Abstract
Purpose
To identify and evaluate textural quantitative imaging signatures (QISes) for tumors occurring within the central gland (CG) and peripheral zone (PZ) of the prostate, respectively, as seen on in vivo 3 Tesla endorectal T2-weighted (T2w) Magnetic Resonance Imaging (MRI).
Materials and Methods
This study utilized 22 pre-operative prostate MRI datasets (16 PZ, 6 CG) acquired from men with confirmed prostate cancer (CaP) and scheduled for radical prostatectomy (RP). The prostate region-of-interest (ROI) was automatically delineated on T2w MRI, following which it was corrected for intensity-based acquisition artifacts. An expert pathologist manually delineated the dominant tumor regions on ex vivo sectioned and stained RP specimens as well as identified each of the studies as either a CG or PZ CaP. A non-linear registration scheme was employed to spatially align and then map CaP extent from the ex vivo RP sections onto the corresponding MRI slices. 110 texture features were then extracted on a per-voxel basis from all T2w MRI datasets. An information theoretic feature selection procedure was then applied to identify QISes comprising T2w MRI textural features specific to CG and PZ CaP, respectively. The QISes for CG and PZ CaP were evaluated via Quadratic Discriminant Analysis (QDA) on a per-voxel basis against the ground truth for CaP on T2w MRI, mapped from corresponding histology.
Results
The QDA classifier yielded an area under the Receiver Operating characteristic curve of 0.86 for the CG CaP studies, and 0.73 for the PZ CaP studies over 25 runs of randomized 3-fold cross-validation. By comparison, the accuracy of the QDA classifier was significantly lower when (a) using all 110 texture features (with no feature selection applied), as well as (b) a randomly selected combination of texture features.
Conclusion
CG and PZ prostate cancers have significantly differing textural quantitative imaging signatures on T2w endorectal in vivo MRI.
Keywords: quantitative imaging signatures, prostate cancer, T2-weighted, magnetic resonance imaging, texture analysis, classification, central gland, peripheral zone
INTRODUCTION
Prostatic adenocarcinoma (CaP) will affect one in 6 men in the United States during his lifetime, and one in 36 will die as a result of it (1). Early detection and staging of this disease offers a 100% 5 year survival rate (1) based on improved treatment options. Over the last decade, there has been significant interest in the use of endorectal magnetic resonance imaging (MRI) to identify prostate cancer in vivo (2,3), allowing for improved detection and staging accuracy compared to ultrasound imaging (4). Recent surveys of the literature (5) have reported a joint sensitivity and specificity of 71%–74% when using a 1.5 Tesla (T) endorectal T2-weighted MRI protocol. While initial enthusiasm for MRI was on account of its utility as a staging modality (6), recent developments in image-guided treatments (such as intensity-modulated radiotherapy, high-focused ultrasound therapy) as well as risk assessment, suggest that there is a growing interest in the use of MRI for accurate identification of prostate cancer stage, presence, and extent in vivo (7).
T2-weighted (T2w) MRI makes use of long echo times (TE) and repetition times (TR) during acquisition, thereby providing excellent contrast for different tissue structures based on fluid content (8). McNeal (9) described four distinct histological regions within the prostate, which are visually differentiable on an endorectal T2w MR image based on their signal intensity (SI) (10) characteristics (see Figure 1): the outer peripheral zone, the inner central and transitional zones, and the anterior fibro-muscular stroma. Other structures such as the prostatic capsule, the neurovascular bundle, the seminal vesicles, and the Denonvilliers fascia (rectal wall) can also be seen in significant detail (10). On account of age, the central and transitional zone regions may not always be easily differentiable on T2w MRI; typically these regions are jointly termed the “central gland” (CG) region (11).
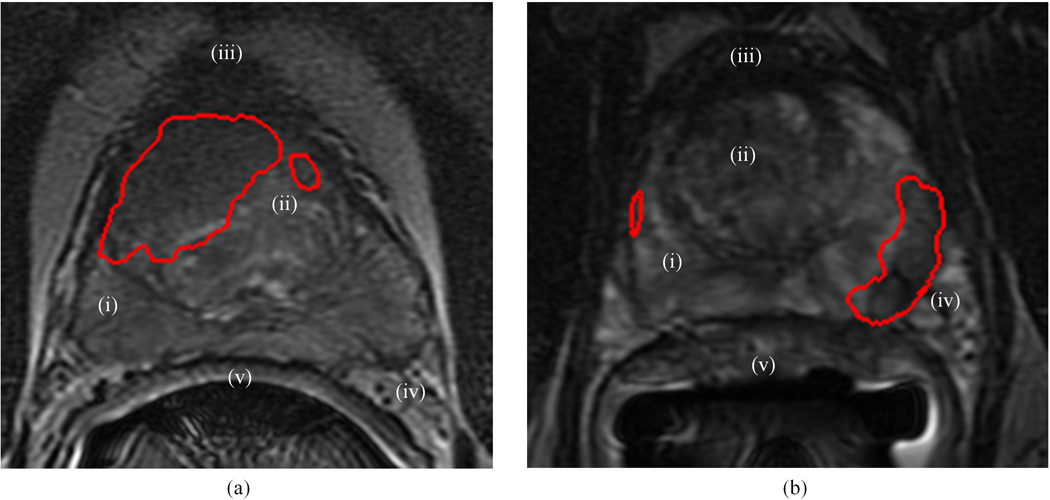
Figure 1.
Depicting visible structures of the prostate on T2w MRI for representative slices with (a) CG CaP, and (b) PZ CaP. On (a) and (b), the red outline shows the delineation of the mapped extent of CaP presence (obtained via registration with corresponding whole-mount histology sections). Structures have been numbered on the images as follows: (i) peripheral zone (PZ), (ii) central and transitional zones jointly termed “central gland” (CG), (iii) anterior fibro-muscular stroma, (iv) seminal vesicles, (v) Denonvilliers fascia (rectal wall).
Due to the lack of epithelial structures in the fibro-muscular region, CaP has typically been found to occur in either the peripheral zone (PZ) or the CG, and its appearance has been found to vary as a function of its spatial location in the gland (12). Within the hyper-intense appearance of PZ on T2w MRI, CaP nodules appear as a region of low SI with incomplete stromal septations within its focus (13). By comparison, CaP nodules in the CG are discerned as a region of purely homogeneous low SI with ill-defined margins and a lenticular (lens-like) shape. This is in contrast to the generally heterogeneous appearance of the CG region on T2w MRI due to nodular areas of varying SI (11,12). While a large majority (~75%) of prostate cancers occur in the PZ (13), accurate localization of CaP within the CG or the PZ is extremely important as CG cancers tend to have lower Gleason scores, suggesting that patients with CG tumors might be candidates for less aggressive therapy and/or active surveillance (14). Additionally, accurate knowledge of the location of CaP within the gland can help in surgical planning by identifying accurate surgical margins (15), targeting biopsies in a more directed fashion (7), as well as in radiation and focal ablation therapy planning to minimize treatment to benign areas (6).
Local staging of CaP in either the CG or PZ of the prostate via T2w MRI has shown significant variability in terms of accuracy (54%–93%) as well as inter-observer agreement (5). For example, the general appearance of an area of low SI in the PZ may be confounded by the presence of non-malignant disease such as prostatitis, fibro-muscular benign prostatic hyperplasia (BPH), or post-biopsy hemorrhagic change (8,10). A significant CaP confounder in the CG particularly is BPH, which has significant overlap with CaP in terms of SI characteristics on T2w MRI. Some researchers have shown that CG and PZ carcinomas may exhibit differences in their apparent diffusion coefficient (16) and ex vivo metabolic profiles (17). However, to our knowledge, no-one has been able to demonstrate significant differences in T2w SI between CG and PZ CaP.
Visual characterization of CaP on T2w MRI has been found to be a function of the resolution and contrast of the imaging technique (5). Bloch et al (18) demonstrated improved signal to noise ratio (SNR) and high spatial resolution using 3 T endorectal MRI, allowing visualization of microscopic and pathologic details that were invisible at 1.5 T. However, the introduction of a bias field in regions close to the endorectal probe (19) significantly affects visual quality of endorectal T2w MRI; an acquisition artifact that is exacerbated by increasing magnetic field strength (11). Other acquisition artifacts which can affect expert assessment of endorectal T2w MRI include “ghosting” (due to patient motion) as well as ambient noise (19).
Based on the differences in SI and appearance between CG and PZ CaP, it is reasonable to assume that there may exist quantitative imaging features (and hence a quantitative imaging signature (QIS)) which are specific to CaP appearance within each of the prostatic zones. The goal of this study is thus to identify unique textural QISes for CaP within each of the PZ and CG. We further posit that these zone-specific QISes will be sufficiently different from each other to allow for building of computerized image-based classifiers for detecting tumor occurrence (on a per voxel basis) in the CG and PZ respectively. To our knowledge, there has been no previous work quantitatively examining the textural appearance of CaP in a manner specific to the zonal location of disease within the prostate. Previous related work in computerized decision support and computer aided detection of CaP from MRI (20,21,22,23) have not considered differences in spatial location of disease within the gland in attempting to identify CaP presence (i.e. the classifiers are monolithic since they do not attempt to distinguish between the spatially distinct types of CaP). We will show that building zone-specific classifiers result in an improved detection of CaP presence on MRI compared to monolithic classifiers.
In order to define zone-specific QISes, we will utilize texture-based characterization of T2w MRI data. The underlying hypothesis for our approach is that the CaP appearance within each of the CG and PZ has distinct textural signatures, i.e. we are attempting to quantify visual characteristics of regions with incomplete stromal septations (corresponding to PZ CaP) or those with homogeneous appearance (corresponding to CG CaP). The result of applying texture analysis algorithms (24,25,26,27) to prostate T2w MRI is that every voxel in a dataset will now be associated with a set of numerical values (reflecting corresponding textural attributes) which describe the local properties and relationships between the SI at the voxel under consideration and its surrounding local neighborhood voxels. Further, given annotations of CaP presence (labels) for each voxel within a dataset, we can train an automated classifier to utilize these textural signatures (and labels) to give us a per-voxel classification for CaP presence in a new, unseen test dataset. In this work, we will construct separate QISes for each of CG and PZ CaP. These QISes will then be used to train independent classifiers to detect CaP in the CG and PZ, respectively. Our hypothesis is that these zone-specific texture-based classifiers will be more accurate in identifying CaP compared to either (a) an intensity-based approach (which would use only the SI at every voxel to classify for CaP presence), or (b) a zone-agnostic monolithic approach (which would not consider zonal differences when constructing the classifier).
Previously, Seltzer et al (28) showed that the combination of a radiologist and a computerized decision support system significantly improves the accuracy of CaP staging on 1.5 T endorectal T2w MRI. Chan et al (20) considered a combination of co-occurrence based (25), discrete cosine transform, and co-ordinate features within an automated classification scheme to obtain an accurate statistical map for CaP presence on in vivo 1.5 T endorectal multi-protocol (line-scan diffusion, T2-mapping, and T2w) MRI. Madabhushi et al (29) presented a machine learning scheme which intelligently combined 3D texture features (1st order statistical (24), Haralick co-occurence (25), Gabor wavelet (26)) to analyze 4 T ex vivo T2w prostate MRI for CaP presence. The system was found to compare favorably to expert radiologists in terms of CaP detection accuracy, with improved reproducibility. A visual study of fractal texture on prostate MRI was conducted by Lv et al (30), who determined that fractal features showed statistically significantly different values for CaP compared to benign regions on 1.5 T T2w MRI. More recently, Lopes et al (22) showed that a machine learning classifier employing fractal and multi-fractal features from 1.5 T T2w prostate MRI was more accurate in detecting CaP compared to more traditional texture features (Haralick co-occurence (25), Gabor (26), and Daubechies (27) wavelet features). Vos et al (23) and Ozer et al (21) developed classifiers for CaP detection using 1.5 T endorectal multi-protocol (dynamic-contrast enhanced, diffusion, and T2w) MRI. These classifiers, however, only employed T2w MRI signal intensity, as opposed to using any textural representations of the original image data. Note that none of these schemes (20,21,22,23) attempted to account for spatial location of disease or zonal anatomy in the classifier construction.
In this work, we will identify distinct zone-specific quantitative textural models of CaP using 3 T endorectal T2w MRI, in order to construct QISes which allow for accurate discrimination between CG and PZ CaP. The overarching application of our work is to build an automated decision support system for improved detection and localization of CaP presence at high-resolution (per-voxel) using T2w MRI; the use of zone-specific models is expected to significantly improve the sensitivity and specificity of CaP detection of such a system. The endorectal 3 T T2w MRI data used in this work was acquired from patients identified with CaP on needle biopsy, who later underwent radical prostatectomies. After pre-processing of both MRI (segmentation (31), acquisition artifact correction (32,33)) and prostatectomy specimens (sectioning, staining, annotation), CaP disease extent is mapped from 2D whole-mount histological prostatectomy sections onto corresponding T2w MRI, via an automated non-linear registration scheme (34). These mapped annotations are considered to constitute the "ground truth" for CaP extent on MRI. A number of different texture features (including Gabor (26) and Haar (27) wavelet, as well as 1st (24) and 2nd order (25) statistical) are then extracted on a per-voxel basis from T2w MRI. An information-theoretic feature selection procedure (35) is applied to identify the most discriminatory features for each of CG and PZ CaP (QISes). The CG and PZ CaP QISes are then evaluated via an automated Quadratic Discriminant Analysis (QDA) classifier (36); corresponding classification results being evaluated on a per-voxel basis against ground truth CaP annotations on MRI (mapped from corresponding histology).
MATERIALS AND METHODS
Data
The data cohort of 22 patients considered in this study was obtained from a previously conducted prospective study approved by the Institutional Review Board at the Beth Israel Deaconess Medical Center. This prospective study considered patients who had first been confirmed to have CaP via positive core needle biopsies and were scheduled for a radical prostatectomy. Prior to surgery, the patients were imaged using a combined torso-phased array and endorectal coil (MedRad, Pittsburgh, PA) using a 3 Tesla whole-body MRI scanner (Genesis Signa LX Excite; GE Medical Systems, Milwaukee, Wisconsin). The parameters for axial T2w MR imaging were TR/TE = 6375/165 msec with a slice thickness of 1.5–2mm (no gap between the slices). Matrix size of acquisition was 320 × 224-192 voxels with a field of view of 12 × 12 cm. Following surgery, the excised glands were sectioned and stained with Haematoxylin and Eosin (H & E), attempting to ensure at all times that the sectioning was done in a plane corresponding to the pre-operative MRI.
The 22 patient studies considered in this work were chosen from a larger cohort of 124 cases, all of which had MR imaging performed prior to a radical prostatectomy procedure. Of the 124 cases, only 65 studies included usable T2w MRI with corresponding digitized whole mount histological sections. Of these, only 25 cases were identified with histological sections on which CaP was visible and could be annotated by a pathologist. An additional 3 cases had to be discarded due to the poor quality of the MR imaging. A pathologist (EMG) and radiologist (NBB) working in unison visually identified 80 corresponding 2D whole-mount histological sections (WMHS) and axial T2w MRI slices from 22 studies. These correspondences were established via anatomical fiducials such as the urethra, veromontanum, as well as prominent BPH nodules that were visually discernible on both the histology and pre-operative MRI.
Based on the recommendations by McNeal (9), a patient study was classified as having CG or PZ CaP if more than 70% of prostate cancer volume was found to be present in a particular zone. Of the 22 datasets, 16 were thus determined as having PZ CaP (50 2D sections), while the remaining 6 were identified as having CaP in the CG (30 2D sections). In order to ensure that the sets of CG and PZ CaP were as distinct from each other as possible, only those sections were included which showed an explicit focus of CaP in either the CG or the PZ.
Notation
We shall denote the set of all samples (voxels on T2w MRI) as C = {c1, c2, …, cn}; note that all analysis in this study has been performed at a per-voxel level as far as possible. After feature extraction (see below), a feature vector Fi = {fi(c1),…, fi(cn)}, i ∈ {1,…, N} is obtained as the collection of feature responses fi(c) for all c ∈ C; hence Fi is a 1 × n vector, where i represents the feature operator. The set of feature vectors corresponding to all operators i is given as F = {F1,…, FN}. Note that n is the total number of voxels considered, while N = 110 is the total number of features extracted. Additionally, every voxel c ∈ C is associated with a label l(c) ∈ {+1, −1}, corresponding to cancer/benign annotations on T2w MRI (obtained via registration with histology, see below); the corresponding label vector is given by L (of size 1 × n).
Pre-processing of T2w MRI
For all datasets considered, the prostate gland was segmented out from the larger field of view of the axial endorectal T2w MRI image using an automated prostate capsule segmentation scheme (31). Briefly, this scheme involves first automatically identifying prostatic voxels within a bounding box around the prostate area in order to obtain a boundary initialization for an Active Shape Model (ASM) scheme. The ASM then transforms a known mean shape of the prostate to the boundary initialization, resulting in a capsule segmentation for the gland. All remaining analysis for CaP presence could then be localized to the prostate region-of-interest alone. We then corrected for known acquisition-based intensity artifacts; bias field inhomogeneity (19) and intensity non-standardness (33). The most significant of these was the effects of bias field on endorectal prostate T2w MRI, occurring due to the usage of an endorectal probe (19). This artifact manifests as a smooth variation of signal intensity across the T2w MR image, and has been shown to significantly affect computerized image analysis algorithms such as the automated classification of tissue regions (29). Bias field artifacts were corrected for via the popular N3 algorithm (32), which incrementally de-convolves smooth bias field estimates from acquired image data, resulting in a bias-field corrected image. A second artifact termed intensity non-standardness (33) refers to the issue of MR image “intensity drift” across different imaging acquisitions; both between different patients as well as for the same patient at different imaging instances. Intensity non-standardness results in MR image intensities lacking tissue-specific numeric meaning within the same MRI protocol, for the same body region, or for images of the same patient obtained on the same scanner (33). This artifact was corrected for via an interactive implementation of the generalized scale algorithm (33). The standardization step allows for alignment of the image intensity histograms across the different patient MRI studies, thus ensuring that MR image intensities have a consistent tissue-specific numeric meaning and interpretation.
Multimodal Elastic Registration of Ex Vivo WMHS and In Vivo MRI to Delineate “Ground Truth” CaP Extent
By aligning ex vivo histologic sections with corresponding in vivo T2w MRI data, ground truth CaP extent can be obtained on MRI. A 2D registration strategy was adopted wherein corresponding 2D WMHS and 2D T2w MRI sections were registered to one another; correspondences between WMHS and T2w MRI sections having previously been established via visual evaluation by clinical experts.
Registration of multi-modal imagery (such as ex vivo WMHS and in vivo MRI) is complicated by differences in both image intensities and shape of the underlying anatomy on scenes corresponding to different modalities and protocols (34). Non-linear differences in the overall shape of the prostate between in vivo MRI and ex vivo WMHS exist due to, (1) the presence of an endorectal coil during MR imaging, and (2) deformations to the histological specimen due to fixation and sectioning. Consequently, achieving correct alignment of such imagery requires elastic transformations to overcome the non-linear shape differences. However, driving such transformations via traditional intensity-based similarity measures (such as mutual information (MI)) has been found to be sub-optimal for robustly registering dissimilar modalities (37). We therefore utilize a non-rigid registration scheme (34) driven by a higher-order variant of MI, which allows handling of images that are very different in terms of intensities (e.g. MRI and WMHS data) as well as deformation characteristics (e.g. in vivo to ex vivo). The spatial alignment of the two modalities is implemented in 2 steps. First, an initial affine alignment of WMHS to the corresponding T2w MRI slice is done. This enables correction of large translations, rotations, and differences in image scale. Then, automated non-rigid registration of the rigidly aligned WMHS and T2w MRI is performed via a fully automated non-linear hierarchical (multi-scale) B-spline mesh grid image warping scheme.
This technique was used to align all 80 corresponding 2D WMHS and T2w MRI slices and subsequently map spatial extent of CaP from WMHS onto corresponding T2w MRI slices, across 22 patient datasets. The spatial extent of CaP mapped onto T2w MRI was examined and manually corrected (as required) by an expert (NBB) using Photoshop® (Adobe Systems Inc, CA). The final result of this procedure was a labeling of each voxel within the prostate as corresponding to CaP or benign. Note that while the image registration was done in 2D, all subsequent feature extraction and classification operations were done in 3D.
Feature Extraction
The goal of our study is to identify unique textural signatures for CaP presence in each of the CG and PZ regions, respectively. It may be expected that some combination of different types of textural features will thus play a role in accurately characterizing CaP appearance in these zones. We have therefore attempted to model CaP appearance via image texture features which have been frequently used in the image processing and computer vision fields (24,38,39), as well as shown to be useful for characterizing the appearance of different pathologies on MRI (40,41,42). These textural descriptors include co-occurrence (25) (which capture spatial greylevel characteristics) and gradient-based features (24) (which capture edge and directional characteristics); these features have previously been shown to be useful in characterizing appearance of CaP on T2w MRI (22,29). A number of different wavelet filter decomposition approaches (43) have also been employed by us as they allow for extraction of fine structural image detail at different orientations and scales, and could prove useful in quantitatively characterizing the micro- and macroscopic visual cues used by radiologists when identifying CaP regions on MRI. Most popular amongst these are the Gabor (continuous) (26) and the Haar (discrete) (27) wavelet transforms. Table 1 summarizes the texture features used in this study as well as the visual significance of such features for identifying CaP on prostate T2w MRI.
Table 1.
Summary of texture features used in this study as well as their significance for localization of CaP on T2w MRI (numbers in brackets signify how many features of each texture category were computed)
| Feature | Implementation | Purpose | Significance for quantifying CaP appearance |
|---|---|---|---|
| Gabor wavelet transform (48) | Modulation of a complex sinusoid by a Gaussian function | Attempt to match localized frequency characteristics at multiple scales and orientations (26) | Quantify visual processing features used by radiologists when examining appearance of the carcinoma |
| Haar wavelet transform (12) | Decomposition coefficients via wavelet decomposition at multiple scales | Attempt decomposition of a signal in the discrete space while offering localization in the time and frequency domains (27) | Differentiate the amorphous nature of the non-CaP regions within foci of low SI |
| Haralick texture feature (36) | Construct joint probability distribution of the occurrence of greylevel intensities in an image (spatial relationship between pixels used to restrict counting of greylevel co-occurrences). Statistical features are then calculated from this distribution | Differentiate between different types of texture excellently due to calculation of 2nd order statistics (which quantify perceptual appearance of image) (25) | Useful in differentiating homogeneous low SI regions (CaP) from more hyper-intense appearance of normal prostate |
| Greylevel statistical features (14) | Mean, standard deviation as well as derivative features such as via convolution with the Sobel and Kirsch operators are calculated | Provide 1st order information, quantifying macroscopic appearance of image e.g. variation of intensities within image (24) etc. | May help localize regions of significant differences on T2w MR image, accurately detect region boundaries |
In all, 110 texture features corresponding to 4 different texture feature classes were extracted on a per-voxel basis from each MRI dataset. All feature extraction methods were implemented using MATLAB® (The Mathworks Inc, MA).
Feature Selection
After extracting texture features, we utilized the minimum Redundancy Maximum Relevance (mRMR) feature selection scheme (35) in order to identify an ensemble of features that will allow for optimal identification of CaP presence on MRI. During feature selection, 2 separate sets of data were considered; one comprising feature and label data corresponding to voxels from the 16 datasets with PZ CaP, and the other comprising feature and label data corresponding to voxels from the 6 datasets with CG CaP. Thus the result of mRMR feature selection was 2 distinct QISes (one corresponding to CG CaP denoted QCG, the other to PZ CaP denoted QPZ); each set comprising texture features which can be considered highly discriminatory for differentiating between CaP and benign regions in the corresponding prostatic zone.
In the following description, the selected subset of features Q is comprised of feature vectors Fi, i ∈ {1,…, |Q|} (note that Q ⊂ F and |Q| < N). The mRMR scheme attempts to simultaneously optimize two distinct criteria. The first is “maximum relevance” which selects features Fi that have the maximal mutual information (MI) with respect to the corresponding label vector L. This is expressed as
The second is “minimum redundancy” which ensures that selected features Fi, Fj ∈ Q, i, j ∈ {1,…, |Q|}, are those which have the minimum MI with respect to each other, given as
Under the second constraint, the selected features will be maximally dissimilar with respect to each other, while under the first, the feature selection will be directed by the similarity with respect to the class labels. There are two major variants of the mRMR scheme: the MI difference (MID, given by U − V) and the MI quotient (MIQ, given by U/V). These represent different techniques to optimize the conditions associated with mRMR feature selection. We evaluated the use of both MID and MIQ for feature selection in this study, as well as determined an optimal number of features comprising each QIS by varying |Q| within the mRMR algorithm.
Automated Supervised Classification
In order to evaluate QCG and QPZ, we utilized a voxel-level QDA classifier (36) to determine their ability to accurately discriminate between CaP and benign regions on T2w MRI data. As an alternative to constructing zone-specific QISes, we also considered the following strategies: (1) utilizing all 110 extracted texture features with no feature selection (F), and (2) a randomly selected subset of texture features (denoted by R). Note that 2 separate voxel-level classification tasks were performed using each of Q, F, R: first to identify regions of CG CaP, and then to identify PZ CaP. The CaP detection accuracy of the QDA classifier in each case was then considered as being reflective of the validity of Q, F, R in characterizing CG and PZ CaP respectively.
Quadratic Discriminant Analysis
The Quadratic Discriminant Analysis (QDA) classifier (36) aims to find a transformation of the input features that is able to optimally discriminate between the classes in the dataset. Given a set of samples C with associated feature set F, QDA solves for Y = FTAF + BTF, where Y = {Y1, Y2,…} denotes the result of QDA.
Based on calculating the means μl(c)=+1, μl(c)=−1, and covariances Σl(c)=+1, Σl(c)=−1, of the 2 classes in the dataset (cancer and benign), we can solve the above equation to calculate the following log likelihood ratio:
The result of QDA classification is a per-voxel probability for CaP presence, given by h(c) ∈ H.
Evaluation
Classification of the T2w MRI data was done on a per-voxel basis, with evaluation of the results against corresponding per-voxel annotations of CaP presence (via registration with histology). Thus, for all samples c ∈ C, we directly compared the probabilistic classifier result h(c) with the label l(c), at different thresholds of h(c). Plotting the true- and false-positive rates as function of varying the threshold of h(c)allowed us to perform Receiver-Operating Characteristic (ROC) curve analysis, with the Area under the ROC curve (AUC) being used as a measure of classifier performance (20,22,29).
In order to ensure robustness of the classifier to training and testing data, a randomized 3 fold cross-validation procedure was implemented. In a single cross-validation run, the datasets being considered (16 in the case of PZ CaP, 6 in the case of CG CaP) were divided into 3 randomized subsets (comprising 6, 5, and 5 studies for the PZ CaP detection problem and 2, 2, and 2 in the case of CG CaP). 2 subsets were considered as training data and the remaining as testing data, following which classification is performed. This was repeated until all 3 subsets were classified, and the entire cross-validation procedure was iterated 25 times. Feature selection and classifier construction were done separately for each set of training data (for all 3 folds over all 25 runs), with corresponding testing data only used for evaluation of classifier performance. All classifications were performed on a per-voxel basis.
Each run of cross-validation yielded an AUC value (cumulatively calculated over all 3 folds); 25 AUC values were therefore calculated for each classification task (identifying CaP presence using different feature sets QCG, QPZ, F, R). The non-parametric pair-wise Wilcoxon rank-sum test was used to test for statistical significance when comparing the CaP detection AUCs of different feature sets (QCG, QPZ, F, R). The Bonferroni correction was employed to address the issue of multiple comparisons, thus correcting the p value used in testing for statistical significance within each individual Wilcoxon test from 0.05 to 1.67E-02. The null hypothesis for the Wilcoxon test in every case is that no statistically significant differences were observed when using QCG (or QPZ) for QDA classification as compared to alternative feature sets (F, R).
RESULTS
Feature Selection to Construct QCG and QPZ
Table 2 summarizes the top 20 features ranked by the mRMR algorithm in terms of their discriminative ability for differentiating between CaP and benign regions within each zone (CG, PZ); these features constitute QCG, QPZ respectively. Note that the combination of features 1 and 2 are considered to be more relevant (while minimizing redundancy) as compared to feature 1 alone, and so on. We examined this property of the QISes in more detail in the experiment below.
Table 2.
Summary of top 20 features selected to accurately identify CG and PZ CaP respectively, obtained by voting of selected features across 25 cross-validation runs. Note that the 2 sets of features are relatively unique. 3 features that were found to be in common have been highlighted in italics (‘w’ stands for window size, an associated parameter setting for the feature).
| Top 20 discriminatory features for CG CaP |
Top 20 discriminatory features for PZ CaP |
|||
|---|---|---|---|---|
| 1 | Gabor | Orient=157.5, Wavelength=5.6569 | Haralick | Difference Average (w=7) |
| 2 | Gabor | Orient=45, Wavelength=2.8284 | Haralick | Sum Entropy (w=7) |
| 3 | Gabor | Orient=0, Wavelength=2.8284 | Haralick | Information Measure (w=5) |
| 4 | Gabor | Orient=135, Wavelength=45.2548 | Haralick | Difference Variance (w=7) |
| 5 | Haralick | Energy (w=5) | Gabor | Orient=0, Wavelength=5.6569' |
| 6 | Haralick | Sum Average (w=5) | Haar | Level 4 Horizontal Coefficient |
| 7 | Gabor | Orient=67.5, Wavelength=5.6569 | Haralick | Entropy (w=7) |
| 8 | Greylevel | Mean (w=5) | Gabor | Orient=157.5, Wavelength=11.3137 |
| 9 | Haralick | Difference Average (w=7) | Gabor | Orient=157.5, Wavelength=8.2 |
| 10 | Haralick | Difference Entropy (w=7) | Gabor | Orient=135, Wavelength=11.3137 |
| 11 | Gabor | Orient=0, Wavelength=45.2548 | Haar | Level 2 Vertical Coefficient |
| 12 | Gabor | Orient=112.5, Wavelength=2.8284 | Haralick | Difference Entropy (w=7) |
| 13 | Haralick | Inverse Difference Moment (w=5) | Gabor | Orient=112.5, Wavelength=45.2548 |
| 14 | Gabor | Orient=90, Wavelength=45.2548 | Gabor | Orient=0, Wavelength=8.2 |
| 15 | Gabor | Orient=157.5, Wavelength=22.6274 | Gabor | Orient=67.5, Wavelength=2.8284 |
| 16 | Gabor | Orient=90, Wavelength=11.3137 | Haralick | Information Measure (w=7) |
| 17 | Gabor | Orient=67.5, Wavelength=2.8284 | Gabor | Orient=0, Wavelength=11.3137 |
| 18 | Greylevel | Standard Deviation (w=5) | Gabor | Orient=112.5, Wavelength=5.6569 |
| 19 | Gabor | Orient=112.5, Wavelength=11.3137 | Greylevel | Kirsch |
| 20 | Gabor | Orient=157.5, Wavelength=45.2548 | Gabor | Orient=67.5, Wavelength=45.2548 |
QCG was seen to largely comprise of Gabor features, while QPZ mainly comprised Haralick texture features. 3 features were found to be in common between QCG, QPZ (highlighted in italics in Table 2). Figure 2 shows representative QCG features derived from a T2w MRI section with CG CaP, while Figure 3 shows representative QPZ feature images corresponding to PZ CaP.
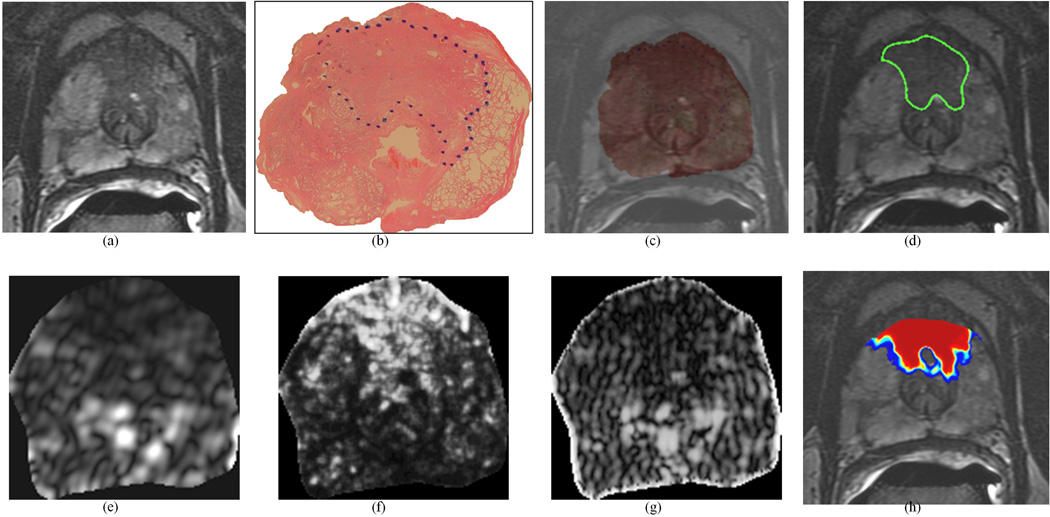
Figure 2.
Representative CG CaP dataset: (a) original 2D T2w MR image, (b) original 2D whole-mount histological section, with CG CaP extent outlined in blue (by a pathologist), (c) overlay of WMHS and T2w MRI after non-linear multi-modal registration, (d) mapped CG CaP extent on T2w MRI (outlined in green, from blue-outlined region in (b)). Representative texture features for this section: (e) Gabor (Orient = 157.5, Wavelength = 5.6569), (f) Haralick (energy, w=5), and (g) Gabor (Orient = 0, Wavelength = 2.8284). Note the improved visual differentiability between CG CaP and benign regions on (e)–(g) texture feature images compared to the (a) original intensity image. (h) shows the probabilistic heatmap of CG CaP presence mapped back onto the image (via automated QDA classification). In (h), red indicates a high probability of CG CaP presence, blue indicates low probabilities of CG CaP presence, and no CaP was detected in the uncolored regions. This study was not used in training the classifier.
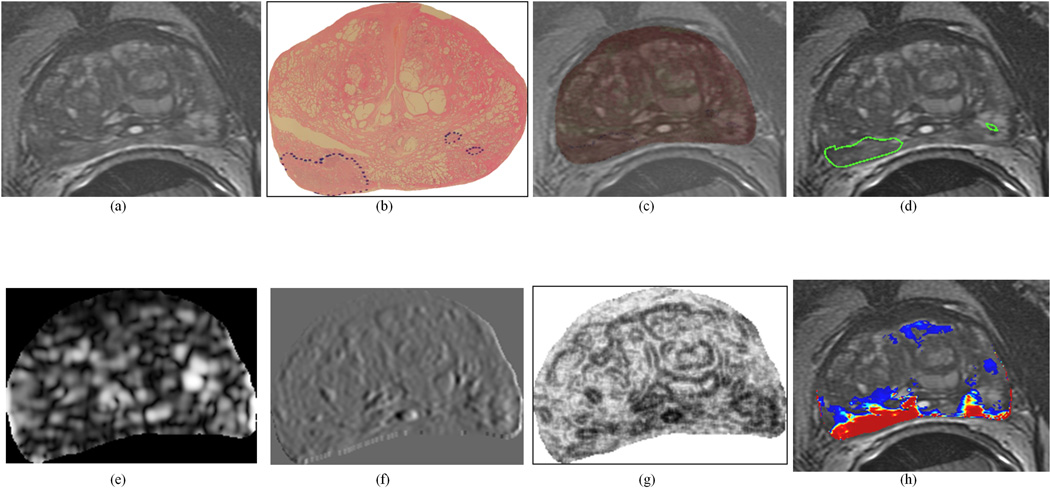
Figure 3.
Representative PZ CaP dataset: (a) original 2D T2w MR image, (b) original 2D whole-mount histological section, with PZ CaP extent outlined in blue (by a pathologist), (c) overlay of WMHS and T2w MRI after non-linear multi-modal registration, (d) mapped PZ CaP extent on T2w MRI (outlined in green, from blue-outlined region in (b)). Representative texture features for this section: (e) Gabor (Orient = 0, Wavelength = 5.6569), (f) Haar (Level 4 vertical coefficient), and (g) Haralick (Information measure, w=5). Note the improved visual differentiability between PZ CaP and benign regions on (e)–(g) texture feature images compared to the (a) original intensity image. (h) shows the probabilistic heatmap of PZ CaP presence mapped back onto the image (via automated QDA classification). In (h), red indicates a high probability of PZ CaP presence, blue indicates low probabilities of PZ CaP presence, and no CaP was detected in the uncolored regions. This study was not used in training the classifier.
Selecting an Optimal Number of Features to Comprise Each of QCG and QPZ
The main free parameter associated with the mRMR scheme is the number of features to be selected (|Q|). We empirically varied the number of features that were selected to comprise QCG and QPZ (from 5 to 50), and evaluated the classification performance of each resulting QCG and QPZ using a QDA classifier. The aim behind this experiment was to identify the minimal number of features required to yield a classifier AUC that most closely approached that obtainable by considering all 110 features. Figures 4 and 5 summarize the results of this evaluation for the CG and PZ, respectively.
Figure 4.
Experiment to determine an optimal number of features to comprise the QIS for CG CaP. Trend in AUC value (Y-axis) of a QDA classifier is plotted as a function of different numbers of features comprising the QIS (X-axis). Note significant improvement when using mRMR feature selection (blue, orange) over using all features (black). The QIS comprising 15 features (selected via mRMR-MIQ, highlighted with a blue polygon) was found to provide the best overall performance for CaP detection compared to any other QIS, as well as compared to using all 110 features (black).
Figure 5.
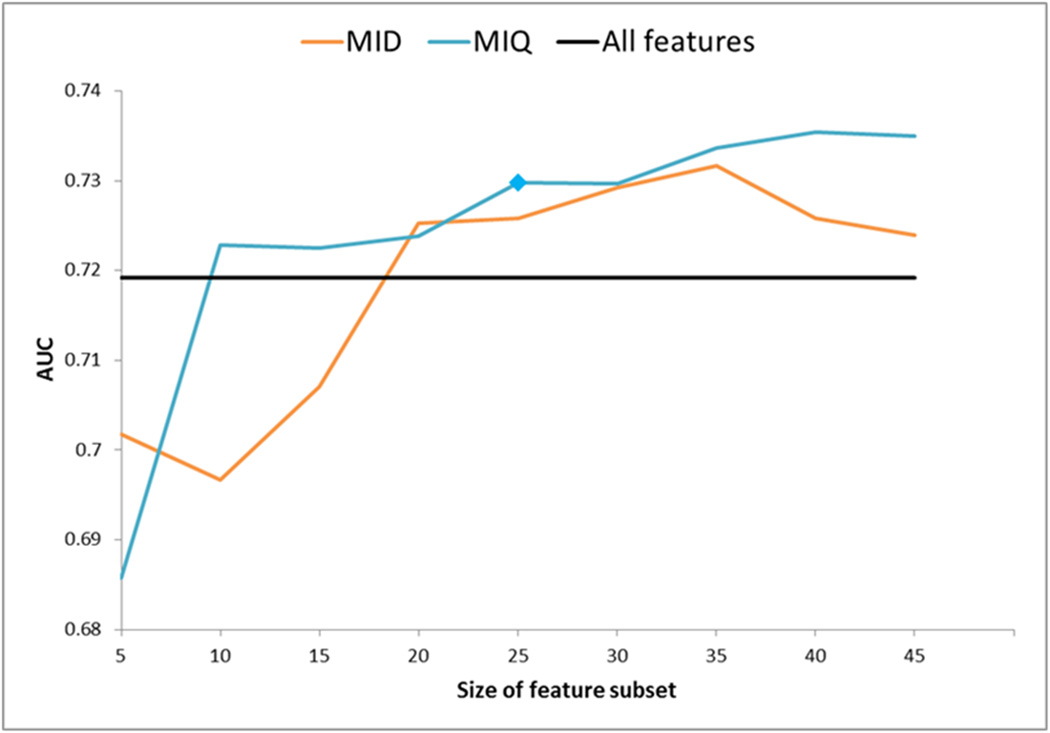
Experiment to determine an optimal number of features to comprise the QIS for PZ CaP. Trend in AUC value (Y-axis) of a QDA classifier is plotted as a function of different numbers of features comprising the QIS (X-axis). Note significant improvement when using mRMR feature selection (blue, orange) over using all features (black). The QIS comprising 25 features (selected via mRMR-MIQ, highlighted with a blue polygon) was found to provide the best overall performance for CaP detection compared to any other QIS, as well as compared to using all 110 features (black).
For the CG, the performance of both variants of the mRMR scheme (MIQ and MID) was consistently found to improve upon using all 110 features (black). Overall, MIQ (blue) was seen to outperform MID (orange). The best performing subset (the most optimal QCG) contained 15 features (selected via MIQ, highlighted with a blue polygon), with an AUC of 0.863 (standard error of 0.002 across 25 cross validation runs). In comparison, using all 110 features (black) yielded a significantly lower AUC of 0.814 (standard error of 0.002, p value = 1.37e-09).
For the PZ, the performance of both variants of the mRMR scheme (MIQ and MID) generally improved upon using all 110 features (black). Overall, MIQ (blue) was seen to outperform MID (orange). The best performing subset (the most optimal QPZ) contained 25 features (selected via MIQ, highlighted with a blue polygon), with an AUC of 0.730 (standard error of 0.004). In comparison, using all 110 features (black) yielded a lower AUC of 0.720 (standard error of 0.003).
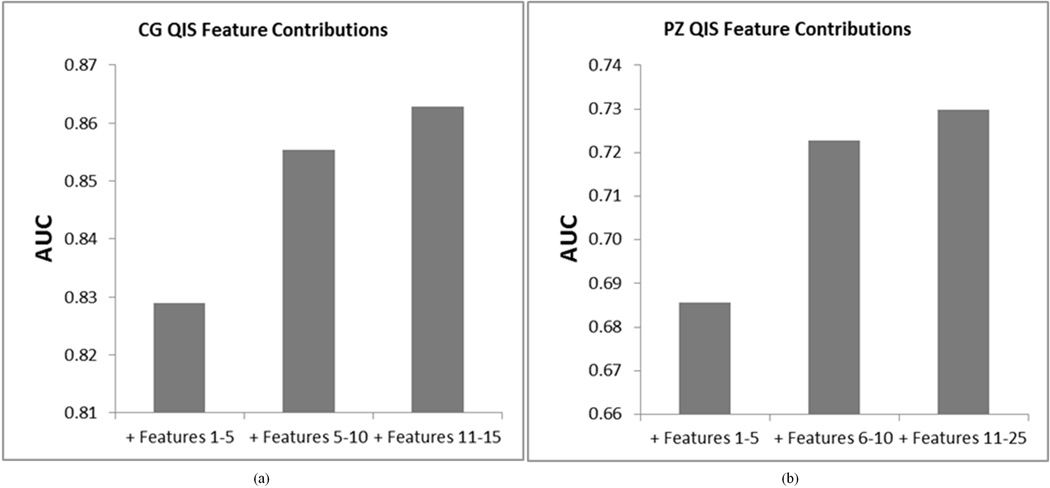
We additionally evaluated the significance as well as the effectiveness (for CaP localization) of each feature Fi ∈ QCG, i ∈ {1,…,15}, and Fj ∈ QPZ, j ∈ {1,…,25}. Figure 6 summarizes the classification performance (in terms of AUC) achieved by incrementally considering features comprising each of QCG and QPZ. For both the CG and PZ, it was seen that the top 5 features contributed most significantly to CaP classification performance. The inclusion of additional features only marginally improved the CaP classification performance over what was obtained by using the first 5 features to be included within QCG and QPZ respectively. However, QCG and QPZ in their entireties yielded the highest AUC values for localizing CaP in the CG (using the top 15 features) and PZ (using the top 25 features).
Figure 6.
Performance of incrementally considering features which comprise CG and PZ QISes. Note the improvement in classification performance with addition of each additional set of features.
Comparing the Use of QCG and QPZ for CaP Detection Against Alternative Feature Sets
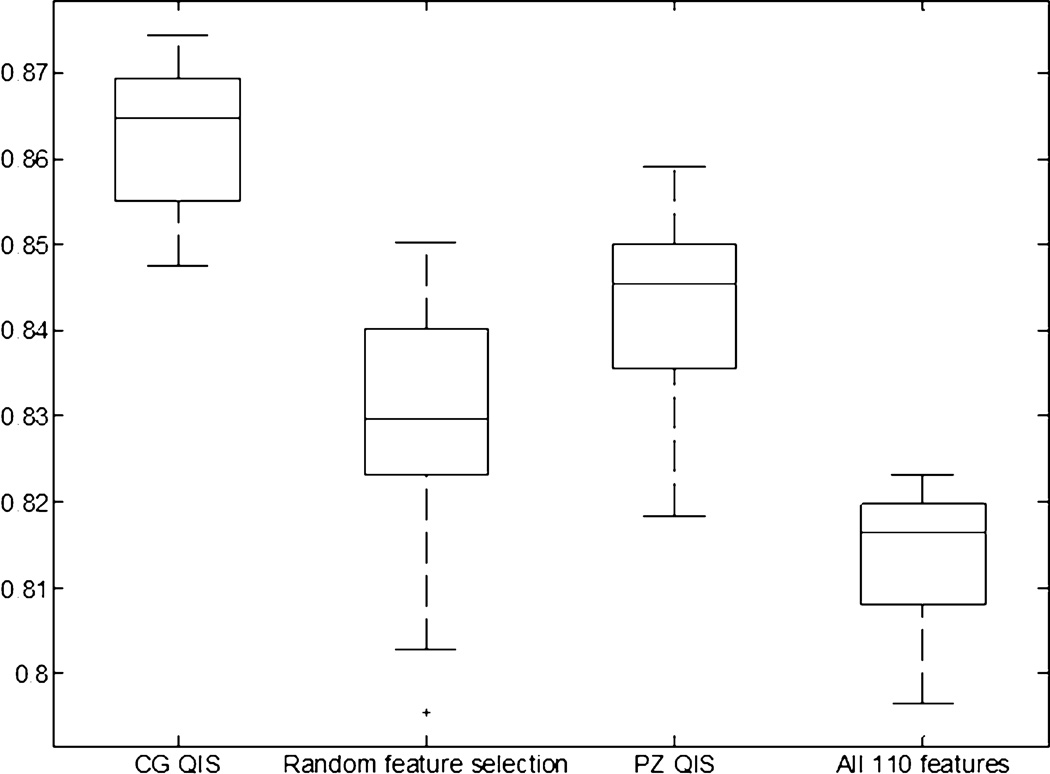
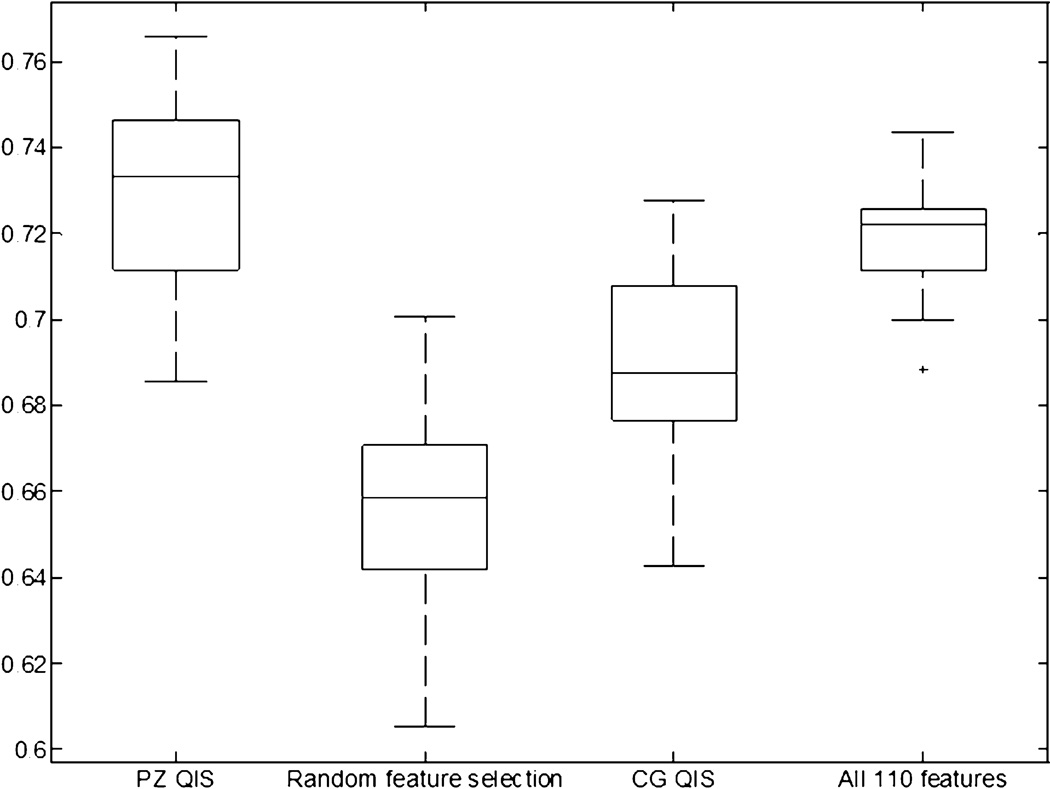
Figures 7 and 8 show box-and-whisker plots reflecting the CaP classification performance (AUC) of QCG and QPZ, as compared to alternate strategies: (1) considering all 110 features (F), (2) a random subset of features R (with the same number of features as the QIS), (3) using QCG to classify for PZ CaP and QPZ to classify for CG CaP. This last experiment is intended to evaluate the specificity of the QIS to the appearance of CaP in a particular zone, as well as highlight the importance of doing a zone-based classification. For the CG, a statistically superior performance (p<1.67E-02) was obtained when using QCG as compared to using F (p=1.37E-09), R (p=5.77E-09), or QPZ (p=1.29E-07). Similarly for the PZ, a statistically superior performance (p<1.67E-02) was obtained when using QPZ as compared to using R (p=2.29E-09) and QCG (p=1.12E-06), but not F (p=7.42E-02).
Figure 7.
Box-and-whisker plot comparing performance of using the CG QIS for CG CaP detection against alternative strategies: (1) using 15 randomly selected features, (2) using the PZ QIS (25 features), (3) using all 110 features (without feature selection/ranking). Using the CG QIS was found to provide a statistically significantly superior performance for CaP detection in the CG, ratifying the effectiveness of the zone-specific textural signature compared to alternative strategies.
Figure 8.
Box-and-whisker plot comparing performance of using the PZ QIS for PZ CaP detection against alternative strategies: (1) using 25 randomly selected features, (2) using the CG QIS (15 features), (3) using all 110 features (without feature selection/ranking). Using the PZ QIS was found to provide a statistically significantly superior performance for CaP detection in the PZ, ratifying the effectiveness of the zone-specific textural signature compared to alternative strategies.
DISCUSSION
Current clinical intuition suggests that the appearance of CG and PZ tumors on endorectal prostate T2w MRI is significantly different (12,13). Given the differing prognoses and outcomes of prostate cancer (CaP) based on its zonal location, there is a significant need for examining quantitative imaging signatures (QISes) specifically targeted to identifying CaP in vivo. The 2 major goals of our study were thus to, (1) define distinct textural signatures specific to CaP appearance in the CG and the PZ, and (2) quantitatively demonstrate the differences in these zone-specific QISes as well as evaluate their effectiveness for detecting CaP on T2w MRI. In this work, we have presented the first attempt at quantitatively defining QISes for CG and PZ tumors on T2w MRI. We believe that this work will allow for the building of targeted classifiers with the ability to incorporate spatial location of the disease into the model. This is a significantly different approach compared to the current trend of building monolithic computer-aided diagnostic models (20,21,22,23), which do not consider the zonal location of CaP in the prostate. Defining and utilizing zone-specific QISes to identify CG and PZ tumors will allow us to build computerized classifiers with improved sensitivity and specificity for CaP detection.
Our results showed that the QISes for each of CG and PZ CaP comprised largely non-overlapping textural attributes. These results appear to confirm current clinical intuition (12,13), which suggests that CG and PZ CaP may have inherently differing appearances on MRI (16). We found that the CG QIS was primarily comprised of the Gabor class of texture features (representative CG QIS images in Figure 2). The multi-scale, multi-orientated Gabor filters appears to be able to accurately model localized frequency characteristics (26) of the hypo-intense, homogeneous appearing CG CaP (12), allowing for discrimination from the heterogeneously appearing normal CG region. By comparison, the PZ CaP QIS was comprised largely of Haralick texture features (representative images in Figure 3). These features, which involve calculating image statistics derived from the co-occurrence of greylevel image intensities, appear to allow for accurate characterization and separation of low SI regions of CaP from hyper-intense normal prostatic regions in the PZ. Features that were found to overlap between the two QISes were high-level macro-resolution features (such as the Haralick energy and difference average features at large window sizes), implying a similarity between the two types of tumors at a gross scale. However, the largely non-overlapping nature of the two QISes at finer scales and resolutions appears to suggest fundamental micro-level textural differences between CG and PZ CaP on T2w MRI.
Significantly improved classification accuracy (via QDA) was achieved when using the zone-specific QISes to detect for CaP presence, as compared to (1) using all 110 features that were extracted, as well as (2) using a random subset of features. More importantly, interchanging the QISes (i.e. using QCG to classify for PZ CaP, and vice versa) also performed significantly worse compared to using the zone-specific QISes (i.e. using QCG to classify for CG CaP). Our findings suggest that CaP presence in different regions of the prostate is characterized by different structural and textural attributes, as captured by the QISes. The relatively high accuracy associated with the zone-specific QISes in detecting CaP (AUC of 0.86 in CG, 0.73 in the PZ) imply that we have largely optimized the classifier for CaP detection on T2w MRI. We expect that further combining the T2w information with DCE or DWI information will allow us to improve on CaP classification accuracy even more.
Our automated classification results are comparable, and in many cases superior, to other computerized decision support schemes in the literature; most of which have only been applied to CaP localization in the PZ alone. When using T2w intensities alone, Chan et al (20) reported an AUC of 0.59±0.14 (11 studies), Vos et al (23) reported an AUC of 0.84 (34 studies), and Ozer et al (21) reported an AUC of 0.7–0.8 (depending on the classifier used). Lopes et al (22) achieved a mean AUC of 0.88 for CaP localization by employing a subset of texture features considered in this work. When examining these results, it is worth noting that the current study differs from previous work in the following ways.
Annotation of CaP on MRI: We have performed automated non-linear registration of T2w MRI and histology (34) in order to map CaP extent onto MRI. This is distinctly more rigorous compared to using either manual annotations (20,21,22) or an approximate affine registration between WMHS and MRI establish CaP extent on MRI (23). The use of more rigorous techniques for mapping of CaP extents onto MRI leads us to have higher confidence in our results.
Resolution of analysis: All of our analysis and evaluation was performed on a per-voxel basis, within an automatically determined ROI (via capsule segmentation (31)), and in 3D. In comparison, classification and evaluation in previous approaches were either done (1) using manually extracted regions-of-interest (20,23) from within the PZ alone, or (2) a per-pixel analysis of representative 2D sections (21,22).
Strength of magnet: To our knowledge, this is the first work to present automated quantitative analysis of 3 Tesla endorectal prostate T2w MRI, compared to the use of 1.5 T endorectal or whole-body MRI in previous work (20,21,22,23).
Our study did however have its limitations. First, our entire study comprised 22 pre-operative patient imaging datasets, 16 with PZ CaP and 6 with CG CaP. However this cohort size is comparable to other CAD studies for prostate cancer detection on MRI: Chan et al (11 datasets) (20), Ozer et al (20 datasets) (21), Lopes et al (27 datasets) (22), Madabhushi et al (5 datasets) (29). To address the issue of the study being under-powered, a randomized cross validation procedure was employed for learning and evaluating the QISes. Additionally, we did not consider textural differences which may exist between different Gleason grades of CaP. We are currently collecting data which will allow us to explore such differences in future work.
Registration of histology and MRI was performed in 2D. Performing the spatial alignment in 3D would have required 3D reconstruction of the histology volume, which was not possible due to (1) an insufficient number of histology slices being retained as part of the clinical protocol, leading to coarse and uneven inter-slice spacing of WMHS, and (2) fixation artifacts such as tissue loss or distortion (caused by prostate sectioning) and inconsistent H & E staining. It is also possible that the candidate zone-specific QISes determined by us in this study were selected based on textural differences between normal CG and PZ tissue, as opposed to the textural differences between CG and PZ CaP. However, an experiment to conclusively demonstrate that there are fundamental textural differences between CG and PZ CaP alone (rather than CG and PZ normal tissue) will require an additional segmentation of the CG and PZ within the prostate ROI. This additional zone-wise segmentation was not performed by us in this study as it is a significantly difficult problem, both for human experts as well as for image segmentation algorithms.
In conclusion, we believe developing quantitative zone-specific models of prostate cancer represent an important step in developing computational image analysis models for improved staging and detection of disease in vivo. Future work will involve further prospective validation of the candidate textural signatures determined in this study on a larger cohort of data, as well as incorporating additional protocols and features into our work, in order to develop a comprehensive multi-functional signature for CaP presence in vivo for use within an automated decision support system. We also intend to study the differences in textural appearance between CaP regions and common confounders for CaP presence (prostatitis, BPH).
Acknowledgments
Grant Support: This work was made possible via grants from the Wallace H. Coulter Foundation, the National Cancer Institute (Grant Nos. R01CA136535-01, R01CA140772-01, and R03CA143991-01), the Cancer Institute of New Jersey, the Department of Defense (W81XWH-09-0072), and the Society for Imaging Informatics in Medicine.
References
- 1.American Cancer Society. Cancer Facts & Figures 2010. Atlanta: American Cancer Society; 2010. [Google Scholar]
- 2.Chelsky MJ, Schnall MD, Seidmon EJ, Pollack HM. Use of endorectal surface coil magnetic resonance imaging for local staging of prostate cancer. J Urol. 1993;150:391–395. doi: 10.1016/s0022-5347(17)35490-3. [DOI] [PubMed] [Google Scholar]
- 3.Jager GJ, Ruijter ET, van de Kaa CA, et al. Local staging of prostate cancer with endorectal MR imaging: correlation with histopathology. AJR Am J Roentgenol. 1996;166:845–852. doi: 10.2214/ajr.166.4.8610561. [DOI] [PubMed] [Google Scholar]
- 4.Bezzi M, Kressel HY, Allen KS, et al. Prostatic carcinoma: staging with MR imaging at 1.5 T. Radiology. 1988;169:339–346. doi: 10.1148/radiology.169.2.3174982. [DOI] [PubMed] [Google Scholar]
- 5.Engelbrecht MR, Jager GJ, Laheij RJ, Verbeek AL, van Lier HJ, Barentsz JO. Local staging of prostate cancer using magnetic resonance imaging: a meta-analysis. Eur Radiol. 2002;12:2294–2302. doi: 10.1007/s00330-002-1389-z. [DOI] [PubMed] [Google Scholar]
- 6.Schiebler ML, Schnall MD, Pollack HM, et al. Current role of MR imaging in the staging of adenocarcinoma of the prostate. Radiology. 1993;189:339–352. doi: 10.1148/radiology.189.2.8210358. [DOI] [PubMed] [Google Scholar]
- 7.Yu KK, Hricak H. Imaging prostate cancer. Radiol Clin North Am. 2000;38:59–85. viii. doi: 10.1016/s0033-8389(05)70150-0. [DOI] [PubMed] [Google Scholar]
- 8.Maio A, Rifkin MD. Magnetic resonance imaging of prostate cancer: update. Top Magn Reson Imaging. 1995;7:54–68. doi: 10.1097/00002142-199500710-00007. [DOI] [PubMed] [Google Scholar]
- 9.McNeal JE. Regional morphology and pathology of the prostate. Am J Clin Pathol. 1968;49:347–357. doi: 10.1093/ajcp/49.3.347. [DOI] [PubMed] [Google Scholar]
- 10.Schiebler ML, Tomaszewski JE, Bezzi M, et al. Prostatic carcinoma and benign prostatic hyperplasia: correlation of high-resolution MR and histopathologic findings. Radiology. 1989;172:131–137. doi: 10.1148/radiology.172.1.2472644. [DOI] [PubMed] [Google Scholar]
- 11.Futterer JJ, Barentsz JO. 3T MRI of prostate cancer. Applied Radiology. 2009;39:25–32. [Google Scholar]
- 12.Akin O, Sala E, Moskowitz CS, et al. Transition zone prostate cancers: features, detection, localization, and staging at endorectal MR imaging. Radiology. 2006;239:784–792. doi: 10.1148/radiol.2392050949. [DOI] [PubMed] [Google Scholar]
- 13.Nicolas V, Beyersdorff D, Mueller-Lise U, Pennekamp W, Heyer C. The Prostate and Seminal Vesicles. MR Imaging of the Abdomen and Pelvis. 2002:220–240. [Google Scholar]
- 14.Shannon BA, McNeal JE, Cohen RJ. Transition zone carcinoma of the prostate gland: a common indolent tumour type that occasionally manifests aggressive behaviour. Pathology. 2003;35:467–471. doi: 10.1080/00313020310001619154. [DOI] [PubMed] [Google Scholar]
- 15.Oto A, Kayhan A, Jiang Y, et al. Prostate cancer: differentiation of central gland cancer from benign prostatic hyperplasia by using diffusion-weighted and dynamic contrast-enhanced MR imaging. Radiology. 257:715–723. doi: 10.1148/radiol.10100021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JH, Kim JK, Park BW, Kim N, Cho KS. Apparent diffusion coefficient: prostate cancer versus noncancerous tissue according to anatomical region. J Magn Reson Imaging. 2008;28:1173–1179. doi: 10.1002/jmri.21513. [DOI] [PubMed] [Google Scholar]
- 17.Swindle P, Ramadan S, Stanwell P, McCredie S, Russell P, Mountford C. Proton magnetic resonance spectroscopy of the central, transition and peripheral zones of the prostate: assignments and correlation with histopathology. MAGMA. 2008;21:423–434. doi: 10.1007/s10334-008-0136-0. [DOI] [PubMed] [Google Scholar]
- 18.Bloch BN, Lenkinski RE, Rofsky NM. The role of magnetic resonance imaging (MRI) in prostate cancer imaging and staging at 1.5 and 3 Tesla: The Beth Israel Deaconess Medical Center (BIDMC) approach. Cancer Biomarkers. 2008;4:251–262. doi: 10.3233/cbm-2008-44-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim CK, Park BK. Update of prostate magnetic resonance imaging at 3 T. J Comput Assist Tomogr. 2008;32:163–172. doi: 10.1097/RCT.0b013e3180683b99. [DOI] [PubMed] [Google Scholar]
- 20.Chan I, Wells W, III, Mulkern RV, et al. Detection of prostate cancer by integration of line-scan diffusion, T2-mapping and T2-weighted magnetic resonance imaging; a multichannel statistical classifier. Medical Physics. 2003;30:2390–2398. doi: 10.1118/1.1593633. [DOI] [PubMed] [Google Scholar]
- 21.Ozer S, Langer DL, Liu X, et al. Supervised and unsupervised methods for prostate cancer segmentation with multispectral MRI. Med Phys. 2010;37:1873–1883. doi: 10.1118/1.3359459. [DOI] [PubMed] [Google Scholar]
- 22.Lopes R, Ayache A, Makni N, et al. Prostate cancer characterization on MR images using fractal features. Med Phys. 2010;38:83–95. doi: 10.1118/1.3521470. [DOI] [PubMed] [Google Scholar]
- 23.Vos PC, Hambrock Thomas, Barenstz Jelle O, Huisman HJ. Computer-assisted analysis of peripheral zone prostate lesions using T2-weighted and dynamic contrast enhanced T1-weighted MRI. Physics in Medicine and Biology. 2010;55:1719. doi: 10.1088/0031-9155/55/6/012. [DOI] [PubMed] [Google Scholar]
- 24.Russ JC. The Image Processing Handbook. CRC Press; 2007. [Google Scholar]
- 25.Haralick RM, Shanmugam K, Dinstein IH. Textural Features for Image Classification. IEEE Transactions on Systems, Man and Cybernetics. 1973;3:610–621. [Google Scholar]
- 26.Bovik AC, Clark M, Geisler WS. Multichannel texture analysis using localized spatial filters. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1990;12:55–73. [Google Scholar]
- 27.Busch C. Wavelet based texture segmentation of multi-modal tomographic images. Computers & Graphics. 1997;21:347–358. [Google Scholar]
- 28.Seltzer SE, Getty DJ, Tempany CM, et al. Staging prostate cancer with MR imaging: a combined radiologist-computer system. Radiology. 1997;202:219–226. doi: 10.1148/radiology.202.1.8988214. [DOI] [PubMed] [Google Scholar]
- 29.Madabhushi A, Feldman MD, Metaxas DN, Tomaszeweski J, Chute D. Automated detection of prostatic adenocarcinoma from high-resolution ex vivo MRI. IEEE Trans Med Imaging. 2005;24:1611–1625. doi: 10.1109/TMI.2005.859208. [DOI] [PubMed] [Google Scholar]
- 30.Lv D, Guo X, Wang X, Zhang J, Fang J. Computerized characterization of prostate cancer by fractal analysis in MR images. Journal of Magnetic Resonance Imaging. 2009;30:161–168. doi: 10.1002/jmri.21819. [DOI] [PubMed] [Google Scholar]
- 31.Bulman J, Toth R, Patel A, et al. Automated Computer-derived Prostate Volumes from MR Imaging Data: Comparison with Radiologist-derived MR Imaging and Pathologic Specimen Volumes. Radiology. 2012;262(1):144–151. doi: 10.1148/radiol.11110266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Transactions on Medical Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 33.Madabhushi A, Udupa JK. New methods of MR image intensity standardization via generalized scale. Med Phys. 2006;33:3426–3434. doi: 10.1118/1.2335487. [DOI] [PubMed] [Google Scholar]
- 34.Chappelow J, Bloch BN, Rofsky N, et al. Elastic registration of multimodal prostate MRI and histology via multiattribute combined mutual information. Med Phys. 2011;38:2005–2018. doi: 10.1118/1.3560879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng H, Long F, Ding C. Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. Pattern Analysis and Machine Intelligence, IEEE Transactions on. 2005;27:1226–1238. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- 36.McLachlan GJ. Discriminant analysis and statistical pattern recognition. Hoboken, N.J: 2004. p. 526. xv. [Google Scholar]
- 37.Pluim JP, Maintz JB, Viergever MA. Image registration by maximization of combined mutual information and gradient information. IEEE Trans Med Imaging. 2000;19:809–814. doi: 10.1109/42.876307. [DOI] [PubMed] [Google Scholar]
- 38.Reed TR, Dubuf JMH. A Review of Recent Texture Segmentation and Feature Extraction Techniques. CVGIP: Image Understanding. 1993;57:359–372. [Google Scholar]
- 39.Haralick RM. Statistical and structural approaches to texture. Proceedings of the IEEE. 1979;67:786–804. [Google Scholar]
- 40.Herlidou S, Rolland Y, Bansard JY, Le Rumeur E, de Certaines JD. Comparison of automated and visual texture analysis in MRI: Characterization of normal and diseased skeletal muscle. Magn Reson Imaging. 1999;17:1393–1397. doi: 10.1016/s0730-725x(99)00066-1. [DOI] [PubMed] [Google Scholar]
- 41.Castellano G, Bonilha L, Li L, Cendes F. Texture analysis of medical images. Clinical Radiology. 2004;59:1061–1069. doi: 10.1016/j.crad.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 42.Kovalev VA, Kruggel F, Gertz HJ, von Cramon DY. Three-dimensional texture analysis of MRI brain datasets. IEEE Transactions on Medical Imaging. 2001;20:424–433. doi: 10.1109/42.925295. [DOI] [PubMed] [Google Scholar]
- 43.Randen T, Husoy JH. Filtering for texture classification: a comparative study. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1999;21:291–310. [Google Scholar]