Abstract
Leishmaniasis is a vector-borne disease, and in the Indian subcontinent the female Phlebotomus argentipes is the vector for Leishmania donovani. However, data on the extent of sand fly infection rates in natural settings using molecular methods have not been extensively reported in India. In this study a PCR technique was applied targeting the 18S rRNA encoding region to determine the prevalence of Leishmania infection in female P. argentipes captured in the field. For this study, sand flies were collected from 897 houses selected from 50 villages endemic for visceral leishmaniasis (VL) in Muzaffarpur district, Bihar state, using CDC miniature light traps and mouth aspirators. A total of 14,585 sand flies were collected of which 449 were female P. argentipes divided into 132 pools. Molecular detection using PCR targeting the 18S rRNA gene was carried out for the identification of P. argentipes and Leishmania. The overall prevalence of infection was 4.90–17.37% for L. donovani in female P. argentipes in endemic regions of Bihar state. In this study no correlation was found between the presence of infected sand flies and the occurrence of clinical VL. This study provides the first report evaluating the prevalence of Leishmania infection in sand flies in a region endemic for VL in India. Sergentomyia species are the most common species of sand fly. Knowledge of the infection rate in female P. argentipes may help in predicting severity of disease and in vector elimination programs.
Key Words: Leishmania donovani, Phlebotomus argentipes, 18S rRNA, Gene, PCR, Sand fly, Visceral leishmaniasis
Introduction
Leishmaniases are among the world's most neglected diseases, affecting mostly developing countries. In all, 350 million people are considered to be at risk of leishmaniasis, and about 2 million new cases occur annually. Leishmaniasis is caused by at least 20 Leishmania species, and is transmitted by the female sand fly of the genus Phlebotomus in the Old World, and Lutzomyia in the New World. In India a total of 33,233 cases were reported in 2008, of which more than 84% were from Bihar state (WHO 2010). In the Indian subcontinent, transmission is anthroponotic, and for visceral leishmaniasis (VL) female P. argentipes is the vector, and L. donovani is the parasite (Desjeux 1996). Female sand flies need blood for their eggs to develop. They become infected with the Leishmania parasites when they suck blood from an infected person or animal (WHO 2010). Monitoring infection in the vector population is an essential tool for surveillance of vector-borne diseases. There are two classical methods for the estimation of infection rates in the reservoir hosts or vectors: microscopic analysis and culture isolation. Both methods are laborious and not feasible for use with large samples. Molecular methods such as PCR are highly sensitive and easy to perform compared to optical microscopy. Using PCR for the detection of Leishmania has proven useful (Cabrera et al. 2002; Bhattarai et al. 2009; Azpurua et al. 2010). This technique would be expensive and highly labor-intensive if individual insects are analyzed, as the number of sand flies collected could be in the thousands. In previous studies of detecting infection rates in the vector population, insects were pooled together before performing PCR (Katholi and Unnasch 2006; Bhattarai et al. 2009). The aim of this study was to assess the prevalence of different species of sand flies, and to find out the infection rate of L. donovani in the natural population using PCR targeting the 18S rRNA of female P. argentipes in VL-endemic regions of Bihar state, India.
Materials and Methods
Study area
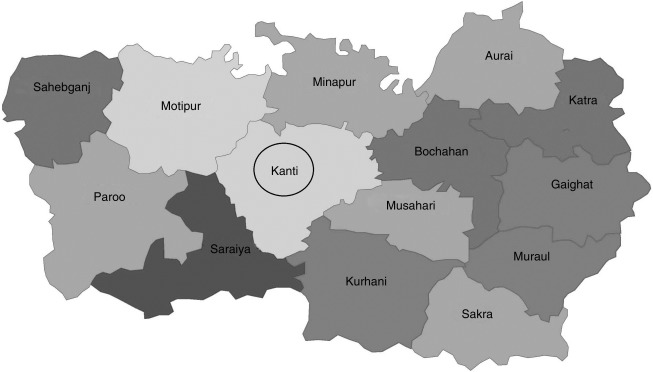
The study area consisted of 50 villages from the endemic regions of Muzaffarpur district, Bihar state, India, situated between 85.163570 and 85.276040 longitude and between 26.126200 and 26.240060 latitude (Fig. 1). For sand fly collection, a total of 897 houses were selected (141 houses with a VL case history, and 282 matched control houses, which included two houses without VL case histories adjacent to each VL-case house, and 474 random houses).
FIG. 1.
Map of the Muzaffarpur district. The black circle shows the area where sand flies were collected for the present study.
This study was approved by the ethics committee of Banaras Hindu University, Varanasi, India, and informed written consent was taken from the head of each household involved in the study.
Sand fly collection
Collection was done in April–June 2009. Insects were collected using Centers for Disease Control and Prevention light traps, and by mouth aspiration. One trap was installed inside each house in the evening and was left there for the whole night (approximately 12 h). The next day, in the early morning the sand flies were collected from the trap, and sand flies resting on the walls were collected by mouth aspiration for 15 min. The collected insects from each house were stored in a Petri dish with chloroform-soaked cotton balls. The collected insects were brought to the entomology laboratory at the Kala Azar Medical Research Centre (KAMRC), Muzaffarpur.
Morphological identification
Field-collected insects were killed for identification after storage at −20°C for 20 min. All the collected insects were differentiated morphologically as sand flies or other insects such as mosquitoes. Further, sand flies were differentiated using sand fly identification keys, such as morphology of the maxillae, and the hairs of the abdominal tergites and terminalia under a stereomicroscope, and identified as P. argentipes, P. papatasi, or Sergentomyia species (Lewis 1982). All sand flies were further differentiated as either male or female on the basis of morphology of the reproductive organs as assessed using the stereomicroscope. Only morphologically-confirmed female P. argentipes were selected for the detection of Leishmania infection, and they were pooled on the basis of the houses from which they were collected. The sand flies were fixed in 70% ethanol and stored at −20°C.
DNA extraction
DNA from each pool of female P. argentipes was extracted using the High Pure PCR Template Preparation Kit (Roche, Mannheim, Germany), following the manufacturer's instructions. DNA was eluted in molecular-biology-grade water, and quantified using a ND-2000 spectrophotometer (Thermo Scientific, Powai, Mumbai, India).
Molecular analysis
Validation of each sand fly pool was done by species-specific PCR. Molecular discrimination of P. argentipes from other sand flies was also done using a species-specific PCR test targeting the 18S rRNA coding gene with forward (5′-TCGAATCTATGGGTGGTGGT-3′) and reverse (5′-CACAATCCCAACCACGAAG-3′) primers that specifically amplified a 399-bp conserved region of phlebotomine sand flies. The reaction was carried out in a volume of 25 μL using the pair of primers (10 pM each) and the Hot Start Taq kit (Qiagen India, New Dehli, India), supplemented with 10× buffer, MgCl2, and 100 ng of sand fly DNA. An initial denaturation of 15 min at 95°C, followed by PCR amplification for 35 cycles of denaturation (95°C for 30 sec), annealing (57°C for 30 sec), and polymerization (72°C for 40 sec), with final extension of 10 min at 72°C. To discriminate P. argentipes from other species, the amplified PCR product was subjected to restriction digestion using a Hae III restriction enzyme (recognition site GGCC; New England Bio Labs, Hitchin, Herts, U.K.) in the supplied reaction buffer supplemented with 1 mg/mL−1 BSA at 37°C for 2 h. The whole reaction was electrophoresed on a 1.5% (w/v) agarose gel and stained with ethidium bromide (Surendran et al. 2005).
A highly sensitive primer specific for the 18S rRNA coding gene of Leishmania was used to identify infection in female P. argentipes (Deborggraeve et al. 2008). The nuclear sequence of the small subunit of the rRNA gene in Leishmania is found as 20–40 copies per cell (van Eys et al. 1992; Meredith et al. 1993; Costa et al. 1996; Osman et al. 1997). Pools of P. argentipes were used for the study of natural infection of Leishmania using PCR specific for the 18S rRNA gene of L. donovani. A forward primer (5′-CGTAGTTGAACTGTGGGCTGTGC-3′) and a reverse primer (5′-ACTCCCGTGTTTCTTGTTTCTTTGAA-3′) were used to amplify a 115-bp sequence within the 18S rRNA gene of L. donovani. The reaction was carried out in a volume of 25 μL using the pair of primers (20 pM each), and a Hot Start Taq kit supplemented with 10× buffer, MgCl2, and 200 ng of template. An initial denaturation was done for 15 min at 95°C, followed by PCR amplification for 45 cycles of denaturation (95°C for 30 sec), annealing (60°C for 30 sec), and polymerization (72°C for 45 sec), with a final extension of 5 min at 72°C (Looker et al. 1988; Deborggraeve et al. 2008; Bhattarai et al. 2009). The DNA of reference strain L. donovani, LEM138 (MHOM/IN/00/DEVI), was used as a positive control, and molecular-biology-grade water was used as the negative control. Amplified products were resolved on 1.5% ethidium bromide-stained agarose gels.
DNA sequencing
Identification of P. argentipes and Leishmania were confirmed by sequencing of amplified PCR products. A few specimens were randomly selected from both groups. The PCR products were extracted from agarose gels using the QIAquick Gel Extraction Kit (Qiagen India). Purified fragments were sequenced using the Big Dye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Carlsbad, CA). The cycle sequencing reaction was stopped according to the manufacturer's instructions, and then analyzed with an ABI 3130 genetic analyzer.
The P. argentipes-specific PCR products of 399 bp were aligned and compared with the available nucleotide sequence of the P. argentipes 18S rRNA gene, clone ARG1 (GenBank accession no. AJ244359.1) (Surendran et al. 2005). The sequence of Leishmania-specific PCR products were aligned and compared with the 18S rRNA sequence of L. donovani (GenBank accession no. X07773.1) (Deborggraeve et al. 2008).
Statistical analysis
For calculating the prevalence of the infection rate of Leishmania parasites in the unequal pools of sand flies, the minimum-maximum infection rate was used. For the minimum infection rate, a pool was considered positive if the pool contained a single infected insect, while for the maximum infection rate, all of the insects were considered to be infected (Katholi and Unnasch 2006).
Results
Sand fly collection
After morphological identification, sand flies were found in 880 out of a total of 897 houses (98%). P. argentipes were detected in 319 houses (Table 1). A total of 14,585 sand flies were collected and their distribution is given in Table 2. For the detection of the Leishmania parasite, only female P. argentipes were selected, and were found in 132 houses (14.7%) from 33 of 50 villages in the study area. The total number of female P. argentipes collected was 449, which were divided into 132 pools, with each pool representing all the female P. argentipes collected from a single house. The number of female P. argentipes in each pool varied from 1 to 22. The total number of pooled samples per village, the total number of sand flies in that pool, the number of pools found to be positive, the total number of sand flies in the positive pool, the percentage of positive samples, and the prevalence of infection (%) are given in Table 3.
Table 1.
The Total Numbers of Houses Positive for Different Types of Sand Flies
| Sand fly species | Male n (%) | Unfed female n (%) | Fed female n (%) | Gravid female n (%) | Total houses |
|---|---|---|---|---|---|
| Sergentomyia spp. | 716 (79.8) | 127 (14.2) | 135 (15.1) | 306 (34.1) | 880 |
| P. argentipes | 276 (30.8) | 122 (13.6) | 72 (8.0) | 27 (3.0) | 319 |
| P. papatasi | 33 (3.7) | 3 (0.3) | 11 (1.2) | 7 (0.8) | 28 |
Table 2.
The Total Number of Sand Flies Collected, Including Males and Females, of All Three Species
| Sand fly species | Male n (%) | Unfed female n (%) | Fed female n (%) | Gravid female n (%) | Total |
|---|---|---|---|---|---|
| Sergentomyia spp. | 5047 (34.6) | 5743 (39.4) | 931 (6.4) | 1060 (7.2) | 12781 (87.6) |
| P. argentipes | 1287 (8.8) | 307 (2.1) | 104 (0.7) | 38 (0.3) | 1736 (11.9) |
| P. papatasi | 43 (0.3) | 3 (0.02) | 14 (0.09) | 8 (0.05) | 68 (0.5) |
| Total | 6377 (43.7) | 6053 (41.5) | 1049 (7.2) | 1106 (7.6) | 14,585 |
Table 3.
Village-Wide Infection Rates of Leishmania donovani in Female Phlebotomus argentipes
| Village | Number of pool | Number of flies | Positive pool | Number of flies in positive pool | Percentage Infection | Minimum infection rate | Maximum infection rate |
|---|---|---|---|---|---|---|---|
| S.C. Devanand | 5 | 17 | 0 | 0 | 0 | 0 | 0 |
| Veerpur East | 6 | 9 | 0 | 0 | 0 | 0 | 0 |
| Veerpur Mid | 9 | 58 | 1 | 15 | 0.76 | 0.22 | 3.34 |
| Veerpur West | 2 | 4 | 0 | 0 | 0 | 0 | 0 |
| Premi Chapra | 4 | 44 | 0 | 0 | 0 | 0 | 0 |
| Bahuara | 3 | 3 | 1 | 1 | 0.76 | 0.22 | 0.22 |
| Sadiquepur | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Bhimalpur | 4 | 13 | 0 | 0 | 0 | 0 | 0 |
| Y. Math | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Bishunpur TP | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| B. Mahanand | 3 | 4 | 3 | 4 | 2.27 | 0.67 | 0.89 |
| B.M. Kachahari | 7 | 28 | 2 | 3 | 1.52 | 0.45 | 0.67 |
| B.A. Bhumihar | 5 | 13 | 2 | 9 | 1.52 | 0.45 | 2.00 |
| B.A. Shekhtolla (E) | 5 | 5 | 1 | 1 | 0.76 | 0.22 | 0.22 |
| B.A. Shekhtolla (W) | 4 | 15 | 0 | 0 | 0 | 0 | 0 |
| B. Thikahan | 7 | 28 | 3 | 6 | 1.51 | 0.45 | 1.34 |
| B. Noonfar | 3 | 4 | 1 | 1 | 0.76 | 0.22 | 0.22 |
| B.G. Bishunpur | 6 | 11 | 0 | 0 | 0 | 0 | 0 |
| Jhitkahi | 6 | 25 | 2 | 13 | 1.52 | 0.45 | 2.9 |
| Madhuban | 3 | 5 | 0 | 0 | 0 | 0 | 0 |
| Gopalpur | 2 | 5 | 1 | 2 | 0.76 | 0.22 | 0.45 |
| Maharatha | 11 | 69 | 2 | 14 | 1.52 | 0.45 | 3.12 |
| Bathnaha | 2 | 6 | 0 | 0 | 0 | 0 | 0 |
| Sin. Phulkahan | 7 | 20 | 1 | 6 | 0.76 | 0.22 | 1.34 |
| Maisahan | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| Salona Bheryahi | 5 | 18 | 0 | 0 | 0 | 0 | 0 |
| G. Phulkahan | 2 | 5 | 0 | 0 | 0 | 0 | 0 |
| M. Chhapra | 5 | 8 | 1 | 1 | 0.76 | 0.22 | 0.22 |
| Panapur Kariyat | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Godai jamal | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Vishwanathpur | 4 | 14 | 0 | 0 | 0 | 0 | 0 |
| Raksha South | 2 | 3 | 1 | 2 | 0.76 | 0.22 | 0.45 |
| Raksha Deah | 2 | 7 | 0 | 0 | 0 | 0 | 0 |
| Total | 132 | 449 | 22 | 78 | 16.67 | 4.90 | 17.37 |
Molecular identification of sand flies
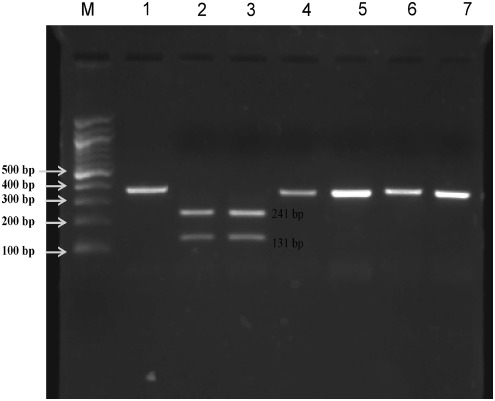
Microscopically-identified female P. argentipes were confirmed using PCR. Using species-specific PCR targeting the 18S rRNA region of phlebotomine sand flies, we amplified a 399-bp region that was further digested with restriction endonuclease to differentiate P. argentipes from other phlebotomine species. The two Hae III cutting sites at positions 27 and 158 in the conserved region of P. argentipes generated three bands of 27 bp (not visible after electrophoresis), of 131 bp and 241 bp, respectively, while the single site in P. papatasi or Sergentomyia species at position 27 generated two bands of 27 bp and 372 bp (Fig. 2).
FIG. 2.
PCR-restriction endonuclease-digested patterns of 399-bp amplified products of the 18S rRNA gene fragment digested with Hae III (lane 1, undigested PCR product; lanes 2 and 3, Hae III restriction digestion patterns of P. argentipes [241-bp and 131-bp fragments]; lanes 4 and 5, P. papatasi; lanes 6 and 7, Sergentomyia species [only one band of 372 bp]; lane M, for 100-bp marker).
Detection of Leishmania infection in sand flies
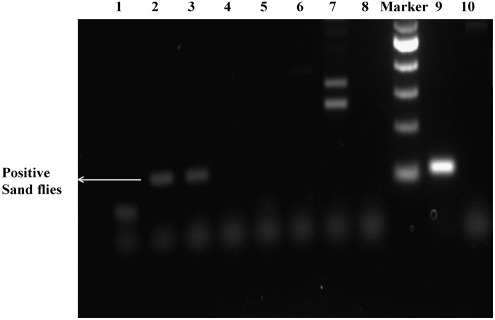
The pools containing P. argentipes collected from 132 houses were used to determine natural infection by L. donovani. This 18S rRNA gene-specific PCR study confirms that only 22 pools, containing a total of 78 sand flies, were infected with L. donovani (Fig. 3). According to the minimum and maximum infection rates, the prevalence of infection of L. donovani in female P. argentipes was 4.90–17.37%, with a minimum infection rate of 22, and a maximum infection rate of 78, out of a total of 449 sand flies considered to be infected.
FIG. 3.
Detection of the L. donovani 18S rRNA gene in female P. argentipes showing amplification of 115 bp (lane 9, positive control; lane 10, negative control; lanes 1, 4, 5, 6, 7, and 8, uninfected samples; lanes 2 and 3, infected samples; lane M, 100-bp marker).
Discussion
The success of vector control programs depends on the vectorial capacity and the understanding of disease transmission by the various vectors. We observed a prevalence rate of sand flies in 98% of houses, with Sergentomyia as the most prevalent genus, followed by P. argentipes. In India, other studies have also done of the distribution of sand flies, but in different endemic regions of Bihar state (Kumar et al. 2009). The prevalence of L. donovani infection in the female P. argentipes population in India has not been studied previously, although the presence of Leishmania DNA in Phlebotomus and Sergentomyia sand flies has previously been reported (Mukherjee et al. 1997). In this study a species-specific primer-based diagnostic PCR was used to detect parasitic nucleic acids in sand flied (Deborggraeve et al. 2008). Another advantage of PCR is that this technique is highly specific and reproducible due to the use of specific primers for conserved regions of P. argentipes and L. donovani, whereas in microscopic dissection studies, there is the possibility of an observer mistaking for other flagellated parasites for L. donovani (Srivastava et al. 2010). In recent years, different methods have been developed to screen pools or clusters of insects, which can efficiently estimate the infection potential of disease spread by a vector species (Yameogo et al. 1999; Martin-Sanchez et al. 2006; Bhattarai et al. 2009; Severinsson, et al. 2010). The rates of sand fly infection varied in accordance with the transmission of the disease, as observed in other studies reported from Panama (Azpurua et al. 2010), Athens (Aransay et al. 2000), and Turkey (Svobodova et al. 2009). The prevalence of infection with Leishmania in the vector female P. argentipes may be exploited as a tool for the surveillance of these infections, and for measuring the success of control programs.
To the best of our knowledge, this is the first report of a molecular-based study in a VL-endemic region of India. Because of the lack of firm evidence and supporting literature regarding the prevalence of infection of sand flies in VL-endemic areas of India, we expected that the prevalence of infection in these sand flies would be low. Large numbers of insects were examined to obtain an accurate estimate of infection levels. The high infection rates observed in this study indicate that a similar type of study can be performed on individual sand flies, which could provide vital information about vector control strategies to help control or eliminate VL in the Indian sub-continent.
Acknowledgments
We thank our entomology team members, KAMRC, and Muzaffarpur for help collecting and differentiating the sand fly samples. The Centre for Genetic Disorders, Banaras Hindu University, provided the ABI 3130 genetic analyzer used in this study. The study was funded by the National Institute of Allergy and Infectious Diseases (NIAID) DMID funding mechanism, Tropical Medicine Research Centre grant number P50AI074321. Puja Tiwary and Dinesh Kumar wish to thank the Council for Scientific and Industrial Research, New Delhi, India, for financial assistance and for providing research fellowships.
Author Disclosure Statement
No competing financial interests exist.
References
- Aransay AM. Scoulica E. Tselentis Y. Detection and identification of Leishmania DNA within naturally infected sand flies by seminested PCR on minicircle kinetoplastic DNA. Appl Environ Microbiol. 2000;66:1933–1938. doi: 10.1128/aem.66.5.1933-1938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpurua J. De La Cruz D. Valderama A, et al. Lutzomyia sand fly diversity and rates of infection by Wolbachia and an exotic Leishmania species on Barro Colorado Island, Panama. PLoS Negl Trop Dis. 2010;4:e627. doi: 10.1371/journal.pntd.0000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai NR. Das ML. Rijal S, et al. Natural infection of Phlebotomus argentipes with Leishmania and other trypanosomatids in a visceral leishmaniasis endemic region of Nepal. Trans R Soc Trop Med Hyg. 2009;103:1087–1092. doi: 10.1016/j.trstmh.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Cabrera OL. Munsterman LE. Cardenas R, et al. [Definition of appropriate temperature and storage conditions in the detection of Leishmania DNA with PCR in phlebotomine flies] Biomedica. 2002;22:296–302. [PubMed] [Google Scholar]
- Costa JM. Durand R. Deniau M, et al. PCR enzyme-linked immunosorbent assay for diagnosis of leishmaniasis in human immunodeficiency virus-infected patients. J Clin Microbiol. 1996;34:1831–1833. doi: 10.1128/jcm.34.7.1831-1833.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deborggraeve S. Boelaert M. Rijal S, et al. Diagnostic accuracy of a new Leishmania PCR for clinical visceral leishmaniasis in Nepal and its role in diagnosis of disease. Trop Med Int Health. 2008;13:1378–1383. doi: 10.1111/j.1365-3156.2008.02154.x. [DOI] [PubMed] [Google Scholar]
- Desjeux P. Leishmaniasis. Public health aspects and control. Clin Dermatol. 1996;14:417–423. doi: 10.1016/0738-081x(96)00057-0. [DOI] [PubMed] [Google Scholar]
- Katholi CR. Unnasch TR. Important experimental parameters for determining infection rates in arthropod vectors using pool screening approaches. Am J Trop Med Hyg. 2006;74:779–785. [PubMed] [Google Scholar]
- Kumar V. Kesari S. Kumar AJ, et al. Vector density and the control of kala-azar in Bihar, India. Mem Inst Oswaldo Cruz. 2009;104:1019–1022. doi: 10.1590/s0074-02762009000700014. [DOI] [PubMed] [Google Scholar]
- Lewis DJ. A taxonomic review of the genus Phlebotomus. Bull Brit Mus Nat Hist Entomol. 1982;45:121–209. [Google Scholar]
- Looker D. Miller LA. Elwood HJ, et al. Primary structure of the Leishmania donovani small subunit ribosomal RNA coding region. Nucleic Acids Res. 1988;16:7198. doi: 10.1093/nar/16.14.7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Sanchez J. Gallego M. Baron S, et al. Pool screen PCR for estimating the prevalence of Leishmania infantum infection in sandflies (Diptera: Nematocera, Phlebotomidae) Trans R Soc Trop Med Hyg. 2006;100:527–532. doi: 10.1016/j.trstmh.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Meredith SE. Zijlstra EE. Schoone GJ, et al. Development and application of the polymerase chain reaction for the detection and identification of Leishmania parasites in clinical material. Arch Inst Pasteur Tunis. 1993;70:419–431. [PubMed] [Google Scholar]
- Mukherjee S. Hassan MQ. Ghosh A, et al. Short report: Leishmania DNA in Phlebotomus and Sergentomyia species during a kala-azar epidemic. Am J Trop Med Hyg. 1997;57:423–425. doi: 10.4269/ajtmh.1997.57.423. [DOI] [PubMed] [Google Scholar]
- Osman OF. Oskam L. Zijlstra EE, et al. Evaluation of PCR for diagnosis of visceral leishmaniasis. J Clin Microbiol. 1997;35:2454–2457. doi: 10.1128/jcm.35.10.2454-2457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severinsson K. Jaenson TG. Pettersson J, et al. Detection and prevalence of Anaplasma phagocytophilum and Rickettsia helvetica in Ixodes ricinus ticks in seven study areas in Sweden. Parasit Vectors. 2010;3:66. doi: 10.1186/1756-3305-3-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava P. Prajapati V. Vanaerschot M, et al. Detection of Leptomonas sp. Parasites in clinical isolates of kala-azar patients from India. Infect Genet Evol. 2010;10:1145–1150. doi: 10.1016/j.meegid.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surendran SN. Karunaratne SH. Adams Z, et al. Molecular and biochemical characterization of a sand fly population from Sri Lanka: evidence for insecticide resistance due to altered esterases and insensitive acetylcholinesterase. Bull Entomol Res. 2005;95:371–380. doi: 10.1079/ber2005368. [DOI] [PubMed] [Google Scholar]
- Svobodova M. Alten B. Zidkova L, et al. Cutaneous leishmaniasis caused by Leishmania infantum transmitted by Phlebotomus tobbi. Int J Parasitol. 2009;39:251–256. doi: 10.1016/j.ijpara.2008.06.016. [DOI] [PubMed] [Google Scholar]
- van Eys GJ. Schoone GJ. Kroon NC, et al. Sequence analysis of small subunit ribosomal RNA genes and its use for detection and identification of Leishmania parasites. Mol Biochem Parasitol. 1992;51:133–142. doi: 10.1016/0166-6851(92)90208-2. [DOI] [PubMed] [Google Scholar]
- World Health Organization (WHO) Control of the leishmaniases. Report of a WHO Expert Committee. World Health Organ Tech Rep Ser. 2010;949:1–200. [Google Scholar]
- Yameogo L. Toe L. Hougard JM, et al. Pool screen polymerase chain reaction for estimating the prevalence of Onchocerca volvulus infection in Simulium damnosum sensu lato: results of a field trial in an area subject to successful vector control. Am J Trop Med Hyg. 1999;60:124–128. doi: 10.4269/ajtmh.1999.60.124. [DOI] [PubMed] [Google Scholar]