Copy number variants (CNVs) are major contributors to genetic disorders1. We have dissected a region on 16p11.2 encompassing 29 genes that confers susceptibility to neurocognitive defects when deleted or duplicated2,3. Overexpression of each human transcript in zebrafish embryos identified KCTD13 as the sole message capable of inducing the microcephaly phenotype associated with the 16p11.2 duplication (dup)2–5, while suppression of the same locus yielded the macrocephalic phenotype associated with the 16p11.2 deletion (del)5,6, capturing the mirror phenotypes of humans. Analyses of zebrafish and mouse embryos suggest that microcephaly is caused by decreased proliferation of neuronal progenitors, with concomitant increase in apoptosis in the developing brain, whereas macrocephaly arises by increased proliferation and no changes in apoptosis. A role for KCTD13 dosage changes is consistent with autism in both a recently reported family with a reduced 16p11.2 deletion and a subject reported here with a complex 16p11.2 rearrangement involving de novo structural alteration of KCTD13. Our data suggest that KCTD13 is a major driver for the neurodevelopmental phenotypes associated with the 16p11.2 CNV, reinforce the notion that one or a small number of transcripts within a CNV can underpin clinical phenotypes, and offer an efficient route to identifying dosage sensitive loci.
Copy number changes have emerged in recent years as a significant source of genetic variation contributing to the human genetic disease risk1. In addition to genomic disorders such as Charcot-Marie Tooth, DiGeorge syndrome and others7,8, in which large deletions and duplications represent penetrant alleles for discrete syndromic phenotypes, recent advances have highlighted the contribution of such genomic events in a broad range of both common and rare traits (DECIPHER Consortium, http://decipher.sanger.co.uk). Systematic surveys of neurodevelopmental disorders have uncovered an especially high incidence of both inherited and de novo CNVs that can confer either causality or susceptibility9–12. For example, deletions in 1q21.1 and 15q13.3 have been associated with schizophrenia (SCZ), whereas duplications in 15q11-q13 and 7q22-q31 have been associated with autism spectrum disorder (ASD; see review1).
A 600kb deletion on 16p11.2, encompassing 29 annotated genes, has been associated significantly and reproducibly with a range of neurocognitive defects, including epilepsy, autism and ASD2, while the reciprocal duplication has been associated with autism and SCZ3. In addition, extended phenotypic analyses of patients with such genomic lesions have revealed strong mirroring co-morbidities: the common 16p11.2 deletion is associated with pediatric neurodevelopmental disorders including autism, diabetes-independent obesity5 and macrocephaly6, while the reciprocal duplication is associated with both autism and SCZ, anorexia and microcephaly2–5. Moreover, a recent post hoc analysis of ASD and SCZ loci has revealed that such co-morbidities might be causally linked to each other, with macrocephaly shown to be associated with ASD and microcephaly associated with SCZ13.
A pervasive challenge in the interpretation of CNV discovery is the transition from the detection of a genomic lesion that can often span large regions encompassing many genes to the identification of the critical loci whose dosage sensitivity drives the phenotype. For some disorders, this has been achievable through the discovery of highly penetrant point mutations at a single locus; for example, mutations in PMP22 are sufficient to cause CMT14, while mutations in RAI1 cause Smith-Magenis syndrome15. In other instances, gene-specific genomic alterations such as chromosomal translocations, inversions, or small coding deletions can narrow the critical region to a single gene (e.g. MBD5 in 2q23.1 microdeletion syndrome16). Alternatively, systematic functional dissection through mouse mutagenesis has yielded strong candidates; ablation of Tbx1 recapitulates the cardiac phenotypes of VCSF17, while knockout of Shank3 captures most of the phenotypes seen in the terminal 22q deletion that causes Phelan-McDermid syndrome18. However, these approaches are significantly more challenging for common phenotypes and genetically heterogeneous disorders: systematic engineering of the mouse genome for each gene in a CNV can be impractical, while rare mutations involved in complex traits are likely to exhibit both reduced penetrance and variable expressivity.
Manipulation of zebrafish embryos represents an attractive alternative towards the discovery of human dosage-sensitive genes, especially when the CNV under investigation has mirrored anatomical phenotypes that are detectable during early development and that can thus be assayed by a combination of gene suppression and overexpression experiments19. Given the association between the 16p11.2 CNV and changes in head size, we hypothesized that a) systematic overexpression of each of the 29 genes in the common duplication might yield a defined, reproducible set of transcripts some of which might cause microcephaly; b) that reciprocal suppression of these genes should yield the macrocephalic phenotype seen in the 16p11.2del. To test these hypotheses, we first queried the zebrafish genome by reciprocal BLAST for each of the 29 target genes (Fig. 1a) and identified 24 orthologs (Suppl. Table 1), with five genes, SPN, QPRT, C16orf54, TMEM219, C16orf92, found only in placental mammals. In a manner akin to the classic Drosophila misexpression experiments, we generated capped mRNA for all 29 human genes and injected zebrafish embryos at the two-cell stage with equimolar pairwise ‘cocktail’ combinations at two dosages of 25 pg and 50 pg; these commonly used ranges20 were selected because they represent >0.25–0.5% of total polyA+ mRNA in a zebrafish embryo21 and are thus likely to achieve significant overexpression above the baseline of any single transcript.
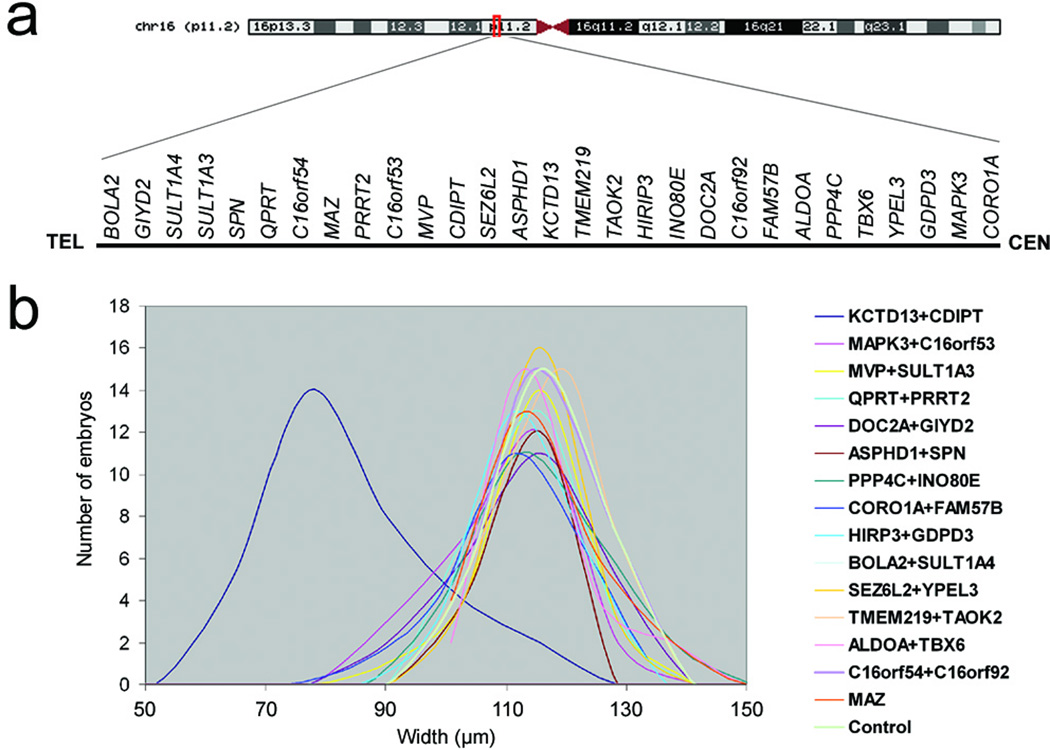
Figure 1. Systematic analysis of 16p11.2 del/dup genes in vivo induces defects in head size.
(a) Schematic of chromosome 16, zoomed in the 16p11.2 CNV, showing gene content (not to scale) above the black line. (b) Plot of head size measurements (in µm) of human mRNA overexpression combinations measured across ~50 embryos/injection cocktail. In all but one injection, both controls and human overexpressed genes result in indistinguishable median head size, with minimal variance. By contrast, embryos injected with the KCTD13/CDIPT mRNAs cocktail show consistent and significant reduction of head size.
One gene, TAOK2, required reduced mRNA dosage to 10 pg because of toxicity. For the remaining 28 genes, we observed no lethality or gross morphological defects at either 25 or 50 pg, while random RT-PCR testing of nine injections including KCTD13 showed persistence of the corresponding human mRNAs up to ~4.25 dpf (Suppl. Fig. 1e). We therefore developed a surrogate measurement for head size at 4.25–4.5 dpf using objective measurements, wherein the distance across the convex tips of the eye cups was recorded in 50 embryos/injection, masked to the injection cocktail (Fig. 1b). Only a single overexpression cocktail containing KCTD13 and CDIPT gave significant changes in head size (two-tailed t-test, p<0.000001). Subsequent single-mRNA injections for the two genes indicated that the phenotype was driven exclusively by the overexpression of KCTD13; injection of KCTD13 at progressive mRNA amounts yielded an increasing percentage of microcephalic embryos (Fig. 2a, b; Suppl. Fig. 1a). In contrast, the head size of embryos injected with CDIPT was indistinguishable from those injected with sham control (data not shown).
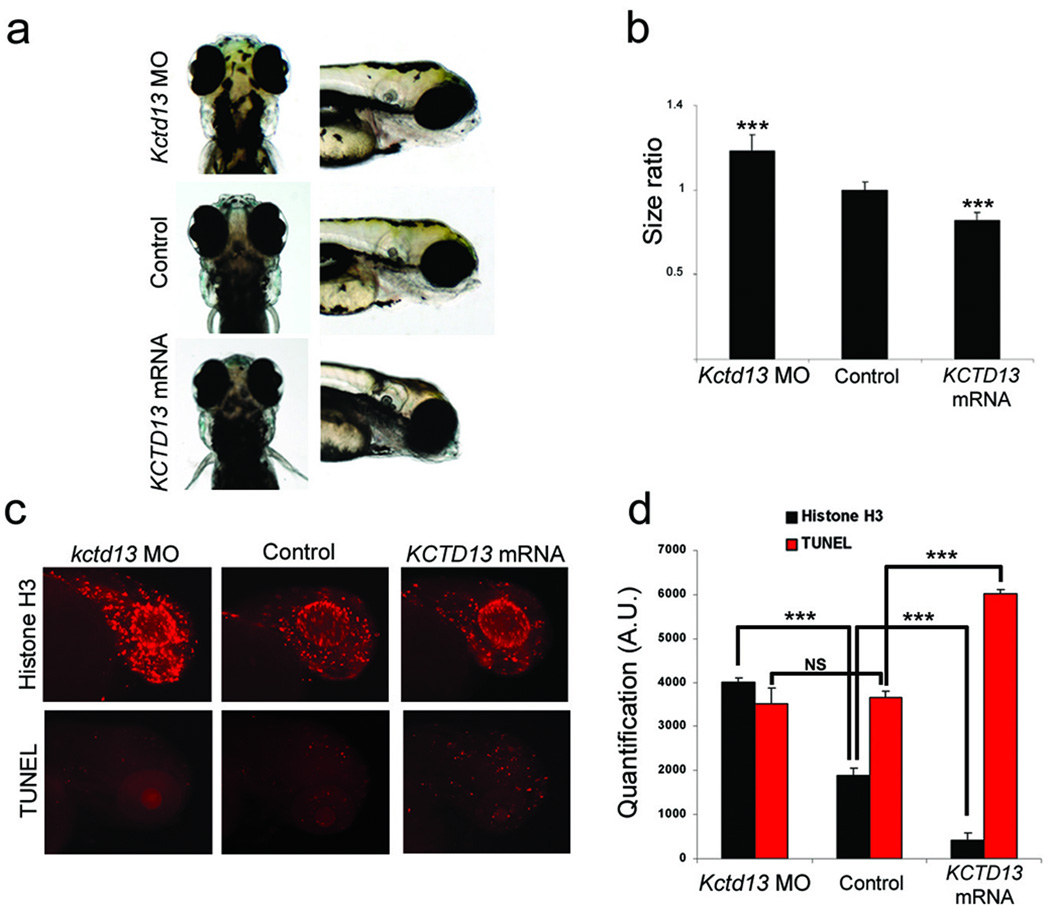
Figure 2. KCTD13 dosage changes lead to head size, proliferation and apoptosis defects.
(a) From top to bottom, dorsal (left) and lateral (right) views of representative embryos injected with Kctd13 MO, control, or KCTD13 mRNA. (b) Graph of the ratio between control and injected embryos head size measures at 4.5 dpf (n=45). (c) Phospho-histone H3 (top panel) and TUNEL (bottom panel) staining for proliferating or apoptotic cells in zebrafish brain at 2 dpf and 3 dpf respectively. From left to right are representative examples of MO-, control- and mRNA-injected embryos. (d) Graph of phospho-histone H3 and TUNEL quantifications from 20 MO-, control and mRNA-injected embryos. Data from three independent experiments are represented as mean ± s.d. *** p<0.00001; two-tailed t-test comparisons between control and either MO-and mRNA-injected embryos.
To validate the specificity of this phenotype and to ask whether we could also simulate the macrocephalic phenotype seen usually in 16p11.2del patients4,13, we designed a splice-blocking morpholino (MO) against the donor site of exon 3 of the sole kctd13 zebrafish ortholog. Injection of 10ng of MO and RT-PCR testing showed ~70% reduction of kctd13 message at 4.5 dpf (Suppl. Fig. 1c). Strikingly, this injection also yielded a significant increase in mean head size (p<0.00001; Fig. 2a, b,Suppl. Fig. 1b). This phenotype is specific to kctd13; a scrambled MO induced no phenotypes, while injection of 10 ng of MO and 50 pg of KCTD13 mRNA rescued both the microcephalic and macrocephalic phenotypes (Suppl. Fig. 1d). Importantly, measurement of the somitic trunk length of scored embryos showed no differences in length (or morphology; Suppl. Fig. 2), indicating that the head size differences are unlikely to be driven by gross developmental delay. We likewise observed no defects in other structures, including the heart and the swim bladder.
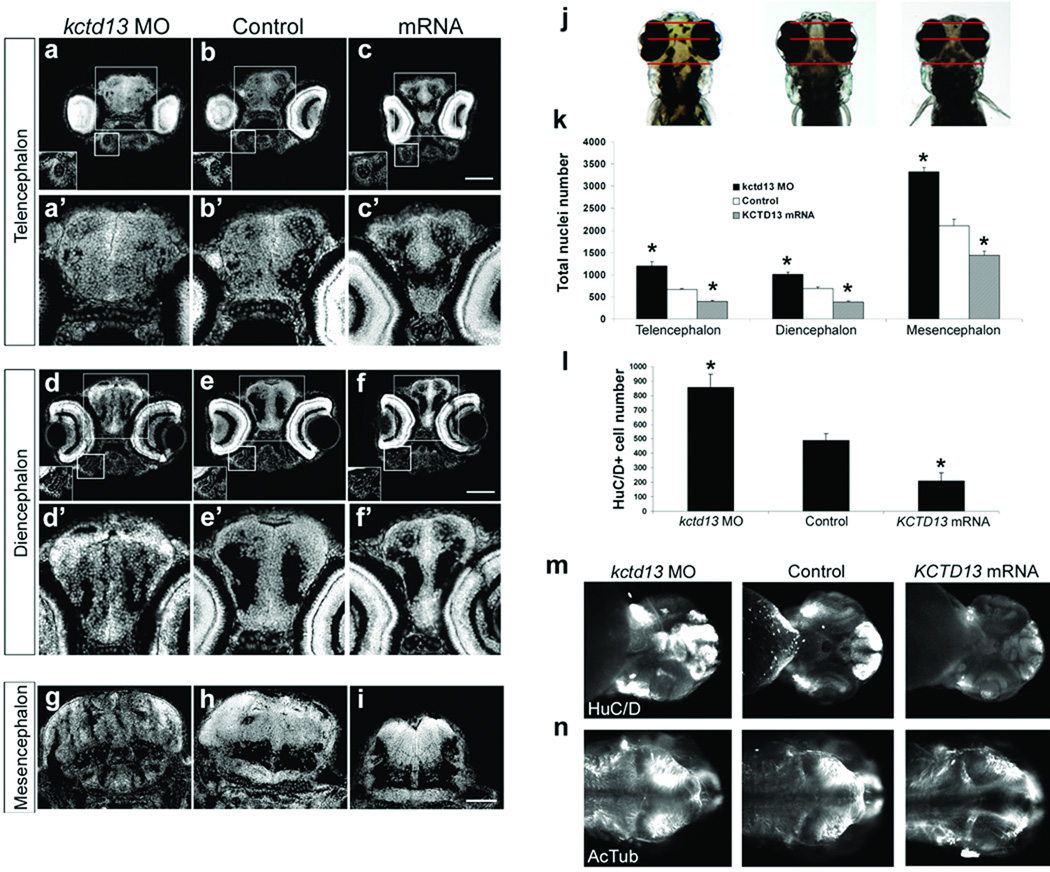
To investigate the mechanism of the head size defects, we examined the developing brain of both macro-and microcephalic embryos. In situ hybridization with an antisense kctd13 probe showed this transcript to be expressed strongly in the developing brain. At 24 hpf, kctd13 is strongly expressed in the anterior forebrain (arrowheads, suppl. Fig. 3a, f), the midbrain, and the hindbrain. In later stages, kctd13 is expressed predominantly in the telencephalon, the diencephalon, and the retina (suppl. Fig. 3b-e). Staining both phenotype classes with terminal deoxynucleotidyl transferase-mediated dUTP nick end-labeling (TUNEL) showed a significant increase in apoptosis exclusively in the microcephalic embryos. At the same time, phospho-histone H3 antibody staining revealed an increase in proliferating cells in the brain of kctd13 morphants and a reciprocal decrease of proliferating cells in overexpressant embryos (Fig. 2 c, d). Detailed analysis of transverse sections from embryos injected with either MO or KCTD13 mRNA confirmed that the observed phenotypes are likely driven by changes in the number of cells in the developing brain, since the architecture and cell content of the Meckel and palatoquadrate pharyngeal cartilages were normal, as was the overall cell content and architecture of the retina (Fig 3a–f). Counting of cell nuclei at 4.5 dpf, when the anatomical measurements were made, showed significant reciprocal changes in the total number of cells in each of the telencephalon, diencephalon, and mesencephalon (Fig 3k). Further, counting of HuC/D-positive cells (a marker for post-mitotic neurons) in the telencephalon recapitulated the differences seen in total cell count but showed no difference in cell circumference (Fig. 3l; Suppl. Fig. 4), indicating that the changes in overall cell numbers and ultimate changes in head size are largely driven by changes in the numbers of mature neurons. These data, collected at 4.5 dpf, predict that the onset of the neuroanatomical defects would precede the manifestation of anatomical micro- or macrocephalic phenotypes. To test this hypothesis, we stained all three classes of embryos with HuC/D and acetylated tubulin at 2 dpf, at which time the head size of kctd13 MOs and overexpressants is indistinguishable from uninjected controls. We observed stark differences in the density and distribution of neurons, particularly in the forebrain, with concomitant loss of organization and bilateral symmetry (Fig. 3m) as well as aberrant distribution of axonal tracks (Fig. 3n).
Figure 3. KCTD13 dosage changes lead to neuroanatomical defects.
DAPI staining on transverse sections of the telencephalon (a, b, c), the diencephalon (d, e, f), and the mesencephalon (g, h, i) of embryos injected with kctd13 MO, control or KCTD13 mRNA at 4.5 dpf. (j) The planes of section are illustrated with red lines on dorsal views of kctd13 MO-, control-, and KCTD13 mRNA-injected embryos (left to right). Higher magnifications of the telencephalon (a–c) and the diencephalon (d–f) are in (a’–f’). The Meckel pharyngeal cartilage (a–c) and the palatoquadrate pharyngeal cartilage (d–f) are in insets. Scale bar, 100µm. (k) Bar graph of the total number of nuclei for the three classes of embryos in the telencephalon, diencephalon and mesencephalon at 4.5 dpf (3 adjacent sections, n=4). (l) Bar graph of the number of HuC/D positive cells in the telencephalon for kctd13 MO-, control- and KCTD13 mRNA-injected embryos at 4.5 dpf (3 adjacent sections, n=4). (m) Ventral and (n) dorsal views of kctd13 MO-, control-, and KCTD13 mRNA-injected embryos at 2 dpf (left to right). Data are represented as mean ± s.d. * p<0.01; two-tailed t-test comparisons between control and either MO- and mRNA-injected embryos.
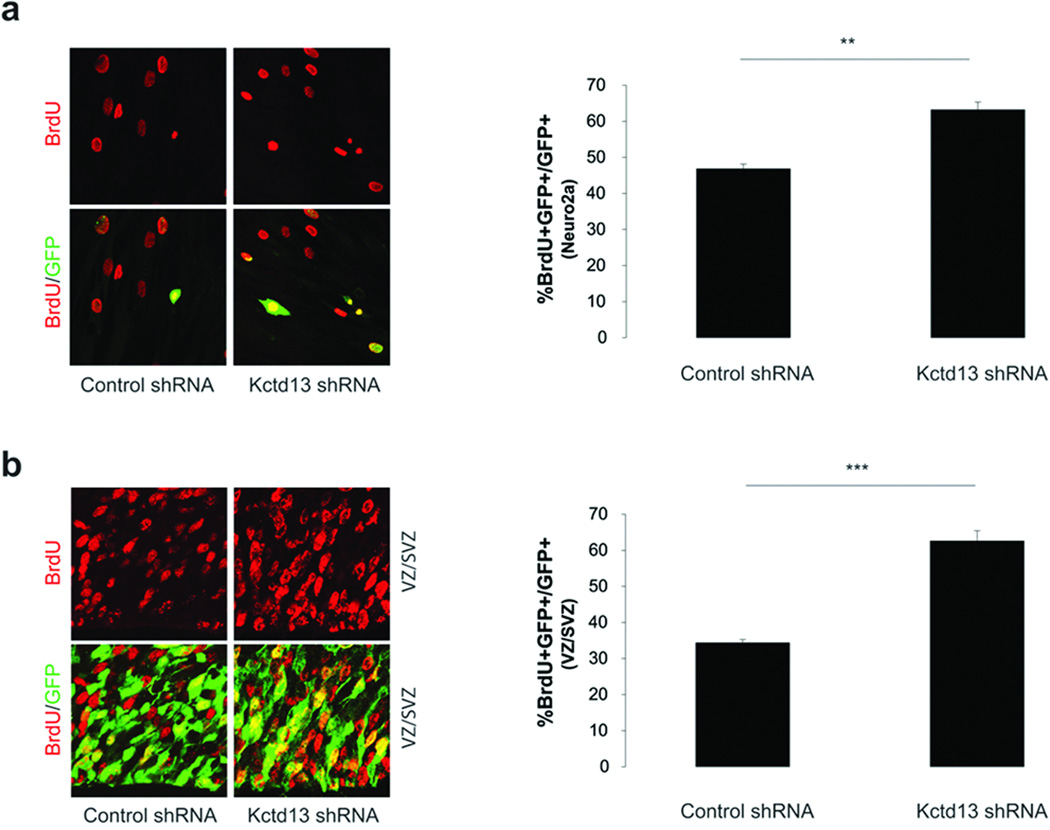
Although the zebrafish brain bears many similarities in terms of developmental programming with the mammalian brain, we sought to test whether our findings might be relevant to cortical development in a mammalian system. Data mining of the Gene Expression Nervous System Atlas (GENSAT; www.gensat.org) and the Allen brain atlas (www.brain-map.org) revealed that human and mouse KCTD13 is expressed throughout development in neurons residing in the cortex, striatum, olfactory tubercle, and hippocampus. We therefore designed shRNAs against murine Kctd13 and tested their efficiency in a cultured mouse neuroblastoma cell line (Neuro-2A). One shRNA, which, similar to the kctd13 MO, downregulated the expression of endogenous message by ~70% (Suppl. Fig. 5), was then co-transfected with a GFP-expressing plasmid into Neuro-2A cells. Two days after transfection, cells were pulsed with 5-bromo-2'-deoxyuridine (BrdU) and analyzed for the effects of Kctd13 knockdown on cellular proliferation. Similar to the proliferation data using zebrafish morphants, depletion of Kctd13 resulted in a 34% increase (p<0.01) in BrdU/GFP labeled cells (Fig 4a). We next injected the Kctd13 shRNA and GFP-expressing plasmid into the ventricular space of wild-type C57BL/6 embryos at embryonic day 13.5 (E13.5) and injected BrdU into pregnant dams two hours before collection of electroporated embryos at E15.5. Knockdown of Kctd13 resulted in a two-fold increase in BrdU/GFP labeling within the ventricular zone (p<0.001; Fig. 4b), suggesting that Kctd13 is required to maintain the proliferative status of cortical progenitors in vivo.
Figure 4. Kctd13 regulates mammalian cell proliferation in vitro and in vivo.
(a) Knockdown of Kctd13 in Neuro-2a cells results in an increase in the number of BrdU+/GFP+ cells relative to control cells. Error bars represent the standard error from two independent experiments.
(b) Analysis of E15.5 mouse cortices injected with either Kctd13 or control shRNA reveal a similar increase in BrdU+/GFP+ cells in knockdown tissue (n=3, error bars represent the standard error). ** p<0.01; ***p<0.001.
The combination of our zebrafish and mouse data suggests that KCTD13 is a major driver of the head size phenotypes associated with the 16p11.2 CNV through the regulation of early neurogenesis. Our data do not exclude the possibility that other loci also have an independent contribution to the 16pdel/dup anatomical phenotypes and cannot inform directly the question whether all of the observed pathology in 16p11.2del/dup patients is driven by dosage changes in KCTD13. However, we were able to ask whether dosage changes of other loci inside the CNV might also be relevant to the head size phenotypes established by KCTD13. Specifically, we performed pairwise overexpressions of KCTD13 with all other 28 transcripts in the region and asked whether we could observe changes in the penetrance (percentile of microcephalic zebrafish in a clutch) or the expressivity (percentile changes in mean head size) of the KCTD13-established phenotype. We observed no changes in penetrance. However, pairwise expression with two other transcripts, MAPK3 and MVP increased significantly the expressivity of the phenotype from 18% for KCTD13 alone to 24% and 22% respectively (Suppl. Fig 6,Suppl. Table 2), predicting that del or dup patients might have a more severe phenotype that individuals with heterozygous loss of function at KCTD13 alone.
Finally, we turned our attention to the question of whether loss of KCDT13 might be sufficient in humans to cause some of the commonly observed phenotypes associated with the 16p11.2del. During our analyses, a submicroscopic ~118 kb deletion in 16p11.2 that segregated with ASD and other neurodevelopmental abnormalities was discovered in a single three-generation pedigree22. This deletion encompasses five genes, MVP, CDIPT1, SEZ6L2, ASPHD1 and KCTD13, in agreement with our hypothesis that haploinsufficiency at KCTD13 might contribute to 16p11.2 phenotypes. We performed multiplex ligation-dependent probe amplification (MLPA; Suppl. Table 3) of this restricted region in 518 subjects that met diagnostic criteria for autism or ASD (Autism Diagnostic Observational Schedule, ADOS10), finding full-segment deletions in eight independent ASD subjects (1.54%; six deletion, two duplication), compared to just five such events from 8,328 controls (0.006%)23. We also observed the deletion of a single probe spanning exon 4 of KCTD13 in one proband with a narrow diagnosis of autism (Suppl. Fig. 7; Suppl. Table 3). The MLPA assay was replicated in the proband and the deletion was confirmed further by qPCR on two independently obtained DNA samples (Suppl. Fig. 7). Identical MLPA analyses, as well as confirmation of paternity by genotyping, were performed in both biological parents, revealing that the deletion arose de novo and was restricted to the KCTD13 coding region, maximally 9 kb and including exons 3, 4, and 5 (Suppl. Fig. 7). However, in an extensive effort to precisely localize the breakpoints by custom tiled array comparative genomic hybridization (aCGH) of the entire 16p11.2 region we discovered the rearrangement to be significantly more complicated. We discovered an additional, atypical ~360 kb deletion distal to the classic 16p11.2 region that was inherited from an asymptomatic mother. The deletion was confirmed by an independent Agilent 24 M feature aCGH, however the precise breakpoints could not be definitely localized as the rearrangement is mediated by a highly complex genomic region of segmental duplication24. The apparent complexity of these co-occurring events, including both inherited and de novo rearrangements, impacts multiple loci in addition to KCTD13, suggesting this phenotype cannot be attributed solely to the KCTD13 alteration.
In aggregate, our data support a major contributory role for KCDT13 in the 16p11.2 CNV through four lines of evidence: a) the in vivo overexpression screen yielded microcephaly in 1/29 genes; b) the reciprocal suppression of this locus mirrored the corresponding human 16p11.2del phenotypes; c) functional analyses established a neurogenic defect that was consistent across species; and d) KCTD13 lies in the putative critical region of ~118 kb region delineated in one family with ASD independent from our study, and, here, we found a complex rearrangement that includes de novo alteration affecting a portion of the coding region in a patient with a narrow diagnosis of autism.
Given that the design of our screen relies on a heterologous system of expression, we do not exclude the possibility that other genes in the 16p11.2 CNV might also be relevant to human pathology but did not trigger phenotypes in zebrafish embryos, especially the five transcripts not present in the zebrafish genome. Moreover, we do not know whether dosage imbalance of KCTD13 might regulate other phenotypes commonly associated with the 16p11.2 CNV, such as obesity and epilepsy4–6. However, it is reasonable to speculate that the neurodevelopmental changes observed upon KCTD13 perturbation could contribute to those phenotypes. We note that loss of function mutations of another member of this family, KCTD7 cause progressive myoclonic epilepsy25, while variants in KCTD15 have been associated with obesity26.
KCTD13 encodes the polymerase delta-interacting protein 1 (PDIP1), that interacts with the Proliferating Cell Nuclear Antigen (PCNA)27 and thus might have a role in the regulation of cell cycle during neurogenesis. Our studies might also inform disease architecture and whether changes in neuronal populations can account for brain overgrowth phenotypes28. Although our data suggest that disregulated KCTD13 levels are sufficient to establish neuroanatomical defects, its genetic interaction with at least one more locus leads us to speculate that the point mutation event that would phenocopy the 16p11.2del might not be represented by single loss of function alleles at KCTD13, but at cis alleles in KCTD13 and MAPK3, as well as MVP, or combined haploinsufficiency of all three. Testing this hypothesis with sufficient statistical power will require the analysis of large cohorts.
Similar to our study, the 16p11.2 CNV has been modeled in mice by chromosomal engineering, where neuroanatomical volumetric changes in regions analogous to our observations (e.g the forebrain and midbrain) have been noted29. Our approach is likely to be particularly useful in resolving a rapidly growing number of genomic regions implicated in a range of human genomic disorder phenotypes30 that a) are involved in both deletion and duplication syndromes such as 7q11.23 CNV10; and b) have mirroring anatomical phenotypes that can be assayed in a physiologically-relevant developmentalsystem (such as 1q21.1, and 3q39 CNVs1). Such analyses will expedite the identification of major dosage-sensitive loci and accelerate our biological understanding of CNVs that, together, account for a significant fraction of the mutational burden of neurodevelopmental disorders1,11.
Methods Summary
Morpholino, in vivo analysis of gene expression and embryo manipulations
Splice-blocking morpholino (MO) against Kctd13 was designed and obtained from Gene Tools, LLC (5’-TCTAAGGGTACACGCCTGACCTGTA-3’). Control MO was the scrambled nucleotide sequence from Gene Tools, LLC (5'-CCTCTTACCTCAGTTACAATTTATA- 3'). MO injection, RNA rescue and overexpression experiments are performed using standard protocol.
Immunostaining and TUNEL assay
Embryos were fixed in 4% PFA or in Dent’s fixative (80% methanol, 20% DMSO) overnight at 4°C. Embryos were incubated in the first antibody solution, 1:750 anti-histone H3 (ser10)-R, (sc-8656-R, Santa Cruz), 1:1000 anti-HuC/D (A21271, Invitrogen), 1:1000 anti-acetylated tubulin (T7451, Sigma-Aldrich). Apoptotic cell death in zebrafish whole-mounts was detected according to a modification of the ApopTag rhodamine In Situ Apoptosis Detection kit (Chemicon,Temecula, CA) protocol. Embryos for cryosectioning were fixed, incubated in PBS-sucrose 30%, embedded in Tissue-Tek O.C.T. Embedding Compound (Sakura Finetek) and sectioned at 7 µm.
In utero microinjection
E13.5 embryos were injected with either control or Kctd13 shRNA constructs and a GFP-expressing vector. Forty-eight hours after electroporation, BrdU (100 mg/kg) was injected intraperitoneally into the pregnant dams and embryos were harvested 2 hours later. Embryo brains were processed and sectioned (20 µm) before staining with a BrdU antibody (Accurate).
Short arm of chromosome 16 custom array-CGH
DNA samples from the proband and both parents were labelled with Cy3 and cohybridized with Cy5-labelled control DNA from an unaffected CEPH individual obtained from Coriell (GM10851) to custom-made Nimblegen arrays. DNA labelling, hybridization and washing were performed according to Nimblegen protocols. Scanning was performed using an Agilent G2565BA Microarray Scanner. Image processing, quality control and data extraction were performed using the Nimblescan software v.2.5.
Methods
Morpholino, in vivo analysis of gene expression and embryo manipulations
Splice-blocking morpholino (MO) against Kctd13 was designed and obtained from Gene Tools, LLC (TCTAAGGGTACACGCCTGACCTGTA). Control MO was the scrambled nucleotide sequence from Gene Tools, LLC (5'-CCTCTTACCTCAGTTACAATTTATA- 3'). We injected 1nl of diluted MO (6, 8, 10 ng) and/or RNA (50, 75, 100 pg) into wildtype zebrafish embryos at the 1-to 2-cell stage. Injected embryos were scored at 4.25 dpf and classified into two groups, i.e. normal and mutant, on the basis of the relative head size compared with age-matched controls from the same clutch. For RNA rescue and overexpression experiments, the human wild type mRNAs was cloned into the pCS2 vector and transcribed in vitro using the SP6 Message Machine kit (Ambion). All the experiments were repeated three times and we ran a t-test to determine the significance of the morphant phenotype. Whole-mount in situ hybridizations were carried out using antisense probes for kctd13 made from clone (Openbiosystems) and following standard protocols. Further processing included clearing in methyl salicylate and flat mounting were performed using standard protocol.
Whole-Mount TUNEL assay
Apoptotic cell death in zebrafish whole-mounts was detected according to a modification of the ApopTag rhodamine In Situ Apoptosis Detection kit (Chemicon,Temecula, CA) protocol. Embryos were fixed in 4% paraformaldehyde (PFA) at 4°C overnight and store in 100% methanol at −20°C. After rehydratation in PBS, embryos were permeabilized by a 5 minutes digestion with proteinase K (10µg/ml) in PBS at room temperature. After two washes in sterile water for 3 minutes each, embryos were postfixed with 4% PFA for 20 minutes, room temperature and then with prechilled ethanol:acetic acid (2:1) for 10 minutes at -20°C. Embryos were washed in PBS-T (PBS 1X, 0.1% Tween-20) for 5 minutes, 3 times at room temperature. The incubation in the equilibration buffer and the further steps were followed according to the standard protocol suggested by the manufacturer. TUNEL staining was quantified by counting positive cells in defined regions of the head and with ImageJ software.
Zebrafish whole-mount and section immunostaining
Embryos were fixed in 4% PFA overnight and stored in 100% methanol at −20°C. For acetylated tubulin staining embryos were fixed in Dent’s fixative (80% methanol, 20% DMSO) overnight at 4°C. The embryos were permeabilized with proteinase K followed by post-fixation with 4% PFA, washed in PBSTX (PBS+0.5% Triton X-100). After rehydration in PBS, PFA-fixed embryos were washed in IF buffer (0.1% Tween-20, 1% BSA in PBS 1X) for 10 minutes at room temperature. The embryos were incubated in the blocking buffer (10% FBS, 1% BSA in PBS1X) for 1hr at room temperature. After two washes in IF Buffer for 10 minutes each, embryos were incubated in the first antibody solution, 1:750 anti-histone H3 (ser10)-R, (sc-8656-R, Santa Cruz), 1:1000 anti-HuC/D (A21271, Invitrogen), 1:1000 anti-acetylated tubulin (T7451, Sigma-Aldrich) in blocking solution, overnight at 4°C. After two washes in IF Buffer for 10 minutes each, embryos were incubated in the secondary antibody solution, 1:1000 Alexa Fluor donkey antirabbit IgG and Alexa Fluor goat anti-mouse IgG (A21207, A11001, Invitrogen) in blocking solution, for 1hr at room temperature. Staining was quantified by counting positive cells in defined regions of the head and with ImageJ software. Embryos for cryosectioning were fixed in 4% PFA overnight at 4°C, washed twice in phosphate-buffered saline (PBS) and incubated in PBS-sucrose 30% at 4°C overnight. Embryos were embedded in Tissue-Tek O.C.T. Embedding Compound (Sakura Finetek). Sections were cut at 7 µm.
Cell Proliferation assay
Neuro-2A cells were seeded in 2 well chamber slides and transfected with control and Kctd13 shRNA constructs and a GFP expressing plasmid. Two days after transfection, cells were pulsed with 10 µM BrdU, fixed and stained with a BrdU antibody (Accurate).
In utero microinjection
E13.5 embryos were injected with either control or Kctd13 shRNA constructs and a GFP-expressing vector. Forty-eight hours after electroporation, BrdU (100 mg/kg) was injected intraperitoneally into the pregnant dams and embryos were harvested 2 hours later. Embryo brains were processed and sectioned (20 µm) before staining with a BrdU antibody (Accurate).
Multiplex ligation-dependent probe amplification (MLPA)
A custom assay with 29 probes was designed to capture candidate genes in the 118 kb 16p11.2 putative critical region (23 probes to the targeted genes KCTD13, MVP, and CDIPT; 2 probes to 16p11.2 microdeletion region genes not within the putative critical region, TAOK2 and TBX6; 4 probes to control genes outside of the region). The assay was performed with 100–150 ng genomic DNA (quantified byPico-Green (Quant-iT, Invitrogen)) according to manufacturer’s instructions. All samples were performed in triplicate. Amplification products from ligated probes were run on an ABI 3730xl DNA Analyzer using Genescan-Rox500 size standards (Applied Biosystems Inc). MLPA peak plots were visualized and analyzed using GeneMarker Software Trial Version 1.91 (SoftGenetics LLC). Peak height and area were compared between control individuals and ASD samples. Values between 0.75 and 1.3 were considered normal.
Real-time Quantitative PCR
Each quantitative real-time PCR was performed in a 96-well plate using 2×LightCycler 480 SYBR Green I Master mix (Roche Applied Science) according to manufacturer's instructions. All products were cycled at: 95°C for 2 min followed by 45–55 cycles of 95°C for 15 s and 60°C for 1 min. Melt temperature analysis was performed at the end of each run to confirm PCR specificity. Relative copy number was determined between the proband (AC02-1467-01) and three control subjects for probes designed within exon 4 of KCTD13 and control probes localized outside of the 16p11.2 microdeletion region.
Short arm of chromosome 16 custom array-CGH
DNA samples from the proband and both parents were labelled with Cy3 and cohybridized with Cy5-labelled control DNA from an unaffected CEPH individual obtained from Coriell (GM10851) to custom-made Nimblegen arrays. These arrays contained 71,000 probes spread across the short arm of chromosome 16 from 22.0 to 32.7 Mb (at a median space of 45 bp between 27.5 and 31.0 Mb) and 1,000 control probes situated in invariable region of the X chromosome. DNA labelling, hybridization and washing were performed according to Nimblegen protocols. Scanning was performed using an Agilent G2565BA Microarray Scanner. Image processing, quality control and data extraction were performed using the Nimblescan software v.2.5.
Supplementary Material
Acknowledgments
We thank Carrie Hanscom for technical assistance. We are also grateful to Joshua Black, Johnathan Whetstine, Kim Brown, and Charles Lee for assistance with the initial and confirmatory aCGH experiments. This work was supported by a Silvo O. Conte center grant from the National Institute of Mental Health, NIH MH-084018 (AS, AK, and NK), grant MH-091230 (AK), grant HD06286 (JFG), the Simon's Foundation, the Autism Consortium of Boston (JFG), the Leenaards Foundation Prize (SJ, DM and AR), the Swiss National Science Foundation (AR, and JSB), a Swiss National Science Foundation Sinergia grant (SJ, JSB and AR). MET was supported by an NIMH National Research Service Award (F32MH087123). SJ is recipient of a « bourse de relève académique de la Faculté de Biologie et Médecine de l’Université de Lausanne ». NK is a Distinguished Brumley Professor.
Footnotes
Author contributions:
CG and NK designed the study and wrote the paper, and all authors approved and commented on it. CG performed the zebrafish studies, immunostaining, TUNEL and sectioning experiments, and counting analyses. JW made the plasmid constructs and capped mRNAs. MET, MS, and JFG performed the human genetic analyses. ECO performed the shRNA silencing, BrdU pulse experiments, and generated mice brain sections. YT, AS, AK performed in utero microinjections. SJ, AR, JSB designed the 16p-specific microarray and shared unpublished data that informed our experimental design.
References
- 1.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 2.Weiss LA, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 3.McCarthy SE, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacquemont S, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478:97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walters RG, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar RA, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 7.Baldini A. Dissecting contiguous gene defects: TBX1. Curr Opin Genet Dev. 2005;15:279–284. doi: 10.1016/j.gde.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Lupski JR, et al. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66:219–232. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- 9.Brunetti-Pierri N, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanders SJ, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stefansson H, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shinawi M, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 2010;47:332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roa BB, et al. Charcot-Marie-Tooth disease type 1A. Association with a spontaneous point mutation in the PMP22 gene. N Engl J Med. 1993;329:96–101. doi: 10.1056/NEJM199307083290205. [DOI] [PubMed] [Google Scholar]
- 15.Slager RE, Newton TL, Vlangos CN, Finucane B, Elsea SH. Mutations in RAI1 associated with Smith-Magenis syndrome. Nat Genet. 2003;33:466–468. doi: 10.1038/ng1126. [DOI] [PubMed] [Google Scholar]
- 16.Talkowski ME, et al. Assessment of 2q23.1 microdeletion syndrome implicates MBD5 as a single causal locus of intellectual disability, epilepsy, and autism spectrum disorder. Am J Hum Genet. 2011;89:551–563. doi: 10.1016/j.ajhg.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindsay EA, et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 18.Peca J, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ceol CJ, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takeda H, Matsuzaki T, Oki T, Miyagawa T, Amanuma H. A novel POU domain gene, zebrafish pou2: expression and roles of two alternatively spliced twin products in early development. Genes Dev. 1994;8:45–59. doi: 10.1101/gad.8.1.45. [DOI] [PubMed] [Google Scholar]
- 21.Detrich HW, 3rd, Westerfield M, Zon LI. Overview of the Zebrafish system. Methods Cell Biol. 1999;59:3–10. doi: 10.1016/s0091-679x(08)61816-6. [DOI] [PubMed] [Google Scholar]
- 22.Crepel A, et al. Narrowing the critical deletion region for autism spectrum disorders on 16p11.2. Am J Med Genet B Neuropsychiatr Genet. 2010;156:243–245. doi: 10.1002/ajmg.b.31163. [DOI] [PubMed] [Google Scholar]
- 23.Cooper GM, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011 doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barge-Schaapveld DQ, Maas SM, Polstra A, Knegt LC, Hennekam RC. The atypical 16p11.2 deletion: a not so atypical microdeletion syndrome? American journal of medical genetics. Part A. 2011;155A:1066–1072. doi: 10.1002/ajmg.a.33991. [DOI] [PubMed] [Google Scholar]
- 25.Azizieh R, et al. Progressive myoclonic epilepsy-associated gene KCTD7 is a regulator of potassium conductance in neurons. Mol Neurobiol. 2011;44:111–121. doi: 10.1007/s12035-011-8194-0. [DOI] [PubMed] [Google Scholar]
- 26.Willer CJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He H, Tan CK, Downey KM, So AG. A tumor necrosis factor alpha- and interleukin 6-inducible protein that interacts with the small subunit of DNA polymerase delta and proliferating cell nuclear antigen. Proc Natl Acad Sci U S A. 2001;98:11979–11984. doi: 10.1073/pnas.221452098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Courchesne E, et al. Neuron number and size in prefrontal cortex of children with autism. Jama. 2011;306:2001–2010. doi: 10.1001/jama.2011.1638. [DOI] [PubMed] [Google Scholar]
- 29.Horev G, et al. Dosage-dependent phenotypes in models of 16p11.2 lesions found in autism. Proc Natl Acad Sci U S A. 2011;108:17076–17081. doi: 10.1073/pnas.1114042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beckmann JS, Estivill X, Antonarakis SE. Copy number variants and genetic traits: closer to the resolution of phenotypic to genotypic variability. Nat Rev Genet. 2007;8:639–646. doi: 10.1038/nrg2149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.