cell Transpl. 2010;1:71–80. doi: 10.3727/215517910X536618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cromer Berman SM, Walczak P, Bulte JW. Tracking stem cells using magnetic nanoparticles. Wiley interdisciplinary reviews. Nanomedicine and nanobiotechnology. 2011;3(4):343–355. doi: 10.1002/wnan.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walczak P, Kedziorek DA, Gilad AA, Barnett BP, Bulte JW. Applicability and limitations of MR tracking of neural stem cells with asymmetric cell division and rapid turnover: the case of the shiverer dysmyelinated mouse brain. Magn Reson Med. 2007;58(2):261–269. doi: 10.1002/mrm.21280. [DOI] [PubMed] [Google Scholar]
- 8.Snyder EY, Deitcher DL, Walsh C, Arnold-Aldea S, Hartwieg EA, Cepko CL. Multipotent neural cell lines can engraft and participate in development of mouse cerebellum. Cell. 1992;68(1):33–51. doi: 10.1016/0092-8674(92)90204-p. [DOI] [PubMed] [Google Scholar]
- 9.Nistor GI, Totoiu MO, Haque N, Carpenter MK, Keirstead HS. Human embryonic stem cells differentiate into oligodendrocytes in high purity and myelinate after spinal cord transplantation. Glia. 2005;49(3):385–396. doi: 10.1002/glia.20127. [DOI] [PubMed] [Google Scholar]
- 10.Huth J, Buchholz M, Kraus JM, Schmucker M, von Wichert G, Krndija D, Seufferlein T, Gress TM, Kestler HA. Significantly improved precision of cell migration analysis in time-lapse video microscopy through use of a fully automated tracking system. BMC Cell Biol. 2010;11:24. doi: 10.1186/1471-2121-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang Y, Shah K, Messerli SM, Snyder E, Breakefield X, Weissleder R. In vivo tracking of neural progenitor cell migration to glioblastomas. Hum Gene Ther. 2003;14(13):1247–1254. doi: 10.1089/104303403767740786. [DOI] [PubMed] [Google Scholar]
- 12.Soenen SJ, Himmelreich U, Nuytten N, De Cuyper M. Cytotoxic effects of iron oxide nanoparticles and implications for safety in cell labelling. Biomaterials. 2011;32(1):195–205. doi: 10.1016/j.biomaterials.2010.08.075. [DOI] [PubMed] [Google Scholar]
- 13.Walls KC, Ghosh AP, Franklin AV, Klocke BJ, Ballestas M, Shacka JJ, Zhang J, Roth KA. Lysosome dysfunction triggers Atg7-dependent neural apoptosis. J Biol Chem. 2010;285(14):10497–10507. doi: 10.1074/jbc.M110.103747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lepore AC, Walczak P, Rao MS, Fischer I, Bulte JW. MR imaging of lineage-restricted neural precursors following transplantation into the adult spinal cord. Experimental neurology. 2006;201(1):49–59. doi: 10.1016/j.expneurol.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 15.Heese O, Disko A, Zirkel D, Westphal M, Lamszus K. Neural stem cell migration toward gliomas in vitro. Neuro Oncol. 2005;7(4):476–484. doi: 10.1215/S1152851704000754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crabbe A, Vandeputte C, Dresselaers T, Sacido AA, Verdugo JM, Eyckmans J, Luyten FP, Van Laere K, Verfaillie CM, Himmelreich U. Effects of MRI contrast agents on the stem cell phenotype. Cell Transplant. 2010;19(8):919–936. doi: 10.3727/096368910X494623. [DOI] [PubMed] [Google Scholar]
- 17.Kedziorek DA, Muja N, Walczak P, Ruiz-Cabello J, Gilad AA, Jie CC, Bulte JW. Gene expression profiling reveals early cellular responses to intracellular magnetic labeling with superparamagnetic iron oxide nanoparticles. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2010;63(4):1031–1043. doi: 10.1002/mrm.22290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khaleghian A, Riazi GH, Ahmadian S, Ghafari M, Rezaie M, Takahashi A, Nakaya Y, Nazari H. Effect of ferric oxide nanoparticles on microtubules organization. Afr Journ Biochem Res. 2010;4(4):99–104. [Google Scholar]
- 19.Soenen SJ, Nuytten N, De Meyer SF, De Smedt SC, De Cuyper M. High intracellular iron oxide nanoparticle concentrations affect cellular cytoskeleton and focal adhesion kinase-mediated signaling. Small. 2010;6(7):832–842. doi: 10.1002/smll.200902084. [DOI] [PubMed] [Google Scholar]
- 20.Verdijk P, Scheenen TW, Lesterhuis WJ, Gambarota G, Veltien AA, Walczak P, Scharenborg NM, Bulte JW, Punt CJ, Heerschap A, Figdor CG, de Vries IJ. Sensitivity of magnetic resonance imaging of dendritic cells for in vivo tracking of cellular cancer vaccines. International journal of cancer Journal international du cancer. 2007;120(5):978–984. doi: 10.1002/ijc.22385. [DOI] [PubMed] [Google Scholar]
- 21.Kostura L, Kraitchman DL, Mackay AM, Pittenger MF, Bulte JW. Feridex labeling of mesenchymal stem cells inhibits chondrogenesis but not adipogenesis or osteogenesis. NMR in biomedicine. 2004;17(7):513–517. doi: 10.1002/nbm.925. [DOI] [PubMed] [Google Scholar]
- 22.Bulte JW, Kraitchman DL, Mackay AM, Pittenger MF. Chondrogenic differentiation of mesenchymal stem cells is inhibited after magnetic labeling with ferumoxides. Blood. 2004;104(10):3410–3412. doi: 10.1182/blood-2004-06-2117. author reply 3412–3413. [DOI] [PubMed] [Google Scholar]
- 23.Henning TD, Sutton EJ, Kim A, Golovko D, Horvai A, Ackerman L, Sennino B, McDonald D, Lotz J, Daldrup-Link HE. The influence of ferucarbotran on the chondrogenesis of human mesenchymal stem cells. Contrast Media Mol Imaging. 2009;4(4):165–173. doi: 10.1002/cmmi.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boddington SE, Sutton EJ, Henning TD, Nedopil AJ, Sennino B, Kim A, Daldrup-Link HE. Labeling human mesenchymal stem cells with fluorescent contrast agents: the biological impact. Molecular imaging and biology: MIB: the official publication of the Academy of Molecular Imaging. 2011;13(1):3–9. doi: 10.1007/s11307-010-0322-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berry CC, Wells S, Charles S, Aitchison G, Curtis AS. Cell response to dextran-derivatised iron oxide nanoparticles post internalisation. Biomaterials. 2004;25(23):5405–5413. doi: 10.1016/j.biomaterials.2003.12.046. [DOI] [PubMed] [Google Scholar]
- 26.Wu X, Tan Y, Mao H, Zhang M. Toxic effects of iron oxide nanoparticles on human umbilical vein endothelial cells. Int J Nanomedicine. 2010;5:385–399. doi: 10.2147/ijn.s10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bright field time-lapse microscopy of Molday labeled C17.2 cells for in vitro motility analysis. The images were acquired at 10X magnification; an image was taken every 5 minutes for 12 hours. The track (red) is marking one cell and the last image shows the entire track taken for that cell over 12 hours.
Bright field time-lapse microscopy 10X of unlabeled C17.2 cells for in vitro motility analysis. The images were acquired at 10x magnification; an image was taken every 5 minutes for 12 hours. The track (blue) is on one cell and the last image shows the entire track taken for that cell over 12 hours.
Fluorescent microscopic time-lapse video of SPIO-labeled C17.2 cell division (red: SPIO; green: C17.2 cells). Images were acquired at 40X magnification and taken every 30 minutes.