Abstract
The pro-apoptotic BH3-only protein, BIK, is widely expressed and although many critical functions in developmental or stress-induced death have been ascribed to this protein, mice lacking Bik display no overt abnormalities. It has been postulated that Bik can serve as a tumour suppressor, on the basis that its deficiency and loss of apoptotic function have been reported in many human cancers, including lymphoid malignancies. Evasion of apoptosis is a major factor contributing to c-Myc-induced tumour development, but despite this, we found that Bik deficiency did not accelerate Eμ-Myc-induced lymphomagenesis. Co-operation between BIK and NOXA, another BH3-only protein, has been previously described, and was attributed to their complementary binding specificities to distinct subsets of pro-survival BCL-2 family proteins. Nevertheless, combined deficiency of Bik and Noxa did not alter the onset of Eμ-Myc transgene induced lymphoma development. Moreover, although p53-mediated induction of Bik has been reported, neither Eμ-Myc/Bik−/− nor Eμ-Myc/Bik−/−Noxa−/− lymphomas were more resistant than control Eμ-Myc lymphomas to killing by DNA damaging drugs, either in vitro or in vivo. These results suggest that Bik, even in combination with Noxa, is not a potent suppressor of c-Myc-driven tumourigenesis or critical for chemotherapeutic drug-induced killing of Myc-driven tumours.
Keywords: Bik, Noxa, Myc, Bcl-2 family, BH3-only, apoptosis
Impaired apoptosis can allow abnormal survival of cells undergoing neoplastic transformation, thereby facilitating the accumulation of further oncogenic mutations.1 In addition to promoting the development of neoplastic disease, defects in the apoptotic machinery can also impair the response of tumour cells to various anti-cancer agents.1
The BCL-2 regulated apoptotic pathway, triggered by various damage signals, is controlled by interactions between the pro- and anti-apoptotic members of the Bcl-2 protein family. The pro-survival members, BCL-2, BCL-XL, BCL-W, MCL-1 and A1, are essential for cell survival. The multi-BH domain pro-apoptotic subfamily members BAX and BAK are the critical effectors of apoptosis, responsible for mitochondrial outer membrane permeabilisation and consequent cellular demolition. The pro-apoptotic BH3-only subfamily members, including BIK/NBK/BLK, BAD, BID, HRK/DP5, BIM, NOXA, BMF and PUMA, are activated by diverse developmental signals and stress stimuli and act as cell-type- and stimulus-specific cell death initiators. Different BH3-only proteins have distinct specificities for binding pro-survival BCL-2 proteins. BIM, BID and PUMA are ‘promiscuous', binding avidly to BCL-2 and all its pro-survival homologues, whereas other BH3-only proteins, such as NOXA and BIK, bind only to subsets of pro-survival proteins.2 According to the ‘direct activation' model, certain BH3-only proteins (called ‘activators'), such as BIM, BID and PUMA, can directly bind and activate BAX/BAK.3 Conversely, the ‘indirect model' posits that cell death is initiated when all pro-survival BCL-2 proteins sequestering BAX/BAK are neutralised by BH3-only proteins, thereby unleashing the pro-apoptotic actions of BAX/BAK.4
BIK/BLK/NBK was identified in a screen for interactors with BCL-2, BCL-XL or the viral analogue E1B19 kD.5, 6 Although Bik is widely expressed, mice lacking Bik display no overt abnormalities.7 Non-transformed hematopoietic cells and fibroblasts from BIK-deficient mice respond normally to apoptotic stimuli, such as treatment with DNA-damaging chemotherapeutic drugs (e.g., etoposide) indicating that BIK may have overlapping functions with other BH3-only proteins.
BIK is proposed to function as a tumour suppressor (see review Chinnadurai et al.8). Deletions in regions of chromosome 22q, encompassing the BIK gene, have been reported in human colorectal as well as head and neck cancers and gliomas. Loss of heterozygosity at the BIK locus has been observed in renal cell carcinomas. Mutations within the BIK gene were found in human B-cell lymphomas and microarray analyses have revealed epigenetic silencing of BIK in multiple myeloma-derived cell lines.
To assess the tumour suppressor potential of Bik, we have examined the impact of Bik deficiency on lymphomagenesis in Eμ-Myc transgenic mice, which develop pre-B or B-cell lymphoma resulting from c-Myc overexpression followed by acquisition of additional oncogenic mutations.9, 10 We found that loss of Bik, even in combination with loss of Noxa (which has a binding specificity complementary to Bik), did not accelerate Eμ-Myc-induced lymphomagenesis.
BIK-induced apoptosis has mainly been demonstrated in epithelial cancer-derived cell lines using in vitro overexpression systems.5, 6, 11, 12, 13, 14, 15, 16 The role of endogenous Bik in chemotherapeutic drug-induced killing of cancer cells has not been examined in detail. Curiously, reports have described both p53-dependent17, 18 and p53–independent19, 20 transcriptional induction of Bik in response to anti-cancer therapeutics in human tumour-derived cell lines but the relevance of these findings is unclear. We have previously shown that Bik loss does not alter sensitivity of Eμ-Myc lymphoma cells with functional p53 to apoptosis induced by etoposide in vitro.21 We show here that neither loss of Bik alone, nor the combined deficiency of Bik and Noxa, affect sensitivity of Eμ-Myc lymphomas to the DNA-damaging chemotherapeutic drug, cyclophosphamide (CTX) in vivo and demonstrate that loss of Bik and Noxa can be adequately compensated for by other BH3-only proteins in the response of lymphoma cells to DNA-damaging chemotherapy.
Results
Bik is expressed in Eμ-Myc lymphoma cells
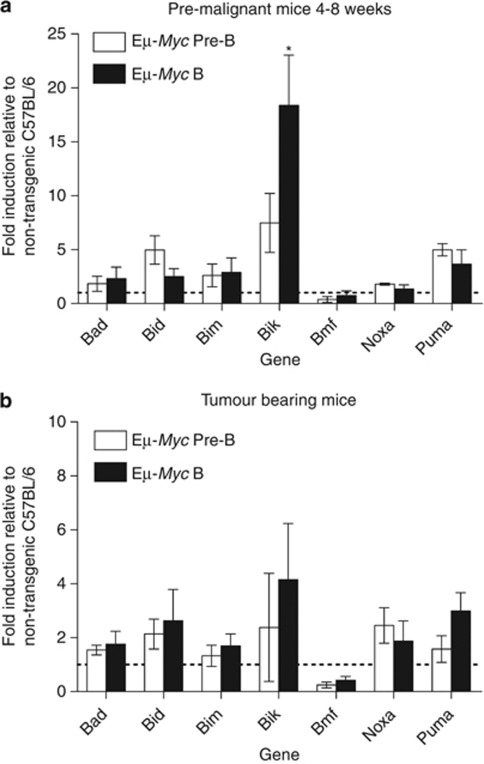
c-Myc overexpression upregulates the expression of the BH3-only genes Noxa, Puma and Bim22 and loss of these proteins was shown to accelerate lymphomagenesis in Eμ-Myc transgenic mice.22, 23, 24, 25 Similarly, pre-leukaemic B220+ sIgM− pre-B and B220+ sIg+ B-lymphoid cells from pre-malignant Eμ-Myc mice (4–6 weeks of age) displayed higher levels of Bik mRNA induction (∼5-fold and ∼17-fold increase, respectively) compared with their counterparts from control (nontransgenic, wt) littermates (Figure 1a). Bik is expressed in a diverse range of haematopoietic cell types,7 however, little has been reported about its levels in neoplastic haematopoietic cells. Higher levels of Bik mRNA were observed in Eμ-Myc pre-B lymphoma (∼2-fold increase) and Eμ-Myc B-lymphoma cells (∼7-fold increase) isolated from sick (tumour-bearing) Eμ-Myc transgenic mice compared with normal pre-B and B-lymphoid cells from healthy C57BL/6 mice (Figure 1b).
Figure 1.
Bik mRNA levels are modestly increased in B-lymphoma cells and pre-malignant lymphoid cells from Eμ-Myc mice compared with control B-lymphoid cells. (a) Bik mRNA expression is increased in pre-neoplastic B-lymphoid cells and (b) in lymphoma cells from sick Eμ-Myc mice. Differences in the levels of mRNA for pro-apoptotic BH3-only genes between FACS-purified pre-B (B220+ sIgM−) and B (B220+ sIgM+) cell subsets obtained from healthy 4–6-week-old Eμ-Myc mice (a) or from sick lymphoma-bearing Eμ-Myc mice (b), compared with the corresponding non-transformed pre-B and B-lymphoid cells from age-matched control C57BL/6 mice. Relative RNA expression levels were calculated by normalising to the β-actin signal in each sample, then dividing the values from the Eμ-Myc transgenic B-lymphoid or -lymphoma cells by the value from the control (nontransgenic) cells (represented by the dotted horizontal line at 1). Data represent mean expression±S.E.M. of cells (lymphoma or control non-transformed) from three individual mice of each genotype analysed in three separate experiments. In pre-malignant Eμ-Myc B220+ sIgM+ B cells, the fold induction of Bik mRNA compared with that observed in control nontransgenic B220+ sIgM+ B cells, was significantly higher (*P<0.05) than the fold induction of Bad, Bid, Bim and Noxa
Neither loss of Bik nor combined loss of Bik and Noxa accelerate Eμ-Myc lymphoma development
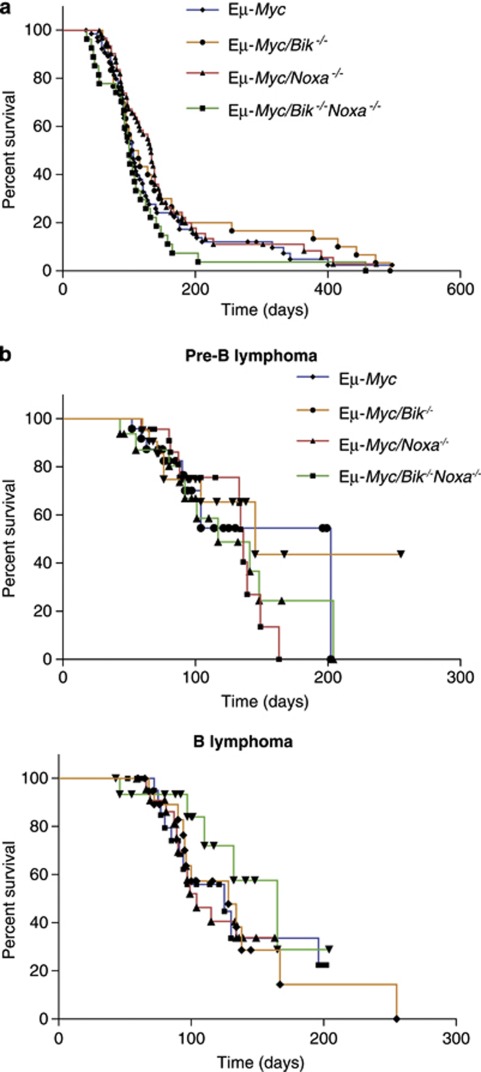
Bik is not essential for normal mouse development and unstressed Bik−/− mice display no haematopoietic abnormalities despite its widespread expression in the haematopoietic compartment.7 We examined the effect of BIK deficiency on Eμ-Myc-induced lymphomagenesis. Eμ-Myc mice (n=68) displayed a median survival of 106 days and Eμ-Myc/Bik−/− (n=30) mice succumbed at a similar rate with a median survival of 109 (Eμ-Myc/Bik−/− vs Eμ-Myc P=0.433) (Figure 2a). Severity of disease manifestation in Eμ-Myc/Bik−/− mice was comparable to that reported in control Eμ-Myc mice9, 10 with conspicuous enlargement of lymph nodes, spleen and thymus.
Figure 2.
Neither loss of Bik, nor combined loss of Bik and Noxa accelerate lymphoma development in Eμ-Myc transgenic mice. (a) Kaplan–Meier survival analysis of mice of the indicated genotypes. Differences in tumour onset between Eμ-Myc (n=68), Eμ-Myc/Noxa−/− (n=64), Eμ-Myc/Bik−/− (n=30) and Eμ-Myc/Bik−/−Noxa−/− (n=27) mice were not significant (P=0.170, 0.433, 0.321, respectively compared with control Eμ-Myc). (b) Loss of BIK alone, or in combination with NOXA loss, in Eμ-Myc mice did not significantly alter the latency or incidence of either pre-B lymphomas (top panel; Eμ-Myc n=10, Eμ-Myc/Bik−/− n=9, Eμ-Myc/Bik−/−Noxa−/− n=10) or B lymphomas (bottom panel; Eμ-Myc n=14, Eμ-Myc/Bik−/− n=11, Eμ-Myc/Bik−/−Noxa−/− n=5) compared with those seen in Eμ-Myc mice
Given the complementary binding patterns of the BH3-only proteins NOXA (MCL-1, A1) and BIK (BCL-2, BCL-XL, BCL-W) to pro-survival BCL-2 family members2, 26 and the report that, when enforcibly expressed, these two BH3-only proteins can cooperate to induce BAX activation, resulting in mitochondrial release of cytochrome c,15 we generated Eμ-Myc mice doubly-deficient for Noxa and Bik. As reported previously,22 loss of Noxa did not significantly affect Eμ-Myc-induced lymphomagenesis with Eμ-Myc/Noxa−/− mice displaying a median survival of 133 days (n=64) compared with a median survival of 106 days in control Eμ-Myc mice (n=68) (Figure 2a). Even the combined loss of both Bik and Noxa did not accelerate Eμ-Myc-induced lymphomagenesis (median survival 100 days; n=27; Figure 2a).
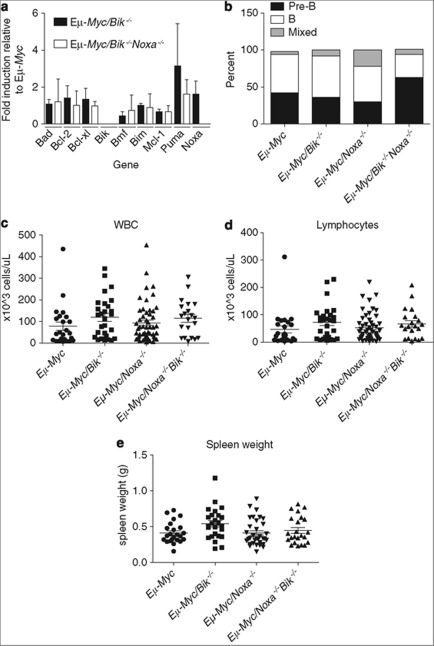
qRT-PCR analysis of Eμ-Myc/Bik−/− and Eμ-Myc/Bik−/−Noxa−/− lymphomas showed no significant differences in expression levels of the pro-apoptotic BH3-only genes Bad, Bim, Bmf, Puma, Noxa, or of the pro-survival Bcl-2 family genes, Bcl-2, Bcl-x, Mcl-1 when compared with control Eμ-Myc lymphoma cells (Figure 3a). These results demonstrate that loss of Bik, or even combined loss of Bik and Noxa, two BH3-only proteins with complementary binding specificity for Bcl-2-like pro-survival proteins, does not cause compensatory upregulation of the other remaining BH3-only genes, or changes in expression of pro-survival Bcl-2 genes at the transcript level.
Figure 3.
Loss of Bik or combined loss of Bik and Noxa does not alter lymphoma subtype or severity of disease in Eμ-Myc transgenic mice. (a) Differences in the levels of mRNA for BCL-2 family members between FACS-purified B220+ lymphoma cells harvested from either sick Eμ-Myc mice lacking Bik, lacking both Bik and Noxa, or from sick control Eμ-Myc mice. qRT-PCR analysis was performed on cDNA and relative expression of RNA was calculated by normalising to the β-actin signal in each sample, then dividing the value from the Eμ-Myc/Bik−/− or Eμ-Myc/Bik−/−Noxa−/− lymphoma cells by the value from the control Eμ-Myc lymphoma cells. Data represent mean expression±S.E.M. of lymphoma cells from three individual mice of each genotype from three separate experiments. (b) Proportions of pre-B (sIgM−), B cell (sIgM+) and mixed pre-B and B-cell lymphomas in sick Eμ-Myc, Eμ-Myc/Bik−/−, Eμ-Myc/Noxa−/− and Eμ-Myc/Bik−/−Noxa−/− mice are shown. (c) Numbers of white blood cells (WBC) and (d) numbers of lymphocytes in the peripheral blood of sick control Eμ-Myc, Eμ-Myc/Bik−/− Eμ-Myc/Noxa−/− and Eμ-Myc/Bik−/−Noxa−/− mice. (e) Spleen weights of sick, lymphoma-bearing Eμ-Myc, Eμ-Myc/Bik−/− Eμ-Myc/Noxa−/− and Eμ-Myc/Bik−/−Noxa−/− mice. For (c–e), each dot represents a single sick (lymphoma-burdened) animal and the mean is represented by a bar. Numbers of Eμ-Myc, Eμ-Myc/Bik−/− and Eμ-Myc/Bik−/−Noxa−/− mice included are 24, 28 and 23, respectively
Loss of Bik alone, or in combination with Noxa loss has no effect on disease severity or lymphoma subtype in Eμ-Myc transgenic mice
Immunophenotyping of primary lymphomas revealed that the majority were either B220+ sIgM− pre-B lymphomas or B220+ sIgM+ B-cell lymphomas, whereas a small minority displayed a mixed population of both sIgM− and sIgM+ lymphoma cells. The overall proportions of pre-B and sIg+ B-cell lymphomas were not significantly altered as a result of Bik deficiency (9/25 Pre-B, 14/25 B) or combined Noxa and Bik deficiency (10/16 pre-B and 5/16 B), compared with control Eμ-Myc (9/21 pre-B, 11/21 B), and Eμ-Myc/Noxa−/− (7/23 pre-B, 11/23 B) mice (Figure 3b).
Differences in tumour onset depending on tumour immunophenotype in Eμ-Myc mice lacking single BH3-only proteins have been documented. Loss of BIM, PUMA, BMF or BAD accelerated Eμ-Myc B-cell lymphoma development while the onset of pre-B lymphomas was not altered.22, 23, 25 These data indicate that BH3-only proteins exert differentiation stage-specific suppressive effects on lymphomagenesis and highlight the necessity of analysing the impact of BH3-only protein deficiencies on tumourigenesis, with respect to immunophenotype. Tumour onset in Eμ-Myc, Eμ-Myc/Bik−/− and Eμ-Myc/Bik−/−Noxa−/− mice was therefore stratified according to pre-B or B lymphoma subtype. No significant differences in survival were observed between Eμ-Myc/Bik−/− mice bearing pre-B (median survival 145 days, n=9) or mature B cell (median survival 128 days, n=11) lymphomas versus control Eμ-Myc mice bearing pre-B (median survival 202 days, n=10) or B cell (median survival 125 days, n=14) lymphomas (Figure 2b). Similarly, the survival of Eμ-Myc/Bik−/−Noxa−/− mice bearing pre-B or mature B-cell lymphomas (median survival 117 days and 165 days, respectively) did not differ significantly from the survival of control Eμ-Myc, Eμ-Myc/Bik−/− or Eμ-Myc/Noxa−/− (median survival pre-B 134 days, B cell 104 days) lymphoma-bearing mice (Figure 2b).
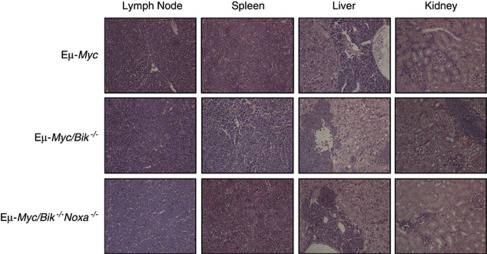
Analysis of peripheral blood of sick Eμ-Myc/Bik−/− and Eμ-Myc/Bik−/−Noxa−/− mice at death revealed no significant differences in leukaemic burden or lymphocyte counts compared with sick control Eμ-Myc or Eμ-Myc/Noxa−/− mice (Figures 3c and d). Spleen weights from Eμ-Myc/Bik−/− mice at autopsy were significantly, albeit modestly (P=0.02), greater than those observed for spleens from control Eμ-Myc mice (Figure 3e). Histological examination of organs from sick Eμ-Myc/Bik−/− and Eμ-Myc/Bik−/−Noxa−/− mice showed similar extent of tumour infiltration into lymph nodes, spleen, thymus, liver, lung and kidney to that seen in control Eμ-Myc mice (Figure 4, Supplementary Table 1).
Figure 4.
Combined loss of Bik and Noxa does not increase the severity of disease in Eμ-Myc transgenic mice. Photomicrographs (magnification × 200) of haematoxylin- and eosin-stained paraffin-embedded sections of lymph nodes, spleen, liver and kidneys from representative lymphoma-burdened sick control Eμ-Myc, Eμ-Myc/Bik−/− and Eμ-Myc/Bik−/−Noxa−/− mice (one mouse per genotype shown)
The incidence of p53 pathway mutations in Eμ-Myc lymphomas is not altered as a result of Bik deficiency or combined deficiency of Noxa and Bik
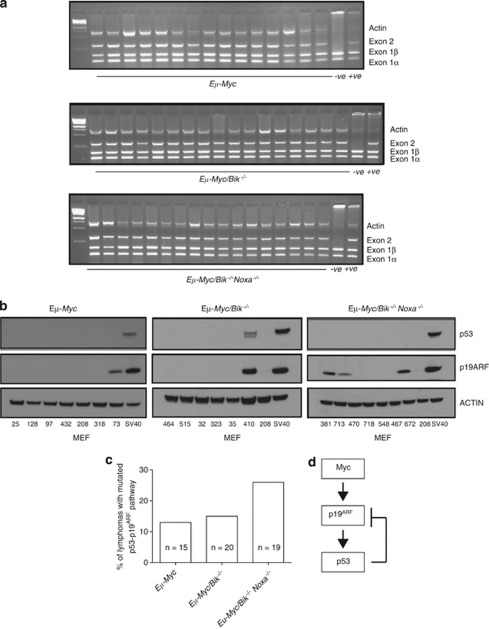
There is significant pressure for the p19ARF – p53 pathway to be compromised during Eμ-Myc-induced lymphoma development.27 Loss of certain BH3-only proteins, such as BIM,23, 28 PUMA22, 24 or BMF25 can alleviate the need for p53 pathway mutations in Eμ-Myc lymphomas. We detected no spontaneous deletions of the p16Ink4a/p19Arf locus by genomic PCR in a panel of randomly selected Eμ-Myc/Bik−/− and Eμ-Myc/Bik−/−Noxa−/− lymphomas (Figure 5a).
Figure 5.
Bik loss, with or without Noxa loss, does not alter the pressure for selection for a mutated p53 pathway during Eμ-Myc-induced lymphoma development. (a) Genomic PCR analysis of the Ink4a/Arf locus revealed that none of the Eμ-Myc/Bik−/− or Eμ-Myc/Bik−/−Noxa−/− lymphomas examined had a deletion in this gene. Controls include an Eμ-Myc lymphoma cell line known to contain a deletion in the Ink4a/Arf locus (denoted as –ve) and an Eμ-Myc lymphoma known to retain the Ink4a/Arf locus (denoted as +ve). (b) Representative western blots of p19ARF, p53 and β-ACTIN (loading control) in randomly selected control Eμ-Myc, Eμ-Myc/Bik−/− and Eμ-Myc/Bik−/−Noxa−/− lymphomas. Extracts from MEF immortalised with SV40 large T antigen were included as a positive control for p19ARF and p53 overexpression. The Eμ-Myc lymphoma #280 was included on each blot as a reference lymphoma. (c) A graphical representation of the proportions of Eμ-Myc, Eμ-Myc/Bik−/− and Eμ-Myc/Bik−/−Noxa−/− lymphomas analysed with a mutated p19ARF–p53 pathway, as determined by western blotting for p53, p19ARF protein. (d) Schematic diagram depicting activation of the p19ARF–p53 pathway by c-MYC and the negative feedback loop that exists between p53 and p19ARF
As p53 normally regulates p19ARF protein expression by a negative feedback loop in healthy cells27 (Figure 5d), the levels of p19ARF as well as p53 protein were examined by western blotting in a panel of randomly selected Eμ-Myc/Bik−/−, Eμ-Myc/Bik−/−Noxa−/− and control Eμ-Myc lymphomas. In these assays high levels of p19ARF indicate loss of p53 function because of impaired negative regulation;29 and high levels of p53 indicate mutant, stabilised p53. High p19ARF and/or p53 protein levels were detected in 3/20 (15%) Eμ-Myc/Bik−/− and in 3/10 (30%) Eμ-Myc/Noxa−/− (Figures 5b and c) lymphomas. This prevalence was comparable to that seen in control Eμ-Myc lymphomas (2/15 (13.3%) as well as to the previously published incidence of 3/10 (30%) in Eμ-Myc/Noxa−/− lymphomas.22
p53-independent Bik induction occurs following DNA-damaging drug treatment in Eμ-Myc lymphomas
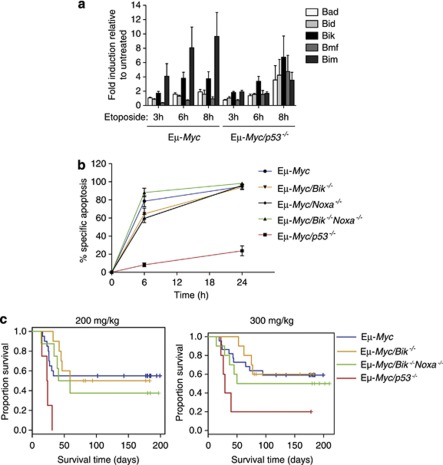
Stable cell lines were generated from lymphomas from sick control Eμ-Myc or p53-deficient Eμ-Myc mice. Upon in vitro treatment of control Eμ-Myc and Eμ-Myc/p53−/− lymphoma cell lines with 0.2 μg/ml etoposide for 3, 6 or 8 h, only negligible induction of Bad, Bid and Bmf was observed (Figure 6a). Bim mRNA, concordant with our previous findings,21 was markedly induced in Eμ-Myc lymphoma cell lines and was significantly less pronounced in Eμ-Myc lymphoma cell lines lacking p53. Similarly, Bik mRNA was upregulated, albeit at slightly lower magnitude than Bim (∼4-fold vs ∼8-fold at 6 h, respectively; Figure 6a). However, in contrast to Bim, Bik mRNA induction (∼3-fold at 6 h and ∼7-fold at 8 h) was also upregulated at similar levels in Eμ-Myc/p53−/− lymphoma cell lines (Figure 6a), demonstrating that this transcriptional induction is p53-independent.
Figure 6.
Bik levels are augmented in Eμ-Myc lymphoma cells in response to DNA damage inducing anti-cancer drugs but loss of Bik does not render these lymphoma cells resistant to drug treatment in vitro or in vivo. (a) Eμ-Myc and Eμ-Myc/p53−/− lymphoma cell lines were treated with 0.2 μg/ml of etoposide for 3, 6 or 8 h in the presence of QVD-OPH (25 μM). Lymphoma cells were harvested at the indicated time points post treatment and total RNA extracted. Differences in the levels of mRNA for the BH3-only genes Bad, Bid, Bik, Bmf and Bim were analysed by qRT-PCR on cDNA. Relative RNA expression levels were calculated by normalising to the β-actin signal in each sample, then dividing the values for etoposide treated lymphoma cells by the values for untreated lymphoma cells. Data represent mean expression±S.E.M. of cells from three individual experiments with three independent lymphoma cell lines for each genotype. (b) Cell lines derived from independent Eμ-Myc lymphomas of the indicated genotypes were treated with etoposide (0.2 μg/ml) for the indicated times in vitro and cell death was assessed by flow cytometry. Data represent mean±S.E.M. from three to seven independent cell lines per genotype, with each cell line analysed in at least three independent experiments. No significant differences were noted, except for the previously reported significant resistance of Eμ-myc/p53−/− cell lines (used here as a control). (c) Response of primary Eμ-Myc/Bik−/− and Eμ-Myc/Bik−/−Noxa−/− lymphomas to CTX (200 or 300 mg/kg body weight) treatment in vivo. Kaplan–Meier survival curves of mice transplanted (day 0) with lymphomas of the indicated genotypes and treated, once lymphomas became palpable at around day 12. Data were pooled from 4–16 independent lymphomas per genotype with 5–10 recipient mice per treatment per independent lymphomas. No significant differences in survival were observed between mice bearing Eμ-Myc/Bik−/−, Eμ-Myc/Bik−/−Noxa−/− or control Eμ-Myc lymphomas
Loss of Bik alone, or in combination with Noxa loss does not render Eμ-Myc lymphoma cells resistant to DNA-damaging drug-induced killing
Although Bik mRNA is induced in Eμ-Myc lymphoma cells following etoposide treatment, we have shown previously that Eμ-Myc/Bik−/− lymphoma cell lines display sensitivity to etoposide, comparable to that of conventional Eμ-Myc lymphoma cell lines.21 The potential for important co-operation between BIK and other BH3-only proteins could not be excluded. We therefore investigated the possible co-operation between BIK and NOXA in DNA damage-induced killing of lymphoma cells. Analysis of stable cell lines from four independent lymphomas arising in Eμ-Myc/Bik−/−Noxa−/− mice revealed that concomitant loss of BIK and NOXA did not afford significant protection from etoposide-induced killing (Figure 6b).
The role of Bik in DNA-damaging drug-induced apoptosis was further assessed in vivo by transplanting primary lymphomas into immuno-competent C57BL/6-recipient mice. Once tumours became palpable, the mice were treated with CTX at either 200 or 300 mg/kg body weight (the maximum tolerated dose21). Kaplan–Meier survival curves for mice bearing control Eμ-Myc (n=17), Eμ-Myc/Bik−/− (n=7), Eμ-Myc/Bik−/−Noxa−/− (n=6) and Eμ-Myc (p53 mutant) (n=4) lymphomas treated with CTX are shown in Figure 6c. For mice bearing control Eμ-Myc lymphomas that were treated with either 200 or 300 mg/kg body weight of CTX, a median survival of 102 or 176 days post treatment was observed, respectively, compared with only 5 days for control Eμ-Myc lymphoma-bearing mice treated with vehicle alone (Supplementary Table 2). A similar prolongation in survival was observed following CTX treatment of mice bearing Eμ-Myc/Bik−/− (80 and 86 days for 200 or 300 mg/kg body weight of CTX, respectively) or Eμ-Myc/Bik−/−Noxa−/− (58 and 175 days for 200 or 300 mg/kg body weight of CTX, respectively) lymphomas, compared with only 2 to 8 days for mice treated with vehicle alone (Supplementary Table 2).
The in vivo survival data were analysed using Cox proportional hazards models, taking into account the correlation between recipient mice. No significant difference post-CTX treatment was observed between the survival of mice bearing control Eμ-Myc (blue curves) and Eμ-Myc/Bik−/− (orange curves) lymphomas (P=0.869 for 200 mg/kg body weight CTX, P=0.336 for 300 mg/kg body weight CTX) or between the survival of mice bearing control Eμ-Myc (blue curves) and Eμ-Myc/Bik−/−Noxa−/− (green curves) lymphomas (P=0.637 for 200 mg/kg body weight CTX; P=0.212 for 300 mg/kg body weight CTX) (Figure 6c). As expected, Eμ-Myc lymphomas with a mutated p53–p19ARF pathway (red curves) displayed profound resistance to CTX and mice bearing such tumours died rapidly following treatment. Consistent with the in vitro data, neither lymphomas lacking Bik alone, nor those lacking both Bik and Noxa, displayed an increase in resistance to CTX treatment in vivo compared with lymphomas with endogenous levels of Bik and Noxa.
Discussion
Bik-deficient mice develop normally with a normal lifespan.7 Non-transformed haematopoietic cells and fibroblasts from these mice respond normally to various apoptotic stimuli, including DNA damage.7 Although these results would suggest functional redundancy between BIK and other pro-apoptotic BH3-only proteins, BIK has been proposed to function as a tumour suppressor in several human cancers, including some lymphoid malignancies.8
Apoptosis triggered by c-Myc overexpression is regulated by the BCL-2 protein family 30, 31 and is thought to impose a major barrier to MYC-induced lymphoma development. Accordingly, overexpression of pro-survival BCL-232 or loss of BH3-only proteins that have potent killing capacity, such as BIM23, 28 and PUMA22, 24 can significantly accelerate MYC-induced lymphomagenesis in mice. A recent study showed that loss of the more ‘selective' binders, and therefore less potent BH3-only proteins, BMF and BAD, can also accelerate Eμ-Myc-driven lymphomagenesis.25 These data are somewhat surprising given that theoretically, the promiscuous BIM or PUMA could adequately compensate for loss of either BMF or BAD. These findings indicate that the role of selective BH3-only proteins in tumour suppression may not simply be redundant to the functions of ‘potent' BH3-only proteins and the control of survival of cells undergoing neoplastic transformation is not solely dictated by the sequestration of pro-survival BCL-2 family proteins.33 We therefore reasoned that BIK might also constrain tumour development in the context of c-MYC overexpression.
Bik mRNA expression levels were modestly elevated in pre-leukaemic, B-lymphoid cells from pre-malignant Eμ-Myc mice compared with those from control (WT) mice. Moreover, lymphoma cells from sick Eμ-Myc mice also expressed readily detectable levels of Bik mRNA. These observations support the notion that BIK may have a role in suppression of Eμ-Myc-induced lymphomagenesis. Induction of apoptosis by c-MYC overexpression is attributable in part to activation of the p19ARF– p53 pathway, in which p19ARF upregulation prevents the degradation of p53 by sequestration of the ubiquitin ligase MDM2.27, 34 The link between Bik levels and p53, however, is controversial. Although putative p53 binding sites have been identified in the promoter of the Bik gene35 and ectopic overexpression of p53 has been shown to robustly induce Bik expression,17 a p53 response could not be detected in promoter/reporter assays.17, 18 c-MYC overexpression can, however, also induce apoptosis independent of p5318, 27 and thus, the elevation of Bik we observed may be critical for apoptosis induction independent of p53.
Despite evidence of loss of BIK expression in human tumours,8 in the Eμ-Myc transgenic mouse model, loss of BIK was insufficient to accelerate lymphoma onset or increase disease severity. Time to tumour morbidity (when animals have to be killed) has been demonstrated to differ according to immunophenotype (sIgM− pre-B or sIgM+ B cell) of lymphomas in Eμ-Myc mice lacking certain BH3-only genes. For example, loss of BIM, PUMA, BMF or BAD accelerated development of Eμ-Myc B-cell lymphoma while the onset of pre-B lymphoma was unaltered.22, 23, 25 Moreover, loss of NOXA delayed the onset of Eμ-Myc pre-B lymphomas but not Eμ-Myc B-cell lymphomas.22 In comparison, BIK deficiency had no impact on pre-B or B-cell Eμ-Myc lymphoma development, implying that BIK may have overlapping functions with other BH3-only proteins. BIK may still be a barrier to tumour development in other cell types, both haematological and nonhaematological, or in tumourigenesis driven by distinct oncogenic lesions (unrelated to c-MYC overexpression). Alternatively, the lack of effect on tumourigenesis may reflect BIK's ability to sequester only select pro-survival BCL-2 family proteins.2, 26 Therefore, the disturbance of the overall balance between pro-apoptotic and anti-apoptotic BCL-2 family members caused by BIK loss would be relatively minor, as loss of BIK can theoretically be compensated for by several other (selective binding) pro-apoptotic BH3-only proteins (such as BMF and BAD) and loss of BIK would not be expected to impact on inhibition of either MCL-1 or A1.
The lack of protection of Eμ-Myc lymphomas from DNA-damaging drug-induced killing conferred by loss of BIK stands in contrast to studies in which ectopic overexpression of BIK was used to potently induce apoptosis in tumour-derived cell lines.5, 6, 11, 12, 14 Overexpression may allow BIK to artificially interact with and inhibit a wider array of pro-survival BCL-2 proteins2, 5, 6 than does endogenous BIK, expressed under physiological conditions. Nonetheless, it is curious that we and others19, 20 have observed a modest induction of Bik following DNA damage, independent of p53. As its loss had little impact on the apoptotic response of lymphoma cells, it is unclear whether Bik induction contributes to apoptosis induction in this setting. The p53-independent upregulation of Bik may function to amplify the response to DNA-damaging drugs observed in Eμ-Myc lymphomas that is elicited by the major p53-dependent apoptosis inducers, PUMA and NOXA.
Enforced co-expression of BIK and NOXA can cooperate potently in killing mouse embryonic fibroblasts (MEFs)2 and in H1299 cells, to synergise in inducing BAX activation and cytochrome c release from mitochondria.15 This cooperative action is presumably due to the complementary nature of their binding abilities to pro-survival BCL-2 family members.2 BIK and NOXA together can theoretically complement one another to allow sufficient neutralisation of all pro-survival BCL-2-like proteins in a cell to promote more potent apoptosis induction than would be possible with sole expression of either protein. We have found, however, that a combined deficiency in BIK and NOXA is not sufficient for acceleration of Eμ-Myc-induced lymphoma development or to increase resistance of Eμ-Myc lymphoma cells to DNA-damaging drug-induced apoptosis indicating that other BH3-only proteins, such as potent family members, PUMA and BIM, can still adequately compensate for their loss. Loss of both BIK and NOXA did not significantly alter the transcription of other pro-apoptotic BH3-only genes, such as Bad, Bim, Puma and Bmf or pro-survival genes, such as Bcl-2, Bcl-xl or Mcl-1, although alterations in the post-transcriptional regulation of these BCL-2 family members cannot be excluded.
Functionally relevant inhibition of the apoptotic signalling pathway downstream of p53, such as that afforded by loss of PUMA or by the combined loss of PUMA and NOXA, have been shown to diminish the pressure for p19ARF– p53 pathway mutations in c-MYC overexpression-driven lymphomas,22, 24 as has loss of BIM23 or BMF (but not loss of BAD).25 Deficiency in BIK alone was not able to diminish the selection for loss of p53 function, indicating that loss of BIK cannot compensate for loss of p53. This indicates that the levels of the more potent (promiscuous binding) BH3-only proteins PUMA and BIM are sufficient in Eμ-Myc transgenic pre-B/B-lymphoid cells to delay neoplastic progression.
Collectively, studies of BH3-only gene deletion in the Eμ-Myc lymphoma model highlight the intricacies of apoptotic regulation in response to oncogene activation and DNA damage. The life or death fate of cells undergoing neoplastic progression or of transformed cancer cells following treatment with anti-cancer drugs must not simply be governed by the overall balance of pro- and anti-apoptotic BCL-2 family members, but instead requires the activation of distinct BH3-only proteins for apoptosis induction in a cell-type- and stimulus-specific manner. Thus Bik, even in combination with Noxa, is not a potent suppressor of c-Myc-driven tumourigenesis or critical for chemotherapeutic drug-induced killing of Myc-driven tumours.
Materials and Methods
Materials
Etoposide and CTX were purchased from Sigma (Castle Hill, NSW, Australia), QVD-OPH (MP Biomedicals, Aurora, OH, USA) was used in experiments at a final concentration of 25 μM.
Experimental animals
All experiments with mice were performed according to the guidelines of The Walter and Eliza Hall Institute of Medical Research Animal Ethics Committee. The Eμ-Myc transgenic mice expressing the c-Myc oncogene under the control of the immunoglobulin heavy chain gene enhancer (Eμ)9, 10 have been backcrossed with C57BL/6 mice for >20 generations. The origins of the Noxa−/−,36 Bik−/−7 and p53−/− 37 mice (all on a C57BL/6 background) have been described. The former two strains were generated on an inbred C57BL/6 background using C57BL/6-derived ES cells; the p53−/− mice were generated on a mixed C57BL/6 × 129SV background, using 129SV-derived ES cells, but were backcrossed with C57BL/6 mice for >10 generations. Eμ-Myc transgenic males were crossed with p53−/− or p53+/− females to generate Eμ-myc/p53+/− offspring and with Noxa−/− or Bik−/− females to produce Eμ-Myc/Noxa+/− or Eμ-Myc/Bik+/− males, which were then crossed with Noxa−/− or Bik−/− females to yield Eμ-Myc/Noxa−/− or Eμ-Myc/Bik−/− progeny, respectively, in separate breedings. Eμ-Myc/Noxa−/− males were crossed with Bik−/−Noxa−/− double knockout females to generate Eμ-Myc/Bik+/−Noxa−/− males, which were then bred to Bik−/−Noxa−/− females to yield Eμ-Myc/Bik−/−Noxa−/− mice.
Eμ-Myc lymphomas and lymphoma cell lines
Eμ-Myc lymphomas were defined by flow cytometry as either pre-B (B220+ sIg−) or B-cell lymphomas (B220+ sIgM+). Lymphoma cells were used for either transplantation into C57BL/6-recipient mice for in vivo drug sensitivity analysis or for in vitro culture to obtain stable lymphoma cell lines. Lymphoma cell lines were generated by preparing single-cell suspensions from spleen, which were cultured at 37 °C in a humidified 10% CO2 incubator in high-glucose Dulbecco's modified Eagle's medium supplemented with 10% foetal calf serum (JRH Biosciences, Brooklyn, VIC, Australia), 50 μM 2-mercaptoethanol (Sigma) and 100 μM asparagine (Sigma) referred to as FMA.
Western blotting
Protein samples were size-fractionated by SDS-PAGE and then blotted onto nitrocellulose (Invitrogen, Grand Island, NY, USA) membranes. Membranes were blocked with 5% non-fat dry milk (Devondale, Melbourne, VIC, Australia) in PBS with 0.1% Tween 20 (Sigma) and then probed with antibodies against p53 (CM5, Novacastra, North Ryde, NSW, Australia), p19ARF (AbCam, Waterloo, NSW, Australia) and β-ACTIN (clone AC-74, Sigma; used as a loading control). Detection was performed with HRP-conjugated secondary antibodies and enhanced chemiluminescence (Amersham Biosciences, Castle Hill, NSW, Australia).
Quantitative reverse transcription (qRT)-PCR
Cells were given a DNA damage insult (etoposide) in the presence of QVD-OPH to prevent terminal cellular destruction, and RNA was isolated at specific time points following treatment. Total RNA was extracted from 0.5–1 × 106 cells using the TRIzol Reagent (Invitrogen) according to the manufacturer's instructions. 0.5–1 μg of total RNA was converted to cDNA using the High Capacity RNA to cDNA kit (Applied Biosystems, Mulgrave, VIC, Australia). For quantitative analysis, the resulting cDNA was subjected to PCR in 10 μl reaction volumes using Taqman Universal PCR Master Mix (Applied Biosystems) with Taqman Gene Expression Assays (Applied Biosystems) and assessed on an ABI-PRISM 7900 thermal cycler (Applied Biosystems). β-Actin served as an endogenous normalisation control. Data analyses were performed by the comparative threshold cycle method.
Cell death assays
Eμ-Myc lymphoma cells were cultured in FMA at a concentration of 25 000–50 000 cells per well in 96-well flat bottom microtiter plates (Falcon, North Ryde, NSW, Australia). Cells were exposed to etoposide/VP16 (Pfizer, West Ryde, NSW, Australia) (1, 0.2 or 0.04 μg/ml) for 6 or 24 h. Cell death was assessed by flow cytometric analysis after staining with propidium iodide (PI; 2 μg/ml) and annexin V-FITC (0.3 μg/ml). The extent of apoptosis induced specifically by treatment with a DNA damage inducing chemotherapeutic drug (percent specific apoptosis) was calculated using the following equation: [% induced apoptosis – (% spontaneous apoptosis/100 – % spontaneous apoptosis)]. Each independent Eμ-Myc lymphoma-derived cell line was analysed in at least three independent experiments.
In vivo lymphoma cell survival analysis post treatment with DNA-damaging drugs
Six- to 8-week-old C57BL/6 female mice were injected (i.v.) on day 0 with 2 × 106 Eμ-Myc lymphoma cells. Mice were treated with CTX (200–300 mg/kg) or vehicle alone when their spleens became palpable (usually 10–16 days post tumour injection). Mice were culled when deemed unwell (lethargy, tremor, hind-leg paralysis, >5% weight loss, palpable tumour) in accordance with ethical guidelines, or in the absence of symptoms of lymphoma at 180 days post treatment (when these experiments were terminated).
Analysis of cdkn2a locus
Multiplex PCR analysis of genomic DNA extracted from lymphoma cells for the cdkn2a (p16Ink4a/p19Arf) locus was performed to reveal gross deletions, using exon-specific primers for α-actin, exons 2, 1α and 1β as previously described.38, 39
Histology
Soft tissues (spleen, lymph nodes, liver, kidney, heart, lung, thymus) and sternum were fixed in 10% buffered formalin. Tissues were embedded in paraffin, sectioned and stained using haematoxylin and eosin.
Statistical analysis
Prism (GraphPad Prism, GraphPad Software Inc., La Jolla, CA, USA), Stata 9.2 (StataCorp., College Station, TX, USA) and R (www.r-project.org) software were used for statistical analysis. Two-group comparisons of leukaemic burden, lymphocyte counts and spleen weights used two-tailed t-tests assuming equal variances.
Survival data were plotted using Kaplan–Meier curves. Differences in survival time to tumour-related deaths of Eμ-Myc mice of various genotypes were tested using log-rank tests (each independent lymphoma analysed separately). P values < 0.05 were considered to indicate statistical significance.
The survival data on lymphoma-burdened mice following in vivo treatment with CTX involved two-levels of biological replication, with many independently derived lymphomas of different genotypes and varying numbers of recipient mice per treatment arm per independent lymphoma (Supplementary Table 2), meaning that the survival times of recipient mice burdened with the same lymphoma were statistically dependent. To produce the Kaplan–Meier plots in Figure 6c, the median survival time was calculated separately for censored and non-censored mice for each independent lymphoma. The resulting Kaplan–Meier curves correspond to independent lymphoma rather than to recipient mice, and hence are statistically independent. To test whether the survival times of the different genotypes were significantly different, Cox proportional hazards regression models (Cox, 1972) were fitted. The correlations between recipient mice were taken into account using a robust sandwich estimator, implemented as method stcox cluster in Stata 9.2, to estimate S.E. from the Cox model.
Acknowledgments
We thank Drs S Cory, JM Adams, P Bouillet, L Coultas, A Villunger, T Jacks and C Vandenberg for genetically modified mice, reagents and advice; ES Jansen for technical assistance; K McKenzie, G Siciliano, K Vella for expert animal husbandry; B Helbert and C Young for genotyping of mice; J Corbin for automated blood analyses; WEHI histology services. This work was supported by the National Health and Medical Research Council of Australia (Programme Grants #461221, #494802 and #257502, Fellowships CLS (#406675), AS (#461299), GKS (#575515)); the Leukaemia and Lymphoma Society (New York; SCOR grant #7015), the National Cancer Institute (NIH, US; CA 80188 and CA 43540) and the Victorian Cancer Council Fellowship CLS (CRF10_20) and scholarships from The Leukaemia Foundation Australia (PhD scholarship to LH), and the University of Melbourne (PhD scholarship to BP). This work was made possible through Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS.
Author Contributions
CLS and AS initiated the studies. LH performed experiments. LH, BP, GKS, AS and CLS analysed the data. LH, AS and CLS wrote the paper.
Glossary
- A1
B-cell lymphoma 2 related protein A1a
- BCL-2
B-cell lymphoma 2
- BCL-W
BCL-2-like 2
- BCL-XL
BCL-2 like 1
- BH3
Bcl-2 homology 3
- BAD
BCL-2-associated agonist of cell death
- BID
BH3-interacting domain death agonist
- BIK
Bcl-2-interacting killer
- BIM
BCL-2-like 11
- BMF
BCL-2 modifying factor
- CDKN2A
cyclin-dependent kindase inhibitor 2A
- cDNA
complementary DNA
- CTX
cyclophosphamide
- DNA
deoxyribose nucleic acid
- HRK
harakiri
- MCL-1
myeloid cell leukaemia sequence 1
- MEF
mouse embryonic fibroblast
- MDM2
p53 E3 ubiquitin ligase (mouse)
- mRNA
messenger RNA
- NOXA
phorbol-12-myristate-13-acetate-induced protein 1
- PUMA
p53 upregulated modulator of apoptosis
- qRT-PCR
quantitative reverse-transcriptase polymerase chain reaction
- QVD-OPH
broad spectrum caspase inhibitor
- RNA
ribonucleic acid
- SDS-PAGE
sodium dodecyl sulphate polyacrylamide gel electrophoresis
- WT
wild-type
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cdd)
Edited by P Salomoni
Supplementary Material
References
- Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of pro-survival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Green DR. How do BCL-2 proteins induce mitochondrial outer membrane permeabilization. Trends Cell Biol. 2008;18:157–164. doi: 10.1016/j.tcb.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotar PE, et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- Hegde R, Srinivasula SM, Ahmad M, Fernandes-Alnemri T, Alnemri ES. Blk a BH3-containing mouse protein that interacts with Bcl-2 and Bcl-xL, is a potent death agonist. J Biol Chem. 1998;273:7783–7786. doi: 10.1074/jbc.273.14.7783. [DOI] [PubMed] [Google Scholar]
- Boyd JM, Gallo GJ, Elangovan B, Houghton AB, Malstrom S, Avery BJ, et al. Bik, a novel death-inducing protein shares a distinct sequence motif with Bcl-2 family proteins and interacts with viral and cellular survival-promoting proteins. Oncogene. 1995;11:1921–1928. [PubMed] [Google Scholar]
- Coultas L, Bouillet P, Stanley EG, Brodnicki TC, Adams JM, Strasser A. Proapoptotic BH3-only Bcl-2 family member Bik/Blk/Nbk is expressed in hemopoietic and endothelial cells but is redundant for their programmed death. Mol Cell Biol. 2004;24:1570–1581. doi: 10.1128/MCB.24.4.1570-1581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnadurai G, Vijayalingam S, Rashmi R. BIK the founding member of the BH3-only family proteins: mechanisms of cell death and role in cancer and pathogenic processes. Oncogene. 2008;27 (Suppl 1):S20–S29. doi: 10.1038/onc.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AW, Pinkert CA, Crawford M, Langdon WY, Brinster RL, Adams JM. The Eμ-myc transgenic mouse: a model for high-incidence spontaneous lymphoma and leukemia of early B cells. J Exp Med. 1988;167:353–371. doi: 10.1084/jem.167.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Han J, Sabbatini P, White E. Induction of apoptosis by human Nbk/Bik, a BH3-containing protein that interacts with E1B 19 K. Mol Cell Biol. 1996;16:5857–5864. doi: 10.1128/mcb.16.10.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain M, Mathai JP, Shore GC. BH-3-only BIK functions at the endoplasmic reticulum to stimulate cytochrome c release from mitochondria. J Biol Chem. 2002;277:18053–18060. doi: 10.1074/jbc.M201235200. [DOI] [PubMed] [Google Scholar]
- Mathai JP, Germain M, Shore GC. BH3-only BIK regulates BAX,BAK-dependent release of Ca2+ from endoplasmic reticulum stores and mitochondrial apoptosis during stress-induced cell death. J Biol Chem. 2005;280:23829–23836. doi: 10.1074/jbc.M500800200. [DOI] [PubMed] [Google Scholar]
- Gillissen B, Essmann F, Graupner V, Starck L, Radetzki S, Dorken B, et al. Induction of cell death by the BH3-only Bcl-2 homolog Nbk/Bik is mediated by an entirely Bax-dependent mitochondrial pathway. EMBO J. 2003;22:3580–3590. doi: 10.1093/emboj/cdg343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain M, Mathai JP, McBride HM, Shore GC. Endoplasmic reticulum BIK initiates DRP1-regulated remodelling of mitochondrial cristae during apoptosis. EMBO J. 2005;24:1546–1556. doi: 10.1038/sj.emboj.7600592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Yang Q, Vater C, Venkatesh LK, Custeau D, Chittenden T, et al. The pro-apoptotic protein, Bik, exhibits potent antitumor activity that is dependent on its BH3 domain. Mol Cancer Ther. 2001;1:95–102. [PubMed] [Google Scholar]
- Mathai JP, Germain M, Marcellus RC, Shore GC. Induction and endoplasmic reticulum location of BIK/NBK in response to apoptotic signaling by E1A and p53. Oncogene. 2002;21:2534–2544. doi: 10.1038/sj.onc.1205340. [DOI] [PubMed] [Google Scholar]
- Hur J, Bell DW, Dean KL, Coser KR, Hilario PC, Okimoto RA, et al. Regulation of expression of BIK proapoptotic protein in human breast cancer cells: p53-dependent induction of BIK mRNA by fulvestrant and proteasomal degradation of BIK protein. Cancer Res. 2006;66:10153–10161. doi: 10.1158/0008-5472.CAN-05-3696. [DOI] [PubMed] [Google Scholar]
- Paquet C, Schmitt E, Beauchemin M, Bertrand R. Activation of multidomain and BH3-only pro-apoptotic Bcl-2 family members in p53-defective cells. Apoptosis. 2004;9:815–831. doi: 10.1023/B:APPT.0000045791.55282.91. [DOI] [PubMed] [Google Scholar]
- Real PJ, Sanz C, Gutierrez O, Pipaon C, Zubiaga AM, Fernandez-Luna JL. Transcriptional activation of the proapoptotic bik gene by E2F proteins in cancer cells. FEBS Lett. 2006;580:5905–5909. doi: 10.1016/j.febslet.2006.08.088. [DOI] [PubMed] [Google Scholar]
- Happo L, Cragg MS, Phipson B, Haga JM, Jansen ES, Herold MJ, et al. Maximal killing of lymphoma cells by DNA-damage inducing therapy requires not only the p53 targets Puma and Noxa but also Bim. Blood. 2010;116:5256–5267. doi: 10.1182/blood-2010-04-280818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak EM, Jansen ES, Happo L, Cragg MS, Tai L, Smyth GK, et al. Puma and to a lesser extent Noxa are suppressors of Myc-induced lymphomagenesis. Cell Death Differ. 2009;16:684–696. doi: 10.1038/cdd.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci USA. 2004;101:6164–6169. doi: 10.1073/pnas.0401471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison SP, Jeffers JR, Yang C, Nilsson JA, Hall MA, Rehg JE, et al. Selection against PUMA gene expression in Myc-driven B-cell lymphomagenesis. Mol Cell Biol. 2008;28:5391–5402. doi: 10.1128/MCB.00907-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenzel A, Labi V, Chmelewskij W, Ploner C, Geley S, Fiegl H, et al. Suppression of B-cell lymphomagenesis by the BH3-only proteins Bmf and Bad. Blood. 2010;115:995–1005. doi: 10.1182/blood-2009-03-212670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, et al. BH3 Domains of BH3-Only Proteins Differentially Regulate Bax-Mediated Mitochondrial Membrane Permeabilization Both Directly and Indirectly. Mol Cell. 2005;17:525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Bric A, Teruya-Feldstein J, Herbst A, Nilsson JA, Cordon-Cardo C, et al. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436:807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- Bissonnette RP, Echeverri F, Mahboubi A, Green DR. Apoptotic cell death induced by c-myc is inhibited by bcl-2. Nature. 1992;359:552–554. doi: 10.1038/359552a0. [DOI] [PubMed] [Google Scholar]
- Fanidi A, Harrington EA, Evan GI. Cooperative interaction between c-myc and bcl-2 proto-oncogenes. Nature. 1992;359:554–556. doi: 10.1038/359554a0. [DOI] [PubMed] [Google Scholar]
- Strasser A, Harris AW, Bath ML, Cory S. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature. 1990;348:331–333. doi: 10.1038/348331a0. [DOI] [PubMed] [Google Scholar]
- Merino D, Strasser A, Bouillet P.Bim must be able to engage all pro-survival Bcl-2 family members for efficient tumor suppression Oncogene 2011. e-pub ahead of print 14 November 2011doi: 10.1038/onc.2011.500 [DOI] [PMC free article] [PubMed]
- Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Budarf ML, Emanuel BS, Chinnadurai G. Structural analysis of the human pro-apoptotic gene Bik: chromosomal localization, genomic organization and localization of promoter sequences. Gene. 2000;254:157–162. doi: 10.1016/s0378-1119(00)00276-6. [DOI] [PubMed] [Google Scholar]
- Villunger A, Michalak EM, Coultas L, Müllauer F, Böck G, Ausserlechner MJ, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins Puma and Noxa. Science. 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 1999;13:2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt CA, Fridman JS, Yang M, Baranov E, Hoffman RM, Lowe SW. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.