Abstract
We analyzed the function of neural progenitors in the developing CNS of Xenopus laevis tadpoles using in vivo time-lapse confocal microscopy to collect images through the tectum at intervals of 2 to 24 hours over 3 days. Neural progenitor cells were labeled with fluorescent protein reporters based on expression of endogenous Sox2 transcription factor. With this construct, we identified Sox2-expressing cells as radial glia and as a component of the progenitor pool of cells in the developing tectum that gives rise to neurons and other radial glia. Lineage analysis of individual radial glia and their progeny demonstrated that less than 10% of radial glia undergo symmetric divisions resulting in two radial glia, while the majority of radial glia divide asymmetrically to generate neurons and radial glia. Time-lapse imaging revealed the direct differentiation of radial glia into neurons. Although radial glia may guide axons as they navigate to superficial tectum, we find no evidence that radial glia function as a scaffold for neuronal migration at early stages of tectal development. Over three days, the number of labeled cells increased 20%, as the fraction of radial glia dropped and the proportion of neuronal progeny increased to approximately 60% of the labeled cells. Tadpoles provided with short-term visual enhancement generated significantly more neurons, with a corresponding decrease in cell proliferation. Together these results demonstrate that radial glial cells are neural progenitors in the developing optic tectum and reveal that visual experience increases the proportion of neurons generated in an intact animal.
Keywords: Radial glial cell, cell division, Sox2, neural progenitor, frog, brain, neuron, cell fate, cell lineage, neurogenesis, experience-dependent
INTRODUCTION
Spatial and temporal regulation of cell proliferation underlies the regionalization and specialization in the brain (Finlay et al., 1998) and is fundamental to proper CNS development (Rubenstein et al., 1994). Over the last decade, the classification of proliferating cells that generate neurons and glia in the brain has changed. Radial glia, a specialized subset of astroglial cells, were once considered to play a predominantly structural role, guiding newly generated neurons in the developing CNS (Costa et al., 2010). Radial glial cells and neurons were thought to arise form distinct lineages until retroviral labeling of progenitors and progeny demonstrated that multipotent precursors generated radial glial cells, neurons and astrocytes (Cepko et al., 1990; Gray and Sanes, 1992; Reid et al., 1995). We now know that that radial glia are themselves progenitor cells of both neurons and other radial glia cells in mammalian neocortex based on observations that isolated radial glia generate neurons in vitro (Malatesta et al., 2003), and live imaging studies that captured radial glial cells undergoing neurogenic division events in slice preparations (Miyata et al., 2001; Miyata et al., 2004; Noctor et al., 2004; Yu et al., 2009). This discovery has revitalized interest in radial glia and the events underlying their proliferation and differentiation.
Radial glia are found across species and throughout developing nervous systems. They were first identified by neuroanatomists more than a century ago in material stained with classical Golgi methods (reviewed in (Alvarez-Buylla et al., 2001; Bentivoglio and Mazzarello, 1999; Rakic, 2003; Sild and Ruthazer, 2011). Radial glia have distinct anatomical features, the most salient of which are the position of the radial glial cell body in the ventricular layer and the extension of a slender radial process from the cell body to the pial surface of the brain, ending in one or more elaborated endfeet (Morest and Silver, 2003). Studies of radial glial progenitors in the CNS of fish, amphibia and reptiles have focused on their role in adult neurogenesis and regeneration (Font et al., 2001; Lopez-Garcia et al., 1992; Margotta et al., 1992; Molowny et al., 1995; Raymond and Easter, 1983; Raymond et al., 1983; Stevenson and Yoon, 1981; Zupanc, 2008) with little attention paid to the potential function of radial glia as progenitors during the development of nervous systems of non-mammalian vertebrates. Retroviral clonal analysis suggests that radial glia may be both the source and migratory scaffold for tectal neurons at relatively late stages of developing chick tectum (Gray and Sanes, 1992), but the function of radial glia in the development of subcortical structures is not yet clear. We address this question in Xenopus tadpoles using in vivo time-lapse imaging to identify and analyze the proliferation and differentiation of radial glial neural progenitors and their progeny in the optic tectum of intact animals.
To identify and follow precursor cells in the tectum with in vivo time-lapse imaging, we employ a cell-type specific, fluorescent reporter system derived from the transcription factor binding element targeted by sex-determining region Y-box 2 (Sox2) and Octamer binding protein 3-4 (Oct3-4) of the fibroblast 4 (FGF4) enhancer. Sox2 and Oct3-4 have well known functions in the coordinated regulation of genes expressed in proliferating cells (Chakravarthy et al., 2008; Rizzino, 2009). In neural precursors, Sox2 and Oct3-4 maintain cell proliferation by inhibiting the expression of proneural genes (Bylund et al., 2003; Graham et al., 2003) and the Sox2 promoter has been used to drive fluorescent reporters (D’Amour and Gage, 2003). We constructed a fluorescent reporter system where repeats of the Sox2/Oct3-4 binding domain were used to drive different fluorescent proteins (FPs). Expression from this construct (henceforth called pSox2-bd::FP) relies on the presence of endogenous Sox2/Oct3-4 proteins and thereby increases cell-type specificity of fluorescent protein expression. Because of the tadpole’s external development, the ease of access to the developing CNS and transparency of the albino tadpole, Xenopus is amenable for direct, in vivo observation of fluorescently labeled proliferating cells and their progeny in the intact animal. Though the morphological development of tectal neurons in Xenopus has been extensively studied using time-lapse methods (Bestman and Cline, 2008; Chen et al., 2010; Chiu et al., 2008; Ewald et al., 2008; Haas et al., 2006; Li et al., 2011; Liu et al., 2009; Sanchez et al., 2006; Schwartz et al., 2009; Shen et al., 2009; Sin et al., 2002; Van Keuren-Jensen and Cline, 2008; Wu and Cline, 2003; Wu et al., 1999; Zou and Cline, 1999), comparably little is known about the cellular origin and lineages that produce these neurons. Using the pSox2-bd::FP construct to drive expression of the photoconvertible Kaede fluorescent protein, or combinations of nucleus-targeted, cytoplasmic or membrane-targeted FPs, we have quantified cell division and differentiation events, and determined the lineages of individual progenitors in the intact optic tectum of living tadpoles.
Retinal ganglion cells synapse onto neurons in the optic tectum and form the major information processing circuit of the visual system in non-mammalian vertebrates. An interesting feature of amphibia and fish is that cell proliferation in the tectum and retina continues throughout developmental stages to generate new neurons that must integrate into the functional circuitry of a visually-responsive and behaving animal (Kaslin et al., 2008; Raymond and Easter, 1983; Straznicky and Gaze, 1972; Straznicky and Gaze, 1971). We studied cell proliferation and differentiation of cells within the Xenopus tectum at developmental stages when the tadpoles are capable of visually-guided, tectum-dependent behaviors (Dong et al., 2008; Schwartz et al., 2011; Shen et al., 2011). At these stages, visual experience modulates the rate of cell proliferation in the Xenopus optic tectum, as determined by incorporation of bromodeoxyuridine derivatives (Sharma and Cline, 2010). In addition, short-term visual enhancement (STVE) increases dendritic arbor development of optic tectal neurons (Sin et al., 2002) and enhances the integration of tectal neurons into the visual circuit (Aizenman and Cline, 2007). Visual activity has also been shown to alter calcium transients and the motility of the fine superficial processes of radial glia in the Xenopus optic tectum (Tremblay et al., 2009). Here, we tested whether short-term visual enhancement affects neurogenesis by imaging radial glial progenitors and their progeny in vivo over 3 days. These results reveal the role of Sox2-expressing radial glia cells as neural progenitors in the developing CNS of Xenopus tadpoles and their response to sensory input.
MATERIALS AND METHODS
Tadpole rearing and tectal cell transfection
Albino Xenopus laevis tadpoles were obtained either by matings of frogs from our colony induced by human chorionic gonadotropin injections or purchased from Nasco (Fort Atkinson, WI). All tadpoles were reared in 12 hr light/12 hr dark cycle incubators at 22-23°C and staged according to (Nieuwkoop and Faber, 1956). Once animal were imaged the first time in a time-lapse series, they were housed singly in a well of a 6-well cell culture plate containing Steinberg’s buffered solution (58.0 mM NaCl, 0.67 mM KCl, 0.34 mM Ca(NO3)2, 0.83 mM MgSO4, 3.0 mM HEPES). The animal protocols were approved by the Institutional Animal Care and Use Committee of the Scripps Research Institute and the Marine Biological Laboratory in accordance with NIH guidelines.
Plasmid constructs
pSox2-bd::FP
The Sox2/Oct3-4 enhancer elements and a minimal promoter from the murine fibroblast growth factor 4 (FGF4) were subcloned from the plasmid described in Figure 1 of (Ambrosetti et al., 2000). The fragment contains six tandem repeats of the 35 bp fragment containing the Sox2/Oct3-4 heterodimer transcription factor binding domain (bd) followed by a 165 bp minimal FGF4 promoter (mFGF4). This plasmid was a gift of Claudio Basilico at New York University. To increase the FP signal, this Sox2/Oct 3-4-mFGF regulatory fragment was moved to the “act-GVP-UG” concatenated plasmid created by Köster and Fraser (2001), which was adapted from the Drosophila Gal4-UAS system (Brand and Perrimon, 1993) and shown to function in Xenopus (Chae et al., 2002; Hartley et al., 2002). We replaced the minimal promoters of the act-GVP-UG plasmid with our Sox2/Oct3-mFGF regulatory fragment to control the expression of the transcriptional activator, Gal4-VP16, which in turn drives the expression of genes controlled by the upstream activating sequence (UAS). The final expression vector (abbreviated as pSox2-bd::FP) contains: 6 repeats of the Sox2/Oct3-4 transcription factor binding domain, mFGF4, Gal4-VP16, a polyadenylation site, 14 repeats of UAS, mFGF4, and a fluorescent protein such as: Kaede (MBL International), turboGFP (Evrogen), turboRFP (Evrogen) alone or tagged with a nuclear localization sequence (nls). We also used the pSox2-bd::FP construct to drive the expression of membrane-targeted GFP (mGFP), a modification of eGFP (Clontech) made by adding the transmembrane domain sequence as described in (Li et al., 2010). The pSox2-bd::FP plasmids were used at 0.5 μg/μl concentration for all electroporations.
Figure 1. Transition from radial glial cells to neurons.
A-B) Xenopus laevis tadpole head (A) and enlarged view of the midbrain (B) with the right lobe of the optic tectum (ot) outlined. The dotted line represents the medial edge of the dorsal tectum. Box width in A = 0.5 mm. C-F) Projections of 2-photon Z-stacks of Timer fluorescent protein expression in the optic tectum. Images were acquired from the same tadpole over time. Immature Fluorescent Timer emits a green fluorescent signal, but as the protein matures the emission spectrum changes to red fluorescent signal (shown as magenta). Coexpression of both immature and mature Timer protein fluorophores is shown as white. Initially the majority of the transfected cells are radial glia, identified by radial processes and pial endfeet (arrows), but over development there are fewer radial glia and most Timer-expressing cells are neurons, identified by elaborate dendritic arbors (final image, F). Asterisks indicate site of intertectal axons in the anterior dorsal commissure. Scale bar = 50 μm.
pCMV::Timer
Fluorescent Timer is a variant of the red fluorescent protein, dsRed (dsRed-E5) that changes its fluorescence emission spectrum from green (500 nm) to red (580 nm; (Terskikh et al., 2000; Verkhusha et al., 2001) as the protein slowly folds and matures. We took the Timer coding domain from the promoterless pTimer plasmid (Clontech) and subcloned it behind the cytomegalovirus promoter of the pEGFP-C1 expression vector (Clontech), replacing the eGFP coding sequence of the pEGFP-C1 plasmid with the 681 bp Fluorescent Timer fragment. The final pCMV::Timer expression vector contains the CMV promoter followed by the Fluorescent Timer coding domain, within the pEGFP-C1 plasmid backbone. The pCMV::Timer plasmid was used at 4-5 μg/μl for all electroporations.
Tectal cell transfection
To transfect cells in the tectum, stage 46 animals were anesthetized in a solution of 0.01% MS-222 (3-Aminobenzoic acid ethyl ester, Sigma-Aldrich, St. Louis, MO), plasmids were injected into the midbrain ventricle, and voltage pulses were applied across the midbrain using platinum electrodes to electroporate cells lining the tectal ventricle (Haas et al., 2002). In tecta where proliferation was quantified, we used three pulses of 35V at each polarity delivered across the midbrain with a Grass SD9 stimulator. This electroporation protocol results in sparsely distributed transfection with an average of 27.3 ± 3.6 pSox2::FP-expressing cells per tectal lobe (Min/max =9-78 cells, median= 21.5), which allowed us to distinguish, count and follow individual radial glial cells and their progeny. Because of the high density of cells in the caudolateral region of the tectum, it is difficult to identify clearly individual cells in this area; therefore, transfection and imaging in this area were avoided.
Tissue processing, immunofluorescence and microscopy
Fixation
Animals were terminally anesthetized in 0.2% MS-222 (Sigma Aldrich). The skin above the brain and the dura mater was peeled back to expose the tectum. The animals were placed in freshly made 4% paraformaldehyde (PFA; Sigma Aldrich) in phosphate buffer for no longer than 10 minutes before the tissue was microwaved in the BioWave (Pelco) at 150W for 1 minute ON, 1 minute OFF, 1 minute ON at 22°C. After microwaving, the tissue was fixed further in 4% PFA for 2 hrs at room temperature or overnight at 4°C.
Antibody Characterization
A list of primary antibodies used in this study is provided in Table 1. The Sox2 antibody (Millipore, AB5603; Journal of Comparative Neurology Database/Neuroinformatics Framework # 570863) is made against a synthetic peptide corresponding to amino acids 249-265 from human Sox2. On western blots of whole cell or nuclear extracts from human NT2/D1 or mouse embryonic stem cells, it recognizes a 34kDa protein not found in cytosolic extracts (Millipore product information). It was also shown to recognize a single band corresponding to endogenous Sox2 and Flag-tagged Sox2 expressed in KH2 ES mouse embryonic stem cells (Kopp et al., 2008). The antibody has been shown to label Sox2-expressing, mitotically active cells in other species (Guo et al., 2010; Mathews et al., 2010; Wang et al., 2006). In Xenopus tadpoles, Agathocleous et al. (2009) showed that Sox2 antibody staining is concentrated in nuclei of proliferating cells in the eye and brain, similar to the expression that we report.
Table 1.
Primary antibodies used in the study.
| Antigen | Immunogen | Manufacturer | Dilution |
|---|---|---|---|
| Sox2 | Synthetic peptide of human Sox2 amino acids 249–265 |
Chemicon/Millipore, rabbit polyclonal, AB5603 | 1:200 |
| Vimentin | Xenopus laevis Vimentin protein | the Developmental Studies Hybridoma Bank | 1:200 |
The Vimentin monoclonal antibody, 14h7, was developed by Michael Klymkowsky, and obtained from the Developmental Studies Hybridoma Bank (Operated under the auspices of the NICHD and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242). This antibody was prepared against the Xenopus protein and recognizes the 55 kDa and 57 kDa isoforms of the vimentin protein on western blots to Xenopus kidney epithelial cell lysate (Dent et al., 1989). Similar to our results, it has been shown to label radial glial cells in the Xenopus laevis tadpole brain (Dent et al., 1989; Tremblay et al., 2009).
Immunolabeling
Fixed brains were mounted in a gelatin/albumin mix, and cut into 30-50 μm vibratome sections. Free-floating sections were washed with PBS+0.3% triton X-100 (PBST; pH 7.2) 3 times for 10 minutes, blocked in 5% normal donkey serum for 30 minutes and then incubated for 48 hrs at 4°C with a mouse anti-Vimentin supernatant or rabbit anti Sox2, each diluted 1:200 in PBS+0.03% triton X-100. After incubation with the primary antibody, the sections were washed in PBST, and incubated in goat anti mouse Alexafluor 633 secondary antibody (1:500; Molecular Probes) overnight at 4°C. The tissue was washed 3 times for 20 minutes with PBS (pH 7.2) before being mounted in Vectashield mounting medium containing DAPI or propidium iodide. Long-term storage degraded the labeling and was avoided.
Microscopy
The images in Figure 1 were acquired using a 2-photon microscope as previously described (Ruthazer et al., 2006). Vibratome sections and in vivo FP-labeled samples were imaged using an Ultraview VoX confocal system using Volocity 5 image acquisition software (Perkin-Elmer). It was composed of a Yokagowa CSU-X1 spinning disk confocal attachment mounted on an Olympus BX61WIF microscope with a XLUMPLFL 20X 0.9 NA or a LUMFLN 60xW 1.1NA objective and piezo microscope stage (ASI, MS-2000). Signals were captured with an EMCCD camera (Hamamatsu, c9100-50). The 20X objective allowed us to capture an entire tectal lobe in a single 180 μm stack (0.5-1 μm Z-steps). DAPI, green fluorescent, red fluorescent and far-red fluorescent signals were excited with 405, 488, 561 and 640 nm laser lines and signals distinguished with the following filters: 445(60), 515(30), 640(120) and 705(90) nm.
In vivo imaging procedures and protocols
For in vivo imaging, tadpoles were anesthetized in 0.01% MS-222 and placed in a custom-built chamber with the coverslip directly on the surface of their heads. Blood flow in the tectum was monitored to ensure that the animals were not compressed.
Daily imaging of pSox2-bd::Kaede-expressing animals
Kaede is easily photoconverted if the tadpoles are exposed to sunlight or halogen light sources. Therefore, animals that were electroporated with pSox2bd::Kaede were kept out of direct light to prevent inadvertent photoconversion of the Kaede fluorophore. Light sources used during the light dark cycle or visual enhancement were wrapped in a flexible plastic UV filter sheet (Edmund Optics, Barrington, NJ) to block wavelengths below ~400 nm. To photoconvert Kaede, tectal lobes were exposed to 405 nm laser excitation through the 20X objective for approximately 20 sec. The extent of the photoconversion was checked by viewing the green fluorescence signal, and if necessary, cells with residual green signal were individually targeted with a brief (~0.5 sec) focal exposure of the cell body to the 405nm laser using the photokinesis unit of the Ultraview system. Within minutes after photoconversion and once daily for the next 2 days, confocal stacks of each tectal lobe were acquired. To minimize exposure to the lasers, confocal stacks were not always taken prior to photoconverting Kaede. To quantify the levels of green to red Kaede fluorescent signal as they change over time, identical imaging conditions (acquisition times, laser and camera settings) were used for each of the 3 days.
Cell division was inhibited using 150 μM aphidicolin and 20 mM hydroxyurea dissolved in 2% dimethyl sulfoxide in Steinberg’s rearing solution. Animals were transferred to the cell division blocker solution immediately after the first image of the time-lapse series and kept in the solution for the duration of the ~48 hour experiment.
Radial glial cell lineage acquisition in pSox2-bd::turboGFP / pSox2-bd::turboRFPnls-expressing animals
24 hours after coelectroporation of pSox2-bd::turboGFP and pSox2-bd::turboRFPnls, anesthetized animals were imaged at time-lapse intervals ranging from every 2 to 19 hours (average interval, 7 hrs; median, 3.5 hrs) over several days (up to 103 hours). All cells expressed both FPs, indicating 100% co-electroporation of the two plasmids. Using these two fluorophores in combination allowed us to resolve the morphology of the cells with cytosolic turboGFP and the nuclear morphology with turboRFPnls. Between imaging sessions, tadpoles were returned to their individual rearing chambers in six well plates.
Image data analysis, quantification and presentation
Image analysis was conducted with Volocity 5 software (Improvision/Perkin Elmer). For the fixed tissue, optical sections were taken at 1 μm and colocalization of Sox2 immunoreactivity with GFP expression was determined using Imaris Software (Bitplane Scientific). The nuclear marker, DAPI, was used to locate cell bodies. Then cell bodies with pSox2-bd::FP expression and endogenous Sox2 immunostaining colocalization were determined. The percent colocalization was determined as the number of Sox2-immunoreactive cells that express pSox2-bd::FP divided by the total number of pSox2-bd::FP-expressing cells. For the time-lapse data, we used the automated functions of Volocity software to identify and outline in 3D the cell bodies of labeled neurons in each tectal lobe. The cell-selection results were verified and modified manually if necessary to improve the single-cell outlines. We assigned neuronal or glia identity to the cells according to morphological criteria based on the three dimensional structure of the cells and our previous work (Wu and Cline, 2003; Wu et al., 1999). The proportions of neurons and glia and the proliferation rates over 3 days were calculated for each tectal lobe. The Volocity software was used to quantify the red and green Kaede fluorescence levels expressed in the cell somata. The total fluorescence intensity for each channel was measured from each identified/outlined cell within the tectal lobe.
The lineages of individual radial glial cells that could be unambiguously followed through the time-lapse series were analyzed. The caudolateral region of tectum was often too densely labeled to allow identification of individual radial glial cells, so the majority of the radial glial lineages were analyzed from cells located midway between the rostrocaudal extent of the tectum. We determined the timing, numbers and sequence of symmetric, proliferative divisions, in which two radial glia were generated, and asymmetric, neurogenic divisions, in which a radial glia and a neuron were generated. We also determined the differentiation state of each cell in the lineage over the time course of the experiment. Criteria to identify cell types are presented in the results.
For the in vivo images presented here, 3D confocal stacks were cropped in XYZ to capture the full extent of the cell’s morphology and 2D projections were made using Volocity software. Because the 2D projections mask the 3D shape of the optic tectum itself as well as the cells within it, we include movies of some Z-stacks in the Supplementary Information. Figures were made with Image J, Adobe Photoshop and Illustrator. Care was taken to minimize saturated pixels during the acquisition of the images. To enhance the appearance of the fainter distal processes of the cells, background levels were subtracted, and the gamma and white levels were lowered. In later time points of Figure 6, images of fine distal processes of the cell were cropped in XYZ, and the white levels were decreased to aid the visualization of the faint branches. These cropped portions of the cell are montaged together.
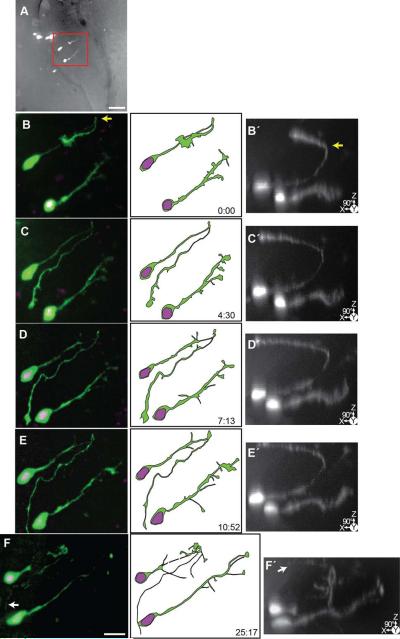
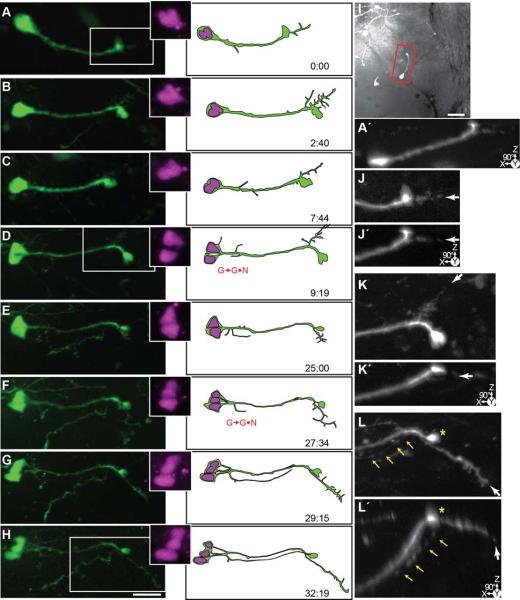
Figure 6. Time-lapse series of a pair of asymmetrically dividing radial glial cells which produce neuronal progeny before differentiating into neurons.
24 hrs after transfection with pSox2-mFGF::turboRFPnls (magenta) and pSox2-bd::turboGFP (green), 16 complete confocal stacks were acquired over 97 hours and 10 minutes. A) A projected confocal stack of the tectal lobe at the first time point with an outline of the tectal lobe. Dotted line indicates ventricular edge of the dorsal tectum. Boxed inset shows the cells followed in the time-lapse series. Scale bar = 50 μm. Bi-ii) Enlarged projection of the glial endfeet at the first time point (boxed regions in B). The confocal stacks in the lower panels are rotated 90° to reveal the Z-depth and the dorsally-projecting pial endfeet. B-Q) Projections of cropped confocal stacks revealing the dividing radial glial cells. Illustrations of the cells are made from the confocal stacks. Insets show an enlargement of the turboRFPnls signal revealing the nuclei. Scale bar = 20 μm. After the radial glial cells divide (marked with G→G•N), the cells become progressively simple before they produce dendrites and other neuronal features (marked with G→N). I-Q) In these later images of the time-lapse, the cells are much fainter. Black and white montaged projections were made to enhance the white levels of the distal processes. H) Asterisk indicates the appearance of an axon from a cell found elsewhere in the tectum. I) Arrow points to the faint axonal process from the cell pair on the right. Inset is the 90°-rotated projection of the boxed area.
Statistical Tests
Mann Whitney U tests were used to make comparisons between groups, and multiple comparisons were made with ANOVA and Tukey-Kramer Honestly Significant Difference (HSD) post-hoc tests. P and F values ≤ 0.05 were considered significant. Graphs show means ± standard error of the mean, and all values for these figures are presented in Tables 2–6.
Table 2.
The ratio of green-Kaede to red-Kaede expression in radial glial cells and neurons over 3 days. This data is presented in Figure 3T.
| DAY 1 | DAY 2 | DAY 3 | Anova/Tukey-Kramer HSD |
|||||
|---|---|---|---|---|---|---|---|---|
| Mean ± SEM (min-max) |
Cell # | Mean ± SEM (min-max) |
Cell # | Mean ± SEM (min-max) |
Cell # | Comparison | P value | |
| Neurons | 0.11±0.01 (0.03-0.29) |
7 | 0.42±0.09 (0.02-6.88) |
155 | 0.52±0.06 (0.03-9.2) |
192 | D1•D2 | 0.003 |
| D2•D3 | 0.44 | |||||||
| D1•D3 | <0.0001 | |||||||
| Radial Glia | 0.14±0.01 (0.01-0.66) |
77 | 0.90±0.33 (0.03-14.6) |
56 | 2.2±0.81 (0.03-30.8) |
53 | D1•D2 | 0.43 |
| D2•D3 | 0.13 | |||||||
| D1•D3 | 0.003 | |||||||
| Mann- Whitney U P value |
0.09 | 0.12 | 0.01 | |||||
| Tectal lobe # |
13 | 13 | 13 | |||||
| Tadpole # | 9 | 9 | 9 | |||||
Table 6.
Proportion of tectal cell types generated in tadpoles exposed to short term visual enhancement (STVE) compared to control animals. This data is presented in Figure 13D.
| DAY 1 12:12 L:D cycle |
DAY 2 12:12 L:D cycle |
DAY 3 12:12 L:D cycle |
DAY 3 STVE |
ANOVA/Tukey-Kramer HSD | ||
|---|---|---|---|---|---|---|
| Comparison | P value | |||||
| Neurons Mean±SEM% |
44.9±3.1 | 59.3±2.7 | 62.6±3.9 | 78.8±3.4 | D1•D2 | 0.003 |
| D2•D3 | 0.91 | |||||
| D1•D3 | 0.0031 | |||||
| D2•D3(STVE) | 0.0007 | |||||
| D3•D3(STVE) | 0.03 | |||||
| Radial Glia Mean±SEM% |
38.8±3.3 | 23.0±2.6 | 14.0±3.4 | 11.0±2.6 | D1•D2 | 0.0006 |
| D2•D3 | 0.23 | |||||
| D1•D3 | <0.0001 | |||||
| D2•D3(STVE) | 0.056 | |||||
| D3•D3(STVE) | 0.95 | |||||
| Unclassified Mean±SEM% |
16.4±2.5 | 17.6±2.2 | 23.4±4.4 | 10.2±3.0 | D1•D2 | 0.98 |
| D2•D3 | 0.53 | |||||
| D1•D3 | 0.35 | |||||
| D2•D3(STVE) | 0.29 | |||||
| D3•D3(STVE) | 0.04 | |||||
| Tectal Lobe # | 27 | 27 | 13 | 14 | ||
RESULTS
Timer Fluorescent protein expression indicates radial glia cells are neuronal progenitors
We used in vivo electroporation of pCMV::Timer to transfect scattered cells along the midbrain ventricle and collected time-lapse in vivo images with a 2 photon laser scanning microscope over six days to obtain an overall evaluation of cell type transitions in the developing optic tectum (Figs. 1A, B). Timer (dsRed-E5) is a fluorescent protein variant of dsRed that changes its fluorescence emission spectrum from green to red as the protein folds and matures (Terskikh et al., 2000; Verkhusha et al., 2001). Timer is a useful reporter for monitoring the dynamics of developmental processes such as cell lineage expansion or the onset of gene expression, because older protein expression can be distinguished from newer protein expression by the relative levels of red and green fluorescence. Figures 1C-F are Z-projections of a time-lapse series of images through the optic tectum. The first image was collected 24 hours after transfection and subsequent images were taken at about 1, 2 and 6 days later (Supplementary Videos 1-4 show the 3D structure of the cells and tectum and Z-series at first and last time points).
While the time-lapse series of the Timer-expression illustrates that transfected cells continue to synthesize the immature green variant of the Timer protein, an increasing proportion of cells express the red-fluorescent signal of the mature Timer in subsequent time points. Colocalization of mature Timer (magenta) with immature Timer (green) appears white in Figures 1D-F. Initially the cells express only the immature green variant of Timer (Fig. 1C). Inspection of these cells indicates that the majority has radial glial morphology and only a few have neuronal morphological features (i. e., dendritic arbors and axons). By the final image of the time-lapse series (Fig. 1F), most cells have neuronal morphologies suggesting that the radial glial cells seen at earlier time-points may have generated neuronal progeny or differentiated into neurons. To explore this idea in detail we conducted a series of experiments in which neural progenitor cells in the optic tectum were targeted for transfection with fluorescent reporters and imaged over time.
Sox2-driven fluorescent protein reporter preferentially labels proliferating radial glial cells
To visualize neural progenitor cells in the tectum, we designed a cell-specific plasmid expression vector based on the endogenous expression of the transcription factor Sox2 (Fig. 2A). Neural progenitor cells express Sox2 and the POU-class transcription factors, Oct3-4, which are required to maintain neural progenitor cell identity (Bylund et al., 2003; Graham et al., 2003; Komitova and Eriksson, 2004). The rationale for our expression vector design is that only cells expressing endogenous Sox2/Oct3-4 transcription factors drive expression from the reporter and this increases cell-type specificity of expression. We refer to this expression vector as pSox2-bd::FP (Fig. 2A). To test whether pSox2-bd::FP transfection results in FP expression in cells expressing endogenous Sox 2, we electroporated tadpoles with pSox2-bd::FP and labeled horizontal sections through the tectum with antibodies to Sox2. Figure 2B shows a DAPI stained horizontal section through the tectum. The boxed area is shown enlarged in Fig. 2C-E. We found that 24 hours after transfection with pSox2-bd::turboRFP tagged with a nuclear localization sequence (turboRFPnls), 95% (47/50) of the RFP positive cells were also Sox2-immunoreactive (Figs. 2C-E).
Figure 2. The pSox2-bd::fluorescent protein (FP) reporter and its expression in the optic tectum.
A) The pSox2-bd::fluorescent protein construct is a single concatenated plasmid containing the Sox2/Oct3-4 and mFGF regulatory fragments controlling the expression of Gal4 and UAS-fluorescent protein. Expression from this construct is dependent upon the binding of endogenous Sox2 transcription factor. B) 30 μm tectal section taken ~100 μm below the dorsal surface of the tectum and stained with DAPI. Box indicates position of the images shown in C-E. Scale bar = 50 μm. C-E) pSox2-bd::turboRFPnls-expressing cells (red, C,E) are Sox2-immunoreactive (D and green, E). DAPI (blue, C, E) was used to visualize the nuclei. In the triple-labeled tectum (E) Sox2-immunoreactive and pSox2-bd::RFPnls-expressing cells appear yellow. Arrows point to pSox2-bd::RFPnls-positive cells. Scale bar = 20 μm. F-K) pSox2-bd::FP expressing cells are Vimentin immunoreactive. F-G) 50 μm section of the right tectal lobe taken ~100 μm below the surface of the tectum stained with DAPI to show nuclei (F). G) The same section as F showing pSox2-bd::GFP-expressing cells (green) and Vimentin immunofluorescence (magenta). The non-specific fluorescence of the embedding material is visible along the right edge of the section. Scale bar = 50 μm. The asterisks indicate the caudolateral region with relatively low Vimentin immunofluorescence. The boxed GFP-expressing radial glial cell in F and G are enlarged in H and I, which shows the GFP-expressing radial glial cell (H) and the radial glial cell together with Vimentin labeling (I). The boxed regions in H and I are expanded in J and K. J, K) Vimentin-positive radial processes are visible and the arrows point toward the GFP-expressing radial process which expresses Vimentin. Scale bar = 10 μm. L-Q) Z-projections of time-lapse confocal stacks showing pSox2-bd::turboGFP expression in the tectum. Starting 24 hrs after electroporation with pSox2-bd::turboGFP, complete confocal stacks were acquired through the same right tectal lobe over 56 hrs. This time-lapse series illustrates the progressive loss of radial glial cells and increase in the number of neurons. Yellow arrows point to glial endfeet and blue arrows indicate examples where glial endfeet present in the preceding time point are now lost. M-Q) The same projections are shown in L and M, but the 108 μm Z-depth is color-coded in 12 μm increments in H. White = most dorsal (d); Blue = most ventral (v). Scale bar = 50 μm.
Cells transfected with pSox2-bd::turboGFP have their cell bodies located within the ventricular layer and extend a radial process tipped with an expanded endfoot to the pial surface of the brain (Figure 2F-Q, and Figures 3-10), characteristic of radial glia described across phyla (Kriegstein and Alvarez-Buylla, 2009). To test whether the pSox2-bd::FP-transfected cells with radial glial morphology express a known marker for radial glia, we electroporated tadpoles with pSox2-bd::turboGFP to produce sparse transfection and labeled horizontal sections through the tectum with antibodies to the intermediate filament protein, Vimentin (Kamei et al., 1998; Noctor et al., 2001; Pinto and Gotz, 2007). In Xenopus, Vimentin immunoreactivity is detected in radial glial cells in the embryo beginning at the time neural tube closure (Stage 19, (Dent et al., 1989), and continues to label radial glia during and after the tadpole stages used in this study (Figs. 2F-K and (Tremblay et al., 2009; Yoshida, 2001). We find that pSox2-bd::turboGFP-expressing radial glial cells are immunoreactive for Vimentin, and specifically that Vimentin immunoreactivity can be detected in GFP-expressing radial processes (Figs. 2F-K). Cells in the caudolateral region of the tectum (asterisks; Figs. F, G) are only faintly immunoreactive for Vimentin. Although cells in this region are highly proliferative (Raymond and Easter, 1983; Schmidt and Roth, 1993; Straznicky and Gaze, 1972), we did not analyze them because they are too densely packed to resolve individual cell lineages. Based on the morphology and expression of radial glial progenitor markers, we identify the pSox2-bd::FP-expressing cells in the tectum to be radial glia.
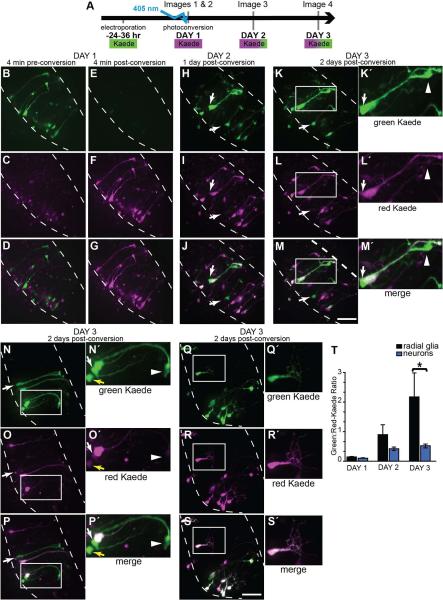
Figure 3. pSox2-bd::Kaede expression reveals newly born cells.
A) Diagram of the time-lapse imaging protocol used. 24 hrs after transfection with pSox2-bd::Kaede, complete confocal stacks were taken immediately before and after photoconversion. Two additional confocal stacks were acquired on each of the subsequent 2 days. Photoconversion of Kaede was achieved with ~20 sec. exposure of the tectal lobe with the 405 nm laser through the microscope objective. In proliferating cells, Kaede continues to be synthesized (appearing green), but cells generated after the photoconversion can be identified because they lack the photoconverted (magenta) Kaede. B-S) Flattened confocal stacks of the right tectal lobe of the unconverted green Kaede (top row), photoconverted red Kaede (magenta, middle row) and merged projections (bottom row). B-D) Kaede expression before and immediately after (E-G) photoconversion. The same tectal lobe on the 2nd (H-J) and 3rd day (K-M) where new, primarily green Kaede-expressing cells have appeared (arrows). K□-M□) Magnified views of boxes in K-M: A radial glial cell expressing high levels of unconverted Kaede. Arrows point to the cell body and the arrowhead at the pial endfoot of a radial glial cell that lacks photoconverted Kaede. It is closely apposed to a neighboring radial glial cell that expresses photoconverted Kaede. N-S) Kaede-positive cells in the right tectal lobe of two additional tadpoles imaged 2 days after photoconversion with an example of a newly-generated radial glial cell (boxed area and N□-P□) and neuron (boxed area and Q□-S□). Yellow arrow indicates the soma of an immature neuron. The white arrow points to the soma of a radial glial cell lacking photoconverted Kaede. Its pial endfoot is shown with the arrowhead. T) Quantification of the red- and green-fluorescent signals from pSox2-bd::Kaede-expressing cells beginning immediately after photoconversion for 3 consecutive days. The increasing green:red ratio of Kaede expression reveals that radial glial cells express significantly greater levels of Sox2 than neurons. MW, p = 0.01. Intensity values for red and green Kaede fluorescence are reported in Table 2.
Figure 10. Time-lapse series of an immature neuron and developing radial glial cell.
24 hrs after transfection with pSox2-bd::turboRFPnls (magenta) and pSox2-bd::turboGFP (green), 5 complete confocal stacks were acquired over 25:17 hrs. A) Projection of the confocal stack of the right tectal lobe superimposed on a brightfield image of the tectal lobe to show tissue edges. Box indicates area of the cell shown in B-F. Scale bar = 50 μm. B-F) Cropped confocal projections of the cells and illustrations. B□-F□) 90° rotated projections of the same confocal stacks as in B-F. This perspective reveals the trajectory of the axon of the immature neuron (left) which grows up to the dorsal surface of the brain (yellow arrow) and then projects medially along of the pial surface. White arrow (F) indicates the tip of the axon. The glial cell (right) projects to the lateral edge of the brain and develops distinct endfeet over the course of the time-lapse (arrows). Scale bar = 20 μm.
To determine the distribution and morphology of pSox2-bd::FP transfected cells in the optic tectum over time, we collected in vivo time-lapse confocal images of FP-expressing cells within each tectal lobe. Figures 2L-Q are examples of confocal projections from a time-lapse series of pSox2-bd::turboGFP-expressing cells in the right tectal lobe over a 56 hour period, beginning 24 hours after electroporation. At the first time point, Figures. 2L-M, the turboGFP-expressing radial glial cells can easily be identified by their radial processes, which extend dorsally and laterally, ending in a pial endfoot (yellow arrows, Figs. 2N-Q; See also Supplementary Video 5 showing the Z-stack). Because the curvature of the tectal lobe, the ventricular layer, indicated with the dotted line in Figures 1B and 2M, extends more medially in deeper sections through the tectum, and more laterally in sections that are more superficial. Therefore, radial glial cell bodies appear along the mediolateral extent of the tectum in the flattened image projections. Similarly, radial glia that extend their processes more dorsally will appear artificially shortened in the Z-stack projection, and their pial endfeet will also appear distributed along the mediolateral extent of the surface of the tectum. These 3-dimensional features of the tectum and the distribution of Timer-expressing and pSox2-bd::FP cells are readily seen in the Z-series movies (Supplementary Data Videos 1-5).
Figures 2L and 2M show the same time point, but in Figure 2L the 108 μm Z-depth of the confocal stack is divided into 12 μm sections and colorized (white is the most dorsal, blue is the most ventral) to demonstrate the three dimensional architecture cells in the tectum. Note that radial glial cell bodies are distributed throughout the dorsoventral (white→pink→yellow→blue) extent of the ventricular layer, and that glial end feet are typically on the dorsal aspect of the tectum.
Once synthesized, the fluorescent protein remains as a marker of a neural progenitor cell and is inherited by its progeny, regardless of the cell identity. This can be seen in the subsequent images of the time-lapse series collected ~2 h, ~9 h, ~23 h and 56 h after the first image, where the number of turboGFP-positive cells with radial glial morphology decreases and the number of turboGFP-positive neurons, identified by their branching dendritic arbors, increases (Figs. 2N-Q). Initially, many radial glial endfeet are visible along the rostral caudal extent of the tectum (yellow arrows, Figs. 2M-Q), but these hallmark radial glial cell features are lost (blue arrows, Figs. 2N-Q) over the progression of the time-lapse. By the final image of the time-lapse series, turboGFP-expressing neurons are found throughout the tectum, suggesting that radial glial progenitors can generate neurons throughout the rostrocaudal and mediolateral axes of the ventricular cell body layer at these stages of development.
pSox2-bd::Kaede expression reveals newly generated cells
The results presented thus far suggest that pSox2-bd-expressing radial glial cells may be neural progenitors: we observed that over time the proportion of FP-labeled radial glia decreases as the proportion of FP-labeled neurons increases. In our previous time-lapse in vivo imaging experiments, individual cells with radial glial morphology appeared to transform into neuronal cells with dendritic arbors (Wu and Cline, 2003). To discriminate parent and progeny cells more clearly, we used the pSox2-bd::FP construct to drive expression of the photoconvertible Kaede fluorophore in the tectal cells. When synthesized by the cell, Kaede emits a green fluorescent signal (peak emission, 518 nm) but can be irreversibly photoconverted to emit red fluorescence (peak emission, 582 nm) by a brief exposure to UV illumination (Ando et al., 2002). The wavelengths required to excite and visualize Kaede in its red or green state are not capable of photoconverting the protein (Ando et al., 2002). Kaede-expressing cells continue to synthesize the green-fluorescent Kaede fluorophore regardless of whether they had been exposed to UV light. Based on these properties, Kaede fusion proteins have been used to identify the time course and cellular distribution of newly generated proteins or organelles (Imai et al., 2010). Furthermore, Kaede has been used to identify newly generated cells over time (Caron et al., 2008; Hatta et al., 2006; McLean and Fetcho, 2009) because cells generated after UV exposure can be distinguished from pre-existing cells based on their expression of green fluorescent Kaede.
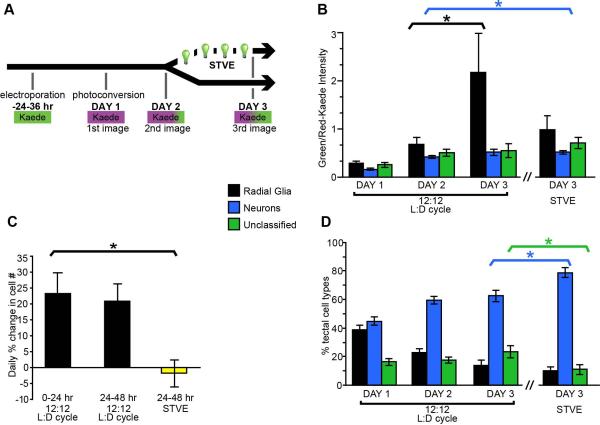
We electroporated tectal cells with pSox2-bd::Kaede, and 24-36 hours later photoconverted the entire population of Kaede-expressing cells so that they emitted the red fluorescent Kaede signal and acquired a confocal stack of the tectum (Fig. 3A). Having marked these potential progenitor cells with the red-Kaede, we then imaged the tectum at daily intervals over the next 48 hours. This protocol (Fig. 3A) revealed newly generated cells expressing relatively high levels of the green fluorescent Kaede. Figures 3B-P show confocal stack projections of a right tectal lobe taken from a single animal over 3 days. The green channel is shown in the top row, the red channel in the middle row (shown as magenta) and the merged projection of the two channels is in the bottom row. At the first time point (Fig. 3B-D), tectum-spanning radial glial cells express only green-fluorescent Kaede (top panel), with background autofluorescence visible in the red channel (middle panel). Immediately after photoconversion (Fig. 3E-G), the Kaede fluorophore is only detectable in the red channel (middle row). This red signal acts as a marker for all the cells present at the time of photoconversion. One day later, newborn (primarily green) cells are detectable (white arrows, Figs. 3H-J) and remain visible 24 hours later (white arrows and box, Figs. 3K-M). Figures 3K’-M’ show an enlarged projection of a newly-generated radial glial cell that is indicated with the boxes in Figures 3K-M. It is closely apposed to a second radial glial cell that was present at the time of the photoconversion based on the presence of red-fluorescent Kaede. The arrows (Figs. 3K’-M’) indicate the position of the cell body of the newly-generated glial cell and arrowhead marks the position of its pial endfoot (Figs. 3K’-M’). Projections of two additional tecta containing Kaede-expressing cells imaged 48 hours after Kaede photoconversion are shown in Figures 3N-P’ and 3Q-S’. Arrows indicate predominately green-Kaede-expressing cells generated after the time of photoconversion. Figures 3N’- P’ are enlarged projections of two cells expressing high levels of green Kaede at the final time point. The white arrow and arrowhead mark a radial glial cell’s soma (arrow) and pial endfoot (arrowhead) and the yellow arrow marks the position of the cell body of an immature neuron with a simple dendritic arbor (Figs. N’-P’). These cells both express relatively low levels of the red-Kaede, indicating they are newly-generated cells that were not present at the time of the photoconversion. In comparison, the insets from Figure 3Q-S’ show an example of a differentiated neuron with a complex dendritic arbor. This cell expresses low levels of green Kaede, consistent with data showing that neurons do not express Sox2 and therefore do not drive FP expression from the pSox2-bd::FP construct.
New green fluorescent Kaede synthesis from the pSox2-bd::Kaede plasmid is limited to proliferating cells expressing the Sox2 and Oct3-4 transcription factors. It should be noted, however, that if red-Kaede-expressing cells were to divide, their Sox2-expressing radial glial progeny would inherit Kaede protein during cytokinesis and therefore express both fluorophores. We predicted that if radial glia are mitotically active and continue to synthesize new green-Kaede, they should have higher ratios of green:red Kaede fluorescence intensities compared to differentiated neurons. To test this, we developed an algorithm to outline the perimeter of all Kaede-expressing cell bodies within the three dimensional confocal stacks and measure the green and red fluorescence levels for each image of the 3 day time-lapse beginning with the first image after photoconversion. Each identified cell was then assigned either radial glial or neuronal identity according to the cell morphology. Over the 3 day period, the green:red ratio increased significantly for both the radial glial cell and neuronal populations (Table 2), indicating that newly-synthesized green-Kaede levels increase over time. The level of new Kaede synthesis in the radial glial population was significantly greater by the 3rd day compared to neurons (2.0±0.8 versus 0.5±0.06; MW, p=0.01) while the average red-Kaede signal across the 3 days was not significantly different for either neurons or glia (ANOVA F>0.05, data not shown). The relatively high green:red-Kaede levels in radial glial cells indicate that they drive expression of pSox2-bd::Kaede after the time of photoconversion on Day 1 (Fig. 3T; Table 2). These data are consistent with the idea that optic tectal radial glia are Sox2-expressing neural progenitor cells in Xenopus CNS.
Cell proliferation blockers prevent an increase in FP-expressing tectal cell number
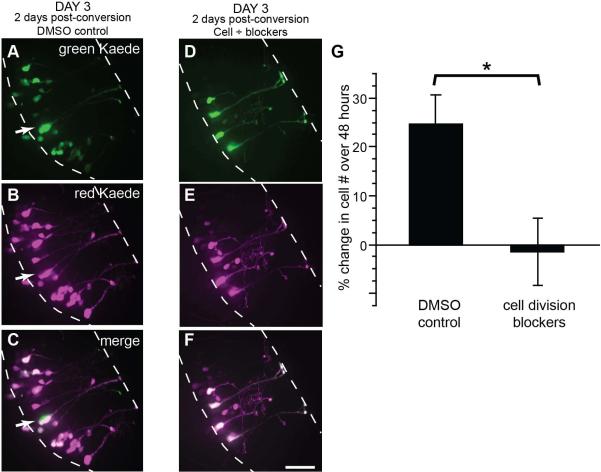
To test whether pSox2-bp::Kaede-expressing radial glia divide and generate additional cells, we counted the numbers of FP-labeled cells over 3 days in control conditions and in the presence of drugs to block cell division. Animals were electroporated with pSox2-bd::Kaede, and after 24-36 hours we photoconverted the Kaede-expressing cells so that all cells expressed the red Kaede fluorophore (magenta in Figs. 4A-F) and imaged the tectal lobes. Immediately after collecting the first image, half the tadpoles were exposed to 150 μM aphidicolin and 20mM hydroxyurea, or the drug vehicle, 2% DMSO in Steinberg’s solution, for the next 48 hours. This drug combination had previously been shown to block cell proliferation in Xenopus (Harris and Hartenstein, 1991). Two days later we collected a second confocal Z-series through the tectal lobes of tadpoles that had been exposed to DMSO alone (Figs. 4A-C) or DMSO with the cell division blockers (Figs. 4D-F). Newly generated cells can be readily recognized on the 3rd day of imaging in control animals by the predominance of green fluorescent protein (arrows, Figs. 3H-P’ and Figs. 4A-C), but no newly generated green fluorescent cells were detected in the drug-treated tadpoles (Figs. 4D-F). We quantified the rates of cell proliferation and found that aphidicolin and hydroxyurea significantly inhibited cell proliferation (Fig. 4G). While control tadpoles generated a 24.5 ± 6.0% increase in cell number over 2 days, drug-treated tadpoles had no detectible increase in cell number (-1.5 ± 6.9% of cell number at Day 1). These data indicate that Sox2-expressing radial glial cells are progenitor cells in the tectum.
Figure 4. Cell division blockers inhibit the generation of tectal cells.
The tectum was transfected with pSox2-bd::Kaede and all cells were photoconverted 24 hrs later at which time tadpoles were placed in a solution of 150 μM aphidicolin and 20 mM hydroxyurea in 2% DMSO or 2% DMSO alone. After 48 hrs, complete confocal stacks were taken of tectal lobes. A-F) Flattened confocal stacks of right tectal lobes of the unconverted green Kaede (top row), photoconverted red Kaede (magenta, middle row) and merged projections (bottom row) taken of tectal lobes 2 days after photoconversion of Kaede. While new cells appeared in the DMSO-control animals (A-C), no examples of cells generated after Kaede photoconversion were detected in animals where cell divisions were inhibited (D-F). Scale bar = 20μm. G) Quantification of cell proliferation between DMSO-treated control tadpoles and those exposed to aphidicolin and hydroxyurea, shows the drugs significantly inhibit cell proliferation in the tectum over 48 hrs. Mann Whitney, p = 0.02. Values are reported in Table 3.
Time-lapse imaging of Sox2-expressing radial glia cell proliferation and differentiation
Previous work suggested that radial glia in subcortical regions like the optic tectum or superior colliculus function as neural progenitor cells during CNS development in chicks (Gray and Sanes, 1992) and in adult fish (Stevenson and Yoon, 1981). To focus on radial glial cells and test their ability to serve as neural progenitors in Xenopus tadpole optic tectum, we imaged cells co-transfected with two plasmids that express fluorophores under pSox2-bd regulatory control: pSox2-bd::turboGFP to reveal cell morphology and pSox2-bd::turboRFPnls to label nuclei. The day after electroporation, we collected confocal Z-stacks through the dorsoventral extent of tectal lobes from 31 tadpoles at time-lapse intervals ranging from every 2 to 19 hours over several days (maximum imaging period: 103 hours).
Experiments in which cells were sparsely transfected with pSox2-bd::GFP and pSox2-bd::RFPnls suggested that about half of the radial glial cells imaged at the first time point appeared to be either in the process of cytokinesis, with one GFP-labeled cell body containing either two nuclei or a lobed nucleus, as visualized with turboRFPnls, or appeared to have recently completed cytokinesis, with two GFP + RFPnls-labeled cell bodies closely apposed to one another (see Figures 6-10). Furthermore, it appeared that the process of cytokinesis, after which two GFP + RFPnls-labeled cell bodies could be clearly distinguished, could be a protracted event that took many hours (see Figures 6-10). It is unlikely that multiple closely apposed radial glial cells were independently transfected, unrelated neighboring cells because the sparse electroporation technique used in this experiment has a low probability of transfecting neighboring cells with both the pSox2-bd::turboGFP and pSox2-bd::turboRFPnls plasmids. Because one of the goals of our study was to determine the lineage of tectal progenitors cells, we wanted to determine whether individual radial glial cells which appeared to have two nuclei were in the process of cell division and whether progeny of a recent division event remained close to one another. To address these questions we co-transfected cells with pSox2-bd::mGFP to express a membrane-localized GFP (mGFP) and pSox2-bd::RFPnls to visualize nuclei (Figure 5).
Figure 5. Prolonged time course of cell division in vivo.
A) A projection of a tectal radial glial cell expressing membrane-targeted GFP (pSox2-bd::mGFP, green signal) and pSox2-bd::RFPnls (magenta) reveals the complex morphology of the pial endfoot, many fine filopodia extending from the radial process and endfoot, and a lobed nucleus. B-C) Enlarged single z-planes through the cell body captured at the same time point as in A. At the first time point, the nucleus is dimpled but not divided (C) and there is no evidence of mGFP-labeled plasma membrane in the center of the cell body between the nuclear lobes (B). D-G) ~6 hours later, the nuclei have separated and mGFP-labeled membrane extends between the nuclei (arrows, F, G). Only one radial process is visible (boxed region of D is enlarged in E). By the third time point, almost 5 hours later (H-M), the nuclei have rotated so they are above and below one another, and remain separated by GFP-labeled membrane (arrows, J-M). Because of the dorso-ventral orientation of the nuclei, two Z-planes separated by ~7 μm are shown: a more dorsal plane (J, K) and ventral plane (L, M). There is no evidence of a second radial process extending from the newly generated cell (boxed region of H is enlarged in I).
A representative radial glial cell labeled with pSox2-bd::mGFP and pSox2-bd::RFPnls imaged over ~11 hours is shown in Figure 5. Figure 5A shows a projection of the cell body with the radial process and elaborate endfoot. Projections of the cell and proximal radial process are shown for images collected 6 and 11 hours after the first image (Figs. 5 D, H). Figures 5B, C, F, G, J-M are single Z-planes through the cell body of the radial glial cell at each designated time point. Because mGFP is targeted to the plasma membrane, it reveals the membrane as it forms during cytokinesis. Initially, the nucleus of the cell is dimpled and appears to have begun to divide, but no mGFP-labeled membrane separates the nuclei (Figs. 5B, C). Six hours later (Figs 5D-G), mGFP-labeled membrane separates the two nuclei (arrows, Figs. 5F, G). Five hours later (Figs 5H-M), at the last time point, the nuclei have rotated around one another slightly and sit partially above and below one another, as shown in images through relatively dorsal (Figs. 5J, K) and ventral (Figs. 5L, M) Z-planes through the somata. Plasma membrane, labeled with mGFP, surrounds each nucleus, indicating that cytokinesis is complete. Throughout the course of cell division, the radial process is maintained and we do not see evidence of a second radial process (Figs. 5E, I). These data demonstrate that the processes of nuclear division and cytokinesis are quite protracted in tectal radial glial progenitors in vivo. Furthermore, the fine structure of the radial glial cells and the nuclei undergo continual rearrangements throughout the period of cell division. These data indicate that the presence of two nuclei within one radial glial cell body and the presence of two closely apposed cells, both of which express GFP and RFPnls, likely report an active or recent proliferative event.
In what follows, we present examples illustrating representative division and differentiation events that we have observed in tectal radial glial cells in vivo. We focused our study on the fate of 48 individual radial glial cells and their progeny because these cells could be unambiguously identified through the time-lapse series. We identified 4 cell fates of radial glial cells labeled by Sox2-bd::FP expression: radial glial cells that remained mitotically inactive, those that divided symmetrically to produce 2 radial glial (G) progeny (designated as G→G•G); those that divided asymmetrically to produce one radial glial cell and one neuronal (N) progeny (designated as G→G•N), or those that appeared to differentiate into a neuron directly with no apparent division event (designated as G→N).
To arrive at these results, we needed to make determinations about the time of cell division and the types of cells generated with the division event based only on the in vivo images. The Z-stacks of images from each time point were rotated in space to determine at which time point two separate nuclei could be resolved; this point of the time-lapse was assigned as the time point of the division. Using only the information in that image of the time-lapse series, we assigned cells either a neuronal or radial glial identity according to the following criteria. Newly generated cells without radial processes were designated as neurons. If the cell retained its radial process and endfoot, but lacked any evidence of dendritic arbor elaboration, it was designated a radial glial cell. When a glial cell directly differentiated into neuron, it was designated a neuron only when the radial process adopted neuronal features including loss of the glial endfoot, formation of axonal or dendritic growth cones, the outgrowth of an axon, and/or the elaboration of dendritic branches. These criteria allow us to account for the situations, for example, where radial glial cells divide, appear to maintain their glial identity for many hours, and only later differentiate into a neuron.
Common to all the examples we show is that the radial glial cells within the tectum, regardless of their eventual fate, exhibit a surprising degrees of morphological rearrangements of their cell bodies, processes and endfeet during the time-lapse imaging periods (Figs. 6-11). Another shared feature across all 42 division events that we captured with time-lapse imaging was that the dividing cell pair, even many hours after the division event, did not migrate away from one another. Although nuclei are very mobile and appear to roll over each other prior to cytokinesis (see Figure 5-10), we find no evidence of pia-directed interkinetic nuclear migration or migration of progeny along radial glial processes. In all cases examined, the cell bodies of the progeny remained within one cell body’s distance from one another, and in most cases remained apposed at the end of the time-lapse. The close association of the parent and progeny cells makes it difficult to pinpoint precisely when cytokinesis is complete.
Figure 11. Time-lapse series of the development of immature neurons.
24 hrs after transfection with pSox2-bd::turboRFPnls (magenta) and pSox2-bd::turboGFP (green), 3 complete confocal stacks were acquired over 42:37 hrs (A-E) and 28:57 hrs (G-J). A) Projection of the uncropped confocal stack of the left tectal lobe superimposed on a brightfield image of the same tectal lobe. Boxed area is enlarged in B-E. Scale bar = 50 μm. B-E) Cropped confocal stacks of a developing neuron projected in Z and rotated 90° to reveal the Z-depth (B□-E□). The cell first sends out an axon and growth cone that reach the dorsal surface of the tectum (arrow) and then extend laterally along the pial surface. The distal process is marked with an arrowhead. In subsequent the time points the neuron elaborates dendritic branches. F) Projection of the uncropped confocal stack superimposed on the brightfield image of the right tectal lobe. Boxed area is enlarged in G-J. Scale bar = 50 μm. G-J) Cropped confocal stack projections and 90° rotate projections (G□-J□) of a developing neuron. The cell send out its axon dorsally where it reaches the pial surface of the tectum (arrow) before extending the axonal processes rostrally. The distal end of the axonal process is marked with the arrowhead and after it reaches the rostral-lateral edge of the tectum, it dives ventrally (arrowhead in J ).
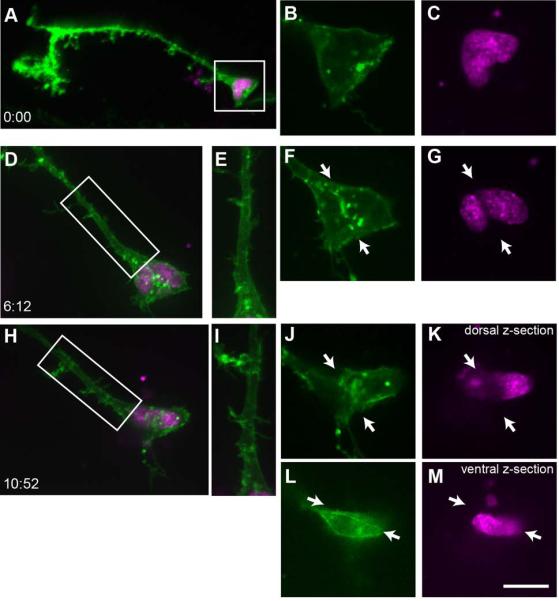
Figure 6 shows the projected confocal images of a pair of radial glial cells that were imaged over a ~97 hour period. Both radial glia divide asymmetrically (G→G•N), and after a delay, the parent cell eventually differentiates into a neuron (G→N). Each image of the time-lapse is grouped with insets of the nuclei as revealed by the pSox2-bd::turboRFPnls signal, and a reconstruction of the cell at that time point. The position of the cells within the tectum is illustrated in Figure 6A. Initially, both cells extend dorsally-projecting processes with endfeet adjacent to the pial surface (boxed region from Fig 6D magnified in Figs. 6B, C), indicating they are radial glia. The features of the pial endfeet can be more easily visualized in the 90° rotated images (Figs. 6B’-C’). Following the cell on the left, we see that its endfoot simplifies progressively over the first 10 hours of imaging, and this cell first shows evidence of a G→G•N division event after ~10 hours (Fig. 6H). At this time point, two nuclei have separated from one another. 48 hours into the time-lapse, with the loss of the pial endfoot, the parent cell has begun to transform into a neuron (G→N; Figure 6N). By the end of the time-lapse, the cell pair on the left is both immature neurons with simple dendritic arbors. The cell pair on the right (Figs. 6D-S) shows a radial glial cell in the process of a G→G•N division. At the first time point in the time-lapse (Fig 6D), the turboRFP reporter indicates the presence of two nuclei, although the cell bodies are so tightly apposed that it is not possible to distinguish the boundaries of two cells. A radial glial process extends from the cell(s), which retains its endfoot. At ~25 hours, there is evidence of an axonal arbor extending from the pial endfoot of the radial glia (arrow, Fig. 6K) which can be seen projecting ventrally along the lateral edge of the tectum from the endfoot (arrows, Fig. 6K arrow and inset). Because the processes of the cell pair are so closely associated it is unclear if this axon-like process is from the parent cell or progeny. The pial endfoot becomes reduced in size and at 48 hours into the time-lapse the parent cell has lost its glial appearance (G→N), and begun to elaborate dendritic branches (Fig. 6N). By the final point of the time-lapse, all four cells have neuronal features.
The ~32 hour time-lapse series presented in Figure 7 shows a radial glial cell in the process of dividing asymmetrically to generate a radial glial cell and a neuronal progeny, an example of the G→G•N lineage. Although two nuclei are present, the cells have not separated enough to distinguish two cell bodies or separate processes. The Z-stack projections show how the radial glial cell projects across the expanse of the tectum with its endfoot along the lateral edge of the pial surface (Fig. 7M). Each confocal projection of the time-lapse in Figures 7A-L is paired with insets showing the turboRFPnls signal and a reconstruction of the cells. Over this time-lapse series, we see the progressive elaboration of the neuronal arbor, and observe dynamic rearrangements of the radial glial cell’s pial endfoot (boxes in Figs. 7A, C, D are expanded in Figures 7N-P with 90 rotated projections in Figures 7N’-P’). At the initial time point (Figs. 7A, N), the endfoot is wide and flat, but over the first 4 hours, it simplifies (Figs. 7C, O), and extends ventrally along the edge of the tectum until it is too deep and faint to resolve (Figs. 7L, P). The transformation and ventrally-oriented growth of the endfoot can be seen in the 90° rotated projections (Figs. 7N’-P’). By the 8th hour, the radial glial cell has lost its pial endfoot and begun to elaborate its axonal process along the edge of the tectum (G→N, Fig. 7E). At this time, both cells have neuronal features, and the cell bodies separate somewhat, although it is difficult to distinguish the growing dendritic arbors of the cells. By the final time point (Fig. 7L), both cells have elaborated entangled dendritic processes and there is no evidence of a radial process with pial endfoot.
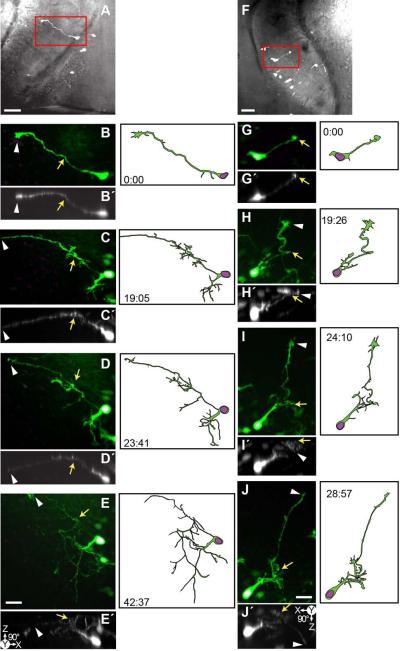
Figure 7. Time-lapse series of an asymmetrically dividing radial glial cell which produces a neuronal progeny before developing into a neuron.
24 hrs after transfection with pSox2-bd::turboRFPnls (magenta) and pSox2-bd::turboGFP (green), 11 complete confocal stacks were acquired over 32 hours and14 minutes. A-L) Projections of confocal stacks cropped to a radial glial cell in the process of dividing to produce a glia and a neuron (a G→G•N lineage) and illustrations of the cells made from the confocal stacks. Insets show an enlargement of the turboRFPnls signal of the nuclei. After 8 hrs the radial glial cell loses its pial endfoot and begins to produce dendrites (G→N). Scale bar = 20 μm. M) Projection of the confocal stack superimposed on a brightfield image of the tectal lobe to show the position of the cells in the tectum. Boxes areas are magnified in N-P show how the glial endfoot simplifies and axon emerges. Enlarged projections Z-stack projections (N-P) of the endfoot indicated in the first, third and last time points (A, C, L) and rotated 90° and projected (N□-P□) show the ventral path and depth of the axon’s projection. Scale bar = 20 μm.
The time-lapse series in Figures 8A-H shows a radial glial cell dividing asymmetrically twice in ~32 hours to generate a radial glial cell and produce two neural progeny, whose cell bodies and processes remain in close contact throughout the time-lapse series. These division events are two examples of the G→G•N lineage. The parent cell’s position in the tectum is shown in Figure 8I. The cell projects a radial process rostrally and the pial endfoot makes contact with the dorsal-medial surface of the tectum. Figure 8A’ is the same confocal stack as Figure 8A rotated 90° and projected to reveal the three-dimensional architecture of the cell in the Z-dimension, particularly the dorsally-projecting radial process and pial endfoot. We also detect a faint axonal process growing out from the endfoot along the dorsal surface in the first time point, which becomes clearer in the next image, 2 h and 40 minutes later. The axon becomes more elaborate over time (Figs. 8C-H). The white arrows point to this axon-like process in expanded boxed regions presented in Figures 8J-L. The same pial endfoot region is rotated 90° and projected in Figures 8J’-L’. Although the turboRFPnls signal in the nucleus appears lobed and could be two closely opposed nuclei in the first time point (inset of turboRFPnls, Fig. 8A), it is not until ~9 hours later that 2 separate cells can be clearly distinguished (inset, Fig. 8D). It is possible that the glial endfoot and axonal processes seen in Figs. 8A-C derive from two separate, closely opposed processes, which appear to emerge from a single cell body and that the nuclear and cell body division evens occur with a delay, seen in Figure 8D. A third cell appears after an additional ~24 hours (inset of nuclei, Fig. 8F). In this series, we see that the daughter cell sends an axonal process out along the same trajectory as the parental glial process (yellow arrows, Figs. 8L and L’). Unlike the cells presented in Figures 6 and 7, the parent glial cell maintains a glial morphology throughout the time-lapse series, determined by the presence of the pial endfoot, labeled with asterisk in Figures 8H-L’L.
Figure 8. Time-lapse series capturing an asymmetrically dividing radial glial cell which produces two neuronal progeny.
24 hrs after transfection with pSox2-bd::turboRFPnls (magenta) and pSox2-bd::turboGFP (green), 8 complete confocal stacks were acquired over 32 hours and 19 minutes. During this period, the parent radial glial cell divides twice to produce two neuronal progeny (marked with G→G•N). A-H) Time-lapse series of cropped confocal stacks of the radial glial cell and its progeny, insets of the turboRFPnls signal of the nuclei and illustrations of the radial glial cell and the newly generated neurons. I) The box outlines the cell’s position within the tectum. Here, a brightfield image is superimposed with the confocal stack of the tectal lobe cropped in the Z-dimension to reveal the radial glial cell in A. Scale bar = 50 μm. A ) Same confocal stack as A, but rotated 90° to show the Z-dimension. J-L) Enlargements of the endfoot from boxed regions of A, D and H; the confocal stacks were rotated 90° and projected (J□-L□) to illustrate the dorsal growth of the axon-like projection (yellow arrows) and the appearance of a second axon growing along the glial process (white arrows, iii and iii ). Asterisk indicates the pial endfoot which is maintained throughout the time series. Scale bar = 20 μm.
The radial glial cell in Figure 9 divides symmetrically to produce two radial glial cell progeny, and is an example of the G→G•G lineage. This parent radial glial cell projects laterally across the width of the tectum (Figs. 9A) and its radial process splits to form multiple endfeet on the dorsal surface of the tectum, seen best when the confocal stacks are rotated 90° to show the Z-axis (Figs. 9B’-9E’). Although the nuclei are not sufficiently separated at the first time point to show that the cell has started to divide (Fig. 9B, inset), there appear to be two separate processes exiting the soma and becoming entwined, and a wispy process extending parallel to the glial process (arrows, Fig. 9B), which may be from the daughter cell. The nuclei have separated at the second time point (Fig. 9C) and, as the time-lapse continues, the pial endfeet of the newly generated glial cell form (Figs. 9D-E). In the final two time points, the two glial processes are separated, however as in the previous examples, the cell bodies of parent and progeny remain in contact throughout the time-lapse series.
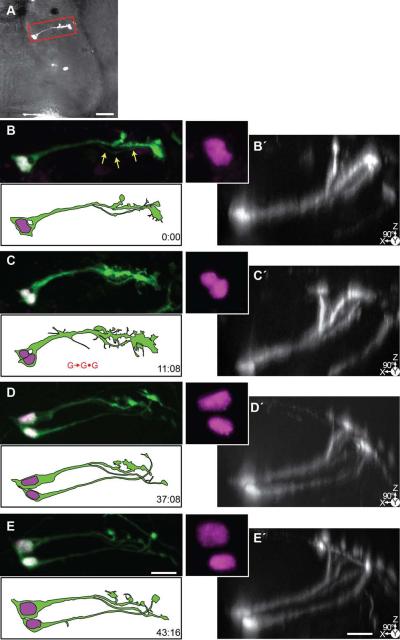
Figure 9. Time-lapse series of a symmetrically dividing radial glial cell.
24 hrs after transfection with pSox2-bd::turboRFPnls (magenta) and pSox2-bd::turboGFP (green), 4 complete confocal stacks were acquired over 43:16 hrs. A) A brightfield image of the tectal lobe overlaid with the projection of the confocal stack of the tectum cropped in Z around the cell shown in B-E. Scale bar = 50 μm. B-E) Projections of the cropped confocal stacks of the radial glial cell, and same stacks (B□-E□) rotated 90° to reveal the dorsal reaching pial endfeet of the glia in the Z-dimension. Note the presence of a faint process of the second cell beginning at t = 0:00 (arrows). The cell divides and nuclei separate (C, marked with G→G•G). At the final time point, two glia, each with endfeet, are visible. Scale bar = 20 μm.
Figure 10 shows two immature cells whose morphology changes significantly over the ~25 hour time-lapse. Both cells project dorsal-laterally across the tectum (Fig. 10A). The cell on the left is a young neuron and the cell on the right is a radial glial cell. At the first time point (Figs. 10B, B’) the neuron on the left has extended an axon tipped with an elaborate growth cone. When the confocal stacks presented in Figures 10B-F are rotated 90° to show the Z-dimension (Figs. 10B’-F’), this rotated perspective shows that the left cell projects a process dorsal-laterally toward the surface of the tectum (arrows, Figs 10 B’), and after reaching the pial surface, the axon-like process extends medially. Over the next 25 hours, the axonal process continues to extend and it becomes more slender and difficult to resolve in the image projections (marked in the final time point of Figs. 10F and F’ with a white arrow). This cell appears to be a radial glial cell that has recently differentiated into a neuron by sending out an axon from the former pial endfoot. By contrast, the cell on the right in Figure 10 is an example of a radial glial cell that exhibits morphological rearrangements throughout the time-lapse series but does not change its fate. Initially the cell’s processes are thicker with greater numbers of fine side branches, but by the final time point (Fig. 10F), the process has become more slender and the pial endfeet have condensed. The 90° XZ projections also illustrate how the radial process projects laterally and dorsally to the pial surface (Figs. 10B’-F’).
Neuronal differentiation in the tectum
Electroporation with pSox2-bd::FP can result in fluorescent protein expression in newly differentiation neurons at the first imaging time point. This provides the opportunity to collect images of neurons at the early stages of differentiation. Time-lapse images of these cells revealed that neuronal progeny of Sox2-expressing progenitors in the tectum follow a predictable developmental program in which the immature neuron extends an axon across the tectal surface as the cell begins to elaborate a dendritic arbor. Examples of this pattern of neural differentiation are shown in Figure 11.
The position of the cells within the tectum is shown in Figure 11A and 11F. At the each time point are projections of confocal stacks, their XZ orthogonal projections (Figs. 11B’-E’ and G’-J’) and reconstructions of the cells. Initially the neuron in the series in Figs 11A-E is quite simple with projecting axon tipped by growth cones. The point at which its axon reaches the pial surface of the tectum is indicated with yellow arrows and the arrowheads mark the distal-most end of the extending axonal process. At the initial time point, the axon of the neuron in Figure 11B has already extended more than 50 μm laterally along the pial surface. The lateral projection of the axon along the curving tectal surface can also be seen in the 90° XZ projections of the confocal stack shown in Figures 11B’. Over the next ~19-24 hours, a cluster of branches has extended from the axon of this neuron near the pial surface and short dendrites have emerged close to its cell body (Figs. 11C, D). By 42.5 hours, both the dendritic and axonal arbors become increasingly elaborate until in the final image of the time-lapse series (Fig. 11E) when the neuron has a mature, wide-branching dendritic arbor. The cell shown in Figures 11F-J begins the time-lapse with the morphological features of a radial glial cell, with the simple radial process and pial endfoot expansion. The ~29 hour time-lapse images reveal the developmental pattern similar to the examples shown in Figures 6, 7 and 10, that after the process reaches the pial surface (marked with the yellow arrow, Figs. 11G-J and 11G’-J’), it turns to elaborate an axon caudally along the dorsal surface of the tectum. The extending axon (the arrowheads mark the distal end of the axon) reaches the rostral-lateral edge of the tectum at the final time point (Fig. 11J). The XZ orthogonal confocal projections show how the axon begins to grow ventrally along the rostral-lateral edge of the surface of the tectum in the later time points (Figs. 11G’-J’). .
Although the neurons in Figures 11A-E and 11F-J develop very different morphological features by the end of their time-lapse series, similar to the neuron on the left side of Figure 10, the initial steps in their development are similar. After the processes of the cells reach the dorsal surface of the tectum (yellow arrows, Fig. 11), the neurons extend their axons adjacent to the pial surface. In contrast to the developmental pattern of these neurons, the radial processes of glia project directly to the brain surface with little torsion (Figs. 6-10). Once a glial cell process reaches the pial surface, its process ends with an endfoot expansion and does not grow adjacent to the pial surface. The meandering and wavy axonal processes of the neurons in Figures 11 are also distinct from glial cell processes (Figs. 6-10).
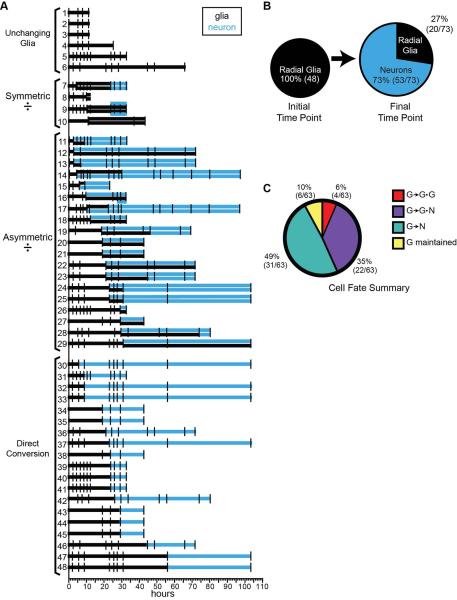
Lineage analysis indicates that most radial glia generate neurons
The division events and fate of 48 radial glial progenitor cells that we could clearly track and re-identify over the course of the time-lapse series, including the examples presented in Figures 6-10, are summarized in Figure 12. We measured the prevalence of the four different cell fates and found: 12.5% (6/48) remained mitotically inactive; 4/48, (8.3%) divided symmetrically to produce 2 radial glial progeny; 39.5% (19/48) divided asymmetrically to produce one radial glial cell and one neuronal progeny; and 19/48 (39.5%) differentiated into a neuron directly with no apparent division event. These data indicate that radial glia cells are an active component of the progenitor pool in the developing tectum that gives rise to neurons and other radial glia. Each horizontal line in Figure 12A is the fate of a single glial cell and its progeny over time, with the image acquisition times indicated with tick marks. The lineage events of each cell and its progeny are marked with changes to the line. When a radial glial cell divides, a second line is added, and the lineage line changes color to indicate a neuronal (blue) or radial glial cell (black) identity. The radial glial cells are grouped along the vertical axis by the type of division/differentiation event that is first identified in each cell’s time-lapse history. Of the 48 radial glia imaged on the first day after transfection, 73% (53/73) of cells at the final time points were neurons and 27% (20/73) were radial glia, producing a ~50% increase in the cell population (Fig. 12B).
Figure. 12. Lineages of radial glial cells revealed with in vivo time-lapse.
Summary of lineage data from 48 radial glial from 31 tadpoles that could be unambiguously reidentified throughout the time-lapse series. A) Lineages of each radial glial cell are presented as a horizontal line, vertical tick marks indicate the time of each confocal stack. Black indicates the cell is a radial glial cell, the addition of an upper or lower line indicates that a division event has occurred. Blue lines represent neurons. Cells with lineages 4, 10, 11,16, 17 are presented in figures 10 (right cell), 9, 7, 6 (right cell), 8 and 6 (left cell), respectively. B) At the final time point, 73% of the cells are neurons, and the total cell count has increased 50%, from 55 to 73 cells. C) Most glia (49%) appeared to differentiate directly into neurons (G→N). 35% divided asymmetrically to generate a neuron and another glial cell (G→G•N), and only 6% displayed a symmetric division generating two glia (G→G•G). 10% of the radial glia that we followed were maintained without a division/differentiation event.
From the lineages of the initial 48 radial glia, we identified 63 division or differentiation events, and the proportions of these events are presented in Figure 12C. The majority (49%, 31/63) of events that we identified were of a radial glial cell undergoing direct differentiation into a neuron (i.e., Figs. 6, 7). Asymmetric divisions which produced a radial glial cell and a neuron constituted 31% (22/63) of the events that we observed (i.e., Figs. 6, 7, 8). Symmetric divisions, in which a radial glial cell generated two radial glial progeny (i.e., example in Fig. 9) were only 6% (4/63) of the events that we observed. Six radial glial cells did not divide or change their fate over the time-lapse period, constituting 10% of the events (i.e., right cell in Fig. 10).
Short-term changes in the visual environment alter cell proliferation and differentiation
Neuronal activity in the brain reportedly modifies rates of cell proliferation (Bonetti and Surace, 2010; LoTurco et al., 1995; Weissman et al., 2004). Our previous work has shown that increasing the activity of the visual circuit by providing short term visual enhancement (STVE) to tadpoles facilitates the development of tectal neurons and their integration into the visual circuit (Aizenman and Cline, 2007; Sin et al., 2002, Hass et al 2006) and that visual deprivation increases the cell proliferation rates in the tectum (Sharma and Cline, 2010). Visual experience also has been shown to produce spontaneous calcium transients in Xenopus tectal radial glial cells and to induce morphological arrangements of the superficial fine processes of radial glial cells (Tremblay et al., 2009). Here, we tested whether increasing the visual experience of tadpoles affects Sox2-expressing radial glial cell proliferation and cell fate in the tectum, using the protocol outlined in Fig. 13A. We electroporated stage 46 tadpole tectum with pSox2-bd::Kaede, waited 24-36 hours to photoconvert all cells, and then acquired confocal stacks through the tectum. Once the first image was acquired, all animals were placed in 12 hr low light/12 hr dark conditions for 24 hours, after which, a second image was taken. Next, half of the animals were provided with the STVE, a simulated motion stimulus, and the other half of the animals were kept in the 12:12 light/dark conditions for the second 24 hour period, after which a third confocal Z-series of images was collected.
Figure 13. Short term visual enhancement decreases proliferation and increases neuronal differentiation.
A) Diagram of the time-lapse imaging protocol used. 24-36 hrs after transfection with pSox2-bd::Kaede, complete confocal stacks were acquired each day for 3 days. Half the animals received a short term visual enhancement (STVE) during the final 24 hr time-lapse interval. Afterward, the cells were identified in the confocal stack and then categorized as neurons, radial glia or undifferentiated cells that could not be assigned either cell identity. B) The ratio of new, green-Kaede expression to photoconverted, red-Kaede expression was calculated from the cell bodies of the pSox2-bd::Kaede-expressing cells in the tectum beginning with the first image after photoconversion of the fluorophore. Radial glial cells continue to express higher levels of green Kaede compared to neurons and ## respond to STVE by significantly lower the level of new Kaede expression. Values given in Table 4. C) The daily change in cell numbers was calculated for each day of the time-lapse. Animals provided with STVE were found to have significantly lower proliferation rates compared to controls Values given in Table 5. D) The proportion of pSox2-bd::Kaede-expressing neurons, radial glia and unclassified cells in each tectal lobe were calculated each day over the 3 day time-lapse series. Animals were exposed to a short term visual enhancement (STVE) instead of control conditions over the last 24 hr period had significantly higher proportion of neurons and fewer unclassifiable cells. Values given in Table 6.
To test whether differences in visual experience affected cell proliferation, we counted the Kaede-expressing cells in images collected on Day 1, Day 2 and Day 3 and determined proliferation rates as the relative change in cell number per day. Animals exposed to the control light:dark conditions had comparable daily proliferation rates over the 2 day period. Cell numbers increased 23.4 ± 6.5 % over the first 24 hrs and 20.9 ± 5.5% over the second 24 hrs (Fig. 13B and Table 4; n=27 tectal lobes from 19 animals). In contrast, cell numbers increased only 1.8 ± 4.3% during the second 24 hour period in tadpoles provided with STVE, significantly lower than the relative increase in cell number over the previous 24 hour period in the same animals (p=0.02; Fig. 13B and Table 4). These data suggest that visual experience decreases cell proliferation in vivo.
Table 4.
Tectal cell proliferation in tadpoles exposed to short term visual enhancement (STVE) compared to control animals. This data is presented in Figure 13C.
| % change in cell # Mean ± SEM |
Tectal lobe # | ANOVA/Tukey-Kramer HSD | ||
|---|---|---|---|---|
| Comparison | P value | |||
| 0-24 hr 12:12 L:D cycle |
23.4±6.5% | 27 | 0-24 hr (12:12)•24-48 hr (12:12) | 0.96 |
| 24-48 hr 12:12 L:D cycle |
20.9±5.5% | 13 | 0-24 hr (12:12)•24-48 hr (STVE) | 0.02 |
| 24-48 hr STVE |
−1.8±4.3%- | 14 | 24-48 hr (12:12)•24-48 hr (STVE) | 0.09 |
Our experiments with Kaede, summarized in Figure 3, showed that after photoconverting Kaede, subsequent expression of green fluorescent Kaede is driven by endogenous Sox2 in radial glial progenitors. To test whether STVE changed the fate of Sox2-expressing radial glial cells, we determined the green:red ratio of Kaede in neurons and glia in animals exposed to STVE or those reared in control light/dark conditions. We find that radial glial cells express significantly higher ratios of green to red Kaede compared to the green to red Kaede ratio in neurons (Fig. 13C, Table 5), consistent with data in Figure 3. Cells that could not be classified as neurons or radial glia by morphological criteria have green:red ratios intermediate between values seen in neurons and radial glia (Table 5). Without STVE, the green:red-Kaede ratio in radial glial cells on Day 3 (2.2±0.81) was significantly higher than on Day 2 (0.68±0.14; Tukey Kramer, p=0.0005; Fig. 13C and Table 5). Again, consistent with data from Figure 3, green-Kaede expression reports the presence of endogenous Sox2 in radial glia. Interestingly, the radial glial cell population in the tadpoles that received STVE had lower green:red Kaede ratios than radial glia from control animals. When the tadpoles were provided with STVE during the final 24 hour period, the average green:red-Kaede ratio measured from the glial cell bodies on Day 3 was only 0.87±0.19, and did not significantly increase from Day 2 levels (Fig. 13B and Table 5). In contrast, the green:red-Kaede ratio was significantly lower in the neuronal population compared to the glial cells across the time points (Fig. 13B and Table 5), and was not altered with STVE (Table 5 and Fig. 13B). These data suggest that the augmented visual experience provided by STVE changes the fate of Sox2-expressing radial glial progenitors so they stop expressing Sox2 and differentiate into neurons.
Table 5.
Ratio of green-Kaede to red-Kaede in tectal cell from tadpoles in control conditions and from animals exposed to short term visual enhancement (STVE). This data is presented in Figure 13B.
| DAY 1 12:12 L:D cycle |
DAY 2 12:12 L:D cycle |
DAY 3 12:12 L:D cycle |
DAY 3 STVE |
ANOVA/Tukey-Kramer HSD | ||
|---|---|---|---|---|---|---|
| Comparison | P value | |||||
| Neurons Mean±SEM (min-max) Cell # |
0.16±0.02 (0.03-3.4) 226 |
0.42±0.03 (0.02-6.9) 370 |
0.52±0.06 (0.03-9.2) 192 |
0.52±0.03 (0.0-4.2) 263 |
D1•D2 | <0.0001 |
| D2•D3 | 0.07 | |||||
| D1•D3 | <0.0001 | |||||
| D2•D3(STVE) | 0.047 | |||||
| D3•D3(STVE) | 0.98 | |||||
| Radial Glia Mean±SEM (min-max) Cell # |
0.29±0.04 (0.01-4.5) 194 |
0.68±0.14 (0.03-14.6) 138 |
2.2±0.81 (0.03-30.8) 53 |
0.87±0.19 (0.07-5.6) 36 |
D1•D2 | 0.45 |
| D2•D3 | 0.0005 | |||||
| D1•D3 | <0.0001 | |||||
| D2•D3(STVE) | 0.97 | |||||
| D3•D3(STVE) | 0.0497 | |||||
| Unclassified Mean±SEM (min-max) Cell # |
0.26±0.05 (0.03-3.0) 80 |
0.51±0.07 (0.02-4.2) 96 |
0.55±0.15 (0.03-8.4) 62 |
0.71±0.11 (0.09-3.5) 37 |
D1•D2 | 0.14 |
| D2•D3 | 0.98 | |||||
| D1•D3 | 0.10 | |||||
| D2•D3(STVE) | 0.54 | |||||
| D3•D3(STVE) | 0.77 | |||||
| ANOVA/Tukey-Kramer HSD | ||||||
| Comparison | P value | |||||
| G•N | 0.01 | 0.02 | 0.0001 | 0.008 | ||
| G•U | 0.80 | 0.36 | 0.003 | 0.28 | ||
| U•N | 0.27 | 0.70 | 0.99 | 0.34 | ||
| Tectal Lobe # | 27 | 27 | 13 | 14 | ||
| Tadpole # | 19 | 19 | 9 | 10 | ||
To test further whether visual experience increased the differentiation of neurons from Sox2-expressing radial glial progenitors, we counted the relative changes in numbers of neurons and radial glia over the 3 days of imaging. Under control light:dark conditions, the proportion of neurons increased from 44.9 ± 3.1% on Day 1 to 62.6 ± 3.9% on Day 3, as the proportion of radial glial cells decreased from 38.3 ± 3.3% on Day 1 to 14.0±3.4% on Day 3 (Fig. 13D, Table 6). The proportion of unclassified cells was stable in the control light:dark conditions (Fig. 13D, Table 6). In tadpoles that received STVE treatment, however, 78.8 ± 3.4% of labeled cells were neurons, a significantly greater proportion than neurons generated in control light:dark conditions (p = 0.03; Fig. 13D and Table 6). Altering the visual experience of the tadpoles did not affect the proportion of radial glia on Day 3 (11.6 ± 3.8% with STVE; Fig. 13D and Table 6), however, STVE significantly decreased the proportion of undifferentiated cells on the 3rd day from 23.4 ± 4.4% in control conditions to 10.2 ± 3.0% in tadpoles receiving the STVE treatment (Fig. 13D, Table 6, TK p = 0.04). These results suggest that the visual experience increases the rate at which neurons are generated in the tadpole optic tectum.
DISCUSSION
Although we know a great deal about the neurons and the neural circuitry of the optic tectum, much less is known about the cellular origins and lineages that give rise to the tectal neurons. To identify the proliferating cells and their progeny, we used in vivo time-lapse imaging over periods lasting up to 103 hrs to visualize Sox2-expressing neural progenitor cell division and differentiation events in the optic tectum of undissected, intact Xenopus tadpoles. Our methods permitted an unbiased retrospective assessment of the proliferative activity of individual cells within the Sox2-expressing progenitor cell population in the tectum. This experimental design also preserves structures and endogenous signaling mechanisms that are likely critical regulators of cell proliferation and differentiation in the intact animal, such as the vascular niche (Javaherian and Kriegstein, 2009; Tavazoie et al., 2008), intercellular signaling mechanisms (Peunova et al., 2001; Putz et al., 2005; Walsh and Reid, 1995), cell-cell interactions (Gao et al., 1991) and activity-dependent mechanisms (Sharma & Cline 2010). With this approach and longitudinal analysis, we analyzed the lineage of neural progenitors in the CNS. We provide direct quantitative evidence that Sox2-expressing radial glia function as neural precursor cells in the optic tectum, both by dividing to generate new neurons and by differentiating directly from radial glia into neurons. Our data also show that providing tadpoles with short-term visual enhancement arrests cell proliferation in the tectum and changes the fate of neural progenitors to increase the proportion of neurons generated.
Evaluation of the pSox2-bd::FP reporters and identification of labeled cell types
Sox2 and Oct3-4 are master regulatory proteins in the cellular pathways that maintain cell proliferation and define stem cells (Chakravarthy et al., 2008). We engineered a reporter plasmid under the regulatory control of the Sox2/Oct3-4 binding domain from the FGF4 promoter, which targets fluorescent protein expression to the Sox2/Oct3-4-expressing proliferating cell population in the brain. By expressing FP in the Sox2-expressing population of radial glial cells, we were able to explore their postulated function as neural progenitor cells in the optic tectum. Furthermore, by imaging fluorescent protein-expressing cells in animals with relatively sparse transfection levels, we were able to collect time-lapse confocal images of individual radial glial cells and their progeny in intact, undissected animals. Over 3 days of imaging, the proportion of pSox2-bd::FP-expressing radial glial cells in the optic tectum decreased as the population of labeled neurons increased, consistent with the role of Sox2-expressing radial glial cells as neural progenitors.
In addition to radial glial cells, Sox2 is also expressed in neuroepithelial cells (Archer et al., 2011; Pevny and Placzek, 2005), the main proliferative cell type found in early development during the formation of the embryonic neuroectoderm and neural tube (Rogers et al., 2009). Radial glial cells, which can be distinguished from neuroepithelial cells based on their Vimentin immunoreactivity (Pinto and Gotz, 2007), are also present at the time of neural tube closure in Xenopus (Dent et al., 1989), consistent with the idea that neuroepithelial cells give rise to or ‘transform into’ radial glial cells, which are then thought to become the principle neural progenitor cell type in the CNS (Farkas and Huttner, 2008(Kriegstein and Alvarez-Buylla, 2009). Because radial glial and neuroepithelial cells are proliferative, and share morphological features and molecular markers (Farkas and Huttner, 2008; Götz and Huttner, 2005), it is often difficult to distinguish them precisely. Based the elongated radial morphology and pial endfeet of the pSox2-bd::FP-expressing cells, the relatively late stage of development, and the observation that pSox2-bd::FP-positive cells express the diagnostic radial glial cell marker, Vimentin (Fig. 2), we identify the cells analyzed in this study as radial glial cells. Columnar shaped cells located in the caudolateral region of the tectum, which have little detectable vimentin immunoreactivity (Fig. 2G), and which have a distinctly squatter morphology compared to radial glial cells, may be neuroepithelial cells.
We also observed fluorescent protein expression in immature neurons (Figs. 10, 11, 13C) at the first imaging time point, which was likely inherited from their radial glial precursors. We began time-lapse imaging 24-36 hours after transfection to allow sufficient production of the fluorescent protein to label the entire morphology of the transfected cells. This interval is long enough for proliferating cells in the tectum to undergo at least one cell division, and so the fluorescent protein detected in neurons may be inherited from their Sox2-expressing radial glial parent cell through cytokinesis. Typically, the labeled neurons we identified at the first imaging time point were morphologically simple and found in close proximity to radial glial cells, suggesting they were the progeny of the radial glial cell. In addition, our lineage analysis indicated that more than half of the cells with neuronal morphology were generated by direct differentiation from cells with radial glial morphology (Fig. 12C). Our time-lapse imaging also demonstrated that the direct differentiation of radial glial cells to neurons could also occur within the interval between transfection and acquisition of the first image. The fluorescent protein expressed by radial glial cells that differentiate directly into neurons is retained in the neurons. Finally, the pSox2-bd::FP construct includes the gal4/UAS amplification system, and so relatively low levels of endogenous Sox2 could drive detectable levels of fluorescent protein expression.
We used pSox2-bd::FP to express Kaede, a fluorophore that can be irreversibly photoconverted from green to red fluorescence. Kaede expression from the neuronal HuC promoter has been used to identify the time-course of neuronal differentiation in the trigeminal ganglion of developing Zebrafish (Caron et al., 2008). An alternate use of Kaede has been to identify the distribution of fusion proteins and labeled organelles over time (Imai et al., 2010). Here, Kaede was driven by the pSox2-bd::FP construct, which allowed us to follow changes in Sox2-expression in tectal progenitor cells and their progeny. After electroporating pSox2-bd::Kaede, we photoconverted Kaede in all Kaede-expressing tectal cells and determined the relative levels of red and green Kaede fluorescence over time in the labeled cell population (Fig. 3, 13). Use of pSox2-bd::Kaede demonstrated that cells with radial glial morphology continue to synthesize Kaede, and therefore have increasing green:red Kaede ratios over 3 days of imaging, whereas cells with neuronal morphology have little or no new Kaede synthesis and their green:red Kaede ratios are relatively stable. We were also able to identify newly generated Sox2-expressing cells based on their predominant expression of green Kaede. The variability in the green:red Kaede ratios (Fig 3E and 13B) likely arose from two sources: 1, newly generated cells inherited variable levels of red Kaede from their parent cells either during cytokinesis, or as a result of direct differentiation of neurons from radial glia, and 2, the differences in synthesis of green Kaede, based on high or low levels of endogenous Sox2 in radial glial cells and neurons.
In vivo time-lapse imaging reveals that tectal radial glia are neural progenitors
Our initial analysis, based on expressing fluorescent protein in a sample of about 20 cells per tectal lobe and collecting images once a day over 3 days, revealed a ~25% increase in total labeled cell number between the first and third day of imaging (Fig. 4G, Table 3). Further analysis of cell type based on morphological criteria showed that the number of radial glia in the tectum decreased and the relative number of neurons increased over this period (Fig. 13D, Table 6). The increase in cell number and shift in the distribution of cell types were readily apparent by comparing the first and last images of the time-lapse series in Figures 1 and 2. The major features of the FP-labeled cells the first day after transfection were their ventricularly positioned tear-drop shaped cell bodies, dorso-laterally directed radial processes and the large flattened endfeet that dotted the pial surface of the tectum. In the final time points in Figures 1 and 2 imaged over three days after transfection, labeled glial endfeet are rare and the neuropil is filled with tangled axonal and dendritic processes.
Table 3.
Quantification of tectal cell proliferation over 48 hours in tadpoles exposed to cell division blockers aphidicolin and hydroxyurea compared to vehicle-control animals. This data is presented in Figure 4G.
| Tadpole Treatment | % change in cell number ± SEM |
Cell# on Day 3 | Tectal Lobe # |
|---|---|---|---|
| 2% DMSO control | +24.6 ± 6.02% | 178 | 8 |
| Cell Division Blockers (150 μM aphidicolin, 20mM hydroxyurea) |
−1.5 ± 6.9% | 172 | 10 |
| Mann-Whitney U P value |
0.02 | ||
Conclusive evidence that Sox2-expressing radial glial cells in the tectum are neural progenitor cells was provided by the lineage analysis of the in vivo time-lapse images of individual radial glial cells as they divided and differentiated (Figs. 6-11). This analysis established that radial glial cell proliferation favors the production of neurons at this developmental stage. From the 48 radial glia imaged on the first day after transfection, 73% (53/73) of the labeled cells at the final time points were neurons. A minority (4/48 or 8%) of the initial radial glia underwent symmetric divisions to generate 2 radial glial progeny (as shown in Fig. 9) while 40% (19/48) of radial glia underwent asymmetric divisions resulting in the production of a neuron and a radial glial cell (Figs. 6-8, 12). By these criteria radial glial cells in the optic tectum are multipotent progenitors that are capable of both symmetric divisions that expand the progenitor pool and asymmetric self-renewing, neurogenic divisions, in which one cell has a restricted neuronal cell fate, and the other retains its multipotent progenitor fate (Kriegstein and Alvarez-Buylla, 2009).
Control of the proportion of asymmetric versus symmetric cell divisions is clearly important for establishing the size of different brain regions and the future proliferation potential of the brain. Live imaging of labeled radial glial cells in slice culture preparations of developing neocortex has revealed similar distributions of symmetric and asymmetric division events to those we observe in tadpole CNS. Noctor et al. (2004) found that the majority of division events were asymmetric, producing only one neuron, and like our results, just ~10% of the mitotically active radial glial cells they imaged divided symmetrically to yield two radial glial progeny. In the developing chick tectum, the majority of retrovirus-labeled clones contained only a single radial glial cell, suggesting that asymmetric neurogenic divisions predominate in that system as well (Gray and Sanes, 1992). The presence of the radial process is often used as a diagnostic feature of radial glia, and could in principle provide information about the asymmetry of the division event. A key element of the in vivo time-lapse imaging data is the opportunity to identify morphological rearrangements of the radial process of progenitors during division events. Although classical studies suggested that the radial process of radial glial progenitors retracted from the pia prior to cell division, recent time-lapse imaging studies have shown that either the radial glial cell or the neuronal progeny can inherit the radial processes during cytokinesis. When the radial glial cell retains the radial process, neuronal progeny appear to bud from the cell body of the radial glial cell (Noctor et al., 2004). This type of cell division is akin to the examples we present in Figures 6 and 7, where the immature neuron begins to elaborate neuronal processes but the parent cell has maintained its radial process. By contrast, it has been reported that the neuron can inherit the radial process (Miyata et al., 2001), in which case the radial glial cell extends a new radial process. There are also examples where the radial process can split and be inherited by both progeny (Kosodo and Huttner, 2009), which may be similar to the time-lapse example we present in Figure 9, where a second process is visible before the nuclei of the presumptive progeny cells have fully separated. Together, these data indicate that the radial process is extremely dynamic during proliferative events and the presence of a radial process alone cannot be used to characterize progeny as neurons or radial glia.
Direct differentiation of radial glia into neurons
The most prevalent neurogenic event we observe was the direct differentiation of radial glia into neurons, which was the fate of half of the radial glial cells imaged (Figs. 6, 7, 10, 12). Furthermore, when radial glia divided asymmetrically to produce a neuron and another radial glial cell (numbered 11-29 in Fig. 12A), the glial cell subsequently differentiated into a neuron in 10/19 or 53% of cases. It is possible that cells identified initially as radial glial cells at the first time-point, which subsequently directly differentiated into neurons (numbered 30-48 in Figure 12A), were the progeny of a recent asymmetric division, similar to radial glial progeny of cells numbered 11-29 in Figure 12A.
Glial cells in general and radial glia in particular have been reported to transform into other cell types (Chanas-Sacre et al., 2000). Recently, convincing evidence of the direct differentiation of radial glial cells into astrocytes has been shown in adult hippocampus (Encinas et al., 2011), consistent with reports that radial glial cells are sparse or absent in the adult hippocampus and cortex in mammals (Hunter and Hatten, 1995; Levitt and Rakic, 1980; Mori et al., 2005; Voigt, 1989). Time-lapse data from cortical slices have shown that as radial glia undergo their morphological transformation into astrocytes, they retract from the ventricle toward the cortical plate and begin elaborating multiple astrocyte processes (Kriegstein and Alvarez-Buylla, 2009). This process of morphological transformation is different from the differentiation of radial glial into neurons that we observe in the tadpole tectum. The first signs of radial glial cell to neuron differentiation that we observed were a thinning of the radial process and transformation of the glial endfoot from a flattened palm-like structure into an axonal process with a filamentous growth cone (Figs. 5, 6, 7, 9). The growth cone then navigates over the surface of the tectum, and the radial process becomes the trailing axonal process. Subsequently, dendritic branches are elaborated from the major process near the cell body. Similar transformation of radial glial endfeet into growth cones has been inferred from Golgi-stained tissue in the chick, teleosts and lizards, where cells of this type are called Type 1 neuroblasts, or Shepherd’s Crook cells (Baez, 2003; Domesick and Morest, 1977; Puelles and Bandela, 1978; Snow and Robson, 1995; Wilm and Fritzsh, 1989).
In a previous study, we labeled cells by DiI iontophoresis and observed with time-lapse imaging that some cells with radial cell morphology differentiate directly into neurons (Wu and Cline, 2003), however, this method does not have the added cell-type specificity provided by the pSox2-bd::FP construct. It is also interesting to note that the DiI-labeled radial glial cells differentiated into neurons by a process in which dendrites are initiated from the cell body prior to extension of the axon (Wu and Cline, 2003). In this study we only occasionally identified this sequence of neuronal differentiation (i. e., Fig. 6), suggesting that two types of morphological differentiation exist for tectal cells. Because most of the cases of direct radial glia to neuronal differentiation that we report here occur in progeny of asymmetric divisions of Sox2-expressing progenitors, it is possible that neurons generated by a plan in which multiple dendrites extend from the cell body prior to axonogenesis derive from a different lineage.
It is noteworthy that in virtually all of the neuronal differentiation events, as soon as we can identify the neuron as a separate cell, we observe the neuron already has an initial polarity, either with respect to the presence of an axon or the asymmetric distribution of dendrites emerging from the cell body. The most common neurogenic event we observe in vivo is one in which the newly differentiated neuron essentially transformed the radial process into the axon. In cases in which a neuronal progeny appears to bud from the parent cell, the first processes extended from the neuron are already biased toward the basal aspect of the cell (for instance, examples in Figure 6, 7,8). Although it is possible that we missed exploratory neurites extending from the apical aspect of the neuron, our data suggest that newly differentiated neurons in vivo are polarized, and there is no evidence of an initial period of morphological symmetry as seen under in vitro conditions (Barnes and Polleux, 2009).
Protracted time course of cytokinesis in vivo
The time-lapse in vivo imaging used in this study has demonstrated that both the cytokinesis of radial glial progenitors and the differentiation of their progeny are protracted events that can take many hours. Using membrane-targeted GFP, we show that the accumulation of plasma membrane to establish two independent cell bodies during cytokinesis can take up to 11 hours. We designate the time at which we can identify two distinct nuclei as the time of cell division and find that progeny can retain radial glial morphology for anywhere from 2 to more than 40 hours before adopting a neuronal fate, determined by morphological criteria (Figure 12). Similarly, for the population of radial glial cells that differentiated directly into neurons, these cells maintained the radial glial cell morphology for imaging periods between 6 and 55 hours before differentiating into neurons. By contrast, in other cases progeny can be recognized as neurons at the same time point as the division event is detected. Of course, with time-lapse data, the division and differentiation events occurred at some point during the inter-imaging interval, so the precise timing of each of these events is unknown. The observation that progeny retain a radial glial morphology for many hours prior to establishing a neuronal phenotype suggests that this interval may be a period during which these cells respond to signals that affect cell fate plasticity. This idea is supported by the experiments in Figure 13 in which we show that short-term visual enhancement increases the proportion of cells with neuronal phenotype. It is likely that signals downstream of the short-term visual enhancement promote the differentiation of radial glial cell progeny into neurons.
Tectal radial glia and neuronal migration
Our time-lapse data show that radial glial cells are not a scaffold for neuronal migration at the developmental stages studied here. It is interesting to note that in embryonic cortex, radial glia are proliferative, but do not function as a migratory scaffold until later stages of cortical development (Rakic, 2003; Sidman and Rakic, 1973) when the cortex becomes laminated. Similarly, in both the optic tectum of older tadpoles after metamorphosis, and in superior colliculus, the mammalian homolog of the tectum, newly generated neurons reportedly migrate past older neurons to superficial tectal layers (Edwards et al., 1986; Schmidt and Roth, 1993; Straznicky and Gaze, 1972). This well-studied “inside-out” radial migration is thought to be an early step in sculpting the “radial unit” which may be relevant to the development of circuitry (Kriegstein and Alvarez-Buylla, 2009; Rakic, 1988; Yu et al., 2009). By contrast, much less is known about cellular and organizational events underlying the earlier stages of development which appear to progress without radial migration of intermediate progenitors and neurons (Rakic, 2003; Sidman and Rakic, 1973). Our data show that the cell bodies of radial glia and their progeny remain in close proximity with one another or are separated by less than one cell body width (~10 μm) throughout the 2-3 day duration of the experiment (Figs. 6-9). Although BrdU-labeled cells that have differentiated into neurons distribute throughout the tectal cell body layer over a period of 2-3 days at comparable stages (Sharma and Cline, 2010), our time-lapse imaging data provide no evidence for organized radial migration of newly generated neurons as they differentiate and mature. Rather, our data are consistent with previous studies indicating that new cells are added at the ventricular surface and ‘push’ older neurons dorso-laterally away from the ventricle and into the mature cell body layers of the tectum (Schmidt and Roth, 1993; Straznicky and Gaze, 1972; Wu et al., 1999).
Newly generated cells in the tadpole tectum are rapidly integrated into a functional retinotectal circuit (Aizenman et al., 2003; Bestman and Cline, 2008; Chiu et al., 2008). Granule neurons generated postnatally in the mammalian hippocampal dentate gyrus also integrate into a pre-existing functional circuit. There are two interesting similarities in the developmental strategies used by newly born neurons in the tadpole optic tectum and mammalian dentate gyrus: in both systems, newly-generated neurons remain in close contact with radial glial cells. Electron microscopy data show that newly generated neurons in the subgranular zone of the dentate gyrus are wrapped in the non-radial processes of the glia (Shapiro et al., 2005). In addition, the immature granule cells first differentiate by sending a slender primary neurite along the neighboring radial glial process to reach the superficial molecular layer, and only later do the granule cells commence their outward migration. Similar to their role in the embryonic development of hamster superior colliculus (Wu et al., 1995), we find that radial glial cells in the tadpole tectum may guide axons into the superficial tectal neuropil (Figs. 6-8). Because of their close association with one another, distinguishing radial glial parent cells and their progeny is difficult (Kosodo and Huttner, 2009) even when aided with GFP proteins that are directed to the membrane (Fig. 5), but our time-lapse images show faint axon-like processes in the superficial tectum that are entwined with the glial cells’ radial fibers (Figs. 6, 7, 8).
Enhanced visual experience decreases rates of proliferation and stimulates neuronal differentiation
Our previous experiments have shown that increasing and decreasing the visual experience during this same developmental period has profound consequences for retinotectal synaptic connectivity. Visual experience enhances the growth and dynamics of tectal cell dendritic arbors (Sin et al., 2002) while consolidating and stabilizing retinal ganglion cell inputs in the tectum (Ruthazer et al., 2006). In addition, Aizenman and Cline (2007) showed that providing short-term visual stimulus for just 4 hours increased retinotectal synaptogenesis and strengthened pre-existing retinotectal synapses. In reciprocal experiments, visual deprivation prevents synaptic maturation and experience-dependent synapse elimination (Li et al., 2011), which is also part of the activity-dependent program of retinotectal circuit development. Our in vivo imaging data reported here show that the rate of cell proliferation in the tectum is lower in tadpoles that receive the short-term visual stimulus, and the animals with visual experience have a higher rate of neuronal differentiation (Fig. 13B). These findings are consistent with our previous study showing that visual deprivation expands the progenitor pool and that visual experience increases the rate at which progenitors exit the cell cycle and express neuronal markers (Sharma and Cline, 2010). These data indicate that visual experience initiates several coincident parallel events in the developing retinotectal system: as a result of increased visual experience radial glial progenitors generate more neurons, differentiated tectal cells both increase their rate of dendritic arbor elaboration and enhance their synaptic connectivity within the retinotectal circuit, and finally retinal axons strengthen their inputs with tectal neuronal dendrites. Together, these data suggest that visual experience increases the generation and integration of new neurons into the visual circuit.
The potential mechanisms by which visual experience affects progenitors are not clear. The membrane potential fluctuates in mitotically active cells and its variations are thought to regulate the transition between phases of the cell cycle (Blackiston et al., 2009). Similarly, the spatial and temporal pattern of calcium signaling in the developing nervous system may control the development and differentiation of neurons (Webb and Miller, 2003). Recently, Trembley et al. (2009) reported that tectal radial glia in the Xenopus tadpole tectum show transient increases in cytosolic calcium in response to visual stimuli similar to that used here. It is tempting to hypothesize that in the development of the tectum, neuronal number can be varied in response to the demands of the tadpole’s visual experience.
Lineage analysis of CNS progenitors
One scheme of CNS progenitor fates is as follows: the development of the CNS in the embryonic stages is thought to be initiated by symmetric division of neurepithelial cells lining the brain ventricle. As the neural tube develops, neurepithelial cells undergo self-regenerating asymmetric divisions to generate a radial glial cell and a neurepithelial cell. The radial glial progenitor cell population is also thought to expand through symmetric divisions and to undergo regenerating asymmetric divisions which generate both a radial glial progeny and neuronal progeny. Finally, radial glia are thought to terminally differentiate into neurons or astrocytes (Kriegstein and Alvarez-Buylla, 2009). Despite the wide-spread support for this scheme, relatively few of the transitional events in lineages have been observed directly. Our lineage analysis, based on in vivo image of individual radial glial cells and their progeny in stage 47 tadpoles, provides evidence for some key aspects of this scheme and adds new insights regarding the dynamic regulation of neural progenitor cells. We find that Sox2-expressing radial glial cells are neural progenitors. They undergo both symmetric divisions and asymmetric neurogenic divisions. About 40% of the radial glial cells divide asymmetrically to generate a neuron and a cell with the radial glial morphology. Although this asymmetric division would be considered regenerative according to the above scheme, if the progeny were phenotyped for instance by antibody labeling shortly after the division, we find that almost half of the radial glial cell progeny in this type of division ultimately differentiate into neurons, after a delay of 4-30 hours. Experiments using immunolabeling to identify cells and determine their phenotype, rather than time-lapse imaging of individual neurons, would be blind to this result and its statistical prevalence.
Under the conditions of our experiments, Sox2-expressing radial glial cells in the stage 47 optic tectum undergo relatively few symmetric divisions that would expand the progenitor pool. This poses a dilemma because it is well-documented that cell proliferation continues in the optic tectum throughout tadpole stages of development (Straznicky and Gaze, 1972). Although it is possible that a population of radial glia that does not express Sox2 is responsible for maintaining an active progenitor pool, our new data suggest that the Sox2-expressing progenitor pool may change its proliferative activity in response to signals from the developing neural circuit. Symmetric divisions by Sox2-expressing radial glial progenitors are more prevalent at earlier stages of development, when overall rates of cell proliferation in the tectum are higher (Sharma and Cline, 2010) and when visually-driven activity in the tectal circuit is lower (Pratt et al., 2008). Furthermore, visual deprivation for 2 days in stage 48 tadpoles expands the progenitor pool, suggesting that activity-dependent signals control the fate of radial glial progeny and thereby control whether divisions are symmetric or asymmetric and neurogenic (Sharma and Cline, 2010). Here we show that visual experience decreases cell proliferation and increases neuronal differentiation. These data indicate that the progenitor pool is plastic and that cell fate decisions are subject to regulation by signals from the developing circuit.
Supplementary Material
Acknowledgements
We thank Melissa Lau for generating the pSox2-bp::mGFP construct and members of the Cline Lab for constructive discussion.
Support: NIH EY011261, Dart Neuroscience, LLC, the Nancy Lurie Marks Family Foundation and an endowment from the Hahn Family Foundation.
Abbreviations
- Sox2
sex-determining region Y-box 2
- Oct3-4
Octamer binding protein 3-4
- FGF4
fibroblast growth factor 4
- FP
fluorescent protein
- nls
nuclear localization signal
- GFP
green fluorescent protein
- mGFP
membrane-targeted GFP
- RFP
red fluorescent protein
- Gal4
regulatory protein Gal4
- UAS
upstream activator sequence
- bd
binding domain
- CMV
cytomegalovirus
- HSD
honestly significant difference
- DiI
1,1′ - Dioctadecyl - 3,3,3′,3′ - tetramethylindocarbocyanine iodide
REFERENCES
- Aizenman CD, Akerman CJ, Jensen KR, Cline HT. Visually driven regulation of intrinsic neuronal excitability improves stimulus detection in vivo. Neuron. 2003;39(5):831–842. doi: 10.1016/s0896-6273(03)00527-0. [DOI] [PubMed] [Google Scholar]
- Aizenman CD, Cline HT. Enhanced visual activity in vivo forms nascent synapses in the developing retinotectal projection. J Neurophysiol. 2007;97(4):2949–2957. doi: 10.1152/jn.00452.2006. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla A, Garcia-Verdugo JM, Tramontin AD. A unified hypothesis on the lineage of neural stem cells. Nat Rev Neurosci. 2001;2(4):287–293. doi: 10.1038/35067582. [DOI] [PubMed] [Google Scholar]
- Ambrosetti D-C, Scholer HR, Dailey L, Basilico C. Modulation of the Activity of Multiple Transcriptional Activation Domains by the DNA Binding Domains Mediates the Synergistic Action of Sox2 and Oct-3 on the Fibroblast Growth Factor-4 Enhancer. J Biol Chem. 2000;275(30):23387–23397. doi: 10.1074/jbc.M000932200. [DOI] [PubMed] [Google Scholar]
- Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci U S A. 2002;99(20):12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer TC, Jin J, Casey ES. Interaction of Sox1, Sox2, Sox3 and Oct4 during primary neurogenesis. Developmental Biology. 2011;350(2):429–440. doi: 10.1016/j.ydbio.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baez J. Neuronal differentiation patterns in the optic tectum of the lizard Gallotia galloti. Brain Research. 2003;975(1-2):48–65. doi: 10.1016/s0006-8993(03)02586-1. [DOI] [PubMed] [Google Scholar]
- Barnes AP, Polleux F. Establishment of axon-dendrite polarity in developing neurons. Annu Rev Neurosci. 2009;32:347–381. doi: 10.1146/annurev.neuro.31.060407.125536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentivoglio M, Mazzarello P. The history of radial glia. Brain Res Bull. 1999;49(5):305–315. doi: 10.1016/s0361-9230(99)00065-9. [DOI] [PubMed] [Google Scholar]
- Bestman JE, Cline HT. The RNA binding protein CPEB regulates dendrite morphogenesis and neuronal circuit assembly in vivo. Proc Natl Acad Sci U S A. 2008;105(51):20494–20499. doi: 10.1073/pnas.0806296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackiston DJ, McLaughlin KA, Levin M. Bioelectric controls of cell proliferation: ion channels, membrane voltage and the cell cycle. Cell Cycle. 2009;8(21):3519–3528. doi: 10.4161/cc.8.21.9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti C, Surace EM. Mouse Embryonic Retina Delivers Information Controlling Cortical Neurogenesis. PLoS One. 2010;5(12):e15211. doi: 10.1371/journal.pone.0015211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1–3 activity. Nature Neuroscience. 2003;6(11):1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- Caron SJ, Prober D, Choy M, Schier AF. In vivo birthdating by BAPTISM reveals that trigeminal sensory neuron diversity depends on early neurogenesis. Development. 2008;135(19):3259–3269. doi: 10.1242/dev.023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepko CL, Austin CP, Walsh C, Ryder EF, Halliday A, Fields-Berry S. Studies of cortical development using retrovirus vectors. Cold Spring Harb Symp Quant Biol. 1990;55:265–278. doi: 10.1101/sqb.1990.055.01.029. [DOI] [PubMed] [Google Scholar]
- Chae J, Zimmerman LB, Grainger RM. Inducible control of tissue-specific transgene expression in Xenopus tropicalis transgenic lines. Mech Dev. 2002;117(1-2):235–241. doi: 10.1016/s0925-4773(02)00219-8. [DOI] [PubMed] [Google Scholar]
- Chakravarthy H, Boer B, Desler M, Mallanna SK, McKeithan TW, Rizzino A. Identification of DPPA4 and other genes as putative Sox2:Oct 3/4 target genes using a combination of in silico analysis and transcription based assays. Journal of Cellular Physiology. 2008;216(3):651–662. doi: 10.1002/jcp.21440. [DOI] [PubMed] [Google Scholar]
- Chanas-Sacre G, Rogister B, Moonen G, Leprince P. Radial glia phenotype: origin, regulation, and transdifferentiation. J Neurosci Res. 2000;61(4):357–363. doi: 10.1002/1097-4547(20000815)61:4<357::AID-JNR1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Chen SX, Tari PK, She K, Haas K. Neurexin-neuroligin cell adhesion complexes contribute to synaptotropic dendritogenesis via growth stabilization mechanisms in vivo. Neuron. 2010;67(6):967–983. doi: 10.1016/j.neuron.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Chiu S-L, Chen C-M, Cline HT. Insulin Receptor Signaling Regulates Synapse Number, Dendritic Plasticity, and Circuit Function In Vivo. Neuron. 2008;58(5):708–719. doi: 10.1016/j.neuron.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MR, Götz M, Berninger B. What determines neurogenic competence in glia? Brain Research Reviews. 2010;63(1-2):47–59. doi: 10.1016/j.brainresrev.2010.01.002. [DOI] [PubMed] [Google Scholar]
- D’Amour KA, Gage FH. Genetic and functional differences between multipotent neural and pluripotent embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(Suppl 1):11866–11872. doi: 10.1073/pnas.1834200100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent JA, Polson AG, Klymkowsky MW. A whole-mount immunocytochemical analysis of the expression of the intermediate filament protein vimentin in Xenopus. Development. 1989;105(1):61–74. doi: 10.1242/dev.105.1.61. [DOI] [PubMed] [Google Scholar]
- Domesick VB, Morest DK. Migration and differentiation of shepherd’s crook cells in the optic tectum of the chick embryo. Neuroscience. 1977;2(3):477–491. doi: 10.1016/0306-4522(77)90012-4. [DOI] [PubMed] [Google Scholar]
- Dong W, Lee RH, Xu H, Yang S, Pratt KG, Cao V, Song YK, Nurmikko A, Aizenman CD. Visual Avoidance in Xenopus Tadpoles Is Correlated With the Maturation of Visual Responses in the Optic Tectum. Journal of Neurophysiology. 2008;101(2):803–815. doi: 10.1152/jn.90848.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MA, Caviness JVS, Schneider GE. Development of the Cell and Fiber Lamination in the Mouse Superior Colliculus. Journal of Comparative Neurology. 1986;248:395–409. doi: 10.1002/cne.902480308. [DOI] [PubMed] [Google Scholar]
- Encinas JM, Michurina TV, Peunova N, Park JH, Tordo J, Peterson DA, Fishell G, Koulakov A, Enikolopov G. Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell. 2011;8(5):566–579. doi: 10.1016/j.stem.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald RC, Van Keuren-Jensen KR, Aizenman CD, Cline HT. Roles of NR2A and NR2B in the development of dendritic arbor morphology in vivo. J Neurosci. 2008;28(4):850–861. doi: 10.1523/JNEUROSCI.5078-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas LM, Huttner WB. The cell biology of neural stem and progenitor cells and its significance for their proliferation versus differentiation during mammalian brain development. Curr Opin Cell Biol. 2008;20(6):707–715. doi: 10.1016/j.ceb.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Finlay BL, Hersman MN, Darlington RB. Patterns of vertebrate neurogenesis and the paths of vertebrate evolution. Brain Behav Evol. 1998;52(4-5):232–242. doi: 10.1159/000006566. [DOI] [PubMed] [Google Scholar]
- Font E, Desfilis E, Perez-Canellas MM, Garcia-Verdugo JM. Neurogenesis and neuronal regeneration in the adult reptilian brain. Brain Behav Evol. 2001;58(5):276–295. doi: 10.1159/000057570. [DOI] [PubMed] [Google Scholar]
- Gao WO, Heintz N, Hatten ME. Cerebellar granule cell neurogenesis is regulated by cell-cell interactions in vitro. Neuron. 1991;6(5):705–715. doi: 10.1016/0896-6273(91)90168-y. [DOI] [PubMed] [Google Scholar]
- Götz M, Huttner WB. The cell biology of neurogenesis. Nature Reviews Molecular Cell Biology. 2005;6(10):777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 Functions to Maintain Neural Progenitor Identity. Neuron. 2003;39(5):749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Gray GE, Sanes JR. Lineage of radial glia in the chicken optic tectum. Development. 1992;114(1):271–283. doi: 10.1242/dev.114.1.271. [DOI] [PubMed] [Google Scholar]
- Guo Z, Packard A, Krolewski RC, Harris MT, Manglapus GL, Schwob JE. Expression of Pax6 and Sox2 in adult olfactory epithelium. The Journal of Comparative Neurology. 2010;518(21):4395–4418. doi: 10.1002/cne.22463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas K, Jensen K, Sin WC, Foa L, Cline HT. Targeted electroporation in Xenopus tadpoles in vivo-from single cells to the entire brain. Differentiation. 2002;70(4-5):148–154. doi: 10.1046/j.1432-0436.2002.700404.x. [DOI] [PubMed] [Google Scholar]
- Haas K, Li J, Cline HT. AMPA receptors regulate experience-dependent dendritic arbor growth in vivo. PNAS. 2006;103(32):12127–12131. doi: 10.1073/pnas.0602670103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris WA, Hartenstein V. Neuronal determination without cell division in Xenopus embryos. Neuron. 1991;6(4):499–515. doi: 10.1016/0896-6273(91)90053-3. [DOI] [PubMed] [Google Scholar]
- Hartley KO, Nutt SL, Amaya E. Targeted gene expression in transgenic Xenopus using the binary Gal4-UAS system. Proc Natl Acad Sci U S A. 2002;99(3):1377–1382. doi: 10.1073/pnas.022646899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta K, Tsujii H, Omura T. Cell tracking using a photoconvertible fluorescent protein. Nat Protoc. 2006;1(2):960–967. doi: 10.1038/nprot.2006.96. [DOI] [PubMed] [Google Scholar]
- Hunter KE, Hatten ME. Radial glial cell transformation to astrocytes is bidirectional: regulation by a diffusible factor in embryonic forebrain. Proc Natl Acad Sci U S A. 1995;92(6):2061–2065. doi: 10.1073/pnas.92.6.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai JH, Wang X, Shi S-H. Kaede-Centrin1 Labeling of Mother and Daughter Centrosomes in Mammalian Neocortical Neural Progenitors. 2010. [DOI] [PMC free article] [PubMed]
- Javaherian A, Kriegstein A. A stem cell niche for intermediate progenitor cells of the embryonic cortex. Cereb Cortex. 2009;19(Suppl 1):i70–77. doi: 10.1093/cercor/bhp029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei Y, Inagaki N, Nishizawa M, Tsutsumi O, Taketani Y, Inagaki M. Visualization of mitotic radial glial lineage cells in the developing rat brain by Cdc2 kinase-phosphorylated vimentin. Glia. 1998;23(3):191–199. doi: 10.1002/(sici)1098-1136(199807)23:3<191::aid-glia2>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Kaslin J, Ganz J, Brand M. Proliferation, neurogenesis and regeneration in the non-mammalian vertebrate brain. Philos Trans R Soc Lond B Biol Sci. 2008;363(1489):101–122. doi: 10.1098/rstb.2006.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komitova M, Eriksson P. Sox-2 is expressed by neural progenitors and astroglia in the adult rat brain. Neuroscience Letters. 2004;369(1):24–27. doi: 10.1016/j.neulet.2004.07.035. [DOI] [PubMed] [Google Scholar]
- Kopp JL, Ormsbee BD, Desler M, Rizzino A. Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. STEM CELLS. 2008;26(4):903–911. doi: 10.1634/stemcells.2007-0951. [DOI] [PubMed] [Google Scholar]
- Kosodo Y, Huttner WB. Basal process and cell divisions of neural progenitors in the developing brain. Dev Growth Differ. 2009;51(3):251–261. doi: 10.1111/j.1440-169X.2009.01101.x. [DOI] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt P, Rakic P. Immunoperoxidase localization of glial fibrillary acidic protein in radial glial cells and astrocytes of the developing rhesus monkey brain. J Comp Neurol. 1980;193(3):815–840. doi: 10.1002/cne.901930316. [DOI] [PubMed] [Google Scholar]
- Li J, Erisir A, Cline H. In Vivo Time-Lapse Imaging and Serial Section Electron Microscopy Reveal Developmental Synaptic Rearrangements. Neuron. 2011;69(2):273–286. doi: 10.1016/j.neuron.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang Y, Chiu SL, Cline HT. Membrane targeted horseradish peroxidase as a marker for correlative fluorescence and electron microscopy studies. Front Neural Circuits. 2010;4:6. doi: 10.3389/neuro.04.006.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XF, Tari PK, Haas K. PKM zeta restricts dendritic arbor growth by filopodial and branch stabilization within the intact and awake developing brain. J Neurosci. 2009;29(39):12229–12235. doi: 10.1523/JNEUROSCI.2842-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Garcia C, Molowny A, Martinez-Guijarro FJ, Blasco-Ibanez JM, Luis de la Iglesia JA, Bernabeu A, Garcia-Verdugo JM. Lesion and regeneration in the medial cerebral cortex of lizards. Histol Histopathol. 1992;7(4):725–746. [PubMed] [Google Scholar]
- LoTurco JJ, Owens DF, Heath MJ, Davis MB, Kriegstein AR. GABA and glutamate depolarize cortical progenitor cells and inhibit DNA synthesis. Neuron. 1995;15(6):1287–1298. doi: 10.1016/0896-6273(95)90008-x. [DOI] [PubMed] [Google Scholar]
- Malatesta P, Hack MA, Hartfuss E, Kettenmann H, Klinkert W, Kirchhoff F, Gotz M. Neuronal or glial progeny: regional differences in radial glia fate. Neuron. 2003;37(5):751–764. doi: 10.1016/s0896-6273(03)00116-8. [DOI] [PubMed] [Google Scholar]
- Margotta V, Filoni S, Del Vecchio P. Degenerative and regenerative phenomena in brain heterotopic homoplastic transplants of adult Triturus carnifex (Urodele Amphibians) J Hirnforsch. 1992;33(1):3–9. [PubMed] [Google Scholar]
- Mathews EA, Morgenstern NA, Piatti VC, Zhao C, Jessberger S, Schinder AF, Gage FH. A Distinctive layering pattern of mouse dentate granule cells is generated by developmental and adult neurogenesis. The Journal of Comparative Neurology. 2010;518(22):4479–4490. doi: 10.1002/cne.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean DL, Fetcho JR. Spinal interneurons differentiate sequentially from those driving the fastest swimming movements in larval zebrafish to those driving the slowest ones. J Neurosci. 2009;29(43):13566–13577. doi: 10.1523/JNEUROSCI.3277-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Okano H, Ogawa M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 2001;31(5):727–741. doi: 10.1016/s0896-6273(01)00420-2. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131(13):3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- Molowny A, Nacher J, Lopez-Garcia C. Reactive neurogenesis during regeneration of the lesioned medial cerebral cortex of lizards. Neuroscience. 1995;68(3):823–836. doi: 10.1016/0306-4522(95)00201-s. [DOI] [PubMed] [Google Scholar]
- Morest DK, Silver J. Precursors of neurons, neuroglia, and ependymal cells in the CNS: What are they? Where are they from? How do they get where they are going? Glia. 2003;43(1):6–18. doi: 10.1002/glia.10238. [DOI] [PubMed] [Google Scholar]
- Mori T, Buffo A, Gotz M. The novel roles of glial cells revisited: the contribution of radial glia and astrocytes to neurogenesis. Curr Top Dev Biol. 2005;69:67–99. doi: 10.1016/S0070-2153(05)69004-7. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin); a systemical and chronological survey of the development from fertilized egg till the end of metamorphosis. North-holland Publishing Co; Amsterdam: 1956. p. 243. 1956. illus. ; 228 cm. p. [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409(6821):714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7(2):136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Peunova N, Scheinker V, Cline H, Enikolopov G. Nitric Oxide Is an Essential Negative Regulator of Cell Proliferation in Xenopus Brain. J Neurosci. 2001;21(22):8809–8818. doi: 10.1523/JNEUROSCI.21-22-08809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny L, Placzek M. SOX genes and neural progenitor identity. Curr Opin Neurobiol. 2005;15(1):7–13. doi: 10.1016/j.conb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Pinto L, Gotz M. Radial glial cell heterogeneity—The source of diverse progeny in the CNS. Progress in Neurobiology. 2007;83(1):2–23. doi: 10.1016/j.pneurobio.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Pratt KG, Dong W, Aizenman CD. Development and spike timing–dependent plasticity of recurrent excitation in the Xenopus optic tectum. Nature Neuroscience. 2008;11(4):467–475. doi: 10.1038/nn2076. [DOI] [PubMed] [Google Scholar]
- Puelles L, Bandela MC. Differentiation of neuroblasts in the chick optic tectum up to eight days of incubation: a golgi study. Neuroscience. 1978;3:307–325. doi: 10.1016/0306-4522(78)90079-9. [DOI] [PubMed] [Google Scholar]
- Putz U, Harwell C, Nedivi E. Soluble CPG15 expressed during early development rescues cortical progenitors from apoptosis. Nat Neurosci. 2005;8(3):322–331. doi: 10.1038/nn1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Specification of cerebral cortical areas. Science. 1988;241(4862):170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- Rakic P. Elusive radial glial cells: Historical and evolutionary perspective. Glia. 2003;43(1):19–32. doi: 10.1002/glia.10244. [DOI] [PubMed] [Google Scholar]
- Raymond PA, Easter SS., Jr. Postembryonic growth of the optic tectum in goldfish. I. Location of germinal cells and numbers of neurons produced. J Neurosci. 1983;3(5):1077–1091. doi: 10.1523/JNEUROSCI.03-05-01077.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond PA, Easter SS, Jr., Burnham JA, Powers MK. Postembryonic growth of the optic tectum in goldfish. II. Modulation of cell proliferation by retinal fiber input. J Neurosci. 1983;3(5):1092–1099. doi: 10.1523/JNEUROSCI.03-05-01092.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid CB, Liang I, Walsh C. Systematic widespread clonal organization in cerebral cortex. Neuron. 1995;15(2):299–310. doi: 10.1016/0896-6273(95)90035-7. [DOI] [PubMed] [Google Scholar]
- Rizzino A. Sox2 and Oct-3/4: a versatile pair of master regulators that orchestrate the self-renewal and pluripotency of embryonic stem cells. Wiley Interdiscip Rev Syst Biol Med. 2009;1(2):228–236. doi: 10.1002/wsbm.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CD, Harafuji N, Archer T, Cunningham DD, Casey ES. Xenopus Sox3 activates sox2 and geminin and indirectly represses Xvent2 expression to induce neural progenitor formation at the expense of non-neural ectodermal derivatives. Mech Dev. 2009;126(1-2):42–55. doi: 10.1016/j.mod.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JL, Martinez S, Shimamura K, Puelles L. The embryonic vertebrate forebrain: the prosomeric model. Science. 1994;266(5185):578–580. doi: 10.1126/science.7939711. [DOI] [PubMed] [Google Scholar]
- Ruthazer ES, Li J, Cline HT. Stabilization of axon branch dynamics by synaptic maturation. J Neurosci. 2006;26(13):3594–3603. doi: 10.1523/JNEUROSCI.0069-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez AL, Matthews BJ, Meynard MM, Hu B, Javed S, Cohen Cory S. BDNF increases synapse density in dendrites of developing tectal neurons in vivo. Development. 2006;133(13):2477–2486. doi: 10.1242/dev.02409. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Roth G. Patterns of cellular proliferation and migration in the developing tectum mesencephali of the frog Rana temporaria and the salamander Pleurodeles waltl. Cell and Tissue Research. 1993;272(2):273–287. doi: 10.1007/BF00302734. [DOI] [PubMed] [Google Scholar]
- Schwartz N, Schohl A, Ruthazer ES. Neural activity regulates synaptic properties and dendritic structure in vivo through calcineurin/NFAT signaling. Neuron. 2009;62(5):655–669. doi: 10.1016/j.neuron.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Schwartz N, Schohl A, Ruthazer ES. Activity-dependent transcription of BDNF enhances visual acuity during development. Neuron. 2011;70(3):455–467. doi: 10.1016/j.neuron.2011.02.055. [DOI] [PubMed] [Google Scholar]
- Shapiro LA, Korn MJ, Shan Z, Ribak CE. GFAP-expressing radial glia-like cell bodies are involved in a one-to-one relationship with doublecortin-immunolabeled newborn neurons in the adult dentate gyrus. Brain Res. 2005;1040(1-2):81–91. doi: 10.1016/j.brainres.2005.01.098. [DOI] [PubMed] [Google Scholar]
- Sharma P, Cline HT. Visual activity regulates neural progenitor cells in developing Xenopus CNS through musashi1. Neuron. 2010;68(3):442–455. doi: 10.1016/j.neuron.2010.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Da Silva JS, He H, Cline HT. Type A GABA-receptor-dependent synaptic transmission sculpts dendritic arbor structure in Xenopus tadpoles in vivo. J Neurosci. 2009;29(15):5032–5043. doi: 10.1523/JNEUROSCI.5331-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, McKeown CR, Demas JA, Cline HT. Inhibition to Excitation Ratio Regulates Visual System Responses and Behavior in vivo. J Neurophysiol. 2011 doi: 10.1152/jn.00641.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidman RL, Rakic P. Neuronal migration, with special reference to developing human brain: a review. Brain Res. 1973;62(1):1–35. doi: 10.1016/0006-8993(73)90617-3. [DOI] [PubMed] [Google Scholar]
- Sild M, Ruthazer ES. Radial glia: progenitor, pathway, and partner. Neuroscientist. 2011;17(3):288–302. doi: 10.1177/1073858410385870. [DOI] [PubMed] [Google Scholar]
- Sin WC, Haas K, Ruthazer ES, Cline HT. Dendrite growth increased by visual activity requires NMDA receptor and Rho GTPases. Nature. 2002;419(6906):475–480. doi: 10.1038/nature00987. [DOI] [PubMed] [Google Scholar]
- Snow RL, Robson JA. Migration and differentiation of neurons in the retina and optic tectum of the chick. Exp Neurol. 1995;134(1):13–24. doi: 10.1006/exnr.1995.1032. [DOI] [PubMed] [Google Scholar]
- Stevenson JA, Yoon MG. Mitosis of radial glial cells in the optic tectum of the adult goldfish. Journal of Neuroscience. 1981;1(8):862–875. doi: 10.1523/JNEUROSCI.01-08-00862.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straznicky K, Gaze R. The development of the tectum in Xenopus laevis: an autoradiographic study. Journal of Embryology and Experimental Morphology. 1972;28(1):87–115. [PubMed] [Google Scholar]
- Straznicky K, Gaze RM. The growth of the retina in Xenopus laevis: an autoradiographic study. J Embryol Exp Morphol. 1971;26(1):67–79. [PubMed] [Google Scholar]
- Tavazoie M, Van der Veken L, Silva-Vargas V, Louissaint M, Colonna L, Zaidi B, Garcia-Verdugo JM, Doetsch F. A specialized vascular niche for adult neural stem cells. Cell Stem Cell. 2008;3(3):279–288. doi: 10.1016/j.stem.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terskikh A, Fradkov A, Ermakova G, Zaraisky A, Tan P, Kajava AV, Zhao X, Lukyanov S, Matz M, Kim S, Weissman I, Siebert P. “Fluorescent timer”: protein that changes color with time. Science. 2000;290(5496):1585–1588. doi: 10.1126/science.290.5496.1585. [DOI] [PubMed] [Google Scholar]
- Tremblay M, Fugere V, Tsui J, Schohl A, Tavakoli A, Travencolo BAN, Costa LdF, Ruthazer ES. Regulation of Radial Glial Motility by Visual Experience. Journal of Neuroscience. 2009;29(45):14066–14076. doi: 10.1523/JNEUROSCI.3542-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Keuren-Jensen KR, Cline HT. Homer proteins shape Xenopus optic tectal cell dendritic arbor development in vivo. Developmental Neurobiology. 2008;68(11):1315–1324. doi: 10.1002/dneu.20659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhusha VV, Otsuna H, Awasaki T, Oda H, Tsukita S, Ito K. An enhanced mutant of red fluorescent protein DsRed for double labeling and developmental timer of neural fiber bundle formation. J Biol Chem. 2001;276(32):29621–29624. doi: 10.1074/jbc.C100200200. [DOI] [PubMed] [Google Scholar]
- Voigt T. Development of glial cells in the cerebral wall of ferrets: direct tracing of their transformation from radial glia into astrocytes. J Comp Neurol. 1989;289(1):74–88. doi: 10.1002/cne.902890106. [DOI] [PubMed] [Google Scholar]
- Walsh C, Reid C. Cell lineage and patterns of migration in the developing cortex. Ciba Found Symp. 1995;193:21–40. doi: 10.1002/9780470514795.ch2. discussion 59-70. [DOI] [PubMed] [Google Scholar]
- Wang TW, Stromberg GP, Whitney JT, Brower NW, Klymkowsky MW, Parent JM. Sox3 expression identifies neural progenitors in persistent neonatal and adult mouse forebrain germinative zones. The Journal of Comparative Neurology. 2006;497(1):88–100. doi: 10.1002/cne.20984. [DOI] [PubMed] [Google Scholar]
- Webb SE, Miller AL. Calcium signalling during embryonic development. Nat Rev Mol Cell Biol. 2003;4(7):539–551. doi: 10.1038/nrm1149. [DOI] [PubMed] [Google Scholar]
- Weissman TA, Riquelme PA, Ivic L, Flint AC, Kriegstein AR. Calcium waves propagate through radial glial cells and modulate proliferation in the developing neocortex. Neuron. 2004;43(5):647–661. doi: 10.1016/j.neuron.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Wilm C, Fritzsh B. Development of tectal neurons in the perciform teleost, Haplochromis burtoni. A Golgi study. Developmental Brain Research. 1989;47:35–52. doi: 10.1016/0165-3806(89)90106-5. [DOI] [PubMed] [Google Scholar]
- Wu DY, Jhaveri S, Schneider GE. Glial environment in the developing superior colliculus of hamsters in relation to the timing of retinal axon ingrowth. J Comp Neurol. 1995;358(2):206–218. doi: 10.1002/cne.903580204. [DOI] [PubMed] [Google Scholar]
- Wu GY, Cline HT. Time-lapse in vivo imaging of the morphological development of Xenopus optic tectal interneurons. J Comp Neurol. 2003;459(4):392–406. doi: 10.1002/cne.10618. [DOI] [PubMed] [Google Scholar]
- Wu GY, Zou DJ, Rajan I, Cline H. Dendritic Dynamics In Vivo Change during Neuronal Maturation. J Neurosci. 1999;19(11):4472–4483. doi: 10.1523/JNEUROSCI.19-11-04472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M. Intermediate filament proteins define different glial subpopulations. J Neurosci Res. 2001;63(3):284–289. doi: 10.1002/1097-4547(20010201)63:3<284::AID-JNR1022>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Yu Y-C, Bultje RS, Wang X, Shi S-H. Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature. 2009;458(7237):501–504. doi: 10.1038/nature07722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D-J, Cline HT. Postsynaptic calcium/calmodulin-dependent protein kinase II is required to limit elaboration of presynaptic and postsynaptic neuronal arbors. Journal of Neuroscience. 1999;19(20):8909–8918. doi: 10.1523/JNEUROSCI.19-20-08909.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupanc G. Adult neurogenesis and neuronal regeneration in the brain of teleost fish. Journal of Physiology-Paris. 2008;102(4-6):357–373. doi: 10.1016/j.jphysparis.2008.10.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.